Bioimpacts. 13(3):191-206.
doi: 10.34172/bi.2022.23813
Original Article
Apoptotic effects of human amniotic fluid mesenchymal stem cells conditioned medium on human MCF-7 breast cancer cell line
Roghiyeh Pashaei-Asl 1  , Maryam Pashaiasl 2, 3, 4, Esmaeil Ebrahimie 5, Maryam Lale Ataei 2, Maliheh Paknejad 1, *
, Maryam Pashaiasl 2, 3, 4, Esmaeil Ebrahimie 5, Maryam Lale Ataei 2, Maliheh Paknejad 1, * 
Author information:
1Department of Clinical Biochemistry, Faculty of Medicine, Tehran University of Medical Sciences, Tehran, Iran
2Department of Anatomical Sciences, School of Medicine, Tabriz University of Medical Sciences
3Stem Cell Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
4Department of Reproductive Biology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
5Genomics Research Platform, School of Life Sciences, College of Science, Health and Engineering, La Trobe University, Melbourne, Victoria 3086, Australia
Abstract
Introduction:
Breast cancer, as the most common malignancy among women, is shown to have a high mortality rate and resistance to chemotherapy. Research has shown the possible inhibitory role of Mesenchymal stem cells in curing cancer. Thus, the present work used human amniotic fluid mesenchymal stem cell-conditioned medium (hAFMSCs-CM) as an apoptotic reagent on the human MCF-7 breast cancer cell line.
Methods:
Conditioned medium (CM) was prepared from hAFMSCs. After treating MCF-7 cells with CM, a number of analytical procedures (MTT, real-time PCR, western blot, and flow cytometry) were recruited to evaluate the cell viability, Bax and Bcl-2 gene expression, P53 protein expression, and apoptosis, respectively. Human fibroblast cells (Hu02) were used as the negative control. In addition, an integrated approach to meta-analysis was performed.
Results:
The MCF-7 cells’ viability was decreased significantly after 24 hours (P < 0.0001) and 72 hours (P < 0.05) of treatment. Compared with the control cells, Bax gene’s mRNA expression increased and Bcl-2’s mRNA expression decreased considerably after treating for 24 hours with 80% hAFMSCs-CM (P = 0.0012, P < 0.0001, respectively); an increasing pattern in P53 protein expression could also be observed. The flow cytometry analysis indicated apoptosis. Results from literature mining and the integrated meta-analysis showed that hAFMSCs-CM is able to activate a molecular network where Bcl2 downregulation stands in harmony with the upregulation of P53, EIF5A, DDB2, and Bax, leading to the activation of apoptosis.
Conclusion:
Our finding demonstrated that hAFMSCs-CM presents apoptotic effect on MCF-7 cells; therefore, the application of hAFMSCs-CM, as a therapeutic reagent, can suppress breast cancer cells’ viabilities and induce apoptosis.
Keywords: MCF-7 cells, hAFMSCs-CM, Bax and Bcl-2 genes, P53, Apoptosis, Meta-analysis
Copyright and License Information
© 2023 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Breast cancer remains as the most common lethal cancer among women around the world.
1
Currently, chemotherapy and surgery are the main approaches in the breast cancer clinical cure. However, the toxicity of chemotherapy agents on normal cells and their resistance to drugs have been considered as the main barrier to proceed with chemotherapy.
2,3
Nowadays, other types of treatments such as hormone replacement therapy and complementary therapies are under clinical consideration, among which targeted therapies, gene therapy, and stem cell therapy have gained considerable attention in the breast cancer research field.
4
In the last decade, stem cell treatment has been considered as a new method for discovering potential therapeutic approaches in cancer therapy.
5-10
In this regard, a great deal of researches have underscored the mesenchymal stem cells’ (MSCs) impact and their related factors on cancer cells.
7,11-13
MSCs are defined as regenerative undifferentiated cells capable of being differentiated into various cell types.
14
Recently, several studies have unveiled MSCs’ potential of suppressing tumors by inhibiting tumor cell proliferation and inducing apoptosis in cancer cells.
6,9,10,15,16
It is argued that the human amniotic stem cells (hAECs) anticancer effect is associated with the endogenous production of growth inhibitors which target tumor growth and progression. Some studies showed hAECs express a range of cytotoxic cytokines, such as IFN-γ, TGF-β, TNF-α and TNF-β as apoptotic inducer substances.
17
Additionally, hAECs secrete various interleukins, including IL-3, IL-4, and IL-2, to promote cytotoxicity in NK cells, the targeting of cancer cells, and the inhibition of tumor formation.
18,19
Furthermore, the ability of MSCs to move to primary tumors could be used to deliver anti-cancer factors to the tumor site.
20,21
Human amniotic fluid mesenchymal stem cell-conditioned medium (hAFMSCs-CM), as effective stem cells in treating a number of human diseases, are achieved from pregnant women at the end or the second trimester of pregnancy using amniocentesis.
22,23
Therefore, not only is the generation of such cell lines considerably easier than human embryonic stem cells (hESCs), they are also not subject to hESCs barriers. Some studies have revealed the inhibitory effects of stem cell conditioned medium (CM) on cancer cells.
24,25
CM has many advantages such as easy production, freezing-thawing competence, and packaging.
26
There is sufficient evidence about hAMSCs ability to produce IFN-γ and CXCL10 as key inhibitors of angiogenesis in the literature.
27
IFN-γ has the potential to hinder a tumor growth and enhance the apoptosis.
28,29
The hAMSCs-CM targets the ratio of cells in S and G2/M phase of PBMC cells leading to apoptosis induction.
30
In addition, hAFMSCs express a number of miRNAs (miR-195-3p, miR-19b-1-5p, miR-20a-5p, miR-20b-3p, miR-26a-1-3p, 708-3p, miR-16-1-3p, 3p, miR-15b-3p, 5p, miR-93-3p, miR204)
31-35
that negatively interact with anti-apoptotic targets.
hAFMSCs are known to have anti-cancer effects by inducing P53 (tumor suppresser) and P21 expression as well as reducing cyclin B1 and D1 after five days of co-culturing with human ovarian cancer cell lines.
6
P21 acts as a P53 transcriptional target, inhibiting cell cycle activity in G1/G2 phases.
36
P53 inhibits the proliferation of abnormal cells by adjusting cell cycle checkpoints in most tissues.
36
Various breast cancers mutate P53, resulting in more aggressive forms of the disease.
37
Bcl-2 inhibition by P53, as a transcriptional factor, is crucial for apoptosis induction. As an anti-apoptotic gene with high expression in most breast cancers, Bcl-2 is known as an effective factor in primary breast cancer prognosis.
38,39
Bax is a pro-apoptotic gene within the Bcl-2 family that presents expression in most breast cancers; a low expression of Bax leads to apoptosis resistance in breast cancer.
40
Based on our literature review and meta-analysis, hardly, we could findany study reporting the effect of the cell-free hAFMSCs conditioned medium on MCF-7 cells viability and the apoptosis. Therefore, the present work aims at assessing the apoptosis and meta-analysis of hAFMSCs-CM on breast cancer cell line.
Materials and Methods
hAFMSCs culture
hAFMSCswere prepared in accordance with previous studies.
22
Cells were plated in 25 cm2 cell culture flasks and DMEM-F12 (Dulbecco's Modified Eagle Medium/ Nutrient Mixture F-12) were supplemented with 15% FBS (Fetal Bovine serum), streptomycin (100 μg/mL), penicillin (100 units/mL), and 10 ng/mL of bFGF (basic fibroblast growth factor). The cells were cultured in an incubator with 5% CO2 humidified gas environment at 37°C.
Preparing conditioned medium
The hAFMSCs were cultured in 75 cm2 flasks toprepare the conditioned media. When the cells reached 70% to 80% confluency, they were washed with phosphate buffer saline (PBS) for 3 times and were kept in DMEM-L (Dulbecco's Modified Eagle Medium-Low Glucose), penicillin (100 units/mL), and streptomycin (100 μg/mL) for 48-72 hours at 37°C in a 5% CO2 humidified environment. Afterward, the media were collected from the flasks and centrifuged at 450 g for 10 minutes to acquire the supernatant and discard the pellet. Passing through a 0.22-μm filter, the media were stored at -80˚C (see Fig. 1).
Fig. 1.
Schematic diagram showing condition medium preparation from hAFMSCs.
.
Schematic diagram showing condition medium preparation from hAFMSCs.
MCF-7 and Hu02 cells culture and treatment
MCF-7 (human breast cancer cell line) and Hu02 (human skin fibroblast cell line) cell lines were obtained from IBRC (Iranian Biological Resource Center). The cells were grown in 25 cm2 flasks with ESCs culture medium (DMEM supplemented with 10% FBS, 100 μg/mL of streptomycin, and 100 units/mL of penicillin). Cells were stored in a humid gas environment with 5% CO2 at 37°C. The media were replaced 3 times per week; 80% (v/v) hAFMSCs-CM was used for the treatment.
Cell viability assay
To determine the effect of hAFMSCs-CM, cell viability was evaluated using MTT (3-(4, 5-dimethylthiazol-2- yl)-2, 5-diphenyltetrazolium bromide) (Sigma, Cas# 298-93-1, USA) assay, as explained elsewhere.
6,41
MCF-7 and Hu02 cells were treated with different percentages of hAFMSCs-CM (20%, 40%, & 80%) for 24, 48, and 72 hours, respectively. In order to determine the cell viability, 0.5 mg/mL of MTT reagent was added to each well and incubated for 4 hours. Then, the MTT solution was removed and 100 μL DMSO (Dimethyl sulfoxide) was added to each well of the 96-well plate to solve formazan crystal. ELISA reader (BioTek, USA) was recruited to measure the absorbance at 570 nm. The untreated cells were considered as the control. To calculate the cell viability, the following formula was used:
Cell viability (%) = (Mean optical absorbance of the treated cells/ Mean optical absorbance of the control cells) × 100
RNA extraction and cDNA synthesis
While the control cells were maintained using the normal media and incubated, MCF-7 and Hu02 cells were treated with 80% of hAFMSCs-CM for total RNA extraction.
After 24 hours, total RNA of MCF-7 was extracted using the RiboEx kit (Gene All, Cat No.301-001, Korea) and the complementary DNA (cDNA) was synthesized from the total RNA using BioFACT kit (BioFACT, Cat No.BR441-096, Korea) based on the manufacture's protocols.
Real-time PCR
To characterize the hAFMSCs-CM effects on the pro-apoptotic (Bax) and the anti-apoptotic (Bcl-2) mRNA expression, Real-time PCR was carried out using SYBER Green (BioFACT, Cat. No. DQ385-40h, Korea) in ABI (Applied Biosystems Step One Plus) detection system in compliance with the manufacture's instruction. Table 1 illustrates the sequence of the primers used in this study; GAPDH (housekeeping gene) was considered as the internal control.
Table 1.
