Bioimpacts. 13(4):313-321.
doi: 10.34172/bi.2023.27490
Original Article
Exosomes are involved in the intercellular transfer of rapamycin resistance in the breast cancer cells
Yuri Yu. Shchegolev Investigation, Visualization, Writing – review & editing, 
Danila V. Sorokin Formal analysis, Methodology, Validation, Writing – review & editing,
Alexander M. Scherbakov Methodology, Resources, Validation, Visualization, Writing – review & editing, , * 
Olga E. Andreeva Investigation, Visualization, Writing – review & editing, 
Diana I. Salnikova Investigation, Writing – review & editing, 
Ekaterina I. Mikhaevich Investigation, Writing – review & editing, 
Margarita V. Gudkova Formal analysis, Writing – review & editing, 
Mikhail A. Krasil’nikov Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, 
Author information:
Department of Experimental Tumor Biology, Institute of Carcinogenesis, N.N. Blokhin National Medical Research Center of Oncology, Kashirskoye Shosse 24, Moscow 115522, Russia
Abstract
Introduction:
Resistance to chemotherapy and/or irradiation remains one of the key features of malignant tumors, which largely limits the efficiency of antitumor therapy. In this work, we studied the progression mechanism of breast cancer cell resistance to target drugs, including mTOR blockers, and in particular, we studied the exosome function in intercellular resistance transfer.
Methods:
The cell viability was assessed by the MTT assay, exosomes were purified by successive centrifugations, immunoblotting was used to evaluate protein expression, AP-1 activity was analyzed using reporter assay.
Results:
In experiments on the MCF-7 cell line (breast cancer) and the MCF-7/Rap subline that is resistant to rapamycin, the capability of resistant cell exosomes to trigger a similar rapamycin resistance in the parent MCF-7 cells was demonstrated. Exosome-induced resistance reproduces the changes revealed in MCF-7/Rap resistant cells, including the activation of ERK/AP-1 signaling, and it remains for a long time, for at least several months, after exosome withdrawal. We have shown that both the MCF-7 subline resistant to rapamycin and cells having exosome-triggered resistance demonstrate a stable decrease in the expression of DNMT3A, the key enzyme responsible for DNA methylation. Knockdown of DNMT3A in MCF-7 cells by siRNA leads to partial cell resistance to rapamycin; thus, the DNMT3A suppression is regarded as one of the necessary elements for the development of acquired rapamycin resistance.
Conclusion:
We propose that DNA demethylation followed by increased expression of key genes may be one of the factors responsible for the progression and maintenance of the resistant cell phenotype that includes exosome-induced resistance.
Keywords: Breast cancer, DNMT3A, Signaling, Rapamycin resistance, Exosomes
Copyright and License Information
© 2023 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Microvesicles, later called "exosomes", were first discovered while studying the process of maturation of reticulocytes. Initially, microvesicles were not considered a mechanism for intercellular communication; they were mainly assigned the function of removing metabolites from cells into the surrounding space.1,2
The investigation of exosome functioning noticeably expanded with the discovery of the exosome capability to comprise and transport its cargo into receiving cells. Proteomic mass spectrometry and western blot analysis provided a real breakthrough in the deciphering of exosome cargo obtained from various sources.3-8 Proteins, which have an increased content in exosomes, according to the online databases of EVs ExoCarta, Vesiclepedia, and EVPedia,9-11 include ribosomal, cytoskeletal (tubulins, actins, mesin, cofilin-1, profilin-1, radixin, ezrin), Rab, cytosolic, plasma membrane and heat shock proteins (Hsp70 and Hsp90), syntenin-1, metabolic enzymes (lactate dehydrogenase, enolases, peroxiredoxins, glyceraldehyde-3-phosphate dehydrogenase, pyruvate kinase), annexins, integrins, Tsg101, tetraspanins (CD81, CD63, CD9), ALIX, and proteins participating in vesicular traffic. During the development of malignant neoplasms, the contents of patient’s exosomes significantly change.12-15 The tumor affects the content (cargo) of proteins and oligonucleotides in the serum exosomes. Moreover, the blood serum of oncologic patients has been used as a source of exosomes with upregulation of several miRNAs. For instance, the considerable upregulation of miR-10a-5p, miR-19b-3p, miR-215-5p, and miR-18a-5p in exosomes of gastric cancer patients in comparison with healthy control samples was revealed by Houman Kahroba et al.13
Recent studies of exosomes have shown that the development of multidrug resistance (MDR) in tumor cells occurs, among other things, with the participation of exosomes, which, due to their nature, can ensure the spread of resistance throughout the pool of tumor cells.12 Thus, in experiments on in vitro cultured breast cancer cells, microvesicles isolated from resistant cells contained an increased amount of P-glycoprotein, one of the main ABC transporters, and the cultivation of sensitive cells with such exosomes leads to the accumulation of exogenous P-glycoprotein in cells and a significant increase in drug resistance.16,17 Further development of these studies has enabled the substantial increase in the amount of groups of biologically active molecules, which are transferred by the microvesicles of resistant cells and are able to trigger drug resistance in receiving cells. First, this statement is true for microRNAs, which control the degree of MDR gene expression. Thus, miR-27a and miR-451 identified with the development of MDR were revealed in exosomes of resistant cell sublines.18 However, microRNAs that mediate exosome-dependent resistance are able to be included implicitly in the control of MDR genes when (anti)apoptotic signaling genes are among their targets. A similar effect was found for miR-222, a broad-spectrum microRNA, one of whose targets is PTEN, a phosphatase that blocks anti-apoptotic PI3K signaling.19 Another microRNA found in exosomes, miR-433, leads to the growth of drug resistance via synthesis suppression of retinoblastoma protein and CDK6 (cyclin-dependent kinase 6) in recipient cells.20
As a result, either the accumulation of ABC transporters or the corresponding microRNAs and mRNAs in exosomes, or, most likely, a combination of these factors, can serve as a decisive factor, which ultimately leads to the formation and maintenance of a resistant phenotype in recipient cells.
Recently, exosomes have been shown to be engaged not only in the expansion of MDR but also in the development of resistance to anticancer medicines, including targeted and hormonal drugs. Thus, the participation of exosomes in the induction of hormonal resistance in recipient cell lines has been shown by us and others. Individual exosomal microRNAs have been identified, the accumulation of which leads to the development of resistance in cells.21-26 The role of epigenomic changes, including methylation of DNA, which is associated with the growth and maintenance of exosome-induced resistance in tumor cell lines, is being actively studied.27-29
In general, the discovery of exosomes, and specifically, their capability to transport biomaterial from one cell to another, has significantly altered current ideas about the mechanism of tumor transformation and progression. The impact of exosomes on the development of a cell phenotype specific to a resistant cell line is one of the best accomplishments in this field. Sensitive cells are certainly able to receive resistance from corresponding cells with the use of exosomes due to a still largely unclear mechanism based on the transport of particular signaling molecules into cells: miRNAs, mRNAs, transporter proteins, etc. Previously, the central place in the development of acquired resistance of tumors was given to the selection of preexisting or emerging de novo cells with pronounced resistance to drugs; however, after the discovery of exosomes, more attention has been paid to the mechanisms of directed changes in tumor cells under the action of exosomes.
