Azam Safary is an Assistant Professor of medical biotechnology at the Connective Tissue Diseases
Research Center (CTDRC) and Research Center for Pharmaceutical Nanotechnology (RCPN) at Tabriz University of Medical Sciences, working on the cellular and molecular mechanisms of immune-mediated inflammatory diseases, new strategies for the detection and prevention of them, nano-formulated therapeutic enzymes, and biotechnological aspects of recombinant proteins.
Professor Yadollah Omidi has a Ph.D. degree in Pharmaceutical Sciences (2003, Cardiff University, UK) and completed a postdoctoral program (2004) at Cardiff University. He is currently working as a full professor at Nova Southeastern University College of Pharmacy, Florida. Prof. Omidi’s research in advanced targeted diagnosis and therapy of diseases have resulted in over 300 published papers in international journals, 27 book chapters, and a few patents. His H-index is 61 and i10-index is 238. He has consecutively been listed
among top 1% highly cited scientists worldwide by WoS-ESI.
Abstract
Epidermal growth factor receptor (EGFR) is a cell surface protein that plays a vital role in regulating cell growth and division. However, certain tumors, such as colorectal cancer (CRC), can exhibit an overexpression of EGFR, resulting in uncontrolled cell growth and tumor progression. To address this issue, therapies targeting and inhibiting EGFR activity have been developed to suppress cancer growth. Nevertheless, resistance to these therapies poses a significant obstacle in cancer treatment. Recent research has focused on comprehending the underlying mechanisms contributing to anti-EGFR resistance and identifying new targets to overcome this striking challenge. Long non-coding RNAs (lncRNAs) are a class of RNA molecules that do not encode proteins but play pivotal roles in gene regulation and cellular processes. Emerging evidence suggests that lncRNAs may participate in modulating resistance to anti-EGFR therapies in CRC. Consequently, combining lncRNA targeting with the existing treatment modalities could potentially yield improved clinical outcomes. Illuminating the involvement of lncRNAs in anti-EGFR resistance mechanisms of cancer cells can provide valuable insights into the development of novel anti-EGFR therapies in several solid tumors.
Keywords: LncRNAs, EGFR, Colorectal cancer, Anti-EGFR resistance, Targeting therapy
Copyright and License Information
© 2024 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Epidermal growth factor receptors (EGFRs) overexpressed by various solid tumors are involved in the initiation, progression, and metastasis of different malignancies such as breast cancer and colorectal cancer (CRC).1,2 EGFRs are considered clinically valid oncomarkers which can be targeted to inhibit/eradicate cancer cells using various advanced treatment modalities such as monoclonal antibodies (mAbs), Ab-armed nanomedicines, Ab-drug nanoconjugates, hybrid Ab scaffolds like bispecific constructs, aptamer-decorated nanosystems, gene therapy, and so forth.1-7 Specific binding of epidermal growth factors (EGF) to EGFRs results in their dimerization and activation of downstream signaling mechanisms such as MAPK/ERK, PI3K/AKT/mTOR pathways leading to the regulation of cancer cell proliferation, invasion, and angiogenesis.8,9 Anti-EGFR mAbs such as cetuximab (CET) were shown to substantially improve the overall survival of metastatic CRC (mCRC) patients with wild-type KRAS genotypes.10 However, in mCRC patients, the therapeutic response to anti-EGFR therapy can be limited due to the development of multiple resistance mechanisms.11 Numerous genetic factors contribute to the development of resistance mechanisms in cancer cells against anti-EGFR mAbs. These factors include:
-
EGFR gene copy number: The number of copies of the EGFR gene can influence the response to anti-EGFR mAbs.
-
Protein expression of EGFR ligands: The levels of ligands that bind to EGFR, such as EGF and TGF-alpha, can impact the efficacy of anti-EGFR mAbs.
-
HER2 and MET gene amplification: Amplification of HER2 and MET genes, which are involved in signaling pathways related to EGFR, can contribute to resistance against anti-EGFR mAbs.