The primers’ sequence used for the Real Time PCR
|
Primer Sequences
|
Gene
|
Forward: 5'- CAAGATCATCAGCAATGCCTCC - 3'
Reverse: 5'- GCCATCACGCCAGTTTCC - 3'
|
GAPDH |
Forward: 5'- GACTCCCCCCGAGAGGTCTT - 3'
Reverse: 5'- ACAGGGCCTTGAGCACCAGTT - 3'
|
BAX |
Forward: 5'- GAGCGTCAACCGGGAGATGTC - 3'
Reverse: 5'- TGCCGGTTCAGGTACTCAGTC - 3'
|
Bcl-2 |
Relative gene expression was calculated using 2-∆∆Ct method based on the following formula
42
:
∆∆Ct = ∆Ct
(treated)
- ∆Ct
(untreated)
= (Ct, Target gene – Ct GAPDH)
(treated)
– (Ct Target gene
– Ct GAPDH)
(untreated)
Western blot analysis
After the hAFMSCs-CM treatment, western blot (WB) analysis was performed to evaluate the amount of P53 protein in MCF-7 and Hu02 cells. While MCF-7 cells were treated with 80% of hAFMSCs-CM, untreated cells were considered as the control. The cells were collected and lysed following a 24-hour incubation, then an electrophoresis was performed when equal amounts of crude protein (50 µg) of sample were loaded in each lane for 10% SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel). The extracted proteins were transferred to a polyvinylidene fluoride (PVDF) membrane where they were blocked with 2% no-fat milk for 1 hour. Next, they were incubated with mouse anti-P53 (Santa Cruz Biotechnology, sc-126, 1:300) and anti-β- actin (sc-47778, 1:300) at 4°C overnight. Afterward, the membrane was incubated with a probed secondary antibody conjugated to HRP (horseradish peroxidase) (Anti-rabbit 1:1000) for 1 hour. An Enhanced Chemiluminescence detection system was employed for detection. Beta-actin was used for normalization and internal control, and ImageJ software was utilized to analyze the image.
Flow cytometry
Apoptotic cells were exposed to phosphatidylserin in their outer plasma membrane, which were identified by fluorescein isothiocyanate (FITC) labeled Annexin-V/PI (propidium iodide) using flow cytometry. Following a 24 hours treatment of cells with 80% hAFMSCs-CM, MCF-7 and Hu02 cells were harvested by trypsin and washed with PBS. After 8 minutes of centrifugation at 1300 rpm, the cells were re-suspended in 100 µL binding buffer (Invitrogen, Lot #4338210) and were mixed with 2 µL Annexin-V (Invitrogen, Lot #1989095); they were then incubated on ice for 20 minutes in a dark place. The cells’ solution was centrifuged at 1300 rpm for 8 min, after which the supernatant was removed and 100 µL binding buffer was added. The sample solution was combined with 1 µL of PI (Invitrogen, Lot #1957465) and was incubated for 20 minutes in a dark place. Flow Jo (7.6.1) software was used to run the flow cytometry analysis on samples utilizing BD FACS Calibur Flow Cytometry (BD Biosciences, NJ, USA).
Finding a possible molecular network underlying the hAFMSCs-CM function using integrated approach of meta-analysis and literature mining
We conducted a literature-mining-based network analysis and employed an integrated approach of meta-analysis of expression data to ascertain the possible regulatory network underlying hAFMSCs-CM function in breast cancer cells.
As presented in Fig. 2, the following steps were performed:
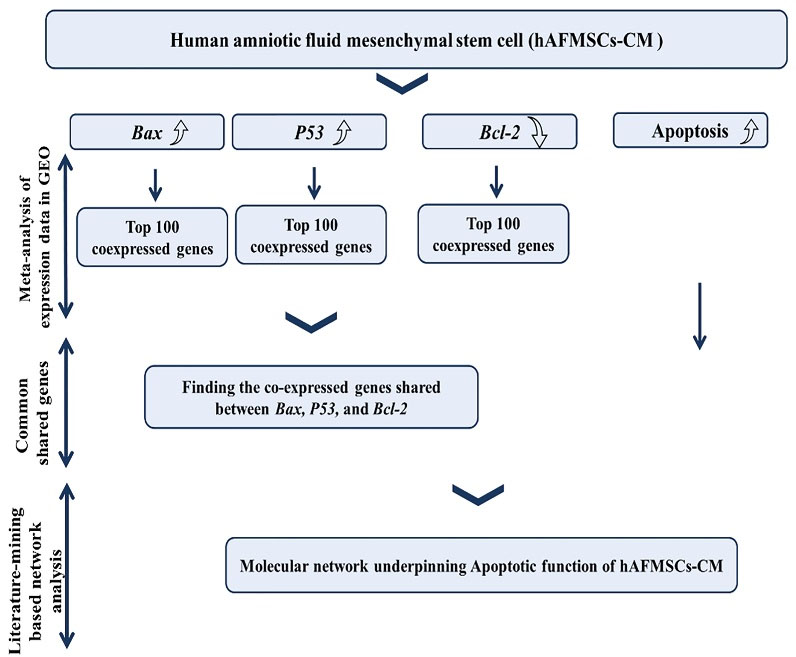
Fig. 2.
Bioinformatics pipeline employed in this study.
.
Bioinformatics pipeline employed in this study.
-
Recruitment of Mutual Ranking (MR) statistics for Co-expression meta-analysis of Bax, Bcl-2, and P53, consulting public transcriptomic data in Gene Expression Omnibus (GEO).
-
Selection of top 100 co-expressed genes with Bax, Bcl-2, and P53.
-
Finding shared genes between co-expressed profiles of Bax, Bcl-2, and P53.
-
Performing Literature-mining based network analysis: Discovery of common targets and regulators with positive interactions with Bax, P53 and apoptosis, and negative interactions with Bcl-2.
Mutual ranking (MR) statistics and Z-transformation of expression data were used for expression data meta-analysis and removal of platform effect, as described elsewhere.
43,44
As compared with the common approach for running the Pearson correlation, MR statistics employs a ranking approach for correlation analysis where it remains unaffected by the experiment. After calculating the rank correlation for each experiment, geometric average of correlation coefficients was ranked in logarithmic manner.
45,46
Correlation rankings were used extensively during the meta-analysis (e.g. Rankprod).
47
The expression data were retrieved from the GEO (NCBI public repository of expression data, https://www.ncbi.nlm.nih.gov/geo/). COXPRESdb v7 tool was performed for analysis.
48
Lower values of MR represent higher level of association where MR value of each gene, including itself, is 0.
Literature mining-based database of Pathway Studio Mammalian (Elsevier)
49,50
was performed, as previously described.
51,52
The database collects data through NLP (Natural Language Processing) algorithm and contains 13 440 356 mined relations from full text published paper and 1 439 833 entities (e.g., proteins/genes, cell process, small molecules, and diseases) (March 2021). The database is enriched with additional inputs from Gene Ontology Consortium for cellular location analysis, MiRbase, and various network construction approaches such as “Common Binding Partner”, “Downstream Target Discovery”, and “Upstream Regulator Discovery”, among others.
Statistical analysis
Each experiment was performed in triplicate. Data were presented as means ± standard error of the mean (SEM). A one-way ANOVA and a t test were conducted to compare the three and the two groups, respectively. Any differences were deemed significant when the P value was smaller than 0.05 (P < 0.05). GraphPad Prism software (La, Jolla, CA) version 8.4.3(686) was utilized to run the statistical analysis.
Results
hAFMSCs-CM effects on MCF-7 cell viability
To investigate the hAFMSCs-CM impact on MCF-7 and Hu02 cell viability, an MTT assay was carried out 3 times (24, 48, and 72 hours) after the treatment. As shown in Fig. 3, hAFMSCs-CM was found to have a cytotoxic effect on MCF-7. Noteworthy, no cytotoxic effects were observed on Hu02 cells. Our data suggest that the cell viability in MCF-7 cells was decreased significantly as a result of CM after the treatments (24, 48, and 72 hours) (see Fig. 3A, 3B, 3C). With 20% of CM no significant effect on cell viability could be observed; however, in 40% and 80% of CMs (after 24 hours), cell growth was inhibited as compared with the control cells. Fig. 3A illustrates the cell viability being declined to 78% (P < 0.0001) when 80% of CM and to 86.99% (P = 0.0027) when 40% of CM were used after the phase 1 of the treatment (24 hours). Upon the completion of the 48-hour and 72-hour incubation (40% CM) phases, the hAFMSCs-CM demonstrated an insignificant effect on the cell viability (P > 0.05). Noteworthy, 80% of the CM was found to have affected the MCF-7 cells considerably (P < 0.05). Although hAFMSCs-CM failed to affect MCF-7’s cell viability, it was found to be capable of promoting the cell viability in Hu02 as normal cells (P = 0.0014, during the 24 hour-treatment).
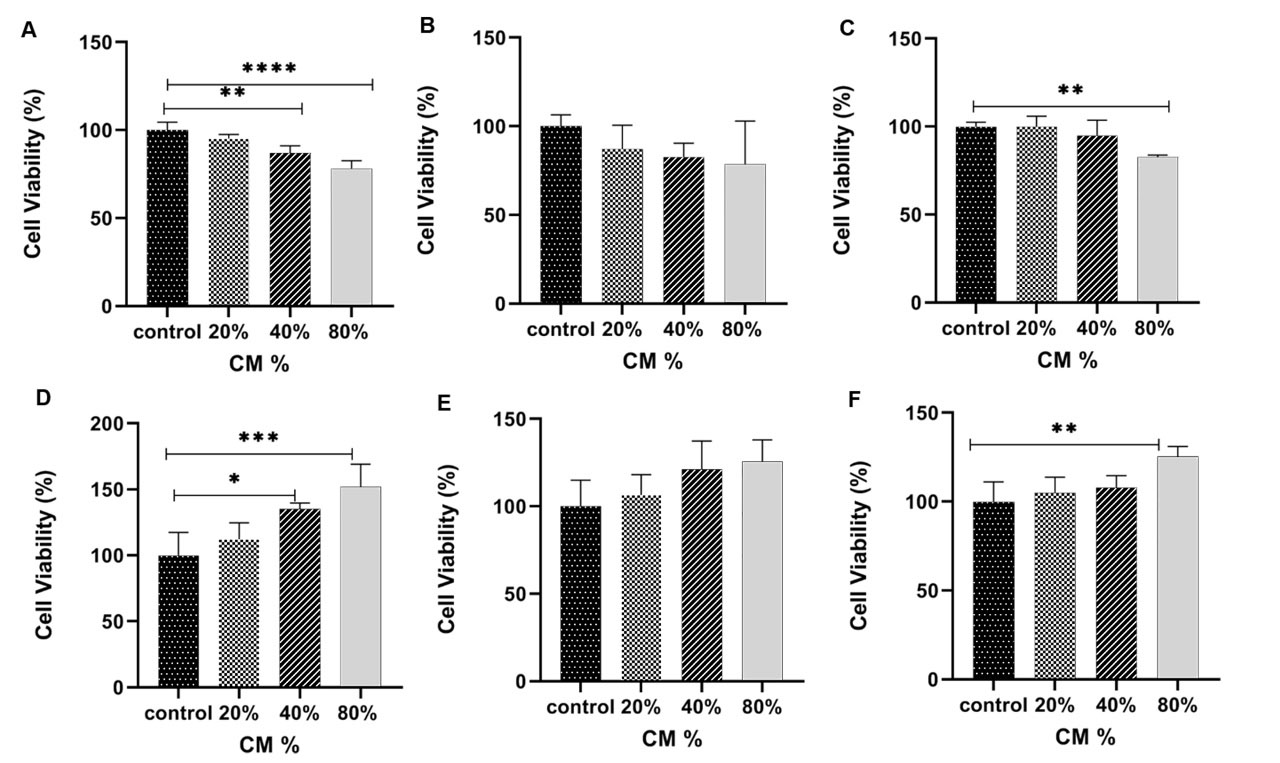
Fig. 3.
The MCF-7 and Hu02 cell viability was assessed by MTT assay for MCF-7 cells within 24 (A), 48 (B) and 72 (C) hours and for and Hu02 within 24 h (D), 48 h (E) and 72 h (F) treatment with hAFMSCs-CM. After 24 h, a significant decrease in MCF-7 cells viability (P<0.0001) and Hu02 cells viability (P<0.005) was observed. The data are presented as mean ± SEM. Significantly different (** P<0.005, ****P<0.0001).
.
