Bioimpacts. 12(3):195-202.
doi: 10.34172/bi.2021.22119
Original Research
Chitosan nanoparticle-mediated effect of antimiRNA-324-5p on decreasing the ovarian cancer cell proliferation by regulation of GLI1 expression
Ysrafil Ysrafil 1, 2, 3, *  , Indwiani Astuti 2
, Indwiani Astuti 2 
Author information:
1Department of Pharmacy, Politeknik Kesehatan Kementerian Kesehatan Gorontalo, Gorontalo, Indonesia
2Department of Pharmacology and Therapy, Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada, Yogyakarta, Indonesia
3Faculty of Medicine, Universitas Palangka Raya, Palangka Raya 73111, Indonesia
Abstract
Introduction:
MicroRNAs (miRNAs) are short-sequence RNAs that regulate gene expression by targeting messenger RNAs (mRNAs). Recent studies reveal that miRNA-324-5p plays an important role in worsening the ovarian cancer prognosis when the expression is very high. This study aimed to develop a miRNA targeted therapy by targeting the miRNA-324-5p function as a miRNA-324-5p inhibitor.
Methods:
Chitosan nanoparticles were used for antimiRNA-324-5p delivery into SKOV3 cell lines formulated by ionic gelation method. Antiproliferative effect of CS-NPs-antimiRNA was assessed by the MTT Assay. A mechanism study assessed the anticancer effect of the formula. In silico analysis used miRTar.Human and StarmiRDB combined with Genecard to predict the target genes of antimiR. Hawkdock web server was used to analyze protein-protein interactions that were further validated by quantitative polymerase chain reaction (qPCR).
Results:
The results of qPCR analysis showed endogenous miRNA-324-5p decreased after 24-hour transfection of antagonist miRNA. Furthermore, the MTT assay results showed that antimiRNA was able to inhibit SKOV3 cell proliferation (80 nM 68.13%, P < 0.05). In silico analysis found miRNA-324-5p can regulate MEN1 and indirectly repress Gli1 mRNA. Validation results confirmed antimiR can decrease GLI1 mRNA expression.
Conclusion:
Our results showed antimiRNA-324-5p can act as a microRNA-based therapy to inhibit ovarian cancer proliferation by the reduction of GLI1 expression.
Keywords: AntimiRNA-324-5p, SKOV3, Ovarian cancer, GLI1, Chitosan nanoparticle
Introduction
Ovarian cancer is the one type of gynecological cancer that causes the most deaths of women worldwide.
1
The high mortality rate of patients with ovarian cancer is commonly caused by aggressive metastasis, recurrence, chemotherapy drug resistance, and late diagnosis. There are no specific manifestations of the early stages in patients, generally causing them to be diagnosed in an advanced stage with only an estimated 5-year survival rate of around 30%.
2-4
Recently, research has demonstrated that cancer progression is related to changes at the molecular level. Moreover, development of more effective diagnostic and therapeutic methods to overcome the treatment challenges are currently receiving attention by cancer researchers.
This newest development refers to small noncoding RNAs (microRNAs/miRNAs) that can regulate the expression of other proteins at the post-transcriptional level.
5,6
MiRNA-mRNA binding can lead to the inhibition of expression or even degradation of the messenger RNA (mRNA) before becoming a protein.
7,8
MiRNA is commonly found dysregulated in various types of cancers including ovarian cancer. The aberrant miRNA expression correlates with the worsening prognosis of patients, so these RNA molecules are often used as biomarkers of certain cancers.
5,9,10
MiRNA has a function as a tumor suppressor (tsmiR) or oncogene (oncomiR).
11
OncomiR is miRNA that suppresses the expression of tsmiR proteins which leads to the dysregulation of signaling in the mechanism of eradicating cancer cells in the body and exacerbating cancer. This process of regulating and/or suppressing cancer cells has attracted significant attention in the development of cancer therapy based on miRNA.
12,13
Previous studies have demonstrated that suppressing the function of oncogenic miRNA that is upregulated in certain cancer cells can suppress the induction of apoptosis, inhibit migration, invasion, proliferation, and metastasis of cancer cells. This strategy can be done by inserting complementary antisense nucleotides of oncomir genes that will inhibit its function.
12,14,15
One characteristic of miRNA is that it is easily degraded in the blood circulation. This makes it difficult to achieve the target action when given systemically in the form of naked miRNA so that it needs an appropriate vector to be able to send microRNA into cells.
7
Chitosan nanoparticles (CS-NPs) are non-viral vectors that are often used in the delivery of miRNA because they are relatively safe, easy to use, and able to protect and deliver into the intracellular compartment of target cells.
16,17
CS-NPs delivery system is proven both in vitro and in vivo in its application for sending miRNA into the cells and mediating the expression of target genes especially in cancer.
18
In this study, we aimed to explore the effects of antimiR-324-5p assisted by CS-NPs in inhibiting the growth of ovarian cancer cells.
Materials and Methods
Materials
AntimiRNA-324-5p were purchased from Integrated DNA Technologies (Danaher, Coralville, USA), while Chitosan medium molecular weight were purchased from Sigma Aldrich (Merck, St. Louis, USA). In this study, we used human epithelial ovarian cancer cell lines SKOV3, obtained from Stem Cell and Cancer Institute Kalbe (Kalbe, Jakarta, Indonesia) that were grown in DMEM High Glucose (Thermo Fisher Scientific, Massachusetts, USA) and supplemented with 10% fetal bovine qualified serum (Thermo Fisher Scientific, Massachusetts, USA), 1% Penicillin-Streptomycin serum (Thermo Fisher Scientific, Massachusetts, USA), and 0.5% amphotericin serum (Thermo Fisher Scientific, Massachusetts, USA). Exiqon miRCURY LNATMUniversal RT microRNA PCR kit, miRCURY RNA Cell and Plant Kit, Universal cDNA synthesis kit II 8-64 rxns were purchased from Exiqon (Qiagen, Hilden, Germany).
Preparation of chitosan nanoparticle-antimiRNA-324-5p
The conjugation of CS-NPs-miRNA was done by an ionic gelation method employing sodium tripolyphosphate (Na TPP) as the crosslinker as described in our previous study.
