BioImpacts. 8(2):129-138.
doi: 10.15171/bi.2018.15
Original Research
Reprogramming of mouse fibroblasts into neural lineage cells using biomaterials
Fahsai Kantawong 1, *, Chanidapa Saksiriwisitkul 1, Chanakan Riyapa 1, Suchalinee Limpakdee 1, Phenphichar Wanachantararak 2, Thasaneeya Kuboki 3
Author information:
1
Department of Medical Technology, Faculty of Associated Medical Sciences, Chiang Mai University, Chiang Mai, Thailand
2
The Dental Research Center, Faculty of Dentistry, Chiang Mai University, Chiang Mai 50200, Thailand
3
Laboratory of Biomedical and Biophysical Chemistry, Institute for Materials Chemistry and Engineering, Kyushu University, Fukuoka,
Japan
Abstract
Introduction:
Induced neural stem cells (iNSCs) have the ability of differentiation into neurons, astrocytes and oligodendrocytes. iNSCs are very useful in terms of research and treatment. The present study offers an idea that biomaterials could be one of the tools that could modulate reprogramming process in the fibroblasts.
Methods:
Gelatin biomaterials were fabricated into 3 types, including (i) gelatin, (ii) gelatin with 1 mg/mL hydroxyapatite, and (iii) gelatin with hydroxyapatite and pig brain. NIH/3T3 fibroblasts were cultured on each type of biomaterial for 7, 9 and 14 days. RT-PCR was performed to investigate the gene expression of the fibroblasts on biomaterials compared to the fibroblasts on tissue culture plates. PI3K/Akt signaling was performed by flow cytometry after 24 hours seeding on the biomaterials. The biomaterials were also tested with the human APCs and PDL cells.
Results:
The fibroblasts exhibited changes in the expression of the reprogramming factor; Klf4 and the neural transcription factors; NFIa, NFIb and Ptbp1 after 9 days culture. The cultivation of fibroblasts on the biomaterials for 7 days showed a higher expression of the transcription factor SOX9. The expression of epigenetic genes; Kat2a and HDAC3 were changed upon the cultivation on the biomaterials for 9 days. The fibroblasts cultured on the biomaterials showed an activation of PI3K/Akt signaling. The human APCs and human PDL cells developed mineralization process on biomaterials
Conclusion:
Changes in the expression of Klf4, NFIa, NFIb, Ptbp1 and SOX9 indicated that fibroblasts were differentiated into an astrocytic lineage. It is possible that the well-designed biomaterials could work as powerful tools in the reprogramming process of fibroblasts into iNSCs.
Keywords: Fibroblasts, Induced neuronal stem cells, Cell reprogramming
Copyright and License Information
© 2018 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
The world population is entering an aging society and soon the health problems of the elderly will inevitably increase. One of the major health problems of the elderly is neurodegenerative disorder due to the deterioration of brain tissue. Neurodegenerative disorder will be more severe as the patient ages increase. Alzheimer's disease (AD), Parkinson's disease and multiple sclerosis are considered as the neurodegenerative disorders, while there exists no therapeutic method to slow down or stop the neuronal damage. The treatment is only to alleviate the symptoms that occur.
A number of researchers have proposed a concept for using stem cells to treat the neurodegenerative disorders.
1-3
However, the use of stem cells in the elderly is limited in both quantity and quality. Recently, researchers have turned their attention to a new technology, so-called induced pluripotent stem cells (iPSCs).
4-6
The iPSCs are very useful in terms of research
5
but the use of iPSCs in the treatment of diseases is also questionable as iPSCs can cause cancer because reprogramming process might result in telomere length
7
which is similar to some types of cancer cells.
8,9
It is reported that the length of the telomere is associated with glioma.
8
Based on the iPSC's technology, there is an expanded field of research called transdifferentiation, which refers to the irreversible conversion of one cell type to another by forcing the expression of lineage-related transcription factor.
10-13
The examples include the conversion of fibroblasts into the chondrocytes by forcing the fibroblasts to express reprogramming factors c-Myc and Klf4 together with the chondrogenic factor SOX9
14
or the conversion of fibroblasts to astrocytes by forcing fibroblasts to express NFIa, NFIb and SOX9 transcription factors.
11
However, new cell types derived from the transdifferentiation process showed a less potential for the cell proliferation compared to stem cells. It has been suggested that the conversion of fibroblasts to the induced neuronal stem cells (iNSCs) would be a better way because it is believed to be more secure as the iNSCs showed a lower incidence of tumorigenicity than the iPSCs and also the iNSCs still have multipotent ability to differentiate into astrocytes, neurons and oligodendrocytes. The method of conversion of fibroblasts into the iNSCs was performed by forcing the fibroblasts to have the expression of transcription factors that were specific to the neural stem cells which could be achieved by transfection of viral vectors into the fibroblasts. This method may cause random integration in the fibroblast genome, in which non-viral methods are preferred.
15,16
Thus, the idea that forcing fibroblasts to express the neural transcription factors by transfection of the viral vector into fibroblasts may not be the safest way.
Biomaterials possibly play an important role in the cellular reprogramming of fibroblasts into the neural cell type. In a previous study, it was shown that the photocurable hydrogel successfully induced a neural differentiation in mesenchymal stem cells (HMSCs).
17,18
The study by Tanum et al presented that the addition of hydroxyapatite (HA) into 10% gelatin and crosslinking by glutaraldehyde (GA) could be used in the HMSCs cultivation.
19
Furthermore, it was shown that the biomaterials from the study of Tanum et al
19
could induce a neural differentiation in the HMSCs.
20
The present study aimed to demonstrate that the gelatin-hydroxyapatite-pig brain biomaterials could induce the expression of the reprogramming factor Klf4 and nerve transcription factors, such as NFIa, NFIb, and SOX9 in fibroblasts. Thus, we hypothesized that biomaterials could be used effectively and economically as a tool to generate iNSCs from fibroblasts. The iNSCs derived from the transdifferentiation process which still have multipotent ability might be useful as a tool in research for finding therapeutic targets, modeling for studying the pathogenesis process, drug screening and ultimately for autologous cell therapy.
Materials and Methods
Materials
Gelatin was obtained from HiMedia Laboratories Pvt. Ltd., Mumbai, India. Nano-hydroxyapatite (HA), particle size <200 nm was from Sigma-Aldrich; Saint Louis, MO 63103, USA. Glutaraldehyde (25%) was from AppliChem Inc.; Saint Louis, USA. Dulbecco's Modified Eagle Medium (DMEM), Trypsin-EDTA (1X), fetal bovine serum (FBS) and Pen-Strep were purchased from Gibco Life technology, Nucleospin Kit was from MACHEREY-NAGEL (Düren, Germany). cDNA synthesis kit was from ReverTra Ace-α-® - Toyobo; Japan. SYBR Green Mastermix was from SensiFAST SYBR® No-ROX Kit-Bioline (Taunton, USA). PI3K Activation Dual Detection Kit was from Merck-Millipore (Burlington, USA).
Brain extract
Pig brain was purchased from the local market and preserved in frozen condition before use. Frozen pig brain was thawed at room temperature then 30 g of pig brain was washed in sterile water before blending in a blender with 100 mL of sterile phosphate buffer saline (PBS) using the liquidify mode for 1 minute. Then, the liquidified brain was spun at 2500 rpm (2320 ×g) for 10 minutes in the centrifuge (KUBOTA Model 5200, KUBOTA CORP., Japan). The supernatant was discarded and the white brain precipitant was kept at -20°C until used.
Biomaterial preparation
Gelatin 10 wt.% in distilled water was incubated in water bath at 50°C for 45 minutes until gelatin was dissolved. Then gelatin solution was stirred at 50°C for 30 minutes. The gelatin solution was stirred for 15 minutes at room temperature before adding 10 mg of nanoHA and continued stirring for 30 minutes. For gel+HA+brain, add 1 mL of brain extract together with HA then 25% glutaraldehyde 5-6 µL/mL was added and mixed together before pouring into 6-well plate. The biomaterial was prepared well-by-well using 2 mL of solution per well. The solution was left overnight then polymerized gelatin was washed with sterile distilled water three times for 15 minutes each. After that polymerized gelatin was soaked in sterile distilled water overnight. Water was discarded and polymerized gelatin was then gently tapped on tissue paper to get rid of the rest of water on the surface of gelatin. Polymerized gelatin was frozen at -20°C before freeze-drying in the lyophilizer (LYOLAB; Lyophilization Systems, Inc., Kingston, USA) at 9 mTorr with condenser temperature 84.1οC for 24 hours.
Preconditioning of biomaterials
Biomaterials were immerged in 70% ethanol for 30 minutes. After 70% ethanol was discarded, biomaterials were immersed in sterile water for 30 minutes 3 times. Complete DMEM (3 mL) was added to each biomaterial and incubated in 5% CO2 incubator at 37 °C for at least 24 hours. Then the biomaterials were ready for cell culture.
NIH/3T3 culture
The NIH/3T3 fibroblasts (passage 8th) were seeded onto preconditioned biomaterials at the density 1 × 105 cells per gel. The cells were cultured in an incubator with 5% CO2 at 37 °C for the desired periods.
