Bioimpacts. 9(2):115-121.
doi: 10.15171/bi.2019.15
Mini Review
Spectroscopic overview of quercetin and its Cu(II) complex interaction with serum albumins
Prasenjit Mondal , Adity Bose * 
Author information:
Department of Chemistry, Presidency University, 86/1 College Street, Kolkata 700073, India
Abstract
Introduction:
Flavonoids are widely used as dietary supplements, and thus, play a significant role in the research field. In recent time, the interaction of flavonoid-metal complexes with serum albumin (SA) has widely been studied since the complexation poses a significant impact on biological activities. Additionally, the binding nature of flavonoids with SA gets modified in the presence of metal ions.
Methods:
In the present review, we studied the interaction of quercetin (Qu), a well-known flavonoid, and its Cu2+ complexes with SA to provide sufficient information about the beneficial role of metal-flavonoid complexes over free flavonoids.
Results:
Complexation with Cu(II) ion may alter the mode of binding of Qu with SAs. The strength of binding might be increased in the presence of Cu(II) as evidenced by the binding constant calculation. However, the drug binding site in bovine serum albumin (BSA) and human serum albumin (HSA) are not altered during the complexation process.
Conclusion:
To enhance the pharmaceutical outcomes of Qu molecules, one may use Qu-Cu(II) complex for the development and delivery of the small molecules.
Keywords: Copper, Flavonoids, Quercetin, Serum albumin
Copyright and License Information
© 2019 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Flavonoids, a subgroup of polyphenolic compounds, are generated in plants, common fruits, vegetables, tea, wine, etc. In recent decades, flavonoids have widely been used in many biological activities, including cardiovascular diseases, cancers or age-related disorders.
1
Due to the variety of activities, the impacts of flavonoids have widely been considered on various biological processes.
2,3
Further, flavonoids can form chelates with the metal ions, which may change their antioxidant activity and biological effects.
4
Quercetin (Qu), as shown in Fig. 1, is one of the most common bioactive and dietary flavonoids, which can be found in the flowers, leaves, and fruits of many plants.
5
Qu can form complexes with transition metal ions, such as Cu2+, Mn2+, Fe2+, etc. Among them, the stability of Qu-Cu(II) complex is reported to be the highest.
5
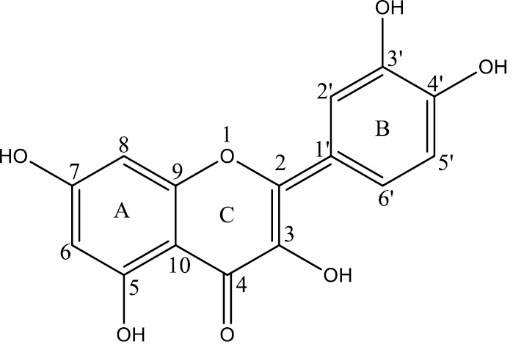
Fig. 1.
Structure of Quercetin (Qu).
.
Structure of Quercetin (Qu).
Cu(II) is one of the most abundant trace minerals in the human body. In the blood, it can often act as a cofactor for angiogenesis.
6
In some recent researches, it has been established that naturally occurring polyphenols can prevent the emergence of angiogenesis. Further, the catalytic role of transition metal ions such as Fe(III) and Cu(II) is well-known in the formation of hydroxyl radical (•OH) via metal-catalyzed Haber–Weiss reaction or Fenton reaction. Metal ion chelation by flavonoids is known to inhibit the aforementioned oxidative damage caused by metal ions. Also, the biological activities such as anti-oxidant effect, anti-bacterial and various kinds of enzymatic functions of free flavonoids get affected as a result of the complex formation.
7,8
Therefore, it is important to understand the effect of Cu(II) on the interactions between flavonoids and SAs. According to available literature, Qu-metal complexes seem to produce better biological activities than the bare Qu itself.
8,9
In keeping with the above facts, in this review, we have studied the ultimate behavior of Qu and Qu-Cu(II) complex with SAs. SAs are the most abundant transport proteins in the blood plasma.
9-11
It carries most of the exogenous and endogenous compounds. The higher conformational adaptability of bovine serum albumin (BSA) and human serum albumin (HAS) to the ligands attracts the researchers to study the ligand-serum albumin interactions.
12-14
It has been found that free flavonoids were less active than their metal complexes, in large part because metal-flavonoid has greater biological efficacy than free flavonoids.
