Bioimpacts. 7(1):41-47.
doi: 10.15171/bi.2017.06
Original Article
Time dependency of morphological remodeling of endothelial cells in response to substrate stiffness
Zahra Goli-Malekabadi 1, Mohammad Tafazzoli-Shadpour 1, *, Ali Tamayol 2, *, Ehsan Seyedjafari 3
Author information:
1Biomedical Engineering Department, Amirkabir University of Technology, Tehran, Iran
2Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA, USA
3Department of Biotechnology, College of Science, University of Tehran, Tehran, Iran
Abstract
Introduction:
Substrate stiffness regulates cellular behavior as cells experience different stiffness values of tissues in the body. For example, endothelial cells (ECs) covering the inner layer of blood vessels are exposed to different stiffness values due to various pathologic and physiologic conditions. Despite numerous studies, cells by time span sense mechanical properties of the substrate, but the response is not well understood. We hypothesized that time is a major determinant influencing the behavior of cells seeded on substrates of varying stiffness.
Methods:
We monitored cell spreading, internal structure, 3D topography, and the viability of ECs over 24 hours of culture on polydimethylsiloxane (PDMS) substrates with two different degrees of elastic modulus.
Results:
Despite significant differences in cell spreading after cell seeding, cells showed a similar shape and internal structure after 24 hours of culture on both soft and stiff substrates. However, 3D topographical images confirmed existence of rich lamellipodia and filopodia around the cells cultured on stiffer PDMS substrates.
Conclusion:
It was concluded that the response of ECs to the substrate stiffness was time dependent with initial enhanced cellular spreading and viability on stiffer substrates. Results can provide a better comprehension of cell mechanotransduction for tissue engineering applications.
Keywords: 3D topography, HUVECs, Morphology, Substrate stiffness, Time dependency
Copyright and License Information
© 2017 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Biological cells are capable of sensing and reacting to their environmental conditions such as substrate stiffness. The nature of adherent cells allows them to feel various physical signals via ligand-receptor interactions when they spread on a surface.1 This process occurs by force exertion and elastic reaction of the substrate, and subsequently translation of mechanical signals into biochemical signals that is called mechanotransduction. 2 Within the human body, there are various hard and soft tissues with a wide range of stiffness moduli, from 10 GPa for bone to 3 kPa for liver.3,4 Hence, cells experience in vivo environments with diverse stiffness values that directly influence mechanotransductive events.
Endothelial cells (ECs) form a lining on the wall of blood vessels, regulate the diffusion of compounds within blood to the tissues, and inhibit blood clotting. Endothelial dysfunction is a major source of cardiovascular diseases which are the main cause of human mortality.5 Within the circulatory system, ECs are exposed to different stiffness values in physiological and pathological conditions that directly influence their functionality.6 Some cardiovascular diseases such as atherosclerosis and hypertension are correlated with stiffening of blood vessels.7,8 Furthermore, blood vessels stiffen with aging.6 It has been reported that endothelial function and morphology can change in response to variations of the intimal membrane.5 In vitro experiments also demonstrated the change in endothelial function cultured on substrates with different stiffness values suggesting the possibility of using such models for studying disease pathology and its effect on ECs formed barrier.9,10
To examine effects of substrate stiffness on cell behavior, there are several materials as culture substrates such as polyacrylamide11 and collagen.12 Polydimethylsiloxane (PDMS) is a widely used material, which is nontoxic, biocompatible, and offers tunable stiffness, surface plasticity, transparency, and flexibility.13-15 The elastic modulus of PDMS can be easily tuned from 0.1 kPa to 2 MPa by manipulation of base monomer: cross-linker ratio. As other chemical and physical properties of the surface including the roughness and hydrophobicity can also change cell behavior, results have confirmed using this method just change elastic modulus of PDMS.11,16,17 Hence, different studies have used PDMS substrates to study effects of substrate stiffness on cellular behavior. Surface stiffness can influence multiple cell behavioral parameters such as morphology,18-20 proliferation,16,19,20 migration,20 viability,18 attachment16 and cell differentiation.20 It has been indicated that when cells were seeded on soft and stiff substrates, the actin filaments were often observed to be thick and thin respectively. On the other hand, cells appeared small and rounded, and lost most of their stress fibers on softer substrates.17-19 Study of cancerous cells cultured on PDMS substrates with two distinct elastic moduli demonstrated changes in the morphological parameters by substrate rigidity, while other chemical and physical properties of the substrates were kept the same.21
Despite, many investigations regarding effects of substrate stiffness on cell behaviors, the time dependent mechanism by which cells sense and respond to their substrate stiffness has not been well studied. Here, we hypothesized that endothelial responses to the substrate stiffness are time dependent. To test this hypothesis, we investigated morphological alterations of ECs in response to the rigid and soft PDMS substrates in specific time points of 24-hour culture. The elongation of cells due to altered substrate stiffness is an important morphological parameter that can affect major cell behavioral parameters such as adhesion and motility.22 This morphological feature is the primary kinetic process following cell attachment events when cells sit on the substrate.23
Methods and Materials
Materials
Monomer and Cross-linking agent (Sylgard 184®), high-glucose Modified Eagle’s Medium, Fetal Bovine Serum, Fibronectin, Triton X-100, Alexa Fluor 488® phalloidin, 4’-6-Diamidino-2-phenylindole (DAPI) and LIVE/DEAD® viability/cytotoxicity kit were obtained from Dow Corning (Midland, Michigan, USA), Gibco (New York, NY, USA), Gibco (New York, NY, USA), Sigma (St. Louis, Missouri, USA), Merck (Darmstadt, Germany), Invitrogen (Waltham, Massachusetts, USA), Invitrogen (Waltham, Massachusetts, USA) and Invitrogen (Waltham, Massachusetts, USA) respectively. Human umbilical vein endothelial cells (HUVECs) were provided from National Cell Bank of Iran, Pasteur Institute of Iran.
