Bioimpacts. 12(2):139-146.
doi: 10.34172/bi.2021.23378
Original Research
The active lung microbiota landscape of COVID-19 patients through the metatranscriptome data analysis
Yang Han 1, 2, #  , Zhilong Jia 1, 2, , # *
, Zhilong Jia 1, 2, , # *  , Jinlong Shi 1, 2, Weidong Wang 1, 2, *, Kunlun He 1, 2, *
, Jinlong Shi 1, 2, Weidong Wang 1, 2, *, Kunlun He 1, 2, *
Author information:
1Key Laboratory of Biomedical Engineering and Translational Medicine, Ministry of Industry and Information Technology, Medical Innovation Research Division of Chinese PLA General Hospital, Beijing, China
2Beijing Key Laboratory for Precision Medicine of Chronic Heart Failure, Medical Innovation Research Division of Chinese PLA General Hospital, Beijing, China
#These authors contributed equally.
Abstract
Introduction:
With the outbreak of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the interaction between the host and SARS-CoV-2 was widely studied. However, it is unclear whether and how SARS-CoV-2 infection affects lung microflora, which contribute to COVID-19 complications.
Methods:
Here, we analyzed the metatranscriptomic data of bronchoalveolar lavage fluid (BALF) of 19 COVID-19 patients and 23 healthy controls from 6 independent projects and detailed the active microbiota landscape in both healthy individuals and COVID-19 patients.
Results:
The infection of SARS-CoV-2 could deeply change the lung microbiota, evidenced by the α-diversity, β-diversity, and species composition analysis based on bacterial microbiota and virome. Pathogens (e.g., Klebsiella oxytoca causing pneumonia as well), immunomodulatory probiotics (e.g., lactic acid bacteria and Faecalibacterium prausnitzii, a butyrate producer), and Tobacco mosaic virus (TMV) were enriched in the COVID-19 group, suggesting a severe microbiota dysbiosis. The significant correlation between Rothia mucilaginosa, TMV, and SARS-CoV-2 revealed drastic inflammatory battles between the host, SARS-CoV-2, and other microbes in the lungs. Notably, TMV only existed in the COVID-19 group, while human respirovirus 3 (HRV 3) only existed in the healthy group. Our study provides insights into the active microbiota in the lungs of COVID-19 patients and would contribute to the understanding of the infection mechanism of SARS-CoV-2 and the treatment of the disease and complications.
Conclusion:
SARS-COV-2 infection deeply altered the lung microbiota of COVID-19 patients. The enrichment of several other pathogens, immunomodulatory probiotics (lactic acid or butyrate producers), and TMV in the COVID-19 group suggests a complex and active lung microbiota disorder.
Keywords: Microbiota, SARS-CoV-2, COVID-19, Lactic acid bacteria, Faecalibacterium prausnitzii
Copyright and License Information
© 2022 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
With the outbreak of severe acute respiratory syndrome coronavirus 2, SARS-CoV-2,
1
leading to coronavirus disease 2019 (COVID-19), many studies on clinical characteristics, three-dimensional structure
2
and interactions between host and virus
3
were reported. Moreover, immunodominant regions of the target antigen was identified through the immunoinformatics approaches.
4
The symptoms of COVID-19 patients (COV) usually include fever, vomiting, diarrhea.
5
As bacterial or viral infections could also induce several symptoms among them, it is necessary to profile the microbiota of COVID-19 patients to effectively relieve their symptoms.
6,7
The bronchoalveolar lavage fluid (BALF), containing microenvironment information on bronchioles and lung alveoli from the lower respiratory tract, is one of key sample types for characterizing the host inflammatory response and microbiota of COVID-19 patients, as lung is one of the main organs infected with SARS-CoV-2.
8,9
The human microbiota in BALF, including bacterial microbiota and virome, is a diverse microbial ecosystem associated with beneficial or deleterious physiological functions, as well as disease etiologies, including COVID-19.
10
It was reported that multiple common respiratory pathogens were co-infected with SARS-CoV-2 in COVID-19 patients.
11
Moreover, the role of commensal viruses in human lungs is poorly understood, particularly in the respiratory tract.
12
Besides, there are close interactions between microorganisms including symbiosis and competition.
13,14
Therefore, the relationship between SARS-CoV-2 and other microorganisms in the body is of great significance for studying its infection mechanism and developing effective treatments. Shen et al briefly reported that the bacterial microbiota in COVID-19 patients was dominated by the pathogens and oral and upper respiratory commensal bacteria.
15
However, there is no systematic microbiota landscape in BALF samples from COVID-19 patients, as well as the healthy individuals.
In this study, we systematically profiled the transcriptionally active microbiota landscape in BALF from COVID-19 patients and healthy individuals, identified microorganism composition in healthy individuals and COVID-19 patients, found disease-specific active microbes in the COVID-19 patient group, and revealed the interaction between several bacteria or viruses and SARS-CoV-2. The systematic microbiota landscape provided crucial insight into understanding the mechanism of SARS-CoV-2 infection and its treatment.
Materials and Methods
Data collection and preprocessing
The raw metatranscriptomic data of 42 BALF samples, consisting 19 COVID-19 patients and 23 healthy controls and two negative controls (NC), were downloaded in FASTQ format from Sequence Read Archive, EMBL-EBI ArrayExpress and National Genomics Data Center, China with project IDs, PRJCA002202, PRJNA601736, PRJCA002326, PRJNA605983, PRJNA615032, and PRJNA434133. All publicly available transcriptome data sets of the COVID-19, as far as we knew, were included in our study. The basic information of these samples is described in Supplementary file 1. The raw fastq data were analyzed using IDSeq web-server
16
(Pipeline v4.9) to obtain the raw read count of each bacterial and viral species. Briefly, the raw reads underwent removal of host reads (Hg38 reference genome) via STAR (Spliced Transcripts Alignment to a Reference), a series of quality control, consisting removal of low-quality, low-complexity, and redundant sequences via paired-read iterative contig extension (PRICE), CD-HIT-DUP and Lempel-Ziv-Welch (LZW) compression score, respectively.
16
After quality control, the assignment of taxonomic IDs was implemented by aligning to the NCBI nucleotide (nt) and non-redundant protein (nr) databases via GSNAPL and RAPsearch,
16
respectively. Then the read counts of nt and reads per million (rPM) of nt at the species level were obtained. To guarantee the quality and credibility of the lung microbiota due to the low biomass, species with rPM value less than 50 in NC were used for downstream analysis. Moreover, bacteria detected in at least two study projects were kept to largely eliminate the reagent and laboratory contamination.