15-18
Additionally, metal ions can bind with SAs
19
and its complexity can influence the binding affinities of other compounds. It has also been reported that in the interaction between SAs and a drug molecule, such as flavonoids,
20,21
vitamins,
22
the presence of a metal ion can change the binding constant values between drugs and SAs. Due to the widespread use of Qu, it is worthwhile to review the result of this flavonoid-albumin complex formation inferred through various techniques to understand the details of the interaction mechanism at the molecular level.
Technical aspects of the interaction of quercetin and its Cu(II) complex with SAs
UV–vis spectroscopy
UV-vis spectroscopy is the simplest analytical method used for the quantitative estimation of different analytes and biological macromolecules. When complexation takes place between biological macromolecules with any ligand/analyte, the association constant of the complex formation can be estimated by the equation (Eq. 1).
7,23
Eq. (1)
Where, is the extinction coefficient, the subscripts b, f, and T denote bound, free and total ligand, respectively. LT is the total Qu or Qu-Cu(II) complex concentration and ∆A is the change in absorbance at a particular wavelength. The association constant (Ka) of complex formation can be obtained from the ratio of the intercept to the slope of a double reciprocal plot of absorbance change against SA concentration.
Fluorescence quenching
Fluorescence quenching of SA can be described by the Stern–Volmer equation (Eg. 2).
24,25
Eq. (2)
Where, F0 and F are respectively the fluorescence intensities before and after the addition of the quencher; kq is the bimolecular quenching constant; and τ0 is the lifetime of the fluorophore in the absence of the quencher. [Q] is the concentration of the quencher and Ksv is the Stern–Volmer quenching constant. Hence, Eq. (2) was used to determine the Ksv value from the linear regression plot of F0/F against [Q]. The results were also analyzed by the modified Stern-Volmer equation given as Eq. (3).
Eq. (3)
Where, ΔF=F0-F; fa is the fraction of fluorophore accessible to the quencher.
For the simultaneous static and dynamic quenching Eq. 4 and Eq. 5 are used.
24
Eq. (4)
Eq. (5)
Where, KS and KD denote the static and dynamic quenching constant, respectively.
Binding constants
Binding parameters are helpful in the study of pharmacokinetics. Calculation of binding constant by different authors can be performed utilizing Eqs. (6), (7) and (8).
25-28
Eq. (6)
Eq. (7)
Eq. (8)
Where, kb is the binding constant and n is the number of binding sites per protein molecule.
Circular dichroism measurement
The CD analysis is a well-known method to estimate the fraction of a biomolecule (e.g., protein) that is in α-helix, β-sheet conformation, beta-turn, and random coil conformation. The CD spectrum of HSA exhibits two negative bands: one at ̴ 208 and another at ̴ 222 nm, typical of a α-helix structure of the protein. When the binding of a molecule to HSA leads to a decrease in negative ellipticity at all wavelengths of the far-UV CD without any remarkable shift of the peaks, it indicates changes in the protein secondary structure, as well as a decrease in the α-helical content of the protein. The percentage of the α-helix is calculated using Eq. (9)
29
:
Eq. (9)
Where, MRE208 is the observed mean residue ellipticity (MRE) value at 208 nm, 4000 is the MRE of the beta-form and random coil conformation cross at 208 nm, and 33 000 is the MRE value of the pure α-helix at 208 nm.
Thermodynamic parameter calculation
The interaction between a drug and a biomolecule generally involve various forces, including hydrophobic forces, electrostatic interactions, van der Waals interactions, and hydrogen bonds. Thermodynamic parameters such as free energy changes (ΔG0), enthalpy changes (ΔH0) and entropy changes (ΔS0) of interactions are essential to interpret the binding mode of interaction between the molecules. In order to elucidate the interaction of a molecule with BSA, the thermodynamic parameters can be calculated from Eqs. (10), (11) and (12). If the temperature does not vary significantly, the enthalpy change (ΔH0) can be regarded as constant. The free energy change (ΔG0) can be estimated from the following equation, based on the binding constants at different temperatures. The thermodynamic parameters can be calculated from the following equations.
Where, R denotes the gas constant, ΔH0 is enthalpy change, ΔS0is entropy change, Kb is the binding constants at the experiment temperature Tly.
According to the literature, the following pattern can be deduced.
30
-
ΔH
0
> 0 and ΔS0> 0indicates the hydrophobic interaction,
-
ΔH
0
< 0 and ΔS0< 0indicates the hydrogen bonding and van der Waals interactions, and
-
ΔH
0
< 0 and ΔS0> 0indicates the electrostatic interactions.
