Bioimpacts. 12(6):549-559.
doi: 10.34172/bi.2022.23489
Original Research
Inhibitory effects of gallic acid on the activity of exosomal secretory pathway in breast cancer cell lines: A possible anticancer impact
Nasrollah Jabbari 1  , Maryam Feghhi 1, Omid Esnaashari 2, Hamid Soraya 3, Jafar Rezaie 1, *
, Maryam Feghhi 1, Omid Esnaashari 2, Hamid Soraya 3, Jafar Rezaie 1, * 
Author information:
1Solid Tumor Research Center, Cellular and Molecular Medicine Institute, Urmia University of Medical Sciences, Urmia, Iran
2Omid Research and Treatment Center, Urmia, Iran
3Department of Pharmacology Toxicology, School of Pharmacy, Urmia University of Medical Sciences, Urmia, Iran
Abstract
Introduction:
Breast cancer cells produce exosomes that promote tumorigenesis. The anticancer properties of gallic acid have been reported. However, the mechanism underlying its anticancer effect on the exosomal secretory pathway is still unclear. We investigated the effect of gallic acid on exosome biogenesis in breast cancer cell lines.
Methods:
The cytotoxic effect of gallic acid on MCF-10a, MCF-7, and MDA-MD-231 cells was measured by MTT assay after 48 hours treatment. Expression of miRNAs including miRNA-21, -155, and 182 as well as exosomal genes such as Rab27a, b, Rab11, Alix, and CD63; along with HSP-70 (autophagy gene), was determined using Q-PCR. The subcellular distribution of it was monitored by flow cytometry analysis. Isolated exosomes were characterized by transmission and scanning electron microscopes and flow cytometry. Acetylcholinesterase activity is used to measure the number of exosomes in supernatants. In addition, autophagy markers including LC3 and P62 were measured by ELISA.
Results:
Data showed that gallic acid was cytotoxic to cells (P < 0.05). Gallic acid modulated expression of miRNAs and down-regulated transcript levels of exosomal genes and up-regulated the HSP-70 gene in three cell lines (P < 0.05). The surface CD63/total CD63 ratio as well as acetylcholinesterase activity decreased in treated cells (P < 0.05). The protein level of LC3 was increased in three cell lines, while the expression of P62 increased in MCF-7 and MDA-MB-231 cancer cell lines.
Conclusion:
Together, gallic acid decreased the activity of the exosomal secretory pathway in breast cancer cell lines, providing evidence for its anti-cancer effects.
Keywords: Breast cancer, Exosomes, Gallic acid, Autophagy, MCF-7 cells
Copyright and License Information
© 2022 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Breast cancer, prevalent cancer, is the second cause of death among women worldwide.
1
According to global cancer statistics, more than2 million new cases of breast cancer patients were diagnosed in 2018.
2
Currently, different treatment methods including radiotherapy, chemotherapy, hormone therapy, and surgery are available
3,4
; unfortunately, some patients suffer from resistance to therapies. Therefore, breast tumor is one of the aggressive and therapy-resistant tumors that release more exosomes with distinct cargo upon exposure to different therapies such as radiotherapy,
5,6
and chemotherapy.
7
Exosomes are extracellular vesicles (30-120 nm) that originate from multivesicular bodies (MVBs) via intrigue mechanisms and are secreted out of cells following MVB-the plasma membrane fusion.
8
Several vital mechanisms are involved in exosome formation and release upon cells' responses to stimuli, despite the fact, that our knowledge of this scenario is still insufficient.
9,10
Different molecules such as CD63 and Alix play pivotal roles in exosomes biogenesis; and Rab proteins (Rab27, Rab11) regulate intracellular trafficking and secretion of MVB/exosomes.
8
Various miRNAs such as miRNA-21, miRNA-155, and miRNA-182 have been shown to regulate tumorigenesis.
11,12
These miRNAs are non-coding RNAs with about 22 nucleotides that target the expression of exosomal genes post-transcriptionally.
13,14
In the tumor environment, tumor cells communicate with each other and adjacent stromal cells through exosomes containing biological components.
15-17
Tumor cells derived exosomes containing different types of biological molecules such as proteins, nucleic acid, and lipids that contribute to promoting proliferation, invasion, angiogenesis, metastasis, and also resistance to therapies.
18
Previous studies indicated that gallic acid, a yellowish-white crystalline phenolic compound, represents broad-spectrum therapeutic properties like anti-carcinogenic, anti-inflammatory, anti-fungal, anti-bacterial, anti-tyrosinase, and anti-diabetic.
19-21
Besides, due to its scavenging capacity against free radicals, gallic acid acts as an ideal antioxidant agent and is frequently used as a constituent of various cosmetic products. It can also protect cells from damage induced by ionizing radiation.
22
Most studies have only focused on the cytotoxic properties of gallic acid on different cancer cells and declared that it is a useful additive with vitamins and as a nutritional complement to inhibit cancer metastasis and relapse.
23-25
Furthermore, it was suggested that gallic acid and its derivatives could be further developed as hopeful lead molecules for new drug development.
25
Given an excellent useful profile, gallic acid is an attractive candidate as a possible anticancer agent. Nevertheless, there remains limited knowledge regarding its effect on the exosomal secretory pathway. In the present study, therefore, we studied the effects of gallic acid on the exosomal and autophagy pathways in MCF-7 and MDA-MB-231 cells as breast cancer cell lines and MCF-10a cells as normal breast epithelial cells; clarifying its effects on the viability of cells and the exosomal secretory pathway. Our findings present new evidence for the potential effect of gallic acid on exosome biogenesis and secretion in breast cancer cells as well as in non-cancerous cells.
Materials and Methods
Reagents
All chemicals and reagents were used as received. The human breast cell lines such as MCF-7, MDA-MB-231, and MCF-10A cells were purchased from the Pasteur Institute of Iran (Tehran, Iran). Gallic acid, Thiazolyl blue tetrazolium bromide (MTT), TRIzol reagent, chloroform, isopropanol, dimethyl sulfoxide (DMSO), BSA, Chromogen substrate 1,2,4,5‐ tetramethyl benzene (TMB), ethanol, and PBS were purchased from Sigma-Aldrich Company (Munich, Germany). RPMI-1640, FBS, penicillin, and Trypsin-EDTA were received from Gibco Company (USA). Exosome isolation kit obtained from Cell Guidance Systems Ltd Company (UK). Primary rabbit LC3 antibody and primary P62 antibody purchased from Cell Signaling Company (USA). SYBR Green High ROX Master mix and miRNA First-Strand cDNA Synthesis Kit obtained from Stem Cell Technology Research Center (Tehran, Iran). cDNA synthesis kit and SYBR Green PCR Master Mix were obtained from Yektatajhiz Company (Tehran, Iran), Cholinesterase kit was provided from Pars-Azmun Company (Tehran, Iran). Secondary anti-mouse IG-FITC was obtained from Biolegend Company (UK). Triton X-100 solution and H2SO4 solution were purchased from Merck Millipore Company (USA). Primary anti-CD63 antibody was obtained from Sunta Cruz Biotechnology Inc (Germany). Horseradish peroxidase (HRP)‐conjugated goat anti-mouse secondary antibody was received from DNAbiotech Company (Tehran, Iran).
Cell culture and treatment
The human breast cancer cell lines MCF-7 and MDA-MB-231 cells, and a normal epithelial cell line MCF-10A were cultured in RPMI-1640, supplemented with 10% FBS and streptomycin, and penicillin (100 μg/100 U ml-1), and incubated at 37°C in a 5% CO2 incubator. About 1 × 106 cells were cultured in each flask and grown until confluence was reached. Every 2-3 days, the supernatants were replaced by a fresh complete medium. We used cells between the second and sixth passages for experiments. Gallic acid was dissolved in double-distilled water (ddH2O) and stored at a concentration of 1000 mM at 4°C.
Cell viability assay
Cell viability was detected by the classical MTT assay based on the manufacturer’s directions. For example, 5000 cells were seeded per well in 96-well cell culture plates and kept at 37°C for 24 hours, to grow monolayer cells. After treatment of cells with serial dilutions of gallic acid (0, 10, 20, 40, 80, 160, 320 mM) for 48 hours, the medium was removed and 20 µL MTT solution (5 mg/mL) was added to each well of a 96-well plate and kept at 37°C and 5% CO2 for 4 hours. Next, the medium in the wells was replaced with 100 μL DMSO for solubilizing the formazan crystals. Using a microplate reader (Biotek), optical density (OD) was recorded at 570 nm. The percentage of cell viability was evaluated by the following equation: Viability (%) = (OD570 of treated cells) / (OD570 of control cells) ×100
Cellular cytotoxicity was assessed by the calculation of the inhibitory concentration of the gallic acid required to cause 50% of cells death recognized as the half-maximal inhibitory concentration (IC50). IC50 was calculated by GraphPad software version 8. For downstream experiments, cells were co-cultured with IC50 gallic acid for 48 hours.