Microbial diversity and differential analysis
Microbial diversity was analyzed by R package vegan (v2.5-6). Shannon diversity index was calculated to evaluate α-diversity. Bacterial or viral loads were computed based on rPM of microorganism. β-diversity was studied by Principal coordinate analysis, PCoA, using R package ape (v5.3) based on Bray-Curtis distance. Differential species were analyzed by DEseq2 (v1.24.0). To avoid the loss of information while visualizing as many species as possible clearly, we used different Log2(Fold Change) thresholds for bacteria and virus. Differential bacteria with a statistical significance (q-value) < 1e-11 and absolute value of log2(Fold Change) > 3, virus with a statistical significance (q-value) < 0.01 and absolute value of log2(Fold Change) > 1 were retained. Student’s t test and Mann-Whitney rank test were used in variables with normal and without normal distribution, respectively. The Benjamini-Hochberg correction was used to obtain FDR adjusted P values (q-values) for multiple hypothesis testing.
Microbiota correlation analysis in BALF
Correlation analysis was implemented by Spearman’s rank correlation. Only bacteria and viruses retained in the differential analysis were used for correlation analysis. Bacteria correlations with a coefficient (r) > 0.8 and a statistical significance (q-value) < 1e-13 were considered for further analysis and visualization. Virus correlations with a coefficient (r) > 0.5 and a statistical significance (q-value) <0.01 were considered for further analysis and visualization. Bacteria and virus correlations with a coefficient (r) > 0.7 and a statistical significance (q-value) <5e-8 were considered for further analysis and visualization. Correlation network was visualized by Cytoscape 3.7.2.
17
Results
We collected the metatranscriptomic data of 42 BALF samples, consisting 19 COVID-19 patients and 23 healthy controls (HC), from six studies.
1,15,18-21
The basic information of the samples is shown in Supplementary file 1. COVID-19 patients were from five different studies, representing individual sample source of the samples. In total, 12413 bacteria and 359 viruses at the species level, which were transcriptionally active, were identified.
Differential microbial diversity in the COVID-19 and healthy groups
The diversity analysis of bacteria and virus, including α- and β-diversity, showed significant differences between two groups. The COVID-19 group had higher Shannon's Diversity Index of bacteria than healthy group (P value = 0.0268, Student’s t test, Fig. 1A). In addition, bacterial loads of COVID-19 group were much higher than healthy group (P value = 9e-4, Mann-Whitney rank test, Fig. 1B). For virome, COVID-19 patients had much higher viral loads than healthy controls (P value < 1e-4, Mann-Whitney rank test) while Shannon's Diversity Index showed no statistical difference (Fig. 1C-D).
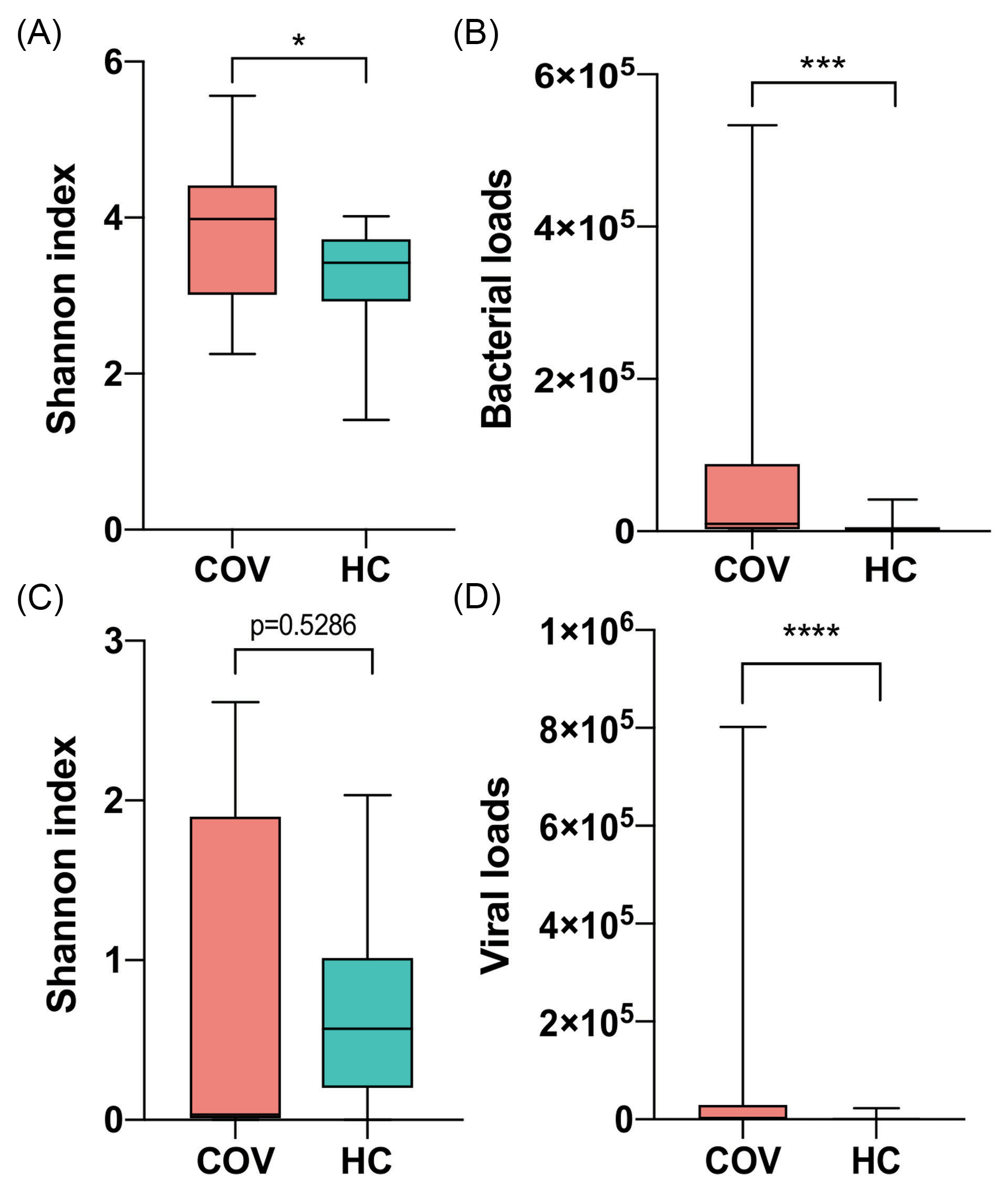
Figure 1.
