BioImpacts. 10(2):65-72.
doi: 10.34172/bi.2020.09
Original Research
Lactobacillus plantarum induces apoptosis in gastric cancer cells via modulation of signaling pathways in Helicobacter pylori
Hadi Maleki-Kakelar 1  , Jaber Dehghani 1, Abolfazl Barzegari 1, Jaleh Barar 1, 2
, Jaber Dehghani 1, Abolfazl Barzegari 1, Jaleh Barar 1, 2  , Masoud Shirmohamadi 3, Javid Sadeghi 4, Yadollah Omidi 1, 2, *
, Masoud Shirmohamadi 3, Javid Sadeghi 4, Yadollah Omidi 1, 2, * 
Author information:
1Research Center for Pharmaceutical Nanotechnology, Biomedicine Institute, Tabriz University of Medical Sciences, Tabriz, Iran
2Department of Pharmaceutics, Faculty of Pharmacy, Tabriz University of Medical Sciences, Tabriz, Iran
3Liver and Gastrointestinal Diseases Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
4Department of Microbiology, School of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
Abstract
Introduction:
Gastric cancer is considered the second prevalent cause of death around the world. This type of cancer is generally induced by Helicobacter pylori which could colonize within the gastric mucosa of the infected cases. To date, triple antibiotic therapy has routinely been utilized for controlling the H. pylori- induced infection. However, this strategy has been unsuccessful, in large part because of issues such as occurring point mutations in the H. pylori genome that can induce resistance to the antibiotics administered. Recently, it has been shown that different probiotics may have strong anti-cancer effects, in which they are capable of inhibiting H. pylori by both immunological and non-immunological mechanisms. Here, we aimed at finding possible anti-cancer impacts of the probiotic bacterium Lactobacillus plantarum on gastric cancer, AGS cells.
Methods:
The anti-cancer effects of the conditioned media of the locally isolated L. plantarum on the AGS cells were evaluated by different analyses such as flow cytometry, DNA ladder assay, DAPI staining, and RT-PCR.
Results:
Our findings showed that the conditioned media of L. plantarum can inhibit both H. pylori and AGS cells through up-/down-regulation of PTEN, Bax, TLR4, and AKT genes. The exudates of the probiotic L. plantarum bacteria can increase the expression of PTEN, Bax, and TLR4, and also decrease the expression of AKT gene.
Conclusion:
In agreement with different reports, our results proved the anti-cancer effects of the locally isolated L. plantarum through some immunological cell signaling pathways. Accordingly, it seems the probiotics could be considered as at least a complementary treatment for different types of malignancies.
Keywords: Gastric cancer, Helicobacter pylori, Lactobacillus plantarum, Apoptosis, PTEN, AKT
Copyright and License Information
© 2020 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Gastric cancer induced by Helicobacter pylori is the second leading cause of death among infected individuals worldwide.
1
Annually, about 700 000 patients are surrendered via cancer induced by the H. pylori.
1,2
This issue is more disastrous in the Middle East region, where the prevalence of H. pylori is very high. Based on different reports, the outbreak of H. pylori infection is respectively ~90%, 58.4%, and 51% in some regions of Iran, Saudi Arabia, and Turkey.
3-6
Unfortunately, the treatment of H. pylori infection still remains unsuccessful, in large part because of its complex biology and adaptive mechanisms.
7
Currently, antibiotic therapy is the only approach for controlling the infection, in which clarithromycin, amoxicillin, or metronidazole can somewhat inhibit the H. pylori infection.
8
However, it is believed that the triple antibiotic therapy may be failed in 10%–23% of patients.
9
In reality, the emergence of point mutations in the H. pylori genome can lead to the resistance of the bacterium to the clarithromycin and metronidazole.
10
Accordingly, exploring new approaches for controlling or treatment of the H. pylori infection are intensely demanded.
1
Although a vaccine that induces a humoral immune response and Th2 cells will be a suitable platform to inhibit the H. pylori bacteria, there is still no effective vaccine(s) available to control the infection.
11
Various groups in the world work to construct efficient vaccine(s) against the H. pylori infection and several of these deigned constructs are being evaluated in different clinical trial phases.
12,13
More recently, probiotics have been considered as a novel tool for controlling H. pylori infections.
1,14
Based on the definition of the Food and Agriculture Organization (FAO) and the World Health Organization (WHO), probiotics are live microorganisms that can confer beneficial health effects on their hosts if they are administered in suitable amounts.
15
Probiotics such as lactobacilli are capable of inhibiting H. pylori by various immunological and non-immunological mechanisms.
16
In general, probiotics can inhibit the proliferation of the bacterium through competing with H. pylori for host surface receptors, and subsequently disrupt its adhesion to the epithelial cells.
17
Moreover, probiotics can counteract with spiral bacteria by secreting some antibacterial compounds including lactic acid, bacteriocins, hydrogen peroxide, short-chain fatty acids, and antibiotic-like substances such as reuterin and reutericyclin.
18
More importantly, lactic acid is probably effective on the H. pylori growth via lowering the pH and inhibiting its urease enzyme.
19
It should be noted that probiotics can modify the immunologic responses including neutrophils, lymphocytes, plasma cells, and macrophages and then trigger the inflammatory responses against H. pylori.
20
Thus, the levels of pro-inflammatory cytokines like IL-1β, IL-2, IL-6, IL-8, and tumor necrosis factor α are significantly increased in the gastric mucosa. However, the precise mechanism of signaling pathways induced by H. pylori is yet to be fully determined in the host cells.
21
It has recently been identified that H. pylori can induce TLR4 expression in the gastric cancer cell line (AGS) cell line, which leads to activating the NF-kB and MAPK depended on pathways and also producing the pro-inflammatory cytokines.
22,23
Furthermore, H. pylori by the phosphorylation of the phosphatase and tensin homolog (PTEN) tumor suppressor at Ser380/Thr382/Thr383 and subsequently inactivating the gene can induce gastric carcinogenesis.
24
Preliminary reports showed that H. pylori bacterium is effective on the expression level of an important apoptosis-regulatory gene known as Bax. Accordingly, a significant level of the Bax protein is accumulated in the H. pylori -infected cells.
25
Moreover, several studies have confirmed different effects of H. pylori on the other apoptosis-involved genes such as AKT, Tp53, and IL-6.
26-28
Here, we evaluated the anti-apoptotic effects of the locally isolated probiotic bacterium L. plantarum on the AGS in the presence of H. pylori. We also evaluated the expression levels of apoptotic and anti-apoptotic related signaling pathways in the presence of L. plantarum, H. pylori, and co-culture of two mentioned bacteria on the AGS and Huvec cells to study the apoptotic and anti-apoptotic signaling pathways in H. pylori.
