Bioimpacts. 12(2):127-138.
doi: 10.34172/bi.2021.22178
Original Research
Evaluating the presence of deregulated tumoral onco-microRNAs in serum-derived exosomes of gastric cancer patients as noninvasive diagnostic biomarkers
Houman Kahroba 1, 2, *  , Nasser Samadi 1, 2, 3, Mostafa Mostafazadeh 3, Mohamad Saied Hejazi 1, 2, 4, *
, Nasser Samadi 1, 2, 3, Mostafa Mostafazadeh 3, Mohamad Saied Hejazi 1, 2, 4, *  , Mohammad Reza Sadeghi 1, 5, Shahryar Hashemzadeh 6, Amir Taher Eftekhar Sadat 7, Abbas Karimi 2
, Mohammad Reza Sadeghi 1, 5, Shahryar Hashemzadeh 6, Amir Taher Eftekhar Sadat 7, Abbas Karimi 2
Author information:
1Department of Molecular Medicine, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
2Molecular Medicine Research Center, Biomedicine Institute, Tabriz University of Medical Sciences, Tabriz, Iran
3Department of Biochemistry, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
4Department of Pharmaceutical Biotechnology, Faculty of Pharmacy, Tabriz University of Medical Sciences, Tabriz, Iran
5Research Center for Pharmaceutical Nanotechnology, Biomedicine Institute, Tabriz University of Medical Sciences, Tabriz, Iran
6Liver and Gastrointestinal Diseases Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
7Department of General and Vascular Surgery, Imam Reza Educational Hospital, School of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
Abstract
Introduction:
Exosomal microRNAs (miRNAs) are emerging diagnostic biomarkers for different types of cancers. We aim to detect gastric cancer (GC)-specific miRNAs in serum exosomes with diagnostic potential.
Methods:
A pair of 43 tumor and tumor-adjacent tissue biopsies obtained from GC patients, also 5 mL peripheral blood (following 12h fasting) were collected from the same patients and healthy controls (HCs). QIAGEN miRCURY LNA miRNA Focus PCR Panel applied to screen differentially expressed onco-miRNAs. The candidate miRNAs with the highest fold changes proceeded for validation by qRT-PCR in individuals.
Results:
We identified that exosomal miR-10a-5p, miR-19b-3p, miR-215-5p, and miR-18a-5p were significantly upregulated in GC patient’s exosomes in contrast to HCs exosomes, Roc curve analysis indicated area under the ROC curve (AUC) of 0.801, 0.721, 0.780 and 0.736 respectively. The Roc curve analysis for the combined signature of four exosomal miRNAs indicated AUC of 0.813. Also, Spearman's correlation coefficients indicated that the miRNA expression is highly correlated between tumor and exosome.
Conclusion:
Herein, we specifically identified four miRNAs in serum exosomes of GC patients for a diagnostic purpose which are directly associated with tumoral miRNA expression profile.
Keywords: Liquid Biopsy, ExomiR, OncomiR, Stem Loop RT-PCR, Stomach
Copyright and License Information
© 2022 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Gastric cancer (GC) is the fourth frequent malignancy, also GC is known as the second leading cause of cancer-derived death worldwide.
1
Almost 90% of all diagnosed gastric tumors are characterized as malignant, and the remains are benign (10%). Approximately 95% of diagnosed malignant tumors are adenocarcinoma, and %5 are squamous cell carcinoma.
2,3
Carcinoembryonic antigens (CEA) and carbohydrate antigen 19-9 (CA19-9) are blood-based biomarkers, which are broadly applied in the clinic to detect GC; however, these biomarkers do not meet the minimum sensitivity and specificity for supporting the early diagnosis of GC.
4,5
Upper endoscopy (gastroscopy), as an invasive approach, is still the main criteria for the screening and diagnosis of GC.
6
Surgical and endoscopic resections are the main therapeutical strategies at the early stages of the GC,
7,8
However, for the advanced tumors, systemic chemotherapies are applied which fails in more than 95% of the cases.
8,9
The poor prognosis of GC, the 5-year survival rate is 5‐15%, is mainly due to advanced tumors at the moment of the diagnosis.
10,11
Therefore, the development of novel diagnostic approaches is an urgent demand in GC studies.
MicroRNAs (miRNAs) are endogenous non-coding-RNAs with about 22 nucleotides in length, which post-transcriptionally orchestrate the gene expression pattern. miRNAs can completely block the expression of the target mRNA(s) or transiently decrease the translation rate. It is reported that these biomolecules can act as oncogenes or tumor-suppressors in cancer.
12
MiRNAs expression patterns are the more accurate hallmark than mRNAs pattern to detect the origin of the unknown tissues, even for tumors.
13
This pattern is tumor-specific and can be used to distinguish between normal- and tumoral-tissues (also in GC).
13,14
However, miRNAs are stable in circulation, but these molecules are persistently under the influence of the different pathophysiological conditions and circulating ribonucleases.
15
The levels of the cell-free miRNAs may be affected by sources other than the tumor cells such as infections, circulating blood cells, hypoxia, diets, and exercise; therefore, the levels of circulating miRNAs may not represent a direct correlation to the cancer tissues.
16,17
Exosomes are spherical and/cup-shaped extracellular nanovesicles that span 40-150 nm in diameter, these vesicles have a bilayer lipid structure for the outer membrane. A multitude of cells releases exosomes into the different body fluids such as blood. Exosomes carry out various biomolecules, including proteins, lipids, DNAs, and RNAs in a multi-directional and conserved manner through the cells which recommends them as possible liquid biopsy-based biomarkers.
1,18
Exosomes improve the stability of the circulating miRNAs by protecting them from ribonuclease activities.
19
Furthermore, exosomes reflect not only the physio-pathological condition of the originating cells but also confirm the upregulated systemic circulation of the exosomes in most cancers.
20
Herein, we screened the onco-miRNA expression profile in GC tumors, tumor margin, and GC patient’s serum-derived exosomes by QIAGEN miRCURY LNA miRNA Focus PCR Panels and validated our findings in GC patients and healthy controls (HCs) by qRT-PCR.
Materials and Methods
Materials
In exosome characterization, uranyl acetate solution (TAAB, England) was used for exosome staining. For the western blotting, the protein lysis buffer (St. Louis, USA), bicinchoninic acid (BCA)-assay (Cat No: DB9684-50ml, DNAbiotech Co. Tehran, Iran), polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, Massachusetts, United States), antibodies from Santa Cruz Biotechnology including anti- CD9 (sc-13118, Dallas, USA), anti-CD81(sc-166029, Dallas, USA) and anti-CD63 (sc-5275, Dallas, USA), anti-Calnexin (sc-23954, Dallas, USA) and anti-Mouse IgG (Goat), HRP Conjugated (1: 10 000; Santa Cruz, sc-2005). For RNA isolation, TRIzol (RiboEx.LS, Seoul, South Korea), isopropyl alcohol, and ethanol from Merck, Germany. The miRCURY LNA RT Kit (Cat No./ID: 339340, Hilden, Germany), miRCURY LNA miRNA Focus PCR Panel (Cat. no. YAHS-102Y, Hilden, Germany), cel-miR-39 Spike-in (Norgen Biotek, Thorold, Canada), RevertAid Reverse Transcriptase (Thermo Scientific, Kaunas, Lithuania), SYBR Green master mix (RealQ Plus 2x Master Mix Green, Ampliqon, Denmark) and primers purchased from (Macrogen, Seoul, South Korea).
Sampling and specimen preparation
A pair of 43 primary adenocarcinoma GC tissue samples (non-cardia and without adjuvant/chemotherapy) with corresponding adjacent non-malignant counterparts were obtained through tumor resection. Also, 5ml of peripheral blood, following 12 hours fasting, was collected from the same patients and 40 HCs at the first affiliated Hospital of Tabriz University of Medical Sciences (Tabriz, Iran). Patients with cardia tumors, adjuvant therapy, radiotherapy and chemotherapy, and previous history of malignancies were excluded from this study. Also, HCs with a history of autoimmune diseases, chronic infections and allergy, and a history of malignancies were excluded. The demographic and clinical information of the GC patients is represented in Table 1. Histopathological data were collected from the patients’ medical records.
Table 1.
Gastric cancer patients' demographic and clinical information
|
Clinical pathological data
|
No. of patients (%) tumor
|
No. of patients (%) exosome
|
| Age |
|
|
| < 65 years |
33 (76.74) |
31 (77.5) |
| ≥65 years |
10 (23.25) |
9 (22.5) |
| Gender |
|
|
| Male |
18 (41.86) |
17 (42.5) |
| Female |
25 (58.13) |
23 (57.49) |
| Tumor size |
|
|
| <5 cm |
19 (44.18) |
19 (47.5) |
| ≥5 cm |
24 (55.81) |
21 (52.5) |
| Histological grade |
|
|
| Poor-differentiated |
11 (25.58) |
11 (27.5) |
| Moderately-differentiated |
21 (48.83) |
18 (45) |
| Well-differentiated |
11 (25.58) |
11 (27.5) |
| Lymph node metastasis |
|
|
| Positive |
26 (60.46) |
26 (65) |
| Negative |
17 (39.53) |
14 (35) |
| Distant metastasis |
|
|
| Positive |
13 (30.23) |
13 (32.5) |
| Negative |
30 (69.76) |
27 (67.5) |
| Pathological variants |
|
|
| Intestinal |
36 (83.72) |
33 (82.5) |
| Diffuse |
7 (16.27) |
7 (17.5) |
Sample preparation
The obtained tissue samples were immediately directed into the liquid nitrogen tank, then homogenized in Trizol and stored at -70°C until RNA extraction (maximum a month). The 5 mL of peripheral blood was obtained in serum clot-activator tubes and centrifuged at 500 g for 15 minutes at room temperature to isolate serum. Subsequently, 1.5-2 mL aliquots of the serum were stored in Eppendorf tubes and proceeded to the -70°C to store for exosome isolation and further RNA extraction.
Exosome isolation
Exosomes were isolated from 1ml of serum by serial ultracentrifugation. Briefly, 1 mL of the serum was centrifuged at 500g for 30 minutes (4˚C) to remove cell debris, and 95% of the supernatant (to avoid debris contamination) proceeded for ultracentrifugation at 30 000 g for 2 hours (4˚C) to remove contaminant microvesicles and apoptotic bodies. Subsequently, 95% of the supernatant was collected and filtered through a membrane filter (0.22 µm) and redirected to the ultracentrifugation at 110 000 g (4˚C) for 2 hours by TLA 100.2 rotor (Beckman Coulter, USA) to isolate the exosomes. The exosome pellets were washed by 1x PBS and proceeded to the final ultracentrifugation step at 110 000 g (4˚C) to reduce the possible protein contamination. The isolated exosomes were resuspended in 1x PBS or distilled water (for dynamic light scattering [DLS] and transmission electron microscopy [TEM]) to store at -70˚C for the next step.
