Bioimpacts. 14(1):26438.
doi: 10.34172/bi.2023.26438
Original Article
Exploratory data analysis of physicochemical parameters of natural antimicrobial and anticancer peptides: Unraveling the patterns and trends for the rational design of novel peptides
Sandeep Saini Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing, 1, 2 
Aayushi Rathore Formal analysis, 3 
Sheetal Sharma Writing – review & editing, 1 
Avneet Saini Conceptualization, Methodology, Supervision, Writing – review & editing, 1, * 
Author information:
1Department of Biophysics, Panjab University, Sector 25, Chandigarh 160014, India
2Department of Bioinformatics, Goswami Ganesh Dutta Sanatan Dharma College, Sector 32-C, Chandigarh 160030, India
3Institute of Bioinformatics and Applied Biotechnology, Biotech Park, Bengaluru 560100, India
Abstract
Introduction:
Peptide-based research has attained new avenues in the antibiotics and cancer drug resistance era. The basis of peptide design research lies in playing with or altering physicochemical parameters. Here in this work, we have done exploratory data analysis (EDA) of physicochemical parameters of antimicrobial peptides (AMPs) and anticancer peptides (ACPs), two promising therapeutics for microbial and cancer drug resistance to deduce patterns and trends.
Methods:
Briefly, we have captured the natural AMPs and ACPs data from the APD3 database. After cleaning the data manually and by CD-HIT web server, further data analysis has been done using Python-based packages, modlAMP and Pandas. We have extracted the descriptive statistics of 10 physicochemical parameters of AMPs and ACPs to build a comprehensive dataset containing all major parameters. The global analysis of datasets has been done using modlAMP to find the initial patterns in global data. The subsets of AMPs and ACPs were curated based on the length of the peptides and were analyzed by Pandas package to deduce the graphical profile of AMPs and ACPs.
Results:
EDA of AMPs and ACPs shows selectivity in the length and amino acid compositions. The distribution of physicochemical parameters in defined quartile ranges was observed in the descriptive statistical and graphical analysis. The preferred length range of AMPs and ACPs was found to be 21-30 amino acids, whereas few outliers in each parameter were evident after EDA analysis.
Conclusion:
The derived patterns from natural AMPs and ACPs can be used for the rational design of novel peptides. The statistical and graphical data distribution findings will help in combining the different parameters for potent design of novel AMPs and ACPs.
Keywords: Antimicrobial peptide, Anticancer peptide, Data analysis, Rational design, Peptide properties, Patterns and trends
Copyright and License Information
© 2024 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
According to the World Health Organization (WHO), antimicrobial resistance (AMR) and cancer are severe threats to human health.1 Recently, global AMR and use surveillance system (GLASS) reported laboratory-confirmed AMR cases in 3 106 602 patients in 70 countries in 2019.2 In the era of antibiotic or multidrug resistance (MDR), there is a need to look for alternative and stable treatment options beyond these small molecules.3 Amongst non-communicable diseases, cancer is the leading cause of death that decreases life expectancy in every country globally. According to the international agency for research on cancer (IARC) GLOBOCAN (2020) database statistics, there were an estimated 19.3 million new cancer cases, and 10 million cancer deaths reported worldwide in 2020.4 Traditional anticancer therapeutics involve surgery, radiation therapy, and chemotherapy as the major treatment options for primary tumors to extensive metastases. However, these traditional therapeutic options suffer from serious problems of drug resistance and adverse side-effects; for instance, data from a clinical study of patients suggest that above 80% of cancer patients acquired single or multiple drug resistance.5
Given the rising prevalence of microbial and cancer drug resistance, there is an essential need to look for alternative therapeutics. Therapeutic peptides (THPs) such as antimicrobial peptides (AMPs) and anticancer peptides (ACPs) are being seen as new arsenals in the era of microbial and cancer drug resistance, respectively.6,7 These peptides provide many advantages over traditional therapeutics drugs because of their better safety.8,9 AMPs are short, cationic, amphiphilic molecules of host defense produced by almost all life forms as components of the innate immune response. They display a broad spectrum of antimicrobial activity against Gram-negative, Gram-positive bacteria, fungi, viruses, and parasites.10 Besides antimicrobial activity, the immunomodulatory role of AMPs in mammals to stimulate pro or anti-inflammatory response by activating cells of the immune system (macrophages and mast cells) and anticancer or antitumor activities in various cancer cell lines or mice models are well established.11
The potential of AMPs as safe, effective, and highly selective drugs against several different types of cancers can be exploited to design novel ACPs as potential drugs.12 ACPs share most of the characteristics with AMPs, such as both possess high hydrophobicity (H), net positive charge, and fold into a well-defined alpha helix or beta-sheet structure upon interaction with cell membranes. However, despite sharing common characteristics, there is still enough uncertainty in the physicochemical parameters that determine the activity of some AMPs against cancer cells.13
The current challenges in peptide therapeutics such as low oral bioavailability, sensitivity to host protease, hemolysis and cytotoxicity, and short half-life hinder the development of successful AMPs or ACPs candidate.14,15 Furthermore, a lack of understanding of rational design approaches further increased the snag in therapeutics peptides development.6 Several previous efforts to explore the physicochemical parameters from the datasets of AMPs or ACPs were mainly made during the curation of peptide databases.16-19 Though, these efforts explored a few physicochemical parameters of the peptides but lack sufficient statistical analysis. Furthermore, a combination of synthetic and natural peptide datasets was used in these studies that may have prevented the overall representation of physicochemical parameters of natural peptides.
The challenges in AMPs or ACPs development and design can be better solved by understanding the underlying principles of designing natural peptides, as recently stated by Wang.20 Additionally, the study of physicochemical parameters of natural AMPs or ACPs may prove advantageous to the clinical peptide candidates as most of these candidate peptides are the analogs or modified derivatives of natural peptides.21,22 Therefore,compared to synthetic peptides, the analysis of physicochemical parameters of natural AMPs and ACPs can decode design principles better which can help in the designing of novel agents.20 Furthermore, several natural or synthetic AMPs and ACPs have been collected from the literature to curate peptide databases.16-19,23-30 The peptide datasets from these databases can provide insights into the overall design parameters for the potent design of novel AMPs and ACPs.22,31
In the current age of data science, exploratory data analysis (EDA) is the process of deriving hidden and unknown information from datasets to discover new patterns and trends in the data.32 Graphical representation of dataset analysis is the main aim of EDA, through which hidden patterns and facts can be easily detected.33 EDA helps gather instant intelligence about the data through visual inspection of graphs, plots, or images that the human brain can easily interpret.34 The statistical analysis during EDA provides only a summary of the data and may miss crucial patterns in the datasets. In contrast, the graphical analysis in EDA displays hidden patterns and facts. EDA prefers multiple plots compared to a single plot because there is no single “best plot” but rather, each different plot helps to identify a unique feature of the dataset.35,36 Recently, the EDA approaches have been used on datasets of different domains to explore hidden information and facts. The datasets analyzed for deducing statistical patterns and different graphical representations were also plotted for visual analysis of data.37-43
EDA of natural AMPs and ACPs datasets can provide insight into the design parameters. In addition, the uncertainty in physicochemical parameters of some AMPs that have shown anticancer activity can also be inferred to develop new potent AMPs or ACPs. Thus, EDA will not only provide a statistical description of physicochemical parameters for rational-based peptide design but can also contribute to the understanding of peptide data for machine learning based model-building.
Hence, here in this work, we have used a new methodology for EDA of physicochemical parameters of natural AMPs and ACPs. The methodology approach was implemented using Python based packages to decipher the patterns and trends in peptide datasets. EDA was performed on a complete dataset termed as global dataset here and subsets (partitioned based on length interval) of both AMPs and ACPs.
Materials and Methods
Dataset preparation
Natural AMPs and ACPs were retrieved from the APD3 database (https://aps.unmc.edu/).30 APD3 contains mostly natural peptides from literature sources with only a few synthetic peptides as derivatives of natural AMPs.20,30 We used “anti-Gram+/Gram- bacteria” and “anticancer” filters to retrieved AMPs and ACPs, respectively. The peptide sequences and physicochemical parameters such as length, charge, Bowman index, and structure and activity types were captured from the APD3.
Dataset preprocessing
By default, the retrieved AMPs also contain ACPs; therefore, we remove ACPs to get AMPs exclusively for analysis. Furthermore, both datasets were checked for the presence of any synthetic peptides by manual checking the annotated name of each peptide. Redundancy in the dataset was checked using the CD-HIT (http://weizhong-lab.ucsd.edu/cdhit-web-server/cgi-bin/index.cgi?cmd=cd-hit-2d) web server.44 Both datasets were cleaned to obtained >99% non-reductant AMPs and ACPs datasets.
Extraction of additional physicochemical parameters
In addition to basic physicochemical parameters captured from the APD3 database, we also calculated other important peptide parameters like molecular weight, isoelectric point (pI), instability index, aromaticity, and aliphatic index. We used modlAMP version 4.2.3(molecular design laboratory’s antimicrobial peptides package), a Python-based package for peptide data analysis to extract additional physicochemical parameters.45 Globaldecsriptor class of descriptors module of modlAMP was used for this purpose. We installed and used modlAMP using Spyder IDE (integrated development environment) of the Anaconda platform.46 Sequence data of AMPs and ACPs were converted into CSV (comma separated values) files to be read by Globaldecsriptor. The output of each parameter was stored in columns in a separate out.csv file. The collected (from APD3) and extracted (from modlAMP) physicochemical parameters of AMPs and ACPs were stored in excel spreadsheets (Supplementary files 1 and 2) for EDA.
Global analysis of AMPs and ACPs
EDA was performed on Spyder, a Python IDE available for data analysis on the Anaconda platform. Python’s pandas version 1.2.2 package47 and Globalanalysis class of the modlAMP analysis module were used for EDA. Datasets were checked for null values, dimensions, and variable types using standard commands of pandas. Descriptive statistics of datasets were calculated using the “describe ()” function of the pandas package.
To analyze the initially hidden pattern and trends graphically in basic physicochemical parameters of global datasets of AMPs and ACPs, we used Globalanalysis class of modlAMP. The sequences of AMPs and ACPs were stored in CSV formats to be read by Globalanalysis. Furthermore, box plots and heatmaps were generated for all physicochemical parameters using the pandas package for graphical analysis.
Subset analysis
Due to the unequal size of the datasets, we avoided a direct comparison of AMPs and ACPs. However, in order to gain insight into the influence of length on other physicochemical parameters, we partitioned each dataset into a length interval of 10, as described in the previous studies.48,49 Usually, the basis of partition depended on the significance of the length parameter in the peptide designing. The number of amino acid (aa) residues in the peptides influences other physicochemical parameters and even the activity of the designed peptides.50
The different subsets of peptides datasets were also analyzed for descriptive statistics using the “describe ()” function. The graphical analysis (boxplot and heatmaps) of subsets was done using the pandas package. Furthermore, we also calculated the amino acid compositions of subsets by using “aa.freq ()” function of modlAMP. The overall methodology used in the study is summarized in Fig. 1.