Preparation of periodontal ligament cells and apical pulp cells
Preparation of periodontal ligament (PDL) cells and apical pulp cells (APCs) were received from patients coming for a routine tooth extraction. Primary PDL cells and APCs were obtained as previously reported.
21
Caries-free third molar that was extracted for orthodontic treatment purposes was used. PDL cells were derived from 2 different donors (N1 and N2). The periodontal ligament and the apical tissues were removed from human third molars and minced into small pieces with a sharp blade. The tissue pieces were then cultured in tissue culture dishes. After becoming confluent, the cells were washed with PBS and trypsinized with trypsin-EDTA to generate the next passage. The PDL cells (passages 6th) or APCs (passages 5th) at a seeding density of 1.0 × 104 cells were cultured onto preconditioned biomaterials and maintained for 1 or 2 weeks in complete DMEM at 37°C with 5% CO2. Cell morphology was examined under an inverted microscope (Nikon Eclipse TS100; Nikon Instruments Inc., Melville, USA).
Real-time PCR
RNA extraction was performed using Nucleospin kit and cDNA synthesis was performed using ReverTra Ace cDNA synthesis kit following the protocols of manufacturers. The cDNA sample was diluted to the desired dilution, then 4.0 µL of the cDNA was mixed with SYBR Green Mastermix and the primer at a final concentration of 0.25 µM in the total volume of 10 µL. Real time-PCR was performed in LightCycler® 480. The PCR program contain 3 steps: (i) pre-incubation step at 95°C for 2 minutes, (ii) real time-PCR step (denature at 95°C, annealing at 60°C and extension at 72°C) for 40 cycles, and (iii) the melting curve analysis at 95, 65 and 97°C. The list of primers used in this study was shown in Table S1 in supplementary data. The experiments were divided into the following groups, including control (cells cultured on tissue culture plate), cells cultured on gelatin (gel), cells cultured on gelatin with 1 mg/mL HA (gel+HA) and cells cultured on gelatin with 1 mg/mL HA and brain (gel+HA+brain). The experiments were repeated at least 3 times. Target genes were normalized with reference genes GAPDH and 18s rRNA using LightCyCler 4.0 software.
PI3 Kinase pathway/AKt assay
NIH/3T3 cells or APCs at a seeding density of 2.0 × 105 cells were cultivated onto each biomaterial in complete DMEM and incubated in 5% CO2 incubator at 37°C for 24 hours. The cells cultured just on tissue culture plates were used as the control. Each type of biomaterial was tested 3 times independently. After 24 hours, the cells were trypsinized with trypsin-EDTA and cell counting was performed. The NIH/3T3 or APCs (7.5 × 104 cells) were used in the analysis. Cells were spun down at 300 ×g for 5 minutes and the media was discarded. The cells were resuspended by adding 200 µL of 1x assay buffer, then 200 µL of fixation buffer was added to the cell suspension and incubated for 5 minutes on ice. The cells were spun down at 300 ×g for 5 minutes and the supernatant was discarded. The cells were permeabilized by adding 400 µL ice-cold permeabilization buffer and incubated on ice for 5 minutes. The cells were spun down at 300 ×g for 5 minutes and the supernatant was discarded, then resuspended in 90 µL 1X assay buffer. Then, 10 µL of the antibody working cocktail solution was added to each microcentrifuge tube containing the cell suspension and incubated for 30 minutes in the dark at room temperature. Following incubation step, 100 µL of 1x assay buffer was added to each microcentrifuge and centrifuged at 300 ×g for 5 minutes on a tabletop centrifuge. The supernatant was discarded and the cells were resuspended in each microcentrifuge tube with 200 µL of 1x Assay Buffer. Samples were assayed on the Muse™ Cell Analyzer using the onscreen instructions.
Data analysis
One-way ANOVA followed by a post hoc analysis was used to test the difference between each type of biomaterials and treated cells. Afterward, the unpaired t-test was used to determine the difference between the test groups and control groups that seemed to be different but showed insignificant statistics by one-way ANOVA followed by a post hoc analysis. For PDL cells, one-way ANOVA with post hoc analysis was used to test the difference between each type of biomaterial. Then, Wilcoxon-Mann-Whitney test was used to determine the difference between the test groups and control groups that seemed to be different but showed insignificant statistics by one-way ANOVA with post hoc analysis.
Results
Biomaterials
It was found that the appropriate amount of GA used in this study was 5-6 µL/mL. The amount of GA less than 5 µL/mL was not enough for polymerization and biomaterials dissolved in the washing process. Too much GA (more than 6 µL) accelerated the cross-linking process and caused heterogeneous polymerization. The surface of all types of biomaterial was ruffled as shown in graphical abstract.
NIH/3T3 fibroblast culture
In this study, NIH/3T3 fibroblasts could be cultured on all types of the biomaterials used. The morphology of NIH/3T3 fibroblasts cultured for 24 hours is shown in Fig. S1 (Supplementary Data). The morphology of NIH/3T3 fibroblasts cultured for 2 weeks is shown in Fig. S2 (Supplementary Data) and the morphology of NIH/3T3 fibroblasts cultured for 9 days is shown in Fig. 1.
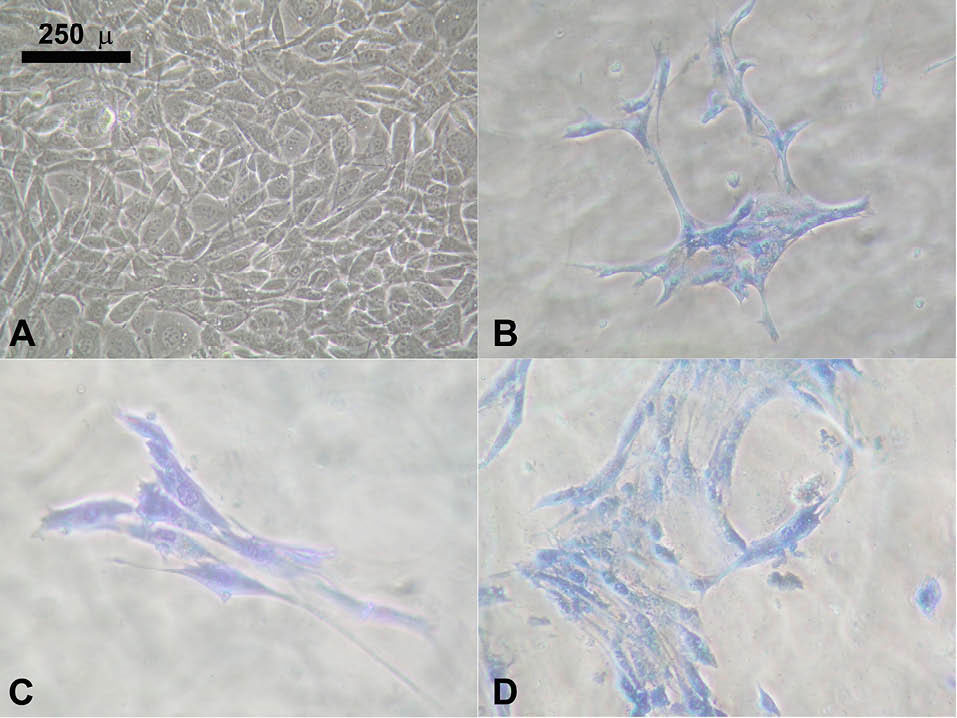
Fig. 1.
NIH/3T3 9 days. A) The control cells (NIH/3T3 cultured on tissue culture plate). B) NIH/3T3 cultured on the gel. C) NIH/3T3 cultured on the gel+HA. D) NIH/3T3 cultured on the gel+HA+brain.
.
NIH/3T3 9 days. A) The control cells (NIH/3T3 cultured on tissue culture plate). B) NIH/3T3 cultured on the gel. C) NIH/3T3 cultured on the gel+HA. D) NIH/3T3 cultured on the gel+HA+brain.
NIH/3T3 gene expression of transcription factors
Klf4 and NFIb were detected at day-7 and day-9 but both genes were significantly upregulated at day-9 especially in gel+HA+brain. The expression of Klf4 is shown in Fig. 2 and the expression of NFIb is shown in Fig. 3. NFIa and Ptbp1 were exclusively detected at day-9 especially in gel+HA+brain (Fig. 4). SOX9 was detectable at day-7 and day-14 but it was significantly upregulated at day-7 (Fig. 5). The summary of transcription factor gene expression in NIH/3T3 is shown in Table S2.
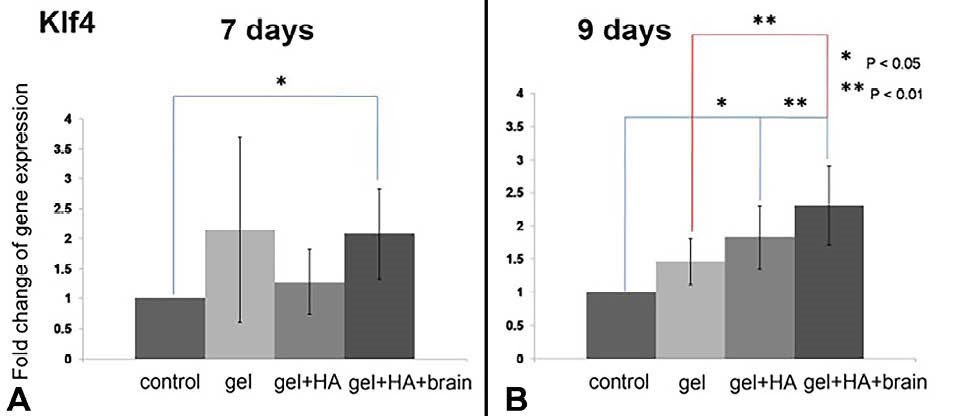
Fig. 2.
