Bioimpacts. 14(4):27640.
doi: 10.34172/bi.2023.27640
Original Article
Telomerase and mitochondria inhibition promote apoptosis and TET2 and ANMT3a expression in triple negative breast cancer cell lines
Zeinab Mazloumi Data curation, Formal analysis, Investigation, Resources, Software, Writing – original draft, 1, 2 
Ali Rafat Formal analysis, Investigation, Resources, Writing – review & editing, 3
Khadijeh Dizaji Asl Investigation, Writing – review & editing, 4
Mohammad Karimipour Conceptualization, Writing – original draft, 5
Dariush Shanehbandi Methodology, 6
Mehdi Talebi Methodology, 1
Majid Montazer Visualization, 7
Ali Akbar Movassaghpour Visualization, 8
Alireza Dehnad Visualization, 9
Raheleh Farahzadi Methodology, 8
Hojjatollah Nozad Charoudeh Conceptualization, Data curation, Funding acquisition, Investigation, Project administration, Resources, Supervision, Validation, Writing – review & editing, 8, * 
Author information:
1Department of Applied Cell Sciences, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
2Student Research Committee, Tabriz University of Medical Sciences, Tabriz, Iran
3Anatomical Sciences Research Center, Institute for Basic Sciences, Kashan University of Medical Sciences, Kashan, Iran
4Department of Histopathology and Anatomy, Faculty of Medical Sciences, Tabriz Medical Sciences, Islamic Azad University, Tabriz, Iran
5Department of Anatomical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
6Immunology Research Center, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
7Department of Cardiovascular Surgery, Imam Reza Hospital, Tabriz University of Medical Sciences, Tabriz, Iran
8Stem Cell Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
9Department of Bacterial Disease Research, Razi Vaccine, and Serum Research Institute, AREEO, Tabriz, Iran
Abstract
Introduction:
High metastasis, resistance to common treatments, and high mortality rate, has made triple-negative breast cancer (TNBC) to be the most invasive type of breast cancer. High telomerase activity and mitochondrial biogenesis are involved in breast cancer tumorigenesis. The catalytic subunit of telomerase, telomerase reverse transcriptase (hTERT), plays a role in telomere lengthening and extra-biological functions such as gene expression, mitochondria function, and apoptosis. In this study, it has been aimed to evaluate intrinsic-, extrinsic-apoptosis and DNMT3a and TET2 expression following the inhibition of telomerase and mitochondria respiration in TNBC cell lines.
Methods:
TNBC cells were treated with IC50 levels of BIBR1532, tigecycline, and also their combination. Then, telomere length, and DNMT3a, TET2, and hTERT expression were evaluated. Finally, apoptosis rate, apoptosis-related proteins, and genes were analyzed.
Results:
The present results showed that IC50 level of telomerase and inhibition of mitochondria respiration induced apoptosis but did not leave any significant effect on telomere length. The results also indicated that telomerase inhibition induced extrinsic-apoptosis in MDA-MB-231 and caused intrinsic- apoptosis in MDA-MB-468 cells. Furthermore, it was found that the expression of p53 decreased and was ineffective in cell apoptosis. The expressions of DNMT3a and TET2 increased in cells. In addition, combination treatment was better than BIBR1532 and tigecycline alone.
Conclusion:
The inhibition of telomerase and mitochondria respiration caused intrinsic- and extrinsic- apoptosis and increased DNMT3a and TET2 expression and it could be utilized in breast cancer treatment.
Keywords: Cancer stem cell, Telomerase, Mitochondria, Apoptosis, DNMT3a, TET2, Triple negative breast cancer
Copyright and License Information
© 2024 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Triple-negative breast cancer (TNBC) is an invasive subtype of breast cancer (containing 15–20%) that do not express Her-2, Estrogen, and Progesterone receptors.1 TNBC cells have cancer stem cells’ (CSCs’) origin with less differentiated property and high proliferation levels.2,3 TNBC patients are resistant to conventional therapy and display higher metastasis, disease recurrence, and mortality rate than other subtypes of breast cancer.4,5 According to the studies, telomerase activity is high in over 90% of breast cancers.6
Telomerase is a reverse transcriptase enzyme that protects telomeres (the ends of chromosomes) from erosion and makes genomic stability.7 Telomerase consists of two subunits including hTERT (catalytic unit) and TERC (RNA template).8,9 Telomerase activity is associated with the expression of hTERT and causes poor prognosis of breast cancer.6,10 In addition, hTERT is involved in extra-biological functions such as gene expression and cell proliferation.8 hTERT translocates to the mitochondria due to exogenous stress, resulting in protecting cells from DNA damage and apoptosis.8,11 Furthermore, high mitochondrial biogenesis causes stemness, proliferation, tumorigenesis, and metastasis in TNBC, and eventually increases tumor growth and chemotherapy resistance.12-15
There are associations between hTERT expression and methylation in breast cancer cells.16 Methylation, as an epigenetic mechanism, involves cellular functions, the regulation of gene expression, and tumor progression.17 Demethylation and methylation of DNA is catalyzed by ten-eleven translocation proteins (TET1,2,3) and DNA methyltransferases (DNMT1,3A,3B), respectively.17,18 Decreased expression of TET and DNMT3a in breast cancer causes metastasis and tumorigenesis.19,20
It has been shown that TNBC is resistant to common treatments. In general, telomerase and mitochondria are essential for the progression of cancers and cancer cell survival.4,14,21 Thus, a better understanding of the effect of their inhibition might help develop effective anti-cancer therapies. In addition, the combination treatment is highly efficient strategy than a single treatment in treating cancer.22,23
BIBR1532 is a nonpeptidic, non-nucleoside small molecule that inhibits telomerase activity by binding specifically to the active site of hTERT. It is indicated that BIBR1532 had potential anti-tumor activity in different cancers by triggering replicative senescence and apoptosis.24,25 BIBR1532 also displays potential efficacy in combination with other anti-cancer agents in breast cancer.26
Tigecycline is an antibiotic, with similar structure to Tetracyclines. It inhibits protein translation by strong binding to the 30S ribosomal subunit. Tigecycline has an anti-cancer effect in human acute myeloid leukemia (AML) and various solid tumors including TNBC. In addition, combinations of tigecycline with chemotherapeutic drugs are for cancer treatment.27,28 Tigecycline has been used in clinical trials for the treatment of acute myeloid leukemia (Clinical Trial No. NCT01332786) and chronic myeloid leukemia (Clinical Trial No. NCT02883036).
In the current study, telomerase and mitochondria respiration in TNBC was inhibited and the expression of apoptosis-related genes, proteins, DNMT3a, and TET 2 were evaluated.
Materials and Methods
Cell lines
MDA-MB-468 and MDA-MB-231 cells (TNBC cell lines) obtained from Pasteur Institute, Tehran, Iran (National Cell Bank). The cells were cultured in Roswell Park Memorial Institute 1640 (RPMI 1640) medium complemented with 1% penicillin/streptomycin and 10% fetal bovine serum (FBS) at 37 °C and 5% CO2. After 3 passages, MDA-MB-231and MDA-MB-468 cells contained (94.6%) CD44+ CD24- (Fig. 1A) and (81%) CD44+ CD24+ (Fig. 1B), respectively.