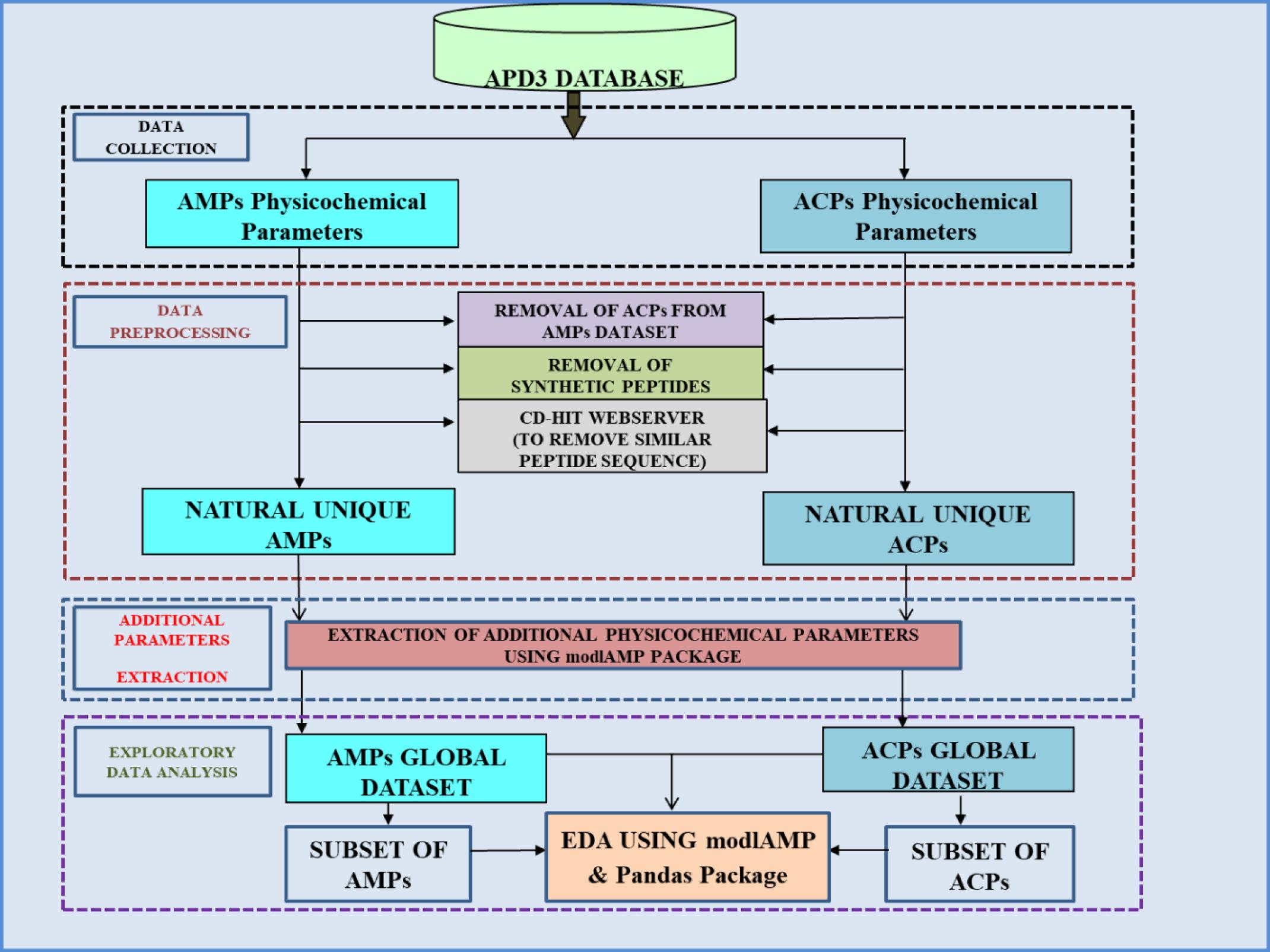
Fig. 1.
Overall methodology used for exploratory data analysis of physicochemical parameters of natural AMPs and ACPs.
.
Overall methodology used for exploratory data analysis of physicochemical parameters of natural AMPs and ACPs.
Results
Dataset characterization
A total of 2680 & 226 natural AMPs and ACPs were retrieved from APD3. Both datasets contain peptides with natural amino acids. Any synthetic peptides in the datasets were removed manually to get precisely natural AMPs and ACPs. After screening AMPs datasets for anticancer or antitumor active peptides, we finally got 2384 AMPs, whereas the ACPs dataset contains 226 ACPs. A total of ten physicochemical parameters of peptides were taken into account for the analysis in which five parameters (such as length, charge, Bowman index, hydrophobicity, and structure type) were retrieved from APD3 and the remaining five (molecular weight, isoelectric point (pI), instability index, aromaticity, and aliphatic index) were calculated using Globaldecsriptor. Out of ten parameters, structure type was the only object data type, and the remaining were of the variable data types. The complete datasets (global and subsets) used in the study can be found in Supplementary file 1 (AMPs dataset) and Supplementary file 2(ACPs dataset).
Trends and patterns in global analysis
To identify trends and patterns in the global AMP dataset, we have performed EDA in two steps. First, we calculated descriptive statistics of all physicochemical parameters to get the data’s statistical pattern. The anomaly observed in the statistical pattern of basic physicochemical parameters was visually inspected by the modlAMP. In the second step, we performed the graphical analysis of all physicochemical parameters using pandas to find the data patterns in each parameter.
Descriptive statistics of AMPs global dataset
The descriptive statistics of the AMPs dataset gave mean, minimum, interquartile range (Q1 (25%), Q2 (50%, median), and Q3 (75%)) and maximum value of all physicochemical parameters along with standard deviation (Table1). As previously mentioned in section 2.5, the length of AMP played an important role in determining the peptide activity. Additionally, the length also affects the mode of action, structure type, and cytotoxicity against red blood cells.50 The descriptive statistical analysis results showed that the mean or average length of AMPs is 34.12 aa, whereas the length of 2 aa and 183 aa are minimum and maximum, respectively. The mean length of 34.12 aa can relate to the mechanism of action of AMPs required for traversing the lipid bilayers. Furthermore, a recent study also suggests the approved peptides in this category range.49 Thus an AMP designed with this length can be of potent activity. The interquartile length range of AMPs was from 20 to 40 aa with a median length of 28 aa. So, designing de-novo or template-based AMPs within these length ranges may provide a potent active AMP.
Table 1.
Descriptive statistics of AMPs global dataset calculated using pandas package
|
Physicochemical parameters
|
Mean
|
SD
|
Min
|
Q1
|
Median, Q2
|
Q3
|
Max
|
| Length |
34.12 |
23.14 |
2 |
20 |
28 |
40 |
183 |
| Charge |
3.86 |
3.39 |
-12 |
2 |
3 |
5 |
30 |
| Hydrophobicity (%) |
42 |
13 |
0 |
34 |
42 |
51 |
87 |
| Boman index (kcal/mol) |
0.84 |
1.54 |
-3.35 |
-0.24 |
0.80 |
1.90 |
8.72 |
| Molecular weight (Da) |
3724.09 |
2538.84 |
294.35 |
2114.28 |
3081.69 |
4323.66 |
19842.55 |
| Isoelectric point (pI) |
9.54 |
1.89 |
2.42 |
8.55 |
10.02 |
10.70 |
13.53 |
| Instability index |
27.95 |
27.61 |
-43.43 |
8.28 |
24.52 |
43.18 |
190.38 |
| Aromaticity |
0.08 |
0.06 |
0 |
0.04 |
0.07 |
0.12 |
0.50 |
| Aliphatic Index |
92.00 |
45.88 |
0 |
56.66 |
90.67 |
121.21 |
256.25 |
SD, Standard deviation; Min, Minimum; Q1, First quartile or 25%; Q2, Second quartile or 50%; Q3, Third quartile or 75%; Max, Maximum.
The net charge is defined as the sum of all charges of ionizable groups of the peptide and is an essential parameter for AMP activity. The net charge of AMPs can be negative or positive, which can be altered during peptide design to change antimicrobial or hemolytic activity.50 We found that the natural AMPs contain an average charge of +3.86; this observation was consistent with the previous finding of Wang.20 The net cationic charge is required for the initial electrostatic attraction of AMPs to negatively charged phospholipid membranes.6 But a minimum and a maximum charge of -12 and +30 respectively were also evident from the analysis in the dataset. The interquartile range of charge lies between +2 to +5, with +3 being the median charge. These findings suggest that natural AMPs preferred a cationic charge range from +2 to +5. Therefore, modulation of AMPs towards this charge range may result in better interaction with negatively charged phospholipid membranes.
Hydrophobicity is the percentage of hydrophobic residues present in the peptide. Previous studies showed that most AMPs were 50% hydrophobic. This parameter plays a key role in the AMP-membrane interaction and also modulates the activity.51 We found mean hydrophobicity of 42% in the global dataset of AMP. Surprisingly, the minimum hydrophobicity in the dataset was found to be 0% and a maximum of 87%. The interquartile range of hydrophobicity spans 34% to 42%, with a median value of 51%. These results reflect the presence of variable hydrophobic amino acid residues in the natural peptides.
The Boman index was proposed by Boman to calculate the potential of peptides towards binding to other proteins (such as receptors) or membranes.52 A peptide having a calculated Boman index value of >2.48 kcal/mol is supposed to have a high binding potential.53 Furthermore, peptides having a high Boman index suggest their multiple roles in the cell due to their tendency to interact with different types of proteins.54 Surprisingly, we found a significantly lower mean value of 0.84 kcal/mol for AMPs. The minimum value was -3.35 kcal/mol, but the maximum value of 8.72 kcal/mol exceeds the threshold value of 2.48 kcal/mol. The interquartile range falls between -0.24 kcal/mol to 1.90 kcal/mol. Despite the active AMPs dataset, the statistical pattern of the Boman index value looks impertinent.
AMPs are usually low molecular weight (<10000 Da) peptides.55 Previous studies have demonstrated that the activity of peptides also relies on the molecular weight,56 which is the sum of the molecular weight of amino acid residues of the peptides.53 The peptide weight often limits its therapeutic value compared to small drug molecules; for example, a 5000 Da peptide production cost increases 10 fold compared to a 500 Da small molecule.57 Hence, understanding the natural peptides weight pattern along with other physicochemical parameters may help in cost-effective and efficient design. In our study, we found a mean weight of 3724.09 Da for AMPs. Active AMPs with a minimum weight of 294.35 Da and a maximum weight of 19842.55 Da were observed from the dataset. The weight of AMPs was found to be in the interquartile range of 2114.8 to 4323.66 Da. The pattern observed shows that an active AMP must not be of large molecular weight.
The pI is defined as the pH at which the net charge of the protein or peptide is equal to 0; this physicochemical parameter affects the solubility of peptides at different pH. The peptide becomes inactive if the pH of the solvent medium is equal to the pI of the peptide.53 The mean pI of AMPs is found to be 9.54 that suggests a preference for basic pH. In contrast to a basic mean value of pI, an acidic pI value of 2.42 and a higher basic pI value of 13.53 were also found from the dataset. Except for these extreme pI values, the interquartile range (8.55 to 10.70) revealed a preference for basic pH in most AMPs. The observed basic pH may be attributable to the high frequency of positive charge basic amino acid residues in the AMPs.
The instability index parameter was given by Guruprasad et al. This parameter is used to predict the stability of a protein in the in-vivo environment based on its amino acid composition. An index value of less than 40 for a peptide reflects the stability of the peptide.53,58 We found a mean value of 27.95 for the AMPs dataset with -43.43 and 190.38 as a minimum and maximum value, respectively. The interquartile range of 8.28 to 43.18 with a median of 24.52 also indicated that most AMPs are stable in-vivo.