The Klf4 expression in the NIH/3T3 cells cultured on the biomaterials. A) The Klf4 overexpression in the cells cultured on the gel+HA+brain on day 7. B) The Klf4 overexpression in the cells cultured on the gel+HA and gel+HA+brain on day 9. * P < 0.05 and ** P < 0.01.
.
The Klf4 expression in the NIH/3T3 cells cultured on the biomaterials. A) The Klf4 overexpression in the cells cultured on the gel+HA+brain on day 7. B) The Klf4 overexpression in the cells cultured on the gel+HA and gel+HA+brain on day 9. * P < 0.05 and ** P < 0.01.
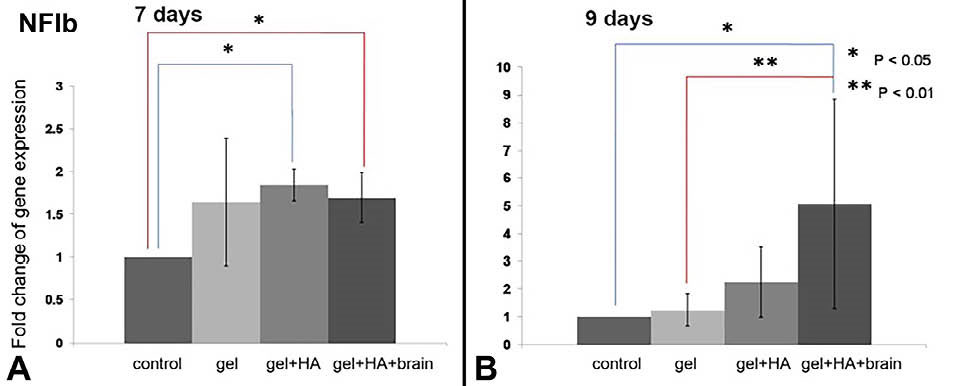
Fig. 3.
The NFIb expression in the NIH/3T3 cells cultured on the biomaterials. A) The NFIb overexpression in the cells cultured on the gel+HA and gel+HA+brain on day 7. B) The NFIb overexpression in the cells cultured on the gel+HA+brain on day 9. * P < 0.05 and ** P < 0.01.
.
The NFIb expression in the NIH/3T3 cells cultured on the biomaterials. A) The NFIb overexpression in the cells cultured on the gel+HA and gel+HA+brain on day 7. B) The NFIb overexpression in the cells cultured on the gel+HA+brain on day 9. * P < 0.05 and ** P < 0.01.
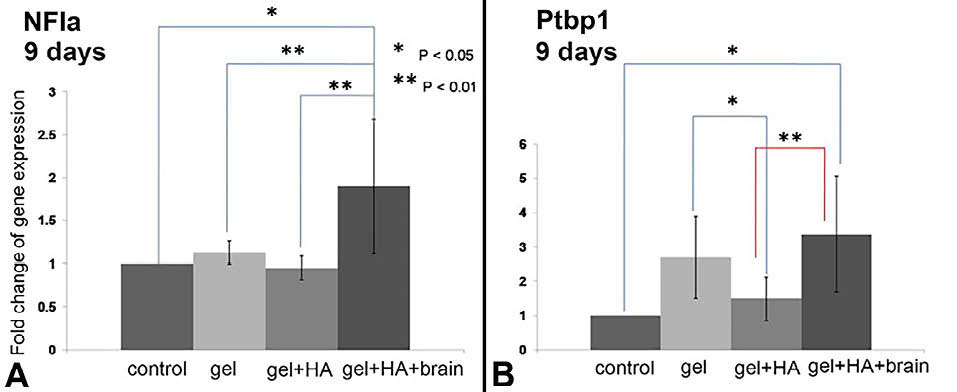
Fig. 4.
The Nfia and Ptbp1 expression in the NIH/3T3 cells cultured on the biomaterials after 9 days. A) The NFIa overexpression in the cells cultured on the gel+HA+brain. B) The Ptbp1 overexpression in the cells cultured on the gel+HA+brain. * P < 0.05 and ** P < 0.01.
.
The Nfia and Ptbp1 expression in the NIH/3T3 cells cultured on the biomaterials after 9 days. A) The NFIa overexpression in the cells cultured on the gel+HA+brain. B) The Ptbp1 overexpression in the cells cultured on the gel+HA+brain. * P < 0.05 and ** P < 0.01.
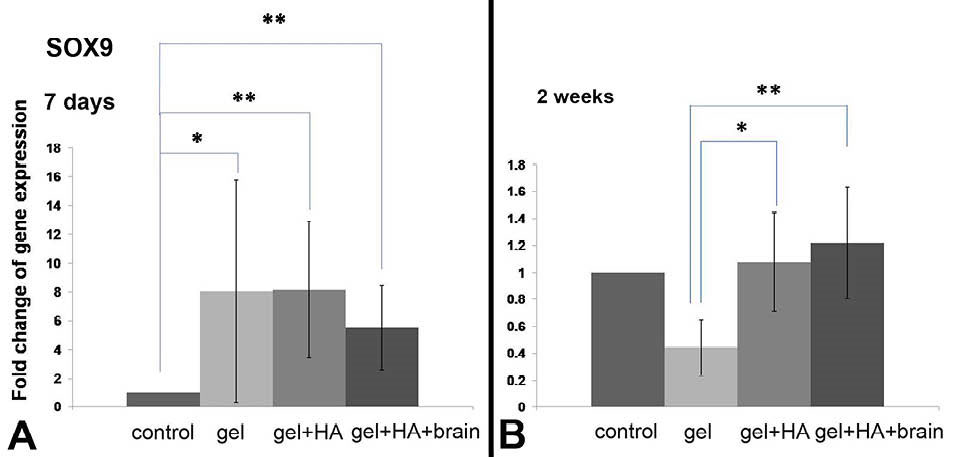
Fig. 5.
The SOX9 expression in the NIH/3T3 cells cultured on the biomaterials. A) The SOX9 overexpression in the cells on day 7. B) The SOX9 expression in the cells after two weeks cultivation. * P < 0.05 and ** P < 0.01.
.
The SOX9 expression in the NIH/3T3 cells cultured on the biomaterials. A) The SOX9 overexpression in the cells on day 7. B) The SOX9 expression in the cells after two weeks cultivation. * P < 0.05 and ** P < 0.01.
NIH/3T3 gene expression of differentiation markers
MAP2 gene was insignificantly detected only at day-9 (Fig. 6). CD44 gene was detected at day-9 and day-14 but the significant upregulation was found in gel and gel+HA+brain at day-9 when compared to control (Fig. 6). Class III β-tubulin, neurofilament light polypeptide, GFAP, nestin and prominin-1 were undetectable at all time points.
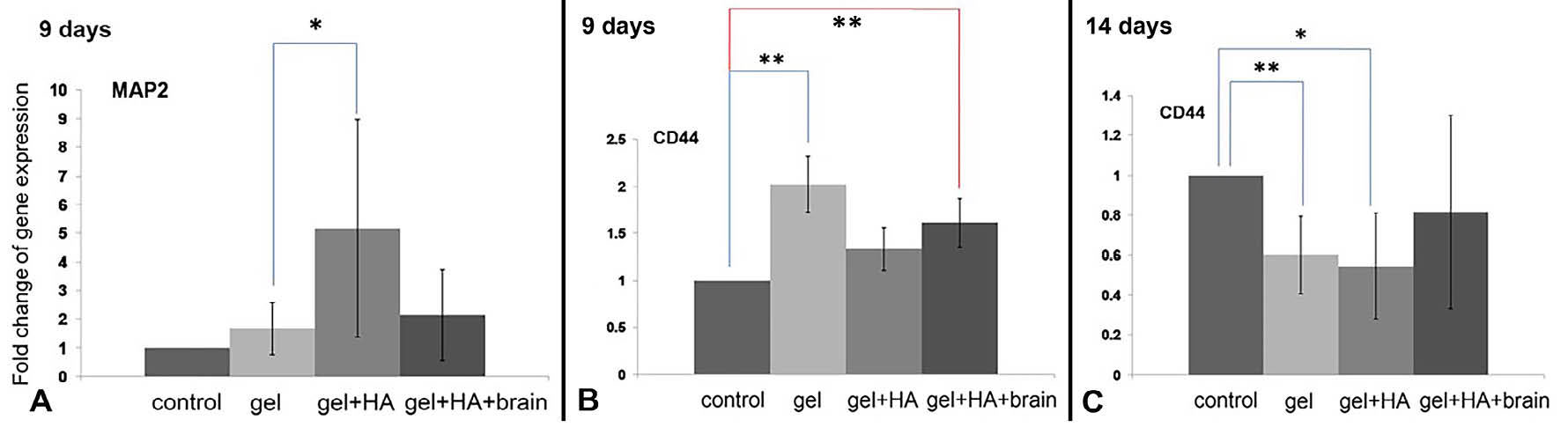
Fig. 6.
