Bioimpacts. 12(3):203-210.
doi: 10.34172/bi.2021.23219
Original Research
Expression of functional eGFP-fused antigen-binding fragment of ranibizumab in Pichia pastoris
Shirin Movaghar Asareh , #  , Tahereh Savei , #
, Tahereh Savei , #  , Sareh Arjmand , *
, Sareh Arjmand , *  , Seyed Omid Ranaei Siadat , *
, Seyed Omid Ranaei Siadat , *  , Fataneh Fatemi , Mehrab Pourmadadi , Javad Shabani Shayeh
, Fataneh Fatemi , Mehrab Pourmadadi , Javad Shabani Shayeh
Author information:
Protein Research Center, Shahid Beheshti University, Tehran, Iran
#These authors are equally contributed to work.
Abstract
Introduction:
Ranibizumab is a mouse monoclonal antibody fragment antigen-binding (Fab) against human vascular endothelial growth factor-A (VEGF-A), inhibiting angiogenesis. This antibody is commercially produced in Escherichia coli host and used to treat wet age-related macular degeneration (AMD).
Methods:
In this study, the heavy and light chains of ranibizumab were expressed in Pichia pastoris. The expressed chains were incubated overnight at 4°C for interaction. The formation of an active structure was evaluated based on the interaction with substrate VEGF-A using an indirect ELISA, and an electrochemical setup. Furthermore, reconstruction of split enhanced green fluorescent protein (eGFP) reporter, chimerized at the C-terminus of the heavy and light chains, was used to characterize chains’ interaction.
Results:
P. pastoris efficiently expressed designed constructs and secreted them into the culture medium. The anti-Fab antibody detected the constructed Fab structure in western blot analysis. Reconstruction of the split reporter confirmed the interaction between heavy and light chains. The designed ELISA and electrochemical setup results verified the binding activity of the recombinant Fab structure against VEGF-A.
Conclusion:
In this work, we indicated that the heavy and light chains of ranibizumab Fab fragments (with or without linkage to split parts of eGFP protein) were produced in P. pastoris. The fluorescence of reconstructed eGFP was detected after incubating the equal ratio of chimeric-heavy and light chains. Immunoassay and electrochemical tests verified the bioactivity of constructed Fab. The data suggested that P. pastoris could be considered a potential efficient eukaryotic host for ranibizumab production.
Keywords: Ranibizumab, Fab fragment, VEGF-A, Pichia pastoris, Split reporter, eGFP
Introduction
Uncontrolled expression of vascular endothelial growth factor (VEGF-A) is the key contributor to diabetic macular edema (DME) and age-related macular degeneration (AMD), which are the most common causes of vision loss in diabetic patients and aged individual.
1-3
Several anti-VEGF-A agents have been approved for the treatment of visual impairment due to macular edema and degeneration that improve the visual function in the majority of patients.
4,5
Ranibizumab is a recombinant humanized monoclonal antibody fragment (IgG kappa isotype) that binds to human VEGF-A and prevents its interaction with VEGF-A receptors on the surface of endothelial cells. This antibody is produced in Escherichia coli and marketed as Lucentis® to treat patients with wet AMD and diabetic macular edema.
3,6,7
Ranibizumab was established as the fragment antigen-binding (Fab fragment) because it was thought the smaller size enhanced penetration through all retina layers.
8
Structurally, ranibizumab contains ten cysteine residues that lead to the four intrachain and one interchain disulfide bonds. Each ranibizumab molecule has one binding site for VEGF-A, compared to two binding sites for its parental full structure antibody. The high-resolution structure of the complex between VEGF-A and ranibizumab is available (Fig. 1, PDB ID: 1CZ8).
9
Due to its production in the prokaryotic host, ranibizumab does not carry any N-glycosylation.
10
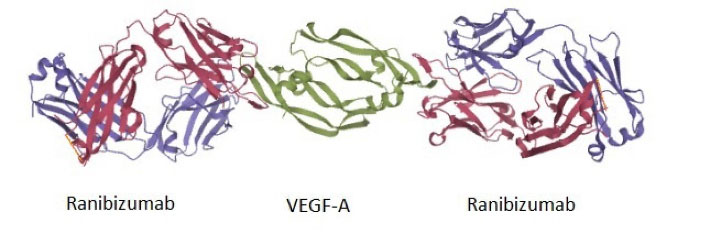
Fig. 1.
The X-ray coordinates from the PDB for VEGF-A engagement of ranibizumab (PDB ID: 1CZ8). Two molecules of ranibizumab bind to one VEGF dimer (green structure). The heavy and light chains of ranibizumab are shown in dark pink and purple colors, respectively. The only disulfide bond between the C-terminus of two chains in each Ab is visible as orange lines.
.
The X-ray coordinates from the PDB for VEGF-A engagement of ranibizumab (PDB ID: 1CZ8). Two molecules of ranibizumab bind to one VEGF dimer (green structure). The heavy and light chains of ranibizumab are shown in dark pink and purple colors, respectively. The only disulfide bond between the C-terminus of two chains in each Ab is visible as orange lines.
Pichia pastoris (currently known as Komagataella phaffii) is a eukaryote microorganism that offers additional advantages over E. coli. Incorporating post-translational modifications, including disulfide-bond formation, facilitates proper folding of recombinant protein produced in this host. Furthermore, lack of endotoxin, high productivity, ease of genetic manipulation, and rapid growth to high cell densities make P. pastoris a favorable host for the cost-effective expression of various recombinant proteins. Simplified purification of recombinant secretory proteins, owing to relatively low amounts of endogenous protein in the extracellular medium, can positively affect the cost price. These advantages make this host a potentially compelling alternative to other hosts which are used currently for the production of expensive drugs, including antibodies and antibody fragments.
