Bioimpacts. 14(4):28870.
doi: 10.34172/bi.2023.28870
Original Article
Integrating rapamycin with novel PI3K/Akt/mTOR inhibitor microRNAs on NOTCH1-driven T-cell acute lymphoblastic leukemia (T-ALL)
Fateme Arjmand Formal analysis, Investigation, Software, Visualization, Writing – original draft, Writing – review & editing, 1 
Samaneh Shojaei Formal analysis, Investigation, Software, 2
Mitra Khalili Data curation, Methodology, Writing – review & editing, 3, 1
Hossein Dinmohammadi Data curation, Methodology, Writing – review & editing, 1
Behzad Poopak Resources, 4
Samira Mohammadi-Yeganeh Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Writing – review & editing, 2, 5, * 
Yousef Mortazavi Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Writing – review & editing, 3, 6, * 
Author information:
1Department of Medical Genetics and Molecular medicine, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran
2Cellular and Molecular Biology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
3Cancer Gene Therapy Research center, Zanjan University of Medical Sciences, Zanjan, Iran
4DCLS PhD. Associate Professor of Hematology Owner & Lab. Director Payvand Clinical & Specialty Laboratory, CEO Amir Payvand Research & Development Co.
5Department of Medical Biotechnology, School of Advanced Technologies in Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
6Department of Medical Biotechnology, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran
Abstract
Introduction:
The PI3K/AKT/mTOR signaling pathway plays a significant role in the development of T-cell acute lymphoblastic leukemia (T-ALL). Rapamycin is a potential therapeutic strategy for hematological malignancies due to its ability to suppress mTOR activity. Additionally, microRNAs (miRNAs) have emerged as key regulators in T-ALL pathophysiology and treatment. This study aimed to investigate the combined effects of rapamycin and miRNAs in inhibiting the PI3K/AKT/mTOR pathway in T-ALL cells.
Methods:
Bioinformatic algorithms were used to find miRNAs that inhibit the PI3K/AKT/mTOR pathway. Twenty-five bone marrow samples were collected from T-ALL patients, alongside five control bone marrow samples from non-leukemia patients. The Jurkat cell line was chosen as a representative model for T-ALL. Gene and miRNA expression levels were assessed using quantitative real-time PCR (qRT-PCR). Two miRNAs exhibiting down-regulation in both clinical samples and Jurkat cells were transfected to the Jurkat cell line to investigate their impact on target gene expression. Furthermore, in order to evaluate the potential of combination therapy involving miRNAs and rapamycin, apoptosis and cell cycle assays were carried out.
Results:
Six miRNAs (miR-3143, miR-3182, miR-99a/100, miR-155, miR-576-5p, and miR-501- 3p) were predicted as inhibitors of PI3K/AKT/mTOR pathway. The expression analysis of both clinical samples and the Jurkat cell line revealed a simultaneous downregulation of miR-3143 and miR-3182. Transfection investigation demonstrated that the exogenous overexpression of miR-3143 and miR-3182 can effectively inhibit PI3K/AKT/mTOR signaling in the Jurkat cell line. Moreover, when used as a dual inhibitor along with rapamycin, miR-3143 and miR-3182 significantly increased apoptosis and caused cell cycle arrest in the Jurkat cell line.
Conclusion:
These preliminary results highlight the potential for improving T-ALL treatment through multi-targeted therapeutic strategies involving rapamycin and miR-3143/miR-3182.