PDMS fabrication
PDMS films of varying Young’s moduli were synthesized following manufacturer's protocol. Briefly, monomer and cross-linking agent were thoroughly blended at the ratios of 10:1 and 50:1. The mixtures were kept in a desiccator for 1 hour for removal of the trapped air bubbles. Then, films were cured at 80°C for 1 hour.
Characterization of elastic modulus of substrates
For the evaluation of the stiffness, uniaxial tensile tests were performed on three standard PDMS sheets of each group in dimension of 100 mm, 20 mm and 0.5 mm for sheet’s length, width and thickness respectively. Briefly, each sample was clamped with tensile device’s clips and stretched in the longitudinal direction. The force-displacement data were acquired and considering the dimensions of samples, stress (F/A) – strain (L/∆L) data were calculated. Here, F, A, L and ∆L are force, cross-section area (width * thickness), original length and the change in length respectively.
Cell culture
HUVECs were cultured in high-glucose modified Eagle’s medium enriched by 10% fetal bovine serum. Cells were passaged once the culture was less than 70%. Cells were used with the passage number of less than 6 throughout the experiments.
Fibronectin coating and cell seeding
To follow up the morphology of HUVECs during 24 hours of culture, a region of the elastic membrane was specified by markers for corresponding image capturing during time intervals. This region was coated by 2 µg/mL fibronectin and incubated at 37°C for 1 hour for surface modification. After removing redundant fibronectin, cells were cultured with the density of 104 cells/cm2.
Image processing and cell elongation evaluation
To quantify alterations in the cell elongation on substrates with different elastic moduli, cell images were captured and processed during culture period. At least 5 images were taken from each sample every 4 hours using an optical phase-contrast inverted microscope. Image J (v1.43e) software was utilized for calculation of morphological parameters. The image processing steps included conversion of captured images to gray scale, separation of cells from their background, and finally generation of binary images. Then, the processed images were used to quantify alterations in the cell circularity (CC) determined for each cell according to Eq. (1),24
(1)
Where P indicates cell perimeter and S describes the area of the cell. The magnitude of circularity parameter is within the range of 0 to 1 representing a line and a circle respectively. When cells become elongated their corresponding circularity value decreases. The cells of each image were clustered in four groups based on cell circularity named spindle (CC: 0.1 to 0.299), semi-spindle (CC: 0.3 to 0.499), semi-round (CC: 0.5 to 0.699) and round (CC: 0.7 to 0.999).
Actin staining
After 24 hours of cell seeding, the internal cytoskeletal structure of cells was stained and displayed by an inverted fluorescence microscope. Cells were washed twice with phosphate buffered saline (PBS) and rinsed with 4% paraformaldehyde diluted in PBS for the fixation. After 10 minutes, cells were washed by PBS again and permeabilized by 0.1% Triton X-100 in PBS for 15 minutes. Cells were then further washed with PBS carefully. For blocking process, samples were incubated for 1 hour with 1% BSA in PBS. Then, F-actin fibers were stained with 1/40 dilution of Alexa Fluor 488® phalloidin in PBS for 45 minutes. Finally, cells were washed with PBS twice. In order to visualize cell nuclei, cells were rinsed in 1/1000 diluted 4’-6-Diamidino-2-phenylindole (DAPI) for 5 minutes. Cell images were then captured by an inverted fluorescence microscope (Nikon TE 2000-U, Nikon instruments Inc., USA).
Fractal dimension calculation
Fractal dimension is an indicator of morphological complexity. This quantitative measurement provides an estimation of cell structure properties, especially their alignment.25,26 The lower fractal dimension indicates higher arrangement.The arrangement of actin filaments was examined using fractal analysis tool of Image J software (v1.43e). Initially, actin staining images were processed by the software as mentioned above. Then, fractal dimension of each image was calculated using ‘‘box counting” method as reported by other studies.24
Scanning electron microscopy
A 3D morphological topography of cells was implemented using SEM (SERON TECHNOLOGIES, AIS2100). Initially, cells were rinsed in 2.5% glutaraldehyde for 20 minutes at room temperature. Samples were then dehydrated in different dilution series of ethanol from 50 to 100%, each step 10 minutes. Subsequently, the substrates were coated by 20 nm of gold for making them electrically conductive and avoiding electric charging (20 kV) during imaging.
MTT assay
The metabolic activity of cells seeded on both substrates was quantified by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium (MTT) assay. Briefly, cells were incubated at 37◦C with a 0.5 mg/mL concentration of MTT solution for 4 hours. Then, the solution was removed and cells were rinsed in 150 µL of dimethylsulphoxide (DMSO) solution for up to 15 minutes. Finally, the absorbance was obtained at 570 nm by an ALISA reader.20,27
Experimental protocol
HUVECs were cultured on two PDMS membranes with stiffness moduli of 2MPa and 50KPa as stiff and soft substrates and their morphology, cytoskeleton, topography, and cell viability of two groups were examined. Cell imaging was performed 4, 8, 12 and 24 hours after culture on specified regions. For each sample, five images were captured and analyzed. All experiments were repeated at least three times. T-test analysis was conducted with significance level set at P= .05 for comparing parameters between both groups.