11,12
Several therapeutic antibodies are produced in P. pastoris expression system, such as two recombinant therapeutic antibody fragments that are already in the clinical development process (Nanobody® ALX-0061, and Nanobody® ALX00171),
13
and eptinezumab that is a fully-humanized IgG1 antibody, approved by the FDA in February 2020 for the preventive treatment of migraine headaches.
14
The recombinant production of therapeutic antibodies requires accurate synthesis, correct folding, and dimerization of two heavy and light chains.
15
There is a broad range of analytical techniques to characterize the dimerization and correct structural formation of proteins, including antibodies. Having the proper functionality implies the correct folding and/or di(poly)merization of a protein or protein complex in an indirect manner.
16,17
Protein complementation assay (PCA), also called the split system, is one of the approaches that are well suited to detect protein interaction in vitro and particularly in vivo. This method is based on the tagging of two (or more) proteins with the rationally designed fragments of a reporter protein.
18
The interaction between reporter protein fragments brings them close enough to enable their non-covalent reassembly, which was recognized by gaining its activity (enzymatic activity or fluorescence emission).
19
Reconstructing the split-fluorescent protein fragments is one of the simplest methods in PCA, which requires no substrate for the production of fluorescence.
20
The main aim of the present study is to produce the ranibizumab Fab fragment in P. pastoris. For this purpose, different constructs were designed to express the heavy and light chains of the Fab fragment. The two chains were expressed individually and incubated for interaction. Reconstruction of enhanced green fluorescent protein (eGFP) reporter was utilized to characterize the interaction of chains. The binding activity of the produced Fab was examined by designing an indirect ELISA experiment and electrochemical biosensor to detect its interaction with VEGF-A.
Materials and Methods
Bacterial and yeast strains, vector, media, and culture condition
The pPINKαHC plasmid was used for gene cloning and expression. E. coli DH5α was used for recombinant plasmid propagation. P. pastoris PichiaPinkTM expression system strain 4 (ade2, pep2, pep4) was used as the host for the expression of target genes. Luria-Bertani (LB) medium (0.5% (w/v) yeast extract, 1% (w/v) tryptone, 1% (w/v) NaCl), supplemented with 100 µg/mL ampicillin, was prepared for cultivation of recombinant E. coli at appropriate condition (37°C with 200 rpm shaking). P. pastoris was grown in buffered glycerol-complex medium (BMGY) (1% (w/v) yeast extract, 2% (w/v) peptone, 100 mM potassium phosphate, 1.34% (w/v) yeast nitrogen base (YNB), 4×10−5% (w/v) biotin, 1% (v/v) glycerol, pH 6.0) and induced in buffered methanol-complex medium (BMMY) (same as the BMGY medium except with 0.5% (v/v) methanol instead of the glycerol). The yeast culture condition was 28-30°C and 250 rpm shaking.
Preparation of target constructs and cloning in P. pastoris
The following constructs were designed for the expression of heavy and light chains of ranibizumab and evaluation of their interaction: I) the heavy chain of ranibizumab, II) the light chain of ranibizumab, III) eGFP (GenBank: AAF62891.1), IV) the heavy chain of ranibizumab linked from its C-terminal to the N-terminal of eGFP (amino acids 1-157) via (G4S)2 Linker, and V) the light chain of ranibizumab linked from its C-terminal to the C-terminal of eGFP (amino acids 158-238) via (G4S)2 Linker. The sequences coding for desired constructs were artificially synthesized according to the codon preference of P. pastoris (Shanghai Generay Biotech Co.). All constructs were cloned in the pPink-αHC plasmid vector under the control of the AOX1 promoter, in-frame with the α-MF signal peptide, and using the XhoI and KpnI restriction enzymes. Transformation of the recombinant plasmid into competent E. coli cells was performed using a heat shock method (90 seconds at 42°C) and screened according to the resistance to ampicillin antibiotic. The accuracy of cloning was confirmed using PCR with specific primers and sequencing.
The recombinant plasmids were linearized using the BsptI restriction enzyme and transformed to the P. pastoris cells using the electroporation apparatus (Gene Pulser, Bio-Rad, USA) according to the Easy Select TM Pichia Expression Kit instructions (Invitrogen, USA). The transformed mixture was spread on PAD plates and incubated at 30°C for ten days. The empty pPink-αHC plasmid was transformed and used as the negative control. Each clone was verified using PCR and sequencing. The obtained verified recombinant yeasts were maintained at -80°C, in 20% (v/v) glycerol solution, for further applications.
Expression of recombinant constructs
For each construct, three recombinant clones were cultivated in the BMGY medium. 50 ml of each culture (~OD600 = 30) were centrifuged at 1500 g for 5 minutes, and the pelleted cells were re-suspended in 50 mL BMMY for induction of constructs expression under the control of AOX1 promoter. The induction was continued for the next four days with 1% methanol. The supernatants of the cultures containing the secreted recombinant proteins were collected for the subsequent experiments. For constructing the Fab fragment structure, the supernatants of two yeast cultures producing heavy and light chains were mixed with equal ratios and incubated overnight at 4°C. The non-recombinant P. pastoris was cultivated as the negative control.
Denaturing SDS-PAGE and native PAGE
The supernatants of cell cultures were mixed with a 2X loading buffer containing (0.2 M Tris, 20% glycerol (v/v), 10% SDS (w/v), 0.05% bromophenol blue (w/v), pH 6.8), heated for 10 min in boiling water, and subjected to denaturing SDS-PAGE. The preparation of electrophoresis was performed according to the standard Laemmli method.
21
The concentration of the separating gel was 12%, and the stacking gel was 5%. The native gel, which minimizes the proteins’ denaturation, was applied to study heavy and light chains’ interaction in the absence of denaturant agents. For native-PAGE, the samples were mixed with a 2x loading buffer (without SDS) and heated for one min. The gel was prepared as the same as SDS-PAGE but without the addition of SDS. Proteins were visualized by silver staining as described by Chevallet et al.
