Bioimpacts. 14(2):27618.
doi: 10.34172/bi.2023.27618
Original Article
A novel imidazo[1,2-a]pyridine derivative and its co-administration with curcumin exert anti-inflammatory effects by modulating the STAT3/NF-κB/iNOS/COX-2 signaling pathway in breast and ovarian cancer cell lines
Havva Afshari Investigation, Writing – original draft, Writing – review & editing, 1 
Shokoofe Noori Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Supervision, 1, * 
Mitra Nourbakhsh Writing – review & editing, 2, 3
Azam Daraei Methodology, 1
Mahsa Azami Movahed Methodology, 4
Afshin Zarghi Methodology, 4
Author information:
1Department of Clinical Biochemistry, Faculty of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
2Finetech in Medicine Research Center, Iran University of Medical Sciences, Tehran, Iran
3Department of Biochemistry, School of Medicine, Iran University of Medical Sciences, Tehran, Iran
4Department of Pharmaceutical Chemistry, School of Pharmacy, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Abstract
Introduction:
Imidazo[1,2-a]pyridine derivatives with diverse pharmacological properties and curcumin, as a potential natural anti-inflammatory compound, are promising compounds for cancer treatment. This study aimed to synthesize a novel imidazo[1,2-a]pyridine derivative, (MIA), and evaluate its anti-inflammatory activity and effects on nuclear factor-κB (NF-κB) and signal transducer and activator of transcription 3 (STAT3) pathways, and their target genes, alone and in combination with curcumin, in MDA-MB-231 and SKOV3 cell lines.
Methods:
We evaluated the interaction between imidazo[1,2-a]pyridine ligand, curcumin, and NF-κB p50 protein, using molecular docking studies. MTT assay was used to investigate the impacts of compounds on cell viability. To evaluate the NF-κB DNA binding activity and the level of inflammatory cytokines in response to the compounds, ELISA-based methods were performed. In addition, quantitative polymerase chain reaction (qPCR) and western blotting were carried out to analyze the expression of genes and investigate NF-κB and STAT3 signaling pathways.
Results:
Molecular docking studies showed that MIA docked into the NF-κB p50 subunit, and curcumin augmented its binding. The MTT assay results indicated that MIA and its combination with curcumin reduced cell viability. According to the results of the ELISA-based methods, MIA lowered the levels of inflammatory cytokines and suppressed NF-κB activity. In addition, real-time PCR and Griess test results showed that the expression of cyclooxygenase-2 (COX-2) and inducible NO synthase (iNOS) genes, and nitrite production were reduced by MIA. Furthermore, the western blotting analysis demonstrated that MIA increased the expression of inhibitory κB (IκBα) and B-cell lymphoma 2 (Bcl-2)-associated X proteins (BAX), and suppressed the STAT3 phosphorylation, and Bcl-2 expression. Our findings revealed that curcumin had a potentiating role and enhanced all the anti-inflammatory effects of MIA.
Conclusion:
This study indicated that the anti-inflammatory activity of MIA is exerted by suppressing the NF-κB and STAT3 signaling pathways in MDA-MB-231 and SKOV3 cancer cell lines.
Keywords: Imidazo[1,2-a]pyridine, Curcumin, NF-κB, Breast cancer, Inflammation, Ovarian cancer
Copyright and License Information
© 2024 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Breast cancer is the most prevalent malignancy with the greatest fatality rate, and ovarian cancer is the third most common cancer among women globally according to the statistics of 2020.1,2 Chemotherapy has been conducted for the treatment of cancer patients: however, it has several significant drawbacks, such as high dosage requirements, serious side effects, and multiple drug resistance.3 Consequently, the development of unique and effective compounds and strategies, such as novel combination therapies for cancer treatment is essential.
In oncology, the medical significance of natural products is well recognized, especially as a substitute for the disadvantages of routine chemotherapy.4-6 Curcumin, the active ingredient in turmeric extracted from the Curcuma longa plant, is a member of the polyphenol superfamily with anti-cancer and anti-inflammatory activities.7 Many investigations have shown that curcumin, either alone or in combination with other agents, acts as a potent compound for cancer treatment by modulating signaling pathways, inflammatory cytokines, and transcription factors.8-11
Imidazo[1,2-a]pyridine(IP),a promising bicyclic 5–6 heterocyclic ring, has recently attracted much attention due to its wide variety of medicinal chemistry applications, including anti-cancer and anti-inflammatory properties. Regarding remarkable inhibitory activity of IP-based compounds against various cancer cell characteristics such as proliferation and migration, they have gained substantial consideration as possible drugs against breast cancer.12 Modifications in the structure of IPs, make them potent substances to arrest cell cycle, induce apoptosis, modulate signal transduction pathways and cause DNA damage.13,14 For example, IP-based compounds, such as IP-Se-05, HS-106, HS-104, HS-173, IP-6, and IPD-196 are able to inhibit the proliferation of breast cancer cells as well as PI3K/Akt/mTOR signaling pathway, even at low concentrations.15-17 Despite efforts that have been made to modify the scaffold structure of IPs to develop new IP-based compounds, none of them have been approved as anti-cancer therapeutics.14,18-20
Inflammation has been shown to be associated with the development of breast and ovarian cancers.21,22 The nuclear factor-κB (NF-κB), as a key inflammatory response mediator, controls the expression of crucial regulatory genes such as immunity and inflammation.23,24 The NF-κB controls the expression of many genes, including the cyclooxygenase-2 (COX-2), inducible nitric oxide (iNOS), and inflammatory cytokines, such as the tumor necrosis factor alpha (TNF)-α, interleukin (IL)-6, and IL-1β.25,26 Studies have shown that inflammatory stimuli, such as lipopolysaccharide (LPS), activate the NF-κB signaling pathway.27 Additionally, transforming growth factor beta (TGF-β) triggers the NF-κB pathway by sequentially controlling the TGF-β-activated kinase 1 (TAK1) and IkappaB kinase.28 Due to the importance of the NF-κB function, its activity is strictly controlled and in unstimulated cells, inhibitory kappa B (IκB) keeps the NF-κB inactive in the cytoplasm.29
The signal transducer and activator of transcription 3 (STAT3) control the transcription of numerous genes, such as inflammatory cytokines, COX-2, iNOS, B-cell lymphoma 2 (Bcl-2), and Bcl-2-associated X protein (BAX).30-32 Phosphorylation of the tyrosine residue (Tyr 705) activates STAT3 and leads to STAT3 dimerization.33 Collaboration between NF-κB and Signal transducers and activators of transcription 3 (STAT3) has an important role in regulating the communication between inflammatory and cancer cells.34,35 The IL-6 has a significant role in cancer and inflammation and acts primarily through the IL-6/STAT3 pathway. The IL-6 is one of the targets of NF-κB. Thus when STAT3 and NF-κB are activated simultaneously, the IL-6 STAT3 axis induces a positive feedback loop for NF-κB activation.36
In the current study, we synthesized a novel imidazo[1,2-a]pyridine derivative, chemically named 8-methyl-2-(4-(methylsulfonyl)phenyl)-N-(p-tolyl)imidazo[1,2- a ]pyridin-3-amine, which we abbreviated as MIA for simplicity, and assessed its possible anti-inflammatory activity in breast and ovarian cancer cell lines.
Materials and Methods
Materials
Cell culture materials, including culture media and supplements, were provided by Thermo Fisher Scientific (Waltham, MA, USA). Plates, flasks and consumables were obtained from SPL Life Sciences, Gyeonggi-do, Korea. Each of the specific kits and reagents is mentioned in their relevant section. All the other chemicals were purchased from Sigma/Aldrich (St. Louis, MO, USA).
Synthesis of 8-methyl-2-(4-(methylsulfonyl)phenyl)-N-(p tolyl)imidazo[1,2- a]pyridin-3-amine
For this synthesis procedure, first α-bromo-4-(methylsulfonyl)acetophenone was prepared according to the literature procedure.37 Then, α-bromo-4-(methylsulfonyl)acetophenone was reacted with p-toluidinein the presence of NaHCO3 in anhydrous methanol (MeOH) to produce 1-(4-(methylsulfonylphenyl-2-(p-tolylamino))ethan-1-one. Subsequently, 1-(4-(methylsulfonylphenyl-2-(p-tolylamino))ethan-1-one underwent a condensation reaction with 2-amino-3-methylpyridine in isopropylalcohol (i-PrOH) at 80 °C to yield 8-methyl-2-(4-(methylsulfonyl)phenyl)-N-(p tolyl)imidazo[1,2- a]pyridin-3-amine (MIA) as the final product, which was used for the subsequent cellular treatments. The synthesis process is depicted in Scheme 1.
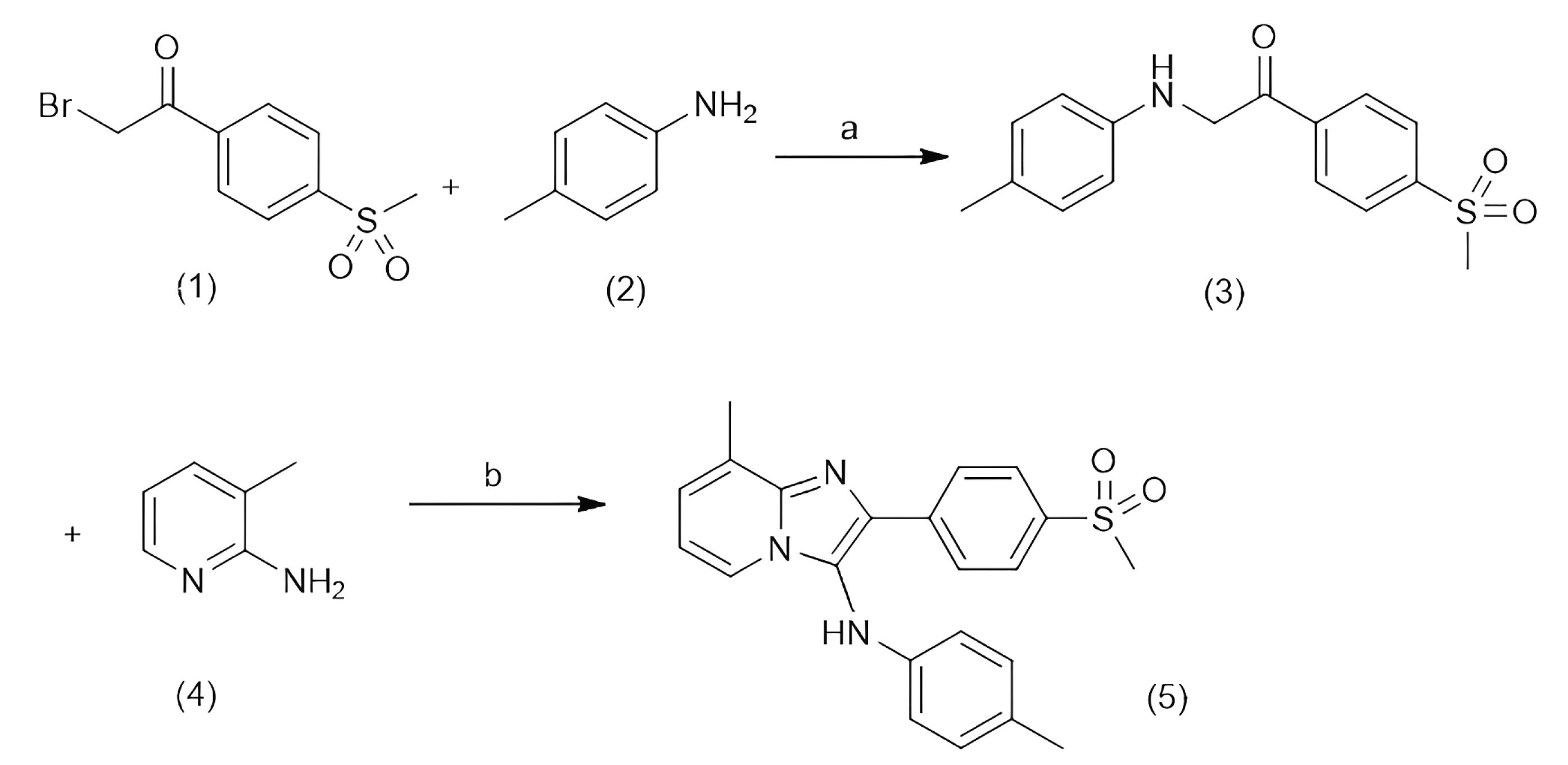
Scheme 1.
The process of the synthesis of 8-methyl-2-(4-(methylsulfonyl)phenyl)-N-(p tolyl)imidazo[1,2- a]pyridin-3-amine (MIA). The first row shows the reaction of 1) α-bromo-4-(methylsulfonyl)acetophenone with 2) p-toluidinein the presence ofa) NaHCO3 in anhydrous MeOH, at room tempreture, producting 3) 1-(4-(methylsulfonylphenyl-2-(p-tolylamino))ethan-1-one. The second row shows the condensation of the product from the first reaction with 4) 2-amino-3-methylpyridine in the presence of b) ZnI2 (30 mol %), air, 4 Å MS, i-PrOH, at 80°C, producing 5) 8-methyl-2-(4-(methylsulfonyl)phenyl)-N-(p tolyl)imidazo[1,2- a]pyridin-3-amine (MIA) as the final product.
.