Results
Elastic modulus of substrates
Fig. 1 shows stress-strain graphs for one sample of each group of substrates. The slope of the line representing stress-strain data determines elastic modulus of the sample and has direct relation to the stiffness of the substrate. Softer samples show lower slopes describing higher extensibility under the same applied force. For all test samples, straight lines were well fitted by the data (with correlation values higher than 0.99) and the corresponding average values of elastic moduli were 2000 ± 0.31 kPa and 50 ± 6 kPa for stiff and soft substrates respectively.
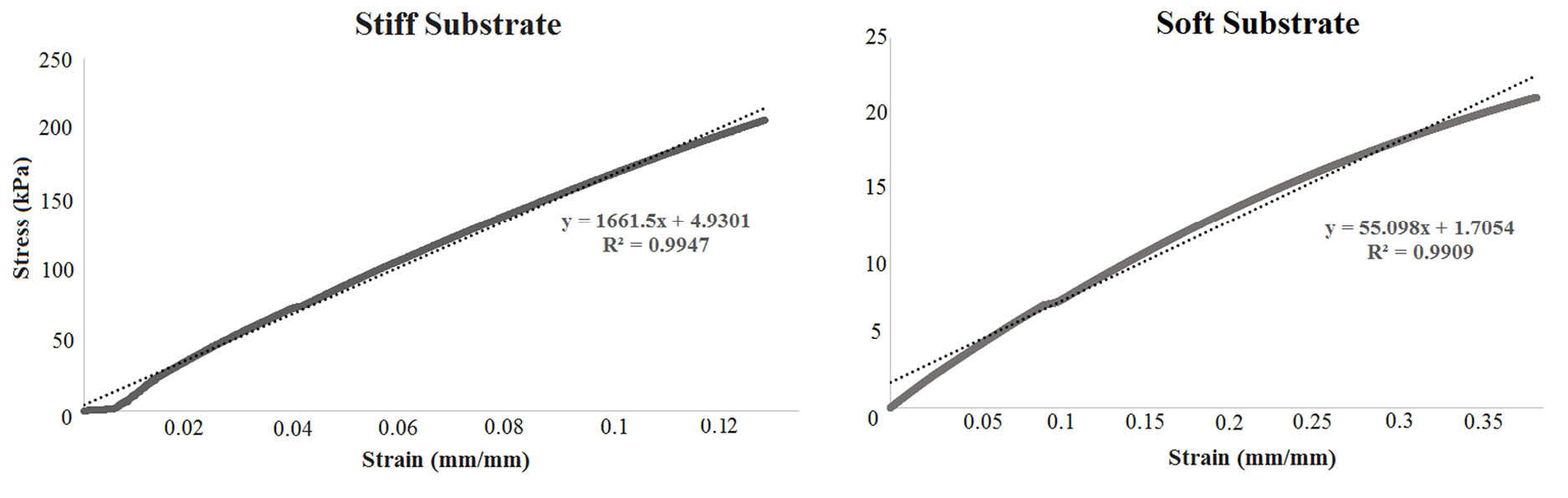
Fig. 1.
The stress-strain data for typical samples of stiff (left) and soft (right) substrates. The slope of the fitted line is higher for the stiff substrate, indicating higher elastic modulus. Conversely, soft substrate has less slop and elastic modulus.
.
The stress-strain data for typical samples of stiff (left) and soft (right) substrates. The slope of the fitted line is higher for the stiff substrate, indicating higher elastic modulus. Conversely, soft substrate has less slop and elastic modulus.
Cell circularity
Fig. 2A shows morphology of cells after 8 and 24 hours of culture on stiff and soft substrates. Qualitatively more cells were elongating on stiffer surface, unlike cells on the softer substrate that mostly remained round. Quantitative cell morphological clustering, i.e., the percentage of each morphological group, is presented in Fig. 2B. Results demonstrated that despite various magnitudes of cell circularity in early hours of cell seeding for two test groups, after 24 hours almost 50% of cells were spindle and there was no significant difference between total percentages of semi-spindle and semi-round cells on 2 types of substrates (P> 0.05). Virtually no cells remained in round shape after 24 hours within 2 groups.
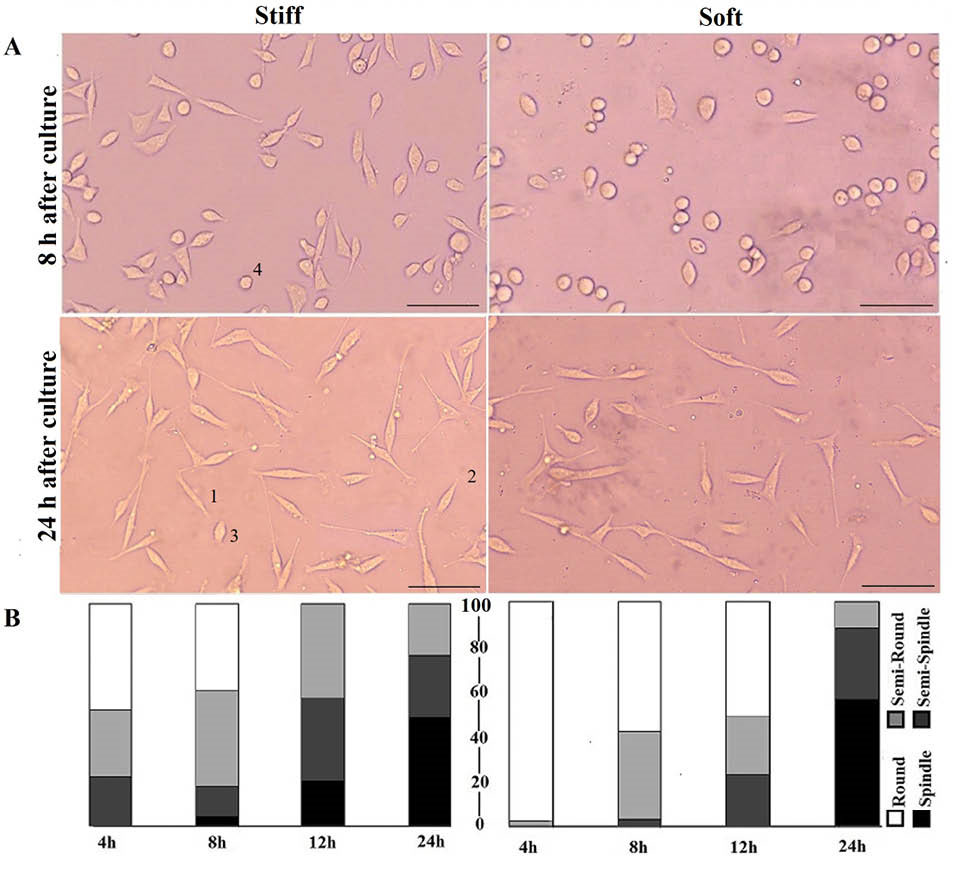
Fig. 2.
