BioImpacts. 10(2):97-104.
doi: 10.34172/bi.2020.12
Original Research
Identification and ranking of important bio-elements in drug-drug interaction by Market Basket Analysis
Reza Ferdousi 1, 2  , Ali Akbar Jamali 3
, Ali Akbar Jamali 3  , Reza Safdari 4, *
, Reza Safdari 4, * 
Author information:
1Department of Health Information Technology, School of Management and Medical Informatics, Tabriz University of Medical Sciences, Tabriz, Iran
2Research Center for Pharmaceutical Nanotechnology, Biomedicine Institute, Tabriz University of Medical Sciences, Tabriz, Iran
3Division of Biomedical Engineering, University of Saskatchewan, Saskatoon, Saskatchewan, Canada
4Department of Health Care Management, Tehran University of Medical Sciences, Tehran, Iran
Abstract
Introduction:
Drug-drug interactions (DDIs) are the main causes of the adverse drug reactions and the nature of the functional and molecular complexity of drugs behavior in the human body make DDIs hard to prevent and threat. With the aid of new technologies derived from mathematical and computational science, the DDI problems can be addressed with a minimum cost and effort. The Market Basket Analysis (MBA) is known as a powerful method for the identification of co-occurrence of matters for the discovery of patterns and the frequency of the elements involved.
Methods:
In this research, we used the MBA method to identify important bio-elements in the occurrence of DDIs. For this, we collected all known DDIs from DrugBank. Then, the obtained data were analyzed by MBA method. All drug-enzyme, drug-carrier, drug-transporter and drug-target associations were investigated. The extracted rules were evaluated in terms of the confidence and support to determine the importance of the extracted bio-elements.
Results:
The analyses of over 45000 known DDIs revealed over 300 important rules from 22 085 drug interactions that can be used in the identification of DDIs. Further, the cytochrome P450 (CYP) enzyme family was the most frequent shared bio-element. The extracted rules from MBA were applied over 2000000 unknown drug pairs (obtained from FDA approved drugs list), which resulted in the identification of over 200000 potential DDIs.
Conclusion:
The discovery of the underlying mechanisms behind the DDI phenomena can help predict and prevent the inadvertent occurrence of DDIs. Ranking of the extracted rules based on their association can be a supportive tool to predict the outcome of unknown DDIs.
Keywords: Drug-drug interaction, Market Basket Analysis, Rule discovery, Biological targets
Copyright and License Information
© 2020 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
The recent advancements in genomics and proteomics have significantly amplified our understanding of the molecular basis for functions of drugs in the human body that has led to the discovery of de novo drugs. It has also led to the identification of the biological targets involved in various and crucial signaling pathways. Since the networks of drug-disease are extremely complicated, the co-administration of designated drugs in the treatment of chronic diseases is an inevitable issue.
1-3
In fact, one may daily consume a variety of therapeutic compounds, including nutritional supplements, medications, and other chemical materials. In most cases, if the drugs are concurrently consumed, they may show somewhat interaction with each other at molecular levels in the body.
4-7
Drug-drug interaction (DDI) refers to a reaction or situation, in which the presence of one drug affects the activity or functionality of another drug, resulting in some medical issues such as toxicity, serious complications, and failure of treatment modality. Under such circumstances, not only a multidrug therapy regime fails to show the desired treatment impacts but also may induce inadvertent adverse reactions. Therefore, it is undeniably important to recognize DDIs prior to their administration.
8-10
Furthermore, the identification of DDIs is considered as vital information for physicians, in large part because of the prescription of various multi-drug therapies with different orders. Although in some cases for existing and well-known drugs, physicians based on their experiential knowledge avoid those drugs, which can interact together. The identification of DDIs can be a crucial issue when the prescribed drugs are not enough well known or they have been designed and introduced recently. It seems that the prediction of drug-pair interactions is limited because of (a) lack of detailed information on drugs’ behavior(s) in the human body at different levels, (b) the low outcome of experimental and laboratory techniques, (c) needs for much more advanced technologies for the detection of DDIs, and (d) the high cost. In addition, sometimes results of in vitro experiments cannot be correlated with the outcomes of in vivo investigations. Hence, there are huge demands for robust and sound methodologies to overcome such shortcomings.
11-15
Unlike the experimental methods, the computational approaches show high degrees of flexibility and result in high-throughput outcomes, providing an opportunity to study and analyze clusters of old and new data to discover novel knowledge.
16-19
These mathematical models can disclose the hidden conditions in which the interactions occur. As a result, new approaches need to be implemented for the prediction of DDIs with high precision in order to provide key information for the promotion of the health by preventing any side effects of administered multi-drug therapy and hence adverse reactions.
17,20
Considering the fact that analogous structures and properties can suggest similar functions,
21,22
we incorporated the biological basis of drug interaction with the mathematical foundation of the MBA approach. Here, we considered any interaction between the pair of drugs as a transaction that is governed by particular rules or items. In such model, common enzymes of drug pairs, their transporters, carriers, and targets were considered as items of our model which caused an interaction between two drugs. Based on these rules, we predicted potential interactions among drugs that might be applied in clinical level.
Materials and Methods
In this work, we applied computational procedure based on association rules discovery algorithm. Market Basket Analysis (MBA) is considered as one of the most reliable and popular approaches that discover co-occurrences of one phenomenon with another one within the same event or transaction. In this scenario, mathematical models search for the association rules resulting in the occurrence of a specific event. Such rules demonstrate that according to the history of all the transactions, if item X is found in a transaction, there is a strong inclination for the occurrence of an item Y within the same transaction.
23-25
In the field of economy and mathematics, items and transactions refer to rules and events, respectively. This methodology has widely been used in the field of biology and medicine.
26,27
For instance, in the field of pharmacology, this procedure has been used to identify the associations of drugs’ features in traditional Chinese medicine drug pairs.