22
Western blot
The incubated mixture of heavy and light chains (24 hours at 4°C) was subjected to the native-PAGE. Western blotting was performed according to the previously described method.
23
Briefly, the resolved proteins were electro-blotted from the native-PAGE to polyvinylidene difluoride (PVDF) membrane (Millipore, USA), in transferring buffer (0.025 MTris, 0.19 Mglycine, and 20% (v/v) methanol), overnight at 20 V/4°C. The membrane was blocked with PBS-T-BSA (PBS, 0.1% (v/v) Tween 20, 1% (w/v) BSA) for 2 hours. 500-fold diluted goat anti-human IgG (Fab specific)-peroxidase antibody (Sigma, A0293) was used to detect Fab structure on the gel. The protein band related to the Fab structures were visualized using DAB substrate solution (1.5 mM 3,3′-diaminobenzidine, 0.06 % (v/v) H 2O2 in PBS, pH 7.2).
Binding activity of ranibizumab to the substrate
Two studies were designed to monitor the interaction between ranibizumab and its target molecule, VEGF-A. In the first study, an indirect ELISA assay with immobilized VEGF-A molecules was used, and in the latter approach, an electrochemical biosensor setup was planned and employed.
ELISA
An indirect ELISA assay was developed to detect either the formation of Fab structure and interaction of the structured Fab with VEGF-A substrate. For this purpose, the supernatants of two yeast cultures producing heavy and light chains were mixed in a 1:1 volume ratio and incubated overnight at 4°C. The supernatants of non-recombinant yeast and heavy chain alone were used as the negative controls, and Lucentis® (Novartis) was used as standard. The recombinant human VEGF-A protein (R&D systems, 293-VE) was coated onto the plate wells in 1 µg/mL concentration and incubated for 6 hours. The wells were washed three times with PBST buffer (PBS containing 0.1% tween 20) and blocked for 2 hours at room temperature with PBS containing 2% skim milk. The washing step was repeated, and the mixtures diluted with PBS (pH 7.4) were used in the volume of 100 µL per well. The plate was incubated overnight at 4°C. 100 µL of anti-human IgG (Fab specific)-peroxidase antibody, diluted 1: 14000 in PBS, were added to the samples, and the plate was incubated at room temperature for 2 hours. After washing, the substrate (0.1 mg/mL TMB, phosphate citrate buffer 0.1 M pH 5.0, 0.02% hydrogen peroxide) was added to each well and incubated for 20 minutes at room temperature. The reaction was stopped using sulfuric acid 2 N, and the plate was read at 450 nm. The PBS (pH 7.4) and bovine serum albumin (1 µg/mL) were used as negative controls. All the standards and samples were run in triplicate.
Electrochemical assay of binding activity
The previously designed electrochemical biosensor composed of reduced graphene oxide (RGO) and gold nanoparticles (AuNPs), and with excellent selectivity for VEGF-A was reconstructed.
24
Synthesis of RGO/AuNPs composite and preparation of nanoprobe was conducted as described earlier.
25
Recombinant heavy and light chains individually, and then incubated mixture of them (24 hours at 4°C), were immobilized covalently on the surface of RGO/AuNPs. Four microliters of RGO/AuNPs-Fab was dropped on the electrode’s surface and placed directly under a normal lamp light to dry. The unbounded RGO/AuNPs-Fab was washed with 100 mM PBS (pH 7.4). The experiment was equally conducted for the heavy and light chains of ranibizumab linked to N- and C-terminals of eGFP and their incubated mixture. Ten microliters of VEGF-A with different concentrations (from 0.002 to 20 µg/mL) was added to the biosensor cell with the scan rate of 50 mV/s in the presence of 0.1 M hexacyanoferrate solution, and then the volumetric responses were recorded. The bare electrode and the experiment without the addition of VEGF-A were used as the controls. Electrochemical measurements were performed using IviumStat XR (a potentiostat/galvanostat with frequency response analyzer), and cyclic voltammetry (CV) test was used to illustrate the performance of materials.
Spectrofluorimetry
The eGFP split reporter was used to study the recombinant ranibizumab heavy and light chain interaction. Two complementary segments of eGFP were fused via a linker to the recombinant heavy and light chains. To analyze the complementation of the segments, the equal amounts of supernatants of two yeast cultures producing heavy and light chains with complementary parts of eGFP were mixed and incubated for 24 hours at 4°C. In another experiment, the cited mixture was incubated with substrate VEGF-A. The supernatants of yeast cultures producing non-chimeric heavy and light chains and the mixture of them were evaluated as well. The empty BMGY medium and the supernatant of eGFP producing yeasts were negative and positive controls, respectively. The intensity of fluorescence was measured using the spectrofluorometer (Shimadzu RF-5000, Japan) with 470 nm excitation and 520 nm emission.
Results
Construction of recombinant yeasts and expression of designed constructs
The correct construction of recombinant plasmid pPICZA containing the five designed constructs (Fig. 2) was confirmed by PCR, using specific primers and sequencing.