The process of the synthesis of 8-methyl-2-(4-(methylsulfonyl)phenyl)-N-(p tolyl)imidazo[1,2- a]pyridin-3-amine (MIA). The first row shows the reaction of 1) α-bromo-4-(methylsulfonyl)acetophenone with 2) p-toluidinein the presence ofa) NaHCO3 in anhydrous MeOH, at room tempreture, producting 3) 1-(4-(methylsulfonylphenyl-2-(p-tolylamino))ethan-1-one. The second row shows the condensation of the product from the first reaction with 4) 2-amino-3-methylpyridine in the presence of b) ZnI2 (30 mol %), air, 4 Å MS, i-PrOH, at 80°C, producing 5) 8-methyl-2-(4-(methylsulfonyl)phenyl)-N-(p tolyl)imidazo[1,2- a]pyridin-3-amine (MIA) as the final product.
Molecular docking studies
Docking studies were performed using AutoDock software version 4.0 to predict the interactions between imidazo[1,2-a]pyridine ligand, curcumin, and protein NF-κB p50. These investigations were conducted using the high-resolution complex NF-κB-DNA (PDB ID: 1NFK) retrieved from Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank. The cocrystallized DNA macromolecule and the protein's water molecules were eliminated, nonpolar hydrogens were combined, Kollman charges were added, and the AutoDock 4 atom type was chosen to create the protein's PDBQT format. Using AutoDock tools, the ligand structure was first minimized in HyperChem8.0 (MM+ technique) and then converted to PDBQT file format. The docking grid box was built from the set of active site residues involved in hydrogen bonds (30×30×30). The docking run was set to 100 and the Lamarckian genetic search algorithm was used. For improved efficiency, protein residues from the docking box with atom count larger than 6.0 Å were deleted.
Cell culture
The cell lines MDA-MB-231 and SKOV3 were provided by the Pasteur Institute of Iran. The MDA-MB-231 cells were grown in DMEM cell culture medium containing streptomycin (100 μg mL-1), penicillin (100 U mL-1), and 10% fetal bovine serum (FBS). The SKOV3 cells were cultured in RPMI-1640 medium containing 1% penicillin/streptomycin and 10% FBS. The cell flasks were then incubated at 37 °C in a humid environment with 5% CO2.
MTT assay
The MTT test was performed to evaluate the impacts of MIA alone and combined with curcumin on the viability of MDA-MB-231 and SKOV3 cell lines. Cell treatment was performed with various concentrations of MIA (10-50 μM), alone and together with curcumin (10 μM), and incubated for 24 hours. The chemicals were solubilized in DMSO, followed by dilution in the cell culture media. A group of cells received only the solvent (DMSO) and no chemical and served as the negative control. Cyclophosphamide (Cpm), a cytotoxic drug against breast and ovarian cancer cells, was also used as the positive control. Afterwards, each well received 5 mg/mL MTT solution (Thermo Fischer Scientific, Waltham, MA, USA) and then was incubated for 4 hours. Finally, the absorbance of formazan was calculated by an ELISA reader at 570 nm. Curcumin was purchased from Sigma-Aldrich (St. Louis, MO, USA) and dissolved in ethanol.
NF-κB activity assay
The p65 and p50 subunits form the NF-κB heterodimer. In inflammatory conditions and during cell development, NF-κB p65 subunits move to the cell nucleus.29 In this study, the effects of MIA alone or in combination with curcumin on NF-κB translocation in LPS-stimulated cells were investigated by the NF-κB p65 Transcription Factor Assay Kit (Abcam, Cambridge, UK) based on enzyme-linked immunosorbent assay (ELISA) method, using a double-stranded DNA sequence that contained the NF-κB response element. Using a nuclear extraction solution containing protease and phosphatase inhibitors, cells were harvested, and nuclear extracts were made. The nuclear extracts were added to the immobilized DNA that contains the NF-κB response element. The NF-κB primary antibody was used for detection, followed by the horseradish peroxidase (HRP)-conjugated secondary antibody. After incubation and washing, the absorbance was calculated by a plate reader at 450 nm. The absorbance rate of each treatment group and the control group were compared.
Measurement of cytokines
The levels of released cytokines (TGF-β1, IL-6, TNF-α, and IL-1β) were measured in LPS-induced cells in response to MIA alone or in combination with curcumin. Using ELISA kits (Sigma-Aldrich, St. Louis, MO, USA), cytokines were quantified in the cell culture supernatants according to the guidelines provided by the manufacturer. An ELISA plate reader (PerkinElmer, USA) calculated the absorbance at 540 nm.
Gene expression analysis
qPCR technique was used to assess COX-2 and iNOS genes expression in LPS-stimulated cells in response to MIA and co-treatment with curcumin. The RNeasy mini kit (Qiagen, Hilden, Germany) was used to extract RNA, and the first-strand cDNA synthesis kit (Bio FACT, Daejeon, South Korea) reverse transcribed the RNA into cDNA. The SYBR Green Master Mix kit (Ampliqon, Odense, Denmark) and the specific primers (Table 1) were used to perform real-time PCR. The GAPDH gene served as a reference gene, and the outcomes were analyzed using the 2-ΔΔCt method using the formula ΔΔCT = ΔCT(target sample)−ΔCT(control sample).
Table 1.
Primers sequences in real-time PCR
|
Gene name
|
Sequence (5'->3')
|
Product length
|
Tm
|
COX-2
NM_000963.4 |
F 5′-TTCAAATGAGATTGTGGGAAAAT-3′
R 5′- AGATCATCTCTGCCTGAGTATCTT -3′ |
305 |
55.01
58.60 |
iNOS
NM_000625.4 |
F 5′-GTTCTCAAGGCACAGGTCTC-3′
R 5′-GCAGGTCACTTATGTCACTTATC-3′ |
127 |
58.20
57.02 |
GAPDH
NM_001256799.3 |
F 5′-CAAATTCCATGGCACCGTCAAG-3′
R 5′-AGAGATGATGACCCTTTTGGCT-3′ |
205
|
60.67
59.42 |
Griess test
NO has a short half-life and its direct measurement is challenging. Therefore, the amounts of nitrate and nitrite, which are stable end products of NO breakdown, are measured to assess NO generation.38 For this purpose, a nitrite assay kit (Sigma-Aldrich, Munich, Germany) based on the Griess Reagent was used to assess the effects of MIA and its combination with curcumin on nitric production in LPS-induced cells. The cells were given a 24-hour incubation period with LPS (10 ng/mL)39 in the presence of different doses of MIA alone and in combination with curcumin (10 μM). According to the instructions, the procedures were carried out. Finally, the absorbance was calculated at 540 nm by an ELISA plate reader (PerkinElmer, USA).
Western blotting
To assess the effects of compounds on IκBα protein expression, cells were treated with the substances for 30 min before exposure to LPS (1 μg/mL) for one hour.40 Also, to investigate the expression of STAT3, p-STAT3, BAX, and Bcl-2 proteins, cells received IL-6 (50 ng/mL) for four hours after treatment with compounds.41 The supernatant of centrifuged lysates was used for western blot analysis. The protein content was evaluated using a BCA test kit (Thermo Fisher Scientific, Oxford, UK). The samples were electroblotted onto a polyvinylidene fluoride (PVDF) membrane (Millipore, St. Louis Missouri, USA) after loading on 10% SDS-PAGE gels. After membrane blocking, the membranes were coated with the primary antibodies at 1:1000 dilution against BAX, Bcl-2, IκBα, STAT3, p-STAT3, and β-actin (Sigma-Aldrich, Munich, Germany) overnight at 4 °C. Afterward, the membranes were then coated with HRP-conjugated anti-mouse immunoglobulin G secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, California, USA). Eventually, enhanced chemiluminescence (ECL) (SuperSignal, Thermo Fisher Scientific, Oxford, UK) was used for detection.
Analytical statistics
GraphPad Prism was used for data analysis. After comparing experimental groups using a one-way analysis of variance (ANOVA), Tukey's post-Hoc analysis was done. P < 0.05 was considered statistically significant, and data were displayed as mean ± standard deviation (SD).
Results
Chemistry
The target compound, 8-methyl-2-(4-(methylsulfonyl)phenyl)-N-(p-tolyl)imidazo[1,2-a]pyridine-3-amine was synthesized and characterized using infrared (IR), liquid chromatography–mass spectrometry (LC-MS),hydrogen-1 nuclear magnetic resonance(1H NMR), and carbon-13 nuclear magnetic resonance (13C NMR). The 1H NMR and 13C NMR results are presented in Fig. 1.
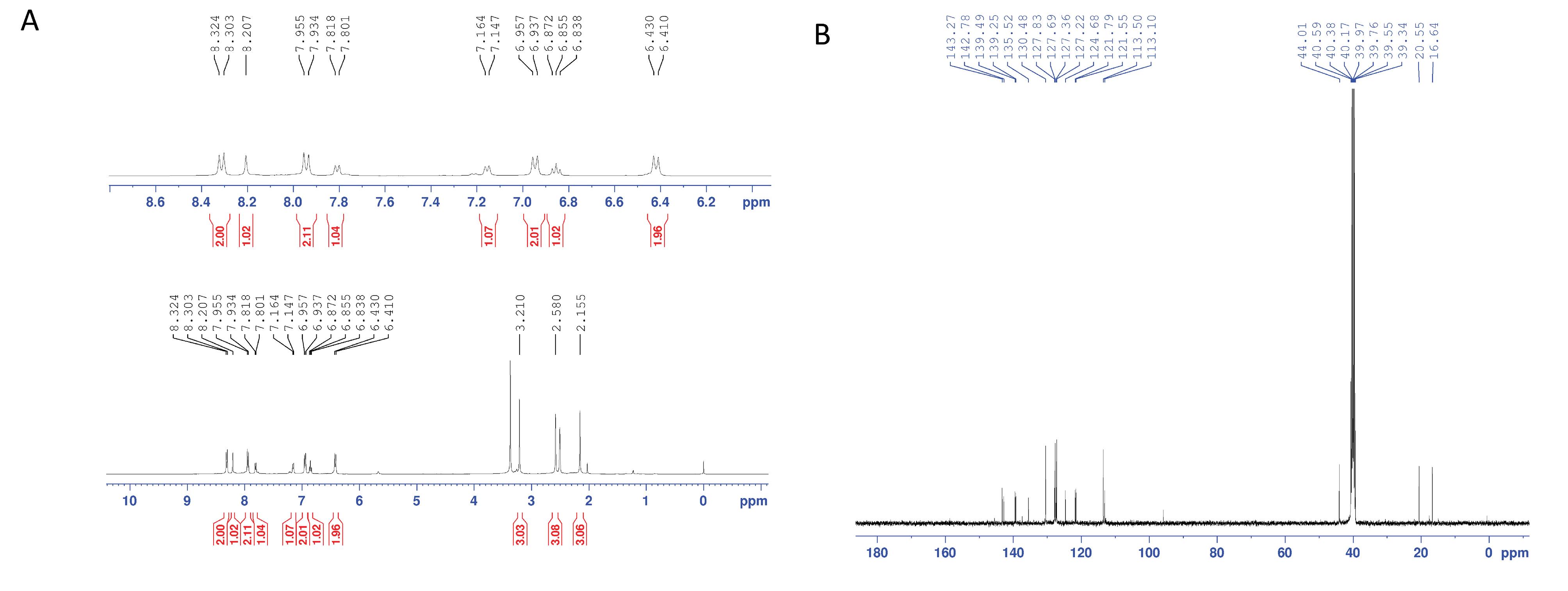
Fig. 1.
The 1H NMR (A) and 13C NMR (B) of 8-methyl-2-(4-(methylsulfonyl)phenyl)-N-(p-tolyl) imidazo[1,2-a]pyridine-3-amine (MIA).
.
The 1H NMR (A) and 13C NMR (B) of 8-methyl-2-(4-(methylsulfonyl)phenyl)-N-(p-tolyl) imidazo[1,2-a]pyridine-3-amine (MIA).
Yield, 59%; dark yellow powder; mp: 200-202 °C; IR (KBr disk): νcm-1 1156, 1315 (SO2), 1632 (C=N), 3370; 1H NMR (DMSO-d6): δ ppm 2.15 (s, 3H, 4-CH3), 2.58 (s, 3H, 8-CH3), 3.21 (s, 3H, SO2Me), 6.41-6.43 (d, 2H, J = 8.0 Hz, phenyl H3 & H5), 6.84-6.87 (t, 1H, J = 6.8 Hz, imidazopyridine H6), 6.94-6.96 (d, 2H, J = 8.0 Hz, phenyl H2 & H6), 7.14-7.16 (d, 1H, J = 6.8 Hz, imidazopyridine H7), 7.80-7.82 (d, 1H, J = 6.8 Hz, imidazopyridine H5), 7.93-7.95 (d, 1H, J = 8.4 Hz, methylsulfonylphenyl H2 & H6), 8.20 (s, 1H, NH), 8.30-8.32 (d, 1H, J = 8.4 Hz, methylsulfonylphenyl H3 & H5); 13C NMR (DMSO-d6): δ ppm 16.64, 20.55, 44.01, 113.10, 113.50, 121.55, 121.79, 124.68, 127.22, 127.36, 127.69, 127.83, 130.48, 135.52, 139.25, 139.49, 142.78, 143.27; LC-MS ESI m/z: 392 ([M+H]+, 100).
