miR-155 effectively induces apoptosis in K562 Philadelphia positive cell line through upregulation of p27kip1
Bioimpacts, 7(2), 109-114; DOI:10.15171/bi.2017.14
Original Research
miR-155 effectively induces apoptosis in K562 Philadelphia positive cell line through upregulation of p27kip1
Mahdi Edalati Fathabad1,
Morteza Karimipoor2,*,
Shaban Alizadeh1,*,
Asghar Abdoli3,
Amir Atashi4,
Mahtab Sayadi4
1
Hematology Department, School of Allied Medicine, Tehran University of Medical Sciences, Tehran, Iran
2
Molecular Medicine Department, Biotechnology Research Center, Pasteur Institute of Iran, Tehran, Iran
3
Department of Hepatitis and AIDS, Pasteur Institute of Iran, Tehran, Iran
4
Cancer Prevention Research Center, Shahroud University of Medical Sciences, Shahroud, Iran
*Corresponding authors: Shaban Alizadeh, Email: alizadeh1982@gmail.com; Morteza Karimipoor, Email: mortezakarimi@pasteur.ac.ir
© 2017 The Author(s). This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by-nc/4.0). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Abstract
Introduction:
Chronic myelogenous leukemia (CML) is a myeloproliferative disorder caused by the Philadelphia chromosome translocation, at (9; 22), which results in BCR-ABL fusion tyrosine kinase oncoprotein. This fusion induces down-regulation of miR-155. Upregulation of miR-155 can influence cell fate via the effect on p27kip1 and apoptosis. The aim of this study was to induce apoptosis in K562 CML cell line by overexpression of miR-155.
Methods:
The K562 cell line was transfected with pLenti-III-pre mir155-GFP constructs through electroporation. Then, overexpression of miR-155 as well as the expression level of p27kip1 and c-Myc was analyzed by quantitative PCR (qPCR). The level of p27 (Kip1) protein expression was measured by Western blot and the Annexin V method was carried out to investigate apoptosis.
Results:
Flow cytometric analysis results of K562 cells transfected with pLenti-III-pre mir155-GFP construct showed a significant increase in cell apoptosis. Gene expression and protein level of p27kip1 were upregulated. However, there was no change in c-Myc expression profile.
Conclusion:
miR-155 could be a promising approach to aid in the treatment of CML. However, further studies are required in this respect.
Keywords: Apoptosis, K562, miR-155, Philadelphia, p27kip1
Introduction
Chronic myelogenous leukemia (CML) is a disorder of bone marrow hematopoietic stem cells associated with increased and unregulated growth of myeloid cells in bone marrow and peripheral blood.1 The incidence of CML is 1 or 2 cases per 100 000 people every year with a median age range of around 65 years.2
Philadelphia chromosome or Philadelphia translocation between chromosomes 9 and 22 [t (9; 22) (q34; q11)] is the major cytogenetic abnormality in CML. This translocation results in the formation of a hybrid BCR-ABL oncogene on chromosome 22. Consequently, deregulated tyrosine kinase leads to uncontrolled cell proliferation and reduced apoptosis.3,4 Tyrosine-kinase inhibitors (TKIs) are the main approved chemotherapy drug groups for CML. Imatinib mesylate (IM) is a TKI as first line therapy of Ph-positive CML.5-7 Despite the many advantages of IM it confers several adverse effects. For example, heart failure related to chemotherapy is a major clinical challenge and cardiotoxicity of IM has been reported in patients.8 Furthermore, IM administration does not result in complete remission of the CML. It is noteworthy that due to Bcr-Abl translocation sophisticated molecular mechanisms occur, especially in blast crisis stage, which prevents the induction of apoptosis and interferes with IM effect.9,10
Therefore, a new therapeutic strategy for eradication of CML has been considered for both basic and clinical scientists. miRNAs gene therapy could be a promising therapeutic approach to replace chemotherapy in future.