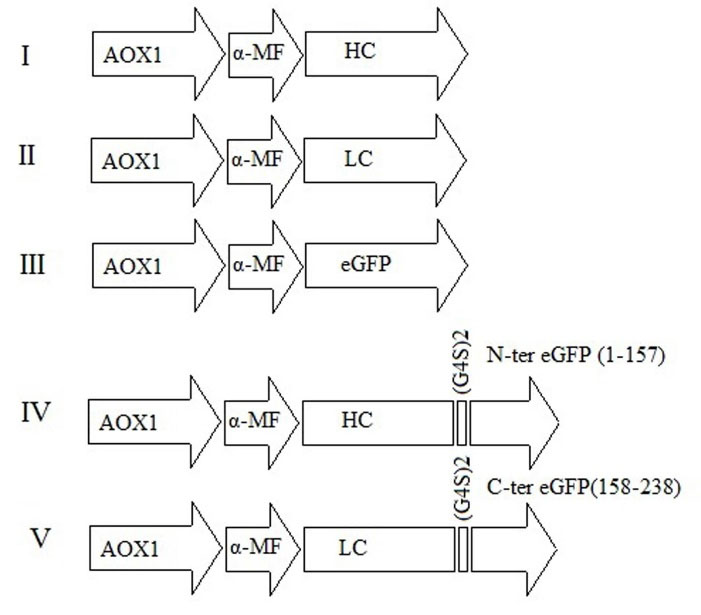
Fig. 2.
Design of five constructs for expression of heavy and light chains of ranibizumab. All constructs are under the control of the AOX1 promoter and in-frame with α-MF, which directs the secretion of recombinant protein.
.
Design of five constructs for expression of heavy and light chains of ranibizumab. All constructs are under the control of the AOX1 promoter and in-frame with α-MF, which directs the secretion of recombinant protein.
The transformant P. pastoris clones, which incorporated the recombinant constructs in its genome (under the control of AOX1 promoter), was used for their expression in a secretory manner. After 96 hours of induction with methanol, the supernatants of recombinant yeasts were subjected to denaturing and native SDS-PAGE. The results showed that P. pastoris efficiently produces all the designed constructs and secrete them using the α-MF signal sequence. The construction of the Fab fragment was studied in native PAGE. The western blot results on native gel indicated that anti-human IgG (Fab specific) could detect the Fab structure on the native gel for the heavy or light chains when subjected to the gel individually and when the mixture was used. The Fab-specific antibody could not recognize the Fab structure fabricated from the mixture of heavy and light chains linked to eGFP split reporter (Fig. 3).
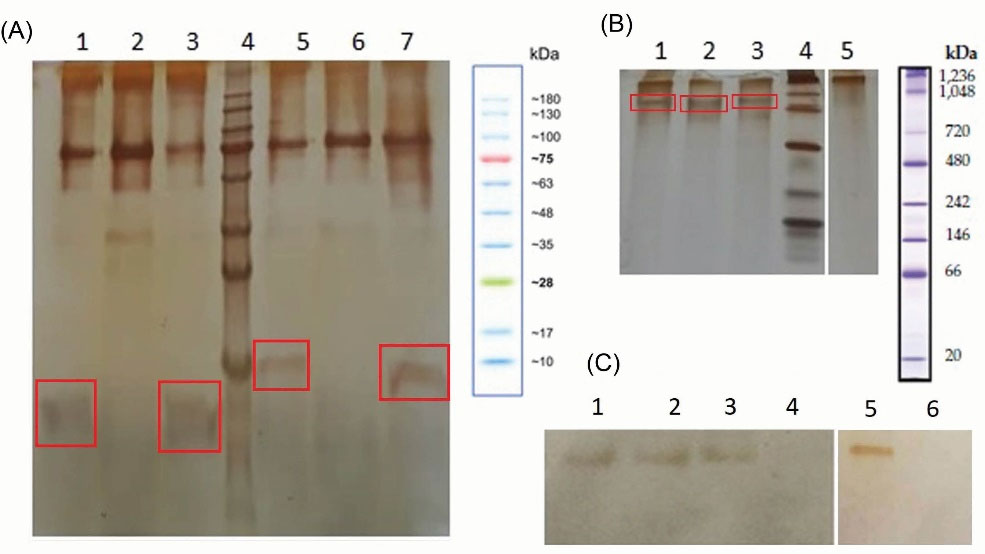
Fig. 3.
A) Denaturing SDS-PAGE of recombinant ranibizumab chains. Lanes 1, 3; heavy and light chains (~ 23 kDa), lane 5 and 7; heavy and light chains of ranibizumab linked to N-, and C-terminals of eGFP (~ 28 kDa), lanes 2 and 6; negative controls, and lane 4 is protein marker. The target proteins are shown in the square boxes. B) Native PAGE of recombinant ranibizumab chains. Lane 1; the incubated mixture of heavy and light chains, lane 2 and 3; heavy and light chains, lane 4; native gel protein marker, and lane 5; negative control. C) Western blot of native gel. Lane 1; heavy chain, lane 2; light chain, lane 3 and 5; the incubated mixture of heavy and light chains, lane 4; negative control, lane 6; the incubated mixture of heavy and light chains, chimerized with fragments of eGFP.
.
A) Denaturing SDS-PAGE of recombinant ranibizumab chains. Lanes 1, 3; heavy and light chains (~ 23 kDa), lane 5 and 7; heavy and light chains of ranibizumab linked to N-, and C-terminals of eGFP (~ 28 kDa), lanes 2 and 6; negative controls, and lane 4 is protein marker. The target proteins are shown in the square boxes. B) Native PAGE of recombinant ranibizumab chains. Lane 1; the incubated mixture of heavy and light chains, lane 2 and 3; heavy and light chains, lane 4; native gel protein marker, and lane 5; negative control. C) Western blot of native gel. Lane 1; heavy chain, lane 2; light chain, lane 3 and 5; the incubated mixture of heavy and light chains, lane 4; negative control, lane 6; the incubated mixture of heavy and light chains, chimerized with fragments of eGFP.
Binding activity of constructed Fab
An indirect ELISA experiment was conducted to detect the interaction of fabricated Fab with the ranibizumab substrate, VEGF-A. The dilution series of Lucentis® was used to generate the standard curve (Fig. S1, Supplementary file 1). According to the results, the incubated mixture of heavy and light chains can bind to VEGF-A, and the measured quantity for the fabricated Fab was ~ 30 µg/mL. No binding activity was detected for heavy and light chains alone. Furthermore, the Fab structure’s binding activity fabricated from the eGFP split reporter-linked chains was not detected. No signal was founded for negative controls.