19
In the first step, chitosan powder was dissolved in 1% acetic acid and a pH of 5.5 was sufficient using NaOH. Chitosan solution was first diluted using acetate buffer to obtain 0.02% chitosan solution. Before it was conjugated with miRNA, the solution was mixed with Na.TPP at a ratio of 5:1 (Fig. 1).
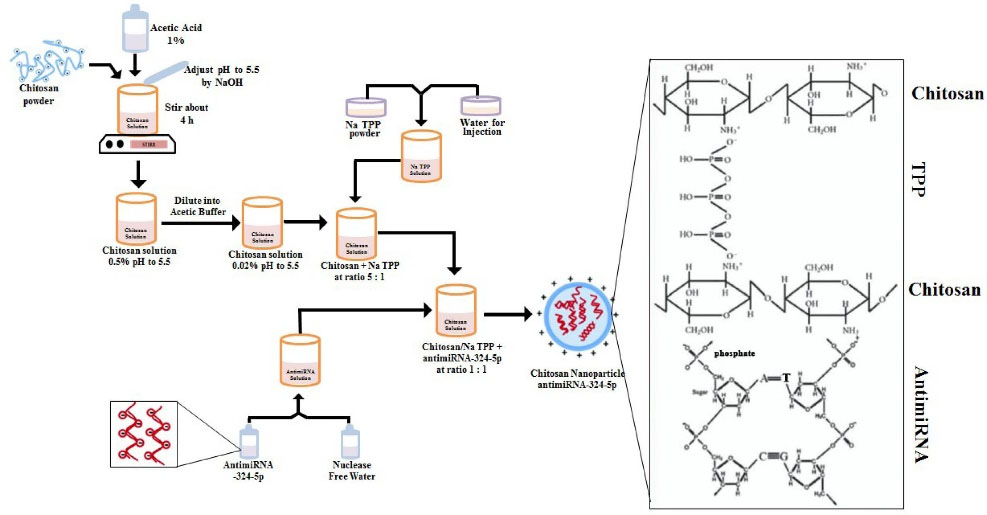
Fig. 1.
Schematic preparation and chitosan nanoparticle antimiRNA formation by crosslinking.
.
Schematic preparation and chitosan nanoparticle antimiRNA formation by crosslinking.
Gel electrophoresis assay
The formation of nanocomplex (CS-NPs-miRNA) in the preparation was assessed by electrophoresis test. The CS-NPs-miRNA were run on 2% electrophoresis gel with 50 V of voltage, 400 mA for 20 minutes (Kaban et al
20
modification).
Loading efficiency
Efficiency of antimiRNA-324-5p encapsulation by CS-NPs was assessed by measuring the free concentration antimiR-324-5p of total antimiRNA-324-5p in the preparations. Prepared CS-NPs-miRNA were centrifuged for 15 minutes at 13,000 g. Then, supernatant was measured by 260/280 nm of wavelength using NANO-Quant (SPARK TECAN). Its absorption efficiency (%) was obtained from the percentage of encapsulated miRNAs (total miRNA-free miRNA) compared to the total RNA.
Particle size and zeta potential of CS-NPs-miRNA
Particle size and zeta potential of CS-NPs-miRNA formed were measured using Nanoparticles Size Analyzer, Horiba Sz-100 with 24.9°C. Previously, samples were prepared with nuclease free water.
Cell viability assay
MTT assay method was used to assess the ability of CS-NPs of antimiRNA to inhibit ovarian cancer cell proliferation that was further compared to naked antimiRNA. A total of 6 × 104 cells/100µL SKOV3 in DMEM HG medium were grown on 96-well plate medium for 24 hours at 37°C in a 5% CO2 incubator. Cells that had been grown were treated with several series of concentrations of CS-NPs-antimiRNA-324-5p or naked antimiRNA-324-5p (30,40, 50, 60, 70, and 80 nM) and were diluted in DMEM without serum. After 24 hours of incubation, the test solution was removed and replaced with 100 µL of MTT reagent in each well. Dehydrogenase enzyme activity was seen from the formazan crystals formed after the addition of stop solution (SDS 10%, and 0.01 mol/L HCl) after 4 hours incubation and the plates were read immediately in a microplate reader at 595 nm wavelength using a Micro Plate Reader (Bio-Rad Model 680 XR).
Bioinformatic analysis to predict target gene of miRNA-324-5p
Bioinformatic analysis were done to predict target genes of miRNA-324-5p using miRTar.Human (http://mirtar.mbc.nctu.edu.tw/human/) and StarMirDB (http://sfold.wadsworth.org/starmirDB.php) to determine the most likely complementary bindings of miRNA-mRNA that lead to the repression of gene expression.
Molecular docking of MEN1-GLI1 Interaction
Data of MEN1 and GLI1 proteins for molecular docking were obtained from protein data bank RCSB PDB of 3U84 and 2GLI, respectively. The protein-protein complex was docked using the Hawkdock web server (http://cadd.zju.edu.cn/hawkdock/) and visualized by UCSF Chimera and Ez Mol 2.1. Hawkdock web server was also used for Molecular Mechanics/Generalized Born Surface Area (MM-GBSA) study.
RNA extraction, cDNA synthesis, and quantitative real time-polymerase chain reaction ( qRT -PCR)
Total RNA of cell treatment was extracted using miRCURY
TM
RNA Isolation Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. To confirm the purified and quantified isolation of RNA, we measured the absorbance at 260 nm and 280 nm wavelengths. Synthesis of cDNA was done by Universal cDNA synthesis kit II 8-64 rxns and conducted according to the kit’s guide with 10 ng RNA. qRT-PCR was done using two different kits.
Quantification of miRNA-324-5p was done using the ExiLent SYBR Green master mix kit, 2.5 mL (Qiagen, Hilden, Germany) with running conditions as follows: 95°C for 10 minutes; 40 cycles of 95°C for 10 seconds, 60°C for 10 seconds, and 60°C for 5 seconds; followed by 95°C for 50 seconds. We used miRNA-16-5p (5'-UAGCAGCACGU AAAUAUUGGCG-3 ') as an endogenous control.
