BioImpacts. 8(3):167-176.
doi: 10.15171/bi.2018.19
Original Research
Expression, purification and DNA-binding properties of zinc finger domains of DOF proteins from Arabidopsis thaliana
Hakimeh Moghaddas Sani 1, 2, Maryam Hamzeh-Mivehroud 2, 3, Ana P. Silva 4, James L. Walshe 4, S. Abolghasem Mohammadi 5, Mahdyieh Rahbar-Shahrouziasl 2, Milad Abbasi 2, Omid Jamshidi 2, Jason KK Low 4, Siavoush Dastmalchi 2, 3, 6, *, Joel P. Mackay 4
Author information:
1Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
2Biotechnology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
3School of Pharmacy, Tabriz University of Medical Sciences, Tabriz, Iran
4School of Life and Environmental Sciences, The University of Sydney, NSW 2006, Australia
5School of Agriculture, University of Tabriz, Tabriz, Iran
6Faculty of Pharmacy, Near East University, POBOX:99138, Nicosia, North Cyprus, Mersin 10, Turkey
Abstract
Introduction:
DOF proteins are a family of plant-specific transcription factors with a conserved zinc finger (ZF) DNA-binding domain. Although several studies have demonstrated their specific DNA binding, quantitative affinity data is not available for the binding of DOF domains to their binding sites.
Methods:
ZF domains of DOF2.1, DOF3.4, and DOF5.8 from Arabidopsis thaliana were expressed and purified. Their DNA binding affinities were assessed using gel retardation assays and microscale thermophoresis with two different oligonucleotide probes containing one and two copies of recognition sequence AAAG.
Results:
DOF zinc finger domains (DOF-ZFs) were shown to form independently folded structures. Assessments using microscale thermophoresis demonstrated that DOF-ZFs interact more tightly (~ 100 fold) with double-motif probe than the single-motif probe. The overall Kd values for the DOF3.4-ZF and DOF5.8-ZF to the double-motif probe were ~2.3±1 and 2.5±1 µM, respectively.
Conclusion:
Studied DOF-ZF domains formed stable complexes with the double-motif probe. Although DOF3.4-ZF and DOF5.8-ZF do not dimerize with an appreciable affinity in the absence of DNA (judging from size-exclusion and multiangle laser light scattering data), it is possible that these ZFs form protein-protein contacts when bound to this oligonucleotide, consistent with previous reports that DOF proteins can homo- and hetero-dimerize.
Keywords: DOF zinc finger domain, DNA binding affinity, Gel retardation assay, Microscale thermophoresis
Copyright and License Information
© 2018 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Transcription factors (TFs) are central control points in gene regulatory networks. The DNA-binding domains of TFs can be divided into a number of structural families and recognize specific sequences of target genes.
1
Zinc finger (ZF) proteins are one of the largest TF super-families in plants.
2
Members of this super-family contain domains that are stabilized by the presence of one or more zinc ions, and several families of ZF domains have been identified, based on their structure and zinc coordination pattern.
3
The DNA-binding protein with one finger (DOF) family is a plant-specific multigene family of TFs and since the discovery of the first member of the family in maize,
4
numerous putative coding genes for DOF proteins have been detected in various species of plants ranging from lower plants such as the green alga Chlamydomonas reinhardtii, which has a single CrDOF-domain containing protein, and the moss Physcomitrella patens (19 PpDOF proteins)
5
to the higher plants, including both angiosperms and gymnosperms such as potato (35 StDOF proteins),
6
tomato (34 SlDOF proteins),
7
barley (26 HvDOF proteins),
8
and rice (30 OsDOF proteins).
9
Phylogenetic analysis has shown that the number of DOF protein-coding genes is directly related to the complexity of the organism.
8
In Arabidopsis thaliana, the 37 DOF coding genes comprise about 10% of all ZF-coding genes in that organism.
10
The DOF protein family is mostly expanded during the evolution of the vascular plants through recurrent duplication of the ancestor genes after the divergence of green algae and the ancestors of terrestrial plants.
5
Unlike many other ZF proteins, which contain several ZFs that cooperate in DNA and protein binding, DOF proteins exclusively harbor only one ZF. This domain is characterized by a highly conserved DNA-binding domain at the N-terminal region of the protein, consisting of 52 amino acids and defined by a C-x(2)-C-x(7)-[CS]-x(13)-C-x(2)-C-x-R-x-W-T-x-G-G motif (where x is any amino acid).
11
Despite the similarity of the DOF-ZF consensus to ZFs in steroid hormone receptors and metazoan GATA ZFs (Fig. 1), the DOF-ZF has a longer loop separating the putative zinc-coordinating cysteine pairs (the C-X2-C units). It has been shown that shortening of this loop in the DOF-ZF domain of ascorbate oxidase binding protein (AOBP) by deleting seven or more residues or replacement of the loop with that of zinc-binding unit 1 of estrogen receptor abolishes DNA binding activity.
12
Substitution of the conserved cysteine residue near the center of the loop region of ZF domain of pumpkin AOBP improves DNA binding properties while substitution of the tryptophan residue close to the end of the same ZF domain (Fig. 1) reduces DNA binding.
13
So far, all reported DOF proteins for which DNA-binding data are available to recognize promoters containing cis-regulatory elements with the AAAG motif, except for pumpkin AOBP for which the recognition site is AAGT.
11

Fig. 1.
Sequence alignment of the DOF domains of AOBP, AtDOF3.4, and NtBBF1 with two zinc binding fingers of GATA1 (F1 and F2) and unit1 (F1) of the human androgen receptor (ANDR) and the glucocorticoid receptor (GLU). Conserved Cys residues proposed to be involved in DNA binding are highlighted in gray and the two conserved aromatic residues are boxed. The conserved Cys near at the center of the DOF-ZF sequence is shown by an asterisk (*).The UniProt ID of each protein sequence is given inside the bracket.
.
Sequence alignment of the DOF domains of AOBP, AtDOF3.4, and NtBBF1 with two zinc binding fingers of GATA1 (F1 and F2) and unit1 (F1) of the human androgen receptor (ANDR) and the glucocorticoid receptor (GLU). Conserved Cys residues proposed to be involved in DNA binding are highlighted in gray and the two conserved aromatic residues are boxed. The conserved Cys near at the center of the DOF-ZF sequence is shown by an asterisk (*).The UniProt ID of each protein sequence is given inside the bracket.
DOF proteins are essential for the growth and development of plants and interestingly they mostly control biological processes that are exclusive to plants, such as vascular development, phytochrome signaling, seed germination, nitrogen assimilation, photosynthetic process and resistance to abiotic stresses.
11
Although DOF proteins vary in sequence and function, their DOF-ZF domains show significant sequence similarity (Fig. S1). High similarity among the amino acid sequences of DOF-ZF domains suggests the similarity in their DNA binding specificity. However, although features of the interaction between DOF3.4-ZF from A. thaliana and its cognate DNA recognition sequence have been proposed based on molecular models built with GATA1 ZF proteins as templates, no experimental structural information is available either on DOF-ZF domains themselves or on their interaction with DNA.
14
In the current study, a number of DOF-ZF domains from A. thaliana, namely DOF2.1, 3.4 and 5.8, were cloned, expressed, and purified in a bacterial expression system. One-dimensional NMR and fluorescence spectra confirmed that these recombinant domains are able to form stably folded structures and the binding ability of these domains to double-stranded oligonucleotides containing one and 2 copies of the consensus sequence AAAG was investigated. To the best of our knowledge, this is the first experimental report describing the quantitative measurement of the sequence-specific DNA binding affinity of DOF domains.