Here, we have demonstrated the ability of exosomes from rapamycin-resistant derivatives of the MCF-7 breast cancer cell line to induce a similar resistance in the parent cells. Exosome-induced resistance reproduces changes similar to those in resistant cells, including the activation of ERK/AP-1 signaling and stable suppression of DNMT3A expression. Knockdown of DNMT3A in parent MCF-7 cells by siRNAs leads to the partial cell resistance to rapamycin, showing the involvement of DNA demethylation in the generation of the resistant cell phenotype.
Materials and Methods
Reagents
High glucose-containing (4.5 g/L) DMEM medium was provided by PanEco. Fetal bovine serum (FBS) was purchased from HyClone. The MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] reagent and dimethyl sulfoxide (DMSO) were obtained from AppliChem. Primary antibodies against PTEN, ERK 1/2, p-ERK 1/2, estrogen receptor α (ERα), mTOR, p-mTOR, DNMT1, DNMT3A, and Bcl-2 were received from Cell Signaling Technology. The secondary antibodies were obtained from Jackson ImmunoResearch. Scrambled nonspecific siRNA and DNMT3A specific siRNA in addition to the appropriate antisense RNA oligonucleotides were acquired from Syntol. Lipofectamine 2000 from Thermo Fisher Scientific Inc was used for transfection. The Milli-Q water purification system (Millipore) was used to produce ultrapure water.
Cell cultures and obtaining the drug-resistant subline
The MCF-7 (ATCC HTB-22TM) human breast cancer cell line was acquired from ATCC. Using Gordiz (http://gordiz.ru/), the cells were authenticated by morphology and STR profiling. We purchased rapamycin and metformin from Merck.
The development of the MCF-7/Rap rapamycin-resistant subline from the original MCF-7 cells was achieved by long-term cell incubation with rapamycin as described earlier.30 All of the following experiments were performed on the MCF-7/Rap line within at least 2 months after drug removal. High glucose-containing (4.5 g/L) DMEM medium with the addition of 7% FBS (HyClone) was used for the MCF-7 and MCF-7/Rap cell cultivation under the conditions of 37 °C and 5% CO2. To identify the cell reaction to rapamycin, the cell cultures were exposed to rapamycin within 72 hours in the abovementioned DMEM medium containing 7% FBS; then, cell viability was assessed. The MTT assay with modifications31 was applied to estimate cellular growth, as characterized previously.32 DMSO was used for formazan crystal dissolution.
Transient transfection and measurement of reporter gene activity
Plasmids containing the luciferase reporter gene controlled by the AP-1-responsive element were used for cellular transfection to assess the transcriptional activity of AP-1. The duration of transfection was 4 hours. Cells were transfected at 37 °C in the presence of Lipofectamine 2000. For this, complex formation between 0.8 µL of Lipofectamine 2000 and 0.4 µg of DNA was performed for transfection of one well (24-well plate, Corning). The β-galactosidase plasmid was also used for cell transfection for regulation of the efficiency and possible toxicity.33 After 24 hours of transfection, the following tests were conducted. The luciferase activity was quantitatively evaluated in compliance with a regular protocol (Promega) using Tecan Infinite M200 Pro and expressed in conditional units as the ratio of the luciferase/galactosidase activity, as described previously.33
Cell treatment with exosome preparations
Exosomes were separated from the conditioned medium using differential ultracentrifugation. Immunoblotting with antibodies against CD9 (marker of exosomes) was applied to characterize exosomes, as we have previously reported.21 The nanoparticle tracking analysis (NTA) using the Nanosight LM10 HS-BF instrument (Nanosight, Ltd.) was used to measure the size and concentration of the purified exosomes.
The addition of exosomes to cells was performed before cell attachment to the 24-well plate. The amount of exosomes was defined by NTA, and the protein level was estimated by Bradford reagent (Merck). Exosomes were added in PBS to 1.5 mL of cell suspension to make an ultimate concentration of 1.7 μg/mL of exosomal protein or CI95 = (5.5 ± 0.3) × 109 vesicles/mL once every 72 hours during cell seeding for 2 weeks.
Western blot analysis
During incubation, the cell cultures reached 80% of the monolayer phase, and 150 μL of lysis buffer was applied for dissolving. The buffer contained Tris-HCl at pH 7.4 (50 mM), NaCl (150 mM), Igepal CA-630 (1%), dithiothreitol (1 mM), ethylenediamine tetraacetate (1 mM), pepstatin, leupeptin, aprotinin (1 μg/mL), sodium orthovanadate, and sodium fluoride (1 mM). Cooling of the samples on ice for 20 minutes was performed before centrifugation. The conditions of the procedure included 10 000 × g for 10 minutes at 4 °C. The Bradford method was used to determine the total protein level and normalization of loading. Polyacrylamide gel at a concentration of 10% was utilized for protein electrophoresis (SDS-PAGE). Cell lysates containing 40 µg of the protein were separated under reducing conditions in accordance with the standard protocol. Of note, the study of exosome samples included non-reducing conditions in comparison with cell analysis, and the sample buffer did not include mercaptoethanol. Then, the proteins were transferred to a nitrocellulose membrane (SantaCruz) for immunoblotting.
The treatment of membranes was performed with 5% nonfat milk (AppliChem) solution in TBS buffer at a pH of 7.5 containing Tris (20 mM), NaCl (500 mM), and 0.1% Tween-20. Furthermore, the incubation of membranes with primary antibodies was performed overnight at 4 °C.
Primary antibodies against PTEN, ERK 1/2, p-ERK 1/2, ERα, mTOR, p-mTOR, DNMT1, DNMT3A, and Bcl-2 were used. The antibodies to GAPDH were used as a control for sample loading. The secondary antibodies were presented by the respective IgGs (Jackson ImmunoResearch) conjugated with horseradish peroxidase. The ECL reagent was prepared in compliance with Mruk’s protocol34 and was utilized for signal identification. An ImageQuant LAS4000 system was used for chemiluminescence detection (GE HealthCare).
The immunoblotting of exosomes or cell samples containing 10 μg of protein was performed in accordance with the protocol from our recent work.21 Primary antibodies to CD9 (Millipore), were used for exosome detection. Densitometry for the tested proteins was carried out using ImageJ software.