21
Exosome characterization
Size and morphology
DLS (Zetasizer Nano ZS, Malvern Panalytical) was employed to measure the average size and size distribution of isolated exosomes. Briefly, the isolated exosomes were resuspended in distilled water and diluted (1:10) to obtain better results. TEM was applied to visualize the morphology and size of the exosomes. The isolated exosomes were resuspended in 250 μL of distilled water, and a small aliquot of them was fixed on a 300 mesh copper grid under the ambient condition for 5 minutes. For negative staining, exosome-grids were stained with 1.5% (wt/v) uranyl acetate solution (TAAB, England) for 2 minutes and coated by carbon film to avoid degradation by electron beams. Grids were dried with a filter paper to remove stain residue and stored overnight at room temperature to visualize by LEO 906 Zeiss instrument (Freiburg, Switzerland) with an accelerating voltage of 80 kV.
Western blotting
Western blotting was performed by lysing of exosome pellet in 250 μL of 1x protein lysis buffer (St. Louis, USA) and followed by thrice sonication for 5min with cool vortex-mixing in between. The protein concentration was measured by bicinchoninic acid (BCA)-assay (Cat No: DB9684-50 mL, DNAbiotech Co. Tehran, I.R. Iran) according to the manufacture's instruction. 20-30 µg of extracted protein incubated with loading buffer containing β-mercaptoethanol) at 70˚C for 10 minutes to run on 12% SDS-polyacrylamide gel (SDS-PAGE). A 5% skim milk solution was applied to block the polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, Massachusetts, United States). The CD9, CD81, and CD63 protein markers were detected by Santa Cruz Biotechnology mouse monoclonal anti- CD9 (sc-13118, Dallas, USA), anti-CD81(sc-166029, Dallas, USA), and anti-CD63 (sc-5275, Dallas, U.S.A.) (1: 1000). Also, mouse monoclonal anti-Calnexin (sc-23954, Dallas, USA) (1: 1000) was used against Calnexin as the negative control for exosomes.
22
This step was followed by incubation with anti-Mouse IgG (Goat), HRP Conjugated (1: 10 000; Santa Cruz, sc-2005) antibody, and the protein bands detected by Chemi-Lumi One Super (Product No. 02230, Kyoto, Japan).
RNA isolation
Total RNA isolated from tissues and exosomes by the TRIzol method (RiboEx.LS, Seoul, South Korea). Briefly, the samples were lysed by 750 μL of the RiboEx and 200 μL chloroform added and stored for 2 minutes at room temperature. The samples were centrifuged at 12 000 g for 20 minutes (4oC) and an aqueous phase was collected and transferred to a new collection tube and incubated with 1 volume of isopropyl alcohol for about 90 minutes at -20. Subsequently, the samples were centrifuged at 12 000 g for 1 hour (4oC) to precipitate the total RNA. The RNA was washed with 1 ml of 75% ethanol alcohol by centrifugation at 12 000 g for 20 minutes (4oC), and the RNA pellet dissolved in 20 λ DEPC-treated water (Cinnagen, Tehran, Iran).
Reverse transcription and Human Cancer Focus Panel (screening phase)
Six samples were used to make pooled samples for each of the tumor, tumor margin, and serum-derived exosomes groups. About 15 ng/μL of total RNA was used to make a pool from each sample. The pooled RNA was reverse transcribed by miRCURY LNA RT Kit (Cat No./ID: 339340, Hilden, Germany) on PeQlab thermocycler (Fareham, United Kingdom) and quantitate PCR (qPCR) ran on Human Cancer Focus, miRCURY LNA miRNA Focus PCR Panel (Cat. no. YAHS-102Y, Hilden, Germany) by Roche light cycler (LightCycler 96 Real-Time PCR Cycler, Indianapolis, USA) to screen 86 cancerous miRNAs expression profile in three distinct groups. UniSp6 spike-in was used as a predefined references gene in the miRNA Focus PCR Panel to evaluate the expression analysis by a qPCR panel. UniSp6 was spiked in each sample at the time of RNA extraction. Also, melting curve analysis was performed at the end of the PCR cycles. The relative expression levels of miRNAs among different groups were calculated by the 2-ΔΔCt method by GenEx software version 6.0.
Reverse transcription and qPCR of candidate miRNAs (validation phase)
Differentially expressed miRNAs (according to the most significant fold change in contrast to the margin group) were selected for the validation phase. The sequences of the miRNAs were obtained from https://www.mirbase.org and used to design stem-loop reverse transcriptase (RT)-PCR specific primers by sRNAPrimerDB.
23
The primer sequences were evaluated by Oligo7 (version: 7.60) (Table 2).
Table 2.
Sequences of the Stem-loop reverse transcriptase-PCR and quantitative real-time PCR primers for candidate miRNAs
|
miRNA
|
Reverse transcriptase-specific-primer (5'-3')
|
Forward-primer (5'-3')
|
Reverse-primer (5'-3')
|
| miR-18a-5p |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTATCT |
AACACGCTAAGGTGCATCTAGT |
GTCGTATCCAGTGCAGGGT |
| miR-19b-3p |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAGTT |
AACAAGTGTGCAAATCCATGC |
| miR-10a-5p |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCACAAA |
AACACGCTACCCTGTAGATCC |
| miR-215-5p |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGTCTGT |
AGCCAGCGATGACCTATGAAT |
| cel-mir-39-3p |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAAGCT |
AACAGTGTCACCGGGTGTAAA |
Also, cel-miR-39 Spike-in (Norgen Biotek, Thorold, Canada) was used as a reference gene in all templates. 5 μL of synthetic cel-mir-39-3p was spiked into each sample after the addition of denaturing solution for normalization of the sample-to-sample variation. RevertAid Reverse Transcriptase (Thermo Scientific, Kaunas, Lithuania) was used for reverse transcription of the candidate miRNAs according to the company instruction. All the experiments were performed as duplicate, the qPCR program was as the following: PCR initial heat activation on 95oC for 15 minutes, 70 cycles of denaturation on 95oC for 10 seconds, and annealing/extension on 60oC for 1 minute (5 μL SYBR Green master mix (RealQ Plus 2x Master Mix Green, Ampliqon, Denmark), 2 μL cDNA, 0.5 μL primer pair mix (5 pmol/μL each primer), and 3 μL H2O).
24
Statistics
All statistical analyses for miRNAs expression levels through GC patients and NCs were performed using the GenEx software (version 6), SPSS 18.0 software, MedCalc Statistical Software version 16.4.3 (MedCalc Software bv, Ostend, Belgium, 2016), and the graphs were generated using GraphPad Prism 6.0 (GraphPad Software, San Diego, CA, USA). MiRNAs expression levels through GC tumors and exosomes by unpaired two-sample Mann-Whitney U test. The association between miRNAs and the clinical characteristics was assessed by the χ2 test. One-way ANOVA with the Tukey-Kramer post-hoc test was applied to test the differences in miRNAs expression levels through different histological grades. Receiver operating characteristic (ROC) curves, the area under the ROC curve (AUC), negative predictive value (NPV), positive predictive value (PPV), and likelihood ratios (LRs) used to estimate the diagnostic value of the candidate miRNAs for GC. Also, the Spearman correlation test was used to calculate any correlation between the miRNA’s expression levels. For all the tests P-value, less than 0.05 was assumed as a statistically significant difference.
Results
Exosome characterization
DLS measurement for exosome indicated 100-200 nm mean size distribution as illustrated in Fig. 1A. The expected size for exosomes is about 30-150nm, the right-skewed distribution in DLS results is due to artificial cup-shaped morphology and hydrodynamic radius which are reported previously by certain studies.
25,26
TEM confirmed cup-shaped structure for exosomes with a size of 30-150 nm. Also, the exosomes were positive for CD9, CD81, and CD63 markers and negative for calnexin (Fig. 1).
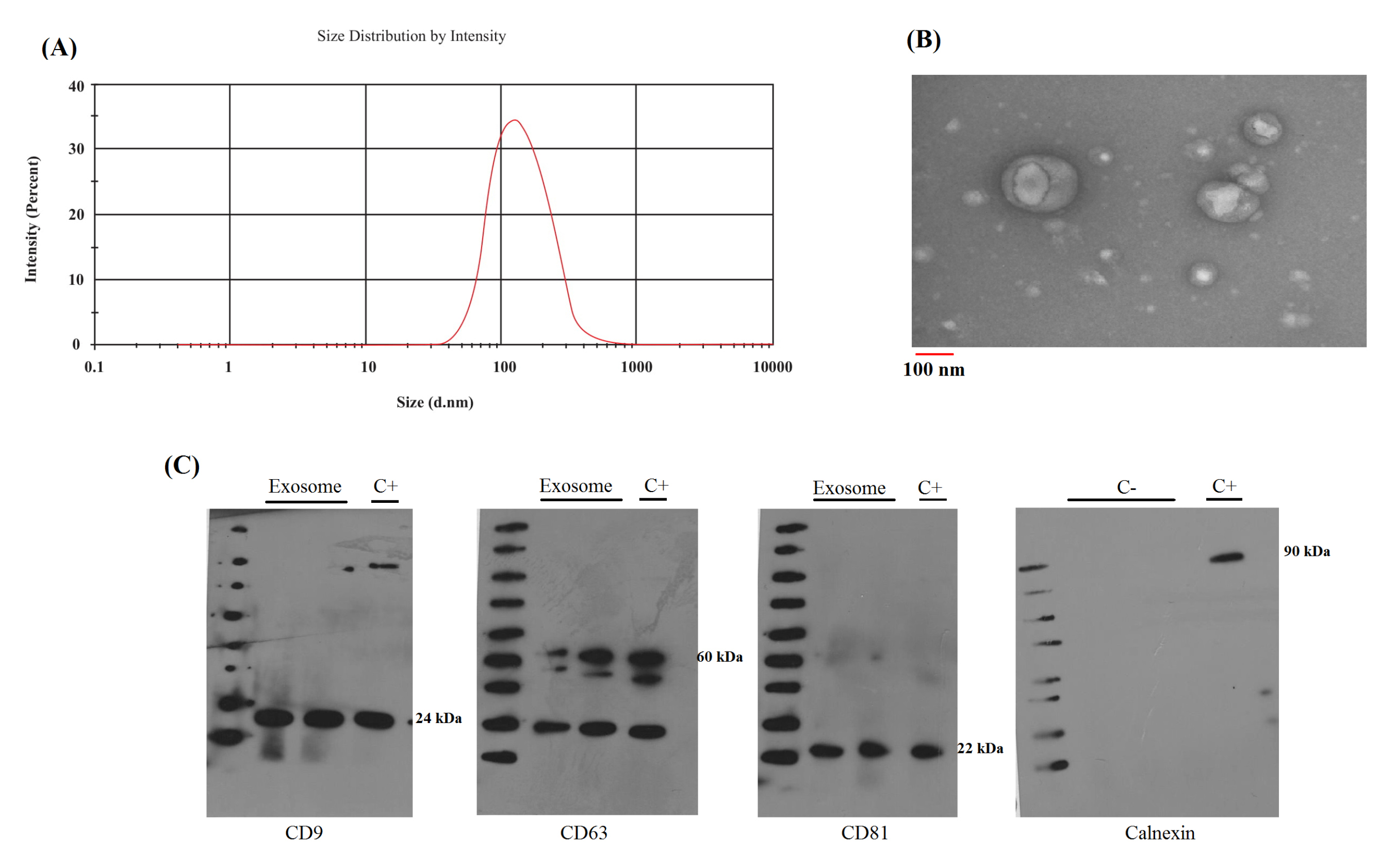
Figure 1.
