Bioimpacts. 13(2):133-144.
doi: 10.34172/bi.2022.23876
Original Article
Targeted gene delivery to the brain using CDX-modified chitosan nanoparticles
Tina Sepasi Resources, 1 
Farhad Bani Formal analysis, 1, 2
Reza Rahbarghazi Formal analysis, Writing – original draft, 3, 4
Abbas Ebrahimi-Kalan Methodology, 5
Mohammad-Reza Sadeghi Data curation, Investigation, 6
Seyedeh Zahra Alamolhoda Investigation, 7
Amir Zarebkohan Conceptualization, Supervision, 1, 2, * 
Tahereh Ghadiri Supervision, 5
Huile Gao Writing – review & editing, 8
Author information:
1Department of Medical Nanotechnology, Advanced Faculty of Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
2Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
3Stem Cell Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
4Department of Applied Cell Sciences, Advanced Faculty of Medical Sciences, Tabriz University of Medical, Tabriz, Iran
5Department of Neuroscience and Cognitive, Advanced Faculty of Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
6Department of Molecular Medicine, Advanced Faculty of Medical Sciences, Tabriz University of Medical, Tabriz, Iran
7Department of Medical Biotechnology, Advanced Faculty of Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
8Key Laboratory of Drug-Targeting and Drug Delivery System of the Education Ministry, Sichuan Engineering Laboratory for Plant-Sourced Drug and Sichuan Research Center for Drug Precision Industrial Technology, West China School of Pharmacy, Sichuan University, Chengdu, 610064, P. R. China
Abstract
Introduction:
Blood-brain barrier with strictly controlled activity participates in a coordinated transfer of bioactive molecules from the blood to the brain. Among different delivery approaches, gene delivery is touted as a promising strategy for the treatment of several nervous system disorders. The transfer of exogenous genetic elements is limited by the paucity of suitable carriers. As a correlate, designing high-efficiency biocarriers for gene delivery is challenging. This study aimed to deliver pEGFP-N1 plasmid into the brain parenchyma using CDX-modified chitosan (CS) nanoparticles (NPs).
Methods:
Herein, we attached CDX, a 16 amino acids peptide, to the CS polymer using bifunctional polyethylene glycol (PEG) formulated with sodium tripolyphosphate (TPP), by ionic gelation method. Developed NPs and their nanocomplexes with pEGFP-N1 (CS-PEG-CDX/pEGFP) were characterized using DLS, NMR, FTIR, and TEM analyses. For in vitro assays, a rat C6 glioma cell line was used for cell internalization efficiency. The biodistribution and brain localization of nanocomplexes were studied in a mouse model after intraperitoneal injection using in vivo imaging and fluorescent microscopy.
Results:
Our results showed that CS-PEG-CDX/pEGFP NPs were uptaken by glioma cells in a dose-dependent manner. In vivo imaging revealed successful entry into the brain parenchyma indicated with the expression of green fluorescent protein (GFP) as a reporter protein. However, the biodistribution of developed NPs was also evident in other organs especially the spleen, liver, heart, and kidneys.
Conclusion:
Based on our results, CS-PEG-CDX NPs can provide a safe and effective nanocarrier for brain gene delivery into the central nervous system (CNS).
Keywords: Targeted gene delivery, Brain, CDX, Chitosan, Nanoparticles
Copyright and License Information
© 2023 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
During the past decades, several modalities and approaches have been used for the delivery of certain biomolecules to the target organs.1 One of the challenges in this area is the existence of natural barriers inside the body restricting the easy access of certain molecules to the target sites.2 The brain provides an impermeable interface namely the blood-brain barrier (BBB) which substantially limits the entrance of hazardous agents, bioactive molecules from the blood to the nervous system.3,4 In this regard, gene delivery is one of the available approaches to the transfer of exogenous genetic elements into the brain parenchyma. This approach per se is a powerful tool to achieve apparent therapeutic effects stand-alone or in combination with modalities.5 Since 1989, more than 1400 clinical trials have been performed around gene therapy whereas very limited numbers are approved by the Food and Drug Administration (FDA).1,2,6 The success and efficiency of gene therapy correlate with vector features.7 Despite encouraging results achieved by viral-based vectors, the stimulation of immune response, fusion possibility with wild-type viruses, spontaneous mutations, and the very high cost of engineered viral vector production are serious concerns.8 As a matter of fact, the selection of certain administration routes is another bottleneck in the field. For instance, most current gene vectors are administrated via craniotomy or direct intracerebral injection, which are highly invasive and have topical effects.9 Considering the involvement of different brain regions during most pathologies, the gene delivery system should be capable of widespread distribution throughout the central nervous system (CNS) post-systemic administration.10,11 During the recent two decades, researchers have focused on the development of targeted non-viral vectors to circumvent the above-mentioned drawbacks.
Chitosan (CS) is a biodegradable, biocompatible, and non-immunogenic polymer derived from chitin. Owing to its cationic nature, CS is commonly used as a promising non-viral vector for gene delivery purposes.12,13 Generally, the formation of highly stable polyplexes between negatively charged genetic materials (e.g., pDNA=plasmid DNA) and cationic structures like CS is electrostatically achievable.14 Of note, inherent lysosomal escape occurs during BBB transfer of several bioactive molecules. As a correlate, receptor-mediated transcytosis (RMT) systems are the best option for brain-targeted gene delivery.15 To date, different types of ligands like transferrin, leptin-derived peptides, angiopep-2, platelet endothelial cell adhesion molecule-1 (PECAM), and SRL peptides have been used for gene delivery into the brain parenchyma.16-18 CDX is a 16-amino acid peptide sequence isolated from snake venom known as candoxin. This peptide can identify the α7 subunit of acetylcholine nicotinic receptors (nAchR) on the surface of brain capillary endothelial cells.19-22 Commensurate with these descriptions, CDX has been used for the release of drugs into the brain alone or combined with other peptides (e.g., RGD) for dual targeting purposes.23-25
So far, this is the first report which shows CDX-modified CS nanoparticles (NPs) for in vivo gene delivery purposes. Here, we aimed to evaluate the potency of CDX-modified CS NPs (CS-PEG-CDX) and their combination with pEGFP-N1 (CS-PEG-CDX/pEGFP) in the delivery of certain genetic elements into the brain in in-vivo conditions.
Materials and Methods
Materials
Plasmid pEGFP-N1 (Clontech, Palo Alto, CA, USA) was purified using QIAGEN Plasmid Mega Kit (Qiagen GmbH, Hilden, Germany). Cysteine terminated CDX peptide was purchased from Biomatic Co. (Wilmington, USA) with a purity percentage of 99%. Bifunctional PEG (NHS-PEG-MAL, MW 2000) was purchased from Nanocs Inc. (Boston, USA). Fluorescein was purchased from Molecular Probes (Eugene, Oregon, United States). The drug Fingolimod was purchased from (AdooQ Bioscience, Centerstone Plaza, Irvine, USA). Other purchased materials were as follows: CS (Chitolytic, 17 Carlaw Ave, Toronto, ON M4M 2R7, Canada), Acetic acid (Sigma–Aldrich, St. Louis, Missouri, USA), cell culture flasks and plates (SPL, Geumgang-ro, Pocheon-si, Gyeonggi-do, Korea), tripolyphosphate (TPP) (Sigma–Aldrich, St. Louis, Missouri, USA), cellulose dialysis bag (cut off 12-14 kDa) (Sigma–Aldrich, St. Louis, Missouri, USA), agarose powder (Bio-Rad, Hercules, California, USA), SYBRTM Green (Thermo Fisher, Waltham, Massachusetts, USA), TNBSA assay (Thermo Fisher, Waltham, Massachusetts, USA), Ellman reagent (Thermo Fisher, Waltham, Massachusetts, USA), DMEM/HG cell medium (Sigma–Aldrich, St. Louis, Missouri, USA), fetal bovine serum (GibcoTM, Thermo Fisher, Waltham, Massachusetts, USA), Pen-Strep, and Trypsin-EDTA (Sigma–Aldrich, St. Louis, Missouri, USA).
Synthesis of CS derivatives
Preparation of CS-PEG-CDX polymers
To prepare the CS solution, 3 mg low molecular weight CS, ranging 50–150 kDa, were dispersed in 1% acetic acid and stirred continuously for 2 hours followed by incubation at 4˚C overnight. For covalent attachment of NHS-PEG2000-MAL (in 10 M excess) to CS polymers, pH was adjusted to 6 using 1 M NaOH. The reaction was preceded for 3 hours at room temperature. Then, the pH of the solution was adjusted to 7 and stirred overnight under similar conditions to obtain CS-PEG conjugates. To eliminate unreacted PEG molecules, we used a cellulose dialysis bag (cut off 12-14 kDa). In this phase, the N-hydroxysuccinimide (NHS) groups of bifunctional PEG reacted with NH2 groups on the CS polymer backbone. For the best reactivity of NHS groups and the instability of NHS esters at higher pH, we adjusted the reaction pH to 7-9. While this value was set to about 6.5 to 7.5 for the maleimide (MAL) groups and 6.5 for the CS polymers. The pH of peptide binding to CS reaction was about 7. We then added cysteine-terminated CDX peptide in a 1:1 molar ratio (CDX: PEG), and the solution was stirred for the next 12 hours. Again, the cellulose dialysis bag (cut off 12-14 kD) was used for discharging unreacted CDX. Finally, CS-PEG-CDX polymers were used for the synthesis of CDX-modified CS NPs.