Transfection of small interfering RNA oligonucleotides
Scrambled nonspecific siRNA and DNMT3A specific siRNA in addition to the appropriate antisense RNA oligonucleotides were applied. The annealing buffer at a pH of 7.5 containing Tris-HCl (10 mM), EDTA (1 mM), and NaCl (50 mM) was used to dissolve these RNAs to a concentration of 100 µM. Equimolar mixtures of oligonucleotides were annealed at 95°C followed by cooling to 25 °C. Lipofectamine 2000 was used for the RNA oligonucleotide transfection to achieve a final RNA concentration of 50 nM.
Sequences used for siRNA treatment: scrambled sense UUCUCCGAACGUGUCACGUTT, scrambled anti-sense ACGUGACACGUUCGGAGAATT; DNMT3A sense GCCAAGGUCAUUGCAGGAATT, DNMT3A anti-sense UUCCUGCAAUGACCUUGGCTT.
Statistical analysis
Three technical replicates were necessary for all experiments, which were repeated three times. Microsoft Excel and GraphPad were used for statistical analysis. If not stated explicitly, results are presented as the mean ± standard deviation (SD). Student's test and ANOVA were applied for statistical comparisons. A P value less than 0.05 is regarded as statistically reliable.
Results
Development and characteristics of the rapamycin-resistant MCF-7 subline
MCF-7 breast cancer cell culture was used for in vitro experiments. MCF-7/Rap subline resistant to rapamycin was established under prolonged incubation of the MCF-7 cells with the appropriate drug, as described previously.30 Briefly, the MCF-7 cells were cultured with the addition of 1–4 μM rapamycin over 3 months, and the cell reaction to rapamycin was assessed by MTT assays. The selected cell subline was characterized by the acquired resistance to the anti-proliferative rapamycin effect that was preserved within at least 6 months after rapamycin withdrawal (Fig. 1A). Furthermore, MCF-7/Rap cells demonstrated cross-resistance to metformin, which is another inhibitor of mTOR/S6K signaling (Fig. 1B).
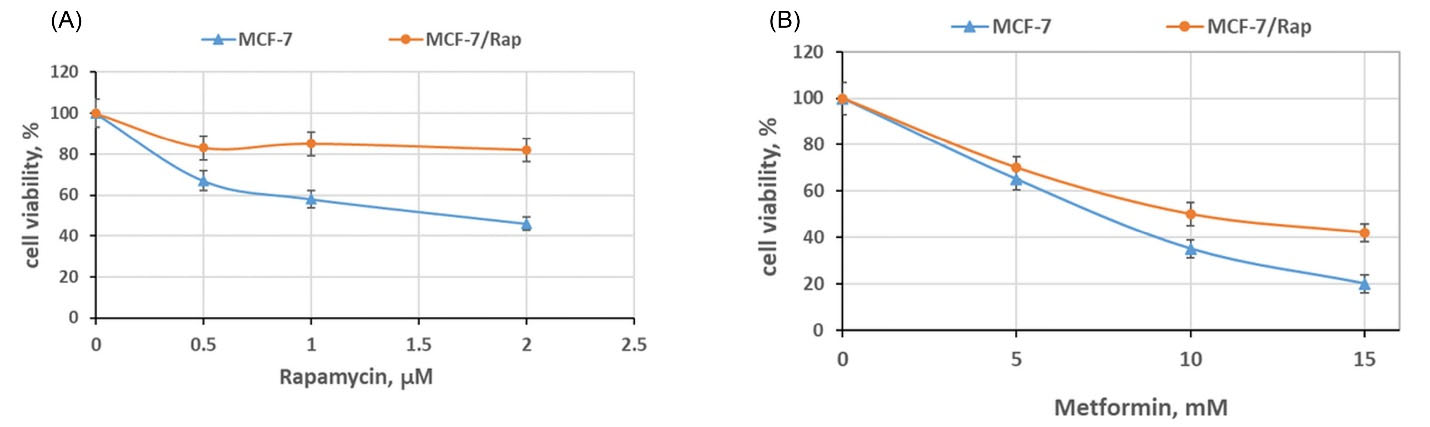
Fig. 1.
The sensitivity of the rapamycin-resistant MCF-7/Rap subline to rapamycin and metformin. The MCF-7 and MCF-7/Rap cells were treated with the indicated doses of rapamycin (A) or metformin (B) for 3 days, and the cell viability was assessed by MTT assay. Data represent the mean ± S.D. of three independent experiments. Then, 100% was set as the viability of cells treated with the vehicle control; P < 0.05 MCF-7 versus MCF-7/Rap cells.
.
The sensitivity of the rapamycin-resistant MCF-7/Rap subline to rapamycin and metformin. The MCF-7 and MCF-7/Rap cells were treated with the indicated doses of rapamycin (A) or metformin (B) for 3 days, and the cell viability was assessed by MTT assay. Data represent the mean ± S.D. of three independent experiments. Then, 100% was set as the viability of cells treated with the vehicle control; P < 0.05 MCF-7 versus MCF-7/Rap cells.
Purification and description of the exosomes
The MCF-7 and MCF-7/Rap-conditioned medium was the source for exosome isolation by differential ultracentrifugation. The relevant controls were involved in determining the exosomal functions upon recommendation of ISEV35 and Takov et al.36 The NTA was used to study the exosomes and has demonstrated that the typical range of the exosome size varied from 50 to 300 nm (Fig. 2A).
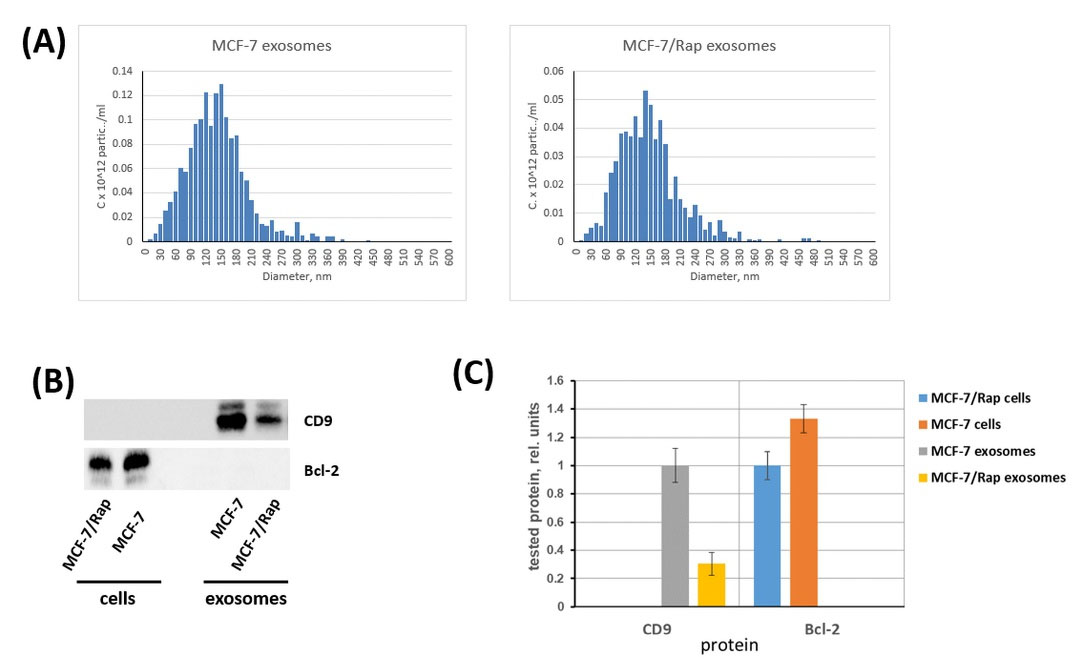
Fig. 2.