The identification of serum exosomes. (A) DLS result for size distribution of serum exosomes indicates that the distribution is around 100 nm. (B) TEM for morphology and size of serum exosomes. TEM confirms cup-shape for exosomes and indicates 100 nm diameter for these vesicles. (C) Specific protein markers for exosomes. Serum exosomes were positive for CD9, CD81, CD63 and negative for Calnexin. Also, exosome supernatant applied as negative control (C-) and cell lysate applied as positive control (C+).
.
The identification of serum exosomes. (A) DLS result for size distribution of serum exosomes indicates that the distribution is around 100 nm. (B) TEM for morphology and size of serum exosomes. TEM confirms cup-shape for exosomes and indicates 100 nm diameter for these vesicles. (C) Specific protein markers for exosomes. Serum exosomes were positive for CD9, CD81, CD63 and negative for Calnexin. Also, exosome supernatant applied as negative control (C-) and cell lysate applied as positive control (C+).
Human Cancer Focus Panel
The result of the Human Cancer miRNA Focus PCR Panel used for fold change analysis with the 2-ΔΔCt method by GenEx software (version 6), and UniSp6 used as a reference gene for data normalization. The tumor margin was assumed as a control group. The miRNA expression heat map is illustrated as single-linkage clustering in Fig. 2. The data comprises two main categories: (a) miRNAs which are upregulated in both exosome and tumor samples in contrast to tumor margin, and (b) miRNAs which are upregulated in exosome and downregulated in tumor samples. Four miRNAs (MiR-10a-5p, miR-19b-3p, miR-215-5p, and miR-18a-5p) indicated the highest dysregulation in exosome and tumor samples, these candidate miRNAs were proceeded to the validation phase for evaluating the possibility to serve as GC-specific diagnostic biomarkers. The selected miRNAs were significantly upregulated in gastric tumors and the patient’s exosomes. The accurate quantification of the downregulated miRNA expression level is challenging. In terms of measurement, upregulation of miRNAs is preferred to downregulation due to ease of quantification in contrast to non-pathological conditions.
27
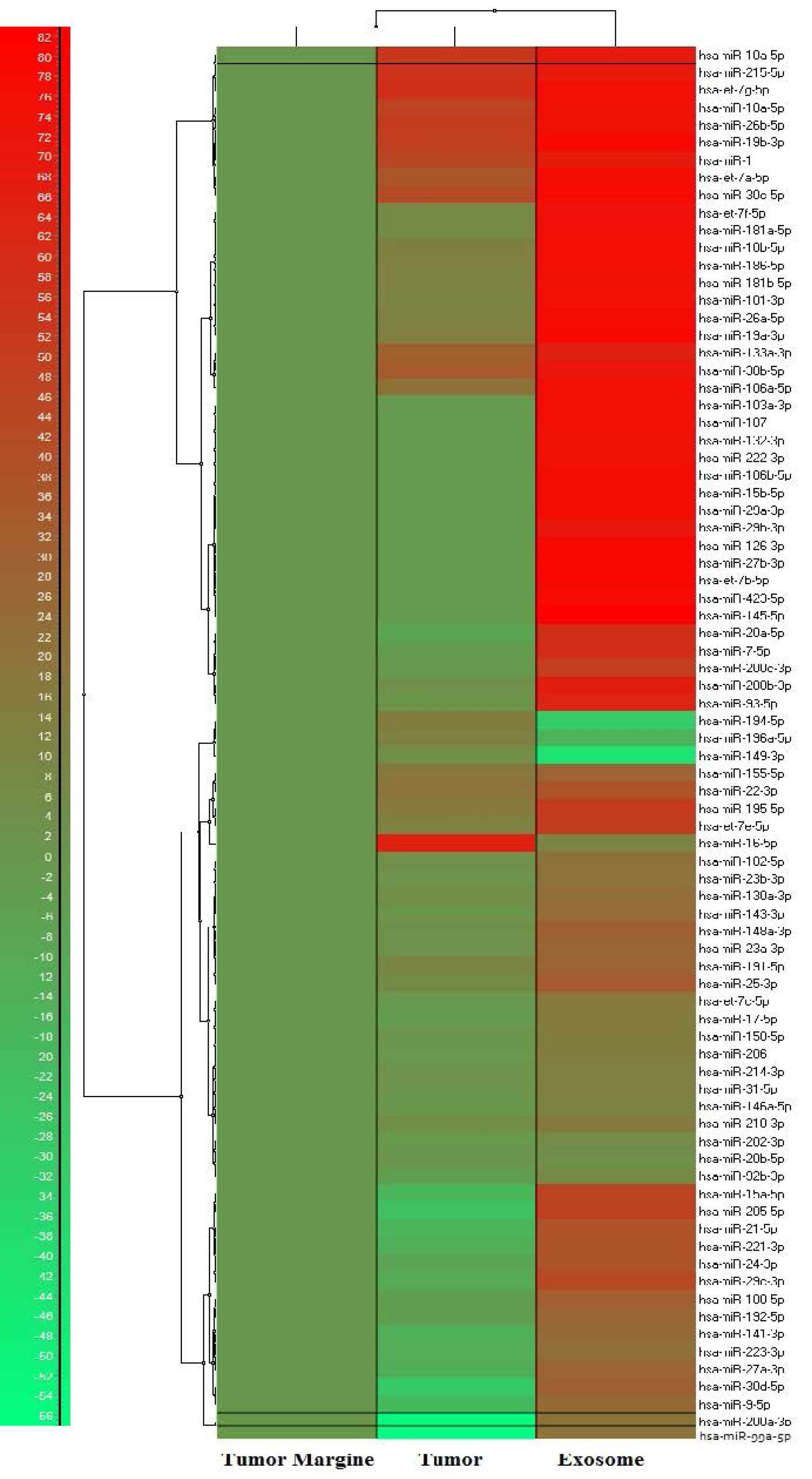
Figure 2.
Human Cancer Focus PCR Panel heat map for miRNA expression in tumor, tumor adjacent non-cancerous tissue and GC patient’s serum exosome. The represented data is illustrated as single-linkage clustering according to fold change of the miRNAs and tumor margin is assumed as control group.
.
Human Cancer Focus PCR Panel heat map for miRNA expression in tumor, tumor adjacent non-cancerous tissue and GC patient’s serum exosome. The represented data is illustrated as single-linkage clustering according to fold change of the miRNAs and tumor margin is assumed as control group.
Candidate miRNAs expression in tumor and exosome
The candidate miRNAs were evaluated by q-RT PCR in two groups including (i) tumor with paired cancer-adjacent non-tumorous tissues (ii) GC patient’s serum exosomes with HCs serum exosomes. The 2-ΔΔCt method was used for data analyzing and cel-mir-39-3p was used as a reference gene for data normalization. We explored the expression levels of the four candidate miRNAs in the GC patient’s tumor and tumor-adjacent tissues (n = 43) by qRT-PCR. We found that miR-10a-5p, miR-19b-3p, miR-215-5p and miR-18a-5p were significantly upregulated in tumors in contrast to tumor-adjacent tissues (Fig. 3). Although we determined the expression levels of these miRNAs in the GC patient’s serum exosomes in contrast to HCs serum exosomes (n = 40). The results confirmed the miR-10a-5p (P < 0.001), miR-19b-3p (P < 0.001), miR-215-5p (P < 0.001) and miR-18a-5p (P < 0.005) upregulation in contrast to HCs exosomes (Fig. 4).