Docking studies
To confirm the binding site and explain the relationship between the structure of the interacting molecule and the function of a protein from a theoretical viewpoint, docking can be used as a helpful tool.
Interaction of Quercetin and its Cu(II) complex with SAs
UV-Vis studies
Being an efficient tool to observe the ground state interaction of proteins with drugs, UV-Vis spectroscopy is often utilized by researchers. Roy et al and Zhang et al have exploited this tool to observe the ground state interaction of Qu and its Cu(II) complex with SAs but in a different manner.
7,31
Roy et al
7
synthesized the Qu-Cu(II) complex and observed the effect of the addition of the complex on the absorption spectra of SAs, while Zhang et al
31
have observed the changes in the absorption spectra of BSA (incubating with or without Cu) with increasing amounts of Qu. It has been reported that Qu can exhibit two absorption maxima in the UV-vis spectra, including (i) at ~261 nm (Benzoyl ring A) and (ii) at ~376 nm (cinnamoyl ring B). According to Roy et al, the Qu-Cu(II) synthesized complex can exhibit an absorbance peak at 438 nm (due to strong π-π*transition) and with the addition of SAs intensity of the absorption peak decreases with a blue shift due to the increase in energy gap between the π-π* electronic levels.
7,32,33
However, in contrast, when SAs were added to free Qu, a red-shift from 376 nm to 400 nm with a decrease in the absorbance intensity can be observed. Hence, a difference in the interaction mode can be discerned. According to Zhang et al,
31
with the gradual addition of Qu (in the absence of Cu(II)), the absorption of BSA (280 nm) as well as the characteristic peak of Qu (376 nm) gradually increased. Slight blue shift of the peak at 280 nm was observed, which was explained as an evidence of ground state complex formation between BSA and Qu.Similar observation was also found in the presence of Cu(II), however, the intensity around 376 nm totally disappeared while a new absorption peak appeared at 328 nm, which can symbolize the formation of Qu–Cu(II) complex.
34
Thus, from their observations, it is evident that a complexation between Qu and BSA can occur.
Fluorescence spectroscopic studies: quenching, binding, and thermodynamic parameters
Both HSA and BSA exhibit similar kind of fluorescence emission based on excitation at 295 nm due to the presence of tryptophan residue (Trp 214 in HSA and Trp 134/Trp 213 in BSA).
7
The fluorescence of BSA/HSA was observed to be quenched in the presence of both Qu and its Cu(II) complex. In the opinion of Roy et al, the addition of Qu-Cu(II) complex to SAs resulted in a non-linear Stern-Volmer plot which can symbolize the simultaneous occurrence of static and dynamic quenching process. However, the authors have ruled out the possibility of dynamic quenching by evaluating the higher value of bimolecular quenching constant (kq).
24
The values of KSV and kq have given in Table 1. Additionally Roy et al have observed the effect of temperature on the binding of Qu-Cu(II) with SAs. With the increase in temperature, the binding constant values were found to increase (Table 2) while the fraction accessibility (fa) decreased. Thus, the protein structure was concluded to be less accessible to the complex on heating.
Table 1.
Stern-Volmer quenching constants (K
sv) and bimolecular quenching rate constants (k
q) for the binding of the Qu-Cu(II) complex with HSA and BSA
7
|
|
Temperature (K)
|
10
5
×
K
sv
( M1)
|
10
13
×
K
q
( M1 S-1)
|
| HSA |
299
306
312
|
2.15 ± 0.10 (R2= 0.99)
2.21 ± 0.09 (R2= 0.98)
2.27 ± 0.01 (R2= 0.99)
|
4.31 ± 0.10
4.41 ± 0.09
4.55 ± 0.01
|
BSA
|
299
306
312
|
2.02 ± 0.10 (R2= 0.97)
2.11 ± 0.03 (R2= 0.98)
2.29 ± 0.03 (R2= 0.96)
|
4.05 ± 0.10
4.23 ± 0.03
4.59 ± 0.03
|
Table 2.
