Bioimpacts. 12(4):315-324.
doi: 10.34172/bi.2021.40
Original Research
A fuzzy logic-based computational method for the repurposing of drugs against COVID-19
Yosef Masoudi-Sobhanzadeh 1, *  , Hosein Esmaeili 2
, Hosein Esmaeili 2  , Ali Masoudi-Nejad 3, *
, Ali Masoudi-Nejad 3, * 
Author information:
1Research Center for Pharmaceutical Nanotechnology, Biomedicine Institute, Tabriz University of Medical Sciences, Tabriz, Iran
2Institute of Biochemistry and Biophysics, University of Tehran, Tehran, Iran
3Laboratory of Systems Biology and Bioinformatics (LBB), Institute of Biochemistry and Biophysics, University of Tehran, Tehran, Iran
Abstract
Introduction:
COVID-19 has spread out all around the world and seriously interrupted human activities. Being a newfound disease, not only many aspects of the disease are unknown, but also there is not an effective medication to cure the disease. Besides, designing a drug is a time-consuming process and needs large investment. Hence, drug repurposing techniques, employed to discover the hidden benefits of the existing drugs, maybe a useful option for treating COVID-19.
Methods:
The present study exploits the drug repositioning concepts and introduces some candidate drugs which may be effective in controlling COVID-19. The suggested method consists of three main steps. First, the required data such as the amino acid sequences of targets and drug-target interactions are extracted from the public databases. Second, the similarity score between the targets (protein/enzymes) and genome of SARS-COV-2 is computed using the proposed fuzzy logic-based method. Since the classical approaches yield outcomes which may not be useful for the real-world applications, the fuzzy technique can address the issue. Third, after ranking targets based on the obtained scores, the usefulness of drugs affecting them is examined for managing COVID-19.
Results:
The results indicate that antiviral medicines, designed for curing hepatitis C, may also cure COVID-19. According to the findings, ribavirin, simeprevir, danoprevir, and XTL-6865 may be helpful in controlling the disease.
Conclusion:
It can be concluded that the similarity-based drug repurposing techniques may be the most suitable option for managing emerging diseases such as COVID-19 and can be applied to a wide range of data. Also, fuzzy logic-based scoring methods can produce outcomes which are more consistent with the real-world biological applications than others.
Keywords: Computational method, COVID-19, Drug repurposing, Fuzzy logic, Hepatitis
Copyright and License Information
© 2022 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Since the emergence of coronavirus disease 2019 (COVID-19), researchers have tried to discover its biological behavior and its mechanism of action.
1
The disease is caused by the severe acute respiratory syndrome corona virus-2 (SARS-CoV-2) which is a new member of the beta coronavirus subfamily.
2,3
The virus has a single-stranded positive-sense RNA genome that is 79% similar to SARS-CoV and 50% similar to Middle East respiratory syndrome coronavirus (MERS-CoV) in term of genomic sequence.
4
The virus complete genome was published on January 12, 2020, in the GISAD database.
5
Analysis of the virus encoding sequences shows that its 30 kb genome encodes 14 open reading frames (ORFs). ORF1a and ORF1b, on the 5'-end of the genome, encode a polyprotein that converts to 16 nonstructural proteins (NSPs 1-16) by autoproteolytic degradation to form the replicase-transcriptase complex, whose members of this complex are involved in the virus transcription and replication. This complex includes Papain-like protease (NSP3), Main protease (NSP5), and primary RNA-dependent RNA polymerase (RdRp or NSP12), which are considered drug targets for inhibiting virus replication activity.
6
The four main ORFs on the 3'-end of the genome of the virus also encode the structural proteins spike glycoprotein (S), envelope (E), membrane (M), and nucleocapsid (N) proteins.
7
Currently, countries all around the world are faced with a big challenge and are trying to come up with a suitable treatment plan.
8
Whereas some aspects of the virus are unknown, producing an efficient medication may require a long time. In addition, the process of developing a drug is a very time-consuming procedure and needs a huge investment.
9
Therefore, researchers are testing alternative approaches such as drug repositioning, which aims to investigate the effectiveness of the existing drugs on COVID-19.
10
Regardless of cost reduction, the drug repurposing technique the trims development time of drug development projects and may play a critical role in curing the disease.
11
Moreover, some drug development projects, which have failed due to various reasons such as safety, low performance, side effect issues, bioavailability, etc., may be revived and used for treating COVID-19.
12
To address the mentioned challenge, this study offers a fuzzy logic-based drug repurposing method which contains two main contributions. First, a fuzzy logic-based alignment technique is proposed to compute the similarity score between the amino acid sequences of the existing protein targets and genome of SARS-CoV-2. Although there is the main challenge for scoring matches, insertions, and deletions between the sequences, the fuzzy logic can address such the limitation. This principle can obtain results which are more consistent with real-world applications than classical methods. Second, potential drugs, which may help treat COVID-19, are detected based on the calculated scores. The medications, which affect the targets sharing a higher value of sequence similarity score with SARS-CoV-2, may have an application in curing the disease. The proposed approach utilizes information existing in the sequences of the targets (proteins or enzymes as well as the genome of SARS-CoV-2). The amino acid sequence of a protein is considered the primary structure of that protein and determines the secondary, tertiary, and quaternary structures of a protein. Proteins can play a structural or functional role through their tertiary or quaternary structures. Each protein can form unique structural and functional motifs due to its specific amino acid sequence. Thus, the amino acid sequences of a protein and being directly involved in its function can also indirectly affect its function through its effect on forming the protein's spatial structure. Hence, proteins with similar amino acid sequences are more likely to have the same structure and function.
Since the COVID-19 outbreak, different efforts have been made to obtain vaccines worldwide to increase the body's immunity to the SARS-CoV-2 and reduce the mortality rate. Fortunately, vaccines for the virus were developed quickly, and according to the World Health Organization (WHO) reports, up to now (February 12, 2021), 66 vaccines are in clinical trials. Some of these vaccines included in the clinical trials have been approved and licensed for utilization, such as Pfizer-BioNTech, Moderna, Comirnaty, AstraZeneca, and Sputnik V.
The main contributions of this study are listed as follows:
simple
-
i) Introducing a novel fuzzy logic-based sequence alignment algorithm
-
ii) Proposing some candidate medications for combating COVID-19.
Related works
For the repurposing of drugs, researchers have proposed various approaches such as in silico, in vitro, and in vivo techniques.
13
Computational methods are at the center of the attention for researchers since they reduce time and produce helpful solutions.
14,15
In this study, the computational techniques have been divided into several categories as follows:
i) Molecular docking techniques: most of the carried out studies fall into this class of computational methods and consider the 3D structure of SARS-CoV-2’s proteins and existing drugs.
16
Usually, two classes of docking methods exist. The first class determines whether two biological components complement each other or not in terms of the total number of bonds and their properties such as the length of the bond. The second class calculates the existing energy between elements.
17
The lower the potential energy; the tighter the interaction between two biological components.
13,18
The two above-mentioned approaches have been frequently used, which each of them face some advantages and disadvantages. The docking methods are very time-consuming processes and are not practical to use on larger scales. For instance, it is not possible to calculate the score between all the existing drugs and components of SARS-CoV-2. In addition, if the 3D structure of a protein is not available, these approaches cannot be employed. Unlike these techniques, our proposed method can be used on a large scale and does not rely on the 3D structure of a protein.
ii) Machine learning (ML) techniques: these methods create machines that mine relationships between inputs and outputs.