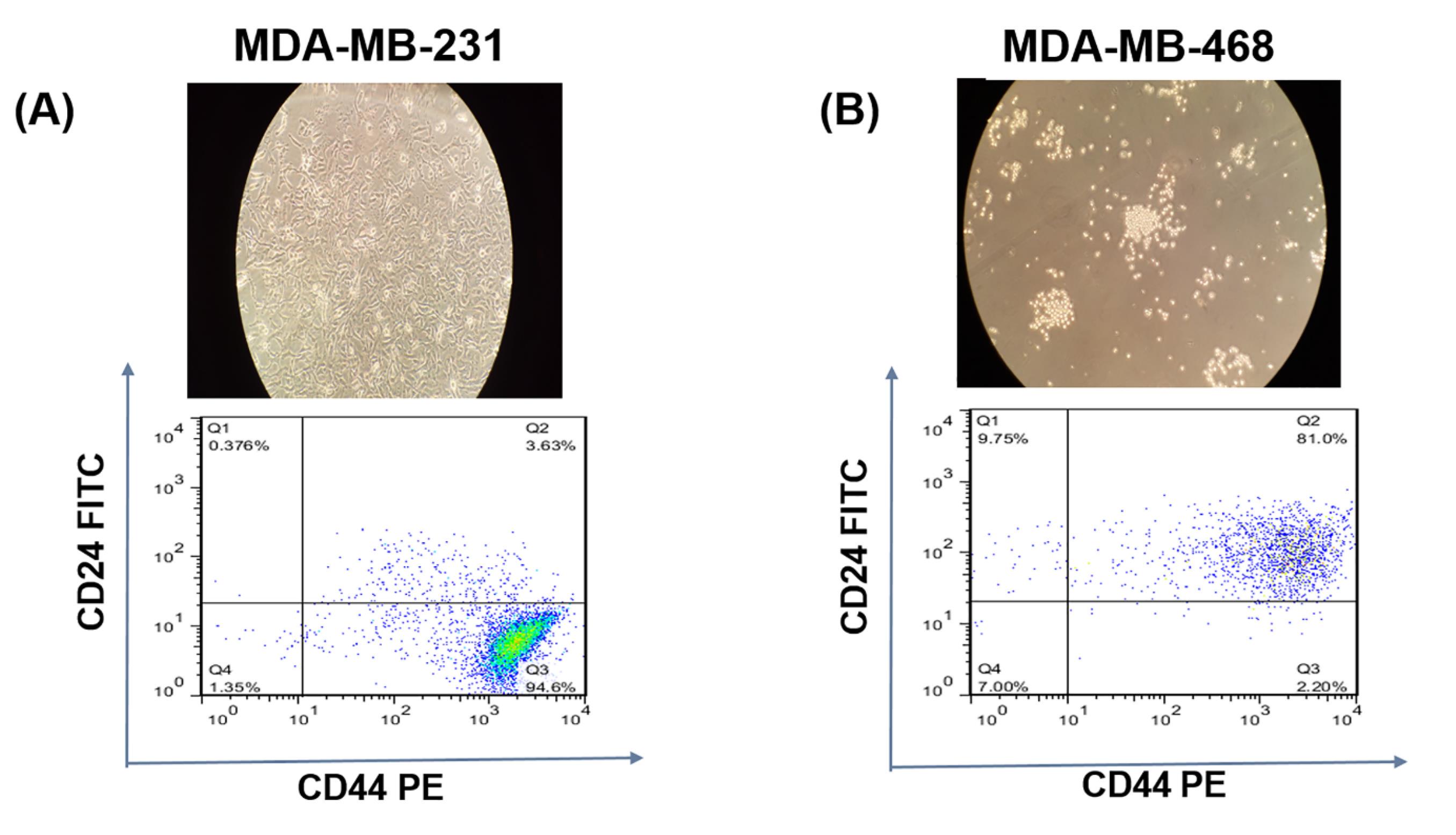
Fig. 1.
Characteristics of cells. MDA-MB-231(A) and MDA-MB-468 (B).
.
Characteristics of cells. MDA-MB-231(A) and MDA-MB-468 (B).
MTT assay
The telomerase inhibitor (BIBR1532) and mitochondria inhibitor (tigecycline) were bought from (Cayman Chemical, MI, USA) and (TCI), respectively. BIBR1532 and tigecycline were prepared in different concentrations during use. As a control group, the cells were cultured without inhibitors.
MDA-MB-231 and MDA-MB-468 cells were seeded in 96‐well at 1×103 cells/wells and incubated for overnight. After 24 hours, the cells were treated with BIBR1532 (7.5, 15, 30, 60,120) µM and tigecycline (1, 2.5, 5, 10, 20, 40) µM for 24, 48, and 72 hours. After supernatant removal, 30 µL of MTT [5 mg/mL in phosphate-buffered saline (PBS)] (ROTH) solution was added to each well and incubated for 4 hours. Then, the medium was removed and 100 µL dimethyl sulfoxide (DMSO) was added. Finally, the absorbance of each well (OD) was measured at 570 nm by a microplate reader (BioTek ELx808, USA). Half-maximal inhibitory concentration (IC50) of BIBR1532 and tigecycline were calculated using the software GraphPad Prism (6.01).
Apoptosis assay
To evaluate apoptosis in TNBC cells, an Annexin V/7AAD assay was carried out. Briefly, TNBC cells were seeded in a 12-well plate with a density of 5×104 and incubated for 24 hours. Then, they were treated with IC50 levels of BIBR1532 and tigecycline for 48 hours. The treated and untreated cells were separated by trypsin (Gibco, UK) and washed with PBS. The cells were suspended in 100 µL of 1X binding buffer (BioLegend). 5 µL annexin V antibody plus 5 µL 7AAD were added and incubated in dark and on ice for 25 minutes. Finally, the cells were washed and suspended in 500 µL binding buffer and evaluated using FACS (BD Bioscience). The results were evaluated using FlowJo software (version X.0.7).
RNA extraction and real-time PCR
IC50 levels of BIBR1532 and tigecycline were used to treat the cells. Then, the cells were collected and RNA extraction was performed using kit Yekta Tajhiz Azma, Iran. The purity of RNA samples was measured by the Nanodrop instrument (NanoDrop1000; Thermo Fisher Scientific). cDNA synthesis was performed according to the manufacturer’s instructions cDNA kit (Yekta Tajhiz Azma, Iran). qPCR was done in 20 µL solution which mixed 10 µL SYBR Green (Yekta Tajhiz Azma, Iran), 2 µL of cDNA, 0.5 µL of each reverse andforward primer, and 7 µL of water without nuclease. The following PCR reactions were performed using a Roche Molecular diagnostic. The PCR reaction started with a 10-minute denaturing step at 94 °C, followed by 40 cycles at 94 °C for 10 seconds, annealing at 56 °C (hTERT, Bax, Bad, Bid, Bcl-2, p53, TET2, DNMT3a and β-actin) and 58 °C (Bcl-xl) for 1 minute, 1 minute at 72 °C, and finally 10 minutes of extension at 72 °C. LightCycler® 96 software was utilized to achieve CT values. For normalization, b-actin was used and the data were analyzed by the 2-ΔΔCT method. The sequences of primer are depicted in Table 1.
Table 1.
The details of primers
|
Gene
|
Forward primer (5′-3′)
|
Reverse primer (5′-3′)
|
Size (bp)
|
| b-actin |
AAACTGGAACGGTGAAGGTG |
TATAGAGAAGTGGGGTGGCT |
174 |
| hTERT |
CAGCAAGTTTGGAAGAACCC |
GACATCCCTGCGTTCTTGG |
234 |
| Bax |
TCACTGAAGCGACTGATGTCC |
CTCCCGCCACAAAGATGGTC |
194 |
| bad |
ACTTCCTCGCCCGAAGAGC |
CTTCCCCTGCCCAAGTTCC |
198 |
| bcl-2 |
AGTGAACATTTCGGTGACTT |
CTTCCAGACATTCGGAGACC |
208 |
| Bcl-xl |
ATCCCAGCTCCACATCACC |
CGATCCGACTCACCAATACC |
202 |
| Bid |
AATCAGAGAAGGAACATACCC |
ACTTCCCATCATTTGAGTGC |
154 |
| P53 |
TCAGTCTACCTCCCGCCATAA |
AGTGGGGAACAAGAAGTGGAG |
176 |
| DNMT3a |
GAAGACCCCTGGAACTGC |
GTCAAATTCCTGGTCGTGG |
123 |
| TET2 |
TGGCTGACAAACTACTGG |
CTTCTGGCAAACTTACATCC |
190 |
DNMT3a: DNA methyltransferases 3A, TET2: Ten-eleven translocation proteins.
DNA extraction for telomerase length assay
Genomic DNA was extracted using Yekta Tajhiz Azma (Iran) DNA extraction kit. In brief, the harvested cells were added to 20 µL of proteinase K and 200uL FABG buffer and were incubated for 15 minutes at 60 ˚C. 200 µL of 96% ethanol was added, vortexed, passed to the BG column and then centrifuged at 5000×g for 1 minute. Afterward, 400 µL washing buffer 1 was added and centrifuged for the 30 seconds at 14 000×g. Next, 750 µL washing buffer 2 was added, and centrifuged for the 30 seconds at 14000×g. Finally, 50~200 µL elution buffer or ddH2O (pH 7.5~9.0) was added to the BG column and centrifuged for three minutes at 14 000×g. Extracted DNA was stored at -20 ˚C until it being used to measure telomere length. Standard curves and quantitative PCR for measuring telomere length before defined by Farahzadi et al.29,30
Protein analysis by western blotting
The harvested cells were lysed with lysis buffer (0.01gr SDS, 0.025gr Sodium Deoxycholate, 0.08 g NaCl, 0.003 gr EDTA, 500 µL, PH=8 Tris-HCL, 1 tablet protease inhibitor cocktail, 10 µL Triton 1%) and were centrifuged at 400×g for 10 minutes at 4 °C. The quantity of protein was determined by Bradford assay. First, lysates were resolved on SDS-page gels, transferred to polyvinylidene difluoride (PVDF) membranes and then incubated at room temperature for 90 minutes in blocking buffer, (composed of 5% non-fat milk powder in Tris-buffered saline with 0.1% Tween (TBS-T). The membranes were pre-incubated with primary antibodies for 15 hours, rinsed with TBS-T buffer (Tris-buffered saline with 0.1% Tween), and then incubated with the secondary conjugated antibody for 1h. Finally, the membrane was washed 3 times with TBS-T buffer. The protein was detected with a chemiluminescence (ECL) kit (Roche, UK). The bands were evaluated by NIH ImageJ 1.6 program. For normalization of protein loading, b-actin was utilized.