Cell elongation on stiff and soft substrates. A) The morphology of cells 8 and 24 h after seeding. Cells labeled by 1, 2, 3 and 4 are spindle, semi-spindle, semi-round and round respectively. (Scale bar: 50 µm), B) Quantitative presentation of cell circularity (CC) at four time periods; the numbers are an average of CC of each cluster. Despite statistical significance between clusters of two groups at initial hours, there is no marked difference after 24 h of culture. Vertical axis shows percentage of each cluster. The numerical data for different clusters are shown in Table 1.
.
Cell elongation on stiff and soft substrates. A) The morphology of cells 8 and 24 h after seeding. Cells labeled by 1, 2, 3 and 4 are spindle, semi-spindle, semi-round and round respectively. (Scale bar: 50 µm), B) Quantitative presentation of cell circularity (CC) at four time periods; the numbers are an average of CC of each cluster. Despite statistical significance between clusters of two groups at initial hours, there is no marked difference after 24 h of culture. Vertical axis shows percentage of each cluster. The numerical data for different clusters are shown in Table 1.
Our results suggested that cells on both stiff and soft surfaces had a tendency to become elongated during seeding time though with different rates. Cells on stiffer substrate showed a higher rate of elongation in initial hours of culture, while in later hours of culture elongation of cells on soft substrates was accelerated with higher rate compared to stiffer substrates. Although stiffer substrates initially had fewer round cells, it was observed that cells on soft surfaces had a tendency of rapid elongation after 8 hours of culture compared to cells on stiff substrate. In other words, the response of cells on stiff substrates through morphological remodeling started earlier, however cell remodeling on soft substrates started later and progressed more rapidly. The CC results are presented in Table 1 by numbers.
Table 1.
Quantitative presentation of cell circularity (CC) at 4 time pointsa
|
Time points
|
Round (%)
|
Semi-round (%)
|
Semi-spindle (%)
|
Spindle (%)
|
| 4 h (Stiff) |
47.5±4 |
30±4 |
22.5±3 |
0 |
| 4 h (Soft) |
97.8±2 |
2.2±1 |
0 |
0 |
| 8 h (Stiff) |
38.6±5 |
43±2 |
14±2 |
4.4±2 |
| 8 h (Soft) |
57.7±8 |
39.4±5 |
2.8±1 |
0 |
| 12 h (Stiff) |
0 |
42.1±4 |
36.8±6 |
21±4 |
| 12 h (Soft) |
51±3 |
25.5±4 |
23.5±5 |
0 |
| 24 h (Stiff) |
0 |
23.1±2 |
27.7±1 |
49.2±5 |
| 24 h (Soft) |
0 |
11.7±3 |
31.7±5 |
56.7±8 |
aThe sign ± indicates standard deviation of each cluster.
Actin arrangements
The internal structure of cells cultured on 2 substrates was observed 24 hours after seeding. As described in Fig. 2, cells remodeled themselves in response to seeding on substrates with 2 elastic moduli by elongation on the surfaces differently during first day of culture. After 24 hours of culture, cells reached almost similar percentages of elongated morphology for both types of substrates. Results of the actin staining confirmed such similarity through comparable internal structure arrangement of cells among 2 test groups (Fig. 3A). The number of actin fibers and bundles of stress fibers remained virtually the same among semi-round and spindle cells (Fig. 3A). As Fig. 3B shows, the fractal dimension of different clusters is the same between 2 groups. In other words, actin structure of the cells has the same complexity after 24 hours of culture and the substrate stiffness did not affect them.
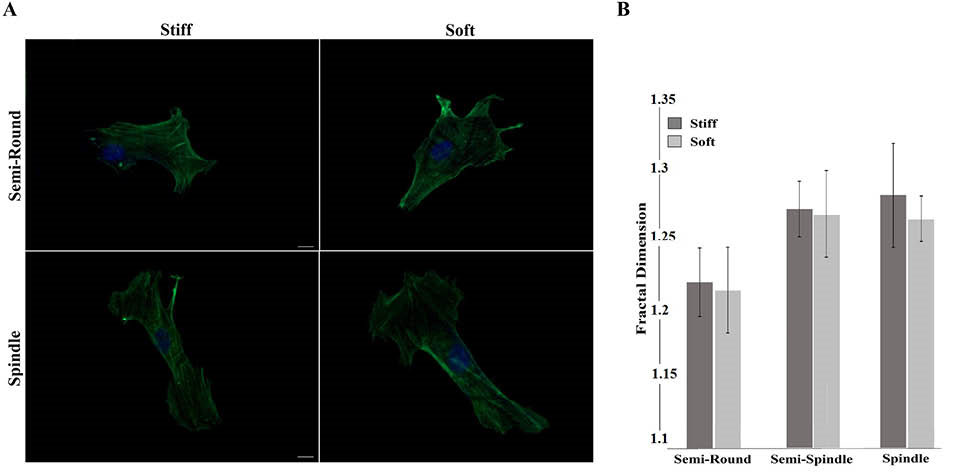
Fig. 3.