Furthermore, quantification of mRNA GLI1 expression was done with the SensiFASTTM SYBR® kit (Bioline, London, UK). We used GLI1 primers purchased from IDT: (GLI1/forward; 3’-TCTGCCCCCATT GCCCACTTG-5’ GLI1/reverse; 3’ TACATAGCCCC CAGCCCATACCTC-5’) and β-actin (forward; 5'-GGGAATTCAAAACTGGA ACGGTGA AGG-3'; and reverse; 5'-GGAAGCTTATCA AAGTCCTCGGCCACA-3') was employed as the reference gene. qRT-PCR running conditions were set as follows: 95°C for 2 minutes; 40 cycles of 95°C for 5 seconds, 58°C for 10 seconds, and 60°C for 5 seconds; followed by 95°C for 50 seconds. All measurements were done by qRT-PCR Bio-Rad CFX 96 C.1000 quantitative PCR machine. The relative gene expressions of miRNA and mRNA were analyzed by 2 -ΔΔCT method that was included on the GenEx version 6.1 software.
Statistical analysis
The data were measured as the mean ± standard deviation and the statistical analysis was performed using SPSS 16 software (IBM Corp., Chicago). The independent samples t test was used to compare mean values of the independent groups.Additionally, the correlations of the reduction of GLI1 and miRNA-324-5p expressions were analyzed by Spearman correlation tests. All the data results were considered statistically significant if P <0.05.
Results
Characterization of CS-NPs-miRNA
CS-NPs were prepared by ionic gelation method and sodium tripolyphosphate (Na TPP) was employed as the cross linker. As seen in Fig. 2a in well B, the closely complete nanocomplex formation with antimiRNA were formed. The nanochitosan-antimiRNA complex was retained within the well while the naked miRNA (Fig. 2a in well A) was able to migrate to the agarose gel. Furthermore, loading efficiency was about 65.2% that suggested antimiRNA were entrapped in matrix chitosan-TPP nanoparticles (Fig. 2b). The particle size and zeta potential of CS-NPs-antimiRNA complexes were evaluated by dynamic light scattering (Table 1) including 326.4 ± 3.041 nm with polydispersity index (PDI) lower than 0.7 that revealed the normality of particle size.
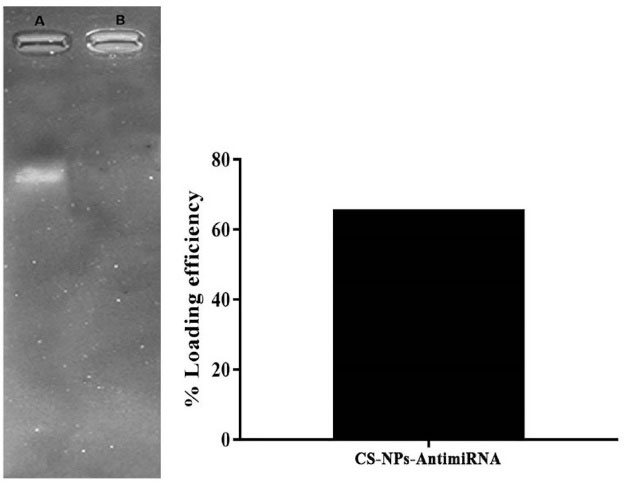
Fig. 2.
Characterization of CS-NPs/antimiRNA-324-5p (a) Agarose gel electrophoresis of nanoplexes [well (A) naked miRNA, well (B) CS-NPs miRNA complex] (b) loading efficiency of NPs-CS-antimiRNA-324-5p.
.
Characterization of CS-NPs/antimiRNA-324-5p (a) Agarose gel electrophoresis of nanoplexes [well (A) naked miRNA, well (B) CS-NPs miRNA complex] (b) loading efficiency of NPs-CS-antimiRNA-324-5p.
Table 1.
Particle size, polydisperse index, and zeta potential of CS-NPs-antimiRNA-324-5p
|
|
Particle Size (nm)
|
Polydisperse Index
b
|
Zeta Potential
b
(mV)
|
| CS-NPs/antimiR-324-5p |
326.4 ± 3.041 |
0.455 ± 0.015 |
47.267 ± 0.961 |
The average particle charge was positively consistent above 30 mV (+47.267 ± 0.961 and +47.2 ± 0.529) indicating that the nanoparticle was stable and would not lead to aggregation.
Differential expression of miRNA-324-5p in treated and untreated SKOV3 cell lines by antisense miRNA-324-5p
In this study, we evaluated the effects of antimiRNA-324-5p transfection to suppress the overexpression of miRNA-324-5p in ovarian cancer cells. As a vector, CS-NPs assisted inhibition of the function of miRNA-324-5p. The results showed significant downregulation of endogenous miRNA-324-5p when SKOV3 cell were treated with antimiR that was loaded by CS-NPs (Fig. 3). This finding supports the premise that through targeting the miRNA, blocking the function of the miRNA can lead to the reduction of endogenous miRNA-324-5p.
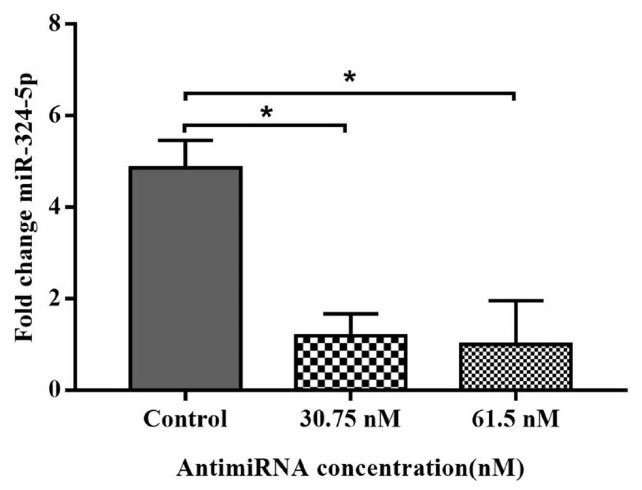
Fig. 3.