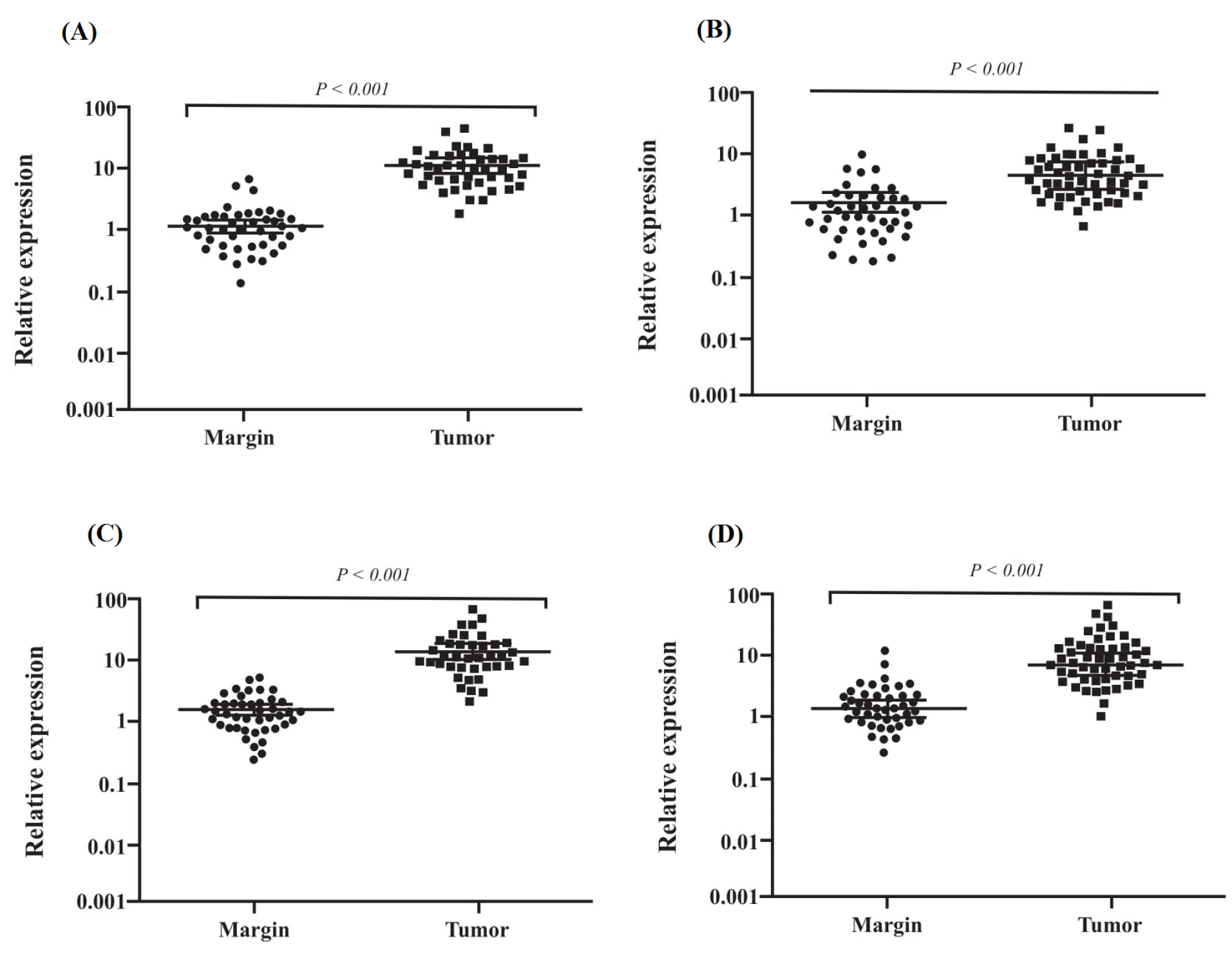
Figure 3.
Tumoral miRNAs expression levels. (A) mir-10a-5p expression, (B) mir-18a-5p, (C) mir-19b-3p expression and (D) mir-215-5p. A p-value less than 0.05 was assumed as statistically significant difference.
.
Tumoral miRNAs expression levels. (A) mir-10a-5p expression, (B) mir-18a-5p, (C) mir-19b-3p expression and (D) mir-215-5p. A p-value less than 0.05 was assumed as statistically significant difference.
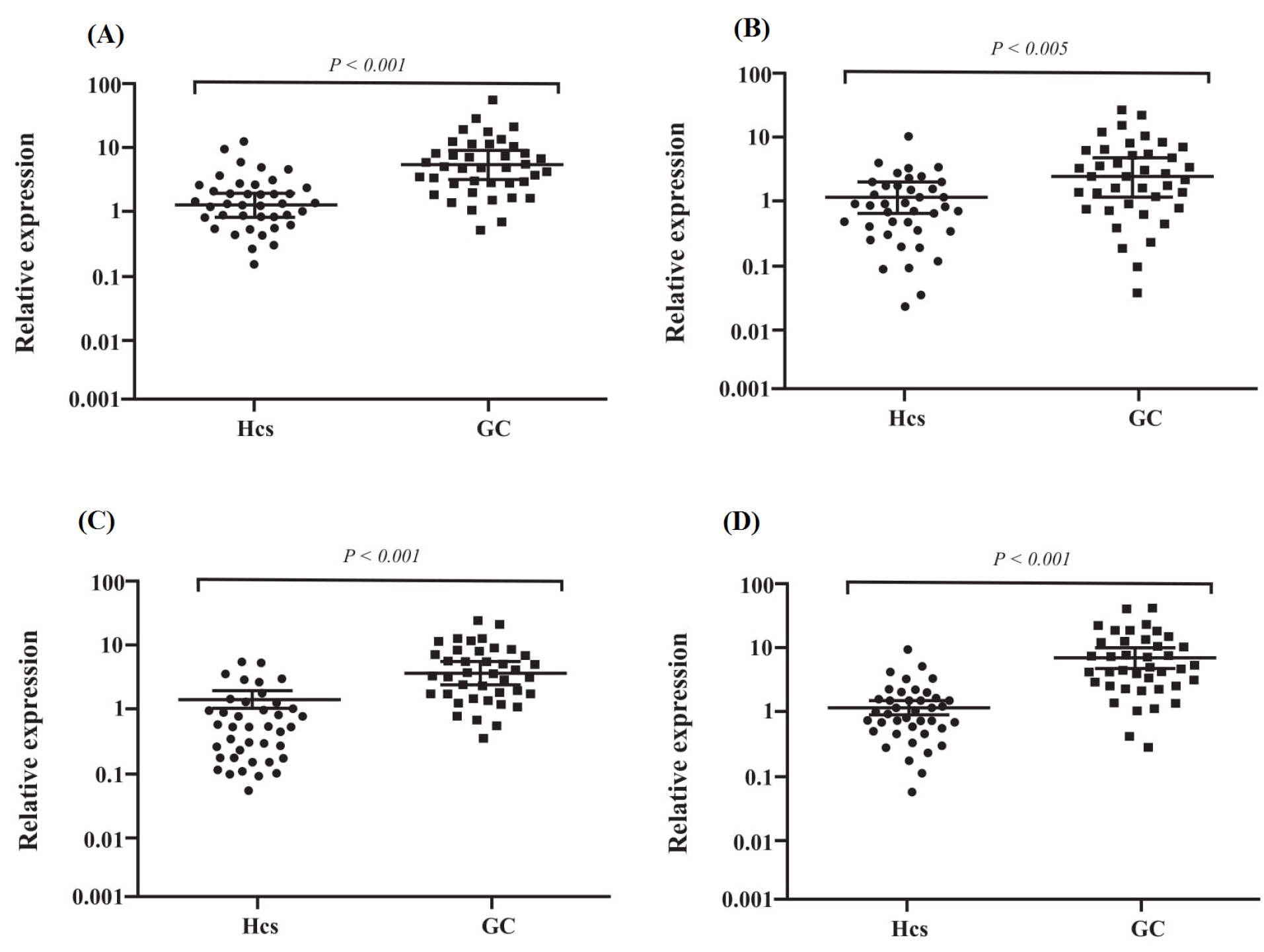
Figure 4.
Exosomal miRNAs expression level. (A) Serum exosomal (Ser-Exo) mir-10a-5p, (B) Ser-Exo mir-18a-5p, (C) Ser-Exo mir-19b-3p expression, (D) Ser-Exo mir-215-5p. A p-value less than 0.05 was assumed as statistically significant difference.
.
Exosomal miRNAs expression level. (A) Serum exosomal (Ser-Exo) mir-10a-5p, (B) Ser-Exo mir-18a-5p, (C) Ser-Exo mir-19b-3p expression, (D) Ser-Exo mir-215-5p. A p-value less than 0.05 was assumed as statistically significant difference.
Diagnostic value of the candidate miRNAs in tumor and exosomes
Generation of ROC curves and the AUC of the four candidate miRNAs was performed to explore the diagnostic value of these molecules in discriminating the gastric tumor from non-cancerous adjacent tissue, which is indicated in Fig. 5 and Fig. 6 for tumoral and exosomal miRNAs, respectively. Also, the overall diagnostic values for tumoral and exosomal miRNAs, including positive likelihood ratio (+LR), negative likelihood ratio (-LR), PPVs, NPVs, and accuracy of the test are calculated and represented in Table 3. Our results showed relatively high sensitivity and specificity for tumoral MiR-10a-5p, miR-19b-3p, miR-215-5p and miR-18a-5p for diagnosis of GC. The combined signature of the four tumoral miRNAs indicated higher diagnostic values for discriminating the gastric tumor from adjacent noncancerous tissue in contrast to individual miRNAs, as indicated in Table 3.
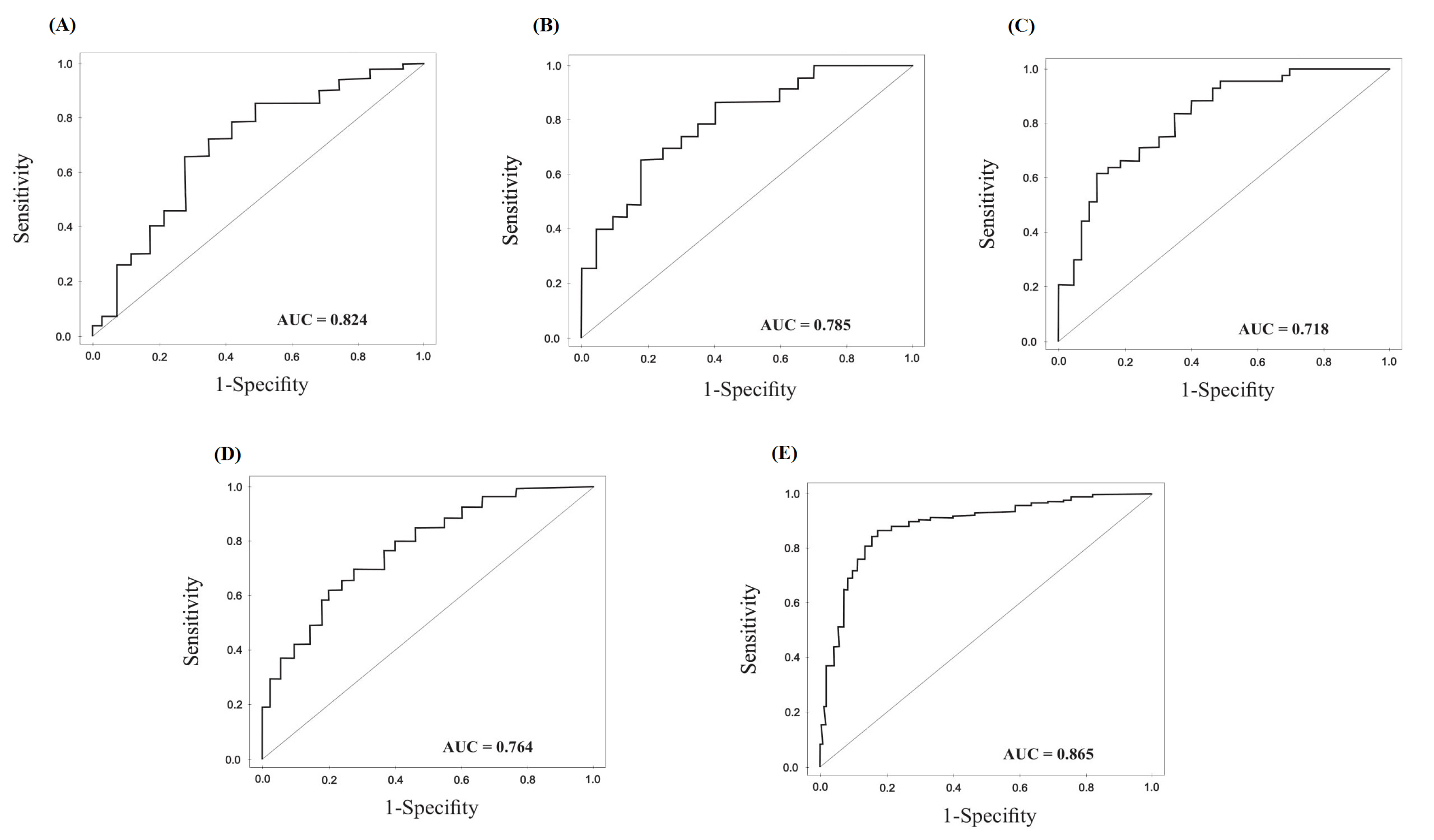
Figure 5.