Comparison of microbial α-diversity and loads between COVID-19 patients (COV) and healthy controls (HC). (A) and (C) Shannon index of bacteria and virus respectively. (B) and (D) Bacterial and viral loads respectively. Boxes represent the interquartile ranges (IQRs) between the first and third quartiles, and the line inside the box represents the median; whiskers represent the lowest or highest values within 1.5 times IQR from the first or third quartiles. * P value < 0.05, ** P value < 0.01, *** P value <0.001, **** P value <0.0001.
.
Comparison of microbial α-diversity and loads between COVID-19 patients (COV) and healthy controls (HC). (A) and (C) Shannon index of bacteria and virus respectively. (B) and (D) Bacterial and viral loads respectively. Boxes represent the interquartile ranges (IQRs) between the first and third quartiles, and the line inside the box represents the median; whiskers represent the lowest or highest values within 1.5 times IQR from the first or third quartiles. * P value < 0.05, ** P value < 0.01, *** P value <0.001, **** P value <0.0001.
Furthermore, principal coordinates analysis (PCoA) with Bray-Curtis distance was applied to evaluate the β-diversity in BALF microbiota across groups. The PCoA results of bacterial microbiome showed there was a significant difference of β-diversity between two groups (P value < 1e-3, R = 0.217, ANOSIM analysis) (Fig. 2A). Moreover, similar results for virome were found (P value < 1e-3, R = 0.541, ANOSIM analysis) (Fig. 2B), indicating a heterogeneous community diversity in the patient and control groups. The diversity analysis revealed that the infection of SARS-CoV-2 probably caused a different lung microbiota composition in the COVID-19 patient group compared with the healthy group.
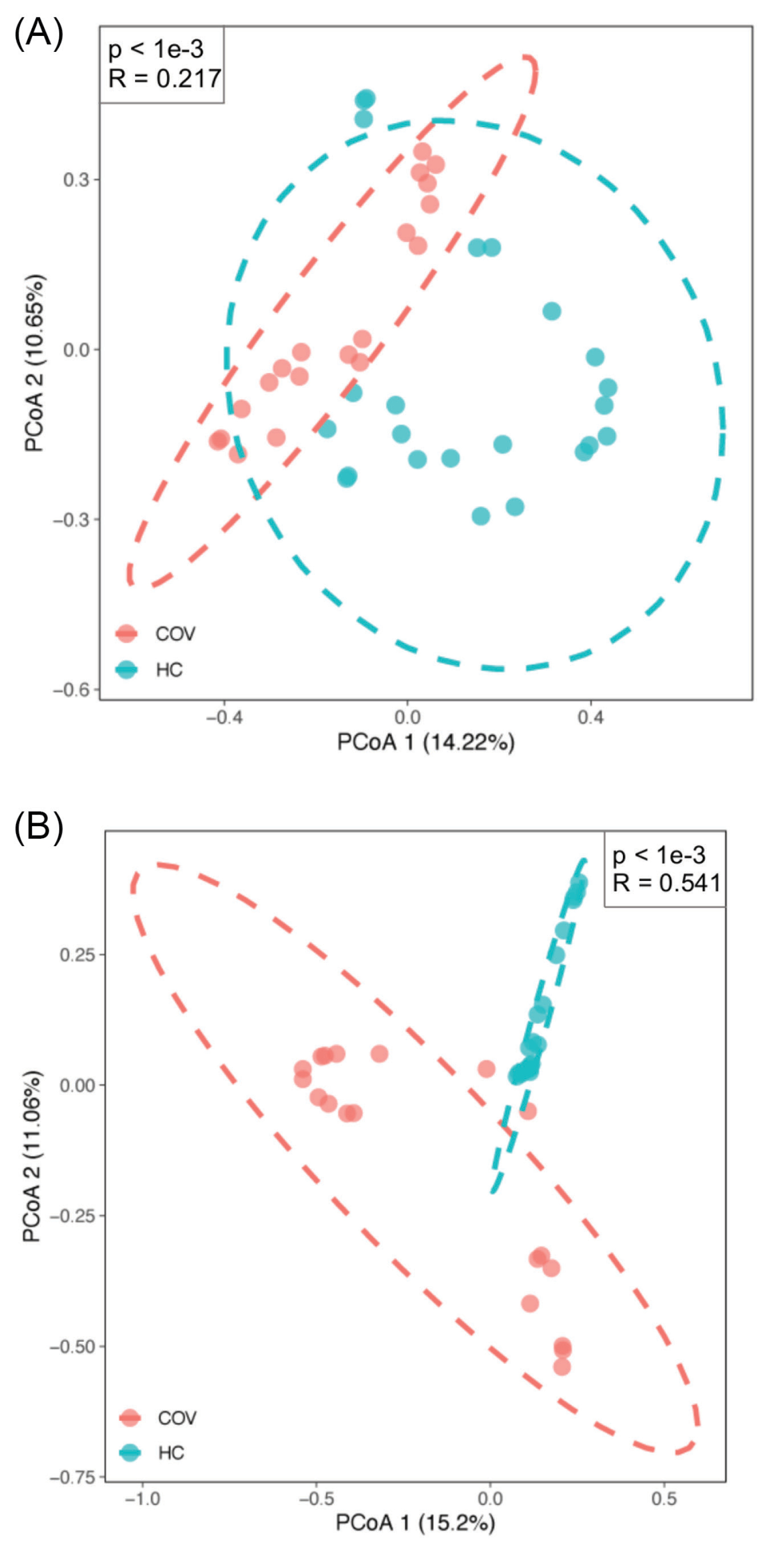
Figure 2.
β-Diversity of bacteria and viruses between COVID-19 patients (COV) and healthy controls (HC). (A) and (B) Principal coordinate analysis plot of bacteria and virus respectively based on Bray-Curtis distance. The pink dot indicates COVID-19 patients and the light green indicates healthy controls. R and P value are the results of ANOSIM test.
.
β-Diversity of bacteria and viruses between COVID-19 patients (COV) and healthy controls (HC). (A) and (B) Principal coordinate analysis plot of bacteria and virus respectively based on Bray-Curtis distance. The pink dot indicates COVID-19 patients and the light green indicates healthy controls. R and P value are the results of ANOSIM test.