Materials and Methods
Materials
RPMI 1640 medium and Trypsin–EDTA (1X), antibiotics (penicillin and streptomycin), and DAPI staining solution (4,6-diamidino-2-phenylindole) were purchased from Sigma-Aldrich Co. (Poole, UK). Fetal bovine serum (FBS) was obtained from Gibco Life technology, (Paisley, UK). TRIzol® reagent and SYBRTM Green master mix were obtained from Thermo Fisher Scientific (Waltham, MA, USA). Primers were designed by oligo software version 7 and synthesized via Bioneer Company (Daejeon, Korea). The solutions for RNA extraction and cDNA synthesis were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Annexin V-FITC apoptosis detection kit was purchased from EMD Chemicals (Gibbstown, USA).
Microbial strain and molecular identification
The probiotic bacterium L. plantarum was isolated from traditional dairy products and subsequently, their bile acid resistance, and antibiotic susceptibilities were examined based on the following work.
29
Furthermore, molecular identification of the bacteria was performed by 16S rRNA gene sequencing. For this purpose, bacterial DNA was extracted and then the 16S rRNA gene was amplified by the specific primers (Table 1). The polymerase chain reaction (PCR) was performed in 25 μL volumes, containing 20 ng genomic DNA, 50 ng of each primer, Taq master mix (Ampliqon Co., Odense, Denmark), and deionized water. Gene amplifications were achieved by Peqlab thermal cycler (Wilmington, USA) as follows: initial denaturation at 94°C for 5 minutes, 32 cycles of initiation at 94°C for 1 minute, annealing at 57°C for 1 minute, and extension at 72°C for 2 minutes, with a final extension at 72°C for 10 minutes. The PCR products were monitored using agarose gel (1%).
Cell culture and treatment
The human gastric (AGS, CRL-1739) and umbilical vein/vascular endothelium cells (Huvec, ATCC: CRL-1730) were obtained from the National Cell Bank of Iran (Pasteur Institute, Tehran, Iran).
30
The mentioned cells were cultured at a seeding density of 1.0×104 cells/cm2 in RPMI 1640 medium containing 10 % FBS, 0.1 μg/μL concentration of penicillin/streptomycin and then incubated at 37°C and 5% CO2. After reaching the confluency of 40%-50%, the cultivated cells were treated with the conditioned-media composed of the co-cultured cells and bacteria for 4-6 hours. In more detail, the bacteria firstly were cultured in the MRS broth at 37°C during the overnight. Subsequently, the bacteria were harvested at 2800 ×g for 5 minutes and the bacterial pellets were washed two times with PBS (1X, 137mM Sodium Chloride, 10mM phosphate, 2.7mM Potassium Chloride; pH: 7.4). Afterward, 0.5 McFarland concentration of bacteria was co-cultured with AGS and Huvec cell line for 4-6 hours. After harvesting and filtering the co-cultured condition media, the supernatant solutions were introduced to a different passage of the cultivated cells with 40%-50% confluency for 2, 6, and 24 hours. Finally, the treated cells were subjected to different biological analyses.
31
RNA extraction and RT-PCR
Total RNA was extracted from the treated AGS and Huvec cell lines using the TRIzol® reagent (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. The purity of RNAs was assessed via a NanoDrop ND®-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The cDNA synthesis was performed by the RevertAid Reverse Transcriptase kit (Thermo Fisher Scientific, Waltham, MA, USA) based on the manufacturer’s instructions. Real-time PCR reactions were performed to measure the expression levels of phosphatase and tensin homolog (PTEN), Bcl-2-associated X (Bax), Toll-like receptor 4 (TLR4), and protein kinase B (AKT) genes using a power SYBRTM Green master mix on a real-time PCR using Bio-Rad IQ5 system (Bio-Rad CO., Hercules, CA, USA) with the specific primers (Table 1). Moreover, the GAPDH as a house-keeping gene was applied as a reference gene to standardize the Ct of target genes. The data were analyzed by GraphPad Prism software and the Heat Map analysis of the genes expression patterns in different conditions was performed by CIMminer (https://discover.nci.nih.gov/cimminer/home.do).
Table 1.
The sequences of primers used for detecting specific RNAs using RT-PCR
|
Gene symbol
|
Gene name
|
Gene ID
|
Primer sequences [5
´
→ 3
´
]
|
Amplicon length (bp)
|
R
2
|
| GAPDH |
Glyceraldehyde 3-phosphate dehydrogenase |
NM_002046.3 |
For 5´ AAGCTCATTTCCTGGTATGACAACG 3´
Rev 5´ TCTTCCTCTTGTGCTCTTGCTGG 3´
|
126 |
0.999 |
| AKT |
Protein kinase B |
NM_181690 |
For 5´ CGCAGTGCCAGCTGATGAAG 3´
Rev 5´ GTCCATCTCCTCCTCCTCCTG 3´
|
185 |
0.997 |
| BAX |
bcl-2-like protein 4 |
NM_001291428 |
For 5´ GATGCGTCCACCAAGAAG 3´
Rev 5´ AGTTGAAGTTGCCGTCAG 3´
|
163 |
0.995 |
| TLR4 |
Toll-like receptor 4 |
NM_138557 |
For 5´ AAGTTATTGTGGTGGTGTCTAG 3´
Rev 5´ GAGGTAGGTGTTTCTGCTAAG 3´
|
191 |
0.996 |
| PTEN |
Phosphatase and tensin homolog |
NM_000314 |
For 5´ TCCCAGTCAGAGGCGCTATG 3´
Rev 5´ CACAAACTGAGGATTGCAAGTTC 3´
|
203 |
0.993 |
Construction of protein/gene-protein/gene interaction networks
A protein/gene-protein/gene interaction network analysis was performed on the identified, differentially-expressed proteins. We utilized String software (http://string-db.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) to search for possible protein/gene-protein/gene interaction networks (https://www.genome.jp/kegg/).
DNA ladder assay for detection of apoptotic cells
The treated cells with ~70 % confluency were harvested from the culture flask using 0.05 % trypsin/EDTA solution. Then, the genomic DNA of the cell lines was extracted and monitored by 1 % agarose gel on the gel documentation instrument to detect the level of apoptosis.
32
DAPI staining assay
DAPI (4,6-diamidino- 2-phenylindole) staining technique was used to monitor the morphology of the nucleus in the apoptotic cells. The AGS cells were cultivated at a seeding density of 1.0×104 cells/cm2 in the plates and then the probiotic bacterium was added to the cells and incubated overnight (O/N). Next, the cells were fixed in 4% paraformaldehyde, washed with PBS and permeabilized with 0.1% Triton X-100 for 15 minutes. Finally, the microscopic observations were performed with the invert fluorescence microscope, IX51 (Olympus, Hamburg, Germany).
