Bioimpacts. 14(2):27778.
doi: 10.34172/bi.2023.27778
Original Article
Comparative molecular docking and toxicity between carbon-capped metal oxide nanoparticles and standard drugs in cancer and bacterial infections
Navid Mohammadjani Conceptualization, Data curation, Formal analysis, Validation, Visualization, Writing – original draft, Writing – review & editing, 1 
Sahand Karimi Data curation, Formal analysis, Validation, Visualization, Writing – original draft, 1
Musa Moetasam Zorab Formal analysis, Validation, Visualization, Writing – original draft, 2 
Morahem Ashengroph Conceptualization, Data curation, Formal analysis, Project administration, Supervision, Validation, Visualization, Writing – review & editing, 1, * 
Mehran Alavi Conceptualization, Data curation, Formal analysis, Project administration, Supervision, Validation, Visualization, Writing – review & editing, 1, 3, * 
Author information:
1Department of Biological Science, Faculty of Science, University of Kurdistan, Sanandaj, Kurdistan, Iran
2Department of Physics, University of Halabja, Kurdistan Region, Iraq
3Nanobiotechnology Department, Faculty of Innovative Science and Technology, Razi University, Kermanshah, Iran
Abstract
Introduction:
Nanoparticles (NPs) are of great interest in the design of various drugs due to their high surface-to-volume ratio, which result from their unique physicochemical properties. Because of the importance of examining the interactions between newly designed particles with different targets in the case of various diseases, techniques for examining the interactions between these particles with different targets, many of which are proteins, are now very common.
Methods:
In this study, the interactions between metal oxide nanoparticles (MONPs) covered with a carbon layer (Ag2O3, CdO, CuO, Fe2O3, FeO, MgO, MnO, and ZnO NPs) and standard drugs related to the targets of Cancer and bacterial infections were investigated using the molecular docking technique with AutoDock 4.2.6 software tool. Finally, the PRO TOX-II online tool was used to compare the toxicity (LD50) and molecular weight of these MONPs to standard drugs.
Results:
According to the data obtained from the semi flexible molecular docking process, MgO and Fe2O3 NPs performed better than standard drugs in several cases. MONPs typically have a lower 50% lethal dose (LD50) and a higher molecular weight than standard drugs. MONPs have shown a minor difference in binding energy for different targets in three diseases, which probably can be attributed to the specific physicochemical and pharmacophoric properties of MONPs.
Conclusion:
The toxicity of MONPs is one of the major challenges in the development of drugs based on them. According to the results of these molecular docking studies, MgO and Fe2O3 NPs had the highest efficiency among the investigated MONPs.
Keywords: Metal oxide nanoparticle, Molecular docking, Virtual screening, Computer-aided drug design, Bacterial infection
Copyright and License Information
© 2024 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Nowadays, one of the most important techniques in computer-aided drug design is molecular docking, which is used in drug design projects to investigate and measure the interactions between target proteins and drugs, synthetic or natural compounds with potentially therapeutic properties. AutoDock 4.2.6 is a critically important and widely utilized software in the field of molecular docking for various research studies,1-3 which is a potential capability in molecular docking due to the ability to define the parameters of metal atoms that cannot be identified by much other software in the field of molecular docking.4 Nanotechnology has provided new options to treat various diseases in recent decades; however, many applications of this science, particularly in the design of drugs and medical diagnostic systems, have yet to be realized at the clinical level.5 The lack of biocompatibility and proper stability and toxicity of nanoparticles (NPs) produced by chemical and physical techniques, as well as the inappropriate scalability of NPs, are among the reasons for this. Fortunately, nanobiotechnology is helping to solve many of these issues.6,7 In fact, microorganisms and other living organisms, including animals and plants, have a unique ability in the biosynthesis of NPs with caps, which significantly decreases toxicity, increases biocompatibility, and stability.8,9 The binding of nanomaterials to various proteins can cause physiological and pathological changes, such as the tendency of proteins to bind or the activation or inhibition.10,11 Silver oxide NPs are increasingly being used in food packaging, medical equipment, and food supplements because of their excellent antimicrobial properties.12,13 Iron oxide NPs have also been approved by food and drug regulatory organizations around the world for anemia treatment, and some studies have reported their antiviral ability.14 Furthermore, copper oxide NPs also have antimicrobial properties due to their high surface-to-volume ratio as well as specific morphological properties.15 Other NPs also have shown their potential ability in the production of reactive oxygen species (ROS) and the destruction of cancer cells, microbials cells, etc. ZnO NPs have a number of benefits, including lower toxicity and simple absorption by humans. ZnO NPs are regarded by the FDA as generally recognized as safe (GRAS) when compared to other MONPs.16 Multidrug-resistant bacterial (MDR) infections, as one of the emerging challenges, have caused the development of new antibiotics and the use of innovative approaches in antibacterial drug design.17-19 The dihydropteroate synthase enzyme plays an important role in the synthesis of folic acid in Escherichia coli.20 Penicillin-binding protein, in particular its 2X type, is a protein responsible for beta-lactam antibiotic group resistance, and it is a major contributor to the development of antibiotic resistance in a variety of bacteria against beta-lactam-based antibiotics.21 Aquaporin Z is another antibacterial target that functions in water exchange in the bacterial cell membrane.22 Bacterial DNA gyrase plays a key role in the translation and transcription of bacterial DNA because of its role in breaking two strands of DNA by the process of catalysis of the negative supercoil; therefore, it is one of the major targets of antibacterial pharmaceutical inhibitors.23-25 Filamenting temperature-sensitive mutant Z, or FtsZ, has been introduced as one of the new antibacterial therapeutic targets for the development of new antibiotics because of its important role in bacterial cell division and also conserved across different bacterial species. One of the primary benefits of targeting this protein is that it is absent in more advanced organisms, including humans, implying that antibiotics targeting this protein do not harm human cells.26 The DNA molecule has been a primary and traditional target for the development of anti-cancer drugs. these drugs can play a significant role in disabling cancer cells by creating bonds with the DNA structure and being placed inside the structure.27 The hypoxia-inducible factor I, or HIF-1, is one of the transcription factors whose increase in the tumor microenvironment in response to hypoxia. This protein is one of the therapeutic targets in the treatment of certain types of cancer, such as liver cancer.28 Several studies have shown that human leukemia inhibitory factor, or LIF, plays a key role in the development of pancreatic ductal adenocarcinoma tumors.29 Caspases, such as caspase-3 and caspase-9, are cysteine proteases that play a key role in cancer cell apoptosis; therefore, the regulators of these proteins can be significant in anti-tumor functions.30,31 Serum and glucocorticoid-regulated kinase 1 (SGK1), also known as one of the SGK isoforms and a member of the AGC family, is one of the therapeutic targets because it has a favorable impact on tumor cell survival, increases tumor invasiveness and adhesion, and stimulates tumor growth.32 Cell division protein kinase 2 (CDK2) is another type of protein whose overexpression, promotes cancer cell growth.33 The X-linked inhibitor of apoptosis protein (XIAP) is one of the proteins whose increased expression disrupts the cell apoptosis system and, as a result, the development of cancer; thus, protein inhibitors can play a role in cancer treatment via inhibition this protein.34 Telomerase overactivity has been observed in a variety of human cancers, suggesting that telomerase reverse transcriptase inhibitors could be used as anti-tumor therapeutic targets.35 Based on CRISPR dropout screens performed by Gao et al., the 3-hydroxy-3-methyl-glutaryl-coenzyme A enzyme or (HMG-CoA) reductase has been introduced as a therapeutic target for the anti-tumor activity of colon cancer stem cells, and inhibitors of this enzyme may have an anti-tumor role in this field.36
In this study, the molecular docking technique was used to investigate the way introduced drugs interact with antibacterial and anticancer drug targets compared to MONPs with a conceptual carbon cap. Considering the need to develop new drugs for various diseases, as well as several studies that have pointed to the medicinal role of MONPs in the diseases mentioned above, this study used the semi flexible molecular docking process to obtain the conceptual and theoretical feasibility of the possible medicinal role of these MONPs. This study used MONPs capped with carbon layer to simulate MONPs that biosynthesized with different organisms. Because such caps are very diverse and it is not possible to introduce a specific cap as a representative of all such caps, only a carbon cap layer was used in this study. To further investigate the performance of MONPs, the toxicity of these particles was compared to standard drugs.
Materials and Methods
Preparation of required files
The RCSB Protein Data Bank (https://www.rcsb.org/) was used to obtain the crystal structure files for the targets in this article. Antibacterial targets included 1AJ0, 1QME, 1RC2, 4URO, and 5V68, while 1BNA, 1EMR, 1H2N, 1RE1, 2AR9, 3HDN, 3IG7, 5CQG, 5OQW, 8DJM were anticancer targets.
All files downloaded from the RCSB PDB were in PDB format. The Drug Bank database (https://go.drugbank.com/) was then used to identify the main drugs of the targets. In this manner, the search in Drug Bank was performed based on the type and title of the desired target, as well as the type of organism, and then the drug files in PDB format were saved. After storing all of the target files and standard drugs, it was time to prepare and design the MONPs. At this stage, 8 metal oxide nanostructures with dimensions X × Y × Z= 5 × 5 × 5 angstroms were produced by Materials Studio 2020 software, including Ag2O3, CdO, CuO, Fe2O3, FeO, MgO, MnO, and ZnO with a carbon layer cap. After the nanostructures were created, the PDB format file for each nanostructure was saved. It should be noted that the Open Babel GUI 3.1.1 software is used in the process of performing the steps if the file format needs to be changed.
Preparation of targets and prediction of their binding sites
The UCSF Chimera 1.16 software was used to remove extra atoms (such as water, ions, etc) from the PDB files, as well as chains and other extra parts from the target files, before saving the file in mol2 format. The process of adding hydrogen to the target structure and then calculating and adding the Gasteiger charge was then completed using the software AutoDock tools 1.5.7 (ADT). The targets were saved in the pdbqt format file at this point. The Discovery Studio 2021 software was used at this point to predict the possible binding sites of all targets, and the XYZ coordinates of the best ones (number one) according to natural ligand-protein interactions recorded in PDB, were chose for the molecular docking process.
Carbon capped-MONPs and drugs
UCSF Chimera 1.16 was used to add hydrogen and then Gasteiger charge added to the received drugs from DrugBank, after which the file was saved in mol2 format and read by ADT 1.5.7 software, then Torsion Tree was calculated with this software, and the bonds that could be rotated were confirmed.
Molecular docking process
The docking software that used in this study was AutoDock 4.2.6.4 The type of docking is semi flexible in this study.4 The GPF (Grid parameter file) and DPF (docking parameter file) were created in the first stage of the docking process using a genetic algorithm with the ADT 1.5.7. The grid box size was 120 × 120 × 120 angstroms. Also, in the case of NPs, parameter and bond files was accompanied by a few changes, and the parameter of metal atoms used in the study were added. Before producing the GPF files and DPF, the new parameter and bond files, which contained the parameter information of the new metal atoms, were defined for the ADT software in each case. At this point, all of the files required to carry out the molecular docking procedure were ready.
Analysis of molecular docking results
The results from AutoDock 4.2.6 were analyzed by ADT 1.5.7 and Discovery Studio 2021 software. To evaluate the results of drug docking with their targets, ADT software first saved the final virtual screening pdbqt file of the best binding pose run, and then the interactions between drugs and key residues and their types were analysed by ADT and Discovery Studio software using 3D and 2D visualizations. In addition, to validate the nanoparticle docking results with targets, ADT software and discovery studio checked the key residues involved in the NP-target interaction, and then Discovery Studio and ADT software evaluated the types of interactions.
Toxicity of MONPs compared with standard drugs
We predicted the toxicity level of drugs and NPs using the online tool PRO TOX-II (https://tox-new.charite.de/protox_II/).37,38 This server predicted the LD50 (Lethal dose of 50%) of drugs and NPs using input files in mol format.