Statistical analysis
In order to assess data, GraphPad Prism software (6.01) was utilized. The results are reported as mean ± SD. All tests were carried out three times. One-way and two-way analysis of variance (ANOVA) trailed by Bonferroni's and Tukey's multiple comparisons test were used for the significance of experimental variables between different groups. The significance values were defined as *P ≤ 0.05.
Results
Cytotoxicity assay of telomerase and mitochondria respiration inhibition inTNBC
IC50 values of telomerase inhibitor (BIBR1532) and mitochondria inhibitor (tigecycline) were determined in the TNBC cell lines by the MTT assay.25,31 The IC50 rates of the BIBR1532 in MDA-MB-231 and MDA-MB-468 cells were respectively 36 µM and 10 µM at 48 hours (Fig. 2A, B). Furthermore, the IC50 level of the tigecycline was 3 µM in MDA-MB-231 cells and 5 µM in MDA-MB-468 cells (Fig. 2C, D). The MDA-MB-231 cells showed higher IC50 when compared to BIBR1532- treated MDA-MB-468 cells (Fig. 2A). This may be due to the nature of MDA-MB-231, which contains the highest BCSCs rate and is an aggressive breast cancer cell line.32
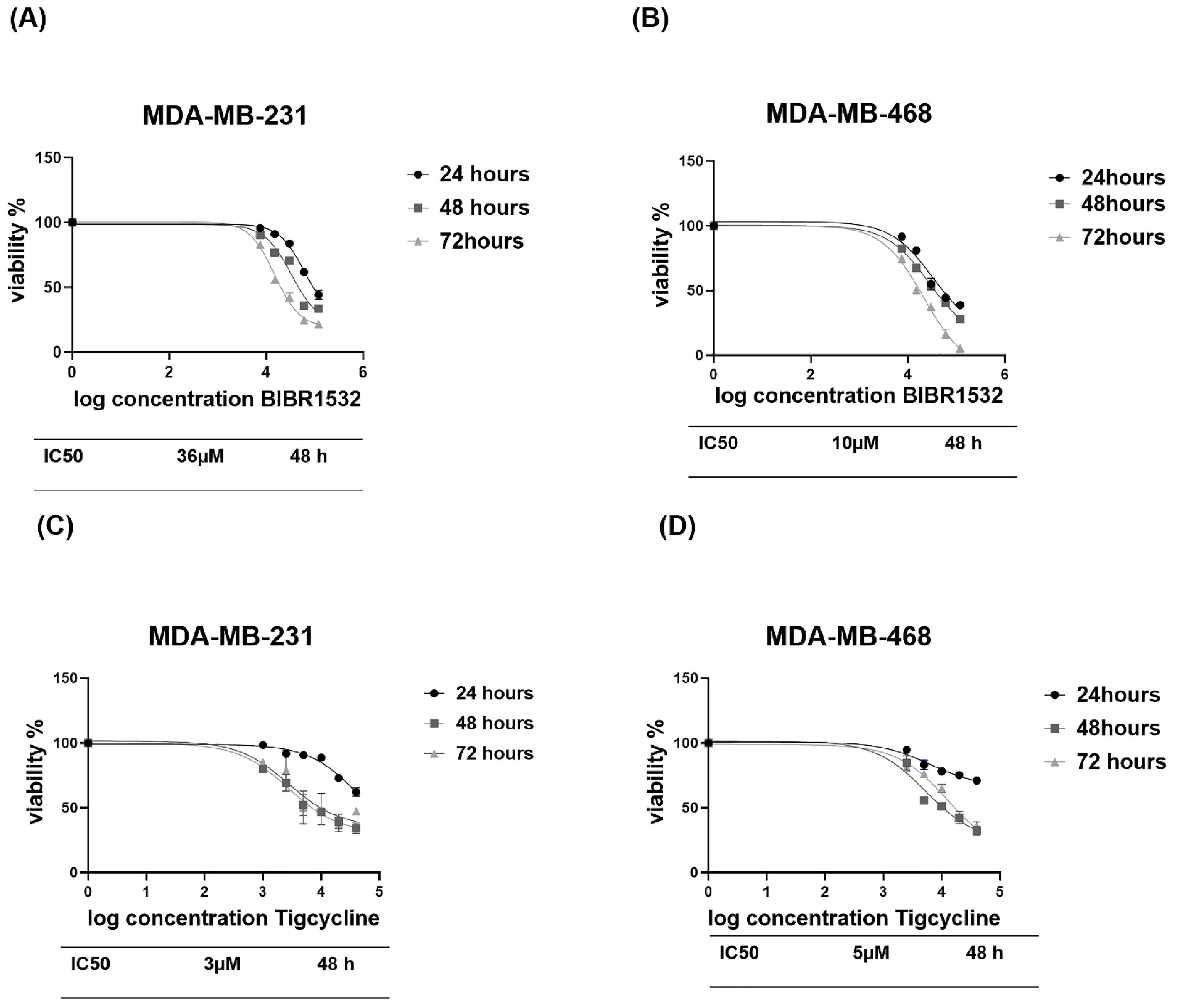
Fig. 2.
MTT assay of BIBR1532 and tigecycline in TNBC cell lines. MTT assay of BIBR1532 in MDA-MB-231(A) and MDA-MB-468(b) and for tigecycline in MDA-MB-231(C) and MDA-MB-468 (D).
.
MTT assay of BIBR1532 and tigecycline in TNBC cell lines. MTT assay of BIBR1532 in MDA-MB-231(A) and MDA-MB-468(b) and for tigecycline in MDA-MB-231(C) and MDA-MB-468 (D).
Telomerase and mitochondria respiration inhibition reduced telomerase activity but not changed telomere length in TNBC cell lines
The expression of hTERT and telomere length in cell lines after treatment with IC50 rate (36 µM and10 µM) of BIBR1532 and (3 µM and 5µM) tigecycline was measured in the current. hTERT mRNA levels decreased significantly in cell lines when compared to untreated cells (control) (Fig. 3A, C) and the highest reduction was noted in the combination group. No significant difference in telomere length in both cells (Fig. 3B, D) was noticed. Telomere length in untreated cells of MDA-MB-231 and MDA-MB-468 were 5.9 kbp and 5 kbp, respectively.