Binding and thermodynamic parameters for the interaction of the Qu-Cu(II) complex with HSA and BSA
7
|
|
Temperature (K)
|
10-5×Kb (M1)
|
N
|
ΔG
0
(KJ mol-1 )
|
ΔH
0
(KJ mol-1 )
|
ΔS
0
(KJ mol-1 )
|
| HSA |
299
306
312
|
2.11 ± 0.18
2.30 ± 0.31
2.41 ± 0.22
|
1.24
1.11
1.01
|
-(31.64 ± 1.05)
-(32.36 ± 1.23)
-(33.04 ± 1.32)
|
+(7.87 ± 1.92) |
+(128.27 ± 2.07) |
| BSA |
299
306
312
|
2.03 ± 0.20
2.19 ± 0.18
2.42 ± 0.32
|
1.23
1.22
1.24
|
-(31.18 ± 1.25)
-(32.05 ± 1.32)
-(33.98 ± 1.22)
|
+(10.30 ± 2.17) |
+(138.72 ± 3.83) |
On the other hand, Zhang et al conducted their experiments in a different manner by adding Cu(II) into the BSA solution first, and then, Qu was added into that same solution. According to them, in the absence of Cu(II), the quenching of BSA fluorescence occurred with a slight blue shift with the addition of Qu. This observation was indicative of hydrophobic interactions, which occurred mainly via hydrogen bonds. The above observations were also supported by Xiao and coworkers. Based on their findings, the hydroxyl groups of Qu played an important role in the binding of Qu to BSA due to the formation of hydrogen bonding between Qu and the polar groups at the BSA surface.
35,36
They have also considered the double bond between C2 and C3 position of Qu to influence the binding affinity of Qu to BSA. The involvement of hydrophobic interactions was also confirmed by Roy et al with the help of thermodynamic parameters (Table 2).
Next, Zhang et al reported that, in the absence and presence of Cu(II), the nature of quenching plots was the same but the values of quenching constant (Ksv) were different. In both cases, the involved quenching mechanism(s) was predicted to be static with the help of kq values (Table 3). A similar observation was also found by Roy et al as mentioned above. Further, the presence of Cu(II) somehow increased the KSV value probably due to a competitive binding effect of Qu and Cu(II) with BSA or a conformational change of BSA or may be complex formation between Qu with Cu(II) ion. Hence, the Cu(II) ion was found to play a major role in the binding of Qu with BSA. Zhang and his co-workers also established that the presence of Cu(II) decreased the binding affinity of Qu to BSA which has the capability to enhance the biological activity of Qu (Table 4).
31
Table 3.
Stern-Volmer quenching constants (
Ksv) and bimolecular quenching rate constants (
kq) for the interactions of Qu with BSA with and without Cu(II) at 298 K
31
|
K
sv
/10
4
M
-1
|
K
q
/10
12
M
1
.S
-1
|
|
Absence
|
Presence
|
Absence
|
Presence
|
| 4.38 |
5.63 |
8.76 |
11.26 |
Table 4.
The static binding constants (K in M
-1) and binding sites (n) for the interaction of Qu with BSA in the presence and absence of Cu(II) at 298 K
31
|
Absence
|
Presence
|
|
logk
b
|
K
b
(10
5
M1)
|
N
|
R
|
logk
b
|
K
b
(105
M1)
|
N
|
R
|
| 5.86 |
7.24 |
1.24 |
0.9984 |
5.49 |
3.09 |
1.15 |
0.9989 |
In the experiments conducted by Roy et al, ΔG0values were similar for both BSA and HSA (Table 2) but the values of ΔH0 and ΔS0 were different which indicated the change in the mode of interaction of Qu-Cu(II) complex with SAs. According to Roy et al the binding interaction of Qu with SAs occurred via electrostatic interaction and the positive ΔS0 value indicated that the interaction can become hydrophobic which was also evident from fluorescence studies.
7,37,38
According to the available literature, Qu binds to site 1 (subdomain IIA) which is the hydrophobic site of BSA and HSA.
39
Hence, it was concluded that like Qu, the Qu-Cu(II) complex also binds to the hydrophobic site 1 (subdomain IIA) of BSA/HSA.
7,40
Circular dichroism studies
The typical CD spectra of both BSA and HSA generally shows two negative bands at ~208 nm and ~220 nm, which are attributed to the α-helix structure of proteins. Roy et al observed that in the far UV CD spectra, the band intensity slightly increased without any significant peak shift when Qu or Qu–Cu(II) complex were made to interact with HSA/BSA. Additionally, the α-helical content of both the SAs changed when it was bound with Qu–Cu(II) complex, but for BSA the change was to some extent greater than that of HSA. This study indicated the conformational change in the SAs while interacting with Qu and Qu-Cu(II) complex. In the experiment carried out by Zhang et al, the percentage α-helical content of BSA decreased in the presence of Cu(II) with respect to only BSA.
31
In contrast, the degree of change of α-helical content in the presence of Cu(II) had a greater effect than in the absence of Cu(II). Hence, Qu and Cu(II) were found to change the secondary structure of BSA.