Results
Docking results of anti-bacterial targets
The binding energy in kcal/mol and inhibition constant in micromolar were calculated after molecular docking for bacterial targets with PDB IDs 1AJ0, 1QME, 1RC2, 4URO, and 5V68 (Table 1). Sulfaphenazole had the best binding energy with -8.51 kcal/mol for the Dihydropteroate synthase (1AJ0) target, as shown in Table 1. Also, Fe2O3 had the best MONP with -6.27 kcal/mol. For second bacterial target, penicillin-binding protein 2X (1QME), the best drug and NP, were cloxacillin, and MgO, with binding energies of -8.50 and -7.87 kcal/mol respectively (Fig. 1). The best drug and NP also were found to bind to the aquaporin Z (1RC2) protein as third bacterial target. Octyl beta-D-galactopyranoside and MgO had binding energies of 3.58 and -7.97 kcal/mol, respectively. Novobiocin is the only standard drug that has been introduced for the DNA gyrase subunit B (4URO) target. It has a binding energy of -4.73 kcal/mol and also the best NP is Fe2O3 with binding energy of -6.17 kcal/mol (Fig. 2). The protein involved in cell division, Ftsz (5V68), is the final bacterial target investigated in this study. The best drug is 5'-guanosine-diphosphate-monothiophosphate, which has a binding energy of -5.03 kcal/mol, and the best NP is MgO, which has a binding energy of -5.27 kcal/mol (Fig. 3). Figs. 1 to 3 clearly summarizes the pattern of interaction between antibacterial targets and drugs or NPs/MONPs. The key residues of the investigated targets in interactions with MONPs and drugs are also summarized in Table 1.
Table 1.
The results of the molecular docking process of drugs and MONPs with a carbon layer cap with anti-bacterial targets
Target protein (PDB ID) (organism)
(Predicted binding site)
|
Drugs/MONPs
|
Binding energy
(kcal/mol)
|
Ki (µM) Inhibition constant
|
Dihydropteroate synthase (1AJ0) (E. coli)
(41.899, 1.026, 9.232) |
Drugs: |
|
|
| 1. Acetyl sulfisoxazole |
-6.39 |
20.80 |
| 2. Sulfacetamide |
-5.95 |
43.55 |
| 3. Sulfacytine |
-6.59 |
14.71 |
| 4. Sulfamerazine |
-6.43 |
19.30 |
| 5. Sulfameter |
-6.49 |
17.48 |
| 6. Sulfamethazine |
-6.99 |
7.51 |
| 7. Sulfamethizole |
-6.59 |
14.80 |
| 8. Sulfamethoxazole |
-6.90 |
8.69 |
| 9. Sulfanilamide |
-5.61 |
77.20 |
| 10. Sulfaphenazole |
-8.51 |
579.59nM |
| 11. Sulfisoxazole |
-7.12 |
5.99 |
| NPs: |
|
|
| 1. Ag2O3 |
-5.84 |
52.59 |
| 2. CdO |
-4.82 |
295.44 |
| 3. CuO |
-4.37 |
627.30 |
| 4. Fe2O3 |
-6.27 |
25.37 |
| 5. FeO |
-5.54 |
86.71 |
| 6. MgO |
-5.87 |
49.88
105.97 |
| 7. MnO |
-5.42 |
| 8. ZnO |
-5.36 |
117.61 |
Penicillin-binding protein 2X (1QME)
(Streptococcus penumoniae)
(95.173, 57.183, 53.895) |
Drugs: |
|
|
| 1. 6-o-Capryloylsucrose |
-4.59 |
433.94 |
| 2. Cloxacillin |
-8.50 |
589.12nM |
| NPs: |
|
|
| 1. Ag2O3 |
-5.85 |
51.32 |
| 2. CdO |
-7.39 |
3.84 |
| 3. CuO |
-5.38 |
114.71 |
| 4. Fe2O3 |
-6.25 |
26.31 |
| 5. FeO |
-7.57 |
2.82 |
| 6. MgO |
-7.87 |
1.71 |
| 7. MnO |
-7.59 |
2.72 |
| 8. ZnO |
-6.07 |
35.59 |
Aquaporin Z
(1RC2) (E. coli)
(79.830, 7.067, -26.468) |
Drugs: |
|
|
| 1. C19H36O10P - |
-2.66 |
11.19mM |
| 2. B-2-Octylglucoside |
-3.59 |
2.33mM |
| 3. octyl alpha-L-altropyranoside |
-3.18 |
4.69mM |
| 4. octyl beta-D galactopyranoside |
-3.85 |
1.50mM |
| NPs: |
|
|
| 1. Ag2O3 |
-6.20 |
28.34 |
| 2. CdO |
-7.24 |
4.93 |
| 3. CuO |
-5.19 |
156.46 |
| 4. Fe2O3 |
-6.23 |
27.16 |
| 5. FeO |
-7.52 |
3.06 |
| 6. MgO |
-7.97 |
1.43 |
| 7. MnO |
-7.48 |
3.30 |
| 8. ZnO |
-5.71 |
65.62 |
| DNA gyrase subunit B (4URO) (Staphylococcus aureus) (35.214, -12.539, 8.265) |
Drugs: |
|
|
| 1. Novobiocin |
-4.73 |
343.28 |
| NPs: |
|
|
| 1. Ag2O3 |
-5.85 |
51.08 |
| 2. CdO |
-5.41 |
108.69 |
| 3. CuO |
-4.87 |
270.64 |
| 4. Fe2O3 |
-6.17 |
30.09 |
| 5. FeO |
-5.59 |
80.45 |
| 6. MgO |
-5.84 |
52.41 |
| 7. MnO |
-5.57 |
82.62 |
| 8. ZnO |
-5.46 |
98.70 |
Filamenting temperature-sensitive mutant Z or Ftsz (5V68)
(Mycobacterium tuberculosis)
(-9.737, 73.159, 18.073) |
Drugs:
|
|
|
| 1. 5’-Guanosine-diphosphate-monothiophosphate |
-5.03 |
204.28 |
| 2. Guanosine-5’-diphosphate |
-2.79 |
8.98mM |
|
NPs:
|
|
|
| 1. Ag2O3 |
-5.05 |
199.36 |
| 2. CdO |
-4.70 |
356.10 |
| 3. CuO |
-4.26 |
749.81 |
| 4. Fe2O3 |
-5.05 |
197.93 |
| 5. FeO |
-5.03 |
204.31 |
| 6. MgO |
-5.27 |
136.79 |
| 7. MnO |
-4.93 |
242.46 |
| 8. ZnO |
-4.86 |
272.09 |
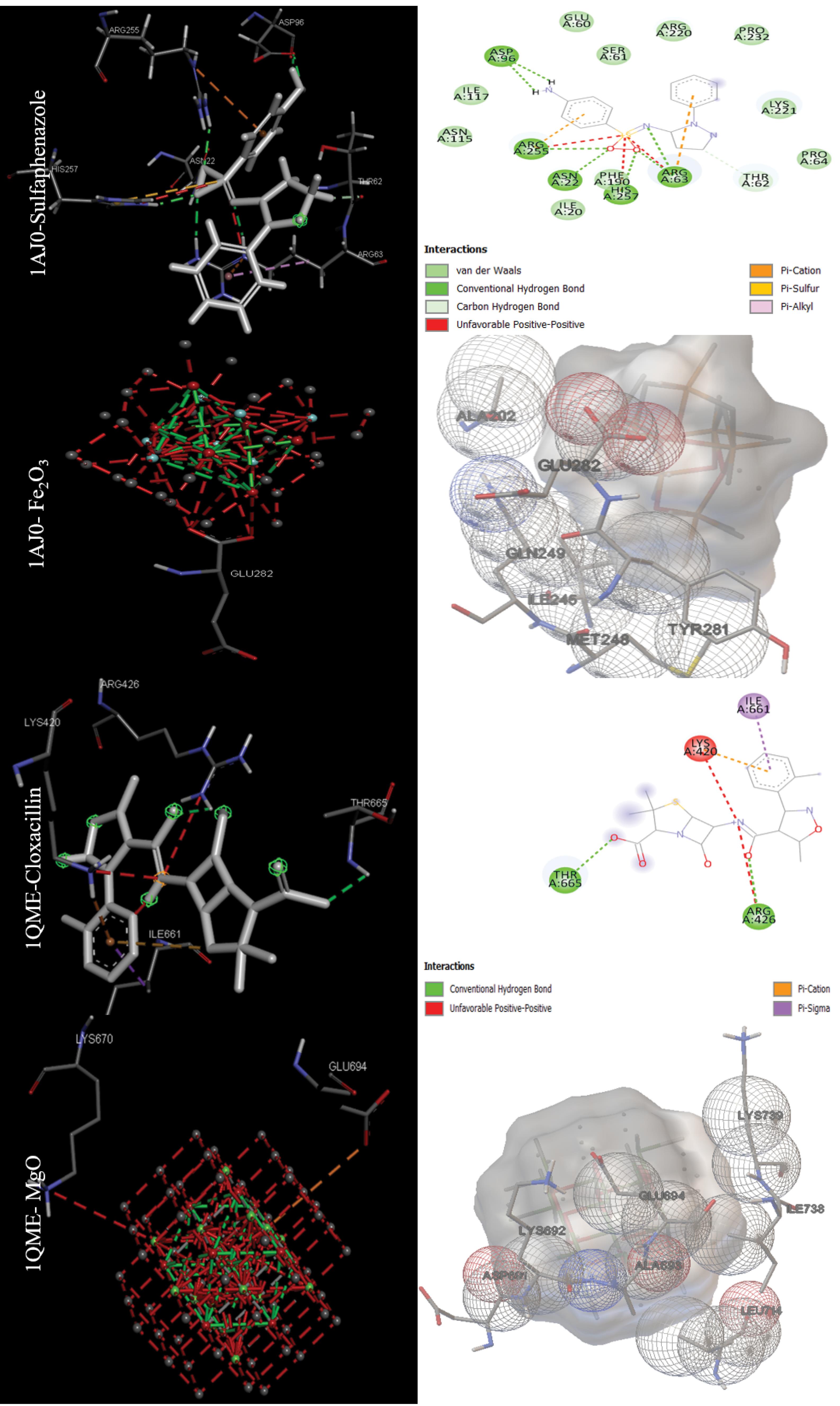
Fig. 1.
Docking positions for the main drugs and NPs with antibacterial receptors (Dihydropteroate synthase (1AJ0) and Penicillin-binding protein 2X (1QME)) with lowest binding energy (visualized were obtained using ADT 1.5.7 and discovery studio 2021).
.
Docking positions for the main drugs and NPs with antibacterial receptors (Dihydropteroate synthase (1AJ0) and Penicillin-binding protein 2X (1QME)) with lowest binding energy (visualized were obtained using ADT 1.5.7 and discovery studio 2021).
