Bioimpacts. 14(5):27846.
doi: 10.34172/bi.2024.27846
Original Article
A novel therapeutic multiepitope vaccine based on oncoprotein E6 and E7 of HPV 16 and 18: An in silico approach
Sari Eka Pratiwi Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Writing – original draft, 1, * 
Ysrafil Ysrafil Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Writing – original draft, 2 
Mardhia Mardhia Funding acquisition, Investigation, Writing – original draft, 3 
Mahyarudin Mahyarudin Validation, Writing – review & editing, 3 
Muhammad Inam Ilmiawan Writing – review & editing, 1
Heru Fajar Trianto Writing – review & editing, 1, 4
Delima Fajar Liana Writing – review & editing, 3
Yuri Amia Data curation, Software, 5
Author information:
1Department of Biology and Pathobiology, Faculty of Medicine, Universitas Tanjungpura, Pontianak, Indonesia
2Department of Pharmacology, Faculty of Medicine, Universitas Palangka Raya, Palangka Raya, Indonesia
3Department of Microbiology, Faculty of Medicine, Universitas Tanjungpura, Pontianak, Indonesia
4Department of Histology, Faculty of Medicine, Universitas Tanjungpura, Pontianak, Indonesia
5Medical School, Faculty of Medicine, Universitas Tanjungpura, Pontianak, Indonesia
Abstract
Introduction:
The current vaccine strategies to prevent cervical cancer are effective only for individuals unexposed to HPV, lacking therapeutic effects against pre-existing infections. Multiepitope vaccines, using an immunoinformatic approach, are promising against tumors and viral infections because of their high specificity, safety, and stability, as well as the cheap cost of development.
Methods:
This study employed computer-based immunoinformatic analysis to design therapeutic multiepitope vaccines against cervical cancer using oncoproteins E6 and E7 of HPV 16 and 18. Several immunoinformatic tools were applied to analyze potential vaccine constructs capable of stimulating immune responses against both oncoproteins.
Results:
The constructed vaccine exhibited antigenic, immunogenic, nonallergenic, nontoxic, stable, and soluble characteristics. Additionally, it effectively interacted with TLR2 and TLR4, showing high binding capacity. Computational analysis indicated the vaccine could induce immune responses through the elevation of cytokine levels after the third injection, antibody production, activation of memory B and T cells, and promotion of increased dendritic cell counts.
Conclusion:
The novel multiepitope vaccine based on E6 and E7 presented as a promising candidate for combating HPV infections and associated cervical cancer. Further in vitro and in vivo studies were essential to validate the efficacy and safety of the vaccine.
Keywords: Multiepitope vaccine, HPV vaccine, Therapeutic vaccine, Immunoinformatic, Reverse vaccinology
Copyright and License Information
© 2024 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
Not applicable.
Introduction
Cervical cancer is caused by persistent human papillomavirus (HPV) infection. In 2020, there were 604 127 new cases of cervical cancer, positioning it as the fourth most prevalent cancer among women globally, following breast, colorectal, and lung cancer. In Indonesia, cervical cancer stands as the second most common type of malignant condition affecting women after breast cancer. The projected figures for 2020 anticipated 36 633 (17.2%) diagnosed cases, with an incidence rate of 24.4 per 100 000 Indonesian women.1,2
HPV is a small virus containing 8 kb circular double-stranded DNA. Furthermore, its genome is partitioned into three segments, namely the early region (E1, E2, E4, E5, E6, and E7), responsible for cell replication, transcription, and transformation; the late region (L1, L2) which encodes capsid proteins; and the long control region (LCR), governing E6 and E7 transcription.3 HPV belongs to the Papillomaviridae family and consists of five main genera, including alpha, beta, gamma, mu, and nu. The alpha, divided into low-risk and high-risk HPV groups, is the most critical genus harboring oncogenic transformation potential.4,5
High-risk HPV responsible for 87% of cervical cancer cases in Indonesia is strains 16 and 18, known as the most carcinogenic types (1). Their oncogenic potential is closely related to the E6 and E7 oncoproteins. Upon integration of HPV with the squamous epithelial cell genome, the oncoprotein genes become expressed, promoting malignant transformation.2 E6 and E7 trigger genomic instability in both the host DNA and viral genome, leading to the integration of viral DNA into the genome of the host. The ensuing genomic instability facilitates HPV-induced oncogenesis alongside viral DNA-host genome fusion. E6 disrupts the apoptosis pathway by blocking the Fas/Fas ligand, tumor necrosis factor (TNF), BCL2 antagonist, and Bak proteins in infected cells, while E7 induces cell proliferation through pRB protein targeting.6 E6 and E7 become therapeutic targets due to their dominant roles in oncogene-induced cervical carcinogenesis, driving the development of genome-targeted techniques and vaccine strategies.7
Presently, bivalent, quadrivalent, and nonavalent prophylactic HPV vaccines, including Cervarix® (targeting HPV strains 16 and 18), Gardasil® (addressing 6, 11, 16, and 18), and Gardasil 9® (combating 6, 11, 16, 18, 31, 33, 45, 52, and 58), offer protection primarily to HPV-unexposed individuals.8,9-11 However, they lack therapeutic effects against pre-existing infections.10,12
To holistically address HPV, a vaccine capable of both preventing infections and treating different forms of tumors is imperative. The E6 and E7 genes of HPV 16 and 18 are ideal targets for therapeutic vaccine development, considering their roles in the cell cycle and tumorigenesis.13,14 Effective elimination of viral infections necessitates a cell-mediated immune response, involving vaccine-induced type 1 helper T cell and cytotoxic T cell, which can kill infected and malignant cell. This type of response correlated with spontaneous HPV clearance and inhibition of progression.13,15,16
Popularly available therapeutic vaccines include live vectors, proteins or peptides, nucleic acids, and cell-based types. This study focuses on a protein recombinant vaccine consisting of E6 and E7 antigen fragments of HPV 16 and 18. Despite being safe and easily producible, this vaccine lacks immunogenicity, necessitating the addition of adjuvants for potency augmentation. The interaction of the vaccine with antigen-presenting cells through major histocompatibility complex (MHC) class I and II molecules stimulates an immune response by activating CD4+ and CD8+ effector T cells. For this to be successful, the vaccine protein must attach to the proteasome, become processed, bind to the TAP transporter, and then enter the APC cellular process.14-17
Current technological advancements should be integrated into vaccine development procedure.12 Bioinformatics serves as a crucial tool in the progression of cancer immunotherapy by aiding in the design of cancer neoantigen-based vaccines, detection of mutations, subsequent prediction of potential epitopes, and evaluation of peptide immunogenicity arising from these mutations.18 Additionally, this system facilitates T cell epitope mapping across HLA Class I and II, selection of antigens based on their cytotoxic T cell or T helper epitope content, and immunogen design to improve antigenicity, mitigate immune tolerance, and minimize cross-reactivity with self-epitopes.19
Immunoinformatics, a fusion of bioinformatics and immunology, enables vaccine discovery through the reverse vaccinology approach. This innovative technique involves using the genome of a pathogenic microorganism to identify antigens and predicting the epitopes (the lowest portions of the antigen capable of inducing an immune response) of T and B cells through several algorithms. Subsequently, the identified epitopes are linked appropriately for vaccine design. Multiepitope vaccines are promising agents against tumors and viral infections due to their high specificity, safety, and stability, as well as cheap development costs.12,20
Previous studies showed that vaccines designed with an in silico approach incorporating E5 and E7 for high-risk HPV strains could induce robust immune responses when tested in vivo.21 The combination of in silico and in vivo techniques holds the propensity to produce effective HPV vaccines.Therefore, this study aims to design a therapeutic multiepitope vaccine against cervical cancer, by employing oncoproteins E6 and E7 of HPV 16 and 18, using computer-based immunoinformatics analysis as a primary developmental step.
Materials and Methods
Retrieval of E6 and E7 protein sequences, antigenicity evaluation, and physicochemical characterization
The E6 and E7 proteins were obtained from the National Center for Biotechnology Information (NCBI) repository with corresponding accession numbers E6 and E7 protein of HPV16 (RefSeq: NC_001526.4) and HPV18 (GenBank: LC509006.1). The antigenicity and physiochemical properties of the selected proteins were analyzed using online tools VaxiJen v2.0 webserver (threshold value 0.40) and Protparam (http://web.expasy.org/protparam/), respectively.
Prediction of cytotoxic T lymphocyte (CTL) epitopes
To identify 9-mer amino acid sequences, epitopes for CTL were predicted.22 This prediction was conducted through the NetMHCpan EL 4.0 method using the Immune Epitopes Database (IEDB) tools. The analysis encompassed 12 different alleles of human leukocyte antigens (HLA) references, including HLA-A*01:01, HLA-A*02:01, HLA-A*03:01, HLA-A*24:02, HLA-A*26:01, HLA-B*07:02, HLA-B*08:01, HLA-B*27:05, HLA-B*39:01, HLA-B*40:01, HLA-B*58:01, and HLA-B*15:01. Subsequently, epitopes with characteristics of percentile lower than 2 were analyzed for their immunogenicity using IEDB test (http://tools.iedb.org/immunogenicity/), and those yielding positive scores were considered as immunogenic.
Prediction of helper T lymphocyte (HTL) Epitopes
For the peptide vaccine, the prediction of helper T lymphocyte (HTL) epitopes was accomplished using the Immune Epitopes Database (IEDB) tools, aiming to identify 15-mer amino acid sequences.23 In this study, 27 or a complete set of different alleles of MHC-II were employed, namely HLA-DRB1*01:01, HLA-DRB1*03:01, HLA-DRB1*04:01, HLA-DRB1*04:05, HLA- DRB1*07:01, HLA-DRB1*08:02, HLA-DRB1*09:01, HLA-DRB1*11:01, HLA-DRB1*12:01, HLA-DRB1*13:02, HLA-DRB1*15:01, HLA-DRB3*01:01, HLA-DRB3*02:02, HLA- DRB4*01:01, HLA-DRB5*01:01, HLA-DQA1*05:01/DQB1*02:01, HLA-DQA1*05:01/DQB1*03:01, HLA-DQA1*03:01/DQB1*03:02, HLA- DQA1*04:01/DQB1*04:02, HLA-DQA1*01:01/DQB1*05:01, HLA-DQA1*01:02/DQB1*06:02, HLA- DPA1*02:01/DPB1*01:01, HLA-DPA1*01:03/DPB1*02:01, HLA-DPA1*01:03/DPB1*04:01, HLA-DPA1*03:01/DPB1*04:02, HLA-DPA1*02:01/DPB1*05:01, and HLA DPA1*02:01/DPB1*14:01. The resultant epitopes were selected based on percentile ranks lower than 2.