Microbial composition of BALF samples
To get an insight into the microbial composition of BALF, we screened out the 10 most abundant bacteria at the species level in healthy controls and COVID-19 patients (Fig. 3A), respectively. Concerning the bacterial community, Porphyromonas gingivalis, often found in the oral cavity where it is implicated in periodontal diseases,
22
had a relatively higher abundance in some healthy individuals. The abundance of Lautropia mirabilis,which was commonly found in the human oral cavity and the upper respiratory tract,
23
and Enterobacter kobei, leading to nosocomial infections,
24
were increased in a few COVID-19 patients. Notably, Variovoraxaccount for a very high proportion in 6 patients. Interestingly, Staphylococcus xylosus and Staphylococcus simulans had high abundances in three healthy controls (H21, H22, and H23).
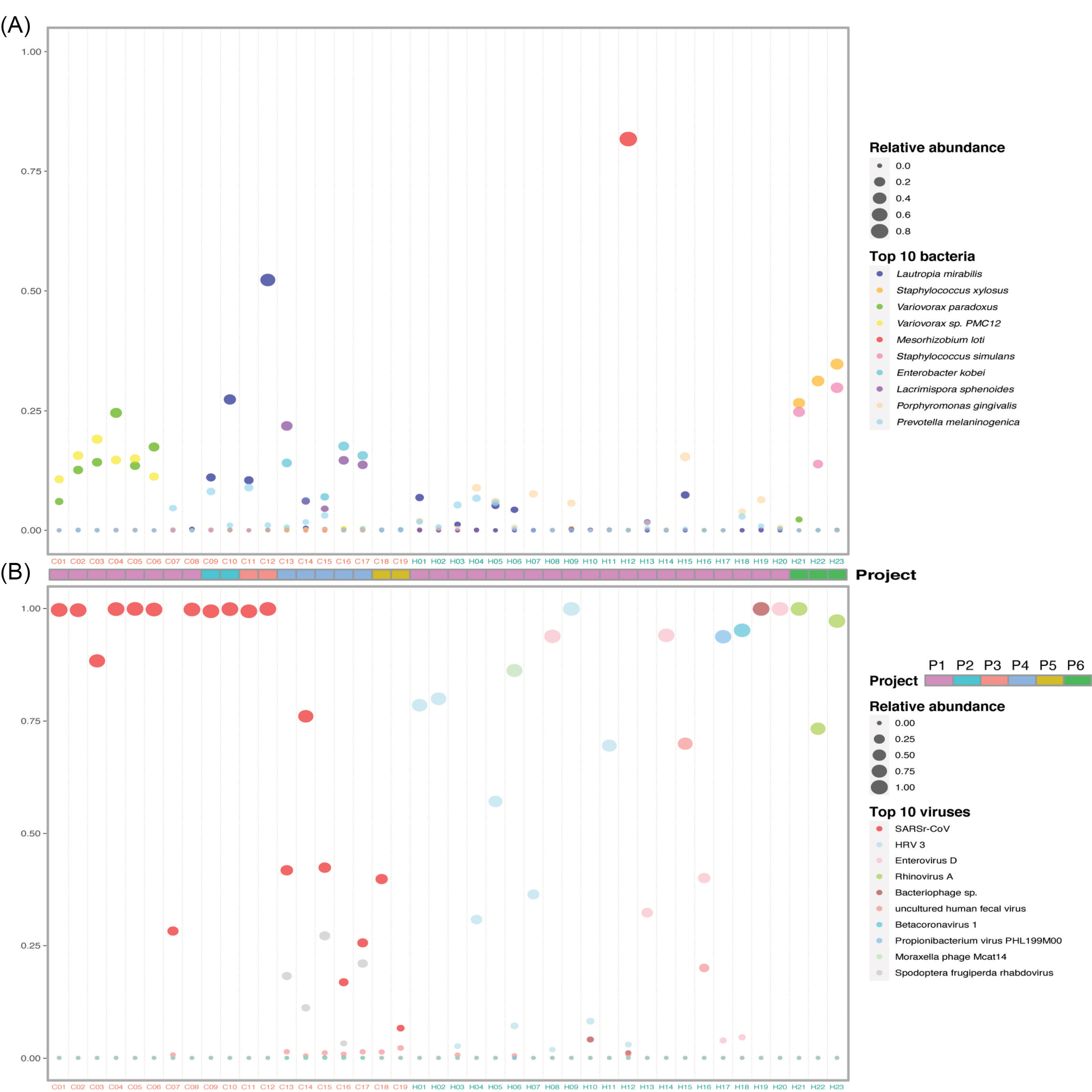
Figure 3.
Composition of the top 10 species in COVID-19 patients and healthy controls. (A) and (B) Relative abundance of the top 10 bacteria and viruses respectively in each sample. The size of the point corresponds to the relative abundance value. The color of the sample names represents the sample group, red represents the COVID-19 patient group, and blue represents the healthy control group. The order of the species in the legend from top to bottom represents the relative abundance from high to low. Project bar represents the sample sources (P1, PRJCA002202; P2, PRJNA601736; P3, PRJCA002326; P4, PRJNA605983; P5, PRJNA615032; P6, PRJNA434133).
.
Composition of the top 10 species in COVID-19 patients and healthy controls. (A) and (B) Relative abundance of the top 10 bacteria and viruses respectively in each sample. The size of the point corresponds to the relative abundance value. The color of the sample names represents the sample group, red represents the COVID-19 patient group, and blue represents the healthy control group. The order of the species in the legend from top to bottom represents the relative abundance from high to low. Project bar represents the sample sources (P1, PRJCA002202; P2, PRJNA601736; P3, PRJCA002326; P4, PRJNA605983; P5, PRJNA615032; P6, PRJNA434133).
The virome composition analysis showed that human respirovirus 3 (HRV 3) and enterovirus D were higher in a few healthy samples (Fig. 3b). Except severe acute respiratory syndrome-related coronavirus (SARSr-CoV, NCBI Taxonomy ID: 694009), spodoptera frugiperda rhabdovirus was significantly enriched in five patient samples (Fig. 3B). SARSr-CoV includes severe acute respiratory syndrome coronavirus (SARS-CoV) and SARS-CoV-2. The top abundant species showed that the infection of SARS-CoV-2 changed the bacterial and viral community in the lung of COVID-19 patients.