The characterization of the exosomes. (A) Size and concentration of purified exosomes were detected via nanoparticle tracking analysis (NTA) using the Nanosight LM10 HS-BF instrument (Nanosight, Ltd.). (B) Immunoblotting of exosomal marker CD9 in the MCF-7 and MCF-7/Rap exosomes versus MCF-7 and MCF-7/Rap cell lysates. As a non-exosomal marker, the Bcl-2 protein was chosen. (C) Densitometry for immunoblotting data (n=3) was performed using ImageJ software (Wayne Rasband, NIH). The protocol for analysis was provided by The University of Queensland with the recommendations from Taylor et al.37
.
The characterization of the exosomes. (A) Size and concentration of purified exosomes were detected via nanoparticle tracking analysis (NTA) using the Nanosight LM10 HS-BF instrument (Nanosight, Ltd.). (B) Immunoblotting of exosomal marker CD9 in the MCF-7 and MCF-7/Rap exosomes versus MCF-7 and MCF-7/Rap cell lysates. As a non-exosomal marker, the Bcl-2 protein was chosen. (C) Densitometry for immunoblotting data (n=3) was performed using ImageJ software (Wayne Rasband, NIH). The protocol for analysis was provided by The University of Queensland with the recommendations from Taylor et al.37
The exosome samples were investigated by Western blotting, which revealed the main marker of exosomes CD9 in all preparations. Bcl-2, a non-exosomal (cell) marker, was applied to the studied cell lines to determine the preparation purity (Fig. 2B,C), as previously recommended.35
Effect of the exosomes on the cell reaction to rapamycin
Differential centrifugation of the MCF-7 and MCF-7/Rap-conditioned media after 72 hours of cell incubation was used for exosome preparation, as described in the Methods section. To modulate the conditions that were used earlier to select the MCF-7/Rap subline that was resistant to rapamycin, the MCF-7 cell culture was treated with exosomes for at least 30 days. Exosomes were dispersed in PBS and added to 1.5 mL of cellular suspension to achieve a final concentration of 3.5 μg/mL once every 72 hours during cell seeding. After a month of the cell treatment with exosomes isolated from MCF-7 and MCF-7/Rap cell cultures, the recipient cells (named MCF-7/exoC and MCF-7/exoR) were placed in a standard medium without the exosomes. The cell growth was maintained over a month; then, the MTT assay was used to evaluate the sensitivity of the cell cultures to the drugs metformin and rapamycin. We found that prolonged cultivation of the breast cancer cell line with exosomes from the MCF-7/Rap cells led to partial resistance to both rapamycin and metformin (MCF-7/exoR subline) compared with the cells treated with the exosomes that were derived from the original MCF-7 cell culture (MCF-7/exoC subline) (Fig. 3A,B).

Fig. 3.
The influence of the exosome treatment on the cell sensitivity to rapamycin and metformin. The MCF-7 cells were cultured with exosomes from MCF-7 and MCF-7/Rap cells within 30 days. Afterwards, the recipient cells (named as MCF-7/exoC and MCF-7/exoR) were transferred to a standard exosome-free medium, and cell growth was maintained for 30 days; then, the cell sensitivity to rapamycin (A) and metformin (B) was evaluated by MTT assay. Data represent the mean ± SD of three independent experiments. Then, 100% was set as the viability of cells treated with the vehicle control. P < 0.05 MCF-7 versus MCF-7/Rap cells.
.
The influence of the exosome treatment on the cell sensitivity to rapamycin and metformin. The MCF-7 cells were cultured with exosomes from MCF-7 and MCF-7/Rap cells within 30 days. Afterwards, the recipient cells (named as MCF-7/exoC and MCF-7/exoR) were transferred to a standard exosome-free medium, and cell growth was maintained for 30 days; then, the cell sensitivity to rapamycin (A) and metformin (B) was evaluated by MTT assay. Data represent the mean ± SD of three independent experiments. Then, 100% was set as the viability of cells treated with the vehicle control. P < 0.05 MCF-7 versus MCF-7/Rap cells.
Cell signaling and rapamycin resistance
To further compare the features of the cell signaling associated with acquired rapamycin resistance or exosome-induced resistance, the expression of the key signaling proteins was analyzed. We revealed no changes in the level of the proteins of ERα, PTEN, or mTOR signaling, while a noticeable increase in the ERK activation (phosphorylation) in both variants of the resistant cells was found (Fig. 4A). The transcription factors of the Maf, ATF, Fos, and Jun subfamilies are related to the activator protein-1 (AP-1) family and support proliferation and cell viability.38,39 The luciferase-based gene reporter analysis was used to estimate the transcriptional activity of AP-1. The reporter analysis identified an increase in the transcriptional activity of AP-1 in MCF-7 cells treated with exosomes that were derived from control and resistant samples. AP-1 transcriptional activity in MCF-7/Rap and MCF-7/exoR cells was higher compared to that in the corresponding control samples (Fig. 4B). This observation supports the key role of the ERKs/AP-1 pathway in resistance development. Of note, the revealed protein changes were maintained for at least 6 months after withdrawal of rapamycin or exosome treatment. This verifies the idea of probable epigenetics participation in the progression of the stable resistant subline.