Site-specific and docking studies
The binding interaction of Qu and Qu-Cu(II) complex with SAs were also verified using the docking study. Roy et al confirmed that both Qu and its Cu(II) complex can bind to the hydrophobic pocket (subdomain IIA) of SAs.
Anti-oxidant studies
Though very few reports are available on the anti-oxidant behavior of Qu-Cu(II) complex, some authors have found that the radical scavenging activity of Qu-Cu(II) complex was greater than that of free Qu.
41-43
According to the authors, in the case of Qu-Cu(II) complex, the Cu(II) binds with the aforesaid oxygen atoms of 3′ and 4′ position. This newly formed metal center provided extra scavenging activity by stabilizing the semiquinone complex formed after H-abstraction from Qu-Cu(II) complex.
Conclusions
Flavonoids are naturally occurring compounds that exhibit various biological and pharmacological potentials. The biological behaviors of flavonoids are known to alter in the presence of metal ions. In most of the cases, the metal-flavonoid complexes being more anti-oxidant than that of free flavonoids. In recent years, many researchers are interested to introduce different flavonoids as potent metal chelating agents to inhibit metal-induced free radical generation. In this study, we have focused on the interaction of Qu and its Cu(II) complex with serum albumins (BSA and HSA) and the effect of metal ion chelation on the anti-oxidant activity of Qu.
The binding affinity of BSA with Qu was found to decrease significantly in the presence of Cu(II) due to the bulkiness of Qu-Cu(II). But a group of researchers found that with an increase in temperature, the binding affinity was remarkably increased in the case of both BSA and HAS (Table 5). The thermodynamic parameter indicated that the interaction of Qu-Cu(II) complex with both BSA and HSA are hydrophobic in nature but in the case of Qu, nature of the interaction was hydrophobic as well as electrostatic in nature. The site-marker and molecular docking analysis confirmed that both Qu and Qu-Cu(II) complex binds to the Trp-214 of HSA and Trp-213 of BSA which is located at the site I of subdomain IIA of the proteins. The CD spectra revealed that the secondary structure of BSA was altered in the presence of Qu and Qu-Cu(II) complex. Thus, the aforementioned results reveal the different interacting nature of Qu and Qu-Cu(II) complex with serum albumins which may affect their roles in the biological system. Further, Qu can be introduced as an excellent Cu(II) chelating agent so as to inhibit Cu(II) induced •OH generation and the overall complex will also behave as a potential anti-oxidant agent. Taken all, Qu can be introduced as an anti-angiogenesis drug, which may reduce the concentration of Cu(II) in the blood by forming a chelate complex and it may also show some unique biological properties that can be utilized in the pharmaceutical industry.
Table 5.
Stern-Volmer quenching constants (
Ksv), bimolecular quenching rate constants (
kq), binding and thermodynamic parameters for the interaction of the Qu-Cu(II) complex with HSA and BSA.
7,31
|
|
Temp. (K)
|
10
5
×
K
sv
( M1)
|
10
13
× K
q
(M1 S-1)
|
10-5×
K
b (M1)
|
N
|
ΔG
0
(KJ mol-1 )
|
ΔH
0
(KJ mol-1 )
|
ΔS
0
(KJ mol-1 )
|
| HSA |
2997
3067
3127
|
2.15 ± 0.10 (R2= 0.99)
2.21 ± 0.09 (R2= 0.98)
2.27 ± 0.01 (R2= 0.99)
|
4.31 ± 0.10
4.41 ± 0.09
4.55 ± 0.01
|
2.11 ± 0.18
2.30 ± 0.31
2.41 ± 0.22
|
1.24
1.11
1.01
|
-(31.64 ± 1.05)
-(32.36 ± 1.23)
-(33.04 ± 1.32)
|
+(7.87 ± 1.92) |
+(128.27 ± 2.07) |
| BSA |
2997
29831
3067
3127
|
2.02 ± 0.10 (R2= 0.97)
0.563
2.11 ± 0.03 (R2= 0.98)
2.29 ± 0.03 (R2= 0.96)
|
4.05 ± 0.10
-
4.23 ± 0.03
4.59 ± 0.03
|
2.03 ± 0.20
3.09
2.19 ± 0.18
2.42 ± 0.32
|
1.23
1.15
1.22
1.24
|
-(31.18 ± 1.25)
-
-(32.05 ± 1.32)
-(33.98 ± 1.22)
|
+(10.3 ± 2.17)
-
|
+(138.72 ± 3.83)
-
|
Acknowledgments
PM thanks Mrs. Priti Sengupta for her help and suggestions.