MicroRNAs (miRNAs) are 19–25 nucleotide noncoding RNAs that play key roles in regulation of the output of many protein-coding genes.11-14 The maturation process of miRNAs occurs inside the cells and matured miRNAs recognize their target mRNAs by matching nucleotides 2 and 8 of the miRNA to the 3′-untranslated region (3′-UTR) of mRNAs.15
It has been shown that BCR-ABL kinase activity induces the down-regulation of miR-31, miR-155, and miR-564 in CML and K562 cell line with BCR-ABL translocation.3
Recently, several studies have focused on miR-155 as a new versatile molecule in cancers. The MIR155 gene is located at chromosome 21q21.3 and encodes for miR-155. It stimulates cell apoptosis by c-Myc and p27 cell signaling in human FLT3-wildtype AML, demonstrating the anti-leukemic role of this microRNA.16 P27Kip1 is a member of the Cip/Kip family of cyclin-dependent kinase (CDK) inhibitors that can arrest the cell cycle in G1 stage. Overexpression of this protein in A549, HeLa, RKO, human melanoma SK-MEL 110, human lung fibroblast (IMR90) and in the rat fibroblast cells resulted in apoptotic cell death.16-18 In support of the p27kip1 function in apoptosis induction, depletion of PCTAIRE1, a cyclin-dependent kinase family protein, stimulates apoptosis of melanoma cells via p27kip1 accumulation.19
In this work, we aimed to overexpress miR-155 in K562 Philadelphia positive CML cell line as an alternative potential approach to aid in the treatment of CML through investigation of its role on the expression of c-MYC and p27 genes.
Materials and Methods
Cell lines and culture
The K562 BCR-ABL positive cell line (obtained from the Pasteur Institute of Iran) was cultured in RPMI-1640 (Gibco-BRL, Eggenstein, Germany) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM glutamine, 1% penicillin and streptomycin and cultured at 37°C in a humidified incubator with 5% CO2.
Plasmids construct and extraction
The pLenti-III-pre mir155-GFP expression vector construct and pLenti-III-blank-GFP (mock) were purchased from ABM Inc. (Applied Biological Materials, Richmond, BC, Canada). E. coli Stbl4 strain harboring the vectors was cultured in LB broth medium with 25 µg/mL kanamycin. The plasmid was extracted with Qiagen plasmid extraction kit (Qiagen, Hilden, Germany).
Transient transfection
Fresh K562 cells were maintained between 0.5–1.5 × 106 cells/mL. The cells were subcultured and 2.0 × 106 cells were transfected with 4 µg pLentiIII-pre mir155-GFP and mock vector using electroporation by Amaxa® Cell Line Nucleofector® Kit V and Lonza Device II (Lonza/Amaxa Biosystems, Walkersville, MD, USA) according to Amaxa™ Optimized Protocol.
RNA extraction and cDNA synthesis
Twenty-four hours after transfection, total RNA was extracted from K562 cell lines by Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Then the quality of RNA was determined by electrophoresis. cDNA of total RNA was synthesized by Fermentaze cDNA Synthesis kit (Fermentaze, Massachusetts, USA).
qPCR for c-Myc and p27kip1 genes expression
c-Myc and p27kip1 were determined as direct and indirect targets of miR-155 respectively via miRNAs target prediction site (http://mirmap.ezlab.org) and according to the study of Palma et al.16 Then, primers of c-Myc genes and p27 gene were designed by Oligo7 and Gen Runner software and GAPDH was used as reference gene (Table 1). Quantitative PCR (qPCR) was performed for genes using Takara SYBR green PCR kit (Takara Bio Inc., Shiga, Japan). Relative expression was calculated for target genes using the ∆∆CT method (n=3).
| Table 1. The list of primers used in qPCR analyses |
|
Primers
|
|
| miR-155 |
Stem-loop RT: GTCGTATGCAGAGCAGGGTCCGAGGTATTC
GCACTGCATACGACACCCCT
Forward: 5´-CGGTTTAATGCTAATCGTGA-3´
Reverse: 5´-GAGCAGGGTCCGAGGT-3´ |
| P27kip1 |
Forward: 5´-GGAGAAGCACTGCAGAGACA-3´
Reverse: 5´-CTCTTGCCACTCGTACTTGC-3´ |
| c-Myc |
Forward: 5´-CGTCTCCACACATCAGCACAA-3´
Reverse: 5´-TCTTGGCAGCAGGATAGTCCTT-3´ |
| GAPDH |
Forward: 5´-TCCACCACCCTGTTGCTGTAG-3´
Reverse: 5´-ACACCCACTCCTCCACCTTTG-3´ |
| Snord47 |
Stem-loop RT: GTCGTATGCAGAGCAGGGTCCGAGGTAT
TCGCACTGCATACGACAACCTC
Forward: 5´-ATCACTGTAAAACCGTTCCA-3´
Reverse: 5´-GAGCAGGGTCCGAGGT-3´ |
cDNA synthesis and stem-loop qPCR of miR-155
Stem-loop primer as well as forward and reverse primers were designed to synthesize cDNA of miR-155 according to Chen et al20 and for qPCR, respectively. Snord47 was selected as reference gene (Table 1). Relative expression was evaluated by the ∆∆CT method (n=3).