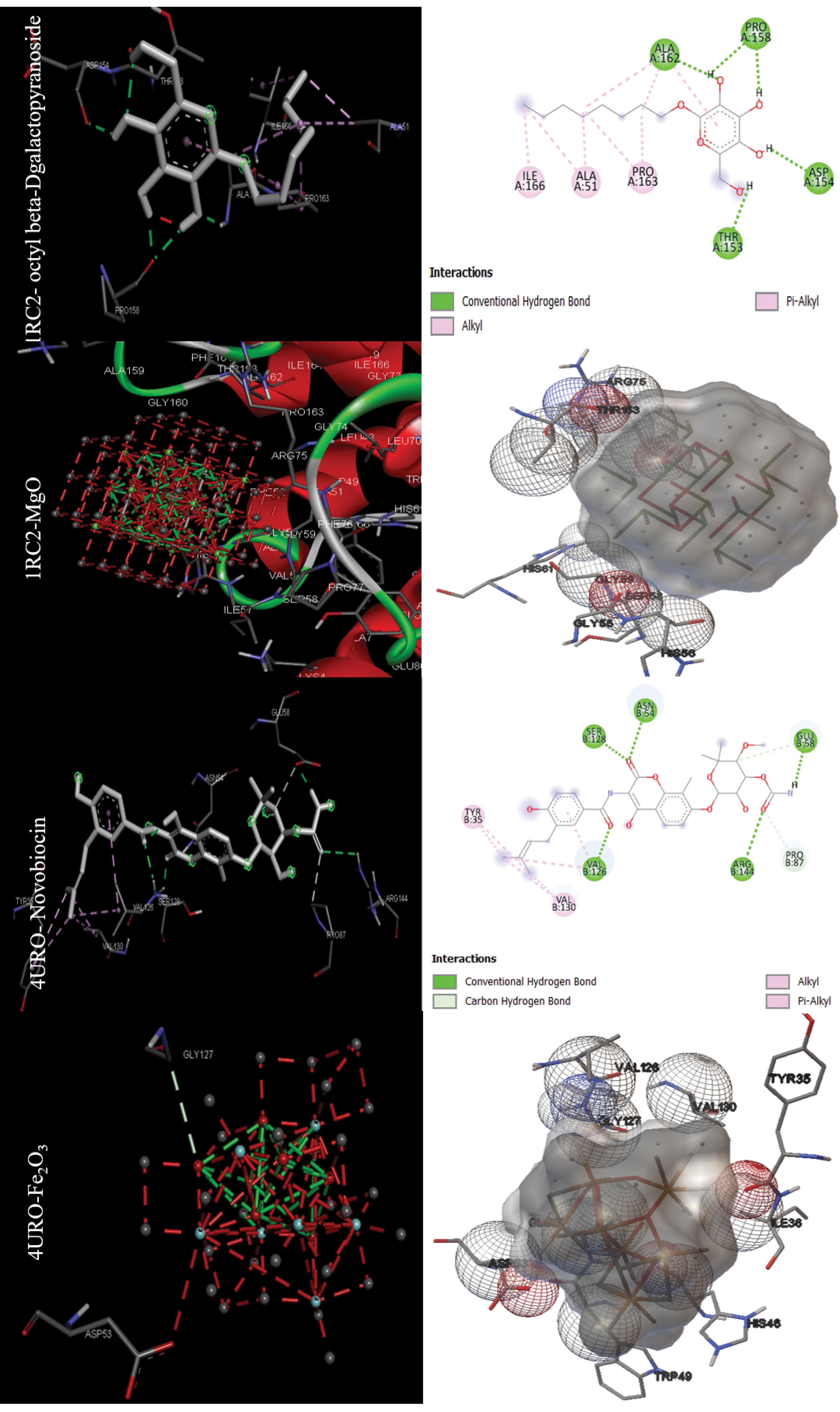
Fig. 2.
Docking positions for the main drugs and NPs with antibacterial receptors (Aquaporin Z (1RC2) and DNA gyrase subunit B (4URO)) with lowest binding energy (visualized were obtained using ADT 1.5.7 and discovery studio 2021).
.
Docking positions for the main drugs and NPs with antibacterial receptors (Aquaporin Z (1RC2) and DNA gyrase subunit B (4URO)) with lowest binding energy (visualized were obtained using ADT 1.5.7 and discovery studio 2021).
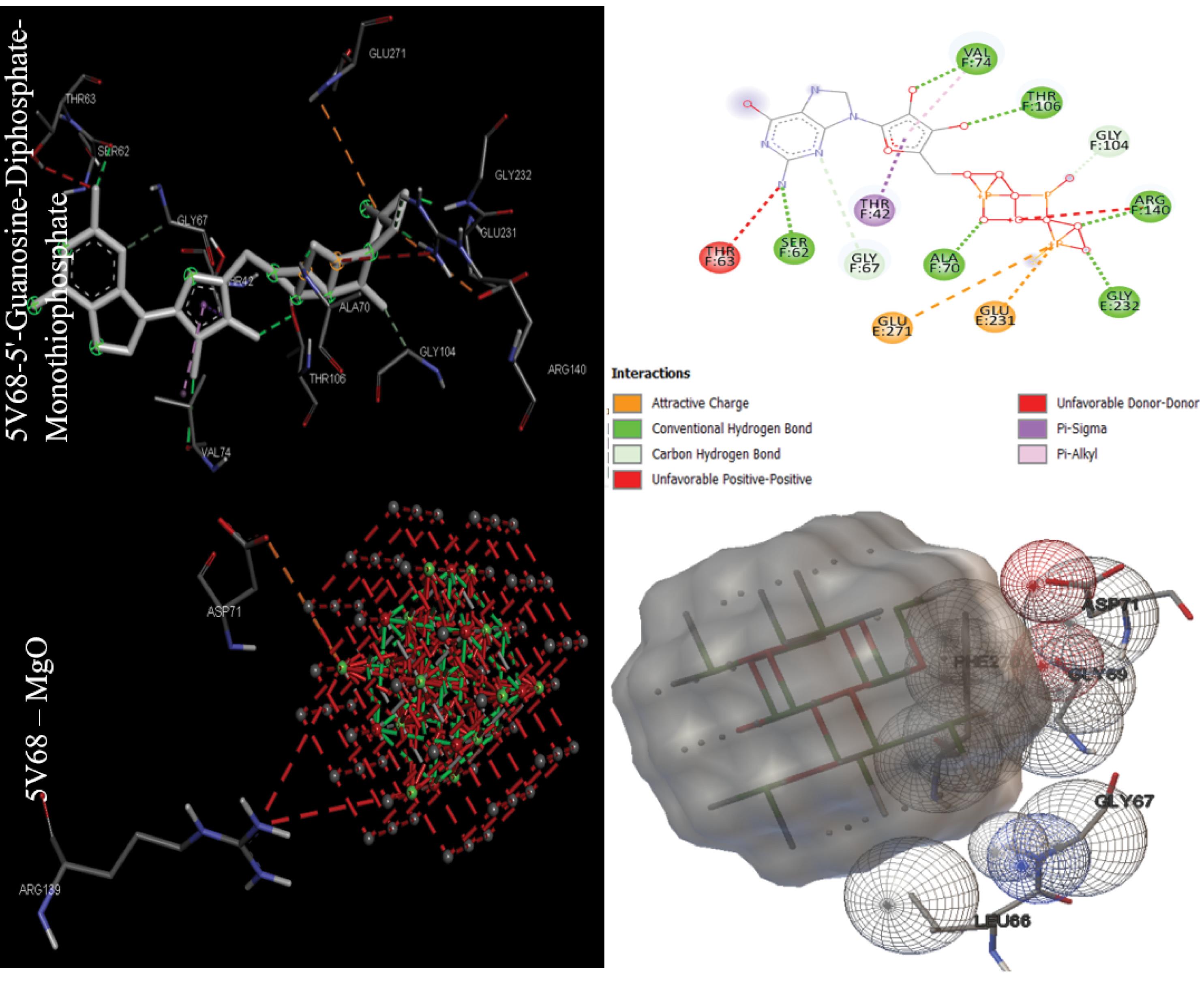
Fig. 3.
Docking positions for the main drugs and NPs with antibacterial receptor (Filamenting temperature-sensitive mutant Z or Ftsz (5V68) ) with lowest binding energy (visualized were obtained using ADT 1.5.7 and discovery studio 2021).
.
Docking positions for the main drugs and NPs with antibacterial receptor (Filamenting temperature-sensitive mutant Z or Ftsz (5V68) ) with lowest binding energy (visualized were obtained using ADT 1.5.7 and discovery studio 2021).
Docking results of anti-cancer targets
The data in Table 2 were obtained by performing molecular docking on anticancer targets with PDB IDs 1BNA, 1EMR, 1H2N, 1RE1, 2AR9, 3HDN, 3IG7, 5CQG, 5OQW, and 8DJM using AutoDock 4.2.6 software. Gentian Violet (-12.23 kcal/mol) was the best drug in terms of binding energy for the first target, human B-DNA (1BNA). The best MNOPs docked at the first target was MgO, with a binding energy of -6.41 kcal/mol. For the second target, human leukemia inhibitory factor (1EMR), doxorubicin and MgO with binding energies of -6.09 and -5.36, respectively, were found to be the best drugs and MONPs (Fig. 4). The best drug, 2-Methoxyestradiol, had a binding energy of -8.19 kcal/mol for the third anti-cancer target, hypoxia-inducible factor I (1H2N), and the best nanoparticle, MgO, had a binding energy of -7.19 kcal/mol. The fourth anti-cancer target, caspase3 (1RE1), was then studied. In terms of binding energy, glycyrrhizic acid (-14.26) and MgO (-6.98 kcal/mol) were obtained for the fourth target (Fig. 5). The only drug introduced for the fifth target, caspase9 (2AR9), had a binding energy of -6.35 kcal/mol, and the best MONP was also Fe2O3 with a binding energy of -6.89. The best drug for the sixth anti-cancer target, serum and glucocorticoid-regulated kinase 1 (3HDN), was Fostamatinib, with a binding energy of -10.43 kcal/mol, and the best MONP was Fe2O3, with a binding energy of -6.53 (Fig. 6). Following the molecular docking process, other drugs and nanoparticles with other anticancer targets were investigated. The only introduced drug, doxorubicin, had a binding energy of -8.46 kcal/mol for the seventh target, cell division protein kinase 2 (3IG7), and the best MONP was identified as MgO with a binding energy of -7.24 kcal/mol. The eighth target is also its only drug, mebendazole, which has a binding energy of -7.49 kcal/mol, and the best MONP is MgO, which has a binding energy of -8.03 kcal/mol (Fig. 7). Regarding the ninth target (XIAP), with PDB ID 5OQW, the best drug is N-Methylalanyl-3-methylvalyl-4-phenoxy-N-(1,2,3,4-tetrahydronaphthalen-1-YL)prolinamide (C31H42N4O4)with a binding energy of -9.77 kcal/mol, and the best MONP is Fe2O3 with a binding energy of -7.40 kcal/mol. The best drug and MONP for the 10th anticancer target, the HMG CoA reductase enzyme (8DJM), were Fluvastatin and MgO, with binding energies of -8.58 and -6.46 kcal/mol, respectively (Fig. 8). Table 2 shows complementary results of how the best drugs and MONPs bind. Figs. 4 to 8 clearly shows the key residues that play a role in the interactions of targets with drugs or MONPs in a summary form and with a specified pattern.
Table 2.