26,28
This section describes our approach in terms of mining DDIs based on the biomedical data. Fig. 1 summarizes the architecture of this study.
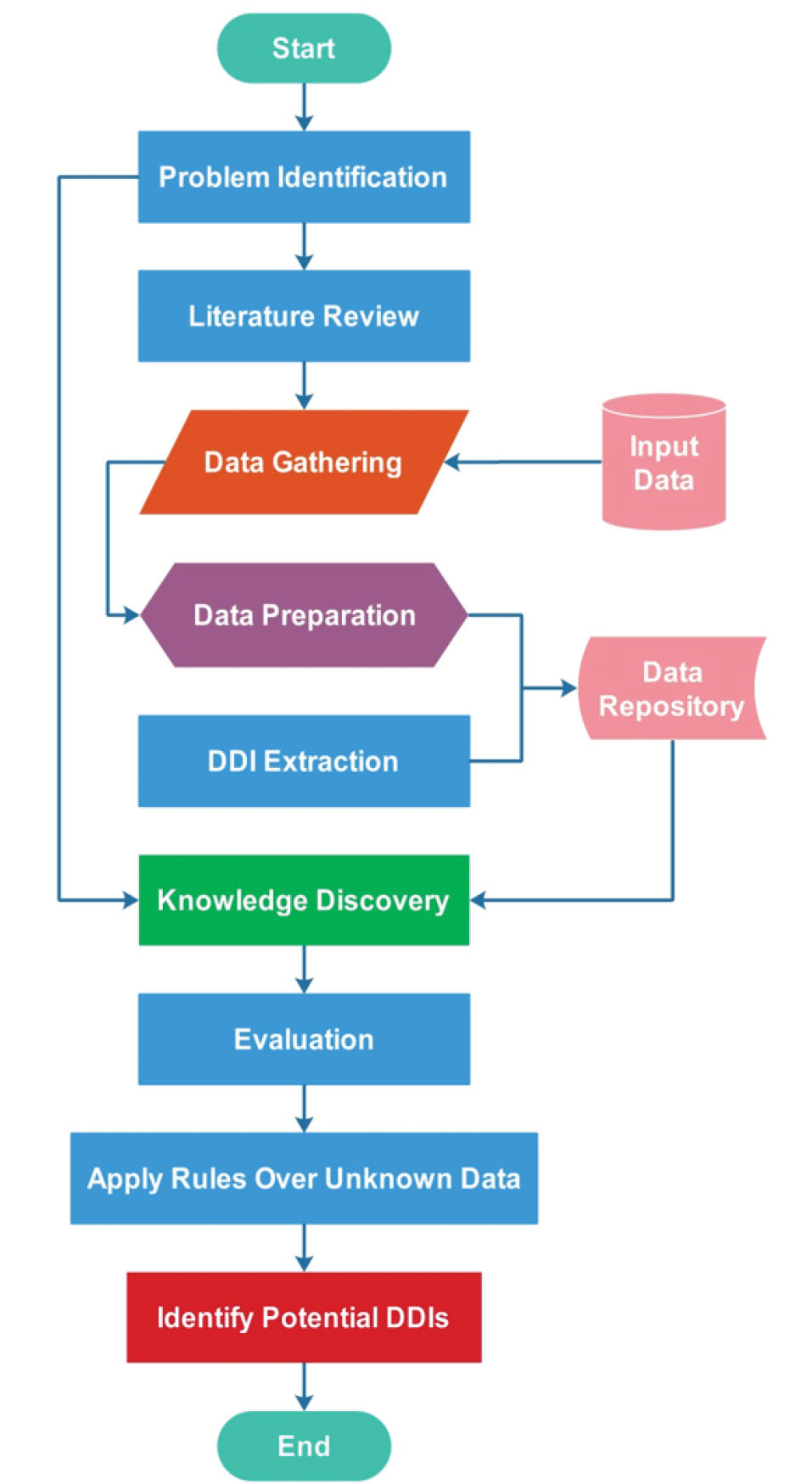
Fig. 1.
Schematic representation of the proposed Market Basket Analysis approaches for the identification of important bio elements in DDI occurrence.
.
Schematic representation of the proposed Market Basket Analysis approaches for the identification of important bio elements in DDI occurrence.
Data acquisition and preparation
It has been reported that there are 2186 drugs approved by the US FDA.
7
The aim of this study was to predict and identify possible interactions between each pair of these drugs. To develop such predictor, experimentally validated data are needed. These data were retrieved from the DrugBank dataset as drug interactions (version 4.3).
29,30
Since there was no list of drug pairs interacting together, we extracted these pairs from stored data in this dataset, where about 45 530 approved drug-pair interactions were found. Since we needed the interactions which resemble the aforementioned items, such filtering decreased the number of these interactions to 22 085 DDIs. Therefore, these interactions were used as the training dataset.
Market Basket Analysis, items and transactions
As mentioned above, the MBA is an approach in data mining, which focuses on the identification of events occurring at the same time on each transaction. As the input data belongs to pharmacology, the output of this method is a set of rules governing the interaction of two drugs. Moreover, these rules are considered as association rules that declare "where event X happens, then the event Y occurs".
23
In this approach, a time-effective analysis of large data is possible via apriori algorithm and its succeeding adjustments.
In this study, we considered any interaction between a pair of drugs as a transaction where items of these transactions refer to the common targets, including drug pairs, their enzymes, carriers, and transporters. Furthermore, different subsets of these items provide a situation, in which one interaction occurs. In such model, each rule is accompanied with a support level, which designates the number of interactions containing items X, Y (Fig. 2) and confidence level that represents the accuracy of association rules. In addition, each rule is equipped with a lift and expected confidence.