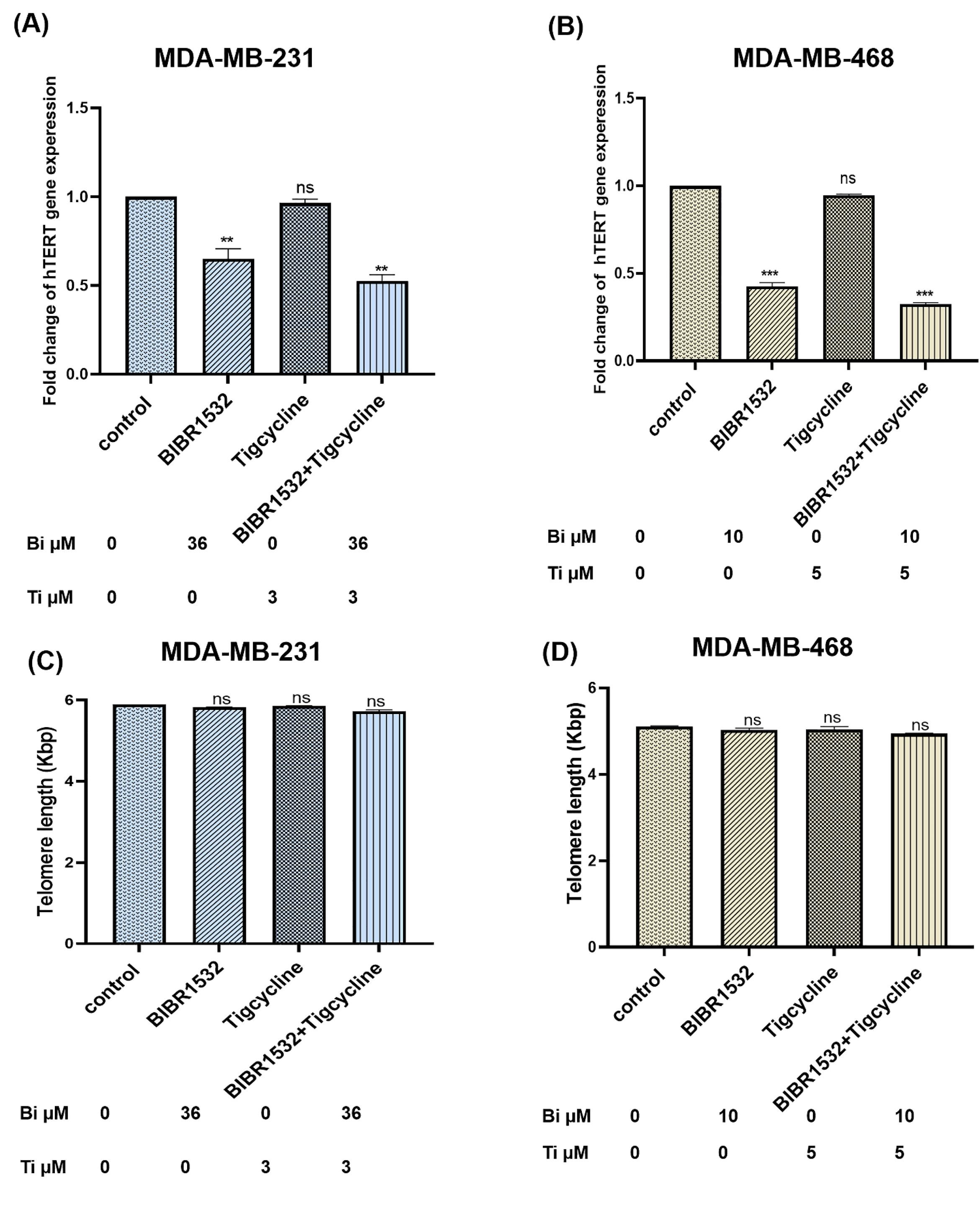
Fig. 3.
hTERT expression and telomere length in TNBC cell lines. The expression of hTERT and telomere length is shown in MDA-MB-231(A, B) and MDA-MB-468 (C, D) following BIBR1532, Tigecycline and in combination treatment. The data are normalized against b-actin and indicated as mean ± SD in each group in three independent tests **P < 0.001 and ***P < 0.001 vs. the untreated cells.
.
hTERT expression and telomere length in TNBC cell lines. The expression of hTERT and telomere length is shown in MDA-MB-231(A, B) and MDA-MB-468 (C, D) following BIBR1532, Tigecycline and in combination treatment. The data are normalized against b-actin and indicated as mean ± SD in each group in three independent tests **P < 0.001 and ***P < 0.001 vs. the untreated cells.
Telomerase and mitochondria respiration inhibition increased apoptosis in TNBC cell lines
The rate of apoptosis was assessed with the IC50 level of BIBR1532 and tigecycline at the 48 hours in cells using Annexin V (Fig. 4). The results displayed a significant increase in the total percentage of apoptotic cells with BIBR1532, tigecycline, and in combination treatment of MDA-MB-231 (Fig. 4A, B) and MDA-MB-468 (Fig. 4C, D) at 48 hours. The apoptosis rate in combination treatment was significantly higher than BIBR1532 and tigecycline alone in cells.
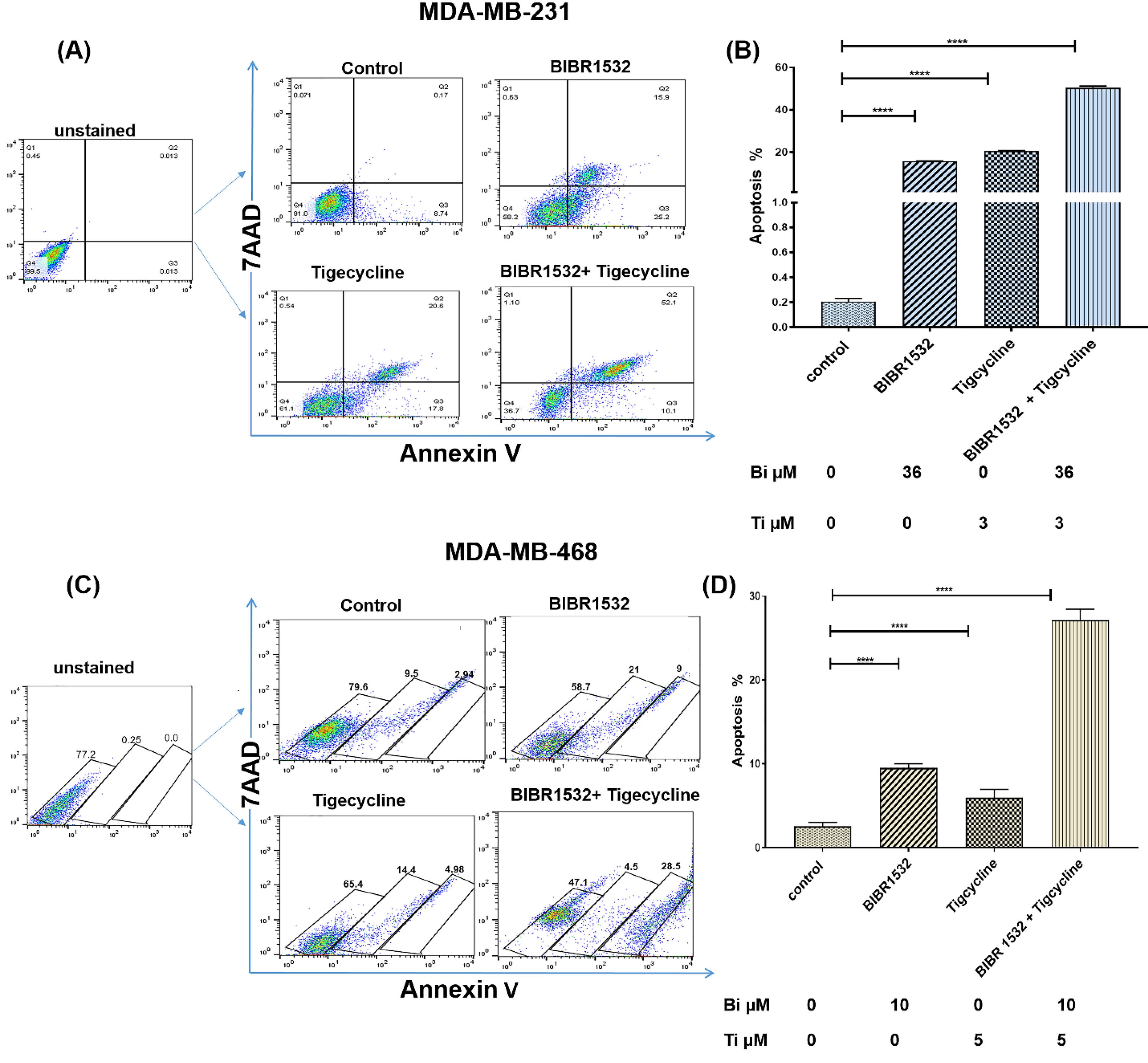
Fig. 4.
Apoptosis rate in TNBC cell lines. Flow cytometry results of MDA-MB-231 (A, B) and MDA-MB-468 cells (C, D) cells following treatment with IC50 level of BIBR1532, tigecycline, combination, and untreated control group. The results are indicated as mean ± SD in each group in three independent experiments ****P < 0.0001 vs the untreated cells.
.
Apoptosis rate in TNBC cell lines. Flow cytometry results of MDA-MB-231 (A, B) and MDA-MB-468 cells (C, D) cells following treatment with IC50 level of BIBR1532, tigecycline, combination, and untreated control group. The results are indicated as mean ± SD in each group in three independent experiments ****P < 0.0001 vs the untreated cells.
The apoptotic genes (Bax, Bad, Bid, Bcl-2, Bcl-xl, and p53) were evaluated following the treatment with IC50 level of BIBR1532 and tigecycline in MDA-MB-231(Fig. 5A) and MDA-MB-468 (Fig. 5D) cells. The expressions of Bax, bad, and bid were significantly upregulated in treated group when compared to the control group. The expressions of Bcl-2 and Bcl-xl were significantly downregulated in tigecycline and combination treatment but did not change in response to BIBR1532 treatment in MDA-MB-231 cells (Fig. 5A and D). Furthermore, p53 was significantly reduced in MDA-MB-231 and MDA-MB-468 cells (Fig. 5A, D) and the highest reduction was for combination treatment.