Quantitative and qualitative presentation of the actin fibers arrangement 24 h after seeding on stiff and soft substrates A) Fluorescent images, And B) Fractal dimension calculation of actin structure. As the images show, there is no statistical difference among the clusters in each group (Scale bar: 5 µm).
.
Quantitative and qualitative presentation of the actin fibers arrangement 24 h after seeding on stiff and soft substrates A) Fluorescent images, And B) Fractal dimension calculation of actin structure. As the images show, there is no statistical difference among the clusters in each group (Scale bar: 5 µm).
Scanning electron microscopy
To visualize 3D cellular topography, cell images were taken by SEM after 24 hours of seeding. Fig. 4 shows 3D images of semi-round and spindle cells after 24 hours of seeding on soft and stiff substrates. Unlike internal structure shown by actin staining (Fig. 3), there were clear differences between 3D images of cells cultured on PDMS of varying stiffness. Although HUVECs became elongated after 24 hours of culture (no significant difference between CC values of cells on 2 substrates after 24 hours), cells on stiff substrate had many lamellipodia and filopodia that tightly connected them to the substrate. Soft matrix did not enable cells to anchor to the substrate firmly.
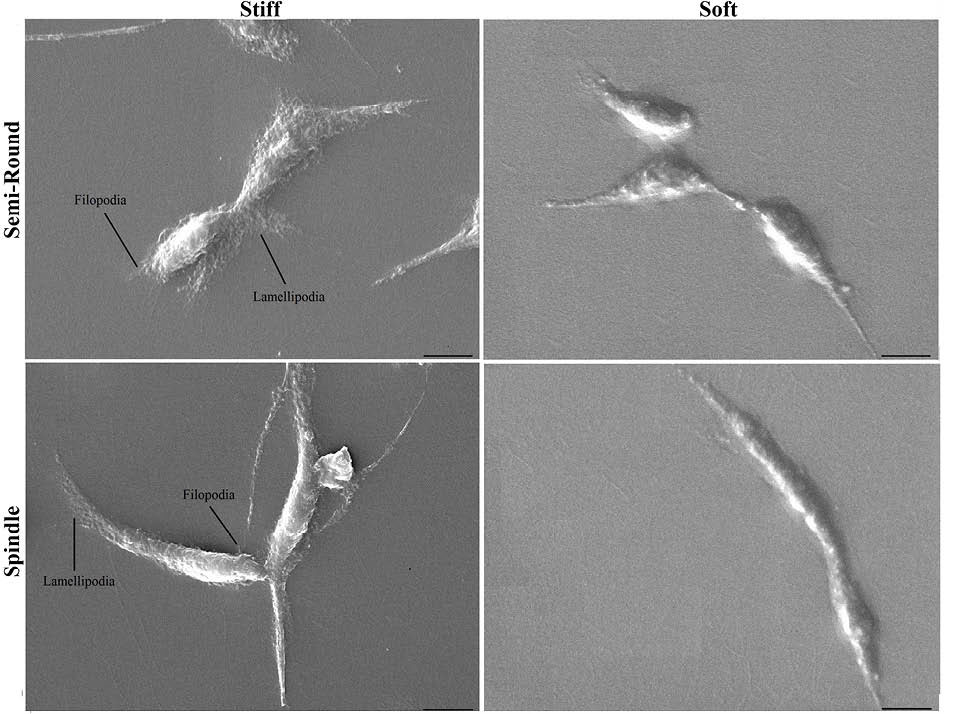
Fig. 4.
Three dimensional topography of cells by SEM after 24 h of culture on stiff and soft substrates for semi-round and spindle cell shapes. (Scale bar: 10 µm).
.
Three dimensional topography of cells by SEM after 24 h of culture on stiff and soft substrates for semi-round and spindle cell shapes. (Scale bar: 10 µm).
Cell viability
To better quantify the function of cells cultured on different substrates, the metabolic activity of cells was further measured using MTT assay. The metabolic activity of cells cultured on stiffer substrates was higher, in agreement with the live/dead assay data (Fig. 5).