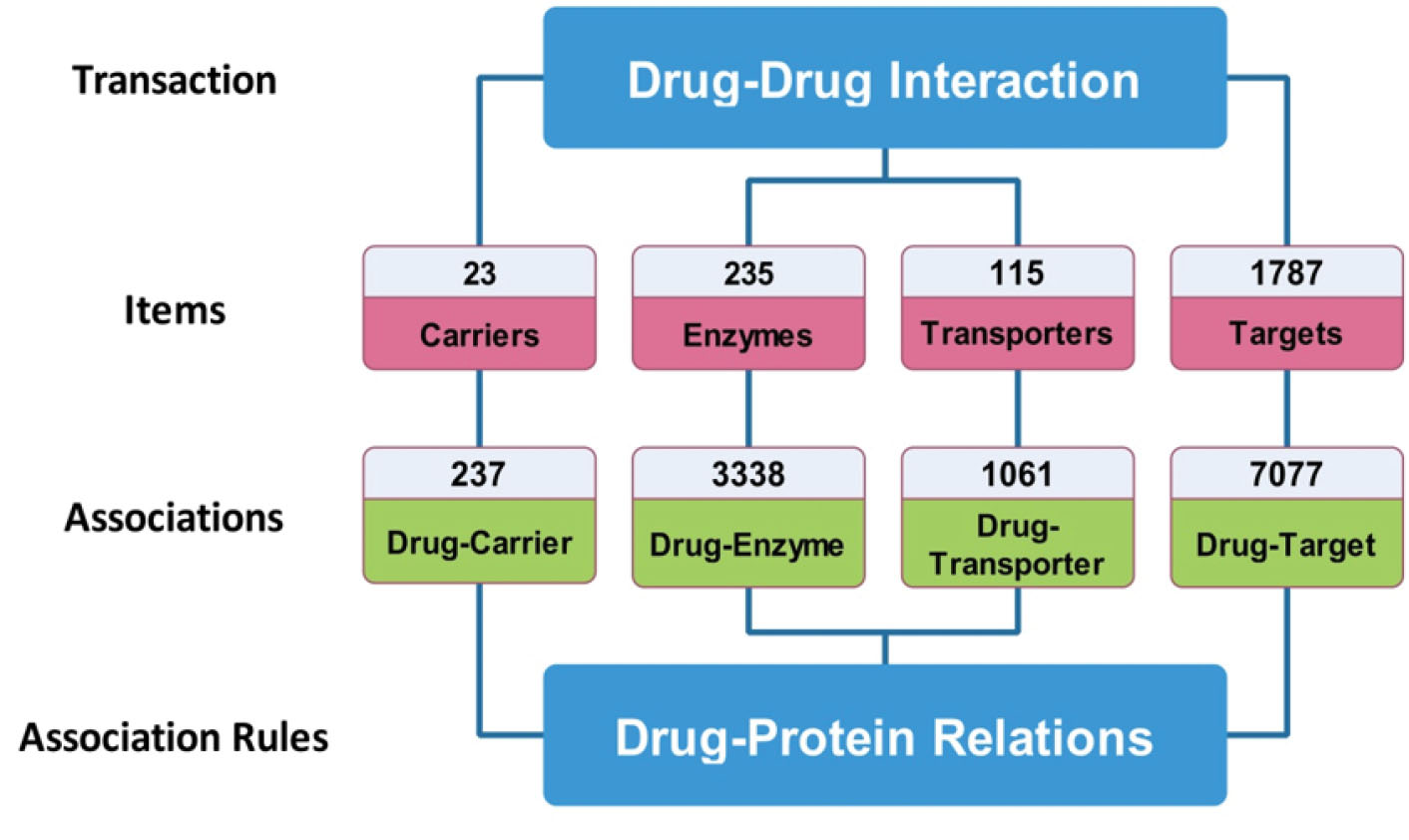
Fig. 2.
The integration of “Market Basket Analysis” with “Drug Interactions”. Reported DDIs were considered as transactions. All the carriers, enzymes, transporters, and targets were considered as items and their interactions with drugs were considered as associations and finally the rules were extracted.
.
The integration of “Market Basket Analysis” with “Drug Interactions”. Reported DDIs were considered as transactions. All the carriers, enzymes, transporters, and targets were considered as items and their interactions with drugs were considered as associations and finally the rules were extracted.
I= {i1, i2, … , im} – items are considered as binary factors
-
Different subsets of X ⊂I are called itemset, then individual subset with k elements: k-itemset.
-
D = {(id1,T1), (id2,T2),...,(idn,Tn)} transaction database,
where, for any j idj ⊂TID is a specific transaction identifier, and for any j Tj ⊂I is a set of occurred interaction.
-
s(X) is the number of interactions which contain s set of X.
-
Association rule is called: if X then Y: X àY
Where, X ⊂I, Y ⊂I, and X ∩ Y = Ø.
Assessment of mathematical model
In all computational procedures, it is undeniably crucial to evaluate the performance and accuracy of the proposed model to decide whether it is reliable or not.
31
When the performance is confirmed, the results can be used in the experimental tasks with a minimum doubt and uncertainty. There are different parameters to evaluate and judge how MBA method works properly and how the results are trustworthy.
If n stands for the number of interactions, X ⊂I, Y ⊂I, and X ∩ Y = Ø, then parameters accompanying the rules, as follows
32
:
Support: Number of interactions containing X & Y/Number of interactions
Confidence: Number of interactions containing X & Y/Number of interactions containing X
Expected Confidence: Number of interactions containing Y/Number of interactions
Lift: Confidence/Expected confidence
In addition, two factors should be considered, including min_conf, and min_sup. They refer to the minimum support and the minimum confidence, describing searched association rules. Their proper distribution can help to find only the appropriate implications.
There are two steps to construct association rules, as follows:
First, it is necessary to find all frequent sets X ⊂I it means
> min_sup
Second, establishing association rules is required according to the frequent sets X ⊂I.