The CV is an electrochemical test and approved method to show the binding affinity. The recorded volumetric responses are shown in Fig. 4. According to the obtained results, after immobilization of the incubated mixture of heavy and light chains (with or without linkage to split parts of eGFP protein) on the electrode’s surface, the electrochemically active surface area was decreased. The addition of different concentrations of VEGF-A to the prepared electrode led to further decreases in the surface area that has a negative ratio with the VEGF-A concentration and confirmed the binding activity. The immobilization of heavy and light chains individually showed no interaction with VEGF-A and was overplotted with the Ab line. As it is clear, the loading of the electrode with reconstructed Fab linked with the split parts of eGFP led to more reduction (~15%) in the electrochemically active area, compared to the non-chimeric Fab, which can be attributed to its heavier weight.
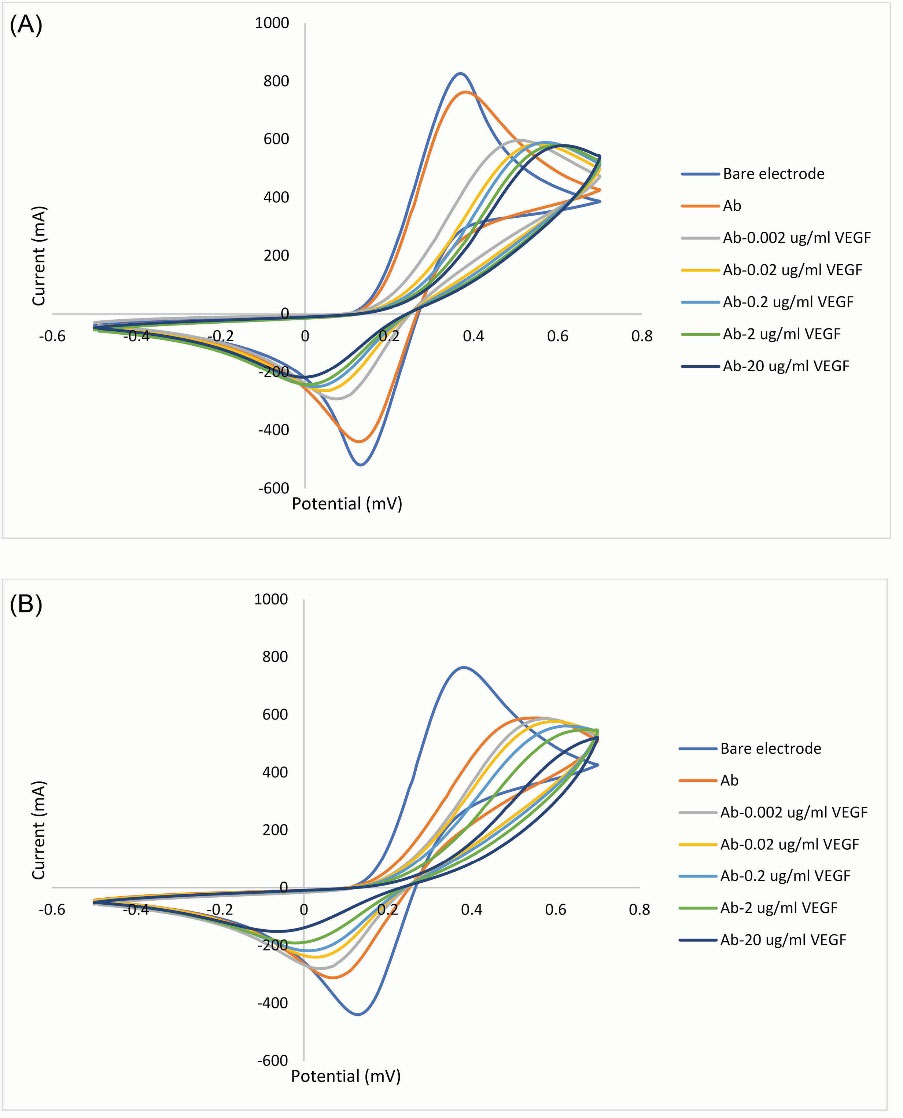
Fig. 4.
Electrochemical CV curves RGO/AuNPs electrode in the presence of 0.1 M Hexacyanoferrate solution and at the scan rate of 50 mV/s. Plots A and B are related to the structured Fab without and with the reconstituted eGFP split reporter, respectively. Bare RGO/AuNPs electrode; (A) after immobilization of incubated mixture of heavy and light chains; (B) after the addition of serial dilutions of VEGF-A. The redox peak of CV curves is related to the electrochemical reaction of Fe+3/+2 ions. The step-down reduction in the electrochemically active surface area confirmed the binding activity of structured Fab.
.
Electrochemical CV curves RGO/AuNPs electrode in the presence of 0.1 M Hexacyanoferrate solution and at the scan rate of 50 mV/s. Plots A and B are related to the structured Fab without and with the reconstituted eGFP split reporter, respectively. Bare RGO/AuNPs electrode; (A) after immobilization of incubated mixture of heavy and light chains; (B) after the addition of serial dilutions of VEGF-A. The redox peak of CV curves is related to the electrochemical reaction of Fe+3/+2 ions. The step-down reduction in the electrochemically active surface area confirmed the binding activity of structured Fab.