Keywords: miRNA, PI3K/Akt/mTOR, miR-3143, miR-3182, T-cell acute lymphoblastic leukemia, Rapamycin
Copyright and License Information
© 2024 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) diagnosis and treatment have made considerable strides, but those who have refractory or recurrent leukemia, still face negative side effects and poor prognosis.1,2 Defective maturation processes are linked to differentiation arrest, hyper-proliferation, and accumulation of leukemia cells in the bone marrow and extra-medullary locations.3 Anemia, granulocytopenia, thrombocytopenia, and the suppression of physiologic hematopoiesis are among the clinical complications of this type of malignancy.4,5 The current standard of care for pediatric instances is combination chemotherapy; however, these powerful therapies frequently result in protracted side effects over time.6
The interaction between oncogenic signals and T cell growth signals is the most conspicuous feature of T-ALL. The proliferation of T cell progenitors is primarily controlled by the phosphatidyl 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signaling pathway, which plays a vital role in hematopoiesis.7 As thymocytes develop, NOTCH1 activates the PI3K/AKT/mTOR signaling pathway to regulate cell growth and proliferation.1,8 In more than 60% of T-ALL cases, gain-of-function (oncogenic) mutations activates NOTCH1, which results in the expansion, proliferation, and self-renewal of T cells through binding to the distal enhancer region of MYC and activating the PI3K/AKT/mTOR pathways in a feed-forward loop.9-11 Furthermore, PTEN inhibition by NOTCH1 promotes the development of T-ALL by preventing the activation of AKT signaling.12
The inhibition of PI3K/AKT/mTOR signaling has been shown to be effective in reducing the proliferation of T-ALL cells in vitro and increasing the survival rate of most T-ALL patients with activated NOTCH1 pathway.13 Research on clinical models and trials has demonstrated the anti-cancer properties of rapamycin, one of the mTOR inhibitors currently available.14 Molecular studies have revealed that rapamycin can induce G1 arrest and reduce S-phase entry in T-ALL cell lines, as well as decreasing cell proliferation and downregulating Cyclin D3 expression.15,16 Rapamycin may have limited anticancer effect as a monotherapy, while studies suggest that it can be highly beneficial in combination with other therapies, such as miRNA.17,18
Research has consistently demonstrated that microRNAs (miRNAs) play a critical role in the initiation and development of cancer.19,20 The fact that more than half of miRNA genes are located in "fragile sites" or genomic regions associated with cancer, indicates their profound stimulatory and inhibitory effects on tumors.19 Transferring mimic or inhibitory miRNAs, which are based on the manipulation of miRNA function, is one of the currently known treatment strategies for cancer.20-22
The effect of combination therapy strategies using rapamycin concurrently with PI3K/AKT/mTOR signaling pathway-inhibiting miRNAs in apoptosis induction in NOTCH1 overexpressed T-ALL cells was investigated in this study.
Materials and Methods
Bioinformatic analysis to predict miRNAs targeting main genes of mTOR signaling pathway
Utilizing bioinformatic tools, we predicted miRNAs that potentially target the mTOR signaling pathway in T-ALL. Initially, expression data on T-ALL was retrieved from the Gene Expression Omnibus (GEO) database, with the accession number GSE225751. GEO2R analysis was employed to identify the expression of mTOR, PI3K, AKT, and S6KB1 mRNAs after Everolimus23 (a rapamycin analog) treatment of Jurkat cells. The TargetScan was used for initial miRNA target prediction, followed by the use of DIANA tools,24 miRanda,25 and miRWalk.26 Ultimately, the most frequently predicted miRNAs in different tools with the highest scores were investigated in target validated tools like MiRTarBase.27
Primer design
To evaluate the expression of target genes (mTOR, PI3K, AKT, S6KB1) and candidate miRNAs ( miR-99a/100, miR-501-3p, miR-576-5p, miR-155, miR-3143, and miR-3182), the primers were designed using the Allele ID 7.0 and Oligo7 28 software. The sequences of primer pairs are shown in Table 1.
Table 1.
The sequences of primer-set in this study
|
miRNAs
|
Forward sequence (5ʹ → 3ʹ)
|
|
| miR-99a/100 |
CGCAACCCGTAGATCCGAA |
|
| miR-501-3p |
CAATGCACCCGGGCAAG |
|
| miR-576-5p |
GACCGCATTCTAATTTCTCCAC |
|
| miR-155 |
GCCGTTAATGCTAATCGTGATAG |
|
| miR-3143 |
CATAGCACGCGGGCTAG |
|
| miR-3182 |
GACCGGATTACATTCTATAGGC |
|
| U6 |
AAGGATGACACGCAAATTC |
|
| Universal reverse primer |
GACGTGGGACGGAGGT |
|
| Genes |
Forward sequence (5ʹ → 3ʹ)
|
Reverse sequence (5ʹ → 3ʹ)
|
| NOTCH1 |
CGAGCGGCCCTACGTG |
GTACAGTACTGACCTGTCCAC |
| AKT |
TGAGGAGCGGGAGGAGTG |
GAGGATCTTCATGGCGTAGTAGC |
| PI3K |
CCACGACCATCATCAGGTGAA |
CCTCACGGAGGCATTCTAAAGT |
| mTOR |
CAATGCTATGGAGGTTACAGG |
ATCGCTTGTTGCCTTTGG |
| S6KB1 |
CTTCCGAGACAGGGAAGCTG |
TCACAATGTTCCATGCCAAGTT |
| ACTB (β-Actin) |
CTTCCTTCCTTCCTGGGCATG |
GTCTTTGCGGATGTCCAC |
U6: Housekeeping non-coding RNA, ACTB (β-actin): housekeeping gene.