Prediction of linear B lymphocyte (LBL) Epitopes
The linear B cell epitopes were predicted with ABCpred web tools and their length was established at a maximum of 16 peptides. The selection threshold for this LBL epitope prediction was set at 0.5, hence, any peptide with a percentile value ≥ 0.5 was selected for subsequent analysis.
Evaluation of antigenicity, allergenicity, and toxicity of CTL, HTL, and LBL epitopes
All the selected epitopes for CTL, HTL, and LBL were further evaluated for antigenicity, allergenicity, and toxicity. The antigenicity assessments were conducted using VaxiJen v2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) with a specific tumor model and a threshold value of 0.40. During the evaluation of allergenicity, two online tools renowned for their accuracy were employed, namely AllergenFP v.1.0 (https://ddg-pharmfac.net/AllergenFP/) and AllerTOP v.2.0 (https://www.ddg-pharmfac.net/AllerTOP/). Meanwhile, toxicity was analyzed through ToxinPred (http://crdd.osdd.net/raghava/toxinpred/) running in the default setting. Eventually, only epitopes demonstrating good antigenicity, non-allergenic properties, and nontoxic attributes were incorporated into the vaccine construction.
Population coverage of T cell epitopes
The assessment of population coverage in vaccine design was used to predict the efficacy globally by evaluating the prevalence of HLA alleles related to the selected MHC-I and MHC-II epitopes. To accomplish this, the Immune Epitopes Database (IEDB) population coverage tool was employed (http://tools.iedb.org/population/). The assessed HLA alleles were all suitable for CTL and HTL epitope prediction, corresponding to the epitopes selected for vaccine construction (with percentile values lower than 2, showing good antigenicity, non-allergenic, and nontoxic properties).24
Construction of multiepitope vaccine
The multiepitope vaccine comprised the selected CTL, HTL, and LBL epitopes arranged in an organized manner. A modification of the Ysrafil et al construction approach presented in Fig. 1 was employed in joining all the epitopes. This process was conducted using linkers such as AAY, GPGPG, and KK respectively for epitopes of CTL, HTL and HTL to CTL connections, and individual LBL or LBL paired with HTL. To augment vaccine efficacy, essential sequences including adjuvant (such as 50S ribosome) were incorporated and joined through an EAAAK linker. Furthermore, the C terminal of the vaccine was finalized by adding a 6xHis-tag.
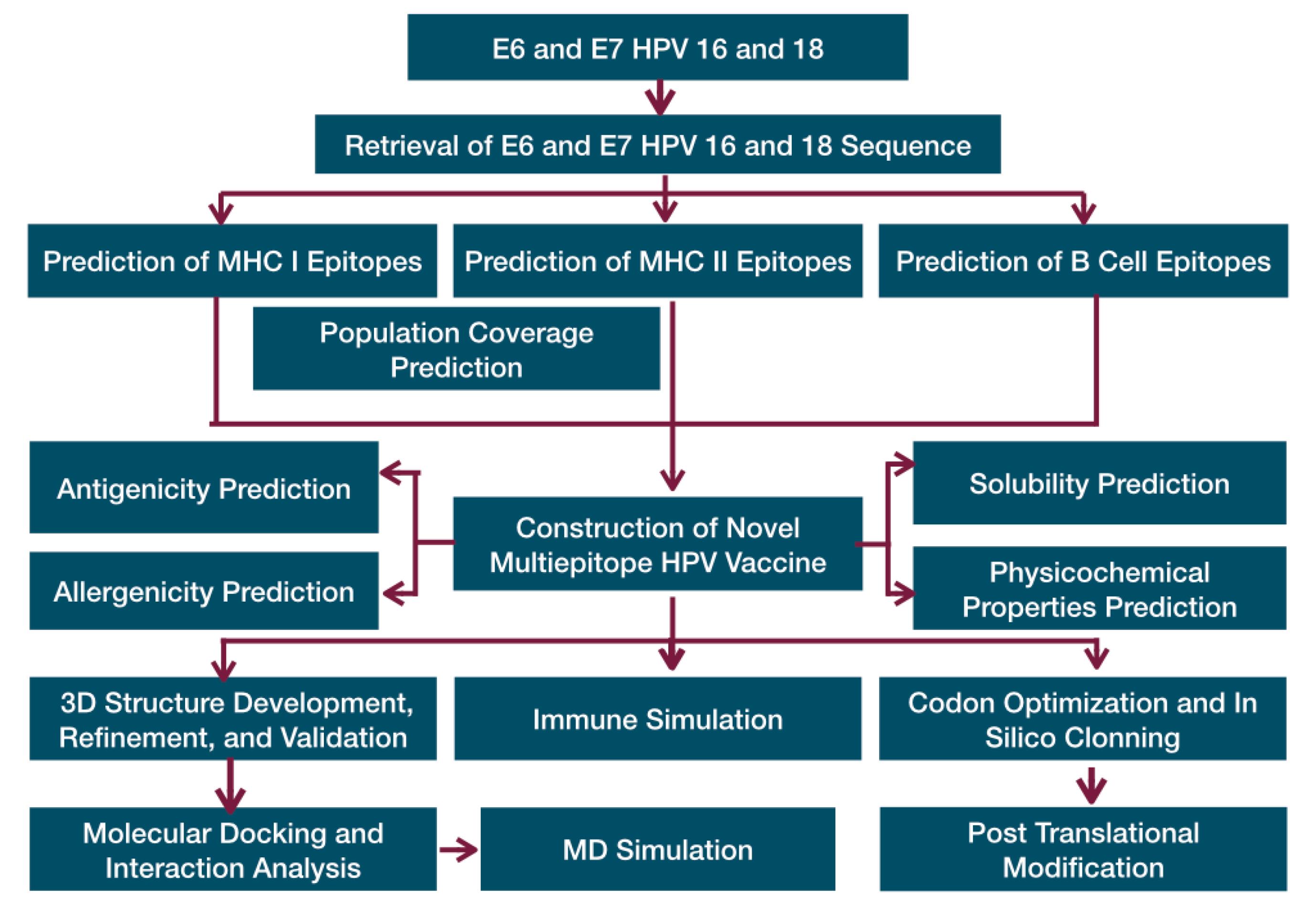
Fig. 1.
The flow chart of novel therapeutic multiepitope vaccine designing.
.
The flow chart of novel therapeutic multiepitope vaccine designing.
Analysis of physicochemical properties, solubility, antigenicity, and allergenicity of multiepitope vaccine
The multiepitope vaccine construct was comprehensively characterized for physicochemical properties, solubility, antigenicity, and allergenicity. The physicochemical characterization was conducted with ProtParam tools (https://web.expasy.org/protparam/), providing information about the molecular weight, isoelectric point (pI), extension coefficient, thermostability (aliphatic index), instability index, and hydrophobicity value (GRAVY). Additionally, the vaccine solubility was evaluated through the SOLpro webserver. The antigenicity and allergenicity evaluations were executed using VaxiJen v2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html), AllergenFP v.1.0 (https://ddg-pharmfac.net/AllergenFP/), and AllerTOP v.2.0 (https://www.ddg-pharmfac.net/AllerTOP/), respectively.
TAP transporter binding analysis
An analysis of the binding affinity of the vaccine to a TAP transporter was conducted using the TAPPred web tool (https://webs.iiitd.edu.in/cgibin/tappred/tappred.pl). This aimed to determine the ability of the vaccine to interact with TAP in the CTL stimulation mechanism involving MHC class I binding.
Predicting secondary and tertiary structure of multiepitope vaccine
The secondary structure of the constructed vaccine was determined with the servers of PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) and SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html). Meanwhile, the tertiary structure was generated through the online trRosetta tool (transform-restrained Restta) (https://yanglab.nankai.edu.cn/trRosetta/), producing a 3D model of the linear vaccine construct, which was visualized using PyMOL.
Refinement, validation, and flexibility evaluation of multiepitope vaccine
The final 3D structure of the vaccine construct was subsequently refined and validated. The refinement process was performed using online refinement tools, such as the GalaxyRefine server (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE), generating five best-refined results for further selection. Validation of the selected vaccine protein structure was executed with freely accessible online tools, including ProSA (https://prosa.services.came.sbg.ac.at/prosa.php), ERRAT (https://servicesn.mbi.ucla.edu/ERRAT/), and PROCHECK (https://servicesn.mbi.ucla.edu/PROCHECK/). Furthermore, CABS-Flex 2.0 server was employed during the evaluation of the flexibility property.
Molecular docking of multiepitope vaccine with host receptors
The refined 3D structure of the vaccine construct (ligand) was subjected to molecular docking with several antigen-presenting cell (APC) receptors to validate its capability to induce immune responses through the APC pathway. This process involved TLR2 (PDB ID:6nig) and TLR4 (PDB ID:4g8a) as receptors retrieved from the RCSB PDB protein data bank (https://www.rcsb.org/). Multiple online docking tools, including the ClusPro server and HDOCK, were employed to analyze the interactions of the vaccine with receptors, and the results were visualized using PyMOL.
Molecular dynamic simulation and estimation of MM/GB-PBSA binding energy
The molecular docking results were subjected to molecular dynamic analysis and MM/GB-PBSA binding energy estimation. The molecular dynamic analysis was conducted with an iMOD server (iMODS) (http://imods.chaconlab.org), while the binding energy was estimated using HawkDock (http://cadd.zju.edu.cn/hawkdock/).
Codon optimization and in silico cloning for genetic expression of the multiepitope vaccine in Pichia pastoris
The expression of the vaccine construct was simulated within selected host organisms. The process began by conducting codon optimization, specifically tailored for humans as the host organism. To accomplish this, the Codon Optimization Tool from Integrated DNA Technologies was used (https://www.idtdna.com/pages/tools/codon-optimization-tool). Subsequently, the vaccine genetic sequence targeted for cloning was simulated in Pichia pastoris. The resulting nucleotide sequence which had been adapted for optimal expression was employed for in silico cloning into the pUC19 expression vector, a popular plasmid DNA. This entire cloning procedure was executed with SnapGene v4.2 software.
Prediction of post-translational modifications in multiepitope vaccine
Post-translational modification (PTM) was analyzed with the same approach described by Ysrafil et al. This involved using the MusiteDeep online deep-learning framework and tool for protein PTM site prediction (https://www.musite.net/). A comprehensive exploration of all potential PTM within the vaccine construct was carried out through simulations.