Preparation of CDX-modified CS NPs
The ionic gelation method was used for the synthesis of CDX-modified CS NPs. Briefly, the TPP was separately prepared by dissolving in 3 mL of distilled water (TPP-to-CS ratio was 1:5 w/w) and slowly added into the polymer solution at 900 rpm stirring speed. The cloudy solution was stirred for 2 hours to yield a homogeneous solution.
Preparation of CS-PEG-CDX/pEGFPN1 polyplexes
Considering the electrostatic interactions of cationic NPs with the negatively charged plasmid, the pH of the CS-PEG-CDX solution was set to 6.5. In this pH, unoccupied NPs amine (NH2) groups were protonated and changed into NH3+. Finally, 5 µL of pEGFPN1 was mixed with 12 µL of the pH-adjusted CS-PEG-CDX NPs solution. Values such as zeta potential, morphology, and real size of NPs measured by transmission electron microscopy (TEM), and nanocomplexes formation and stability are shown in Figure 3A-D,whilethehydrodynamic diameter of NPs and nanocomplexes, and their polydispersity index (PDI) measured by dynamic light scattering (DLS) are respectively shown in Table 1.
Table 1.
The size distribution and zeta potential of CS-derived polymers and NPs
|
Formulation
|
Size (nm)
|
Zeta potential (mV)
|
|
DLS
|
TEM
|
| CS NP |
80 |
50 |
+28.2 |
| CS-PEG NP |
120.3 |
NA |
-1.76 |
| CS-PEG-CDX NP |
162.4 |
70 |
0.91 |
CS-PEG-CDX:pEGFP-N1
N:P ratio |
5:1 |
283.8 |
NA |
-23.0 |
| 10:1 |
174.6 |
120 |
-16.9 |
| 20:1 |
391.7 |
NA |
-14.2 |
CS; chitosan, NP; nanoparticle, NA; not assessed.
Electrophoresis of CS-PEG-CDX/pEGFP nanocomplex
Different ratios of NP/plasmid (N/P; mol/mol) were prepared as follows: 1: 20, 1: 10, 1: 5, 5: 1, 10: 1, and 20: 1. Thereafter, the stability and mobility of synthesized CS-PEG-CDX/pEGFPN1 complexes were evaluated in 0.7% agarose gel electrophoresis. Electrophoresis was carried out for 30 minutes under 100 V/cm and the bands were visualized and recorded using the Gel Doc system.
Characterization of synthesized polymers and NPs
NMR, FTIR, DLS, and zeta sizer were used to measure the physicochemical characteristics of the synthesized NPs. To analyze NMR, CS-PEG-MAL and CS-PEG-CDX solutions were initially turned into powder and dissolved in D2O. The solutions were further analyzed in an NMR system (Bruker Avance II 400 MHz spectrometer). The exact amount of PEG and Cys-CDX substitution on the polymer constructs was estimated using TNBSA and Ellman's reagent assays, respectively. Size and the zeta potential of NPs were studied using the DLS method and Zeta Plus Analyzer (Zetasizer Nano ZS, Malvern; UK). The peaks related to CS-PEG and CS-PEG-CDX polymers were confirmed by FTIR (SpectrumTM 100 Optica system). Data were separately collected in the form of the spectrum ranging from 500 to 4000 cm-1 for each CS, PEG, CDX, CS-PEG, and CS-PEG-CDX polymers. After the completion of NMR, FTIR, DLS, and zeta sizer analyses, the size and morphology of the NPs, as well as NP–gene complex, were visualized using TEM (Digital Instrument Inc., Santa Barbara, USA).
Degree of substitution (DS)
Quantifying free amine groups: TNBSA assay
To quantify the unmodified free amine groups on the CS polymer, we used a TNBSA assay.26 The mixture of CS with NHS-PEG-maleimide was dialyzed through a cellulose dialysis bag (cutoff 12-14 kDa). Afterward, the filtered solution was freeze-dried. One milligram of CS-PEG powder was quantified based on the TNBSA reagent protocol.26 The DS of PEG on CS was calculated using a standard curve (R2 = 0.9959) using 8 different concentrations of unmodified CS according to the following equation:
DS amine = [(C - CP)/C] × 100% (1)
where “C” and “CP” stand for amine group contents in CS and CS-PEG polymers, respectively.
Quantifying free sulfhydryl groups: Ellman’s assay
To determine the number of free cysteine terminates in the CS-PEG-CDX solution, we performed Ellman’s assay.26 In brief, the reaction buffer containing 0.1 M sodium phosphate and 1 mM ethylenediaminetetraacetic acid (EDTA) was prepared at pH 8.0. Two hundred and fifty microliter of sample solution was added to a test tube containing a mix of 50 µL Ellman’s reagent and 2.5 mL reaction buffer. Then, the mixture was incubated at 37°C for 15 minutes, and the absorbance was measured at 412 nm. Data were compared to a standard curve prepared based on different serial dilutions of cysteine hydrochloride monohydrate (n=8 and R2 = 0.9743). The free sulfhydryl group content was quantified according to the following equation:
DS thiol = [(T - CPT)/T] × 100%,(2)
Where “T” and “CPT” stand for sulfhydryl group contents in CDX peptide and CS-PEG-CDX polymer, respectively.
Cell culture and expansion protocol
Rat C6 glioma cells were purchased from the National Cell Bank of Iran (NCBI Code: C575; Tehran; Iran). The cells were cultured in high-glucose content Dulbecco's Modified Eagle Medium (DMEM/HG). The medium was supplemented with 10% fetal bovine serum (FBS), and 1% Pen-Strep solution. Cells were maintained at 37˚C in a humidified incubator with 5% CO2. Cells were passaged at 70-80% confluence using 0.25% Trypsin-EDTA. Rat C6 glioma cells at passages 3-6 were used for different analyses.
Measuring CS-PEG-CDX/pEGFP uptake by C6 glioma cells can express high levels of α7 subunit of nAchR.27 The ethidium bromide (EtBr)-labeled pEGFP solution was used to prepare NPs by the same method as the above-mentioned. To this end, C6 glioma cells were seeded at an initial density of 2 ×104 cells/well in 24-well plates and allowed to reach 70-80% confluence. Then, different concentrations (5, 10, 20, and 40 ppm) of EtBr-labeled pEGFP/CS-PEG-CDX nano-complexes were added to the culture medium and maintained for 3 hours at standard condition.28,29 To avoid the possible effects of the protein corona, cells were incubated with EtBr-labeled pEGFP/CS-PEG-CDX nano-complexes in a culture medium containing 2% FBS. After completion of the incubation period, the supernatant was removed, and cells were washed with phosphate-buffered saline (PBS) three times. The number and intensity of fluorescent cells were checked by fluorescence microscopy (Olympus, Osaka, Japan).
In vivo imaging analysis
Around 4–5-week-old male Balb/c mice (n=6), weighing 20–25 g were purchased from Razi Vaccine and Serum Research Institute (Karaj, Iran). The animal was kept in standard cages with unrestricted access to food and water under standard 12-hour light/dark cycles. Animals were treated according to the Guide for the Care and Use of Laboratory Animals (NIH, 1986), and all phases of this study were approved by the Local Ethics Committee of Tabriz University of Medical Sciences. To assess the BBB transfer of EtBr-labeled pEGFP/CS-PEG-CDX into the brain, polyplexes containing pEGFP (10:1, CS-PEG-CDX to DNA, w/w) with a dose of 50 µg DNA/mice were injected intraperitoneally once a day for 5 consecutive days. After this period, the mice were imaged using CRi Maestro (Fluorescence) Imaging (Kodak Fx Pro, New York, USA).18
Measuring gene expression using fluorescence imaging ex vivo
The mice were humanely euthanized using an overdose of Xylazine and Ketamine. Then, brain samples were maintained in OCT for 24 hours at -20oC. For fluorescent imaging, 20-µm thick sections were prepared from samples embedded in OCT using Cryostat (Leica, CM 1900, Wetzlar, Germany). For nuclear staining, slides were stained with 300 nM DAPI. After two-time PBS washes, the slides were visualized using fluorescence microscopy.
Statistical analysis
Experimental results are expressed as means ± SD. Statistical analysis was carried out by SPSS software (ver. 17.0; IBM; USA) using the general linear model and Tukey’s post hoc test. P< 0.05 was considered statistically significant.
Results
Characterization of CS derived NPs
FTIR results
The mechanism of synthesis of CDX-modified CS/pEGFP-N1 nanocomplexes is illustrated in Fig 1.