Literature reviews showed that the activation of the PI3K/AKT/mTOR pathways is a consequence of NOTCH1 over-activation. Twenty-five bone marrow samples of patients diagnosed with T-ALL were obtained from Peyvand Laboratory in Tehran, Iran. Informed consent was obtained from all the patients to participate in this study. The diagnosis of T-ALL was confirmed via combination of immuno-phenotyping, immuno-cytogenetic analysis, and cellular morphology. None of the patients had undergone chemotherapy or radiation therapy prior to sampling. The expression of NOTCH1 was investigated in clinical samples, and finally, 15 out of 25 samples that were confirmed to be NOTCH1overexpressed were recruited for further analysis. Five control bone marrow samples from non-leukemia patients were also provided.
RNA extraction, cDNA synthesis, and quantitative RT-PCR for miRNAs and their target genes expression analysis
To investigate the expression of miRNAs and target genes, TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., USA) was used to extract total RNA in accordance with the manufacturer's instructions. The spectrophotometer (MaestroNano Pro, Hong Kong) was employed to evaluate the quality and quantity of RNA. Reverse transcription kit (Thermo Fisher Scientific, USA) was used to create cDNA in accordance with the manufacturer's instructions. Stem-loop based cDNA synthesis kit for miRNAs’ expression analysis was purchased from AnaCell company (Tehran, Iran) and applied as previously described.28 Quantitative real-time PCR (qRT-PCR) was carried out to evaluate gene expression using SYBR Green-based method in StepOneTM instrument (ABI, USA). U6 and β-actin served as internal controls for miRNA and gene expression analyses, respectively. The relative expression levels of genes and miRNAs were calculated using comparative threshold cycles (∆∆Ct).
Cell culture
Jurkat cells (T-cell acute lymphoblastic leukemia cell lines) carrying early activating mutations in NOTCH129 was acquired from the Iranian Biological Resource Center in Tehran, Iran. The cells were cultured in RPMI-1640 medium (PAN biotech, Germany), supplemented with 10% fetal bovine serum (Gibco, USA), at 37 °C in a humidified atmosphere containing 5% carbon dioxide.
Evaluating the inhibitory concentration (IC50) of rapamycin in Jurkat cells using MTT assay
To evaluate the IC50 of rapamycin in Jurkat cells, the cells were seeded (50 000 cells/well) in 96 well plates and treated with the increasing concentrations of rapamycin for 24, 48, and 72 hours (10, 20, 40, 60, and 80 µM). After the treatment period, 100 µL of MTT dye (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, Sigma, USA) was added to each well. The plates were incubated in the dark for 4 hours to allow formazan crystal formation. Then, 100 µL of dimethyl sulfoxide (DMSO, Sigma, USA) was added to dissolve the formazan crystals. Finally, the absorbance of the solution was measured using a plate reader at 570 nm, with a reference wavelength of 630 nm.
Transfection of FAM-labeled scramble into the Jurkat cells
To achieve the best transfection efficiency in Jurkat cells, which are difficult to transfect, FAM-labeled control (Scramble) was obtained from EXIQON (Denmark). Jurkat cells were cultured in 6-well plates with a cell density of 250 000 per well and then incubated at 37 °C. These cells were subsequently transfected with 50 nM of FAM-labeled Scramble using TurboFectTM transfection reagent (Thermo Fisher, USA) for 48 hours. The effectiveness of oligo transfection was confirmed by examining the cells using a Nikon TE2000 fluorescent microscope 48 hours after the procedure. To further ensure transfection efficiency, FAM-labeled cells were counted using a flow cytometer (Beckman Coulter Inc. USA). The transfection protocol followed specific guidelines, and all subsequent tests were conducted under the same conditions for consistency.
microRNAs transfection and rapamycin treatment
MiR-3143 and miR-3182 mimics were purchased from EXIQON (Denmark), and utilized to evaluate miRNAs’ effects on cellular response. Two groups were transfected with each mimic using TurboFectTM for 48 hours at a concentration of 50 nM. To investigate the impact of rapamycin on the cells, one group was subjected to rapamycin treatment alone at its IC50 concentration, another two groups received the same treatment procedure while being transfected with each mimic, whereas the other group received a combination of all three drugs. Finally, a control group was included that underwent identical conditions to the experimental groups without any treatment.