Downregulation of endogenous miRNA-324-5p expression in SKOV3 cell treated with antimiRNA-324-5p encapsulated in CS-NPs (Data are mean ± SD. *P < 0.05).
.
Downregulation of endogenous miRNA-324-5p expression in SKOV3 cell treated with antimiRNA-324-5p encapsulated in CS-NPs (Data are mean ± SD. *P < 0.05).
AntimiRNA-324-5p blocks miRNA-324-5p function and leads to inhibition of proliferation of ovarian cancer cell line SKOV3
Antiproliferation activity tests were done to ensure the inhibition of ovarian cancer cell proliferation after that downregulation of oncomiRNA-324-5p (Fig. 4) was caused by the blocking function of antisense miRNA. Assessment of antiproliferative activity was done by confirming the mitochondrial dehydrogenase performance in reducing the yellow tetrazole to purple formazan or commonly known as the MTT assay.
This test involved several variations of antimiRNA concentrations (30 to 80 nM) to see the effect of antisense concentrations in initiating SKOV3 ovarian cancer cell death. The results showed that the increase in the concentration of antisense was proportional to the antiproliferative effect (Fig. 4).
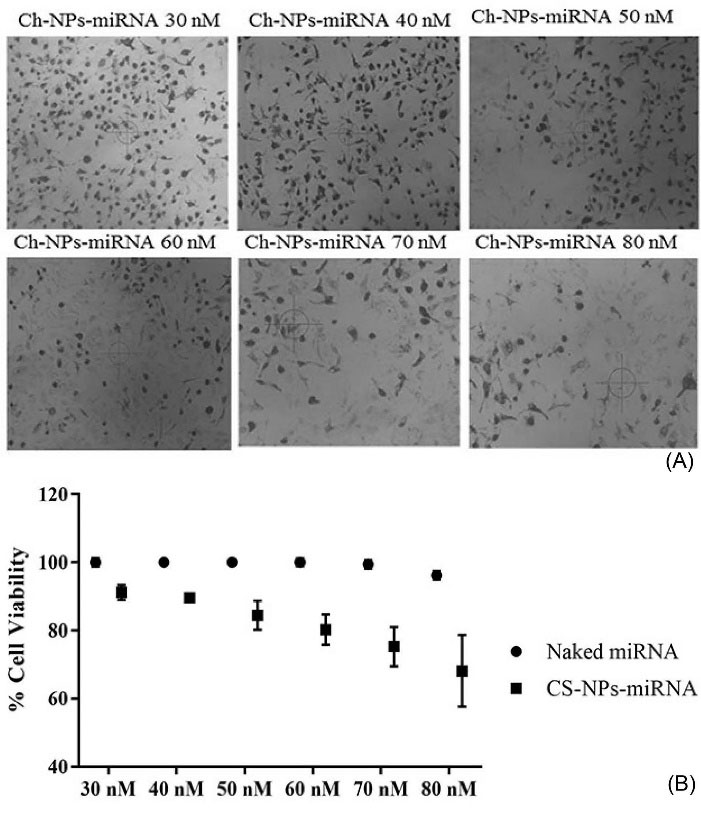
Fig. 4.
Inhibition of cell viability effect of CS-NPs/antimiRNA-324-5p: (A) Phase contrast microscopic image of SKOV3 cells treated with antimiR-324-5p, (B) Percentage of cell viability treated with various concentrations of antimiR-324-5p compared to naked antimiRNA-324-5p (data are mean of three different experiments).
.
Inhibition of cell viability effect of CS-NPs/antimiRNA-324-5p: (A) Phase contrast microscopic image of SKOV3 cells treated with antimiR-324-5p, (B) Percentage of cell viability treated with various concentrations of antimiR-324-5p compared to naked antimiRNA-324-5p (data are mean of three different experiments).
MiRNA-324-5p target gene prediction with bioinformatic analysis
We used miRTar.Human and StarMirDB to generate target genes that are regulated by miRNA-324-5p. According to our findings by Genecard, the hsa-miRNA-324-5p precursor gene is located on chromosome 17p13.1 and it is predicted to regulate more than 800 target genes. Furthermore, Menin-1 (MEN1) is a tumor suppressor gene that is highly regulated by this miRNA. It was proven by the interaction of both miRNA-324-5p-MEN1 that occurred in 2331-2342 base, generating the logistic probability of 0.765 and ΔGtotal -15 kcal/mol. The interaction also resulted in minimum free energy -21 and alignment score 162 (Fig. 5) that indicate high interaction.
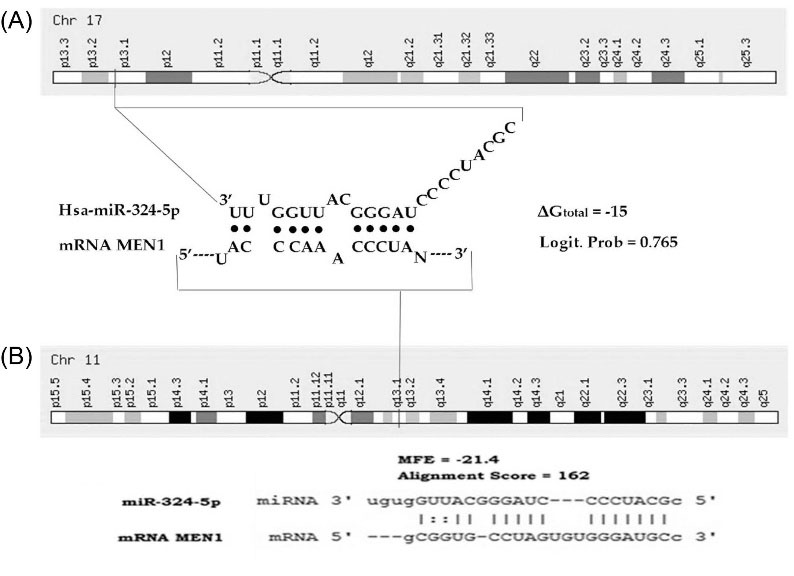
Fig. 5.
In silico gene target prediction for recognition and interaction of miRNA-324-5p with 3’UTR of MEN1 (direct target of miRNA-324-5p) by online software packages StarMirDB (A) and miRTar.Human (B).