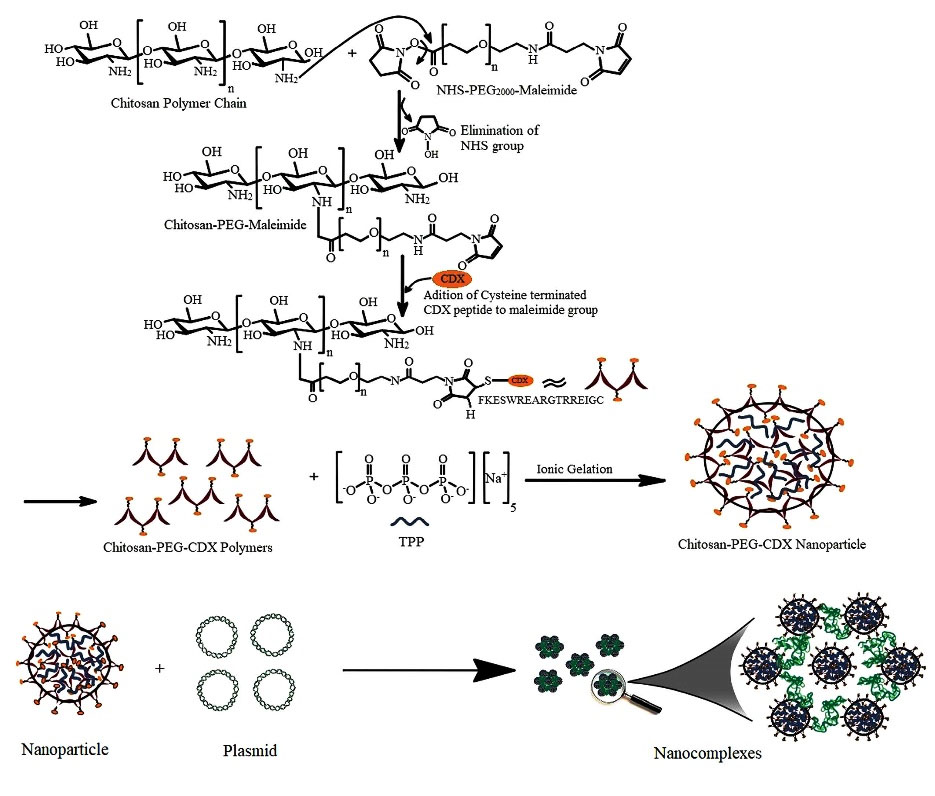
Fig. 1.
Schematic representation of CS-PEG-CDX/pEFGN1 nanocomplexes preparation methods. The important steps of synthesis have been illustrated schematically.
.
Schematic representation of CS-PEG-CDX/pEFGN1 nanocomplexes preparation methods. The important steps of synthesis have been illustrated schematically.
FTIR spectroscopy was used to characterize the covalent attachment of reaction components to each other. Fig. 2A-E depicts the characteristic spectra of functional groups and bonds in CS, NHS-PEG2000-MAL, CDX, CS-PEG, and CS-PEG-CDX. The characteristic peak of CS at 3441 cm-1 is related to the overlapping tensile vibration of N-H and O-H (Fig. 2B). The peak at 2879 and 1111 cm-1 are associated with the traction of C=O and C-H that appeared after the binding of PEG. These peaks appeared strongly in grafted materials compared to the CS polymer. Another characteristic peak of PEG, 1728 cm-1, is related to C-O-C tensile vibration and can also be associated with esterification (Fig. 2C and 2E). An increase in the vibration of the alkyl was observed at 1072 cm-1, which represents the tensile vibrations of the C-O bond of the glucosamine ring. The peak at 1580 cm-1 indicates the N-H bond of the primary amines of CS polymer, in which the amide I peak was removed after the binding of PEG. The tensile vibrations of the glycoside C-O-C linkage are observed at 896 cm-1 and the CH2 flexural bond at 1420 cm-1 (Fig. 2B).
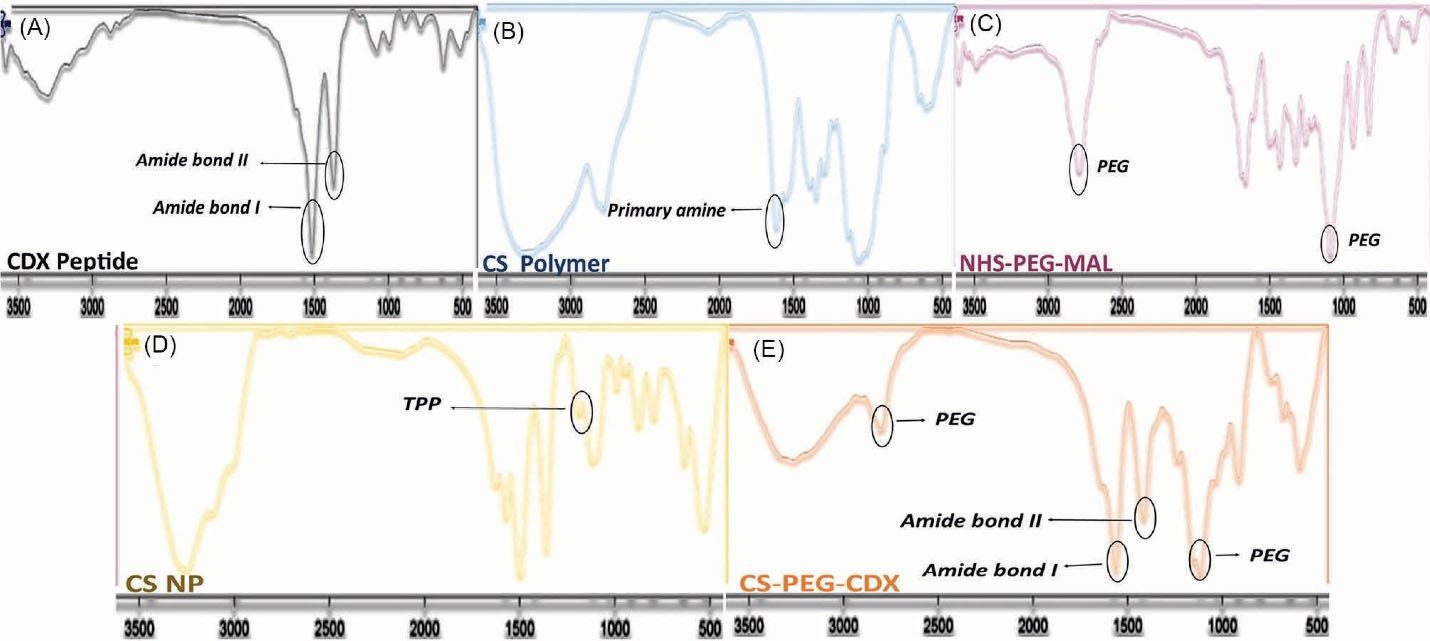
Fig. 2.
FTIR spectrum of (A) CDX peptide, (B) CS polymer, (C) NHS-PEG2000-MAL, (D) CS NP, and (E) CS-PEG-CDX polymer. FTIR: using KBr disc.
.
FTIR spectrum of (A) CDX peptide, (B) CS polymer, (C) NHS-PEG2000-MAL, (D) CS NP, and (E) CS-PEG-CDX polymer. FTIR: using KBr disc.
The peak at 1240 cm-1 is related to the binding of the thiol peptide group to the maleimide group of the PEG molecule (Fig. 2A).30,31 Two characteristic peaks of the attached peptide at 1645 cm-1 and 1565 cm-1 were associated with amide I and amide II bands, respectively.32 Finally, the electrostatic interactions between the amine groups of CS and the oxygen group of TPP were shown at 1215 cm-1 (Fig. 2E).33 We evaluated also the created characteristic peaks after the reaction of CS, bifunctional PEG, and CDX peptide with each other using 1HNMR (Supplementary data).
Consistent with FTIR and NMR data, the TNBSA and Ellman assays confirmed the concise attachment of PEG and peptide molecules to the CS polymeric backbone. The measured concentration of free amines with TNBSA assay for CS, CS-PEG, and CS-PEG-CDX were 468.56, 132.01, and 94.79 μM, respectively. These data show that the degree of amine substitution was approximately %71.82. Following the reaction of the CDX with the CS-PEG solution, all CDX contents were attached to the PEG MAL groups in the CS-PEG polymer structure. These findings are in agreement with the zeta potential of prepared NPs, in which the zeta potential of CS, CS-PEG, and CS-PEG-CDX NPs reached +28, -1.76, and +0.9mV, respectively (Table 1, Fig. 3C).