The MCF-7 and Hu02 cell viability was assessed by MTT assay for MCF-7 cells within 24 (A), 48 (B) and 72 (C) hours and for and Hu02 within 24 h (D), 48 h (E) and 72 h (F) treatment with hAFMSCs-CM. After 24 h, a significant decrease in MCF-7 cells viability (P<0.0001) and Hu02 cells viability (P<0.005) was observed. The data are presented as mean ± SEM. Significantly different (** P<0.005, ****P<0.0001).
hAFMSCs-CM effects on Bax and Bcl-2 genes expression and P53 protein expression
Following the hAFMSCs-CM 24-hour treatment, the Bax and Bcl-2 mRNA level expressions were analyzed. The genes’ Ct values were normalized against the GAPDH mRNA level (the housekeeping gene). Notably, as illustrated in Fig. 4A, the pro-apoptotic Bax gene’s expression level increased significantly as compared with the control group (P < 0.0001). On the other hand, the anti-apoptotic Bcl-2 gene’s mRNA level decreased considerably when cells were treated with 80% hAFMSCs-CM for 24 hours (P = 0.0012). Nevertheless, as Fig. 4C shows, in normal cells (Hu02), the level of the Bax gene declined and Bcl2 increased after the hAFMSCs-CM treatment.
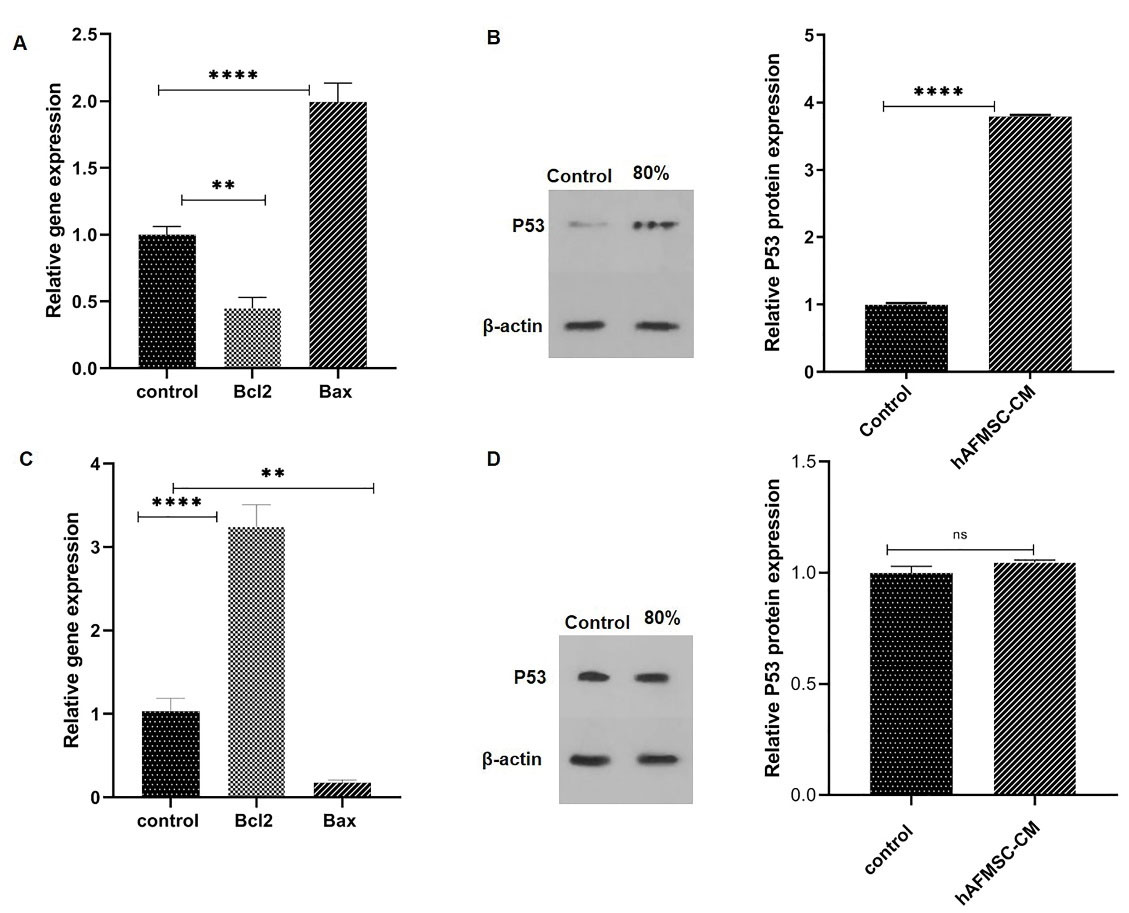
Fig. 4.
Real-time PCR and western blot analysis were used to assess hAFMSCs-CM effect on MCF-7 and Hu02 cells. (A) pro-apoptotic Bax and anti-apoptotic Bcl-2 genes of MCF-7 were treated with 80% hAFMSCs-CM in 24 h. (B) WB analysis and P53 protein quantification were used to evaluate 80% hAFMSCs-CM effect on P53 protein expression compared with the control in MCF-7 cells. (C) Hu02’s Bax and Bcl-2 genes were treated with 80% hAFMSCs-CM within 24 h. (D) WB analysis and P53 protein quantification to evaluate the 80% hAFMSCs-CM effect on P53 protein expression compared with the control in Hu02 cells. P53 protein level was determined by ImageJ analysis. The data are presented as mean ± SEM. (****P<0.0001, ***P<0.005).
.
Real-time PCR and western blot analysis were used to assess hAFMSCs-CM effect on MCF-7 and Hu02 cells. (A) pro-apoptotic Bax and anti-apoptotic Bcl-2 genes of MCF-7 were treated with 80% hAFMSCs-CM in 24 h. (B) WB analysis and P53 protein quantification were used to evaluate 80% hAFMSCs-CM effect on P53 protein expression compared with the control in MCF-7 cells. (C) Hu02’s Bax and Bcl-2 genes were treated with 80% hAFMSCs-CM within 24 h. (D) WB analysis and P53 protein quantification to evaluate the 80% hAFMSCs-CM effect on P53 protein expression compared with the control in Hu02 cells. P53 protein level was determined by ImageJ analysis. The data are presented as mean ± SEM. (****P<0.0001, ***P<0.005).
Fig. 4B illustrates the WB analysis of P53 protein expression, demonstrating a significant (P < 0.0001) increase (about 3.7 fold) after the hAFMSCs-CM treatment, as compared with control (untreated) cells. However, we could not observe meaningful differences in P53 expression in Hu02 cells (P > 0.05) (Fig. 4D).
hAFMSCs-CM effects on apoptosis
Apoptosis was measured using a flow cytometry assay via annexin V and PI staining of the cells. As demonstrated in Fig. 5, apoptosis was induced in the female human breast cancer cells by hAFMSCs-CM. The flow cytometry analysis of MCF-7 cells, treated with 80% hAFMSCs-CM for 24 hours, showed early apoptosis (annexin V+ PI-) of nearly 22.7%, whereas the control cells’ apoptotic functions were about 6.2%. Despite insignificant differences among normal cells (P > 0.05), no considerable apoptosis could be observed in Hu02 cells (P > 0.05) (Fig. 5F).
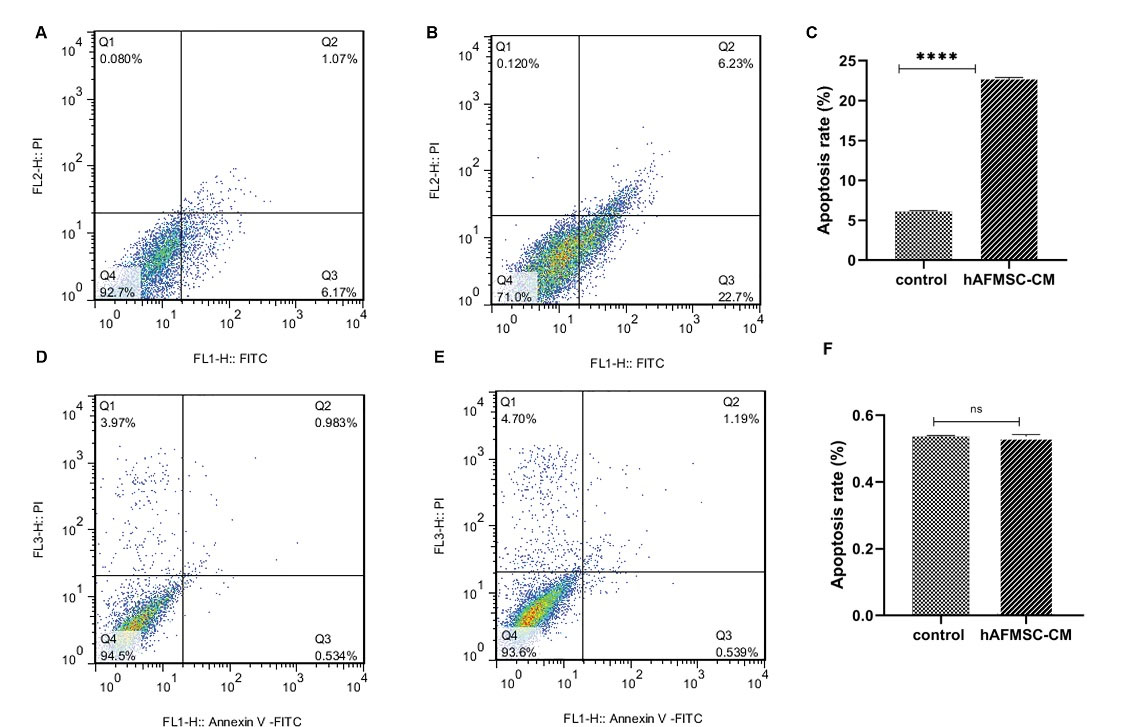
Fig. 5.
Apoptotic evaluation using the flow cytometry via annexin V and PI staining. (A) Control MCF-7 cells’ (untreated cells) apoptosis. (B) The effect of 80% hAFMSCs-CM on MCF-7 cells after 24 h -- hAFMSCs-CM induces apoptosis in MCF-7 cells. (C) Quantification of apoptosis in MCF-7 cells. (D) Control Hu02 cells (untreated cells) apoptosis. (E) The effect of 80% hAFMSCs-CM on Hu02 cells after 24 h. hAFMSCs-CM do not affect apoptosis in Hu02 cells. (F) Quantification of apoptosis in Hu02 cells. Fig. 5. represents viable cells (annexin V-PI-) population, early apoptosis (annexin V+ PI-), late apoptosis (annexin V+PI+), and necrotic cells (annexin V-PI Flow cytometry analysis was performed for samples using BD FACS Calibur flow cytometry [BD Biosciences, NJ, USA]). Flow Jo. (7.6.1) software was used to analyze the data.
.
Apoptotic evaluation using the flow cytometry via annexin V and PI staining. (A) Control MCF-7 cells’ (untreated cells) apoptosis. (B) The effect of 80% hAFMSCs-CM on MCF-7 cells after 24 h -- hAFMSCs-CM induces apoptosis in MCF-7 cells. (C) Quantification of apoptosis in MCF-7 cells. (D) Control Hu02 cells (untreated cells) apoptosis. (E) The effect of 80% hAFMSCs-CM on Hu02 cells after 24 h. hAFMSCs-CM do not affect apoptosis in Hu02 cells. (F) Quantification of apoptosis in Hu02 cells. Fig. 5. represents viable cells (annexin V-PI-) population, early apoptosis (annexin V+ PI-), late apoptosis (annexin V+PI+), and necrotic cells (annexin V-PI Flow cytometry analysis was performed for samples using BD FACS Calibur flow cytometry [BD Biosciences, NJ, USA]). Flow Jo. (7.6.1) software was used to analyze the data.