Computational approaches are not able to evaluate all rules quickly. Application of the priori algorithm could improve the search process and let the detection of the most reliable and significant associations. By application of MBA, one can choose thresholds min_conf and min_sup. At this step, it is possible to decide whether the developed model can be applied to typical data to predict potential DDIs.
Prediction of potential DDIs
Since there are 2189 FDA approved drugs, considering equation (Eq.) 1, it can mathematically be assumed to have 2 394 766 possible interactions of pair-drug.
P ( n,r ) = P
r
n
= (n)
r Eq. 1
All the 2189 drugs were used as the input for our predictor to reveal whether a pair of drugs can interact with one another.
Results
Item classification
Considering the methodology of MBA, the classification on the items and common components are essential. It is clear that dominance and range of the items alter the performance of the prediction machine. Therefore, it is crucial to determine which items are effective in the interaction of two drugs. As discussed in section 2.1, we classified the interactions according to the occurrence of items. Fig. 3A and Table 1 show the distribution of these interactions considering their mutual items. This suggests that the high number of shared items might guarantee the occurrence of the interaction between pairs of drugs.
Table 1.
Top 20 Known DDIs with a number of the shared biological itemsa
|
Drug Pair ( DrugBank ID)
|
No. of shared items (fired rules)
|
| Amoxapine, Olanzapine (DB00543,DB00334) |
42 |
| Olanzapine, Ergoloid mesylate (DB00334,DB01049) |
34 |
| Amitriptyline, Olanzapine (DB00321,DB00334) |
29 |
| Aripiprazole, Olanzapine (DB01238,DB00334) |
27 |
| Olanzapine, Trimipramine (DB00334,DB00726) |
26 |
| Nortriptyline, Olanzapine (DB00540,DB00334) |
26 |
| Doxepin, Olanzapine (DB01142,DB00334) |
26 |
| Aripiprazole, Clozapine (DB01238,DB00363) |
26 |
| Amoxapine, Loxapine (DB00543,DB00408) |
26 |
| Amitriptyline, Clozapine (DB00321,DB00363) |
25 |
| Amoxapine, Quetiapine (DB00543,DB01224) |
24 |
| Amoxapine, Clozapine (DB00543,DB00363) |
24 |
| Primidone, Ethanol (DB00794,DB00898) |
23 |
| Desipramine, Olanzapine (DB01151,DB00334) |
23 |
| Clozapine, Doxepin (DB00363,DB01142) |
23 |
| Amoxapine, Ziprasidone (DB00543,DB00246) |
23 |
| Amoxapine, Aripiprazole (DB00543,DB01238) |
23 |
| Pentobarbital, Primidone (DB00312,DB00794) |
22 |
| Trimipramine, Loxapine (DB00726,DB00408) |
22 |
| Quetiapine, Trimipramine (DB01224,DB00726) |
22 |
ae.g. there is 42 common items between amoxapine and olanzapine.
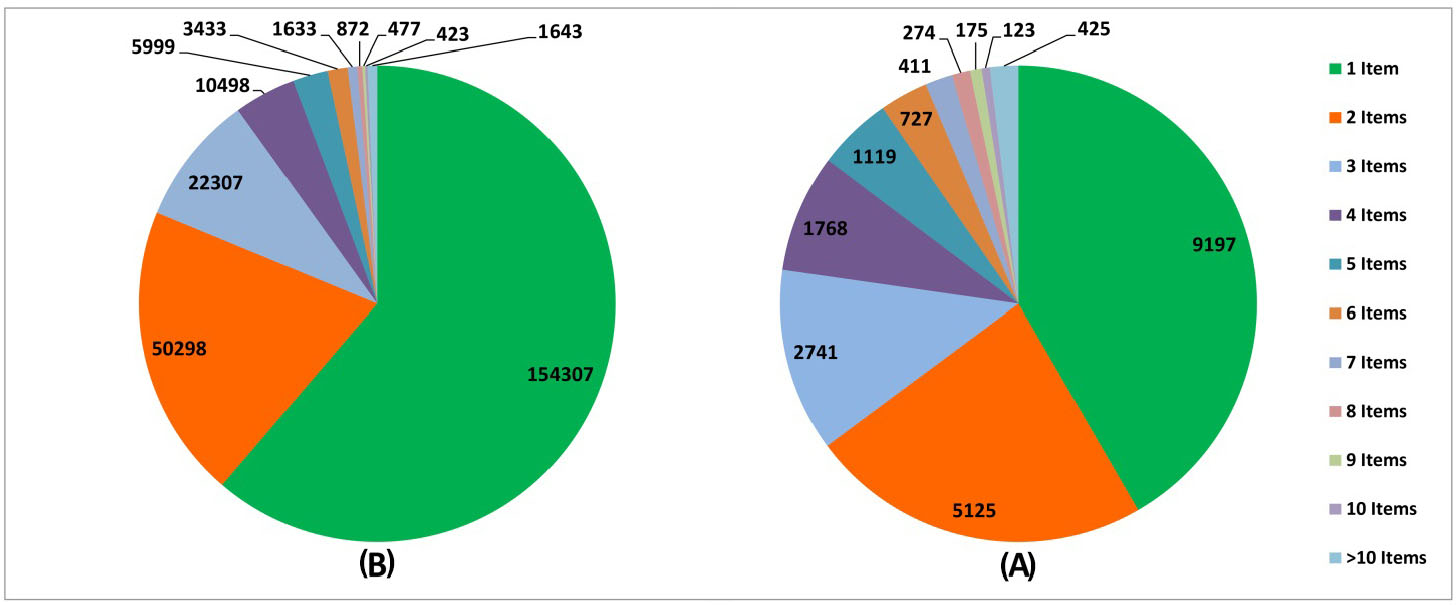
Fig. 3.