19
For example, they can be used to discover whether a drug and a target may interact or not.
20
In this field, many types of learners, such as support vector machine, decision tree, artificial neural network, and many other approaches are used.
21,22
Although ML methods can produce a model in an acceptable time and lead to desired results, they face particular challenges in generating a model. There is also another type of artificial neural networks which is known as deep neural networks and has a higher prediction capacity.
23
For example, using deep learning approaches, an attempt to design potential inhibitors of SARS-CoV-2 has been carried out.
24
The ML-based approaches face a great challenge in invalidating the predicted outcomes. To this end, researchers usually compare their outcomes with other experimentally reported data or use docking methods to score the interactions between the predicted drug and target. ML methods also require an adequate volume of data to generate an acceptable model. Although our fuzzy logic-based proposed method needs secondary validation techniques such as molecular docking, it only depends on the amino acid or nucleotide sequence of the targets and viruses and, unlike the ML methods, can be used in the early stage of a disease pandemic.
c) Network-based techniques: biological elements are part of a complex system, which can be modeled by the graph theory concepts.
25
By doing this, a network of different biological components and their relationships is formed.
26
Then, graph analyzing algorithms are applied to discover hidden information and introduce effective therapeutic plans against COVID-19.
27,28
Drug repurposing using a generative network method, is an approach that researchers have introduced to identify potential drugs for curing COVID-19. For this purpose, associated genes with SARS-CoV-2 have been considered as the seeds of biological communications, and the network has then been extended by them. In other words, the interactions between the elements of SARS-CoV-2 (including genes or proteins) and the host cell are constituted. After analysis, the desired targets have been specified, and some potential drugs have been proposed for controlling them.
29
Since some aspects of SARS-CoV-2 are unknown, it is essential to produce data of interest by experimental techniques such as mass spectrometry. Hence, as the ML approaches, this category of the methods cannot be used in the early stages of a pandemic whereas the method proposed in this study can address this limitation.
d) Hybrid techniques: the main idea of the hybrid methods is to utilize the advantages of single approaches and obtain an efficient method with higher reliability capabilities.
30
For example, researchers have proposed a two-step technique for the repurposing of drugs against COVID-19. To this end, first, the candidate drugs have been chosen using a computer-aided method. Second, the docking techniques have been applied to select drugs that may have a proper functionality in treating COVID-19.
31,32
Besides, another study has combined the statistical and ML methods and has generated an efficient model for predicting drugs, which may be useful in curing COVID-19.
33
To validate the approach, the researchers have used medications exploited for healing SARS and MERS diseases. Although these techniques utilize the benefits of each of the single methods used in the combined procedure, they may suffer from their synthetic disadvantages.
34
The proposed method of this study is a similarity-based technique, which considers the likeness between the amino acid sequence of targets (proteins and enzymes) and the genome of SARS-CoV-2. The approach uses fuzzy logic to score the insertions/deletions and the matches in the sequences and aims to reduce the false-positive rate.
Methods
To discover potential drugs which may be useful in curing COVID-19, the present study suggests a fuzzy-based computational approach. Fig. 1 depicts the framework of the proposed method. Fig. 1 includes three main parts as follows:
-
Collecting data: the amino acid and nucleotide sequences of the existing enzymes and proteins have been acquired from DrugR+,
35
which is a relational version of DrugBank
36
and consists of some information about the KEGG
37
database. The database also includes drugs-targets interaction data and different chemical properties of drugs. Besides, the genome of SARS-CoV-2 has been extracted from PDB.
38
The extracted data consist of 4893 drugs and more than 18 000 drug-target interactions.
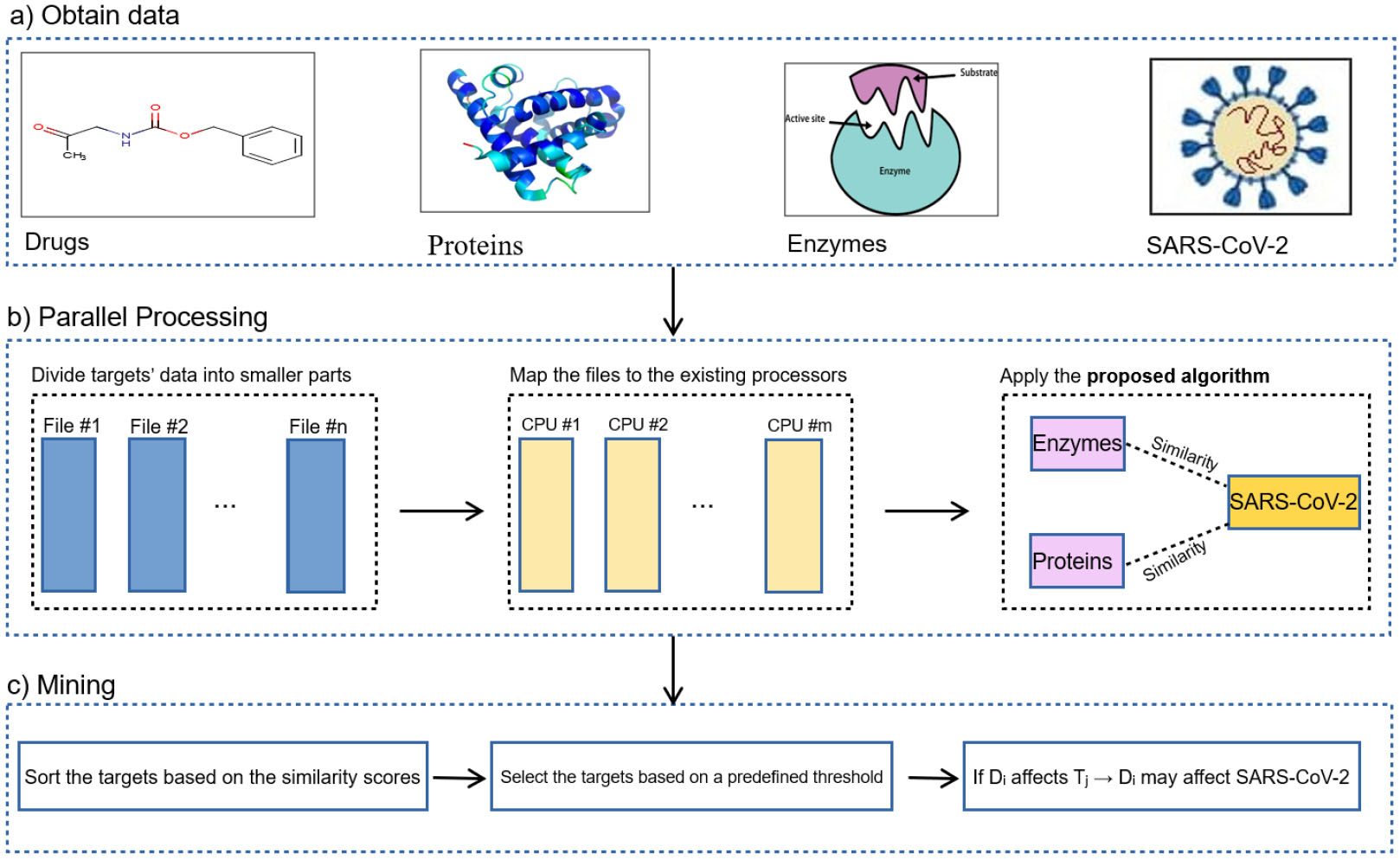
Fig. 1.