Flow cytometry assay
The flow cytometry analysis was carried out to monitor the occurrence of apoptosis and/or necrosis in the treated cells with L. plantarum. For this purpose, the co-cultured cells were harvested, washed with PBS, re-suspended in the binding buffer and stained with annexin V-FITC for 15 minutes. Subsequently, the stained cells were incubated at room temperature. Finally, the cells were re-suspended in the PI binding buffer and prepared for the monitoring of apoptosis. To this end, we capitalized on the detection of FITC and PI respectively in the FL-1 and FL-2 channels employing BD FACSCalibur™ (BD Biosciences, San Jose, CA, USA).
Statistical analysis
Data were presented as mean values ± standard deviation (SD) from three independent experiments. Between two groups, comparisons of continuous numerical variables were performed using the independent-sample t- test. One-way ANOVA, followed by the multiple comparison Tukey post hoc test, was used to test for the statistical differences. Data represented the mean values and standard deviations of the replication of at least three independent experiments. A P value less than 0.05 was considered statistically significant.
Results
Probiotics bacteria identification
The amplification of the 16S rRNA gene by PCR showed approximately 1700 bp band on the electrophoresis gel (Fig. 1A). Sequencing and blasting the sequences with the NCBI database revealed that our locally isolated bacterium is considered as L. plantarum. Furthermore, the sequence was submitted to the NCBI database (AC: MG708111).
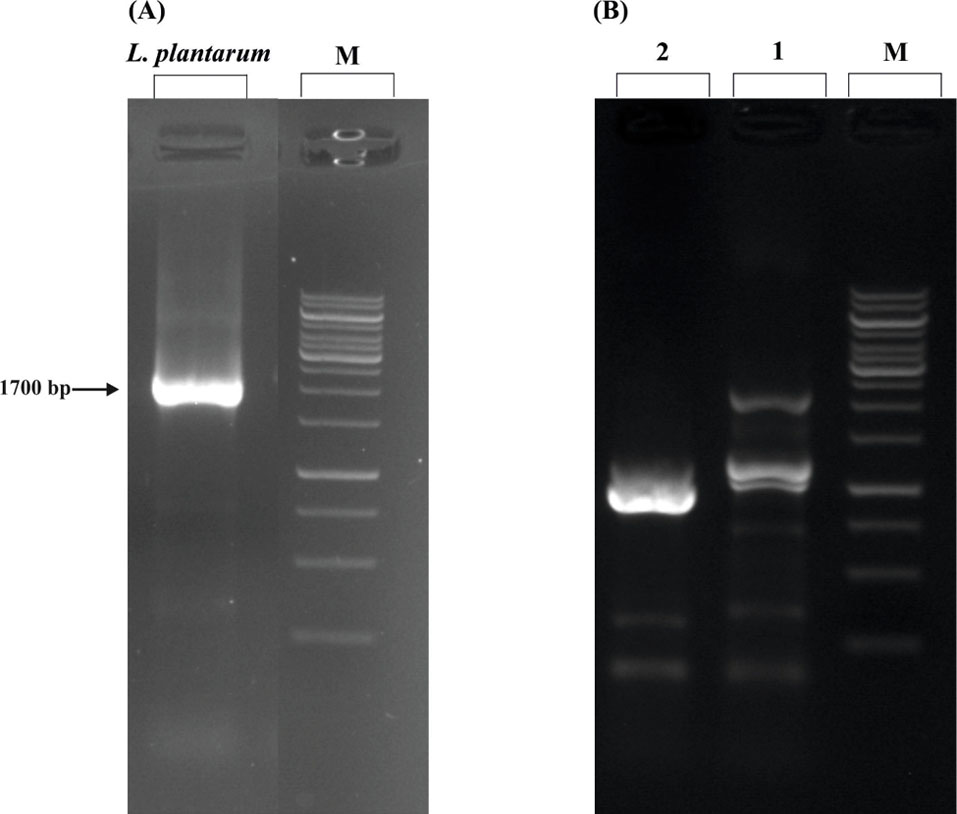
Fig. 1.
The molecular structure of 16S rDNA marker and DNA ladder assay. (A) Molecular identification of the locally isolated probiotic by 16S rDNA marker. The PCR results showed a ~1700 bp amplicon in the gel electrophoresis. On the basis of the sequencing and subsequent blasting in the NCBI server, the isolated probiotic is identified as one strain of Lactobacillus plantarum. (B) DNA ladder assay. The conditioned media of the locally isolated L. plantarum induced apoptosis in the AGS cells shown as fragmented DNA bands in the gel electrophoresis. M: 1kb ladder, 1: treated AGS cells, and 2: untreated AGS cells.
.
The molecular structure of 16S rDNA marker and DNA ladder assay. (A) Molecular identification of the locally isolated probiotic by 16S rDNA marker. The PCR results showed a ~1700 bp amplicon in the gel electrophoresis. On the basis of the sequencing and subsequent blasting in the NCBI server, the isolated probiotic is identified as one strain of Lactobacillus plantarum. (B) DNA ladder assay. The conditioned media of the locally isolated L. plantarum induced apoptosis in the AGS cells shown as fragmented DNA bands in the gel electrophoresis. M: 1kb ladder, 1: treated AGS cells, and 2: untreated AGS cells.
AKT, PTEN, BAX, and TLR4 genes expression
The RT-PCR data showed that the conditioned media of L. plantarum could change the expression levels of some apoptosis-involved genes. In detail, the treatment of the AGS cell line with the conditioned media for six hours caused a significant increase in PTEN (P <0.05) expression level and a decrease in the expression level of AKT gene (Figs. 2A, B and C and Fig. 3). Interestingly, the conditioned media of mixed H. pylori and L. plantarum in 12 hours can relatively increase the expression level of PTEN and decrease the expression level of AKT gene (Figs. 2A, B and C). Indeed, the AGS cells that were treated with the conditioned media for 12 hours, the down-regulation of AKT and up-regulation of PTEN, BAX, and TLR4 were not significant in them (Figs. 2A, B and C and Fig. 3).