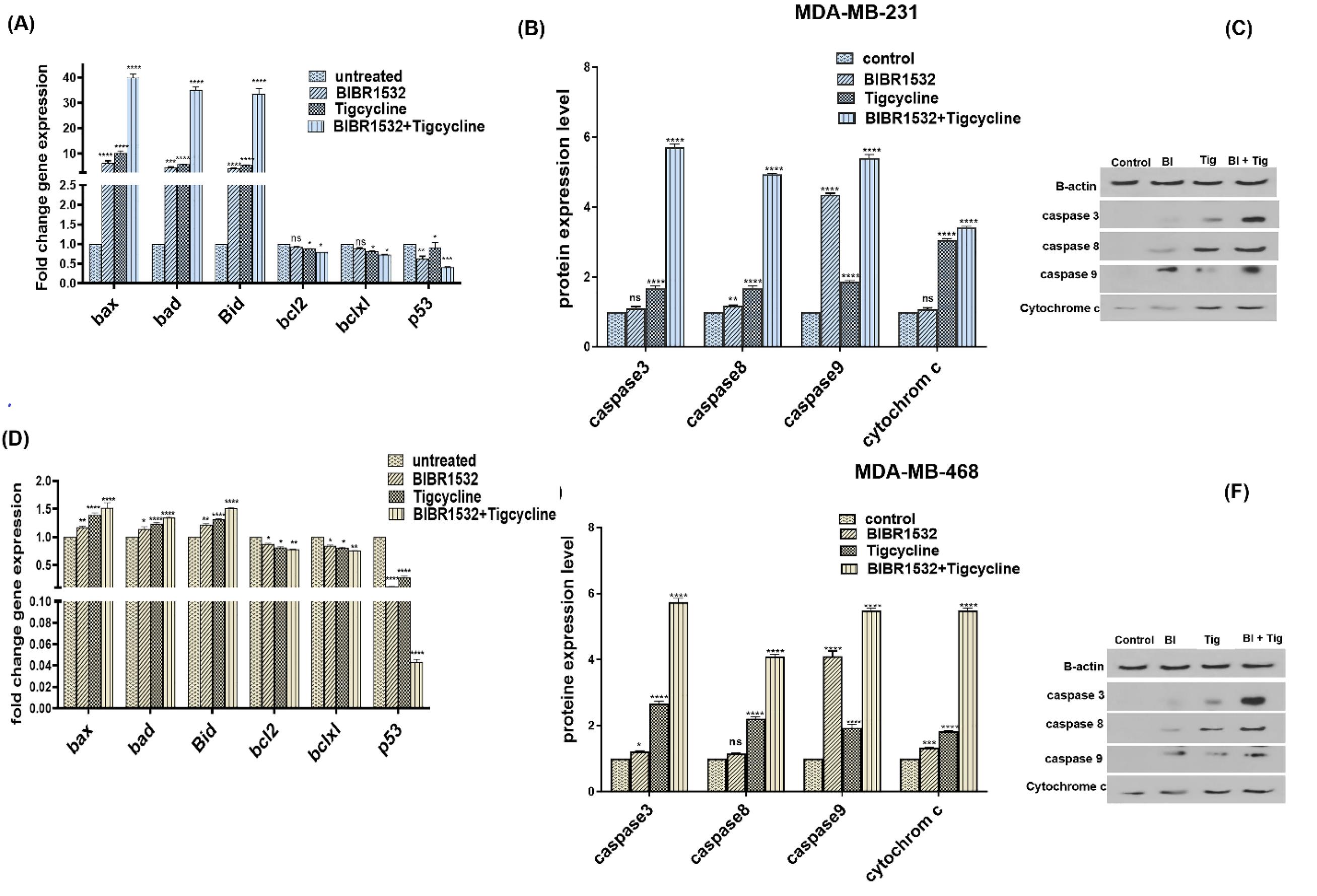
Fig. 5.
Expression of apoptotic genes and proteins in TNBC cell lines. The relative mRNA expression of Bax, Bad, Bid Bcl-2, Bcl-xL, and p53 (A) and proteins (B, C) are shown in MDA-MB-231 following IC50 level of BIBR1532, Tigecycline, in combination treatment, and untreated control group. Furthermore, expression of Bax, Bad, Bid Bcl-2, Bcl-xL, and p53 and proteins of MDA-MB-468 are shown in (D) and (E, F) respectively. The data are normalized against b-actin and indicated as mean ± SD in each group in three independent investigations, *P < 0.05, **P < 0.01, ***P < 0.001 ****P<0.0001 vs the untreated cells.
.
Expression of apoptotic genes and proteins in TNBC cell lines. The relative mRNA expression of Bax, Bad, Bid Bcl-2, Bcl-xL, and p53 (A) and proteins (B, C) are shown in MDA-MB-231 following IC50 level of BIBR1532, Tigecycline, in combination treatment, and untreated control group. Furthermore, expression of Bax, Bad, Bid Bcl-2, Bcl-xL, and p53 and proteins of MDA-MB-468 are shown in (D) and (E, F) respectively. The data are normalized against b-actin and indicated as mean ± SD in each group in three independent investigations, *P < 0.05, **P < 0.01, ***P < 0.001 ****P<0.0001 vs the untreated cells.
To understand the apoptosis pathway, the protein level of cytochrome C and caspases (9, 8, 3) were measured in MDA-MB-231(Fig. 5B, C) and MDA-MB-468 (Fig. 5E, F). The protein levels of cytochrome C and caspases (9, 8, 3) were significantly increased in combination treatment in cells. It is while, cytochrome C and caspase 9 partially remained unchanged in MDA-MB-231 in response to BIBR1532 treatment and caspase 8 was not changed in MDA-MB-468 with tigecycline treatment.
Telomerase and mitochondria respiration inhibition changed the expression of DNMT3a and TET2
DNMT3a mRNA levels were increased in tigecycline and combination treatment in MDA-MB-231 (Fig. 6A) and MDA-MB-468 (Fig. 6B) cells. However, there was no difference in DNMT3a expression in BIBR1532 treatment in cells. Furthermore, TET2 expression was increased in tigecycline and combination treatment in MDA-MB-468 (Fig. 6B) cells. While no difference in TET2 expression in some treated groups in MDA-MB-231 and MDA-MB-468 cells was found.
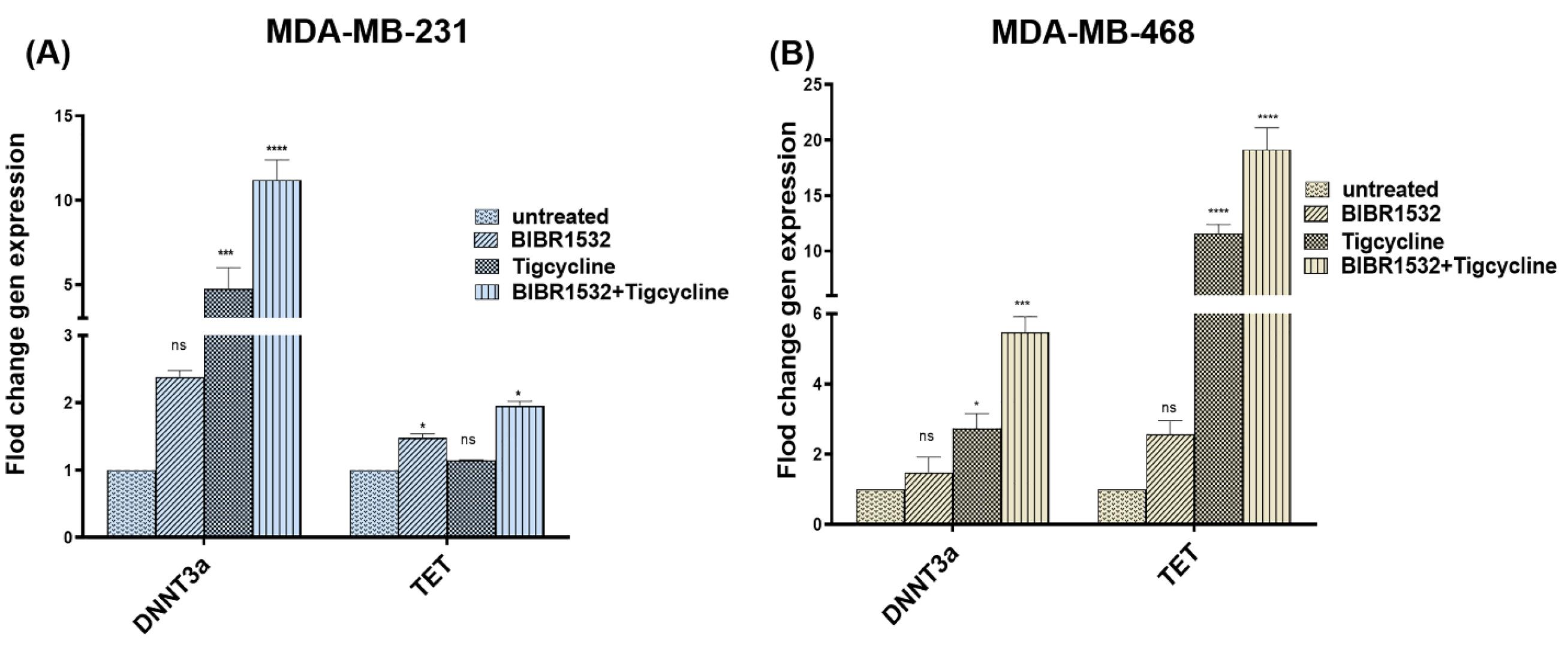
Fig. 6.