Materials and Methods
Reagents
Qiagen RNeasy, and RevertAid cDNA synthesis kits were purchased from Qiagen (Hilden, Germany). Gel purification and plasmid mini extraction kits were obtained from Bioneer (South Korea). Primers were supplied from Bioron (Ludwigshafen, Germany). Glutathione Sepharose 4B was purchased from GE Healthcare Life Sciences (Daejeon, Sweden). 5’-Fluorescein-labelled and unlabelled oligonucleotides were purchased as synthetic oligonucleotides from Integrated DNA Technologies (NSW, Australia). Deuterium oxide (D2O, >99.96% purity) was from Sigma (Missouri, USA) and 2, 2-dimethyl-2-silapentane-5-sulfonate sodium salt (DSS) was obtained from Fluka A.G. (Buchs, Switzerland).
Plant
Arabidopsis thaliana (ecotype: Columbia) seeds were obtained from the Faculty of Agriculture, Tabriz University, Iran. The seed preparation and growing were all carried out in a greenhouse at Tabriz University. The protocol for planting and growing A. thaliana was that used by the Arabidopsis Biological Resource Center with minor modifications (http://abrc.osu.edu/seed-handling).
Construction of the expression vector
To construct the vectors used for DOF-ZF domain expression, total RNA from 3-week old A. thaliana was utilized. Nested PCR amplification was performed using 2 sets of primers (Table 1) designed based on sequence information for DOF2.1 (AT2G28510), DOF3.4 (At3g50410) and DOF5.8 (At5g66940) available at NCBI. Plasmids to express the DOF-ZF domains N-terminally fused to glutathione S-transferase were constructed by inserting BamHI and EcoRI double digested PCR products between the same restriction sites in the pGEX-6p-1 vector. The accuracy of the constructs was verified by Sanger sequencing.
Table 1.
Oligonucleotide primers used for amplification of DOF ZF domains
|
DOF proteins
|
|
Outer primers
|
Inner primers
|
| DOF2.1 |
Forward |
5'GTGCAGGAAATCTCAAACGAGAC3' |
5'AAAGGATCCAGAGGAGAATTAGGAGG3' |
|
|
Reverse |
5'TGATGCTTTTGGAGAGTAGCGA3' |
5'GAGGAGAATTCTCATCGACGGCAACCA3' |
| DOF3.4 |
Forward |
5'CCAAATTCTCACTCTCTCATACCCT3' |
5'GCGTAGGATCCCCGATTTCTGACC3' |
|
|
Reverse |
5'GAGGGAAGAGAACAGGCGTC3' |
5'GAACGGAATTCTCATTTACGAGTACC3' |
| DOF5.8 |
Forward |
5'ACGGCCAAGGAGGATCTGTTGC3' |
5'TCTGGATCCATTCCGACGGATCAACAA3' |
|
|
Reverse |
5'CCGTCGTGATACCGCCGTTGG3' |
5'GAACGGAATTCTCATTTACGGGAAAC3' |
Expression and purification of recombinant DOF-ZF proteins
Plasmids containing GST-DOF-ZF coding sequences were transformed and expressed in Escherichia. coli BL21 (DE3) cells. Bacteria were grown at 37°C in one liter of LB broth containing 100 µg/mL ampicillin to an optical density of 0.7 and expression of the GST-DOF protein induced by addition of 0.4 mM of isopropyl-1-thio-ß-D-galactopyranoside (IPTG) followed by overnight incubation at 20°C. Cells were harvested and resuspended in lysis buffer (50 mM Tris-HCl pH 7.2, 500 mM NaCl, 10% Triton-X100, 1.4 mM phenyl methyl sulfonyl fluoride (PMSF), 0.1% beta-mercaptoethanol (2-ME), 0.1 mg/mL lysosyme, 10 μg/mL DNaseI, and 10 µM ZnSO4). Cell disruption was induced by 5 rounds of sonication pulses (30% amplitude) for 30 seconds with a pause interval of 30 seconds. The samples were cooled to 4°C prior to sonication and kept on ice during sonication. The bacterial lysate was centrifuged at 10 000 rpm for 20 minutes at 4°C. PEI (polyethyleneimine, 0.8% v/v) was used to precipitate nucleic acid. The supernatant was subjected to affinity chromatography by incubation with glutathione-Sepharose 4B beads for 1.5 hours at 4°C. Subsequently, the beads were washed with 5 column volumes of wash buffer (150 mM NaCl, 50 mM Tris pH 7.2, and 1 mM DTT). Then, the GST-DOF-ZF was eluted using 20 mM Tris-HCl pH 7.2, 10 mM glutathione, 150 mM NaCl, 1 mM DTT. Fractions containing GST-DOF-ZF were collected and used in gel retardation assays. To prepare recombinant protein free from GST, the eluted GST-DOF-ZF protein was incubated with PreScission protease in cleavage buffer (150 mM NaCl, 50 mM Tris pH 7.2, 1 mM DTT) at 4°C overnight. The cleaved DOF-ZFs (which have 5 non-native amino acids at their N-terminus, of which 3 are derived from the cleavage site and 2 from the translation of the BamHI site) was purified using UNO SI cation exchange chromatography (IEC) (BioRad). Proteins at each step of the expression and purification were subjected to SDS-PAGE analysis. Protein concentration was measured on a Nanodrop® ND-1000 UV-Vis spectrophotometer (ThermoFisher Scientific, Wilmington DE, USA) at 280 nm, using extinction coefficients calculated from the amino acid sequence.
Atomic absorption spectroscopy
The zinc content of the GST-DOF-ZF samples was determined by flame atomic absorption spectroscopy on a CTA3000 spectrometer (ChemTech Analytical Instruments Limited, UK). A protein sample was applied into the atomic absorption spectrophotometer and the signal monitored at the absorption maximum for Zn2+ (213.5 nm). Concentrations were determined in reference to a standard curve constructed using standard Zn2+ solutions (1000 mg/L in 0.5 mol/L nitric acid; BDH, stock solution diluted to final concentrations of 1-10 ppm). Two determinations were performed for each GST-DOF-ZF and the error value for the determination was calculated based on the error of the calibration curve.
Folding assessment of the proteins
Protein samples (100 µM) were prepared in 10 mM potassium phosphate buffer pH 7.2, containing 100 mM NaCl, 1 mM DTT, in 5% (v/v) D2O and 2 μM 2, 2-dimethyl-2-silapentane-5-sulfonate sodium salt (DSS). One-dimensional (1D) 1H NMR spectra were collected on a Bruker AvanceIII 600-MHz spectrometer and processed by TOPSPIN3 (Bruker, Karlsruhe, Germany). To record the fluorescence emission spectra of DOF-ZFs, protein samples were prepared at 2 μM concentration in a buffer comprising of 10 mM potassium phosphate buffer pH 7.2, 100 mM NaCl, and 0.1% 2-ME. The fluorescence intensity was recorded with a Jasco FP-750 spectrofluorimeter (Jasco Corporation, Tokyo, Japan) at 25°C, before and following the addition of guanidinium hydrochloride to a final concentration of up to 6 M. An excitation wavelength of 280 nm was used and the excitation slit width was set to 5 nm.
Oligonucleotide preparation
The lyophilized 5’-fluorescein-labeled and unlabeled oligonucleotides were reconstituted to a concentration of 1 mM in DEPC water. For the preparation of dsDNA, forward and reverse oligonucleotides were mixed in a 1:1 molar ratio. The mixture was heated at 90°C for 10 min and then was cooled slowly to room temperature. The resulting double-stranded DNA was purified using a Superose™ 12 HR 10/30 column using an AKTA FPLC system (GE Healthcare Life Sciences, Silverwater, NSW). The concentration of each oligomer was determined by UV absorbance using their calculated extinction coefficient.
Gel retardation assay
Single- and double-motif probes (Table 2) were derived from the promoter sequence of the GST6 and DOF2.3 genes in A. thaliana. GST-DOF-ZF proteins (10 µM) were incubated with each dsDNA fragment (at 3 µM) in a total volume of 10 µL in a reaction buffer comprising 10 mM HEPES pH 7.8, 50 mM KCl, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT and 5% glycerol. After a 30-minute incubation at 4°C, the samples were analyzed by 6% native polyacrylamide gel electrophoresis and DNA bands were visualized using ethidium bromide staining.
Table 2.