According to Lobry and Gautier, aromaticity is the relative frequency of aromatic amino acids (F, W, and Y) in the protein or peptide sequence.59 Previous research works on the role of aromaticity described its importance in membrane interaction and structural integrity, which are critical to AMP activity.60,61 In another study, the role of aromatic interactions in the identification of biomolecules was highlighted, which may help in biomaterial research and molecular recognition.62 We found a mean value of 0.08 equivalent to 8% from the global dataset, but a minimum value of 0 reflects the lack of aromatic residues in some AMPs, whereas the interquartile range of 0.04 to 0.12 shows the presence of few aromatic residues.
Ikai purposed the aliphatic index parameter to measure the thermal stability of proteins by calculating the relative volume occupied by aliphatic side chains of amino acid residues, A, V, I, and L.63 The higher value of this parameter means higher heat stability.64 Descriptive statistics showed higher values of instability index in the range of 56.66 to 121.21. The mean value was found to be 92.
modlAMP analysis of AMPs global dataset
The initial descriptive statistics gave insight into the statistical pattern of physicochemical parameters but revealed unusual trends in the data (Table 1) compared to known patterns about physicochemical parameters.50,65 For example, the maximum length of 183 aa and the maximum charge of 30 are lesser-known facts when considering literature that shows AMPs length up to 100 aa and net charge in the range of +2 to +9.65 Therefore, to uncover the hidden pattern, we used the modlAMP package that contains Globalanalysis class for plotting the peptide dataset's basic physicochemical parameters. Moreover, it also gave amino acids frequency distribution in the peptide dataset.
The amino acid composition pattern observed in the AMPs dataset revealed the preference for G, K, L, I, and A (Fig. 2A) amino acids over other amino acids. Furthermore, the parameter length (Fig. 2C) was found to contain outliers in the dataset that may have resulted in unusual trends during the descriptive statistics. The graphical representations by modlAMP (Figs. 2A, 2B, 2C, 2D, 2E and 2F) uncover some of the unusual patterns observed in descriptive statistics. Additionally, it also provided the most favorable patterns for the basic physicochemical parameters; for instance, the global charge tends to accumulate around +2 to +5 (Fig. 2B) with the most frequent value of +4. The length of AMPs distributed around 20-40 aa (Fig. 2C) is consistent with descriptive statistics.
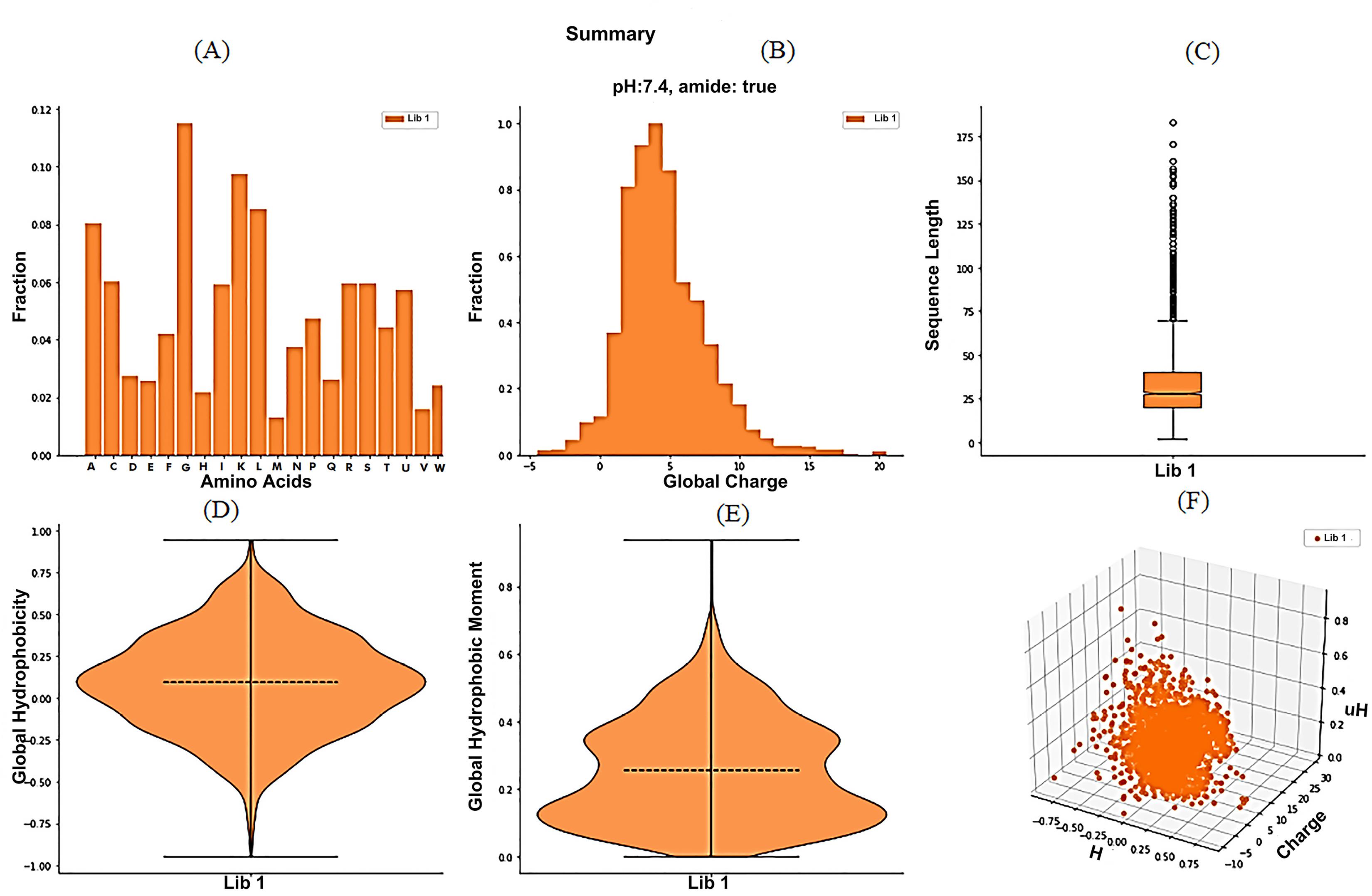
Fig. 2.
Global analysis of physicochemical parameters of AMPs global dataset plotted using modlAMP. The global summary plot showing the distribution of basic physicochemical parameters of AMPs like the relative abundance of individual amino acids (A), global charge (B), sequence length (C), global hydrophobicity (D), hydrophobic moment (E) and 3D scatter plot (F) that shows the correlation.
.
Global analysis of physicochemical parameters of AMPs global dataset plotted using modlAMP. The global summary plot showing the distribution of basic physicochemical parameters of AMPs like the relative abundance of individual amino acids (A), global charge (B), sequence length (C), global hydrophobicity (D), hydrophobic moment (E) and 3D scatter plot (F) that shows the correlation.
Interestingly, a graphical global analysis summary of the AMPs dataset by modlAMP shows that the data points in the hydrophobicity (Fig. 2D) and hydrophobic moment, µH (a measure of the helix amphipathicity)) (Fig. 2E) are uniformly distributed in the violin plot. Moreover, a 3D scatterplot (Fig. 2F) in the global summary also revealed a good correlation pattern among the hydrophobicity (H), charge, and hydrophobic moment (µH).
Graphical profile analysis of global AMPs dataset
The patterns observed above in the global summary plot of basic physicochemical parameters were not evident in the descriptive statistical analysis; thus, it prompted us to visually inspect each physicochemical parameter of the global AMPs dataset. This task constituted our second stage of global data analysis, where we have used the Python pandas package to plot the distribution of AMPs data points graphically. Box plots were obtained for each parameter to analyze the distribution pattern further and trends in AMPs global data set. Only object data type, i.e., structure type, was also considered in this analysis for which descriptive statistic was not possible. To explore the correlation among each physicochemical parameter, we plotted a heatmap. The box plots obtained by the pandas package for each physicochemical parameter were compiled with heatmap and amino acid composition patterns to form a graphical profile of the AMPs global dataset, as shown in Figs. 3A-L.
The graphical profile of the AMPs global data set helps to find the unusual pattern observed in descriptive statistical analysis. For instance, the boxplot of the most parameter contains outlier beyond the Q3 (Figs. 3A, 3B, 3C, 3D, 3E, 3G, 3H, 3I, 3J, 3K and 3L) except pI (Fig. 3F) in which outliers were detected below Q1. The correlation pattern in the parameters was found using a heatmap. The dark blue color shows a more positive correlation, whereas lighter blue to white colors shows a decrease or a negative correlation among the two variables. A value of 1 represents the highest correlation, whereas -1 shows a negative correlation. We found that parameters such as hydrophobicity, aromaticity, aliphatic index, and pI show a negative correlation with the length of AMPs (Fig. 3J). The charge was positively related to pI, whereas a negative correlation was found with the aliphatic index and hydrophobicity (Fig. 3J). This correlation analysis of natural AMPs will be helpful in the rational design of novel peptides. By analyzing the relationships between two variables in the natural dataset, we can better modulate the template peptides for activity.
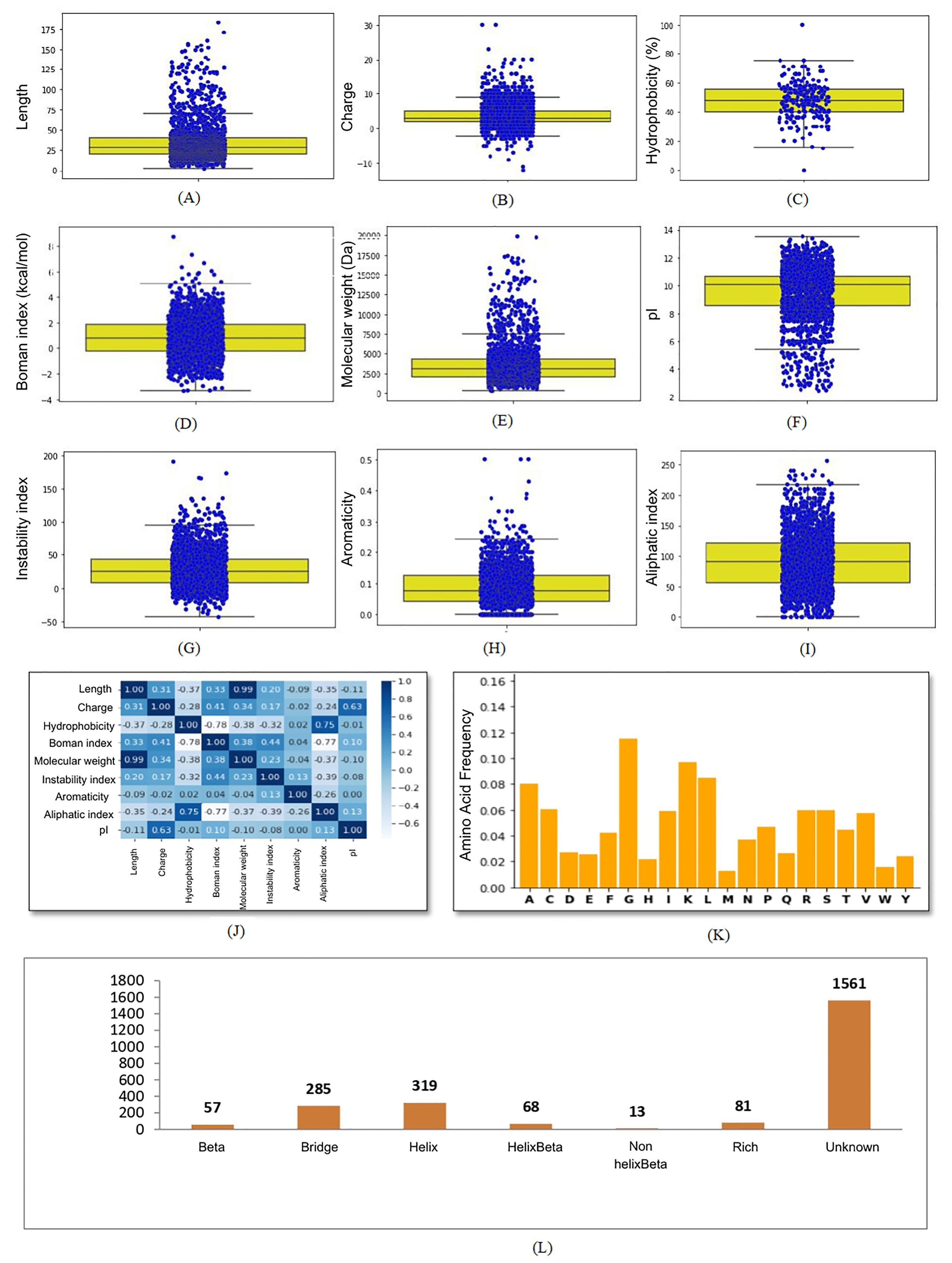
Fig. 3.