The framework of the proposed method. First, the data, including proteins and enzymes, are divided into smaller parts, and they are mapped to the existing CPUs. Second, a fuzzy-based similarity algorithm is applied to compute the similarity between the amino acid sequence of targets and SARS-COV-2.
.
The framework of the proposed method. First, the data, including proteins and enzymes, are divided into smaller parts, and they are mapped to the existing CPUs. Second, a fuzzy-based similarity algorithm is applied to compute the similarity between the amino acid sequence of targets and SARS-COV-2.
-
Applying parallel processing techniques: due to the sheer volume of data, it is essential to utilize parallel processing approaches. For every target, a file, including the profile of the targets (such as the drugs affecting them, their amino acid and nucleotide sequences, the genes coding them, etc.), is generated. Then, they are assigned to the existing CPUs to compute the similarity between the genome of SARS-CoV-2 and the extracted sequences of proteins and enzymes. To this end, different types of algorithms such as Smith-Waterman, longest common substring, and some others can be used as well.
39
Since finding a suitable similarity strategy is a challenging problem in computational biology, fuzzy logic, which may produce results consistent with the biological real-world facts, can be a proper option. To implement the fuzzy logic, three steps are considered, as follows:
(1) Fuzzification: the classical inputs are turned into fuzzy data. For this purpose, the inputs are mapped to the membership functions, and it is determined how much a value belongs to a specific group. The inputs incorporate two main components:
First, the total number of matched amino acids between the sequence of a given target and the genome of SARS-CoV-2 is calculated. To this end, an R×C matrix, named M, is constituted and is completed using Eq. 1. R and C are the length of the first and the second sequences, respectively.
(1)
The maximum value of M (MAX) shows the total number of the matched amino acids, and MAX divided by the length of the given sequence represents the match percentage between two sequences.
Second, the percentage of insertions and deletions (INDELS), which is computed by Eq. 2, is specified. R indicates the length of the sequence.
Based on the calculated percentage of the matches and INDELS, the final similarity score is determined. For the inputs, three fuzzy sets including low, medium, and high sets have been considered. Fig. 2A-C depicts the coverage range of the sets. To determine the ranges of the sets, a phylogenetic tree between different organisms were considered, and, based on the distance among them, the ranges were specified. For instance, for some entities having the farthest distance, 50% of similarity was observed. Hence, the [0, 50] range was selected for the low setting.
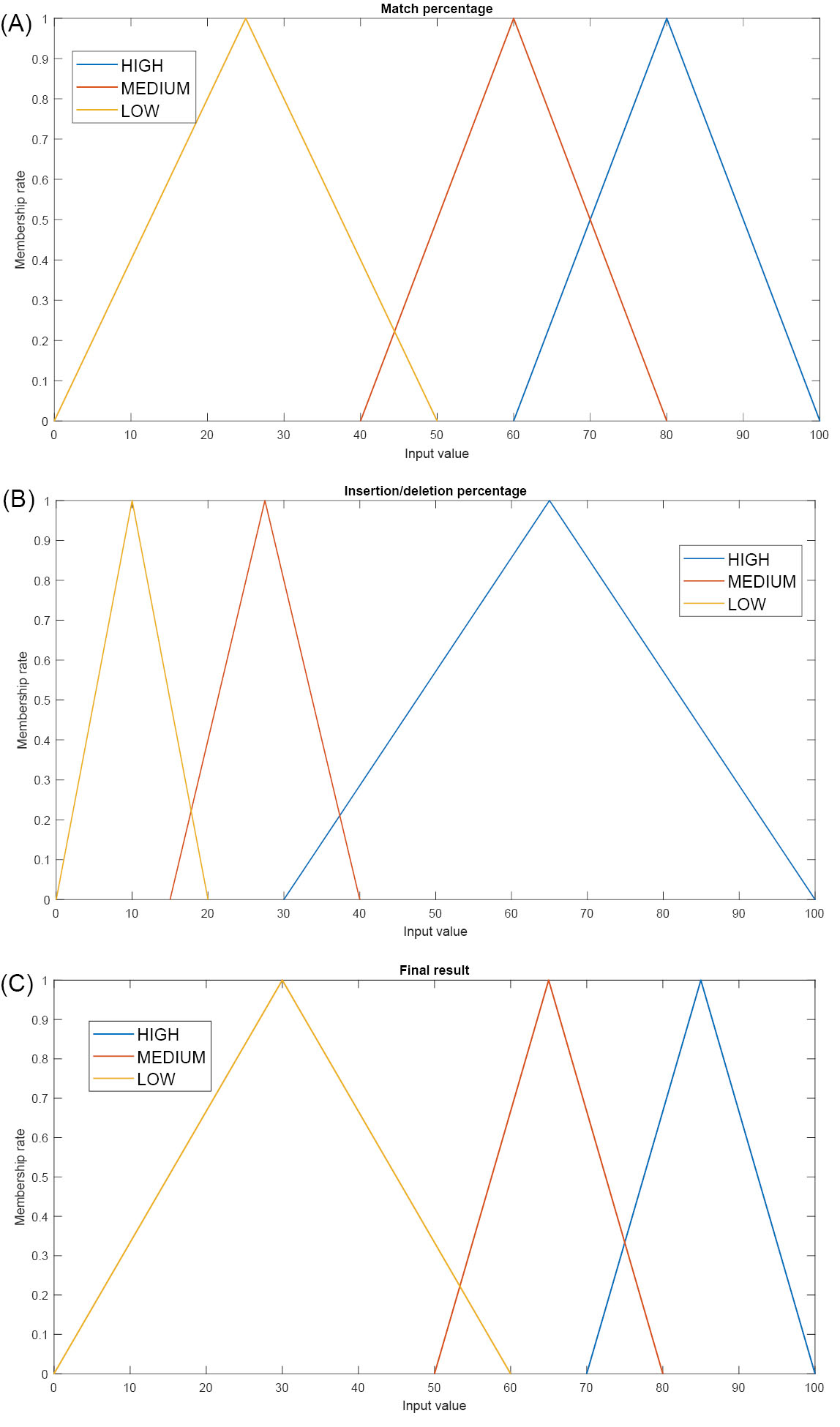
Fig. 2.
Fuzzy membership function for the (A) matched elements, (B) unmatched elements and (C) final similarity.
.
Fuzzy membership function for the (A) matched elements, (B) unmatched elements and (C) final similarity.
(2) Fuzzy inference step: This process combines the fuzzy membership functions and the defined rules. Given the three fuzzy sets for the inputs, nine rules, shown in Table 1, have been defined.
Table 1.