Oligonucleotide sequences used for gel retardation assays
|
Probe name
|
Sequence
|
Gene
|
Reference
|
| Single-motif probe |
AATCCAAAAGTGTAGAGGAG
|
GST6 promoter |
Chen,1996 |
| Double-motif probe |
AAATAATCATAAAGTATTAAAGTAATATATAC
|
DOF2.3 promoter |
Skirycz, 2008 |
| Control for single-motif probe |
AATCCAGCGATGTAGAGGAG
|
|
|
| Control for double-motif probe |
AAATAATCATGCGATATTGCGAAATATATAAC
|
|
|
Microscale thermophoresis
The affinity of DOF-ZF domains for target oligonucleotides was assessed using microscale thermophoresis. A constant quantity of fluorescently-labeled DNA (100 nM) was incubated with a range of concentrations of protein at room temperature for 15 minutes in 20 μL reaction buffer (10 mM potassium phosphate pH 7.2, 100 mM NaCl, 1 mM DTT, 5 mM MgCl2, 0.05% Tween 20). Samples were aspirated into the standard treated capillaries and scanned using a blue excitation laser on a Monolith™ NT.115 instrument (NanoTemper Technologies GmbH, München, Germany). The assessment was carried out using 50% LED power and 20% IR-laser power. The normalized fluorescence values were fitted to a simple 1:1 Langmuir binding isotherm in the Nanotemper software. The data from 3 individual measurements for each sample were used for the analyses.
Protein molecular weight determination using SEC and multiangle laser light scattering (SEC-MALLS)
Purified proteins were subjected to size-exclusion chromatography using a Superose™ 12 HR 10/30 size-exclusion column (GE Healthcare, Parramatta, NSW) with an in-line MiniDawn MALLS detector with a laser source at 690 nm (Wyatt Technology, Santa Barbara, CA, USA) and Wyatt Refractometer. Proteins were eluted in 20 mM Tris pH 8.0, 150 mM NaCl, 1mM DTT using a flow rate of 0.4 mL/min. The weight-average molecular weight was calculated using the intensity of scattered light at 90° in combination with the change in refractive index. Protein concentration at the detector was determined by the change in refractive index.
Results
DOF domains are bona fide ZFs that form stable, folded structures
To investigate the interaction of DOF-ZF domains with DNA, the DOF domains from 3 DOF proteins from the model organism A. thaliana, namely DOF2.1, DOF3.4 and DOF5.8, were cloned into the pGEX-6p-1 bacterial expression vector. The constructs used here were 60 amino acids in length to cover the coding sequence of the predicted DOF-ZF domain. The similarity of these 3 DOF-ZF domains to other members of the family from different plant species is high, as is shown in supplemental Fig. S1. All 3 DOF-ZF domains were expressed at high levels in E. coli BL21 (DE3) as a fusion protein with a GST tag at the amino terminus and the GST-tagged DOF-ZF proteins were purified using glutathione affinity chromatography (Fig. 2A). In order to assess whether these polypeptides are in fact zinc-binding domains, each of the 3 purified proteins was subjected to atomic absorption spectroscopy. The 3 proteins yielded Zn: protein ratios of 0.74±0.16 (DOF2.1-ZF), 1.07±0.08 (DOF3.4-ZF) and 1.07±0.23 (DOF5.8-ZF). Thus, these data are consistent with each DOF domain binding a single Zn2+ ion, most likely via the sidechains of the 4 cysteine residues highlighted in grey in Fig. 1.
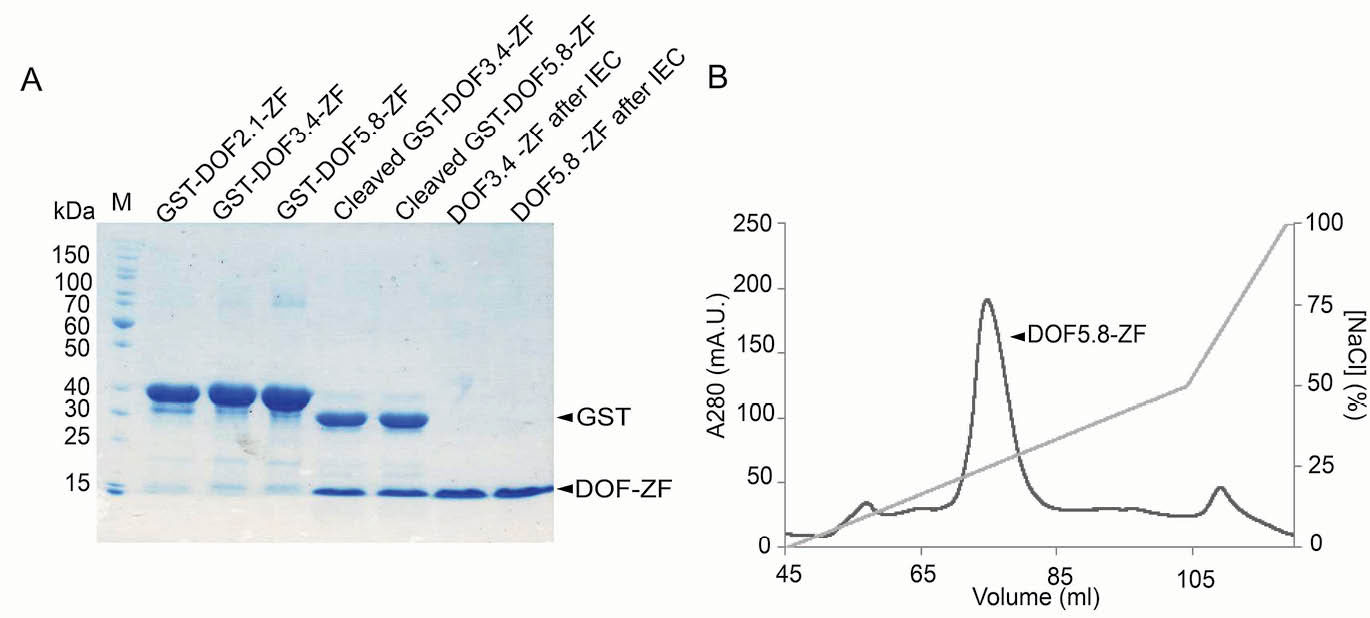
Fig. 2.
Expression and purification of A. thaliana DOF-ZF domains. (A) SDS-PAGE analysis of the proteins after cleavage and purification. M: protein molecular weight marker. (B) A representative cation exchange chromatogram for purification of DOF5.8-ZF after cleaving the GST tag.
.
Expression and purification of A. thaliana DOF-ZF domains. (A) SDS-PAGE analysis of the proteins after cleavage and purification. M: protein molecular weight marker. (B) A representative cation exchange chromatogram for purification of DOF5.8-ZF after cleaving the GST tag.
To allow further studies of these domains, the GST tag was removed by enzymatic cleavage and the DOF-ZF domains of DOF3.4 and DOF5.8 were successfully purified by cation exchange chromatography (Fig. 2A). In contrast, DOF2.1-ZF underwent significant precipitation following cleavage and could not be obtained in appreciable amounts. As shown in Fig. 2B, DOF3.4-ZF and DOF5.8-ZF were able to be prepared to a high level of purity (95%, as assessed by the image processing program ImageJ).
To assess the structural properties of these purified recombinant DOF-ZFs, we first recorded one-dimensional 1H NMR spectra for the DOF3.4-ZF and DOF5.8-ZF. The sharp and well-dispersed 1H NMR signals illustrated in Fig. 3A suggest stably folded structures for both proteins. We also recorded fluorescence emission spectra of DOF3.4-ZF and DOF5.8-ZF to monitor the physical environment of the single tryptophan residue. Fig. 3B and 3C show the variation in the intrinsic fluorescence emission spectra of both DOF3.4-ZF and DOF5.8-ZF, respectively, upon the addition of guanidinium hydrochloride (GndHCl) at concentrations ranging from 1 to 6 M. A significant redshift of λmax (from 331 to 356 nm) was observed following the addition of the GndHCl, consistent with an increase in solvent exposure for the tryptophan. In 5 and 6 M GndHCl the spectra were identical with that of pure tryptophan (not shown), indicating complete denaturation of the protein. Therefore, it can be concluded that each domain takes up a well-ordered structure in phosphate buffer.
