Bioimpacts. 12(3):219-231.
doi: 10.34172/bi.2021.43
Original Research
Evaluation of in vitro fibroblast migration by electrospun triple-layered PU-CA/gelatin.PRGF/PU-CA scaffold using an AAVS1 targeted EGFP reporter cell line
Forough Shams 1  , Hamideh Moravvej 2
, Hamideh Moravvej 2  , Simzar Hosseinzadeh 3, 4, Bahram Kazemi 1, Masoumrh Rajabibazl 5, Azam Rahimpour 3, 4, *
, Simzar Hosseinzadeh 3, 4, Bahram Kazemi 1, Masoumrh Rajabibazl 5, Azam Rahimpour 3, 4, * 
Author information:
1Department of Medical Biotechnology, School of Advanced Technologies in Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
2Skin Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
3Medical Nano-Technology and Tissue Engineering Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
4Department of Tissue Engineering and Applied Cell Sciences, School of Advanced Technologies in Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
5Department of Clinical Biochemistry, Faculty of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Abstract
Introduction:
Migration of fibroblast cells in wound areas is a critical aspect of the wound healing process. Employment of enhanced green fluorescent protein (EGFP) labeled fibroblast cells facilitates real-time monitoring and functional evaluation of these cells in both in vitro and in vivo settings. Plasma rich in growth factor (PRGF) is a potent accelerator of wound healing; therefore, in this study, a novel method to fabricate an electrospun bioactive scaffold containing PRGF was employed to induce in vitro cell proliferation and migration.
Methods:
First, the EGFP reporter gene was integrated into the AAVS1 locus of fibroblast cells using CRISPR/Cas9 system. Then, PRGF was obtained from platelet-rich plasma, and a multi-layered scaffold was fabricated using polyurethane-cellulose acetate (PU-CA) fibers as the outer layers and PRGF-containing gelatin fibers were located in the internal layer like a central strip. Scanning electron microscopy (SEM), tensile, water contact angle, and FTIR tests were performed to assess the characteristics of the scaffolds. The EGFP targeted cells were cultured on scaffolds with or without PRGF to investigate their viability, toxicity, and migration pattern in response to the release profile.
Results:
Fluorescence images showed that the number of migrating cells on scaffold containing PRGF was more significant than PU-CA scaffold up to day 6. Increased expression of SGPL1, DDR2, and VEGF genes was also observed on the scaffold containing PRGF compared to PU-CA using real-time polymerase chain reaction (PCR) analysis with around 3-, 2-, and 2-fold enhancement, respectively.
Conclusion:
The current scaffold provides the appropriate template for cell attachment and migration. In addition, the present results highlight the potential of reporter gene targeting for the in vitro analysis of biological processes such as migration.
Keywords: Gene targeting, Fibroblast migration, Polyurethane-cellulose acetate, Platelet-rich plasma, AAVS1, Enhanced green fluorescent protein
Introduction
Wound healing consists of well-orchestrated cellular responses such as migration, differentiation, extracellular matrix (ECM) synthesis, and proliferation. The migration of fibroblast cells plays a critical role in this process.
1,2
Fibroblast cells are summoned into the wound area to increase and advance several critical activities under the tight regulation of injury-mediated factors. Therefore, migration is vital for changing the wound environment at the end state of the wound healing. Following migration, fibroblast cells are induced by growth factors and cytokines accessible in the serum. This process is mediated by different growth factors such as epidermal growth factor (EGF), fibroblast growth factor (FGF), platelet-derived growth factor (PDGF), transforming growth factor (TGF), nerve growth factor (NGF), and vascular endothelial growth factor (VEGF). Plasma rich in growth factor (PRGF) is a platelet-enriched plasma which can influence and promote biological processes of fibroblast cells such as migration, proliferation, and differentiation.
3
Studies have indicated that PRGF contains several bioactive proteins and growth factors that facilitate the process of tissue regeneration and wound healing.
4,5
Therefore, the transport of growth factors and cells to the wound area is vital in the wound healing process. The application of PRGF in the scaffold along with fibrillar and cellular components is highly beneficial for the generation of elastic, dense, and haemostatic fibrin during the re-epithelialization phase of soft tissue repair.
6
Since the migration of fibroblast cells is critical during repair, investigating the pattern of migration in the presence of growth factors provides more comprehensive understanding of the wound healing process. In this regard, nanofibers are favorable candidates to create a simple system for in vitro experiments. Electrospinning is a simple, versatile, scalable, and cost-effective technique to generate fibers with a wide range of diameters from a few micrometers to nanometers in various forms, such as core-shell structures, pure/composite nanofibrous patches, and multi-layered scaffolds.
7
The bioactive dressing materials that have been widely studied, are influenced by many factors, such as polymer type, as well as their adhesion and solubility. According to most studies, applying bilayer or multi-layered scaffolds with inherent similarity to multi-layered skin could be highly beneficial in mechanical characteristics, degradation, and drug release.
8
Moreover, due to the large surface-to-volume ratio of fibers, these scaffolds provide a proper substrate for improved attachment, proliferation, and delivery of biomolecules. Given the significance of multi-layered scaffolds and the role of growth factors in skin tissue engineering, we designed a novel electrospun triple-layered scaffold.
5
The external layers contain polyurethane (PU) and cellulose acetate (CA). An electrospun tape layer containing PRGF-gelatin nanofibers with a length of 20 cm and a width of 1 cm was placed in the middle of the scaffold as an internal layer. Gelatin which is obtained from dehydration of denatured collagen offers reduced immunogenicity. In addition, gelatin has been considered as a biodegradable, hydrophilic, and non-toxic material commonly used in such scaffolds. Cellulose acetate is a widely used biopolymer due to its unique features such as liquid transport, water absorption, and hydrophilicity. The acetate ester of cellulose offers several advantages such as biocompatibility, biodegradability, adequate flexural, tensile strength, high affinity with other substances, and high modulus, which promotes its utility for the generation of nanofiber mats.
9
However, CA cannot be used alone as a biomedical material in the clinic due to its poor resistance, strain, and low breaking stress. On the other hand, some researchers have reported that semi-permeable dressings like PU, enhance wound healing rate due to their oxygen permeability and unique barrier properties.
9,10
Polyurethanes include three basic components: (1) the polyol, (2) the diisocyanate, and (3) the chain extender. Polyol is a soft segment that is terminated by hydroxyl (-OH) groups. The diisocyanate constitutes a part of the low molecular weight that reacts with either the chain extender or polyol. The chain extender includes a small molecule with either amine or hydroxyl end groups
11
. Thermoplastic polyurethane (TPU) is a class of PU that has linear molecular chains and can be easily developed through melt methods or solutions
12
. TPU is regarded as biodegradable materials because it is synthesized using either degradable chain developer or polyester diols as soft segments. The unique biological and elastomechanical properties of TPU including air permeability and blood compatibility make it an attractive material in biomedical applications. As a result, TPU has been extensively applied as foams or films in wound-dressings.
13
Tegaderm and OpSite are PU-based wound dressing commercial products that soak up a limited amount of ulcer exudation because of their dense structure.
11
However, extensive application of PU, as a soft and hydrophobic material, is associated with a few drawbacks including the displacement of the dressing and prevention of fluid exuding from the wound area. Therefore, the combination of PU-CA along with PRGF would be an ideal wound dressing material. This study evaluated the effects of a PU-CA scaffold containing PRGF-gelatin on the migration pattern of fibroblast cells.
An enhanced green fluorescent protein (EGFP) reporter gene was targeted in the adeno-associated virus integration site 1 (AAVS1) safe harbor locus using CRISPR/Cas9 system to facilitate fibroblast cells monitoring during the migration. CRISPR/Cas9 system has gained tremendous attention as a precise and effective tool for gene targeting in mammalian cells.
14,15
AAVS1 locus which is located on human chromosome 19 (position 19q13.42) has served as a safe harbor locus for targeted integration of the exogenous DNA. Studies have shown that disruption of the phosphatase 1 regulatory subunit 12C (PPP1R12C) gene located in this locus is not associated with any adverse cellular effects. In addition, it has been indicated that AAVS1 supports the long-term and stable expression of exogenous genes in a wide variety of cells.
16,17
This study examined the potential of EGFP targeted fibroblast cells as an in vitro model to investigate the migration pattern of fibroblast cells on a triple-layered scaffold containing PRGF-gelatin.
Materials and Methods
Vector construction
To target the AAVS1 locus in fibroblast cells, the AAVS1_Puro_PGK1_3xFLAG_Twin cassette (Addgene #68375) assembled donor vector, and the eSpCas9 (1.1) No_FLAG_AAVS1_T2 vector (Addgene #79888) which expresses the AAVS1 T2 gRNA in combination with eSpCas9 (1.1) were purchased from Addgene. To construct the AAVS1-EGFP donor vector, the EGFP fragment (720 bp) was first amplified from pEGFP-N1 vector, cloned into an intermediate plasmid (pTG19-T, Vivantis, Malaysia), and sequenced. Subsequently, the EGFP fragment was sub-cloned in KpnI restriction site of the AAVS1 donor vector.