Analysis of eGFP reconstitution
The spectroscopy results revealed the reconstitution of split fragments and confirmed the interaction between heavy and light chains and the Fab fragment construction. As it is shown in Fig. 5 the culture medium has an intrinsic fluorescence. The measured fluorescence for the supernatant of non-recombinant clones and the other clones that secret the chimeric and non-chimeric heavy or light chains, and the mixture incubated of non-chimeric heavy and light chains were at the base level of empty medium. A significant increase was detected in the fluorescence of the incubated mixture of chimeric heavy and light chains (with split fragments), which was measured with the same quantity when incubated with VEGF-A as well. The clone of P. pastoris producing recombinant eGFP was used as the positive control.
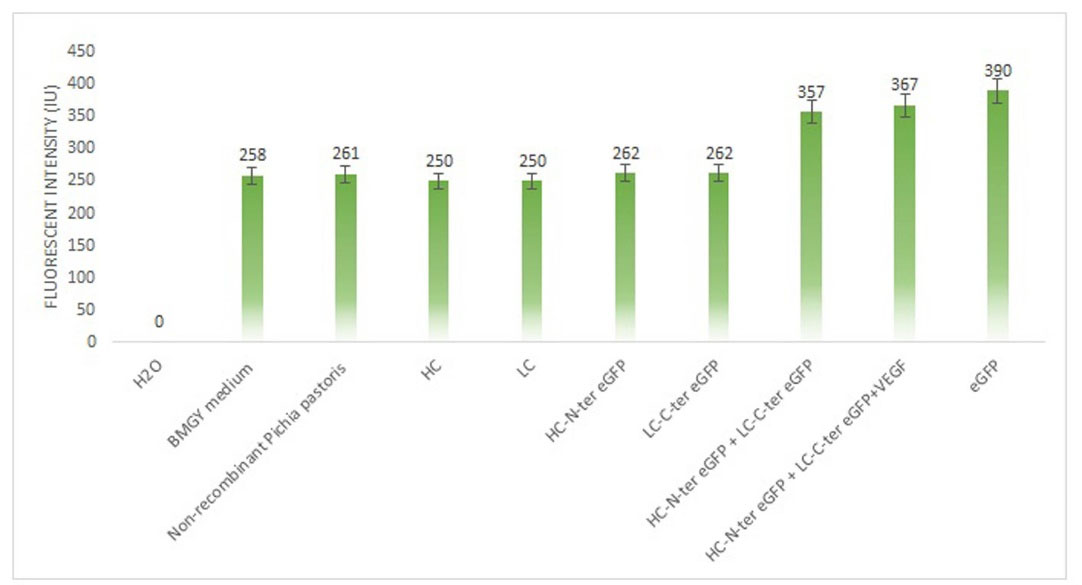
Fig. 5.
Study the heavy and light chain interaction using split-eGFP. Incubation of heavy and light chains (chimerized with fragments of eGFP), with and without VEGF-A substrate, led to a significant increase in the measured fluorescence. The clones producing intact eGFP protein was used as the positive control. HC: heavy chain; LC: light chain.
.
Study the heavy and light chain interaction using split-eGFP. Incubation of heavy and light chains (chimerized with fragments of eGFP), with and without VEGF-A substrate, led to a significant increase in the measured fluorescence. The clones producing intact eGFP protein was used as the positive control. HC: heavy chain; LC: light chain.
Discussion
Ranibizumab is an effective but expensive drug for DME and AMD, which is commercially produced in E. coli. Host change is a promising strategy to improve recombinant proteins’ production and making cost objects more efficient.
26
Different expression systems have been introduced to produce Fab fragments, but none is regarded as optimal.
27
The study scope herein described was to produce the ranibizumab Fab fragments in the new host, P. pastoris. This host’s advantages, importantly easy scale-up and reducing the secretary protein purification efforts, make it an attractive expression system for large-scale production of heterologous proteins with therapeutic use.
28
Additionally, the successful production of functional mAbs and Fab fragments by yeast P. pastoris suggested this non-conventional yeast strain for the upcoming project.
29-31
The heavy and light chain constructs were cloned and expressed separately, and their interaction to generate Fab fragments was evaluated by the construction of split reporter eGFP protein. eGFP is a mutant variant of wild-type GFP with brighter fluorescence intensity that makes the detection much more sensitive.
32
The eGFP was cleaved into two fragments between residues 157 and 158 and were fused to the heavy and light chains. After expression, equal amounts of supernatant from the media of recombinant clones were mixed and incubated for 24 hours at 4°C. The measured fluorescence intensity from the mixture of two chimeric chains was comparable with the intact eGFP, expressed as secretary protein. After incubation of the mixture with the VEGF-A, similar fluorescence intensity was taken. Neither the incubation of non-chimeric nor the intact heavy or light chains alone showed an increase in the fluorescence signal. The results suggested that dissected eGFP parts complement each other after appropriate interaction between heavy and light chains and Fab reconstruction. Since rich media, such as BMGY, tends to has high-background fluorescence,
33
a basic level of fluorescence has been detected in the negative controls.
The construction of the Fab structure was tested by anti-Fab specific Ab in western blot, as well. The bands appeared in the lanes related to the individual and mixture of non-chimeric heavy and light chains. It seems that heavy and light chains can interact individually, and the generated structures were distinguishable by anti-Fab Ab. The absence of signal for the mixture of chimeric chains suggested that anti-Fab detected the Fab structure’s N-terminal, here occupied by cleaved eGFP fragments.
Besides, the anti-Fab specific Ab was used to confirm the molecular interaction between the constructed Fab and the VEGF-A substrate in an indirect ELISA, designed for this study. The VEGF-A was coated on the plate, and the signal can only be obtained if the Fab structure interacts with it. The absence of signals in the plate wells that contained only heavy chains or only light chains indicated that although each chain molecule may non-specifically interact with each other and make Fab-likes, these structures cannot detect the substrate. In other words, the antigen-binding site at the N-terminal is not constructed except that the interaction happens between heavy and light chains.