Meta-analysis based co-expressed genes with Bax, Bcl-2, and P53
Tables 2, 3, and 4 show the genes that were found to be co-expressed with Bax, Bcl-2, and P53 after a meta-analysis. Fig. 6 also presents the shared genes found within the meta-analysis derived co-expressed profiles of Bax, Bcl-2, and P53. Notably, Bax and P53 were found to be co-expressed. DDB2 (Damage specific DNA Binding protein 2) is among the top 3 co-expressed genes with Bax that co-expresses with P53. In the same vein, EIF5A (Eukaryotic Translation Initiation Factor 5A) is an important protein that co-expresses with Bax and P53.
Table 2.
Meta-analysis-derived co-expressed genes with Bax
|
|
Gene
|
Function
|
Entrez Gene ID
|
| 0 |
BAX |
BCL2 associated X: apoptosis regulator |
581 |
| 1 |
FDXR |
Ferredoxin reductase |
2232 |
| 2 |
AP1S1 |
Adaptor related protein complex 1 sigma 1 subunit |
1174 |
| 3 |
DDB2 |
Damage specific DNA binding protein 2 |
1643 |
| 4 |
RPS27L |
Ribosomal protein S27 like |
51065 |
| 5 |
BBC3 |
BCL2 binding component 3 |
27113 |
| 6 |
MDM2 |
MDM2 proto-oncogene |
4193 |
| 7 |
ZMAT3 |
Zinc finger matrin-type 3 |
64393 |
| 8 |
LINC01759 |
Long intergenic non-protein coding RNA 1759 |
1.03E+08 |
| 9 |
PHLDA3 |
Pleckstrin homology like domain family A member 3 |
23612 |
| 10 |
PDAP1 |
PDGFA associated protein 1 |
11333 |
| 11 |
AEN |
Apoptosis enhancing nuclease |
64782 |
| 12 |
PFDN6 |
Prefoldin subunit 6 |
10471 |
| 13 |
TIGAR |
TP53 induced glycolysis regulatory phosphatase |
57103 |
| 14 |
PFN1 |
Profilin 1 |
5216 |
| 15 |
CDKN1A |
Cyclin dependent kinase inhibitor 1A |
1026 |
| 16 |
PRMT1 |
Protein arginine methyltransferase 1 |
3276 |
| 17 |
APH1A |
Aph-1 homolog A: gamma-secretase subunit |
51107 |
| 18 |
TRIAP1 |
TP53 regulated inhibitor of apoptosis 1 |
51499 |
| 19 |
PHPT1 |
Phosphohistidine phosphatase 1 |
29085 |
| 20 |
ZNF428 |
Zinc finger protein 428 |
126299 |
| 21 |
HNRNPUL2 |
Heterogeneous nuclear ribonucleoprotein U like 2 |
221092 |
| 22 |
PSENEN |
Presenilin enhancer gamma-secretase subunit |
55851 |
| 23 |
EML2 |
Echinoderm microtubule associated protein like 2 |
24139 |
| 24 |
EDA2R |
Ectodysplasin A2 receptor |
60401 |
| 25 |
ISOC2 |
Isochorismatase domain containing 2 |
79763 |
| 26 |
MIR34AHG |
MIR34A host gene |
1.07E+08 |
| 27 |
AP2S1 |
Adaptor related protein complex 2 sigma 1 subunit |
1175 |
| 28 |
ARPC4 |
Actin related protein 2/3 complex subunit 4 |
10093 |
| 29 |
ARPC1B |
Actin related protein 2/3 complex subunit 1B |
10095 |
| 30 |
RPS19 |
Ribosomal protein S19 |
6223 |
| 31 |
RFXANK |
Regulatory factor X associated ankyrin containing protein |
8625 |
| 32 |
GTF2H4 |
General transcription factor IIH subunit 4 |
2968 |
| 33 |
LAMTOR2 |
Late endosomal/lysosomal adaptor: MAPK and MTOR activator 2 |
28956 |
| 34 |
THOC6 |
THO complex 6 |
79228 |
| 35 |
SYMPK |
Symplekin |
8189 |
| 36 |
TMEM160 |
Transmembrane protein 160 |
54958 |
| 37 |
TP53I3 |
Tumor protein p53 inducible protein 3 |
9540 |
| 38 |
PPP4C |
Protein phosphatase 4 catalytic subunit |
5531 |
| 39 |
PNKP |
Polynucleotide kinase 3'-phosphatase |
11284 |
| 40 |
GPX1 |
Glutathione peroxidase 1 |
2876 |
| 41 |
ITPA |
Inosine triphosphatase |
3704 |
| 42 |
SPATA18 |
Spermatogenesis associated 18 |
132671 |
| 43 |
MPDU1 |
Mannose-P-dolichol utilization defect 1 |
9526 |
| 44 |
EIF5A |
Eukaryotic translation initiation factor 5A |
1984 |
| 45 |
PURPL |
p53 upregulated regulator of p53 levels |
643401 |
| 46 |
TFPT |
TCF3 fusion partner |
29844 |
| 47 |
IRF3 |
Interferon regulatory factor 3 |
3661 |
| 48 |
CHMP4B |
Charged multivesicular body protein 4B |
128866 |
| 49 |
GDF15 |
Growth differentiation factor 15 |
9518 |
| 50 |
CYBA |
Cytochrome b-245 alpha chain |
1535 |
| 51 |
PGLS |
6-Phosphogluconolactonase |
25796 |
| 52 |
DLST |
Dihydrolipoamide S-succinyltransferase |
1743 |
| 53 |
FBXO22 |
F-box protein 22 |
26263 |
| 54 |
PIDD1 |
p53-induced death domain protein 1 |
55367 |
| 55 |
GEMIN7 |
Gem nuclear organelle associated protein 7 |
79760 |
| 56 |
TNFRSF10B |
TNF receptor superfamily member 10b |
8795 |
| 57 |
U2AF2 |
U2 small nuclear RNA auxiliary factor 2 |
11338 |
| 58 |
UBE2M |
Ubiquitin conjugating enzyme E2 M |
9040 |
| 59 |
TMED9 |
Transmembrane p24 trafficking protein 9 |
54732 |
| 60 |
CTSZ |
Cathepsin Z |
1522 |
| 61 |
APOBEC3H |
Apolipoprotein B mRNA editing enzyme catalytic subunit 3H |
164668 |
| 62 |
LAMTOR4 |
Late endosomal/lysosomal adaptor: MAPK and MTOR activator 4 |
389541 |
| 63 |
POLH |
DNA polymerase eta |
5429 |
| 64 |
QTRT1 |
queuine tRNA-ribosyltransferase catalytic subunit 1 |
81890 |
| 65 |
PTCHD4 |
Patched domain containing 4 |
442213 |
| 66 |
HIRA |
histone cell cycle regulator |
7290 |
| 67 |
RCC1 |
Regulator of chromosome condensation 1 |
1104 |
| 68 |
COMMD4 |
COMM domain containing 4 |
54939 |
| 69 |
TRAPPC1 |
Trafficking protein particle complex 1 |
58485 |
| 70 |
ARF5 |
ADP ribosylation factor 5 |
381 |
| 71 |
BAK1 |
BCL2 antagonist/killer 1 |
578 |
| 72 |
RAB35 |
RAB35: member RAS oncogene family |
11021 |
| 73 |
STK16 |
Serine/threonine kinase 16 |
8576 |
| 74 |
FTL |
Ferritin light chain |
2512 |
| 75 |
PIH1D1 |
PIH1 domain containing 1 |
55011 |
| 76 |
SHKBP1 |
SH3KBP1 binding protein 1 |
92799 |
| 77 |
KDELR1 |
KDEL endoplasmic reticulum protein retention receptor 1 |
10945 |
| 78 |
RANGRF |
RAN guanine nucleotide release factor |
29098 |
| 79 |
TP53 |
Tumor protein p53 |
7157 |
| 80 |
PBX2 |
PBX homeobox 2 |
5089 |
| 81 |
SDHC |
Succinate dehydrogenase complex subunit C |
6391 |
| 82 |
MMP14 |
Matrix metallopeptidase 14 |
4323 |
| 83 |
DPP3 |
dipeptidyl peptidase 3 |
10072 |
| 84 |
TNFRSF10C |
TNF receptor superfamily member 10c |
8794 |
| 85 |
PPP1CA |
Protein phosphatase 1 catalytic subunit alpha |
5499 |
| 86 |
PPP2R1A |
Protein phosphatase 2 scaffold subunit Aalpha |
5518 |
| 87 |
FUS |
FUS RNA binding protein |
2521 |
| 88 |
CFL1 |
Cofilin 1 |
1072 |
| 89 |
TM7SF3 |
Transmembrane 7 superfamily member 3 |
51768 |
| 90 |
WDR83 |
WD repeat domain 83 |
84292 |
| 91 |
FBXL19 |
F-box and leucine rich repeat protein 19 |
54620 |
| 92 |
KPTN |
Kaptin: actin binding protein |
11133 |
| 93 |
ALYREF |
Aly/REF export factor |
10189 |
| 94 |
TAX1BP3 |
Tax1 binding protein 3 |
30851 |
| 95 |
VASP |
Vasodilator stimulated phosphoprotein |
7408 |
| 96 |
MRPS12 |
Mitochondrial ribosomal protein S12 |
6183 |
| 97 |
LINC02051 |
Long intergenic non-protein coding RNA 2051 |
1.08E+08 |
| 98 |
SRSF9 |
Serine and arginine rich splicing factor 9 |
8683 |
| 99 |
TWF2 |
Twinfilin actin binding protein 2 |
11344 |
| 100 |
DRG2 |
Developmentally regulated GTP binding protein 2 |
1819 |
Table 3.