Apoptosis assay
K562 cells were transfected with pLentiIII-pre mir155-GFP expression construct and mock vector and cultured for 72 hours in 12 well plates. Annexin V assay was performed by Annexin V-PE/7-AAD eBioscience kit (eBioscience, Inc., CA, USA). The cells were harvested and washed with PBS. Then, 2.5 μL Annexin V-PE was added to 100 μL of the cells suspended in binding buffer and incubated for 15 minutes in dark at room temperature. Next, the cells were washed in binding buffer and 2.5 μL of the 7-AAD solution was added to the cell suspension. Finally, the treated cells were analyzed versus untreated fresh cells by flow cytometry (Partec, Münster, Germany) and FlowMax software (Partec) (n=3).
Western blotting analysis
After 48 hours, K562 cells transfected by pLentiIII-pre mir155-GFP expression vector and mock vector were harvested and washed three times with PBS. Then, cellular proteins were extracted by cell lysis buffer containing 40 mM Tris-HCl (pH 7.4), 7M Urea, 2M Thiourea, 4% CHAPS, 0.2% Biolyte, 50 mM dithiothreitol (DTT), 200 µM PMSF and 1 × protease inhibitor cocktail (Roche, Mannheim, Germany). The cell extracts were run on 12% SDS–polyacrylamide gel and transferred to PVDF membrane (Life Science, Amersham, Braunschweig, Germany). The membrane was blocked overnight in Blocking Buffer at 4°C (2.5% skim milk, 2.5% glycerol and 0.05% tween 20 in TBS buffer). Next, the membrane was incubated overnight with 1:1000 dilution of p27kip1 rabbit primary antibody (Abcam, Inc., Cambridge, MA, USA) at 4°C. 1:3000 dilution of goat secondary antibody (Abcam) was added to membrane and incubated for 1h at room temperature. The p27kip1 protein band was visualized by ECL (Kodak Image Station; New Haven, CT, USA). Densitometry of bands was analysed by ImageJ software (http://rsb.info.nih.gov/ij) (n=3).
Statistical analysis
Statistical data analysis was done using student t test by GraphPad Prism software version 6.01 (La Jolla, California, USA). P < 0.05 was considered statistically significant. All the experiments of this study were carried out in triplicate (n = 3).
Results
Overexpression of miR-155 in K562 cells
To investigate the effects of miR-155 on regulation of c-Myc, p27kip1 and finally on apoptosis in K562 cells, the cells were first transfected with pLenti-III-premiR155-GFP expression vector construct and mock vector by electroporation followed by detection of apoptosis. Fluorescent microscopy confirmed transfection efficacy after 24 hours. Approximately 50% of cells were transfected (data not shown). qPCR shows significantly increased expression of miR-155 in the cells after 24 hours of transfection as shown in Fig. 1. The expression of miR-155 was approximately 20-fold higher relative to mock vector transfected cells (P < 0.001).