Immune response post multiepitope vaccine administration
The immune response following the administration of the Multiepitope Vaccine was simulated using the C-ImmSim server 10.1 (http://www.cbs.dtu.dk/services/C-ImmSim-10.1/). The simulation encompassed a minimum of three vaccine injections at intervals of 0, 28, and 56 days. The obtained results were subsequently analyzed through comparison with existing literature.
Results
Retrieval of oncoproteins E6 and E7
Genomic data sourced from the NCBI website provided comprehensive genomic information, including the amino acid sequences of oncoproteins E6 and E7 for each HPV type, presented in Table S1. Antigenicity prediction and physicochemical characterization of the selected protein were carried out, showing a high antigenic potential with an antigenic value >0.4.
CTL epitope prediction
CTL epitope prediction centered on identifying MHC-I-binding epitopes, followed by assessments for antigenicity, immunogenicity, non-toxicity, and non-allergenicity to determine the optimal candidates for vaccine construction. This study successfully identified 10 CTL epitopes associated with HLA alleles including HLA-A*02:01, HLA-A*03:01, HLA-A*24:02, HLA-A*26:01, HLA-B*07:02, HLA-B*08:01, HLA-B*15:01, HLA-B*39:01, and HLA-B*40:01. This selection encompassed 2, 2, 5, and 1 epitopes from E6 of HPV 16, E7 of HPV 16, E6 of HPV 18, and E7 of HPV18. All selected epitopes demonstrated antigenic, immunogenic, non-allergenic, and non-toxic characteristics (Table S2).
Among the ten optimal epitopes, 49RAHYNIVTF57 from oncoprotein E7 of HPV 16 exhibited binding to four alleles, specifically HLA-A*24:02, HLA-A*26:01, HLA-B*07:02, and HLA-B*08:01. 126RFHNIAGHY134 from E6 of HPV 18 bound to three alleles, namely HLA-A*03:01, HLA-A*26:01, and HLA-B*15:01, while others attached to one or two alleles. The ability of these epitopes to bind with diverse HLA alleles influenced broader population coverage and vaccine effectiveness.
Helper T lymphocyte epitope prediction
To identify HTL epitopes, 15-mer amino acids were selected from the four oncoproteins using 27 MHC-II alleles. According to Table S3, six selected peptides as epitopes met the antigenicity criteria with an exhibition of optimal characteristics for vaccine candidacy, and each was bound to a minimum of one HLA allele. 44VFEFAFKDLFVVYRD58 was the only epitope attached to three alleles, namely HLA-DPA1*01:03/DPB1*04:01, HLA-DQA1*01:01/DQB1*05:01, and HLA-DPA1*02:01/DPB1*05:01, while others were linked to one or two alleles.
Four of the six epitopes stimulated the release of three cytokines, IFN-γ, IL-4, and IL10. These included 49VYDFAFRDLCIVYRD63 and 84RHYCYSLYGTTLEQQ98 from E6 of HPV 16, as well as 44VFEFAFKDLFVVYRD58 and 45FEFAFKDLFVVYRDS59 from E6 of HPV 18. Meanwhile, E7 epitopes from both HPV 16 and 18 induced a single cytokine response, as indicated in Table S3. This cytokine-inducing ability was found to be essential for activating the cellular and humoral immune systems.
Population coverage of T cell epitopes
The population coverage of T cell epitopes was assessed using the IEDB Population Coverage web tool. T cell epitopes that met the vaccine design criteria (10 CTL and 6 HTL epitopes) were submitted alongside their respective alleles. This study unveiled a global population coverage of 99.3% for the 16 T cell epitopes, as presented in Fig. 2. Regions with the highest population coverage reaching 100% included the Cook Islands, Mexico, and Sweden, while the lowest at 2.5% was in Rwanda. Up to 92.28% was recorded in Southeast Asia, featuring 71.21% in Indonesia, while the highest range at 99.51% was found in Japan.
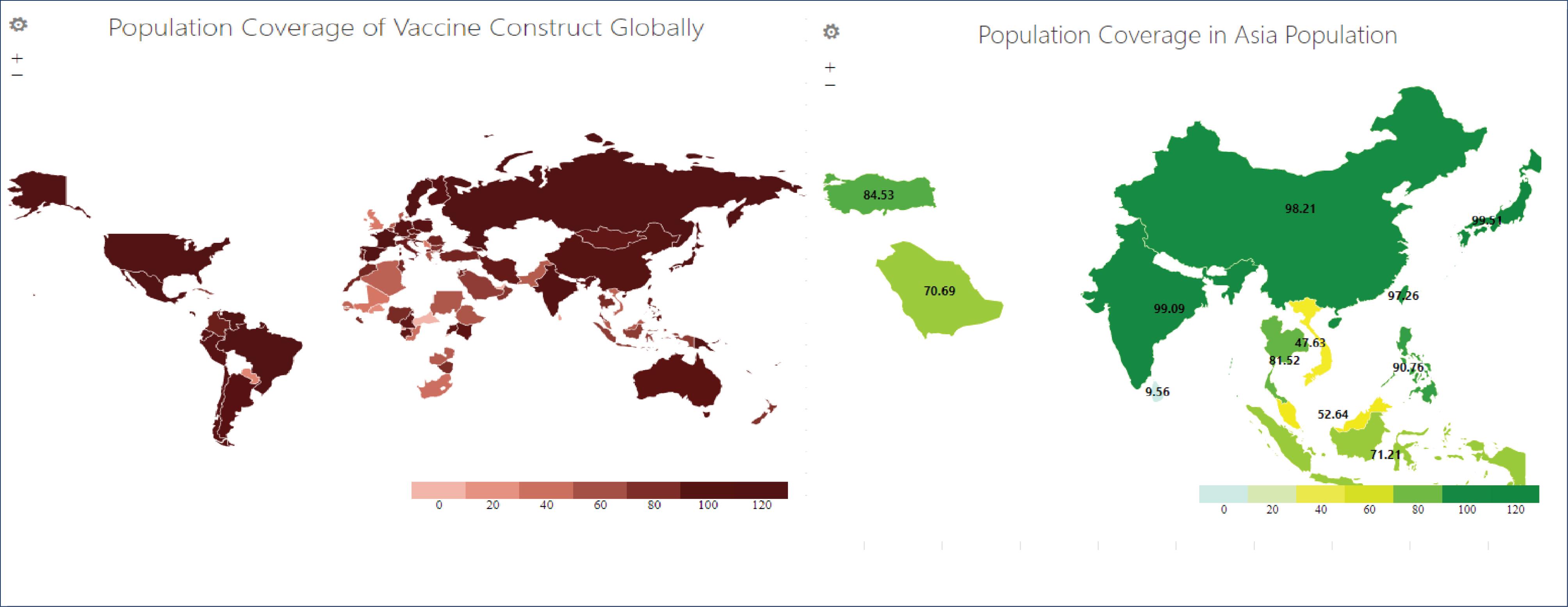
Fig. 2.
Population coverage of vaccine construct in global and Asia populations.
.
Population coverage of vaccine construct in global and Asia populations.
B cell lymphocyte (LBL) epitope prediction
LBL epitope prediction was performed with the ABCpred web tool, selecting 16-mer peptides as preferred epitopes. The selected LBL epitopes were scrutinized for their antigenicity, allergenicity, toxicity, and ability to induce immunoglobulins (Ig).25 Only antigenic, non-allergenic, and non-toxic epitopes were retained for vaccine design, while the capability to stimulate Ig production was not a prerequisite. A total of 12 peptides were identified as LBL epitopes, among which 5, 2, 3, and another 2 originated from HPV 16 E6, HPV 16 E7, HPV 18 E6, and HPV 18 E7, respectively. Only three of these functioned as IgG epitopes, namely 2 from HPV 16 E7 and 1 from HPV 18 E7 (See Supplementary file 1, Table S4).
Construction of vaccine from selected epitopes and evaluation of the allergenicity, antigenicity, and physicochemical characteristics
A total of 28 epitopes, including 10 CTL, 6 HTL, and 12 LBL epitopes, were integrated to form a multiepitope vaccine. According to Fig. 3, the construction process incorporated 50S ribosomal protein L7/L12 (Locus RL7_MYCTU)from Mycobacterium tuberculosis as an adjuvant, along with linkers binding the epitopes. Six histidines (6xHis-tag) were tagged to both ends of the vaccine following the procedure described by Ysrafil et al,23 Yang et al,26 Bhattacharya et al27, and Chukwudozie et al.28
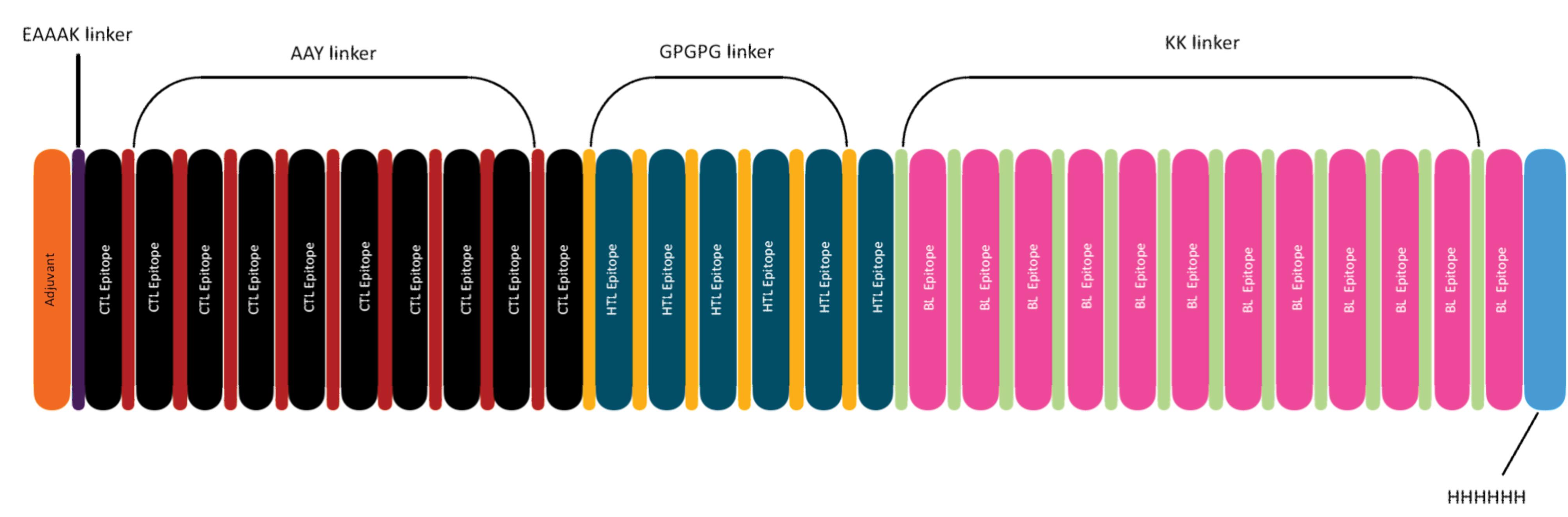
Fig. 3.