Graphical profile of global dataset of natural AMPs. (A) Distribution of length (B) charge (C) hydrophobicity (D) Boman index (E) molecular weight (F) pI (G) instability index (H) aromaticity (I) aliphatic index (J) correlation heatmap (K) amino acid frequency, and (L) structure type in global dataset of natural AMPs.
.
Graphical profile of global dataset of natural AMPs. (A) Distribution of length (B) charge (C) hydrophobicity (D) Boman index (E) molecular weight (F) pI (G) instability index (H) aromaticity (I) aliphatic index (J) correlation heatmap (K) amino acid frequency, and (L) structure type in global dataset of natural AMPs.
AMPs can adopt different secondary structures, such as α-helix, β-sheet, extended or mixed structures. α-helix peptides are unstructured in an aqueous solution, whereas β-sheet peptides are more ordered. The α-helical AMPs are usually more active against microbes due to their ability to undergo conformation change upon interaction with membrane.57,66 We found that secondary structures of the large number (1561 AMPs) of natural AMPs were unknown, and structure type α-helix (319) dominates over other structure types, as shown in Fig. 3L.
Descriptive statistics of ACPs global dataset
The same procedure which has been followed in AMPs dataset analysis was used for the ACPs dataset. The descriptive statistics of the ACPs dataset are summarized in Table 2 also revealed some unusual patterns and trends; for example, the maximum and minimum length of ACPs in the dataset is 111 aa and 5 aa, respectively. The mean or average length was found to be 26.47 aa. The maximum length of 111 aa among ACPs is an unusual pattern observed compared to the known facts.13 Therefore, the modlAMP analysis was conducted to explore the hidden information in the dataset.
Table 2.
Descriptive statistics of ACPs global dataset calculated using Pandas package
|
Physicochemical parameters
|
Mean
|
SD
|
Min
|
Q1
|
Median, Q2
|
Q3
|
Max
|
| Length |
26.47 |
14.44 |
5 |
17 |
25 |
31 |
111 |
| Charge |
3.09 |
3.11 |
-6 |
1 |
3 |
4.75 |
16 |
| Hydrophobicity (%) |
47 |
12 |
0 |
40 |
48 |
56 |
100 |
| Boman index (kcal/mol) |
0.44 |
1.55 |
-3.82 |
-0.74 |
0.25 |
1.28 |
8.33 |
| Molecular weight (Da) |
2871.14 |
1596.92 |
407.44 |
1744.84 |
2618.24 |
3279.91 |
12251.12 |
| Isoelectric point |
9.08 |
2.27 |
2.55 |
7.82 |
9.83 |
10.70 |
12.80 |
| Instability index |
26.61 |
27.77 |
-31.77 |
10.91 |
24.16 |
41.20 |
141.26 |
| Aromaticity |
0.076 |
0.059 |
0 |
0.037 |
0.065 |
0.107 |
0.307 |
| Aliphatic Index |
105.35 |
52.54 |
0 |
61.75 |
97.73 |
142.39 |
264.28 |
SD, Standard deviation; Min, Minimum; Q1, First quartile or 25%; Q2, Second quartile or 50%; Q3, Third quartile or 75%; Max, Maximum.
modlAMP analysis of ACPs global dataset
The amino acid composition of the ACPs dataset shows the preference of G, K, L, I, A, and C amino acids over others as shown in Fig. 4A. The most frequent charge on ACPs was +2 (Fig. 4B), and the most preferred charge range was +1 to +5. We observed outliers above the length of 50 aa (Fig. 4C), containing a peptide of 111 aa (largest ACP) as also depicted in descriptive statistics. Most ACPs were present in the length range of 17-31 aa as shown in the Fig. 4C. Global hydrophobicity and moment (µH) were also distributed uniformly as depicted in Fig. 4D and 4E, respectively. However, the correlation pattern in the 3D scatter plot (Fig. 4F) between charge, hydrophobicity, and moment (µH) shows minimal correlations compared to the AMPs dataset.
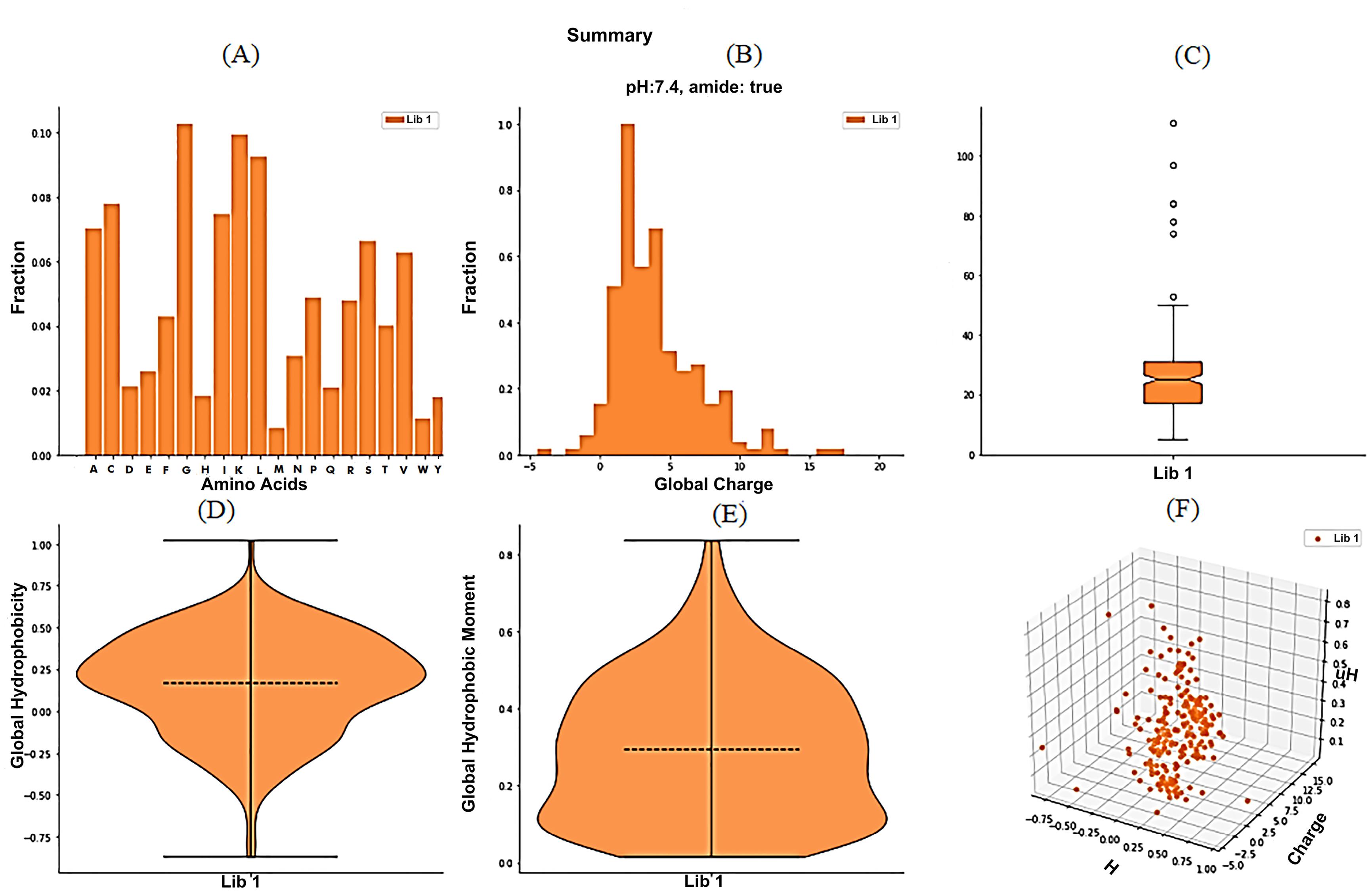
Fig. 4.
Global analysis of physicochemical parameters of ACPs global dataset plotted using modlAMP. The global summary plot showing the distribution of basic physicochemical parameters of AMPs like the relative abundance of individual amino acids (A), global charge (B), sequence length (C), global hydrophobicity (D), hydrophobic moment (E) and 3D scatter plot (F) that shows the correlation.
.
Global analysis of physicochemical parameters of ACPs global dataset plotted using modlAMP. The global summary plot showing the distribution of basic physicochemical parameters of AMPs like the relative abundance of individual amino acids (A), global charge (B), sequence length (C), global hydrophobicity (D), hydrophobic moment (E) and 3D scatter plot (F) that shows the correlation.
Graphical profile analysis of global ACPs dataset
Graphical profile of ACPs global dataset revealed the distribution of ACPs data points on the box plots. It was observed that compared to AMPs data points distribution, the numbers of outliers in ACPs are very few (Fig. 5A-5L). This observed pattern is may be due to the small set of ACPs dataset. But as observed in the AMPs dataset, the number of outliers for each parameter in the ACPs dataset also contains outlier beyond the Q3 except pI. We found the same correlation (dark brown color shows a more positive correlation, whereas lighter brown color shows a decrease or a negative correlation) among the different parameters in ACPs dataset as was present in the AMPs dataset but with different magnitude (Fig. 5J). The analysis of the structure type object variable in the ACPs dataset shows that the secondary structure type, α-helix was the most preferred conformation adopted by ACPs, as shown in Fig. 5L. Fig. 5A-5L shows thegraphical profile of the ACPs global dataset.