Meta-analysis-derived co-expressed genes with Bcl-2
|
|
Gene
|
Function
|
Entrez Gene ID
|
| 0 |
BCL2 |
BCL2: apoptosis regulator |
596 |
| 1 |
BACH2 |
BTB domain and CNC homolog 2 |
60468 |
| 2 |
IKZF1 |
IKAROS family zinc finger 1 |
10320 |
| 3 |
MDFIC |
MyoD family inhibitor domain containing |
29969 |
| 4 |
ITPKB |
inositol-trisphosphate 3-kinase B |
3707 |
| 5 |
KIAA1328 |
KIAA1328 |
57536 |
| 6 |
NFATC1 |
Nuclear factor of activated T cells 1 |
4772 |
| 7 |
NSD3 |
Nuclear receptor binding SET domain protein 3 |
54904 |
| 8 |
KDSR |
3-Ketodihydrosphingosine reductase |
2531 |
| 9 |
LDLRAD4 |
Low density lipoprotein receptor class A domain containing 4 |
753 |
| 10 |
NEMP2 |
Nuclear envelope integral membrane protein 2 |
1E+08 |
| 11 |
ANKRD44 |
Ankyrin repeat domain 44 |
91526 |
| 12 |
MYB |
MYB proto-oncogene: transcription factor |
4602 |
| 13 |
RAPGEF6 |
Rap guanine nucleotide exchange factor 6 |
51735 |
| 14 |
HNRNPA0 |
Heterogeneous nuclear ribonucleoprotein A0 |
10949 |
| 15 |
ZXDA |
Zinc finger: X-linked: duplicated A |
7789 |
| 16 |
AFF3 |
AF4/FMR2 family member 3 |
3899 |
| 17 |
LOC374443 |
C-type lectin domain family 2 member D pseudogene |
374443 |
| 18 |
KIAA1147 |
KIAA1147 |
57189 |
| 19 |
TNFRSF13B |
TNF receptor superfamily member 13B |
23495 |
| 20 |
PTGER4 |
Prostaglandin E receptor 4 |
5734 |
| 21 |
BMI1 |
BMI1 proto-oncogene: polycomb ring finger |
648 |
| 22 |
ITPR1 |
Inositol 1:4:5-trisphosphate receptor type 1 |
3708 |
| 23 |
LOC100509088 |
Hypothetical LOC100509088 |
1.01E+08 |
| 24 |
RHOH |
Ras homolog family member H |
399 |
| 25 |
KCNQ5 |
Potassium voltage-gated channel subfamily Q member 5 |
56479 |
| 26 |
ESR1 |
Estrogen receptor 1 |
2099 |
| 27 |
SLC38A1 |
Solute carrier family 38 member 1 |
81539 |
| 28 |
ELP2 |
Elongator acetyltransferase complex subunit 2 |
55250 |
| 29 |
GPR174 |
G protein-coupled receptor 174 |
84636 |
| 30 |
TAF4B |
TATA-box binding protein associated factor 4b |
6875 |
| 31 |
RCSD1 |
RCSD domain containing 1 |
92241 |
| 32 |
SETBP1 |
SET binding protein 1 |
26040 |
| 33 |
NOP53 |
NOP53 ribosome biogenesis factor |
29997 |
| 34 |
LINC00341 |
Long intergenic non-protein coding RNA 341 |
79686 |
| 35 |
LTA |
Lymphotoxin alpha |
4049 |
| 36 |
CEP68 |
Centrosomal protein 68 |
23177 |
| 37 |
DDHD2 |
DDHD domain containing 2 |
23259 |
| 38 |
LRCH1 |
Leucine rich repeats and calponin homology domain containing 1 |
23143 |
| 39 |
ZNF24 |
Zinc finger protein 24 |
7572 |
| 40 |
TARSL2 |
Threonyl-tRNA synthetase like 2 |
123283 |
| 41 |
LRRC8C-DT |
LRRC8C divergent transcript |
400761 |
| 42 |
PPM1K |
Protein phosphatase: Mg2+/Mn2+ dependent 1K |
152926 |
| 43 |
SMAD2 |
SMAD family member 2 |
4087 |
| 44 |
RGS1 |
Regulator of G protein signaling 1 |
5996 |
| 45 |
FMNL3 |
Formin like 3 |
91010 |
| 46 |
CD69 |
CD69 molecule |
969 |
| 47 |
C21orf2 |
Chromosome 21 open reading frame 2 |
755 |
| 48 |
ZNF407 |
Zinc finger protein 407 |
55628 |
| 49 |
GNA13 |
G protein subunit alpha 13 |
10672 |
| 50 |
XYLT1 |
xylosyltransferase 1 |
64131 |
| 51 |
SP4 |
Sp4 transcription factor |
6671 |
| 52 |
RBBP6 |
RB binding protein 6: ubiquitin ligase |
5930 |
| 53 |
LINC00909 |
Long intergenic non-protein coding RNA 909 |
400657 |
| 54 |
IRF4 |
Interferon regulatory factor 4 |
3662 |
| 55 |
WDR7 |
WD repeat domain 7 |
23335 |
| 56 |
POU6F1 |
POU class 6 homeobox 1 |
5463 |
| 57 |
PPP3CC |
Protein phosphatase 3 catalytic subunit gamma |
5533 |
| 58 |
NTRK2 |
Neurotrophic receptor tyrosine kinase 2 |
4915 |
| 59 |
TSHZ1 |
Teashirt zinc finger homeobox 1 |
10194 |
| 60 |
PM20D2 |
Peptidase M20 domain containing 2 |
135293 |
| 61 |
PRKCE |
Protein kinase C epsilon |
5581 |
| 62 |
MSI2 |
Musashi RNA binding protein 2 |
124540 |
| 63 |
SLC39A6 |
Solute carrier family 39 member 6 |
25800 |
| 64 |
RSBN1 |
Round spermatid basic protein 1 |
54665 |
| 65 |
ZBTB32 |
Zinc finger and BTB domain containing 32 |
27033 |
| 66 |
EPM2A |
EPM2A: laforin glucan phosphatase |
7957 |
| 67 |
RFTN1 |
Raftlin: lipid raft linker 1 |
23180 |
| 68 |
NFATC2 |
Nuclear factor of activated T cells 2 |
4773 |
| 69 |
N4BP2L1 |
NEDD4 binding protein 2 like 1 |
90634 |
| 70 |
FOXN3 |
Forkhead box N3 |
1112 |
| 71 |
LOC107985690 |
Uncharacterized LOC107985690 |
1.08E+08 |
| 72 |
ZADH2 |
Zinc binding alcohol dehydrogenase domain containing 2 |
284273 |
| 73 |
ARMC5 |
Armadillo repeat containing 5 |
79798 |
| 74 |
ANKRD33B |
Ankyrin repeat domain 33B |
651746 |
| 75 |
LEF1 |
Lymphoid enhancer binding factor 1 |
51176 |
| 76 |
PRDM8 |
PR/SET domain 8 |
56978 |
| 77 |
STAP1 |
Signal transducing adaptor family member 1 |
26228 |
| 78 |
JADE2 |
Jade family PHD finger 2 |
23338 |
| 79 |
MIR155HG |
MIR155 host gene |
114614 |
| 80 |
RABEP1 |
Rabaptin: RAB GTPase binding effector protein 1 |
9135 |
| 81 |
P2RY10 |
P2Y receptor family member 10 |
27334 |
| 82 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor 6 |
9459 |
| 83 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
55188 |
| 84 |
SYNE3 |
Spectrin repeat containing nuclear envelope family member 3 |
161176 |
| 85 |
ABCD2 |
ATP binding cassette subfamily D member 2 |
225 |
| 86 |
SPNS3 |
Sphingolipid transporter 3 (putative) |
201305 |
| 87 |
FBXL17 |
F-box and leucine rich repeat protein 17 |
64839 |
| 88 |
LNPEP |
Leucyl and cystinyl aminopeptidase |
4012 |
| 89 |
GRASP |
General receptor for phosphoinositides 1 associated scaffold protein |
160622 |
| 90 |
LINC00938 |
Long intergenic non-protein coding RNA 938 |
400027 |
| 91 |
MAST4 |
Microtubule associated serine/threonine kinase family member 4 |
375449 |
| 92 |
RNF157 |
Ring finger protein 157 |
114804 |
| 93 |
SOCS2 |
Suppressor of cytokine signaling 2 |
8835 |
| 94 |
MALT1 |
MALT1 paracaspase |
10892 |
| 95 |
LINC00926 |
Long intergenic non-protein coding RNA 926 |
283663 |
| 96 |
CLECL1 |
C-type lectin like 1 |
160365 |
| 97 |
CLNK |
Cytokine dependent hematopoietic cell linker |
116449 |
| 98 |
RASGRP1 |
RAS guanyl releasing protein 1 |
10125 |
| 99 |
FCMR |
Fc fragment of IgM receptor |
9214 |
| 100 |
SDK2 |
Sidekick cell adhesion molecule 2 |
54549 |
Table 4.
Meta-analysis-derived co-expressed genes with P53
|
|
Gene
|
Function
|
Entrez Gene ID
|
| 0 |
TP53 |
Tumor protein p53 |
7157 |
| 1 |
PFN1 |
Profilin 1 |
5216 |
| 2 |
BANF1 |
Barrier to autointegration factor 1 |
8815 |
| 3 |
YWHAE |
Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon |
7531 |
| 4 |
CDK4 |
Cyclin dependent kinase 4 |
1019 |
| 5 |
THOC6 |
THO complex 6 |
79228 |
| 6 |
RAVER1 |
Ribonucleoprotein: PTB binding 1 |
125950 |
| 7 |
ALDH16A1 |
Aldehyde dehydrogenase 16 family member A1 |
126133 |
| 8 |
APEX1 |
Apurinic/apyrimidinic endodeoxyribonuclease 1 |
328 |
| 9 |
MYBBP1A |
MYB binding protein 1a |
10514 |
| 10 |
SHMT2 |
Serine hydroxymethyltransferase 2 |
6472 |
| 11 |
NONO |
Non-POU domain containing octamer binding |
4841 |
| 12 |
TRIM28 |
Tripartite motif containing 28 |
10155 |
| 13 |
SMARCC1 |
SWI/SNF related: matrix associated: actin dependent regulator of chromatin subfamily c member 1 |
6599 |
| 14 |
TRAPPC1 |
Trafficking protein particle complex 1 |
58485 |
| 15 |
GEMIN4 |
Gem nuclear organelle associated protein 4 |
50628 |
| 16 |
CASP2 |
Caspase 2 |
835 |
| 17 |
SF3B3 |
Splicing factor 3b subunit 3 |
23450 |
| 18 |
DRG2 |
Developmentally regulated GTP binding protein 2 |
1819 |
| 19 |
G3BP1 |
G3BP stress granule assembly factor 1 |
10146 |
| 20 |
BTBD2 |
BTB domain containing 2 |
55643 |
| 21 |
SF3B4 |
Splicing factor 3b subunit 4 |
10262 |
| 22 |
PELP1 |
Proline: glutamate and leucine rich protein 1 |
27043 |
| 23 |
EIF5A |
Eukaryotic translation initiation factor 5A |
1984 |
| 24 |
AAAS |
Aladin WD repeat nucleoporin |
8086 |
| 25 |
HNRNPUL1 |
Heterogeneous nuclear ribonucleoprotein U like 1 |
11100 |
| 26 |
RCC2 |
Regulator of chromosome condensation 2 |
55920 |
| 27 |
PFAS |
Phosphoribosylformylglycinamidine synthase |