Microbial differences between the COVID-19 patient and healthy control groups
To identify the species associated with COVID-19, differential abundance analysis was performed between the two groups. Ninety-nine bacteria at the species level were significantly different between the two groups (q-value < 1e-11, absolute value of log2(Fold Change) > 3) (Supplementary file 2). Meanwhile, 82 species with the maximum relative abundance greater than 0.05% were selected from the 99 bacteria and visualized using heatmap to get a deeper investigation (Fig. 4A).
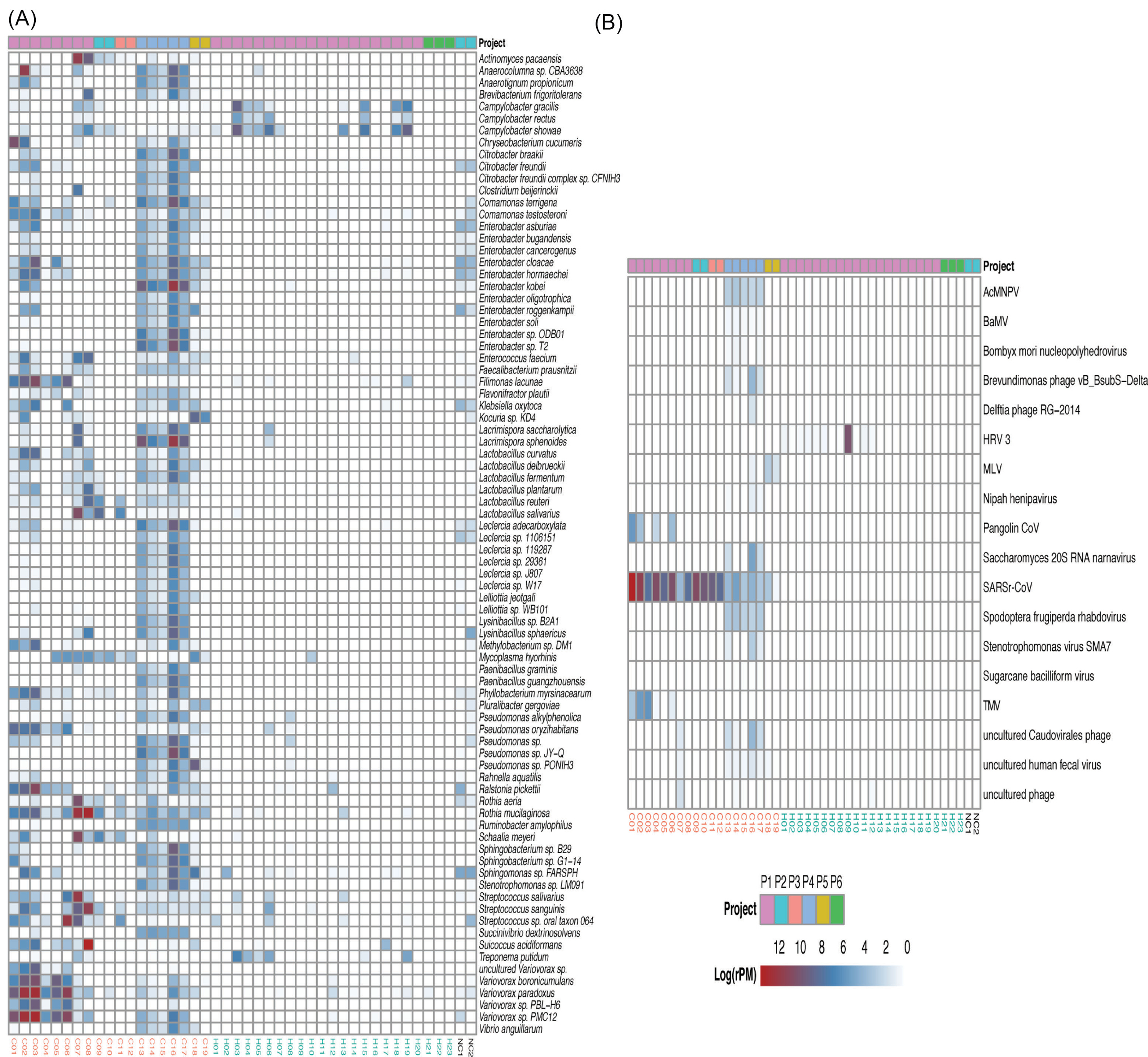
Figure 4.
Heat map of the species with strongly significant differences and important differential pathogens. (A) Heat map of differential bacteria with q-value < 1e-11, absolute value of log2(Fold Change) >3 and the maximum relative abundance greater than 0.05%. (B) Heat map of virus with q-value < 0.01 and absolute value of log2(Fold Change) >1. Project bar represents the sample sources (P1, PRJCA002202; P2, PRJNA601736; P3, PRJCA002326; P4, PRJNA605983; P5, PRJNA615032; P6, PRJNA434133). The color of the sample names represents the sample group, red represents the COVID-19 patient group, and blue represents the healthy control group. NC1 and NC2 are negative controls.
.
Heat map of the species with strongly significant differences and important differential pathogens. (A) Heat map of differential bacteria with q-value < 1e-11, absolute value of log2(Fold Change) >3 and the maximum relative abundance greater than 0.05%. (B) Heat map of virus with q-value < 0.01 and absolute value of log2(Fold Change) >1. Project bar represents the sample sources (P1, PRJCA002202; P2, PRJNA601736; P3, PRJCA002326; P4, PRJNA605983; P5, PRJNA615032; P6, PRJNA434133). The color of the sample names represents the sample group, red represents the COVID-19 patient group, and blue represents the healthy control group. NC1 and NC2 are negative controls.
The abundance of lactic acid bacteria such as Lactobacillus fermentum, Lactobacillus reuteri, Lactobacillus delbrueckii, and Lactobacillus salivarius were higher in the COVID-19 group than that in the healthy group (Supplementary file 2). Of note, Lactobacillus reuteri was detected only in the patient group. Some pathogens were enriched in COVID-19 patients and depleted in healthy controls. For example, Klebsiella oxytoca,leading to pneumonia, colitis and sepsis,
25
and Enterobacter cloacae, positively correlated with COVID-19 severity,
26
were increased in COVID-19. The abundance of some nosocomial infection pathogens such as Enterobacter kobei,
24
Enterobacter cloacae,
27
and Ralstonia pickettii
28
were higher in COVID-19 patients than that in healthy controls. Several gut bacteria like Faecalibacterium prausnitzii,
29
Enterococcus faecium,
30
and Citrobacter freundii,
31
and commensal bacteria residing in the mouth and respiratory tracts such as Rothia mucilaginosa were also enriched in the lung of COVID-19 patients. In addition, a few of pathogens causing severe nausea, vomiting, and diarrhea like Bacillus cereus
32
increased in patients significantly.