MIA docked into the NF-κB p50 subunit, and curcumin reinforced its binding
Docking experiments revealed an interaction between MIA and NF-κB p50 subunit. The findings of the docking experiment were visualized and analyzed using AutoDock4 and PyMOL (open-Source). The docking simulations were performed on p50 monomers (chains A and B). The orientation of the ligand in the active site is shown in Fig. 2. The ligand occupied the active site surrounded by several amino acids (i.e., Arg54, Arg56, Tyr57, Glu60, His141, and Lys241). As demonstrated in Fig. 2A, the oxygen atom of the SO2Me group plays a role as a hydrogen bond acceptor and forms a hydrogen bond with the amine group of Lys241 (distance = 2.52 Å). Moreover, the NH of His141 can interact with the NH group of 3-phenylamino (distance = 2.86 Å). Curcumin fits into the same pocket of the NF-κB p50 active site. Two OH groups of curcumin bond with NH groups of Lys241 as hydrogen bond donors and acceptors (distance = 2.14 and 2.16 Å). Also, the other OH forms a hydrogen bond with the NH group of Ser208 (distance = 2.73 Å). The oxygen atom of carbonyl forms a hydrogen bond with NH of Thr143 amine moiety.
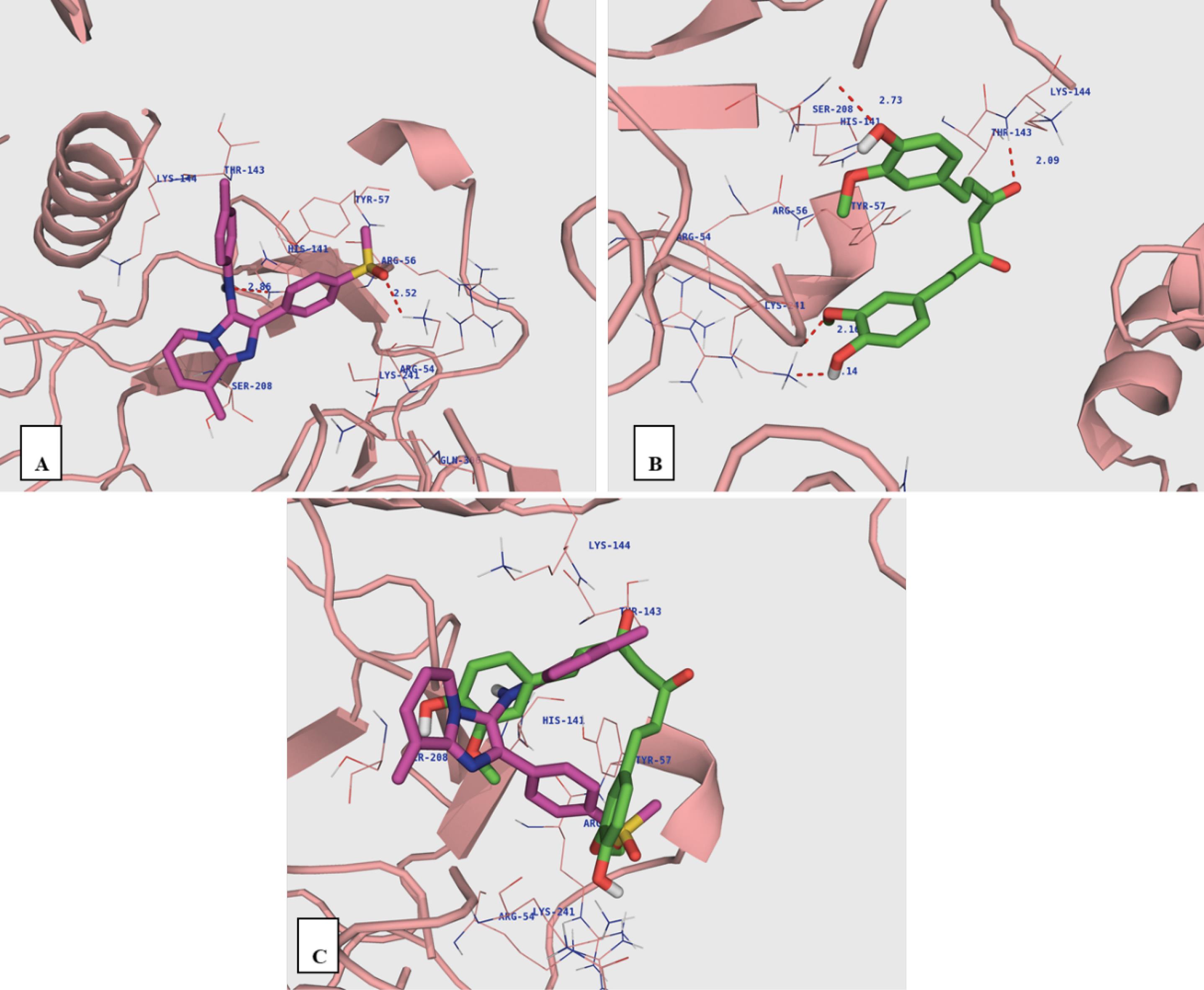
Fig. 2.
The ligands docked into the NF-κB p50 monomer. Imidazo[1,2-a]pyridine (A), curcumin (B) and superimposition of imidazo[1,2-a]pyridineon curcumin (C).
.
The ligands docked into the NF-κB p50 monomer. Imidazo[1,2-a]pyridine (A), curcumin (B) and superimposition of imidazo[1,2-a]pyridineon curcumin (C).
Curcumin enhanced the cytotoxicity of MIA in both cell lines
We assessed the cytotoxic activity of compounds using the MTT test. The MTT assay results demonstrated that MIA reduced cell viability after 24 h in a dose-dependent manner, which was significant at concentrations of 30 μM, 40 μM, and above in MDA-MB-231 and SKOV3 cell lines, respectively (Figs. 3A and 3B). Moreover, the combination of MIA and curcumin caused a greater cytotoxicity compared to MIA treatment alone, and the presence of curcumin decreased the minimum effective concentration of MIA to 20 μM in MDA-MB-231 and 30 μM in SKOV3 cell lines.
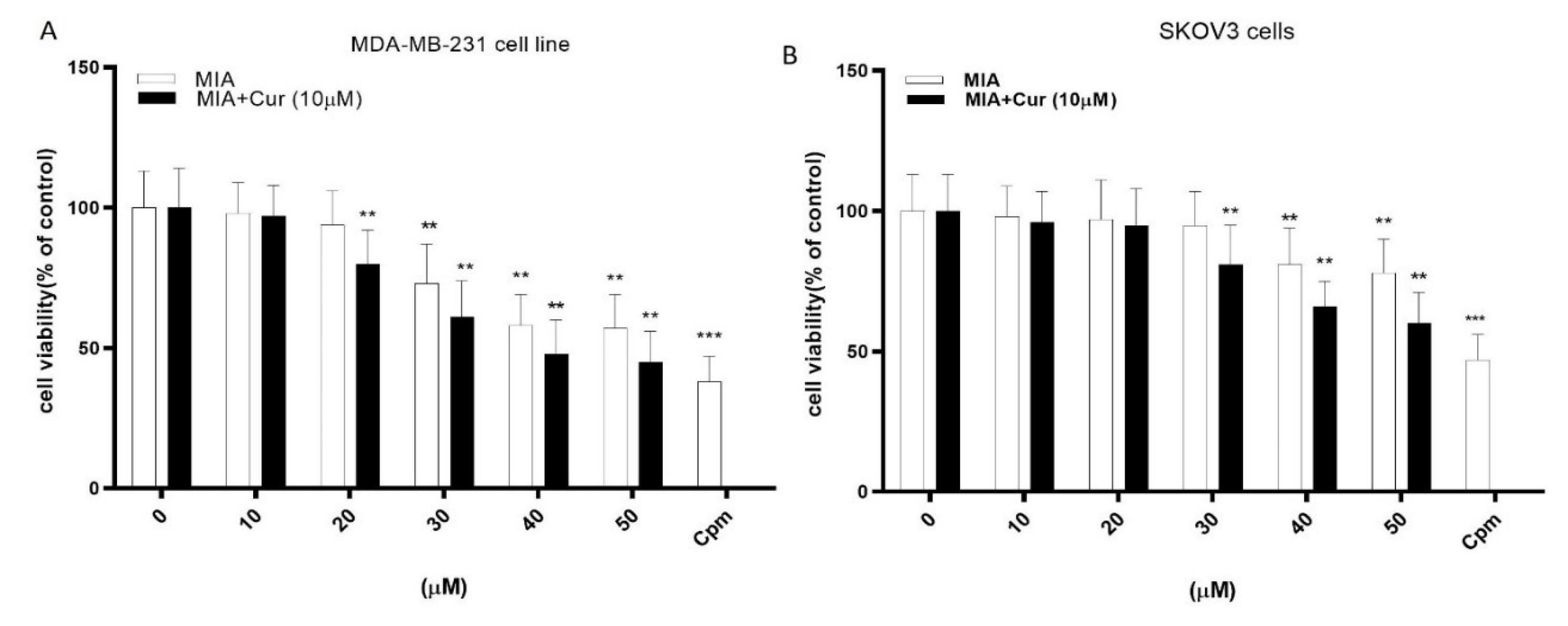
Fig. 3.
MIA and its combination with curcumin reduced the viability of MDA-MB-231 and SKOV3 cells. The effects of different concentrations of MIA (10-50 μM) and its combination with curcumin (10 μM) on the viability of MDA-MB-231 (A) and SKOV3 (B) cells. Negative control cells received only the solvent (0). Cyclophosphamide (Cpm) was used as the positive control. MIA: imidazo[1,2-a]pyridine derivative, Cur: curcumin. The presented data are the mean ± SD of three separate experiments. The values were evaluated in comparison with the control. *P < 0.05, ** P < 0.01 compared with control.
.
MIA and its combination with curcumin reduced the viability of MDA-MB-231 and SKOV3 cells. The effects of different concentrations of MIA (10-50 μM) and its combination with curcumin (10 μM) on the viability of MDA-MB-231 (A) and SKOV3 (B) cells. Negative control cells received only the solvent (0). Cyclophosphamide (Cpm) was used as the positive control. MIA: imidazo[1,2-a]pyridine derivative, Cur: curcumin. The presented data are the mean ± SD of three separate experiments. The values were evaluated in comparison with the control. *P < 0.05, ** P < 0.01 compared with control.
Co-administration of MIA and curcumin was more effective than MIA alone in decreasing NF-κB DNA binding activity
We investigated the NF-κB activity in response to MIA and its combination with curcumin in LPS-stimulated cells using an ELISA-based method. The data showed that LPS alone significantly enhanced the DNA binding activity of NF-κB compared to cells that did not receive LPS (Figs. 4A and 4B). At concentrations of 20 μM, 30 μM, and above, MIA significantly decreased NF-κB DNA binding activity in LPS-induced MDA-MB-231 and SKOV3 cell lines, respectively. Additionally, co-treatment with curcumin boosted the suppressive effect of MIA on NF-κB activity compared to MIA treatment alone in both cell lines, and curcumin decreased the lowest effective concentration of MIA to 10 μM in MDA-MB-231 and 20 μM in SKOV3 cell lines.
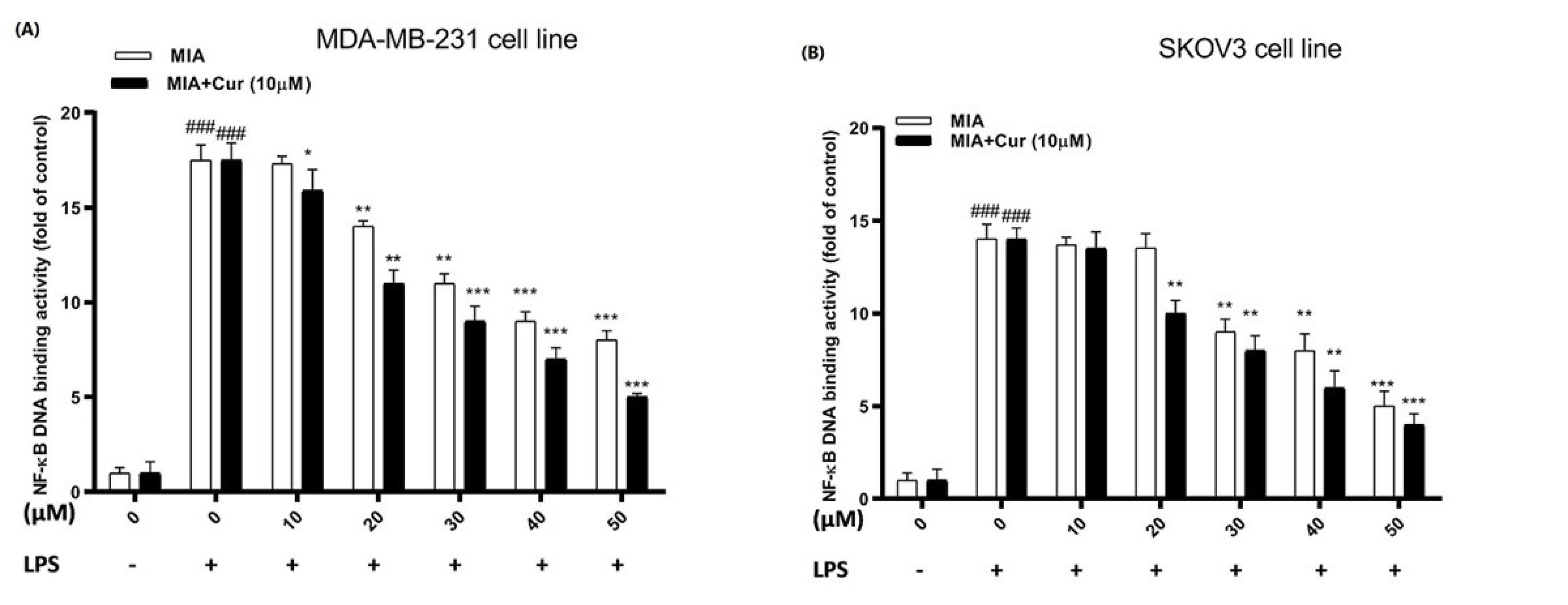
Fig. 4.