Cell culture
Human foreskin fibroblast cells, Hu02, (Iranian Biological Resource Center, I.R. Iran) were grown in Dulbecco’s modified eagle medium (DMEM)/F-12 medium supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine and 1% penicillin/streptomycin (100 μg/mL) (all purchased from Gibco, USA). The cells were seeded in the vent cap flasks, incubated with 5% CO2 and 90% humidity at 37°C, and were sub-cultured at 48 hours intervals. Trypan blue exclusion assay was used to evaluate the cell count and viability.
Gene targeting
One day before transfection, fibroblast cells were cultured at a density of 1 × 105 cells/well in 24- well plates. TurboFect Transfection Reagent (Thermo Fisher Scientific, USA) was applied for the transfection based on the manufacturers' instructions. The cells were co-transfected in duplicates with 500 ng of AAVS1-EGFP targeting vector and Cas9-gRNA pU6 vector in a 1:1 ratio. Seventy-two hours post-transfection, EGFP expression was evaluated using fluorescence microscopy (Nikon Instruments, USA). The cells were then detached, diluted in 1:10 ratio in complete medium and seeded in 6-well plates. A selective medium containing 0.25 µg/mL puromycin (Sigma-Aldrich, USA) was applied to select the cells during 7 days of culture. Cells were then diluted to 10 cells/mL concentration in DMEM-F12 medium with 15% FBS and 1% nonessential amino acid mixture (Thermo Fisher Scientific, USA), and 100 μL of the resulting dilutions were cultured in 96-well plates to achieve 1 cell/well concentration. Following one week of incubation, single-cell EGFP expressing clones were identified using fluorescent microscopy and were subjected to the selective medium for another week. The EGFP positive surviving clones were then expanded in 12-well plates.
Genomic DNA extraction and junction PCR analysis
Genomic DNA of selected EGFP positive clones was extracted using the genomic DNA extraction kit (Qiagen, Germany) following the manufacturer’s instructions. Approximately 200 ng of genomic DNA was used for each polymerase chain reaction (PCR). Forward primer (F1) 5ʹ- ATGGTGAGCAAGGGCGAGGAG -3ʹ located on the EGFP, and reverse primer (R1) 5ʹ- GGTCCAGGCCAAGTAGGTG -3ʹ located on genomic DNA downstream of the right homology arm were used to detect the junction point of integration. PCR cycle parameters were 95°C for 5 minutes, 60°C for 30 seconds, and 72°C for 2 minutes, with a final elongation step of 72°C for 10 minutes. The resulting fragment was then purified and sequenced.
Preparation of the PRGF
Blood was drawn from the voluntary person ulnar vein and put into 9 mL tubes containing 3.8% (wt/vol) sodium citrate. To prepare PRGF, the blood was centrifuged at 200 g for 20 minutes. Subsequently, the plasma was transferred to new tubes and centrifuged again at 3000 g for 15 minutes at 4°C. Two milliliters (mL) of plasma at the bottom of the tube along with the precipitated platelets which constitute the inactivated PRGF was transferred to a new glass tube. To activate PRGF, 50 μL of CaCl2 (10%) was added per 1 mL of the PRGF solution. The mixture was incubated for 1 hour at 37°C, which led to formation of the fibrin clot. Upon PRGF activation, fibrinogen transforms into fibrin through the function of thrombin and the PRGF activator. The solution was then placed at 4°C for 2 days to develop all possible deposits. Eventually, the mixture was centrifuged at 3000 g for 10 minutes, and the supernatant was filtered and stored at -80°C until use.
Preparation of the polymer solution
Two solutions were used to produce the triple-layered scaffold. Solution A contained PU, 4 wt% (Sigma Aldrich, USA) and CA, 18 wt% (Sigma Aldrich, USA). PU was dissolved in the 3:1 ratio of N, N dimethylformamide (Merck, Germany) and tetrahydrofuran (Merck, Germany) at room temperature to acquire a uniform solution overnight. CA was dissolved in the 3:1 ratio of acetic acid (Merck, Germany) and H2O at room temperature for 24 hours. The PU and CA solutions were mixed in 3:1 ratio. To prepare solution B, a 20% (w/v) gelatin type A (Sigma Aldrich, USA) was dissolved in 40% acetic acid and mixed at 37°C for 30 minutes. PRGF was then added and mixed to obtain a 12.5% solution.
Fabrication of the electrospun scaffolds
PU-CA solution was placed in a plastic syringe tube and fibers were developed by the electrospinning device (Nanoazma, Iran) under 18 kV voltage in 17 cm distance with two nozzles at a flow rate of 0.2 mL/hour and drum speed of 300 rpm with a 45° angel between the needles’ tip and the collector. The solution of PU-CA scaffold was electrospun for 4 hours at the conditions mentioned above. For the inner layer, the whole scaffold was covered except a 1 cm strip in the middle of the scaffold. This layer was electrospun by the gelatin/PRGF solution for 4 hours in the identical situation except for the voltage, which was set to 16 kV. Then the cover was removed and PU-CA solution was again electrospun for 4 hours on the inner layer. In order to enhance the mechanical and biological features of the scaffold it was cross-linked through glutaraldehyde (GTA,1%).
Mechanical and morphological characterization of the scaffolds
To measure the surface structure and the size distribution of fibers on PU-CA and PU-CA/gelatin.PRGF/PU-CA scaffolds, scanning electron microscopy (SEM) (Philips XL30; Philips, Netherlands) was used under a 25 kV accelerated voltage after coating with a thin layer of gold on the samples. The fiber diameter was measured using the ImageJ software (National Institute of Health, USA). Mechanical characteristics of PU-CA and PU-CA/gelatin.PRGF/PU-CA (1 × 4 cm) were defined through tensile measurement on an Instron universal testing machine (Model STM‐20, SANTAM, Iran) at a speed of 5 mm/min. The elastic modulus, final tensile strength, and strain at break were measured. Ultimately, the data were plotted as a stress-strain curve. The Contact angle was performed with water to measure the surface hydrophilicity of scaffolds. To obtain a contact angle goniometer (Krüss, Hamburg, Germany), scaffolds were cut 1×1 cm and set on microscope slides. For this analysis, 4 µL of the deionized water was located on the scaffold and the contact angle was measured.
Fourier transform-infrared spectroscopy (FTIR)
The bonding configurations of the specimens were determined by employing FTIR spectroscopy. FTIR spectroscopy was operated in the 400–4000 cm -1 range, with a scanning speed of 32 cm -1 and spectral width 2.0 cm -1. The two samples were read by grinding along with KBr and compressing into the thin pellets.
Protein Release
Two 1 × 1 cm2 punches of PU-CA/gelatin.PRGF/PU-CA and PU-CA scaffolds were placed in a 24-well plate containing 500 μL per well of DW and incubated at 37°C. 100 μL aliquots were taken out at certain times (6, 12, 24, 48, 72, 96, 120, 144, 168, 192, and 216 hours) and stored at -20°C. Simultaneously, 100 μL of DW was added to fix the final volume at 500 μL. Each sample was subjected to protein quantification using a micro BCA assay. The optical density was read at 570 nm by using a microplate reader (BioTek, USA). The absorbance values of the sample scaffold were normalized according to the values obtained in the PU-CA control scaffold. The values were then converted to μg/mL using the calibration curve that was obtained using BSA as a standard. Since 20% of the total proteins were removed from each collection, the amount of cumulated proteins in each collection time were measured according to following equation:
where
is the real amount of cumulative protein in time t, Xnis the amount of absorbed protein at time t, and
is the real amount of cumulative protein in one step time (∆t) before time t.
MTT assay
The proliferation rate of fibroblast cells on scaffolds was measured using MTT (3–(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; Sigma, Germany) assay. A total of 1 × 1 cm2 sections of scaffolds were sterilized in a 24-well plate by UV irradiation. 5000 EGFP positive cells in 25 μL DMEM-F12 supplemented with 5% FBS were cultured on the center of scaffolds and incubated for 1 hour at 37°C, 5% CO2, and 95% humidity. Then 475 μL DMEM-F12 with 10% FBS was added gently. The same conditions were applied for the sample without scaffold, tissue culture polystyrene (TCPS), as a control group. Cell viability assay was examined on days 1, 3, 7, and 14. After removing the culture medium, PBS was used to wash the cells and 50 μI of MTT solution (5 mg/mL in DMEM) was added to each well. The plate was incubated at 37°C for 4 hours to change MTT into formazan crystals by mitochondrial dehydrogenases of living cells. Next, the medium was removed and dimethyl sulfoxide (DMSO) was used to dissolve formazan crystals. Finally, solutions were collected and their absorbance was recorded at 570 nm using a microplate reader (BioTek, USA).