Real-time polymerase chain reaction
For real-time PCR, initially, total RNA was isolated from cells by Trizol reagent based on the manufacturer’s instruction. Briefly, treated and control cells were lysed by Trizol reagent and then the components of lysates were separated by chloroform. Afterward, samples were centrifuged at 12000 g for 15 minutes at 4°C and the colorless upper aqueous phase was obtained, and isopropanol was added for 20 minutes at -20°C. Samples were centrifuged and then the RNA pellet was washed using ethanol (75%) twice. The RNA pellet was dissolved in DEPC-treated water. The concentration and purity of total RNA were calculated using a Nanodrop system (BioTek). A proper amount of total RNA (1000 ng) was converted into cDNA using a cDNA synthesis kit and miRNA First-Strand cDNA Synthesis Kit according to protocols. PCR was done on a MIC Real-Time PCR System (Swiss), with a PCR Master Mix for exosomal mRNA analysis and SYBR Green High ROX Master mix for the relative expression level of miRNAs. Expressions of mRNAs were controlled against GAPDH and the expression of miRNAs was normalized to that of Snord47. Relative expression was evaluated using the 2−ΔΔCt method. Primers were designed by Oligo v7.60 software and evaluated by Primer-Blast tool of NCBI databases (https://www.ncbi.nlm.nih.gov/). The sequences of primers are presented in Tables 1 and 2, and the conditions for real-time PCR are presented in Table 3.
Table 1.
List of miRNA primers
|
Name
|
Sequence (5 3 )
|
Tm (°C)
|
| miRNA-21 |
GGC TTG TCA GAC TGA TG |
60 |
| miRNA-182 |
TTG GCA ATG GTA GAA CT |
60 |
| miRNA-155 |
TAG GCT AAG CGT GAT AG |
60 |
| SNORD47 |
ATC ACT GTA AAA CCG TT |
60 |
Table 2.
List of miRNA primers
|
Genes
|
Sense
|
Antisense
|
Tm (°C)
|
| Rab27a |
AGAGGAGGAAGCCATAGCAC |
CATGACCATTTGATCGCACCAC |
59 |
| Rab27b |
GGAACTGGCTGACAAATATGG |
CAGTATCAGGGATTTGTGTCTT |
59 |
| Rab11 |
CCTCAGCCTCTACGAAGCAAA |
CCGGAAGTTGATCTCCTCCTG |
59 |
| CD63 |
TCCTGAGTCAGACCATAATCC |
GATGGCAAACGTGATCATAAG |
63 |
| Alix |
CTGGAAGGATGCTTTCGATAAAGG |
AGGCTGCACAATTGAACAACAC |
63 |
| HSP-70 |
GCCGAGCATTCTCTGATCCA |
AACACTTTCGGCTGTCTCCT |
59 |
| GAPDH |
CAAGTTCAACGGCACAGTCAAG |
ATACTCAGCACCAGCATCACC |
60 |
Table 3.
The real-time PCR conditions for mRNAs and miRNAs
|
Bio-materials
|
Temperature (C°)
|
Duration
|
Cycles
|
| mRNAs |
95 |
5 min |
1 |
| 95 |
10 s |
| Tm |
35 s |
40 |
| 72 |
20 s |
| miRNAs |
95 |
2 min |
1 |
| 95 |
5 s |
40 |
| 60 |
30 s |
Isolation of exosomes
Exosomes were extracted from the supernatant of treated and control cells.
26
Cells were cultured in FBS-free medium for 48 hours. In brief, according to the exosome isolation kit’s recommendation, cell culture supernatants were cleared of cell debris by continuous centrifuging (300 × g and then 10 000 × g for 10 minutes at 4°C) to remove cellular debris. Then, the supernatant was filtered by a 0.22 µm filter. Supernatants were mixed with isolation buffer and kept overnight at 4°C. Next, samples were centrifuged at 16 000 × g for 1 hour and then precipitate dissolved in 200 µL PBS and moved onto filter columns and centrifuged 670 × g for 1 minute. Exosome pellets were suspended in 100 µL PBS and kept at 4°C for downstream studies.
Quantification of exosomes
The amount of exosome released into supernatants of treated and control cells was evaluated using theacetylcholinesterase activity (AChE activity) assay
27,28
according to the manufacturer’s recommendation for a cholinesterase kit. Briefly, 500 µL R1 buffer was mixed with 100 µL of exosome from each group and incubated for 5 minutes at room temperature. Then, samples were mixed with 20 µL R2 buffer, and absorbance was read at 405 nm at three different intervals with a microplate reader system. AChE activity was calculated accordance with the following formula: Activity (U/l) = ΔAbs/ min × 65,800
Characterization of exosomes
We used TEM (The Netherlands) and SEM (Tescan), and flow cytometry analysis for the characterization of exosomes. For TEM analysis, glutaraldehyde 1% was mixed with 20 μL exosome samples with, and then placed on the carbon grids at room temperature and were allowed to dry. After twice washing, uranyl acetate stain 1% added on samples for 10 minutes at room temperature and photographs were taken at 80 kV by a TEM system. For SEM analysis, briefly, exosome sample (20 μL) was placed on grids and then freeze-dried. After, grids coated by gold and exosomes were visualized by the SEM system at 30 kV. For flow cytometry analysis, the primary anti-CD63 antibody mixed with 200 μL exosome samples for 2 hours at 4°C. Then, samples were incubated with secondary anti-mouse IG-FITC for 1 hour at 37°C. Expression of the CD63 marker was detected by a BD FACSCalibur system (BD Biosciences) and evaluated by FlowJo software (version 7.6.1).
Flow cytometry analysis
The intracellular distribution of exosomal marker CD63 inside treated and control cells analyzed by flow cytometry. Two protocols were planned as follows: protocol I (total CD63): cells firstly were fixed and then permeabilized with 0.1% Triton X-100 solution. Protocol II (surface CD63): cells only fixed. In brief, the primary anti-CD63 antibody) was added to cell suspension and incubated for 2 hours at 4°C. Next, cells were mixed with secondary anti-mouse IG-FITC for 1 hour at 37°C. Finally, exosomal marker CD63 was measured using a BD FACSCalibur system and Flow Jo software (version 7.6.1). The data was presented as a surface CD63/total CD63 ratio.
Enzyme-linked immunosorbent assay (ELISA)
ELISA assay was designed to measure the protein level of the autophagic markers LC3 and P62 in treated and control cells.
13,29
Briefly, primary rabbit LC3 antibody and primary P62 antibody was added into each well of 96‐well plates and kept plates at 4°C overnight. Then, wells were washed twice in PBS and 1% BSA was added to wells for blocking the remaining free activated sites for 1 hour at room temperature. After washing, an equal amount of protein (30 ng) were poured into wells and kept for 40 minutes at room temperature. Next, wells were incubated with either 100 µL of HRP‐conjugated goat anti-mouse secondary antibody and then wells were washed with PBS twice. For 20 min, 50 µL of TMB were added. To end the reaction, 50 µL of H2SO4 solution (5%) was added to each well. The absorbance of wells at 450 nm was recorded by an ELISA reader. The quantity of LC3 and P62 in treated cells was described as the level of protein against the control.
Statistical analysis
All experiments were accomplished in triplicate. Measurements were presented as mean ± standard deviation (SD). A one-way analysis of variance (ANOVA) and a Tukey post hoc test were used to analyses differences between the multiple groups, whereas comparisons between two groups were analyzed by the two-sided Student’s t test. The SPSS Statistics software (version 16.0) used to analyze experimental data.
Results
Gallic acid was cytotoxic to cells
Gallic aciddose-dependentlyreduced the survival rate of MCF-10a, MCF-7, and MDA-MB-231 breast cancer cells after 48 hours as shown in Fig. 1. Compared with the control group, cell viability values decreased in treated MCF-10a cells (P < 0.001) (Fig. 1A). In addition, there was a significant difference between 10 mM treated cells and other treated cells (P < 0.05). We found that the viability of 160 mM and 320 mM treated cells was low as compared with 20 mM, 40 mM, and 80 mM treated cells (P < 0.001; Fig. 1). There was a significant reduction in cell viability at 320 mM (35.34 ± 1.7 %) treated cells compared with that of 160 mM treated cells (43.75 ± 1.54 %) (P < 0.01).