-
Activation of EGFR downstream cascade signaling pathways: Mutations or alterations in downstream signaling pathways of EGFR, such as KRAS, NRAS, BRAF, and PIK3CA, can lead to resistance against anti-EGFR mAbs.
-
Loss of PTEN and STAT3 phosphorylation: Inactivation of the tumor suppressor PTEN and abnormal phosphorylation of STAT3 might be associated with resistance to anti-EGFR mAbs.
-
Epithelial-mesenchymal transition (EMT) occurrence: EMT, a process where epithelial cells acquire mesenchymal characteristics, seems to be linked to resistance against anti-EGFR mAbs in cancer cells.
These factors appear to contribute collectively to the emergence of resistance mechanisms in cancer cells, reducing the effectiveness of anti-EGFR mAbs.12,13 Besides, the role of non-genetic factors is a highly debated issue even though little is known about the exact resistance mechanism of such a phenomenon. All in all, it is necessary to explore new strategies for improving the cytotoxic impacts of anti-EGFR mAbs, enhancing apoptosis in CRC, and overcoming drug resistance mechanisms in mCRC patients.14
In recent studies, the crucial functions of small non-coding RNAs (sncRNAs) and long non-coding RNAs (lncRNAs) have gained significant attention, particularly in relation to tumor progression and the development of resistance mechanisms against anti-EGFR mAbs in CRC.15 Of these, lncRNA biomacromolecules are considered complex ncRNAs structures, which have been indiscriminately described as RNA molecules longer than 200 nucleotides with no translation into proteins.16 These biomacromolecules appear to modulate CRC via altering the expression of genes, triggering chromosomal remodeling, orchestrating transcriptional/post-transcriptional impacts, and self-translation of lncRNAs into polypeptides.17,18 Additionally, lncRNAs can prevent therapeutic-induced cell death, stimulate the EMT phenomenon, and promote non-cell-autonomous resistance mechanisms.19 Table 1 lists some of the aberrant expressions of specific lncRNAs together with their potential biological and clinical relevance during colorectal carcinogenesis.20,21
Table 1.
Long non-coding RNAs and colorectal cancer
|
LncRNAs
|
Expression level
|
Potential function and mechanism
|
References
|
| MALAT1 |
Upregulated |
Promoted proliferation, invasion, and migration through activating PRKA kinase anchor protein 9 (AKAP-9) |
22
|
| H19 |
Upregulated |
Downregulation of its target tumor suppressor retinoblastoma (RB) and hence regulation of the CRC development |
23
|
| CCAT1 |
Upregulated |
Promoted CRC progression by regulating the miR-181a-5p expression |
24
|
| CCAT2 |
Upregulated |
Enhance the proliferation and metastasis of CRC cells by engaging in direct interactions with TAF15, facilitating the transcriptional activation of RAB14, and triggering the AKT/GSK3β signaling pathway. |
25
|
| PANDAR |
Upregulated |
Enhanced CRC progression by EMT pathway |
26
|
| UCA1 |
Upregulated |
Promoted progression of CRC via the miR-143/MYO6 axis |
27
|
| MEG3 |
Downregulated |
SOCS3-mediated repression of the malignant proliferation of colonic stem cells by activation of miR-708 and hence inhibition of CRC progression |