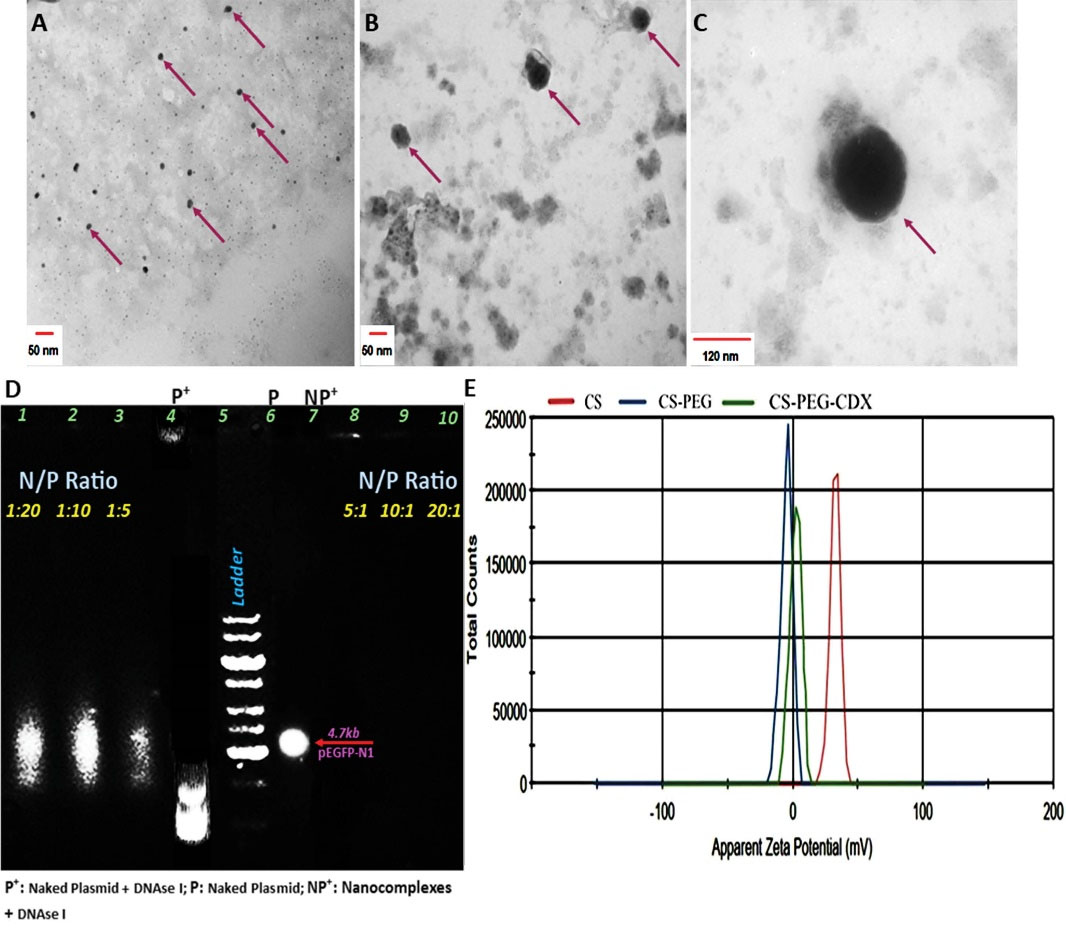
Fig. 3.
(A) TEM image of CS NPs (scale bar: 50 nm), (B) CS-PEG-CDX NPs (scale bar: 50 nm), and (C) CS-PEG-CDX/pEGFP nanocomplexes (scale bar: 120 nm). (D) Agarose gel electrophoresis. The stability of nano-complex in electromagnetic field and enzyme degradation increased by an increase in the concentration of NPs to plasmid (lanes 7-10). (E) Zeta potential of CS, CS-PEG, and CS-PEG-CDX NPs is touted as successful attachment of PEG and CDX to the structure of NPs. Agarose gel concentration: 0.7%, Electrophoresis time: 30 minutes, Electrophoresis voltage: 100 V/cm.
.
(A) TEM image of CS NPs (scale bar: 50 nm), (B) CS-PEG-CDX NPs (scale bar: 50 nm), and (C) CS-PEG-CDX/pEGFP nanocomplexes (scale bar: 120 nm). (D) Agarose gel electrophoresis. The stability of nano-complex in electromagnetic field and enzyme degradation increased by an increase in the concentration of NPs to plasmid (lanes 7-10). (E) Zeta potential of CS, CS-PEG, and CS-PEG-CDX NPs is touted as successful attachment of PEG and CDX to the structure of NPs. Agarose gel concentration: 0.7%, Electrophoresis time: 30 minutes, Electrophoresis voltage: 100 V/cm.
Size, zeta potential, and morphology results
The chemical formation of NPs was performed in a self-assembly manner, in which the complex was formed by adding TPP as an ionic cross-linker. Generally, the CS-PEG-CDX/TPP ratio, stirring speed, and TPP adding speed have a crucial effect on the size and zeta potential of synthesized CS-based NPs. In this regard, we used various CS-PEG-CDX: TPP ratios ranging from 1:1 to 5:1 w/w, with different stirring speeds, and the TPP adding speed. Our results showed that the best ratio of CS-PEG-CDX: TPP and stirring speed for CS-PEG-CDX NP synthesis are 5:1 w/w and 1000 rpm, respectively (Table 2).
Table 2.
Optimizing the CS-PEG-CDX: TPP ratio and stirrer speed
|
CS-PEG-CDX:TPP ratio
|
Speed (RPM)
|
Size (nm)
|
PDI
|
| 1:1 |
1000 |
316.5 |
0.237 |
| 2:1 |
1000 |
204.1 |
0.440 |
| 3:1 |
1000 |
190 |
0.215 |
| 4:1 |
1000 |
107 |
0.181 |
| 5:1 |
1000 |
78.75 |
0.292 |
| 5:1 |
750 |
170.9 |
0.376 |
| 5:1 |
500 |
276.0 |
0.237 |
RPM; Round per minute, CS; Chitosan, CDX; Candoxin derived 14 amino acids sequence, TPP; Sodium tripolyphosphate, PDI; Poly dispersity index, PEG; Poly ethylene glycol.
The zeta potential of CS NPs was +28 mV. This value is due to the presence of numerous amine groups on the CS surface. However, the addition of PEG into the CS NP’s structure reduced the zeta potential to -1.76 mV, indicating the successful reaction of PEG with CS amine groups (Fig. 3C). Finally, the zeta potential of CS-PEG-CDX was +0.9 mV, which is due to the positive charge of CDX peptide at physiological pH (about +2 mV). It is noteworthy to mention that the final positive charge of synthesized NPs is a vital factor in compacting plasmids. DLS data showed that the average hydrodynamic diameter of the CS, CS-PEG, and CS-PEG-CDX NPs is around 73.34, 97.52, and 123 nm, respectively. The real size and morphology of the NPs were also checked using TEM (Fig. 3A-C). Our results showed that the morphology of synthesized NPs is spherical. The mean size of CS, CS-PEG-CDX, and CS-PEG-CDX/pEGFP NPs is around 50, 70, and 120 nm, respectively (Table 1, Fig. 3A-C).
The agarose gel electrophoresis is widely used to evaluate the stability of synthesized NPs like CS-PEG-CDX/pEGFP complexes (Fig. 3B). In this regard, we labeled the pEGFP plasmid with EtBr, and different ratios of N/P were prepared. The complexes were electrophoresed on 0.7% agarose gel at different N/P ratios (1: 20-20: 1). Our results showed that the N/P ratio at 5:1 and higher ratios have good stability and appropriate molecular weights. The N/P ratio of 10:1 was selected for subsequent in vivo analysis based on the proper size and appropriate zeta potential. Moreover, in Fig. 3B (lanes 4, and 7), it is well shown that the plasmid shows different resistance behavior in the presence and absence of DNAse I. Thus, this maintained stability in the presence of the enzyme indicates the enzymatic stability of the synthesized nanocomplexes in the biological environment, which is a crucial factor for a proper gene delivery system.
In-vitro uptake of CS-PEG-CDX/ pEGFP by C6 glioma cells
The cellular uptake of CS-PEG-CDX/pEGFP nanocomplexes was analyzed in the rat C6 glioma cell line using a fluorescence microscope. To evaluate the internalization efficiency, the cells were imaged 3 hours after treatment with different concentrations (5, 10, 20, and 40 ppm) of EtBr-labeled CS-PEG-CDX/pEGFP nanocomplexes. As shown in Fig. 4, the cellular uptake of CS-PEG-CDX/pEGFP nanocomplexes was intensified by increasing the concentration of nanocomplex from 5 ppm (Fig. 4A-C) up to 40 ppm (Fig. 4J-L), indicating a dose-dependent uptake of the synthesized nanocomplexes.