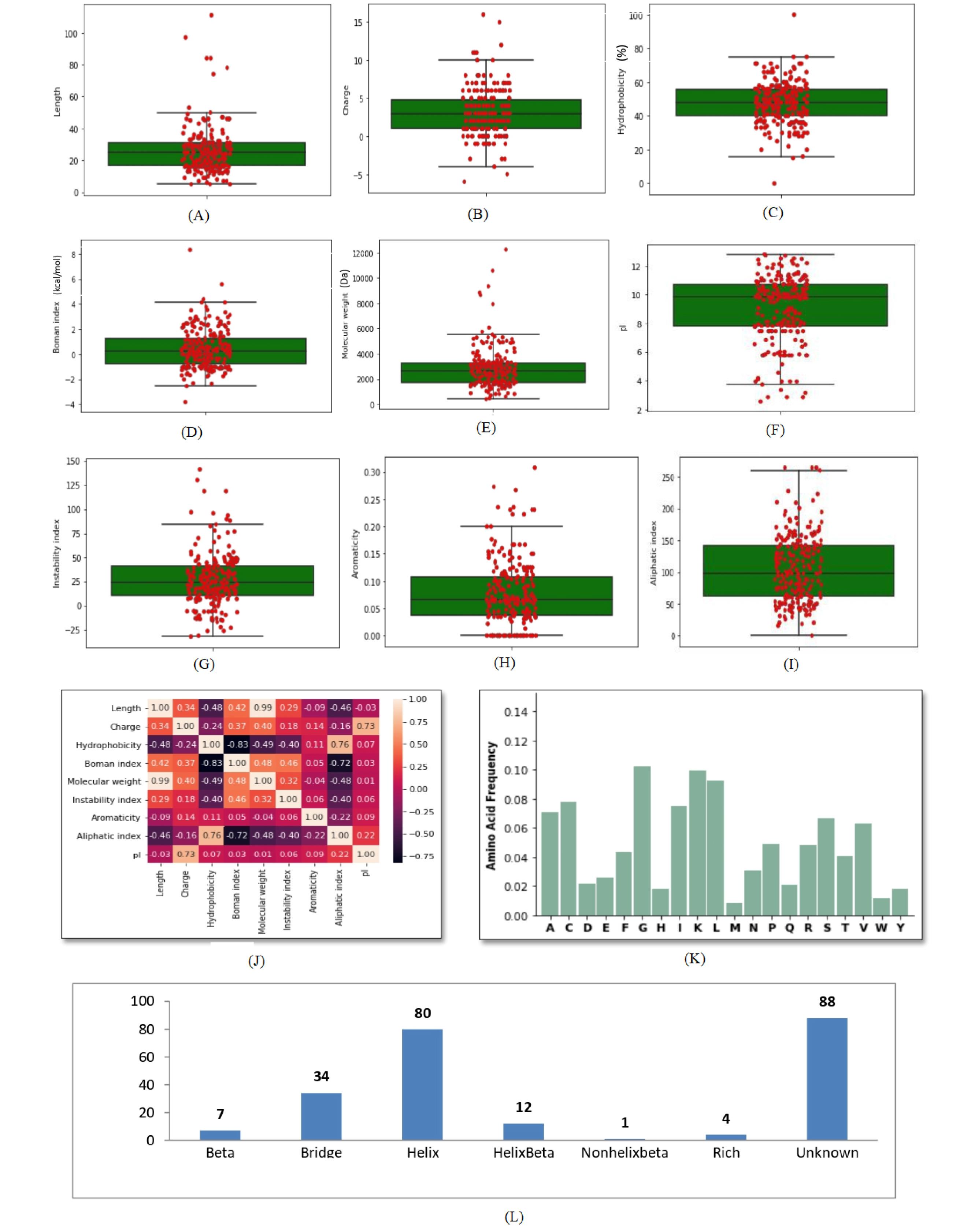
Fig. 5.
Graphical profile of global dataset of natural ACPs. (A) Distribution of length (B) charge (C) hydrophobicity (D) Boman index (E) molecular weight (F) pI (G) instability index (H) aromaticity (I) aliphatic index (J) correlation heatmap (K) amino acid frequency, and (L) structure type in global dataset of natural ACPs.
.
Graphical profile of global dataset of natural ACPs. (A) Distribution of length (B) charge (C) hydrophobicity (D) Boman index (E) molecular weight (F) pI (G) instability index (H) aromaticity (I) aliphatic index (J) correlation heatmap (K) amino acid frequency, and (L) structure type in global dataset of natural ACPs.
Trends and patterns in subsets analysis
Although descriptive statistics, a summary of the modlAMP global analysis, and a graphical profile analysis provide interesting facts about the global data sets, since the parameters length and amino acid composition are important in peptide design, we have created and analyzed subsets of AMPs and ACPs data sets based on lengths. The purpose of the subset analysis was to deduce the parameters of correlation among each parameter based on the length and the relative compositions of amino acids in the peptide. For this purpose, each dataset was divided into a length range of 10 intervals until each length range contains a significant number of peptides (in the case of AMPs >20 aa and ACP >10 aa)
Subsets analysis of AMPs
AMPs dataset was divided into 11 subsets (Supplementary file 1) in which the first 10 sets were of length interval 10 each. The 11th subset contains AMPs that were >100 aa and for which there were less than 20 AMPs in the length range. The subsets formed after splitting the AMPs dataset show that approximately 74% of natural AMPs present in the length range of 11-40. The maximum numbers of AMPs were present in the length range 21-30 (674 AMPs), followed by 11-20 (559 AMPs) and 31-40 (529 AMPs), as shown in Fig. 6. The amino acids distribution pattern in most AMPs subsets were found to be slightly different with the presence of high frequency of C, R, S, T, and V residues as compared to global AMPs datasets. However, the frequency pattern of G, K, L and I residues was found to be relatively consistent throughout all AMPs subsets which highlighted the significance of these amino acid residues even in the longer form of AMPs.
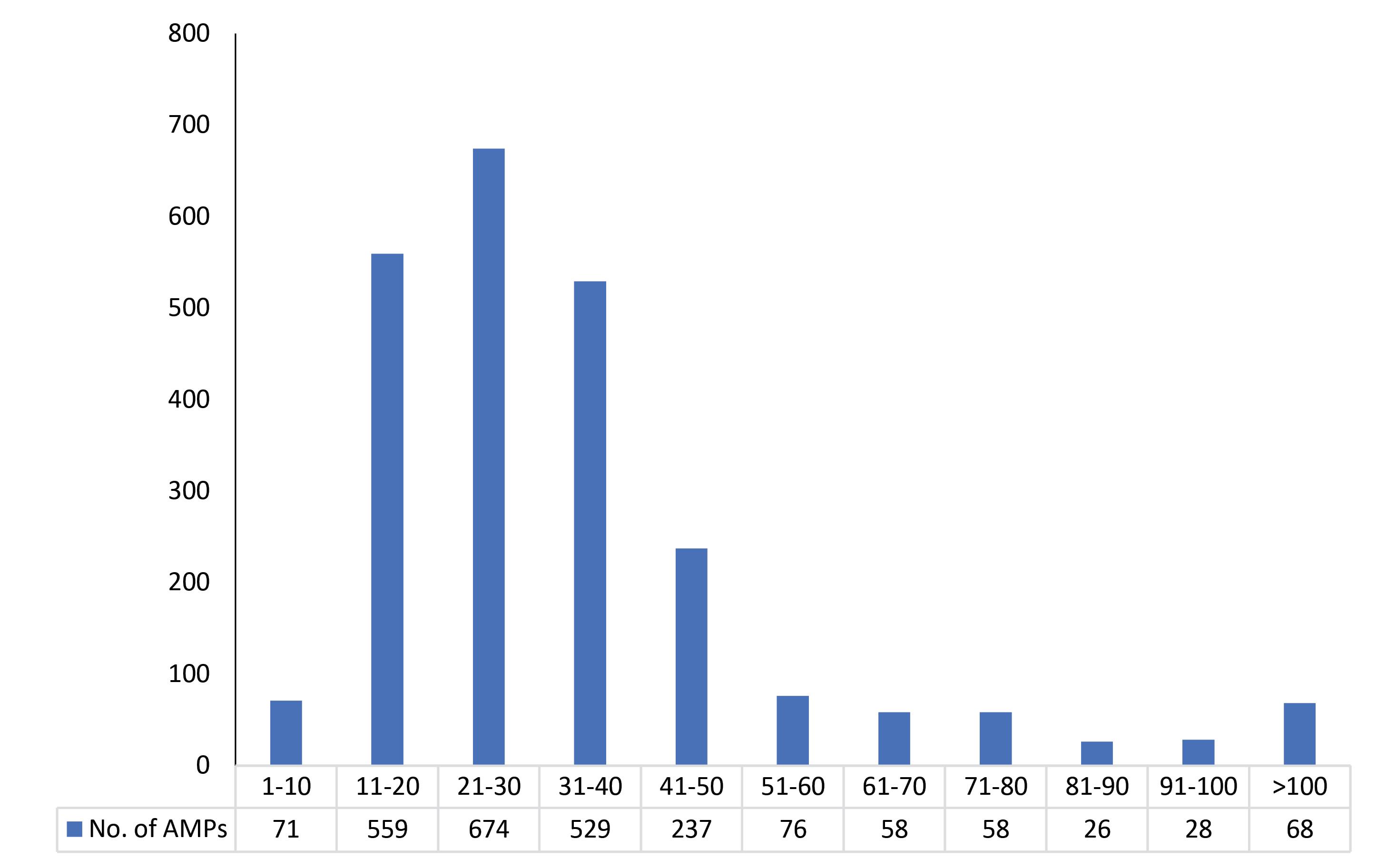
Fig. 6.
Number of AMPs present in each subset. The subset 21-30 contains the most AMPs, followed by subsets 11-20 & 31-40.
.
Number of AMPs present in each subset. The subset 21-30 contains the most AMPs, followed by subsets 11-20 & 31-40.
Similar to the amino acid composition analysis, we analyzed the other physicochemical parameters of each subset by calculating descriptive statistics and graphical profile analysis. The descriptive statistics of each AMPs subset can be found in Table S1, Supplementary file 3 and the graphical profile analysis of AMPs subsets in Figs. S1-S11, Supplementary file 4. The subsets 21-30 and 31-40 containing most of the AMPs and are included in the interquartile range of the global dataset, have been discussed here.
Trends in AMP subset 11-20
The subset 11-20 containing 559 AMPs (as shown in Fig. 6) shows the average length of 16 aa residues; the maximum and minimum lengths of residues were 20 and 11, respectively (Table S1, Supplementary file 3). This subset has 121 AMPs, of which peptides with 13 aa residues were the most frequent. Amino acid composition analysis (Fig. S2-K, Supplementary file 4) shows the same amino acid preferences as the global dataset (Fig. 3K) and subset 21-30 (Fig. S3-K, Supplementary file 4). Similar to subset 21-30, this subset also shows few outliers among other parameters, but the central distribution of AMPs data points in quartile regions was also observed, as shown in Fig. S2 (Panels A-L) (Supplementary file 4).
Trends in AMP subset 21-30
The analysis of subset 21-30 shows that the average lengths of AMPs are ~25 aa (Table S1, Supplementary file 3), less than the average length of the global dataset that is 34.12 aa. The maximum and minimum lengths are 30 aa, and 21 aa respectively (Table S1, Supplementary file 3), and the 134 AMPs were found to be 24 aa residues long, the most frequent count in the subset. The frequency distribution of amino acids in this subset was similar to that of the AMPs global dataset, with K, L, G, I, and A residues as most frequent (Fig. S3-K, Supplementary file 4). These results reflect that this subset largely determined the composition of AMP's global dataset. The other AMPs parameters, though centered in the quartile regions in the box plots, but also contains few outliers, as shown in Fig. S3 (Panels A-L) (Supplementary file 4).