The expression of differentiation markers in NIH/3T3 cells. A) The MAP2 gene overexpression in the cells cultured on the gel+HA on day 9. B) The CD44 overexpression in the cells cultured on the gelatin and gel+HA+brain on day 9. C) The CD44 underexpression in the cells cultured on gelatin and gel+HA on day 14. * P < 0.05 and ** P < 0.01.
.
The expression of differentiation markers in NIH/3T3 cells. A) The MAP2 gene overexpression in the cells cultured on the gel+HA on day 9. B) The CD44 overexpression in the cells cultured on the gelatin and gel+HA+brain on day 9. C) The CD44 underexpression in the cells cultured on gelatin and gel+HA on day 14. * P < 0.05 and ** P < 0.01.
NIH/3T3 gene expression of epigenetic markers
At 9 days culture, gene expression of Kat2a was decreased in all types of biomaterial compared to control, whilst the expression of HAT1 on biomaterials was not different from control. In the meanwhile, HDAC3 expression was increased significantly in gel+HA+brain (Fig. 7).
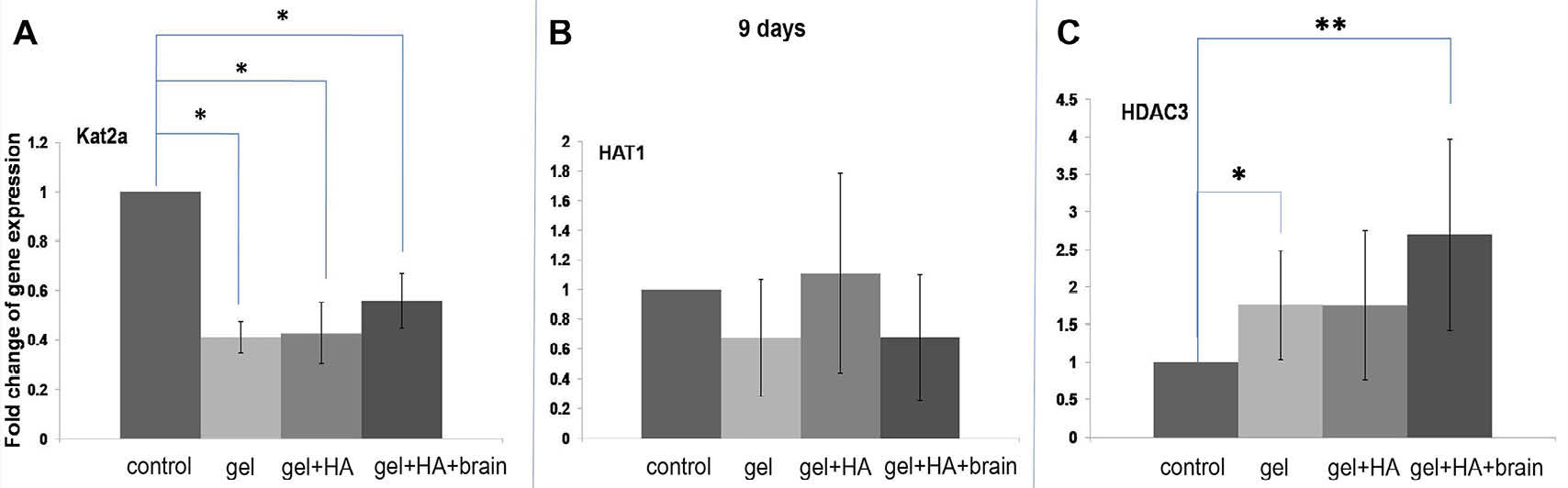
Fig. 7.
Epigenetic markers in the NIH/3T3 cells after 9 days of culture. A) The Kat2a underexpression in the cells cultured on all types of biomaterials. B) The HAT1 expression in the cells cultured on all types of biomaterials. C) The HDAC3 overexpression in the cells cultured on the gelatin and gel+HA+brain. * P < 0.05 and ** P < 0.01.
.
Epigenetic markers in the NIH/3T3 cells after 9 days of culture. A) The Kat2a underexpression in the cells cultured on all types of biomaterials. B) The HAT1 expression in the cells cultured on all types of biomaterials. C) The HDAC3 overexpression in the cells cultured on the gelatin and gel+HA+brain. * P < 0.05 and ** P < 0.01.
Gene expression of human APC and human PDL culture
The morphology of APCs culture on biomaterials are shown in Fig. S4 and PDL cells culture on biomaterials are shown in Fig. S3. The results of gene expression at 1 week and 2 weeks cultures were different from the result of NIH/3T3. At 1 week culture, only SOX9 and OCN were detected in the APCs and PDL cells as shown in Fig. 8. The 1-week result revealed that SOX9 and OCN were highest in the PDL cells cultured on the gel+HA+brain. Then, the PDL cells from another donor (N2) were tested with the gel+HA and gel+HA+brain for 2 weeks to observed the gene expression in long-term culture (Fig. 9). Early bone markers; collagen type I and ALP were upregulated in the gel+HA. Non-collagenous bone matrix proteins; OCN was significantly upregulated in the gel+HA+brain, while OPN was downregulated in all biomaterials compared to the control. Versican was upregulated in the gel+HA with fold-change of the gene expression <2, while aggrecan was significantly upregulated in the gel+HA+brain. The summary of transcription factor gene expression in APCs and PDL cells is shown in Table S3.
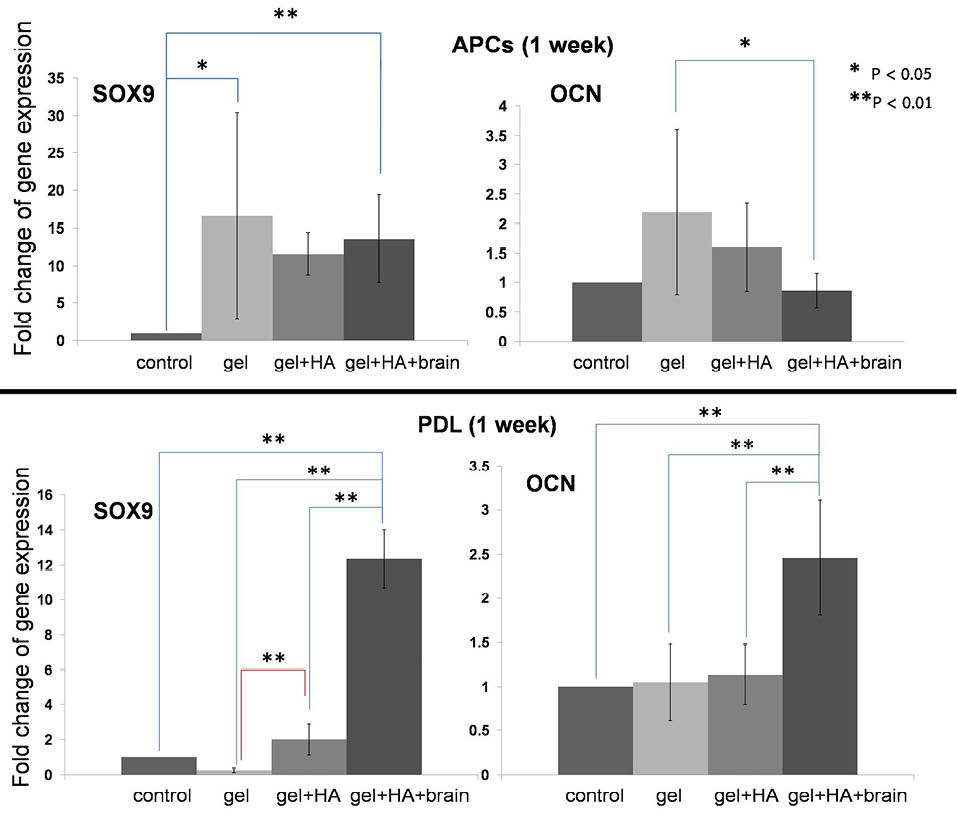
Fig. 8.
The expression of SOX9 and OCN in the APCs and PDL cells cultured for 1 week. A) The SOX9 overexpression in the APCs cultured on the gel and gel+HA+brain. B) The OCN underexpression in the APCs cultured on the gel+HA+brain. C) The SOX9 overexpression in the PDL cells cultured on the gel+HA+brain. D) The OCN overexpression in the PDL cells cultured on the gel+HA+brain. * P < 0.05 and ** P < 0.01.
.
The expression of SOX9 and OCN in the APCs and PDL cells cultured for 1 week. A) The SOX9 overexpression in the APCs cultured on the gel and gel+HA+brain. B) The OCN underexpression in the APCs cultured on the gel+HA+brain. C) The SOX9 overexpression in the PDL cells cultured on the gel+HA+brain. D) The OCN overexpression in the PDL cells cultured on the gel+HA+brain. * P < 0.05 and ** P < 0.01.