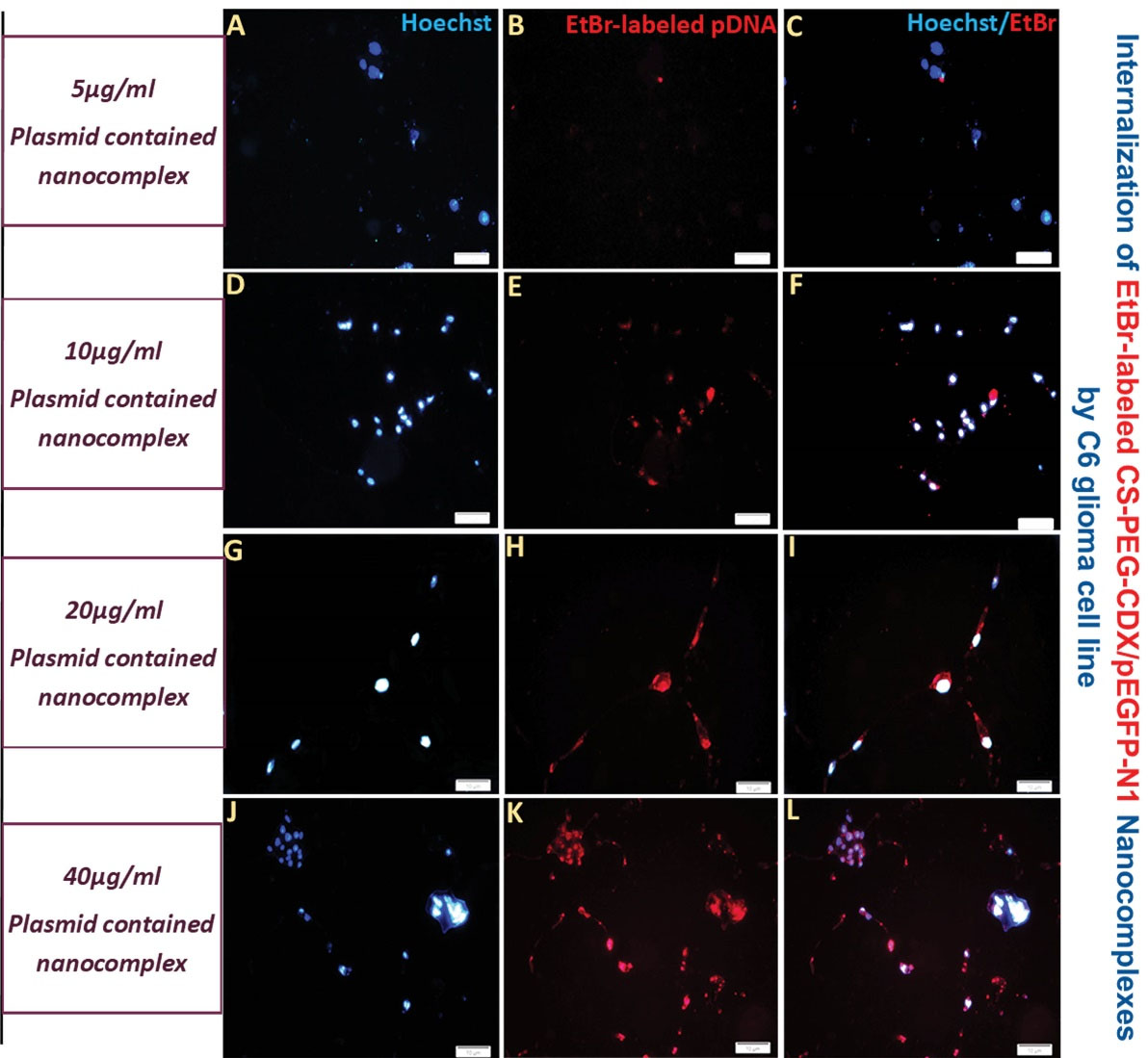
Fig. 4.
Cellular internalization of EtBr-labeled CS–PEG–CDX/pEGFP nanocomplexes in the concentration of 5 (A-C), 10 (D-F), 20 (G-I), and 40 ppm (J-L) by C6 glioma cells, in a concentration-dependent manner. Red: Ethidium bromide, Blue: Hoechst. Scale bar 100 µm.
.
Cellular internalization of EtBr-labeled CS–PEG–CDX/pEGFP nanocomplexes in the concentration of 5 (A-C), 10 (D-F), 20 (G-I), and 40 ppm (J-L) by C6 glioma cells, in a concentration-dependent manner. Red: Ethidium bromide, Blue: Hoechst. Scale bar 100 µm.
Fluorescent Imaging
In vivo imaging analysis
For evaluating the efficiency of synthesized nano-complexes to reach the brain, the EtBr-labeled pEGFP/CS–PEG–CDX and EtBr-labeled pEGFP/CS (10:1, NPs to the plasmid, w/w) nano-complexes were injected intraperitoneally at a dose of 50 μg DNA per mouse once a day for 5 consecutive days. The total volume injection was 200 μl per day.18 To assess the biodistribution nanocomplexes, different organs including the heart, spleen, kidneys, and liver were sampled along with the brain. As shown in Fig. 5A, EtBr-labeled pEGFP can be seen in the brain of the animal that received EtBr-labeled pEGFP/CS–PEG–CDX compared to the control group. The biodistribution of NPs in vital organs like the liver, spleen, heart, and kidneys was also evident in which the spleen was the main target for EtBr-labeled pEGFP/PEG–CDX (Fig. 5B).
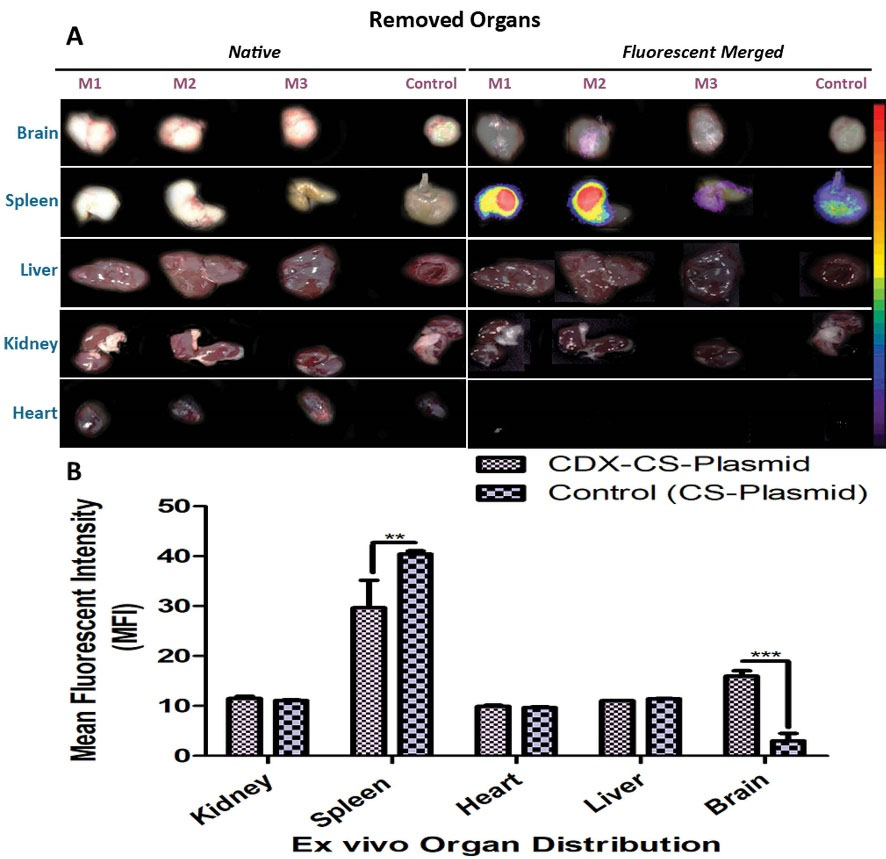
Fig. 5.
Ex vivo imaging of major organs of the mice was treated with EtBr-labeled CS/pEGFP (control) and EtBr-labeled CS–PEG–CDX/pEGFP nanocomplexes. Images were taken 48 h after IP injection of NPs. (A) Organs image (Left), The fluorescent merged images of the same organs (Right). (B) Semi-quantitative fluorescence intensity of the brain and different organs. P value ≤0.05, (n=3, M1-3).
.
Ex vivo imaging of major organs of the mice was treated with EtBr-labeled CS/pEGFP (control) and EtBr-labeled CS–PEG–CDX/pEGFP nanocomplexes. Images were taken 48 h after IP injection of NPs. (A) Organs image (Left), The fluorescent merged images of the same organs (Right). (B) Semi-quantitative fluorescence intensity of the brain and different organs. P value ≤0.05, (n=3, M1-3).
Distribution and GFP expression in the brain
To ascertain the presence of synthesized nanocomplexes in the brain tissue, we checked the red fluorescence of EtBr-labeled nanocomplexes in the mice brain slices using a fluorescent microscope (n=3). Our results showed that the EtBr-labeled pEGFP/CS-PEG-CDX nanocomplexes were efficiently transfected into the cells and expressed across the brain parenchyma (Fig. 6).