Rapamycin and miRNAs’ effects on apoptosis of Jurkat cells
To evaluate the effect of rapamycin treatment and miRNAs on cell apoptosis, the proportion of cells that remained alive following the intended treatment was indicated by staining the cells with Annexin V–FITC (BD Biosciences, Bedford, MA, USA). The assay relies on the ability of Annexin V, a calcium-dependent phospholipid-binding protein, to bind specifically to phosphatidylserine (PS) that is exposed on the outer surface of the plasma membrane during early stages of apoptosis. The assay involves incubating the cells with Annexin V-FITC and a vital dye such as propidium iodide (PI) that stains necrotic cells. The cells are then analyzed by flow cytometry to determine the proportion of Annexin V-positive cells, which are considered apoptotic, and PI-positive cells, which are considered necrotic.
Rapamycin and miRNAs’ effects on cell cycle of Jurkat cells
To evaluate the effect of rapamycin treatment and miRNAs on the cell progression, cell cycle assay was performed. This assay facilitates the identification of alterations in DNA content that occur during G1, S, G2, and M phases of cell cycle. Propidium iodide (PI) dye (BD Biosciences, Bedford, Massachusetts, United States) binds to DNA without sequence specificity. In cell cycle analysis, PI is added to cells subsequent to their fixation and permeabilization, and the resulting fluorescence intensity is directly proportional to the amount of DNA present in each cell.
Statistical analyses
All tests were done in duplicate, and findings were presented as the mean standard deviation. ANOVA and Student’s t-test were utilized to compare the groups with normally-distributed data. GraphPad Prism software was utilized for the statistical analysis (Prism application, GraphPad Software, San Diego, CA.). Statistical significance was defined as P< 0.05.
Results
Selection of candidate miRNAs
GEO2R analysis of GSE225751 showed that the main genes of mTOR signaling pathway were downregulated after Everolimus treatment. Using prediction methods, the miRNAs which target the 3'-UTR of PI3K, Akt, mTOR, and S6KB1 mRNAs were retrieved; finally, the six miRNAs with the highest scores, highest number of repetitions, and highest number of predictions across multiple databases were chosen and listed in Table 2. All putative miRNAs contain at least one binding site on the 3′-UTR of their respective mRNA, as determined by bioinformatics and sequence alignment analyses.
Table 2.
Candidate miRNAs and their target genes from PI3K/AKT/mTOR pathway
|
miRNA
|
Target gene
|
TargetScan
|
miRanda
|
microCosm
|
microPIR
|
miRDB
|
miRSearch
|
miRWalk
|
DIANA-microT
|
PicTar
|
TarBase
|
miRTarBase
|
miRNAMap
|
MirZ
|
SUM
|
|
miR-99a/100
|
mTOR |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
3
|
|
miR-3182
|
mTOR |
1 |
1 |
0 |
1 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
1 |
6
|
|
miR-155
|
S6Kb1 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
3
|
|
miR-3182
|
S6Kb1 |
1 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
4
|
|
miR-3143
|
PIK3CA |
1 |
0 |
0 |
1 |
0 |
1 |
1 |
0 |
1 |
0 |
1 |
0 |
0 |
6
|
|
miR-576-5p
|
PI3K |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
3
|
|
miR-3143
|
AKT |
1 |
1 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
5
|
|
miR-501-3p
|
AKT |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
3
|
Interaction was indicated by a score of 1 and its absence by a score of 0. The SUM displays the total number of prediction/validation of miRNA: mRNA interactions in thirteen bioinformatic tools.
Selection of NOTCH1-overexpressed clinical samples
The PI3K/AKT/mTOR pathway is activated by mutant NOTCH1, which promotes leukemia cell growth and proliferation. To investigate this further, we recruited 15 clinical samples with NOTCH1-overexpression from a total of 25 T-ALL samples. The expression levels of selected candidate miRNAs were evaluated in these samples to determine their potential roles in modulating the downstream effects of NOTCH1 overexpression.