The distribution of interactions based on the number of shared items. (A) Experimentally validated interactions, (B) Predicted interactions.
.
The distribution of interactions based on the number of shared items. (A) Experimentally validated interactions, (B) Predicted interactions.
Model and rules assessment
The output of MBA reveals how regularly items co-occur in any transaction. On the other hand, items might synchronize due to their "nature". Our input data presented the way in which interactions occurred. The mathematical model of these interactions should have been validated with the biological results. Therefore, the accuracy of the proposed method seemed to be crucial. As noted earlier, there are some measures to evaluate the association rules. Considering our rules, these measures assigned a real value to association rules. These parameters are considered as gold standards to evaluate and judge the performance of the machine. Our proposed method possessed a reliable performance in term of support. Based on our results, about 32% standed for this parameter. These results seem to be satisfactory enough to be validated via some relevant experimental approaches. Table 2 summarizes values for these measures.
Table 2.
Distribution of DDIs according to the number of items
|
Number of Items
|
1
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
9
|
10
|
>10
|
| Experimental Interactions |
9197 |
5125 |
2741 |
1768 |
1119 |
727 |
411 |
274 |
175 |
123 |
425 |
| Predicted Interactions |
154307 |
50298 |
22307 |
10498 |
5999 |
3433 |
1633 |
872 |
477 |
423 |
1643 |
DDIs classes
Considering the associations between items and transactions, we classified predicted DDIs based on the items. These interactions are governed by the number of items varying from 1 to 32. Fig. 3B and Table 1 summarize the distribution of interactions according to the items. It is well-established fact that when the numbers of items increase, the number of interactions decrease. Further, not only the number of interactions is dropped, but also the feasibility of interactions is strengthened. The greater the number of supporting items are, the stronger the interactions will be. In such condition, the predictive power of the machine can be strengthened.
Rule discovery
The identification of governing rules in this method can be considered as a vital step since they might affect the occurrence of DDIs. In this study, according to the experimentally validated DDIs, we extracted about 341 rules (see Supplementary file 1, Table S1). These rules, considering their biological importance, prove and confirm the availability of DDIs. All these rules belong to the protein families with an effective role in the pathways of drug metabolism, drug signaling, and drug delivery.
DDI identification
Extracted rules from the previous section were applied over 2 000 000 drug pairs that resulted in the identification of over 200 000 potential DDI (see Supplementary file 1, Table S2).
Discussion
The computational and mathematical approaches provide a great opportunity for the identification of new drug interactions. Considering the high-throughput results obtained by these methods, they have widely been used in the field of pharmacology.
11,33
In this work, we capitalized on the association rule discovery analysis to be applied for the prediction of possible DDIs. This proposed methodology emphasizes different and effective sets of items and their associations in the occurrence of interactions. Utilizing MBA to identify important bio element in DDI occurrence is the key point of this study and the co-occurrence of the events is the main concept of the MBA approaches. In DDI a shared bio element could be considered as a common event. Therefore, MBA could identify shared bio element as DDI triggers. It should be noted that the sets of items affect the interactions and also guarantee the potency of interactions.
34
On the other hand, interactions with more items and involved components seem to be probable and feasible. From the rules viewpoint, our introduced rules can strongly be approved in terms of biology.
26
The strongest rules with the frequency of 14231 and 5443 belong to two isoforms of the CYP family, which is one of the most important enzymes in drug metabolism. The CYP family encompasses several well-known enzymes involved in the metabolism of nutrients, drugs, and toxicants. In addition, CYP plays significant roles in drug metabolism and the incidence of numerous DDIs.
35-37
Considering the variety of drugs interacting with CYP, it has been suggested that these drugs have some issues in common which potentially can result in DDI.
7,38
Therefore, the drugs interacting with CYP, are considered as potential candidates for DDIs and it is necessary to find out the capability and probability of these interactions. Therefore, biologically it is important to design a drug with high specificity to avoid unwanted interactions.
39,40
Moreover, the third rule with the frequency of 5164 has been ascribed to multidrug resistance protein (P-GP), which plays a crucial role in the outward transportation of various drugs.
41,42
Given the undeniable importance of CYP in the biological systems, the following 10 rules belong to this protein family (Fig. 4). The analyses of the rules, considering their biological functions, prove that these rules are meaningful and can be applied in the computational methods. Therefore, discovered association rules in this study seem to be rational enough to be implemented in the prediction machine. By taking all above-mentioned argument into consideration, the predicted potential DDIs based on these rules can reliably be applied. Although this algorithm tries to find potential interactions among drugs with high accuracy, there are some limitations which should be noted. The core assumption of DDIs in this algorithm is established based on mutual interacting compounds as rules including enzymes, carriers, targets, and transporters. It has been suggested that there is a linear association between the number and variety of rules and performance and accuracy of DDIs prediction. There is no doubt that the high number of rules can affect and guarantee the results of prediction. Therefore, if we can provide more training data including more rules, we can strongly confirm the results. This can be performed with a deep analysis of drugs and their mutual interacting elements to empower input data to obtain precise DDI interactions.