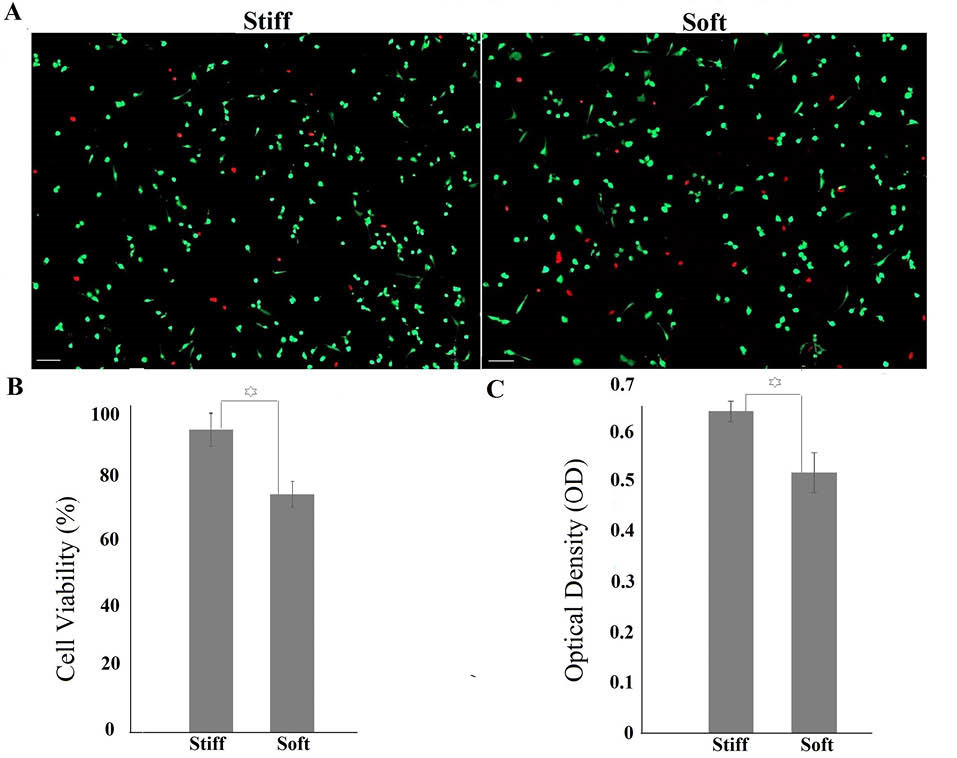
Fig. 5.
Metabolic activity of cells measured by MTT assay. OD number has direct relation to metabolic activity. Increased elastic modulus enhanced metabolic activity of cells. The star sign shows significant difference between two groups.
.
Metabolic activity of cells measured by MTT assay. OD number has direct relation to metabolic activity. Increased elastic modulus enhanced metabolic activity of cells. The star sign shows significant difference between two groups.
Discussion
The interaction between cells and their mechanical environment guides cellular responses that play essential roles in controlling their behavior and is addressed by mechanotransduction.1,2 Elastic modulus, as the major mechanical feature of the cell substrate among in vitro experiments, may simulate the mechanical condition of tissues in which biological cells function in vivo. Hence, in vitro study of cellular responses to the substrate stiffness expands the understanding of cell remodeling due to environmental stimuli and assists in obtaining cells with optimized function for tissue engineering and cell therapy applications. It appears that the opposing yet sustained results between various studies might be due to differences among test conditions and experimental protocols including surface protein coating, hydrophobic surface correction by plasma treatment, fabrication technique, utilized ranges of PDMS elastic moduli, cell type, cell crowdedness and/or proliferation, and checked time points. It seems that strong cell signaling through substrate occurs when suitable culture conditions trigger cell remodeling in specific time span.
As an example, an adequate density of cells (depending on cell phenotype) is required to cause cell signaling between adjacent cells due to substrate stiffness.28 Both highly confluent or disperse cell population reduce effects of proper signaling through substrate.28 Moreover, surface conditioning in terms of method of coating and thickness of surface protein might be another determinant. Thick coating layer might result in lack of sense of substrate stiffness by cells.21 Hence, despite the fact that hydrophobic nature of PDMS requests an intermediate protein for cell attachment,16 in some cases even the usage of PDMS substrate without coating has been suggested to properly investigate effects of bulk stiffness on cell behavior.21 Additionally, it has been argued that some surface modifications such as plasma treatment can create a thin layer with different elastic behavior from the bulk of substrate which might undermine effect of substrate properties on cell behavior.29 Here we coated substrates with a very thin layer of fibronectin to allow cell interact with the substrates and sense their stiffness.
Some studies have used other materials such as polyacrylamide as the cell substrate and concluded that PDMS is not a proper material for study of substrate stiffness since it does not effectively influence cellular behavior.11 However, as a holistic view, it can be hypothesized that each substrate can modulate behavior of cells provided that culture conditions including checked time points, cell density, surface modification and fabrication technique are adequately balanced. Furthermore, studies which concluded no effects of substrate stiffness on cell morphology, did not examine cell elongation among the initial hours of seeding, which is a major determinant of cell behavior after primary attachment.23 The short term culture provides an appropriate time to examine effects of substrate stiffness on cell behavior. Due to cell growth after first day of culture, cell confluence becomes high especially among ECs which naturally have high confluence potential. While during first day of culture cell signaling through the substrate is prominent, by higher cell confluence cells receive strong signals from each other and less from the substrate.
Here, we utilized PDMS substrates with two different elastic moduli for analyzing the morphology of HUVECs at 4, 8, 12 and 24 hours after seeding. Cells responded to PDMS stiffness in term of cell elongation with a time dependent trend. While cells from two test groups experienced different elongation algorithm within the initial hours of culture, after 24 hours they practically reached similar elongation and actin filament arrangement, although SEM images revealed that there were clear differences among two groups of cultured cells, where cells showed better adherence to the stiffer substrate. This was in agreement with viability results showing significantly higher rate of survival for cells attached to stiffer substrates. Previous results have also confirmed that stiffer substrates can increase cells survival rate.30
Time dependency of the mechanosensing still remains an open question. Such a trend has been already observed among epithelial cells cultured on polyacrylamide gels with different elastic moduli. Although cells were round and flat on soft and stiff substrates respectively in first two days after culture, at the fifth day of culture they achieved similar flat morphology on all substrates.31 Furthermore, when fibroblasts were cultured on gels of varying stiffness with the same adhesiveness, the earliest time point at which cells reacted differently was after 2 minutes.32 In general published results on effects of substrate stiffness on cellular behavior have basically shown that substrate stiffness has an elementary effect on cell behavior.33
Conclusion
The response of cells to substrate stiffness was shown to be a time dependent process. In current study the time dependency was within the time span of 24 hours. The cell-substrate interaction through stiffness of substrate diminished after this period based on the general morphology and cytoskeleton of cells. However, clear difference was observed between 3D topographical images of cells in two test groups, describing different patterns of cell anchorage to the substrate. The enhanced anchorage of cells to the stiffer substrate may improve cell adherence and spreading, the possible reason for higher viability of cells on stiffer substrates. In conclusion, stiffer substrates enhance cell viability and attachment through enrichment of lamellipodia and filopodia among ECs that can subsequently affect other cell behaviors. Results of this study may contribute to promotion of our knowledge in achieving functional cells in cell therapy and tissue engineering.