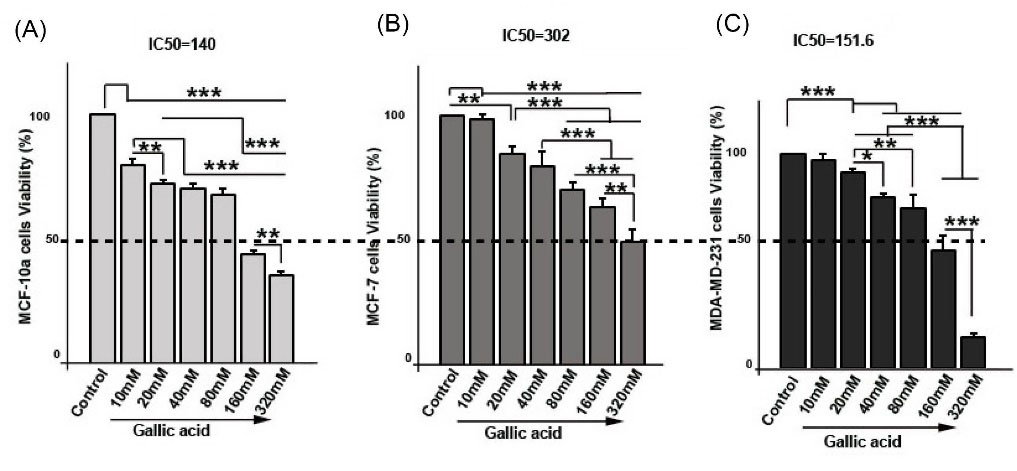
Fig. 1.
The cell survival rate of MCF-10a (A), MCF-7 (B), and MDA-MB-231 (C) was evaluated in response to Gallic acid at different concentrations by MTT assay. IC50 assay was performed after 48 h Gallic acid treatment by GraphPad (Prism) software. MCF-10a (IC50= 140 mM; A), MCF-7 (IC50= 302 mM; B), and MDA-MB-231 (IC50=151.6; C). One-way ANOVA with Tukey test. The data is the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
.
The cell survival rate of MCF-10a (A), MCF-7 (B), and MDA-MB-231 (C) was evaluated in response to Gallic acid at different concentrations by MTT assay. IC50 assay was performed after 48 h Gallic acid treatment by GraphPad (Prism) software. MCF-10a (IC50= 140 mM; A), MCF-7 (IC50= 302 mM; B), and MDA-MB-231 (IC50=151.6; C). One-way ANOVA with Tukey test. The data is the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Similar to MCF-10a cells, gallic acidreduced the survival rate of MCF-7 cells in comparison with control cells (PControl and 10mM vs.20 mM < 0.01;PControl and 10mM vs.40mM, 80mM, 160mM, and 320mM < 0.001, Fig. 1B). As shown in Fig. 1, gallic acid declined the viability of cells in a dose manner (P20mM vs.80mM, 160mM, and 320 mM < 0.001; P40mM vs.160mM and 320mM < 0.001 0; P160mM vs. 320mM < 0.01).
The same results were observed in MDA-MB-231 cells cultured with gallic acid (Fig. 1C). Results pointed out that MDA-MB-231 cells exhibited a significant decrease in percentage of viability than cells of control group (PControl, 10mM and 20mM vs.40mM, 80mM, 160mM, and 320mM < 0.001, Fig. 1). We also found that viability of cells in 40 mM, 80 mM, 160 mM, and 320 mM decreased as compared to 20 mM treated cells (P20mM vs.40mM< 0.05; P 20mM vs. 80mM <0.01; P20mM vs. 160mM and 320mM < 0.001). The same results were observed when 40 mM and 80 mM treated cells were compared with 160 mM and 320 mM treated cells (P < 0.001; Fig. 1). Compared to 160 mM treated cells (58.14 ± 6.9 %), a decreased level of viability in 320 mM cells (14.2 ± 2.19 %) was observed (P < 0.001). Results indicate the viability of MCF-10a, MCF-7, and MDA-MB-231 cells was dose-dependently reduced after 48 hours. Furthermore, results from Graph pad software showed that gallic acid IC50 for MCF-10a (Fig. 1A), MCF-7 (Fig. 1B), and MDA-MB-231 (Fig. 1C) cells were 140 mM, 302 mM, and 151.6 mM for 48 hours respectively.
Gallic acid modulated expression of miRNAs in cells
Fig. 2A shows that, in MCF-10a cells treated with gallic acid, the expression of miRNA-21 was not affected significantly (P> 0.05), however, the expression levels of both miRNA-155 (0.31 ± 0.014 fold change) and miRNA-182 (0.39 ± 0.019 fold change) significantly decreased (P < 0.01). In addition, we found that relative expression of miRNA-21 and miRNA-155 were slightly decreased (P> 0.05), reversely, expression of miRNA-182 was increased in gallic acid treated MCF-7 cells (P < 0.05, Fig. 2B). In MDA-MB-231 cells, we found that expression of miRNA-21 was not affected significantly (0.76 ± 0.05 fold change; P> 0.05), and expression of miRNA-155 was decreased, whereas expression of miRNA-182 was increased (P < 0.05, Fig. 2C).
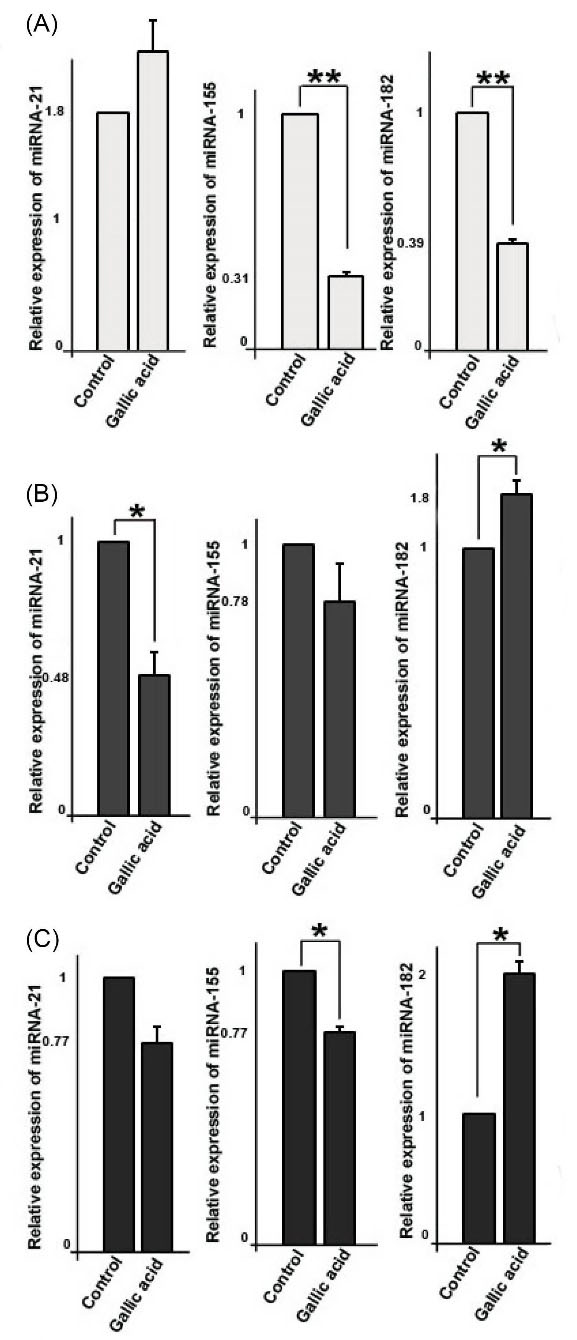
Fig. 2.
Relative quantitative expression of oncogenic miRNAs including miRNA-21, miRNA-155, and miRNA82 in three cell types; MCF-10a (A), MCF-7 (B), and MDA-MB-231(C). Student’s t test. The data represent the mean ± SD from three independent experiments. *P < 0.05, **P < 0.01.
.
Relative quantitative expression of oncogenic miRNAs including miRNA-21, miRNA-155, and miRNA82 in three cell types; MCF-10a (A), MCF-7 (B), and MDA-MB-231(C). Student’s t test. The data represent the mean ± SD from three independent experiments. *P < 0.05, **P < 0.01.