28
|
| PCAT6 |
Upregulated |
Inhibited apoptosis of CRC cells through regulation of anti-apoptotic protein ARC expression via EZH2 |
29
|
| BCAR4 |
Upregulated |
Enhanced CRC progression via activating Wnt/β-catenin signaling |
30
|
| TUSC7 |
Downregulated |
Promote cell migration and invasion in CRC via regulation of miR-23b/PDE7A Axis |
31
|
| MAPKAPK5-AS1 |
Upregulated |
Enhanced CRC progression by cis-regulating the nearby gene MK5 and acting as a let-7f-1-3p sponge |
29,32
|
| RP9P |
Upregulated |
Promote CRC progression by modulating miR-133a-3p/FOXQ1 axis |
33
|
| u50535 |
Upregulated |
Promoted CRC growth and metastasis by regulating CCL20 |
29
|
| PVT1 |
Upregulated |
Promoting CRC tumorigenesis through miR-16-5p stabilization and interaction with the VEGFA/VEGFR1/AKT Axis |
27
|
| NEAT1 |
Upregulated |
Promoted progression of CRC via modulation of the KDM5A/Cul4A and Wnt signaling pathway |
34
|
| FTX |
Upregulated |
Enhanced migration and invasion of CRC cells by miRNA-590-5p/RBPJ axis |
35
|
| XIST |
Upregulated |
Enhanced growth and metastatic potential of colorectal cancer cells by directly affecting miR-486-5p and facilitating the activation of neuropilin-2, a critical regulator of epithelial-mesenchymal transition (EMT) |
36
|
Remarkably, lncRNAs are believed to be potential modulators of genes related to the cancer cells resistance mechanisms, which might (i) influence intracellular drug concentrations, (ii) prompt alternative signaling pathways, and (iii) modify drug efficacy by hindering cell cycle regulation and DNA damage response. All in all, they are probably responsible for developing resistance to anticancer agents, especially anti-EGFR mAbs.31 LncRNAs may induce CET-resistance in mCRC through different mechanisms, including (i) the EGFR mutations and disrupting the CET binding to EGFR (lncRNA POU5F1P4),37 (ii) the mutations of the EGFR downstream pathways (lncRNA CRART16),38 (iii) the activation of Wnt/β-catenin signaling (lncRNA MIR100HG),26 and (iv) the activation of the parallel pathway such as MET (lncRNA UCA1).39 Remarkably, lncRNA CRART16 was reported to elicit CET-resistance in CRC cells most likely by functioning as a miR-371a-5p sponge and thereby enhancing the expression of erythroblasts Leukemia viral oncogene Homolog 3 (ERBB3) through the miR-371a-5p/ERBB3/MAPK pathway.38 The downregulation of lncRNAs LNC00973 seems to improve CET resistance in mCRC cells most likely by regulating the metabolism of glucose.29 Besides, lncRNA HCG18 appears to facilitate the progress of the CRC resulting in CET-resistance through upregulation of PD-L1 and also suppressed CD8+ T lymphocytes cells via sponging miR-20b-5p.28 The upregulation of exosomal lncRNA UCA1,29 and downregulation of lncRNA POU5F1P4 were shown to be involved with the emergence of drug resistance in CET-sensitive CRC cells.37 Recent studies have supported lncRNAs' roles in anti-EGFR drug-resistance inducing based on lncRNAs-mRNAs, or lncRNAs-miRNAs-mRNAs regulatory networks through the EGFR, RAS, and PI3K/AKT signaling pathways.40 Fig. 1 illustrates the possible involvement of lncRNAs on EGFR-related signaling pathways.