The combination of MIA and curcumin attenuated nuclear translocation of the NF-kB p65 subunit in LPS-stimulated cells. NF-κB DNA binding activity in response to different concentrations of MIA (10-50 μM) and MIA+curcumin (10 μM) in LPS-induced MDA-MB-231 (A) and SKOV3 (B) cell lines. Cur: curcumin. MIA: imidazo[1,2-a]pyridine derivative. The standard deviation of three distinct experiments is represented by the error bar. # P < 0.05 compared with untreated unstimulated control cells; * P < 0.05, ** P < 0.01, *** P < 0.001 compared with LPS-stimulated untreated cells.
.
The combination of MIA and curcumin attenuated nuclear translocation of the NF-kB p65 subunit in LPS-stimulated cells. NF-κB DNA binding activity in response to different concentrations of MIA (10-50 μM) and MIA+curcumin (10 μM) in LPS-induced MDA-MB-231 (A) and SKOV3 (B) cell lines. Cur: curcumin. MIA: imidazo[1,2-a]pyridine derivative. The standard deviation of three distinct experiments is represented by the error bar. # P < 0.05 compared with untreated unstimulated control cells; * P < 0.05, ** P < 0.01, *** P < 0.001 compared with LPS-stimulated untreated cells.
Cytokine levels were attenuated by MIA; an effect that was enhanced by curcumin in LPS-stimulated cells
We used an ELISA-based assay to measure the effects of MIA and its combination with curcumin on the inflammatory released cytokines in LPS-induced MDA-MB-231 and SKOV3 cell lines. The results indicated that LPS considerably raised the levels of cytokines (IL-6, TNF-α, IL1-β, TGF-β1) in the extracellular fluids in both cell lines. MIA meaningfully reduced the relative levels of all assessed cytokines stimulated by LPS in both cell lines (Figs. 5A-H). Furthermore, these inhibitory effects were more remarkable in the combined treatment with MIA and curcumin, and curcumin augmented the suppressive impacts of MIA on cytokine production.
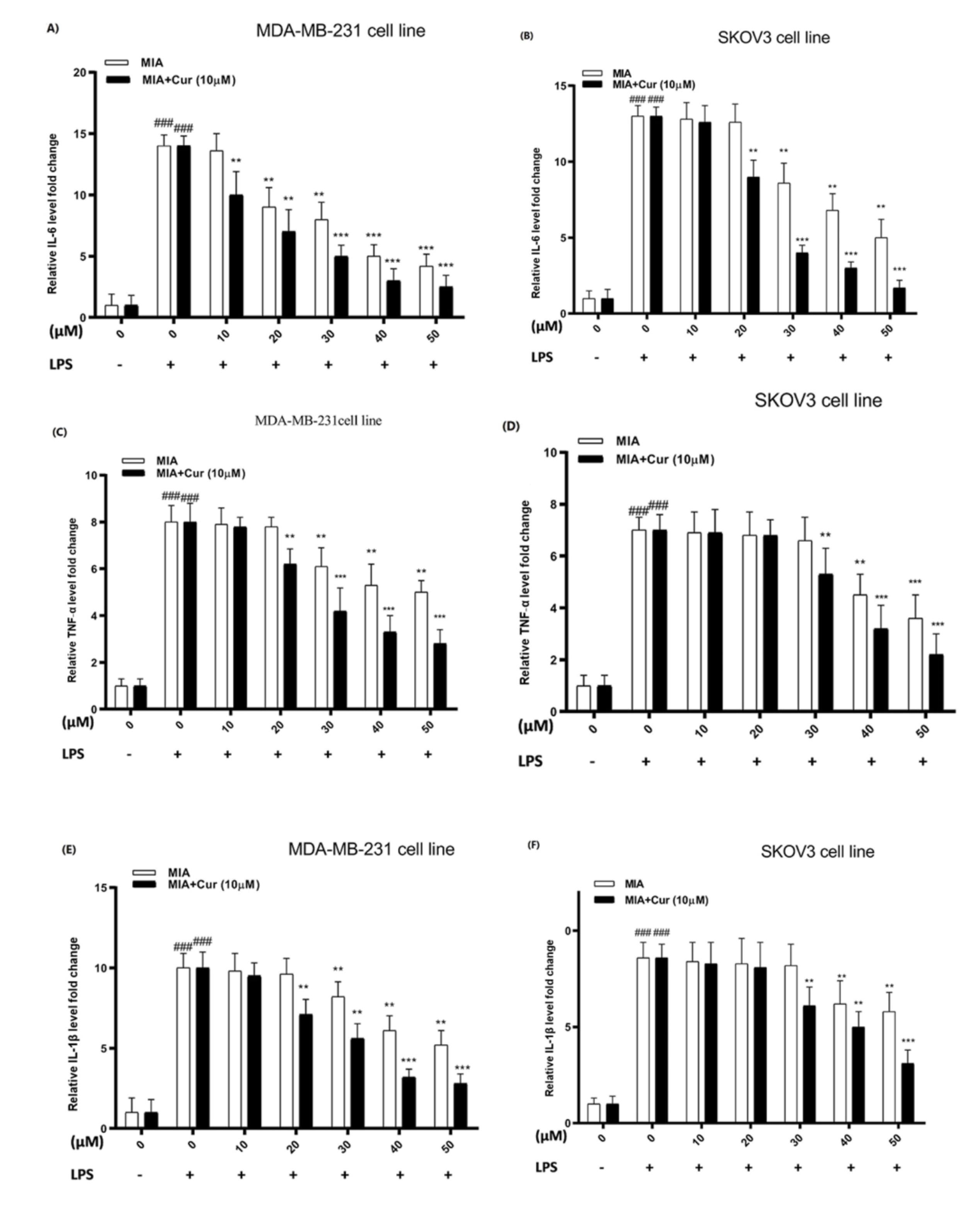
Fig. 5.
Cytokine levels were decreased by MIA and co-treatment with curcumin increased its suppressive effects in LPS-treated cells.The effect of different concentrations of MIA (10-50 μM) and co-treatment with curcumin (10 μM) on IL-6 (A, B), TNF-α (C, D), IL1-β (E, F), and TGF-β1 (G, H) cytokine levels in LPS-induced MDA-MB-231 and SKOV3 cell lines. Cur: curcumin. MIA: imidazo[1,2-a]pyridine derivative. # P < 0.05 compared with untreated unstimulated control cells; * P < 0.05, ** P < 0.01, ** P < 0.001 compared with LPS-stimulated untreated cells
.
Cytokine levels were decreased by MIA and co-treatment with curcumin increased its suppressive effects in LPS-treated cells.The effect of different concentrations of MIA (10-50 μM) and co-treatment with curcumin (10 μM) on IL-6 (A, B), TNF-α (C, D), IL1-β (E, F), and TGF-β1 (G, H) cytokine levels in LPS-induced MDA-MB-231 and SKOV3 cell lines. Cur: curcumin. MIA: imidazo[1,2-a]pyridine derivative. # P < 0.05 compared with untreated unstimulated control cells; * P < 0.05, ** P < 0.01, ** P < 0.001 compared with LPS-stimulated untreated cells
Curcumin increased the suppressive effects of MIA on the COX-2 and iNOS gene expression
As demonstrated in Figs. 6A-D, LPS raised the expression of both genes in breast and ovarian cancer cell lines. On the contrary, MIA downregulated the expression of both genes and counteracted the stimulatory effects of LPS. MIA had significant inhibitory effects on the expression of COX-2 and iNOS genes at concentrations of 30μM, 40μM, and higher in both LPS-stimulated cell lines. Additionally, the expression of genes was further reduced by the combination of MIA and curcumin (10 μM) and was more effective compared to MIA treatment alone.
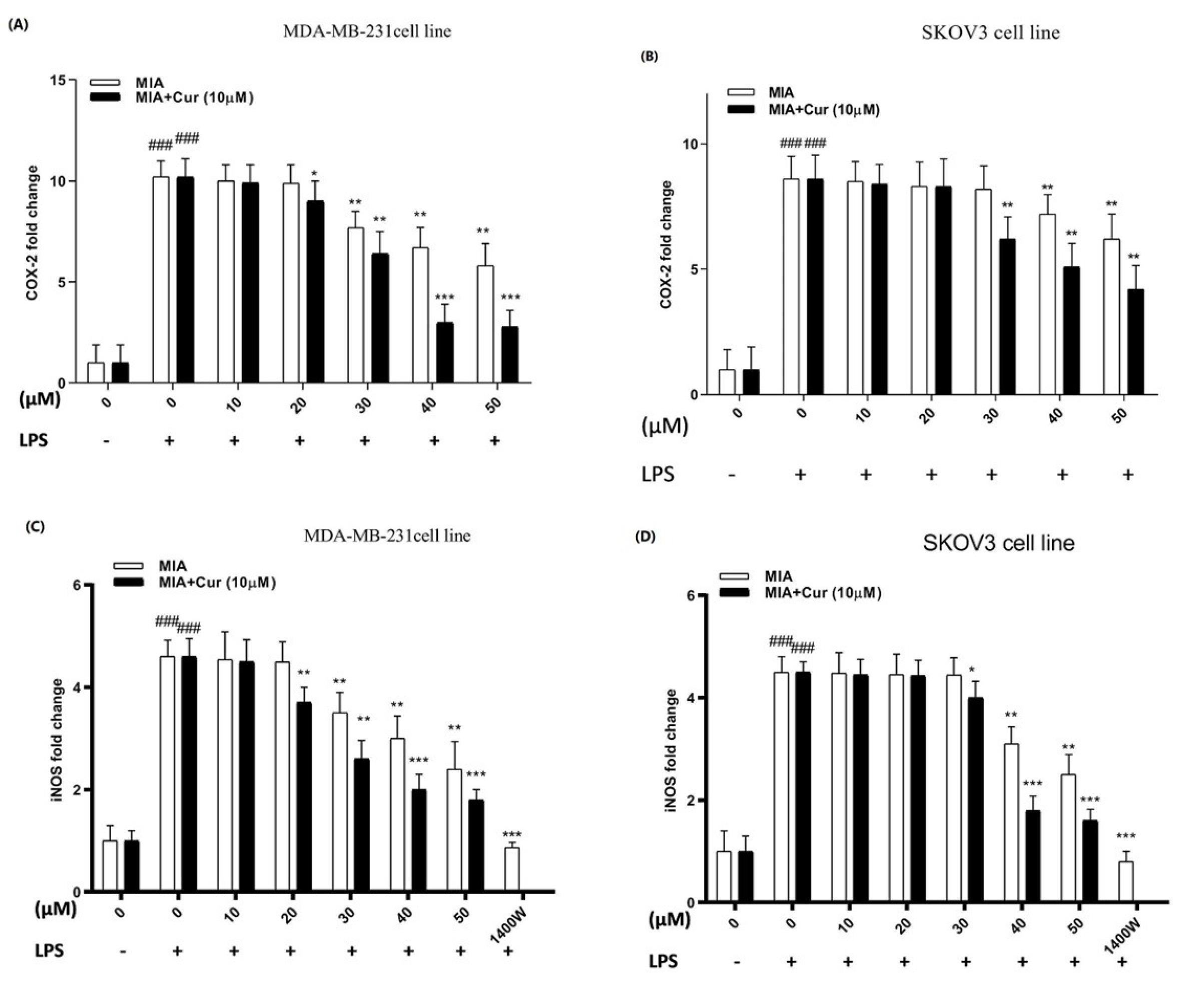
Fig. 6.
MIA alone and together with curcumin diminished COX-2 and iNOS gene expression in LPS-stimulated MDA-MB-231 and SKOV3 cell lines.The fold change levels of COX-2 (A, B) and iNOS (C, D) genes were determined by RT-PCR in response to various concentrations of MIA (10-50 μM) alone or in combination with curcumin (10 μM) in LPS-induced MDA-MB-231 and SKOV3 cells. The presented data are the mean ± SD of at least three separate experiments. # P < 0.05 comparison with untreated unstimulated control cells; * P < 0.05, ** P < 0.01, *** P < 0.001 compared with LPS-stimulated untreated cells.
.
MIA alone and together with curcumin diminished COX-2 and iNOS gene expression in LPS-stimulated MDA-MB-231 and SKOV3 cell lines.The fold change levels of COX-2 (A, B) and iNOS (C, D) genes were determined by RT-PCR in response to various concentrations of MIA (10-50 μM) alone or in combination with curcumin (10 μM) in LPS-induced MDA-MB-231 and SKOV3 cells. The presented data are the mean ± SD of at least three separate experiments. # P < 0.05 comparison with untreated unstimulated control cells; * P < 0.05, ** P < 0.01, *** P < 0.001 compared with LPS-stimulated untreated cells.