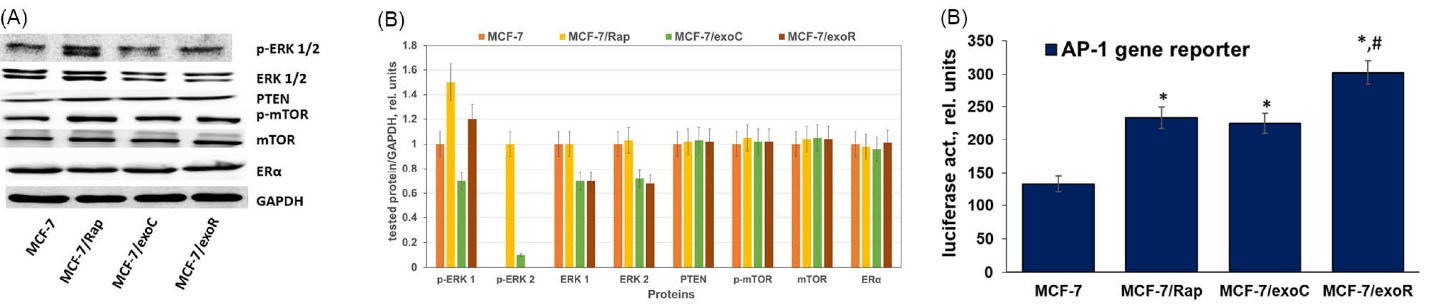
Fig. 4.
Signaling profile of the MCF-7 cells and resistant sublines. (A) Western blotting analysis of total cell extracts. Protein loading was controlled by membrane hybridization with GAPDH Abs. The blot represents the results of one of three similar experiments. (B) Densitometry for immunoblotting data (n=3) was performed using ImageJ software. (C) Reporter analysis of the AP-1 transcriptional activity. The cells were transfected with the AP-1 plasmid containing the luciferase reporter gene under the AP-1-responsive elements and the β-galactosidase plasmid. Then, 24 h after transfection, the luciferase and β-galactosidase activities were determined as described in the Methods section. The relative luciferase activity was calculated in arbitrary units as the ratio of the luciferase to the galactosidase activity. Data represent the mean ± S.D. of three independent experiments. * P < 0.05 versus MCF-7, # P < 0.05 versus MCF-7/Rap and MCF-7/exoC.
.
Signaling profile of the MCF-7 cells and resistant sublines. (A) Western blotting analysis of total cell extracts. Protein loading was controlled by membrane hybridization with GAPDH Abs. The blot represents the results of one of three similar experiments. (B) Densitometry for immunoblotting data (n=3) was performed using ImageJ software. (C) Reporter analysis of the AP-1 transcriptional activity. The cells were transfected with the AP-1 plasmid containing the luciferase reporter gene under the AP-1-responsive elements and the β-galactosidase plasmid. Then, 24 h after transfection, the luciferase and β-galactosidase activities were determined as described in the Methods section. The relative luciferase activity was calculated in arbitrary units as the ratio of the luciferase to the galactosidase activity. Data represent the mean ± S.D. of three independent experiments. * P < 0.05 versus MCF-7, # P < 0.05 versus MCF-7/Rap and MCF-7/exoC.
Changes in the DNMT profile in rapamycin-resistant cells
The analysis of the expression level of the main DNA methylation enzymes, DNMT3A and DNMT1, was performed in the sensitive and resistant cells. While the DNMT1 expression was not changed in all variants of the resistant cells, the DMNT3A expression was considerably decreased in the MCF-7/Rap resistant subline. Moreover, the cell cultures with rapamycin resistance triggered by exosomes (MCF-7/exoR cells) were characterized by a similar tendency, i.e., a decrease in DNMT3A expression (Fig. 5A).
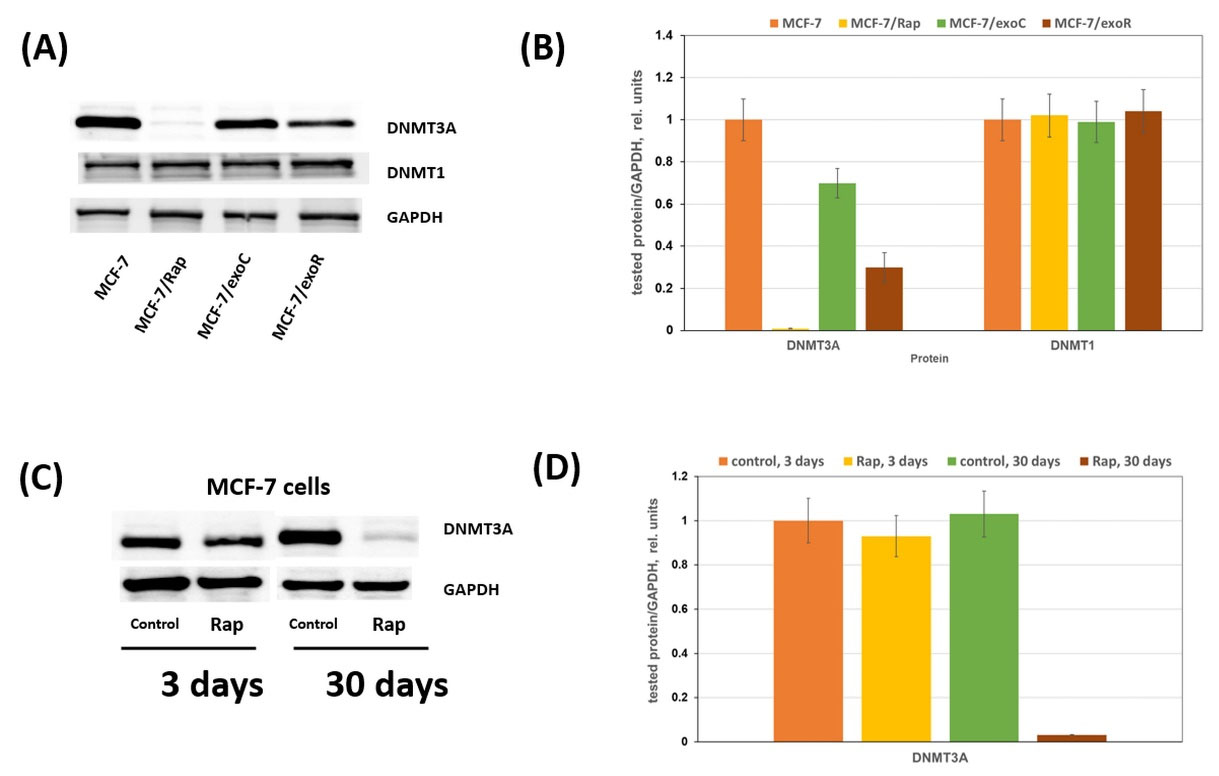
Fig. 5.
Rapamycin resistance and DNMT3A expression. (A) Western blotting analysis of DNMT3A in total cell extracts. Protein loading was controlled by membrane hybridization with GAPDH Abs. The blot represents the results of one of three similar experiments. (C) Rapamycin influence on DNMT3A expression. MCF-7 cells were treated with 2 μM rapamycin for 3 or 30 days with subsequent Western blotting analysis of DNMT3A in cell lysates. (B), (D)Densitometry for corresponding immunoblotting data (n=3).
.