Cell culture for scanning electron microscopy
To evaluate the attachment and growth morphology of cells on the scaffolds, specimens were fixed by glutaraldehyde for SEM. Briefly, fibroblast cells were seeded on both PU-CA and PU-CA/ gelatin.PRGF/PU-CA scaffolds at a final DMEM-F12 volume of 500 μL supplemented with 10% FBS in a 24-wells plate and incubated for 72 hours. Then, scaffolds were fixed using a glutaraldehyde solution (2.5%) for 2 hours and a serial dilution of ethanol was applied for dehydration treatment. Ultimately, scaffolds were coated with gold and evaluated using an SEM at an accelerating voltage of 25 kV.
Migration assay
Chemotactic properties of PRGF released from the triple-layered scaffold were assessed through EGFP positive fibroblast cells. For this purpose, the PU-CA/gelatin.PRGF/PU-CA scaffold was punched with a 3.8 cm diameter that contained a PRGF strip in the middle. Also, the PU-CA scaffold was punched with a 3.8 cm diameter as a control group. Scaffolds were sterilized and placed in a 6-well culture plate. 1.5 × 104 EGFP positive cells in the volume of 15 μL DMEM-F12 were seeded on each side (about 2 mm) of the scaffold, which had the farthest distance from the center. After 1 hour incubation at 37°C and 5% CO2, 2 mL of DMEM-F12 with 10% FBS was gently added to the samples. The sample and control groups were monitored by fluorescence microscopy for 6 days and fluorescent images were taken on days 1, 3, and 6. To evaluate migration on the scaffold, the cells on 8 considered positions were counted using Image J.
Gene expression analysis
Quantitative real-time PCR (qRT-PCR) was employed to investigate the expression level of fibroblast migration markers, including discoidin domain receptor tyrosine kinase 2 (DDR2) and sphingosine-1-phosphate lyase (SGPL1). In addition, vascular endothelial growth factor (VEGF) was evaluated as a proliferation marker. Following 6 days of culture, PU-CA, PU-CA/PRGF scaffolds and TCPS as a control group were removed for RNA extraction (Favorgen, Taiwan) according to the manufacturer’s protocol. Then, cDNA synthesis kit (Vivantis, Malaysia) was used for preparing total cDNA. qRT-PCR was carried out in a StepOnePlusTM system (Applied Biosystems, USA) using a high ROX SYBR Green master mix (Ampliqon, Denmark). The amplification program was set at 95°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds, and 60°C for 1 minute. Relative quantification analysis was carried out using the comparative CT (ΔΔCT) method and β-actin housekeeping gene was selected to normalize the relative expression. Table 1 shows the primers used in this study.
Table 1.
Primers sequences used in real-time RT-PCR analysis
|
Gene
|
Primer sequences 5’-3’
|
| DDR2 |
F: GAGACCAAGGGAGGCAGAC |
| R:CATCACTCGTCGCCTTGTTG |
| SGPL1 |
F:CATCTACCGACTATCAAACCTG |
| R:GCGTGTAGTAATGTGTATGCAG |
| VEGF |
F: CCCATGGCAGAAGGAGGAG |
| R: GGATGGCTTGAAGATGTACTCG |
| β-Actin |
F: TCCTCCTGAGCGCAAGTAC |
| R: CCTGCTTGCTGATCCACATCT |
Statistical analysis
REST 2009 (Qiagen, Germany) and GraphPad online software were employed for the gene expression analysis. Data were reported as means ± standard deviation (SD). Un-paired t test and one-way ANOVA were used to compare the means between two and multiple groups, respectively (Graph Pad Prism 4.0, US). P values <0.05 were considered statistically significant.
Results
Targeted integration of EGFP reporter gene into the AAVS1 locus
Site-specific gene integration of the reporter genes into safe harbor loci facilitates the development of stably labeled cells without affecting the phenotype of the cells. Therefore, CRISPR/Cas9 system was employed to integrate the EGFP expression unit into the first intron of PPP1R12C gene in the AAVS1 locus (Fig. 1A). The EGFP coding sequence was inserted downstream of the hPGK promoter in the AAVS1 donor vector. The vector utilizes a splicing acceptor (SA)-linked promoter-less puromycin resistant gene, which allows selection of AAVS1 targeted cells while restricting the growth of cells harboring randomly integrated vectors in puromycin containing medium. Following transfection and selection, two EGFP positive clones were selected for further analysis. Observation of puromycin-resistant EGFP expressing cells using fluorescent microscopy indicated consistent expression of the reporter gene (Fig. 1B). Furthermore, successful targeting of the donor vector to the AAVS1 locus in selected clones was verified using PCR analysis. As indicated in Fig. 1C a 2.2 kb fragment corresponding to the junction PCR product was observed. Site-specific integration was further verified by sequencing of the resulting fragment (Fig. 1D).


Fig. 1.
CRISPR/Cas9 based insertion of EGFP into AAVS1 locus of fibroblast cells. (A) Schematic illustration of the chromosomal location of the AAVS1 safe harbor site. This locus was targeted using sgRNA in a CRISPR/Cas9 strategy. The inserted cassette consists of promoter-less puromycin-resistant gene (puro), and the EGFP expression unit. Following integration, puro expression is driven by the endogenous promoter of the PPP1R12C gene. EGFP expression is driven by hPGK promoter. This is flanked on each side by about 0.8 kb of homology to the AAVS1 locus. (B) GFP-positive cells were illustrated after colony selection. (C) Primers F1, R1 used to detect the junction point of insertion. The junction PCR amplifies a fragment from intron 1 of the PPP1R12C gene, just downstream of the right homology arm (AAVS1-HA-R), to the EGFP element. While a 2.2-kb fragment was detected in the fibroblast-insertion group, PCR amplification was not observed in the fibroblast group. (D) Sequencing was carried out to confirm the fidelity of 2.2 kb fragment obtained in junction PCR.
.
CRISPR/Cas9 based insertion of EGFP into AAVS1 locus of fibroblast cells. (A) Schematic illustration of the chromosomal location of the AAVS1 safe harbor site. This locus was targeted using sgRNA in a CRISPR/Cas9 strategy. The inserted cassette consists of promoter-less puromycin-resistant gene (puro), and the EGFP expression unit. Following integration, puro expression is driven by the endogenous promoter of the PPP1R12C gene. EGFP expression is driven by hPGK promoter. This is flanked on each side by about 0.8 kb of homology to the AAVS1 locus. (B) GFP-positive cells were illustrated after colony selection. (C) Primers F1, R1 used to detect the junction point of insertion. The junction PCR amplifies a fragment from intron 1 of the PPP1R12C gene, just downstream of the right homology arm (AAVS1-HA-R), to the EGFP element. While a 2.2-kb fragment was detected in the fibroblast-insertion group, PCR amplification was not observed in the fibroblast group. (D) Sequencing was carried out to confirm the fidelity of 2.2 kb fragment obtained in junction PCR.
Physiochemical characteristics of scaffolds
SEM images demonstrated the mean diameter, the overall morphology, and diameter distribution of the scaffolds (Fig. 2A-B). Morphological characterization of both scaffolds based on SEM showed smooth, continuous, randomly aligned and bead-free fibers. The diameter distributions of PU-CA/gelatin.PRGF/PU-CA fibers were as follows: 10% of fibers were less than 300 nm diameter. Forty- three percent of fibers were ranged between 300 to 600 nm diameter and 46% of fibers were more than 600 nm diameter (Fig. 2C). In PU-CA scaffold, 17% of electrospun fibers were less than 300 nm diameter, 70% of fibers were ranged between 300 to 600 nm, and 13% of fibers were more than 600 nm diameter (Fig. 2D). According to these results, the mean diameter of the PU-CA and PU-CA.PRGF fibers were 596.26 and 439.1 nm, respectively. Therefore, there is a noticeable difference between PU-CA and PU-CA/PRGF fibers’ mean diameter. The hydrophilic properties of scaffolds were determined using contact angles (Fig. 3). The contact angle of PU-CA scaffolds with and without PRGF was 55.45° and 61°, respectively. A contact angle under 90° correlates with a hydrophilic surface and both scaffolds maintain hydrophilicity characteristics. Therefore, the presence of PRGF within the PU-CA scaffold was associated with reduced wettability of electrospun fibers.