MIA and its combination with curcumin diminished nitrite production in LPS-induced cells
As shown in Figs. 7A and 7B, the presence of LPS significantly enhanced nitrite production compared to control cells without LPS stimulation. MIA decreased nitrite production, which was significant at concentrations of 30 μM, 40 μM, and above in LPS-induced MDA-MB-231 and SKOV3 cell lines, respectively. Furthermore, these reduction effects were more remarkable in the combined treatment with curcumin compared to when the cells were treated with MIA alone. These findings suggest that the inhibitory impact of MIA on LPS-stimulated nitrite generation might be due to the suppression of iNOS mRNA expression. 1400W (N-(3-(aminomethyl)benzyl)acetamidine), which a slow and irreversible inhibitor of human iNOS,42 was used as positive control.
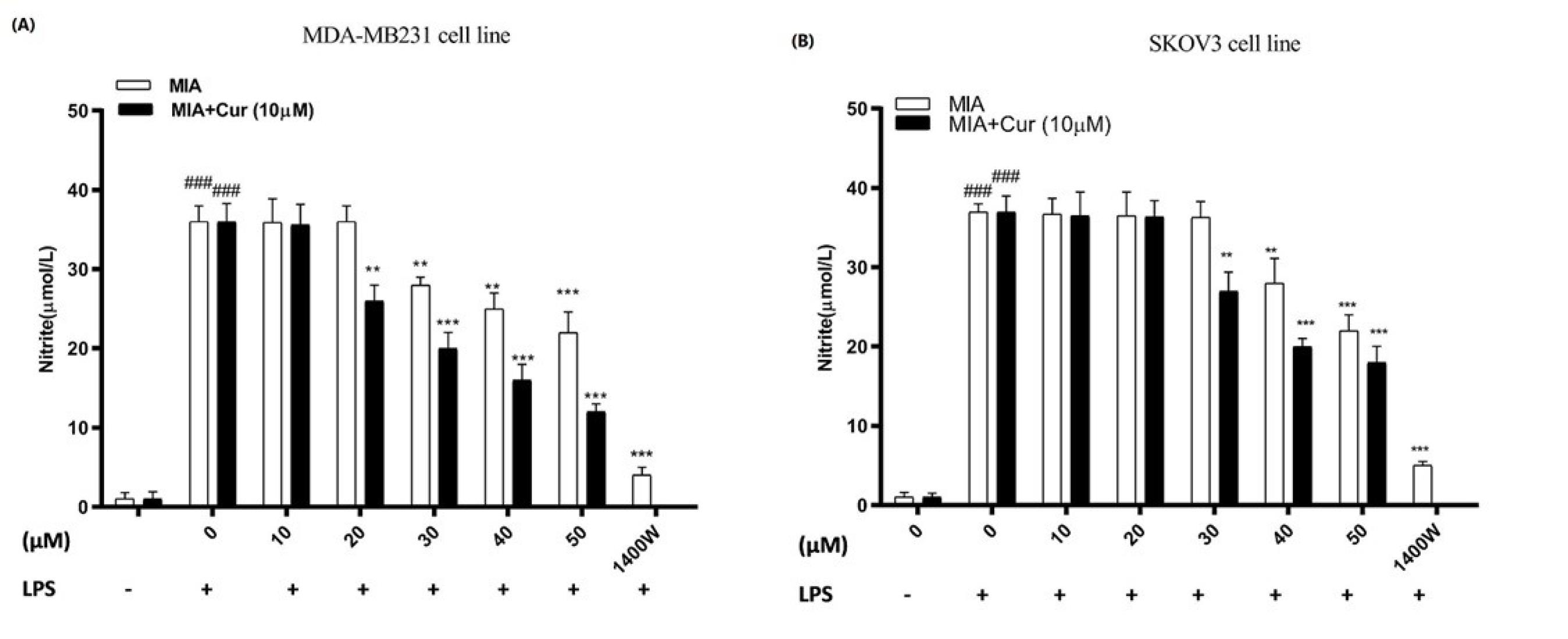
Fig. 7.
MIA lowered nitrite production in LPS-induced MDA-MB-231 and SKOV3 cell lines.The effects of various concentrations of MIA and its combination with curcumin (10 μM) on nitrite production were evaluated by Griess reagent in LPS-stimulated MDA-MB-231 (A) and SKOV3 cells (B). An iNOS inhibitor, called 1400W was used as positive control. # P < 0.05 compared to untreated, unstimulated control cells. * P < 0.05, ** P < 0.01, *** P < 0.001 compared with untreated control cells. At least three times, the experiment was carried out separately.
.
MIA lowered nitrite production in LPS-induced MDA-MB-231 and SKOV3 cell lines.The effects of various concentrations of MIA and its combination with curcumin (10 μM) on nitrite production were evaluated by Griess reagent in LPS-stimulated MDA-MB-231 (A) and SKOV3 cells (B). An iNOS inhibitor, called 1400W was used as positive control. # P < 0.05 compared to untreated, unstimulated control cells. * P < 0.05, ** P < 0.01, *** P < 0.001 compared with untreated control cells. At least three times, the experiment was carried out separately.
MIA and its co-treatment with curcumin enhanced IκBα protein expression in LPS-stimulated cells
We examined the effects of MIA, either alone or combined with curcumin, on IκBα protein expression in LPS-induced cells using western blotting analysis. The results indicated that LPS caused a remarkable reduction in IκBα expression compared to the control in breast and ovarian cancer cell lines (Figs. 8 A and B). In contrast, MIA boosted the IκBα expression and blocked IκBα degradation by LPS in both cell lines. Treatment with MIA and curcumin further increased the IκBα protein expression compared to treatment with MIA alone.
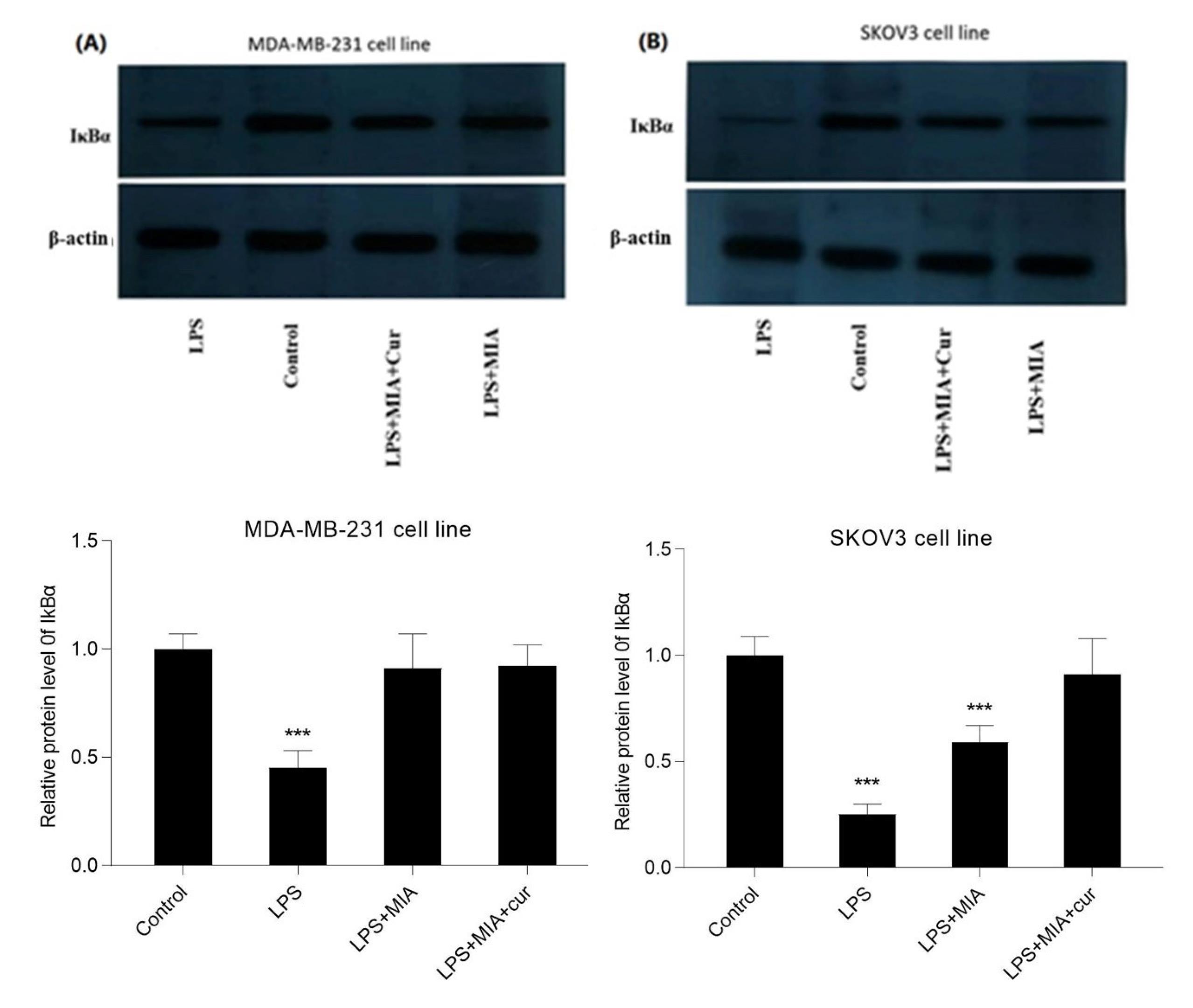
Fig. 8.
MIA and co-treatment with curcumin boosted the IκBα expression in LPS-induced cells.The effects of LPS, MIA, and MIA+curcumin on IκBα protein expression in MDA-MB-231 (A) and SKOV3 (B) cell lines. The cells were treated with MIA (30 μM for MDA-MB-231 and 40 μM for SKOV3 cells) alone and in combination with curcumin (10 μM) for 30 min before exposure to LPS (1 μg/ml) for 1 h. The results of western blotting showed that MIA reduced IκBα degradation and curcumin enhanced its effects. LPS served as the negative control.
.
MIA and co-treatment with curcumin boosted the IκBα expression in LPS-induced cells.The effects of LPS, MIA, and MIA+curcumin on IκBα protein expression in MDA-MB-231 (A) and SKOV3 (B) cell lines. The cells were treated with MIA (30 μM for MDA-MB-231 and 40 μM for SKOV3 cells) alone and in combination with curcumin (10 μM) for 30 min before exposure to LPS (1 μg/ml) for 1 h. The results of western blotting showed that MIA reduced IκBα degradation and curcumin enhanced its effects. LPS served as the negative control.
MIA and its combination with curcumin suppressed p-STAT3 and Bcl-2 and enhanced BAX protein expression in IL-6-stimulated cells
The effects of MIA, either alone or in combination with curcumin, on STAT3, p-STAT3, Bcl-2, and BAX protein expression were evaluated in IL-6-induced cells by western blot analysis. Western blotting results revealed that IL-6 as an activator of the STAT3 signaling pathway enhanced STAT3 phosphorylation at Tyr705 (Fig. 9) in both MDA-MB-231 and SKOV3 cells, without affecting the total STAT3 level. Treatment of cells with MIA caused a significant decline in STAT3 phosphorylation. IL-6 also remarkably enhanced Bcl-2 expression and reduced BAX expression in MDA-MB-231 and SKOV3 cell lines (Fig. 10). On the other hand, MIA suppressed IL-6-induced Bcl-2 protein expression and up-regulated BAX protein expression in both studied cell lines. Furthermore, combined treatment with curcumin augmented the effects of MIA on protein expression compared to MIA treatment alone. These findings suggest that MIA may induce apoptosis by suppressing the IL-6/STAT3 signaling pathway, and curcumin potentiates the induction of apoptosis by MIA. However, this study does not specify the mechanism of apoptosis induction.