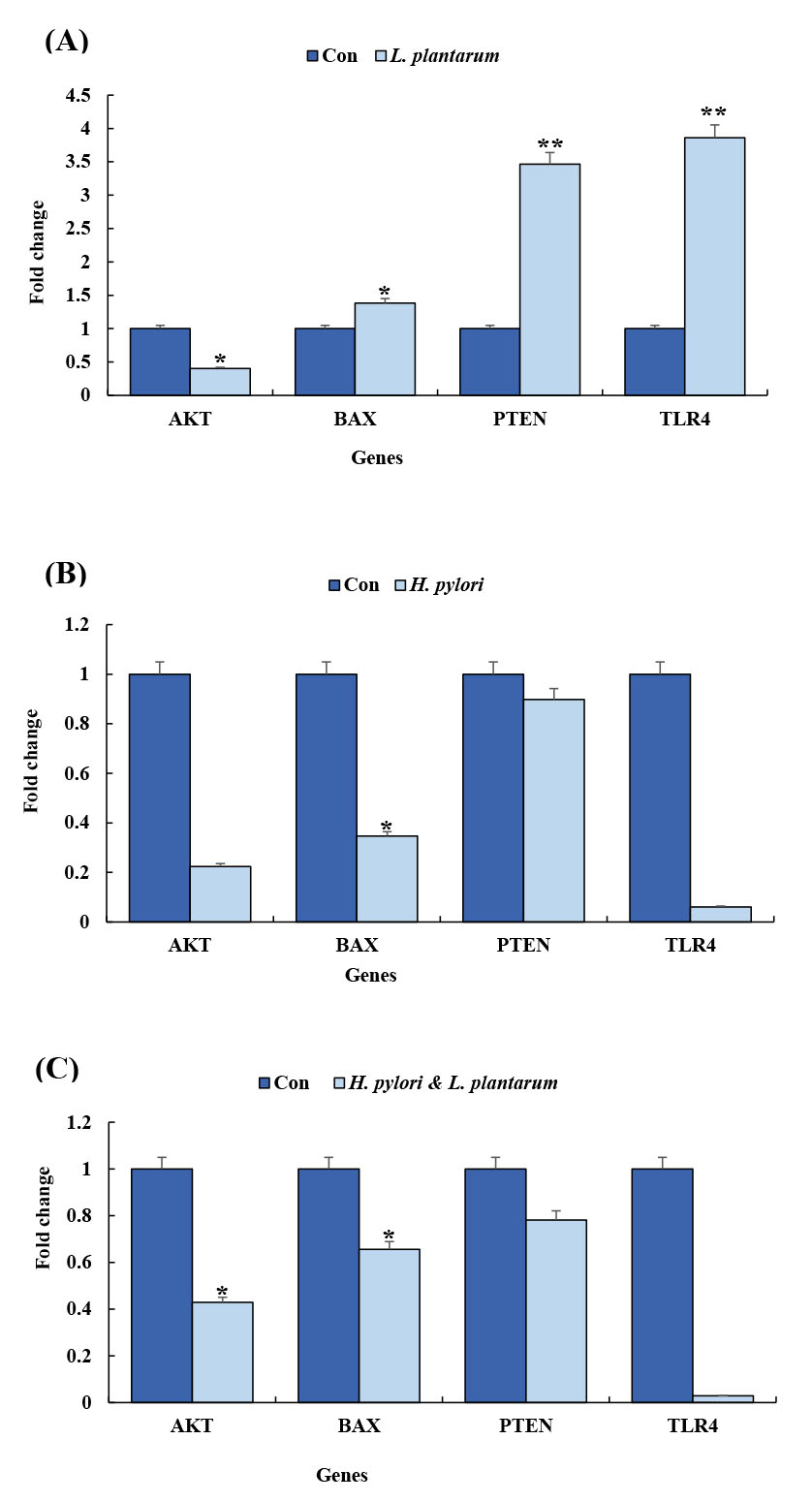
Fig. 2.
The RT-PCR analysis of the expression of PTEN, Bax, TLR4, and AKT genes. Panels A, B, and C represent the quantitative gene expression analyses at different conditions for L. plantarum, H. pylori, and H. pylori & L. plantarum, respectively.
.
The RT-PCR analysis of the expression of PTEN, Bax, TLR4, and AKT genes. Panels A, B, and C represent the quantitative gene expression analyses at different conditions for L. plantarum, H. pylori, and H. pylori & L. plantarum, respectively.
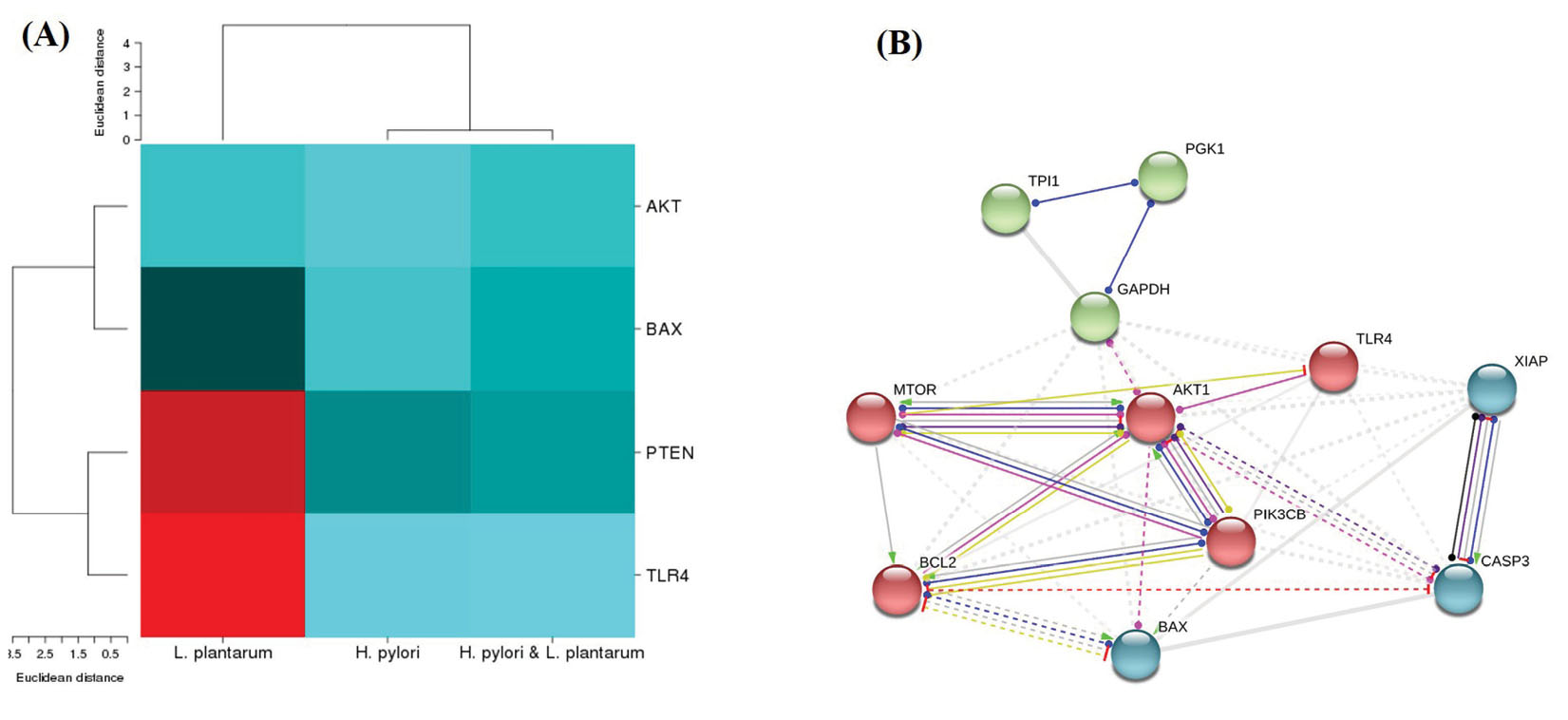
Fig. 3.