The defined rules
|
No.
|
Rule
|
| 1 |
If the similarity percentage is HIGH and the INS is LOW then the similarity is HIGH |
| 2 |
If the similarity percentage is HIGH and the INS is MEDIUM then the similarity is MEDIUM |
| 3 |
If the similarity percentage is HIGH and the INS is HIGH then the similarity is LOW |
| 4 |
If the similarity percentage is MEDIUM and the INS is LOW then the similarity is MEDIUM |
| 5 |
If the similarity percentage is MEDIUM and the INS is MEDIUM then the similarity is LOW |
| 6 |
If the similarity percentage is MEDIUM and the INS is HIGH then the similarity is LOW |
| 7 |
If the similarity percentage is LOW and the INS is LOW then the similarity is LOW |
| 8 |
If the similarity percentage is LOW and the INS is MEDIUM then the similarity is LOW |
| 9 |
If the similarity percentage is LOW and the INS is HIGH then the similarity is LOW |
(3) Defuzzification: many methods have been proposed for concluding the fuzzy output. Mean of maximum, center of gravity (COG), and height methods are commonly used. Because the outputs of this step are continuous values, COG method, considering all areas of the output, has been used in this study. Eq. 3 presents how the defuzzification process is done.
Where s and x indicate the fuzzy set and the input value of the output membership function, respectively.
-
Mining: the targets are ranked based on their calculated fuzzy similarity scores, and, the list is filtered by a prespecified threshold. Next, a defined association rule is applied to discover the drugs that may help treat COVID-19. A drug (D) that affects a target (T) which is similar to the genome of SARS-CoV-2, may inhibit the virus and show therapeutic properties. The mentioned rule has been shown as follows:
If a drug such as D affects a target such as T then D may be useful in curing COVID-19.
Results
The proposed fuzzy-based method was implemented in the python programming language. As mentioned before, the amino acid sequence of protein and enzyme targets, as well as drug-target interactions, were extracted from DrugR+ database. After applying the proposed method to the data, the targets were sorted by the calculated fuzzy similarity scores. The range of the scores fitted between 0 and 93, among which, the score of 10 targets was more than 65. Table 2 explains the mentioned targets and their properties. The listed targets perform several biological functions, including:
-
ATP binding activity: adenosine triphosphate (ATP) is an organic compound that is responsible for preparing energy for cell activities. Some proteins interact with ATP and transport different substrates within or outside of a cell.
-
Cysteine-type endopeptidase activity: it is an activity in which proteins play the main role in apoptotic cell death.
-
Ion-channel activity: ion-channel proteins move across the cell membrane and transport some ions into the cell.
-
RNA binding activity: a specific type of proteins, named RNA binding proteins, connect to RNA and help to form ribonucleoproteins.
-
RNA helicase activity: helicase enzymes, also known as motor proteins, unpackaged genes of an organism.
-
RNA-directed RNA polymerase activity: this activity is referred to as the specific type of viral enzymes that are responsible for catalyzing RNA replication.
-
Structural molecule activity: the activity is the contribution of a protein or enzyme to form complex molecular components.
Table 2.
The protein targets sharing a higher value of similarity with the SARS-CoV-2’s genome
|
Target ID
|
Description
|
Length
|
Score
|
| BE0001031 |
Basement membrane-specific heparin sulfate proteoglycan core protein |
13492 |
65 |
| BE0001261 |
Genome polyprotein |
10362 |
93 |
| BE0001283 |
Apo lipoprotein(a) |
13900 |
73 |
| BE0001478 |
Genome polyprotein |
9208 |
69 |
| BE0002110 |
Prolow-density lipoprotein receptor-related protein 1 |
13940 |
80 |
| BE0002460 |
E3 ubiquitin-protein ligase UBR4 |
15868 |
78 |
| BE0002813 |
Genome polyprotein |
10368 |
68 |
| BE0003427 |
Genome polyprotein |
9211 |
91 |
| BE0003801 |
Replicase polyprotein 1ab |
21624 |
88 |
| BE0003935 |
Genome polyprotein |
9211 |
79 |
The discovered targets can be inhibited by 23 drugs which have been listed in Table 3. This Table consists of different medicines from the approved and clinical investigation groups. Two classes of drugs exist in Table 3. The first class is the small molecules group which incorporates chemical compounds having less than 900 Dalton molecular weight. Small molecules drugs, which have more therapeutic properties than non-small molecule medicines, can be considered as a tool for probing biological functions.
40
The second category is biotech drugs, which researchers have recently been interested in them. These drugs use biotechnology as a means to manipulate microorganisms such as bacteria. Then, the organisms can perform a specific biological process.
Table 3.
The detected potential medications and their descriptions
|
ID
|
Name
|
Drug type
|
Description
|
| DB00031 |
Tenecteplase |
Biotech |
Tissue plasminogen activator |
| DB00039 |
Palifermin |
Biotech |
Recombinant human keratinocyte growth factor |
| DB00513 |
Aminocaproic acid |
Small molecule |
This drug inhibits plasminogen activator |
| DB00811 |
Ribavirin |
Small molecule |
The drug interferes with the synthesis of viral mRNA. |
| DB02331 |
Phenyl-Propionic acid |
Small molecule |
It is an experimentally validated drug and its application has not been reported |
| DB03605 |
Propionic acid |
Small molecule |
It is an experimentally validated drug and its application has not been reported |
| DB04005 |
Uridine 5-triphosphate |
Small molecule |
The drug consists of three phosphate groups esterified to the sugar moiety. |
| DB04298 |
Dihydro-pyran-2-one |
Small molecule |
It is an experimentally validated drug and its application has not been reported |
| DB04473 |
Alpha-L-fucose |
Small molecule |
It is an experimentally validated drug and its application has not been reported |
| DB04959 |
Verpasep caltespen |
Biotech |
It is a vaccine and is an immune system booster |
| DB05868 |
Ciluprevir |
Small molecule |
It acts against HCV NS3 protease |
| DB06058 |
XTL-6865 |
Small molecule |
The drugs operate against the hepatitis C virus |
| DB06974 |
phenylpyridazin |
Small molecule |
It is an experimentally validated drug and its application has not been reported |
| DB07062 |
methanesulfonamide |
Small molecule |
It is an experimentally validated drug and its application has not been reported |
| DB07293 |
benzyl (2-oxopropyl) carbamate |
Small molecule |
It is an experimentally validated drug and its application has not been reported |
| DB08578 |
oxobutanoic acid |
Small molecule |
It is an experimentally validated drug and its application has not been reported |
| DB08579 |
dimethylpiperidin |
Small molecule |
It is an experimentally validated drug and its application has not been reported |
| DB08580 |
(2-chlorobenzyl) pyrrolidin |
Small molecule |
It is an experimentally validated drug and its application has not been reported |
| DB08581 |
dimethylpiperidin-1-y |
Small molecule |
It is an experimentally validated drug and its application has not been reported |
| DB08582 |
Morpholin-4-yl-4-oxobutanamide |
Small molecule |
It is an experimentally validated drug and its application has not been reported |
| DB08644 |
CARBAMIC ACID |
Small molecule |
It is an experimentally validated drug and its application has not been reported |
| DB08656 |
Amino-2-methyl |
Small molecule |
It is an experimentally validated drug and its application has not been reported |
| DB11779 |
Danoprevir |
Small molecule |
The drug is in the investigation phase and has been developed for treating hepatitis C |
Fig. 3 depicts the relationships between the targets and the drugs affecting them. COVID-19 (the disease induced by SARS-CoV-2) has been shown in the core of communications. The targets, which share an impressive number of sequence individuals with the virus (determined by the proposed fuzzy score), exist in the second level. Also, the drugs, having an interaction with the identified targets, have been shown in the third layer.