Gallic acid down-regulated expression of CD63, Alix, and Rabs
We used quantitative PCR analysis to measure the expression of genes regulating exosome secretion pathways including Rab27a, Rab27b, Rab11, CD63, and Alix. Data showed that mRNA levels of Rab27a in treated MCF-10a cells were not affected significantly (0.7 ± 0.13 fold change; P> 0.05), however, mRNA levels of Rab27b (P < 0.05), Rab11 (P < 0.01), CD63 (P < 0.05), and Alix (P < 0.05) significantly decreased (Fig. 3). In addition, we detected the expression of Rab27a in MCF-7 cells was decreased (0.48 ± 0.02 fold change; P < 0.05). mRNA level of Rab27b (0.56 ± 0.034 fold change) was lower in treated MCF-7 cells, while mRNA level of Rab11 (1.7 ± 0.075 fold change) increased in gallic acid-treated cells as compared with non-treated control cells (P < 0.05). Similar to MCF-10a cells, expression of Alix and CD63 decreased in treated cells (P < 0.01 and P < 0.05 respectively).
As shown in Fig. 3, gallic acid did not affect the expression of Rab27a and Rab11 in MDA-MB-231 cells (P> 0.05), conversely, it decreased the expression of Rab27b, CD63, and Alix (P < 0.05). These results indicate gallic acid could decrease the expression of genes involved in the exosomal secretory pathway.
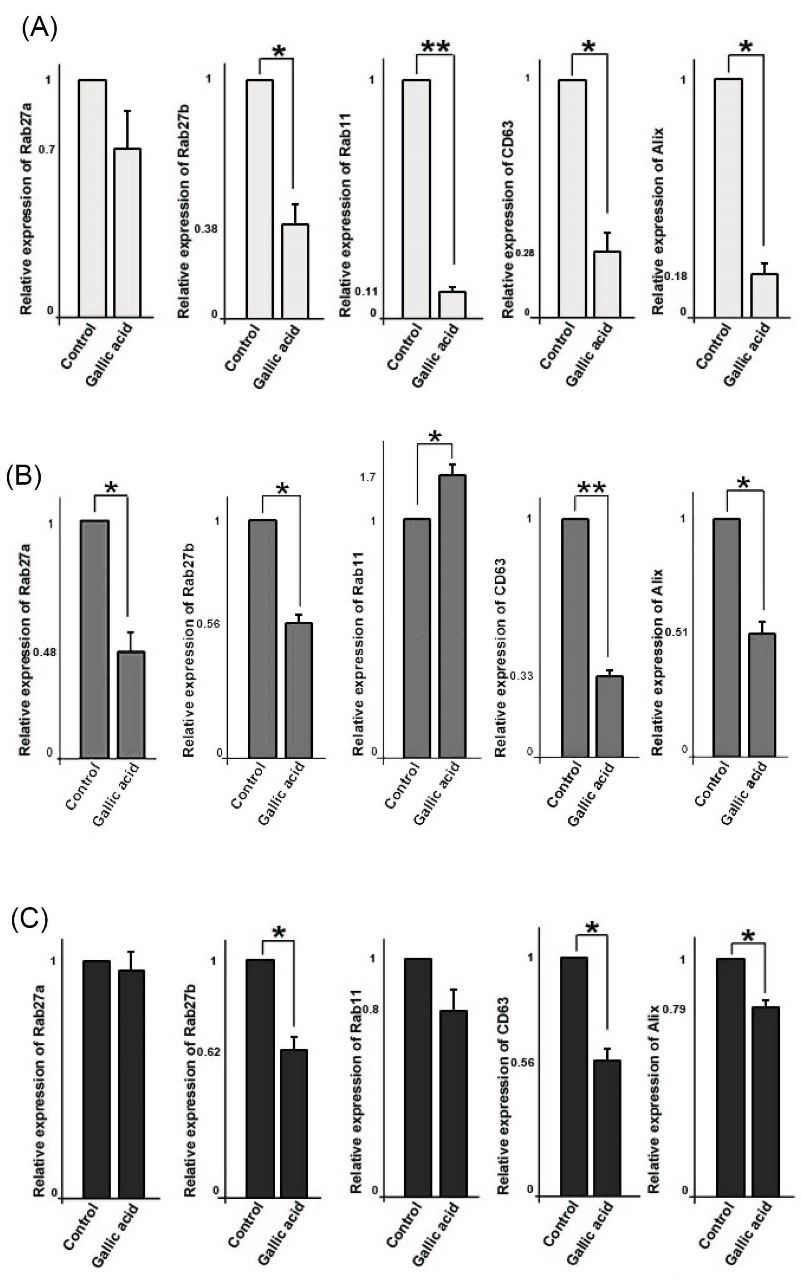
Fig. 3.
Relative quantitative expression of genes involved in exosome biogenesis and secretion such as Rab27a, Rab27b, Rab11, CD63, and Alix in three cell types; MCF-10a (A), MCF-7 (B), and MDA-MB-231(C). Student’s t test. The data shows the mean ± SD. *P < 0.05, **P < 0.01.
.
Relative quantitative expression of genes involved in exosome biogenesis and secretion such as Rab27a, Rab27b, Rab11, CD63, and Alix in three cell types; MCF-10a (A), MCF-7 (B), and MDA-MB-231(C). Student’s t test. The data shows the mean ± SD. *P < 0.05, **P < 0.01.
Gallic acid altered the intercellular distribution of CD63 in treated cells
We used flow cytometry analysis to further monitor the distribution of exosomal marker CD63 inside cells following exposure to gallic acid. For this purpose, the total and surface CD63 were measured by permeabilizing or nonpermeabilizing methods, respectively (Fig. 4A). As compared to the control cells, flow cytometry analysis showed that the surface CD63/total CD63 ratio was considerably decreased in the treated cells (P < 0.05; Figs. 4B, C, D). This observation suggested an increase in the cell-surface CD63 accumulation in gallic acid-treated cells.
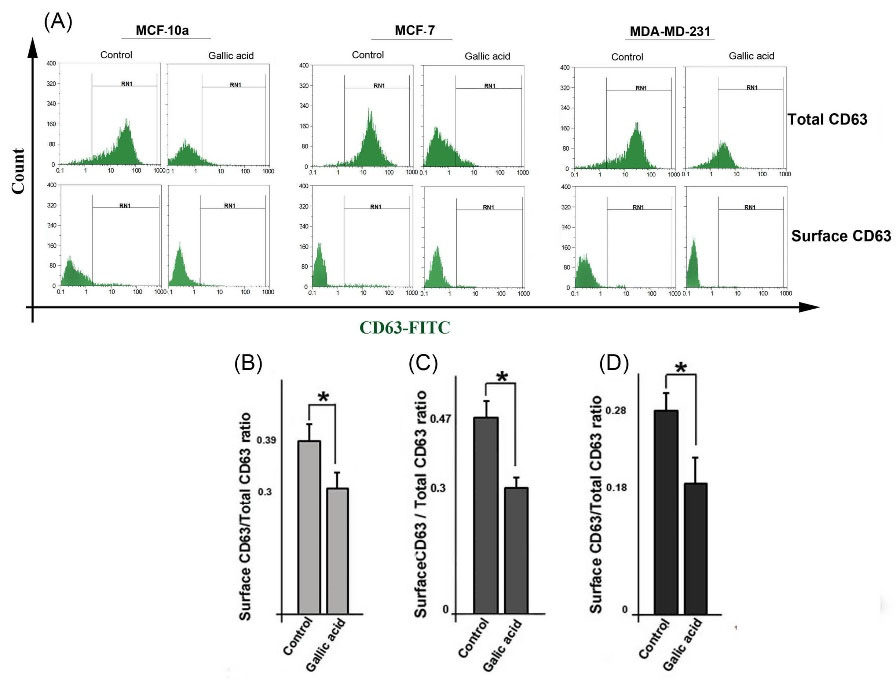
Fig. 4.
Flow cytometry analysis of the subcellular distribution of CD63 in cells. Cells were stained with a CD63-FITC antibody for measuring the percentage of total and surface CD63 protein (A) and analyzed the ratio of surface CD63/ total CD63 for MCF-10a (B), MCF-7 (C), and MDA-MB-231 (D) cells. Student’s t-test. The data represent the mean ± SD. *P < 0.05.
.
Flow cytometry analysis of the subcellular distribution of CD63 in cells. Cells were stained with a CD63-FITC antibody for measuring the percentage of total and surface CD63 protein (A) and analyzed the ratio of surface CD63/ total CD63 for MCF-10a (B), MCF-7 (C), and MDA-MB-231 (D) cells. Student’s t-test. The data represent the mean ± SD. *P < 0.05.