.
In silico gene target prediction for recognition and interaction of miRNA-324-5p with 3’UTR of MEN1 (direct target of miRNA-324-5p) by online software packages StarMirDB (A) and miRTar.Human (B).
Molecular docking of MEN1-GLI1 Interaction
The Hawkdock server generates the 10 best models of complex proteins from the interactions of the two proteins that were ranked according to energy and scores. Lower energy and scores correspond to better scores indicating good interaction of both proteins. As the result, the model 1 is the best model which had score and binding free energy of -6789.54 and -35.98 kcal/mol, respectively (Fig. 6).
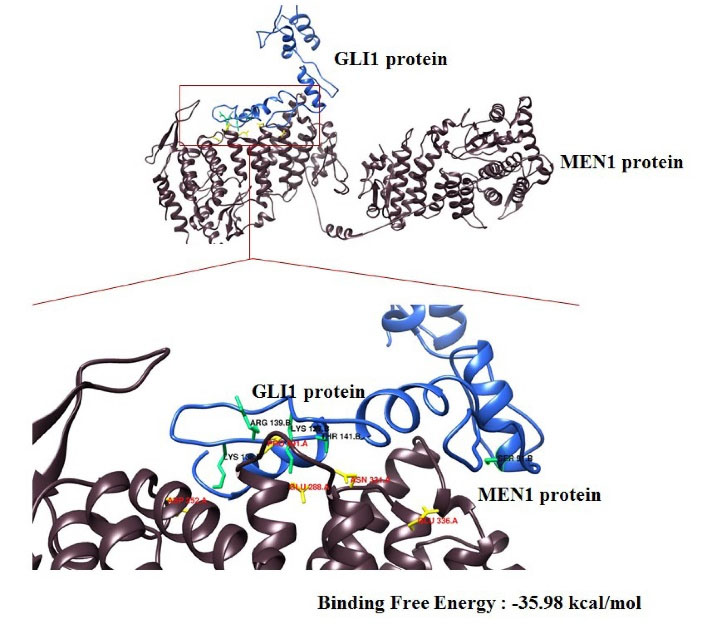
Fig. 6.
The molecular interaction of MEN1-GLI1 docked by Hawkdock server and visualized by UCSF Chimera.
.
The molecular interaction of MEN1-GLI1 docked by Hawkdock server and visualized by UCSF Chimera.
Furthermore, we analyzed the model 1 for the molecular mechanics/generalized born surface area (MM-GBSA) that was generated by the Hawkdock server and this resulted in some residues both of MEN1 and GLI1 which are predicted to be responsible for the interaction of the both proteins (Table 2).
Table 2.
The results of MM/GBSA of MEN1-GLI1 interaction
|
Rank
|
Residue (MEN1)
|
Binding Free Energy (kcal/mol)
|
Residue (GLI1)
|
Binding Free Energy (kcal/mol)
|
|
1
|
Glu.288 |
-4.62 |
Arg.139 |
-6.04 |
|
2
|
Pro.291 |
-3.41 |
Thr.141 |
-2.93 |
|
3
|
Glu.336 |
-2.47 |
Lys.138 |
-2.83 |
|
4
|
Asp.252 |
-2.18 |
Ser.91 |
-2.21 |
|
5
|
Asn.331 |
-2.06 |
Lys.127 |
-2.19 |
Transfection of antimiRNA-324-5p leads to the downregulation of GLI1 mRNA expression
Following the in silico and molecular docking results, we conducted quantification of GLI1 expression to further validate our findings in uptake of antimiRNA. GLI1 is a direct target of MEN1 that plays an important role in Hedgehog signaling. As the result, treatment of CS-NPs-antimiRNA-324-5p in SKOV3 cell line could reduce relatively the GLI1 expression (P < 0.05) (Fig. 7), suggesting that treatment of antimiRNA could cause the miRNA-324-5p expression (Fig. 3) and restore the MEN1 function to directly repress the GLI1 expression as previously hypothesized and proved by molecular docking (Fig. 6) and inhibit the ovarian cancer proliferation (Fig. 4).
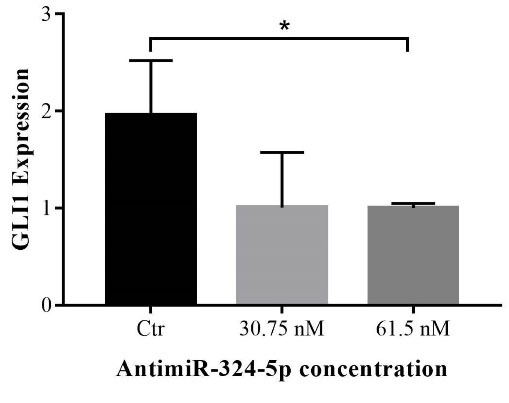
Fig. 7.
qRT-PCR of GLI1 mRNA expression in the treated SKOV3 cell line by antimiRNA-324-5p (All data are presented in mean ± SD *P < 0.05) versus control (Ctr) group (untreated SKOV3 cell line).
.
qRT-PCR of GLI1 mRNA expression in the treated SKOV3 cell line by antimiRNA-324-5p (All data are presented in mean ± SD *P < 0.05) versus control (Ctr) group (untreated SKOV3 cell line).
Discussion
The involvement of miRNAs in the pathogenesis of various diseases has increasingly attracted the attention of researchers. Ovarian cancer is no exception because it is the one type of female reproductive cancer with the highest mortality rate.
21
Upregulated expression of oncogenic miRNA that occurs in ovarian cancer worsens the prognosis of the cancer.
22
This condition is used by some research groups as one of the miRNA-based targeted molecular therapy strategies by restoring the endogenous miRNA balance, one of which uses the blocking function of oncomiRNA by using the antisense of that miRNA.
7
In this study, we used antimiRNA-324-5p to block the function of the miRNA and CS-NPs to assist miRNA delivery in SKOV3 cell lines. CS-NPs are cationic nanocarriers that are proven effective in delivering miRNA into the cells through the mechanism of interaction with negatively charged cell membranes, making it easier to enter cells.