The abundance of 18 viruses significantly differed between the two groups (q-value <0.01, absolute value of log2(Fold Change) > 1) (Fig. 4B, Supplementary file 3). In addition to SARSr-CoV, Surprisingly, 13 out of 18 differential viruses were only detected in the COVID-19 group such as Pangolin coronavirus (Pangolin CoV) and Tobacco mosaic virus (TMV), which were probably closely related to the infection of SARS-CoV-2, though the function of them is unclear. Notably, HRV 3 was only detected in healthy controls (12 of 23 individuals).
Microbiota correlation in BALF
To further investigate whether the altered microbiota interacted with each other, we constructed networks of bacteria and viruses based on Spearman rank correlations, respectively. Network within bacteria (Fig. 5A and Supplementary file 4) showed that lots of bacteria were highly correlated. For example, Enterobacter sp., Pseudomonas sp. and Leclerciasp. were positively correlated to each other. What's more, Anaerotignum propionicum, detected only in the patient group, was positively correlated to Chryseobacterium cucumeris, Ruminococcus gnavus, and Leclerciasp. LSNIH1.

Figure 5.
Correlation network of differential species between the COVID-19 patient and healthy control groups. (A) The correlation network within bacteria with a coefficient (r) > 0.8 and a statistical significance (q-value) < 1e-13. (B) The correlation network within viruses with a coefficient (r) > 0.5 and a statistical significance (q-value) < 0.01. (C) The correlation between bacteria and viruses with a coefficient (r) > 0.7 and a statistical significance (q-value) < 5e-8. Diamonds represent viruses and circles represent bacteria. Blue line represents positive correlations and pink line represents negative correlations. Green represents up-regulation of the abundance of the species and pink represents down-regulation.
.
Correlation network of differential species between the COVID-19 patient and healthy control groups. (A) The correlation network within bacteria with a coefficient (r) > 0.8 and a statistical significance (q-value) < 1e-13. (B) The correlation network within viruses with a coefficient (r) > 0.5 and a statistical significance (q-value) < 0.01. (C) The correlation between bacteria and viruses with a coefficient (r) > 0.7 and a statistical significance (q-value) < 5e-8. Diamonds represent viruses and circles represent bacteria. Blue line represents positive correlations and pink line represents negative correlations. Green represents up-regulation of the abundance of the species and pink represents down-regulation.
Network within viruses (Fig. 5B and Supplementary file 5) showed SARSr-CoV, TMV, and Pangolin CoV were positively correlated to each other, while SARSr-CoV was negatively correlated to HRV 3. The Network between bacteria and viruses (Fig. 5C and Supplementary file 6) presented that SARSr-CoV was positively correlated to Rothia mucilaginosa, Schaalia meyeri, and Phyllobacterium myrsinacearumwhich was also positively related to TMV.
Taken together, the results of correlation analysis suggest that the altered lung microbiota, particularly species from the Lactobacillus and Rothia, Enterobacter, and TMV, actively played a certain role during the infection of SARS-CoV-2. The negative correlation between SARS-CoV-2 and HRV 3 indicates that the former probably had an inhibitory effect on the latter.
Discussion
The transcriptionally active microbiota landscape of BALF in COVID-19 patients and healthy controls could shed light on the infection mechanism of SARS-COV-2. Our study described the microbial composition at the species level in BALF of COVID-19 patients and healthy controls. Although Porphyromonas gingivalis existed widely in healthy lungs and accounted for a high proportion, lower richness and bacterial loads mean it still stays at a low level. In addition, though HRV 3 is one of the most common respiratory viruses, particularly among young children under 5 years of age and immunocompromised patients,
33,34
it was detected in the healthy group with no symptoms.
The infection of SARS-CoV-2 changed the active microbiota in the lungs, characterized by the α-diversity, β-diversity, species composition of bacteria and viruses respectively. The changes should be a result of complex interactions between the host, SARS-CoV-2, bacteria, and other viruses. The enrichment of lactic acid bacteria, and probiotics usually producing lactic acid in the COVID-19 group could probably inhibit the inflammatory response caused by SARS-CoV-2 to some extent, as several species of Lactobacillus could decrease the inflammatory response, and balance the immune response while limiting the potential of pathogens.
35,36
Besides, Lactobacillus could help treat diarrhea. The enrichment of several pathogens such as bacteria (Klebsiella oxytoca, Enterobacter kobei, and Bacillus cereus) and viruses (Pangolin CoV and TMV) in the COVID-19 group probably indicated co-infections or a second infection. Several symptoms, such as gastrointestinal symptoms and extreme fatigue in addition to fever, may be caused by the increase of these pathogens.
Although Rothia mucilaginosawas a part of the normal oropharyngeal flora, it was linked to causing disease, including pneumonia, in immunosuppressed humans.
37
Its enrichment in COVID-19 patients may be the result of the impact of SARS-CoV-2 on the patients’ immune system. In our study, we also found that some intestinal bacteria (e.g., Faecalibacterium prausnitziiand Enterococcus faecium) were enriched in the lung of COVID-19 patients. A study reported that intestinal bacteria could transfer to other organs through the intestinal barrier,
38
so we speculated that the infection of SARS-CoV-2 could probably promote the transfer of intestinal bacteria. Faecalibacterium prausnitzii, a prototypical IL-10 and butyrate producer in the gut,
39
showed a negative correlation with COVID-19 severity.
26
Therefore, it has the potential to be a protector of the host for the infection.
Subjects with COVID-19 have unexpected colonization of the lung by TMV and Pangolin CoV, suggesting that they may play a role in the infection of SARS-CoV-2. It was reported that TMV could enter and persist in mouse lungs, raising questions about the potential interactions between the virus and human host.
40
Accordingly, a further investigation was necessary. Our BALF metatranscriptome results detailed the species in the lungs of COVID-19 patients and shed light on the complex interactions between SARS-CoV-2 and other microbes in the lungs.