5198 |
| 28 |
CHTF8 |
Chromosome transmission fidelity factor 8 |
54921 |
| 29 |
DVL2 |
Dishevelled segment polarity protein 2 |
1856 |
| 30 |
SCAMP4 |
Secretory carrier membrane protein 4 |
113178 |
| 31 |
ASB16-AS1 |
ASB16 antisense RNA 1 |
339201 |
| 32 |
WDR6 |
WD repeat domain 6 |
11180 |
| 33 |
MTA2 |
Metastasis associated 1 family member 2 |
9219 |
| 34 |
CAD |
Carbamoyl-phosphate synthetase 2: aspartate transcarbamylase: and dihydroorotase |
790 |
| 35 |
CHST14 |
Carbohydrate sulfotransferase 14 |
113189 |
| 36 |
HNRNPA0 |
Heterogeneous nuclear ribonucleoprotein A0 |
10949 |
| 37 |
IMPDH2 |
Inosine monophosphate dehydrogenase 2 |
3615 |
| 38 |
SF3A2 |
Splicing factor 3a subunit 2 |
8175 |
| 39 |
G6PC3 |
Glucose-6-phosphatase catalytic subunit 3 |
92579 |
| 40 |
APEX2 |
Apurinic/apyrimidinic endodeoxyribonuclease 2 |
27301 |
| 41 |
APOBEC3C |
Apolipoprotein B mRNA editing enzyme catalytic subunit 3C |
27350 |
| 42 |
PRPF8 |
Pre-mRNA processing factor 8 |
10594 |
| 43 |
DDB2 |
Damage specific DNA binding protein 2 |
1643 |
| 44 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
23399 |
| 45 |
UCP2 |
Uncoupling protein 2 |
7351 |
| 46 |
VARS |
Valyl-tRNA synthetase |
7407 |
| 47 |
SET |
SET nuclear proto-oncogene |
6418 |
| 48 |
PATZ1 |
POZ/BTB and AT hook containing zinc finger 1 |
23598 |
| 49 |
NOB1 |
NIN1/PSMD8 binding protein 1 homolog |
28987 |
| 50 |
SNRPA |
Small nuclear ribonucleoprotein polypeptide A |
6626 |
| 51 |
SLC16A13 |
Solute carrier family 16 member 13 |
201232 |
| 52 |
MRPS27 |
Mitochondrial ribosomal protein S27 |
23107 |
| 53 |
NCOA5 |
Nuclear receptor coactivator 5 |
57727 |
| 54 |
RPA1 |
Replication protein A1 |
6117 |
| 55 |
TGIF2 |
TGFB induced factor homeobox 2 |
60436 |
| 56 |
C17orf49 |
Chromosome 17 open reading frame 49 |
124944 |
| 57 |
MAZ |
MYC associated zinc finger protein |
4150 |
| 58 |
DNAAF5 |
Dynein axonemal assembly factor 5 |
54919 |
| 59 |
GART |
Phosphoribosylglycinamide formyltransferase: phosphoribosylglycinamide synthetase: phosphoribosylaminoimidazole synthetase |
2618 |
| 60 |
C19orf54 |
Chromosome 19 open reading frame 54 |
284325 |
| 61 |
ATIC |
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
471 |
| 62 |
PHF23 |
PHD finger protein 23 |
79142 |
| 63 |
CBX5 |
Chromobox 5 |
23468 |
| 64 |
FAM86C1 |
Family with sequence similarity 86 member C1 |
55199 |
| 65 |
DAXX |
Death domain associated protein |
1616 |
| 66 |
ELAVL1 |
ELAV like RNA binding protein 1 |
1994 |
| 67 |
MTA1 |
Metastasis associated 1 |
9112 |
| 68 |
MEN1 |
Menin 1 |
4221 |
| 69 |
TUBB |
Tubulin beta class I |
203068 |
| 70 |
SIGMAR1 |
Sigma non-opioid intracellular receptor 1 |
10280 |
| 71 |
FAM86B1 |
Family with sequence similarity 86 member B1 |
85002 |
| 72 |
EIF4A1 |
Eukaryotic translation initiation factor 4A1 |
1973 |
| 73 |
ALDH1B1 |
Aldehyde dehydrogenase 1 family member B1 |
219 |
| 74 |
ELAC2 |
elaC ribonuclease Z 2 |
60528 |
| 75 |
PTBP1 |
Polypyrimidine tract binding protein 1 |
5725 |
| 76 |
GLOD4 |
GLyoxalase domain containing 4 |
51031 |
| 77 |
EXOSC5 |
Exosome component 5 |
56915 |
| 78 |
ALDH18A1 |
Aldehyde dehydrogenase 18 family member A1 |
5832 |
| 79 |
RPL22L1 |
RIbosomal protein L22 like 1 |
200916 |
| 80 |
RFX5 |
Regulatory factor X5 |
5993 |
| 81 |
UNG |
Uracil DNA glycosylase |
7374 |
| 82 |
C1QBP |
Complement C1q binding protein |
708 |
| 83 |
BAX |
BCL2 associated X: apoptosis regulator |
581 |
| 84 |
EEFSEC |
Eukaryotic elongation factor: selenocysteine-tRNA specific |
60678 |
| 85 |
METTL16 |
Methyltransferase like 16 |
79066 |
| 86 |
KDELR1 |
KDEL endoplasmic reticulum protein retention receptor 1 |
10945 |
| 87 |
ZNF286A |
Zinc finger protein 286A |
57335 |
| 88 |
APRT |
Adenine phosphoribosyltransferase |
353 |
| 89 |
SLC35A4 |
Solute carrier family 35 member A4 |
113829 |
| 90 |
ZNF740 |
Zinc finger protein 740 |
283337 |
| 91 |
PA2G4 |
Proliferation-associated 2G4 |
5036 |
| 92 |
PRR3 |
Proline rich 3 |
80742 |
| 93 |
ZNF362 |
Zinc finger protein 362 |
149076 |
| 94 |
VPS35L |
VPS35 endosomal protein sorting factor like |
57020 |
| 95 |
CHAMP1 |
Chromosome alignment maintaining phosphoprotein 1 |
283489 |
| 96 |
SENP3 |
SUMO1/sentrin/SMT3 specific peptidase 3 |
26168 |
| 97 |
GANAB |
Glucosidase II alpha subunit |
23193 |
| 98 |
UBTF |
Upstream binding transcription factor: RNA polymerase I |
7343 |
| 99 |
PRKCSH |
Protein kinase C substrate 80K-H |
5589 |
| 100 |
TSR1 |
TSR1: ribosome maturation factor |
55720 |
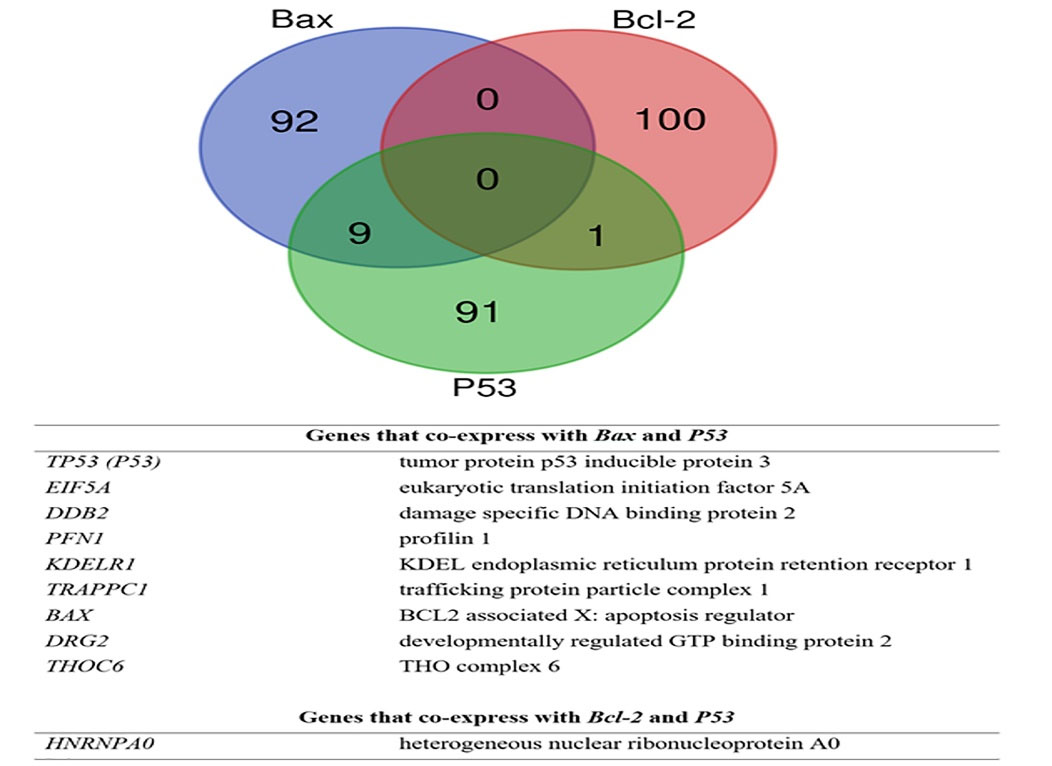
Fig. 6.
Shared genes between meta-analysis derived co-expressed profiles of Bax, Bcl-2, and P53.
.
Shared genes between meta-analysis derived co-expressed profiles of Bax, Bcl-2, and P53.
Molecular network underlying hAFMSCs-CM function in MCF-7 cells
Fig. 7 illustrates the molecular network underlying the apoptotic function of hAFMSCs-CM in MCF-7 cells. Supplementary data represents the underlying relations, mined sentences through literature mining, and the reference publications. P53 (TP53), EIF5A, DDB2, BcL2, and Bax are hubs in the network where BcL2 downregulation stands in harmony with the upregulation of P53, EIF5A, DDB2, and Bax, leading to apoptosis activation.
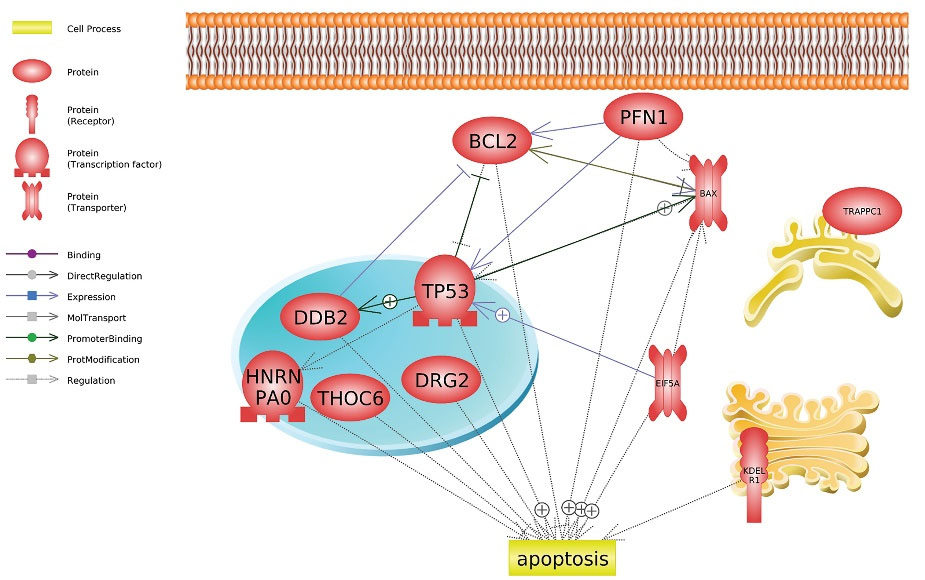
Fig. 7.
Molecular network underlying apoptotic function of hAFMSCs function in MCF-7 cells. The positive sign represents the positive/upregulation and the negative sign represents the negative/downregulation interaction.
.
Molecular network underlying apoptotic function of hAFMSCs function in MCF-7 cells. The positive sign represents the positive/upregulation and the negative sign represents the negative/downregulation interaction.
Discussion
Currently, chemotherapy and surgery are the principal approaches in clinical-base breast cancer treatment. However, the side effects of surgery, the toxicity of chemotherapy agents on normal cells, and drug resistance in cancer cells are undeniable post-treatment problems.
2,3
As a result, other types of breast cancer treatments, such as targeted therapies and gene therapy, have become the focus of recent research.
4
Multiple studies have shown that MSCs can fight cancer, which has led researchers to think about using them as a new treatment.
6,7,11,15,53,54
Nonetheless, the MSCs-CM’s anti-cancer effects, especially hAFMSCs-CM, on breast cancer apoptosis, have not been clearly understood. TROY, TAIL, and Fas Ligand/TNFSF6 were found in the MSCs-CM made from bone marrow.
55
The present work has investigated the apoptotic potential effects of hAFMSCs-CM through cellular and molecular approaches. Our data indicated that MCF-7 cell viability declined as a result of hAFMSCs-CM treatment as compared with control cells. Our findings are consistent with studies that have highlighted the promising aspects of human amniotic-derived MSCs’ effects on cancer inhibition.
6,56
Moreover, we have shown that hAFMSCs-CM induces apoptosis in MCF-7 breast cancer cells due to the increase in Bax gene expression and the decrease in Bcl-2 gene expression. Furthermore, based on the protein analysis and compared with the untreated cells, our data revealed that the level of tumor suppressor protein expression, P53, was enhanced in MCF-7 due to the hAFMSCs-CM treatment (P < 0.0001). There is ample evidence confirming that P53 overexpression in breast cancer downregulates Bcl2 expression, promotes Bax expression, and stimulates Bax function as a result of P53-induced apoptosis.
57-59
Gholizadeh et al stated that hAFMSCs medium could significantly promote p53 expression in the ovarian cancer cell line (P < 0.05).
6
Apoptosis can be caused in breast cancer cells by giving them hAFMSCs-CM, and this could lead to more P53 protein in the cells. Consistently, Kalamegam et al. found that CM from Wharton’s jelly stem cell had inhibitory effects on an ovarian cancer cell line.