Gallic acid reduced AChE activity in exosome
AChE is an enzyme that is associated with exosomes
30
and AChE activity could serve as a sign of the amount of secreted exosomes.
6,31
Data showed that the AChE activity of treated three cell lines decreased as compared to corresponding control cells (P < 0.05, Fig. 5B).
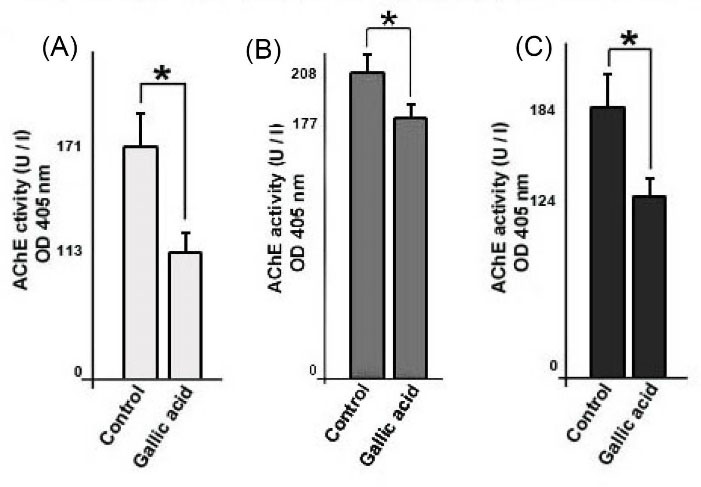
Fig. 5.
Measurement of exosomes in supernatants of cells by acetylcholinesterase (AChE) activity assay. MCF-10a (A), MCF-7 (B), and MDA-MB-231 (C). Student’s t test. The data present the mean ± SD. *P < 0.05.
.
Measurement of exosomes in supernatants of cells by acetylcholinesterase (AChE) activity assay. MCF-10a (A), MCF-7 (B), and MDA-MB-231 (C). Student’s t test. The data present the mean ± SD. *P < 0.05.
Characterization of purified exosomes
We characterized exosomes released from cells according to International Society for Extracellular Vesicles (ISEV) guidelines by SEM, TEM, and flow cytometry techniques. SEM and TEM showed the round shape and nano-scale-sized exosomes derived from MCF-7 cells (Fig. 6A, 6B). Immunophenotyping by flow cytometry confirmed the CD63 marker in isolated exosomes (Fig. 6C).
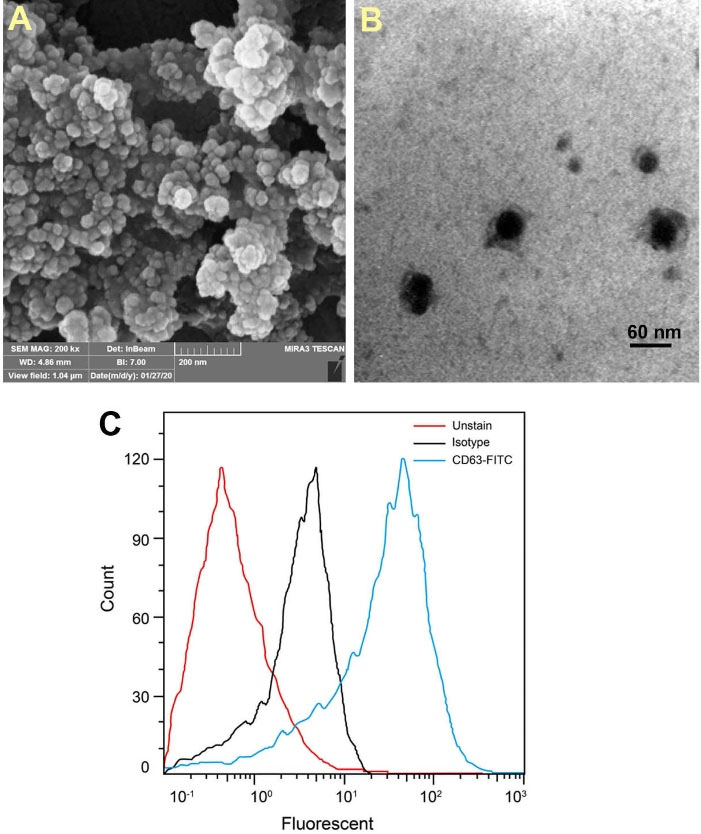
Fig. 6.
Characterizing of purified exosomes from MCF-7 cells using transmission electron micrographs (A), scanning electron micrographs (B), and flow cytometry analysis for CD63 marker (C).
.
Characterizing of purified exosomes from MCF-7 cells using transmission electron micrographs (A), scanning electron micrographs (B), and flow cytometry analysis for CD63 marker (C).
Gallic acid altered mRNA level of HSP-70 gene and protein levels of LC3 and P62 in cells
We further investigated the levels of autophagy markers in gallic acid-treated cells (Fig. 7 I, II). Real time-PCR showed that the mRNA level of HSP-70 increased in treated cells (The >1.32–fold for MCF-10a, the >1.6–fold for MCF-7, the >1.8–fold for MDA-MB-231) (P < 0.05, Fig. 7A, 7B, and 7C). In addition, ELISA showed that levels of LC3 and P62 increased and decreased in treated MCF-10a cells, respectively (P < 0.05; Fig. 7D). Levels of both LC3 and P62 proteins increased in MCF-7 and MDA-MB-231 cells against control cells (P < 0.05, Fig. 7E, 7F). Gallic acid has been identified as a cellular stressor that modulates autophagy in cells.
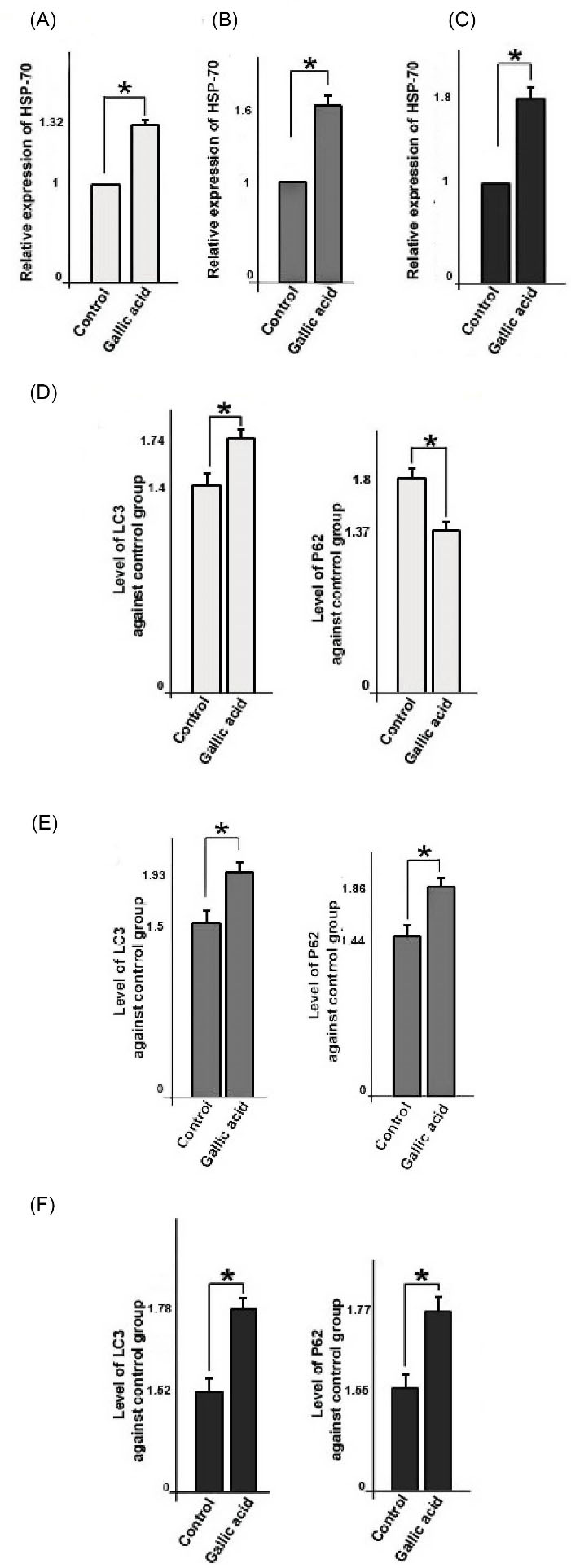
Fig. 7.