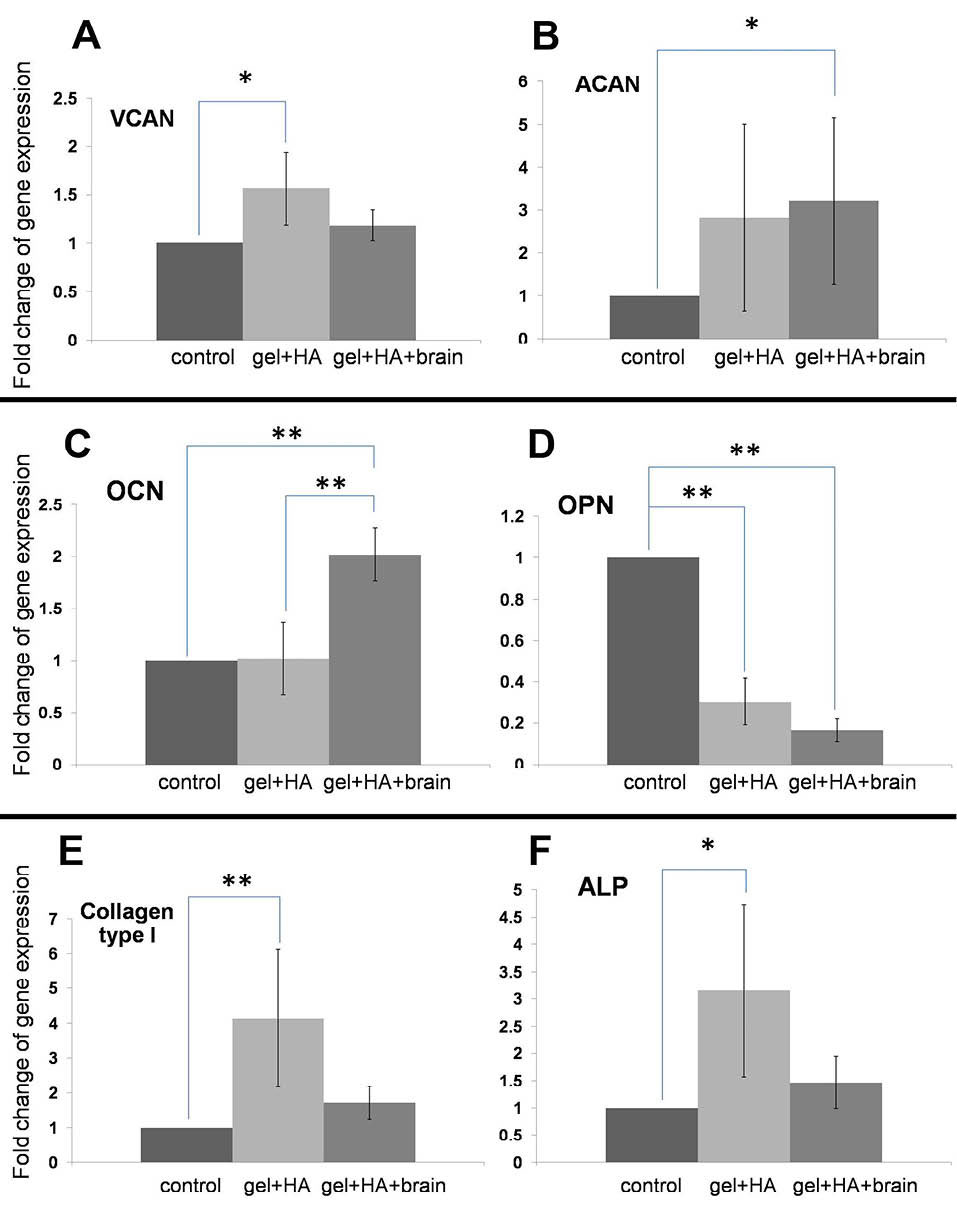
Fig. 9.
Gene expression in the PDL cells cultured for 2 weeks. A) The versican overexpression in the cells cultured on the gel+HA. B) The aggrecan upregulation in the cells cultured on the gel+HA+brain. C) The OCN upregulation in the cells cultured on the gel+HA+brain. D) The OPN downregulation in the cells cultured on all types of the biomaterials. E) The collagen type I upregulation in the cells cultured on the gel+HA. F) The ALP upregulation in the cells cultured on the gel+HA. * P < 0.05 and ** P < 0.01.
.
Gene expression in the PDL cells cultured for 2 weeks. A) The versican overexpression in the cells cultured on the gel+HA. B) The aggrecan upregulation in the cells cultured on the gel+HA+brain. C) The OCN upregulation in the cells cultured on the gel+HA+brain. D) The OPN downregulation in the cells cultured on all types of the biomaterials. E) The collagen type I upregulation in the cells cultured on the gel+HA. F) The ALP upregulation in the cells cultured on the gel+HA. * P < 0.05 and ** P < 0.01.
PI3 Kinase pathway/AKt assay
The NIH/3T3 cells cultured in the gel and gel+HA showed the activation of PI3K/Akt pathway (Fig. 10), while the APCs showed a decreased PI3K/Akt pathway activation when they were cultured on the gel and gel+HA (Fig. 11). However, the PI3K/Akt pathway activation was dominantly increased in the gel+HA+brain (both NIH/3T3 and APCs). For the NIH/3T3 cells, control and each type of biomaterial were repeated 3 times. The mean value and SD of each experimental group were calculated (Fig. 10F), which indicate the reproducibility of the PI3K/Akt activation.
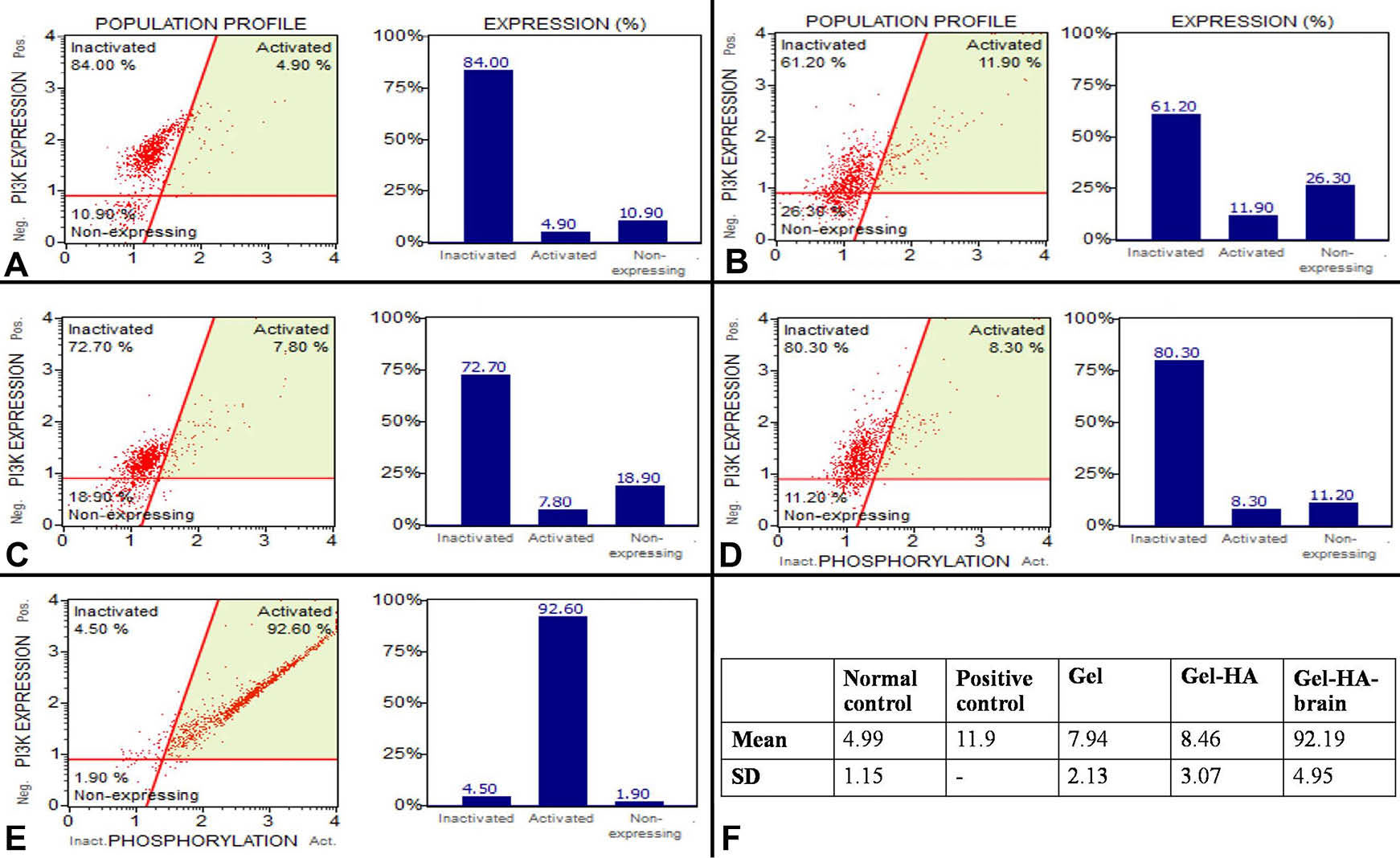
Fig. 10
.