12
Similarly, Serhal et al isolated CM from adipose-derived MSCs and assessed its effect on hepatocellular carcinoma cells. They posited that, after the CM treatment, the apoptosis rate increased due to P53 upregulation and retinoblastoma gene expression. They also highlighted the significant decrease in cell proliferation by dint of hTERE downregulation and c-Myc expression. Likewise, the present study found a noticeable decrease in Bcl-2 mRNA level expression and an increase in Bax mRNA level within the treated cells with hAFMSCs-CM in comparison with the untreated cells (P < 0.005). Consistent with our findings, in 2020, Rahmatizadeh et al showed that indirect hAFMSCs co-culturing with human cervical cancer (HeLa) resulted in an increase in the Bax/Bcl-2 ratio and cells’ sensitivity to apoptosis. They also said that the level of p53 mRNA in Hela cells rose a lot after day 5 of co-culture with indirect hAFMSCs, which is when they were mixed with the cells.
60
In 2018, Rodrigues et al reported that P53 is active in human amniotic fluid stem cells.
61
More importantly, we found that P53’s protein level increased after the hAFMSCs-CM treatment. According to our findings, it seems that hAFMSCs-CM could interfere with the apoptosis signal pathway associated with P53, inhibiting Bcl-2 expression. Consistent with this study, Jiao et al. demonstrated that hAMCs decreased tumor size significantly (P < 0.05) in gliomas by increasing Bax expression and reducing Bcl-2 levels.
62
In addition, Qiao and her colleagues reported that MSCs inhibited hepatoma cancer cell lines by downregulating the levels of Bcl-2, c-Myc, Survivin, PCNA, and β-catenin.
9
Conversely, Farahmand et al showed that bone marrow derived stem cell CM has tumorigenic effects on human breast cancer.
63
In this study, we developed a systems biology analysis approach by integrating the meta-analysis of expression data, using rank correlation and Z standardization, and performing literature mining analysis. The employed systems biology approach led us to an apoptotic-promoting gene interaction network, including P53, EIF5A, DDB2, and Bax, activated by hAFMSCs-CM treatment. More research should be conducted to validate this type of treatment.
Research Highlights
What is the current knowledge?
√ The role of MSCs in clinical application is well researched.
√ Stem cells such as hAFMSCs have anticancer effects in some tumors.
√ hAFMSCs-CM ability to downsize tumors should be investigated.
√ Given that the MSCs is the best among different sources, hAFMSCs-CM could target tumor cells and inhibit their growth rate through expressing apoptotic factors.
What is new here?
√ The current study focused on apoptotic effect of the cell-free hAFMSCs-CM on the cancer cells, especially the breast cancer.
√ This study explained the relationship between hAFMSCs-CM and the apoptotic molecules (antitumor).
√ The meta-analysis study illustrated that an apoptotic-promoting gene interaction network, including P53, EIF5A, DDB2, and Bax, can be activated by hAFMSCs-CM treatment.
Conclusion
The present work revealed that hAFMSCs-CM could promote apoptosis in MCF-7 cells. Our data shown a high level of P53 in MCF-7 cells, but not in normal cells (Hu02). After the treatment, P53 was found competent to downregulate Bcl2 expression and upregulate Bax to induce apoptosis in MCF-7 cells. On the other hand, our data suggest that hAFMSCs-CM has proliferation effects on normal cells but not on p53 expression; thus, we observed a decrease in Bax and an increase in Bcl2 mRNA levels. As per our findings, amniotic fluid-derived stem cells could seemingly target the tumor cells, inhibiting their growth rate by expressing various apoptotic factors. In the end, we suggest that more research be conducted on hAFMSCs’ effects on cancer therapy for stem cell CM.
Acknowledgment
We would like to appreciate Tehran University of Medical Sciences, Tehran, Iran for their financial support and Clinical Research Development Unit of Alzahra Educational, Research and Treatment Center,Tabriz University of Medical Sciences, Tabriz, Iran for their assistance in this research.
Funding
This study was funded by Tehran University of Medical Sciences for a PhD thesis (No. 9611184003).
Ethical Statement
This study was approved by the Ethics Committee of Tehran University of Medical Sciences, Tehran, Iran (ID number IR.TUMS.MEDICINE.REC.1398.690).
Competing Interests
The authors declared no conflict of interest.
Supplementary Materials
Supplementary file 1 contains molecular network relations underlying hAFMSCs function in MCF-7 cells.
(xlsx)
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020; 70:7-30. doi: 10.3322/caac.21590 [Crossref] [ Google Scholar]
- Zhang W, Jiang H, Chen Y, Ren F. Resveratrol chemosensitizes adriamycin-resistant breast cancer cells by modulating miR-122-5p. J Cell Biochem 2019; 120:16283-92. doi: 10.1002/jcb.28910 [Crossref] [ Google Scholar]
- Sadegh-Nejadi S, Afrisham R, Emamgholipour S, Izadi P, Eivazi N, Tahbazlahafi B. Influence of plasma circulating exosomes obtained from obese women on tumorigenesis and tamoxifen resistance in MCF-7 cells. IUBMB Life 2020; 72:1930-40. doi: 10.1002/iub.2305 [Crossref] [ Google Scholar]
- Akram M, Iqbal M, Daniyal M, Khan AU. Awareness and current knowledge of breast cancer. Biol Res 2017; 50:33. doi: 10.1186/s40659-017-0140-9 [Crossref] [ Google Scholar]
- Riedel R, Pérez-Pérez A, Carmona-Fernández A, Jaime M, Casale R, Dueñas JL. Human amniotic membrane conditioned medium inhibits proliferation and modulates related microRNAs expression in hepatocarcinoma cells. Sci Rep 2019; 9:14193. doi: 10.1038/s41598-019-50648-5 [Crossref] [ Google Scholar]
- Gholizadeh-Ghaleh Aziz S, Fardyazar Z, Pashaiasl M. The human amniotic fluid mesenchymal stem cells therapy on, SKOV3, ovarian cancer cell line. Mol Genet Genomic Med 2019; 7:e00726. doi: 10.1002/mgg3.726 [Crossref] [ Google Scholar]
- Kang NH, Hwang KA, Kim SU, Kim YB, Hyun SH, Jeung EB. Potential antitumor therapeutic strategies of human amniotic membrane and amniotic fluid-derived stem cells. Cancer Gene Ther 2012; 19:517-22. doi: 10.1038/cgt.2012.30 [Crossref] [ Google Scholar]
- Ayuzawa R, Doi C, Rachakatla RS, Pyle MM, Maurya DK, Troyer D. Naïve human umbilical cord matrix derived stem cells significantly attenuate growth of human breast cancer cells in vitro and in vivo. Cancer Lett 2009; 280:31-7. doi: 10.1016/j.canlet.2009.02.011 [Crossref] [ Google Scholar]
- Qiao L, Xu Z, Zhao T, Zhao Z, Shi M, Zhao RC. Suppression of tumorigenesis by human mesenchymal stem cells in a hepatoma model. Cell Res 2008; 18:500-7. doi: 10.1038/cr.2008.40 [Crossref] [ Google Scholar]
- Ramasamy R, Lam EW, Soeiro I, Tisato V, Bonnet D, Dazzi F. Mesenchymal stem cells inhibit proliferation and apoptosis of tumor cells: impact on in vivo tumor growth. Leukemia 2007; 21:304-10. doi: 10.1038/sj.leu.2404489 [Crossref] [ Google Scholar]
- Khalil C, Moussa M, Azar A, Tawk J, Habbouche J, Salameh R. Anti-proliferative effects of mesenchymal stem cells (MSCs) derived from multiple sources on ovarian cancer cell lines: an in-vitro experimental study. J Ovarian Res 2019; 12:70. doi: 10.1186/s13048-019-0546-9 [Crossref] [ Google Scholar]
- Kalamegam G, Sait KHW, Anfinan N, Kadam R, Ahmed F, Rasool M. Cytokines secreted by human Wharton's jelly stem cells inhibit the proliferation of ovarian cancer (OVCAR3) cells in vitro. Oncol Lett 2019; 17:4521-31. doi: 10.3892/ol.2019.10094 [Crossref] [ Google Scholar]
- Serhal R, Saliba N, Hilal G, Moussa M, Hassan GS, El Atat O. Effect of adipose-derived mesenchymal stem cells on hepatocellular carcinoma: In vitro inhibition of carcinogenesis. World J Gastroenterol 2019; 25:567-83. doi: 10.3748/wjg.v25.i5.567 [Crossref] [ Google Scholar]
- Rahmatizadeh F, Gholizadeh-Ghaleh Aziz S, Khodadadi K, Lale Ataei M, Ebrahimie E, Soleimani Rad J. Bidirectional and Opposite Effects of Naïve Mesenchymal Stem Cells on Tumor Growth and Progression. Adv Pharm Bull 2019; 9:539-58. doi: 10.15171/apb.2019.063 [Crossref] [ Google Scholar]
- Han KH, Kim AK, Jeong GJ, Jeon HR, Bhang SH, Kim DI. Enhanced Anti-Cancer Effects of Conditioned Medium from Hypoxic Human Umbilical Cord-Derived Mesenchymal Stem Cells. Int J Stem Cells 2019; 12:291-303. doi: 10.15283/ijsc19002 [Crossref] [ Google Scholar]
- Özcan S, Alessio N, Acar MB, Toprak G, Gönen ZB, Peluso G. Myeloma cells can corrupt senescent mesenchymal stromal cells and impair their anti-tumor activity. Oncotarget 2015; 6:39482-92. doi: 10.18632/oncotarget.5430 [Crossref] [ Google Scholar]
- Jafari A, Rezaei-Tavirani M, Farhadihosseinabadi B, Zali H, Niknejad H. Human amniotic mesenchymal stem cells to promote/suppress cancer: two sides of the same coin. Stem Cell Res Ther 2021; 12:126. doi: 10.1186/s13287-021-02196-x [Crossref] [ Google Scholar]
- Kang NH, Yi BR, Lim SY, Hwang KA, Baek YS, Kang KS. Human amniotic membrane-derived epithelial stem cells display anticancer activity in BALB/c female nude mice bearing disseminated breast cancer xenografts. Int J Oncol 2012; 40:2022-8. doi: 10.3892/ijo.2012.1372 [Crossref] [ Google Scholar]
- Kim YS, Hwang KA, Go RE, Kim CW, Choi KC. Gene therapy strategies using engineered stem cells for treating gynecologic and breast cancer patients (Review). Oncol Rep 2015; 33:2107-12. doi: 10.3892/or.2015.3846 [Crossref] [ Google Scholar]
- Aboody KS, Bush RA, Garcia E, Metz MZ, Najbauer J, Justus KA. Development of a tumor-selective approach to treat metastatic cancer. PLoS One 2006; 1:e23. doi: 10.1371/journal.pone.0000023 [Crossref] [ Google Scholar]
- Yi BR, Kang NH, Hwang KA, Kim SU, Jeung EB, Choi KC. Antitumor therapeutic effects of cytosine deaminase and interferon-β against endometrial cancer cells using genetically engineered stem cells in vitro. Anticancer Res 2011; 31:2853-61. [ Google Scholar]