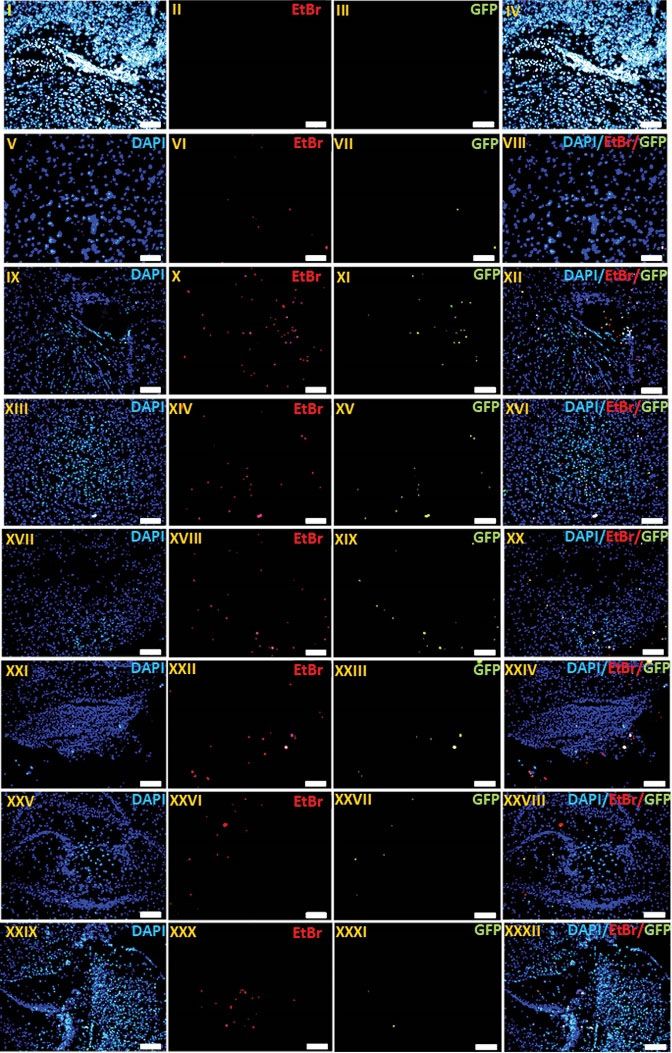
Fig. 6.
The qualitative evaluation of EtBr-labeled CS-PEG-CDX/pEGFP NPs and gene expression in vivo. Profile of labeled pDNA distribution and GFP gene expression in brains of Balb/c mice treated with EtBr-labeled CS/pEGFP (I-IV) and CS–PEG–CDX/pEGFP (n=3) nano-complexes 48 h after last i.p. injection (V-XXXII). Frozen sections (thickness of 20 µm) of the striatum (V-VIII), ventricle (IX-XII), substantia nigra (XIII-XVI), cortical layer (XVII-XX), brain stem (XXI-XXIV), basal ganglia (XXV-XXVIII), and hippocampus (XXIX-XXXII) were observed by fluorescent microscopy. The sections were stained with 300 nM DAPI for 10 min at room temperature. Green: GFP. Blue: DAPI. Red: EtBr. Original magnification: 100.
.
The qualitative evaluation of EtBr-labeled CS-PEG-CDX/pEGFP NPs and gene expression in vivo. Profile of labeled pDNA distribution and GFP gene expression in brains of Balb/c mice treated with EtBr-labeled CS/pEGFP (I-IV) and CS–PEG–CDX/pEGFP (n=3) nano-complexes 48 h after last i.p. injection (V-XXXII). Frozen sections (thickness of 20 µm) of the striatum (V-VIII), ventricle (IX-XII), substantia nigra (XIII-XVI), cortical layer (XVII-XX), brain stem (XXI-XXIV), basal ganglia (XXV-XXVIII), and hippocampus (XXIX-XXXII) were observed by fluorescent microscopy. The sections were stained with 300 nM DAPI for 10 min at room temperature. Green: GFP. Blue: DAPI. Red: EtBr. Original magnification: 100.
Discussion
Given the unique features of CS, including biocompatibility, biodegradability, and mucous attachment, CS-based NPs are one of the promising tools for drug and gene delivery.34 However these NPs suffer from some deficiencies, especially in biological environments. The most important of them is their instability in the physiological pH because of their ionic interactions with TPP. It was shown that in pH above 7.4, the protonation of TPP causes the dissociation of CS polymer from TPP and results in NPs' rapid degradation.35 In alignment with those findings, our results showed that unmodified CS NPs synthesized using the ionic gelation method were stable in the RPMI media with pH 7.4 only for 4 hours. While after modification with bifunctional PEG and CDX peptide, this time increased to around 72 hours (data was not shown). The possible reason behind this enhanced stability might be the more condensation of the CS polymers due to interactions among added molecules to the NP structure. Based on our multi-step characterization, CS NPs, CS-PEG-CDX polymer, and CS-PEG-CDX NPs were successfully synthesized. Further, physicochemical properties of developed NPs like their size and zeta potential are under 100 nm, which are proper features for a nano-drug delivery system (NDDS).36 Our findings established that in the case of functionalized-CS NPs, preparation of polymers, CDX-PEG-CS polymer is the best strategy compared to surface modification after the synthesis of CS NPs. Furthermore, the physicochemical properties of CDX modified Cs NPs complex with pEGFP-N1 confirmed that in the N/P ratio of 10:1, the aforementioned features, condensation, and their stability against DNAse are in the suitable range for in vivo study.18,37 Among different routes for NP entrance to the brain, including adsorption-mediated endocytosis (AME), carrier-mediated endocytosis (CME), and RMT, the latter one is a more suitable route for intact delivery of cargo to the brain parenchyma.38,39 Hence, using the ligands which initiate this biological process is the best option for drug/gene delivery to the brain. Up to now, there have been some ligands discovered with the aforementioned capability, such as Angiopep-2,40 SRL peptide,18 Leptin-derived peptides,41 and CDX peptide.25 CDX is a 16-amino acid peptide sequence isolated from snake venom known as Candoxin.42 This peptide can identify the α7 subunit of nAchR on the surface of brain capillary endothelial cells.19-22 It has been shown that this receptor is highly expressed on the brain capillary endothelial cells with a very high affinity to the CDX peptide and fast transcytosis ability.25
Moreover, as mentioned earlier, the C6 glioma cell line overexpressed α7 nAchRs. An increase in the fluorescence intensity in the present study is due to the high cellular internalization of nanocomplexes into the rat C6 glioma cells via α7 nAchRs, in a dose-dependent manner. Therefore, CS-PEG-CDX/pEGFP nanocomplexes are promising agents for the dual-targeting of glioma. Interestingly, it has been shown that the α7 nAChRs are involved in proliferation, migration,43 tumor formation,44 and resistance to apoptosis induced by different therapeutics.45,46 According to the abovementioned evidence, our developed CS-PEG-CDX/pEGFP nanocomplexes are suitable candidates for the dual-targeting of glioma.47,48 On the other hand, our results showed an interesting outcome, in which the spleen has trapped most of the nanocomplexes in the in vivo study. It seems that the synthesized nanocomplexes were captured by the spleen due to the high levels of IgM adsorbed to the CDX segment of nanocomplexes. It has been shown that the positive net charge of CDX peptide is the main reason for IgM attachment to this peptide.49 Different studies have shown that IgM-dependent fast removal of NPs is completely associated with the activation of the spleen marginal zone B lymphocytes controlled by type 2 T helpers.50 Commensurate with the comments, we proposed that the same mechanism contributed to the splenic accumulation of EtBr-labeled pEGFP/CS-PEG-CDX nanocomplexes. Given that homomeric α7 nAChRs are abundantly expressed in the CNS and spinal cord,51 and they exert neuroprotection activity in microglia,52 and astrocytes,53,54 by reducing the pro-inflammatory cytokines, they can be considered as a promising choice for treating neuroinflammatory diseases.55 In this regard, it was suggested that α7 nAchRs have a vital role in the proliferation and viability of glutamatergic synapses in the hippocampus,56 a site in the brain which is responsible for memory formation and consolidation. Consistent with these data, the pivotal role of α7 nAchRs deficiency in developing a cognitive related disease, especially Alzheimer's disease (AD) has been established.57,58 Concerning our findings (Fig. 6), we proposed that the currently synthesized nanocomplexes could be used for gene therapy for CNS-related cancers and neurodegenerative diseases.
Research Highlights
What is the current knowledge?
√ The therapeutic agents have not been summarized in drugs and small molecules.
√ The genetic materials, including different kinds of microRNAs, siRNAs, and larger ones like plasmids, have shown promising results alone or in combination with drugs for CNS disorders.
√ However, the main drawbacks in usage of these therapeutic agents are their instability in the blood stream and inability in passage from the cell’s membrane.
What is new here?
√ We successfully delivered (pEGFP) to the brain using CDX-modified CS NPs in vivo, by preferred brain distribution according to nAchR expression profile. These findings are promising and applicable for gene delivery to some CNS diseases like Alzheimer, which has a close relationship with the abovementioned receptor expression profile.
Conclusion
For the first time, developed CDX modified CS NPs were demonstrated to be efficient for brain-targeting gene delivery in vivo conditions. The qualitative experiments proved by in vivo imaging system and fluorescent microscopy showed that CS–PEG–CDX/pEGFP-N1 are capable of crossing the BBB and reach the brain parenchyma. Based on the importance of nAchR function in the pathology of glioma and AD, we believe that CS-PEG-CDX NPs have a promising potential for efficient noninvasive targeting gene delivery to mentioned diseases.
Acknowledgment
This work is a part of an MSc thesis of Tina Sepasi in medical nanotechnology at Tabriz University of Medical Sciences (TBZMED). The authors also would like to thank TUMS Preclinical Core Facility for their kind technical support in in-vivo imaging studies.
Funding sources
This work was fully supported by Iran National Science Foundation (INSF) (Grant number: 97002663).
Ethical statement
All phases of this study were approved by the Local Ethics Committee of Tabriz University of Medical Sciences.