The PI3K activation in the NIH/3T3 cells. A) The PI3K activation in the cells cultured on tissue culture plate. B) The PI3K activation, as the positive control, in the cells cultured on tissue culture plate supplemented with 2.5 mM ascorbic acid. C) The PI3K activation in the cells cultured on the gelatin. D) The PI3K activation in the cells cultured on the gel+HA. E) The PI3K activation in the cells cultured on the gel+HA+brain. F) The mean and SD values of PI3K/Akt activation of each group in the replicated the experiments (n=3).
.
The PI3K activation in the NIH/3T3 cells. A) The PI3K activation in the cells cultured on tissue culture plate. B) The PI3K activation, as the positive control, in the cells cultured on tissue culture plate supplemented with 2.5 mM ascorbic acid. C) The PI3K activation in the cells cultured on the gelatin. D) The PI3K activation in the cells cultured on the gel+HA. E) The PI3K activation in the cells cultured on the gel+HA+brain. F) The mean and SD values of PI3K/Akt activation of each group in the replicated the experiments (n=3).
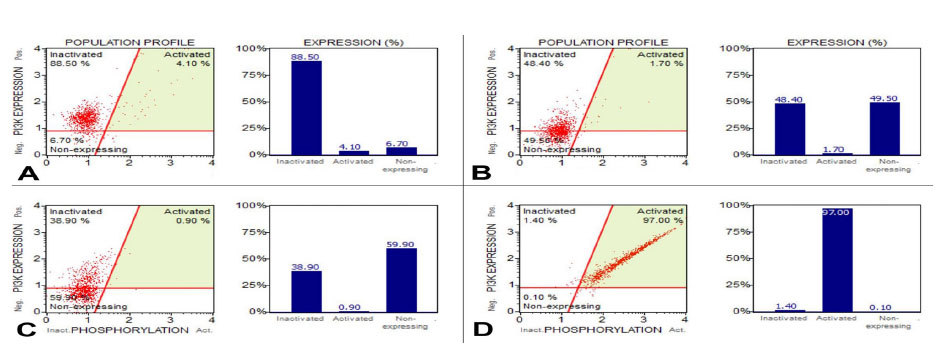
Fig. 11
.
The PI3K activation in the APCs. A) The PI3K activation in the cells cultured on tissue culture plate. B) The PI3K activation in the cells cultured on the gelatin. C) The PI3K activation in the cells cultured on the gel+HA.D) The PI3K activation in the cells cultured on the
gel+HA+brain.
.
The PI3K activation in the APCs. A) The PI3K activation in the cells cultured on tissue culture plate. B) The PI3K activation in the cells cultured on the gelatin. C) The PI3K activation in the cells cultured on the gel+HA.D) The PI3K activation in the cells cultured on the
gel+HA+brain.
Discussion
Some researchers have reported on the transformation of fibroblasts into the neural progenitor cells or neural stem cells (NSCs) using a set of specific transcription factors together with reprogramming agents.
22,23
Many previous studies
24-26
forced the fibroblasts to express the neural transcription factor by infecting fibroblasts with the pMX retroviral vectors. In this study, cells expressed the transcription factors using a non-invasive method by seeding cells on the gelatin biomaterials and growing in conventional medium without infection of viral particles, in which change of epigenetic genes occurred without the addition of any chemical reagents so this point showed the novelty of this work. It is possible that the neurodegenerative diseases could be cured by the induced NSCs produced from the somatic cells. Currently, there were 2 ways to produce iNSCs. The first method was an indirect reprogramming approach, in which fibroblasts were passing through iPSCs as the intermediate stage.
24-29
However, the iPSCs showed the potential to become cancerous. It has been reported that the iNSCs generated by the iPSCs had also potential for tumorigenicity but the efficiency was less than iPSCs.
30
A solution could be achieved by the direct reprogramming of the somatic cells without going through the pluripotent state.
31
The direct transformation of somatic cells into the lineage-committed multipotent stem cells might be an attractive way to generate desired types of cells. This could allow sufficient numbers of cells to be produced for basic researches, drug screening and disease modeling or cell therapy.
23
An increased expression of the reprogramming factor Klf4 suggested a state of reprogramming,
32,33
hence Kim et al showed that transient induction of the four reprogramming factors (i.e., Oct4, Sox2, Klf4, and c-Myc) could efficiently transdifferentiate fibroblasts into the functional neural stem/progenitor cells (NPCs).
28
The Klf4 gene encoded zinc finger protein which was required for a normal development of skin function. Transcription factors NFIa, NFIb and Ptbp1 indicated an astrocytic cell type.
11,34-36
The NFI family played important roles in the development of the central nervous system.
37,38
The study by Deneen et al indicated that NFIa was important in the transition from neurogenesis to gliogenesis.
39
Oligodendrocytes, astrocytes, ependymal cells and microglia were considered as glial cells in the central nervous system. Schwann cells and satellite cells were considered as glial cells in the peripheral nervous system.
40
The oligodendrocyte lineage required the maintenance of Olig2 and Nkx2.2 expression.
41,42
Because Olig2 and Nkx2.2 were undetectable throughout this study thus this evidence indicated that NIH/3T3 did not differentiate to oligodendrocytes. Up-regulation of NFIa was required to promote migration and differentiation of astrocyte precursors.
39
Thus, the up-regulation of NFIa in this study might be an indicative of the development of an astrocytic lineage. In this study, NFIa could be detected on day 9 of culture. It was upregulated significantly in NIH/3T3 cultured in gel+HA+brain. NFIb was found significantly increased only in gel+HA+brain on day 9 of culture. Polypyrimidine tract-binding proteins (PTBPs) were RNA-binding proteins that were extensively characterized as trans-acting splicing repressors.
43
PTBPs gene expressed heterogeneous nuclear ribonucleoproteins (hnRNPs) which bound to heterogeneous nuclear RNA (hnRNA). During neuronal differentiation, the splicing of a large group of exons was reprogrammed by a switch in expression between two types of PTBPs.
44
NPCs expressed PTB and when NPCs differentiated into the neurons, PTB expression was disappeared and the neural PTB was induced. However, during the differentiation of these cells into the astrocytes, they maintained the PTB expression.
44
Ptbp1 was a modulator of pre-mRNA splicing
45
which expressed at high levels in the non-neuronal cells and neuronal precursors. The Ptbp1 expression was reduced by miR-124 during the neuronal differentiation. The reduction of Ptbp1 allowed a neuronal differentiation to happen.
43
We assumed that if Ptbp1 was high so the miR-124 should be low and downregulation of miR-124 suppressed the neuronal differentiation and promoted the astrocyte differentiation.
46
The study by Neo et al suggested that miR-124 is among the most efficient miRNA regulators of the histone H3 Lys-27 histone methyltransferase (Ezh2) expression.
46
The downregulation of miR-124 inhibited the neuronal differentiation and promoted the astrocyte differentiation.
46
Thus, the astrocyte differentiation might happen when the histone H3 Lys-27 histone methyltransferase was increased. Other than Ezh2, the histone deacetylases (HDACs) also regulated miR-124, which implied an important function for miR-124 and its downstream target, Ptbp1, in the generation of epigenetically reprogrammed.
47
These phenomena suggested the relation of epigenetic modification and transcription factors. Thus, the increased gene expression of HDAC3 in the gel+HA+brain on day 9 of the culture might imply that miR-124 was decreased and, according to the work of Neo et al., the astrocyte-specific differentiation was promoted when miR-124 was decreased.
46
The relation between Ptbp1, HDAC3 and astrocyte-specific differentiation is presented in the graphical abstract. SOX9 is involved in differentiation mechanism that INSCs to differentiate from the neurogenesis to the gliogenesis. Stolt et al indicated that SOX9 expressed firstly in the neural stem cells, and later in the glial cells. However, the SOX9 expression in the oligodendrocyte lineage was ended at the onset of terminal differentiation. Then, mature oligodendrocytes expressed SOX10 and SOX8.
48
SOX9 and Olig2 appeared to be important for the oligodendrocyte identity, while SOX9 seemed to be essential for the glial identity and Nkx2.2 and Olig2 were important for the subtype identity. Without the expression of Nkx2.2 and Olig2, it could be concluded that the upregulation of Ptbp1 in accordance with Klf4, SOX9, NFIa and NFIb might result in the astrocytic lineage differentiation, while the neuronal differentiation might be suppressed.
11
Real time-PCR assays for other transcription factors (i.e., Pou3f2, Pax6, Sox2, Pou5f1, Nanog, Notch1, Foxa2) and other neuronal markers (i.e., CD133, nestin, βIIITUB, GFAP) were undetectable. Only 2 astrocyte differentiation markers (i.e., MAP2 and CD44) were detected, thus it could be possible that the biomaterials in our study played just only partial roles in the astrocyte differentiation process. Further studies using ELISA, immunofluorescent staining and RNA sequencing techniques are now going on in our laboratory to confirm the real time-PCR results. For functional astrocyte differentiation, better-designed biomaterials are needed. The importance of each transcription factors
49
is explained in the diagram in Fig. S5 (Supplementary Data).
The PI3K/Akt pathway was activated during reprogramming.
50
PI3K-Akt signaling was important in the early steps of the reprogramming process.