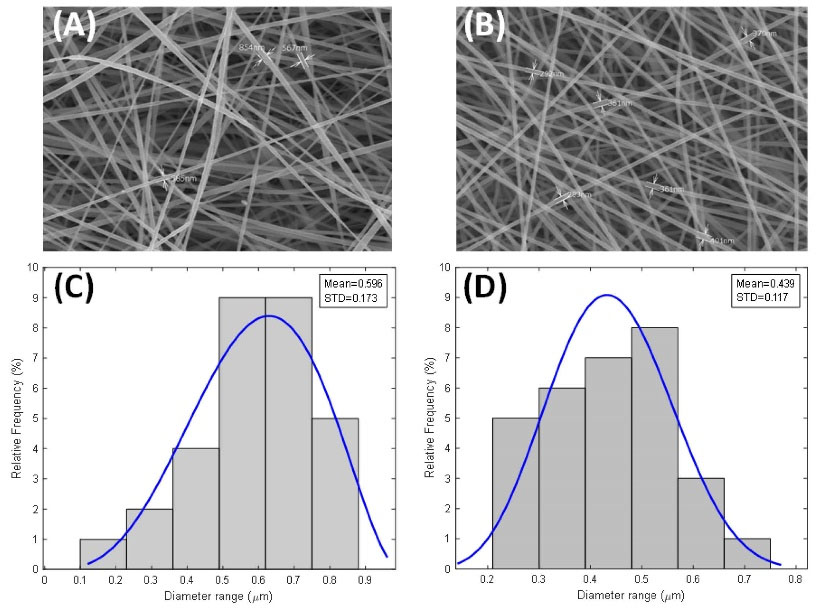
Fig. 2.
Morphology and diameter distribution of PU-CA/gelatin.PRGF/PU-CA (A, C) and PU-CA (B, D) scaffolds.
.
Morphology and diameter distribution of PU-CA/gelatin.PRGF/PU-CA (A, C) and PU-CA (B, D) scaffolds.
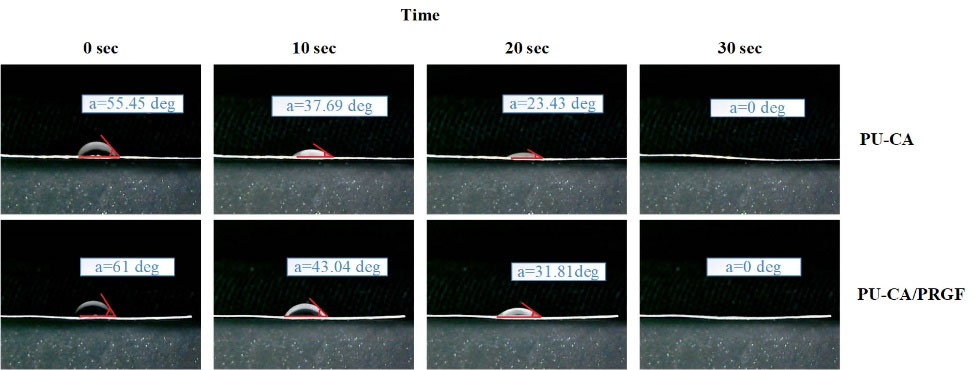
Fig. 3.
Contact angle measurements of PU-CA (A) and PU-CA/gelatin.PRGF/PU-CA (B) scaffolds.
.
Contact angle measurements of PU-CA (A) and PU-CA/gelatin.PRGF/PU-CA (B) scaffolds.
Mechanical properties
The effect of PRGF on diverse mechanical parameters such as young modulus, ultimate tensile strength and strain at break was investigated by obtaining the stress-strain curve of both scaffolds (Fig. 4). Data were summarized in Table 2. A higher strain was observed in PU-CA scaffold (12.708642%) compared to PU-CA/gelatin.PRGF/PU-CA (6.666837%) (P ≤ 0.05). Therefore, the presence of gelatin and PRGF in PU-CA scaffold meaningfully decreased the ultimate tensile strength and strain at the break of the scaffold, while there was no considerable difference in young modulus.
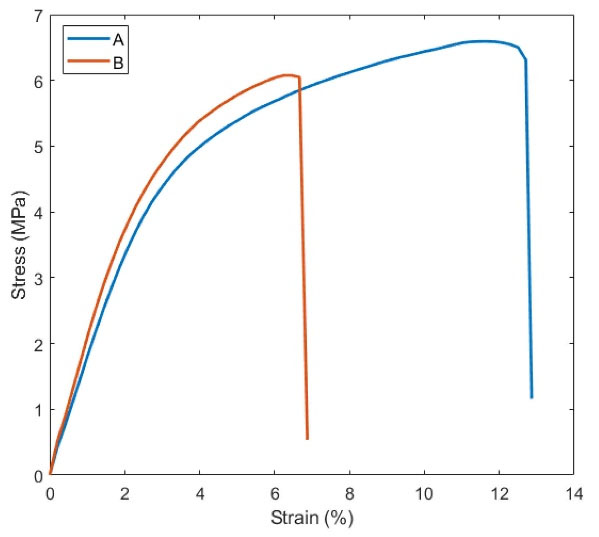
Fig. 4.
Representative stress-strain curves of PU-CA (A) and PU-CA/gelatin.PRGF/PU-CA (B) scaffolds.
.
Representative stress-strain curves of PU-CA (A) and PU-CA/gelatin.PRGF/PU-CA (B) scaffolds.
Table 2.
Different mechanical properties of PU-CA and PU-CA/gelatin.PRGF/PU-CA
|
Gropes
|
Ultimate tensile strength (MPa)
|
Strain %
|
Young modulus
|
| PU-CA |
6.310835 |
12.708642 |
1.79 |
PU-CA /gelatin.PRGF/
PU-CA
|
6.048841 |
6.666837 |
1.58 |
FTIR analysis
FTIR was utilized to determine the intermolecular interactions of the mixture. The spectra of PU–CA and PU–CA/gelatin.PRGF/PU-CA fibers are shown in Fig. 5. The spectroscopy of PU-CA showed the characteristic peaks at 3442, 2954, 1530, 1167, 1060 and 906-605 wave-numbers (ν), which belongs to PU, and represents ν (N-H), ν (C-H), ν (C-C), ν (C-O, ν (C-O), ν (C-H) on replaced benzene, respectively.
18
Also, the important adsorption peaks of CA were at 1733 cm -1 ν (C-O) and 1235 cm -1 ν C(-O-C).
19
FITR spectra of PU-CA/gelatin.PRGF/PU-CA, contained common protein bands in addition to PU and CA bands that were almost revealed at 1650 cm -1 (amide I) and 1532 cm -1 (amide II), corresponding to the stretching vibrations of C–O bond, N–H bond and stretching of C–N bonds. The peak of amide I band at 1650 cm -1 was related to both a random coil and α-helix conformation of gelatin.
20
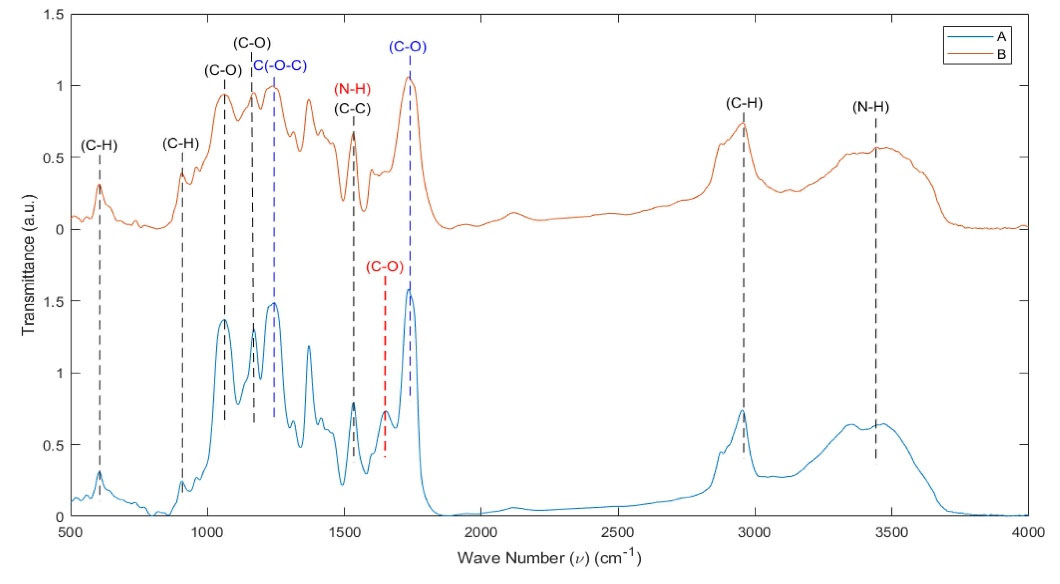
Fig. 5.
FTIR spectra of PU-CA/gelatin.PRGF/PU-CA (blue line) (A) and PU-CA (red line) (B) scaffolds. Black dashed line (PU peaks), Blue dashed line (CA peaks), and Red dashed line (PRGF peaks).
.
FTIR spectra of PU-CA/gelatin.PRGF/PU-CA (blue line) (A) and PU-CA (red line) (B) scaffolds. Black dashed line (PU peaks), Blue dashed line (CA peaks), and Red dashed line (PRGF peaks).