The results of the molecular docking process of drugs and MONPs with anti-cancer targets
Target Protein (PDB ID) (organism)
(Predicted Binding site)
|
Drugs/MONPs
|
Binding energy
(kcal/mol)
|
Ki (µM)
Inhibition constant
|
B DNA (1BNA)
(11.275, 23.032, 7.195) |
Drugs: |
|
|
| 1. Amsacrine |
-9.40 |
129.55 |
| 2. Carmustine |
-5.59 |
80.21 |
| 3. Chlorambucil |
-7.55 |
2.92 |
| 4. Cladribine |
-6.79 |
10.61 |
| 5. Gentian Violet |
-12.23 |
1.08 |
| 6. Mitomycin |
-8.78 |
363.63 |
| 7. Pipobroman |
-7.06 |
6.73 |
| 8. Streptozocin |
-6.17 |
29.80 |
| 9. Tioguanine |
-5.57 |
82.37 |
| Nanoparticles: |
|
|
| 1. Ag2O3 |
-4.89 |
261.46 |
| 2. CdO |
-5.46 |
99.37 |
| 3. CuO |
-4.51 |
491.84 |
| 4. Fe2O3 |
-5.17 |
162.85 |
| 5. FeO |
-6.10 |
33.74 |
| 6. MgO |
-6.41 |
19.86 |
| 7. MnO |
-5.86 |
50.98 |
| 8. ZnO |
-4.96 |
231.90 |
| Human-leukemia inhibitory factor (1EMR) (39.467, -0.948, 23.953) |
Drugs: |
|
|
| 1. Doxorubicin |
-6.09 |
34.12 |
| 2. Fluorouracil |
-4.36 |
639.72 |
| Nanoparticles: |
|
|
| 1. Ag2O3 |
-5.09 |
186.57 |
| 2. CdO |
-4.88 |
266.99 |
| 3. CuO |
-4.53 |
475.73 |
| 4. Fe2O3 |
-5.13 |
172.55 |
| 5. FeO |
-5.09 |
186.28 |
| 6. MgO |
-5.36 |
117.09 |
| 7. MnO |
-5.02 |
208.25 |
| 8. ZnO |
-5.00 |
216.79 |
Hypoxia-inducible factor I
(1H2N) (19.080, 25.679, 33.882) |
Drugs: |
|
|
| 1. 2-Methoxyestradiol |
-8.19 |
995.87 |
| 2. Carvedilol |
-7.29 |
4.52 |
| 3. ENMD-1198 |
-7.32 |
4.29 |
| 4. FG-2216 |
-6.44 |
18.88 |
| 5. Hydralazine |
-5.87 |
49.54 |
| 6. PX-478 |
-6.65 |
13.30 |
| 7. Vadadustat |
-6.52 |
16.76 |
| Nanoparticles: |
|
|
| 1. Ag2O3 |
-6.28 |
25.06 |
| 2. CdO |
-6.06 |
36.18 |
| 3. CuO |
-5.80 |
55.65 |
| 4. Fe2O3 |
-6.27 |
25.25 |
| 5. FeO |
-6.94 |
8.22 |
| 6. MgO |
-7.19 |
5.38 |
| 7. MnO |
-6.86 |
9.30 |
| 8. ZnO |
-5.77 |
59.25 |
Caspase 3 (1RE1)
(36.935, 94.291, 17.839) |
Drug: |
|
|
| 1. C19H13CIN2O6 |
-5.55 |
85.57 |
| 2. C19H14N2O6 |
-6.39 |
20.62 |
| 3. C20H23N3O5 |
-5.08 |
187.70 |
| 4. C20H20N2O5S |
-5.95 |
43.69 |
| 5. C9H11NO5S2 |
-5.12 |
176.61 |
| 6. C17H14N2O6 |
-6.66 |
13.11 |
| 7. C20H20N2O9S |
-5.95 |
43.46 |
| 8. Aspirin |
-4.76 |
325.27 |
| 9. Emricasan |
-6.87 |
9.27 |
| 10. Glycyrrhizic acid |
-14.26 |
35.34 |
| 11. Incadronic acid |
-8.91 |
293.63 |
| 12. Methyl (3S)-3-[(tert-butoxycarbonyl)amino]-4-oxopentanoate |
-4.04 |
1.09 |
| 13. Minocycline |
-6.01 |
39.61 |
| 14. C21H18FN3O6 |
-6.12 |
32.45 |
| 15. Oleandrin |
-8.29 |
831.61 |
| 16. PAC-1 |
-7.25 |
4.82 |
| 17. Pamidronic acid |
-5.87 |
49.63 |
| 18. Tributyrin |
-2.89 |
7.64 |
| Nanoparticles: |
|
|
| 1. Ag2O3 |
-5.87 |
49.68 |
| 2. CdO |
-6.61 |
14.22 |
| 3. CuO |
-5.72 |
64.44 |
| 4. Fe2O3 |
-6.01 |
39.51 |
| 5. FeO |
-6.79 |
10.46 |
| 6. MgO |
-6.98 |
7.61 |
| 7. MnO |
-6.85 |
9.58 |
| 8. ZnO |
-5.35 |
120.60 |
Caspase9 (2AR9)
(27.831, 44.977, 9.946) |
Drugs: |
|
|
| 1. Pamidronic acid |
-6.35 |
22.07 |
| Nanoparticles: |
|
|
| 1. Ag2O3 |
-5.30 |
130.17 |
| 2. CdO |
-5.41 |
108.36 |
| 3. CuO |
-4.97 |
226.43 |
| 4. Fe2O3 |
-6.89 |
8.96 |
| 5. FeO |
-6.38 |
21.01 |
| 6. MgO |
-6.78 |
10.65 |
| 7. MnO |
-6.24 |
26.76 |
| 8. ZnO |
-5.64 |
72.81 |
Serum and glucocorticoid-regulated kinase 1 or Sgk1
(3HDN) (28.185, 34.646, 71.484) |
Drugs: |
|
|
| 1. Dabrafenib |
-10.23 |
31.83 |
| 2. Fostamatinib |
-10.43 |
22.54 |
| Nanoparticles: |
|
|
| 1. Ag2O3 |
-6.08 |
34.92 |
| 2. CdO |
-5.01 |
213.45 |
| 3. CuO |
-5.34 |
121.09 |
| 4. Fe2O3 |
-6.53 |
16.40 |
| 5. FeO |
-5.64 |
73.95 |
| 6. MgO |
-6.41 |
19.88 |
| 7. MnO |
-5.46 |
98.65 |
| 8. ZnO |
-6.16 |
30.63 |
Cell division protein kinase2 or CDK2
(3IG7) (-1.776, 31.688, 10.929) |
Drugs: |
|
|
| 1. Doxorubicin |
-8.46 |
626.37 |
| Nanoparticles: |
|
|
| 1. Ag2O3 |
-5.70 |
66.73 |
| 2. CdO |
-6.79 |
10.56 |
| 3. CuO |
-4.92 |
246.69 |
| 4. Fe2O3 |
-5.61 |
76.83 |
| 5. FeO |
-6.96 |
7.93 |
| 6. MgO |
-7.24 |
4.95 |
| 7. MnO |
-6.98 |
7.61 |
| 8. ZnO |
-5.83 |
53.34 |
Telomerase reverse transcriptase
(5CQG) (-22.471, 31.524, -22.574) |
Drugs: |
|
|
| 1. Mebendazole |
-7.49 |
3.23 |
| Nanoparticles: |
|
|
| 1. Ag2O3 |
-6.39 |
20.56 |
| 2. CdO |
-6.33 |
22.85 |
| 3. CuO |
-4.81 |
296.87 |
| 4. Fe2O3 |
-6.03 |
38.01 |
| 5. FeO |
-6.79 |
10.61 |
| 6. MgO |
-8.03 |
1.29 |
| 7. MnO |
-6.30 |
23.93 |
| 8. ZnO |
-5.28 |
135.93 |
X-linked inhibitor of apoptosis protein or XIAP ( 5OQW)
(40.179, 0.450, -13.666) |
Drugs: |
|
|
| 1. C25H38N4O3 |
-8.48 |
612.46 |
| 2. Dequalinium |
-7.03 |
7.07 |
| 3. C31H42N4O4 |
-9.77 |
69.07 |
| 4. Terpinen-4-ol |
-5.08 |
187.56 |
| Nanoparticles: |
|
|
| 1. Ag2O3 |
-5.55 |
86.17 |
| 2. CdO |
-4.30 |
701.78 |
| 3. CuO |
-6.70 |
12.24 |
| 4. Fe2O3 |
-7.40 |
3.77 |
| 5. FeO |
-4.52 |
484.08 |
| 6. MgO |
-5.06 |
197.00 |
| 7. MnO |
-4.35 |
649.31 |
| 8. ZnO |
-6.81 |
10.11 |
HMG CoA Reductase (8DJM)
(145.724, 111.371, 136.181) |
Drugs: |
|
|
| 1. Atorvastatin |
-7.42 |
3.63
2.24 |
| 2. Cerivastatin |
-7.71 |
| 3. Fluvastatin |
-8.58 |
511.79 |
| 4. Lovastatin |
-5.95 |
43.19 |
| 5. Mevastatin |
-6.00 |
40.33 |
| 6. Pitavastatin |
-7.58 |
2.76 |
| 7. Pravastatin |
-4.74 |
334.93 |
| 8. Rosuvastatin |
-6.68 |
12.64 |
| 9. Simvastatin |
-6.27 |
25.32 |
| Nanoparticles: |
|
|
| 1. Ag2O3 |
-5.83 |
53.62 |
| 2. CdO |
-5.87 |
49.69 |
| 3. CuO |
-4.68 |
372.13 |
| 4. Fe2O3 |
-5.62 |
75.98 |
| 5. FeO |
-6.11 |
33.12 |
| 6. MgO |
-6.46 |
18.39 |
| 7. MnO |
-6.04 |
37.61 |
| 8. ZnO |
-5.59 |
79.72 |
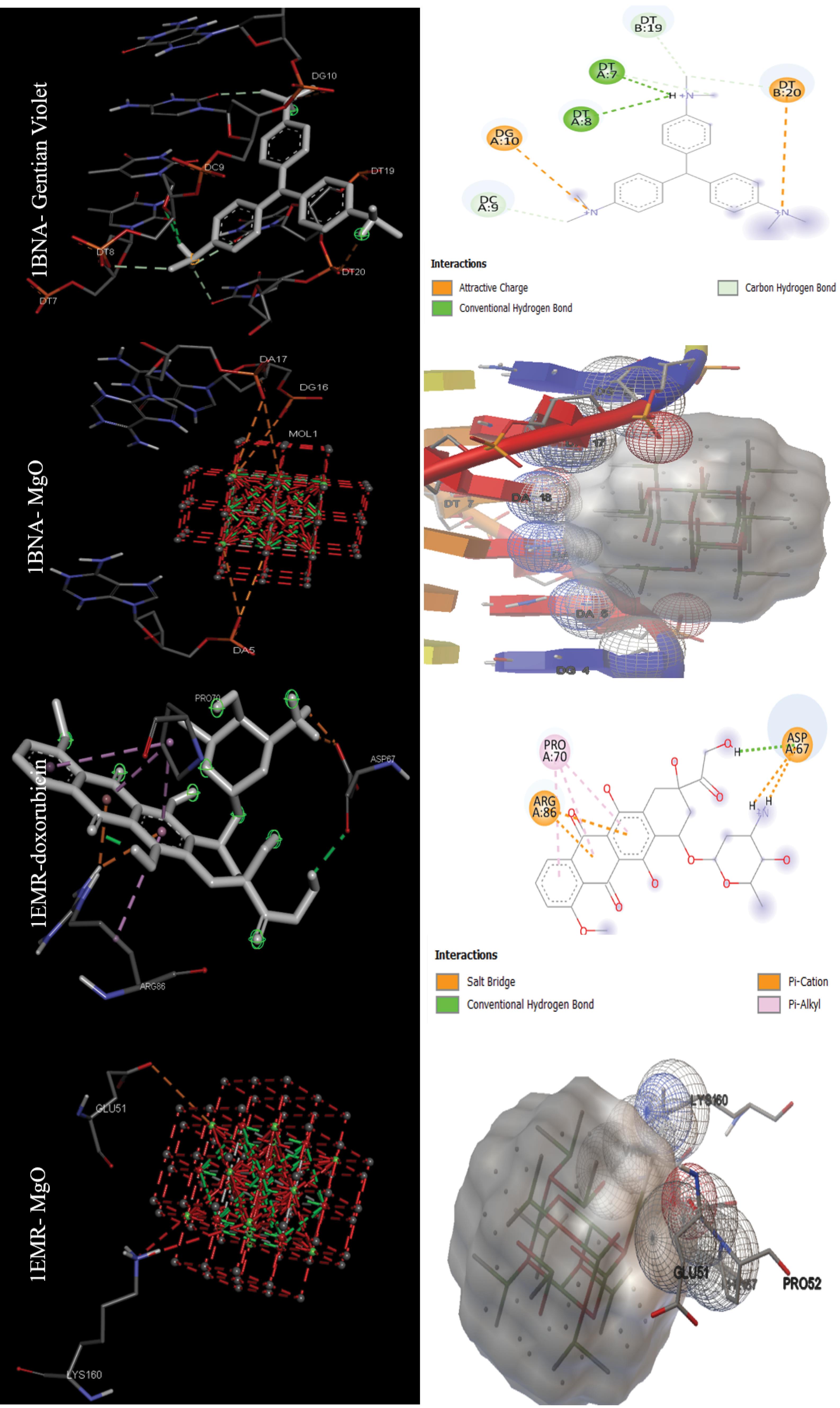
Fig. 4.
Images of the interactions involved between the best drugs and MONPs with anti-cancer targets (B DNA (1BNA) and Human-leukemia inhibitory factor (1EMR)) based on molecular docking results.
.
Images of the interactions involved between the best drugs and MONPs with anti-cancer targets (B DNA (1BNA) and Human-leukemia inhibitory factor (1EMR)) based on molecular docking results.