Funding sources
The authors also thank WBDST (Project No.546 (sanc.)/ST/P/S&T/4G-13/2014), DST (Project no. YSS/2014/000403) and the FRPDF grant from Presidency University for financial support.
Ethical statement
Not applicable.
Competing interests
There is no conflict of interest.
Authors contribution
AB has chosen the theme of the review. Both AB and PM have discussed the review topic. PM has written themanuscript.
Review Highlights
What is the current knowledge?
simple
-
√ Quercetin and its Cu(II) complex binds with serum
albumins.
-
√ The binding of quercetin with serum albumin may be
hydrophobic or hydrophilic.
What is new here?
simple
-
√ We have prepared a detailed review on interaction of
quercetin and its Cu(II) complex with SA’s.
-
√ Quercetin and its Cu(II) complex binds with serum
albumin and the binding affinity of quercetin-Cu(II) complex
is greater than that of bare quercetin.
-
√ The radical scavenging activity of quercetin increases when
its binds with Cu(II).
References
- Fresco P, Borges F, Diniz C, Marques MPM. New insights on the anticancer properties of dietary polyphenols. Med Res 2006; 26:747-766. doi: 10.1002/med.20060 [Crossref] [ Google Scholar]
- Bondžić AM, Lazarević-Pašti TD, Bondžić BP, Čolović MB, Jadraninb MB, Vasić VM. Colovic, Milka B Jadranin, Vesna M Vasic Investigation of reaction between Qu and Au (III) in acidic media: mechanism and identification of reaction products. New J Chem 2013; 37:901-908. doi: 10.1039/C2NJ40742F [Crossref] [ Google Scholar]
- Momic T, Savic JZ, Cernigoj U, Trebse P, Vasic VM. Protolytic equilibria and photodegradation of Qu in aqueous solution. Collect Czech Chem Commun 2007; 72:1447-1460. doi: 10.1135/cccc20071447 [Crossref] [ Google Scholar]
- Kasprzak MM, Erxleben A, Ochocki J. Properties and applications of flavonoid metal complexes. RSC Adv 2015; 5:45853. doi: 10.1039/C5RA05069C [Crossref] [ Google Scholar]
- Liu Y, Guo M. Studies on transition metal-Qu complexes using electrospray ionization tandem mass spectrometry. Molecules 2015; 20:8583-8594. doi: 10.3390/molecules20058583 [Crossref] [ Google Scholar]
- Tripathy DR, Singha Roy A, Dasgupta S. Complex formation of rutin and quercetin with copper alters the mode of inhibition of Ribonuclease A. FEBS Letters 2011; 585:3270-3276. doi: 10.1016/j.febslet.2011.09.005 [Crossref] [ Google Scholar]
- Singha Roy A, Tripathy DR, Ghosh AK, Dasgupta S. An alternate mode of binding of the polyphenol Qu withserum albumins when complexed with Cu (II). Journal of Luminescence 2012; 132:2943-295. [ Google Scholar]
- Mendoza EE, Burd R. Quercetin as a systemic chemopreventative agent: structural and functional mechanisms. Mini Rev Med Chem 2011; 11:1216-1221. doi: 10.2174/13895575111091216 [Crossref] [ Google Scholar]
- Leopoldini M, Russo N, Chiodo S, Toscano M. Iron chelation by the powerful antioxidant flavonoid Qu. J Agric Food Chem 2006; 54:6343-6351. doi: 10.1021/jf060986h [Crossref] [ Google Scholar]
- Geng R, Ma L, Liu L, Xie Y. Influence of bovine serum albumin-flavonoid interaction on the antioxidant activity of dietary flavonoids: new evidence from electrochemical quantification. Molecules 2019; 24(1):70. doi: 10.3390/molecules24010070 [Crossref] [ Google Scholar]
- Liu S, Guo C, Guo Y, Yu H, Greenaway F, Sun MZ. Comparative binding affinities of flavonoid phytochemicals with bovine serum albumin. Iran J Pharm Res 2014; 13(3):1019-28. [ Google Scholar]