Release profile
A total protein released from PU-CA/gelatin.PRGF/PU-CA scaffold was monitored for nine days in DW at 37°C, employing BCA assay. Based on the size of scaffold, which was about 30 × 10 cm2, the time of the electrospinning, flow rate, and percentage of gelatin it was estimated that about 700 µg gelatin exists in each 1 × 1 cm2 section of the scaffold. Fig. 6 illustrates the release profile of PU-CA/gelatin.PRGF/PU-CA scaffold in the form of cumulative release. Given this, about 3.4% of all proteins combined in the scaffold had been released until 216 hours. The release in 24, 48, 72, 96, and 120 hours were 0.18%, 0.6%, 1.14%, 1.8%, and 2.25%, respectively, which were equal to 1.27, 4.1, 8, 12.7 and 15.7 concentrations (µg/mL). Furthermore, the release velocity had been enhanced from 48 to 192 hours and after that, it was reduced. Notably, the rate of cumulative release between days 8 and 9 was only 0.04%. The kinetic release parameters were analyzed with zero-order, first-order, Korsmeyer-Peppas, Higuchi, and Peppas-Sahlin models represented in Table 3.

Fig. 6.
Cumulative release of PU-CA/gelatin.PRGF/PU-CA scaffold.
.
Cumulative release of PU-CA/gelatin.PRGF/PU-CA scaffold.
Table 3.
The release mechanism of PRGF from PU-CA/gelatin.PRGF/PU-CA scaffold
|
Release mechanism
|
|
Zero-order
|
First-order
|
Higuchi
|
Korsmeyer-Peppas
|
Peppas-Sahlin
|
|
K
0
|
R
2
|
K
F
|
R
2
|
K
H
|
R
2
|
K
KP
|
R
2
|
Release exponent (n)
|
K
1
|
K
2
|
R
2
|
Release exponent (n)
|
| 0.017 |
0.977 |
0.0002 |
0.973 |
0.211 |
0.827 |
0.022 |
0.973 |
1.01 |
0.001 |
2 ×10 -7
|
0.999 |
1.551 |
MTT assay
According to Fig. 7, fibroblast cells on PU-CA/gelatin.PRGF/PU-CA scaffold showed the highest proliferation rate until days 3 and 7 compared to TCPS and PU-CA groups. On day one, the number of active cells in the TCPS group exceeded those of the other groups (P ≤ 0.05). Moreover, PU-CA, PU-CA/PRGF, and TCPS groups demonstrated a steady enhancement in cell number until day 7, while the number of active cells reduced on day fourteen. In addition, The number of viable cells in the PU-CA/PRGF group was more than PU-CA and TCPS groups on day fourteen.

Fig. 7.
Viability of cells seeded on scaffolds and TCPS as control during the 14-day culture period. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001
.
Viability of cells seeded on scaffolds and TCPS as control during the 14-day culture period. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001
Cell attachment and growth on scaffolds
SEM and fluorescent images showed proper expansion of the cells (Fig. 8). The cells were cultured on PU-CA/gelatin.PRGF/PU-CA scaffold aligned and directed well after 72 hours compared to those seeded on PU-CA scaffold (Fig. 8A-B). Cell behavior on the PU-CA scaffold was different and showed a random orientation. SEM images indicated that cells on the PU-CA scaffold were located cumulatively. While on the PRGF scaffold, cells were growing in one direction, were protruding well on the substrate, and their movement was much faster. The cell attachment pattern and growth on both scaffolds were also confirmed by fluorescent microscopy (Fig. 8C-D).

Fig. 8.
Expanded cells on PU-CA (A) and PU-CA/gelatin.PRGF/PU-CA (B) scaffolds. Cells could protrude well on both scaffolds especially on PU-CA/PRGF scaffold which cells illustrated acceptable migratory and proliferation morphology 72 hours after seeding (B, D). In the PU-CA scaffold, the cells demonstrated random orientation (A, C) at a certain time. The protrusions and orientation of cells are shown with arrows.
.
Expanded cells on PU-CA (A) and PU-CA/gelatin.PRGF/PU-CA (B) scaffolds. Cells could protrude well on both scaffolds especially on PU-CA/PRGF scaffold which cells illustrated acceptable migratory and proliferation morphology 72 hours after seeding (B, D). In the PU-CA scaffold, the cells demonstrated random orientation (A, C) at a certain time. The protrusions and orientation of cells are shown with arrows.
Migration of EGFP targeted fibroblast on scaffolds
Evaluation of cell migration is essential for validation of novel scaffolds in wound healing studies. In this study, the EGFP targeted fibroblasts cells were employed to investigate the effect of electrospun PRGF on cell migration. A migration test was designed to trace the migrating fibroblast cells towards the center of the scaffold where a strip of gelatin-PRGF was located. Fig. 9 shows the behavior of cultured cells on the PU-CA (control) and PU-CA/gelatin.PRGF/PU-CA scaffolds during different days. On the first day, there was no noticeable difference in the appearance and behavior of cells in both scaffolds. On day 3, migration was quite clear, according to the behavior and orientation of cells. During the following days, most of the cells were observed to migrate toward the center of the scaffold. On day 6, cells were mostly residing in the centre of the scaffold, albeit migration was continuing (Fig. 9K). Cell migration was not observed in the control group during this period despite noticeable cell proliferation. Fig. 10A-B represents the number of migrating cells on days 3 and 6 in eight positions. Interestingly, the number of migrating cells at positions 2, 3 and 6, 7 were higher compared to other regions. The total number of migrating cells was also measured on days 3 and 6 (Fig. 10C). On both days, extensive migration was observed on the PRGF scaffold with a significant difference from the control (P ≤ 0.01).

Fig. 9.
The behavior of cultured cells on PU-CA (control) and PU-CA/gelatin PRGF/PU-CA scaffolds. Migration on PU-CA/PRGF scaffold was clearly observed on days 3 and 6. Also, the proliferation of cells is obvious on both scaffolds.
.
The behavior of cultured cells on PU-CA (control) and PU-CA/gelatin PRGF/PU-CA scaffolds. Migration on PU-CA/PRGF scaffold was clearly observed on days 3 and 6. Also, the proliferation of cells is obvious on both scaffolds.
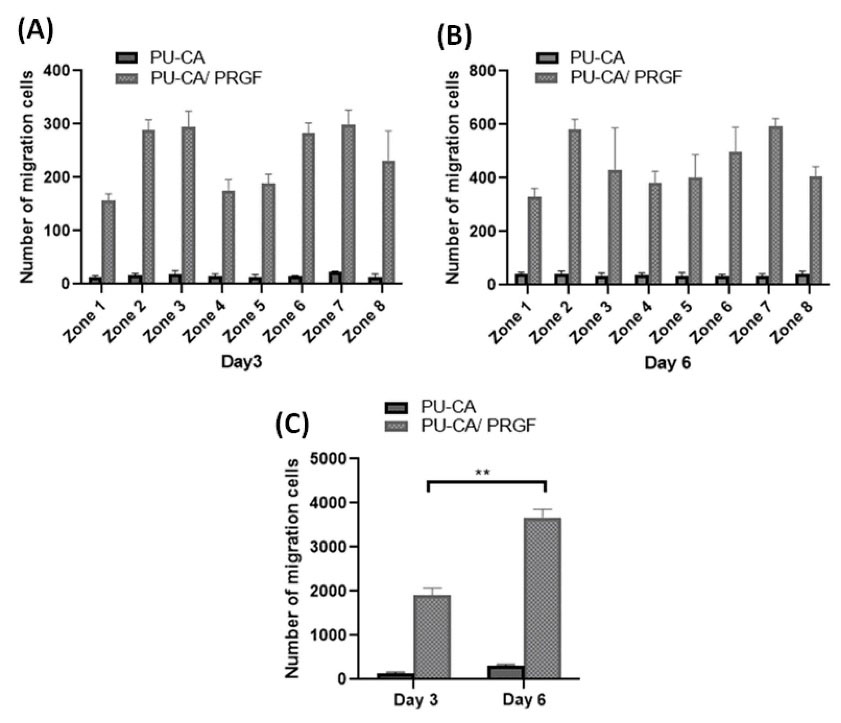
Fig. 10.
The total number of migrating cells at each position of scaffolds, day 3 (A) and day 6 (B). The total number of migrating cells at days 3 and 6(C). **P ≤ 0.01.
.
The total number of migrating cells at each position of scaffolds, day 3 (A) and day 6 (B). The total number of migrating cells at days 3 and 6(C). **P ≤ 0.01.