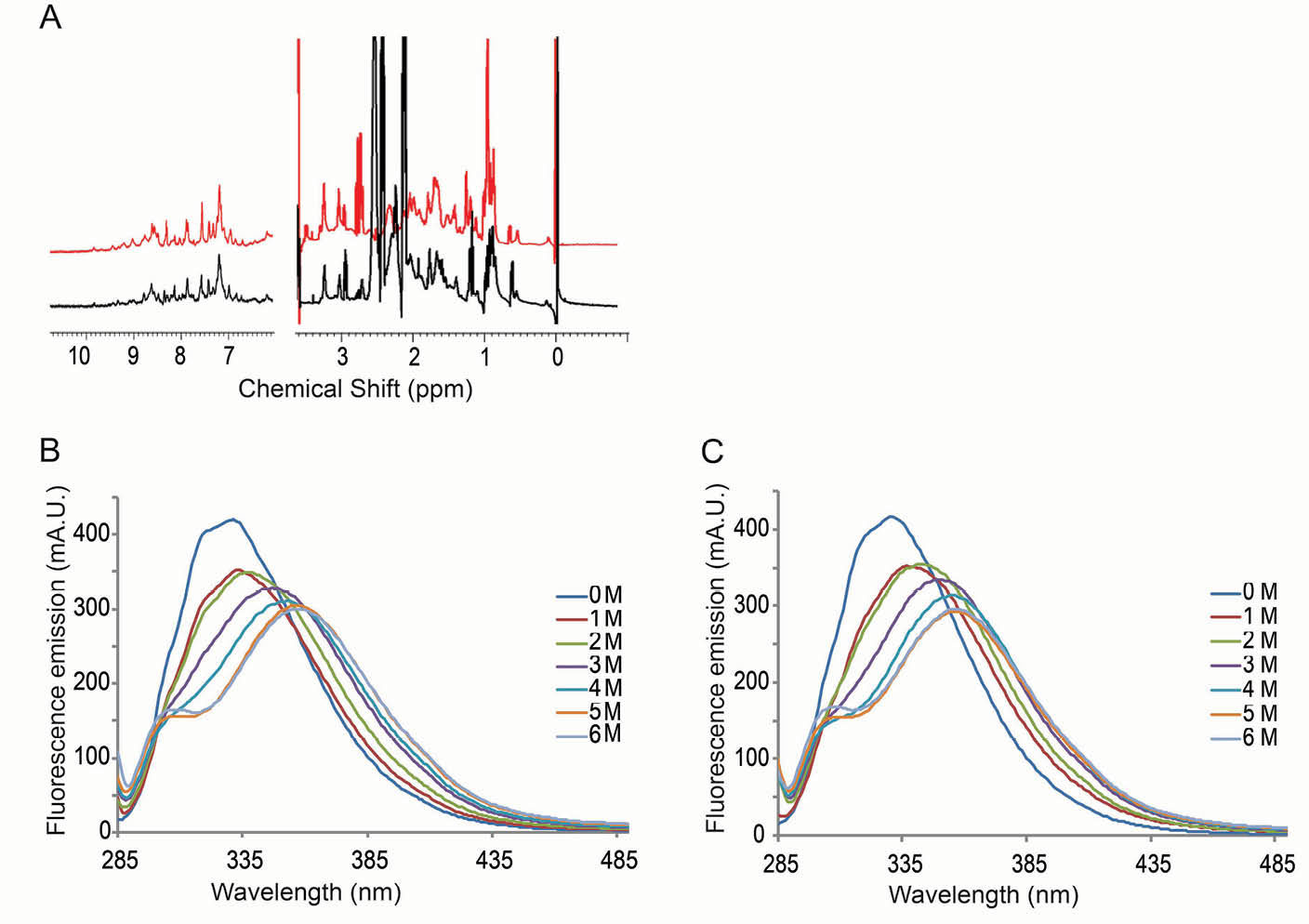
Fig. 3.
Assessment of the folding of DOF-ZFs. (A) Portions of the 1D 1H NMR spectra of DOF5.8-ZF (red) and DOF3.4-ZF (black) (100 µM, 25°C). (B) Fluorescence emission spectra of DOF5.8-ZF and DOF3.4-ZF, respectively, before and after the addition of guanidinium hydrochloride to the indicated concentrations.
.
Assessment of the folding of DOF-ZFs. (A) Portions of the 1D 1H NMR spectra of DOF5.8-ZF (red) and DOF3.4-ZF (black) (100 µM, 25°C). (B) Fluorescence emission spectra of DOF5.8-ZF and DOF3.4-ZF, respectively, before and after the addition of guanidinium hydrochloride to the indicated concentrations.
GST-DOF-ZFs bind AAAG motifs in a sequence-specific manner
We next investigated the ability of GST fused DOF-ZF domains to bind DNA. The promoter of the GST6 gene, a gene involved in environmental stress responses, contains an octopine synthase (ocs) element and a DOF binding site upstream to the ocs element. In vitro gel retardation assays have shown that full-length DOF3.4 stimulates recruitment of OBF4 and OBF5 (ocs binding proteins) to their binding site and also that DOF3.4 binds by itself to a single AAAG sequence derived from the GST6 promoter.
15
We had synthesized a 20-base pair oligonucleotide that contains this single AAAG site from the GST6 promoter (single-motif probe, Table 2). Another study using chromatin immunoprecipitation coupled to quantitative PCR verified the DOF 2.3 gene promoter as another target of DOF3.4.
16
The ability of the full -length DOF3.4 to the DNA fragment derived from the DOF2.3 promoter was demonstrated using an in vitro gel retardation assay. This fragment contains 2 AAAG motifs in a parallel arrangement separated by 4 nucleotides and we selected this sequence for our experiments (double-motif probe, Table 2). We performed the gel retardation assays by titrating both single and double-motif probes with GST-DOF-ZF proteins. As shown in Fig. 4, both probes exhibited interaction with all 3 GST-DOF-ZFs, albeit with significantly different characteristics leading to a decreased amount of free DNA at the bottom of the gel. All 3 double-motif probe-protein mixtures showed a clear retarded DNA band on the gel, whereas for single-motif probe there is no such discrete retarded band. This sharp band appears in a concentration-dependent manner, consistent with the formation of a single complex with a defined stoichiometry. The data suggest that the dissociation constant for formation of a complex with the double-motif probe under these conditions is ~3 µM for GST-DOF-2.1 and ~5 µM for GST-DOF-3.4 and GST-DOF-5.8.