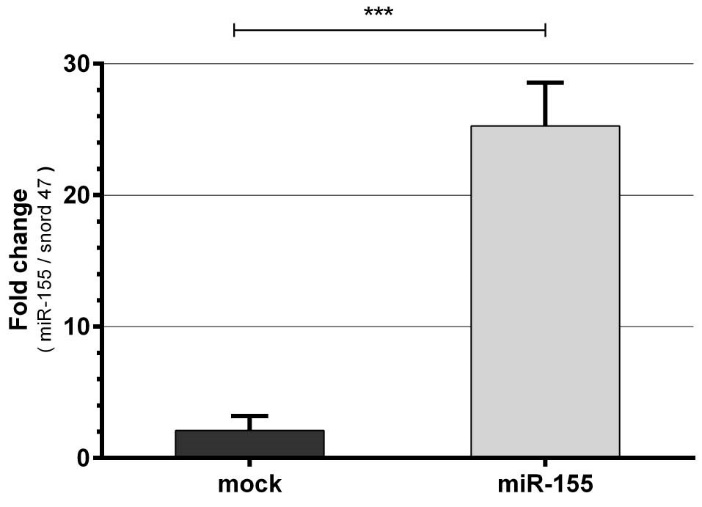
Fig. 1. Confirmation of miR-155 in K562 cells after transfection by qPCR. K562 cells were transfected with plentiIII-pre mir155- GFP expression vector construct or mock. The expression of the miR-155 in the K562 cells transfected with the recombinant vector was considerably higher than mock after 24 hours (Mean ± SD, *** P < 0.001).
miR-155 induces apoptosis in K562 cells
Next, programmed cell death in the K562 cells was evaluated by Annexin V-PE / 7-AAD kit 72 hours post-transfection. As shown in Fig. 2A-2C, the number of apoptotic cells in K562 cell line transfected with pLentiIII-pre mir155-GFP construct was higher than mock transfected cells. The number of apoptotic cells transfected with pLentiIII-pre mir155-GFP construct was about 17% higher than those transfected with mock vector (Fig. 2D).
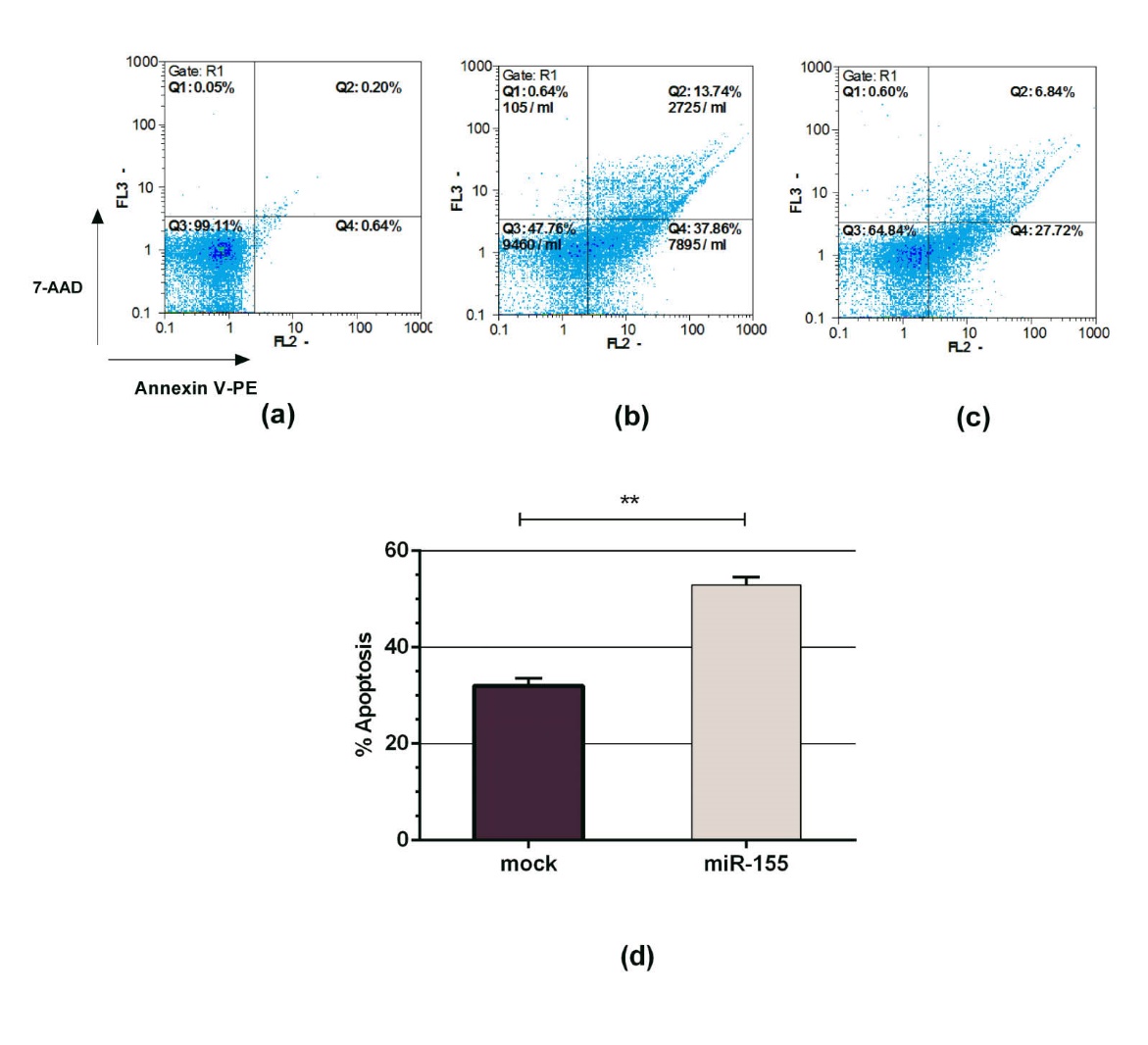
Fig. 2. Effect of miR-155 overexpression on programmed cell death in k562 cells 72 hours post -transfection. The transfected K562 cells were treated with Annexin V-PE/7-AAD and followed by flow cytometry analysis. (A) K562 cells were used as a control. (B) In the K562 cells transfected with plentiIII-pre mir155-GFP construct, approximately 48.6 % of cells became Annexin V-PE / 7-AAD positive. (C) About 34.5 % of K562 transfected cells