- Tang X, Tang P, Liu Liu. Molecular Structure–Affinity Relationship of Flavonoids in Lotus Leaf (Nelumbo nucifera Gaertn) on Binding to Human Serum Albumin and Bovine Serum Albumin by Spectroscopic Method. Molecules 2017; 22:E1036. doi: 10.3390/molecules22071036 [Crossref] [ Google Scholar]
- Jaiswal R, Khan MA, Mussarat J. Photosensitized paraquat-induced structural alterations and free radical-mediated fragmentation of serum albumin. J Photochem Photobiol B 2002; 67:163-170. [ Google Scholar]
- Amorim Porfírioa D, de Queiroz Ferreira R, Renata Malagutti A, Maira Agostini Valle E. Electrochemical study of the increased antioxidant capacity of flavonoids through complexation with iron(II) ions. Electrochimica Acta 2014; 141:33-38. doi: 10.1016/j.electacta.2014.07.046 [Crossref] [ Google Scholar]
- Mishra B, Barik A, Priyadarsini KI, Mohan H. Fluorescence spectroscopic studies on the binding of a flavonoid antioxidant Qu to serum albumins. J Chem Sci 2005; 117:641-647. [ Google Scholar]
- Dolatabadi JE. Molecular aspects on the interaction of quercetin and its metal complexes with DNA. Int J Biol Macromol 2011; 48:227-233. doi: 10.1016/j.ijbiomac.2010.11.012 [Crossref] [ Google Scholar]
- Bravo A, Anacona JR. Metal complexes of the flavonoid quercetin: antibacterial properties. Transit Metal Chem 2001; 26:20-23. doi: 10.1023/A:1007128325639 [Crossref] [ Google Scholar]
- Lo YC, Ko TP, Su WC, Su TL, Wang AHJ. Terpyridine–platinum(II) complexes are effective inhibitors of mammalian topoisomerases and human thioredoxin reductase. J Inorg Biochem 2009; 103:1082-1092. doi: 10.1016/j.jinorgbio.2009.05.006 [Crossref] [ Google Scholar]
- Rusak G, Piantanida I, Bretschneider S, Ludwig-Müller J. Complex formation of Qu with lanthanum enhances binding to plant viral satellite double-stranded RNA. J Inorg Biochem 2009; 103:1597-1601. doi: 10.1016/j.jinorgbio.2009.08.008 [Crossref] [ Google Scholar]
- Sanna D, Garribba E, Micera G.
Interaction of VO2+ ion with human serum transferrin and albumin
. J Inorg Biochem 2009; 103:648-655. doi: 10.1016/j.jinorgbio.2009.01.002 [Crossref] [ Google Scholar]
- Cao SH, Jiang XY, Chen JW. Effect of Zinc (II) on the interactions of bovine serum albumin with flavonols bearing a different number of hydroxyl substituent on B-ring. J Inorg Biochem 2010; 104:146-152. doi: 10.1016/j.jinorgbio.2009.10.014 [Crossref] [ Google Scholar]
- Li DJ, Zhu M, Xu C, Chen JJ, Ji BM.
The effect of Cu2+ or Fe3+ on the noncovalent binding of rutin with bovine serum albumin by spectroscopic analysis
. Spectrochim Acta A Mol Biomol Spectrosc 2011; 78:74-79. doi: 10.1016/j.saa.2010.08.069 [Crossref] [ Google Scholar]
- Shaikh SNT, Seetharamappa J, Kandagal PB, Manjunatha DH. In vitro study on the binding of the anti-coagulant vitamin to bovine serum albumin and the influence of toxic ions and common ions on the binding. Int J Biol Macromol 2007; 41:81-86. doi: 10.1016/j.ijbiomac.2007.01.004 [Crossref] [ Google Scholar]
- Benesi HA, Hildebrand JH. A Spectrophotometric Investigation of the Interaction of Iodine with Aromatic Hydrocarbons. J Am Chem Soc 1949; 71:2703-2707. doi: 10.1021/ja01176a030 [Crossref] [ Google Scholar]
-
Lakowicz JR. Principles of Fluorescence Spectroscopy. 2nd ed. New York: Kluwer Academic/Plenum Publishers; 1999. pp. 237-240.