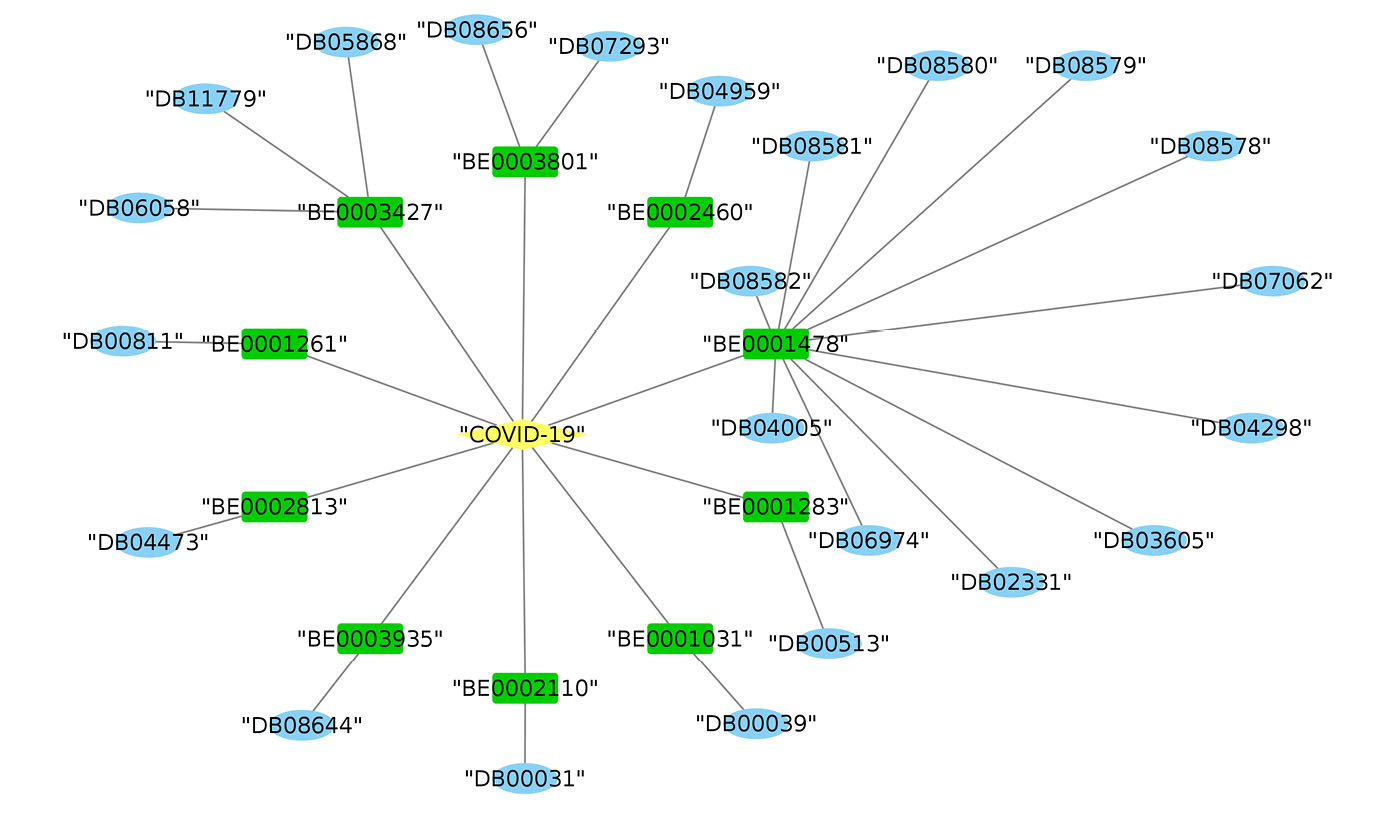
Fig. 3.
The drugs and their similar targets to SARS-COV-2. Yellow, green, and blue colors show the virus related to COVID-19, the targets, and the drugs, respectively.
.
The drugs and their similar targets to SARS-COV-2. Yellow, green, and blue colors show the virus related to COVID-19, the targets, and the drugs, respectively.
Table 2 consists of 4 medications (ribavirin, ciluprevir, XTL-6865, and danoprevir) whose antiviral applications have been experimentally proved. These medications have been developed for curing hepatitis diseases and maybe a proper plan in treating COVID-19 given their scores. Fig. 4 represents the chemical structures of the drugs except for XTL-6865 whose data was not available in databases.
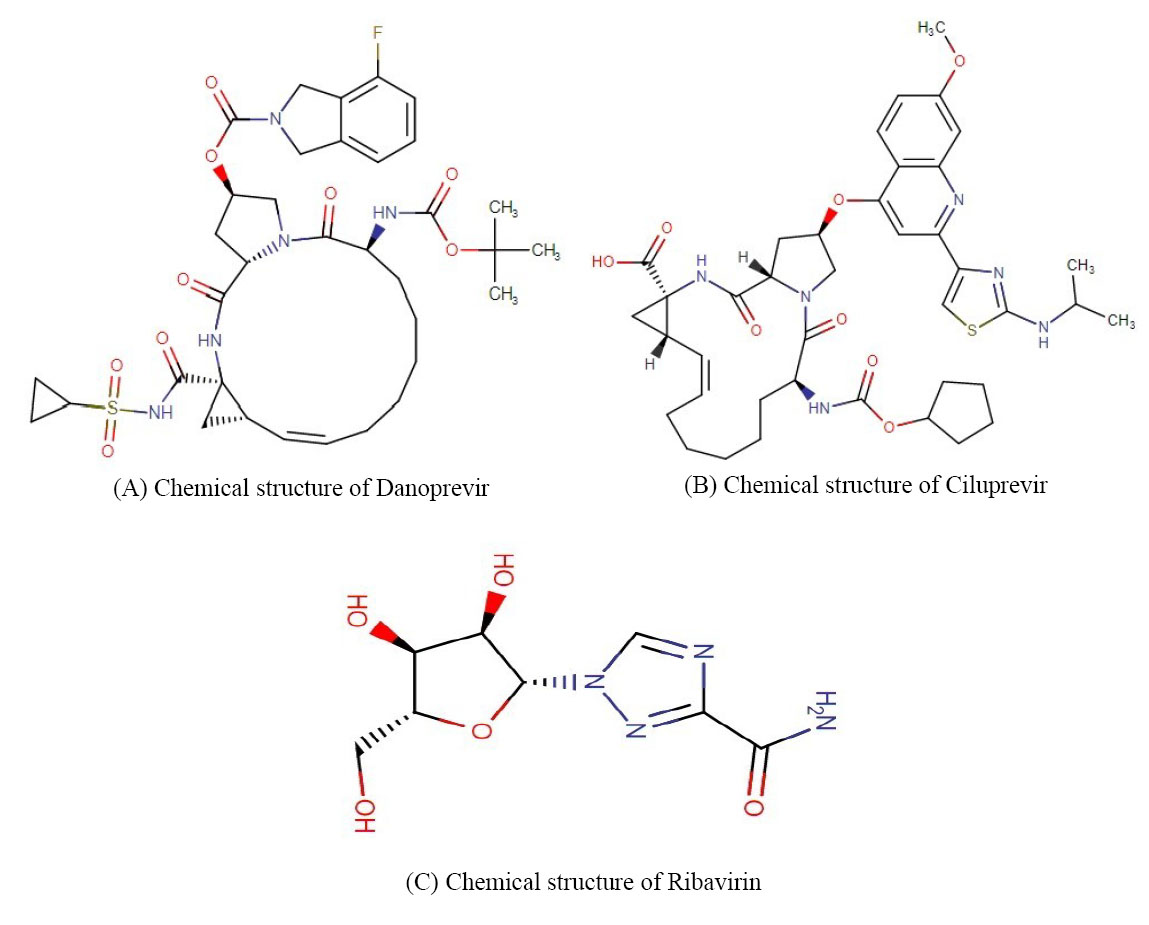
Fig. 4.