Relative quantitative expression of HSP-70 genes in three cell lines; MCF-10a (A), MCF-7 (B), and MDA-MB-231(C). Detection of autophagy markers like LC3 and P62 in three cell types by ELISA (enzyme-linked immunosorbent assay); MCF-10a (D), MCF-7 (E), and MDA-MB-231(F). Student’s t-test. The data represent the mean ± SD from three independent experiments. *P < 0.05.
.
Relative quantitative expression of HSP-70 genes in three cell lines; MCF-10a (A), MCF-7 (B), and MDA-MB-231(C). Detection of autophagy markers like LC3 and P62 in three cell types by ELISA (enzyme-linked immunosorbent assay); MCF-10a (D), MCF-7 (E), and MDA-MB-231(F). Student’s t-test. The data represent the mean ± SD from three independent experiments. *P < 0.05.
Discussion
The effect of gallic acid on exosome biogenesis and secretion in breast epithelial cells remains elusive. In our study, gallic acid was initially found to be cytotoxic to cells for 48 hours. Gallic acid dose-dependently decreased the viability of cells (Fig. 1), which is in excellent agreement with the previous reports on breast cancer cells.
32-34
Rezaei-Seresht et al showed that gallic acid decreased the proliferation of MCF-7 cells and induced apoptosis through the intrinsic apoptotic signaling pathway.
35
In addition, gallic acid has been shown to inhibit cancer cell survival via suppressing NFκB pathways like MEK1, JNK1/2, and p90RSK.
36
Based on the IC50 data (Fig. 1), gallic acid is preferentially cytotoxic to cells, therefore, it seems that breast cancer cells (302 mM and 151.6 mM) are more resistant to gallic acid rather than MCF-10a (IC50:140mM) cells after 48 hours treatment. miRNAs participate in several cellular activities including proliferation, metastasis, differentiation, and chemoresistance.
37,38
We propose that gallic acid may directly affect oncogenic miRNAs such as miRNA-155, miRNA-21, and miRNA-182. We found that expression of miRNA-21 was not affected in both treated MCF-10a and MDA-MB-231 cells but decreased in MCF-7 cells. Besides, the expression of miRNA-155 decreased in MCF-10a and MDA-MB-231 cells, while was not altered in MCF-7 cells. The expression of miRNA-182 was increased in MCF-7 cells and MDA-MB-231 cells but was decreased in MCF-10a cells (Fig. 2). These miRNAs are oncogenic,
39
chemoresistances,
12,40
and angiogenic
41
molecules that are implicated in promoting tumorigenesis. Most likely, this is the first report and their expression patterns may be related to cell type. Therefore, further scrutiny is essential to elucidate the underlying mechanisms behind the diverse expression patterns of these miRNAs in different breast cancer cells. Since stress conditions such as exposure to the exogenous compound are capable of affecting the exosome signaling pathway, we seek to monitor the possible effect of gallic acid on exosome biogenesis and secretion.
We found that gallic acid treatment down-regulated key genes involved in exosome biogenesis and secretion. Tumor cells produce abundant exosomes to communicate with other cells and promote tumorigenesis and resistance to therapies.
15
Different Rab-GTPase proteins are involved in the intracellular trafficking of MVB/exosomes.
15
For example, Rab27b mediates the movement of MVB/exosomes from the prenuclear area to the plasma membrane,
42
while Rab27a and Rab11 regulate the plasma membrane and MVB fusion.
42,43
As shown in Fig. 3, expression of Rab27a was not affected in treated MDA-MB-231 and MCF-10a cells however were decreased in MCf-7 cells. Rab27b gene was down-regulated in all three cells, however, Rab11 decreased in MCF-10a cells and increased in MCF-7 cells, and was not affected in MDA-MB-231 cells. It may be assumed that the expression pattern of Rab proteins depended on cell type. We found that the transcript levels of CD63 and Alix genes simultaneously with CD63 protein levels were decreased in treated cells. It was confirmed that CD63, a tetraspanin protein located on MVB and exosome membranes,
44
and Alix
45
are involved in exosome loading and biogenesis. A decrease in expression of these genes may indicate a reduction in exosome formation, loading, and secretion. Commensurate with these findings, we observed a decrease in surface CD63/total CD63 fraction in cells incubated with gallic acid (Fig. 4), representing the reduction of MVB/exosomes fusion with the plasma membrane upon exposure to gallic acid. Moreover, the activity of AChE, an exosome-associated enzyme, was reduced (Fig. 5), demonstrating a decreased exosome secretion rate in treated cells. Overall, these data support the idea that gallic acid is able to inhibit the biogenesis and abscission of exosomes in cells.
Furthermore, we found that gallic acid-induced autophagy in cells by modulating the expression of HSP-70, LC3, and P62 molecules (Fig. 7). Similarly, previous studies indicated that gallic acid increased autophagy both in vitro and in animal models.
46,47
We found that the transcript level of the HSP-70 gene increased in treated cells (Fig. 7). Abdelwahab et al demonstrated that gallic acid modulated expression of the HSP-70 in rat gastric lesion model.
48
HSP-70, heat shock protein, facilitates the degradation of irreversibly denatured proteins, which accumulate inside cells in response to stress conditions.
49
Hsp-70 mediates suitable assembly, conformation, and transport of proteins; and autophagy eliminates proteins and organelles that are no longer useful or necessary and recycles their components, thus, autophagy and HSP-70 under stress conditions complement each other to maintain cell homeostasis.
50
Autophagy is a multi-step process and is known to play a pivotal role in conserving the homeostasis of tumor cells under different stress conditions, like nutrient insufficiency, chemotherapy, and radiotherapy.
51-53
In the initiation step, LC3 in the conjugation system contributes to inducing autophagosome biogenesis.
54
Thus, LC3 is considered a marker of an early stage of autophagy.
55
P62, known as sequestosome 1 (SQSTM1), is involved in the final step of autophagy and indirectly mediates the fusion of the autophagosome with the lysosome.
56
In fact, the P62 protein directly cooperates with GABARAP family proteins and also LC3, and transfers ubiquitinated molecules into the lysosomes.
56
In keeping, P62 is hydrolyzed by autophagy and then is accumulated when autophagy is suppressed and its decrease is correlated with autophagy initiation step.
55
The preliminary results of our study showed that gallic acid promoted autophagy flux in MCF-10a cells. However in MCF-7 and MDA-MB-231 cells, gallic acid induced the early step of autophagy by up-regulation of LC3 protein, while the lysosome-autophagosome, which is the final step of autophagy, was inhibited as shown by increase of P62 after 48 hours treatment. Nevertheless, these findings are preliminary and further studies are essential.
In summary, by studying gallic acid-treated three cells, we shed light on the cellular mechanism of exosome biogenesis inside breast cancer cells and non-cancerous cells. We showed that gallic acid treatment decreased the activity of the exosomal secretory pathway as well as exosome secretion. Presumably, these features could be beneficial in using gallic acid as a tool for reducing side effects and increasing the efficacy of therapies as radiotherapy and chemotherapy through inhibiting exosome biogenesis and secretion from cancer cells. In addition, as we know that autophagy facilitates the degradation of damaged molecules and organelles,
57
and represents crosstalk with exosome biogenesis, and synergically promotes cellular homeostasis through removing damaged biomolecules.
58
Further, the coordination between autophagy flux and the exosomal secretory pathway is a synchronized route, therefore, deficiency in exosome biogenesis is compensated by autophagy flux and vice versa.
59
We would assume that gallic acid decreased activity of the exosomal secretory pathway and blocked autophagy after 48 h of treatment.
Research Highlights
What is the current knowledge?
√ Breast cancer is an aggressive and therapy-resistant tumor.
√ Breast cancer cells abundantly produce exosomes that support tumor growth.
√ Gallic acid has anti-cancer properties.
What is new here?
√ Gallic acid IC50 values of breast cancer cell lines are more than normal breast cancer cell line
√ Gallic acid inhibits the activity of the exosomal pathway and decreases exosome secretion in cells.
√ Gallic acid affected miRNAs expression patterns in cells.
√ Gallic acid regulated autophagy in cells.
√ A decrease in the activity of the exosomal pathway may correlate to the anticancer effects of gallic acid.
Conclusion
Our findings suggest that gallic acid can decrease exosome biogenesis and secretion in all three cell lines. In addition, it was capable of blocking autophagy in both MCF-7 and MDA-MB-231 cancer cells. These results thus highlight its favorable features for designing supplementary novel cancer treatment approaches. Further studies, which take in vitro and in vivo examinations into account, especially those combined with common therapies, will need to be undertaken.