Gene expression analysis
qRT-PCR analysis showed a significant increase in selected genes at mRNA level in cells seeded on PU-CA/gelatin.PRGF/PU-CA scaffold compared to other groups (Fig. 11). In this group, the expression of the SGPL1 gene at the mRNA level was about 3-fold higher than other groups (P ≤ 0.001). Also, a significant increase was shown in the expression of the DDR2 in comparison to PU-CA and TCPS groups (P ≤ 0.01). However, the cells seeded on the PU-CA scaffold did not show a significant difference in the expression of migration markers compared to the other groups. The expression level of the VEGF gene in both scaffolds was increased significantly compared to TCPS as the control. Furthermore, the expression rate of the VEGF gene in the PU-CA/PRGF scaffold was about 2-fold higher than PU-CA (P ≤ 0.01).
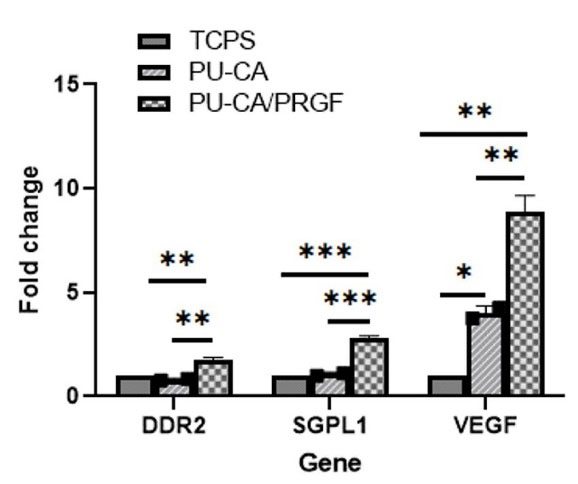
Fig. 11.
DDR2, SGPL1, and VEGF gene expression at mRNA level. After 6 days of cell seeding on PU-CA, PU-CA/PRGF scaffolds, and TCPS the cells were tested by Real-Time PCR. The cells which were seeded on PU-CA/PRGF scaffold showed more expression in these genes compared to the other groups. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001
.
DDR2, SGPL1, and VEGF gene expression at mRNA level. After 6 days of cell seeding on PU-CA, PU-CA/PRGF scaffolds, and TCPS the cells were tested by Real-Time PCR. The cells which were seeded on PU-CA/PRGF scaffold showed more expression in these genes compared to the other groups. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001
Discussion
Proliferation and migration of cells to the damaged area are vital events during the wound healing process. Migration of fibroblast cells to the injured area is particularly crucial in chronic wounds where the natural wound healing process is impaired. Furthermore, fibroblast cells produce and deposit matrix proteins, which are essential for the migration of inflammatory cells to the wound area.
21
Electrospinning is regarded as a practical procedure for generating a physical construct for wound dressing. The multi-layered scaffolds, similar to skin architecture, are employed as potential engineered skin substitutes applicable to wound care.
5
Efficient genome engineering tools and reliable reporter cell lines are invaluable tools to investigate the role and behavior of cells during the treatment processes. Site-specific gene integration at pre-defined safe harbour loci facilitates rapid generation of stable cell lines with limited transgene silencing and few detrimental effects. The AAVS1 locus which supports persistent transgene expression is a favorite choice for the targeted integration of exogenous DNA.
22
Robust expression of EGFP at the AAVS1 locus will facilitate in vitro and in vivo imaging to evaluate the localization, proliferation, survival and migration of cells in experimental and preclinical studies.
23
Here, we demonstrated successful targeting of the AAVS1 locus with the genetically encoded EGFP cassette using a CRISPR/Cas9 system. In accordance with the previous reports, our results showed stable expression of the EGFP from the AAVS1 locus.
16,24
In the current study, the multi-layered scaffold was synthesized using PU-CA as the outer layers and gelatin type A combined with PRGF as the inner layer with optimized morphological and mechanical attributes as a wound dresser. Based on mechanical results, the percentage of strain was reduced due to the lower interfacial interaction between PU-CA chains as a function of gelatin and PRGF presence. In fact, incorporation of the protein into the fibers leads to a reduction of tensile strength. However, the young modulus and elastic behavior of scaffolds were not reduced.
25
In this line, other studies indicated increasing PRGF was associated with an enhancement in fiber diameter/pore area, which can in turn accelerate cellular infiltration while diminishing regression of mechanical properties.
5,26
Reduction in mechanical properties of the scaffold containing PRGF was also found in our study which could have induced rapid migration. Hence, scaffold with PRGF content displayed reduced mechanical attributes that facilitated cell migration more rapidly. The FTIR spectra recorded on both scaffolds demonstrated that the whole peaks of PU-CA were visible in PU–CA/gelatin.PRGF/PU–CA composite fibers and some peaks were being overlapped. This measurement confirmed the presence of the gelatin, PRGF, and other polymers in the fabricated scaffolds. This finding indicates successful incorporation of PRGF in the electrospun scaffolds.
10,27
Since mathematical functions are very useful for describing the release profile, suitable kinetics models were selected to predict the release mechanism according to the R2 values.
28
Based on the evaluated R2 values presented in Table 3, zero-, first-order and Korsmeyer-Peppas model predicted experimental data properly, while; the Peppas-Sahlin kinetics model had the best adaption to experimental results. Peppas-Sahlin model calculates the two contribution mechanisms (diffusional and relaxational) in the release process of the swellable scaffold. This model accounts the effect of the Fickian diffusion and polymer Case II relaxation release in the following form:
Where Mt and M∞ are the amount of the drug released at the time t and the amount released at the infinite time, respectively. Also, K1, K2 and n are constants. According to Table 3, Case II relaxation constant (K2) is negligible respect to Fickian constant (K1) which indicates that the Fickian process has the main contribution to the release mechanism during the incubation time. Fickian diffusion is characterized by solvent high diffusion velocity to the scaffold and low velocity of polymeric relaxation. This behavior induces a solvent penetration gradient in the scaffold.
29
The higher proportion of components released after the first 48 hours, validated the time required for facilitating higher swelling and polymer relaxation. During this time, the lower water sorption was another point for lower component release, thereby, the release was related to time and zero-order kinetic was confirmed.
28
Hence, based on the kinetics of the release profile, very small amounts of PRGF were released into the medium during the first 48 hours. Furthermore, the correlation between gene expression, release profile, and cell behavior displayed that the scaffold containing PRGF can be potentially used as a wound dresser.
5
MTT results showed an increase in the number of active cells on days 1, 3, and 7 in the TCPS group, while reducing viable cells was observed after day 7. In addition, the cells showed a proper proliferation with reasonable speed on these scaffolds until day 14. Our results showed that fabricated scaffolds had appropriate biocompatibility and were not toxic for the EGFP positive cells as indicated by other reports.
30,31
In several studies, CA has been reported to be biodegradable and its degradation rate depends on the degree of acetyl substitution.
31,32
Furthermore, utilization of different biodegradable TPUs as tissue engineering materials for soft tissues, bone tissues, blood vessels, cardiovascular tissues, and wound dressings has been reported recently.
13,33,34
Although most studies have evaluated degradation of PU in the absence of cells and plasma enzymes, this process is expected to accelerate in vivo environment or in the presence of cells in in vitro settings.
35,36
In this line, Unnithan et al
10
reported that PU-CA fibers have reasonable biocompatibility and mechanical properties for dressing wounds.
10
The operation of the scaffold in the presence of water is an important criterion that could be beneficial for the cell attachment. Measurement of contact angle indicated a hydrophilic behavior for both scaffolds and improvement in cell proliferation and adhesion, as reported by other groups.
10,30
Moreover, the arrangement and orientation of cells on PU-CA/gelatin.PRGF scaffold indicates the unique growth pattern in response to PRGF release. PRGF-containing scaffolds stimulate cell migration into the wound area. In addition, improved healing potential has been shown in triple-layered scaffold with PRGF continuous release.
5
According to Fig. 10C, the total number of migrating cells on day 6 compared to day 3 was quite significant, which can be attributed to cell behavior on the scaffold and the amount of PRGF release. Based on migration (Fig. 10) and gene expression results (Fig. 11), minimal migration was observed on the PU-CA scaffold. This conclusion is consistent with the findings reported by Piran et al,
37
which showed the positive impact of PDGF-BB released from the scaffold at a low concentration (even less than 1 ng), on migration and proliferation of fibroblast cells compared to the control group which was subjected to a constant concentration of PDGF in the cultured media. In the study mentioned above, inflammation was induced to further elucidate the underlying mechanism of migration to the wound in the presence of PDGF-BB using under agarose migration assay.
37
In another study, Piranet al
5
demonstrated that the scaffold containing continuously released PRGF at a low rate, maintained its healing potential until day 5. They fabricated a scaffold containing PRGF as a wound dresser to induce fibroblast migration toward the injury site in the agarose migration test.