Declined expression of miR-3143 and miR-3182 in NOTCH1-overexpressed T-ALL samples
The expression levels of miR-155, miR-576-5p, miR-501-3p, miR-3143, miR-3182, and miR-99a/100 were investigated in 15 NOTCH1-overexpressed clinical samples compared with healthy samples. The results showed a significant decrease in the expression of miR-3143, miR-3182, and miR-501-3p by approximately a thousand-fold when compared to the control group (****P<0.0001) (Fig. 1B). These findings suggest potential roles for these miRNAs in regulating the downstream effects of mutant NOTCH1 and warrant further investigation into their therapeutic potential for leukemia treatment.
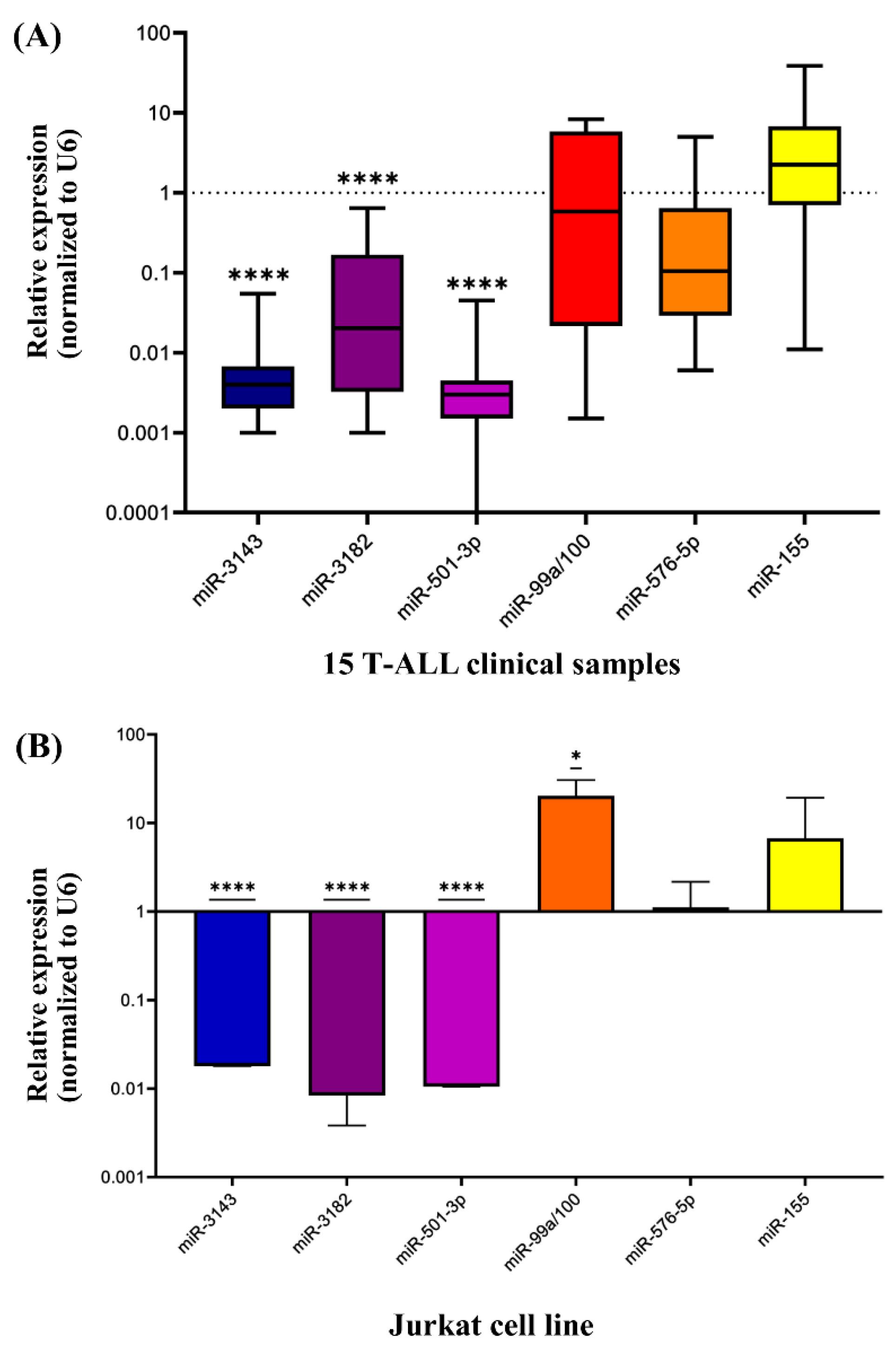
Fig. 1.