Diagnostic values of tumoral miRNAs. (A) ROC curves of miR-10a-3p, (B) miR-215-5p, (C) miR-18a-5p, (D) miR-19b-3p in tumoral and tumor adjacent noncancerous biopsies from the GC patients. (E) ROC analysis for the four combined miRNA signature.
.
Diagnostic values of tumoral miRNAs. (A) ROC curves of miR-10a-3p, (B) miR-215-5p, (C) miR-18a-5p, (D) miR-19b-3p in tumoral and tumor adjacent noncancerous biopsies from the GC patients. (E) ROC analysis for the four combined miRNA signature.
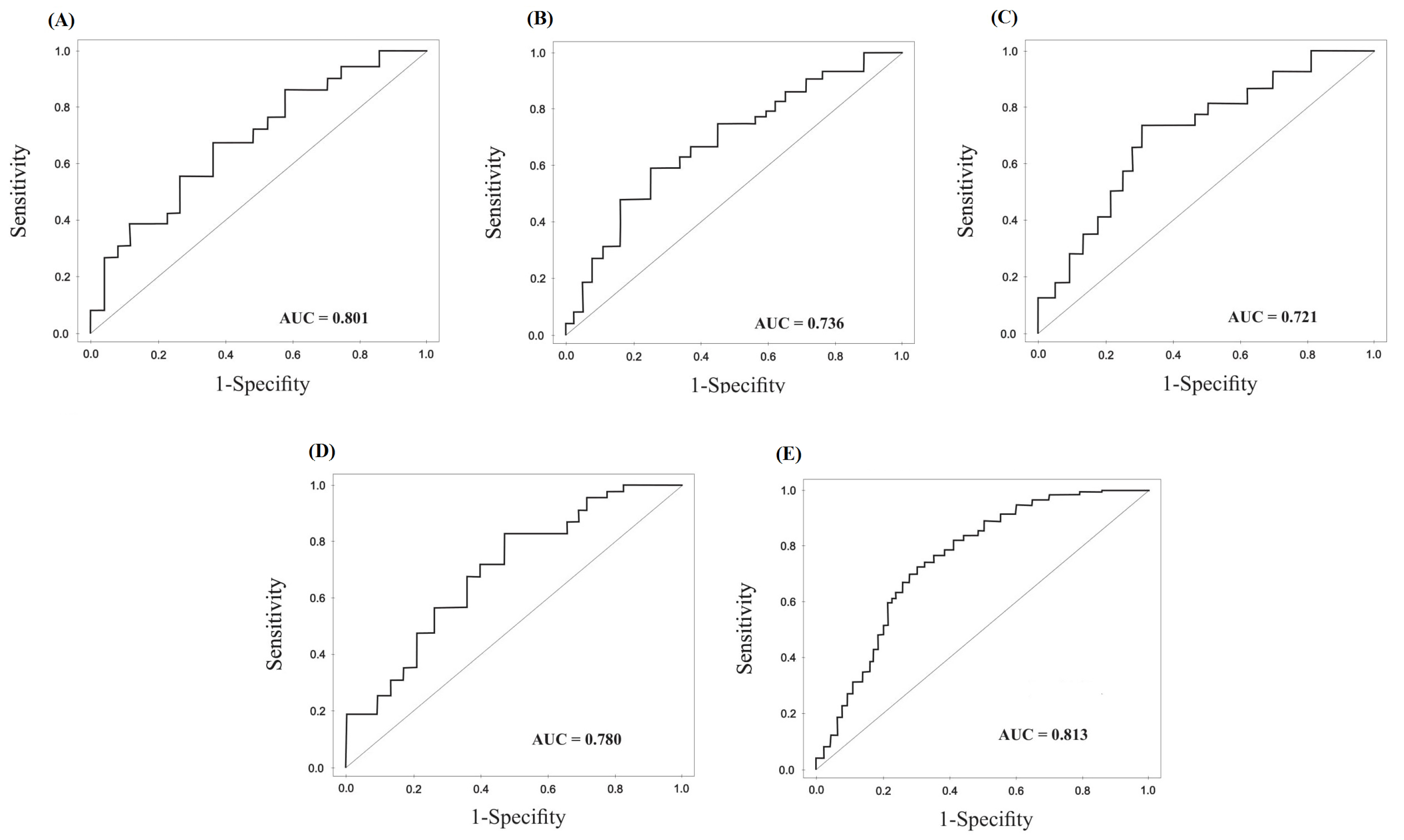
Figure 6.
Diagnostic values of exosomal miRNAs. (A) miR-10a-3p, (B) miR-215-5p, (C) miR-18a-5p, (D)miR-19b-3p in GC patients and HCs. (E) ROC curves analysis for the four combined serum exosomal miRNA signature.
.
Diagnostic values of exosomal miRNAs. (A) miR-10a-3p, (B) miR-215-5p, (C) miR-18a-5p, (D)miR-19b-3p in GC patients and HCs. (E) ROC curves analysis for the four combined serum exosomal miRNA signature.
Table 3.
Diagnostic evaluation of the tumoral and exosomal miRNAs in gastric cancer (GC) patients
|
Tumor
|
miR-10a-5p
|
miR-18a-5p
|
miR-19b-3p
|
miR-215-5p
|
Combined signature
|
| AUC |
0.824
(0.780–0.868)
|
0.718
(0.630 –0.806)
|
0.764
(0.670–0.858)
|
0.785
(0.722–0.848)
|
0.865
(0.788–0.824)
|
| Sensitivity |
85.00%
(70.16%-94.29%)
|
76.32%
(59.76%-88.56%)
|
84.38%
(67.21%-94.72%)
|
83.78%
(67.99%-93.81%)
|
82.89%
(75.95%-88.51%)
|
| Specificity |
80.43%
(66.09%-90.64%)
|
69.39%
(54.58%-81.75%)
|
70.37%
(56.39%-82.02%)
|
75.51%
(61.13%-86.66%)
|
75.26%
(68.57%-81.16%)
|
| +LR |
4.34
(2.38-7.92)
|
2.49
(1.58-3.94)
|
2.85(1.84-4.41) |
3.42
(2.05-5.71)
|
3.35
(2.59-4.33)
|
| -LR |
0.19
(0.09-0.40)
|
0.34
(0.19-0.62)
|
0.22(0.10-0.51) |
0.21
(0.10-0.45)
|
0.23
(0.16-0.33)
|
| PPV |
79.07%
(67.46%-87.32%)
|
65.91%
(55.03%-75.33%)
|
62.79%
(52.15%-72.32%)
|
72.09%
(60.76%-81.17%)
|
72.41%
(67.02%-77.22%)
|
| NPV |
86.05%
(74.42%-92.89%)
|
79.07%
(67.46%-87.32%)
|
88.37%
(76.93%-94.54%)
|
86.05%
(74.45%-92.88%)
|
84.88%
(79.68%-88.94%)
|
| Accuracy |
82.56%
(72.87%-89.90%)
|
72.41%
(61.79%-81.46%)
|
75.58%
(65.13%-84.20%)
|
79.07%
(68.95%-87.10%)
|
78.61%
(73.91%-82.82%)
|
| Exosome |
Exo-miR-10a-5p |
Exo-miR-18a-5p |
Exo-miR-19b-3p |
Exo-miR-215-5p |
Combined signature |
| AUC |
0.801
(0.747–0.855)
|
0.721
(0.609-0.833)
|
0.780
(0.709– 0.851)
|
0.736
(0.634–0.838)
|
0.813
(0.722–0.812)
|
| Sensitivity |
76.32%
(59.76%-88.56%)
|
71.79%
(55.13%-85.00%)
|
74.29%
(56.74%-87.51%)
|
68.42%
(51.35%-82.50%)
|
73.38%
(65.66%-80.17%)
|
| Specificity |
73.81%
(57.96%-86.14%)
|
70.73%
(54.46%-83.87%)
|
68.89%
(53.35%-81.83%)
|
66.67%
(50.45%-80.43%)
|
71.69%
(64.18%-78.40%)
|
| +LR |
2.91
(1.70-4.99)
|
2.45
(1.47-4.11)
|
2.39
(1.48-3.85)
|
2.05
(1.27-3.31)
|
2.59
(2.00-3.36)
|
| -LR |
0.32
(0.18-0.58)
|
0.40
(0.23-0.68)
|
0.37
(0.21-0.68)
|
0.47
(0.28-0.79)
|
0.37
(0.28-0.49)
|
| PPV |
72.50%
(60.63%-81.86%)
|
70.00%
(58.23%-79.61%)
|
65.00%
(53.56%-74.94%)
|
65.00%
(53.49%-74.99%)
|
70.62%
(64.96%-75.72%)
|
| NPV |
77.50%
(65.44%-86.24%)
|
72.50%
(60.62%-81.87%)
|
77.50%
(65.48%-86.21%)
|
70.00%
(58.24%-79.61%)
|
74.38%
(68.71%-79.33%)
|
| Accuracy |
75.00%
(64.06%-84.01%)
|
71.25%
(60.05%-80.82%)
|
70.39%
(61.11%-79.25%)
|
67.50%
(56.11%-77.55%)
|
72.50%
(67.26%-77.32%)
|
AUC is the area under the ROC curve, +LR is positive likelihood ratio, -LR is negative likelihood ratio, PPV is positive predictive value, and NPV is negative predictive values. The 95% confidence interval (95%CI) was calculated for all statistical analyses.