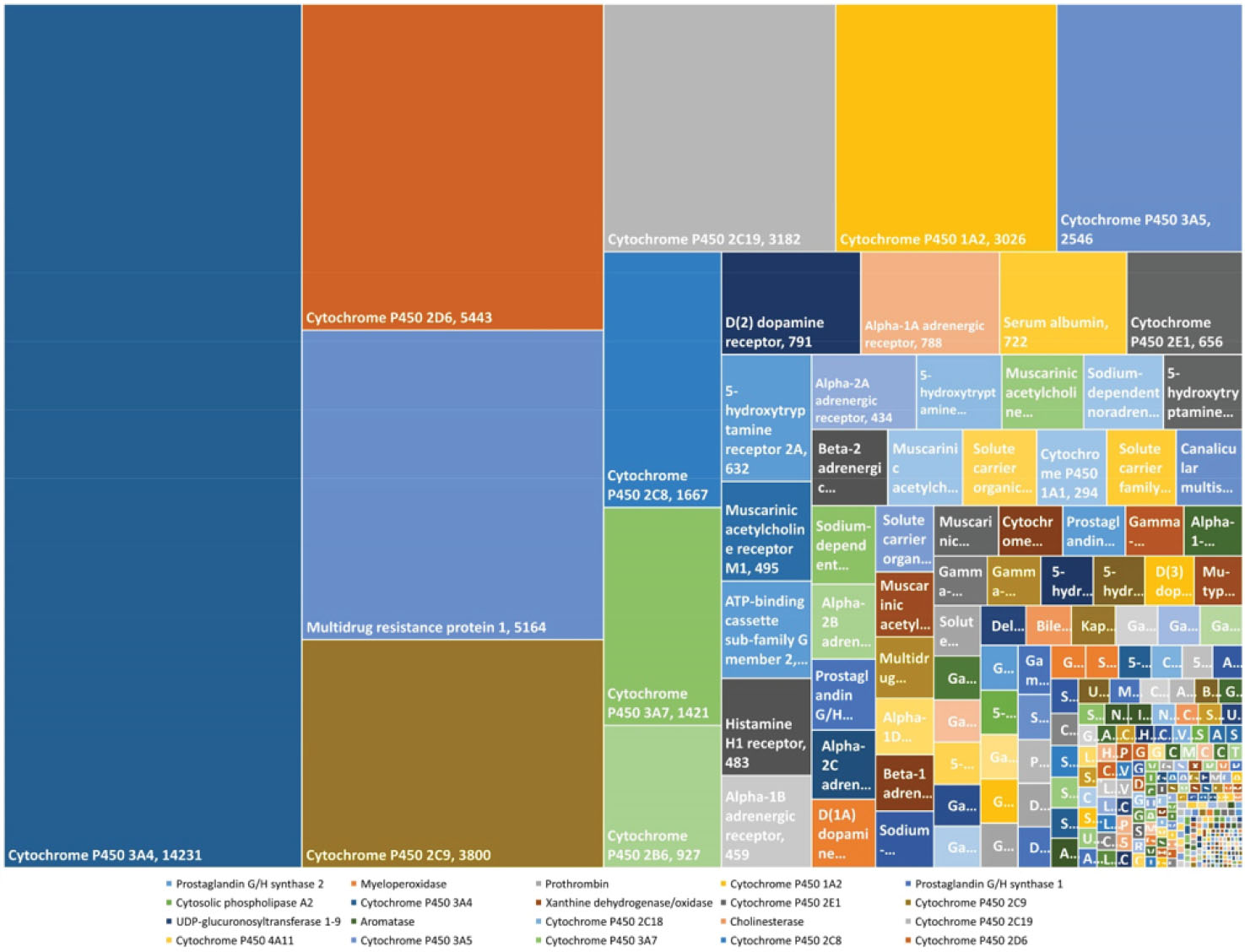
Fig. 4.
Frequency of important biological elements in DDIs. The size of the rectangle represents the role of corresponding bio-element in DDI occurrence.
.
Frequency of important biological elements in DDIs. The size of the rectangle represents the role of corresponding bio-element in DDI occurrence.
Conclusion
The identification of DDIs is considered as the vitally key information, in large part because of their importance in multi-drug therapy. It can be performed via both experimentally and computational approaches.
43,44
Of these, the laboratory-based approaches are costly and time-demanding, which also need the implementation of a variety of equipment. On the contrary, the computational techniques have been considered as a method of choice by many researchers, in large part due to their efficiency and time- and cost-effectiveness. These techniques can simplify the difficulty of the DDIs prediction. In fact, the results of in silico DDIs identification seem to be promising for the clinical practice.
45
That being noted, it is necessary to highlight that these methodologies are not the alternatives but complementary methodologies to fulfill the prediction of DDIs.
11
Acknowledgments
Authors like to acknowledge the Research Center for Pharmaceutical Nanotechnology at Tabriz University of Medical Sciences for the financial support (grant No. RCPN-97006).
Funding sources
This work funded by research center for pharmaceutical nanotechnology, biomedicine Institute, Tabriz University of Medical Sciences.
Ethical statement
The present study was approved by the Ethics Committee of Tabriz University of Medical Sciences (ethical code No. IR.TBZMED.REC.1398.1085).
Competing interests
There are no competing interests to declare.
Authors’ contribution
The conception and the design of the study were performed by RF. The data collection and data analysis carried out by RF and AAJ and RS. The project supervised by RS. The manuscript was drafted by AAJ and RF and revised by RF and RS. All authors participated in manuscript in the critical review process of the manuscript and approval of the final version.
Supplementary Materials
Supplementary file 1 contains Table S1 and Table S2.
(xlsx)
Research Highlights
What is the current knowledge?
simple
-
√ Case reports and research papers are the main source of
DDIs.
-
√ There are some databases for DDIs.
-
√ Computational techniques used to predict possible DDIs.
-
√ Costly Wet lab studies can reveal the interaction.
What is new here?
simple
-
√ A Market Basket Analysis-based method is proposed for
identification of the drug interaction rules.
-
√ DDIs may occur based on common biological elements.
Shared biological elements were employed to identify unseen
DDIs.
-
√ Over 45 000 known DDIs investigated and over 300
important rules, that can be used in the identification of
DDIs, were extracted.