Acknowledgments
The authors acknowledge the Research Vice-Chancellor (VCR) at Urmia University of Medical Sciences (Urmia, Iran) for the endowment.
Funding sources
The Research Committee, Urmia University of Medical Sciences supported this study (IR.UMSU.REC.1398.332).
Ethical statement
All procedures in the present work were permitted by the ethics committee of Urmia University of Medical Sciences (Ethical code: IR.UMSU.REC.1398.332).
Competing interests
The authors declare that they have no conflict of interest.
Authors’ contribution
JR and NJ contributed to the study conception and design. Material preparation and data collection were performed by MF and OE. The first draft of the manuscript was written by MF and HS. JR performed supervision, data analysis, and presentation. NJ contributed to writing and reviewing. All authors read and approved the final manuscript.
References
- Scott LC, Mobley LR, Kuo T-M, Il’yasova D. Update on triple-negative breast cancer disparities for the United States: A population-based study from the United States Cancer Statistics database, 2010 through 2014. Cancer 2019; 125:3412-3417. doi: 10.1002/cncr.32207 [Crossref] [ Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394-424. doi: 10.3322/caac.21492 [Crossref] [ Google Scholar]
- Jabbari N, Zarei L, Esmaeili Govarchin Galeh H, Mansori Motlagh B. Assessment of synergistic effect of combining hyperthermia with irradiation and calcium carbonate nanoparticles on proliferation of human breast adenocarcinoma cell line (MCF-7 cells). Artif Cells NanomedBiotechnol 2018; 46:364-372. doi: 10.1080/21691401.2018.1457537 [Crossref] [ Google Scholar]
- Tutt A, Yarnold J. Radiobiology of breast cancer. Clin Oncol (R Coll Radiol) 2006; 18:166-78. doi: 10.1016/j.clon.2005.11.011 [Crossref] [ Google Scholar]
- Jelonek K, Wojakowska A, Marczak L, Muer A, Tinhofer-Keilholz I, Lysek-Gladysinska M. Ionizing radiation affects protein composition of exosomes secreted in vitro from head and neck squamous cell carcinoma. Acta Biochim Pol 2015; 62:265-72. doi: 10.18388/abp.2015_970 [Crossref] [ Google Scholar]
- Jabbari N, Nawaz M, Rezaie J. Ionizing Radiation Increases the Activity of Exosomal Secretory Pathway in MCF-7 Human Breast Cancer Cells: A Possible Way to Communicate Resistance against Radiotherapy. Int J Mol Sci 2019; 20:3649. doi: 10.3390/ijms20153649 [Crossref] [ Google Scholar]
- Dong X, Bai X, Ni J, Zhang H, Duan W, Graham P. Exosomes and breast cancer drug resistance. Cell Death Dis 2020; 11:987. doi: 10.1038/s41419-020-03189-z [Crossref] [ Google Scholar]
- Jabbari N, Karimipour M, Khaksar M, Akbariazar E, Heidarzadeh M, Mojarad B. Tumor-derived extracellular vesicles: insights into bystander effects of exosomes after irradiation. Lasers Med Sci 2020; 35:531-545. doi: 10.1007/s10103-019-02880-8 [Crossref] [ Google Scholar]
- Alenquer M, Amorim M. Exosome biogenesis, regulation, and function in viral infection. Viruses 2015; 7:5066-83. doi: 10.3390/v7092862 [Crossref] [ Google Scholar]
- Khaksar M, Sayyari M, Rezaie J, Pouyafar A, Montazersaheb S, Rahbarghazi R. High glucose condition limited the angiogenic/cardiogenic capacity of murine cardiac progenitor cells in in vitro and in vivo milieu. Cell BiochemFunct 2018; 36:346-356. doi: 10.1002/cbf.3354 [Crossref] [ Google Scholar]
- Feng Y-H, Tsao C-J. Emerging role of microRNA-21 in cancer. Biomed Rep 2016; 5:395-402. doi: 10.3892/br.2016.747 [Crossref] [ Google Scholar]
- Jia L, Luo S, Ren X, Li Y, Hu J, Liu B. miR-182 and miR-135b mediate the tumorigenesis and invasiveness of colorectal cancer cells via targeting ST6GALNAC2 and PI3K/AKT pathway. Dig Dis Sci 2017; 62:3447-59. doi: 10.1007/s10620-017-4755-z [Crossref] [ Google Scholar]
-
Soraya H, Sani NA, Jabbari N, Rezaie J. Metformin Increases Exosome Biogenesis and Secretion in U87 MG Human Glioblastoma Cells: A Possible Mechanism of Therapeutic Resistance. Arch Med Res 2020; S0188-4409(20)30377-5. 10.1016/j.arcmed.2020.10.007.
- Fabian MR, Sundermeier TR, Sonenberg N. Understanding how miRNAs post-transcriptionally regulate gene expression. Prog Mol Subcell Biol 2010; 50:1-20. doi: 10.1007/978-3-642-03103-8_1 [Crossref] [ Google Scholar]
- Nawaz M, Fatima F, Nazarenko I, Ekström K, Murtaza I, Anees M. Extracellular vesicles in ovarian cancer: applications to tumor biology, immunotherapy and biomarker discovery. Expert Rev Proteomics 2016; 13:395-409. doi: 10.1586/14789450.2016.1165613 [Crossref] [ Google Scholar]
- Fatima F, Nawaz M. Vesiculated long non-coding RNAs: offshore packages deciphering trans-regulation between cells, cancer progression and resistance to therapies. Noncoding RNA 2017; 3:10. doi: 10.3390/ncrna3010010 [Crossref] [ Google Scholar]
- Tukmechi A, Rezaee J, Nejati V, Sheikhzadeh N. Effect of acute and chronic toxicity of paraquat on immune system and growth performance in rainbow trout, Oncorhynchus mykiss. Aquac Res 2014; 45:1737-43. doi: 10.1111/are.12118 [Crossref] [ Google Scholar]
- Whiteside TL. Tumor-derived exosomes and their role in cancer progression. Adv Clin Chem: Elsevier 2016; 74:103-41. doi: 10.1016/bs.acc.2015.12.005 [Crossref] [ Google Scholar]
- Fazary AE, Taha M, Ju Y-H. Iron complexation studies of gallic acid. Int J Chem Eng Data 2009; 54:35-42. doi: 10.1021/je800441u [Crossref] [ Google Scholar]
- Werner RA, Rossmann A, Schwarz C, Bacher A, Schmidt H-L, Eisenreich W. Biosynthesis of gallic acid in Rhus typhina: discrimination between alternative pathways from natural oxygen isotope abundance. Phytochemistry 2004; 65:2809-13. doi: 10.1016/j.phytochem.2004.08.020 [Crossref] [ Google Scholar]
- You BR, Moon HJ, Han YH, Park WH. Gallic acid inhibits the growth of HeLa cervical cancer cells via apoptosis and/or necrosis. Food Chem Toxicol 2010; 48:1334-40. doi: 10.1016/j.fct.2010.02.034 [Crossref] [ Google Scholar]
- Beniwal V, Kumar A, Sharma J, Chhokar V. Recent advances in industrial application of tannases: a review. Recent Pat Biotechnol 2013; 7:228-33. doi: 10.2174/18722083113076660013 [Crossref] [ Google Scholar]
- Otani K, Naito Y, Sakaguchi Y, Seo Y, Takahashi Y, Kikuta J. Cell-cycle-controlled radiation therapy was effective for treating a murine malignant melanoma cell line in vitro and in vivo. Sci Rep 2016; 6:30689. doi: 10.1038/srep30689 [Crossref] [ Google Scholar]
- Wang K, Zhu X, Zhang K, Zhu L, Zhou F. Investigation of gallic acid induced anticancer effect in human breast carcinoma mcf‐7 cells. J Biochem Mol Toxicol 2014; 28:387-93. doi: 10.1002/jbt.21575 [Crossref] [ Google Scholar]
- Verma S, Singh A, Mishra A. Gallic acid: molecular rival of cancer. Environ ToxicolPharmacol 2013; 35:473-85. doi: 10.1016/j.etap.2013.02.011 [Crossref] [ Google Scholar]
- Feghhi M, Rezaie J, Akbari A, Jabbari N, Jafari H, Seidi F. Effect of multi-functional polyhydroxylated polyhedral oligomeric silsesquioxane (POSS) nanoparticles on the angiogenesis and exosome biogenesis in human umbilical vein endothelial cells (HUVECs). Mater Des 2020; 197:109227. doi: 10.1016/j.matdes.2020.109227 [Crossref] [ Google Scholar]