Novel multiepitope HPV-vaccine construct.
.
Novel multiepitope HPV-vaccine construct.
Characterization of the vaccine encompassed various attributes, such as antigenicity, allergenicity, amino acids content, molecular weight, isoelectric point (pI), extension coefficient, thermostability (aliphatic index), index instability, and hydrophobicity value (GRAVY), for quality determination. Antigenicity assessment conducted using the Vaxijen 2.0 server and antigenPRO yielded values of 0.6998 and 0.763696, respectively, confirming the antigenic nature of the vaccine. Additionally, the allergenicity analysis performed with AllergeFP and AllerTOP established the non-allergenic status.
Subsequent characteristic analysis with the Protparam tool showed that the vaccine consisted of 593 amino acids, weighed 66.85 kDa (40-110 kDa), and exhibited a theoretical pI value of 6.27, indicating stability at an acidic pH of 6.27. The half-life of the vaccine was estimated to be 30 hours in mammalian reticulocytes in vitro, >20 hours in yeast in vivo, and >10 hours in Escherichia coli in vivo. Stability evaluation indicated a score of 37.15, meaning the vaccine was stable, and the SOLpro web server showed a score of 0.97, suggesting high solubility.
TAP transporter binding analysis
From the full construct analysis on 593 amino acids (AA), 236 sequences showed binding affinity to the TAP transporter for CTL, with 41 categorized as high affinity and 195 as intermediate affinity. This indicated the ability of the constructed vaccine to attach to the TAP transporter, thereby facilitating MHC class I-binding within the endoplasmic reticulum.
Analysis of the secondary structure of the vaccine
Secondary structure analysis of the vaccine was performed using PSIPRED and SOPMA servers, as depicted in Fig. 4. PSIPRED detected 44.69% (265) alpha helix, 17.20% (102) beta strands, and 38.11% (226) coil forms among the 593 AA. Meanwhile, SOPMA showed 46.88% alpha helix, 18.04% extended strand, 6.91% β-turns, and 28.16% random coil. These results collectively suggested the identified secondary structure to be good.
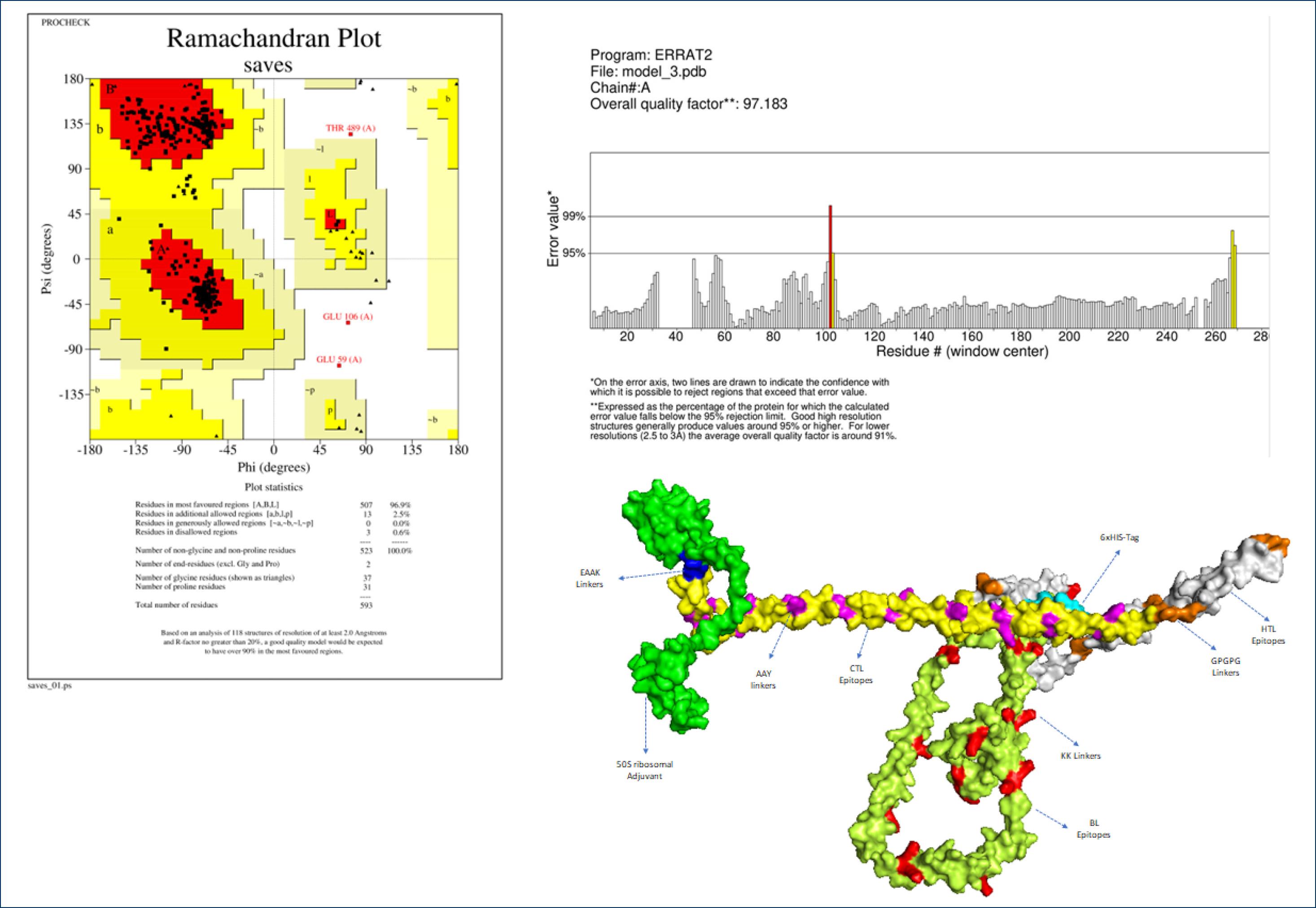
Fig. 4.
3D Structure of vaccine and its analysis.
.
3D Structure of vaccine and its analysis.
Three-dimensional structure building, refinement, and validation
Through the application of the trRosetta web tool, the tertiary (three-dimensional) structure of the vaccine was constructed and subsequently refined with the Galaxy refine server. Among five structural models generated, the fourth identified with the best characteristics was subjected to further analysis. Based on the results, 96.9% of the residues resided within favored regions, accompanied by a high-quality factor of 97.183 according to ERRAT and Ramachandran servers (Fig. 5), indicating that the model possessed good structural integrity.
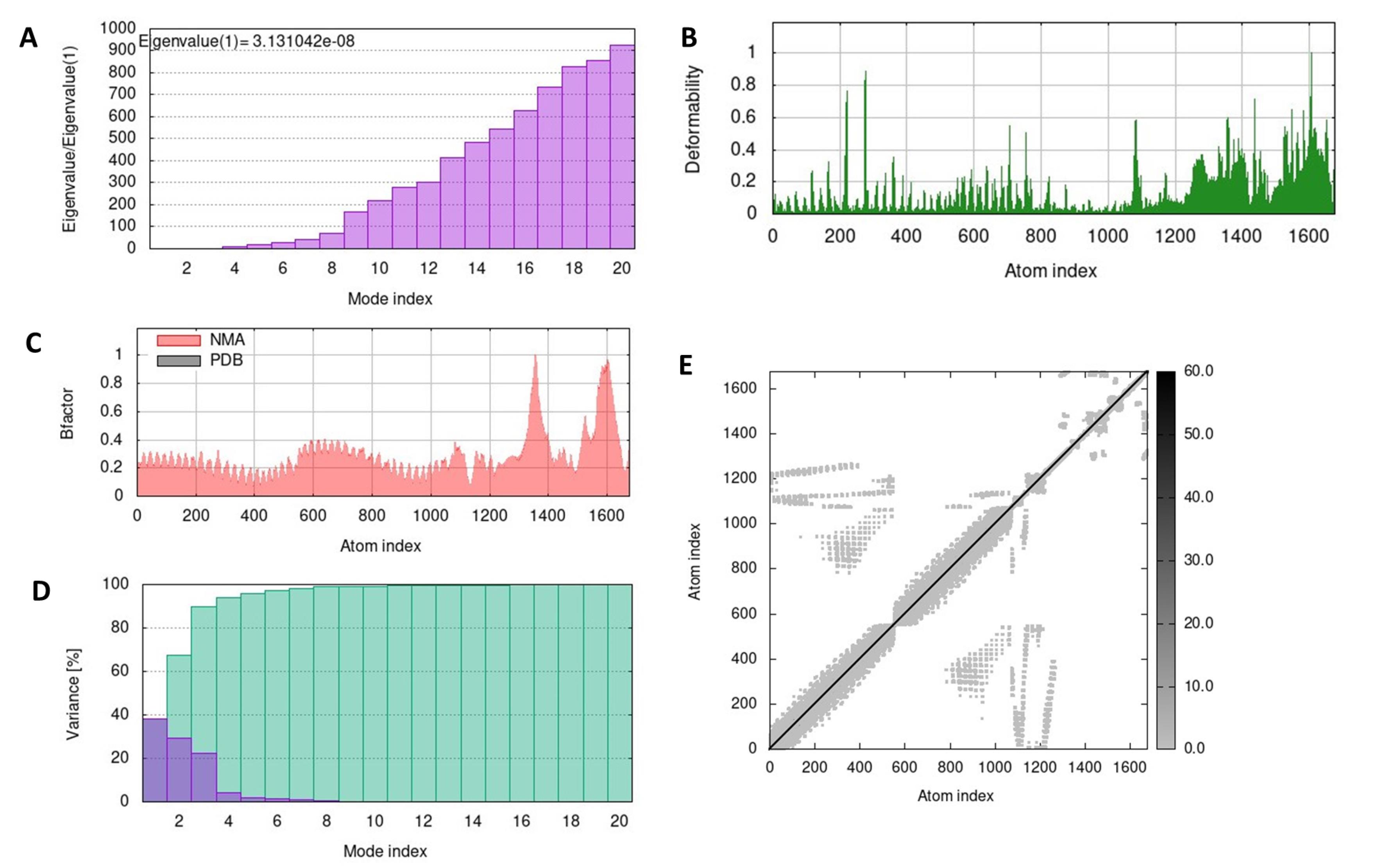
Fig. 5.