Rapamycin resistance and DNMT3A expression. (A) Western blotting analysis of DNMT3A in total cell extracts. Protein loading was controlled by membrane hybridization with GAPDH Abs. The blot represents the results of one of three similar experiments. (C) Rapamycin influence on DNMT3A expression. MCF-7 cells were treated with 2 μM rapamycin for 3 or 30 days with subsequent Western blotting analysis of DNMT3A in cell lysates. (B), (D)Densitometry for corresponding immunoblotting data (n=3).
Is a single rapamycin treatment of cells sufficient for DNMT3A suppression or is continuous rapamycin treatment necessary for irreversible inhibition of DNMT3A? The study of the effect of single or prolonged rapamycin treatment on the DNMT3A level showed no changes after a single treatment, whereas continuous rapamycin treatment resulted in marked DNMT3A suppression (Fig. 5B).
For subsequent investigation of the function of DNMT3A for cell sensitivity regulation, transfection of the initial MCF-7 cell line was performed using the siRNA against DNMT3A. The obtained results showed marked suppression of DNMT3A expression correlated with an increase in the rapamycin resistance after siRNA transfection (Fig. 6A,B). Consequently, the DNMT3A suppression is likely one of the necessary steps in the development of acquired rapamycin resistance.

Fig. 6.
Influence of DNMT3A knockdown on the cell sensitivity to rapamycin. MCF-7 cells were transfected with the scrambled nonspecific siRNA or DNMT3A specific siRNA as described in the Methods section. Western blot analysis of DNMT3A (A) and MTT assay of cell sensitivity to rapamycin (C) were performed as described above. Data represent the mean ± SD of three independent experiments. * P < 0.05 versus control, # P < 0.05 versus MCF-7/scr cells treated with rapamycin. (B) Densitometry for immunoblotting data (n=3) was performed using ImageJ software.
.
Influence of DNMT3A knockdown on the cell sensitivity to rapamycin. MCF-7 cells were transfected with the scrambled nonspecific siRNA or DNMT3A specific siRNA as described in the Methods section. Western blot analysis of DNMT3A (A) and MTT assay of cell sensitivity to rapamycin (C) were performed as described above. Data represent the mean ± SD of three independent experiments. * P < 0.05 versus control, # P < 0.05 versus MCF-7/scr cells treated with rapamycin. (B) Densitometry for immunoblotting data (n=3) was performed using ImageJ software.
Discussion
The development of resistance of malignant tumors to chemotherapy drugs is one of the main factors that diminish the efficiency of antitumor therapy. This fully applies to targeted therapy, the resistance to which develops rather quickly after treatment initiation. If resistance in relation to xenobiotic drug compounds is based mainly on the involvement of ABC transporters and, as a result, on the rapid drug efflux from the tumor cell, resistance to target drugs is based on the rearrangement of signaling pathways aimed at overcoming the proliferation block. Among these targeted drugs are rapamycin and its analogues, rapalogues, which are direct mTOR inhibitors. The mechanism of development of tumor resistance to these drugs is being actively studied, and signaling pathways in which the reactivation can lead to acquired resistance of tumor cells to mTOR inhibitors have been identified. Thus, the development of MCF-7 cell resistance to rapamycin is caused by the involvement of Akt signaling,40 overexpression of MYC,41 activation of the MAP cascade,42,43 overexpression of Survivin, which is a representative of the inhibitor of apoptosis protein family,44 and activation of the integrin/FAK/IGFR pathway.45 However, the mechanism of progression and expansion of acquired resistance in tumor cells and the role of intercellular interactions in this process remain largely unclear.
This work aimed to determine the tumor exosome participation in the development of resistance to mTOR inhibitors. The investigation was performed using MCF-7 breast cancer cells and MCF-7/Rap resistant cell culture, which was obtained from long-term cultivation of parent cells with the mTOR inhibitor, rapamycin. The resulting subline was cross-resistant to rapamycin and metformin, an indirect mTOR inhibitor belonging to the AMPK activator family, and was used, along with parent cells, as a source of exosomes.
We found that long-term (30 days) cultivation of the breast cancer line in the presence of MCF-7/Rap cell exosomes enabled the progression of partial resistance of recipient cells to rapamycin and metformin, in contrast to exosomes of their own (parental) cells, which do not cause this effect. Exosome-induced resistance was quite stable and persisted in cells for at least 30 days after exosome withdrawal.
Analysis of the main functional proteins of sensitive breast cancer cell lines in comparison with the resistant cell proteins did not reveal significant differences in the expression of mTOR signaling proteins but showed marked activation of p42/44 MAP kinases and an increase in the transcriptional activity of AP-1 in resistant cells. Parallel analysis of proteins after exosome treatment revealed similar changes: activation of MAP kinases and AP-1 by the exosomes from resistant but not parental cells. Thus, the exosomes of resistant cells, when interacting with recipient cells, practically reproduce the features of the phenotype of resistant cells, including the activation of the MAP kinase cascade.
As mentioned above, the rapamycin resistance induced by exosomes and the corresponding changes in the protein profile are maintained in cells for a long time, at least for several months, after exosome withdrawal. What is the mechanism for retention of exosome-induced changes in recipient cells? At the final stage of the experiments, we analyzed the level of DNA methyltransferases, the key enzymes responsible for the development and maintenance of epigenetic changes, in resistant and parental cells. The MCF-7/Rap resistant subline is characterized by a stable decrease in the expression of DNA methyltransferase 3α (DNMT3A). A similar decrease occurred in MCF-7 cells after the cell treatment with the “resistant” exosomes, supporting the importance of the DNMT3A suppression in the progression of tumor resistance.
Conclusion
In this study, we discovered novel mechanisms of resistance transfer between breast cancer cells. In rapamycin-resistant cells, the upregulated ERK and AP-1 signaling pathways supported resistance to the antiproliferative effect of the drug. Such “aggressive” changes can be transmitted by exosomes originating from resistant cells. Naive cells that have received exosomes from resistant cells may acquire rapamycin resistance. We propose that DNA demethylation followed by increased expression of key genes may be one of the factors responsible for the progression and maintenance of the resistant cell phenotype.
Research Highlights
What is the current knowledge?
√ Cancer is a leading cause of death worldwide.
√ Resistance limits the efficiency of antitumor therapy.
√ Recent studies have shown that exosomes are involved in the development of resistance to hormonal drugs.
What is new here?
√ Exosomes of resistant cells induce the resistance to rapamycin in the parent MCF-7 cells.
√ Exosome-induced resistance reproduces the changes revealed in the rapamycin-resistant cells, including the activation of ERK/AP-1.
√ DNA demethylation followed by increased expression of key genes may be one of the factors responsible for the resistance.
Acknowledgments
Graphical abstract was created using Servier Medical Art templates. Original templates are licensed under a Creative Commons Attribution 3.0 Unported License (https://smart.servier.com/). Authors thank Gleb Skryabin and Elena Tchevkina for their help in the NTA.