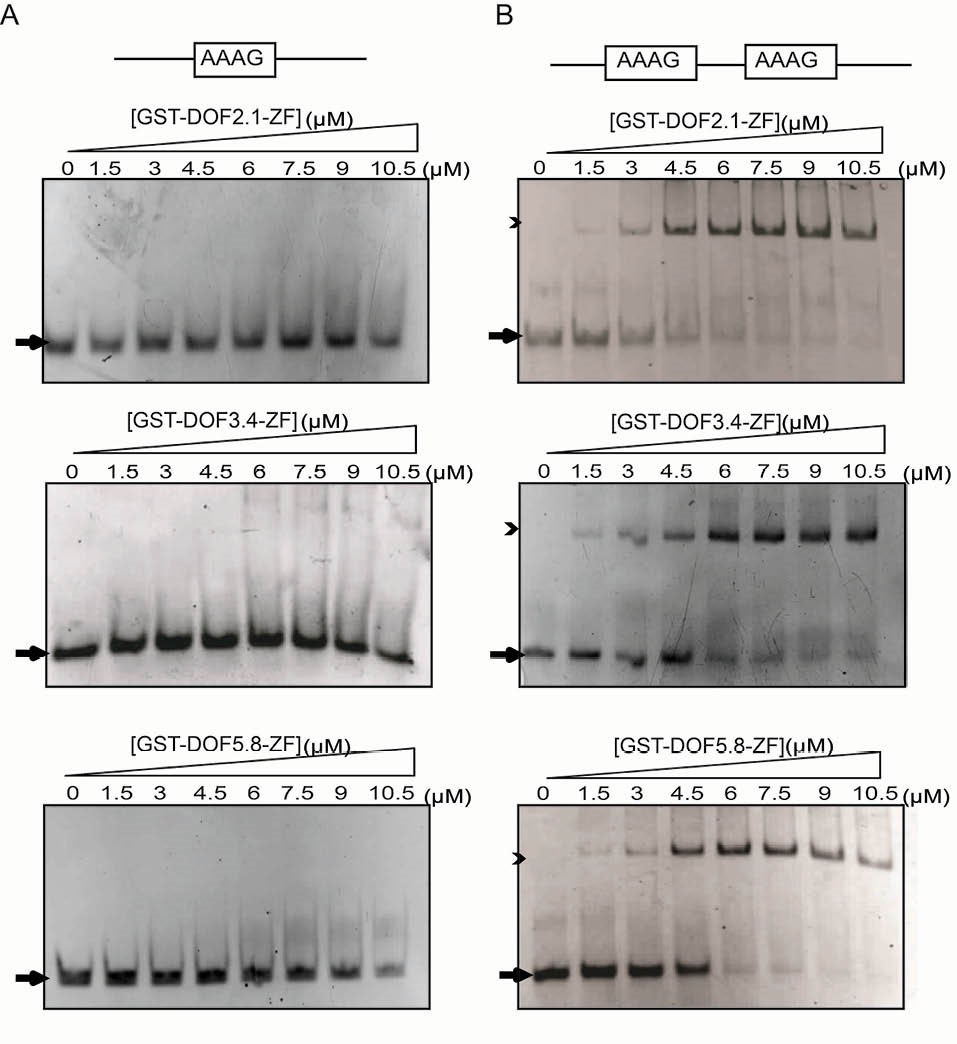
Fig. 4.
DNA binding analysis of GST-DOF-ZFs using gel retardation assays. Interactions are shown between increasing amounts of three GST-DOF-ZFs with (A) single-motif probe (3 µM), and (B) double-motif probe (3 µM) on native polyacrylamide gels stained with ethidium bromide. Arrows and arrow heads show free and bound DNA, respectively. Protein concentrations are shown above the gel.
.
DNA binding analysis of GST-DOF-ZFs using gel retardation assays. Interactions are shown between increasing amounts of three GST-DOF-ZFs with (A) single-motif probe (3 µM), and (B) double-motif probe (3 µM) on native polyacrylamide gels stained with ethidium bromide. Arrows and arrow heads show free and bound DNA, respectively. Protein concentrations are shown above the gel.
To examine the possible contribution of GST dimerization to the observed bands, the DNA retardation assays were repeated for cleaved DOF3.4-ZF and DOF5.8-ZF using both single and double-motif probes (Fig. 5). For the double-motif probe, binding is still clearly observed as a discrete band, although the shifted band is not as well defined as for the GST-fusion proteins. Inspection of the concentration dependence of binding indicates that the apparent dissociation constant is ~9 µM for both domains, which is within a factor of 2 of the estimated dissociation constants for the GST-fusion proteins. These data suggest that the presence of GST have a measurable but relatively small effect on binding of the DOF domains to DNA. The clearer shifted bands might indicate that the GST is somewhat decreasing the off-rate of the complex, increasing the lifetime of the complex as it runs through the gel.
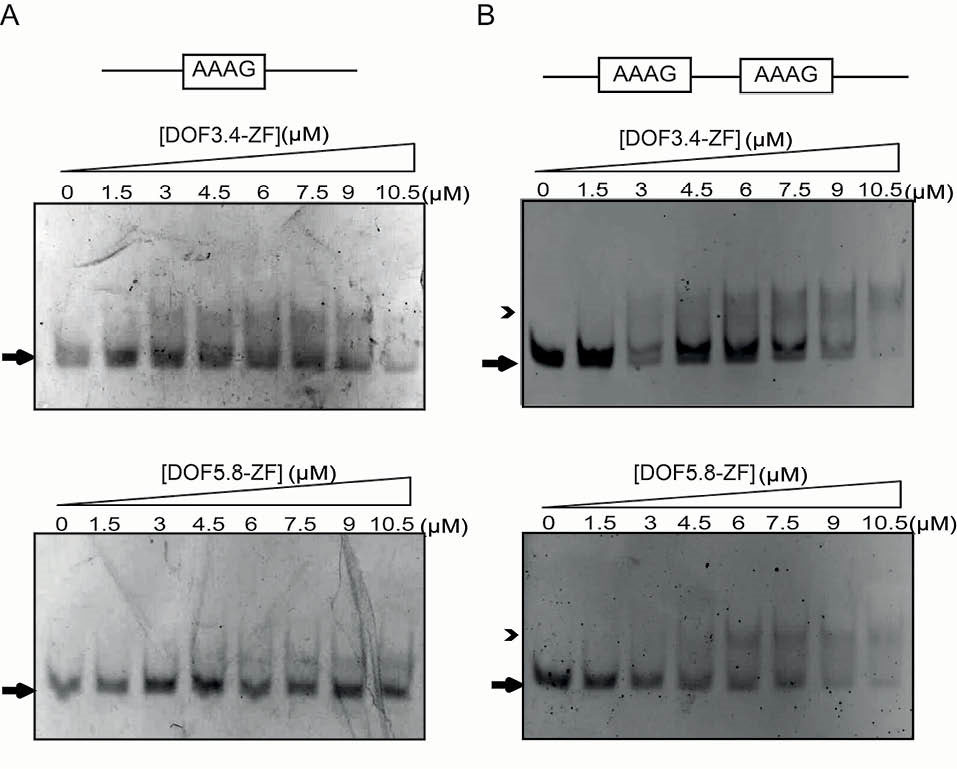
Fig. 5.
DNA binding analysis of DOF-ZFs using gel retardation assays. Interactions are shown between increasing amounts of DOF3.4-ZF and DOF5.8-ZF with (A) single-motif probe (3 µM), and (B) double-motif probe (3 µM) on native polyacrylamide gels. Arrows and arrow heads show free and bound DNA, respectively. Protein concentrations are shown above the gel.
.
DNA binding analysis of DOF-ZFs using gel retardation assays. Interactions are shown between increasing amounts of DOF3.4-ZF and DOF5.8-ZF with (A) single-motif probe (3 µM), and (B) double-motif probe (3 µM) on native polyacrylamide gels. Arrows and arrow heads show free and bound DNA, respectively. Protein concentrations are shown above the gel.
In the case of the single AAAG probe, binding is also noticeably weaker although a smear is still visible, suggesting the formation of a complex with a faster off-rate. Moreover, to begin to assess the DNA-binding specificity of each domain, we also tested the binding of each DOF-ZF proteins with and without GST tag to oligonucleotide probes in which we had mutated AAAG motifs to GCGA (Table 2). None of the 3 DOF domains bound to the control oligonucleotides (data not shown), indicating that these domains are indeed targeting the AAAG motif.
To corroborate these binding data, we turned to microscale thermophoresis. Analysis of the interaction between 5’-fluorescein-labeled single-motif probe and either DOF5.8-ZF or DOF3.4-ZF revealed overall Kd values of greater than 100 µM, which is considered very weak for a DNA-binding ZF domain (Fig. 6A). However, the DNA-binding affinity of DOF3.4-ZF and DOF5.8-ZF to the double-motif probe under the same conditions showed significantly tighter binding with overall Kd values of 2.3±1 µM and 2.5±1 µM, respectively (Fig. 6B). Encouragingly, these values closely resemble the Kd estimates obtained from our gel retardation assays. To assess the oligomerization states of DOF-ZFs in solution, we used size exclusion chromatography coupled to multi-angle laser light scattering (SEC-MALLS). DOF3.4-ZF eluted from SEC-MALLS as a single species with a molecular weight of 6.9±4 kDa (theoretical Mw for the monomer = 7.3 kDa) (Fig. 7A). As shown in Fig. 7B, the retention time of DOF5.8 is very similar to that of DOF3.4, indicating that this domain has the same self-association state as DOF3.4. These results show that both DOF domains are most likely in a monomeric state in solution in the absence of DNA.