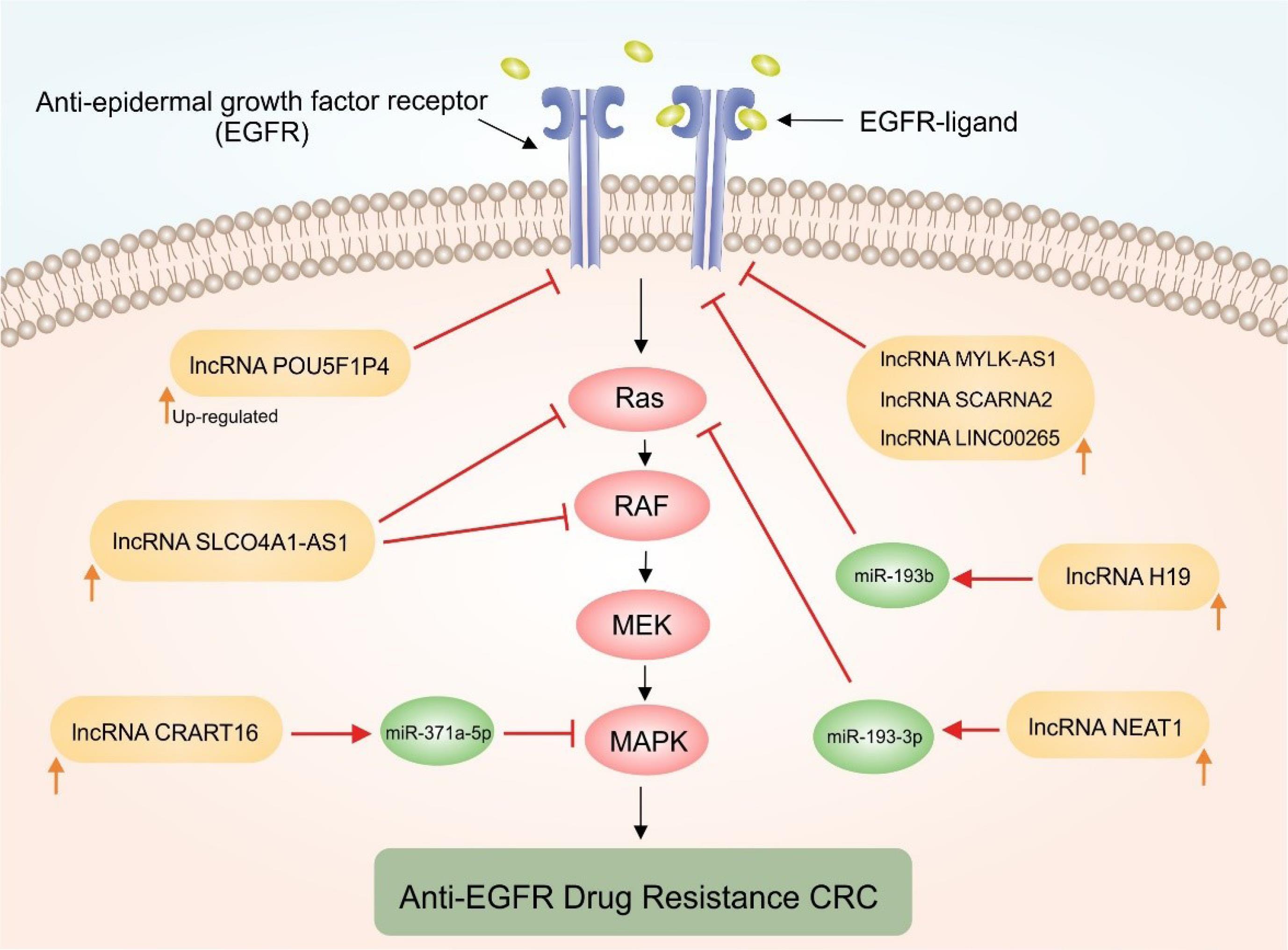
Fig. 1.
Schematic representation of the upregulation impacts of certain lncRNAs on the EGFR-related signaling pathways. Epidermal growth factor receptor (EGFR), as a cell surface receptor, regulates cell growth, proliferation, and survival. Dysregulation of EGFR signaling is commonly observed in cancer, leading to uncontrolled cell growth and tumor progression. A complex network of genes and miRNAs can be modulated, including those associated with the RAS/RAF/MEK/ERK pathways. LncRNA CRART16 upregulates ERBB3 expression through miR-371a-5p, lncRNA NEAT1 downregulates miR-193a-3p, and lncRNA H19 attenuates miR-193b-mediated inhibition. These functions activate the EGFR/RAS/RAF/MAPK pathways and contribute to anti-EGFR resistance in CRC.
.