The genes expression pattern and protein-protein networks. (A) Heat map analysis of the genes expression patterns in different conditions such as L. plantarum, H. pylori & L. plantarum and H.pylori. (B) Network interactions between proteins related to apoptosis and mitochondrial membrane integrity. The network was constructed using the STRING tool.
.
The genes expression pattern and protein-protein networks. (A) Heat map analysis of the genes expression patterns in different conditions such as L. plantarum, H. pylori & L. plantarum and H.pylori. (B) Network interactions between proteins related to apoptosis and mitochondrial membrane integrity. The network was constructed using the STRING tool.
Analysis of protein/gene-protein/gene interactions
The String software was utilized to explore protein/gene-protein/gene interaction network analysis. Then, an interaction network diagram was constructed for four differentially expressed proteins and genes. The AKT, PTEN, and mTOR (mammalian target of rapamycin) proteins/genes were located in the most central area of the network, while cysteine-aspartic acid protease (caspase3), BCL2 -associated X (Bax), and B-cell lymphoma-extralarge (Bcl-xL) were located in the outermost part of the network (Fig. 3B).
DNA ladder assay
Because of the division of DNA, a standard agarose electrophoresis gel could be utilized to detect apoptosis as a ladder pattern of 180-200 bp through the activation of a nuclear endonuclease. Thus, by inducing apoptosis in the AGS cell line, it could be detected the formation of DNA ladder in the gel electrophoresis (Fig. 1B).
DAPI staining
Microscopic analysis of the stained cells with DAPI assay also examined the induction of apoptosis in AGS cells. As shown in Fig. 4, the untreated AGS cells possessed normal morphology, whilst the treated cells exhibited the changed morphological features, including condensed chromatin and nuclear fragmentation.
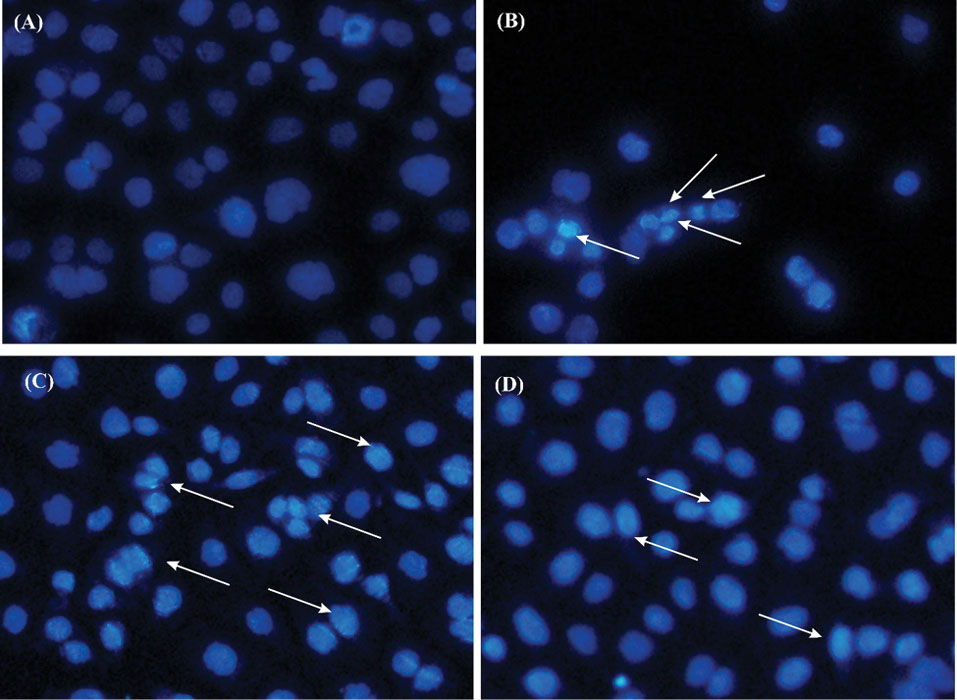
Fig. 4.
The morphological changes in the AGS cells treated with conditioned media (CM) of the probiotic L. plantarum bacteria. Panels A and B respectively represent the untreated cells, and the cells treated with 5% DMSO as the positive control. Panels C and D represent the cells treated with the CM of L. plantarum, and the CM of H.pylori, respectively. The arrows show the condensation/breakage of the nucleus of the affected cells.
.
The morphological changes in the AGS cells treated with conditioned media (CM) of the probiotic L. plantarum bacteria. Panels A and B respectively represent the untreated cells, and the cells treated with 5% DMSO as the positive control. Panels C and D represent the cells treated with the CM of L. plantarum, and the CM of H.pylori, respectively. The arrows show the condensation/breakage of the nucleus of the affected cells.
Flow cytometry assay
Flow cytometry assay was applied to detect the apoptotic and necrotic percentage in the AGS cells. Apoptosis includes the removal of the inner side of the plasma membrane of the membrane of phosphatidylserine from the cell surface. Annexin V has a high affinity to phosphatidylserine, where the cells are stained with Annexin V-FITC/PI. Moreover, the Ca
2+
phospholipid-binding protein showed a high affinity for the apoptotic cells. Our results showed that the percentage of apoptotic cells significantly increased (55.47 %) when the AGS cells were treated with the conditioned media of L. plantarum and also with the conditioned media of mixed H. pylori and L. plantarum (Fig. 5).