5
In addition, Zhao et al
38
investigated the potential of PDGF-BB on the migration of MC3T3 preosteoblastic cells using an agarose cell droplet migration assay. They demonstrated that the incorporation of PDGF with PLGA/PEG-PLA scaffold could induce a cell migration process which serves as the primary step of wound healing.
38
In the final step, the expression profile of genes involved in migration and proliferation was evaluated. DDR2 and SGPL1 are specific genes in fibroblast migration, as Herrera et al
39
showed that silencing of DDR2 with siRNA inhibited human fibroblast migration and Lovric et al
40
demonstrated knock-down of SGPL1 by siRNA inhibited fibroblast migration in ichthyosis and adrenal insufficiency. qRT-PCR showed increased expression of SGPL1 and DDR2 genes in cells seeded on PU-CA/gelatin.PRGF/PU-CA compared to TCPS and PU-CA groups. These results reflect the bioactivity of released PRGF. Moreover, a 1.6-fold increase was observed in SGPL1 mRNA expression compared with DDR2. These findings were in agreement with those obtained by Piran et al
37
which indicated enhanced expression of Arp 2 and PDGFRbgenes, which were involved in fibroblast migration, on the scaffold contained PDGF compared to TCPS and PDGF free scaffold.
37
The VEGF gene plays an important role in cell proliferation and the wound healing process. Our results demonstrated a significant increase in VEGF expression in cells seeded on the PU-CA/gelatin.PRGF/PU-CA scaffold in comparison with the PU-CA and TCPS groups. Although VEGF expression was observed in cells applied to both scaffolds, a 2-fold increase was shown in PU-CA/PRGF scaffold compared to the PU-CA group. These findings reflect the bioactivity of the released PRGF. Enhanced expression of VEGF mRNA in bone marrow stem cells and the primary bladder smooth muscle cells has also been reported in different types of scaffolds.
41,42
Based on our results, PU-CA scaffold did not support proper migration of fibroblast cells; however, dense cell populations formed using this scaffold. Substantial growth and accelerated migration of fibroblast cells were observed in one direction when PRFG was included in the scaffold.
Research Highlights
What is the current knowledge?
√ Migration of fibroblast cells plays a critical role in the wound healing process.
√ PRGF can accelerate the wound healing process.
√ EGFP reporter fibroblast cells facilitates real-time monitoring of cell behaviour on the scaffold.
√
AAVS1 site serves as a safe harbor locus for targeted knock-in of exogenous reporter genes.
What is new here?
√ Triple-layered scaffold along with PRGF was be obtained using the electrospinning process.
√
EGFP reporter gene was targeted in the AAVS1 safe harbor locus to facilitate monitoring of fibroblast cells using CRISPR/Cas9 system.
√ The current scaffold provides the appropriate template for cell attachment and migration.
Conclusion
EGFP targeted fibroblast cells employed in this study facilitated in vitro cell tracking due to the robust and persistent fluorescent gene expression on scaffolds. Since various scaffolds are used in wound healing to create optimal support and accelerate healing, studying the behavior of cells could be beneficial in establishing efficient wound management strategies. In this study, migration of EGFP targetted fibroblast cells on gelatin.PRGF scaffold was observed clearly. This strategy indicated that the release of PRGF from the PU-CA scaffold with mentioned release profile can induce proliferation and migration of fibroblast cells.
Acknowledgments
The authors wish to express their gratitude for financial support from the School of Advanced Technologies in Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran (grants No. 10461 and 145).
Ethical statement
Blood was drawn from a healthy volunteer after obtaining the informed consent according to the ethical requirement of Shahid Beheshti University of Medical Sciences.
Competing interests
None declared.
Authors’ contribution
FS: conceptualization, data handling, experiments design, data analysis, writing and reviewing; HM: conceptualization, experiments design, project administration; SH: experiments design, data analysis, study validation; BK: experiments design, study consultation; MR: experiments design, study consultation; AR: conceptualization, experiments design, data analysis, supervision, project administration.
References
- Anitua E, Sánchez M, Zalduendo MM, de la Fuente M, Prado R, Orive G. Fibroblastic response to treatment with different preparations rich in growth factors. Cell Prolif 2009; 42:162-70. doi: 10.1111/j.1365-2184.2009.00583.x [Crossref] [ Google Scholar]
- Choi WI, Yameen B, Vilos C, Sahu A, Jo S-M, Sung D. Optimization of fibrin gelation for enhanced cell seeding and proliferation in regenerative medicine applications. Polym Adv Technol 2017; 28:124-9. doi: 10.1002/pat.3866 [Crossref] [ Google Scholar]
- Anitua E, Sanchez M, Merayo-Lloves J, De la Fuente M, Muruzabal F, Orive G. Plasma Rich in Growth Factors (PRGF-Endoret) Stimulates Proliferation and Migration of Primary Keratocytes and Conjunctival Fibroblasts and Inhibits and Reverts TGF-β1–Induced Myodifferentiation. Invest Ophthalmol Vis Sci 2011; 52:6066-73. doi: 10.1167/iovs.11-7302 [Crossref] [ Google Scholar]
- Anitua E, Pino A, Orive G. Plasma rich in growth factors promotes dermal fibroblast proliferation, migration and biosynthetic activity. J Wound Care 2016; 25:680-7. doi: 10.12968/jowc.2016.25.11.680 [Crossref] [ Google Scholar]
- Piran M, Shiri M, Soufi Zomorrod M, Esmaeili E, Soufi Zomorrod M, Vazifeh Shiran N. Electrospun triple-layered PLLA/gelatin. PRGF/PLLA scaffold induces fibroblast migration. J Cell Biochem 2019. doi: 10.1002/jcb.28422 [Crossref]
- Anitua E, Sanchez M, Nurden AT, Zalduendo M, de la Fuente M, Orive G. Autologous fibrin matrices: a potential source of biological mediators that modulate tendon cell activities. J Biomed Mater Res A 2006; 77:285-93. doi: 10.1002/jbm.a.30585 [Crossref] [ Google Scholar]
- Ranjbarvan P, Mahmoudifard M, Kehtari M, Babaie A, Hamedi S, Mirzaei S. Natural Compounds for Skin Tissue Engineering by Electrospinning of Nylon-Beta Vulgaris. Asaio j 2018; 64:261-9. doi: 10.1097/mat.0000000000000611 [Crossref] [ Google Scholar]
- Chen S, Liu B, Carlson MA, Gombart AF, Reilly DA, Xie J. Recent advances in electrospun nanofibers for wound healing. Nanomedicine (Lond) 2017; 12:1335-52. doi: 10.2217/nnm-2017-0017 [Crossref] [ Google Scholar]
- Unnithan AR, Barakat NA, Pichiah PB, Gnanasekaran G, Nirmala R, Cha YS. Wound-dressing materials with antibacterial activity from electrospun polyurethane-dextran nanofiber mats containing ciprofloxacin HCl. CarbohydrPolym 2012; 90:1786-93. doi: 10.1016/j.carbpol.2012.07.071 [Crossref] [ Google Scholar]
- Unnithan AR, Gnanasekaran G, Sathishkumar Y, Lee YS, Kim CS. Electrospun antibacterial polyurethane–cellulose acetate–zein composite mats for wound dressing. Carbohydrate Polymers 2014; 102:884-92. doi: 10.1016/j.carbpol.2013.10.070 [Crossref] [ Google Scholar]
- Hacker C, Karahaliloglu Z, Seide G, Denkbas EB, Gries T. Functionally modified, melt-electrospun thermoplastic polyurethane mats for wound-dressing applications. J Appl Polym Sci 2014; 131. doi: 10.1002/app.40132 [Crossref]
- Mi H-Y, Salick MR, Jing X, Jacques BR, Crone WC, Peng X-F.