Research Highlights
What is the current knowledge?
√ Substrate stiffness can affect cellular activities as cells experience different tissues’ stiffness in the body.
√ Substrate stiffness modulates endothelial cell behaviors such as proliferation, migration and apoptosis.
√ Despite numerous studies, the timeline by which cells sense their substrate and respond to its properties are not well understood.
What is new here?
√ Effectiveness of PDMS stiffness on endothelial cell elongation is time dependent.
√ Cells on stiffer substrates showed better spreading and higher ratio of cell viability.
√ Cells cultured on soft substrates lacked the rich lamellipodia and filopodia.
Ethical approval
None to be declared.
Competing interests
Authors declare no conflict of interests.
Acknowledgements
Authors acknowledge the personnel of central laboratories, Amirkabir University of Technology, and Tissue Engineering Laboratory of Biotechnology Department, Tehran University, Tehran, Iran.
References
- Giannone G, Dubin-Thaler BJ, Döbereiner H, Kieffer N, Bresnick AR, Sheetz MP. Periodic lamellipodial contractions correlate with rearward actin waves. Cell 2004; 116:431-443. doi: 10.1016/S0092-8674(04)00058-3 [Crossref] [ Google Scholar]
- Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J 2000; 79:144-152. doi: 10.1016/S0006-3495(00)76279-5 [Crossref] [ Google Scholar]
- T van Eijden TM, van Ruijven LJ, Giesen EB. Bone tissue stiffness in the mandibular condyle is dependent on the direction and density of the cancellous structure. Calcif Tissue Int2004. 75: 502-508. doi: 10.1007/s00223-004-0295-6.
- Georges PC, Hui JJ, Gombos Z, McCormick ME, Wang AY. Increased stiffness of the rat liver precedes matrix deposition: implications for fibrosis. Am J Physiol Gastrointest Liver Physiol 2007; 293:1147-1154. doi: 10.1152/ajpgi.00032.2007 [Crossref] [ Google Scholar]
- Lee K, Forudi F, Saidel GM, Penn MS. Alterations in internal elastic lamina permeability as a function of age and anatomical site precede lesion development in apolipoprotein E-null mice. Circ Res 2005; 97:450-6. doi: 10.1161/01.RES.0000181026.94390.c9 [Crossref] [ Google Scholar]
- McEniery CM, Yasmin Yasmin, Hall IR, Qasem A, Wilkinson IB, Cockcroft JR. Normal vascular aging: differential effects on wave reflection and aortic pulse wave velocity: the Anglo-Cardiff Collaborative Trial (ACCT). J Am Coll Cardiol 2005; 43:1753-60. doi: 10.1016/j.jacc.2005.07.037 [Crossref] [ Google Scholar]
- Fernandes VR, Polak JF, Cheng S, Rosen BD, Carvalho B. Arterial stiffness is associated with regional ventricular systolic and diastolic dysfunction: the Multi-Ethnic Study of Atherosclerosis. Arterioscler Thromb Vasc Biol 2008; 28:194-201. doi: 10.1161/ATVBAHA.107.156950 [Crossref] [ Google Scholar]
- Laurent S, Boutouyrie P. Recent advances in arterial stiffness and wave reflection in human hypertension. Hypertension 2007; 49:1202-6. doi: 10.1161/HYPERTENSIONAHA.106.076166 [Crossref] [ Google Scholar]
- Masona BN, Starchenko A, Williams RM, Bonassar LJ, Reinhart-King Reinhart-King. Tuning three-dimensional collagen matrix stiffness independently of collagen concentration modulates endothelial cell behavior. Acta Biomater 2013; 9:4635-44. doi: 10.1016/j.actbio.2012.08.007 [Crossref] [ Google Scholar]
- Birukova AA, Tian X, Cokic I, Beckham Y, Gardel ML, Birukov KG. Endothelial barrier disruption and recovery is controlled by substrate stiffness. Microvasc Res 2013; 87:50-7. doi: 10.1016/j.mvr.2012.12.006 [Crossref] [ Google Scholar]
- Trappmann B, Gautrot JE, Connelly JT, Strange DGT, Li Y. Extracellular-matrix tethering regulates stem-cell fate. Nature Mat 2012; 11:642-649. doi: 10.1016/j.jmbbm.2011.10.003 [Crossref] [ Google Scholar]
- Sridharan I, Kim T, Wang R. Adapting collagen/CNT matrix in directing hESC differentiation. Biochem Biophys Res Commun 2009; 381:508-512. doi: 10.1016/j.bbrc.2009.02.072 [Crossref] [ Google Scholar]
- Murrell M, Kamm RD, Matsudaira PT. Substrate viscosity enhances correlation in epithelial cell motion. Biophys J 2011; 101:297-306. doi: 10.1016/j.bpj.2011.05.048 [Crossref] [ Google Scholar]
- Lee JN, Jiang X, Ryan D, Whitesides GM. Compatibility of mammalian cells on surfaces of polydimethylsiloxane. Langmuir 2004; 20:11684-91. doi: 10.1021/la048562+ [Crossref] [ Google Scholar]