18,19,23,24
The dissolution of chitosan in 1% acetic acid makes its amine group protonated to the acidic condition and forms NH3. The addition of acidic chitosan solution (pH = 5.5) into the alkali phase of TPP (pH 7-9) could initiate the formation of complex nanosized particles (Table 1). This complex is formed from electrostatic interactions that form an inter- and intra-molecular bridge that connect the amine group of chitosan and phosphate of TPP.
25-27
The conjugation of chitosan-TPP nanocomplex with miRNA allows the encapsulation of miRNA into the matrix polymer (Figs. 1 and 2). The encapsulation process occurs by two main interaction mechanisms: binding of NH3+ group of chitosan to phosphate of NA TPP and the interaction between the amine of chitosan and phosphate group of oligonucleotides and TPP. In addition, this conjugation also allows physical absorption due to the induction of cross-linking of TPP to miRNA.
27,28
Homogenization of particle size (lower than 500 nm and polydisperse index below 0.7) with the positive charge of formed nanocomplex could facilitate its interaction with the negatively charged cell membrane and lead to an increase in the efficiency of the transfection. Furthermore, the positive charge of the nanoparticles promotes the electrostatic interaction with the negative charged ions of the cell membrane to induce antimiR release in cytoplasm.
29
It was further proven by the demonstrated results of endogenous antimiR-324 that simultaneously caused downregulation (Fig. 3). This condition allows it to lose or reduce its function to suppress the tumor suppressor gene for the survival of the cancer cells. We noticed a significant decreasing of SKOV3 cell viability after treatment with antisense miRNA (Fig. 4). This finding is similar to previous results found by Ananta et al who transfected antimiRNA-21 and antimiRNA-10b using PLGA nanoparticle vectors. The second transfection of antimiRNA was shown to reduce endogenous miRNA and co-inhibit glioblastoma cells treated with Temozolomide.
30
Additionally, the blocking function of miRNA-324-5p uses its antisense through molecular mechanisms so that cancer cell proliferation is inhibited. As an oncomiRNA, a miRNA can regulate many tumor suppressor genes to maintain the life of ovarian cancer cells.
22
Based on our analysis in computational tools, miRNA-324-5p is predicted to be able to recognize and interact with 3'UTR of MEN1 on the nucleotide base 2331-2342 (Fig. 5). This interaction produces a logistical probability of 0.765 and ΔGtotal -15 kcal/mol, each of which implies the highest confidence in the predicted and structural accessibility at the target site.
31
MEN1 is one of the highlighted target genes regulated by miRNA-324-5p. The gene is a tumor suppressor that plays an important role in ovarian cancer. Lack of MEN1 expression leads to the invasion and migration of the ovarian cancer cell line.
5
The existence of MEN1 can inhibit and suppress the expression of protein oncogenes such as GLI1 (the protein involved in Hh signaling). MEN1 can also regulate the expression of GLI1 directly or indirectly.
32,33
As found by our molecular docking, MEN1 can bond and regulate GLI1 directly. This binding produces the lowest score and binding of free energy at -6789.54 and -35.98 kcal/mol, respectively, indicating the best interaction of both proteins. From the MM/GBSA results of the best model in docking, we concluded that there were 5 amino acids responsible for this interaction, namely Glu.288, Pro.291, Glu.336, Asp.252, and Asn.331 of MEN1 protein. While, in GLI1 there were Arg.139, Thr.141, Lys.138, Ser.91, and Lys.127. This interaction can further affect the expression of Gli1.
Based on the qPCR analysis, the results showed that giving antimiRNA treatment to SKOV3 can significantly downregulate the GLI1 mRNA expression (P < 0.05) (Fig. 6). We proposed it occurs through some mechanisms by blocking the function of miRNA-324-5p (by antisense miRNA) and causing the increased ability of MEN1 to repress oncogenic protein leading to poor cell viability (Fig. 4). MEN1 directly represses GLI1 expression by recruitment of PRMT5.
32
Furthermore, as a tumor suppressor, MEN1 can interact with the p65 subunit of nuclear factor kappa beta (NFκB) and recruit Sirt1 so that NFκB-dependent transcription and NFκB-mediated transactivation are inhibited. NFκB is a transcription factor for pro-inflammatory proteins such as tumor necrosis factor-alpha and Interleukin-1 beta that can induce the expression of GLI1 in an HH-dependent and non-canonical, HH/SMO independent manner leading to cancer cell proliferation.
33-35
Moreover, MEN1 has the ability to suppress the Wnt/β-catenin signaling pathway. Upregulation of MEN1 can reduce the nuclear accumulation of β-catenin and its transcriptional activity. β-catenin is a human protein encoded by CTNNB1 that plays an important role in the Wnt signaling pathway and leads to metastases.
5
This protein can interact with GLI1. Maeda et al demonstrated that the presence of exogenous β-catenin leads to the increased activity of the GLI-responsive reporter in luciferase assay.
36
Furthermore, other studies found that the interaction of both of the genes regulates proliferation, migration, and tumor growth.
37,38
Research Highlights
What is the current knowledge?
√ Currently, researchers recognize microRNA as a biomarker for diagnosis and prognosis of ovarian cancer.
What is new here?
√ microRNA-based therapeutic approach inhibited ovarian cancer proliferation.
√ Using antimiR-324-5p repressed oncomiRNA role in ovarian cancer.
√ Using CS-NPs as a non-viral vector delivered anti-miR-324-5p to ovarian cancer cell line.
Conclusion
According to the findings, we suggest that transfected antimiRNA-324-5p could mediate the inhibition of ovarian SKOV3 cell line by blocking the function of oncomiRNA-324-5p. The proposed blocking causes the balancing of MEN1 levels and its function leads to killing of ovarian cancer cells by repression of GLI1 expression.
Acknowledgments
The authors are grateful for the Master in Biomedical Science Program, Department of Pharmacology and Therapy and Klinik Bahasa Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada, Farmako Yogyakarta Indonesia.