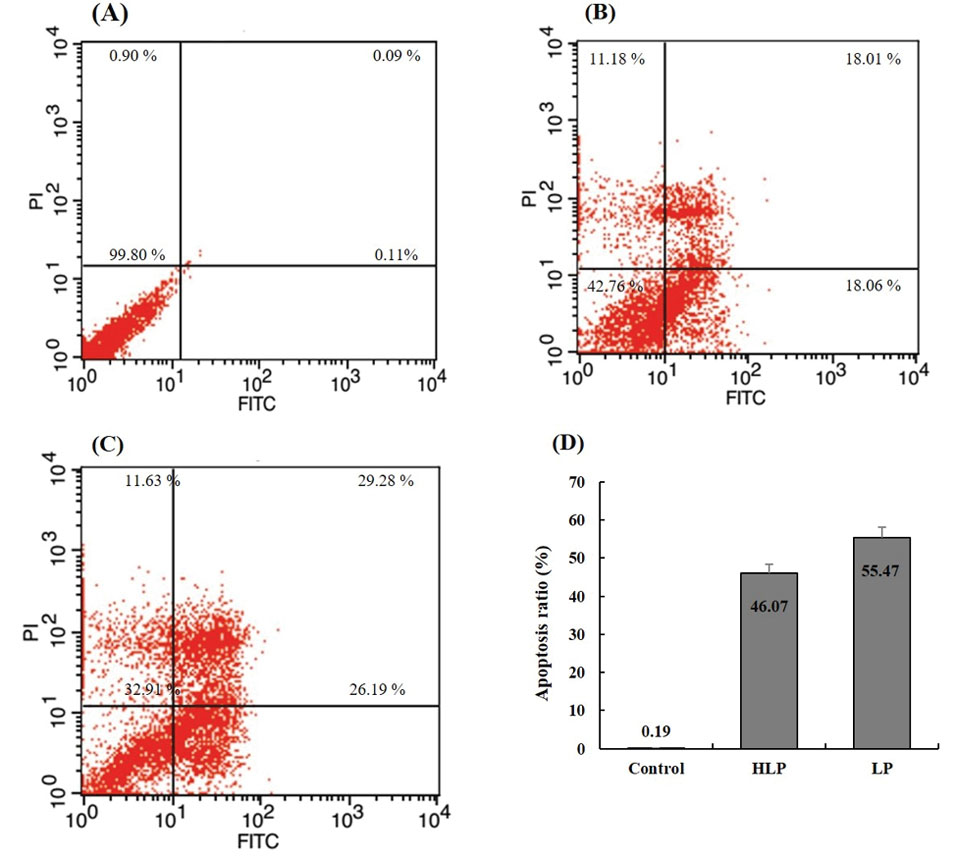
Fig. 5.
The flow cytometry analyses. (A) The ratio of apoptotic cells in the untreated cells. (B) The cells treated with H. pylori &L. plantarum conditioned-media. (C) The cells treated with L. plantarum conditioned-media. The cells were treated for 24 hours and evaluated by Annexin V-FITC/PI double-staining analysis. (D) Quantitative analysis of cell apoptosis by flow cytometry after staining with annexin V and PI. *P < .05, compared with the control experiment.
.
The flow cytometry analyses. (A) The ratio of apoptotic cells in the untreated cells. (B) The cells treated with H. pylori &L. plantarum conditioned-media. (C) The cells treated with L. plantarum conditioned-media. The cells were treated for 24 hours and evaluated by Annexin V-FITC/PI double-staining analysis. (D) Quantitative analysis of cell apoptosis by flow cytometry after staining with annexin V and PI. *P < .05, compared with the control experiment.
Discussion
Despite the recent developments in cancer therapy, this issue is considered as the second cause of death worldwide. Nowadays, probiotics have attracted the attention of many researchers because of their anti-cancer properties.
1
It is believed that probiotics are capable of inhibiting the cancers by the suppression of the growth mutagens and carcinogen microorganisms, protecting the host genome from oxidative damage, and the regulation of the immune system.
33
Moreover, these microorganisms can change the expression of different genes that participate in various biological phenomena, including cell proliferation, apoptosis, metastasis processes, and cell cycle.
34
However, the precise molecular mechanisms of probiotic impacts on the cancer cells are yet to be fully understood. Accordingly, we aimed to explore some molecular mechanisms of the probiotic L. plantarum on the expression of the several important genes (i.e., AKT, PTEN, BAX, and TLR4 genes) involved in the apoptosis and anti-apoptosis signaling pathways in the AGS cell line as a gastric cancer cell. Different analyses such as DNA ladder assay, flow cytometry, DAPI staining, and q-PCR confirmed the L. plantarum and showed positive potential in inducing the apoptosis in the gastric cancer cell line. The latter data showed that L. plantarum by decreasing and increasing the expression of AKT and PTEN genes, respectively, can inhibit the viability of cancer cells. It should be noted that similar results are achieved from different researches which are discussed briefly below.
Based on some cellular and molecular approaches, it was confirmed that L. plantarum could inhibit human oral cancer cells. Similarly, the authors reported that the probiotic bacterium could increase the expression level of the PTEN gene and subsequently diminish the cancer cells.
35
Moreover, it has been reported that the inactivation of PTEN in the head and neck squamous cell carcinoma cancer cell line can increase the expression of AKT gene and suppress the p53 gene. Indeed, over-expression of the PTEN gene can significantly inhibit the growth of the tumor.
36
Furthermore, it has been reported that the probiotic bacterium L. mesenteroides could inhibit the inflammation and cell survival in colon cancer by modulating the NF-kappaB/AKT/PTEN/MAPK pathways, along with miRNA-21 and miRNA-200b. Moreover, another study showed that L. amylovorus could pose an important effect in the inhibition of intestinal infection induced by Escherichia coli K88 through the suppression of TLR4 gene.
37
The up/down-regulation of Bax gene is also effective in controlling the cancer cells as it is confirmed seven species of probiotics could hinder the growth of human colonic carcinoma via modulating the Bax/Bcl-2 pathway.
38
Inconsistent with the other reports, our data represented that the expression level of PTEN significantly increased compared to the untreated control cells when the AGSs were treated with the conditioned media of L. plantarum.
Based on our findings, the treatment of AGS cells with the conditioned media of L. plantarum could significantly increase the PTEN mRNA expression and also markedly decrease the expression of AKT gene. As a result, we speculated that the probiotics especially L. plantarum might play an important role in the inhibition of H. pylori -related gastric cancer through the up-/down-regulation of the PTEN/AKT signaling pathways. Considering that the imposition of an unsuitable dose of encapsulated non-indigenous probiotics may change the normal pattern of the commensal microbiome in the body, probiotics therapy should be considered a supplementary treatment.
39
It should be noted that probiotics similar to many of microalgae (e.g., Dunaliella and Spirulina species) may be generally regarded as safe for use in animal and human systems.
29,40,41
Taken all, these organisms could be assigned as an important vehicle for oral delivery of different biologics such as edible vaccines.
42
Conclusion
Nowadays, using the probiotics (e.g., L. plantarum ) in different aspects of the food industry, pharmacology, and medicine are daily increased. Regarding the anti-cancer effects of probiotics, we aimed at investigating the impacts of the probiotic bacterium L. plantarum on the gastric cancer AGS cells. Our results showed that the conditioned media of the L. plantarum could inhibit cancer through up-/down-regulation of some important apoptosis and anti-apoptosis related genes, including PTEN, Bax, TLR4, and AKT. Taking the aforementioned results into consideration, it seems that the microorganisms could be utilized as a novel therapeutic approach in the prevention of H. pylori -induced gastric cancer. However, this platform should be clinically examined before translation into clinical practice.
Acknowledgments
Authors gratefully appreciate the Research Center for Pharmaceutical Nanotechnology at Tabriz University of Medical Sciences for supporting this work.