Subsets analysis of ACPs
The global data set of ACPs (226 ACPs) was divided into 6 subsets (Supplementary file 2). The first five subsets were of length interval of 10, whereas the sixth subset (>50 residues) contains ACPs, which cannot be partitioned into subsets because of a smaller number of peptides (<10 ACPs). Segregating the ACPs global dataset into subsets unraveled the preferable ranges of length parameter. As shown in Fig. 7, most of the ACPs were found in the residue range 21-30 (83 ACPs), followed by 11-20 (71 ACPs) and 31-40 (34 ACPs). Though the number of ACPs in the global dataset (Supplementary file 2) is lesser than AMPs (Supplementary file 1), the preferred length range is similar in both types of peptides. Both therapeutic agents preferred the length range 21-30, followed by 11-20 and 31-40 as shown in Fig. 6 and Fig. 7, respectively.
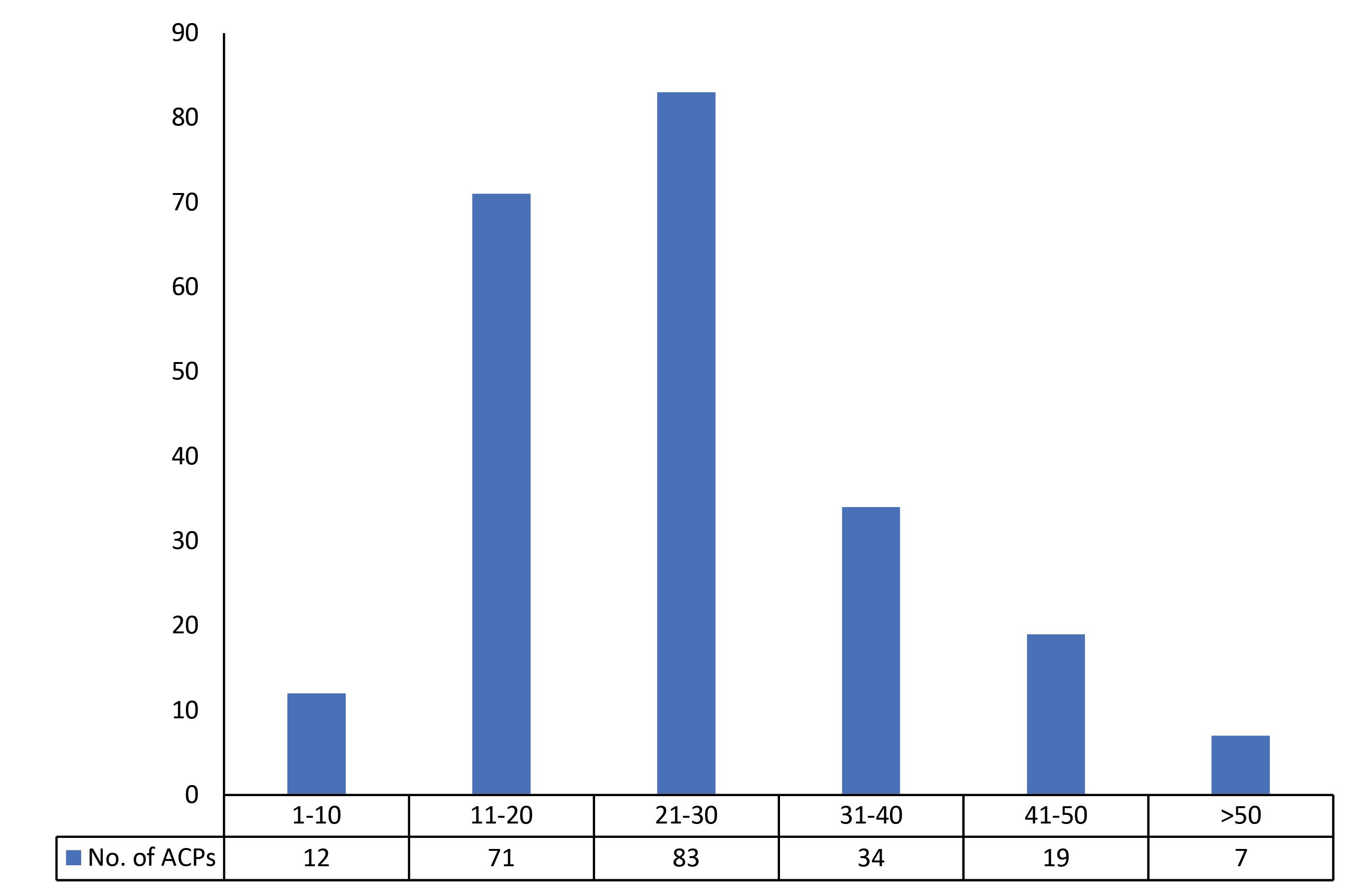
Fig. 7.
Number of ACPs present in each subset. The subset 21-30 contains the most AMPs, followed by subsets 11-20 & 31-40.
.
Number of ACPs present in each subset. The subset 21-30 contains the most AMPs, followed by subsets 11-20 & 31-40.
The analysis of amino acid patterns in ACP subsets revealed the dominance of the G, K, L, A and C as in the global ACPs dataset but the high frequencies of the R and S as compared to I was also seen in most of the subsets. The other physicochemical parameters of all six subsets (Supplementary file 2) of ACPs were analyzed for descriptive statistics, outlier detection by boxplot, and correlation analysis by plotting heat maps using the Pandas package. The subset 21-30 that contains most of the ACPs was discussed here. The remaining subset analysis was given in Table S2 (Supplementary file 3) and graphical profiles in Figs. S1-S6 (Supplementary file 5).
Trends in ACP subset 21-30
The analysis of subset 21-30 shows that the average lengths of ACPs are ~26.33 aa residues (Table S2, Supplementary file 3), similar to the average length of AMPs preferred subset 21-30 (Table S1, Supplementary file 3). The maximum and minimum lengths were found to be 30 aa and 21 aa residues, respectively (Table S2, Supplementary file 3). The frequency distribution of amino acids in this subset of ACPs was similar to that of the AMPs global dataset and subset 21-30, with K, L, G, I, and A residues as the most frequent ones as shown in Fig. S3-K, Supplementary file 5. But the presence of C and P were also observed. The other ACPs parameters are centered in the quartile regions in the box plots but also contain few outliers, as shown in Fig. S3 (Panels A-L) (Supplementary file 5).
Discussion
In the era of antibiotics and cancer drug resistance, the research community has put tremendous effort into designing safe and reliable AMPs and ACPs, respectively, which is evident from number of published literature over the last few years.57 Many research groups have collected the published peptide sequences and physicochemical parameters data to curate peptide databases, servers and machine learning algorithms like AntiCP 2.0, MLCPP 2.0, and xDeep-AcPEP, etc.16,17,24,29,67-71 Several previous efforts have used the peptide datasets for designing novel peptides. For instance, Mishra and Wang used the DFT (database filtering technique) approach, in which they exploited the most likely physicochemical parameters derived from APD3, to design novel potent peptides against Staphylococcus aureus.72 In another study by Pearson et al. (2016), potent peptides were designed against Mycobacterium tuberculosis using database-derived peptide physicochemical parameters.73
The evidence from the above studies shows that the pattern information of physicochemical parameters derived from datasets can be proved significant in designing potent novel peptides. However, there is still a need to quantitatively examine these peptides' physicochemical parameters that can help in future design and application.15 In this context, natural peptides that serve as a template for most of the designed synthetic AMPs and ACPs and for which a vast amount of data is available in the peptide databases can be exploited to retrieve quantitative information.6,20,66
Hence in the present study, we have done EDA of the physicochemical parameters of natural AMPs and ACPs retrieved from the APD3 database. To the best of our knowledge, this is the first attempt to perform EDA on natural AMPs and ACPs datasets. Furthermore, the study's uniqueness lies in the fact that both AMPs and ACPs datasets contain only natural peptides. Additionally, anticancer peptides were removed from the AMPs dataset to get unique AMPs. The major limitation of the study was the unequal size of AMPs and ACPs datasets. Hence, we avoided the direct comparison of descriptive statistical analysis results of the two datasets and emphasized the independent interpretation of these derived parameters.
The descriptive statistical analysis of global AMPs and ACPs datasets revealed a uniform pattern of interquartile ranges (Q1-Q3) with some unobvious trends in the data. For instance, in AMPs global dataset each parameter shows some extreme lower and upper values (length (min = 2 aa, max = 183 aa) charge (min = -12, max =30), detailed analysis is shown in Table 1. The extreme lower and upper trends were also observed in the ACPs global dataset (length (min = 5 aa, max = 111 aa) charge (min = -6, max =16), detailed analysis is shown in Table 2. This quantitative information derived from statistical analysis of each parameter can be used for rational-based peptide design. In short, the mean, median, and Q1, Q2 and Q3 values of the different peptide parameters can be combined to design novel peptides that may likely have more stability and activity. For instance, an AMP or ACP can be designed with a selection of either minimum, maximum, Q1-Q3, or mean length combining with the minimum, maximum, Q1-Q3, or mean observations of other parameters like charge, hydrophobicity, Boman index, isoelectric point, and instability index etc.
In the era of AMR, the computer-aided peptide design approaches such as template and de-novo based has been widely used to design the novel optimized peptide analogs.74 Both of these approaches rely on the pattern information of amino acid frequency that plays a crucial role in selecting an individual or group of amino acids residues for the substitutions in selected template or a seed fragment for new peptide analogs. The amino acid frequency-based pattern information assisted the substitutions of residues during the computational design of new analogs with improve activity in many previous works.75,76 Hence the amino acid composition information derived from the natural AMPs and ACPs datasets in this study could be used for the selection of the most probable amino acids for substitutions in design of template or de-novo based computational approaches.
The frequency pattern of amino acid composition in AMPs and ACPs is similar (G, K, L, I, and A), with the exception of C in the ACPs. A recent study pointed to the role of cysteine in the stabilization of the extracellular domain or motif structure during the interaction of ACP on the cell surface receptor.9 Thus, these most probable amino acids could be used for the substitution during computer-aided design of more potent AMPs and ACPs. The EDA of the AMPs subgroups revealed the high probability of C, R, S, T, and V residues compared to the global dataset pattern whereas the R and S were dominant as compared to I in the ACPs subsets. These patterns of amino acid frequency observed in subsets of natural AMPs and ACPs in our study could be helpful for the length specific computer-aided peptide design by the substitutions of amino acids which are more frequent in the particular range length of AMPs and ACPs datasets.
Moreover, these observed frequency patterns of amino acid compositions can be used in combination with the quantitative statistical values of other physicochemical parameters such as length, charge, hydrophobicity etc for the computer aided rational design of AMPs and ACPs with improve activity.
Given the significance of the length parameter in the peptide therapeutics,77 we formed subsets and analyzed each subset of AMPs and ACPs. The partition of AMPs and ACPs in the subsets shows that both follow a similar pattern. For instance, it has been observed that subset 21-30 contains most of AMPs and ACPs followed by 11-20 and 31-40 as compared to other AMPs and ACPs subsets. Moreover, due to the significance of subsets information in the computer aided design of peptides as shown by few recent studies78 we also extracted descriptive statistics (Table S1 & S2, Supplementary file 3) and prepared a graphical profile (Figs. S1-S11, Supplementary file 4 & Figs. S1-S6, Supplementary file 5) of each subset that can be used in the computer-aided peptide design. Different researchers working on AMPs and ACPs can use the pattern observed among the different parameters to design novel peptide agents.