References
- Berg EL. Systems biology in drug discovery and development. Drug Discov Today 2014; 19:113-25. doi: 10.1016/j.drudis.2013.10.003 [Crossref] [ Google Scholar]
- Sun PG. The human Drug–Disease–Gene Network. Information Sciences 2015; 306:70-80. doi: 10.1016/j.ins.2015.01.036 [Crossref] [ Google Scholar]
- Leung CON, Tong M, Chung KPS, Zhou L, Che N, Tang KH. Overriding adaptive resistance to sorafenib via combination therapy with SHP2 blockade in hepatocellular carcinoma. Hepatology 2019. doi: 10.1002/hep.30989 [Crossref]
- Zheng W, Lin H, Zhao Z, Xu B, Zhang Y, Yang Z. A graph kernel based on context vectors for extracting drug–drug interactions. J Biomed Inform 2016; 61:34-43. doi: 10.1016/j.jbi.2016.03.014 [Crossref] [ Google Scholar]
- Shaked I, Oberhardt Matthew A, Atias N, Sharan R, Ruppin E. Metabolic network prediction of drug side effects. Cell Syst 2016; 2:209-13. doi: 10.1016/j.cels.2016.03.001 [Crossref] [ Google Scholar]
- Safdari R, Ferdousi R, Aziziheris K, Niakan-Kalhori SR, Omidi Y. Computerized techniques pave the way for drug-drug interaction prediction and interpretation. Bioimpacts 2016; 6:71-8. doi: 10.15171/bi.2016.10 [Crossref] [ Google Scholar]
- Ferdousi R, Safdari R, Omidi Y. Computational prediction of drug-drug interactions based on drugs functional similarities. J Biomed Inform 2017; 70:54-64. doi: 10.1016/j.jbi.2017.04.021 [Crossref] [ Google Scholar]
- Andersson ML, Böttiger Y, Lindh JD, Wettermark B, Eiermann B. Impact of the drug-drug interaction database SFINX on prevalence of potentially serious drug-drug interactions in primary health care. Eur J Clin Pharmacol 2012; 69:565-71. doi: 10.1007/s00228-012-1338-y [Crossref] [ Google Scholar]
- Murtaza G, Khan MYG, Azhar S, Khan SA, Khan TM. Assessment of potential drug–drug interactions and its associated factors in the hospitalized cardiac patients. Saudi Pharm J 2016; 24:220-5. doi: 10.1016/j.jsps.2015.03.009 [Crossref] [ Google Scholar]
- Witt J-A, Elger CE, Helmstaedter C. Which drug-induced side effects would be tolerated in the prospect of seizure control?. Epilepsy Behav 2013; 29:141-3. doi: 10.1016/j.yebeh.2013.07.013 [Crossref] [ Google Scholar]
- Jamali AA, Ferdousi R, Razzaghi S, Li J, Safdari R, Ebrahimie E. DrugMiner: comparative analysis of machine learning algorithms for prediction of potential druggable proteins. Drug Discov Today 2016. doi: 10.1016/j.drudis.2016.01.007 [Crossref]
- Chen L, Li B-Q, Zheng M-Y, Zhang J, Feng K-Y, Cai Y-D. Prediction of Effective Drug Combinations by Chemical Interaction, Protein Interaction and Target Enrichment of KEGG Pathways. Biomed Res Int 2013; 2013:10. doi: 10.1155/2013/723780 [Crossref] [ Google Scholar]
- Kovacevic M, Vezmar Kovacevic S, Radovanovic S, Stevanovic P, Miljkovic B. Adverse drug reactions caused by drug-drug interactions in cardiovascular disease patients: introduction of a simple prediction tool using electronic screening database items. Curr Med Res Opin 2019; 1. doi: 10.1080/03007995.2019.1647021 [Crossref]
- Min JS, Bae SK. Prediction of drug-drug interaction potential using physiologically based pharmacokinetic modeling. Arch Pharm Res 2017; 40:1356-79. doi: 10.1007/s12272-017-0976-0 [Crossref] [ Google Scholar]
- Rohani N, Eslahchi C. Drug-Drug Interaction Predicting by Neural Network Using Integrated Similarity. Sci Rep 2019; 9:13645. doi: 10.1038/s41598-019-50121-3 [Crossref] [ Google Scholar]
- Katsila T, Spyroulias GA, Patrinos GP, Matsoukas M-T. Computational approaches in target identification and drug discovery. Comput Struct Biotechnol J 2016; 14:177-84. doi: 10.1016/j.csbj.2016.04.004 [Crossref] [ Google Scholar]
- Sliwoski G, Kothiwale S, Meiler J, Lowe EW. Computational Methods in Drug Discovery. Pharmacol Rev 2014; 66:334-95. doi: 10.1124/pr.112.007336 [Crossref] [ Google Scholar]
- Raef B, Ferdousi R. A Review of Machine Learning Approaches in Assisted Reproductive Technologies. Acta Inform Med 2019; 27:205-11. doi: 10.5455/aim.2019.27.205-211 [Crossref] [ Google Scholar]
- Rezaei-Hachesu P, Oliyaee A, Safaie N, Ferdousi R. Comparison of coronary artery disease guidelines with extracted knowledge from data mining. J Cardiovasc Thorac Res 2017; 9:95. doi: 10.15171/jcvtr.2017.16 [Crossref] [ Google Scholar]
- Wiwanitkit S, Wiwanitkit V. Computational intelligence in tropical medicine. Asian Pac J Trop Biomed 2016; 6:350-2. doi: 10.1016/j.apjtb.2015.11.009 [Crossref] [ Google Scholar]
- Cha K, Kim M-S, Oh K, Shin H, Yi G-S. Drug Similarity Search Based on Combined Signatures in Gene Expression Profiles. Healthc Inform Res 2014; 20:52-60. doi: 10.4258/hir.2014.20.1.52 [Crossref] [ Google Scholar]
- Vyas VK, Ukawala RD, Ghate M, Chintha C. Homology Modeling a Fast Tool for Drug Discovery: Current Perspectives. Indian J Pharm Sci 2012; 74:1-17. doi: 10.4103/0250-474X.102537 [Crossref] [ Google Scholar]
-
Agrawal R, Imielinski T, Swami A, editors. Mining Association Rules between Sets of Items in Large Databases. Proceedings of the ACM SIGMOD International Conference on Management of Data; 1993; Washington, DC.