Molecular dynamic of complex vaccine-TLR2 analysis by iMODS server. (A) Eigenvalue, (B) Deformability, (C) B-factor values, (D) The covariance matrix, and (E) The elastic network.
.
Molecular dynamic of complex vaccine-TLR2 analysis by iMODS server. (A) Eigenvalue, (B) Deformability, (C) B-factor values, (D) The covariance matrix, and (E) The elastic network.
Docking of HPV-vaccine constructs with TLRs
The established tertiary structure of the vaccine was subjected to docking experiments with TLR2 and TLR4 receptors found on antigen-presenting cells (APCs). This assessment aimed to confirm and identify its ability to bind with APC receptors and potentially induce an adaptive immune response. The ClusPro, PatchDock, and HDock servers were employed for this analysis. The results showed strong binding interactions of the HPV vaccine with both TLR2 and TLR4 receptors, featuring confidence scores exceeding 0.7 (0.96 and 0.98, respectively), indicative of a highly possible binding scenario.
Molecular dynamic simulation and estimation of MM/GB-PBSA binding energy
The molecular dynamics of the vaccine-TLR2 and TLR4 complexes were evaluated using the iMODS server, a reliable tool for assessing protein flexibility, rigidity, and deformability through normal model analysis (NMA). Eigenvalues calculated from the structure were 3.131042e-08 and 1.142735e-07, corresponding to the TLR2 and TLR4 receptor components as presented in Figs. 5A and 6A. Lower eigenvalues indicated easier deformation of the complex, and they gradually increased in each mode. Additionally, the deformability showed a similar pattern for the vaccine-TLR2 and vaccine-TLR4, reflecting low degrees of deformation in all residues within the complexes (Figs. 5B and 6B). Their β-factor graphs in Figs. 5C and 6C presented a comparison of NMA results to the root mean square and the calculated uncertainty of each atom. Variance plot analysis revealed a moderate decrease in the individual variance of each successive mode for both complexes (Figs. 5D and 6D). Moreover, the iMODS analysis indicated decreased mobility and deformability, signifying the stability of vaccine-TLR3. An elastic network analysis demonstrated the relationship between atoms and springs, where stiffer springs were visually indicated by darker coloration.
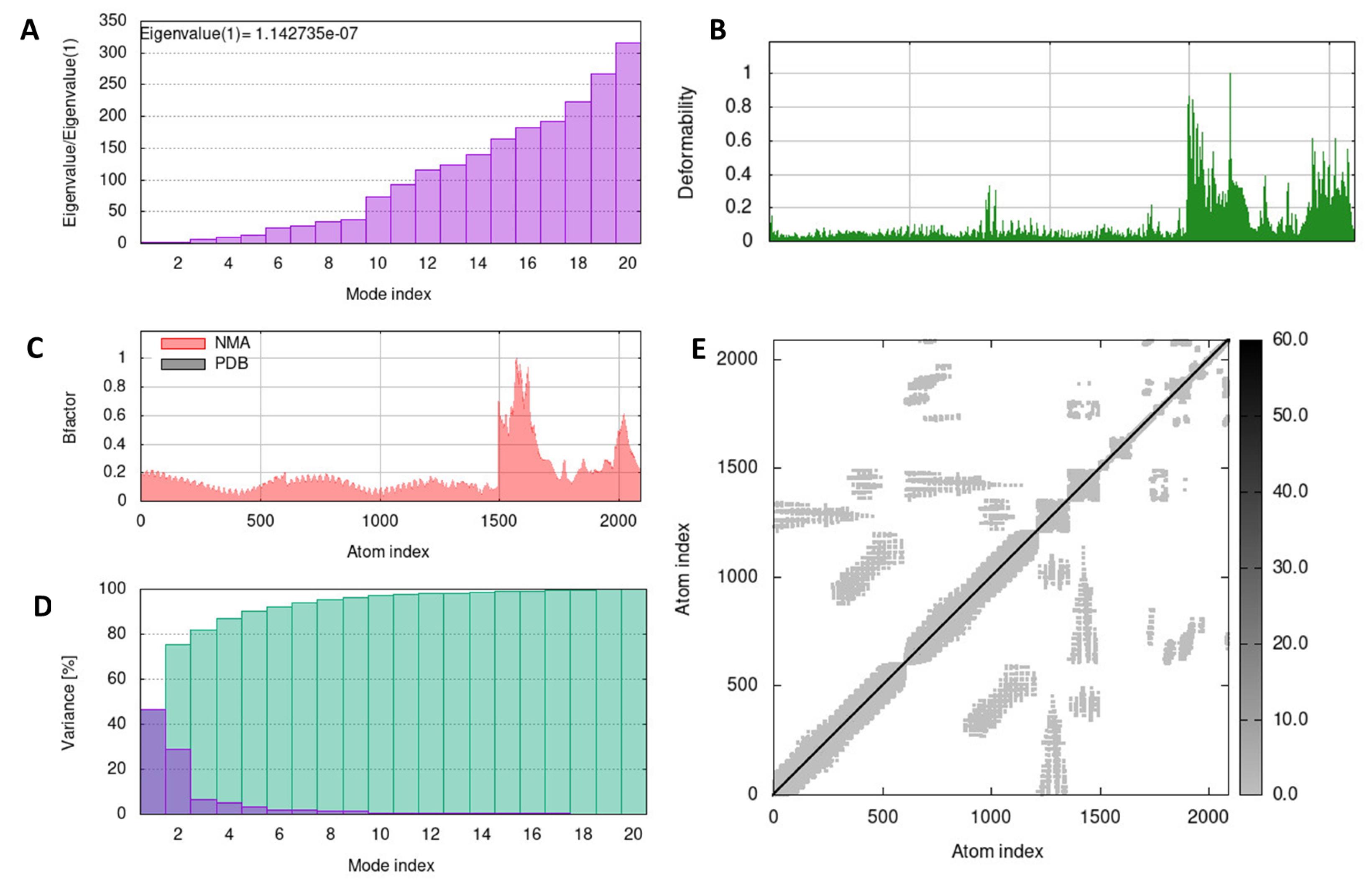
Fig. 6.
Molecular dynamic of complex vaccine-TLR4 analysis by iMODS. (A) Eigenvalue, (B) Deformability, (C) B-factor values, (D) The covariance matrix, and (E) The elastic network.
.
Molecular dynamic of complex vaccine-TLR4 analysis by iMODS. (A) Eigenvalue, (B) Deformability, (C) B-factor values, (D) The covariance matrix, and (E) The elastic network.
Codon adaptation and in silico cloning of the vaccine
Before in silico cloning, the codon optimization process carried out reversed the amino acid sequence of the vaccine to form nucleotides that were inserted into the plasmid during simulation. This process, which was performed using the IDT server, transformed 593 amino acids into a 1779-bp nucleotide sequence, with a total complexity score of 1.6 (<7), indicating acceptability.
During the insertion process, the identification of a suitable restriction enzyme site within the palindromic region was necessary to facilitate plasmid cleavage. The plasmid used for the in vitro vaccine propagation and production stage was pUC19, which also served as the gene expression vector in Pichia pastoris. Among the enzymes compatible with this expression system, BamHI and KpnI were selected as they would not disrupt the vaccine sequence. Consequently, palindromic sequences were added at the 5' and 3' ends of the vaccine to be inserted into the plasmid. The process of in silico cloning are presented in Fig. 7.
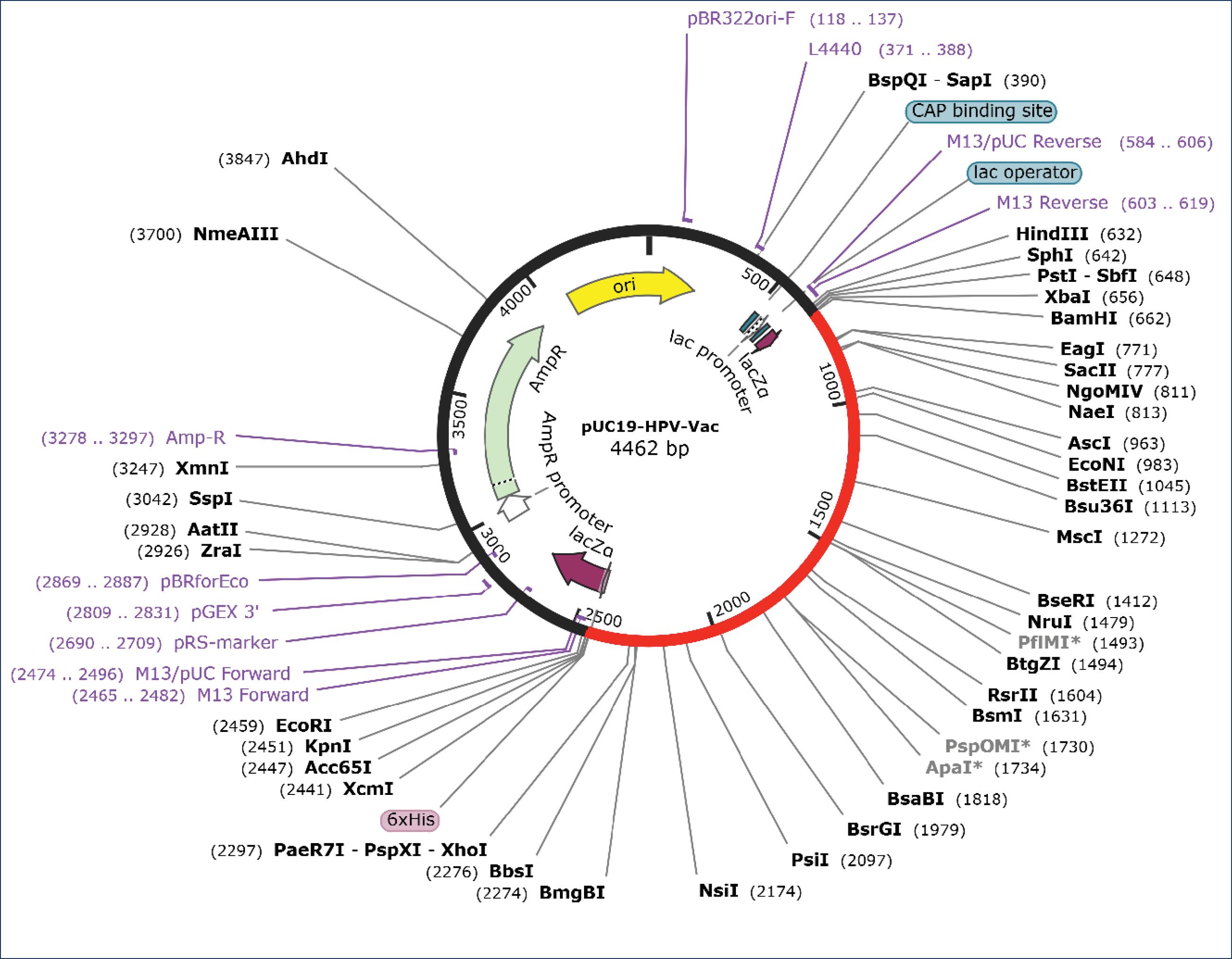
Fig. 7.