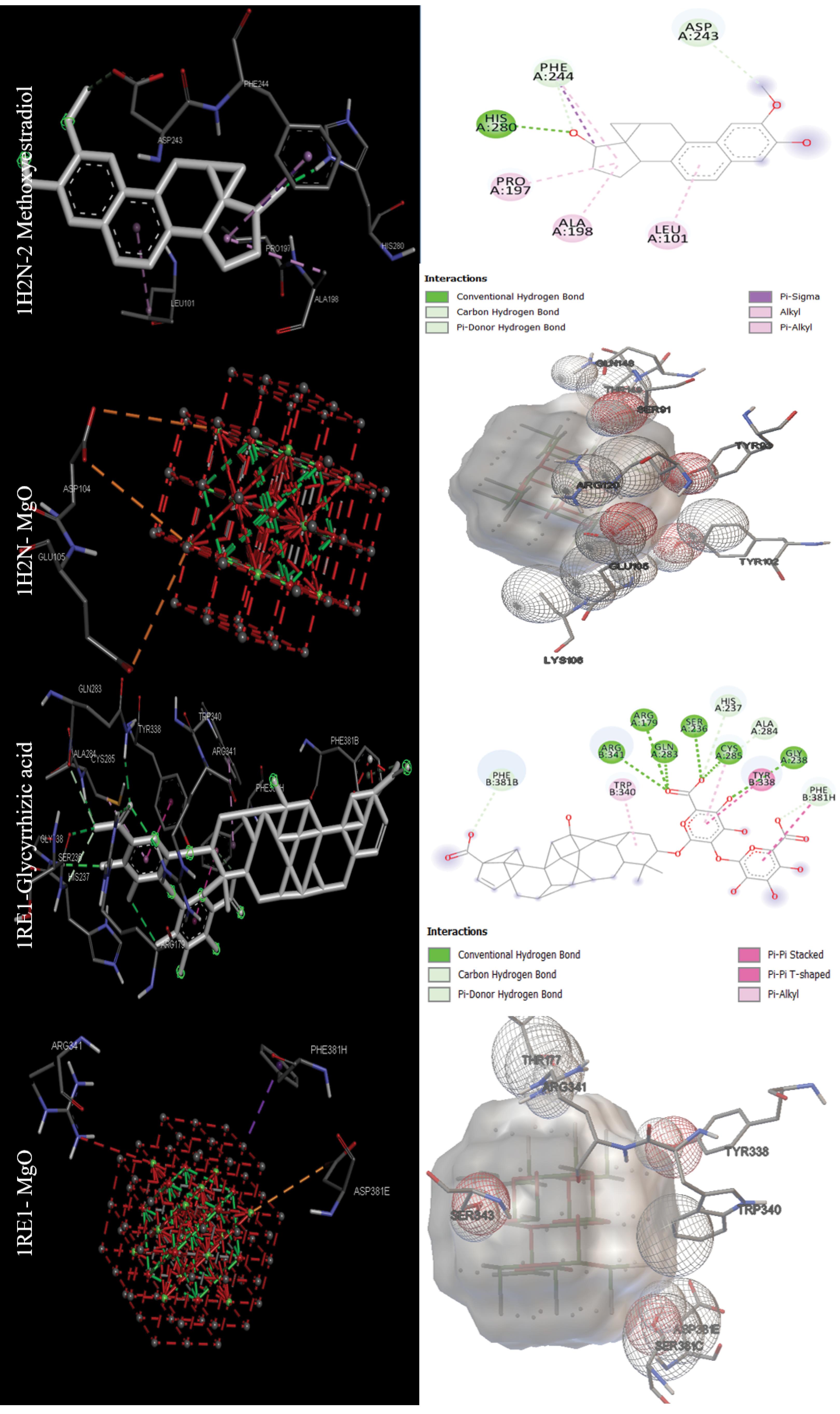
Fig. 5.
Images of the interactions involved between the best drugs and MONPs with anti-cancer targets (Hypoxia-inducible factor I (1H2N) and Caspase 3 (1RE1)) based on molecular docking results.
.
Images of the interactions involved between the best drugs and MONPs with anti-cancer targets (Hypoxia-inducible factor I (1H2N) and Caspase 3 (1RE1)) based on molecular docking results.
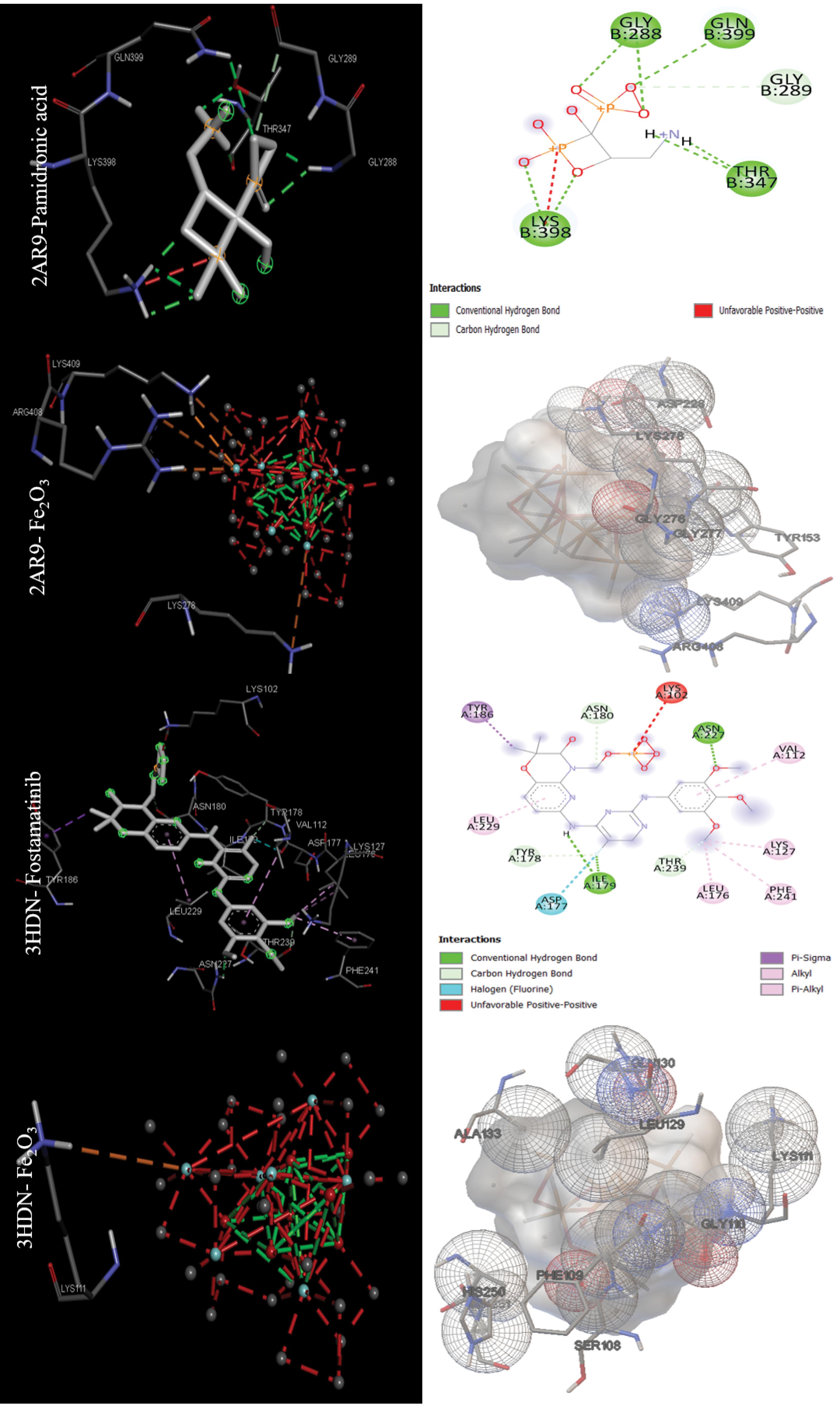
Fig. 6.
Images of the interactions involved between the best drugs and MONPs with anti-cancer targets (Caspase9 (2AR9) and Serum and glucocorticoid-regulated kinase 1 or Sgk1(3HDN)) based on molecular docking results.
.
Images of the interactions involved between the best drugs and MONPs with anti-cancer targets (Caspase9 (2AR9) and Serum and glucocorticoid-regulated kinase 1 or Sgk1(3HDN)) based on molecular docking results.
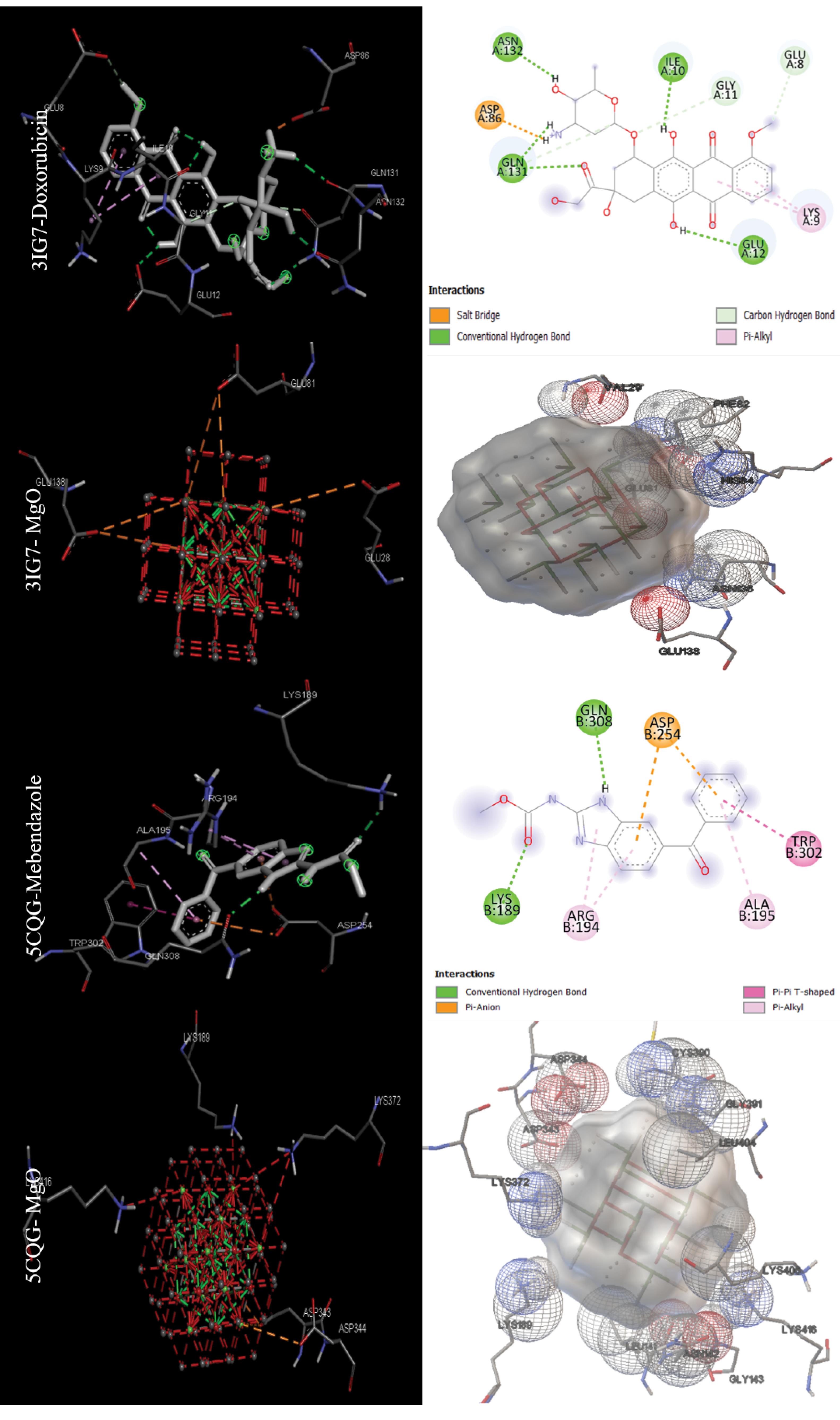
Fig. 7.
Images of the interactions involved between the best drugs and MONPs with anti-cancer targets (Cell division protein kinase2 or CDK2 (3IG7) and Telomerase reverse transcriptase (5CQG)) based on molecular docking results.
.
Images of the interactions involved between the best drugs and MONPs with anti-cancer targets (Cell division protein kinase2 or CDK2 (3IG7) and Telomerase reverse transcriptase (5CQG)) based on molecular docking results.