- Bhagat A, Jothimuthu P, Papautsky I. Photodefinable polydimethylsiloxane (PDMS) for rapid lab-on-a-chip prototyping. Lab Chip 2007; 7:1192-7. doi: 10.1039/b704946c [Crossref] [ Google Scholar]
- Brown QX, Ookawa K, Wong JY. Evaluation of polydimethylsiloxane scaffolds with physiologically relevant elastic moduli: interplay of substrate mechanics and surface chemistry effects on vascular smooth muscle cell response. Biomaterials 2005; 26:3123-9. doi: 10.1016/j.biomaterials.2004.08.009 [Crossref] [ Google Scholar]
- Palchesko RN, Zhang L, Sun Y, Feinberg AW. Development of polydimethylsiloxane substrates with tunable elastic modulus to study cell mechanobiology in muscle and nerve. PLoS One 2012; 7:e51499. doi: 10.1371/journal.pone.0051499 [Crossref] [ Google Scholar]
- Park JY, Yoo SJ, Lee E, Lee DH, Kim JY, Lee S. Increased poly(dimethylsiloxane) stiffness improves viability and morphology of mouse fibroblast cells. BioChip J 2010; 4:230-236. doi: 10.1016/j.biomaterials.2004.08.009 [Crossref] [ Google Scholar]
- Cheng C, LeDuc PR, Lin Y. Localized bimodal response of neurite extensions and structural proteins in dorsal-root ganglion neurons with controlled polydimethylsiloxane substrate stiffness. J Biomech 2011; 44:856-862. doi: 10.1016/j.jbiomech.2010.12.006 [Crossref] [ Google Scholar]
- Wang Y, Wang G, Luo X, Qiu J, Tang C. Substrate stiffness regulates the proliferation, migration and differentiation of epidermal cells. Burns 2012; 38:414-20. doi: 10.1016/j.burns.2011.09.002 [Crossref] [ Google Scholar]
- Prauzner-Bechcicki S, Raczkowska J, Madej E, Pabijan J, Lukes J. PDMS substrate stiffness affects the morphology and growth profiles of cancerous prostate and melanoma cells. J Mech Behav Biomed Mater 2015; 41:13-22. doi: 10.1016/j.jmbbm.2014.09.020 [Crossref] [ Google Scholar]
- Dubin-Thaler BJ, Giannone G, Do¨bereiner HG, Sheetz MP. Nanometer analysis of cell spreading on matrix-coated surfaces reveals two distinct cell states and STEPs. Biophy J 2004; 86:1794-1806. doi: 10.1016/S0006-3495(04)74246-0 [Crossref] [ Google Scholar]
- Li J, Han D, Zhao Y. Kinetic behavior of the cells touching substrate: the interfacial stiffness guides cell spreading. Sci Rep 2014; 4:e5139. doi: 10.1038/srep05139 [Crossref] [ Google Scholar]
- Goli-Malekabadi Z, Tafazzoli-Shadpour M, Rabbani M, Janmaleki M. Effect of uniaxial stretch on morphology and cytoskeleton of human mesenchymal stem cells: static vs dynamic loading. Biomed Tech 2011; 56:259-265. doi: 10.1515/BMT.2011.109 [Crossref] [ Google Scholar]
- Borodinsky LN, Fiszman ML. A single-cell model to study fractal dimension. Methods 2001; 24:341-345. doi: 10.1006/meth.2001.1204 [Crossref] [ Google Scholar]
- Behar TN. Analysis of FD of O2A glial cells differentiating in vitro. Methods 2001; 24:331-9. doi: 10.1006/meth.2001.1203 [Crossref] [ Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983; 65:55-63. doi: 10.1016/0022-1759(83)90303-4 [Crossref] [ Google Scholar]
- Previtera ML, Langhammer CG, Firestein BL. Effects of substrate stiffness and cell density on primary hippocampal cultures. J Biosci Bioeng 2010; 110:459-470. doi: 10.1016/j.jbiosc.2010.04.004 [Crossref] [ Google Scholar]
- Bartalena G, Loosli Y, Zambelli T, Snedeker JG. Biomaterial surface modifications can dominate cell–substrate mechanics: the impact of PDMS plasma treatment on a quantitative assay of cell stiffness. Soft matter 2012; 8:673-681. doi: 10.1039/C1SM06250F [Crossref] [ Google Scholar]
- Sazonova OV, Lee KL, Isenberg BC, Rich CB, Nugent MA, Wong JY. Cell-Cell Interactions Mediate the Response of Vascular Smooth Muscle Cells to Substrate Stiffness. Biophys J 2011; 101:622-30. doi: 10.1016/j.bpj.2011.06.051 [Crossref] [ Google Scholar]
- Eisenberg JL, Safi A, Wei X, Espinosa HD, Budinger GS, Takawira D, Hopkinson SB, Jones JC. Substrate stiffness regulates ECM deposition by alveolar epithelial cells. Res Rep Biol 2011; 2:1-12. doi: 10.2147/RRB.S13178 [Crossref] [ Google Scholar]
- Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton 2005; 60:24-34. doi: 10.1002/cm.20041 [Crossref] [ Google Scholar]
- Petersen A, Joly P, Bergmann C, Korus G, Duda GN. The impact of substrate stiffness and mechanical loading on fibroblast-induced scaffold remodeling. Tissue Eng Part A 2012; 18:1804-17. doi: 10.1089/ten.TEA.2011.0514 [Crossref] [ Google Scholar]