The AUC for exosomal miRNAs is calculated and indicated in Fig. 6. Also, the overall diagnostic values for exosomal miRNAs are represented in Table 3. The exosomal miRNAs indicate lower diagnostic values for GC in contrast to tumoral miRNAs. However, the combined signature of the exosomal miRNAs indicates acceptable diagnostic values for GC in contrast to individual exosomal miRNAs.
Correlation through tumoral and exosomal miRNA expression
The correlation between tumoral and exosomal miR-10a-5p, miR-19b-3p, miR-215-5p, and miR-18a-5p in the samples of the same patients was analyzed by Spearman's correlation coefficients test. Our findings indicated a statistically significant correlation through these miRNAs in tumor and exosome, as represented in Fig. 7. The highest correlation is calculated for mir-10a-5p with an R-squared of 0.563, and the lowest correlation is calculated for mir-215-5p with R2 of 0.191 as indicated in Table 4.
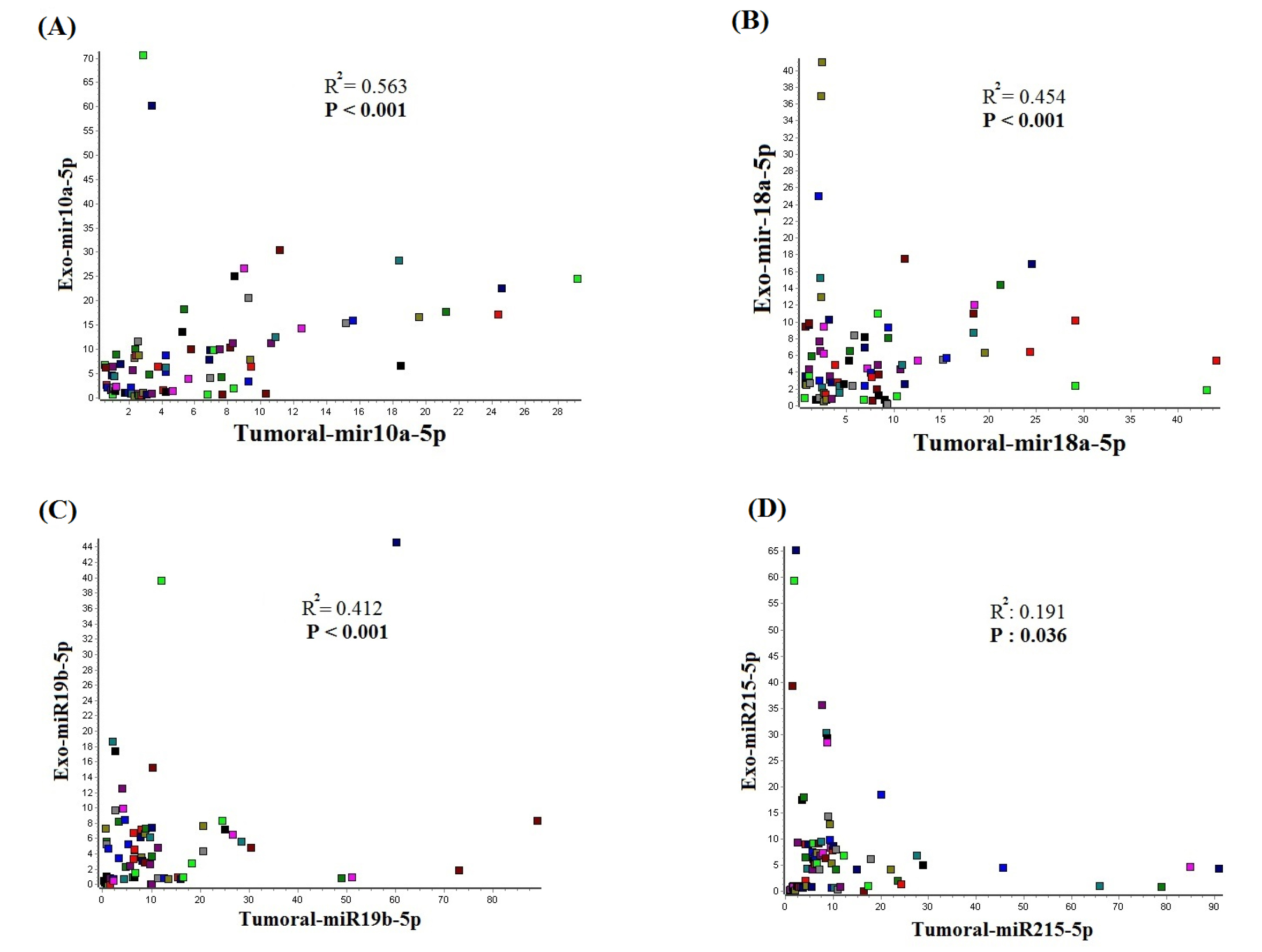
Figure 7.
Correlation analysis of exosomal and tumoral miRNAs. A p-value less than 0.05 was assumed as statistically significant difference.
.
Correlation analysis of exosomal and tumoral miRNAs. A p-value less than 0.05 was assumed as statistically significant difference.
Table 4.
Correlation between the tumoral and serum-exosomal miRNAs in gastric cancer patients
|
Tissue
|
Exosome
|
Correlation Coefficient
|
P
value
|
| Tumoral mir-10a-5p |
Exosomal mir-10a-5p |
0.563 |
<0.001 |
| Tumoral mir-18a-5p |
Exosomal mir-18a-5p |
0.454 |
<0.001 |
| Tumoral mir-19b-5p |
Exosomal mir-19b-5p |
0.412 |
<0.001 |
| Tumoral mir-215-5p |
Exosomal mir-215-5p |
0.191 |
0.036 |
Association of the expression levels of candidate miRNAs with clinic-pathological characteristics of GC patients
The association of the candidate miRNAs expression levels with demographic data of the GC patients were evaluated. There was no significant association among the age, gender, pathological variation and histological with candidate miRNAs expression level. The miR-10a-5p in tumor and serum exosome indicated significant association with tumor size (Ptumor = 0.007, Pserum exosome = 0.036 ), lymph node metastasis (Ptumor = 0.028, Pserum exosome = 0.031) and distant metastasis (Ptumor = 0.025, Pserum exosome = 0.028). Also, miR-19b-3p in tumor and serum exosome indicated significant association with tumor size (Ptumor = 0.029, Pserum exosome = 0.031 ), lymph node metastasis (Ptumor = 0.022) and distant metastasis (Ptumor = 0.027, Pserum exosome = 0.032). The miR-215-5p in tumor and serum exosome indicated significant association with tumor size (Ptumor = 0.026, Pserum exosome = 0.038 ), lymph node metastasis (Ptumor = 0.019, Pserum exosome = 0.028) and distant metastasis (Ptumor = 0.034, Pserum exosome = 0.023). The miR-18a-5p in tumor and serum exosome indicated significant association with tumor size (Ptumor = 0.016 ), lymph node metastasis (Ptumor = 0.014, Pserum exosome = 0.031) and distant metastasis (Ptumor = 0.017, Pserum exosome = 0.032). The whole association analysis for exploring the value of the exosomal and tissue derived miRNAs are indicated in Table 5 with mean fold changes and P value.
Table 5.
Association of the miRNAs expression levels with clinic-pathological variables
|
Demographic data
|
Characteristics
|
Sample
|
No. of patients
|
miR-10a-5p
Fold change
(mean ± SD)
|
P
value
|
miR-19b-3p
Fold change
(mean ± SD)
|
P
value
|
miR-215-5p
Fold change
(mean ± SD)
|
P
value
|
miR-18a-5p
Fold change
(mean ± SD)
|
P
value
|
| Age |
< 65 years
≥65 years
|
Tumor |
33
10
|
10.48 ± 0.55
11.91 ± 0.67
|
0.511 |
15.21 ± 0.29
16.37 ± 0.34
|
0.610 |
8.25 ± 0.61
9.11 ± 0.42
|
0.660 |
8.24 ± 0.32
8.13 ± 0.65
|
0.615 |
| S-Exo |
31
9
|
8.68 ± 0.71
7.19 ± 0.85
|
0.559 |
6.49 ± 0.89
7.28 ± 0.78
|
0.662 |
7.87 ± 0.71
8.09 ± 0.03
|
0.708 |
5.66 ± 0.77
5.28 ± 0.81
|
0.675 |
| Gender |
Male
Female
|
Tumor |
18
25
|
11.25 ± 0.69
10.48 ± 0.57
|
0.692 |
16.31 ± 044
15.56 ± 0.59
|
0.585 |
8.36 ± 0.61
8.22 ± 0.77
|
0.621 |
8.22 ± 0.45
8.55 ± 0.38
|
0.685 |
| S-Exo |
17
23
|
8.36 ± 0.87
8.61 ± 0.81
|
0.736 |
6.23 ± 0.49
6.31 ± 0.49
|
0.613 |
7.59 ± 0.31
6.11 ± 1.26
|
0.719 |
5.28 ± 0.76
5.78 ± 0.82
|
0.713 |
| Tumor size |
< 5 cm
≥5 cm
|
Tumor |
19
24
|
8.22 ± 0.56
10.89 ± 0.54
|
0.007
|
14.11 ± 0.29
18.59 ± 0.36
|
0.029
|
6.57 ± 0.55
8.27 ± 0.27
|
0.026
|
6.24 ± 0.36
8.76 ± 0.41
|
0.016
|
| S-Exo |
19
21
|
6.22 ± 0.81
8.74 ± 0.85
|
0.036
|
4.62 ± 0.82
7.48 ± 0.65
|
0.031
|
7.91 ± 0.64
9.39 ± 0.53
|
0.038
|
4.76 ± 0.65
6.36 ± 0.75
|
0.023
|
| Histological grade |
P-differentiated
M-differentiated
W-differentiated
|
Tumor |
11
21
11
|
10.62 ± 0.61
10.76 ± 0.58
11.46 ± 0.47
|
0.238 |
15.25 ± 0.24
15.37 ± 0.11
14.64 ± 0.31
|
0.243 |
8.25 ± 0.68
8.05 ± 0.71
8.33 ± 0.48
|
0.478 |
8.34 ± 0.21
7.83 ± 0.56
8.41 ± 0.25
|
0.421 |
| S-Exo |
11
18
11
|
8.25 ± 0.54
7.76 ± 0.58
8.46 ± 0.42
|
0.375 |
6.32 ± 0.67
6.17 ± 0.73
5.74 ± 0.61
|
0.428 |
7.52 ± 0.81
6.93 ± 0.92
7.77 ± 0.15
|
0.552 |
5.33 ± 0.58
4.25 ± 0.55
5.71 ± 0.61
|
0.596 |
| Lymph node metastasis |
Positive
Negative
|
Tumor |
26
17
|
10.76 ±0.43
8.85 ± 0.45
|
0.028
|
17.73 ± 0.63
14.29 ± 0.41
|
0.022
|
9.13 ± 0.45
7.49 ± 0.29
|
0.019
|
8.49 ± 0.17
5.3 ± 0.45
|
0.014
|
| S-Exo |
24
16
|
8.45 ± 0.75
6.32 ± 0.63
|
0.031
|
6.24 ±0.43
7.18 ±0.37
|
0.234 |
8.99 ± 0.35
6.08 ± 0.15
|
0.028
|
6.51 ± 1.07
4.24 ± 0.65
|
0.031
|
| Distant metastasis |
Positive
Negative
|
Tumor |
13
30
|
11.35 ± 0.48
9.73 ± 0.29
|
0.025
|
17.25 ± 0.33
15.34 ± 0.13
|
0.027
|
10.68 ± 0.51
8.25 ± 0.22
|
0.034
|
8.64 ± 0.52
6.26 ± 0.35
|
0.017
|
| S-Exo |
13
27
|
9.25 ± 0.81
7.36 ± 0.73
|
0.028
|
8.76 ±0.47
6.56 ±0.35
|
0.032
|
8.18 ± 0.15
6.36 ± 0.36
|
0.023
|
6.45 ± 0.85
4.73 ± 0.93
|
0.032
|
| pathological variants |
Intestinal
diffuse
|
Tumor |
36
7
|
10.86 ± 0.51
9.51 ± 0.63
|
0.456 |
16.31 ± 0.53
16.69 ± 0.63
|
0.489 |
9.27 ± 0.34
9.57 ± 0.25
|
0.316 |
8.69 ± 0.27
8.21 ± 0.65
|
0.445 |
| S-Exo |
33
7
|
7.73 ± 0.71
8.21 ± 0.43
|
0.541 |
7.35 ± 0.54
7.64 ± 0.83
|
0.454 |
8.07 ± 0.85
7.72 ± 0.66
|
0.357 |
4.19 ± 0.94
4.24 ± 0.75
|
0.521 |
The bold characters indicate that the P value is less than 0.05, which has statistical significance. S-Exo: serum-derived exosome. SD: Standard deviation. Poorly-differentiated: p-differentiated, Moderately-differentiated: M-differentiated, Well-differentiated: W-differentiated.