Except for XTL-6865, the chemical structure of the proposed drugs has been depicted.
.
Except for XTL-6865, the chemical structure of the proposed drugs has been depicted.
Discussion
This study proposed a novel fuzzy logic-based method for calculating similarity scores between the amino acid sequence of the existing targets and the genome of SARS-CoV-2, which utilized parallel processing techniques due to the huge volume of the existing data. Although different algorithms have been introduced for computing the similarity score between sequences,
41
they confront some challenging issues such as scoring matches and gaps of the given sequences and may increase the false-positive rate of the predictions. In other words, they may produce outcomes, which are not consistent with real-world biological applications. The proposed fuzzy logic-based method can address this limitation and can be considered as a straightforward therapeutic plan for curing emerging infectious diseases because of its capabilities in discovering potential medicines based on the rudimentary information on the genome sequence of the virus. Also, in the early stages of an infectious disease in which many aspects of its pathogenesis are unknown (e.g., there is not enough information on the proteins of the virus), the proposed fuzzy logic-based computational method can be helpful and may propose candidate drugs for treating the emerged disease.
The final output of the proposed approach is 23 drugs, which have a higher sequence similarity score with the genome of SARS-CoV-2 than the others. Among them, 4 medications (ribavirin, ciluprevir, XTL-6865, and danoprevir) have antiviral traits that have been clinically validated. The drugs have been designed for managing hepatitis patients.
42
Hepatitis, having several types and be induced by different agents, is an inflammation of an area between the liver tissues. Viruses are one of the most prevalent reasons for hepatitis difficulties and can seriously threaten the life of patients.
The outcomes of the proposed methods consist of three hepatitis medications: Ciluprevir, danoprevir, and ribavirin. Ciluprevir is an antiviral medication that is used for controlling hepatitis type C. The experimental studies have shown that this medicine is an inhibitor of nonstructural protein 4A (NS4A) and nonstructural protein 3 (NSP3). NS4A is a viral protein found in the hepatitis C virus (HCV) and is a cofactor of the NSP3 enzyme. Like NS4A, NSP3 is a viral NSP and plays a role as a serine protease, helicase, or nucleoside triphosphatase. Furthermore, a portion of its amino acids is a cofactor domain for NS2. Ciluprevir has been tested on humans in the clinical phase and has shown a remarkable effect on the patients. However, its development has been halted due to the observation that it has a higher value of toxicity in animals. The ideas obtained from developing ciluprevir led to the development of simeprevir and danoprevir that have shown impressive antiviral activities.
43
Simeprevir significantly reduces the viral load when combined with remdesivir. It acts by inhibiting the Mpro (or NSP5) and RNA-dependent RNA polymerase (RdRp).
44
Concomitant use of ribavirin and Simeprevir is likely to reduce the effects of COVID-19. Unlike simeprevir, ciluprevir has higher toxicity, but it may be a suitable option for combination therapy. Using a minimum dosage of medications, combination therapy may enhance the efficacy of drugs and can revive many drugs that halted during different drug development phases.
45,46
Danoprevir, inspired by the ciluprevir project, is also a 15-membered macrocyclic peptidomimetic inhibitor of nonstructural proteins (NS4A/NSP3). From a pharmaceutical perspective, the half-maximum inhibitory concentration (IC50) and effective concentration (EC50) of danoprevir are 0.2-0.4 nM and 1.6 nM, respectively. These parameters state that the medication has a very favorable profile and is an interesting option for curing viral diseases such as hepatitis C.
47
Ribavirin, playing a critical role in blocking viral RNA synthesis, is used for curing hepatitis C patients and is a very helpful agent in managing hemorrhagic fevers and hantavirus infections. This drug, as a guanosine analog, has several effects on the various RNA and DNA viruses and acts as a competitive inhibitor of the inosine monophosphate dehydrogenase enzyme, which is a critical enzyme in the synthesis of de novo purine nucleotides.
48
By inhibiting this enzyme, the amount of intracellular GTP storage is reduced, indirectly reducing the viral polymerase activity. Also, ribavirin is involved in inhibiting viral polymerase's catalytic activity and interfering with the mRNA capping process, thereby preventing virus replication. Clinical trials have also shown that ribavirin inhibits SARS-CoV-2's replication at micromolar concentrations.
49
Although this medicine has shown a very desired effect on patients, it has several side effects. Hence, ribavirin may be combined with newer antiviral drugs developed for treating hepatitis C.
50
Besides, a combination of ciluprevir and Ribavirin may show a synergistic effect in curing COVID-19 and reduce the toxicity of ciluprevir and the side effects of ribavirin. In some studies, the helpfulness of ribavirin has been evaluated and discussed. For instance, in a docking-based computational approach, it has been indicated that ribavirin, Sofosbuvir, and Remdesivir can bind to the SARS-CoV-2 RdRp enzyme.
51
Like the above-mentioned drugs, XTL-6865 is an antiviral agent which is used for curing viral diseases.
52
This drug, which is a combination of two human antibodies ab68 and ab65, is a small molecule, which tries to prevent virus entry into a cell. Researchers have followed two main goals in developing XTL-6865. First, they aim to hinder viral re-infection after doing a liver transplant. Second, they aim to introduce an effective drug for curing chronic HCV. Since the drug is in the investigational group of medications, its different properties have not been reported.
Research Highlights
What is the current knowledge?
√ A similarity-based method has been introduced for the repurposing of drugs against COVID-19.
√ Fuzzy logic may yield to the outcomes which are more consistent with the real-world biological applications.
What is new here?
√ The proposed fuzzy logic-based method investigates huge volume of the existing data and proposes some medications for curing COVID-19.
Conclusion
COVID-19 is an emerging disease, and there is an urgent need for detecting a proper treatment plan for inhibiting it. In order to cut down on the time, computational-based drug repurposing approaches may be a helpful plan and can propose potential drugs in a short time. This study introduced a fuzzy logic-based computational method for the repurposing of drugs against COVID-19. The proposed approach calculated the sequence similarity scores in such a way that they are more compatible with real-world biological concepts. The outcomes showed that hepatitis C antiviral drugs can be useful in treating COVID-19. Although some drugs have a suitable profile in treatment, some others have a higher value of toxicity and a large number of side effects. Therefore, combination therapy can alleviate such limitations and can yield an efficient treatment plan for combating COVID-19.