(A) Expression of miR-155, miR-576-5p, miR-501-3p, miR-3143, miR-3182, miR-99a/100 in 15 NOTCH1 overexpressed T-ALL clinical samples. The expression of miR-3143, miR-3182 and miR-501-3p showed a significant decrease in comparison with healthy normal samples by about a thousand-fold (****P<0.0001). Two independent experiments were performed in duplicates (n=2). (B) Expression of miR-155, miR-576-5p, miR-501-3p, miR-3143, miR-3182, miR-99a/100 in Jurkat cells. The expression of miR-3143, miR-3182, and miR-501-3p indicated a significant decrease compared to healthy normal samples (**** P<0.0001). Three independent experiments were performed in triplicates (n=3).
.
(A) Expression of miR-155, miR-576-5p, miR-501-3p, miR-3143, miR-3182, miR-99a/100 in 15 NOTCH1 overexpressed T-ALL clinical samples. The expression of miR-3143, miR-3182 and miR-501-3p showed a significant decrease in comparison with healthy normal samples by about a thousand-fold (****P<0.0001). Two independent experiments were performed in duplicates (n=2). (B) Expression of miR-155, miR-576-5p, miR-501-3p, miR-3143, miR-3182, miR-99a/100 in Jurkat cells. The expression of miR-3143, miR-3182, and miR-501-3p indicated a significant decrease compared to healthy normal samples (**** P<0.0001). Three independent experiments were performed in triplicates (n=3).
Loss of miR-3143 and miR-3182 in T-ALL cell line ( Jurkat cells)
The expression pattern of miR-155, miR-576-5p, miR-501-3p, miR-3143, miR-3182, and miR-99a/100 in T-ALL cell line were confirmed against healthy controls. As shown in Fig. 1B, miR-3143, miR-3182, and miR-501-3p exhibited a significant decrease in expression levels by almost a thousand-fold when compared to the normal control (****P<0.0001). Notably, this expression pattern closely resembled their expression levels in clinical samples (Fig. 1A), suggesting potential roles for these miRNAs in leukemia development. Due to their ability to target and regulate multiple key genes in the PI3K/AKT/mTOR pathway, which is activated by mutant NOTCH1 and plays a critical role in leukemia cell growth and proliferation, miR-3143 and miR-3182 were chosen for transfection analysis.
Overexpression of PI3K, AKT, mTOR, S6KB1 genes by QRT-PCR in Jurkat cell line
Relative expression levels of target genes (PI3K, AKT, mTOR, and S6KB1) in Jurkat cells were compared with those of healthy control samples. The results obtained from qRT-PCR analysis showed a significant increase in the expression levels of PI3K, AKT, and mTOR genes in the Jurkat cell line when compared to the normal controls (*P <0.05) (Fig. 2).