In silico cloning of vaccine using pUC19 expression vector. The red line is the inserted vaccine.
.
In silico cloning of vaccine using pUC19 expression vector. The red line is the inserted vaccine.
PTM analysis
PTM analysis showed potential modifications to the vaccine construct in the post-translational in vitro phase (production stage). Several modifications including phosphorylation (P), glycosylation (gl), ubiquitination (ub), acetylation (ac), methylation (me), and hydroxylation (hy) were identified. Phosphorylation was the most predominant, appearing seven times at S5, T6, S61, S368, Y477, S572, and S574, followed by ubiquitination which was found in six amino acids, namely K3, K73, K110, K117, K119, and K391.
Simulation of the immune responses triggered by administered vaccine in the human body
This phase assessed the potential of the vaccine to stimulate an immune response in silico, specifically adaptive immune responses encompassing cellular and humoral reactions, as well as memory cell formation. A successful induction was indicated by the activation of B and T cytotoxic memory cells. Additionally, innate immunity provided by macrophages, and particularly APCs such as dendritic cells, was considered due to its role in the release of various cytokines, including IFN-γ.
Based on Fig. 8, the administration of three doses of the HPV vaccine at four-week intervals showed the ability to induce multiple responses. These included elevated IFN-γ levels, increased antibody production, higher active cytotoxic T and helper T cell counts, augmented activation of B and B memory cells, as well as a noticeable rise in the number of dendritic cells and macrophages. These results indicated that injection of the vaccine could stimulate a robust immune response in silico.
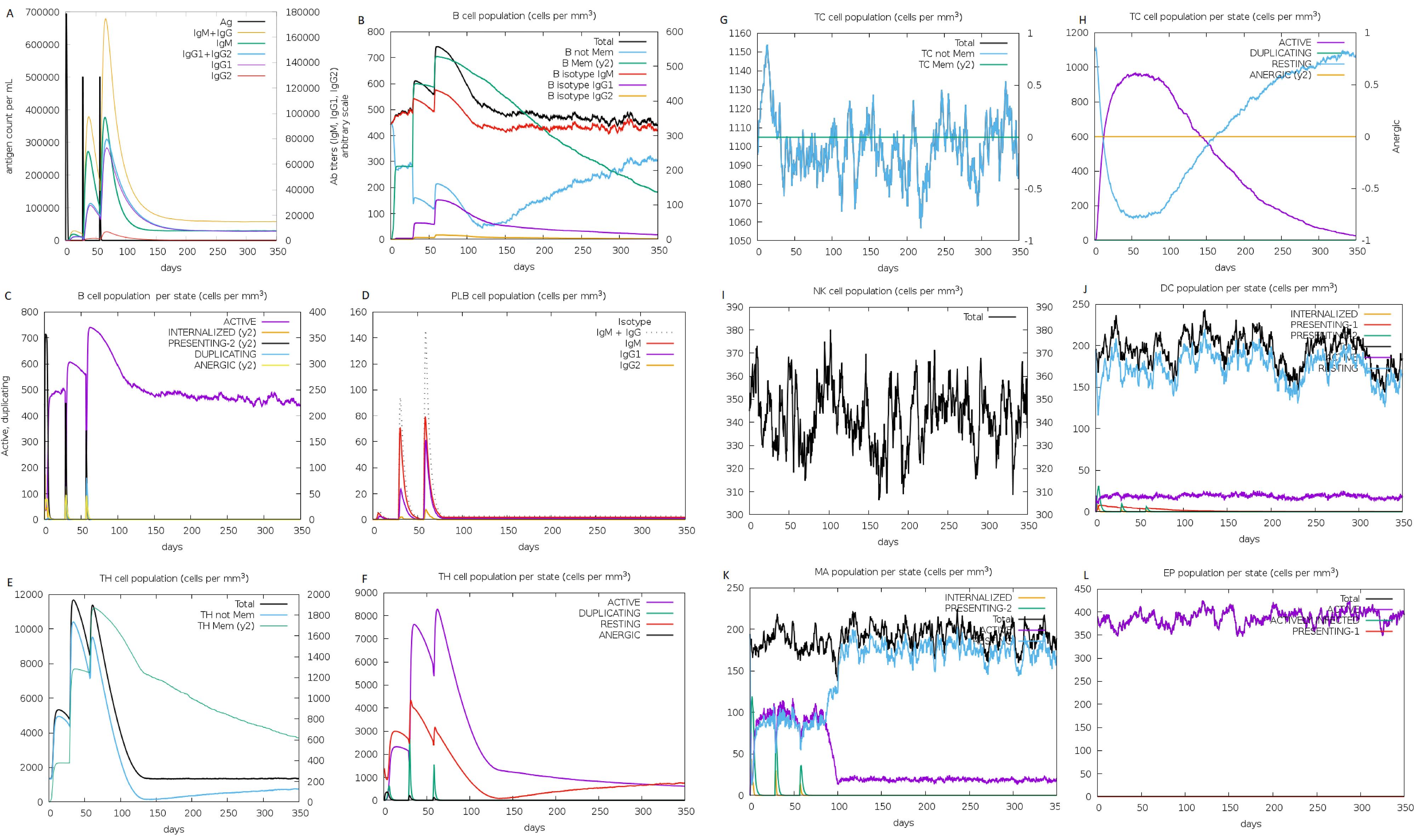
Fig. 8.
Immune simulation of multiepitope vaccine after three-dose injections at 0, 28, and 56 days. (A) Antigen concentration and antibody responses. Vaccines can induce antibody responses after being injected. (B-D) B cells population counts. (F-H) T cell population counts and activated state. (I) Natural Killer (NK) cell population activated. (J) Dendritic cells (DCs) population. (K) macrophages population.
.
Immune simulation of multiepitope vaccine after three-dose injections at 0, 28, and 56 days. (A) Antigen concentration and antibody responses. Vaccines can induce antibody responses after being injected. (B-D) B cells population counts. (F-H) T cell population counts and activated state. (I) Natural Killer (NK) cell population activated. (J) Dendritic cells (DCs) population. (K) macrophages population.
Discussion
Vaccination is one of the primary prevention approaches for infectious diseases, specifically those caused by viruses. Currently, the HPV prophylactic vaccine is in clinical use, targeting individuals presumed to be free of HPV infection. This vaccine mainly triggers the development of humoral immune responses.8,29 However, therapeutic cancer vaccines are expected to stimulate both humoral and cellular immune responses to eradicate tumor cells.30,31 This increases the need for a vaccine that can eliminate HPV infection while concurrently shrinking tumor size within cervical epithelium by engaging the innate, cellular, and humoral immune systems. To induce the described immune responses, an antigen capable of binding to MHC class I and II is required.31,32
MHC class I generally binds to peptides originating from the endogenous pathway, which are associated with the activity of CTL. Interactions involving MHC class I peptides are essential in antiviral and anticancer responses. Meanwhile, MHC class II binds to peptides derived from extracellular sources, playing a significant role in activating HTL, regulating antibody responses, and enhancing cytotoxic responses.22,33,34 In this study, vaccines based on oncoproteins E6 and E7 for HPV 16 and 18 were developed by assembling a construct consisting of proteins that match CTL, HTL, and B cells. The construct encompassed 593 AA, comprising peptides as MHC class I and II epitopes, along with B cell epitopes, adjuvants, linkers, and 6xHis-tag.
Differences in antigen origin and processing also dictate the involvement of distinct pathways. The antigen peptides presented by MHC class II molecules originate from endocytosis and become degraded before encountering these molecules in the late endosome. However, MHC class I molecules exhibit peptides originating from the cytosol. These peptides are acquired through proteasome cleavage and transferred to the endoplasmic reticulum (ER) by the TAP transporter to bind to MHC class I in the ER.22 This study analyzed peptide interactions with the TAP transporter, known as one of the natural HPV immune escape pathways.35 Out of the complete constructs analyzed (593 AA), 236 sequences exhibited binding affinity towards the TAP transporter for CTL, comprising 41 high and 195 intermediate affinities. The evaluated vaccine characteristics showed its potential antigenicity, non-allergenicity, stability, and solubility.
HLA, the MHC in humans, is a highly polymorphic molecule. Specific HLA alleles are expressed differently among various ethnic groups. Consequently, the design and selection of the T cell epitopes for a vaccine based on specific HLAs will determine its population coverage.33 This study employed HLA-A*02:01, HLA-A*03:01, HLA-A*24:02, HLA-A*26:01, HLA-B*07:02, HLA-B*08:01, HLA-B*15:01, HLA-B*39:01, and HLA-B*40:01, for MHC-I, and HLA-DQA1*01:01, HLA-DQA1*03:01, HLA-DQA1*05:01, HLA-DPA1*01:03, and HLA-DPA1*02:01 for MHC-II, to predict the population coverage of the selected epitopes. The results showed a population coverage of over 90% on a global scale and within Southeast Asia, while the data varied across specific ethnic groups in different regions.