- Gholizadeh-Ghaleh Aziz S, Pashaei-Asl F, Fardyazar Z, Pashaiasl M. Isolation, Characterization, Cryopreservation of Human Amniotic Stem Cells and Differentiation to Osteogenic and Adipogenic Cells. PLoS One 2016; 11:e0158281. doi: 10.1371/journal.pone.0158281 [Crossref] [ Google Scholar]
- Loukogeorgakis SP, De Coppi P. Concise Review: Amniotic Fluid Stem Cells: The Known, the Unknown, and Potential Regenerative Medicine Applications. Stem Cells 2017; 35:1663-73. doi: 10.1002/stem.2553 [Crossref] [ Google Scholar]
- Cho JA, Park H, Kim HK, Lim EH, Seo SW, Choi JS. Hyperthermia-treated mesenchymal stem cells exert antitumor effects on human carcinoma cell line. Cancer 2009; 115:311-23. doi: 10.1002/cncr.24032 [Crossref] [ Google Scholar]
- Seyhoun I, Hajighasemlou S, Ai J, Hosseinzadeh F, Mirmoghtadaei M, Seyhoun SM. Novel Combination of Mesenchymal Stem Cell-Conditioned Medium with Sorafenib Have Synergistic Antitumor Effect of Hepatocellular Carcinoma Cells. Asian Pac J Cancer Prev 2019; 20:263-7. doi: 10.31557/apjcp.2019.20.1.263 [Crossref] [ Google Scholar]
- Joseph A, Baiju I, Bhat IA, Pandey S, Bharti M, Verma M. Mesenchymal stem cell-conditioned media: A novel alternative of stem cell therapy for quality wound healing. J Cell Physiol 2020; 235:5555-69. doi: 10.1002/jcp.29486 [Crossref] [ Google Scholar]
- Spitzhorn LS, Rahman MS, Schwindt L, Ho HT, Wruck W, Bohndorf M. Isolation and Molecular Characterization of Amniotic Fluid-Derived Mesenchymal Stem Cells Obtained from Caesarean Sections. Stem Cells Int 2017; 2017:5932706. doi: 10.1155/2017/5932706 [Crossref] [ Google Scholar]
- Rakshit S, Chandrasekar BS, Saha B, Victor ES, Majumdar S, Nandi D. Interferon-gamma induced cell death: Regulation and contributions of nitric oxide, cJun N-terminal kinase, reactive oxygen species and peroxynitrite. Biochim Biophys Acta 2014; 1843:2645-61. doi: 10.1016/j.bbamcr.2014.06.014 [Crossref] [ Google Scholar]
- Detjen KM, Farwig K, Welzel M, Wiedenmann B, Rosewicz S. Interferon gamma inhibits growth of human pancreatic carcinoma cells via caspase-1 dependent induction of apoptosis. Gut 2001; 49:251-62. doi: 10.1136/gut.49.2.251 [Crossref] [ Google Scholar]
- Beretti F, Zavatti M, Casciaro F, Comitini G, Franchi F, Barbieri V. Amniotic fluid stem cell exosomes: Therapeutic perspective. Biofactors 2018; 44:158-67. doi: 10.1002/biof.1407 [Crossref] [ Google Scholar]
- Lazzarini R, Sorgentoni G, Caffarini M, Sayeed MA, Olivieri F, Di Primio R. New miRNAs network in human mesenchymal stem cells derived from skin and amniotic fluid. Int J Immunopathol Pharmacol 2016; 29:523-8. doi: 10.1177/0394632015610228 [Crossref] [ Google Scholar]
- Shen SQ, Huang LS, Xiao XL, Zhu XF, Xiong DD, Cao XM. miR-204 regulates the biological behavior of breast cancer MCF-7 cells by directly targeting FOXA1. Oncol Rep 2017; 38:368-76. doi: 10.3892/or.2017.5644 [Crossref] [ Google Scholar]
- Shirjang S, Mansoori B, Asghari S, Duijf PHG, Mohammadi A, Gjerstorff M. MicroRNAs in cancer cell death pathways: Apoptosis and necroptosis. Free Radic Biol Med 2019; 139:1-15. doi: 10.1016/j.freeradbiomed.2019.05.017 [Crossref] [ Google Scholar]
- Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta 2011; 1812:592-601. doi: 10.1016/j.bbadis.2011.02.002 [Crossref] [ Google Scholar]
- Lazzarini R, Olivieri F, Ferretti C, Mattioli-Belmonte M, Di Primio R, Orciani M. mRNAs and miRNAs profiling of mesenchymal stem cells derived from amniotic fluid and skin: the double face of the coin. Cell Tissue Res 2014; 355:121-30. doi: 10.1007/s00441-013-1725-4 [Crossref] [ Google Scholar]
- Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene 2001; 20:1803-15. doi: 10.1038/sj.onc.1204252 [Crossref] [ Google Scholar]
- Gasco M, Shami S, Crook T. The p53 pathway in breast cancer. Breast Cancer Res 2002; 4:70-6. doi: 10.1186/bcr426 [Crossref] [ Google Scholar]
- Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol 2007; 35:495-516. doi: 10.1080/01926230701320337 [Crossref] [ Google Scholar]
- Dawson SJ, Makretsov N, Blows FM, Driver KE, Provenzano E, Le Quesne J. BCL2 in breast cancer: a favourable prognostic marker across molecular subtypes and independent of adjuvant therapy received. Br J Cancer 2010; 103:668-75. doi: 10.1038/sj.bjc.6605736 [Crossref] [ Google Scholar]
- Sakakura C, Sweeney EA, Shirahama T, Igarashi Y, Hakomori S, Nakatani H. Overexpression of bax sensitizes human breast cancer MCF-7 cells to radiation-induced apoptosis. Int J Cancer 1996; 67:101-5. doi: 10.1002/(sici)1097-0215(19960703)67:1<101::aid-ijc17>3.0.co;2-h [Crossref] [ Google Scholar]
- Ahmadian N, Pashaei-Asl R, Samadi N, Rahmati-Yamchi M, Rashidi MR, Ahmadian M. Hesa-A Effects on Cell Cycle Signaling in Esophageal Carcinoma Cell Line. Middle East J Dig Dis 2016; 8:297-302. doi: 10.15171/mejdd.2016.39 [Crossref] [ Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402-8. doi: 10.1006/meth.2001.1262 [Crossref] [ Google Scholar]
- Deihimi T, Niazi A, Ebrahimi M, Kajbaf K, Fanaee S, Bakhtiarizadeh MR. Finding the undiscovered roles of genes: an approach using mutual ranking of coexpressed genes and promoter architecture-case study: dual roles of thaumatin like proteins in biotic and abiotic stresses. Springerplus 2012; 1:30. doi: 10.1186/2193-1801-1-30 [Crossref] [ Google Scholar]
- Pashaei-Asl R, Pashaei-Asl F, Mostafa Gharabaghi P, Khodadadi K, Ebrahimi M, Ebrahimie E. The Inhibitory Effect of Ginger Extract on Ovarian Cancer Cell Line; Application of Systems Biology. Adv Pharm Bull 2017; 7:241-9. doi: 10.15171/apb.2017.029 [Crossref] [ Google Scholar]
- Obayashi T, Kinoshita K. Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res 2009; 16:249-60. doi: 10.1093/dnares/dsp016 [Crossref] [ Google Scholar]
- Kinoshita K, Obayashi T. Multi-dimensional correlations for gene coexpression and application to the large-scale data of Arabidopsis. Bioinformatics 2009; 25:2677-84. doi: 10.1093/bioinformatics/btp442 [Crossref] [ Google Scholar]
- Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 2006; 22:2825-7. doi: 10.1093/bioinformatics/btl476 [Crossref] [ Google Scholar]
- Obayashi T, Kagaya Y, Aoki Y, Tadaka S, Kinoshita K. COXPRESdb v7: a gene coexpression database for 11 animal species supported by 23 coexpression platforms for technical evaluation and evolutionary inference. Nucleic Acids Res 2019; 47:D55-d62. doi: 10.1093/nar/gky1155 [Crossref] [ Google Scholar]
- Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio--the analysis and navigation of molecular networks. Bioinformatics 2003; 19:2155-7. doi: 10.1093/bioinformatics/btg290 [Crossref] [ Google Scholar]
- Yuryev A, Kotelnikova E, Daraselia N. Ariadne's ChemEffect and Pathway Studio knowledge base. Expert Opin Drug Discov 2009; 4:1307-18. doi: 10.1517/17460440903413488 [Crossref] [ Google Scholar]
- Pashaei-Asl F, Pashaei-Asl R, Khodadadi K, Akbarzadeh A, Ebrahimie E, Pashaiasl M. Enhancement of anticancer activity by silibinin and paclitaxel combination on the ovarian cancer. Artif Cells Nanomed Biotechnol 2018; 46:1483-7. doi: 10.1080/21691401.2017.1374281 [Crossref] [ Google Scholar]
- Pashaiasl M, Ebrahimi M, Ebrahimie E. Identification of the key regulating genes of diminished ovarian reserve (DOR) by network and gene ontology analysis. Mol Biol Rep 2016; 43:923-37. doi: 10.1007/s11033-016-4025-8 [Crossref] [ Google Scholar]
- Reza A, Choi YJ, Yasuda H, Kim JH. Human adipose mesenchymal stem cell-derived exosomal-miRNAs are critical factors for inducing anti-proliferation signalling to A2780 and SKOV-3 ovarian cancer cells. Sci Rep 2016; 6:38498. doi: 10.1038/srep38498 [Crossref] [ Google Scholar]
- He N, Kong Y, Lei X, Liu Y, Wang J, Xu C. MSCs inhibit tumor progression and enhance radiosensitivity of breast cancer cells by down-regulating Stat3 signaling pathway. Cell Death Dis 2018; 9:1026. doi: 10.1038/s41419-018-0949-3 [Crossref] [ Google Scholar]
- Cantinieaux D, Quertainmont R, Blacher S, Rossi L, Wanet T, Noël A. Conditioned medium from bone marrow-derived mesenchymal stem cells improves recovery after spinal cord injury in rats: an original strategy to avoid cell transplantation. PLoS One 2013; 8:e69515. doi: 10.1371/journal.pone.0069515 [Crossref] [ Google Scholar]
- Niknejad H, Khayat-Khoei M, Peirovi H, Abolghasemi H. Human amniotic epithelial cells induce apoptosis of cancer cells: a new anti-tumor therapeutic strategy. Cytotherapy 2014; 16:33-40. doi: 10.1016/j.jcyt.2013.07.005 [Crossref] [ Google Scholar]
- Basu A, Haldar S. The relationship between BcI2, Bax and p53: consequences for cell cycle progression and cell death. Mol Hum Reprod 1998; 4:1099-109. doi: 10.1093/molehr/4.12.1099 [Crossref] [ Google Scholar]
-
Campbell KJ, Tait SWG. Targeting BCL-2 regulated apoptosis in cancer. Open Biol 2018; 8. 10.1098/rsob.180002.
- Haldar S, Negrini M, Monne M, Sabbioni S, Croce CM. Down-regulation of bcl-2 by p53 in breast cancer cells. Cancer Res 1994; 54:2095-7. [ Google Scholar]
- Rahmatizadeh F, Pashaei-Asl F, Dehcheshmeh MM, Rahbar S, LaleAtaei M, Aziz SG-G. Reduction in the Viability of Human Cervical Cancer HeLa Cell Line via Indirect Co-culture With Amniotic Fluid-Derived Mesenchymal Stem Cells. Int J Womens Health Reprod Sci 2020; 8:319-27. doi: 10.15296/ijwhr.2020.51 [Crossref] [ Google Scholar]
- Rodrigues M, Antonucci I, Elabd S, Kancherla S, Marchisio M, Blattner C. p53 Is Active in Human Amniotic Fluid Stem Cells. Stem Cells Dev 2018; 27:1507-17. doi: 10.1089/scd.2017.0254 [Crossref] [ Google Scholar]
- Jiao H, Guan F, Yang B, Li J, Song L, Hu X. Human amniotic membrane derived-mesenchymal stem cells induce C6 glioma apoptosis in vivo through the Bcl-2/caspase pathways. Mol Biol Rep 2012; 39:467-73. doi: 10.1007/s11033-011-0760-z [Crossref] [ Google Scholar]
- Farahmand L, Esmaeili R, Eini L, Majidzadeh AK. The effect of mesenchymal stem cell-conditioned medium on proliferation and apoptosis of breast cancer cell line. J Cancer Res Ther 2018; 14:341-4. doi: 10.4103/0973-1482.177213 [Crossref] [ Google Scholar]