Conflict of interests
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript.
References
- Ginn SL, Amaya AK, Alexander IE, Edelstein M, Abedi MR. Gene therapy clinical trials worldwide to 2017: An update. J Gene Med 2018; 20:e3015. doi: 10.1002/jgm.3015 [Crossref] [ Google Scholar]
- Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet 2003; 4:346-58. doi: 10.1038/nrg1066 [Crossref] [ Google Scholar]
- Sharma G, Sharma AR, Lee S-S, Bhattacharya M, Nam J-S, Chakraborty C. Advances in nanocarriers enabled brain targeted drug delivery across blood brain barrier. Int J Pharm 2019; 559:360-72. doi: 10.1016/j.ijpharm.2019.01.056 [Crossref] [ Google Scholar]
- Kim J, Ahn SI, Kim Y. Nanotherapeutics engineered to cross the blood-brain barrier for advanced drug delivery to the central nervous system. J Ind Eng Chem 2019; 73:8-18. doi: 10.1016/j.jiec.2019.01.021 [Crossref] [ Google Scholar]
- Mijanović O, Branković A, Borovjagin AV, Butnaru DV, Bezrukov EA, Sukhanov RB. Battling Neurodegenerative Diseases with Adeno-Associated Virus-Based Approaches. Viruses 2020; 12:460. doi: 10.3390/v12040460 [Crossref] [ Google Scholar]
- Senior M. After Glybera's withdrawal, what's next for gene therapy?. Nat Biotechnol 2017; 35:491-492. doi: 10.1038/nbt0617-491 [Crossref] [ Google Scholar]
- Mohammadinejad R, Dehshahri A, Sagar Madamsetty V, Zahmatkeshan M, Tavakol S, Makvandi P. In vivo gene delivery mediated by non-viral vectors for cancer therapy. J Con Rel 2020; 325:249-75. doi: 10.1016/j.jconrel.2020.06.038 [Crossref] [ Google Scholar]
- Mohammadinejad R, Dehshahri A, Madamsetty VS, Zahmatkeshan M, Tavakol S, Makvandi P. In vivo gene delivery mediated by non-viral vectors for cancer therapy. J Con Rel 2020; 325:249-75. doi: 10.1016/j.jconrel.2020.06.038 [Crossref] [ Google Scholar]
- Pardridge WM. Drug and gene delivery to the brain: the vascular route. Neuron 2002; 36:555-8. doi: 10.1016/s0896-6273(02)01054-1 [Crossref] [ Google Scholar]
- Ingusci S, Verlengia G, Soukupova M, Zucchini S, Simonato M. Gene therapy tools for brain diseases. Front Pharm 2019; 10:724. doi: 10.3389/fphar.2019.00724 [Crossref] [ Google Scholar]
- Huang R-Q, Qu Y-H, Ke W-L, Zhu J-H, Pei Y-Y, Jiang C. Efficient gene delivery targeted to the brain using a transferrin‐conjugated polyethyleneglycol‐modified polyamidoamine dendrimer. FASEB J 2007; 21:1117-25. doi: 10.1096/fj.06-7380com [Crossref] [ Google Scholar]
- Gu J, Al-Bayati K, Ho EA. Development of antibody-modified CS NPs for the targeted delivery of siRNA across the blood-brain barrier as a strategy for inhibiting HIV replication in astrocytes. Drug DelivTransl Res 2017; 7:497-506. doi: 10.1007/s13346-017-0368-5 [Crossref] [ Google Scholar]
- Apirakaramwong A, Sunthornphan S, Pasuthawong N, Intaravicha T, Ruangthai N, Rojanarata T, editors editors. The Effect of Spermidine and Spermine on CS-Mediated Gene Delivery. Key Eng Mat 2020; 859:113-119. doi: 10.4028/www.scientific.net/KEM.859.113 [Crossref] [ Google Scholar]
- Santos-Carballal B, Fernández Fernández E, Goycoolea FM. CS in non-viral gene delivery: Role of structure, characterization methods, and insights in cancer and rare diseases therapies. Polymers 2018; 10:444. doi: 10.3390/polym10040444 [Crossref] [ Google Scholar]
- Praveena R. A Complete Review-NPs Targeting to Brain. A J Res Pharml Sci 2020; 10:119-203. doi: 10.5958/2231-5659.2020.00038.7 [Crossref] [ Google Scholar]
- Behzadi S, Serpooshan V, Tao W, Hamaly MA, Alkawareek MY, Dreaden EC. Cellular uptake of NPs: journey inside the cell. Chem Soc Rev 2017; 46:4218-44. doi: 10.1039/C6CS00636A [Crossref] [ Google Scholar]
- Glassman PM, Myerson JW, Ferguson LT, Kiseleva RY, Shuvaev VV, Brenner JS. Targeting drug delivery in the vascular system: Focus on endothelium. Adv Drug Del Rev 2020; 157:96-117. doi: 10.1016/j.addr.2020.06.013 [Crossref] [ Google Scholar]
- Zarebkohan A, Najafi F, Moghimi HR, Hemmati M, Deevband MR, Kazemi B. Synthesis and characterization of a PAMAM dendrimer nanocarrier functionalized by SRL peptide for targeted gene delivery to the brain. Eur J Pharm Sci 2015; 78:19-30. doi: 10.1016/j.ejps.2015.06.024 [Crossref] [ Google Scholar]
- Wei X, Zhan C, Shen Q, Fu W, Xie C, Gao J. A D‐peptide ligand of nicotine acetylcholine receptors for brain‐targeted drug delivery. Angew Chem 2015; 127:3066-70. doi: 10.1002/ange.201411226 [Crossref] [ Google Scholar]
- Thompson EG, Sontheimer H. Acetylcholine receptor activation as a modulator of glioblastoma invasion. Cells 2019; 8:1203. doi: 10.3390/cells8101203 [Crossref] [ Google Scholar]
- Niranjan R, Nath C, Shukla R. Melatonin attenuated mediators of neuroinflammation and alpha-7 nicotinic acetylcholine receptor mRNA expression in lipopolysaccharide (LPS) stimulated rat astrocytoma cells, C6. Free Radical Res 2012; 46:1167-77. doi: 10.3109/10715762.2012.697626 [Crossref] [ Google Scholar]
- Terpinskaya TI, Osipov AV, Kryukova EV, Kudryavtsev DS, Kopylova NV, Yanchanka TL. α-Conotoxins and α-Cobratoxin Promote, while Lipoxygenase and Cyclooxygenase Inhibitors Suppress the Proliferation of Glioma C6 Cells. Marine Drugs 2021; 19:118. doi: 10.3390/md19020118 [Crossref] [ Google Scholar]
- Dai T, Jiang K, Lu W. Liposomes and lipid disks traverse the BBB and BBTB as intact forms as revealed by two-step Förster resonance energy transfer imaging. Acta Pharma Sin B 2018; 8:261-71. doi: 10.1016/j.apsb.2018.01.004 [Crossref] [ Google Scholar]
- Zhan C, Li B, Hu L, Wei X, Feng L, Fu W. Micelle-based brain-targeted drug delivery enabled by a nicotine acetylcholine receptor ligand. Angew Chem Int Ed Engl 2011; 50:5482-5. doi: 10.1002/anie.201100875 [Crossref] [ Google Scholar]
- Wei X, Zhan C, Shen Q, Fu W, Xie C, Gao J. A D-peptide ligand of nicotine acetylcholine receptors for brain-targeted drug delivery. Angew Chem 2015; 54:3023-7. doi: 10.1002/anie.201411226 [Crossref] [ Google Scholar]
- Malhotra M, Tomaro-Duchesneau C, Saha S, Kahouli I, Prakash S. Development and characterization of CS-PEG-TAT NPs for the intracellular delivery of siRNA. Int J Nanomed 2013; 8:2041-52. doi: 10.2147/IJN.S43683 [Crossref] [ Google Scholar]
- Zheng Z, Zhang J, Jiang J, He Y, Zhang W, Mo X, et al. Remodeling tumor immune microenvironment (TIME) for glioma therapy using multi-targeting liposomal codelivery. J Immun Cancer 2020; 8. 10.1136/jitc-2019-000207.