with pLentiIII-blank-GFP (mock) showed Annexin V-PE/7-AAD positive. (D) Overexpression of miR-155 in K562 cells significantly increased the apoptotic cell death compared to mock (Mean ±SD, ** P < 0.01).
miR-155 upregulates p27kip1 but does not alter c-Myc expression
Twenty-four hours after transfection, the expression level of p27kip1 and c-Myc genes expression were assessed by qPCR. The results indicated that the expression of p27kip1 in K562 cells transfected with pLentiIII-pre mir155-GFP construct was about 1.7-fold higher than the cells transfected with mock vector (P < 0.001) while the expression level of c-Myc was not altered (Fig. 3).
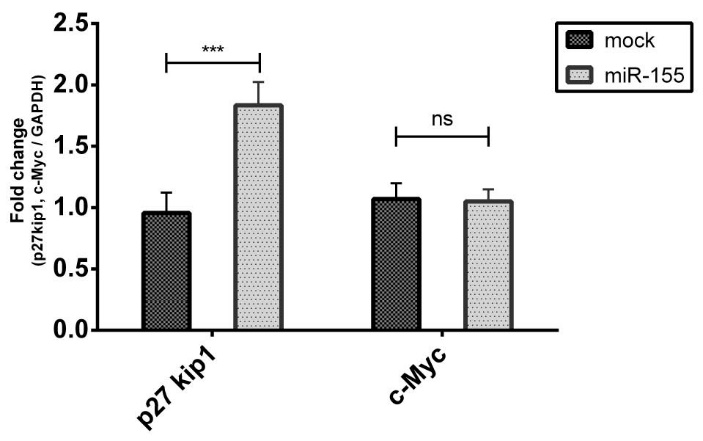
Fig. 3. Expression of p27kip1 and c-Myc genes by qPCR. K562 cells transfected either with the plentiIII-premir155-GFP construct or mock vector followed by expression evaluation of p27kip1 and c-Myc genes 24 hours after transfection. Overexpressed miR-155 resulted in upregulation of p27kip1 expression but caused no alteration in c-Myc expression. (Mean ± SD, *** P < 0.001)
Next, the expression of p27kip1 was evaluated by western blot analysis at protein level 48h post transfection. As shown in Fig. 4A and 4B the expression of p27kip1was upregulated in K562 cells transfected with pLentiIII-premir155-GFP expression vector construct.