The designed electrochemical method has shown the ability to generate an electrochemical signal in response to specific target binding. The results proved a high affinity of the immobilized constructed Fab fragments to VEGF-A by showing the decrease in the maximum peak of CV. Chimerization with the splitted fragments of eGFP showed no adverse effects on antigen-binding of the constructed Fab fragment.
Research Highlights
What is the current knowledge?
√ Ranibizumab is a monoclonal antibody Fab fragment that binds to human VEGF-A, and inhibits angiogenesis.
√ Ranibizumab is commercially produced in E. coli.
What is new here?
√
Pichia pastoris successfully express the heavy and light chains of ranibizumab.
√ Reconstruction of split eGFP reporter, chimerized at the C-terminus of the heavy and light chains, verified chains’ interaction.
√ The interaction of constructed Fab structure and VEGF-A can be verified using electrochemical measurement.
Conclusion
Our data shows that the yeast host P. pastoris could be considered a potential new host for ranibizumab production. The heavy and light chains of this monoclonal Ab were produced in a secretory manner, and the Fab structure was obtained easily by incubation of the equal ratio of the chains at 4°C. The appropriate interaction of chains was confirmed using a split reporter system based on the eGFP protein. The results of immunoassay and electrochemical tests verified the potential of constructed Fab for inhibition of VEGF-A. The studies for appropriate purification of recombinant heavy and light chains, determination of the kinetics of chain interaction, and antigen-binding activity remain to be conducted. Also, further optimizations for protein productivity are undoubtedly possible.
Acknowledgments
The authors would like to thank the Sobhan Recombinant Protein Co. for its material and technical support.
Funding sources
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethical statement
Not applicable.
Competing interests
The authors declare there is no conflict of interest.
Authors’ contribution
TS, SMA: Performing the molecular and biochemical tests, data handling. SA: Conceptualization, experiments design, data analysis, study validation, writing, and reviewing. SORA: Supervision, study validation. FF: Consultation, study validation. MP, JSS: performing the electrochemical test.
Supplementary Materials
Supplementary file 1 contains Figure S1.
(pdf)
References
- Kauppinen A, Paterno JJ, Blasiak J, Salminen A, Kaarniranta K. Inflammation and its role in age-related macular degeneration. Cell Mol Life Sci 2016; 73:1765-86. doi: 10.1007/s00018-016-2147-8 [Crossref] [ Google Scholar]
- Al-Zamil WM, Yassin SA. Recent developments in age-related macular degeneration: a review. Clin Interv Aging 2017; 12:1313-30. doi: 10.2147/CIA.S143508 [Crossref] [ Google Scholar]
- Krispel C, Rodrigues M, Xin X, Sodhi A. Ranibizumab in diabetic macular edema. World J Diabetes 2013; 4:310-8. doi: 10.4239/wjd.v4.i6.310 [Crossref] [ Google Scholar]
- Stefanini FR, Badaro E, Falabella P, Koss M, Farah ME, Maia M. Anti-VEGF for the management of diabetic macular edema. J Immunol Res 2014; 2014:632307. doi: 10.1155/2014/632307 [Crossref] [ Google Scholar]
- Schmidt-Erfurth U, Chong V, Loewenstein A, Larsen M, Souied E, Schlingemann R. Guidelines for the management of neovascular age-related macular degeneration by the European Society of Retina Specialists (EURETINA). Br J Ophthalmol 2014; 98:1144-67. doi: 10.1136/bjophthalmol-2014-305702 [Crossref] [ Google Scholar]
- Ziemssen F, Wachtlin J, Kuehlewein L, Gamulescu MA, Bertelmann T, Feucht N. Intravitreal Ranibizumab Therapy for Diabetic Macular Edema in Routine Practice: Two-Year Real-Life Data from a Non-interventional, Multicenter Study in Germany. Diabetes Ther 2018; 9:2271-89. doi: 10.1007/s13300-018-0513-2 [Crossref] [ Google Scholar]
- Mayor R, Agarwal M, Singh S, Venkatesh R. Ranibizumab for diabetic macular edema. Ophthalmology 2013; 120:221. doi: 10.1016/j.ophtha.2012.07.069 [Crossref] [ Google Scholar]
- Ferrara N, Damico L, Shams N, Lowman H, Kim R. Development of ranibizumab, an anti-vascular endothelial growth factor antigen binding fragment, as therapy for neovascular age-related macular degeneration. Retina 2006; 26:859-70. doi: 10.1097/01.iae.0000242842.14624.e7 [Crossref] [ Google Scholar]
- Walker A, Chung C-W, Neu M, Burman M, Batuwangala T, Jones G. Novel interaction mechanism of a domain antibody-based inhibitor of human vascular endothelial growth factor with greater potency than ranibizumab and bevacizumab and improved capacity over aflibercept. J Biol Chem 2016; 291:5500-11. doi: 10.1074/jbc.M115.691162 [Crossref] [ Google Scholar]
- Zou L, Lai H, Zhou Q, Xiao F. Lasting controversy on ranibizumab and bevacizumab. Theranostics 2011; 1:395-402. [ Google Scholar]
- Weinacker D, Rabert C, Zepeda AB, Figueroa CA, Pessoa A, Farias JG. Applications of recombinant Pichia pastoris in the healthcare industry. Braz J Microbiol 2013; 44:1043-8. doi: 10.1590/s1517-83822013000400004 [Crossref] [ Google Scholar]