Funding sources
This research was fully funded by grants from Ministry of Research Technology, and Higher Education of Republic of Indonesia PTUPT (1818/UNI/DITLIT/DIT-LIT/LT/2018) and (1987/UNI/DITLIT/DIT-LIT/LT/2018).
Ethical statement
The experimental protocol was approved by the Institutional Review Board of the Faculty of Medicine, Public Health and Nursing, Gadjah Mada University (KE/FK/0949/EC/2018).
Competing Interests
None declared.
Authors’ contribution
YY designed and performed the experiment and contributed to manuscript writing. IA supervised the study and contributed to manuscript writing.
References
- Biamonte F, Santamaria G, Sacco A, Perrone FM, Di Cello A, Battaglia AM. MicroRNA let-7g acts as tumor suppressor and predictive biomarker for chemoresistance in human epithelial ovarian cancer. Sci Rep 2019; 9:5668. doi: 10.1038/s41598-019-42221-x [Crossref] [ Google Scholar]
- Fang G, Liu J, Wang Q, Huang X, Yang R, Pang Y. MicroRNA-223-3p regulates ovarian cancer cell proliferation and invasion by targeting SOX11 expression. Int J Mol Sci 2017; 18:1208. doi: 10.3390/ijms18061208 [Crossref] [ Google Scholar]
- Chen S-N, Chang R, Lin L-T, Chern C-U, Tsai H-W, Wen Z-H. MicroRNA in ovarian cancer: biology, pathogenesis, and therapeutic opportunities. Int J Environ Res Public Health 2019; 16:1510. doi: 10.3390/ijerph16091510 [Crossref] [ Google Scholar]
- Eoh KJ, Lee SH, Kim HJ, Lee J-Y, Kim S, Kim SW. MicroRNA-630 inhibitor sensitizes chemoresistant ovarian cancer to chemotherapy by enhancing apoptosis. BiochemBiophys Res Commun 2018; 497:513-20. doi: 10.1016/j.bbrc.2018.02.062 [Crossref] [ Google Scholar]
- Hou R, Yang Z, Wang S, Chu D, Liu Q, Liu J. miR-762 can negatively regulate menin in ovarian cancer. Onco Targets Ther 2017; 10:2127. doi: 10.2147/OTT.S127872 [Crossref] [ Google Scholar]
- Dahiya N, Morin PJ. MicroRNAs in ovarian carcinomas. EndocrRelat Cancer 2010; 17:F77-F89. doi: 10.1677/ERC-09-0203 [Crossref] [ Google Scholar]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017; 16:203. doi: 10.1038/nrd.2016.246 [Crossref] [ Google Scholar]
- Astuti I, Torizal G, Sa’adah N, Oktriani R, Wardana T, Aryandono T. Resistance to doxorubicin correlated with dysregulation of microRNA-451 and P-glyoprotein, caspase 3, estrogen receptor on breast cancer cell line. J Med Sci 2019; 51:282-91. doi: 10.19106/JMedSci005104201901 [Crossref] [ Google Scholar]
- Wei J, Zhang L, Li J, Zhu S, Tai M, Mason CW. MicroRNA-205 promotes cell invasion by repressing TCF21 in human ovarian cancer. J Ovarian Res 2017; 10:33. doi: 10.1186/s13048-017-0328-1 [Crossref] [ Google Scholar]
- Qu K, Zhang X, Lin T, Liu T, Wang Z, Liu S. Circulating miRNA-21-5p as a diagnostic biomarker for pancreatic cancer: evidence from comprehensive miRNA expression profiling analysis and clinical validation. Sci Rep 2017; 7:1692. doi: 10.1038/s41598-017-01904-z [Crossref] [ Google Scholar]
- Deng X, Cao M, Zhang J, Hu K, Yin Z, Zhou Z. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials 2014; 35:4333-44. doi: 10.1016/j.biomaterials.2014.02.006 [Crossref] [ Google Scholar]
- Rui M, Qu Y, Gao T, Ge Y, Feng C, Xu X. Simultaneous delivery of anti-miR21 with doxorubicin prodrug by mimetic lipoprotein nanoparticles for synergistic effect against drug resistance in cancer cells. Int J Nanomedicine 2017; 12:217. doi: 10.2147/IJN.S122171 [Crossref] [ Google Scholar]
- Chen Y, Liu W, Chao T, Zhang Y, Yan X, Gong Y. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett 2008; 272:197-205. doi: 10.1016/j.canlet.2008.06.034 [Crossref] [ Google Scholar]
- Devulapally R, Sekar TV, Paulmurugan R. Formulation of anti-miR-21 and 4-hydroxytamoxifen co-loaded biodegradable polymer nanoparticles and their antiproliferative effect on breast cancer cells. Mol Pharm 2015; 12:2080-92. doi: 10.1021/mp500852s [Crossref] [ Google Scholar]
- Ohyagi-Hara C, Sawada K, Kamiura S, Tomita Y, Isobe A, Hashimoto K. miR-92a inhibits peritoneal dissemination of ovarian cancer cells by inhibiting integrin α5 expression. Am J Pathol 2013; 182:1876-89. doi: 10.1016/j.ajpath.2013.01.039 [Crossref] [ Google Scholar]
- Martien R, Loretz B, Sandbichler AM, Schnuerch AB. Thiolated chitosan nanoparticles: transfection study in the Caco-2 differentiated cell culture. Nanotechnology 2008; 19:045101. doi: 10.1088/0957-4484/19/04/045101 [Crossref] [ Google Scholar]
- Raemdonck K, Martens TF, Braeckmans K, Demeester J, De Smedt SC. Polysaccharide-based nucleic acid nanoformulations. Adv Drug Deliv Rev 2013; 65:1123-47. doi: 10.1016/j.addr.2013.05.002 [Crossref] [ Google Scholar]
- Chen X, Gu S, Chen B-F, Shen W-L, Yin Z, Xu G-W. Nanoparticle delivery of stable miR-199a-5p agomir improves the osteogenesis of human mesenchymal stem cells via the HIF1a pathway. Biomaterials 2015; 53:239-50. doi: 10.1016/j.biomaterials.2015.02.071 [Crossref] [ Google Scholar]