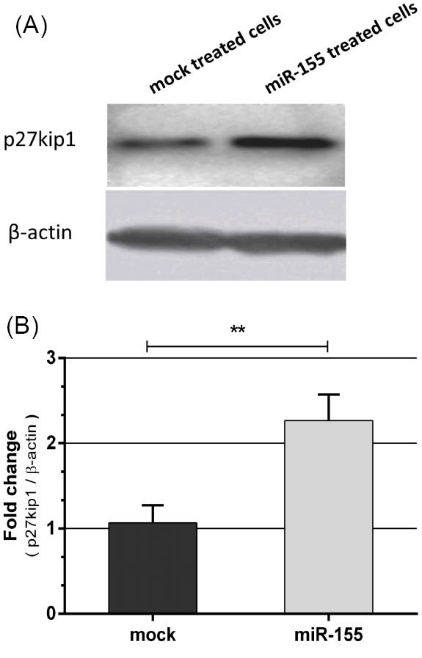
Fig. 4. (A) Western blot analysis of p27kip1 protein expression in K562 cells transfected by either plentiIII-premir155-GFP construct or mock vector 48 hours post-transfection. miR-155 upregulated p27kip1 protein. (B) Densitometry analysis of bands by ImageJ software indicated upregulation of nearly 2.4-fold. β-actin was used as loading control ( Mean ±SD, **P < 0.01).
Discussion
High proliferation and reduced apoptosis are well-known features of CML cells. The apoptotic defect is a major challenge, and limits the efficacy of cancer chemotherapy.3,9 On the other hand, side effects of chemotherapeutic agents are other concerns.21 The mi-RNAs like miR-155 play a crucial role in cell fate as small potent molecules with different targets. Rokah et al demonstrated that Philadelphia chromosome in CML cells can downregulate the expression of miR-155. In another study, Palma et al showed that the upregulation of this microRNA in AML cells results in apoptosis through interfering in the regulation of c-Myc and p27kip1profiles.3,16 In this study, we evaluated the potential anti-cancer effect of miR-155 as a promising approach to aid in the treatment of CML.
Electroporation technique, which is a versatile method in terms of efficiency of transfection, was used to transfect K562 cells.22 Our results revealed that about 50% of the transfected cells express GFP and thereby dramatically induce miR-155.
However, one of the disadvantages of electroporation method could be cytotoxic effects.23
c-Myc is one of the direct targets of miR-155 (http://mirmap.ezlab.org ) and upregulation of miR-155 in CML cells leads to downregulation of c-Myc and it is followed by upregulation of p27kip1 and apoptosis.16
Our finidings revealed that apoptosis was significantly induced in K562 cells following overexpression of miR-155. However, the number of apoptotic cells in the K562 cell line transfected with mock vector was also high, which may be due to electroporation method.
Our results also showed that overexpression of miR-155 resulted in upregulation of p27kip1.
Palma et al assessed the miR-155 expression pattern in AML cells. They reported that, the patients with Fms-like tyrosine kinase 3 (FLT3)-wild type AML had miR-155 expression levels like normal bone marrow. Administration of cytarabine arabinoside and 1, 23-dihydroxy vitamin D3 in FLT3-wild type AML cells induced apoptosis and myelomonocytic differentiation, respectively. Both drugs led to increased miR-155 expression level. A growing body of evidence suggests the anti-leukaemic role for miR-155 in AML cells. Palma et al also reported that overexpression of miR-155 led to downregulation of c-Myc and MEIS1/GFI1 and finally to phosphorylation of JNK or the activation of p27/kip1.16 Interestingly, consistent with the findings reported by Palma et al, our results indicated that the apoptotic role of miR-155 is in contrast with its oncogenic effects.24,25 In our study, contradicting Palma et al’s findings (Fig. 5), the overexpression of miR-155 did not alter the expression level of c-Myc.
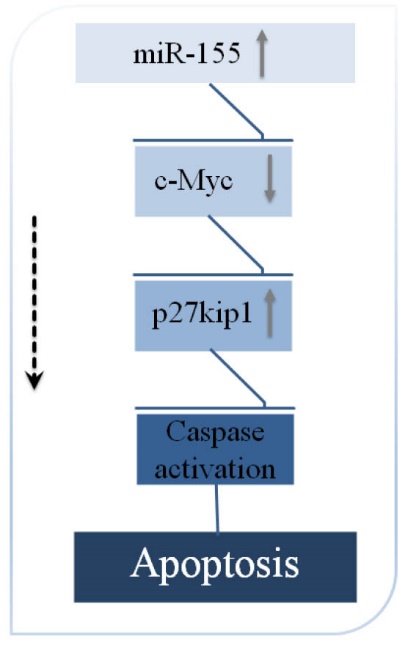
Fig. 5. Upregulation of miR-155 caused downregulation of c-Myc. This potentially leads to upregulation of p27/kip1 and finally activation of the apoptosis cascade.