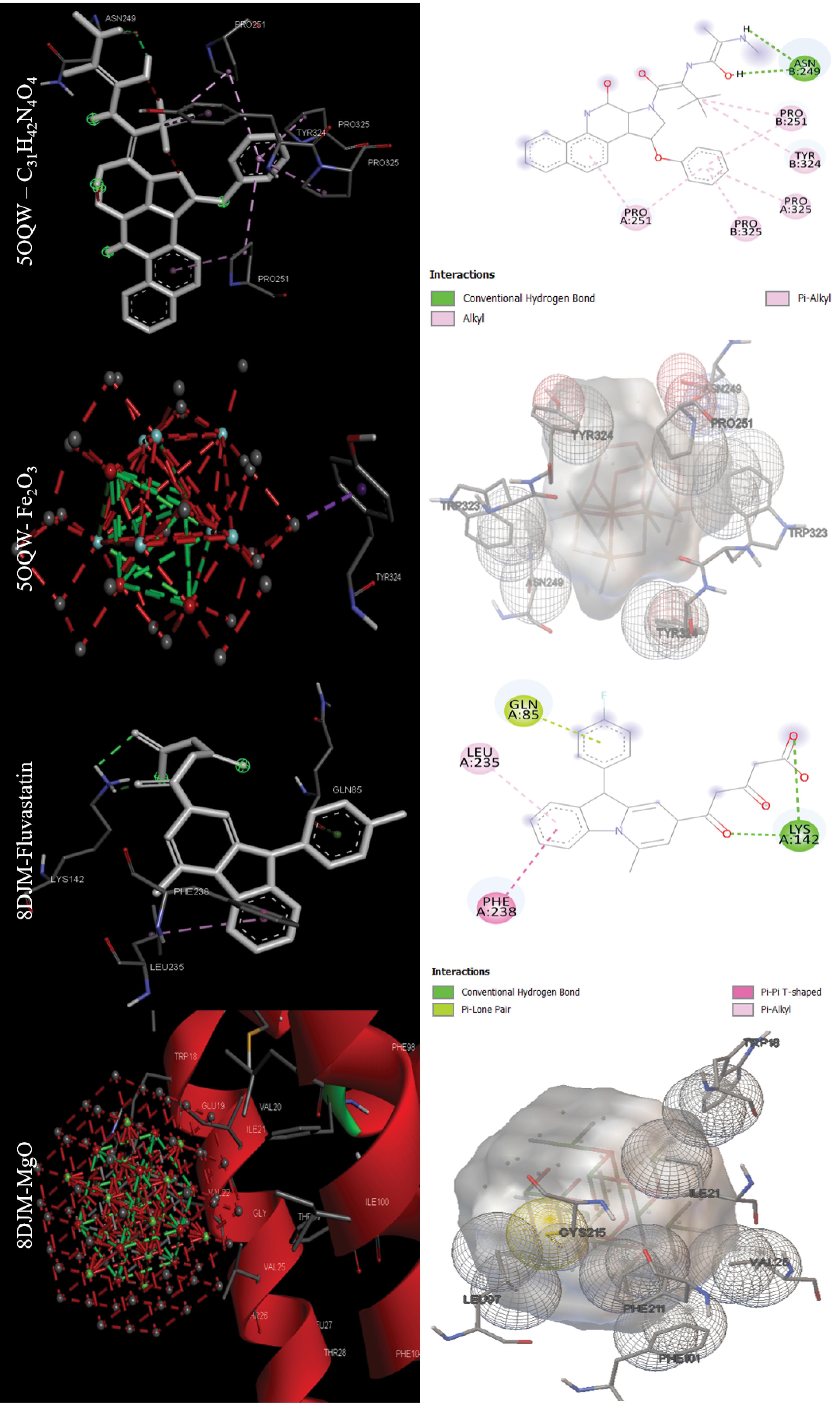
Fig. 8.
Images of the interactions involved between the best drugs and MONPs with anti-cancer targets (Cell division protein kinase2 or CDK2 (3IG7) and Telomerase reverse transcriptase (5CQG)) based on molecular docking results.
.
Images of the interactions involved between the best drugs and MONPs with anti-cancer targets (Cell division protein kinase2 or CDK2 (3IG7) and Telomerase reverse transcriptase (5CQG)) based on molecular docking results.
The efficiency of standard drugs and carbon capped-MONPs
The antibacterial properties of the five targets under study were investigated in this study, and in the case of two targets, 1AJ0 and 1QME, the target’s standard drug had the highest binding energy and the lowest inhibition constant and performed better than MONPs, but other targets, namely 1RC2, 4URO, 5V68, MgO, and Fe2O3 MONPs, performed better. By targeting Aquaporin Z, DNA gyrase subunit B, and FtsZ, these two nanoparticles showed good antibacterial properties. Also, when compared to standard drugs, MONPs have different binding to target proteins, as shown in Table 2, only two MONPs of Fe2O3 and MgO, outperformed conventional drugs in the case of the anti-cancer targets, out of the 10 investigated targets. Both the telomerase reverse transcriptase (5CQG) and caspase-9 (2AR9) were more effectively targeted by Fe2O3 NP and MgO NP, respectively. However, in the eight other anticancer targets studied, the standard drugs performed better than MONPs. As illustrated in Figs. 9 A and B, the binding energy distribution of MONPs for all the targets investigated in this study was between -4 and -8 kcal/mol, whereas the distribution of drugs is between -2.5 and -14.5 kcal/mol. MONPs have shown a minor difference in binding energy for different targets in these diseases, which is likely because of the unique physicochemical and pharmacophoric properties of MONPs.39,40 These properties have resulted in NPs performing similarly to various targets.
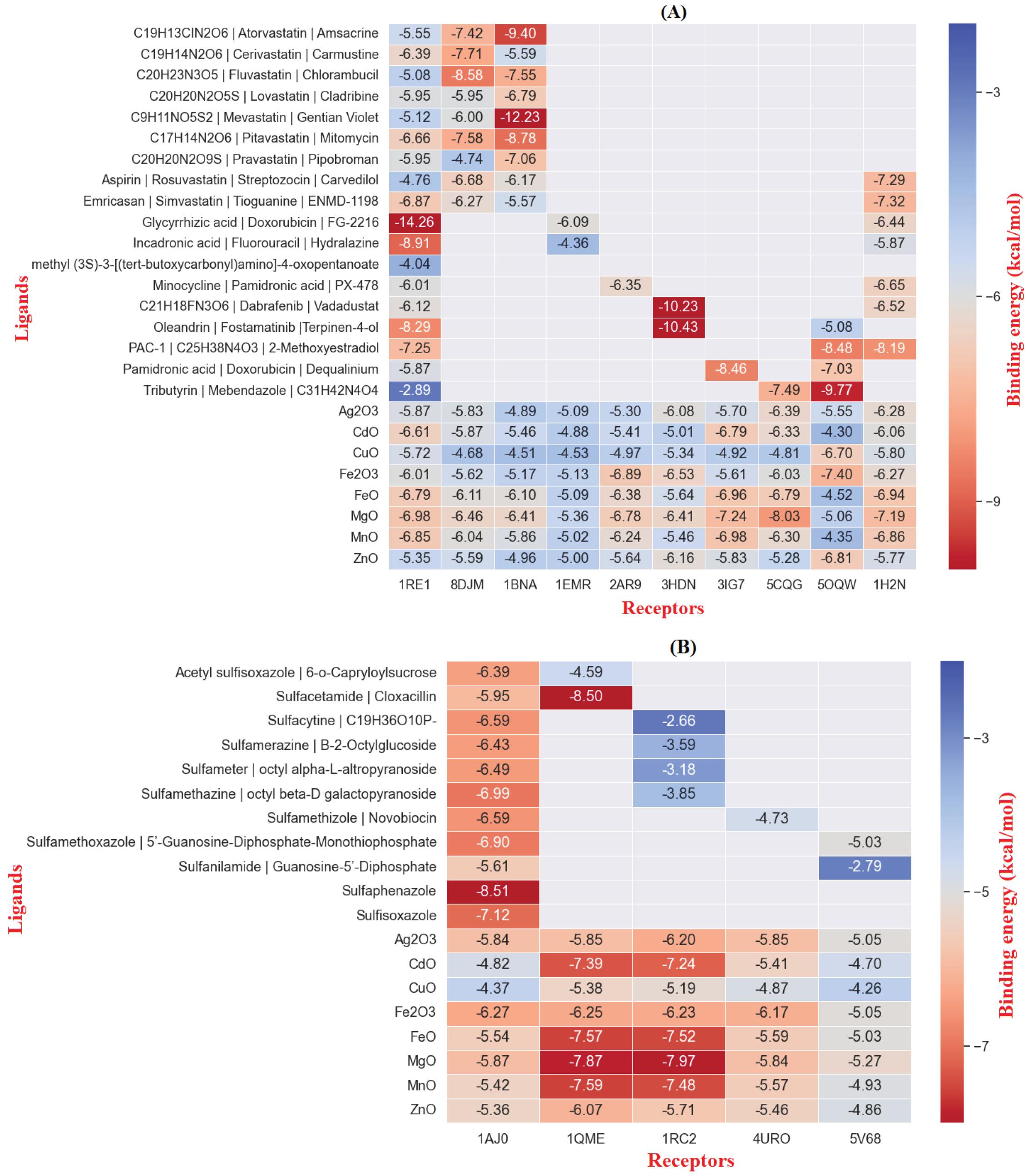
Fig. 9.
Comparison of anti-Cancer (A) anti-Bacterial (B) molecular docking results
.
Comparison of anti-Cancer (A) anti-Bacterial (B) molecular docking results
The toxicity of standard drugs and carbon-capped MONPs
MONPs were more toxic than other drugs in comparison studies between NPs and standard drugs, as shown in Table 3 and Fig. 10 (LD50, or lethal dose 50%, is the concentration of a compound that causes the death of 50% of test samples, especially in the case of cancer cells and bacteria). Another issue is that MONPs have a high molecular weight when compared to standard drugs. Because of their high molecular weight, these NPs are likely to be toxic to various organs of the human body if taken orally. However, such problems can be avoided by adjusting the dosage and application of biocompatible natural materials such as herbal metabolites so that the lowest one has a level of toxicity for the organs of the human body.41-43
Table 3.
The predicted toxicity of MONPs by PRO TOX-II
|
MONPs
|
LD50 (mg/kg)
|
MW (g/mol)
|
| Ag2O3 |
900 |
1109 |
| CdO |
200 |
2617 |
| CuO |
2000 |
873.92 |
| Fe2O3 |
68 |
879.44 |
| FeO |
305 |
1774.47 |
| MgO |
1000 |
1311.75 |
| MnO |
68 |
1637.8 |
| ZnO |
1000 |
993.73 |
MONPs: Metal oxide NPs; LD50: Lethal dose 50%; MW: Molecular weight
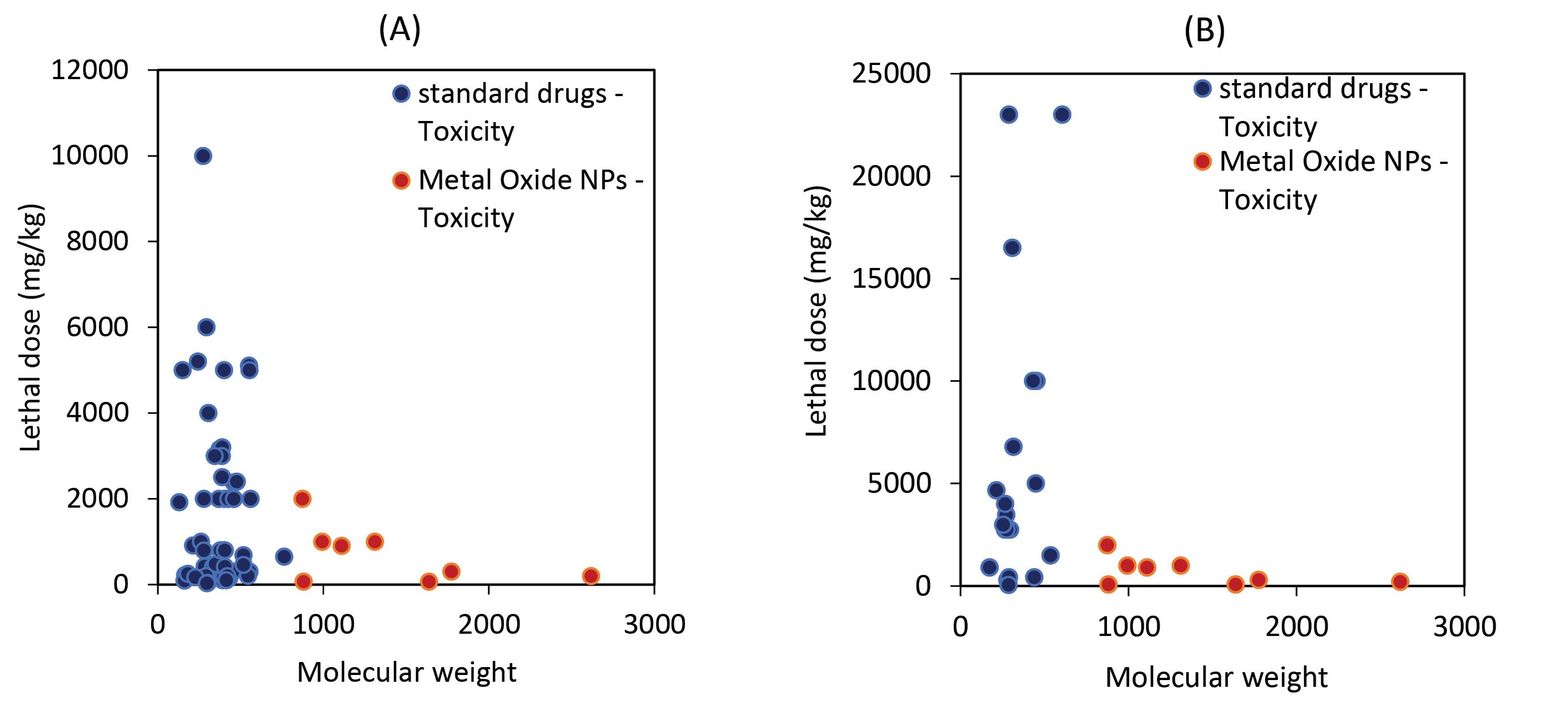
Fig. 10.