Funding sources
Not applicable.
Ethical statement
Not applicable.
Competing interests
None.
Authors’ contribution
YMS: Conceptualization, data preparing, formal analysis, investigation, writing-manuscript. AMN: Conceptualization, Supervision, project administration, editing the manuscript. Both the authors have read and approved the manuscript.
References
- Zhou D, Dai SM, Tong Q. COVID-19: a recommendation to examine the effect of hydroxychloroquine in preventing infection and progression. J Antimicrob Chemother 2020; 75:1667-70. doi: 10.1093/jac/dkaa114 [Crossref] [ Google Scholar]
- Salemi A, Pourseif MM, Omidi Y. Next-generation vaccines and the impacts of state-of-the-art in-silico technologies. Biologicals 2020 2020; 69:83-85. doi: 10.1016/j.biologicals.2020.10.002 [Crossref] [ Google Scholar]
- Bojkova D, Klann K, Koch B, Widera M, Krause D, Ciesek S. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 2020; 583:469-72. doi: 10.1038/s41586-020-2332-7 [Crossref] [ Google Scholar]
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020; 395:565-74. doi: 10.1016/s0140-6736(20)30251-8 [Crossref] [ Google Scholar]
-
Gralinski LE, Menachery VD. Return of the Coronavirus: 2019-nCoV. Viruses 2020; In Press. 10.3390/v12020135
-
Lotfi F, Akbarzadeh-Khiavi M, Lotfi Z, Rahbarnia L, Safary A, Zarredar H, et al. Micronutrient therapy and effective immune response: a promising approach for management of COVID-19. Infection 2021;In Press. 10.1007/s15010-021-01644-3
- Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol 2015; 1282:1-23. doi: 10.1007/978-1-4939-2438-7_1 [Crossref] [ Google Scholar]
- Pourseif MM, Parvizpour S, Jafari B, Dehghani J, Naghili B, Omidi Y. A domain-based vaccine construct against SARS-CoV-2, the causative agent of COVID-19 pandemic: development of self-amplifying mRNA and peptide vaccines. BioImpacts 2021; 11:65-84. doi: 10.34172/bi.2021.11 [Crossref] [ Google Scholar]
- Masoudi-Sobhanzadeh Y, Omidi Y, Amanlou M, Masoudi-Nejad A. Trader as a new optimization algorithm predicts drug-target interactions efficiently. Sci Rep 2019; 9:9348. doi: 10.1038/s41598-019-45814-8 [Crossref] [ Google Scholar]
- Devaux CA, Rolain JM, Colson P, Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19?. Int J Antimicrob Agents 2020; 55:105938. doi: 10.1016/j.ijantimicag.2020.105938 [Crossref] [ Google Scholar]
- Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov 2019; 18:41-58. doi: 10.1038/nrd.2018.168 [Crossref] [ Google Scholar]
- Rosa SGV, Santos WC. Clinical trials on drug repositioning for COVID-19 treatment. Rev PanamSalud Publica 2020; 44:e40. doi: 10.26633/RPSP.2020.40 [Crossref] [ Google Scholar]
-
Masoudi-Sobhanzadeh Y, Salemi A, Pourseif MM, Jafari B, Omidi Y, Masoudi-Nejad A. Structure-based drug repurposing against COVID-19 and emerging infectious diseases: methods, resources and discoveries. Brief Bioinform 2021. 10.1093/bib/bbab113
- Khosravi A, Kouhsar M, Goliaei B, Jayaram B, Masoudi-Nejad A. Systematic analysis of genes and diseases using PheWAS-Associated networks. Comput Biol Med 2019; 109:311-21. doi: 10.1016/j.compbiomed.2019.04.037 [Crossref] [ Google Scholar]
- Masoudi-Sobhanzadeh Y. Computational-based drug repurposing methods in COVID-19. Bioimpacts 2020; 10:205-6. doi: 10.34172/bi.2020.25 [Crossref] [ Google Scholar]
- Muralidharan N, Sakthivel R, Velmurugan D, Gromiha MM. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J Biomol Struct Dyn 2020; 39:2673-78. doi: 10.1080/07391102.2020.1752802 [Crossref] [ Google Scholar]
- Marinho EM, Batista de Andrade Neto J, Silva J, Rocha da Silva C, Cavalcanti BC, Marinho ES. Virtual screening based on molecular docking of possible inhibitors of Covid-19 main protease. MicrobPathog 2020; 148:104365. doi: 10.1016/j.micpath.2020.104365 [Crossref] [ Google Scholar]
- Krishnan DA, Sangeetha G, Vajravijayan S, Nandhagopal N, Gunasekaran K. Structure-based drug designing towards the identification of potential anti-viral for COVID-19 by targeting endoribonuclease NSP15. Inform Med Unlocked 2020; 20:100392. doi: 10.1016/j.imu.2020.100392 [Crossref] [ Google Scholar]
- Beck BR, Shin B, Choi Y, Park S, Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J 2020; 18:784-90. doi: 10.1016/j.csbj.2020.03.025 [Crossref] [ Google Scholar]
- Hooshmand SA, Ghobadi MZ, Hooshmand SE, Jamalkandi SA, Alavi SM, Masoudi-Nejad A. A multimodal deep learning-based drug repurposing approach for treatment of COVID-19. Mol Divers 2020; 25:1717-30. [ Google Scholar]
- Zhao K, So HC. Using Drug Expression Profiles and Machine Learning Approach for Drug Repurposing. Methods Mol Biol 2019; 1903:219-37. doi: 10.1007/978-1-4939-8955-3_13 [Crossref] [ Google Scholar]
- Burdick H, Lam C, Mataraso S, Siefkas A, Braden G, Dellinger RP. Prediction of respiratory decompensation in Covid-19 patients using machine learning: The READY trial. Comput Biol Med 2020; 124:103949. doi: 10.1016/j.compbiomed.2020.103949 [Crossref] [ Google Scholar]
- Abbasi K, Poso A, Ghasemi J, Amanlou M, Masoudi-Nejad A. Deep Transferable Compound Representation across Domains and Tasks for Low Data Drug Discovery. J Chem Inf Model 2019; 59:4528-39. doi: 10.1021/acs.jcim.9b00626 [Crossref] [ Google Scholar]
-
Choi Y, Shin B, Kang K, Park S, Beck BR. Target-Centered Drug Repurposing Predictions of Human Angiotensin-Converting Enzyme 2 (ACE2) and Transmembrane Protease Serine Subtype 2 (TMPRSS2) Interacting Approved Drugs for Coronavirus Disease 2019 (COVID-19) Treatment through a Drug-Target Interaction Deep Learning Model. Viruses 2020; In Press. 10.3390/v12111325
- Taz TA, Ahmed K, Paul BK, Kawsar M, Aktar N, Mahmud SMH. Network-based identification genetic effect of SARS-CoV-2 infections to Idiopathic pulmonary fibrosis (IPF) patients. Brief Bioinform 2020; 22:1254-66. doi: 10.1093/bib/bbaa235 [Crossref] [ Google Scholar]
- Lo YC, Rensi SE, Torng W, Altman RB. Machine learning in chemoinformatics and drug discovery. Drug Discov Today 2018; 23:1538-46. doi: 10.1016/j.drudis.2018.05.010 [Crossref] [ Google Scholar]