Rokah et al identified miRNAs expression profile of CML cell lines by using miRNA microarrays and miRNA qPCR. Their results revealed that miR-31, miR-155 and miR-564 were downregulated in CML cells.3
Li et al reported that the upregulation of cell cycle-dependent kinase inhibitors (CDKIs) p21 and p27 in K562 cell line through overexpression of miR-29b suppressed cell growth and colony formation ability and induced apoptosis. They had previously shown that the expression of miR-29b in CML patient samples was considerably low.26
Lu et al investigated the miRNAs expression in human monocytes, mature dendritic cells, and immature dendritic cells. They showed that the downregulation of miR-221 and the overexpression of miR-155 could lead to the upregulation of p27kip1 and resulted in DCs apoptosis.27
Conclusion
In this study, we propose a new potential anticancer agent in CML cell line, i.e. miR -155. We showed that overexpression of miR-155 can induce apoptosis in k562 cell line via upregulation of p27Kip1. Although signaling for apoptosis occurs through multiple independent pathways, our focus was on c-Myc/p27 kip1 signaling pathway. However, further and complementary studies are required to address some other aspects, including in vivo studies and safety issues.
Authors' contribution
SA and MK contributed essential reagents or tools, designed the research study, revised and approved the submitted and final versions. AsA and AmA contributed essential reagents or tools, drafted and approved the submitted and final versions. MEF performed the research, designed the research study, analyzed the data, wrote the paper, and approved the submitted and final versions. MS designed the research study, drafted and revised the paper and approved the submitted and final versions.
Competing interests
The authors declare that there are no competing interests.
Ethical approval
There is no ethical approval for the present study.
Acknowledgment
This study was a Ph.D. thesis financially supported by Tehran University of Medical Sciences (grant number: 93-04-31-27576). Also, the authors would like to acknowledge the cooperation of Pasteur Institute of Iran, which supported all parts of the experiments.
References
- Pasternak G, Hochhaus A, Schultheis B, Hehlmann R. Chronic myelogenous leukemia: molecular and cellular aspects. J Cancer Res Clin Oncol 1998; 124: 643-60.
- Hehlmann R, Hochhaus A, Baccarani M. Chronic myeloid leukaemia. Lancet 2007; 370: 342-50. doi:10.1016/S0140-6736(07)61165-9. [Crossref]
- Rokah OH, Granot G, Ovcharenko A, Modai S, Pasmanik-Chor M, Toren A, et al. Downregulation of miR-31, miR-155, and miR-564 in chronic myeloid leukemia cells. PloS One 2012; 7: e35501. doi:10.1371/journal.pone.0035501. [Crossref]
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl j Med 2001; 2001: 1031-7. doi:10.1056/NEJM200104053441401. [Crossref]
- Novartis Pharma A. Gleevec (imatinib mesylate) tablets prescribing information. East Hanover, NJ, USA 2006.
- Druker BJ, O’Brien SG, Cortes J, Radich J. Chronic myelogenous leukemia. ASH Education Program Book 2002; 2002: 111-35.
- Baccarani M, Deininger MW, Rosti G, Hochhaus A, Soverini S, Apperley JF, et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood 2013; 122: 872-84. doi:10.1182/blood-2013-05-501569. [Crossref]
- Maharsy W, Aries A, Mansour O, Komati H, Nemer M. Ageing is a risk factor in imatinib mesylate cardiotoxicity. Eur J Heart Fail 2014; 16: 367-76. doi:10.1002/ejhf.58. [Crossref]
- Nimmanapalli R, Fuino L, Bali P, Gasparetto M, Glozak M, Tao J, et al. Histone deacetylase inhibitor LAQ824 both lowers expression and promotes proteasomal degradation of Bcr-Abl and induces apoptosis of imatinib mesylate-sensitive or-refractory chronic myelogenous leukemia-blast crisis cells. Cancer Res 2003; 63: 5126-35.
- Alizadeh S, Azizi SG, Soleimani M, Farshi Y. The Role of MicroRNAs in Myeloproliferative Neoplasia. Int J Hematol Oncol Stem Cell Res 2016.