The identification of B cell epitopes is essential for various medical applications, particularly in vaccine development, and diverse web tools are available for this stage. In this study, ABCpred was used to determine36 12 peptides as LBL epitopes, three of which acted as IgG epitopes. Besides antibody production, B cells could modulate immune cell functions through cytokine secretion, co-stimulation, and antigen presentation, indicating the relevance in cancer vaccines. The epitopes were found to be capable of enhancing vaccine peptide uptake by B cells, subsequent presentation to CD4+ lymphocytes, and activation of CTL.37
Immunodominant epitopes are often selected from CTL, HTL, or B cells, but peptide-based vaccines generally exhibit weak immunogenicity. Therefore, adjuvants are needed to enhance and trigger the immune response.31 In this study, the addition of 50S ribosomal protein L7/L12 (Locus RL7_MYCTU)from Mycobacterium tuberculosis was used as an adjuvant, known to elevate vaccine immunogenicity.12,38
The assessment of the ability of a vaccine to induce subsequent immune responses involves its interaction with Toll-like receptors (TLRs) on APCs. TLRs, being pattern recognition receptors, are situated prominently on the surface or endogenous regions of APCs. The interactions between the vaccine and TLRs initiate the activation of both innate and adaptive immune responses.23
TLR2 is an innate immune system receptor expressed by APCs and mucosal epithelium, contributing to mucosal immune responses.39 Both TLR2 and TLR4 are present on cell surfaces and intracellularly within dendritic, epithelial, and endothelial cells. These receptors can effectively detect viral coat proteins, triggering antiviral immune responses.40 Additionally, TLRs binding may play a significant role in antitumor activity. The previous study by Sher et al. reported the induction of antitumor immunity in animal models through a therapeutic HPV vaccine developed with a recombinant lapidated HPV16E7 mutant, which inactivated E7 oncogenic function by binding to TLR2 on APCs.41 Moreover, TLR4 receptors exhibit heightened expression in cervical cancer HeLa cells than other TLRs, this expression strongly correlates with cancer progression.12 In this study, a comprehensive analysis of vaccine docking onto TLR2 and TLR4 was conducted. The results showed a robust binding interaction of the novel multiepitope therapeutic HPV vaccine with both TLR2 and TLR4 receptors. This binding is promising for vaccine development as it is expected to induce the desired immune response.
During the protein production process, various PTM usually occur, such as ubiquitination, phosphorylation, and glycosylation which can impact the effectiveness of cancer vaccines. Ubiquitination facilitates proteasome binding, degradation, and epitope presentation, thereby enhancing antigen entry into the MHC class I pathway and boosting the CTL response. Similarly, phosphorylation can promote the process of ubiquitination and proteasomal degradation of antigens. Glycosylation is capable of augmenting the attachment of epitopes to both MHC classes.37 In this study, phosphorylation was the most frequently observed modification, occurring seven times, followed by ubiquitination in six amino acids. In summary, the vaccine construct exhibited appropriate PTM that enhanced its immunogenicity and efficacy.
This current study presented the simulation of an in silico vaccine administration performed in three doses at four-week intervals. The simulation successfully generated innate, cellular, and humoral immune responses, while also activating immune and memory cells, along with associated cytokines. The three-dose administration strategy aligned with various investigations on the clinical administration of prophylactic vaccines and in silico simulation of immune response modification.8,23 However, in vivo experimentation should be conducted to determine the optimal dosing frequency and quantity, to reduce tumor size and HPV infection within the cervical epithelium.
The transition to in vivo testing involves recombinant production and purification of the constructed vaccine, as outlined by Safavi et al.42 Furthermore, conjugation with non-viral vectors, including lipid nanoparticles, polymeric nanoparticles, and liposome-polymer hybrid nanoparticles, can facilitate effective vaccine delivery in the human body.43,44 Nanoparticle-based vectors, being non-viral agents, are capable of delivering vaccines to their target and inducing immune responses. The study by Zhao et al44 showed that conjugating a multiepitope vaccine with LNPs led to the acquisition of desirable characteristics and increased vaccine antigen expression in L-02 cells. The intramuscular injection was expected to elevate cellular and humoral immune responses in experimental animals. Similarly, Hassan et al45 reported effective protection against virus transmission upon the intramuscular injection of the multiepitope hepatitis conjugate adenovirus vaccine.
Multiple studies and the development of therapeutic vaccines for HPV infection and cervical cancer are still needed, particularly in developing countries experiencing high morbidity rates associated with these diseases. While traditional vaccine development procedures are time-consuming and costly, reverse vaccinology offers a solution. Due to limitations in computational tool databases, designs tested in silico must be confirmed by sequencing the construct and assessing immune response induction in both in vitro and in vivo settings. Moreover, another significant limitation of this vaccine design approach is its lack of comprehensive consideration for most epitope prediction tools, specifically antigen processing sites involved in the prediction and presentation of epitopes. This discrepancy arises from antigen processing mechanisms which vary based on proinflammatory signals and may differ among distinct cell types.46
Conclusion
In conclusion, the multiepitope vaccine developed in this study tended to be a potential therapeutic intervention for HPV infection and HPV-related cancers. The assessments conducted confirmed its antigenic, immunogenic, non-allergenic, non-toxic, soluble, and stable properties. Computational analysis showed that this vaccine could bind strongly to TLR2 and TLR4 and stimulate an adequate immune response. Administration of three doses at intervals of 28 days led to increased levels of IgG and IgM antibodies, activation of T and B cells, memory cell formation, and induction of phagocytic activity and dendritic cells functioning as APCs. However, this study was exploratory in nature, and additional investigations would be necessary to validate the efficacy of the constructed vaccine. In vitro studies should be conducted to substantiate the vaccine concept, followed by animal experiments to ascertain appropriate dosing and administration frequency. Subsequent clinical trials would be essential to establish the effectiveness of the vaccine in humans, aimed at developing a potent therapeutic HPV vaccine.
Research Highlights
What is the current knowledge?
√ Cervical cancer ranks as the fourth most common cancer among women worldwide.
√ There is a growing need for more effective vaccines to cure HPV infection and its associated cancers, including cervical cancer.
√ Vaccines can be developed through Immunoinformatics approaches using HPV oncoproteins.
What is new here?
√ A vaccine can be designed through immunoinformatics approaches using HPV oncoproteins E6 and E7 from HPV 16 and 18.
√ The multiepitope vaccine is antigenic and nonallergenic, exhibiting good characteristics.
√ The vaccine construct demonstrates strong interactions with human receptors including TLR2 and TLR4.
√ The vaccine construct elicits both humoral and cellular immune responses, crucial for curbing HPV infection and combating cancer.
Acknowledgments
The authors are grateful to the Faculty of Medicine, Universitas Tanjungpura, Pontianak, Indonesia, for the invaluable support offered to facilitate this study.
Competing Interests
None declared.
Ethical Statement
Not applicable.
Supplementary files
Supplementary file 1 contains Tables S1-S4.
(pdf)
References
- Bruni L, Albero G, Serrano B, Mena M, Collado J, Gómez D, et al. ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre). Human Papillomavirus and Related Diseases in Indonesia. Summary Report 10 March 2023. 2023 [cited 10 March 2023 10 March 2023].
- Wild C, Wederpass E, Stewart B. World Cancer Report; Cancer Research For Cancer Prevention. International Agency for Research on Cancer; 2020.
- Dai S, Li C, Yan Z, Zhou Z, Wang X, Wang J. Association of Human Papillomavirus Type 16 Long Control Region Variations with Cervical Cancer in a Han Chinese Population. Int J Med Sci 2020; 17:931-8. doi: 10.7150/ijms.43030 [Crossref] [ Google Scholar]
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology 2004; 324:17-27. doi: 10.1016/j.virol.2004.03.033 [Crossref] [ Google Scholar]
- Kombe Kombe AJ, Li B, Zahid A, Mengist HM, Bounda GA, Zhou Y. Epidemiology and Burden of Human Papillomavirus and Related Diseases, Molecular Pathogenesis, and Vaccine Evaluation. Front Public Health 2020; 8:552028. doi: 10.3389/fpubh.2020.552028 [Crossref] [ Google Scholar]
- Estêvão D, Costa NR, Gil da Costa RM, Medeiros R. Hallmarks of HPV carcinogenesis: The role of E6, E7 and E5 oncoproteins in cellular malignancy. BiochimBiophys Acta Gene Regul Mech 2019; 1862:153-62. doi: 10.1016/j.bbagrm.2019.01.001 [Crossref] [ Google Scholar]
- Pal A, Kundu R. Human Papillomavirus E6 and E7: The Cervical Cancer Hallmarks and Targets for Therapy. Front Microbiol 2019; 10:3116. doi: 10.3389/fmicb.2019.03116 [Crossref] [ Google Scholar]