51
Flow cytometry showed that PI3K/Akt activation was increased in the NIH/3T3 cells cultured using all types of biomaterials, especially on the gel+HA+brain. The PI3K/Akt result was concordant with real time-PCR results because most of the gene expression increased significantly in the gel+HA+brain compared to the control. PI3K/Akt activation was decreased in APCs cultured on the gel and gel+HA but PI3K/Akt activation was increased in APCs cultured on the gel+HA+brain. It was possible that the extremely high PI3K activation could happen because of the interference from pig brain particles in the gel+HA+brain. Kat2a was found significantly downregulated in all types of biomaterial compared to the control. Recently, Tapias et al reviewed the roles of lysine acetylation and deacetylation in the brain development
52
and they mentioned that the KAT activity was necessary for the neuronal progenitor differentiation.
52
When human periodontal ligament cell-like fibroblast
53
and human APCs were cultured on the biomaterials for 1 week, it was found that the human PDL cells and APCs could survive but the profile of gene expression was totally different from the NIH/3T3 cells after 1 week culture. In 1 week culture, only OCN and SOX9 genes were detected in both APCs and PDL cells. Significant upregulation of SOX9 was found in both APCs and the PDL cells. PDL cells from another patient (N2) were cultured on the gel+HA and gel+HA+brain for 2 weeks and results of the gene expression suggested dental tissue mineralization
54-57
instead of the neural differentiation. Both types of biomaterials might induce mineralization in different amplitudes. Such finding might indicate that the adult PDL cells strongly maintained the characteristics of mesenchymal lineage, while NIN/3T3 cells present more plasticity.
58
This phenomenon was similar to the study of Caiazzo et al, who capitalized on a set of transcription factors to convert mouse fibroblasts into the functional astrocytes. However, when the method was applied to the human fibroblasts, the result was less successful as compared to the mouse fibroblasts.
11
Conclusion
The NIH/3T3 cells cultured on the gel+HA+brain expressed a set of transcription factors (i.e., NFIa, NFIb, Ptbp1 and SOX9), indicating that the cells were differentiated into an astrocytic lineage. The cells might be passed through the neural stem cell (neural progenitor) state because the NIH/3T3 cells expressed the Klf4, a pluripotency marker which was significantly upregulated on the gelatin+HA+brain during the first week of the cell culture. The PI3K/Akt activation was involved in the early steps of the reprogramming process as it was increased in NIH/3T3 cells cultured using biomaterials especially on the gelatin+HA+brain. These results demonstrate that the chemical and mechanical properties of biomaterials could initiate reprogramming process in the NIH/3T3 fibroblasts in a non-invasive manner. However, the properties of biomaterials are needed to be improved for the clinical applications.
Ethical approval
Ethical approval was obtained for the collection of APCs and PDL cells from third molar or premolar of the patients who had orthodontic treatment at the Faculty of Dentistry, Chiang Mai University.
Competing interests
There is no conflict of interests to be reported.
Acknowledgments
We are grateful to Prof. Kidoaki, Institute for Materials Chemistry and Engineering, Kyushu University, for NIH/3T3 fibroblasts. We would like to give special thanks to Dr. Khajornsak Tragoolpua for all of his help that supported this work.
Funding
We would also like to acknowledge the support from the National Research Council of Thailand (NRCT), [grant number 170327].
Supplementary Data
Supplementary file 1 contains Tables S1-S3 and Figs. S1-S5.
(pdf)
Research Highlights
What is current knowledge?
simple
-
√ Proper biomaterials could be used to initiate cellular
reprogramming which involved with epigenetic modification.
-
√ The reprogramming initiated by the biomaterials could not
drive the complete reprogramming process and might need
combination of other growth factors.
What is new here?
simple
-
√ Biomaterials could induce upregulation of a set of
transcription factors that specific for astrocyte differentiation.
-
√ Biomaterials might impose some impacts on the epigenetic
pattern, indicating biomaterials application as a tool for the
cellular reprogramming or transdifferentiation in the future.
References
- Dantuma E, Merchant S, Sugaya K. Stem cells for the treatment of neurodegenerative diseases. Stem Cell Res Ther 2010; 1:37. doi: 10.1186/scrt37 [Crossref] [ Google Scholar]
- Ho TJ, Chan TM, Ho LI, Lai CY, Lin CH, Macdonald I. The possible role of stem cells in acupuncture treatment for neurodegenerative diseases: a literature review of basic studies. Cell Transplant 2014; 23:559-66. doi: 10.3727/096368914X678463 [Crossref] [ Google Scholar]
- Tanna T, Sachan V. Mesenchymal stem cells: potential in treatment of neurodegenerative diseases. Curr Stem Cell Res Ther 2014; 9:513-21. doi: 10.2174/1574888X09666140923101110 [Crossref] [ Google Scholar]
- Gao A, Peng Y, Deng Y, Qing H. Potential therapeutic applications of differentiated induced pluripotent stem cells (iPSCs) in the treatment of neurodegenerative diseases. Neuroscience 2013; 228:47-59. doi: 10.1016/j.neuroscience.2012.09.076 [Crossref] [ Google Scholar]
- Jung YW, Hysolli E, Kim KY, Tanaka Y, Park IH. Human induced pluripotent stem cells and neurodegenerative disease: prospects for novel therapies. Curr Opin Neurol 2012; 25:125-30. doi: 10.1097/WCO.0b013e3283518226 [Crossref] [ Google Scholar]
- Xie N, Tang B. The Application of Human iPSCs in Neurological Diseases: From Bench to Bedside. Stem Cells Int 2016; 2016:6484713. doi: 10.1155/2016/6484713 [Crossref] [ Google Scholar]
- Allsopp R. Telomere length and iPSC re-programming: survival of the longest. Cell Res 2012; 22:614-5. doi: 10.1038/cr.2012.6 [Crossref] [ Google Scholar]
- Walsh KM, Wiencke JK, Lachance DH, Wiemels JL, Molinaro AM, Eckel-Passow JE. Telomere maintenance and the etiology of adult glioma. Neuro Oncol 2015; 17:1445-52. doi: 10.1093/neuonc/nov082 [Crossref] [ Google Scholar]
- Haycock PC, Burgess S, Nounu A, Zheng J, Okoli GN, Bowden J. Association between telomere length and risk of cancer and non-neoplastic diseases: a mendelian randomization study. JAMA Oncol 2017; 3:636-51. doi: 10.1001/jamaoncol.2016.5945 [Crossref] [ Google Scholar]
- Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A 2011; 108:10343-8. doi: 10.1073/pnas.1105135108 [Crossref] [ Google Scholar]
- Caiazzo M, Giannelli S, Valente P, Lignani G, Carissimo A, Sessa A. Direct conversion of fibroblasts into functional astrocytes by defined transcription factors. Stem Cell Reports 2015; 4:25-36. doi: 10.1016/j.stemcr.2014.12.002 [Crossref] [ Google Scholar]
- Thoma EC, Merkl C, Heckel T, Haab R, Knoflach F, Nowaczyk C. Chemical conversion of human fibroblasts into functional Schwann cells. Stem Cell Reports 2014; 3:539-47. doi: 10.1016/j.stemcr.2014.07.014 [Crossref] [ Google Scholar]
- Shahbazi E, Moradi S, Nemati S, Satarian L, Basiri M, Gourabi H. Conversion of Human Fibroblasts to Stably Self-Renewing Neural Stem Cells with a Single Zinc-Finger Transcription Factor. Stem Cell Reports 2016; 6:539-51. doi: 10.1016/j.stemcr.2016.02.013 [Crossref] [ Google Scholar]
- Outani H, Okada M, Yamashita A, Nakagawa K, Yoshikawa H, Tsumaki N. Direct induction of chondrogenic cells from human dermal fibroblast culture by defined factors. PLoS One 2013; 8:e77365. doi: 10.1371/journal.pone.0077365 [Crossref] [ Google Scholar]
- Deng X, Wang H, Wang T, Fang X, Zou L, Li Z. Non-viral methods for generating integration-free, induced pluripotent stem cells. Current Stem Cell Research & Therapy 2015; 10:153-8. doi: 10.2174/1574888X09666140923101914 [Crossref] [ Google Scholar]
- Zhou YY, Zeng F. Integration-free methods for generating induced pluripotent stem cells. Genomics Proteomics Bioinformatics 2013; 11:284-7. doi: 10.1016/j.gpb.2013.09.008 [Crossref] [ Google Scholar]
- Kantawong F, Kuboki T, Kidoaki S. Redox gene expression of adipose-derived stem cells in response to soft hydrogel. Turk J Biol 2015; 39:682-91. doi: 10.3906/biy-1412-29 [Crossref] [ Google Scholar]