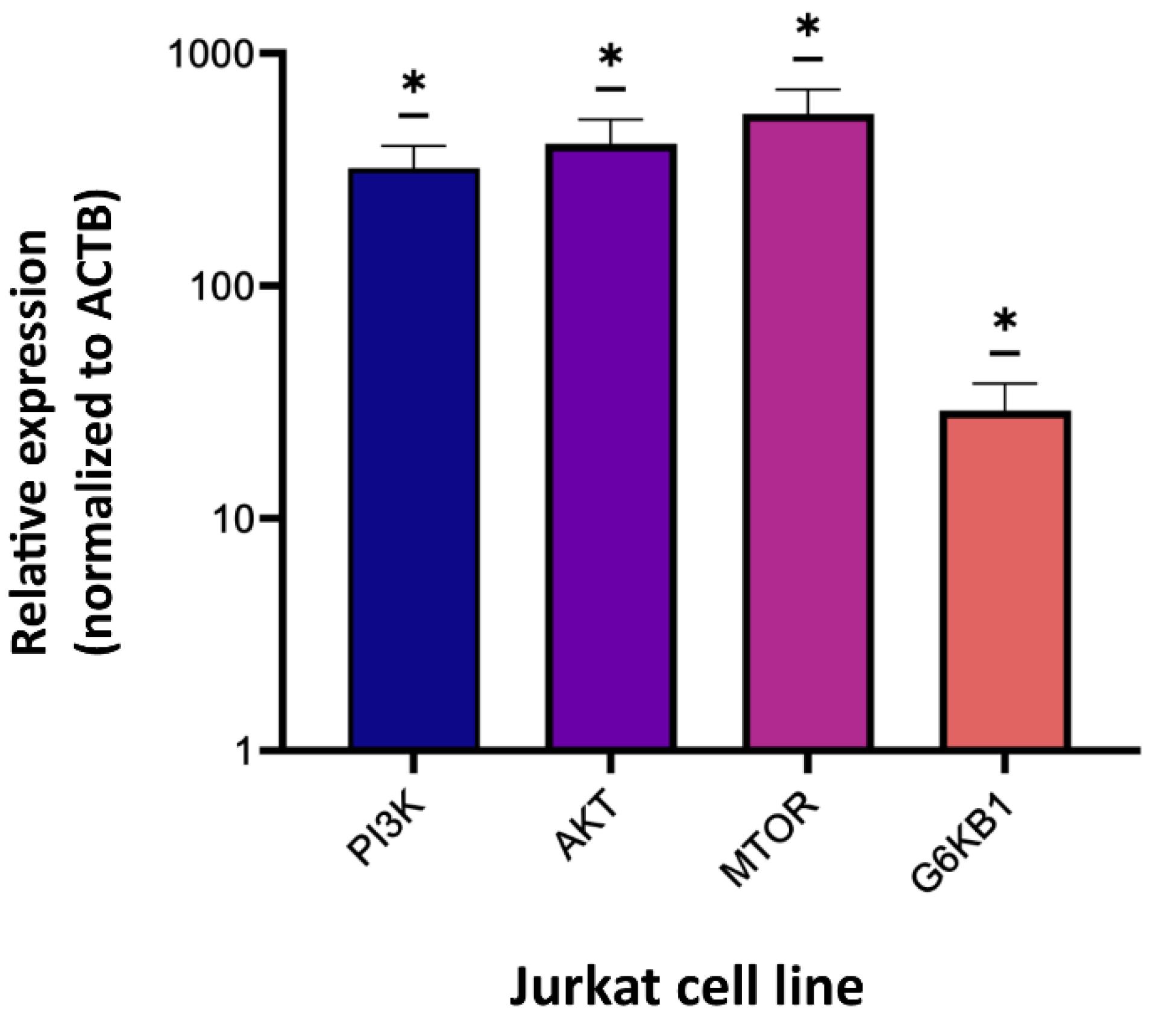
Fig. 2.
Expression of PI3K, AKT, mTOR, and S6KB1 genes in Jurkat cells. The expression of PI3K, AKT, mTOR, and S6KB1 genes in Jurkat cells indicated a significant increase compared to control samples (*P<0.05). Three independent experiments were done in triplicates (n=3).
.
Expression of PI3K, AKT, mTOR, and S6KB1 genes in Jurkat cells. The expression of PI3K, AKT, mTOR, and S6KB1 genes in Jurkat cells indicated a significant increase compared to control samples (*P<0.05). Three independent experiments were done in triplicates (n=3).
Anti-proliferative effect of rapamycin on Jurkat cells
Jurkat cells were treated with varying concentrations of rapamycin (5-80 µM) for 24, 48, or 72 hours, and cell viability was assessed using the MTT assay. The results showed that after exposure to rapamycin, the viability of Jurkat cells was significantly reduced in a dose- and time-dependent manner (Fig. 3A). Notably, after 48 hours of treatment, the concentration of rapamycin at 13 µM had a 50% inhibitory effect, resulting in the calculation of 13 µM as the IC50 of rapamycin (Fig. 3B and 4C). Accordingly, this concentration was chosen for further investigations. To ensure the accuracy and reliability of our findings, we conducted two independent experiments in quadruplicate (n=2).