- Goncalves AM, Pedro AQ, Maia C, Sousa F, Queiroz JA, Passarinha LA. Pichia pastoris: a recombinant microfactory for antibodies and human membrane proteins. J Microbiol Biotechnol 2013; 23:587-601. doi: 10.4014/jmb.1210.10063 [Crossref] [ Google Scholar]
- Kim H, Yoo SJ, Kang HA. Yeast synthetic biology for the production of recombinant therapeutic proteins. FEMS Yeast Res 2015; 15:1-16. doi: 10.1111/1567-1364.12195 [Crossref] [ Google Scholar]
- Dhillon S. Eptinezumab: First Approval. Drugs 2020; 80:733-9. [ Google Scholar]
- Gil D, Schrum AG. Strategies to stabilize compact folding and minimize aggregation of antibody-based fragments. Adv BiosciBiotechnol 2013; 4:73-84. doi: 10.4236/abb.2013.44A011 [Crossref] [ Google Scholar]
- Deperalta G, Alvarez M, Bechtel C, Dong K, McDonald R, Ling V. Structural analysis of a therapeutic monoclonal antibody dimer by hydroxyl radical footprinting. MAbs 2013; 5:86-101. doi: 10.4161/mabs.22964 [Crossref] [ Google Scholar]
- Gell DA, Grant RP, Mackay JP. The detection and quantitation of protein oligomerization. Adv Exp Med Biol 2012; 747:19-41. doi: 10.1007/978-1-4614-3229-6_2 [Crossref] [ Google Scholar]
- Michnick SW, Remy I, Campbell-Valois FX, Vallee-Belisle A, Pelletier JN. Detection of protein-protein interactions by protein fragment complementation strategies. Methods Enzymol 2000; 328:208-30. doi: 10.1016/s0076-6879(00)28399-7 [Crossref] [ Google Scholar]
- Morell M, Ventura S, Aviles FX. Protein complementation assays: approaches for the in vivo analysis of protein interactions. FEBS Lett 2009; 583:1684-91. doi: 10.1016/j.febslet.2009.03.002 [Crossref] [ Google Scholar]
- Moustaqil M, Bhumkar A, Gonzalez L, Raoul L, Hunter DJB, Carrive P. A Split-Luciferase Reporter Recognizing GFP and mCherry Tags to Facilitate Studies of Protein-Protein Interactions. Int J Mol Sci 2017; 18. doi: 10.3390/ijms18122681 [Crossref]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227:680-5. doi: 10.1038/227680a0 [Crossref] [ Google Scholar]
- Chevallet M, Luche S, Rabilloud T. Silver staining of proteins in polyacrylamide gels. Nat Protoc 2006; 1:1852-8. doi: 10.1038/nprot.2006.288 [Crossref] [ Google Scholar]
- Arjmand S, Bidram E, Lotfi AS, Shamsara M, Mowla SJ. Expression and Purification of Functionally Active Recombinant Human Alpha 1-Antitrypsin in Methylotrophic Yeast Pichia pastoris. Avicenna J Med Biotechnol 2011; 3:127-34. [ Google Scholar]
- Arjmand S, Tavasoli Z, Ranaei Siadat SO, Saeidi B, Tavana H. Enhancing chimeric hydrophobin II-vascular endothelial growth factor A165 expression in Pichia pastoris and its efficient purification using hydrophobin counterpart. Int J Biol Macromol 2019; 139:1028-34. doi: 10.1016/j.ijbiomac.2019.08.080 [Crossref] [ Google Scholar]
- Pourmadadi M, Shayeh JS, Arjmand S, Omidi M, Fatemi F. An electrochemical sandwich immunosensor of vascular endothelial growth factor based on reduced graphene oxide/gold nanoparticle composites. Microchemical Journal 2020; 159:105476. doi: 10.1016/j.microc.2020.105476 [Crossref] [ Google Scholar]
- Jafarian V, Bagheri K, Zarei J, Karami S, Ghanavatian P. Improved expression of recombinant sweet-tasting brazzein using codon optimization and host change as new strategies. Food Biotechnol 2020; 34:62-76. [ Google Scholar]
- Lebozec K, Jandrot-Perrus M, Avenard G, Favre-Bulle O, Billiald P. Quality and cost assessment of a recombinant antibody fragment produced from mammalian, yeast and prokaryotic host cells: A case study prior to pharmaceutical development. New Biotechnol 2018; 44:31-40. doi: 10.1016/j.nbt.2018.04.006 [Crossref] [ Google Scholar]
- Baeshen MN, Bouback TA, Alzubaidi MA, Bora RS, Alotaibi MA, Alabbas OT. Expression and purification of C-peptide containing insulin using Pichia pastoris expression system. BioMed Res Int 2016; 2016:3423685. doi: 10.1155/2016/3423685 [Crossref] [ Google Scholar]
- Shah KA, Clark JJ, Goods BA, Politano TJ, Mozdzierz NJ, Zimnisky RM. Automated pipeline for rapid production and screening of HIV-specific monoclonal antibodies using Pichia pastoris. BiotechnolBioeng 2015; 112:2624-9. doi: 10.1002/bit.25663 [Crossref] [ Google Scholar]
- Jiang Y, Li F, Zha D, Potgieter TI, Mitchell T, Moore R. Purification process development of a recombinant monoclonal antibody expressed in glycoengineered Pichia pastoris. Protein Expr Purif 2011; 76:7-14. doi: 10.1016/j.pep.2010.11.004 [Crossref] [ Google Scholar]
- Ohkuri T, Murase E, Sun SL, Sugitani J, Ueda T. Characterization of deamidation at Asn138 in L-chain of recombinant humanized Fab expressed from Pichia pastoris. J Biochem 2013; 154:333-40. doi: 10.1093/jb/mvt061 [Crossref] [ Google Scholar]
- Yang TT, Cheng L, Kain SR. Optimized codon usage and chromophore mutations provide enhanced sensitivity with the green fluorescent protein. Nucleic Acids Res 1996; 24:4592-3. doi: 10.1093/nar/24.22.4592 [Crossref] [ Google Scholar]
- Chalfie M, Kain SR. Green Fluorescent Protein: Properties, Applications and Protocols: John Wiley & Sons; 2005.