- Ke W, Shao K, Huang R, Han L, Liu Y, Li J. Gene delivery targeted to the brain using an Angiopep-conjugated polyethyleneglycol-modified polyamidoamine dendrimer. Biomaterials 2009; 30:6976-85. doi: 10.1016/j.biomaterials.2009.08.049 [Crossref] [ Google Scholar]
- Najafi F, Moghimi HR, Hemmati M, Deevband MR, Kazemi B. SRL-coated PAMAM dendrimer nano-carrier for targeted gene delivery to the glioma cells and competitive inhibition by lactoferrin. Iranian journal of pharmaceutical research: IJPR 2016; 15:629. [ Google Scholar]
- Papadimitriou SA, Achilias DS, Bikiaris DN. CS-g-PEG NPs ionically crosslinked with poly (glutamic acid) and tripolyphosphate as protein delivery systems. Int J Pharm 2012; 430:318-27. doi: 10.1016/j.ijpharm.2012.04.004 [Crossref] [ Google Scholar]
- Zeng Z, Mo X-m, He C, Morsi Y, El-Hamshary H, El-Newehy M. An in situ forming tissue adhesive based on poly (ethylene glycol)-dimethacrylate and thiolated CS through the Michael reaction. J Mat Chem B 2016; 4:5585-92. doi: 10.1039/C6TB01475E [Crossref] [ Google Scholar]
- Du J, Hsieh Y-L. PEGylation of CS for improved solubility and fiber formation via electrospinning. Cellulose 2007; 14:543-52. doi: 10.1007/s10570-007-9122-3 [Crossref] [ Google Scholar]
- Mazancová P, Némethová V, Treľová D, Kleščíková L, Lacík I, Rázga F. Dissociation of CS/tripolyphosphate complexes into separate components upon pH elevation. Carbo pol 2018; 192:104-10. doi: 10.1016/j.carbpol.2018.03.030 [Crossref] [ Google Scholar]
- Divya K, Jisha MS. CS NPs preparation and applications. Env Chem Let 2018; 16:101-12. doi: 10.1007/s10311-017-0670-y [Crossref] [ Google Scholar]
- Huang Y, Cai Y, Lapitsky Y. Factors affecting the stability of CS/tripolyphosphate micro- and nanogels: resolving the opposing findings. J Mat Chem B 2015; 3:5957-70. doi: 10.1039/C5TB00431D [Crossref] [ Google Scholar]
- Tiwari G, Tiwari R, Sriwastawa B, Bhati L, Pandey S, Pandey P. Drug delivery systems: An updated review. Int J pharm Invest 2012; 2:2-11. doi: 10.4103/2230-973X.96920 [Crossref] [ Google Scholar]
- Huang RQ, Qu YH, Ke WL, Zhu JH, Pei YY, Jiang C. Efficient gene delivery targeted to the brain using a transferrin-conjugated polyethyleneglycol-modified polyamidoamine dendrimer. FASEB j 2007; 21:1117-25. doi: 10.1096/fj.06-7380com [Crossref] [ Google Scholar]
- Patel MM, Patel BM. Crossing the Blood-Brain Barrier: Recent Advances in Drug Delivery to the Brain. CNS Drugs 2017; 31:109-33. doi: 10.1007/s40263-016-0405-9 [Crossref] [ Google Scholar]
- Pandey PK, Sharma AK, Gupta U. Blood brain barrier: An overview on strategies in drug delivery, realistic in vitro modeling and in vivo live tracking. Tissue Barriers 2016; 4:e1129476. doi: 10.1080/21688370.2015.1129476 [Crossref] [ Google Scholar]
- Shi X-X, Miao W-M, Pang D-W, Wu J-S, Tong Q-S, Li J-X. Angiopep-2 conjugated NPs loaded with doxorubicin for the treatment of primary central nervous system lymphoma. Biomat Sci 2020; 8:1290-7. doi: 10.1039/C9BM01750J [Crossref] [ Google Scholar]
- Liu Y, Li J, Shao K, Huang R, Ye L, Lou J. A leptin derived 30-amino-acid peptide modified pegylated poly-L-lysine dendrigraft for brain targeted gene delivery. Biomaterials 2010; 31:5246-57. doi: 10.1016/j.biomaterials.2010.03.011 [Crossref] [ Google Scholar]
- Zhan C, Li B, Hu L, Wei X, Feng L, Fu W. Micelle-Based Brain-Targeted Drug Delivery Enabled by a Nicotine Acetylcholine Receptor Ligand. Angew Chem 2011; 50:5482-5. doi: 10.1002/anie.201100875 [Crossref] [ Google Scholar]
- Nguyen HD, Liao Y-C, Ho Y-S, Chen L-C, Chang H-W, Cheng T-C. The α9 nicotinic acetylcholine receptor mediates nicotine-induced PD-L1 expression and regulates melanoma cell proliferation and migration. Cancers 2019; 11:1991. doi: 10.3390/cancers11121991 [Crossref] [ Google Scholar]
- Fararjeh AFS, Tu SH, Chen LC, Cheng TC, Liu YR, Chang HL. Long‐term exposure to extremely low‐dose of nicotine and 4‐(methylnitrosamino)‐1‐(3‐pyridyl)‐1‐butanone (NNK) induce non‐malignant breast epithelial cell transformation through activation of the a9‐nicotinic acetylcholine receptor‐mediated signaling pathway. Env Toxicol 2019; 34:73-82. doi: 10.1002/tox.22659 [Crossref] [ Google Scholar]
- Wang YY, Liu Y, Ni XY, Bai ZH, Chen QY, Zhang Y. Nicotine promotes cell proliferation and induces resistance to cisplatin by α7 nicotinic acetylcholine receptor-mediated activation in Raw264. 7 and El4 cells. Onco Rep 2014; 31:1480-8. doi: 10.3892/or.2013.2962 [Crossref] [ Google Scholar]
- Wang C, Niu W, Chen H, Shi N, He D, Zhang M. Nicotine suppresses apoptosis by regulating α7nAChR/Prx1 axis in oral precancerous lesions. Oncotarget 2017; 8:75065. doi: 10.18632/oncotarget.20506 [Crossref] [ Google Scholar]
- Wessler I, Kirkpatrick C. Acetylcholine beyond neurons: the non‐neuronal cholinergic system in humans. Bri J Pharmacol 2008; 154:1558-71. doi: 10.1038/bjp.2008.185 [Crossref] [ Google Scholar]
- Gotti C, Clementi F. Neuronal nicotinic receptors: from structure to pathology. Progress in Neurobiol 2004; 74:363-96. doi: 10.1016/j.pneurobio.2004.09.006 [Crossref] [ Google Scholar]
- Guan J, Shen Q, Zhang Z, Jiang Z, Yang Y, Lou M. Enhanced immunocompatibility of ligand-targeted liposomes by attenuating natural IgM absorption. Nat Comm 2018; 9:1-11. doi: 10.1038/s41467-018-05384-1 [Crossref] [ Google Scholar]
- Cataldi M, Vigliotti C, Mosca T, Cammarota M, Capone D. Emerging role of the spleen in the pharmacokinetics of monoclonal antibodies, NPs and exosomes. Int J Mol Sci 2017; 18:1249. doi: 10.3390/ijms18061249 [Crossref] [ Google Scholar]
- Tribollet E, Bertrand D, Marguerat A, Raggenbass M. Comparative distribution of nicotinic receptor subtypes during development, adulthood and aging: an autoradiographic study in the rat brain. Neurosci 2004; 124:405-20. doi: 10.1016/j.neuroscience.2003.09.028 [Crossref] [ Google Scholar]
- Suzuki T, Hide I, Matsubara A, Hama C, Harada K, Miyano K. Microglial alpha7 nicotinic acetylcholine receptors drive a phospholipase C/IP3 pathway and modulate the cell activation toward a neuroprotective role. J Neurosci Res 2006; 83:1461-70. doi: 10.1002/jnr.20850 [Crossref] [ Google Scholar]
- Papouin T, Dunphy JM, Tolman M, Dineley KT, Haydon PG. Septal Cholinergic Neuromodulation Tunes the Astrocyte-Dependent Gating of Hippocampal NMDA Receptors to Wakefulness. Neuron 2017; 94: 840-54.e7. 10.1016/j.neuron.2017.04.021.
- Broide RS, Winzer-Serhan UH, Chen Y, Leslie FM. Distribution of α7 nicotinic acetylcholine receptor subunit mRNA in the developing mouse. Front Neuroanatomy 2019; 13:76. doi: 10.3389/fnana.2019.00076 [Crossref] [ Google Scholar]
- Yue Y, Liu R, Cheng W, Hu Y, Li J, Pan X. GTS-21 attenuates lipopolysaccharide-induced inflammatory cytokine production in vitro by modulating the Akt and NF-κB signaling pathway through the α7 nicotinic acetylcholine receptor. Int Immunopharmacol 2015; 29:504-12. doi: 10.1016/j.intimp.2015.10.005 [Crossref] [ Google Scholar]
- Lozada AF, Wang X, Gounko NV, Massey KA, Duan J, Liu Z. Glutamatergic synapse formation is promoted by α7-containing nicotinic acetylcholine receptors. J Neurosci 2012; 32:7651-61. doi: 10.1523/JNEUROSCI.6246-11.2012 [Crossref] [ Google Scholar]
- Stoiljkovic M, Kelley C, Nagy D, Hurst R, Hajós M. Activation of α7 nicotinic acetylcholine receptors facilitates long-term potentiation at the hippocampal-prefrontal cortex synapses in vivo. Eur Neuropsychopharmacol 2016; 26:2018-23. doi: 10.1016/j.euroneuro.2016.11.003 [Crossref] [ Google Scholar]
- Inestrosa NC, Godoy JA, Vargas JY, Arrazola MS, Rios JA, Carvajal FJ. Nicotine prevents synaptic impairment induced by amyloid-β oligomers through α7-nicotinic acetylcholine receptor activation. Neuromol Med 2013; 15:549-69. doi: 10.1007/s12017-013-8242-1 [Crossref] [ Google Scholar]