- Sidorkiewicz M, Grek M, Jozwiak B, Majda-Stanislawska E, Piekarska A, Bartkowiak J. Expression of microRNA-155 precursor in peripheral blood mononuclear cells from Hepatitis C patients after antiviral treatment. Acta Virol 2010; 54: 75-8.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281-97.
- Tang B, Xiao B, Liu Z, Li N, Zhu E-D, Li B-S, et al. Identification of MyD88 as a novel target of miR‐155, involved in negative regulation of Helicobacter pylori‐induced inflammation. FEBS Lett 2010; 584: 1481-6. doi:10.1016/j.febslet.2010.02.063. [Crossref]
- Kouhkan F, Hafizi M, Mobarra N, Mossahebi-Mohammadi M, Mohammadi S, Behmanesh M, et al. miRNAs: a new method for erythroid differentiation of hematopoietic stem cells without the presence of growth factors. Appl Biochem Biotechnol 2014; 172: 2055-69. doi:10.1007/s12010-013-0633-0. [Crossref]
- Elton TS, Selemon H, Elton SM, Parinandi NL. Regulation of the MIR155 host gene in physiological and pathological processes. Gene 2013; 532: 1-12. doi:10.1016/j.gene.2012.12.009. [Crossref]
- Palma CA, Al Sheikha D, Lim TK, Bryant A, Vu TT, Jayaswal V, et al. MicroRNA-155 as an inducer of apoptosis and cell differentiation in Acute Myeloid Leukaemia. Mol Cancer 2014; 13: 1. doi:10.1186/1476-4598-13-79. [Crossref]
- Woltman AM, van der Kooij SW, Coffer PJ, Offringa R, Daha MR, van Kooten C. Rapamycin specifically interferes with GM-CSF signaling in human dendritic cells, leading to apoptosis via increased p27KIP1 expression. Blood 2003; 101: 1439-45. doi:10.1182/blood-2002-06-1688. [Crossref]
- Wang X, Gorospe M, Huang Y, Holbrook NJ. p27Kip1 overexpression causes apoptotic death of mammalian cells. Oncogene 1997; 15: 2991-7. doi:10.1038/sj.onc.1201450. [Crossref]
- Yanagi T, Reed J, Matsuzawa SI. PCTAIRE1 regulates p27 stability, apoptosis and tumor growth in malignant melanoma. Oncoscience 2014; 1: 624. doi:10.18632/oncoscience.86. [Crossref]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res 2005; 33: e179-e. doi:10.1093/nar/gni178. [Crossref]
- Gurung S, Pandey RA. Perception of Side Effects of Chemotherapy among Cancer Patients in BP Koirala Memorial Cancer Hospital Bharatpur, Nepal. Journal of College of Medical Sciences-Nepal 2016; 11: 14-9. doi:10.3126/jcmsn.v11i4.14319. [Crossref]
- Hamm A, Krott N, Breibach I, Blindt R, Bosserhoff AK. Efficient transfection method for primary cells. Tissue Eng 2002; 8: 235-45. doi:10.1089/107632702753725003. [Crossref]
- Lepik D, Jaks V, Kadaja L, Värv S, Maimets T. Electroporation and carrier DNA cause p53 activation, cell cycle arrest, and apoptosis. Anal Biochem 2003; 318: 52-9. doi:10.1016/S0003-2697(03)00135-0. [Crossref]
- Sandhu SK, Volinia S, Costinean S, Galasso M, Neinast R, Santhanam R, et al. miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Eµ-miR-155 transgenic mouse model. Proc Natl Acad Sci U S A 2012; 109: 20047-52. doi:10.1073/pnas.1213764109. [Crossref]
- O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med 2008; 205: 585-94. doi:10.1084/jem.20072108. [Crossref]
- Li Y, Wang H, Tao K, Xiao Q, Huang Z, Zhong L, et al. miR-29b suppresses CML cell proliferation and induces apoptosis via regulation of BCR/ABL1 protein. Exp Cell Res 2013; 319: 1094-101. doi:10.1016/j.yexcr.2013.02.002. [Crossref]
- Lu C, Huang X, Zhang X, Roensch K, Cao Q, Nakayama KI, et al. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood 2011; 117: 4293-303. doi:10.1182/blood-2010-12-322503. [Crossref]