Schematic representation of the upregulation impacts of certain lncRNAs on the EGFR-related signaling pathways. Epidermal growth factor receptor (EGFR), as a cell surface receptor, regulates cell growth, proliferation, and survival. Dysregulation of EGFR signaling is commonly observed in cancer, leading to uncontrolled cell growth and tumor progression. A complex network of genes and miRNAs can be modulated, including those associated with the RAS/RAF/MEK/ERK pathways. LncRNA CRART16 upregulates ERBB3 expression through miR-371a-5p, lncRNA NEAT1 downregulates miR-193a-3p, and lncRNA H19 attenuates miR-193b-mediated inhibition. These functions activate the EGFR/RAS/RAF/MAPK pathways and contribute to anti-EGFR resistance in CRC.
Collectively, the mechanisms underlying the CRC resistance to anti-EGFR therapy are the most complicated issue, and the lncRNAs spectrum associated with this resistance mechanism remained largely unknown due to the paucity of lncRNAs-specific microarray/RNA sequencing analysis. Upon some published data, lncRNAs can affect the therapeutic efficacy of anti-EGFR mAbs mainly through intracellular signaling even though their specific mechanisms are yet to be fully addressed. Notably, the interaction between ncRNAs and their crosstalk with anti-EGFR resistance-related pathways needs to be fully understood. To specifically target and inhibit the overexpressed lncRNAs, various therapeutic approaches can be developed. First, antisense oligonucleotides (ASOs), antagomirs, small interfering RNAs (siRNAs), short hairpin RNAs (shRNAs), miRNA sponges, and CRISPR/Cas9-based genome editing technique, which can be used to directly interfere with the expression or function of the overexpressed lncRNAs, leading to their inhibition or degradation. Second, treatment strategies that involve the use of tumor suppressor lncRNAs. Some lncRNAs have the ability to suppress tumor growth and progression. By introducing or enhancing the expression of tumor suppressor lncRNAs, it may be possible to regulate the functional expression of oncomiRs and restore normal cellular functions. Third, small molecule inhibitors, which specifically target the functional domains or binding sites of overexpressed lncRNAs, can be developed. Fourth, screening natural compounds and extracts for their ability to inhibit overexpressed lncRNAs can be another approach. Finally, nanoscale formulations such as Ab-drug nanoconjugates can be employed. These involve coupling therapeutic agents, such as small molecule drugs or antibodies, to nanoparticles with potential to specifically target and suppress the cells or tissues expressing the lncRNAs. Altogether, therapeutic strategies for targeting overexpressed lncRNAs include the use of antisense oligonucleotides, RNA interference, genome editing, exploitation of tumor suppressor lncRNAs, and nanoscale treatment modalities like Ab-drug nanoconjugates. These approaches hold promise in combating cancers associated with aberrant lncRNA expression. Despite the recent progress in ncRNA-based therapeutics, understanding the precise role of lncRNAs and related molecular mechanisms in anti-EGFR therapy and CET resistance in mCRC requires deep insights. In this regard, mismatched base pairing to non-target mRNAs, unexpected effects on normal tissue, and especially off-target effects are still tremendous challenges that need to be addressed. Thus, further evaluations are required to improve therapeutics' specificity, delivery, and tolerability using lncRNAs. Moreover, lncRNAs can be applied in monitoring and forecasting treatment response and resistance to personalized treatments to improve clinical outcomes.
Competing interests
None to be stated.
Ethical statement
The present study was approved by the Ethics Committee of Tabriz University of Medical Sciences (Ethical No. IR.TBZMED.REC.1401.338).
Funding sources
The support provided by the Liver and Gastrointestinal Diseases Research Center at Tabriz University of Medical Sciences (grant #69709) to MAK is acknowledged.