Funding sources
This study was financially supported by the Research Center for Pharmaceutical Nanotechnology (RCPN) at Tabriz University of Medical Sciences (grant No. 94010). This project is part of a Ph.D. thesis (No. 94/010/156/3) conducted at RCPN.
Ethical statement
Not applicable.
Competing interests
The authors declare there is no conflict of interests.
Authors’ contribution
All authors conceived and planned the project, the main conceptual ideas, and design of the experiments. HMK, JD, and YO wrote the draft and JB and YO finalized the latest version of the manuscript.
Research Highlights
What is the current knowledge?
simple
-
√ The locally isolated L. plantarum showed a significant antigastric
cancer activity.
-
The conditioned media of the probiotic bacterium could be
used for the treatment of gastric cancer induced by H. pylori.
What is new here?
simple
-
√ The probiotic bacterium L. plantarum can induce apoptosis
in the AGS cells and hence inhibit their growth significantly.
-
√ By up/down regulation of PTEN and AKT genes
respectively, L. plantarum can inhibit the proliferation of
AGS cells and H. pylori bacterium.
References
- Maleki Kakelar H, Barzegari A, Dehghani J, Hanifian S, Saeedi N, Barar J. Pathogenicity of Helicobacter pylori in cancer development and impacts of vaccination. Gastric Cancer 2019; 22:23-36. doi: 10.1007/s10120-018-0867-1 [Crossref] [ Google Scholar]
- Sabbagh P, Javanian M, Koppolu V, Vasigala VR, Ebrahimpour S. Helicobacter pylori infection in children: an overview of diagnostic methods. Eur J Clin Microbiol Infect Dis 2019. doi: 10.1007/s10096-019-03502-5 [Crossref]
- Malekzadeh R, Sotoudeh M, Derakhshan MH, Mikaeli J, Yazdanbod A, Merat S. Prevalence of gastric precancerous lesions in Ardabil, a high incidence province for gastric adenocarcinoma in the northwest of Iran. J Clin Pathol 2004; 57:37-42. doi: 10.1136/jcp.57.1.37 [Crossref] [ Google Scholar]
- Sotoudeh M, Derakhshan MH, Abedi-Ardakani B, Nouraie M, Yazdanbod A, Tavangar SM. Critical role of Helicobacter pylori in the pattern of gastritis and carditis in residents of an area with high prevalence of gastric cardia cancer. Dig Dis Sci 2008; 53:27-33. doi: 10.1007/s10620-007-9817-1 [Crossref] [ Google Scholar]
- Abasiyanik MF, Tunc M, Salih BA. Enzyme immunoassay and immunoblotting analysis of Helicobacter pylori infection in Turkish asymptomatic subjects. Diagn Microbiol Infect Dis 2004; 50:173-7. doi: 10.1016/j.diagmicrobio.2004.07.005 [Crossref] [ Google Scholar]
- Khan MA, Ghazi HO. Helicobacter pylori infection in asymptomatic subjects in Makkah, Saudi Arabia. J Pak Med Assoc 2007; 57:114-7. [ Google Scholar]
- Harris A. Treatment of Helicobacter pylori. World J Gastroenterol 2001; 7:303-7. doi: 10.3748/wjg.v7.i3.303 [Crossref] [ Google Scholar]
- Goderska K, Agudo Pena S, Alarcon T. Helicobacter pylori treatment: antibiotics or probiotics. Appl Microbiol Biotechnol 2018; 102:1-7. doi: 10.1007/s00253-017-8535-7 [Crossref] [ Google Scholar]
- Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev 2006; 19:449-90. doi: 10.1128/cmr.00054-05 [Crossref] [ Google Scholar]
- Megraud F. Basis for the management of drug-resistant Helicobacter pylori infection. Drugs 2004; 64:1893-904. doi: 10.2165/00003495-200464170-00003 [Crossref] [ Google Scholar]
- Blosse A, Lehours P, Wilson KT, Gobert AP. Helicobacter: Inflammation, immunology, and vaccines. Helicobacter 2018; 23 Suppl 1:e12517. doi: 10.1111/hel.12517 [Crossref] [ Google Scholar]
- Sutton P, Boag JM. Status of vaccine research and development for Helicobacter pylori. Vaccine 2018. doi: 10.1016/j.vaccine.2018.01.001 [Crossref]
- Talebi Bezmin Abadi A. Vaccine against Helicobacter pylori: Inevitable approach. World J Gastroenterol 2016; 22:3150-7. doi: 10.3748/wjg.v22.i11.3150 [Crossref] [ Google Scholar]
- Qureshi N, Li P, Gu Q. Probiotic therapy in Helicobacter pylori infection: a potential strategy against a serious pathogen?. Appl Microbiol Biotechnol 2019. doi: 10.1007/s00253-018-09580-3 [Crossref]
- Reid G. Probiotics: definition, scope and mechanisms of action. Best Pract Res Clin Gastroenterol 2016; 30:17-25. doi: 10.1016/j.bpg.2015.12.001 [Crossref] [ Google Scholar]
- Bruno G, Rocco G, Zaccari P, Porowska B, Mascellino MT, Severi C. Helicobacter pylori infection and gastric dysbiosis: can probiotics administration be useful to treat this condition?. Can J Infect Dis Med Microbiol 2018; 2018:6237239. doi: 10.1155/2018/6237239 [Crossref] [ Google Scholar]
- Mukai T, Asasaka T, Sato E, Mori K, Matsumoto M, Ohori H. Inhibition of binding of Helicobacter pylori to the glycolipid receptors by probiotic Lactobacillus reuteri. FEMS Immunol Med Microbiol 2002; 32:105-10. doi: 10.1111/j.1574-695X.2002.tb00541.x [Crossref] [ Google Scholar]
- Foligne B, Nutten S, Grangette C, Dennin V, Goudercourt D, Poiret S. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J Gastroenterol 2007; 13:236-43. doi: 10.3748/wjg.v13.i2.236 [Crossref] [ Google Scholar]
- Lin WH, Wu CR, Fang TJ, Guo JT, Huang SY, Lee MS. Anti-Helicobacter pylori activity of fermented milk with lactic acid bacteria. J Sci Food Agric 2011; 91:1424-31. doi: 10.1002/jsfa.4327 [Crossref] [ Google Scholar]
- Homan M, Orel R. Are probiotics useful in Helicobacter pylori eradication?. World J Gastroenterol 2015; 21:10644-53. doi: 10.3748/wjg.v21.i37.10644 [Crossref] [ Google Scholar]