- Ysrafil Y, Astuti I, Anwar SL, Martien R, Sumadi FAN, Wardhana T. MicroRNA -155-5p Diminishes in Vitro Ovarian Cancer Viabiity by Tergeting HIF1α Expression. Adv Pharm Bull 2020; in press. doi: 10.15171/apb.2020.076 [Crossref]
- Kaban K, Salva E, Akbuga J. In vitro dose studies on chitosan nanoplexes for microRNA delivery in breast cancer cells. Nucleic Acid Ther 2017; 27:45-55. doi: 10.1089/nat.2016.0633 [Crossref] [ Google Scholar]
- Yokoi A, Matsuzaki J, Yamamoto Y, Yoneoka Y, Takahashi K, Shimizu H. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat Commun 2018; 9:1-10. doi: 10.1038/s41467-018-06434-4 [Crossref] [ Google Scholar]
- Zhao X-G, Hu J-Y, Tang J, Yi W, Zhang M-Y, Deng R. miR-665 expression predicts poor survival and promotes tumor metastasis by targeting NR4A3 in breast cancer. Cell Death Dis 2019; 10:1-21. doi: 10.1038/s41419-019-1705-z [Crossref] [ Google Scholar]
- Gaur S, Wen Y, Song JH, Parikh NU, Mangala LS, Blessing AM. Chitosan nanoparticle-mediated delivery of miRNA-34a decreases prostate tumor growth in the bone and its expression induces non-canonical autophagy. Oncotarget 2015; 6:29161. doi: 10.18632/oncotarget.4971 [Crossref] [ Google Scholar]
- Rudzinski WE, Aminabhavi TM. Chitosan as a carrier for targeted delivery of small interfering RNA. Int J Pharm 2010; 399:1-11. doi: 10.1016/j.ijpharm.2010.08.022 [Crossref] [ Google Scholar]
- Katas H, Alpar HO. Development and characterisation of chitosan nanoparticles for siRNA delivery. J Control Release 2006; 115:216-25. doi: 10.1016/j.jconrel.2006.07.021 [Crossref] [ Google Scholar]
- Karimi M, Avci P, Ahi M, Gazori T, Hamblin MR, Naderi-Manesh H. Evaluation of chitosan-tripolyphosphate nanoparticles as a p-shRNA delivery vector: formulation, optimization and cellular uptake study. J Nanopharmaceutics Drug Delivery 2013; 1:266-78. doi: 10.1166/jnd.2013.1027 [Crossref] [ Google Scholar]
- Elgadir M, Uddin M, Ferdosh S, Adam A, Chowdhury AJK, Sarker MI. Impact of chitosan composites and chitosan nanoparticle composites on various drug delivery systems: a review. J Food Drug Anal 2015; 23:619. doi: 10.1016/j.jfda.2014.10.008 [Crossref] [ Google Scholar]
- Csaba N, Köping-Höggård M, Alonso MJ. Ionically crosslinked chitosan/tripolyphosphate nanoparticles for oligonucleotide and plasmid DNA delivery. Int J Pharm 2009; 382:205-14. doi: 10.1016/j.ijpharm.2009.07.028 [Crossref] [ Google Scholar]
- Kharia A, Singhai A, Verma R. Formulation and evaluation of polymeric nanoparticles of an antiviral drug for gastroretention. Int J Pharm Sci Nanotech 2012; 4:1557-62. [ Google Scholar]
- Ananta JS, Paulmurugan R, Massoud TF. Tailored nanoparticle codelivery of antimiR-21 and antimiR-10b augments glioblastoma cell kill by temozolomide: toward a “personalized” anti-microRNA therapy. Mol Pharm 2016; 13:3164-75. doi: 10.1021/acs.molpharmaceut.6b00388 [Crossref] [ Google Scholar]
- Rennie W, Kanoria S, Liu C, Mallick B, Long D, Wolenc A. STarMirDB: a database of microRNA binding sites. RNA Biol 2016; 13:554-60. doi: 10.1080/15476286.2016.1182279 [Crossref] [ Google Scholar]
- Gurung B, Feng Z, Hua X. Menin directly represses Gli1 expression independent of canonical Hedgehog signaling. Mol Cancer Res 2013; 11:1215-22. doi: 10.1158/1541-7786.MCR-13-0170 [Crossref] [ Google Scholar]
- Matkar S, Thiel A, Hua X. Menin: a scaffold protein that controls gene expression and cell signaling. Trends Biochem Sci 2013; 38:394-402. doi: 10.1016/j.tibs.2013.05.005 [Crossref] [ Google Scholar]
- Grund-Gröschke S, Stockmaier G, Aberger F. Hedgehog/GLI signaling in tumor immunity-new therapeutic opportunities and clinical implications. Cell Commun Signal 2019; 17:1-9. doi: 10.1186/s12964-019-0459-7 [Crossref] [ Google Scholar]
- Gang D, Hongwei H, Hedai L, Ming Z, Qian H, Zhijun L. The tumor suppressor protein menin inhibits NF-κB-mediated transactivation through recruitment of Sirt1 in hepatocellular carcinoma. Mol Biol Rep 2013; 40:2461-6. doi: 10.1007/s11033-012-2326-0 [Crossref] [ Google Scholar]
- Maeda O, Kondo M, Fujita T, Usami N, Fukui T, Shimokata K. Enhancement of GLI1-transcriptional activity by β-catenin in human cancer cells. Oncol Rep 2006; 16:91-6. doi: 10.3892/or.16.1.91 [Crossref] [ Google Scholar]
- Zinke J, Schneider FT, Harter PN, Thom S, Ziegler N, Toftgård R. β-Catenin-Gli1 interaction regulates proliferation and tumor growth in medulloblastoma. Mol Cancer 2015; 14:17. doi: 10.1186/s12943-015-0294-4 [Crossref] [ Google Scholar]
- Wang Y, Lin P, Wang Q, Zheng M, Pang L. Wnt3a-regulated TCF4/β-catenin complex directly activates the key Hedgehog signalling genes Smo and Gli1. Exp Ther Med 2018; 16:2101-7. doi: 10.3892/etm.2018.6379 [Crossref] [ Google Scholar]