- Kuboki T, Kantawong F, Burchmore R, Dalby MJ, Kidoaki S. 2D-DIGE proteomic analysis of mesenchymal stem cell cultured on the elasticity-tunable hydrogels. Cell Struct Funct 2012; 37:127-39. doi: 10.1247/csf.12012 [Crossref] [ Google Scholar]
- Tanum J, Udomsom S, Wattanutchariya W, Sooksaen P, Kantawong F. Characterization of gelatin composite with low content hydroxyapatite. Key Eng Mater 2016; 675-676:473-6. doi: 10.4028/www.scientific.net/KEM.675-676.473 [Crossref] [ Google Scholar]
- Kantawong F, Tanum J, Wattanutchariya W, Sooksaen P. Variation of Hydroxyapatite Content in Soft Gelatin Affects Mesenchymal Stem Cell Differentiation. BABT 2016:59. doi: 10.3906/biy-1505-29 [Crossref]
- Kantawong F, Thaweenan P, Mungkala S, Tamang S, Manaphan R, Wanachantararak P. Mucus of Achatina fulica stimulates mineralization and inflammatory response in dental pulp cells. Turk J Biol 2016; 40:353-9. doi: 10.3906/biy-1505-29 [Crossref] [ Google Scholar]
- Wang L, Huang W, Su H, Xue Y, Su Z, Liao B. Generation of integration-free neural progenitor cells from cells in human urine. Nat Methods 2013; 10:84-9. doi: 10.1038/nmeth.2283 [Crossref] [ Google Scholar]
- Miura T, Sugawara T, Fukuda A, Tamoto R, Kawasaki T, Umezawa A. Generation of primitive neural stem cells from human fibroblasts using a defined set of factors. Biol Open 2015; 4:1595-607. doi: 10.1242/bio.013151 [Crossref] [ Google Scholar]
- Han DW, Tapia N, Hermann A, Hemmer K, Höing S, Araúzo-Bravo MJ. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell 2012; 10:465-72. doi: 10.1016/j.stem.2012.02.021 [Crossref] [ Google Scholar]
- Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, Li G. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell 2012; 11:100-9. doi: 10.1016/j.stem.2012.05.018 [Crossref] [ Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010; 463:1035-41. doi: 10.1038/nature08797 [Crossref] [ Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ. Induction of human neuronal cells by defined transcription factors. Nature 2011; 476:220-3. doi: 10.1038/nature10202 [Crossref] [ Google Scholar]
- Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A 2011; 108:7838-43. doi: 10.1073/pnas.1103113108 [Crossref] [ Google Scholar]
- Kim SM, Flaßkamp H, Hermann A, Araúzo-Bravo MJ, Lee SC, Lee SH. Direct conversion of mouse fibroblasts into induced neural stem cells. Nat Protoc 2014; 9:871-81. doi: 10.1038/nprot.2014.056 [Crossref] [ Google Scholar]
- Hemmer K, Zhang M, van Wüllen T, Sakalem M, Tapia N, Baumuratov A. Induced neural stem cells achieve long-term survival and functional integration in the adult mouse brain. Stem Cell Reports 2014; 3:423-31. doi: 10.1016/j.stemcr.2014.06.017 [Crossref] [ Google Scholar]
- Zhu S, Wang H, Ding S. Reprogramming fibroblasts toward cardiomyocytes, neural stem cells and hepatocytes by cell activation and signaling-directed lineage conversion. Nat Protoc 2015; 10:959-73. doi: 10.1038/nprot.2015.059 [Crossref] [ Google Scholar]
- Nishimura K, Kato T, Chen C, Oinam L, Shiomitsu E, Ayakawa D. Manipulation of KLF4 expression generates iPSCs paused at successive stages of reprogramming. Stem Cell Reports 2014; 3:915-29. doi: 10.1016/j.stemcr.2014.08.014 [Crossref] [ Google Scholar]
- Schmidt R, Plath K. The roles of the reprogramming factors Oct4, Sox2 and Klf4 in resetting the somatic cell epigenome during induced pluripotent stem cell generation. Genome Biol 2012; 13:251. doi: 10.1186/gb-2012-13-10-251 [Crossref] [ Google Scholar]
- Petersen GF, Strappe PM. Generation of diverse neural cell types through direct conversion. World J Stem Cells 2016; 8:32-46. doi: 10.4252/wjsc.v8.i2.32 [Crossref] [ Google Scholar]
- Linares AJ, Lin CH, Damianov A, Adams KL, Novitch BG, Black DL. The splicing regulator PTBP1 controls the activity of the transcription factor Pbx1 during neuronal differentiation. Elife 2015; 4:e09268. doi: 10.7554/eLife.09268 [Crossref] [ Google Scholar]
- Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit Rev Biochem Mol Biol 2012; 47:360-78. doi: 10.3109/10409238.2012.691456 [Crossref] [ Google Scholar]
- Mason S, Piper M, Gronostajski RM, Richards LJ. Nuclear factor one transcription factors in CNS development. Mol Neurobiol 2009; 39:10-23. doi: 10.1007/s12035-008-8048-6 [Crossref] [ Google Scholar]
- Heng YH, Barry G, Richards LJ, Piper M. Nuclear factor I genes regulate neuronal migration. Neurosignals 2012; 20:159-67. doi: 10.1159/000330651 [Crossref] [ Google Scholar]
- Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 2006; 52:953-68. doi: 10.1016/j.neuron.2006.11.019 [Crossref] [ Google Scholar]
- Rasband MN. Glial Contributions to Neural Function and Disease. Mol Cell Proteomics 2016; 15:355-61. doi: 10.1074/mcp.R115.053744 [Crossref] [ Google Scholar]
- Zhou Q, Choi G, Anderson D. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx22. Neuron 2001; 31:791-807. doi: 10.1016/S0896-6273(01)00414-7 [Crossref] [ Google Scholar]
- Ligon K, Alberta J, Kho A, Weiss J, Kwaan M, Nutt C. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol 2004; 63:499-509. [ Google Scholar]
- Ling JP, Chhabra R, Merran JD, Schaughency PM, Wheelan SJ, Corden JL. PTBP1 and PTBP2 Repress Nonconserved Cryptic Exons. Cell Rep 2016; 17:104-13. doi: 10.1016/j.celrep.2016.08.071 [Crossref] [ Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev 2007; 21:1636-52. doi: 10.1101/gad.1558107 [Crossref] [ Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 2007; 27:435-48. doi: 10.1016/j.molcel.2007.07.015 [Crossref] [ Google Scholar]
- Neo WH, Yap K, Lee SH, Looi LS, Khandelia P, Neo SX. MicroRNA miR-124 controls the choice between neuronal and astrocyte differentiation by fine-tuning Ezh2 expression. J Biol Chem 2014; 289:20788-801. doi: 10.1074/jbc.M113.525493 [Crossref] [ Google Scholar]
- Wang D, Zhang H, Li M, Frid MG, Flockton AR, McKeon BA. MicroRNA-124 controls the proliferative, migratory, and inflammatory phenotype of pulmonary vascular fibroblasts. Circ Res 2014; 114:67-78. doi: 10.1161/CIRCRESAHA.114.301633 [Crossref] [ Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev 2003; 17:1677-89. doi: 10.1101/gad.259003 [Crossref] [ Google Scholar]
- Gopalakrishnan S, Hor P, Ichida JK. New approaches for direct conversion of patient fibroblasts into neural cells. Brain Res 2017; 1656:2-13. doi: 10.1016/j.brainres.2015.10.012 [Crossref] [ Google Scholar]
- Hawkins K, Joy S, McKay T. Cell signalling pathways underlying induced pluripotent stem cell reprogramming. World J Stem Cells 2014; 6:620-8. doi: 10.4252/wjsc.v6.i5.620 [Crossref] [ Google Scholar]
- Hossini AM, Quast AS, Plötz M, Grauel K, Exner T, Küchler J. PI3K/AKT Signaling Pathway Is Essential for Survival of Induced Pluripotent Stem Cells. PLoS One 2016; 11:e0154770. doi: 10.1371/journal.pone.0154770 [Crossref] [ Google Scholar]
- Tapias A, Wang ZQ. Lysine Acetylation and Deacetylation in Brain Development and Neuropathies. Genomics Proteomics Bioinformatics 2017; 15:19-36. doi: 10.1016/j.gpb.2016.09.002 [Crossref] [ Google Scholar]
- Jönsson D, Nebel D, Bratthall G, Nilsson BO. The human periodontal ligament cell: a fibroblast-like cell acting as an immune cell. J Periodontal Res 2011; 46:153-7. doi: 10.1111/j.1600-0765.2010.01331.x [Crossref] [ Google Scholar]
- Orimo H. The mechanism of mineralization and the role of alkaline phosphatase in health and disease. J Nippon Med Sch 2010; 77:4-12. doi: 10.1272/jnms.77.4 [Crossref] [ Google Scholar]
- Goldberg M, Septier D, Lécolle S, Chardin H, Quintana MA, Acevedo AC. Dental mineralization. Int J Dev Biol 1995; 39:93-110. [ Google Scholar]
- Goldberg M, Kulkarni AB, Young M, Boskey A. Dentin: structure, composition and mineralization. Front Biosci (Elite Ed) 2011; 3:711-35. [ Google Scholar]
- McKee MD, Hoac B, Addison WN, Barros NM, Millán JL, Chaussain C. Extracellular matrix mineralization in periodontal tissues: Noncollagenous matrix proteins, enzymes, and relationship to hypophosphatasia and X-linked hypophosphatemia. Periodontol 2000 2013; 63:102-22. doi: 10.1111/prd.12029 [Crossref] [ Google Scholar]
- Dadheech N, Srivastava A, Belani M, Gupta S, Pal R, Bhonde RR. Basal expression of pluripotency-associated genes can contribute to stemness property and differentiation potential. Stem Cells Dev 2013; 22:1802-17. doi: 10.1089/scd.2012.0261 [Crossref] [ Google Scholar]