References
- Omidi Y, Mobasher M, Castejon AM, Mahmoudi M. Recent advances in nanoscale targeted therapy of HER2-positive breast cancer. J Drug Target 2022; 30:687-708. doi: 10.1080/1061186X.2022.2055045 [Crossref] [ Google Scholar]
- Akbarzadeh Khiavi M, Safary A, Barar J, Ajoolabady A, Somi MH, Omidi Y. Multifunctional nanomedicines for targeting epidermal growth factor receptor in colorectal cancer. Cell Mol Life Sci 2020; 77:997-1019. doi: 10.1007/s00018-019-03305-z [Crossref] [ Google Scholar]
- Bakhtiary Z, Barar J, Aghanejad A, Saei AA, Nemati E, Ezzati Nazhad Dolatabadi J. Microparticles containing erlotinib-loaded solid lipid nanoparticles for treatment of non-small cell lung cancer. Drug Dev Ind Pharm 2017; 43:1244-53. doi: 10.1080/03639045.2017.1310223 [Crossref] [ Google Scholar]
- Baradaran B, Majidi J, Farajnia S, Barar J, Omidi Y. Targeted therapy of solid tumors by monoclonal antibody specific to epidermal growth factor receptor. Hum Antibodies 2014; 23:13-20. doi: 10.3233/HAB-140278 [Crossref] [ Google Scholar]
- Najar AG, Pashaei-Asl R, Omidi Y, Farajnia S, Nourazarian AR. EGFR antisense oligonucleotides encapsulated with nanoparticles decrease EGFR, MAPK1 and STAT5 expression in a human colon cancer cell line. Asian Pac J Cancer Prev 2013; 14:495-8. doi: 10.7314/apjcp.2013.14.1.495 [Crossref] [ Google Scholar]
- Nourazarian AR, Pashaei-Asl R, Omidi Y, Najar AG. c-Src antisense complexed with PAMAM denderimes decreases of c-Src expression and EGFR-dependent downstream genes in the human HT-29 colon cancer cell line. Asian Pac J Cancer Prev 2012; 13:2235-40. doi: 10.7314/apjcp.2012.13.5.2235 [Crossref] [ Google Scholar]
- Nourazarian AR, Najar AG, Farajnia S, Khosroushahi AY, Pashaei-Asl R, Omidi Y. Combined EGFR and c-Src antisense oligodeoxynucleotides encapsulated with PAMAM Denderimers inhibit HT-29 colon cancer cell proliferation. Asian Pac J Cancer Prev 2012; 13:4751-6. doi: 10.7314/apjcp.2012.13.9.4751 [Crossref] [ Google Scholar]
- Bianco R, Gelardi T, Damiano V, Ciardiello F, Tortora G. Rational bases for the development of EGFR inhibitors for cancer treatment. Int J Biochem Cell Biol 2007; 39:1416-31. doi: 10.1016/j.biocel.2007.05.008 [Crossref] [ Google Scholar]
- Akbarzadeh Khiavi M, Safary A, Somi MH. Recent advances in targeted therapy of colorectal cancer: impacts of monoclonal antibodies nanoconjugates. Bioimpacts 2019; 9:123-7. doi: 10.15171/bi.2019.16 [Crossref] [ Google Scholar]
- Li QH, Wang YZ, Tu J, Liu CW, Yuan YJ, Lin R. Anti-EGFR therapy in metastatic colorectal cancer: mechanisms and potential regimens of drug resistance. Gastroenterol Rep (Oxf) 2020; 8:179-91. doi: 10.1093/gastro/goaa026 [Crossref] [ Google Scholar]
- Zhou J, Ji Q, Li Q. Resistance to anti-EGFR therapies in metastatic colorectal cancer: underlying mechanisms and reversal strategies. J Exp Clin Cancer Res 2021; 40:328. doi: 10.1186/s13046-021-02130-2 [Crossref] [ Google Scholar]
- Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/beta-catenin signaling. Nat Med 2017; 23:1331-41. doi: 10.1038/nm.4424 [Crossref] [ Google Scholar]
- Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov 2014; 4:1269-80. doi: 10.1158/2159-8290.CD-14-0462 [Crossref] [ Google Scholar]
- Stintzing S, Modest DP, Rossius L, Lerch MM, von Weikersthal LF, Decker T. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab for metastatic colorectal cancer (FIRE-3): a post-hoc analysis of tumour dynamics in the final RAS wild-type subgroup of this randomised open-label phase 3 trial. Lancet Oncol 2016; 17:1426-34. doi: 10.1016/S1470-2045(16)30269-8 [Crossref] [ Google Scholar]
- Sana J, Faltejskova P, Svoboda M, Slaby O. Novel classes of non-coding RNAs and cancer. J Transl Med 2012; 10:103. doi: 10.1186/1479-5876-10-103 [Crossref] [ Google Scholar]
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer 2018; 18:5-18. doi: 10.1038/nrc.2017.99 [Crossref] [ Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012; 81:145-66. doi: 10.1146/annurev-biochem-051410-092902 [Crossref] [ Google Scholar]
- Tang Y, Cheung BB, Atmadibrata B, Marshall GM, Dinger ME, Liu PY. The regulatory role of long noncoding RNAs in cancer. Cancer Lett 2017; 391:12-9. doi: 10.1016/j.canlet.2017.01.010 [Crossref] [ Google Scholar]
- Hahne JC, Valeri N. Non-Coding RNAs and Resistance to Anticancer Drugs in Gastrointestinal Tumors. Front Oncol 2018; 8:226. doi: 10.3389/fonc.2018.00226 [Crossref] [ Google Scholar]
- Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res 2013; 23:1446-61. doi: 10.1101/gr.152942.112 [Crossref] [ Google Scholar]
- Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. Int J Oncol 2011; 39:169-75. doi: 10.3892/ijo.2011.1007 [Crossref] [ Google Scholar]
- Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. BiochimBiophys Acta 2015; 1852:166-74. doi: 10.1016/j.bbadis.2014.11.013 [Crossref] [ Google Scholar]
- Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2010; 31:350-8. doi: 10.1093/carcin/bgp181 [Crossref] [ Google Scholar]
- Shang A, Wang W, Gu C, Chen W, Lu W, Sun Z. Long non-coding RNA CCAT1 promotes colorectal cancer progression by regulating miR-181a-5p expression. Aging (Albany NY) 2020; 12:8301-20. doi: 10.18632/aging.103139 [Crossref] [ Google Scholar]
- Wang D, Li Z, Yin H. Long Non-Coding RNA CCAT2 Activates RAB14 and Acts as an Oncogene in Colorectal Cancer. Front Oncol 2021; 11:751903. doi: 10.3389/fonc.2021.751903 [Crossref] [ Google Scholar]
- Lu M, Liu Z, Li B, Wang G, Li D, Zhu Y. The high expression of long non-coding RNA PANDAR indicates a poor prognosis for colorectal cancer and promotes metastasis by EMT pathway. J Cancer Res Clin Oncol 2017; 143:71-81. doi: 10.1007/s00432-016-2252-y [Crossref] [ Google Scholar]
- Luan Y, Li X, Luan Y, Zhao R, Li Y, Liu L. Circulating lncRNA UCA1 Promotes Malignancy of Colorectal Cancer via the miR-143/MYO6 Axis. Mol Ther Nucleic Acids 2020; 19:790-803. doi: 10.1016/j.omtn.2019.12.009 [Crossref] [ Google Scholar]