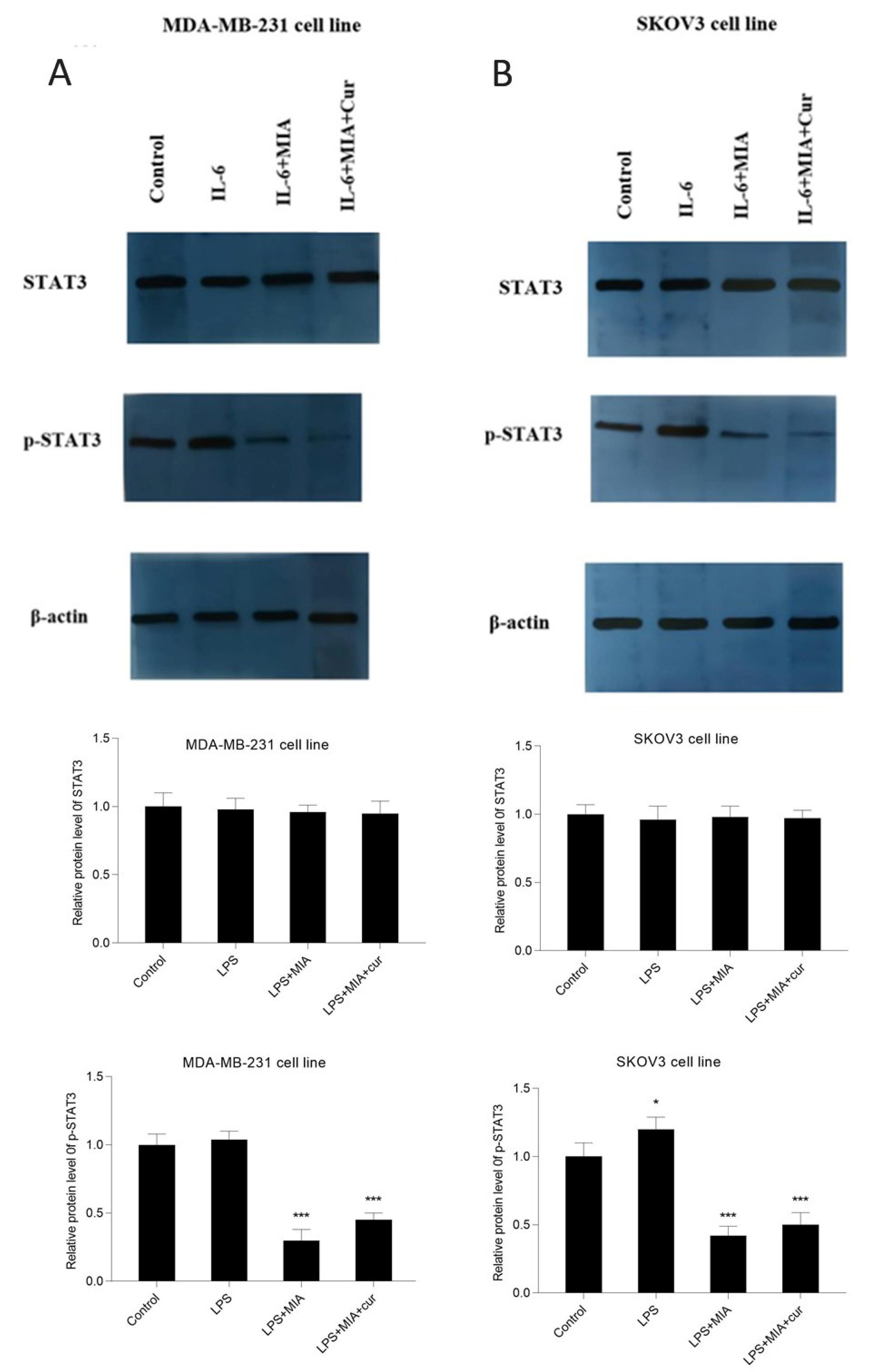
Fig. 9.
MIA and its combination with curcumin suppressed the IL-6-induced phosphorylation of STAT3. The effects of IL-6, MIA, and MIA+curcumin on the protein expression of STAT3 and its phosphorylated form, p-STAT3 in MDA-MB-231 (A) and SKOV3 (B) cells. The densitometric analysis of each blot is presented below the blot image. The cells received IL-6 (50 ng/mL) for 4 h after treatment with MIA (30 μM for MDA-MB-231 and 40 μM for SKOV3 cells) and curcumin (10 μM). Western blotting data indicated that MIA reduced the phosphorylation of STAT3 without affecting STAT3 total proteini level. * P < 0.05, *** P < 0.001 compared with untreated control cells.
.
MIA and its combination with curcumin suppressed the IL-6-induced phosphorylation of STAT3. The effects of IL-6, MIA, and MIA+curcumin on the protein expression of STAT3 and its phosphorylated form, p-STAT3 in MDA-MB-231 (A) and SKOV3 (B) cells. The densitometric analysis of each blot is presented below the blot image. The cells received IL-6 (50 ng/mL) for 4 h after treatment with MIA (30 μM for MDA-MB-231 and 40 μM for SKOV3 cells) and curcumin (10 μM). Western blotting data indicated that MIA reduced the phosphorylation of STAT3 without affecting STAT3 total proteini level. * P < 0.05, *** P < 0.001 compared with untreated control cells.
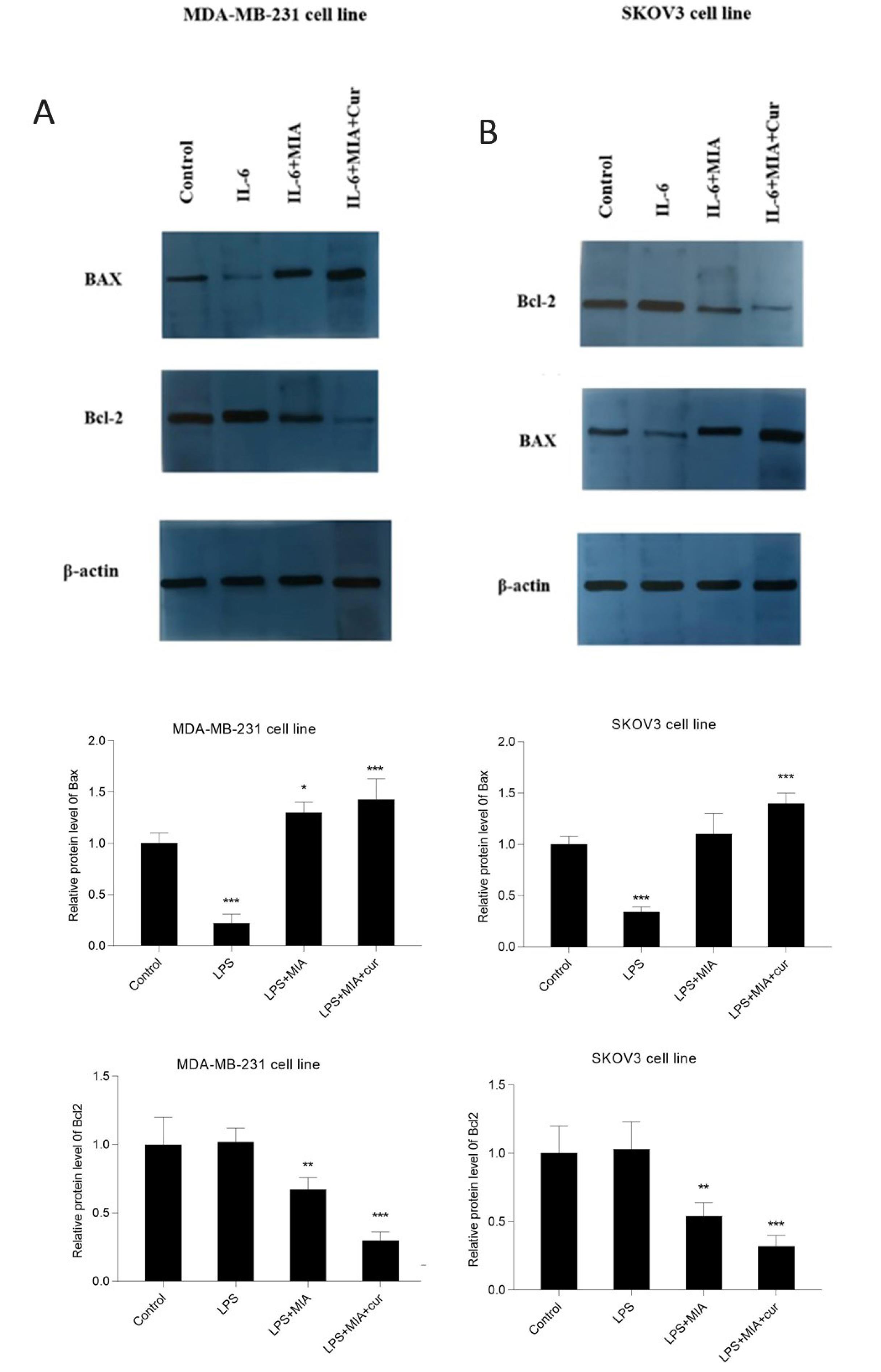
Fig. 10.
MIA either alone or with curcumin repressed the expression of Bcl-2 and enhanced BAX protein in IL-6-stimulated cell lines. The effects of IL-6, MIA, and MIA+curcumin on the protein expression of Bcl-2 and BAX in MDA-MB-231 (A) and SKOV3 (B) cell lines. The cells received IL-6 (50 ng/mL) for 4 h after treatment with MIA (30 μM for MDA-MB-231 and 40 μM for SKOV3 cells) and curcumin (10 μM). The densitometric analysis of blots are presented under each blotting image. Western blotting data indicated that MIA reduced the expression Bcl-2 and up-regulated BAX expression. * P < 0.05, ** P < 0.01, *** P < 0.001 compared with untreated control cells.
.
MIA either alone or with curcumin repressed the expression of Bcl-2 and enhanced BAX protein in IL-6-stimulated cell lines. The effects of IL-6, MIA, and MIA+curcumin on the protein expression of Bcl-2 and BAX in MDA-MB-231 (A) and SKOV3 (B) cell lines. The cells received IL-6 (50 ng/mL) for 4 h after treatment with MIA (30 μM for MDA-MB-231 and 40 μM for SKOV3 cells) and curcumin (10 μM). The densitometric analysis of blots are presented under each blotting image. Western blotting data indicated that MIA reduced the expression Bcl-2 and up-regulated BAX expression. * P < 0.05, ** P < 0.01, *** P < 0.001 compared with untreated control cells.
Discussion
Breast and ovarian cancers are still the most common cancers in women.43 Despite the advances in cancer treatment, there are still many problems and limitations, such as serious side effects of chemotherapy drugs and the emergence of drug resistance.44 Therefore, developing innovative therapeutic approaches are required to effectively control different carcinogenic processes. Compounds with the imidazo[1,2-a]pyridine core have demonstrated remarkable anti-inflammatory activity.45 In this study, we evaluated the anti-inflammatory activity of 8-methyl-2-(4-(methylsulfonyl)phenyl)-N-(p-tolyl) imidazo[1,2-a]pyridine -3-amine, MIA, as a novel synthetic imidazo[1,2-a]pyridine derivative, alone and in combination with curcumin in breast and ovarian cancer cell lines, particularly in regards to its effects on STAT3 and NF-κB pathways. We first evaluated the cytotoxic effects of compounds, the results of the MTT assay indicated that MIA had dose-dependent cytotoxic effects and reduced cell viability in breast and ovarian cancer cells.Previous studies have shown thatimidazo[1,2-a]pyridine derivatives are able to cause cytotoxicity in breast and ovarian cancer cell lines.46,47
Several studies have revealed that curcumin has anti-cancer and anti-inflammatory activities against breast and ovarian malignancies by promoting apoptosis and inhibiting cell growth.48,49 Curcumin has shown beneficial effects, alone or in combination with other drugs.50,51 Novel imidazo[1,2-a]pyridine analogs, inspired by curcumin structure, demonstrated cytotoxicity and antiproliferative activities against several cell lines, including breast and cervical cells.52 In the current investigation, we used curcumin and a new imidazo[1,2-a]pyridine derivative as a novel combination treatment. We provided the first evidence that curcumin can augment the cytotoxic effects of the synthetic imidazo[1,2-a]pyridine derivative, MIA, against MDA-MB-231 and SKOV3 cell lines.
Uncontrolled or aberrant NF-κB activity has been associated with breast and ovarian cancer.53,54 Kamala K. Vasu et al have demonstrated that a series of new imidazo[1,2-a]pyridine possess NF-κB inhibitory activity.55 In this study, our findings showed that MIA diminished the DNA-binding activity of NF-κB and reduced the effects of LPS on NF-κB in breast and ovarian cancer cells.Previous studies have shown that curcumin could be a promising treatment for ovarian and breast cancer by suppressing NF-κB activity.56,57 Furthermore, curcumin was able to enhance the inhibitory effects of MIA on NF-κB activity. Docking studies indicated that both MIA and curcumin could be inserted in the same pocket in the NF-κB p50 monomer active site and form hydrogen bonds. Curcumin formed four strong hydrogen bonds, whereas MIA participated in two hydrogen bonds. Therefore, the docking results could explain that curcumin could enhance the inhibitory effect of this new imidazo[1,2-a]pyridine derivative.
Studies have shown that the direct interaction between NF-κB and STAT3 is linked to inflammation.58 Recently, the cooperation of these two transcription factors has been demonstrated in breast and ovarian cancers,59,60 STAT3 is a critical activator of NF-κB that regulates the expression of pro-inflammatory cytokine, iNOS, COX-2, and TGF-β1 genes.32,61,62 Additionally, the significant role of activated STAT3 has been demonstrated in the nuclear retention of NF-κB.63 IκB keeps the NF-κB transcription factor inactive in the cytoplasm.29 The effect of imidazo[1,2-a]pyridine derivatives on IκBα protein expression has not been investigated yet. We investigated for the first time the effects of MIA, as a synthetic derivative of imidazo[1,2-a]pyridine, on IκBα protein expression in LPS-induced cells. Western blot results revealed that MIA increased IκBα protein expression and counteracted the suppressive effects of LPS. Moreover, curcumin potentiated the effect of MIA on IκBα expression and reduced IκBα degradation in LPS-induced MDA-MB-231 and SKOV3 cells. Proinflammatory cytokines, including IL-1β, TNF-α, and IL-6are related to chronic and acute inflammation. Rether et al reported that TNF-α expression was suppressed by imidazo[1,2-a]pyridine derivatives in T cells.64
Our findings showed that MIA, as a new synthetic imidazo[1,2-a]pyridine derivative, reducedinflammatory cytokines in breast and ovarian cancer cell lines and curcumin enhanced its inhibitory effects. Many studies have demonstrated that increased iNOS expression and NO production are associated with tumor development, such as breast and ovarian carcinoma.65 COX-2 is overexpressed in different cancers and exerts its effects through modulating immune response and inflammation.66 Previous studies have demonstrated that novel synthetic imidazo[1,2-a]pyridine derivatives act as COX-2 blockers.37,67 Gafner et al have shown that curcumin and its derivatives reduce LPS-induced iNOS and COX-2 gene expression in a mouse model.68 Until now, the effects on imidazo[1,2-a]pyridine derivatives have not been evaluated. We investigated for the first time the impacts of a novelimidazo[1,2-a]pyridine derivative on iNOS gene expression. The real-time PCR and Griess test results demonstrated that our new imidazo[1,2-a]pyridine derivative, suppressed the expression of COX-2, iNOS genes, and NO production and curcumin enhanced its inhibitory effects in breast and ovarian cancer cells.