Expression of DNMT3a and TET2 in TNBC cell lines. DNMT3a and TET2 mRNA expression are shown in MDA-MB-231(a) and MDA-MB-468 (b) after IC50 level of BIBR1532, tigecycline, in combination treatment, and untreated control group. The data are normalized against b-actin and indicated as mean ± SD in each group in three independent investigations, *P < 0.05, ***P < 0.001, and ****P < 0.0001 vs the untreated cells.
.
Expression of DNMT3a and TET2 in TNBC cell lines. DNMT3a and TET2 mRNA expression are shown in MDA-MB-231(a) and MDA-MB-468 (b) after IC50 level of BIBR1532, tigecycline, in combination treatment, and untreated control group. The data are normalized against b-actin and indicated as mean ± SD in each group in three independent investigations, *P < 0.05, ***P < 0.001, and ****P < 0.0001 vs the untreated cells.
Discussion
Considering that, TNBC patients treatment remained a challenge and hTERT involves in the progression of cancer and inhibits mitochondrial apoptosis.21 In addition, combination therapy is a more effective strategy than monotherapy for improving cancer.22 Therefore, we inhibited telomerase and mitochondria respiration and evaluated apoptosis and expression of DNMT3a and TET 2.
According to the former studies, different breast cancer cell lines respond differently to BIBR1532 and tigecycline treatment.31,33 The present results indicated that BIBR1532 exposure significantly diminished the expression of hTERT but did not leave a significant effect on telomere length. BIBR1532 treatment reduced hTERT expression in breast cancer cell lines.26,33,34 The inhibition of telomerase often has a long lag phase prior to telomere shortening.35 A study indicated that long-term exposure with low doses of BIBR 1532 in different malignancy cells induced telomere shortening while, EL-Daly showed that telomere length was not changed with short-term exposure of BIBR 1532 in cancer cells.25,36,37 Furthermore, hTERT inhibitor did not affect telomere length in breast cancer.38 Therefore, telomere shortening may be time and dose-dependent with BIBR1532 treatment.
The overexpression of hTERT inhibits mitochondrial apoptosis due to the inhibition of cytochrome c release, the reduction of mitochondrial potential, transmission the Bax to mitochondria.39-41 Also, hTERT inhibit extrinsic cell death. According to Zhang et al, TERT hampers the induction of TRAIL-mediated cell death.42,43 Moreover, tigecycline induces intrinsic apoptosis by activating the release of cytochrome c, and cleavage of caspase-9/caspase-3/caspase-7.27,44 Based on the current findings, the inhibition of telomerase and mitochondria respiration increased apoptosis in TNBC cells. It was found that the inhibition of telomerase and mitochondria respiration up-regulated the expression levels of pro-apoptotic Bax, Bad, Bid genes and caspase 3, 8, 9, and cytochrome c proteins, and it downregulates the expression levels of anti-apoptotic Bcl-2 and Bcl-xl and cause intrinsic-, extrinsic- mediated apoptosis in TNBC cells. These results are consistent with the role of the mitochondrial pathway in apoptosis. Bax accelerated the opening of voltage-dependent anion channels at the outer mitochondrial membrane with the subsequent release of cytochrome c that interacts with the apoptotic protease activating factor-1 (Apaf-1) and activates the initiator caspase 9. Bad also blocks the anti-apoptotic function of Bcl-xl and Bcl-2 and induces apoptosis.45-48 Mitochondrial- and receptor-dependent pathways cooperate through Bid to enhance apoptosis. Bid, a pro-apoptotic protein, is cleaved and activated by caspase 8 and led to the mitochondrial permeability transition.49 The expressions of Bcl-2 and Bcl-xl remain unchanged in hTERT inhibition of MDA-MB-231 cells. It suggests that post-translational modification including methylation modifications can reduce the effect of Bcl-2.50 Our result showed that telomerase inhibition increased caspase 8 and 3 and induced extrinsic-apoptosis in MDA-MB-231. It is consistent with the result of Rubis et al who showed that the reduction of telomerase activity stimulated the expression of caspase-8, Fas, and FasL genes and activated the death receptor apoptosis in MDA-MB-231 cells.38 It is while, mitochondria pathway plays a vital role in apoptosis in the inhibition of telomerase in MDA-MB-468 cells. p53 is a DNA-binding transcription factor that is increased in response to cell stress, and plays role in DNA damage repair, apoptosis, and other cellular processes.51,52 p53 mutant accumulates in TNBC, disturbs DNA repair complex, and causes tumor progression with apoptosis resistance and cancer invasion.53,54 Based on studies tigecycline acts independently from p53.55 BIBR1532 and tigecycline therapy down-regulates p53 expression in TNBC cells and induces apoptosis.
The current results suggest that the expressions of DNMT3a and TET2 increased. Epigenetic markers (DNMT3a and TET2) have tumor-suppression functions.56,57 DNMT3a depletion enhances self-renewal, cancer cell progression, and breast cancer metastasis and decreases TET2 mRNA leading to poor prognosis in breast cancer patients.19,20,58 DNMT3a andTET deficiency happens in increased telomere length.59,60
Conclusion
Inhibition of telomerase and mitochondria respiration caused intrinsic and extrinsic apoptosis and increased DNMT3a and TET2 expression and could be utilized in breast cancer future treatments.
Research Highlights
What is the current knowledge?
√ Triple-negative breast cancers (TNBCs) exhibit the properties of cancer stem cells (CSCs)
√ CSCs are resistant to conventional therapy
√ hTERT, subunit of telomerase, displays several functions such as the effect on mitochondria, gene expression, and apoptosis
√ Mitochondria is involved in breast cancer tumorigenesis.
What is new here?
√ Telomerase and mitochondria respiration inhibition decreased hTERT expression but no change in telomere length in TNBC cell lines.
√ The inhibition of telomerase and mitochondria respiration induced apoptosis in TNBC cell lines.
√ The inhibition of telomerase and mitochondria respiration affected the expression of DNAT3a and TET2 in TNBC cell lines.
Acknowledgments
This is a report of the database from a PhD thesis registered at Tabriz University of Medical Sciences.
Competing Interests
None of the authors claimed to have any competing interests.
Ethics Statement
This study was approved by the Academic Research Ethics Committee of Tabriz University of Medical Sciences (code of ethics, IR.TBZMED.REC.1398.785).
Funding
This study is supported by the faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences (Grant code. 62836) as the PhD thesis of Zeinab Mazloumi.