Discussion
Early diagnosis can increase the survival rates of cancer patients through timely treatment. Currently, GC diagnosis is mainly based on invasive and non-invasive approaches which do not meet the minimum sensitivity and specificity for supporting the early detection of gastric tumors in many cases.
4,5
Therefore, most of the patients are diagnosed with advanced tumors and suffer poor prognosis due to metastasis.
10,11
Signature of miRNAs can utilize to detect the origin of the unknown tissues, also tumor tissues, but the instability of the cell-free miRNAs is the main obstacle for accurate detection approaches.
13
The main problem seems to be the lack of concordance for miRNA signatures reported from over 154 studies by different research groups that leads to a lack of a single diagnostic signature.
16
Also, the other problem is the lack of correlation between expression levels of cell-free miRNAs and tumoral tissues.
17
Recent studies have confirmed the secretion of exosomes by tumor cells into the body fluids, exosomes can mirror the origin cell functional status and exosomal miRNAs are considerable sources for biomarker studies.
13,19
Tumor cells are able to release a greater amount of exosomes into the circulation in contrast to normal cells which is due to improved access to the vascular system, and potentially increasing the signal-to-noise ratio of cancer biomarkers that can be applied for the early detection and screening of cancer.
28
Hence, this study was conducted to study gastric tumor-specific dysregulated miRNAs in serum exosomes to evaluate the possibility of clinical application of these markers in diagnosis.
In this study, we applied QIAGEN miRCURY LNA miRNA Focus PCR Panels to explore the differential expression profiling of miRNAs in 6 gastric tumors, 6 non-cancerous adjacent tissue and 6 GC patient’s serum exosome as pooled samples for the screening phase and proceeded with validation of candidate miRNAs by qRT-PCR. The candidate miRNAs from the screening phase (miRNAs with the highest fold change) were evaluated between different groups including gastric tumors with tumor-adjacent non-cancerous tissue and GC patient’s serum exosomes with HCs serum exosomes. The serum was collected following 12h fasting from the GC patients and HCs. We found that miR-10a-5p, miR-19b-3p, miR-215-5p and miR-18a-5p were significantly upregulated in serum exosomes. We proposed a model including four miRNAs (miR-10a-5p, miR-19b-3p, miR-215-5p, and miR-18a-5p) in serum exosome which can use to diagnose the GC with an AUCs of 0.813.
MiR-10a-5p can modulate the progression of the cell cycle, cell proliferation, metastasis and apoptosis in the benefit of cancerization in the gastric cells.
29
In vitro studies indicated that miR-10a-5p is upregulated in exosomes of five GC cell lines (MKN45, SGC7901, NCI-N87, and AGS) in contrast to normal gastric cell line (GES-1).
30
Also, it is reported that miR-10a-5p can suppress the expression of the MAPK8IP1 which is the main mechanism for metastasis of GC.
31
Also, miR-10a can promote migration and invasion in a gastric cell line (BGC823), the proposed mechanism for this phenomenon is targeting of RPL23.
32
These data support the direct role of miR-10a-5p in gastric tumors which confirms the specify of the serum exosomal miR-10a-5p as a diagnostic marker for GC. MiR-19b-3p upregulation in serum exosomes of the GC patients is reported previously.
19,33
There is doubt about the oncogenic or tumor-suppressive role of the miR-19b-3p in GC.
34
MiR-215 is reported to be downregulated in a number of the cancers, however, this molecule is preferentially overexpressed in GC, also miR-215 can promote the development of GC by targeting the RUNX1.
35
MiR-215-5p is able to establish cancerization in gastric cells.
29
There are a few studies about the role of the exosomal miR-215-5p in GC, we indicated that miR-215-5p is over-expressed in serum exosome of the GC patients. MiR-18a is reported to be over-expressed in gastric tumors in contrast to normal tissues,
36
also plasma levels of miR-18a are higher in GC patients comparing to plasma of HCs.
37
MiR-18a can modify the expression of P53 in GC by suppressing IRF2 and therefore can modulate the apoptosis in GC.
36
However, there is no study until now about the role of the exosomal miR-18a-5p in the diagnosis of the GC. Our results indicated that miR-18a-5p is upregulated in serum exosomes. GC patient’s serum exosomal miRNA indicates promising power for GC diagnosis.
The expression pattern of the identified miRNAs in serum exosomes was in concordance with tumoral miRNA expression profile and indicated a direct association with gastric tumors expression profile. The candidate miRNAs association with clinicopathological features of the patients were analyzed. The results indicated a significant association with tumor size, lymph node metastasis and distance metastasis. however, there was no significant association with age, gender, histological grade and pathological variants.
Exosomes exhibit significant potential for application as biomarkers in cancer studies but there are certain limitations for their application. Exosome isolation is one of the most challenging issues, also most scientists utilize ultracentrifugation as a common method for exosome isolation but there are certain limitations including co-isolation of non-exosomal impurities and low reproducibility of this method.
38
It has been reported that variety in the exosome isolation method could change the final miRNA profile of these vesicles. Therefore, the application of an appropriate method for exosome isolation from body fluids is still the most critical aspect.
39
Furthermore, it is reported that 21% to 99% of vesicle-free miRNAs (depending on the individual miRNA) can precipitate during exosome isolation by precipitation-based extracellular vesicle isolation methods.
40
Therefore, the main hurdle for application of exosomes and their compartment in the clinic and early diagnosis of the cancers is lack of robust and reproducible technique besides the presence of contaminants such as similarly sized micro-vesicles of different origins, proteins and cell-free RNAs.
41
Also, it is not clear that what amount of the exosomes can support the extrapolation for early detection and screening of the different malignancies.
42
To enable exosome-based miRNA biomarkers to be beneficial in the clinical diagnosis, the reproducibility and performance must be evidently examined. Application of the universally accepted standard sets can efficiently improve the reproducibility of the various laboratories result. Furthermore, clinical validation with relatively large sample sizes and a sufficient quantity of specimens is an obligatory approach to produce reliable results among the different researchers.
27
We believe that there is a great demand for further studies on the exosomal miRNAs as emerging biomarkers for establishing an accurate isolation method with a high reproducibility outcome.
Conclusion
In this study, we could identify the upregulated onco-miRNAs in gastric tumors in contrast to non-cancerous tumor-adjacent tissue. We found that these miRNAs also were upregulated in serum-derived exosomes which introduce them as possible non-invasive biomarkers for early diagnosis of gastric cancer by liquid sampling, however, lots of studies are needed for clinical application.
Acknowledgments
Authors gratefully appreciate the Molecular Medicine Research Center and Department of Molecular Medicine, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, for supporting this work.
Funding sources
This study was financially supported by the Molecular Medicine Research Center at Tabriz University of Medical Sciences (grant No. IR.TBZMED.REC.1396.910).
Ethical statement
The current research was carried out according to The Code of Ethics of the World Medical Association (Declaration of Helsinki), and informed consent was obtained. This study was approved by the Institutional Board Review and Ethics Committee of Tabriz University of Medical Sciences (IR.TBZMED.REC.1396.910).
Competing interests
The authors declare there is no conflict of interest.
Authors’ contribution
Author contributions are as follows: H.K and N.S designed and performed the study. A.K contributed to sample size calculation. H.K, M.M, SH.H and A.E collected the patient data. H.K performed the laboratory tests and data analysis. H.K drafted the manuscript, prepared the tables and figures, and N.S and MRS critically read the manuscript. N.S, MRS and MS.H revised the manuscript. All authors have read the journal's authorship agreement and the manuscript has been reviewed by and approved by all named authors.
Research Highlights
What is the current knowledge?
√ Exosomes are emerging nano-vesicles with a significant role in cancer development and metastasis
What is new here?
√ Exosomal miRNAs are capable molecules for developing non-invasive approaches for the early diagnosis of gastric cancer. Application of the serum-derived exosomes can reduce the time for early diagnosis with high accuracy in contrast to current blood-based biomarkers.