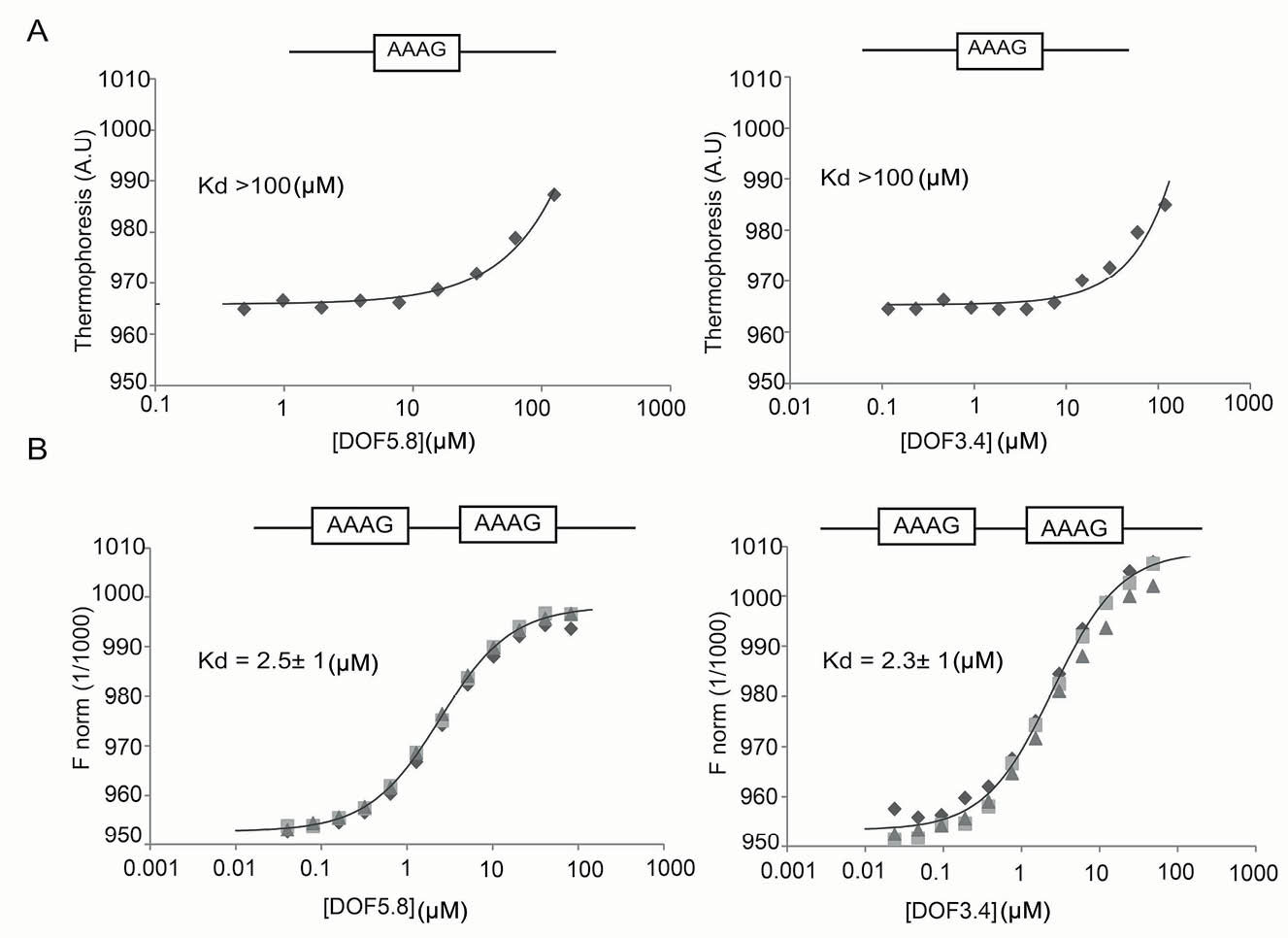
Fig. 6.
Quantitative assessment of DNA binding by DOF-ZFs using MST. Binding of the ZF domains of DOF5.8 and DOF3.4 to fluorescently labeled single-motif probe (A) and double-motif (B) probes is shown. Data points from independent titrations are fitted to a 1:2 binding model, assuming that the two binding sites are equivalent.
.
Quantitative assessment of DNA binding by DOF-ZFs using MST. Binding of the ZF domains of DOF5.8 and DOF3.4 to fluorescently labeled single-motif probe (A) and double-motif (B) probes is shown. Data points from independent titrations are fitted to a 1:2 binding model, assuming that the two binding sites are equivalent.
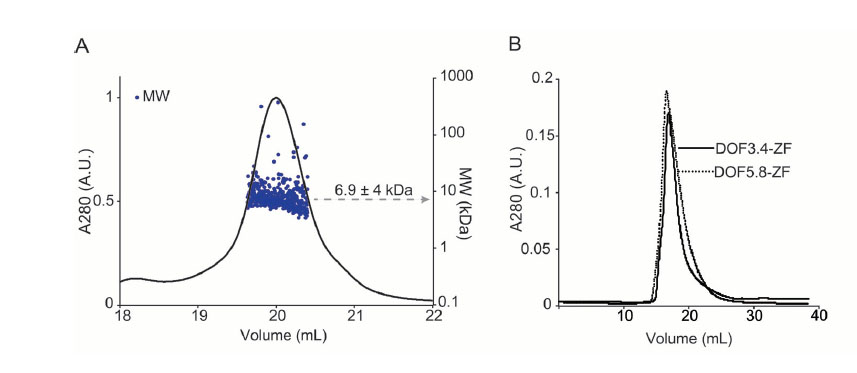
Fig. 7.
Protein molecular weight determination. A) SEC-MALLS result on DOF3.4-ZF. The solid curve corresponds to the UV absorbance profile at 280 nm and the scatter plot represents the mass estimated from light scattering profile. A major peak on SEC-MALLS was detected with molecular weight of 6.9±4 kDa. B) SEC chromatograms of the purified DOF3.4-ZF and DOF5.8-ZF.
.
Protein molecular weight determination. A) SEC-MALLS result on DOF3.4-ZF. The solid curve corresponds to the UV absorbance profile at 280 nm and the scatter plot represents the mass estimated from light scattering profile. A major peak on SEC-MALLS was detected with molecular weight of 6.9±4 kDa. B) SEC chromatograms of the purified DOF3.4-ZF and DOF5.8-ZF.
Discussion
Previous in vivo and in vitro studies on DOF proteins have shown the ability of DOF-domain proteins to bind to specific promoters. Baumann et al showed that NtBBF1, a DOF protein from Nicotiana tabacum, binds to ACTTTA site in the rolB promoter and activates the expression of the beta-glucuronidase reporter gene in the apical meristem and vascular system of transgenic tobacco.
17
In another study, full-length StDOF1 from potato was shown in gel retardation assays to bind to a DNA fragment containing 3 TAAAG repeats that is found in the promoter of the KST1 gene (which encodes a K+ influx channel that is expressed predominantly in guard cells.
18
However, until now there have been no reports in the literature of the quantitation of direct DNA binding of isolated DOF-ZF domains.
In this work, N-terminally GST-tagged polypeptides encoding the putative DOF-ZF domains from A. thaliana DOF2.1, DOF3.4, and DOF5.8 were expressed in E. coli and purified either as fusion proteins or as isolated DOF domains. The final proteins were purified to ~95% judged based on SDS-PAGE analysis. Examination of the DOF-ZF domains by 1H NMR spectroscopy revealed that the DOF-ZF domains were able to form a well-defined 3-dimensional structure and atomic absorption spectrometry confirmed that all of the DOF domains bind the expected one molar equivalent of Zn2+.