References
- Huang L, Li A, Liao G, Yang F, Yang J, Chen X. Curcumol triggers apoptosis of p53 mutant triple-negative human breast cancer MDA-MB 231 cells via activation of p73 and PUMA. Oncol Lett 2017; 14:1080-8. doi: 10.3892/ol.2017.6273 [Crossref] [ Google Scholar]
- Nedeljković M, Damjanović A. Mechanisms of Chemotherapy Resistance in Triple-Negative Breast Cancer-How We Can Rise to the Challenge. Cells 2019; 8:957. doi: 10.3390/cells8090957 [Crossref] [ Google Scholar]
- Dizaji Asl K, Rafat A, Mazloumi Z, Valipour B, Movassaghpour A, Talebi M. Cord blood stem cell-generated KIR(+)NK cells effectively target leukemia cell lines. Hum Immunol 2023; 84:98-105. doi: 10.1016/j.humimm.2022.10.010 [Crossref] [ Google Scholar]
- Kamalabadi-Farahani M, H Najafabadi MR, Jabbarpour Z. Apoptotic Resistance of Metastatic Tumor Cells in Triple Negative Breast Cancer: Roles of Death Receptor-5. Asian Pac J Cancer Prev 2019; 20:1743-8. doi: 10.31557/APJCP.2019.20.6.1743 [Crossref] [ Google Scholar]
- Park S-Y, Choi J-H, Nam J-S. Targeting Cancer Stem Cells in Triple-Negative Breast Cancer. Cancers 2019; 11:965. doi: 10.3390/cancers11070965 [Crossref] [ Google Scholar]
- Herbert BS, Wright WE, Shay JW. Telomerase and breast cancer. Breast Cancer Res 2001; 3:146-9. doi: 10.1186/bcr288 [Crossref] [ Google Scholar]
- Rafat A, Dizaji Asl K, Mazloumi Z, Movassaghpour AA, Farahzadi R, Nejati B. Telomerase-based therapies in haematological malignancies. Cell Biochem Funct 2022; 40:127-40. doi: 10.1002/cbf.3687 [Crossref] [ Google Scholar]
- Singhapol C, Pal D, Czapiewski R, Porika M, Nelson G, Saretzki GC. Mitochondrial Telomerase Protects Cancer Cells from Nuclear DNA Damage and Apoptosis. PLoS One 2013; 8:e52989. doi: 10.1371/journal.pone.0052989 [Crossref] [ Google Scholar]
- Rafat A, Dizaji Asl K, Mazloumi Z, Movassaghpour AA, Talebi M, Shanehbandi D. Telomerase inhibition on acute myeloid leukemia stem cell induced apoptosis with both intrinsic and extrinsic pathways. Life Sci 2022; 295:120402. doi: 10.1016/j.lfs.2022.120402 [Crossref] [ Google Scholar]
- Kang Y, Wan L, Wang Q, Yin Y, Liu J, Liu L. Long noncoding RNA SNHG1 promotes TERT expression by sponging miR-18b-5p in breast cancer. Cell Biosci 2021; 11:169. doi: 10.1186/s13578-021-00675-5 [Crossref] [ Google Scholar]
- Wu L, Fidan K, Um J-Y, Ahn KS. Telomerase: Key regulator of inflammation and cancer. Pharmacol Res 2020; 155:104726. doi: 10.1016/j.phrs.2020.104726 [Crossref] [ Google Scholar]
- Martinez-Outschoorn UE, Pavlides S, Sotgia F, Lisanti MP. Mitochondrial biogenesis drives tumor cell proliferation. Am J Pathol 2011; 178:1949-52. doi: 10.1016/j.ajpath.2011.03.002 [Crossref] [ Google Scholar]
- Weiner-Gorzel K, Murphy M. Mitochondrial dynamics, a new therapeutic target for Triple Negative Breast Cancer. Biochim Biophys Acta Rev Cancer 2021; 1875:188518. doi: 10.1016/j.bbcan.2021.188518 [Crossref] [ Google Scholar]
- García-Heredia JM, Carnero A. Role of Mitochondria in Cancer Stem Cell Resistance. Cells 2020; 9:1693. doi: 10.3390/cells9071693 [Crossref] [ Google Scholar]
- Patel PS, Castelow C, Patel DS, Bhattacharya SK, Kuscu C, Kuscu C. Mitochondrial Role in Oncogenesis and Potential Chemotherapeutic Strategy of Mitochondrial Infusion in Breast Cancer. Int J Mol Sci 2022; 23:12993. doi: 10.3390/ijms232112993 [Crossref] [ Google Scholar]
- Goldgar S, Heerboth S, Byler S, Rosenthal S, Sarkar S, editors. Epigenetic Regulation of hTERT Expression in Breast Cancer Cells: Analysis of Methylation Status, Expression Levels, and Apoptosis with HDACi and Calpeptin Combination Therapy 2012.
- Mazloumi Z, Farahzadi R, Rafat A, Dizaji Asl K, Karimipour M, Montazer M. Effect of aberrant DNA methylation on cancer stem cell properties. Exp Mol Pathol 2022; 125:104757. doi: 10.1016/j.yexmp.2022.104757 [Crossref] [ Google Scholar]
- Cakouros D, Hemming S, Gronthos K, Liu R, Zannettino A, Shi S. Specific functions of TET1 and TET2 in regulating mesenchymal cell lineage determination. Epigenetics Chromatin 2019; 12:3. doi: 10.1186/s13072-018-0247-4 [Crossref] [ Google Scholar]
- Yang L, Yu S-J, Hong Q, Yang Y, Shao Z-M. Reduced Expression of TET1, TET2, TET3 and TDG mRNAs Are Associated with Poor Prognosis of Patients with Early Breast Cancer. PloS One 2015; 10:e0133896. doi: 10.1371/journal.pone.0133896 [Crossref] [ Google Scholar]
- Deivendran S, Marzook H, Santhoshkumar TR, Kumar R, Pillai MR. Metastasis-associated protein 1 is an upstream regulator of DNMT3a and stimulator of insulin-growth factor binding protein-3 in breast cancer. Sci Rep 2017; 7:44225. doi: 10.1038/srep44225 [Crossref] [ Google Scholar]
- Li Y, Tergaonkar V. Noncanonical Functions of Telomerase: Implications in Telomerase-Targeted Cancer Therapies. Cancer Res 2014; 74:1639-44. doi: 10.1158/0008-5472.can-13-3568 [Crossref] [ Google Scholar]
- Chen Q, He L, Li X, Xu L, Chen T. Ruthenium complexes boost NK cell immunotherapy via sensitizing triple-negative breast cancer and shaping immuno-microenvironment. Biomaterials 2022; 281:121371. doi: 10.1016/j.biomaterials.2022.121371 [Crossref] [ Google Scholar]
- Mazloumi Z, Rafat A, Dizaji Asl K, Nozad Charoudeh H. A combination of telomerase inhibition and NK cell therapy increased breast cancer cell line apoptosis. Biochem Biophys Res Commun 2023; 640:50-5. doi: 10.1016/j.bbrc.2022.11.090 [Crossref] [ Google Scholar]
- Altamura G, Degli Uberti B, Galiero G, De Luca G, Power K, Licenziato L. The Small Molecule BIBR1532 Exerts Potential Anti-cancer Activities in Preclinical Models of Feline Oral Squamous Cell Carcinoma Through Inhibition of Telomerase Activity and Down-Regulation of TERT. Front Vet Sci 2020; 7:620776. doi: 10.3389/fvets.2020.620776 [Crossref] [ Google Scholar]
- Lavanya C, Venkataswamy MM, Sibin MK, Srinivas Bharath MM, Chetan GK. Down regulation of human telomerase reverse transcriptase (hTERT) expression by BIBR1532 in human glioblastoma LN18 cells. Cytotechnology 2018; 70:1143-54. doi: 10.1007/s10616-018-0205-9 [Crossref] [ Google Scholar]
- Nasrollahzadeh A, Bashash D, Kabuli M, Zandi Z, Kashani B, Zaghal A. Arsenic trioxide and BIBR1532 synergistically inhibit breast cancer cell proliferation through attenuation of NF-κB signaling pathway. Life Sci 2020; 257:118060. doi: 10.1016/j.lfs.2020.118060 [Crossref] [ Google Scholar]
- Dong Z, Abbas MN, Kausar S, Yang J, Li L, Tan L. Biological Functions and Molecular Mechanisms of Antibiotic Tigecycline in the Treatment of Cancers. Int J Mol Sci 2019; 20:3577. doi: 10.3390/ijms20143577 [Crossref] [ Google Scholar]
- Reed GA, Schiller GJ, Kambhampati S, Tallman MS, Douer D, Minden MD. A Phase 1 study of intravenous infusions of tigecycline in patients with acute myeloid leukemia. Cancer Med 2016; 5:3031-40. doi: 10.1002/cam4.845 [Crossref] [ Google Scholar]
- Farahzadi R, Fathi E, Mesbah-Namin SA, Zarghami N. Zinc sulfate contributes to promote telomere length extension via increasing telomerase gene expression, telomerase activity and change in the TERT gene promoter CpG island methylation status of human adipose-derived mesenchymal stem cells. PLoS One 2017; 12:e0188052. doi: 10.1371/journal.pone.0188052 [Crossref] [ Google Scholar]