Competing Interests
The authors declare no conflict of interest.
Ethical Statement
None to be declared.
Funding
The study was supported by the Russian Science Foundation, project #19-15-00245.
References
- Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem 1987; 262:9412-20. [ Google Scholar]
- Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol 1985; 101:942-8. doi: 10.1083/jcb.101.3.942 [Crossref] [ Google Scholar]
- Choi DS, Kim DK, Kim YK, Gho YS. Proteomics of extracellular vesicles: Exosomes and ectosomes. Mass Spectrom Rev 2015; 34:474-90. doi: 10.1002/mas.21420 [Crossref] [ Google Scholar]
- Banfi C, Brioschi M, Wait R, Begum S, Gianazza E, Pirillo A. Proteome of endothelial cell-derived procoagulant microparticles. Proteomics 2005; 5:4443-55. doi: 10.1002/pmic.200402017 [Crossref] [ Google Scholar]
- Bard MP, Hegmans JP, Hemmes A, Luider TM, Willemsen R, Severijnen LA. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am J Respir Cell Mol Biol 2004; 31:114-21. doi: 10.1165/rcmb.2003-0238OC [Crossref] [ Google Scholar]
- Choi DS, Choi DY, Hong BS, Jang SC, Kim DK, Lee J, et al. Quantitative proteomics of extracellular vesicles derived from human primary and metastatic colorectal cancer cells. J Extracell Vesicles 2012; 1. 10.3402/jev.v1i0.18704.
- Pashoutan Sarvar D, Shamsasenjan K, Akbarzadehlaleh P. Mesenchymal Stem Cell-Derived Exosomes: New Opportunity in Cell-Free Therapy. Adv Pharm Bull 2016; 6:293-9. doi: 10.15171/apb.2016.041 [Crossref] [ Google Scholar]
- Moradi-Chaleshtori M, Koochaki A, Shojaei S, Paryan M, Safarzadeh M, Hashemi SM. Overexpression of Pigment epithelium-derived factor in breast cancer cell-derived exosomes induces M1 polarization in macrophages. Immunol Lett 2022; 248:31-36. doi: 10.1016/j.imlet.2022.05.005 [Crossref] [ Google Scholar]
- Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol 2012; 10:e1001450. doi: 10.1371/journal.pbio.1001450 [Crossref] [ Google Scholar]
- Kim DK, Lee J, Kim SR, Choi DS, Yoon YJ, Kim JH. EVpedia: a community web portal for extracellular vesicles research. Bioinformatics 2015; 31:933-9. doi: 10.1093/bioinformatics/btu741 [Crossref] [ Google Scholar]
- Keerthikumar S, Chisanga D, Ariyaratne D, Al Saffar H, Anand S, Zhao K. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J Mol Biol 2016; 428:688-92. doi: 10.1016/j.jmb.2015.09.019 [Crossref] [ Google Scholar]
- Rahbar Saadat Y, Barar J. Exosomes as versatile nanoscaled biocompartments in cancer therapy and/or resistance. Bioimpacts 2022; 12:87-8. doi: 10.34172/bi.2022.24253 [Crossref] [ Google Scholar]
- Kahroba H, Samadi N, Mostafazadeh M, Hejazi MS, Sadeghi MR, Hashemzadeh S. Evaluating the presence of deregulated tumoral onco-microRNAs in serum-derived exosomes of gastric cancer patients as noninvasive diagnostic biomarkers. Bioimpacts 2022; 12:127-38. doi: 10.34172/bi.2021.22178 [Crossref] [ Google Scholar]
- Mousavi SM, Amin Mahdian SM, Ebrahimi MS, Taghizadieh M, Vosough M, Sadri Nahand J. Microfluidics for detection of exosomes and microRNAs in cancer: State of the art. Mol Ther Nucleic Acids 2022; 28:758-91. doi: 10.1016/j.omtn.2022.04.011 [Crossref] [ Google Scholar]
- Zokaei E, Darbeheshti F, Rezaei N. Prospect of exosomal circular RNAs in breast Cancer: presents and future. Mol Biol Rep 2022; 49:6997-7011. doi: 10.1007/s11033-022-07472-4 [Crossref] [ Google Scholar]
- Jaiswal R, Luk F, Dalla PV, Grau GE, Bebawy M. Breast cancer-derived microparticles display tissue selectivity in the transfer of resistance proteins to cells. PLoS One 2013; 8:e61515. doi: 10.1371/journal.pone.0061515 [Crossref] [ Google Scholar]
- Pasquier J, Galas L, Boulangé-Lecomte C, Rioult D, Bultelle F, Magal P. Different modalities of intercellular membrane exchanges mediate cell-to-cell p-glycoprotein transfers in MCF-7 breast cancer cells. J Biol Chem 2012; 287:7374-87. doi: 10.1074/jbc.M111.312157 [Crossref] [ Google Scholar]
- de Souza PS, Cruz AL, Viola JP, Maia RC. Microparticles induce multifactorial resistance through oncogenic pathways independently of cancer cell type. Cancer Sci 2015; 106:60-8. doi: 10.1111/cas.12566 [Crossref] [ Google Scholar]
- Chen WX, Liu XM, Lv MM, Chen L, Zhao JH, Zhong SL. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS One 2014; 9:e95240. doi: 10.1371/journal.pone.0095240 [Crossref] [ Google Scholar]
- Weiner-Gorzel K, Dempsey E, Milewska M, McGoldrick A, Toh V, Walsh A. Overexpression of the microRNA miR-433 promotes resistance to paclitaxel through the induction of cellular senescence in ovarian cancer cells. Cancer Med 2015; 4:745-58. doi: 10.1002/cam4.409 [Crossref] [ Google Scholar]
- Semina SE, Scherbakov AM, Vnukova AA, Bagrov DV, Evtushenko EG, Safronova VM, et al. Exosome-Mediated Transfer of Cancer Cell Resistance to Antiestrogen Drugs. Molecules 2018; 23. 10.3390/molecules23040829.