We went on to carry out gel retardation assays using (a) a single-motif DNA probe, from the GST6 promoter, which contains a single AAAG motif, and (b) a double-motif probe derived from the DOF2.3 promoter, which contains 2 AAAG motifs. All 3 GST-DOF-ZF proteins interacted with both oligonucleotides. However, only the complexes formed with the double-motif probe were observed as well-defined bands in the gels; the single-motif probe complexes were highly smeared, suggesting that the double AAAG motif forms higher affinity complexes.
To assess these interactions in more detail, we used MST to measure the dissociation constants for the binding of the double-motif probe to the (GST-free) ZF domains of DOF3.4 and DOF5.8; in both cases, a Kd of ~2 µM was obtained. In contrast, single-motif probe displayed very weak binding to both DOF domains (>100 µM). Collectively, the results of gel retardation assay and MST experiments suggest the presence of only a very weak interaction between the single-motif probe and DOF-ZFs furthermore, the DOF-ZF domain affinity for DNA is significantly stronger (at least 100-fold) when the target DNA contains 2 repeats of the AAAG motif. This conclusion is in agreement with gel retardation data for full-length His6-DOF3.4 binding to the promoter of DOF2.3 from A. thaliana.
16
The stronger interaction observed for the double-motif probe could arise from one of 2 possible scenarios. First, one DOF-ZF could bind to each site and the 2 ZFs could contact each other, stabilizing their interaction with DNA. Although no appreciable self-association is observed for DOF3.4 or DF5.8 ZFs in solution, it is quite plausible that they could make contacts when localized by their binding to the same DNA probe. The second possibility is that the context of the AAAG motifs in the double-site probe is sufficiently different from the single-motif probe that they account for the ~100-fold increase in binding affinity. Overall, we consider the first scenario to be more likely (and more consistent with published data on the homo- and heterodimerization of DOF proteins; see below), and additional confirmation awaits detailed structural studies of these proteins bound to DNA.
There are numerous reports describing nanomolar (or stronger) affinities of ZF proteins for DNA.
19,20
Generally, an array of 3 or more ZFs is required for tight, specific binding of this type, although all ZFs in a protein may not be equally involved in protein or DNA interactions.
21
However, there are examples of ZF proteins that tightly interact with their cognate DNA through a single ZF domain. For example, the DNA affinity of the one-ZF GAGA protein from Drosophila melanogaster is in the nanomolar range. However, in this case, 2 basic regions N-terminal to the GAGA ZF domain are thought to make interactions with the DNA.
22
Several other one-ZF DNA-binding proteins make use of an adjacent region that has some affinity for nucleic acids. For example, the TFs NIT2 from Neurospora crassa and AreA from Aspergillus nidulans each bear a single Cys4 ZF and display a C-terminally extended basic region that contributes to DNA binding (Fig. 8).
23,24
The affinity of DOF-ZF domains for the double-motif probe measured in this study is comparable to the affinity of the minimal ZF domain of AreA (lacking the basic region) for its cognate DNA site, in contrast to the sub-nanomolar affinity measured for a C-terminally extended version of the domain. These extended regions can potentially contribute to DNA-binding affinity through direct interactions (either specific or nonspecific) with DNA or by stabilizing the structures – such as ZF domains – that are directly involved in the binding.
24
The constructs used in the current study did not include the extended C-terminal basic region in the DOF-ZF sequences.

Fig. 8.
Alignment of single ZF domain and basic flanking regions of GAGA, AreA, NIT2 and three DOF proteins from A. thaliana. Underlined sequences show basic regions. The UniProt ID of each protein sequence is given inside the bracket.
.
Alignment of single ZF domain and basic flanking regions of GAGA, AreA, NIT2 and three DOF proteins from A. thaliana. Underlined sequences show basic regions. The UniProt ID of each protein sequence is given inside the bracket.
Genome-wide analysis of A. thaliana reveals a high frequency of repeated DOF binding sequences in the same or inverted orientations, with the most common arrangement being AAAG-N7-CTTT. In this study, it was shown that the predominant bases separating AAAG sequences in AAAG-Nn-AAAG pattern are A and G (N denotes any nucleotide bases).
25
Such repeats are found in the promoters of the ANAC069 (seven copies in both orientation) and MYB60 (a cluster of DOF motifs in both orientation) genes, which encode a member of NAC family of plant-specific TFs (NTM2) and a guard-cell specific protein, respectively.
26,27
This feature is not limited to the A. thaliana as these repeated elements are abundantly seen in the promoters of genes controlled by DOF proteins in other plants. Guard cell specific KST1, ADP-glucose pyrophosphorylase, and PEP carboxylase genes in potato each contain multiple copies of such a motif.
18
The position of these motifs in the promoter region and the number of repetitions and of intervening nucleotides differ between genes, but these repeated motifs might give rise to higher affinity and/or higher specificity DNA binding.
Interactions between TFs are important in the regulation of gene expression. The interaction of DOF proteins with each other and with different TFs has been observed previously. A previous gel retardation analysis of DOF1 and DOF2, 2 maize DOF proteins, demonstrated that these proteins are able to self-associate and create homomeric and heteromeric complexes.
28
The non-tagged DOF-ZF domain of DOF1 was shown to specifically interact with DNA probes containing one and 2 binding sites (separated by 7 nucleotides). The DOF1-ZF formed higher order complexes with both DNA probes as its concentration increases. This could be due to the binding of dimerized DOF1-ZF to the binding site.
28
Other studies have shown that particular DOF proteins need an ‘assistant’ protein for optimum binding to DNA. In barley, in vivo studies revealed the physical interaction between HvWRKY38 and BPBF (a DOF protein), as a repressor, in regulating the gibberellin induced Amy32b α-amylase protein. The binding sites of these 2 TFs are only 14 bp apart.
29
Finally, in vitro experiments using gel retardation assays indicated that the maize HMGB protein stimulates the binding of the DOF-ZF domain of the maize TF, DOF2, to a probe with a single AAAG site. In the absence of the HMGB protein, the amount of complex formed between DOF2-ZF and DNA reduced significantly.
30
Overall, it can be concluded that self-association and formation of the protein complexes are likely to be mechanisms that DOF proteins use for tight binding to their target sites. In the present study, the binding affinity of DOF3.4-ZF and DOF5.8-ZF to the probe with a single binding site was very weak compared to their affinity to the probe with 2 binding sites. Since the binding sites are only 4 bp apart, tighter binding for 2 sites possibly indicates that proteins are able to physically contact each other and bind as a dimer to the DNA.
Conclusion
In summary, this work presents experimental evidence that DOF-ZF domains are bona fide ZF domains and that the DOF-ZF domains in isolation bind to DNA fragments containing AAAG sequence. Quantitative assessment of the binding is also performed. According to our results, the binding affinity of DOF-ZF domains to an oligonucleotide containing 2 binding sites is ~100-fold higher than that for a similar oligonucleotide with one binding site. This observation provides a possible explanation for the presence of repeated numbers of AAAG in the promoter regions of DOF TF target genes.
Ethical approval
None to be declared.
Competing interests
There is no conflict of interests to be reported.
Supplementary Materials
Supplementary file 1 contains Fig. S1.
(pdf)
Acknowledgment
The authors would like to thank the Research Office of Tabriz University of Medical Sciences for providing financial support under the Postgraduate Research Grant scheme toward the Ph.D. thesis No: 92/4-5/2. The support of School of Life and Environmental Sciences, The University of Sydney is also gratefully acknowledged.
Research Highlights
What is current knowledge?
simple
-
√ DOF proteins are plant-specific TFs.
-
√ Zinc finger domains of DOF proteins from A. thaliana bind
to their specific AAAG cognate DNA with unknown affinity.