Huang et al have reported that anew imidazo[1,2-a]pyridine derivativecan inhibit STAT3 phosphorylation at Tyr705 in TNBC cell lines.69 In this study, the western blot analysis indicated that MIA suppressed the STAT3 phosphorylation (Tyr705) without altering STAT3 level in IL-6-stimulated breast and ovarian cancer cells, and it was more remarkable with the combination of MIA and curcumin.Taken together, considering the interaction of NF-κB and STAT3 in inflammation and the commonality of target genes (COX-2, iNOS, and inflammatory cytokines), we hypothesize that the anti-inflammatory activity of MIA is mediated via suppression of STAT3/NF-κB/iNOS/COX-2 signaling pathway.
The IL-6 has been demonstrated to activate STAT3 to promote inflammatory responses in breast and ovarian cancers.70-72 Activated STAT3 controls the expression of target genes, such as the Bcl-2 family.73 Previous study indicated that a new imidazo[1,2-a]pyridine compound induced apoptosis via the p53/Bax pathway in HeLa cells.74 Also, Bhavya et al have shown that novel imidazo [1,2-a] pyridine derivatives enhance pro-apoptotic BAX expression, and reduce Bcl-2 expression in A549 cells.75 In this investigation, the impacts of MIA on the BAX and Bcl-2 protein expression was assessed in IL-6-induced breast and ovarian cancer cells. The western blot results revealed that MIA suppressed Bcl-2 expression while boosting BAX protein expression. Although this study does not clarify the mechanism of apoptosis induction by MIA, considering the suppression of STAT3 phosphorylation by MIA, we suppose that MIA may induce apoptosis through inhibiting the IL-6/STAT3 signaling pathway.
Conclusion
Taken together, this study suggests that the anti-inflammatory activity of MIA, a new synthetic derivative of imidazo[1,2-a]pyridine, may be mediated by the suppression of the STAT3/NF-κB/iNOS/COX-2 pathwayin MDA-MB-231 and SKOV3 cell lines. Furthermore, curcumin effectively potentiated the anti-inflammatory effects of MIA in breast and ovarian cancer cells. Therefore, MIA might be a potential candidate with anti-inflammatory activities alongside curcumin for cancer treatment. Nevertheless, the therapeutic value of these compounds must first be determined through additional pharmacokinetic and in vivo investigations.
Research Highlights
What is the current knowledge?
√ Curcumin, either alone or in combination with other agents, exerts significant anti-cancer and anti-inflammatory impacts on various cancer cell lines.
√ Imidazo[1,2-a]pyridine derivatives have a broad pharmaceutical activity, such as anti-inflammatory and anti-cancer effects.
√ STAT3 and NF-κB pathways have a significant collaboration in inflammation and cancer.
What is new here?
√ MIA as a new imidazo[1,2-a]pyridine derivative is able to reduce cell viability, trigger apoptosis and suppress inflammation in breast and ovarian cancer cells.
√ Curcumin-imidazo[1,2-a]pyridine derivative as a novel combination therapy exerts anti-inflammatory effects in breast and ovarian cancer cells.
√ The anti-inflammatory activity of MIA is exerted by modulating the STAT3/NF-κB/iNOS/COX-2 signaling pathway in MDA-MB-231 and SKOV3 cell lines.
√ Curcumin enhances the anti-inflammatory effects of MIA.
Competing Interests
The authors declare that there is no conflict of interest.
Ethical Statement
Not applicable.
Funding
This study was financially supported by Iran National Science Foundation, grant number 90003943 and Shahid Beheshti University of Medical Sciences grant number 43002565.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021; 71:209-49. doi: 10.3322/caac.21660 [Crossref] [ Google Scholar]
- Huang J, Chan WC, Ngai CH, Lok V, Zhang L, Lucero-Prisno DE. Worldwide Burden, Risk Factors, and Temporal Trends of Ovarian Cancer: A Global Study. Cancers (Basel) 2022; 14:2230. doi: 10.3390/cancers14092230 [Crossref] [ Google Scholar]
- Senapati S, Mahanta AK, Kumar S, Maiti P. Controlled drug delivery vehicles for cancer treatment and their performance. Signal Transduct Target Ther 2018. 3: 1-19. 10.1038/s41392-017-0004-3.
- Dehelean CA, Marcovici I, Soica C, Mioc M, Coricovac D, Iurciuc S. Plant-derived anticancer compounds as new perspectives in drug discovery and alternative therapy. Molecules 2021; 26:1109. doi: 10.3390/molecules26041109 [Crossref] [ Google Scholar]
- Dinic J, Podolski-Renic A, Stankovic T, Bankovic J, Pesic M. New approaches with natural product drugs for overcoming multidrug resistance in cancer. Curr Pharm Des 2015; 21:5589-604. doi: 10.2174/1381612821666151002113546 [Crossref] [ Google Scholar]
- Huang M, Lu J-J, Ding J. Natural products in cancer therapy: past, present and future. Nat Prod Bioprospect 2021; 11:5-13. doi: 10.1007/s13659-020-00293-7 [Crossref] [ Google Scholar]
- Tomeh MA, Hadianamrei R, Zhao X. A review of curcumin and its derivatives as anticancer agents. Int J Mol Sci 2019; 20:1033. doi: 10.3390/ijms20051033 [Crossref] [ Google Scholar]
- Giordano A, Tommonaro G. Curcumin and cancer. Nutrients 2019; 11:2376. doi: 10.3390/nu11102376 [Crossref] [ Google Scholar]
- Liu D, Chen Z. The effect of curcumin on breast cancer cells. J Breast Cancer 2013; 16:133-7. doi: 10.4048/jbc.2013.16.2.133 [Crossref] [ Google Scholar]
- Maiti P, Plemmons A, Dunbar GL. Combination treatment of berberine and solid lipid curcumin particles increased cell death and inhibited PI3K/Akt/mTOR pathway of human cultured glioblastoma cells more effectively than did individual treatments. PloS One 2019; 14:e0225660. doi: 10.1371/journal.pone.0225660 [Crossref] [ Google Scholar]
- DiMarco‐Crook C, Rakariyatham K, Li Z, Du Z, Zheng J, Wu X. Synergistic anticancer effects of curcumin and 3', 4'‐didemethylnobiletin in combination on colon cancer cells. J Food Sci 2020; 85:1292-301. doi: 10.1111/1750-3841.15073 [Crossref] [ Google Scholar]
- Kim YB, Kang CW, Ranatunga S, Yang H, Sebti SM, Del Valle JR. Imidazo[1,2-a]pyridine-based peptidomimetics as inhibitors of Akt. Bioorg Med Chem Lett 2014; 24:4650-3. doi: 10.1016/j.bmcl.2014.08.040 [Crossref] [ Google Scholar]
- Almeida GM, Rafique J, Saba S, Siminski T, Mota N, Filho DW. Novel selenylated imidazo[1,2-a]pyridines for breast cancer chemotherapy: Inhibition of cell proliferation by Akt-mediated regulation, DNA cleavage and apoptosis. Biochem Biophys Res Commun 2018; 503:1291-7. doi: 10.1016/j.bbrc.2018.07.039 [Crossref] [ Google Scholar]
- Altaher AM, Adris MA, Aliwaini SH, Awadallah AM, Morjan RY. The Anticancer Effects of Novel Imidazo[1,2-a]Pyridine Compounds against HCC1937 Breast Cancer Cells. Asian Pac J Cancer Prev 2022; 23:2943-51. doi: 10.31557/apjcp.2022.23.9.2943 [Crossref] [ Google Scholar]
- Kim O, Jeong Y, Lee H, Hong S-S, Hong S. Design and Synthesis of Imidazopyridine Analogues as Inhibitors of Phosphoinositide 3-Kinase Signaling and Angiogenesis. J Med Chem 2011; 54:2455-66. doi: 10.1021/jm101582z [Crossref] [ Google Scholar]
- Li GY, Jung KH, Lee H, Son MK, Seo J, Hong SW. A novel imidazopyridine derivative, HS-106, induces apoptosis of breast cancer cells and represses angiogenesis by targeting the PI3K/mTOR pathway. Cancer Lett 2013; 329:59-67. doi: 10.1016/j.canlet.2012.10.013 [Crossref] [ Google Scholar]
- Lee H, Jung KH, Jeong Y, Hong S, Hong SS. HS-173, a novel phosphatidylinositol 3-kinase (PI3K) inhibitor, has anti-tumor activity through promoting apoptosis and inhibiting angiogenesis. Cancer Lett 2013; 328:152-9. doi: 10.1016/j.canlet.2012.08.020 [Crossref] [ Google Scholar]
- Deep A, Kaur Bhatia R, Kaur R, Kumar S, Kumar Jain U, Singh H. Imidazo [1, 2-a] pyridine scaffold as prospective therapeutic agents. Curr Top Med Chem 2017; 17:238-50. doi: 10.2174/1568026616666160530153233 [Crossref] [ Google Scholar]
- Aliwaini S, Awadallah AM, Morjan RY, Ghunaim M, Alqaddi H, Abuhamad AY. Novel imidazo [1, 2-a] pyridine inhibits AKT/mTOR pathway and induces cell cycle arrest and apoptosis in melanoma and cervical cancer cells. Oncol Lett 2019; 18:830-7. doi: 10.3892/ol.2019.10341 [Crossref] [ Google Scholar]
- Kwong HC, Kumar C, Mah SH, Mah YL, Chia TS, Quah CK. Crystal Correlation of Heterocyclic Imidazo [1, 2-a] pyridine Analogues and their Anticholinesterase potential evaluation. Sci Rep 2019; 9:1-15. doi: 10.1038/s41598-018-37486-7 [Crossref] [ Google Scholar]
- Macciò A, Madeddu C. Inflammation and ovarian cancer. Cytokine 2012; 58:133-47. doi: 10.1016/j.cyto.2012.01.015 [Crossref] [ Google Scholar]
- Danforth DN. The role of chronic inflammation in the development of breast cancer. Cancers (Basel) 2021; 13:3918. doi: 10.3390/cancers13153918 [Crossref] [ Google Scholar]
- Zinatizadeh MR, Schock B, Chalbatani GM, Zarandi PK, Jalali SA, Miri SR. The Nuclear Factor Kappa B (NF-kB) signaling in cancer development and immune diseases. Genes Dis 2021; 8:287-97. doi: 10.1016/j.gendis.2020.06.005 [Crossref] [ Google Scholar]
- Xia Y, Shen S, Verma IM. NF-κB, an active player in human cancers. Cancer Immunol Res 2014; 2:823-30. doi: 10.1158/2326-6066.CIR-14-0112 [Crossref] [ Google Scholar]
- Tak PP, Firestein GS. NF-κB: a key role in inflammatory diseases. J Clin Invest 2001; 107:7-11. doi: 10.1172/JCI11830 [Crossref] [ Google Scholar]
- Serasanambati M, Chilakapati SR. Function of nuclear factor kappa B (NF-kB) in human diseases-a review. South Indian J Biol Sci 2016; 2:368-87. doi: 10.22205/sijbs/2016/v2/i4/103443 [Crossref] [ Google Scholar]
- Sharif O, Bolshakov VN, Raines S, Newham P, Perkins ND. Transcriptional profiling of the LPS induced NF-κB response in macrophages. BMC Immunol 2007; 8:1-17. doi: 10.1186/1471-2172-8-1 [Crossref] [ Google Scholar]
- Luo K. Signaling cross talk between TGF-β/Smad and other signaling pathways. Cold Spring Harb Perspect Biol 2017; 9:a022137. doi: 10.1101/cshperspect.a022137 [Crossref] [ Google Scholar]
- Oeckinghaus A, Ghosh S. The NF-κB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol 2009; 1:a000034. doi: 10.1101/cshperspect.a000034 [Crossref] [ Google Scholar]
- Al Zaid Siddiquee K, Turkson J. STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res 2008; 18:254-67. doi: 10.1038/cr.2008.18 [Crossref] [ Google Scholar]
- Fathi N, Rashidi G, Khodadadi A, Shahi S, Sharifi S. STAT3 and apoptosis challenges in cancer. Int J Biol Macromol 2018; 117:993-1001. doi: 10.1016/j.ijbiomac.2018.05.121 [Crossref] [ Google Scholar]
- Carpenter RL, Lo H-W. STAT3 target genes relevant to human cancers. Cancers (Basel) 2014; 6:897-925. doi: 10.3390/cancers6020897 [Crossref] [ Google Scholar]
- Rébé C, Végran F, Berger H, Ghiringhelli F. STAT3 activation: A key factor in tumor immunoescape. JAKSTAT 2013; 2:e23010. doi: 10.4161/jkst.23010 [Crossref] [ Google Scholar]