- Shiokawa Y, Misawa T, Date Y, Kikuchi J. Application of market basket analysis for the visualization of transaction data based on human lifestyle and spectroscopic measurements. Anal Chem 2016; 88:2714-9. doi: 10.1021/acs.analchem.5b04182 [Crossref] [ Google Scholar]
- Raeder T, Chawla NV. Market basket analysis with networks. Social Net Anal and Min 2010; 1:97-113. doi: 10.1007/s13278-010-0003-7 [Crossref] [ Google Scholar]
- Yusof I, Shah F, Hashimoto T, Segall MD, Greene N. Finding the rules for successful drug optimisation. Drug Discov Today 2014; 19:680-7. doi: 10.1016/j.drudis.2014.01.005 [Crossref] [ Google Scholar]
- Agapito G, Guzzi PH, Cannataro M. DMET-Miner: Efficient discovery of association rules from pharmacogenomic data. J Biomed Inform 2015; 56:273-83. doi: 10.1016/j.jbi.2015.06.005 [Crossref] [ Google Scholar]
- Erxin S, Liang Y, Xinsheng F, Yuping T, Jinao D. Discovery of association rules between TCM properties in drug pairs by association mining between datasets and probability tests. World Sci and Tech 2010; 12:377-82. doi: 10.1016/S1876-3553(11)60017-3 [Crossref] [ Google Scholar]
- Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A. DrugBank 30: a comprehensive resource for ‘Omics’ research on drugs. Nucleic Acids Res 2011; 39:D1035-D41. doi: 10.1093/nar/gkq1126 [Crossref] [ Google Scholar]
- Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y. DrugBank 40: shedding new light on drug metabolism. Nucleic Acids Res 2014; 42:D1091-D7. doi: 10.1093/nar/gkt1068 [Crossref] [ Google Scholar]
- Sawant Ms V, Shah K. Performance Evaluation of Distributed Association Rule Mining Algorithms. Procedia Comp Sci 2016; 79:127-34. doi: 10.1016/j.procs.2016.03.017 [Crossref] [ Google Scholar]
- Agrawal R, Mannila H, Srikant R, Toivonen H, Verkamo AI. Fast discovery of association rules. Adv Knowl Discov Data Min 1996; 12:307-28. [ Google Scholar]
- Ivanov AA. Explore Protein-Protein Interactions for Cancer Target Discovery Using the OncoPPi Portal. Methods Mol Biol 2020; 2074:145-64. doi: 10.1007/978-1-4939-9873-9_12 [Crossref] [ Google Scholar]
- Ibrahim H, Saad A, Abdo A, Sharaf Eldin A. Mining association patterns of drug-interactions using post marketing FDA’s spontaneous reporting data. J Biomed Inform 2016; 60:294-308. doi: 10.1016/j.jbi.2016.02.009 [Crossref] [ Google Scholar]
- Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 2013; 138:103-41. doi: 10.1016/j.pharmthera.2012.12.007 [Crossref] [ Google Scholar]
- Ogu CC, Maxa JL. Drug interactions due to cytochrome P450. Proc (Bayl Univ Med Cent) 2000; 13(4):421-3. doi: 10.1080/08998280.2000.11927719 [Crossref] [ Google Scholar]
- Huth F, Gardin A, Umehara K, He H. Prediction of the impact of cytochrome P450 2C9 genotypes on the drug-drug interaction potential of siponimod with physiologically-based pharmacokinetic modeling: a comprehensive approach for drug label recommendations. Clin Pharmacol Ther 2019; 106:1113-24. doi: 10.1002/cpt.1547 [Crossref] [ Google Scholar]
- Akamine Y, Yasui-Furukori N, Uno T. Drug-Drug Interactions of P-gp Substrates Unrelated to CYP Metabolism. Curr Drug Metab 2019; 20:124-9. doi: 10.2174/1389200219666181003142036 [Crossref] [ Google Scholar]
- Raschke RA, Gollihare B, Wunderlich TA. A computer alert system to prevent injury from adverse drug events: Development and evaluation in a community teaching hospital. JAMA 1998; 280:1317-20. doi: 10.1001/jama.280.15.1317 [Crossref] [ Google Scholar]
- Bennett CL, Nebeker JR, Lyons E. The research on adverse drug events and reports (radar) project. JAMA 2005; 293:2131-40. doi: 10.1001/jama.293.17.2131 [Crossref] [ Google Scholar]
-
Keppler D. Multidrug Resistance Proteins (MRPs, ABCCs): Importance for Pathophysiology and Drug Therapy. In: Fromm FM, BR Kim, eds. Drug Transporters. Berlin, Heidelberg: Springer g; 2011. p. 299-323.
- Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters: the multidrug resistance-associated proteins. J Natl Cancer Inst 2000; 92:1295-302. doi: 10.1093/jnci/92.16.1295 [Crossref] [ Google Scholar]
-
Anonymous. Drug-Drug Interactions. In: Stolerman IP, LH Price, ed. Encyclopedia of Psychopharmacology. Berlin, Heidelberg: Springer; 2015. p. 562.
-
Minzenberg M. Drug–Drug Interactions. In: Stolerman IP, editor. Encyclopedia of Psychopharmacology. Berlin, Heidelberg: Springer; 2010. p. 430-1.
- Ivanov SM, Lagunin AA, Poroikov VV. In silico assessment of adverse drug reactions and associated mechanisms. Drug Discov Today 2016; 21:58-71. doi: 10.1016/j.drudis.2015.07.018 [Crossref] [ Google Scholar]