- Brazvan B, Farahzadi R, Mohammadi SM, Montazer Saheb S, Shanehbandi D, Schmied L. Key Immune Cell Cytokines Affects the Telomere Activity of Cord Blood Cells In vitro. Adv Pharm Bull 2016; 6:153-61. doi: 10.15171/apb.2016.022 [Crossref] [ Google Scholar]
- Dong Z, Abbas MN, Kausar S, Yang J, Li L, Tan L. Biological Functions and Molecular Mechanisms of Antibiotic Tigecycline in the Treatment of Cancers. Int J Mol Sci 2019; 20:3577. doi: 10.3390/ijms20143577 [Crossref] [ Google Scholar]
- Xu L, Zhang L, Hu C, Liang S, Fei X, Yan N. WNT pathway inhibitor pyrvinium pamoate inhibits the self-renewal and metastasis of breast cancer stem cells. Int J Oncol 2016; 48:1175-86. doi: 10.3892/ijo.2016.3337 [Crossref] [ Google Scholar]
- Doğan F, Özateş NP, Bağca BG, Abbaszadeh Z, Söğütlü F, Gasımlı R. Investigation of the effect of telomerase inhibitor BIBR1532 on breast cancer and breast cancer stem cells. J Cell Biochem 2019; 120:1282-93. doi: 10.1002/jcb.27089 [Crossref] [ Google Scholar]
- Shi Y, Sun L, Chen G, Zheng D, Li L, Li L. A combination of the telomerase inhibitor, BIBR1532, and paclitaxel synergistically inhibit cell proliferation in breast cancer cell lines. Target Oncol 2015; 10:565-73. doi: 10.1007/s11523-015-0364-y [Crossref] [ Google Scholar]
- Goldblatt EM, Gentry ER, Fox MJ, Gryaznov SM, Shen C, Herbert BS. The telomerase template antagonist GRN163L alters MDA-MB-231 breast cancer cell morphology, inhibits growth, and augments the effects of paclitaxel. Mol Cancer Ther 2009; 8:2027-35. doi: 10.1158/1535-7163.mct-08-1188 [Crossref] [ Google Scholar]
- Celeghin A, Giunco S, Freguja R, Zangrossi M, Nalio S, Dolcetti R. Short-term inhibition of TERT induces telomere length-independent cell cycle arrest and apoptotic response in EBV-immortalized and transformed B cells. Cell Death Dis 2016; 7:e2562. doi: 10.1038/cddis.2016.425 [Crossref] [ Google Scholar]
- El-Daly H, Kull M, Zimmermann S, Pantic M, Waller CF, Martens UM. Selective cytotoxicity and telomere damage in leukemia cells using the telomerase inhibitor BIBR1532. Blood 2005; 105:1742-9. [ Google Scholar]
- Rubis B, Holysz H, Gladych M, Toton E, Paszel A, Lisiak N. Telomerase downregulation induces proapoptotic genes expression and initializes breast cancer cells apoptosis followed by DNA fragmentation in a cell type dependent manner. Mol Biol Rep 2013; 40:4995-5004. doi: 10.1007/s11033-013-2600-9 [Crossref] [ Google Scholar]
- Qin Y, Guo H, Tang B, Yang S-M. The non-reverse transcriptase activity of the human telomerase reverse transcriptase promotes tumor progression (Review). Int J Oncol 2014; 45:525-31. doi: 10.3892/ijo.2014.2470 [Crossref] [ Google Scholar]
- Indran IR, Hande MP, Pervaiz S. hTERT overexpression alleviates intracellular ROS production, improves mitochondrial function, and inhibits ROS-mediated apoptosis in cancer cells. Cancer Res 2011; 71:266-76. [ Google Scholar]
- Del Bufalo D, Rizzo A, Trisciuoglio D, Cardinali G, Torrisi MR, Zangemeister-Wittke U. Involvement of hTERT in apoptosis induced by interference with Bcl-2 expression and function. Cell Death Differ 2005; 12:1429-38. [ Google Scholar]
- Dudognon C, Pendino F, Hillion J, Saumet A, Lanotte M, Segal-Bendirdjian E. Death receptor signaling regulatory function for telomerase: hTERT abolishes TRAIL-induced apoptosis, independently of telomere maintenance. Oncogene 2004; 23:7469-74. [ Google Scholar]
- Zhang R-G, Zhao J-J, Yang L-Q, Yang S-M, Wang R-Q, Chen W-S. RNA interference-mediated hTERT inhibition enhances TRAIL-induced apoptosis in resistant hepatocellular carcinoma cells. Oncol Rep 2010; 23:1013-9. [ Google Scholar]
- D’Andrea A, Gritti I, Nicoli P, Giorgio M, Doni M, Conti A. The mitochondrial translation machinery as a therapeutic target in Myc-driven lymphomas. Oncotarget 2016; 7:72415-30. doi: 10.18632/oncotarget.11719 [Crossref] [ Google Scholar]
- Bergmann A. Survival Signaling Goes BAD. Dev Cell 2002; 3:607-8. doi: 10.1016/S1534-5807(02)00328-3 [Crossref] [ Google Scholar]
- Ottilie S, Diaz J-L, Horne W, Chang J, Wang Y, Wilson G. Dimerization properties of human Bad: Identification of a BH-3 domain and analysis of its binding to mutant Bcl-2 and Bcl-Xl proteins. J Biol Chem 1997; 272:30866-72. [ Google Scholar]
- Pastorino JG, Tafani M, Rothman RJ, Marcineviciute A, Hoek JB, Farber JL. Functional consequences of the sustained or transient activation by Bax of the mitochondrial permeability transition pore. J Biol Chem 1999; 274:31734-9. [ Google Scholar]
- Santucci R, Sinibaldi F, Cozza P, Polticelli F, Fiorucci L. Cytochrome c: An extreme multifunctional protein with a key role in cell fate. J Biol Macromol 2019. 136: 1237-46.
- Schimmer AD, Hedley DW, Penn LZ, Minden MD. Receptor-and mitochondrial-mediated apoptosis in acute leukemia: a translational view. Blood 2001; 98:3541-53. [ Google Scholar]
- Chrysovergis A, Papanikolaou Vs, Tsiambas E, Ragos V, Peschos D, Kyrodimos E. Digital Analysis of BCL2 Expression in Laryngeal Squamous Cell Carcinoma. Anticancer Res 2019; 39:1253-7. doi: 10.21873/anticanres.13235 [Crossref] [ Google Scholar]
- Tutton S, Lieberman PM. A role for p53 in telomere protection. Mol Cell Oncol 2016; 4:e1143078-e. doi: 10.1080/23723556.2016.1143078 [Crossref] [ Google Scholar]
- Hickman ES, Moroni MC, Helin K. The role of p53 and pRB in apoptosis and cancer. Curr Opin Genet Dev 2002; 12:60-6. [ Google Scholar]
- Arora R, Jain S, Rahimi H. Evaluating the efficacy of Tigecycline to target multiple cancer-types: A Review. STEM Fellowship Journal 2018; 4:5-11. doi: 10.17975/sfj-2018-002 [Crossref] [ Google Scholar]
- Wang X, Zhang Y, Pei X, Guo G, Xue B, Duan X. TRIM3 inhibits P53 signaling in breast cancer cells. Cancer Cell Int 2020; 20:559. doi: 10.1186/s12935-020-01630-z [Crossref] [ Google Scholar]
- Saha MN, Qiu L, Chang H. Targeting p53 by small molecules in hematological malignancies. J Hematol Oncol 2013; 6:23. doi: 10.1186/1756-8722-6-23 [Crossref] [ Google Scholar]
- Biray Avci C, Dogan F, Ozates Ay NP, Goker Bagca B, Abbaszadeh Z, Gunduz C. Effects of telomerase inhibitor on epigenetic chromatin modification enzymes in malignancies. J Cell Biochem 2018; 119:9817-24. [ Google Scholar]
- Chou W-C, Chou S-C, Liu C-Y, Chen C-Y, Hou H-A, Kuo Y-Y. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood 2011; 118:3803-10. [ Google Scholar]
- Jeong M, Park HJ, Celik H, Ostrander EL, Reyes JM, Guzman A. Loss of Dnmt3a Immortalizes Hematopoietic Stem Cells In Vivo. Cell Rep 2018; 23:1-10. doi: 10.1016/j.celrep.2018.03.025 [Crossref] [ Google Scholar]
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol 2006; 8:416-24. doi: 10.1038/ncb1386 [Crossref] [ Google Scholar]
- Yang J, Guo R, Wang H, Ye X, Zhou Z, Dan J. Tet Enzymes Regulate Telomere Maintenance and Chromosomal Stability of Mouse ESCs. Cell Rep 2016; 15:1809-21. doi: 10.1016/j.celrep.2016.04.058 [Crossref] [ Google Scholar]