- Semina SE, Scherbakov AM, Kovalev SV, Shevchenko VE, Krasil'nikov MA. Horizontal Transfer of Tamoxifen Resistance in MCF-7 Cell Derivates: Proteome Study. Cancer Invest 2017; 35:506-18. doi: 10.1080/07357907.2017.1368081 [Crossref] [ Google Scholar]
- Andreeva OE, Sorokin DV, Mikhaevich EI, Bure IV, Shchegolev YY, Nemtsova MV. Towards Unravelling the Role of ERα-Targeting miRNAs in the Exosome-Mediated Transferring of the Hormone Resistance. Molecules 2021; 26:6661. doi: 10.3390/molecules26216661 [Crossref] [ Google Scholar]
- Liu J, Zhu S, Tang W, Huang Q, Mei Y, Yang H. Exosomes from tamoxifen-resistant breast cancer cells transmit drug resistance partly by delivering miR-9-5p. Cancer Cell Int 2021; 21:55. doi: 10.1186/s12935-020-01659-0 [Crossref] [ Google Scholar]
- Hu K, Liu X, Li Y, Li Q, Xu Y, Zeng W. Exosomes Mediated Transfer of Circ_UBE2D2 Enhances the Resistance of Breast Cancer to Tamoxifen by Binding to MiR-200a-3p. Med Sci Monit 2020; 26:e922253. doi: 10.12659/msm.922253 [Crossref] [ Google Scholar]
- La Camera G, Gelsomino L, Caruso A, Panza S, Barone I, Bonofiglio D, et al. The Emerging Role of Extracellular Vesicles in Endocrine Resistant Breast Cancer. Cancers (Basel) 2021; 13. 10.3390/cancers13051160.
- Xie W, Sun H, Li X, Lin F, Wang Z, Wang X. Ovarian cancer: epigenetics, drug resistance, and progression. Cancer Cell Int 2021; 21:434. doi: 10.1186/s12935-021-02136-y [Crossref] [ Google Scholar]
- Qian Z, Shen Q, Yang X, Qiu Y, Zhang W. The Role of Extracellular Vesicles: An Epigenetic View of the Cancer Microenvironment. Biomed Res Int 2015; 2015:649161. doi: 10.1155/2015/649161 [Crossref] [ Google Scholar]
- Hayashi T, Hoffman MP. Exosomal microRNA communication between tissues during organogenesis. RNA Biol 2017; 14:1683-9. doi: 10.1080/15476286.2017.1361098 [Crossref] [ Google Scholar]
- Shchegolev Y, Sorokin D, Scherbakov A, Shunaev A, Andreeva O, Mikhaevich E. Upregulation of Akt/Raptor signaling is associated with rapamycin resistance of breast cancer cells. Chem Biol Interact 2020; 330:109243. doi: 10.1016/j.cbi.2020.109243 [Crossref] [ Google Scholar]
- Iselt M, Holtei W, Hilgard P. The tetrazolium dye assay for rapid in vitro assessment of cytotoxicity. Arzneimittelforschung 1989; 39:747-9. [ Google Scholar]
- Volkova YA, Antonov YS, Komkov AV, Scherbakov AM, Shashkov AS, Menchikov LG. Access to steroidal pyridazines via modified thiohydrazides. RSC Advances 2016; 6:42863-8. doi: 10.1039/C6RA06881B [Crossref] [ Google Scholar]
- Scherbakov AM, Komkov AV, Komendantova AS, Yastrebova MA, Andreeva OE, Shirinian VZ. Steroidal Pyrimidines and Dihydrotriazines as Novel Classes of Anticancer Agents against Hormone-Dependent Breast Cancer Cells. Front Pharmacol 2017; 8:979. doi: 10.3389/fphar.2017.00979 [Crossref] [ Google Scholar]
- Mruk DD, Cheng CY. Enhanced chemiluminescence (ECL) for routine immunoblotting: An inexpensive alternative to commercially available kits. Spermatogenesis 2011; 1:121-2. doi: 10.4161/spmg.1.2.16606 [Crossref] [ Google Scholar]
- Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, Gardiner C. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 2014; 3:26913. doi: 10.3402/jev.v3.26913 [Crossref] [ Google Scholar]
- Takov K, Yellon DM, Davidson SM. Confounding factors in vesicle uptake studies using fluorescent lipophilic membrane dyes. J Extracell Vesicles 2017; 6:1388731. doi: 10.1080/20013078.2017.1388731 [Crossref] [ Google Scholar]
- Taylor SC, Berkelman T, Yadav G, Hammond M. A defined methodology for reliable quantification of Western blot data. Mol Biotechnol 2013; 55:217-26. doi: 10.1007/s12033-013-9672-6 [Crossref] [ Google Scholar]
- Wu Z, Nicoll M, Ingham RJ. AP-1 family transcription factors: a diverse family of proteins that regulate varied cellular activities in classical hodgkin lymphoma and ALK+ ALCL. Exp Hematol Oncol 2021; 10:4. doi: 10.1186/s40164-020-00197-9 [Crossref] [ Google Scholar]
-
Chen TK, Smith LM, Gebhardt DK, Birrer MJ, Brown PH. Activation and inhibition of the AP-1 complex in human breast cancer cells. Mol Carcinog 1996; 15: 215-26. 10.1002/(sici)1098-2744(199603)15:3<215::aid-mc7>3.0.co;2-g.
- Sun SY, Rosenberg LM, Wang X, Zhou Z, Yue P, Fu H. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res 2005; 65:7052-8. doi: 10.1158/0008-5472.can-05-0917 [Crossref] [ Google Scholar]
- Bihani T, Ezell SA, Ladd B, Grosskurth SE, Mazzola AM, Pietras M. Resistance to everolimus driven by epigenetic regulation of MYC in ER+ breast cancers. Oncotarget 2015; 6:2407-20. doi: 10.18632/oncotarget.2964 [Crossref] [ Google Scholar]
- Hoang B, Benavides A, Shi Y, Yang Y, Frost P, Gera J. The PP242 mammalian target of rapamycin (mTOR) inhibitor activates extracellular signal-regulated kinase (ERK) in multiple myeloma cells via a target of rapamycin complex 1 (TORC1)/eukaryotic translation initiation factor 4E (eIF-4E)/RAF pathway and activation is a mechanism of resistance. J Biol Chem 2012; 287:21796-805. doi: 10.1074/jbc.M111.304626 [Crossref] [ Google Scholar]
- Soares HP, Ni Y, Kisfalvi K, Sinnett-Smith J, Rozengurt E. Different patterns of Akt and ERK feedback activation in response to rapamycin, active-site mTOR inhibitors and metformin in pancreatic cancer cells. PLoS One 2013; 8:e57289. doi: 10.1371/journal.pone.0057289 [Crossref] [ Google Scholar]
- Koike H, Nitta T, Sekine Y, Arai S, Furuya Y, Nomura M. YM155 reverses rapamycin resistance in renal cancer by decreasing survivin. J Cancer Res Clin Oncol 2014; 140:1705-13. doi: 10.1007/s00432-014-1734-z [Crossref] [ Google Scholar]
- Yoon SO, Shin S, Karreth FA, Buel GR, Jedrychowski MP, Plas DR, et al. Focal Adhesion- and IGF1R-Dependent Survival and Migratory Pathways Mediate Tumor Resistance to mTORC1/2 Inhibition. Mol Cell 2017; 67: 512-27.e4. 10.1016/j.molcel.2017.06.033.