Conclusion
To succeed in designing more effective and stable AMPs & ACPs for therapeutics, the research community now needs to look for the data-based selection of the peptide physicochemical parameters. Here in this work, our study provides a blueprint of physicochemical parameters of natural AMPs and ACPs datasets. Some of the broad conclusions drawn from the results are: most natural AMPs and ACPs were present in the particular length ranges of 21-30 followed by 11-20 and 31-40, the frequency pattern of amino acid composition in natural AMPs and ACPs was found to be similar (G, K, L, I, and A) except the presence of C in the ACPs. However, AMPs and ACPs subsets were found to have high abundance of C, R, S, T, V and R and S amino acid residues respectively as compared to their respective global datasets. The alpha-helix conformation was found to be preferred by both AMPs and ACPs.
The present observations found in global and subset datasets of AMPs and ACPs might help to design more potent and stable peptides. These statistical and graphical profiles of AMPs and ACPs can impact the decision-making while selecting the design parameters for computer-aided design of AMPs and ACPs for instance, preferred length ranges pattern, amino acid compositions among the global and subsets, the correlation pattern as heatmaps, quartile ranges of parameters and the information of which preferred secondary structure types adopted by AMPs and ACPs global and subsets can prove advantageous during tuning the different physicochemical parameters for novel analogs designs. Moreover, the outliers in graphical profiles will help the in detection of anomalies among each parameter.
Furthermore, the methodology used in this work can be used for EDA of other peptide datasets such as anti-allergic, anti-hypertensive, anti-diabetic, anti-inflammatory and immunomodulatory peptides, etc. Additionally, our future work includes designing novel AMPs or ACPs based on the derived parameters.
Research Highlights
What is the current knowledge?
√ Natural antimicrobial and anticancer peptides can be used for the design of novel potent analogs.
√ The pattern information of physicochemical parameters can be used for the computer-aided peptide design.
What is new here?
√ Natural AMPs and ACPs global and subsets were used for deriving the pattern information from the physicochemical parameters.
√ The patterns and trends in physicochemical parameters of AMPs and ACPs were presented using different graphics and descriptive statistics.
Acknowledgements
The authors gratefully acknowledge the APD3 database for the use of raw data.
Competing Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical Statement
Not applicable. This paper does not involve research on humans.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Supplementary Materials
Supplementary file 1 contains AMPs global and subset datasets used in the study.
(xlsx)
Supplementary file 2 contains ACPs global and subset datasets used in the study.
(xlsx)
Supplementary file 3 contains Table S1 and S2.
(doc)
Supplementary file 4 contains figs. S1-S11.
(pdf)
Supplementary file 5 contains figs. S1-S6.
(pdf)
References
- World Health Organization (WHO), Ten threats to global health in 2019. https://www.who.int/vietnam/news/feature-stories/detail/ten-threats-to-global-health-in-2019. Accessed on 15 April, 2022.
- World Health Organization (WHO), Global antimicrobial resistance and use surveillance system (GLASS) report 2021. https://www.who.int/publications/i/item/9789240027336. Accessed on 15 April, 2022.
- Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist 2018; 11:1645-1658. doi: 10.2147/IDR.S173867 [Crossref] [ Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394-424. doi: 10.3322/caac.21492 [Crossref] [ Google Scholar]
- Ke X, Shen L. Molecular targeted therapy of cancer: The progress and future prospect. Frontiers in Laboratory Medicine 2017; 1:69-75. doi: 10.1016/j.flm.2017.06.001 [Crossref] [ Google Scholar]
- Li J, Koh JJ, Liu S, Lakshminarayanan R, Verma CS, Beuerman RW. Membrane active antimicrobial peptides: Translating mechanistic insights to design. Front Neurosci 2017; 11:73. doi: 10.3389/fnins.2017.00073 [Crossref] [ Google Scholar]
- Gabernet G, Gautschi D, Müller AT, Neuhaus CS, Armbrecht L, Dittrich PS. In silico design and optimization of selective membranolytic anticancer peptides. Sci Rep 2019; 9:11282. doi: 10.1038/s41598-019-47568-9 [Crossref] [ Google Scholar]
- Lei J, Sun LC, Huang S, Zhu C, Li P, He J. The antimicrobial peptides and their potential clinical applications. Am J Transl Res 2019; 1 1:3919-3931. [ Google Scholar]
- Chiangjong W, Chutipongtanate S, Hongeng S. Anticancer peptide: Physicochemical prope rty, functional aspect and trend in clinical application (Review). Int J Oncol 2020; 57:678-696. doi: 10.3892/ijo.2020.5099 [Crossref] [ Google Scholar]
- Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol 2006; 24:1551-1557. doi: 10.1038/nbt1267 [Crossref] [ Google Scholar]
- Gaspar D, Veiga AS, Castanho MA. From antimicrobial to anticancer peptidesA review. Front Microbiol 2013; 4:294. doi: 10.3389/fmicb.2013.00294 [Crossref] [ Google Scholar]
- Hoskin DW, Ramamoorthy A. Studies on anticancer activities of antimicrobial peptides. Biochim Biophys Acta 2008; 1778:357-375. doi: 10.1016/j.bbamem.2007.11.008 [Crossref] [ Google Scholar]
- Felício MR, Silva ON, Gonçalves S, Santos NC, Franco OL. Peptides with dual antimicrobial and anticancer activities. Front Chem 2017; 5:5. doi: 10.3389/fchem.2017.00005 [Crossref] [ Google Scholar]
- Torres MDT, Sothiselvam S, Lu TK, de la Fuente-Nunez C. Peptide Design Principles for Antimicrobial Applications. J Mol Biol 2019; 431:3547-3567. doi: 10.1016/j.jmb.2018.12.015 [Crossref] [ Google Scholar]
- Mahlapuu M, Björn C, Ekblom J. Antimicrobial peptides as therapeutic agents: opportunities and challenges. Crit Rev Biotechnol 2020; 40:978-992. doi: 10.1080/07388551.2020.1796576 [Crossref] [ Google Scholar]
- Jhong JH, Chi YH, Li WC, Lin TH, Huang KY, Lee TY. dbAMP: an integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data. Nucleic Acids Res 2019; 47:D285-D297. doi: 10.1093/nar/gky1030 [Crossref] [ Google Scholar]
- Fan L, Sun J, Zhou M, Zhou J, Lao X, Zheng H. DRAMP: a comprehensive data repository of antimicrobial peptides. Sci Rep 2016; 6:24482. doi: 10.1038/srep24482 [Crossref] [ Google Scholar]
- Kapoor P, Singh H, Gautam A, Chaudhary K, Kumar R, Raghava GPS. Tumorhope: a database of tumor homing peptides. PLoS One 2012; 7:e35187. doi: 10.1371/journal.pone.0035187 [Crossref] [ Google Scholar]
- Shi G, Kang X, Dong F, Liu Y, Zhu N, Hu Y. DRAMP 30: an enhanced comprehensive data repository of antimicrobial peptides. Nucleic acids research 2022; 50:D488-D496. doi: 10.1093/nar/gkab651 [Crossref] [ Google Scholar]
- Wang G. The antimicrobial peptide database provides a platform for decoding the design principles of naturally occurring antimicrobial peptides. Protein Sci 2020; 29:8-18. doi: 10.1002/pro.3702 [Crossref] [ Google Scholar]
- Wang, G, Vaisman II, van Hoek ML. Machine Learning Prediction of Antimicrobial Peptides. In: Simonson T (eds). Computational Peptide Science. Methods in Molecular Biology. New York: Humana; 2022. p. 2405:1-37. 10.1007/978-1-0716-1855-4_1.
- Magana M, Pushpanathan M, Santos AL, Leanse L, Fernandez M, Ioannidis A. The value of antimicrobial peptides in the age of resistance. Lancet Infect Dis 2020; 20:e216-e230. doi: 10.1016/S1473-3099(20)30327-3 [Crossref] [ Google Scholar]
- Liu S, Fan L, Sun J, Lao X, Zheng H. Computational resources and tools for antimicrobial peptides. J Pept Sci 2017; 23:4-12. doi: 10.1002/psc.2947 [Crossref] [ Google Scholar]
- Li Y, Chen Z. RAPD: a database of recombinantly-produced antimicrobial peptides. FEMS Microbiol Lett 2008; 289:126-129. doi: 10.1111/j.1574-6968.2008.01357.x [Crossref] [ Google Scholar]
- Waghu FH, Barai RS, Gurung P, Idicula-Thomas S. CAMPR3: a database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res 2016; 44:D1094-D1097. doi: 10.1093/nar/gkv1051 [Crossref] [ Google Scholar]
- Brahmachary M, Krishnan SPT, Koh JLY, Khan AM, Seah SH, Tan TW. ANTIMIC: a database of antimicrobial sequences. Nucleic Acids Res 2004; 32:586-589. doi: 10.1093/nar/gkh032 [Crossref] [ Google Scholar]
- Di Luca M, Maccari G, Maisetta G, Batoni G. BaAMPs: The database of biofilm-active antimicrobial peptides. Biofouling 2015; 31:193-199. doi: 10.1080/08927014.2015.1021340 [Crossref] [ Google Scholar]
- Tyagi A, Tuknait A, Anand P, Gupta S, Sharma M, Mathur D. CancerPPD: A database of anticancer peptides and proteins. Nucleic Acids Res 2015; 43:D837-D843. doi: 10.1093/nar/gku892 [Crossref] [ Google Scholar]
- Piotto SP, Sessa L, Concilio S, Iannelli P. YADAMP: Yet another database of antimicrobial peptides. Int J Antimicrob Agents 2012; 39:346-351. doi: 10.1016/j.ijantimicag.2011.12.003 [Crossref] [ Google Scholar]
- Wang G, Li X, Wang Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res 2016; 44:D1087-D1093. doi: 10.1093/nar/gkv1278 [Crossref] [ Google Scholar]
- Ramazi S, Mohammadi N, Allahverdi A, Khalili E, Abdolmaleki P. A review on antimicrobial peptides databases and the computational tools. Database (Oxford) 2022; 2022:baac011. doi: 10.1093/database/baac011 [Crossref] [ Google Scholar]
- Behrens JT. Principles and Procedures of Exploratory Data Analysis. Psychological Methods 1997; 2:131-160. doi: 10.1037/1082-989X.2.2.131 [Crossref] [ Google Scholar]
- Good IJ. Exploratory data analysis. Joural of Statistical Computation and Simulation 1990; 37:243-245. doi: 10.1080/00949659008811311 [Crossref] [ Google Scholar]
- Vigni M Li, Durante C, Cocchi M. Exploratory Data Analysis. In: Marini F, eds. Data Handling in Science and Technology. Netherlands: Elsevier; 2013. p. 55-126. 10.1016/B978-0-444-59528-7.00003-X.
- Jebb AT, Parrigon S, Woo SE. Exploratory data analysis as a foundation of inductive research. Human Resource Management Review 2017; 27:265-276. doi: 10.1016/j.hrmr.2016.08.003 [Crossref] [ Google Scholar]
- Shreffler, J, Huecker MR. Exploratory Data Analysis: Frequencies, Descriptive Statistics, Histograms, and Boxplots. in StatPearls. StatPearls Publishing, Treasure Island (FL); 2022. Available from: https://www.ncbi.nlm.nih.gov/books/NBK557570/.