Comparison of the toxicities of MONPs and conventional medications with anticancer (A) and antibacterial (B) activities is shown in two graphs. MONPs, as is well known, have a lower LD50 and a higher molecular weight than standard drugs, and NPs exhibit greater toxicity.
.
Comparison of the toxicities of MONPs and conventional medications with anticancer (A) and antibacterial (B) activities is shown in two graphs. MONPs, as is well known, have a lower LD50 and a higher molecular weight than standard drugs, and NPs exhibit greater toxicity.
Discussion
Traditional drug discovery depending on in vitro and in vivo studies has several disadvantages of time-consuming processes, large investments for experimental set-ups, and feasibility only to biopharmaceutical companies. In this case, to save time and resources, screening of repurposable chemical materials relying on modern artificial intelligence-based algorithms and state-of-the-art computational techniques can be applied to discover new drugs.44 Rapid and cost-effective in silico methods based on structure-based drug repurposing have a critical role in targeting specific receptors related to health-threatening diseases such as cancers, and diabetes by natural compounds and NPs.45 Drug/target network-based models, modern artificial intelligence, and structure-based approaches have been used for the viral pandemic. However, there are some limitations to these computational drug repositioning/repurposing/reprofiling approaches. For example, the results of modern artificial intelligence-based networks are less relevant due to the necessity of more data for their application.46 In addition, a low chemical library can lead to the limited efficiency of molecular docking approaches.47 Therefore, in vitro and in vivo studies should be considered to overcome these disadvantages.48
Based on the toxicity property and efficiency of the MONPs tested in this study, it can be concluded that biocompatible MONPs have therapeutic potential for targeting specific receptors. However, the role of these NPs in ROS production should not be underestimated, because previous studies found that many of the antibacterial and anticancer properties of NPs were due to the production of this active species of free radicals.49 For example, MgO NPs caused apoptosis in K-562 lymphoblast cell lines by the production of ROS. Moreover, molecular docking results for this study showed that MgO NPs with a diameter of 1.5 nm can interact with human serum albumin via hydrophobic residues using hydrophobic forces.50 In a comparative study, docking of four nanocomposites including Fe3O4@SiO2, Fe3O4@SiO2@APTS (3-aminopropyl) triethoxysilane), Fe3O4@SiO2@APTS~Schiff base, and Fe3O4@SiO2@APTS~Schiff base-Cu(II) were evaluated against DNA duplex of sequence d (ACCGACGTCGGT)2, ribonucleotide reductase, and topoisomerase II. Highest binding affinity was found for docking of Fe3O4@SiO2@APTS with DNA and Fe3O4@SiO2@APTS~Schiff base-Cu(II) with topoisomerase II by values of –10.85 and –7.94 kcal/mol, respectively.51 In another study, the best binding energy value between AgO NPs and Als3 adhesin was -9.60 kcal/mol. In addition, these NPs exhibited an IC50 value of 36.56 µg/mL against A431 epidermoid carcinoma cell line.52 One of the most important aspects of using nanoparticles as therapeutic agents is their ease of penetration into the structures of proteins or other targets.53 It is worth noting that the toxicity of drug agents is not limited to cancer cells; these agents must also be able to respond appropriately to the tumour microenvironment.54 Introducing MONPs as potential agents in the field of cancer therapy can be effective if the primary characteristic of the disease, namely its complexity, is considered. The agents may have anti-cancer properties against cancer cells in vitro, but they are ineffective in vivo.55 Due to their unique physicochemical properties, nanoparticles have the potential to be among the leading candidates in cancer chemotherapy.
Conclusion
The results of this study indicate that MONPs have therapeutic potential for use as oral medications against a variety of disease-related targets. These findings undoubtedly confirm the anticancer and antibacterial properties reported in earlier studies. One of the major challenges in the development of drugs based on MONPs is their toxicity as measured by LD50, so this important parameter was investigated in this study. The toxicity of MONPs as oral drugs was higher than standard drugs, but this needs to be investigated further in the future. MgO and Fe2O3 NPs have the highest efficiency among the MONPs under investigation, according to the data obtained from these molecular docking studies. One intriguing finding was that these two MONPs had a greater ability to bind to specific targets for cancer, and bacterial infection than other MONPs. It is even greater than the standard drugs for these targets. The toxicity level of MgO (1000 mg/kg) was determined to be much lower than that of Fe2O3 (68 mg/kg). In this context, it is clear that bioinformatics tools for drug development based on MONPs must be developed. In addition, the best MONPs must be developed and designed to have higher efficacy and lower toxicity.
Research Highlights
What is the current knowledge?
√ One of the main challenges in the development of drugs based on MONPs is their toxicity based on LD50.
What is new here?
√ The toxicity of MONPs when used as an oral drug was higher than standard drugs, although their toxicity needs further investigation in the future.
Competing Interests
No potential conflict of interest was reported by the authors.
Ethical Statement
None to be declared.
Funding
There was no any funding source for this study.
References
- Santos-Martins D, Solis-Vasquez L, Tillack AF, Sanner MF, Koch A, Forli S. Accelerating AutoDock4 with GPUs and gradient-based local search. J Chem Theory Comput 2021; 17:1060-73. doi: 10.1021/acs.jctc.0c01006 [Crossref] [ Google Scholar]
- El Gammal RN, Elmansi H, El-Emam AA, Belal F, Hammouda ME. Exploring the molecular interaction of mebendazole with bovine serum albumin using multi-spectroscopic approaches and molecular docking. Sci Rep 2022; 12:1-13. doi: 10.1038/s41598-022-15696-4 [Crossref] [ Google Scholar]
- Alomari FY, Sharfalddin AA, Abdellattif MH, Domyati D, Basaleh AS, Hussien MA. QSAR Modeling, Molecular Docking and Cytotoxic Evaluation for Novel Oxidovanadium (IV) Complexes as Colon Anticancer Agents. Molecules 2022; 27:649. doi: 10.3390/molecules27030649 [Crossref] [ Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 2009; 30:2785-91. doi: 10.1002/jcc.21256 [Crossref] [ Google Scholar]
- Adefegha SA, Salawi A, Bumrungpert A, Khorasani S, Torkaman S, Mozafari MR. Encapsulation of polyphenolic compounds for health promotion and disease prevention: Challenges and opportunities. Nano Micro Biosystems 2023; 1:1-12. doi: 10.22034/NMBJ.2023.163756 [Crossref] [ Google Scholar]
- Rahman MM, Islam MR, Akash S, Harun-Or-Rashid M, Ray TK, Rahaman MS. Recent advancements of nanoparticles application in cancer and neurodegenerative disorders: At a glance. Biomed Pharmacother 2022; 153:113305. doi: 10.1016/j.biopha.2022.113305 [Crossref] [ Google Scholar]
- Alavi M, Aghaie E. Self-assembled nanostructures for anticancer applications: Advances and limitations. Nano Micro Biosystems 2022; 1:27-31. doi: 10.22034/NMBJ.2022.161602 [Crossref] [ Google Scholar]
- Ashengroph M, Khaledi A, Bolbanabad EM. Extracellular biosynthesis of cadmium sulphide quantum dot using cell-free extract of Pseudomonas chlororaphis CHR05 and its antibacterial activity. Process Biochem 2020; 89:63-70. doi: 10.1016/j.procbio.2019.10.028 [Crossref] [ Google Scholar]
- Amraei S, Ahmadi S. Recent studies on antimicrobial and anticancer activities of saponins: a mini-review. Nano Micro Biosystems 2022; 1:22-6. doi: 10.22034/NMBJ.2022.160182 [Crossref] [ Google Scholar]
- Ranjan S, Dasgupta N, Chinnappan S, Ramalingam C, Kumar A. A novel approach to evaluate titanium dioxide nanoparticle–protein interaction through docking: an insight into mechanism of action. Proceedings of the National Academy of Sciences, India Section B: Biological Sciences 2017; 87:937-43. doi: 10.1007/s40011-015-0673-z [Crossref] [ Google Scholar]
- Alencar WLM, da Silva Arouche T, Neto AFG, de Castro Ramalho T, de Carvalho Júnior RN, de Jesus Chaves Neto AM. Interactions of Co, Cu, and non-metal phthalocyanines with external structures of SARS-CoV-2 using docking and molecular dynamics. Sci Rep 2022; 12:1-20. doi: 10.1038/s41598-022-07396-w [Crossref] [ Google Scholar]
- Wasukan N, Kuno M, Maniratanachote R. Molecular Docking as a Promising Predictive Model for Silver Nanoparticle-Mediated Inhibition of Cytochrome P450 Enzymes. J Chem Inf Model 2019; 59:5126-34. doi: 10.1021/acs.jcim.9b00572 [Crossref] [ Google Scholar]
- Al-Khalaf AA, Hassan HM, Alrajhi AM, Mohamed RAEH, Hozzein WN. Anti-Cancer and Anti-Inflammatory Potential of the Green Synthesized Silver Nanoparticles of the Red Sea Sponge Phyllospongia lamellosa Supported by Metabolomics Analysis and Docking Study. Antibiotics 2021; 10:1155. doi: 10.3390/antibiotics10101155 [Crossref] [ Google Scholar]
- Abo-zeid Y, Ismail NSM, McLean GR, Hamdy NM. A molecular docking study repurposes FDA approved iron oxide nanoparticles to treat and control COVID-19 infection. Eur J Pharm Sci 2020; 153:105465. doi: 10.1016/j.ejps.2020.105465 [Crossref] [ Google Scholar]
- Chibber S, Ahmad I. Molecular docking, a tool to determine interaction of CuO and TiO2 nanoparticles with human serum albumin. Biochemistry and Biophysics Reports 2016; 6:63-7. doi: 10.1016/j.bbrep.2016.03.004 [Crossref] [ Google Scholar]
- Anjum S, Hashim M, Malik SA, Khan M, Lorenzo JM, Abbasi BH. Recent Advances in Zinc Oxide Nanoparticles (ZnO NPs) for Cancer Diagnosis, Target Drug Delivery, and Treatment. Cancers (Basel) 2021; 13:4570. doi: 10.3390/cancers13184570 [Crossref] [ Google Scholar]
- Yu H, Shang L, Yang G, Dai Z, Zeng X, Qiao S. Biosynthetic microcin J25 exerts strong antibacterial, anti-inflammatory activities, low cytotoxicity without increasing drug-resistance to bacteria target. Front Immunol 2022; 13:811378. doi: 10.3389/fimmu.2022.811378 [Crossref] [ Google Scholar]
- Amraei S, Eslami G, Taherpour A, Hashemi A. Relationship between MOX genes and antibiotic resistance in Klebsiella pneumoniae strains in nosocomial infections. Micro Nano Bio Aspects 2022; 1:12-7. doi: 10.22034/mnba.2022.155320 [Crossref] [ Google Scholar]
- Amraei S, Eslami G, Taherpour A, Hashemi A. The role of ACT and FOX genes in Klebsiella pneumoniae strains isolated from hospitalized patients. Micro Nano Bio Aspects 2022; 1:18-25. doi: 10.22034/mnba.2022.155447 [Crossref] [ Google Scholar]
- Verma T, Aggarwal A, Singh S, Sharma S, Sarma SJ. Current challenges and advancements towards discovery and resistance of antibiotics. J Mol Struct 2022; 1248:131380. doi: 10.1016/j.molstruc.2021.131380 [Crossref] [ Google Scholar]
- Beres SB, Zhu L, Pruitt L, Olsen RJ, Faili A, Kayal S. Integrative Reverse Genetic Analysis Identifies Polymorphisms Contributing to Decreased Antimicrobial Agent Susceptibility in Streptococcus pyogenes. mBio 2022; 13:e03618-21. doi: 10.1128/mbio.03618-21 [Crossref] [ Google Scholar]
- Priya AK, Gnanasekaran L, Kumar PS, Jalil AA, Hoang TKA, Rajendran S. Recent trends and advancements in nanoporous membranes for water purification. Chemosphere 2022; 303:135205. doi: 10.1016/j.chemosphere.2022.135205 [Crossref] [ Google Scholar]
- Amer HH, Eldrehmy EH, Abdel-Hafez SM, Alghamdi YS, Hassan MY, Alotaibi SH. Antibacterial and molecular docking studies of newly synthesized nucleosides and Schiff bases derived from sulfadimidines. Sci Rep 2021; 11:17953. doi: 10.1038/s41598-021-97297-1 [Crossref] [ Google Scholar]
- Aliye M, Dekebo A, Tesso H, Abdo T, Eswaramoorthy R, Melaku Y. Molecular docking analysis and evaluation of the antibacterial and antioxidant activities of the constituents of Ocimum cufodontii. Sci Rep 2021; 11:10101. doi: 10.1038/s41598-021-89557-x [Crossref] [ Google Scholar]
- Fang Y, Lu Y, Zang X, Wu T, Qi X, Pan S. 3D-QSAR and docking studies of flavonoids as potent Escherichia coli inhibitors. Sci Rep 2016; 6:1-13. doi: 10.1038/srep23634 [Crossref] [ Google Scholar]
- Ur Rahman M, Wang P, Wang N, Chen Y. A key bacterial cytoskeletal cell division protein FtsZ as a novel therapeutic antibacterial drug target. Bosn J Basic Med Sci 2020; 20:310-8. doi: 10.17305/bjbms.2020.4597 [Crossref] [ Google Scholar]
- Ivens E, Cominetti MM, Searcey M. Junctions in DNA: underexplored targets for therapeutic intervention. Biorg Med Chem 2022; 116897. 10.1016/j.bmc.2022.116897.