- Taverna S, Flugy A, Saieva L, Kohn EC, Santoro A, Meraviglia S. Role of exosomes released by chronic myelogenous leukemia cells in angiogenesis. Int J Cancer 2012; 130:2033-43. doi: 10.1002/ijc.26217 [Crossref] [ Google Scholar]
- Savina A, Furlán M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem 2003; 278:20083-90. doi: 10.1074/jbc.M301642200 [Crossref] [ Google Scholar]
- Shokrollahi E, Nourazarian A, Rahbarghazi R, Salimi L, Karbasforush S, Khaksar M. Treatment of human neuroblastoma cell line SH‐SY5Y with HSP27 siRNA tagged‐exosomes decreased differentiation rate into mature neurons. J Cell Physiol 2019; 234:21005-21013. doi: 10.1002/jcp.28704 [Crossref] [ Google Scholar]
- Ge Q, Zhou Y, Lu J, Bai Y, Xie X, Lu Z. miRNA in plasma exosome is stable under different storage conditions. Molecules 2014; 19:1568-75. doi: 10.3390/molecules19021568 [Crossref] [ Google Scholar]
- Martins TS, Catita J, Rosa IM, e Silva OAdC, Henriques AG. Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS One 2018; 13:e0198820. doi: 10.1371/journal.pone.0198820 [Crossref] [ Google Scholar]
- Tor YS, Yazan LS, Foo JB, Wibowo A, Ismail N, Cheah YK. Induction of apoptosis in MCF-7 cells via oxidative stress generation, mitochondria-dependent and caspase-independent pathway by ethyl acetate extract of Dillenia suffruticosa and its chemical profile. PLoS One 2015; 10:e0127441. doi: 10.1371/journal.pone.0127441 [Crossref] [ Google Scholar]
- Hwang EY, Huh J-W, Choi M-M, Choi SY, Hong H-N, Cho S-W. Inhibitory effects of gallic acid and quercetin on UDP-glucose dehydrogenase activity. FEBS Lett 2008; 582:3793-7. doi: 10.1016/j.febslet.2008.10.010 [Crossref] [ Google Scholar]
- Moghtaderi H, Sepehri H, Delphi L, Attari F. Gallic acid and curcumin induce cytotoxicity and apoptosis in human breast cancer cell MDA-MB-231. BioImpacts 2018; 8:185-194. doi: 10.15171/bi.2018.21 [Crossref] [ Google Scholar]
- Rezaei-Seresht H, Cheshomi H, Falanji F, Movahedi-Motlagh F, Hashemian M, Mireskandari E. Cytotoxic activity of caffeic acid and gallic acid against MCF-7 human breast cancer cells: An in silico and in vitro study. Avicenna J Phytomed 2019; 9:574-586. doi: 10.22038/AJP.2019.13475 [Crossref] [ Google Scholar]
- García-Rivera D, Delgado R, Bougarne N, Haegeman G, Berghe WV. Gallic acid indanone and mangiferin xanthone are strong determinants of immunosuppressive anti-tumour effects of Mangifera indica L. bark in MDA-MB231 breast cancer cells. Cancer Lett 2011; 305:21-31. doi: 10.1016/j.canlet.2011.02.011 [Crossref] [ Google Scholar]
- Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduction and Signal Transduct Target Ther 2016; 1:15004. doi: 10.1038/sigtrans.2015.4 [Crossref] [ Google Scholar]
- Si W, Shen J, Zheng H, Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics 2019; 11:25. doi: 10.1186/s13148-018-0587-8 [Crossref] [ Google Scholar]
- Johansson J, Berg T, Kurzejamska E, Pang M-F, Tabor V, Jansson M. MiR-155-mediated loss of C/EBPβ shifts the TGF-β response from growth inhibition to epithelial-mesenchymal transition, invasion and metastasis in breast cancer. Oncogene 2013; 32:5614-24. doi: 10.1038/onc.2013.322 [Crossref] [ Google Scholar]
- Wang YQ, Guo RD, Guo RM, Sheng W, Yin LR. MicroRNA‐182 promotes cell growth, invasion, and chemoresistance by targeting programmed cell death 4 (PDCD4) in human ovarian carcinomas. J Cell Biochem 2013; 114:1464-73. doi: 10.1002/jcb.24488 [Crossref] [ Google Scholar]
- Kong W, He L, Richards E, Challa S, Xu C, Permuth-Wey J. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene 2014; 33:679-89. doi: 10.1038/onc.2012.636 [Crossref] [ Google Scholar]
- Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 2010; 12:19-30; sup pp 1. doi: 10.1038/ncb2000 [Crossref] [ Google Scholar]
- Blanc L, Vidal M. New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases 2018; 9:95-106. doi: 10.1080/21541248.2016.1264352 [Crossref] [ Google Scholar]
- Andreu Z, Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front Immunol 2014; 5:442. doi: 10.3389/fimmu.2014.00442 [Crossref] [ Google Scholar]
- Ghossoub R, Lembo F, Rubio A, Gaillard CB, Bouchet J, Vitale N. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat Commun 2014; 5:3477. doi: 10.1038/ncomms4477 [Crossref] [ Google Scholar]
- Tian S, Wen E, Mi N, Li H. Tian S, Wen E, Mi N, Li HDetermination of three active components in Euphorbia humifusa willdUsing high-performance liquid chromatography with diode-array detection and autophagy and apoptosis analysis of normal rat kidney and HeLa cells. Pharmacogn Mag 2019; 15:348-355. doi: 10.4103/pm.pm_348_18 [Crossref] [ Google Scholar]
- Doan KV, Ko CM, Kinyua AW, Yang DJ, Choi Y-H, Oh IY. Gallic acid regulates body weight and glucose homeostasis through AMPK activation. Endocrinology 2015; 156:157-68. doi: 10.1210/en.2014-1354 [Crossref] [ Google Scholar]
- Abdelwahab SI. Protective mechanism of gallic acid and its novel derivative against ethanol-induced gastric ulcerogenesis: Involvement of immunomodulation markers, Hsp70 and Bcl-2-associated X protein. Int Immunopharmacol 2013; 16:296-305. doi: 10.1016/j.intimp.2013.04.005 [Crossref] [ Google Scholar]
- Witkin SS, Kanninen TT, Sisti G. The role of Hsp70 in the regulation of autophagy in gametogenesis, pregnancy, and parturition. Adv AnatEmbryol Cell Biol 2017; 222:117-127. doi: 10.1007/978-3-319-51409-3_6 [Crossref] [ Google Scholar]
- Fernández-Fernández MR, Gragera M, Ochoa-Ibarrola L, Quintana-Gallardo L, Valpuesta JM. Hsp70 – a master regulator in protein degradation. FEBS Lett 2017; 591:2648-2660. doi: 10.1002/1873-3468.12751 [Crossref] [ Google Scholar]
- Russell RC, Yuan H-X, Guan K-L. Autophagy regulation by nutrient signaling. Cell Res 2014; 24:42-57. doi: 10.1038/cr.2013.166 [Crossref] [ Google Scholar]
- Fang Y, Tan J, Zhang Q. Signaling pathways and mechanisms of hypoxia-induced autophagy in the animal cells. Cell Biol Int 2015; 39:891-8. doi: 10.1002/cbin.10463 [Crossref] [ Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer cell 2006; 10:51-64. doi: 10.1016/j.ccr.2006.06.001 [Crossref] [ Google Scholar]
- Lee Y-K, Lee J-A. Role of the mammalian ATG8/LC3 family in autophagy: differential and compensatory roles in the spatiotemporal regulation of autophagy. BMB Rep 2016; 49:424-30. doi: 10.5483/bmbrep.2016.49.8.081 [Crossref] [ Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2016; 12:1-222. doi: 10.1080/15548627.2015.1100356 [Crossref] [ Google Scholar]
- Rogov V, Dötsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell 2014; 53:167-78. doi: 10.1016/j.molcel.2013.12.014 [Crossref] [ Google Scholar]
-
Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol2010; 221: 3-12. 10.1002/path.2697
- Baixauli F, López-Otín C, Mittelbrunn M. Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front Immunol 2014; 5:403. doi: 10.3389/fimmu.2014.00403 [Crossref] [ Google Scholar]
- Xu J, Camfield R, Gorski SM. The interplay between exosomes and autophagy–partners in crime. J Cell Sci 2018; 131:jcs215210. doi: 10.1242/jcs.215210 [Crossref] [ Google Scholar]