- Liu CG, Li J, Xu Y, Li W, Fang SX, Zhang Q. Long non-coding RNAs and circular RNAs in tumor angiogenesis: From mechanisms to clinical significance. Mol TherOncolytics 2021; 22:336-54. doi: 10.1016/j.omto.2021.07.001 [Crossref] [ Google Scholar]
- Huang W, Su G, Huang X, Zou A, Wu J, Yang Y. Long noncoding RNA PCAT6 inhibits colon cancer cell apoptosis by regulating anti-apoptotic protein ARC expression via EZH2. Cell Cycle 2019; 18:69-83. doi: 10.1080/15384101.2018.1558872 [Crossref] [ Google Scholar]
- Ouyang S, Zheng X, Zhou X, Chen Z, Yang X, Xie M. LncRNA BCAR4 promotes colon cancer progression via activating Wnt/beta-catenin signaling. Oncotarget 2017; 8:92815-26. doi: 10.18632/oncotarget.21590 [Crossref] [ Google Scholar]
- Hao L, Yun Y, Liang R, Yuan G. Long non-coding RNA TUSC7 suppressed colorectal cancer progression via regulation of miR-23b/PDE7A Axis. Clin Invest Med 2020; 43:E35-43. doi: 10.25011/cim.v43i4.34703 [Crossref] [ Google Scholar]
- Yang T, Chen WC, Shi PC, Liu MR, Jiang T, Song H. Long noncoding RNA MAPKAPK5-AS1 promotes colorectal cancer progression by cis-regulating the nearby gene MK5 and acting as a let-7f-1-3p sponge. J Exp Clin Cancer Res 2020; 39:139. doi: 10.1186/s13046-020-01633-8 [Crossref] [ Google Scholar]
- Jin Z, Liu B, Lin B, Yang R, Wu C, Xue W. The Novel lncRNA RP9P Promotes Colorectal Cancer Progression by Modulating miR-133a-3p/FOXQ1 Axis. Front Oncol 2022; 12:843064. doi: 10.3389/fonc.2022.843064 [Crossref] [ Google Scholar]
- Shen X, Ye Z, Wu W, Zhao K, Cheng G, Xu L, et al. lncRNA NEAT1 facilitates the progression of colorectal cancer via the KDM5A/Cul4A and Wnt signaling pathway. Int J Oncol 2021; 59. 10.3892/ijo.2021.5231.
- Chen GQ, Liao ZM, Liu J, Li F, Huang D, Zhou YD. LncRNA FTX Promotes Colorectal Cancer Cells Migration and Invasion by miRNA-590-5p/RBPJ Axis. Biochem Genet 2021; 59:560-73. doi: 10.1007/s10528-020-10017-8 [Crossref] [ Google Scholar]
- Jing C, Ma R, Cao H, Wang Z, Liu S, Chen D. Long noncoding RNA and mRNA profiling in cetuximab-resistant colorectal cancer cells by RNA sequencing analysis. Cancer Med 2019; 8:1641-51. doi: 10.1002/cam4.2004 [Crossref] [ Google Scholar]
- Ji H, Hui B, Wang J, Zhu Y, Tang L, Peng P. Long noncoding RNA MAPKAPK5-AS1 promotes colorectal cancer proliferation by partly silencing p21 expression. Cancer Sci 2019; 110:72-85. doi: 10.1111/cas.13838 [Crossref] [ Google Scholar]
- Wu H, Wei M, Jiang X, Tan J, Xu W, Fan X. lncRNA PVT1 Promotes Tumorigenesis of Colorectal Cancer by Stabilizing miR-16-5p and Interacting with the VEGFA/VEGFR1/AKT Axis. Mol Ther Nucleic Acids 2020; 20:438-50. doi: 10.1016/j.omtn.2020.03.006 [Crossref] [ Google Scholar]
- Yuan HH, Zhang XC, Wei XL, Zhang WJ, Du XX, Huang P. LncRNA UCA1 mediates Cetuximab resistance in Colorectal Cancer via the MiR-495 and HGF/c-MET Pathways. J Cancer 2022; 13:253-67. doi: 10.7150/jca.65687 [Crossref] [ Google Scholar]
- Chu J, Fang X, Sun Z, Gai L, Dai W, Li H. Non-Coding RNAs Regulate the Resistance to Anti-EGFR Therapy in Colorectal Cancer. Front Oncol 2021; 11:801319. doi: 10.3389/fonc.2021.801319 [Crossref] [ Google Scholar]