- McNamara M, Batur P, Walsh JME, Johnson KM. HPV Update: Vaccination, Screening, and Associated Disease. J Gen Intern Med 2016; 31:1360-6. doi: 10.1007/s11606-016-3725-z [Crossref] [ Google Scholar]
- de Martel C, Plummer M, Vignat J, Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer 2017; 141:664-70. doi: 10.1002/ijc.30716 [Crossref] [ Google Scholar]
- Dong D, Zhu Y, Aili Z, Chen Z, Ding J. Bioinformatics analysis of HPV-68 E6 and E7 oncoproteins for designing a therapeutic epitope vaccine against HPV infection. Infect Genet Evol 2020; 81:104266. doi: 10.1016/j.meegid.2020.104266 [Crossref] [ Google Scholar]
- Mastutik G, Alia R, Rahniayu A, Rahaju AS, Kurniasari N, Putra ST. Genotyping of human pappilomavirus in cervical precancerous lesion and squamous cell carcinoma at drSoetomo hospital, Surabaya, Indonesia. Afr J Infect Dis 2018; 12:7-12. doi: 10.21010/Ajid.12v1S.2 [Crossref] [ Google Scholar]
- Sanami S, Azadegan-Dehkordi F, Rafieian-Kopaei M, Salehi M, Ghasemi-Dehnoo M, Mahooti M. Design of a multi-epitope vaccine against cervical cancer using immunoinformatics approaches. Sci Rep 2021; 11:12397. doi: 10.1038/s41598-021-91997-4 [Crossref] [ Google Scholar]
- Chabeda A, Yanez RJR, Lamprecht R, Meyers AE, Rybicki EP, Hitzeroth Hitzeroth, II II. Therapeutic vaccines for high-risk HPV-associated diseases. Papillomavirus Res 2018; 5:46-58. doi: 10.1016/j.pvr.2017.12.006 [Crossref] [ Google Scholar]
- Yang A, Farmer E, Wu TC, Hung CF. Perspectives for therapeutic HPV vaccine development. J Biomed Sci 2016; 23:75. doi: 10.1186/s12929-016-0293-9 [Crossref] [ Google Scholar]
- Yang A, Jeang J, Cheng K, Cheng T, Yang B, Wu TC. Current state in the development of candidate therapeutic HPV vaccines. Expert Rev Vaccines 2016; 15:989-1007. doi: 10.1586/14760584.2016.1157477 [Crossref] [ Google Scholar]
- Zhang J, Fan J, Skwarczynski M, Stephenson RJ, Toth I, Hussein WM. Peptide-Based Nanovaccines in the Treatment of Cervical Cancer: A Review of Recent Advances. Int J Nanomedicine 2022; 17:869-900. doi: 10.2147/ijn.S269986 [Crossref] [ Google Scholar]
- Jabbar B, Rafique S, Salo-Ahen OMH, Ali A, Munir M, Idrees M. Antigenic Peptide Prediction From E6 and E7 Oncoproteins of HPV Types 16 and 18 for Therapeutic Vaccine Design Using Immunoinformatics and MD Simulation Analysis. Front Immunol 2018; 9:3000. doi: 10.3389/fimmu.2018.03000 [Crossref] [ Google Scholar]
- Holtsträter C, Schrörs B, Bukur T, Löwer M. Bioinformatics for Cancer Immunotherapy. Methods Mol Biol 2020; 2120:1-9. doi: 10.1007/978-1-0716-0327-7_1 [Crossref] [ Google Scholar]
- De Groot AS, Moise L, Terry F, Gutierrez AH, Hindocha P, Richard G. Better Epitope Discovery, Precision Immune Engineering, and Accelerated Vaccine Design Using Immunoinformatics Tools. Front Immunol 2020; 11:442. doi: 10.3389/fimmu.2020.00442 [Crossref] [ Google Scholar]
- Taupiqurrohman O, Yusuf M, Nuswantara S, Subroto T. Potensi Gen Oncoprotein Human Papillomavirus Tipe 16 Sebagai Kandidat Vaksin Kanker Serviks. Maj Kedokt Bandung 2016; 48:84-91. doi: 10.15395/mkb.v48n2.761 [Crossref] [ Google Scholar]
- Namvar A, Panahi HA, Agi E, Bolhassani A. Development of HPV(16,18,31,45) E5 and E7 peptides-based vaccines predicted by immunoinformatics tools. Biotechnol Lett 2020; 42:403-18. doi: 10.1007/s10529-020-02792-6 [Crossref] [ Google Scholar]
- Sanchez-Trincado JL, Gomez-Perosanz M, Reche PA. Fundamentals and Methods for T- and B-Cell Epitope Prediction. J Immunol Res 2017; 2017:2680160. doi: 10.1155/2017/2680160 [Crossref] [ Google Scholar]
- Ysrafil Y, Sapiun Z, Astuti I, Anasiru MA, Slamet NS, Hartati H. Designing multi-epitope based peptide vaccine candidates against SARS-CoV-2 using immunoinformatics approach. Bioimpacts 2022; 12:359-70. doi: 10.34172/bi.2022.23769 [Crossref] [ Google Scholar]
- Ysrafil Y, Imran AK, Wicita PS, Kamba V, Mohamad F, Ismail I, et al. Mosaic vaccine design targeting mutational spike protein of SARS-COV-2: an Immunoinformatics Approaches. Bioimpacts 2023; 13. 10.34172/bi.2023.26443.
- Pourseif MM, Moghaddam G, Daghighkia H, Nematollahi A, Omidi Y. A novel B- and helper T-cell epitopes-based prophylactic vaccine against Echinococcus granulosus. Bioimpacts 2018; 8:39-52. doi: 10.15171/bi.2018.06 [Crossref] [ Google Scholar]
- Yang Z, Bogdan P, Nazarian S. An in silico deep learning approach to multi-epitope vaccine design: a SARS-CoV-2 case study. Sci Rep 2021; 11:3238. doi: 10.1038/s41598-021-81749-9 [Crossref] [ Google Scholar]
- Bhattacharya M, Sharma AR, Ghosh P, Lee SS, Chakraborty C. A Next-Generation Vaccine Candidate Using Alternative Epitopes to Protect against Wuhan and All Significant Mutant Variants of SARS-CoV-2: An Immunoinformatics Approach. Aging Dis 2021; 12:2173-95. doi: 10.14336/ad.2021.0518 [Crossref] [ Google Scholar]
- Chukwudozie OS, Gray CM, Fagbayi TA, Chukwuanukwu RC, Oyebanji VO, Bankole TT. Immuno-informatics design of a multimeric epitope peptide based vaccine targeting SARS-CoV-2 spike glycoprotein. PLoS One 2021; 16:e0248061. doi: 10.1371/journal.pone.0248061 [Crossref] [ Google Scholar]
- Gonçalves AK, Cobucci RN, Rodrigues HM, de Melo AG, Giraldo PC. Safety, tolerability and side effects of human papillomavirus vaccines: a systematic quantitative review. Braz J Infect Dis 2014; 18:651-9. doi: 10.1016/j.bjid.2014.02.005 [Crossref] [ Google Scholar]
- Crews DW, Dombroski JA, King MR. Prophylactic Cancer Vaccines Engineered to Elicit Specific Adaptive Immune Response. Front Oncol 2021; 11:626463. doi: 10.3389/fonc.2021.626463 [Crossref] [ Google Scholar]
- Liu J, Fu M, Wang M, Wan D, Wei Y, Wei X. Cancer vaccines as promising immuno-therapeutics: platforms and current progress. J Hematol Oncol 2022; 15:28. doi: 10.1186/s13045-022-01247-x [Crossref] [ Google Scholar]
- Galanis KA, Nastou KC, Papandreou NC, Petichakis GN, Pigis DG, Iconomidou VA. Linear B-Cell Epitope Prediction for In Silico Vaccine Design: A Performance Review of Methods Available via Command-Line Interface. Int J Mol Sci 2021; 22. 10.3390/ijms22063210.
- Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics 2006; 7:153. doi: 10.1186/1471-2105-7-153 [Crossref] [ Google Scholar]
- Comber JD, Philip R. MHC class I antigen presentation and implications for developing a new generation of therapeutic vaccines. Ther Adv Vaccines 2014; 2:77-89. doi: 10.1177/2051013614525375 [Crossref] [ Google Scholar]
- Steinbach A, Riemer AB. Immune evasion mechanisms of human papillomavirus: An update. Int J Cancer 2018; 142:224-9. doi: 10.1002/ijc.31027 [Crossref] [ Google Scholar]
- Jespersen MC, Mahajan S, Peters B, Nielsen M, Marcatili P. Antibody Specific B-Cell Epitope Predictions: Leveraging Information From Antibody-Antigen Protein Complexes. Front Immunol 2019; 10:298. doi: 10.3389/fimmu.2019.00298 [Crossref] [ Google Scholar]
- Safavi A, Kefayat A, Sotoodehnejadnematalahi F, Salehi M, Modarressi MH. In silico analysis of synaptonemal complex protein 1 (SYCP1) and acrosin binding protein (ACRBP) antigens to design novel multiepitope peptide cancer vaccine against breast cancer. Int J Pept Res Ther 2019; 25:1343-59. doi: 10.1007/s10989-018-9780-z [Crossref] [ Google Scholar]
- Lee SJ, Shin SJ, Lee MH, Lee MG, Kang TH, Park WS. A potential protein adjuvant derived from Mycobacterium tuberculosis Rv0652 enhances dendritic cells-based tumor immunotherapy. PLoS One 2014; 9:e104351. doi: 10.1371/journal.pone.0104351 [Crossref] [ Google Scholar]
- Ashhurst AS, Johansen MD, Maxwell JWC, Stockdale S, Ashley CL, Aggarwal A. Mucosal TLR2-activating protein-based vaccination induces potent pulmonary immunity and protection against SARS-CoV-2 in mice. Nat Commun 2022; 13:6972. doi: 10.1038/s41467-022-34297-3 [Crossref] [ Google Scholar]
- Sartorius R, Trovato M, Manco R, D'Apice L, De Berardinis P. Exploiting viral sensing mediated by Toll-like receptors to design innovative vaccines. NPJ Vaccines 2021; 6:127. doi: 10.1038/s41541-021-00391-8 [Crossref] [ Google Scholar]
- Sher YP, Lee C, Liu SY, Chen IH, Lee MH, Chiu FF. A therapeutic vaccine targeting HPV E6/E7 with intrinsic Toll-like receptor 2 agonist activity induces antitumor immunity. Am J Cancer Res 2018; 8:2528-37. [ Google Scholar]
- Safavi A, Kefayat A, Sotoodehnejadnematalahi F, Salehi M, Modarressi MH. Production, purification, and in vivo evaluation of a novel multiepitope peptide vaccine consisted of immunodominant epitopes of SYCP1 and ACRBP antigens as a prophylactic melanoma vaccine. Int Immunopharmacol 2019; 76:105872. doi: 10.1016/j.intimp.2019.105872 [Crossref] [ Google Scholar]
- Pourseif MM, Parvizpour S, Jafari B, Dehghani J, Naghili B, Omidi Y. A domain-based vaccine construct against SARS-CoV-2, the causative agent of COVID-19 pandemic: development of self-amplifying mRNA and peptide vaccines. Bioimpacts 2021; 11:65-84. doi: 10.34172/bi.2021.11 [Crossref] [ Google Scholar]
- Zhao Z, Ma X, Zhang R, Hu F, Zhang T, Liu Y. A novel liposome-polymer hybrid nanoparticles delivering a multi-epitope self-replication DNA vaccine and its preliminary immune evaluation in experimental animals. Nanomedicine 2021; 35:102338. doi: 10.1016/j.nano.2020.102338 [Crossref] [ Google Scholar]
- Hassan AO, Amen O, Sayedahmed EE, Vemula SV, Amoah S, York I. Adenovirus vector-based multi-epitope vaccine provides partial protection against H5, H7, and H9 avian influenza viruses. PLoS One 2017; 12:e0186244. doi: 10.1371/journal.pone.0186244 [Crossref] [ Google Scholar]
- Maleki A, Russo G, Parasiliti Palumbo GA, Pappalardo F. In silico design of recombinant multi-epitope vaccine against influenza A virus. BMC Bioinformatics 2022; 22:617. doi: 10.1186/s12859-022-04581-6 [Crossref] [ Google Scholar]