- Fan Y, Mao R, Yang J. NF-κB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell 2013; 4:176-85. doi: 10.1007/s13238-013-2084-3 [Crossref] [ Google Scholar]
- Grivennikov SI, Karin M. Dangerous liaisons: STAT3 and NF-κB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev 2010; 21:11-9. doi: 10.1016/j.cytogfr.2009.11.005 [Crossref] [ Google Scholar]
- Hirano T. IL-6 in inflammation, autoimmunity and cancer. Int Immunol 2021; 33:127-48. doi: 10.1093/intimm/dxaa078 [Crossref] [ Google Scholar]
- Azami Movahed M, Daraei B, Zarghi A. Synthesis and biological evaluation of new imidazo [1, 2-a] pyridine derivatives as selective cox-2 inhibitors. Lett Drug Des Discov 2016; 13:793-9. doi: 10.2174/1570180813666160613090944 [Crossref] [ Google Scholar]
- Griendling KK, Touyz RM, Zweier JL, Dikalov S, Chilian W, Chen Y-R. Measurement of reactive oxygen species, reactive nitrogen species, and redox-dependent signaling in the cardiovascular system: a scientific statement from the American Heart Association. Circ Res 2016; 119:e39-e75. doi: 10.1161/RES.0000000000000110 [Crossref] [ Google Scholar]
- Shin JS, Lee KG, Lee HH, Lee HJ, An HJ, Nam JH. Shin JS, Lee KG, Lee HH, Lee HJ, An HJ, Nam JH, et alα‐Solanine Isolated From Solanum Tuberosum Lcv Jayoung Abrogates LPS‐Induced Inflammatory Responses Via NF‐κB Inactivation in RAW 2647 Macrophages and Endotoxin‐Induced Shock Model in Mice. J Cell Biochem 2016; 117:2327-39. doi: 10.1002/jcb.25530 [Crossref] [ Google Scholar]
- Gutiérrez-Venegas G, Torras-Ceballos A, Gómez-Mora JA, Fernández-Rojas B. Luteolin, quercetin, genistein and quercetagetin inhibit the effects of lipopolysaccharide obtained from Porphyromonas gingivalis in H9c2 cardiomyoblasts. Cell Mol Biol Lett 2017; 22:1-12. doi: 10.1186/s11658-017-0047-z [Crossref] [ Google Scholar]
- Afshari H, Nourbakhsh M, Salehi N, Mahboubi-Rabbani M, Zarghi A, Noori S. STAT3-mediated apoptotic-enhancing function of sclareol against breast cancer cells and cell sensitization to cyclophosphamide. Iran J Pharm Res 2020; 19:398. doi: 10.22037/ijpr.2020.112587.13843 [Crossref] [ Google Scholar]
- Garvey EP, Oplinger JA, Furfine ES, Kiff RJ, Laszlo F, Whittle BJ. 1400W is a slow, tight binding, and highly selective inhibitor of inducible nitric-oxide synthase in vitro and in vivo. J Biol Chem 1997; 272:4959-63. doi: 10.1074/jbc.272.8.4959 [Crossref] [ Google Scholar]
- Wooster R, Weber BL. Breast and Ovarian Cancer. N Engl J Med 2003; 348:2339-47. doi: 10.1056/NEJMra012284 [Crossref] [ Google Scholar]
- Chakraborty S, Rahman T. The difficulties in cancer treatment. Ecancermedicalscience 2012; 6:ed16. doi: 10.3332/ecancer.2012.ed16 [Crossref] [ Google Scholar]
- Márquez-Flores YK, Campos-Aldrete M, Salgado-Zamora H, Correa-Basurto J, Meléndez-Camargo M. Acute and chronic anti-inflammatory evaluation of imidazo [1, 2-a] pyridine carboxylic acid derivatives and docking analysis. Med Chem Res 2012; 21:3491-8. doi: 10.1007/s00044-011-9870-3 [Crossref] [ Google Scholar]
- Dam J, Ismail Z, Kurebwa T, Gangat N, Harmse L, Marques HM. Synthesis of copper and zinc 2-(pyridin-2-yl) imidazo [1, 2-a] pyridine complexes and their potential anticancer activity. Eur J Med Chem 2017; 126:353-68. doi: 10.1016/j.ejmech.2016.10.041 [Crossref] [ Google Scholar]
- Teng Q-H, Peng X-J, Mo Z-Y, Xu Y-L, Tang H-T, Wang H-S. Transition-metal-free C–N and C–C formation: synthesis of benzo [4, 5] imidazo [1, 2-a] pyridines and 2-pyridones from ynones. Green Chem 2018; 20:2007-12. doi: 10.1039/C8GC00069G [Crossref] [ Google Scholar]
- Hu S, Xu Y, Meng L, Huang L, Sun H. Curcumin inhibits proliferation and promotes apoptosis of breast cancer cells. Exp Ther Med 2018; 16:1266-72. doi: 10.3892/etm.2018.6345 [Crossref] [ Google Scholar]
- Liu L-d, Pang Y-x, Zhao X-r, Li R, Jin C-j, Xue J. Curcumin induces apoptotic cell death and protective autophagy by inhibiting AKT/mTOR/p70S6K pathway in human ovarian cancer cells. Arch Gynecol Obstet 2019; 299:1627-39. doi: 10.1007/s00404-019-05058-3 [Crossref] [ Google Scholar]
- Hu Y, Ran M, Wang B, Lin Y, Cheng Y, Zheng S. Co-delivery of docetaxel and curcumin via nanomicelles for enhancing anti-ovarian cancer treatment. Int J Nanomedicine 2020; 15:9703. doi: 10.2147/IJN.S274083 [Crossref] [ Google Scholar]
- Wang K, Zhang C, Bao J, Jia X, Liang Y, Wang X. Synergistic chemopreventive effects of curcumin and berberine on human breast cancer cells through induction of apoptosis and autophagic cell death. Sci Rep 2016; 6:1-14. doi: 10.1038/srep26064 [Crossref] [ Google Scholar]
- Ramya PS, Guntuku L, Angapelly S, Digwal CS, Lakshmi UJ, Sigalapalli DK. Synthesis and biological evaluation of curcumin inspired imidazo [1, 2-a] pyridine analogues as tubulin polymerization inhibitors. Eur J Med Chem 2018; 143:216-31. doi: 10.1016/j.ejmech.2017.11.010 [Crossref] [ Google Scholar]
- Wu JT, Kral JG. The NF-κB/IκB signaling system: a molecular target in breast cancer therapy. J Surg Res 2005; 123:158-69. doi: 10.1016/j.jss.2004.06.006 [Crossref] [ Google Scholar]
- Harrington BS, Annunziata CM. NF-κB signaling in ovarian cancer. Cancers (Basel) 2019; 11:1182. doi: 10.3390/cancers11081182 [Crossref] [ Google Scholar]
- Vasu KK, Digwal CS, Pandya AN, Pandya DH, Sharma JA, Patel S. Imidazo [1, 2-a] pyridines linked with thiazoles/thiophene motif through keto spacer as potential cytotoxic agents and NF-κB inhibitors. Bioorg Med Chem Lett 2017; 27:5463-6. doi: 10.1016/j.bmcl.2017.10.060 [Crossref] [ Google Scholar]
- Lin YG, Kunnumakkara AB, Nair A, Merritt WM, Han LY, Armaiz-Pena GN. Curcumin inhibits tumor growth and angiogenesis in ovarian carcinoma by targeting the nuclear factor-κB pathway. Clin Cancer Res 2007; 13:3423-30. doi: 10.1158/1078-0432.CCR-06-3072 [Crossref] [ Google Scholar]
- Aggarwal BB, Shishodia S, Takada Y, Banerjee S, Newman RA, Bueso-Ramos CE. Curcumin suppresses the paclitaxel-induced nuclear factor-κB pathway in breast cancer cells and inhibits lung metastasis of human breast cancer in nude mice. Clin Cancer Res 2005; 11:7490-8. doi: 10.1158/1078-0432.CCR-05-1192 [Crossref] [ Google Scholar]
- Lee H, Herrmann A, Deng J-H, Kujawski M, Niu G, Li Z. Persistently activated Stat3 maintains constitutive NF-κB activity in tumors. Cancer Cell 2009; 15:283-93. doi: 10.1016/j.ccr.2009.02.015 [Crossref] [ Google Scholar]
- Chung SS, Aroh C, Vadgama JV. Constitutive activation of STAT3 signaling regulates hTERT and promotes stem cell-like traits in human breast cancer cells. PloS One 2013; 8:e83971. doi: 10.1371/journal.pone.0083971 [Crossref] [ Google Scholar]
- Chen C, You F, Wu F, Luo Y, Xu H, Liu Y. Antiangiogenesis efficacy of ethanol extract from Amomum tsaoko in ovarian Cancer through inducing ER stress to suppress p-STAT3/NF-kB/IL-6 and VEGF loop. Evid Based Complement Alternat Med 2020; 2020:2390125. doi: 10.1155/2020/2390125 [Crossref] [ Google Scholar]
- Bolli R, Dawn B, Xuan Y-T. Role of the JAK–STAT pathway in protection against myocardial ischemia/reperfusion injury. Trends Cardiovasc Med 2003; 13:72-9. doi: 10.1016/s1050-1738(02)00230-x [Crossref] [ Google Scholar]
- Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 2009; 9:798-809. doi: 10.1038/nrc2734 [Crossref] [ Google Scholar]
- McFarland BC, Hong SW, Rajbhandari R, Twitty Jr GB, Gray GK, Yu H. NF-κB-induced IL-6 ensures STAT3 activation and tumor aggressiveness in glioblastoma. PloS One 2013; 8:e78728. doi: 10.1371/journal.pone.0078728 [Crossref] [ Google Scholar]
- Rether J, Erkel G, Anke T, Bajtner J, Sterner O. Imidazo [1, 2-a] pyridine derivatives as inhibitors of TNF-α expression in T cells. Bioorg Med Chem 2008; 16:1236-41. doi: 10.1016/j.bmc.2007.10.074 [Crossref] [ Google Scholar]
- Liao W, Ye T, Liu H. Prognostic value of inducible nitric oxide synthase (iNOS) in human cancer: a systematic review and meta-analysis. Biomed Res Int 2019; 2019:6304851. doi: 10.1155/2019/6304851 [Crossref] [ Google Scholar]
- Xu X-C. COX-2 inhibitors in cancer treatment and prevention, a recent development. Anticancer Drugs 2002; 13:127-37. doi: 10.1097/00001813-200202000-00003 [Crossref] [ Google Scholar]
- Ismael AS, Amin NH, Elsaadi MT, Ali MR, Abdel-Rahman HM. Design, synthesis and biological evaluation of new imidazo [1, 2-a] pyridine derivatives as selective COX-2 inhibitors. J Mol Struct 2022; 1250:131652. doi: 10.1016/j.molstruc.2021.131652 [Crossref] [ Google Scholar]
- Gafner S, Lee S-K, Cuendet M, Barthélémy S, Vergnes L, Labidalle S. Biologic evaluation of curcumin and structural derivatives in cancer chemoprevention model systems. Phytochemistry 2004; 65:2849-59. doi: 10.1016/j.phytochem.2004.08.008 [Crossref] [ Google Scholar]
- Huang Q, Zhong Y, Li B, Ouyang S, Deng L, Mo J. Structure-based discovery of potent and selective small-molecule inhibitors targeting signal transducer and activator of transcription 3 (STAT3). Eur J Med Chem 2021; 221:113525. doi: 10.1016/j.ejmech.2021.113525 [Crossref] [ Google Scholar]
- Neurath MF, Finotto S. IL-6 signaling in autoimmunity, chronic inflammation and inflammation-associated cancer. Cytokine Growth Factor Rev 2011; 22:83-9. doi: 10.1016/j.cytogfr.2011.02.003 [Crossref] [ Google Scholar]
- Browning L, Patel MR, Horvath EB, Tawara K, Jorcyk CL. IL-6 and ovarian cancer: Inflammatory cytokines in promotion of metastasis. Cancer Manag Res 2018; 10:6685. doi: 10.2147/CMAR.S179189 [Crossref] [ Google Scholar]
- Aryappalli P, Al-Qubaisi SS, Attoub S, George JA, Arafat K, Ramadi KB. The IL-6/STAT3 signaling pathway is an early target of manuka honey-induced suppression of human breast cancer cells. Front Oncol 2017; 7:167. doi: 10.3389/fonc.2017.00167 [Crossref] [ Google Scholar]
- Xie Q, Yang Z, Huang X, Zhang Z, Li J, Ju J. Ilamycin C induces apoptosis and inhibits migration and invasion in triple-negative breast cancer by suppressing IL-6/STAT3 pathway. J Hematol Oncol 2019; 12:1-14. doi: 10.1186/s13045-019-0744-3 [Crossref] [ Google Scholar]
- Yu Y, Li Y, Yang X, Deng Q, Xu B, Cao H. A Novel Imidazo [1, 2-a] pyridine Compound Reduces Cell Viability and Induces Apoptosis of HeLa Cells by p53/Bax-Mediated Activation of Mitochondrial Pathway. Anticancer Agents Med Chem 2022; 22:1102-10. doi: 10.2174/1871520621666210805130925 [Crossref] [ Google Scholar]
- Bhavya K, Mantipally M, Roy S, Arora L, Badavath VN, Gangireddy M. Novel imidazo [1, 2-a] pyridine derivatives induce apoptosis and cell cycle arrest in non-small cell lung cancer by activating NADPH oxidase mediated oxidative stress. Life Sci 2022; 294:120334. doi: 10.1016/j.lfs.2022.120334 [Crossref] [ Google Scholar]