- Borruel N, Casellas F, Antolin M, Llopis M, Carol M, Espiin E. Effects of nonpathogenic bacteria on cytokine secretion by human intestinal mucosa. Am J Gastroenterol 2003; 98:865-70. doi: 10.1111/j.1572-0241.2003.07384.x [Crossref] [ Google Scholar]
- Su B, Ceponis PJ, Lebel S, Huynh H, Sherman PM. Helicobacter pylori activates Toll-like receptor 4 expression in gastrointestinal epithelial cells. Infect Immun 2003; 71:3496-502. doi: 10.1128/iai.71.6.3496-3502.2003 [Crossref] [ Google Scholar]
- Allison CC, Kufer TA, Kremmer E, Kaparakis M, Ferrero RL. Helicobacter pylori induces MAPK phosphorylation and AP-1 activation via a NOD1-dependent mechanism. J Immunol 2009; 183:8099-109. doi: 10.4049/jimmunol.0900664 [Crossref] [ Google Scholar]
- Yang Z, Cao X, Xu W, Xie C, Chen J, Zhu Y. Phosphorylation of phosphatase and tensin homolog induced by Helicobacter pylori promotes cell invasion by activation of focal adhesion kinase. Oncol Lett 2018; 15:1051-7. doi: 10.3892/ol.2017.7430 [Crossref] [ Google Scholar]
- Liu HF, Liu WW, Wang GA, Teng XC. Effect of Helicobacter pylori infection on Bax protein expression in patients with gastric precancerous lesions. World J Gastroenterol 2005; 11:5899-901. doi: 10.3748/wjg.v11.i37.5899 [Crossref] [ Google Scholar]
- Yan F, Polk DB. Probiotic bacterium prevents cytokine-induced apoptosis in intestinal epithelial cells. J Biol Chem 2002; 277:50959-65. doi: 10.1074/jbc.M207050200 [Crossref] [ Google Scholar]
- Li N, Xie C, Lu N-H. p53, a potential predictor of Helicobacter pylori infection-associated gastric carcinogenesis?. Oncotarget 2016; 7:66276-86. doi: 10.18632/oncotarget.11414 [Crossref] [ Google Scholar]
- Song H-Y, Zhou L, Liu D-Y, Yao X-J, Li Y. What roles do probiotics play in the eradication of Helicobacter pylori? Current knowledge and ongoing research. Gastroenterol Res Pract 2018; 2018:9379480. doi: 10.1155/2018/9379480 [Crossref] [ Google Scholar]
- Maleki Kakelar H, Barzegari A, Hanifian S, Barar J, Omidi Y. Isolation and molecular identification of Lactobacillus with probiotic potential from abomasums driven rennet. Food Chem 2019; 272:709-14. doi: 10.1016/j.foodchem.2018.08.081 [Crossref] [ Google Scholar]
- Eskandani M, Abdolalizadeh J, Hamishehkar H, Nazemiyeh H, Barar J. Galbanic acid inhibits HIF-1α expression via EGFR/HIF-1α pathway in cancer cells. Fitoterapia 2015; 101:1-11. doi: 10.1016/j.fitote.2014.12.003 [Crossref] [ Google Scholar]
- Hamishehkar H, Khani S, Kashanian S, Ezzati Nazhad Dolatabadi J, Eskandani M. Geno- and cytotoxicity of propyl gallate food additive. Drug Chem Toxicol 2014; 37:241-6. doi: 10.3109/01480545.2013.838776 [Crossref] [ Google Scholar]
- Rahbar Saadat Y, Saeidi N, Zununi Vahed S, Barzegari A, Barar J. An update to DNA ladder assay for apoptosis detection. Bioimpacts 2015; 5:25-8. doi: 10.15171/bi.2015.01 [Crossref] [ Google Scholar]
- Ma EL, Choi YJ, Choi J, Pothoulakis C, Rhee SH, Im E. The anticancer effect of probiotic Bacillus polyfermenticus on human colon cancer cells is mediated through ErbB2 and ErbB3 inhibition. Int J Cancer 2010; 127:780-90. doi: 10.1002/ijc.25011 [Crossref] [ Google Scholar]
- So SS, Wan ML, El-Nezami H. Probiotics-mediated suppression of cancer. Curr Opin Oncol 2017; 29:62-72. doi: 10.1097/cco.0000000000000342 [Crossref] [ Google Scholar]
- Asoudeh-Fard A, Barzegari A, Dehnad A, Bastani S, Golchin A, Omidi Y. Lactobacillus plantarum induces apoptosis in oral cancer KB cells through upregulation of PTEN and downregulation of MAPK signalling pathways. BioImpacts 2017; 7:193-8. doi: 10.15171/bi.2017.22 [Crossref] [ Google Scholar]
- Squarize CH, Castilho RM, Abrahao AC, Molinolo A, Lingen MW, Gutkind JS. PTEN deficiency contributes to the development and progression of head and neck cancer. Neoplasia 2013; 15:461-71. doi: 10.1593/neo.121024 [Crossref] [ Google Scholar]
- Finamore A, Roselli M, Imbinto A, Seeboth J, Oswald IP, Mengheri E. Lactobacillus amylovorus inhibits the TLR4 inflammatory signaling triggered by enterotoxigenic Escherichia coli via modulation of the negative regulators and involvement of TLR2 in intestinal Caco-2 cells and pig explants. PLoS One 2014; 9:e94891. doi: 10.1371/journal.pone.0094891 [Crossref] [ Google Scholar]
- Chen ZY, Hsieh YM, Huang CC, Tsai CC. Inhibitory Effects of probiotic Lactobacillus on the growth of human colonic carcinoma cell line HT-29. Molecules 2017; 22. doi: 10.3390/molecules22010107 [Crossref]
- Zununi Vahed S, Barzegari A, Rahbar Saadat Y, Goreyshi A, Omidi Y. Leuconostoc mesenteroides-derived anticancer pharmaceuticals hinder inflammation and cell survival in colon cancer cells by modulating NF-kappaB/AKT/PTEN/MAPK pathways. Biomed Pharmacother 2017; 94:1094-100. doi: 10.1016/j.biopha.2017.08.033 [Crossref] [ Google Scholar]
- Dehghani J, Adibkia K, Movafeghi A, Barzegari A, Pourseif MM, Maleki Kakelar H. Stable transformation of Spirulina (Arthrospira) platensis: a promising microalga for production of edible vaccines. Appl Microbiol Biotechnol 2018; 102:9267-78. doi: 10.1007/s00253-018-9296-7 [Crossref] [ Google Scholar]
- Dehghani J, Movafeghi A, Barzegari A, Barar J. Efficient and stable transformation of Dunaliella pseudosalina by 3 strains of Agrobacterium tumefaciens. Bioimpacts 2017; 7:247-54. doi: 10.15171/bi.2017.29 [Crossref] [ Google Scholar]
- Amdekar S, Dwivedi D, Roy P, Kushwah S, Singh V. Probiotics: multifarious oral vaccine against infectious traumas. FEMS Immunol Med Microbiol 2010; 58:299-306. doi: 10.1111/j.1574-695X.2009.00630.x [Crossref] [ Google Scholar]