Characterization of thermoplastic polyurethane/polylactic acid
(TPU/PLA) tissue engineering scaffolds fabricated by microcellular injection molding
. Mater Sci Eng C 2013; 33:4767-76. doi: 10.1016/j.msec.2013.07.037 [Crossref] [ Google Scholar]
- Mi H-Y, Jing X, Napiwocki BN, Hagerty BS, Chen G, Turng L-S. Biocompatible, degradable thermoplastic polyurethane based on polycaprolactone-block-polytetrahydrofuran-block-polycaprolactone copolymers for soft tissue engineering. J Mater Chem B 2017; 5:4137-51. doi: 10.1039/C7TB00419B [Crossref] [ Google Scholar]
- Wu M, Wei C, Lian Z, Liu R, Zhu C, Wang H. Rosa26-targeted sheep gene knock-in via CRISPR-Cas9 system. Scientific Reports 2016; 6:24360. doi: 10.1038/srep24360 [Crossref] [ Google Scholar]
- Bayat H, Omidi M, Rajabibazl M, Sabri S, Rahimpour A. The CRISPR Growth Spurt: from Bench to Clinic on Versatile Small RNAs. J Microbiol Biotechnol 2017; 27:207-18. doi: 10.4014/jmb.1607.07005 [Crossref] [ Google Scholar]
- Luo Y, Liu C, Cerbini T, San H, Lin Y, Chen G. Stable enhanced green fluorescent protein expression after differentiation and transplantation of reporter human induced pluripotent stem cells generated by AAVS1 transcription activator‐like effector nucleases. Stem Cells Transl Med 2014; 3:821-35. doi: 10.5966/sctm.2013-0212 [Crossref] [ Google Scholar]
- Oceguera-Yanez F, Kim SI, Matsumoto T, Tan GW, Xiang L, Hatani T. Engineering the AAVS1 locus for consistent and scalable transgene expression in human iPSCs and their differentiated derivatives. Methods 2016; 101:43-55. doi: 10.1016/j.ymeth.2015.12.012 [Crossref] [ Google Scholar]
- Jiang Z, Yuan K-j, Li S-f, Chow WK. [Study of FTIR spectra and thermal analysis of polyurethane]. Guang Pu Xue Yu Guang Pu Fen Xi 2006; 26:624-8. [ Google Scholar]
- Yu D-G, Li X-Y, Wang X, Chian W, Liao Y-Z, Li Y. Zero-order drug release cellulose acetate nanofibers prepared using coaxial electrospinning. Cellulose 2013; 20:379-89. [ Google Scholar]
- Safaeijavan R, Soleimani M, Divsalar A, Eidi A, Ardeshirylajimi A. Biological behavior study of gelatin coated PCL nanofiberous electrospun scaffolds using fibroblasts. J Paramed Sci (JPS) Winter 2014; 5:2008-4978. doi: 10.22037/jps.v5i1.5467 [Crossref] [ Google Scholar]
- Li J, Chen J, Kirsner R. Pathophysiology of acute wound healing. Clin Dermatol 2007; 25:9-18. doi: 10.1016/j.clindermatol.2006.09.007 [Crossref] [ Google Scholar]
- Lyu C, Shen J, Wang R, Gu H, Zhang J, Xue F. Targeted genome engineering in human induced pluripotent stem cells from patients with hemophilia B using the CRISPR-Cas9 system. Stem Cell Res Ther 2018; 9:1-12. doi: 10.1186/s13287-018-0839-8 [Crossref] [ Google Scholar]
- Bhagwan JR, Collins E, Mosqueira D, Bakar M, Johnson BB, Thompson A. Variable expression and silencing of CRISPR-Cas9 targeted transgenes identifies the AAVS1 locus as not an entirely safe harbour. F1000Research 2020; 8:1911. doi: 10.12688/f1000research.19894.2 [Crossref] [ Google Scholar]
- Dalvai M, Loehr J, Jacquet K, Huard CC, Roques C, Herst P. A scalable genome-editing-based approach for mapping multiprotein complexes in human cells. Cell R 2015; 13:621-33. doi: 10.1016/j.celrep.2015.09.009 [Crossref] [ Google Scholar]
- Norouzi M, Shabani I, Ahvaz HH, Soleimani M. PLGA/gelatin hybrid nanofibrous scaffolds encapsulating EGF for skin regeneration. Journal of Biomedical Materials Research Part A 2015; 103:2225-35. doi: 10.1002/jbm.a.35355 [Crossref] [ Google Scholar]
- Sell SA, Wolfe PS, Ericksen JJ, Simpson DG, Bowlin GL. Incorporating platelet-rich plasma into electrospun scaffolds for tissue engineering applications. Tissue Engineering Part A 2011; 17:2723-37. doi: 10.1089/ten.TEA.2010.0663 [Crossref] [ Google Scholar]
- Sadeghianmaryan A, Karimi Y, Naghieh S, Sardroud HA, Gorji M, Chen X. Electrospinning of scaffolds from the polycaprolactone/polyurethane composite with graphene oxide for skin tissue engineering. Appl BiochemBiotechnol 2019; 191:567-578. doi: 10.1007/s12010-019-03192-x [Crossref] [ Google Scholar]
- Singhvi G, Singh M. In-vitro drug release characterization models. Int J Pharm Stud Res 2011; 2:77-84. [ Google Scholar]
- Peppas NA, Sahlin JJ. Peppas NA, Sahlin JJA simple equation for the description of solute releaseIIICoupling of diffusion and relaxation. Int J Pharm 1989; 57:169-72. doi: 10.1016/0378-5173(89)90306-2 [Crossref] [ Google Scholar]
- Esmaeili E, Eslami-Arshaghi T, Hosseinzadeh S, Elahirad E, Jamalpoor Z, Hatamie S. The biomedical potential of cellulose acetate/polyurethane nanofibrous mats containing reduced graphene oxide/silver nanocomposites and curcumin: Antimicrobial performance and cutaneous wound healing. Int J Biol Macromol 2020; 152:418-27. doi: 10.1016/j.ijbiomac.2020.02.295 [Crossref] [ Google Scholar]
- Buchanan CM, Gardner RM, Komarek RJ. Aerobic biodegradation of cellulose acetate. J Appl Polym Sci 1993; 47:1709-19. doi: 10.1002/app.1993.070471001 [Crossref] [ Google Scholar]
- Komarek RJ, Gardner RM, Buchanan CM, Gedon S. Biodegradation of radiolabeled cellulose acetate and cellulose propionate. J Appl Polym Sci 1993; 50:1739-46. doi: 10.1002/app.1993.070501009 [Crossref] [ Google Scholar]
- Karchin A, Simonovsky FI, Ratner BD, Sanders JE. Melt electrospinning of biodegradable polyurethane scaffolds. Acta Biomater 2011; 7:3277-84. doi: 10.1016/j.actbio.2011.05.017 [Crossref] [ Google Scholar]
- Hong Y, Ye SH, Pelinescu AL, Wagner WR. Synthesis, characterization, and paclitaxel release from a biodegradable, elastomeric, poly(ester urethane)urea bearing phosphorylcholine groups for reduced thrombogenicity. Biomacromolecules 2012; 13:3686-94. doi: 10.1021/bm301158j [Crossref] [ Google Scholar]
- Guan J, Fujimoto KL, Sacks MS, Wagner WR. Preparation and characterization of highly porous, biodegradable polyurethane scaffolds for soft tissue applications. Biomaterials 2005; 26:3961-71. doi: 10.1016/j.biomaterials.2004.10.018 [Crossref] [ Google Scholar]
- Guan J, Sacks MS, Beckman EJ, Wagner WR. Biodegradable poly (ether ester urethane) urea elastomers based on poly (ether ester) triblock copolymers and putrescine: synthesis, characterization and cytocompatibility. Biomaterials 2004; 25:85-96. doi: 10.1016/s0142-9612(03)00476-9 [Crossref] [ Google Scholar]
- Piran M, Vakilian S, Piran M, Mohammadi-Sangcheshmeh A, Hosseinzadeh S, Ardeshirylajimi A. In vitro fibroblast migration by sustained release of PDGF-BB loaded in chitosan nanoparticles incorporated in electrospun nanofibers for wound dressing applications. Artif Cells NanomedBiotechnol 2018; 46:511-20. doi: 10.1080/21691401.2018.1430698 [Crossref] [ Google Scholar]
- Zhao X, Hadjiargyrou M. Induction of cell migration in vitro by an electrospun PDGF-BB/PLGA/PEG-PLA nanofibrous scaffold. J Biomed Nanotechnol 2011; 7:823-9. doi: 10.1166/jbn.2011.1342 [Crossref] [ Google Scholar]
- Herrera-Herrera ML, Quezada-Calvillo R. DDR2 plays a role in fibroblast migration independent of adhesion ligand and collagen activated DDR2 tyrosine kinase. BiochemBiophys Res commun 2012; 429:39-44. doi: 10.1016/j.bbrc.2012.10.103 [Crossref] [ Google Scholar]
- Lovric S, Goncalves S, Gee HY, Oskouian B, Srinivas H, Choi W-I. Mutations in sphingosine-1-phosphate lyase cause nephrosis with ichthyosis and adrenal insufficiency. Journal Clin Invest 2017; 127:912-28. doi: 10.1172/JCI89626 [Crossref] [ Google Scholar]
- Liao H-T, Chen Y-Y, Lai Y-T, Hsieh M-F, Jiang C-P. The osteogenesis of bone marrow stem cells on mPEG-PCL-mPEG/hydroxyapatite composite scaffold via solid freeform fabrication. Biomed Res Int 2014; 2014:321549. doi: 10.1155/2014/321549 [Crossref] [ Google Scholar]
- Mokhames Z, Rezaie Z, Ardeshirylajimi A, Basiri A, Taheri M, Omrani MD. VEGF-incorporated PVDF/collagen nanofibrous scaffold for bladder wall regeneration and angiogenesis. Int J Polyme Mater 2020:521-529. doi: 10.1080/00914037.2020.1740985 [Crossref]