- Bowyer C, Lewis AL, Lloyd AW, Phillips GJ, Macfarlane WM. Hypoxia as a target for drug combination therapy of liver cancer. Anticancer Drugs 2017; 28:771-80. doi: 10.1097/cad.0000000000000516 [Crossref] [ Google Scholar]
- Wrona E, Potemski P, Sclafani F, Borowiec M. Leukemia Inhibitory Factor: A Potential Biomarker and Therapeutic Target in Pancreatic Cancer. Arch Immunol Ther Exp (Warsz) 2021; 69:2. doi: 10.1007/s00005-021-00605-w [Crossref] [ Google Scholar]
- Yadav P, Yadav R, Jain S, Vaidya A. Caspase-3: A primary target for natural and synthetic compounds for cancer therapy. Chem Biol Drug Des 2021; 98:144-65. doi: 10.1111/cbdd.13860 [Crossref] [ Google Scholar]
- Kim B, Srivastava SK, Kim S-H. Caspase-9 as a therapeutic target for treating cancer. Expert OpinTher Targets 2015; 19:113-27. doi: 10.1517/14728222.2014.961425 [Crossref] [ Google Scholar]
- Cicenas J, Meskinyte-Kausiliene E, Jukna V, Rimkus A, Simkus J, Soderholm D. SGK1 in Cancer: Biomarker and Drug Target. Cancers (Basel) 2022; 14:2385. doi: 10.3390/cancers14102385 [Crossref] [ Google Scholar]
- Said MA, Abdelrahman MA, Abourehab MAS, Fares M, Eldehna WM. A patent review of anticancer CDK2 inhibitors (2017–present). Expert OpinTher Pat 2022; 32:885-98. doi: 10.1080/13543776.2022.2078193 [Crossref] [ Google Scholar]
- Opo FA, Rahman MM, Ahammad F, Ahmed I, Bhuiyan MA, Asiri AM. Structure based pharmacophore modeling, virtual screening, molecular docking and ADMET approaches for identification of natural anti-cancer agents targeting XIAP protein. Sci Rep 2021; 11:1-17. doi: 10.1038/s41598-021-83626-x [Crossref] [ Google Scholar]
- Gu W, Lin Z, Zhao S, Wang G, Shen Z, Liu W. Research Progress on G-Quadruplexes in Human Telomeres and Human Telomerase Reverse Transcriptase (hTERT) Promoter. Oxid Med Cell Longev 2022; 2022:2905663. doi: 10.1155/2022/2905663 [Crossref] [ Google Scholar]
- Gao S, Soares F, Wang S, Wong CC, Chen H, Yang Z. CRISPR screens identify cholesterol biosynthesis as a therapeutic target on stemness and drug resistance of colon cancer. Oncogene 2021; 40:6601-13. doi: 10.1038/s41388-021-01882-7 [Crossref] [ Google Scholar]
- Banerjee P, Dehnbostel FO, Preissner R. Prediction Is a Balancing Act: Importance of Sampling Methods to Balance Sensitivity and Specificity of Predictive Models Based on Imbalanced Chemical Data Sets. Frontiers in Chemistry 2018; 6. 10.3389/fchem.2018.00362.
- Drwal MN, Banerjee P, Dunkel M, Wettig MR, Preissner R. ProTox: a web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Res 2014; 42:W53-W8. doi: 10.1093/nar/gku401 [Crossref] [ Google Scholar]
- Elrayess R, Darwish KM, Nafie MS, El-Sayyed GS, Said MM, Yassen ASA. Quinoline–hydrazone hybrids as dual mutant EGFR inhibitors with promising metallic nanoparticle loading: rationalized design, synthesis, biological investigation and computational studies. New J Chem 2022; 46:18207-32. doi: 10.1039/D2NJ02962F [Crossref] [ Google Scholar]
- Alavi M, Yarani R. ROS and RNS modulation: the main antimicrobial, anticancer, antidiabetic, and antineurodegenerative mechanisms of metal or metal oxide nanoparticles. Nano Micro Biosystems 2023; 2:22-30. doi: 10.22034/nmbj.2023.382133.1012 [Crossref] [ Google Scholar]
- Tundisi LL, Ataide JA, Costa JSR, Coêlho DdF, Liszbinski RB, Lopes AM. Nanotechnology as a tool to overcome macromolecules delivery issues. Colloids Surf B Biointerfaces 2023; 222:113043. doi: 10.1016/j.colsurfb.2022.113043 [Crossref] [ Google Scholar]
- Alavi M, Yarani R, Sreedharan M, Thomas S. Micro and nanoformulations of catechins for therapeutic applications: recent advances and challenges. Micro Nano Bio Aspects 2023; 2:8-19. doi: 10.22034/mnba.2023.382922.1021 [Crossref] [ Google Scholar]
- Aljelehawy QHA, Mohammadi S, Mohamadian E, Raji Mal Allah O, Mirzaei A, Ghahremanlou M. Antimicrobial, anticancer, antidiabetic, antineurodegenerative, and antirheumatic activities of thymol: clarification of mechanisms. Micro Nano Bio Aspects 2023; 2:1-7. doi: 10.22034/mnba.2023.381107.1019 [Crossref] [ Google Scholar]
- Choudhury C, Arul Murugan N, Priyakumar UD. Structure-based drug repurposing: Traditional and advanced AI/ML-aided methods. Drug Discov Today 2022; 27:1847-61. doi: 10.1016/j.drudis.2022.03.006 [Crossref] [ Google Scholar]
- Aljelehawy Q, Mal Allah OR, Sourazur G. Physicochemical properties, medicinal chemistry, toxicity, and absorption of quercetin and its interaction with spike glycoprotein of SARS-CoV-2: Molecular docking. Nano Micro Biosystems 2022; 1: 32-9. 10.22034/nmbj.2022.163207.
- Dotolo S, Marabotti A, Facchiano A, Tagliaferri R. A review on drug repurposing applicable to COVID-19. Brief Bioinform 2021; 22:726-41. doi: 10.1093/bib/bbaa288 [Crossref] [ Google Scholar]
- Wang G, Zhu W. Molecular docking for drug discovery and development: a widely used approach but far from perfect. Future Med Chem 2016; 8:1707-10. doi: 10.4155/fmc-2016-0143 [Crossref] [ Google Scholar]
- Maroufi Y, Hoseini SR, Alavi M. Antiparasitic Effect of Leaf Extract and Major Metabolites of Pelargonium quercetorumAgnew. against Leishmania Major: In Vitro and In Silico Studies. Journal of Applied Biotechnology Reports 2022; 9:817-30. doi: 10.30491/jabr.2022.325242.1487 [Crossref] [ Google Scholar]
- G K P, Prashanth PA, Singh P, Nagabhushana BM, Shivakumara C, G M K. Effect of doping (with cobalt or nickel) and UV exposure on the antibacterial, anticancer, and ROS generation activities of zinc oxide nanoparticles. Journal of Asian Ceramic Societies 2020; 8:1175-87. doi: 10.1080/21870764.2020.1824328 [Crossref] [ Google Scholar]
- Behzadi E, Sarsharzadeh R, Nouri M, Attar F, Akhtari K, Shahpasand K. Albumin binding and anticancer effect of magnesium oxide nanoparticles. Int J Nanomedicine 2019; 14:257-70. doi: 10.2147/ijn.s186428 [Crossref] [ Google Scholar]
- Eshaghi Malekshah R, Fahimirad B, Aallaei M, Khaleghian A. Synthesis and toxicity assessment of Fe3O4 NPs grafted by ∼ NH2-Schiff base as anticancer drug: modeling and proposed molecular mechanism through docking and molecular dynamic simulation. Drug Deliv 2020; 27:1201-17. doi: 10.1080/10717544.2020.1801890 [Crossref] [ Google Scholar]
- Ganesh Kumar A, Pugazhenthi E, Sankarganesh P, Muthusamy C, Rajasekaran M, Lokesh E, et al. “Cleome rutidosperma leaf extract mediated biosynthesis of silver nanoparticles and anti-candidal, anti-biofilm, anti-cancer, and molecular docking analysis”. Biomass Conversion and Biorefinery 2023. 10.1007/s13399-023-03806-9.
- Waheed S, Li Z, Zhang F, Chiarini A, Armato U, Wu J. Engineering nano-drug biointerface to overcome biological barriers toward precision drug delivery. Journal of Nanobiotechnology 2022; 20:395. doi: 10.1186/s12951-022-01605-4 [Crossref] [ Google Scholar]
- Zughaibi TA, Mirza AA, Suhail M, Jabir NR, Zaidi SK, Wasi S. Evaluation of Anticancer Potential of Biogenic Copper Oxide Nanoparticles (CuO NPs) against Breast Cancer. Journal of Nanomaterials 2022; 2022:5326355. doi: 10.1155/2022/5326355 [Crossref] [ Google Scholar]
- Yee YJ, Benson HAE, Dass CR, Chen Y. Evaluation of novel conjugated resveratrol polymeric nanoparticles in reduction of plasma degradation, hepatic metabolism and its augmentation of anticancer activity in vitro and in vivo. Int J Pharm 2022; 615:121499. doi: 10.1016/j.ijpharm.2022.121499 [Crossref] [ Google Scholar]