- Bose A. Interaction of tea polyphenols with serum albumins: A fluorescence spectroscopic analysis. Journal of Luminescence 2016; 169:220-226. doi: 10.1016/j.jlumin.2015.09.018 [Crossref] [ Google Scholar]
- Bi S, Ding L, Tian Y, Song D, Zhou X, Liu X. Investigation of the interaction between flavonoids and human serum albumin. J Mol Struct 2004; 703:37-45. doi: 10.1016/j.molstruc.2004.05.026 [Crossref] [ Google Scholar]
- Trnkova L, Bousova I, Stankova V, Drsata J. Study on the interaction of catechins with human serum albumin using spectroscopic and electrophoretic techniques. J Mol Struct 2011; 985:243-250. doi: 10.1016/j.molstruc.2010.11.001 [Crossref] [ Google Scholar]
- Cui F, Zhang Q, Yao X, Luo H, Yang Y, Qin L. The investigation of the interaction between 5-Iodouracil and human serum albumin by spectroscopic and modeling methods and determination of protein by synchronous fluorescence technique. Pestic Biochem Physiol 2008; 90:126-134. doi: 10.1016/j.pestbp.2007.11.002 [Crossref] [ Google Scholar]
- Bhogale A, Patel N, Mariam J, Dongre PM, Miotello A, Kothari DC. Comprehensive studies on the interaction of copper nanoparticles with bovine serum albumin using various spectroscopies. Colloids Surf B 2014; 113:276-284. doi: 10.1016/j.colsurfb.2013.09.021 [Crossref] [ Google Scholar]
- Ross PD, Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry 1981; 20:3096-3102. doi: 10.1021/bi00514a017 [Crossref] [ Google Scholar]
- Zhang Y, Shi S, Sun X, Xiong X, Peng M.
The effect of Cu2+ on the interaction between flavonoids with different C-ring substituents and bovine serum albumin: Structure-affinity relationship aspect
. J Inorg Biochem 2011; 105:1529-1537. doi: 10.1016/j.jinorgbio.2011.08.007 [Crossref] [ Google Scholar]
- Chen W, Sun S, Cao W, Liang Y, Song J. Antioxidant property of Qu-Cr(III) complex: The role of Cr(III) ion. J Mol Struct 2009; 918:194-197. doi: 10.1016/j.molstruc.2008.08.008 [Crossref] [ Google Scholar]
- Yang F, Zhang Y, Liang H. Interactive association of drugs binding to human serum albumin. Int J Mol Sci 2014; 15:3580-3595. doi: 10.3390/ijms15033580 [Crossref] [ Google Scholar]
- Yamashita N, Tanemura H, Kawanishi S. Mechanism of oxidative DNA damage induced by Qu in the presence of Cu(II). Mutation Research 1999; 425:107-115. [ Google Scholar]
- Ni YN, Du S, Kobot S. Interaction between Qu–copper(II) complex and DNA with the use of the Neutral Red dye fluorophor probe. Analytica Chimica Acta 2007; 584:19-27. doi: 10.1016/j.aca.2006.11.006 [Crossref] [ Google Scholar]
- Xiao J, Suzuki M, Jiang X, Chen X, Yamamoto K, Ren F. Influence of B-Ring Hydroxylation on Interactions of Flavonols with Bovine Serum Albumin. JJ Agric Food Chem 2008; 56:2350-2356. doi: 10.1021/jf7037295 [Crossref] [ Google Scholar]
- Rubens FV, Souza de, F Wagner, Giovani D. Antioxidant properties of complexes of flavonoids with metal ions. Redox Rep 2004; 9:97-104. doi: 10.1179/135100004225003897 [Crossref] [ Google Scholar]
-
Timaseff SN. Thermodynamics of protein interactions. In: H. Peeters H, ed. Proteins of Biological Fluids. Oxford: Pergamon Press; 1972. p. 511-520.
- Singha Roy A, Pandey NK, Dasgupta S. Preferential binding of fisetin to the native state of bovine serum albumin: spectroscopic and docking studies. Mol Biol Rep 2013; 40:3239-3253. doi: 10.1007/s11033-012-2399-9 [Crossref] [ Google Scholar]
- Hu YJ, Yang YO, Dai CM, Liu Y, Xiao XH. Site-Selective Binding of Human Serum Albumin by Palmatine: Spectroscopic Approach. Biomacromolecules 2010; 11:106-112. doi: 10.1021/bm900961e [Crossref] [ Google Scholar]
- Bukhari SB, Memon S, Mahroof-Tahir M, Bhanger MI. Synthesis, characterization and antioxidant activity copper–quercetin complex. Spectrochimica Acta Part A 2009; 71:1901-1906. doi: 10.1016/j.saa.2008.07.030 [Crossref] [ Google Scholar]
- Said Al Amri F, Amzad Hossain M. Comparison of total phenols, flavonoids and antioxidant potential of local and imported ripe bananas. Egyptian Journal of Basic and Applied Sciences 2018; 5:245-251. doi: 10.1016/j.ejbas.2018.09.002 [Crossref] [ Google Scholar]
- Al-Jadaan SAN, Al-Diwan MA, Mathi AS. Synthesis, characterization and antioxidant activity of novel quercetin derivative. Life Sc Arch 2018; 4:1260-1272. [ Google Scholar]