References
- Kahroba H, Hejazi MS, Samadi N. Exosomes: from carcinogenesis and metastasis to diagnosis and treatment of gastric cancer. Cell Mol Life Sci 2019; 76:1747-58. doi: 10.1007/s00018-019-03035-2 [Crossref] [ Google Scholar]
- Dicken BJ, Bigam DL, Cass C, Mackey JR, Joy AA, Hamilton SM. Gastric adenocarcinoma: review and considerations for future directions. Ann Surg 2005; 241:27-39. doi: 10.1097/01.sla.0000149300.28588.23 [Crossref] [ Google Scholar]
- Gülçiçek OB, Solmaz A, Özdoğan K, Erçetin C, Yavuz E, Yiğitbaş H. Primary squamous cell carcinoma of the stomach. Ulus Cerrahi Derg 2016; 32:221-3. doi: 10.5152/ucd.2015.2811 [Crossref] [ Google Scholar]
- Feng F, Tian Y, Xu G, Liu Z, Liu S, Zheng G. Diagnostic and prognostic value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer 2017; 17:737. doi: 10.1186/s12885-017-3738-y [Crossref] [ Google Scholar]
- Kanda M, Suh YS, Park DJ, Tanaka C, Ahn SH, Kong SH. Serum levels of ANOS1 serve as a diagnostic biomarker of gastric cancer: a prospective multicenter observational study. Gastric Cancer 2020; 23:203-11. doi: 10.1007/s10120-019-00995-z [Crossref] [ Google Scholar]
- Moon JS. Screening Upper Endoscopy for Early Detection of Gastric Cancer. J Korean Med Sci 2018; 33:e190. doi: 10.3346/jkms.2018.33.e190 [Crossref] [ Google Scholar]
- González CA, Agudo A. Carcinogenesis, prevention and early detection of gastric cancer: where we are and where we should go. Int J Cancer 2012; 130:745-53. doi: 10.1002/ijc.26430 [Crossref] [ Google Scholar]
- Maeda O, Ando T, Ohmiya N, Ishiguro K, Watanabe O, Miyahara R. Alteration of gene expression and DNA methylation in drug-resistant gastric cancer. Oncol Rep 2014; 31:1883-90. doi: 10.3892/or.2014.3014 [Crossref] [ Google Scholar]
- An X, Sarmiento C, Tan T, Zhu H. Regulation of multidrug resistance by microRNAs in anti-cancer therapy. Acta Pharm Sin B 2017; 7:38-51. doi: 10.1016/j.apsb.2016.09.002 [Crossref] [ Google Scholar]
- Jung HY, Fattet L, Yang J. Molecular pathways: linking tumor microenvironment to epithelial-mesenchymal transition in metastasis. Clin Cancer Res 2015; 21:962-8. doi: 10.1158/1078-0432.ccr-13-3173 [Crossref] [ Google Scholar]
- Yang W, Ma J, Zhou W, Cao B, Zhou X, Yang Z. Molecular mechanisms and theranostic potential of miRNAs in drug resistance of gastric cancer. Expert Opin Ther Targets 2017; 21:1063-75. doi: 10.1080/14728222.2017.1389900 [Crossref] [ Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97. doi: 10.1016/s0092-8674(04)00045-5 [Crossref] [ Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D. MicroRNA expression profiles classify human cancers. Nature 2005; 435:834-8. doi: 10.1038/nature03702 [Crossref] [ Google Scholar]
- Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol 2010; 11:136-46. doi: 10.1016/s1470-2045(09)70343-2 [Crossref] [ Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008; 105:10513-8. doi: 10.1073/pnas.0804549105 [Crossref] [ Google Scholar]
- Jarry J, Schadendorf D, Greenwood C, Spatz A, van Kempen LC. The validity of circulating microRNAs in oncology: five years of challenges and contradictions. Mol Oncol 2014; 8:819-29. doi: 10.1016/j.molonc.2014.02.009 [Crossref] [ Google Scholar]
- Wang J, Wang Q, Liu H, Hu B, Zhou W, Cheng Y. MicroRNA expression and its implication for the diagnosis and therapeutic strategies of gastric cancer. Cancer Lett 2010; 297:137-43. doi: 10.1016/j.canlet.2010.07.018 [Crossref] [ Google Scholar]
- Aslan C, Maralbashi S, Salari F, Kahroba H, Sigaroodi F, Kazemi T. Tumor-derived exosomes: Implication in angiogenesis and antiangiogenesis cancer therapy. J Cell Physiol 2019; 234:16885-903. doi: 10.1002/jcp.28374 [Crossref] [ Google Scholar]
- Wang N, Wang L, Yang Y, Gong L, Xiao B, Liu X. A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem Biophys Res Commun 2017; 493:1322-8. doi: 10.1016/j.bbrc.2017.10.003 [Crossref] [ Google Scholar]
- Wang J, Liu Y, Sun W, Zhang Q, Gu T, Li G. Plasma exosomes as novel biomarker for the early diagnosis of gastric cancer. Cancer Biomark 2018; 21:805-12. doi: 10.3233/cbm-170738 [Crossref] [ Google Scholar]
- Helwa I, Cai J, Drewry MD, Zimmerman A, Dinkins MB, Khaled ML. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS One 2017; 12:e0170628. doi: 10.1371/journal.pone.0170628 [Crossref] [ Google Scholar]
- Riazifar M, Mohammadi MR, Pone EJ, Yeri A, Lässer C, Segaliny AI. Stem Cell-Derived Exosomes as Nanotherapeutics for Autoimmune and Neurodegenerative Disorders. ACS Nano 2019; 13:6670-88. doi: 10.1021/acsnano.9b01004 [Crossref] [ Google Scholar]
- Xie S, Zhu Q, Qu W, Xu Z, Liu X, Li X. sRNAPrimerDB: comprehensive primer design and search web service for small non-coding RNAs. Bioinformatics 2019; 35:1566-72. doi: 10.1093/bioinformatics/bty852 [Crossref] [ Google Scholar]
- Salone V, Rederstorff M. Stem-loop RT-PCR based quantification of small non-coding RNAs. Methods Mol Biol 2015; 1296:103-8. doi: 10.1007/978-1-4939-2547-6_10 [Crossref] [ Google Scholar]
- Paulaitis M, Agarwal K, Nana-Sinkam P. Dynamic Scaling of Exosome Sizes. Langmuir 2018; 34:9387-93. doi: 10.1021/acs.langmuir.7b04080 [Crossref] [ Google Scholar]
- Petersen KE, Manangon E, Hood JL, Wickline SA, Fernandez DP, Johnson WP. A review of exosome separation techniques and characterization of B16-F10 mouse melanoma exosomes with AF4-UV-MALS-DLS-TEM. Anal Bioanal Chem 2014; 406:7855-66. doi: 10.1007/s00216-014-8040-0 [Crossref] [ Google Scholar]
- Wang W, Young MR, Srivastava S. MicroRNA Biomarkers for Early Detection of Cancer. In: Srivastava S, ed. Biomarkers in Cancer Screening and Early Detection. 2017. p. 27-36.
- Lee J, Ghosh S, Srivastava S. Exosomes: A Valuable Biomedical Tool in Biomarker Discovery and Development. In: Srivastava S, ed. Biomarkers in Cancer Screening and Early Detection. 2017. p. 50-63.
- Pereira AL, Magalhães L, Moreira FC, Reis-das-Mercês L, Vidal AF, Ribeiro-Dos-Santos AM. Epigenetic Field Cancerization in Gastric Cancer: microRNAs as Promising Biomarkers. J Cancer 2019; 10:1560-9. doi: 10.7150/jca.27457 [Crossref] [ Google Scholar]
- Ren J, Zhou Q, Li H, Li J, Pang L, Su L. Characterization of exosomal RNAs derived from human gastric cancer cells by deep sequencing. Tumour Biol 2017; 39:1010428317695012. doi: 10.1177/1010428317695012 [Crossref] [ Google Scholar]
- Lu Y, Wei G, Liu L, Mo Y, Chen Q, Xu L. Direct targeting of MAPK8IP1 by miR-10a-5p is a major mechanism for gastric cancer metastasis. Oncol Lett 2017; 13:1131-6. doi: 10.3892/ol.2016.5544 [Crossref] [ Google Scholar]
- Yang O, Huang J, Lin S. Regulatory effects of miRNA on gastric cancer cells. Oncol Lett 2014; 8:651-6. doi: 10.3892/ol.2014.2232 [Crossref] [ Google Scholar]
- Fu M, Gu J, Jiang P, Qian H, Xu W, Zhang X. Exosomes in gastric cancer: roles, mechanisms, and applications. Mol Cancer 2019; 18:41. doi: 10.1186/s12943-019-1001-7 [Crossref] [ Google Scholar]
- Zhang T, Liu C, Huang S, Ma Y, Fang J, Chen Y. A Downmodulated MicroRNA Profiling in Patients with Gastric Cancer. Gastroenterol Res Prac 2017; 2017:1526981. doi: 10.1155/2017/1526981 [Crossref] [ Google Scholar]
- Li N, Zhang QY, Zou JL, Li ZW, Tian TT, Dong B. miR-215 promotes malignant progression of gastric cancer by targeting RUNX1. Oncotarget 2016; 7:4817-28. doi: 10.18632/oncotarget.6736 [Crossref] [ Google Scholar]
- Chen YJ, Wu H, Zhu JM, Li XD, Luo SW, Dong L. MicroRNA-18a modulates P53 expression by targeting IRF2 in gastric cancer patients. J Gastroenterol Hepatol 2016; 31:155-63. doi: 10.1111/jgh.13041 [Crossref] [ Google Scholar]
- Tsujiura M, Komatsu S, Ichikawa D, Shiozaki A, Konishi H, Takeshita H. Circulating miR-18a in plasma contributes to cancer detection and monitoring in patients with gastric cancer. Gastric Cancer 2015; 18:271-9. doi: 10.1007/s10120-014-0363-1 [Crossref] [ Google Scholar]
- Konoshenko MY, Lekchnov EA, Vlassov AV, Laktionov PP. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed Res Int 2018; 2018:8545347. doi: 10.1155/2018/8545347 [Crossref] [ Google Scholar]
- Ludwig N, Whiteside TL, Reichert TE. Challenges in Exosome Isolation and Analysis in Health and Disease. Int J Mol Sci 2019; 20. doi: 10.3390/ijms20194684 [Crossref]
- Karttunen J, Heiskanen M, Navarro-Ferrandis V, Das Gupta S, Lipponen A, Puhakka N. Precipitation-based extracellular vesicle isolation from rat plasma co-precipitate vesicle-free microRNAs. J Extracell Vesicles 2019; 8:1555410. doi: 10.1080/20013078.2018.1555410 [Crossref] [ Google Scholar]
- Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 2012; 56:293-304. doi: 10.1016/j.ymeth.2012.01.002 [Crossref] [ Google Scholar]
- Zhang HG, Grizzle WE. Exosomes: a novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am J Pathol 2014; 184:28-41. doi: 10.1016/j.ajpath.2013.09.027 [Crossref] [ Google Scholar]