Dr. Jafar Ezzati Nazhad Dolatabadi has received a PhD degree in Pharmaceutical Nanotechnology from Tabriz University of Medical Sciences (TUOMS). He currently works as an assistant professor at TUOMS Research Center for Pharmaceutical Nanotechnology (RCPN). His research activities at RCPN include pharmaceutical nanotechnology, drug delivery and biosensors.
Prof. Miguel de la Guardia is a Full Professor at Valencia University (Department of Analytical Chemistry) from 1991. He has published over 500 papers in journals of the Science Citation Index, 3 books on Green Analytical Chemistry and 1 book on Food analysis additionally more than 8 book chapters. He is member of the Editorial board of Spectroscopy Letters (USA), Ciencia (Venezuela), Journal of the Brazilian Chemical Society (Brazil), Chemical Speciation & Bioavailability (UK) and BioImpacts. He was member of the advisory board of Analytica Chimica Acta (The Netherlands) between 1995 and 2000.
Abstract
Surface plasmon resonance (SPR) technique offers a robust label-free approach applicable in various investigations including binding affinity, specificity and kinetics of biological macromolecules (e.g., peptides, proteins and nucleotidase) and small molecules. SPR provides extremely important data on the kinetics and affinity of substances examined, through which bio-specific interaction(s) can be established by the analysis of adsorption of analyteonto the immobilized ligand(s) on a sensor-based analytical system. Due to SPR wide applications in biomedical laboratories, the aim of this editorial is to highlight the importance of SPR in affinity kinetics and ligand immobilization.
Keywords: Analyte, Kinetic study, Ligand immobilization, Surface plasmon resonance
Copyright and License Information
© 2016 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Recently, much interest has been devoted on surface plasmon resonance (SPR) biosensors that are used for evaluation of binding affinity as real-time in clinical and research laboratories.
1
The basis of the SPR technique is the changes in the interfacial refractive index of a sensor surface due to the high-affinity interaction between an immobilized biomolecule and its biospecific analyte in solution.
2
SPR provides the evaluation of the analyte-immobilized ligand interaction as well as antigen–antibody (Ag-Ab) interaction without any needs for further chemical or biological markers. To acquire a detectable signal in an enzyme-linked immunosorbent assay (ELISA), Abs need to be labeled while SPR permits direct detection in real-time without any labels or additional steps.
1,3
Monitoring of immunoresponses by SPR biosensor can be acquired by either use of designated Ag or Ab as ligands. The use of Abs as ligands is favored for analytes with a molecular weight over 1000 Da. Therefore, the immobilized Ab molecules onto the sensor surface may recognize its complementary Ag molecules in a simple, fast and direct way.
1
The response of SPR is directly related to the mass changes on the sensor surface. Hence, usually the smaller Ags are immobilized and the larger Abs are present as soluble analytes, through which the Ag–Ab interaction can be investigated.
4
There are some tips that should be considered before ligand immobilization: (i) molecular weight, (ii) reactive group, (iii) pH and buffer types, (iv) purity and availability, and (v) control molecule. We discuss some of these hints in details in the following context.
(i) Smaller molecules can change refractive index lower than the larger molecules. Thus, researchers usually immobilize small ligands to obtain high signals when large analytes are bounded.
(ii) Reactive groups are important to assure covalent coupling to the sensor surface, thus, ligands should contain reactive groups such as -NH2, -SH or, -COOH for capturing proteins and oligonucleotides. Streptavidin-biotin capturing methods are also used.
(iii) Buffer composition and pH influence analyte binding to the ligand. The typical buffers that are used in SPR experiments are HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) or PBS (Phosphate buffered saline) at concentration of 10 mM and pH 7.4.
(iv) The purest interactants are usually applied as the ligand while more available compounds should be used as the analyte.
(v) To confirm the successful ligand immobilization, its availability in its proper conformation must be assured.
5
Kinetic parameters give information about the rate of complex formation between two molecules (A and B). Association rate constant (ka) is indicative of AB number formed per second and dissociation rate constant (kd) is demonstrative of complexes fraction which decay per second.
Real-time monitoring of the binding response (i.e., response unit) has been shown in Fig. 1, which presents a way to evaluate binding kinetics.
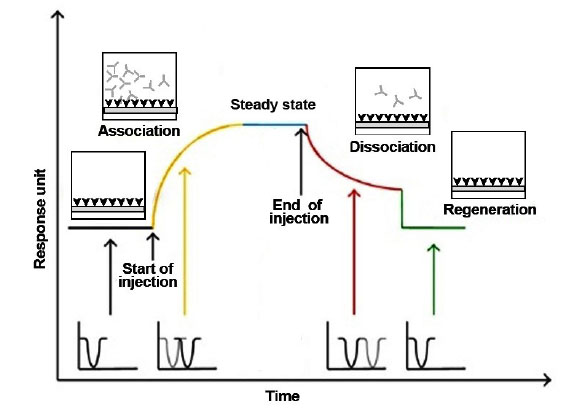
Fig. 1
.
Typical sensorgram of kinetic based on the interaction of a
surface bounded molecule (A) and a dissolved target analyte (B).
.
Typical sensorgram of kinetic based on the interaction of a
surface bounded molecule (A) and a dissolved target analyte (B).
Different molecules can have the same affinity to the target but different kinetics. Affinity shows that how much complex is formed at steady state or equilibrium (when association is balanced with dissociation). Affinity unit depends on equilibrium dissociation constant (KD) and low values of KD indicate a high tendency of association between two materials that can be calculated through the equation (Eq.) 1
6
:
Based on the equilibrium described by equation 2.
Finally, it is noteworthy that SPR is a useful tool for the direct measurement of affinity between small molecules. Not only steady state but also kinetics of the binding process can be efficiently calculated through SPR real-time and label free measurements.
Ethical approval
There is none to be declared.
Competing interests
The authors declare no competing interests.
References
- Mauriz E, García-Fernández MC, Lechuga LM. Towards the design of universal immunosurfaces for SPR-based assays: A review. TrAC Trends in Analytical Chemistry 2016; 79:191-8. doi: 10.1016/j.trac.2016.02.006 [Crossref] [ Google Scholar]
- Chiu N-F, Huang T-Y. Sensitivity and kinetic analysis of graphene oxide-based surface plasmon resonance biosensors. Sensors and Actuators B: Chemical 2014; 197:35-42. doi: 10.1016/j.snb.2014.02.033 [Crossref] [ Google Scholar]
- Souto DEP, Fonseca AM, Barragan JTC, Luz RdCS, Andrade HM, Damos FS. SPR analysis of the interaction between a recombinant protein of unknown function in Leishmania infantum immobilised on dendrimers and antibodies of the visceral leishmaniasis: A potential use in immunodiagnosis. Biosens Bioelectron 2015; 70:275-81. doi: 10.1016/j.bios.2015.03.034 [Crossref] [ Google Scholar]
- England P, Bregegere F, Bedouelle H. Energetic and kinetic contributions of contact residues of antibody D13 in the interaction with lysozyme. Biochemistry 1997; 36:164-72. doi: 10.1021/bi961419y [Crossref] [ Google Scholar]
-
Affinity and kinetics. Available from: http://www.bionavis.com/en/life-science/technology/affinity-and-kinetics/.
- Fathi F, Ezzati Nazhad Dolatanbadi J, Rashidi M-R, Omidi Y. Kinetic studies of bovine serum albumin interaction with PG and TBHQ using surface plasmon resonance. Int J Biol Macromol 2016; 91:1045-50. doi: 10.1016/j.ijbiomac.2016.06.054 [Crossref] [ Google Scholar]