- Masoudi-Sobhanzadeh Y, Omidi Y, Amanlou M, Masoudi-Nejad A. Drug databases and their contributions to drug repurposing. Genomics 2020; 112:1087-95. doi: 10.1016/j.ygeno.2019.06.021 [Crossref] [ Google Scholar]
- Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 2020; 6:14. doi: 10.1038/s41421-020-0153-3 [Crossref] [ Google Scholar]
- Zhou Y, Hou Y, Shen J, Mehra R, Kallianpur A, Culver DA. A network medicine approach to investigation and population-based validation of disease manifestations and drug repurposing for COVID-19. PLoS Biol 2020; 18:e3000970. doi: 10.1371/journal.pbio.3000970 [Crossref] [ Google Scholar]
- MotieGhader H, Gharaghani S, Masoudi-Sobhanzadeh Y, Masoudi-Nejad A. Sequential and Mixed Genetic Algorithm and Learning Automata (SGALA, MGALA) for Feature Selection in QSAR. Iran J Pharm Res 2017; 16:533-53. [ Google Scholar]
- Hakmi M, Bouricha EM, Kandoussi I, Harti JE, Ibrahimi A. Repurposing of known anti-virals as potential inhibitors for SARS-CoV-2 main protease using molecular docking analysis. Bioinformation 2020; 16:301-6. doi: 10.6026/97320630016301 [Crossref] [ Google Scholar]
-
Kumar A, Kumar D, Kumar R, Singh P, Chandra R, Kumari K. DFT and docking studies of designed conjugates of noscapines & repurposing drugs: promising inhibitors of main protease of SARS-CoV-2 and falcipan-2. J Biomol Struct Dyn 2020; In Press. 10.1080/07391102.2020.1841030
- Lu H, Zhang J, Liang Y, Qiao Y, Yang C, He X. Network topology and machine learning analyses reveal microstructural white matter changes underlying Chinese medicine Dengzhan Shengmai treatment on patients with vascular cognitive impairment. Pharmacol Res 2020; 156:104773. doi: 10.1016/j.phrs.2020.104773 [Crossref] [ Google Scholar]
- Masoudi-Sobhanzadeh Y, Motieghader H, Masoudi-Nejad A. FeatureSelect: a software for feature selection based on machine learning approaches. BMC Bioinformatics 2019; 20:170. doi: 10.1186/s12859-019-2754-0 [Crossref] [ Google Scholar]
- Masoudi-Sobhanzadeh Y, Omidi Y, Amanlou M, Masoudi-Nejad A. DrugR+: A comprehensive relational database for drug repurposing, combination therapy, and replacement therapy. Comput Biol Med 2019; 109:254-62. doi: 10.1016/j.compbiomed.2019.05.006 [Crossref] [ Google Scholar]
- Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018; 46:D1074-D82. doi: 10.1093/nar/gkx1037 [Crossref] [ Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 1999; 27:29-34. doi: 10.1093/nar/27.1.29 [Crossref] [ Google Scholar]
- Rose PW, Prlic A, Bi C, Bluhm WF, Christie CH, Dutta S. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res 2015; 43:D345-56. doi: 10.1093/nar/gku1214 [Crossref] [ Google Scholar]
- Okada D, Ino F, Hagihara K. Accelerating the Smith-Waterman algorithm with interpair pruning and band optimization for the all-pairs comparison of base sequences. BMC Bioinformatics 2015; 16:321. doi: 10.1186/s12859-015-0744-4 [Crossref] [ Google Scholar]
-
Hooshmand SA, Jamalkandi SA, Alavi SM, Masoudi-Nejad A. Distinguishing drug/non-drug-like small molecules in drug discovery using deep belief network. Mol Divers 2020: 25: 827-838. 10.1007/s11030-020-10065-7
- Wang M, Kong L. pblat: a multithread blat algorithm speeding up aligning sequences to genomes. BMC Bioinformatics 2019; 20:28. doi: 10.1186/s12859-019-2597-8 [Crossref] [ Google Scholar]
- Gotte M, Feld JJ. Direct-acting antiviral agents for hepatitis C: structural and mechanistic insights. Nat Rev Gastroenterol Hepatol 2016; 13:338-51. doi: 10.1038/nrgastro.2016.60 [Crossref] [ Google Scholar]
- Wei L, Shang J, Ma Y, Xu X, Huang Y, Guan Y. Efficacy and Safety of 12-week Interferon-based Danoprevir Regimen in Patients with Genotype 1 Chronic Hepatitis C. J Clin Transl Hepatol 2019; 7:221-5. doi: 10.14218/JCTH.2019.00018 [Crossref] [ Google Scholar]
- Lo HS, Hui KP, Lai H-M, Khan KS, Kaur S, Li Z, et al. Simeprevir suppresses SARS-CoV-2 replication and synergizes with remdesivir. bioRxiv 2020.
- Pott-Junior H, Bricks G, Grandi G, Figueiredo Senise J, Castelo Filho A. Sofosbuvir in combination with daclatasvir or simeprevir for 12 weeks in noncirrhotic subjects chronically infected with hepatitis C virus genotype 1: a randomized clinical trial. Clin Microbiol Infect 2019; 25:365-71. doi: 10.1016/j.cmi.2018.06.007 [Crossref] [ Google Scholar]
- Masoudi-Sobhanzadeh Y, Masoudi-Nejad A. Synthetic repurposing of drugs against hypertension: a datamining method based on association rules and a novel discrete algorithm. BMC Bioinformatics 2020; 21:313. doi: 10.1186/s12859-020-03644-w [Crossref] [ Google Scholar]
- Jiang Y, Andrews SW, Condroski KR, Buckman B, Serebryany V, Wenglowsky S. Discovery of danoprevir (ITMN-191/R7227), a highly selective and potent inhibitor of hepatitis C virus (HCV) NS3/4A protease. J Med Chem 2014; 57:1753-69. doi: 10.1021/jm400164c [Crossref] [ Google Scholar]
- Carrillo-Bustamante P, Nguyen THT, Oestereich L, Günther S, Guedj J, Graw F. Determining Ribavirin's mechanism of action against Lassa virus infection. Sci Rep 2017; 7:11693. doi: 10.1038/s41598-017-10198-0 [Crossref] [ Google Scholar]
- Lam S, Lombardi A, Ouanounou A. COVID-19: A review of the proposed pharmacological treatments. Eur J Pharmacol 2020; 886:173451. doi: 10.1016/j.ejphar.2020.173451 [Crossref] [ Google Scholar]
- Charlton M, Gane E, Manns MP, Brown RS, Jr Jr, Curry MP, Kwo PY. Sofosbuvir and ribavirin for treatment of compensated recurrent hepatitis C virus infection after liver transplantation. Gastroenterology 2015; 148:108-17. doi: 10.1053/j.gastro.2014.10.001 [Crossref] [ Google Scholar]
- Elfiky AA. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci 2020; 248:117477. doi: 10.1016/j.lfs.2020.117477 [Crossref] [ Google Scholar]
- McCarthy B. Antivirals--an increasingly healthy investment. Nat Biotechnol 2007; 25:1390-3. doi: 10.1038/nbt1207-1390 [Crossref] [ Google Scholar]