- Dsouza J, Senthil Velan S. Using Exploratory Data Analysis for Generating Inferences on the Correlation of COVID-19 cases. 2020 11th International Conference on Computing, Communication and Networking Technologies. (ICCCNT); 2020. 10.1109/ICCCNT49239.2020.9225621.
- Pérez C, Claveria O. Natural resources and human development: Evidence from mineral-dependent African countries using exploratory graphical analysis. Resources Policy 2020; 65:101535. doi: 10.1016/j.resourpol.2019.101535 [Crossref] [ Google Scholar]
- Tummers J, Catal C, Tobi H, Tekinerdogan B, Leusink G. Coronaviruses and people with intellectual disability: an exploratory data analysis. J Intellect Disabil Res 2020; 64:475-481. doi: 10.1111/jir.12730 [Crossref] [ Google Scholar]
- Bondu R, Cloutier V, Rosa E, Roy M. 2020An exploratory data analysis approach for assessing the sources and distribution of naturally occurring contaminants (F, Ba, Mn, As) in groundwater from southern Quebec (Canada). Applied Geochemistry 2020; 114:104500. doi: 10.1016/j.apgeochem.2019.104500 [Crossref] [ Google Scholar]
- Hamed MM, Al-Eideh BM. An exploratory analysis of traffic accidents and vehicle ownership decisions using a random parameters logit model with heterogeneity in means. Anal Methods Accid Res 2020; 25:100116. doi: 10.1016/j.amar.2020.100116 [Crossref] [ Google Scholar]
- Satwika MV, Sushma DS, Jaiswal V, Asha S, Pal T. The Role of Advanced Technologies Supplemented with Traditional Methods in Pharmacovigilance Sciences. Recent Pat Biotechnol 2021; 15:34-50. doi: 10.2174/1872208314666201021162704 [Crossref] [ Google Scholar]
- Zeiss R, Hafner S, Schönfeldt-Lecuona C, Connemann BJ, Gahr M. Drug-Associated Liver Injury Related to Antipsychotics: Exploratory Analysis of Pharmacovigilance Data. J Clin Psychopharmacol 2022. 10.1097/JCP.0000000000001576.
- Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010; 26:680-682. doi: 10.1093/bioinformatics/btq003 [Crossref] [ Google Scholar]
- Müller AT, Gabernet G, Hiss JA, Schneider G. modlAMP: Python for antimicrobial peptides. Bioinformatics 2017; 33:2753-2755. doi: 10.1093/bioinformatics/btx285 [Crossref] [ Google Scholar]
- Anaconda Inc. Anaconda Guide. 2019. Available from: https://www.anaconda.com/distribution/.
- McKinney W, Team PD. Pandas - Powerful Python Data Analysis Toolkit. 2015.
- Shoombuatong W, Schaduangrat N, Nantasenamat C. Unraveling the bioactivity of anticancer peptides as deduced from machine learning. EXCLI J 2018; 17:734-752. doi: 10.17179/excli2018-1447 [Crossref] [ Google Scholar]
- Lau JL, Dunn MK. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg Med Chem 2018; 26:2700-2707. doi: 10.1016/j.bmc.2017.06.052 [Crossref] [ Google Scholar]
- Bahar AA, Ren D. Antimicrobial peptides. Pharmaceuticals (Basel) 2013; 6:1543-1575. doi: 10.3390/ph6121543 [Crossref] [ Google Scholar]
- Nakajima Y. Mode of action and resistance mechanisms of antimicrobial macrolides. In: Omura S, ed. Macrolide Antibiotics, Second Edition: Chemistry, Biology, and Practice. San Diego: Academic Press; 2003. p. 453-499. 10.1016/B978-012526451-8/50011-4.
- Wang X, Mishra B, Lushnikova T, Narayana JL, Wang G. Amino Acid Composition Determines Peptide Activity Spectrum and Hot-Spot-Based Design of Merecidin. Adv Biosyst 2018; 2:1700259. doi: 10.1002/adbi.201700259 [Crossref] [ Google Scholar]
- Osorio D, Rondon-Villarreal P, Torres R. Peptides: A package for data mining of antimicrobial peptides. The R Journal 2015. 7: 4-14.
- Azad MA, Huttunen-Hennelly HEK, Friedman CR. Bioactivity and the first transmission electron microscopy immunogold studies of short de novo-designed antimicrobial peptides. Antimicrob Agents Chemother 2011; 55:2137-2145. doi: 10.1128/AAC.01148-10 [Crossref] [ Google Scholar]
- Jakubczyk A, Karas M, Rybczynska-Tkaczyk K, Zielinska E, Zielinski D. Current trends of bioactive peptides - New sources and therapeutic effect. Foods 2020; 9:846. doi: 10.3390/foods9070846 [Crossref] [ Google Scholar]
- Fjell CD, Hiss JA, Hancock REW, Schneider G. Designing antimicrobial peptides: Form follows function. Nat Rev Drug Discov 2012; 11:37-51. doi: 10.1038/nrd3591 [Crossref] [ Google Scholar]
- Mahlapuu M, Håkansson J, Ringstad L, Björn C. Antimicrobial peptides: An emerging category of therapeutic agents. Front Cell Infect Microbiol 2016; 6:194. doi: 10.3389/fcimb.2016.00194 [Crossref] [ Google Scholar]
- Guruprasad K, Reddy BV, Pandit MW. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng 1990; 4:155-161. doi: 10.1093/protein/4.2.155 [Crossref] [ Google Scholar]
- Lobry JR, Gautier C. Hydrophobicity, expressivity and aromaticity are the major trends of amino-acid usage in 999 escherichia coli chromosome-encoded genes. Nucleic Acids Res 1994; 22:3174-3180. doi: 10.1093/nar/22.15.3174 [Crossref] [ Google Scholar]
- Datta A, Bhattacharyya D, Singh S, Ghosh A, Schmidtchen A, Malmsten M. Role of aromatic amino acids in lipopolysaccharide and membrane interactions of antimicrobial peptides for use in plant disease control. J Biol Chem 2016; 291:13301-13317. doi: 10.1074/jbc.M116.719575 [Crossref] [ Google Scholar]
- Lee E, Kim JK, Jeon D, Jeong KW, Shin A, Kim Y. Functional roles of aromatic residues and helices of papiliocin in its antimicrobial and anti-inflammatory activities. Sci Rep 2015; 5:12048. doi: 10.1038/srep12048 [Crossref] [ Google Scholar]
- Waters ML. Aromatic interactions in peptides: Impact on structure and function. Biopolymers 2004; 76:435-445. doi: 10.1002/bip.20144 [Crossref] [ Google Scholar]
- Ikai A. Thermostability and aliphatic index of globular proteins. J Biochem 1980; 88:1895-1898. doi: 10.1093/oxfordjournals.jbchem.a133168 [Crossref] [ Google Scholar]
- Li RF, Lu ZF, Sun YN, Chen SH, Yi YJ, Zhang HR. Molecular Design, Structural Analysis and Antifungal Activity of Derivatives of Peptide CGA-N46. Interdiscip Sci 2016; 8:319-326. doi: 10.1007/s12539-016-0163-x [Crossref] [ Google Scholar]
- Kang SJ, Kim DH, Mishig-Ochir T, Lee BJ. Antimicrobial peptides: Their physicochemical properties and therapeutic application. Arch Pharm Res 2012; 35 409-413. 10.1007/s12272-012-0302-9.
- Barreto-Santamaría A, Patarroyo ME, Curtidor H. Designing and optimizing new antimicrobial peptides: all targets are not the same. Crit Rev Clin Lab Sci 2019; 56:351-373. doi: 10.1080/10408363.2019.1631249 [Crossref] [ Google Scholar]
- Agrawal P, Bhagat D, Mahalwal M, Sharma N, Raghava G. AntiCP 20: an updated model for predicting anticancer peptides. Brief Bioinform 2021; 22:bbaa153. doi: 10.1093/bib/bbaa153 [Crossref] [ Google Scholar]
- Jaiswal V, Negi A, Pal T. A review on current advances in machine learning based diabetes prediction. Prim Care Diabetes 2021; 15:435-443. doi: 10.1016/j.pcd.2021.02.005 [Crossref] [ Google Scholar]
- Rathore A, Saini A, Kaur N, Singh A, Dutta O, Bamhotra M et al. Amino Acid Composition and Charge Based Prediction of Antisepsis Peptides by Random Forest Machine Learning Algorithm. bioRxiv 2021: 461860. 10.1101/2021.09.26.461860.
- Manavalan B, Patra MC. Manavalan B, Patra MCMLCPP 20: An Updated Cell-penetrating Peptides and Their Uptake Efficiency Predictor. J Mol Biol 2022; 434:167604. doi: 10.1016/j.jmb.2022.167604 [Crossref] [ Google Scholar]
- Chen J, Cheong HH, Siu S. xDeep-AcPEP: Deep Learning Method for Anticancer Peptide Activity Prediction Based on Convolutional Neural Network and Multitask Learning. J Chem Inf Model 2021; 61:3789-3803. doi: 10.1021/acs.jcim.1c00181 [Crossref] [ Google Scholar]
- Mishra B, Wang G. Ab initio design of potent anti-MRSA peptides based on database filtering technology. J Am Chem Soc 2012; 134:12426-12429. doi: 10.1021/ja305644e [Crossref] [ Google Scholar]
- Pearson CS, Kloos Z, Murray B, Tabe E, Gupta M. Combined bioinformatic and rational design approach to develop antimicrobial peptides against Mycobacterium tuberculosis. Antimicrob Agents Chemother 2016; 60:2757-2764. doi: 10.1128/AAC.00940-15 [Crossref] [ Google Scholar]
- Farhadi T, Hashemian SM. Computer-aided design of amino acid-based therapeutics: a review. Drug Des Devel Ther 2018; 12:1239-1254. doi: 10.2147/DDDT.S159767 [Crossref] [ Google Scholar]
- Madanchi H, Akbari S, Shabani AA, Sardari S, Farmahini Farahani Y. Alignment-based design and synthesis of new antimicrobial Aurein-derived peptides with improved activity against Gram-negative bacteria and evaluation of their toxicity on human cells. Drug Dev Res 2019; 80:162-170. doi: 10.1002/ddr.21503 [Crossref] [ Google Scholar]
- Bobde SS, Alsaab FM, Wang G, Van Hoek ML. Ab initio Designed Antimicrobial Peptides Against Gram-Negative Bacteria. Front Microbiol 2021; 12:715246. doi: 10.3389/fmicb.2021.715246 [Crossref] [ Google Scholar]
- Liscano Y, Oñate-Garzón J, Delgado JP. Peptides with dual antimicrobial–anticancer activity: Strategies to overcome peptide limitations and rational design of anticancer peptides. Molecules 2020; 25:4245. doi: 10.3390/molecules25184245 [Crossref] [ Google Scholar]
- Ripperda T, Yu Y, Verma A, Klug E, Thurman M. Improved Database Filtering Technology Enables More Efficient Ab Initio Design of Potent Peptides against Ebola Viruses. Pharmaceuticals (Basel) 2022; 15:521. doi: 10.3390/ph15050521 [Crossref] [ Google Scholar]