What is new here?
simple
-
√ DOF-ZFs are structurally folded in the monomeric state
in the solution.
-
√ The binding affinity of DOF-ZF domains to their cognate
DNA sequence was determined using biophysical methods.
-
√ A mechanism for binding of DOF-ZFs to DNA was
proposed.
References
-
Hughes TR. A Handbook of Transcription Factors. Netherlands: Springer; 2011.
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 2000; 290:2105-10. doi: 10.1126/science.290.5499.2105 [Crossref] [ Google Scholar]
- Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci 2007; 32:63-70. doi: 10.1016/j.tibs.2006.12.007 [Crossref] [ Google Scholar]
- Yanagisawa S. A novel DNA-binding domain that may form a single zinc finger motif. Nucleic Acids Res 1995; 23:3403-10. [ Google Scholar]
- Shigyo M, Tabei N, Yoneyama T, Yanagisawa S. Evolutionary processes during the formation of the plant-specific Dof transcription factor family. Plant Cell Physiol 2007; 48:179-85. doi: 10.1093/pcp/pcl044 [Crossref] [ Google Scholar]
- Venkatesh J, Park SW. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol Biochem 2015; 94:73-85. doi: 10.1016/j.plaphy.2015.05.010 [Crossref] [ Google Scholar]
- Cai X, Zhang Y, Zhang C, Zhang T, Hu T, Ye J. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J Integr Plant Biol 2013; 55:552-66. doi: 10.1111/jipb.12043 [Crossref] [ Google Scholar]
- Moreno-Risueno MA, Martinez M, Vicente-Carbajosa J, Carbonero P. The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol Genet Genomics 2007; 277:379-90. doi: 10.1007/s00438-006-0186-9 [Crossref] [ Google Scholar]
- Lijavetzky D, Carbonero P, Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol 2003; 3:17. doi: 10.1186/1471-2148-3-17 [Crossref] [ Google Scholar]
- Riechmann JL, Ratcliffe OJ. A genomic perspective on plant transcription factors. Curr Opin Plant Biol 2000; 3:423-34. doi: 10.1016/S1369-5266(00)00107-2 [Crossref] [ Google Scholar]
- Noguero M, Atif RM, Ochatt S, Thompson RD. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci 2013; 209:32-45. doi: 10.1016/j.plantsci.2013.03.016 [Crossref] [ Google Scholar]
- Umemura Y, Ishiduka T, Yamamoto R, Esaka M. The Dof domain, a zinc finger DNA-binding domain conserved only in higher plants, truly functions as a Cys2/Cys2 Zn finger domain. Plant J 2004; 37:741-9. doi: 10.1111/j.1365-313X.2003.01997.x [Crossref] [ Google Scholar]
- Shimofurutani N, Kisu Y, Suzuki M, Esaka M. Functional analyses of the Dof domain, a zinc finger DNA-binding domain, in a pumpkin DNA-binding protein AOBP. FEBS Lett 1998; 430:251-6. doi: 10.1016/S0014-5793(98)00670-X [Crossref] [ Google Scholar]
- Hamzeh-Mivehroud M, Moghaddas-Sani H, Rahbar-Shahrouziasl M, Dastmalchi S. Identifying key interactions stabilizing DOF zinc finger-DNA complexes using in silico approaches. J Theor Biol 2015; 382:150-9. doi: 10.1016/j.jtbi.2015.06.013 [Crossref] [ Google Scholar]
- Chen W, Chao G, Singh KB. The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J 1996; 10:955-66. doi: 10.1046/j.1365-313X.1996.10060955.x [Crossref] [ Google Scholar]
- Skirycz A, Radziejwoski A, Busch W, Hannah MA, Czeszejko J, Kwasniewski M. The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J 2008; 56:779-92. doi: 10.1111/j.1365-313X.2008.03641.x [Crossref] [ Google Scholar]
- Baumann K, De Paolis A, Costantino P, Gualberti G. The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell 1999; 11:323-34. [ Google Scholar]
- Plesch G, Ehrhardt T, Mueller-Roeber B. Involvement of TAAAG elements suggests a role for Dof transcription factors in guard cell-specific gene expression. Plant J 2001; 28:455-64. doi: 10.1046/j.1365-313X.2001.01166.x [Crossref] [ Google Scholar]
- Eom KS, Cheong JS, Lee SJ. Structural Analyses of Zinc Finger Domains for Specific Interactions with DNA. J Microbiol Biotechnol 2016; 26:2019-29. doi: 10.4014/jmb.1609.09021 [Crossref] [ Google Scholar]
- Wai DC, Shihab M, Low JK, Mackay JP. The zinc fingers of YY1 bind single-stranded RNA with low sequence specificity. Nucleic Acids Res 2016; 44:9153-65. doi: 10.1093/nar/gkw590 [Crossref] [ Google Scholar]
- Gamsjaeger R, Swanton MK, Kobus FJ, Lehtomaki E, Lowry JA, Kwan AH. Structural and biophysical analysis of the DNA binding properties of myelin transcription factor 1. J Biol Chem 2008; 283:5158-67. doi: 10.1074/jbc.M703772200 [Crossref] [ Google Scholar]
- Pedone PV, Ghirlando R, Clore GM, Gronenborn AM, Felsenfeld G, Omichinski JG. The single Cys2-His2 zinc finger domain of the GAGA protein flanked by basic residues is sufficient for high-affinity specific DNA binding. Proc Natl Acad Sci U S A 1996; 93:2822-6. doi: 10.1073/pnas.93.7.2822 [Crossref] [ Google Scholar]
- Fu YH, Marzluf GA. Site-directed mutagenesis of the 'zinc finger' DNA-binding domain of the nitrogen-regulatory protein NIT2 of Neurospora. Mol Microbiol 1990; 4:1847-52. doi: 10.1111/j.1365-2958.1990.tb02033.x [Crossref] [ Google Scholar]
- Manfield IW, Reynolds LA, Gittins J, Kneale GG. The DNA-binding domain of the gene regulatory protein AreA extends beyond the minimal zinc-finger region conserved between GATA proteins. Biochim Biophys Acta 2000; 1493:325-32. doi: 10.1016/S0167-4781(00)00197-4 [Crossref] [ Google Scholar]
- Mehrotra R, Jain V, Shekhar C, Mehrotra S. Genome wide analysis of Arabidopsis thaliana reveals high frequency of AAAGN7CTTT motif. Meta Gene 2014; 2:606-15. doi: 10.1016/j.mgene.2014.05.003 [Crossref] [ Google Scholar]
- Cominelli E, Galbiati M, Albertini A, Fornara F, Conti L, Coupland G. DOF-binding sites additively contribute to guard cell-specificity of AtMYB60 promoter. BMC Plant Biol 2011; 11:162. doi: 10.1186/1471-2229-11-162 [Crossref] [ Google Scholar]
- He L, Su C, Wang Y, Wei Z. ATDOF58 protein is the upstream regulator of ANAC069 and is responsive to abiotic stress. Biochimie 2015; 110:17-24. doi: 10.1016/j.biochi.2014.12.017 [Crossref] [ Google Scholar]
- Yanagisawa S. Dof DNA-binding domains of plant transcription factors contribute to multiple protein-protein interactions. Eur J Biochem 1997; 250:403-10. doi: 10.1111/j.1432-1033.1997.0403a.x [Crossref] [ Google Scholar]
- Zou X, Neuman D, Shen QJ. Interactions of two transcriptional repressors and two transcriptional activators in modulating gibberellin signaling in aleurone cells. Plant Physiol 2008; 148:176-86. doi: 10.1104/pp.108.123653 [Crossref] [ Google Scholar]
- Krohn NM, Yanagisawa S, Grasser KD. Specificity of the stimulatory interaction between chromosomal HMGB proteins and the transcription factor Dof2 and its negative regulation by protein kinase CK2-mediated phosphorylation. J Biol Chem 2002; 277:32438-44. doi: 10.1074/jbc.M203814200 [Crossref] [ Google Scholar]