Conclusion
The metatranscriptome of human BALF can reveal transcriptionally active microbiota in the lungs. The microbes in the lungs have a great influence on human health and disease. We detailed the active microbiota landscape in both healthy individuals and COVID-19 patients and found the infection of SARS-CoV-2 could deeply change the microbiota in the lungs. This is the first detailed description of lung colonization in COVID-19 patients. The enrichment of several microbes, Lactic Acid Bacteria, Faecalibacterium prausnitzii, and TMV, the significant correlation between Rothia, TMV and SARS-CoV-2 reveals a microbiota disorder and drastic battles between the host, SARS-CoV-2, and other microbes in the lungs. Our study provides insights into the active microbiota in the lungs of COVID-19 patients and would contribute to the understanding of the infection mechanism of SARS-CoV-2 and the treatment of the disease and complications.
Availability of data
The raw datasets analyzed for this study were downloaded from Sequence Read Archive, EMBL-EBI ArrayExpress and National Genomics Data Center, China with project IDs, PRJCA002202, PRJNA601736, PRJCA002326, PRJNA605983, PRJNA615032, and PRJNA434133.
Funding sources
This work was supported by the National Natural Science Foundation of China [grant number: 31701155] and National Key Research and Development Program of China [grant number: 2017YFC0908403].
Ethical statement
None to be declared.
Competing interests
The authors declared no conflict of interests.
Authors’ contribution
Conceptualization: ZJ, KH and WW; Data analysis: YH, ZJ, JS; Writing and Editing: YH and ZJ. All authors read and approved the final manuscript.
Supplementary Materials
Supplementary file 1. Sample information.
(xlsx)
Supplementary file 2. Differential bacteria.
(xlsx)
Supplementary file 3. Differential viruses.
(xlsx)
Supplementary file 4. Bacterial correlations.
(xlsx)
Supplementary file 5. Viral correlations.
(xlsx)
Supplementary file 6. Correlations between Bacteria tand viruses.
(xlsx)
Research Highlights
What is the current knowledge?
√ The interaction between the host and SARS-CoV-2 was widely studied. However, it is unclear whether and how SARS-CoV-2 infection affects lung microflora, which contributes to COVID-19 complications.
What is new here?
√ SARS-COV-2 infection could deeply alter the lung microbiota, proved by the α-diversity, β-diversity and species composition based on bacterial microbiota and virome, respectively.
√ Pathogens (e.g., Klebsiella oxytoca, causing pneumonia as well), immunomodulatory probiotics (e.g., lactic acid bacteria and Faecalibacterium prausnitzii, a butyrate producer), and Tobacco mosaic virus were significantly enriched in the COVID-19 group, while depleted in the healthy group, suggesting a complex and active microbiota disorder in the lungs of COVID-19 patients.
√ The significant correlation between Rothia, TMV, and SARS-CoV-2 revealed drastic inflammatory battles between the host, SARS-CoV-2, and other microbes in the lungs.
√ TMV was only identified in 9 out of 19 patients, but in none of the healthy.
References
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579:270-3. doi: 10.1038/s41586-020-2012-7 [Crossref] [ Google Scholar]
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020; 581:215-20. doi: 10.1038/s41586-020-2180-5 [Crossref] [ Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020; 181:271-80. doi: 10.1016/j.cell.2020.02.052 [Crossref] [ Google Scholar]
- Pourseif MM, Parvizpour S, Jafari B, Dehghani J, Naghili B, Omidi Y. A domain-based vaccine construct against SARS-CoV-2, the causative agent of COVID-19 pandemic: development of self-amplifying mRNA and peptide vaccines. Bioimpacts 2021; 11(1):65-84. doi: 10.34172/bi.2021.11 [Crossref] [ Google Scholar]
- Li LQ, Huang T, Wang YQ, Wang ZP, Liang Y, Huang TB. COVID-19 patients' clinical characteristics, discharge rate, and fatality rate of meta-analysis. J Med Virol 2020. doi: 10.1002/jmv.25757 [Crossref]
- Zhang H, Ai J-W, Yang W, Zhou X, He F, Xie S. Metatranscriptomic Characterization of COVID-19 Identified A Host Transcriptional Classifier Associated With Immune Signaling. Clinical Infectious Diseases 2020; 73:376-385. doi: 10.1093/cid/ciaa663 [Crossref] [ Google Scholar]
- Fan J, Li X, Gao Y, Zhou J, Wang S, Huang B. The lung tissue microbiota features of 20 deceased patients with COVID-19. Journal of Infection 2020; 91:e64-e67. doi: 10.1016/j.jinf.2020.06.047 [Crossref] [ Google Scholar]
- Yatera K, Noguchi S, Mukae H. The microbiome in the lower respiratory tract. Respir Investig 2018; 56:432-9. doi: 10.1016/j.resinv.2018.08.003 [Crossref] [ Google Scholar]
- Marsh RL, Kaestli M, Chang AB, Binks MJ, Pope CE, Hoffman LR. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome 2016; 4:37. doi: 10.1186/s40168-016-0182-1 [Crossref] [ Google Scholar]
- Matijašić M, Meštrović T, Paljetak H, Perić M, Barešić A, Verbanac D. Gut Microbiota beyond Bacteria-Mycobiome, Virome, Archaeome, and Eukaryotic Parasites in IBD. Int J Mol Sci 2020; 21:2668. doi: 10.3390/ijms21082668 [Crossref] [ Google Scholar]
- Kim D, Quinn J, Pinsky B, Shah NH, Brown I. Rates of Co-infection Between SARS-CoV-2 and Other Respiratory Pathogens. Jama 2020; 323:2085-6. doi: 10.1001/jama.2020.6266 [Crossref] [ Google Scholar]
- Ren L, Zhang R, Rao J, Xiao Y, Zhang Z, Yang B. Transcriptionally Active Lung Microbiome and Its Association with Bacterial Biomass and Host Inflammatory Status. mSystems 2018; 3:e00199-18. doi: 10.1128/mSystems.00199-18 [Crossref] [ Google Scholar]
- Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunological Reviews 2017; 279:70-89. doi: 10.1111/imr.12567 [Crossref] [ Google Scholar]
-
Zuo T, Liu Q, Zhang F, Lui GC-Y, Tso EY, Yeoh YK, et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut 2020; gutjnl-2020-322294. 10.1136/gutjnl-2020-322294
-
Shen Z, Xiao Y, Kang L, Ma W, Shi L, Zhang L, et al. Genomic diversity of SARS-CoV-2 in Coronavirus Disease 2019 patients. Clin Infect Dis 2020. 10.1093/cid/ciaa203
- Kalantar KL, Carvalho T, de Bourcy CFA, Dimitrov B, Dingle G, Egger R. IDseq – An Open Source Cloud-based Pipeline and Analysis Service for Metagenomic Pathogen Detection and Monitoring. bioRxiv 2020. doi: 10.1101/2020.04.07.030551 [Crossref]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13:2498-504. doi: 10.1101/gr.1239303 [Crossref] [ Google Scholar]
- Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect 2020; 9:761-70. doi: 10.1080/22221751.2020.1747363 [Crossref] [ Google Scholar]
- Chen L, Liu W, Zhang Q, Xu K, Ye G, Wu W. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect 2020; 9:313-9. doi: 10.1080/22221751.2020.1725399 [Crossref] [ Google Scholar]
- Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Møller R. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020; 181:1036-45. doi: 10.1016/j.cell.2020.04.026 [Crossref] [ Google Scholar]
- Michalovich D, Rodriguez-Perez N, Smolinska S, Pirozynski M, Mayhew D, Uddin S. Obesity and disease severity magnify disturbed microbiome-immune interactions in asthma patients. Nat Commun 2019; 10:5711. doi: 10.1038/s41467-019-13751-9 [Crossref] [ Google Scholar]
- Naito M, Hirakawa H, Yamashita A, Ohara N, Shoji M, Yukitake H. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P gingivalis. DNA Res 2008; 15:215-25. doi: 10.1093/dnares/dsn013 [Crossref] [ Google Scholar]
- Gerner-Smidt P, Keiser-Nielsen H, Dorsch M, Stackebrandt E, Ursing J, Blom J. Lautropia mirabilis gen nov, sp nov, a gram-negative motile coccus with unusual morphology isolated from the human mouth. Microbiology 1994; 140:1787-97. doi: 10.1099/13500872-140-7-1787 [Crossref] [ Google Scholar]
- Hoffmann H, Schmoldt S, Trülzsch K, Stumpf A, Bengsch S, Blankenstein T. Nosocomial urosepsis caused by Enterobacter kobei with aberrant phenotype. Diagn Microbiol Infect Dis 2005; 53:143-7. doi: 10.1016/j.diagmicrobio.2005.06.008 [Crossref] [ Google Scholar]
- Högenauer C, Langner C, Beubler E, Lippe IT, Schicho R, Gorkiewicz G. Klebsiella oxytoca as a causative organism of antibiotic-associated hemorrhagic colitis. N Engl J Med 2006; 355:2418-26. doi: 10.1056/NEJMoa054765 [Crossref] [ Google Scholar]
- Zuo T, Zhang F, Lui GCY, Yeoh YK, Li AYL, Zhan H. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology 2020; 159:944-955. doi: 10.1053/j.gastro.2020.05.048 [Crossref] [ Google Scholar]
- Keller R, Pedroso MZ, Ritchmann R, Silva RM. Occurrence of virulence-associated properties in Enterobacter cloacae. Infect Immun 1998; 66:645-9. [ Google Scholar]
- Ryan MP, Pembroke JT, Adley CC. Ralstonia pickettii: a persistent gram-negative nosocomial infectious organism. J Hosp Infect 2006; 62:278-84. doi: 10.1016/j.jhin.2005.08.015 [Crossref] [ Google Scholar]
- Bag S, Ghosh TS, Das B. Complete Genome Sequence of Faecalibacterium prausnitzii Isolated from the Gut of a Healthy Indian Adult. Genome Announc 2017; 5. doi: 10.1128/genomeA.01286-17 [Crossref]
- McArthur DB. Emerging Infectious Diseases. Nurs Clin North Am 2019; 54:297-311. doi: 10.1016/j.cnur.2019.02.006 [Crossref] [ Google Scholar]
- Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A. Genomic variation landscape of the human gut microbiome. Nature 2013; 493:45-50. doi: 10.1038/nature11711 [Crossref] [ Google Scholar]
- Kotiranta A, Lounatmaa K, Haapasalo M. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect 2000; 2:189-98. doi: 10.1016/s1286-4579(00)00269-0 [Crossref] [ Google Scholar]
- Reed G, Jewett PH, Thompson J, Tollefson S, Wright PF. Epidemiology and clinical impact of parainfluenza virus infections in otherwise healthy infants and young children < 5 years old. J Infect Dis 1997; 175:807-13. doi: 10.1086/513975 [Crossref] [ Google Scholar]
- Liu WK, Liu Q, Chen DH, Liang HX, Chen XK, Huang WB. Epidemiology and clinical presentation of the four human parainfluenza virus types. BMC Infect Dis 2013; 13:28. doi: 10.1186/1471-2334-13-28 [Crossref] [ Google Scholar]
- Llopis M, Antolin M, Carol M, Borruel N, Casellas F, Martinez C. Lactobacillus casei downregulates commensals' inflammatory signals in Crohn's disease mucosa. Inflamm Bowel Dis 2009; 15:275-83. doi: 10.1002/ibd.20736 [Crossref] [ Google Scholar]
- Oksaharju A, Kankainen M, Kekkonen RA, Lindstedt KA, Kovanen PT, Korpela R. Probiotic Lactobacillus rhamnosus downregulates FCER1 and HRH4 expression in human mast cells. World J Gastroenterol 2011; 17:750-9. doi: 10.3748/wjg.v17.i6.750 [Crossref] [ Google Scholar]
- Yuan Z, Panchal D, Syed MA, Mehta H, Joo M, Hadid W. Induction of cyclooxygenase-2 signaling by Stomatococcus mucilaginosus highlights the pathogenic potential of an oral commensal. J Immunol 2013; 191:3810-7. doi: 10.4049/jimmunol.1300883 [Crossref] [ Google Scholar]
- Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018; 359:1156-61. doi: 10.1126/science.aar7201 [Crossref] [ Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008; 105:16731-6. doi: 10.1073/pnas.0804812105 [Crossref] [ Google Scholar]
- Balique F, Colson P, Barry AO, Nappez C, Ferretti A, Moussawi KA. Tobacco mosaic virus in the lungs of mice following intra-tracheal inoculation. PLoS One 2013; 8:e54993. doi: 10.1371/journal.pone.0054993 [Crossref] [ Google Scholar]