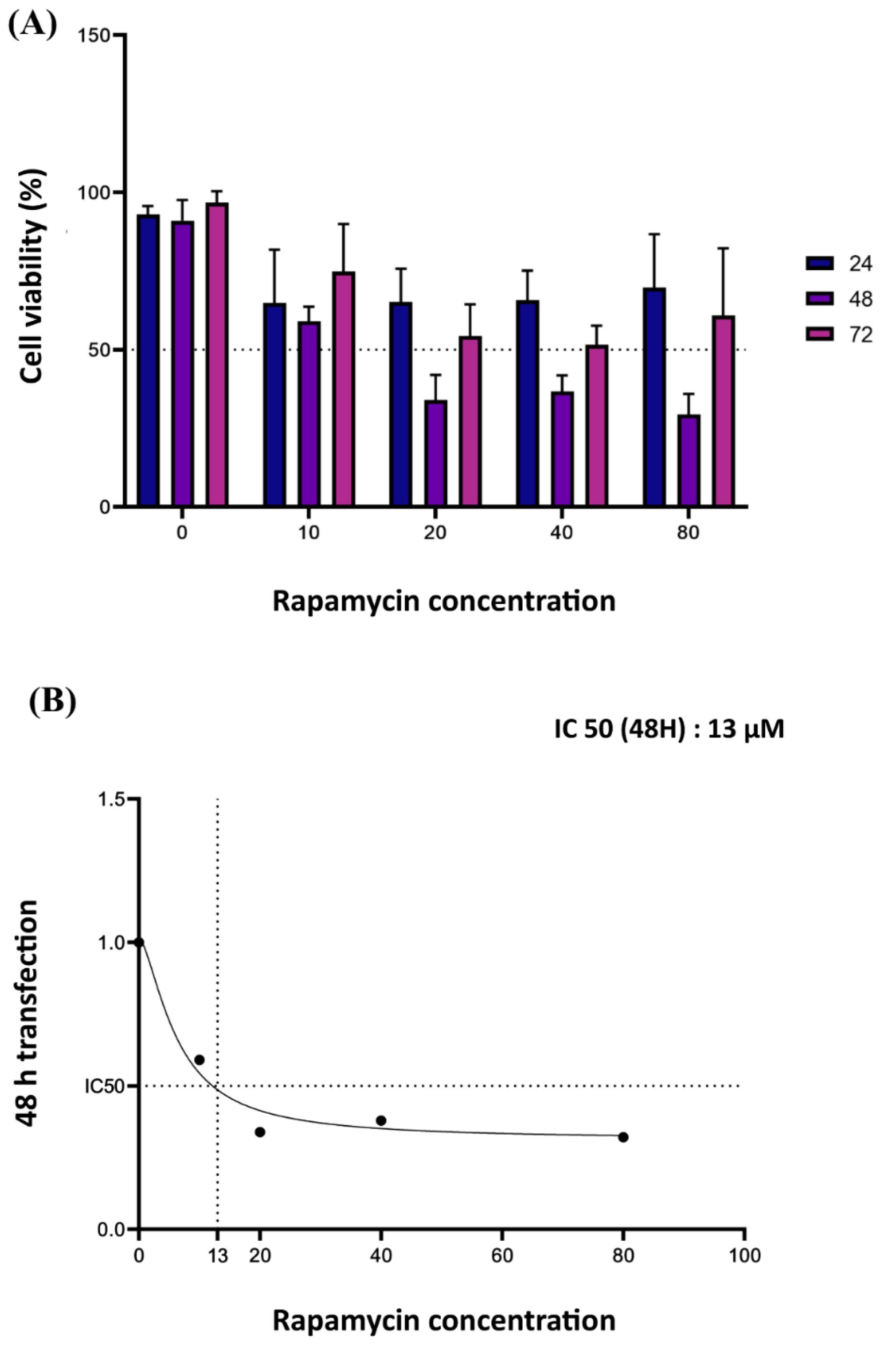
Fig. 3.
(A) MTT assay on Jurkat cells showed that the viability of Jurkat cells was significantly reduced after exposure to rapamycin in a dose- and time-dependent manner. (B) The colorimetric assays indicated that rapamycin with a concentration of 13 μM can inhibit cell proliferation by 50% (IC50) 48 hours after treatment. Two independent experiments were done in quadruplicates (n=2).
.
(A) MTT assay on Jurkat cells showed that the viability of Jurkat cells was significantly reduced after exposure to rapamycin in a dose- and time-dependent manner. (B) The colorimetric assays indicated that rapamycin with a concentration of 13 μM can inhibit cell proliferation by 50% (IC50) 48 hours after treatment. Two independent experiments were done in quadruplicates (n=2).
Efficient transfection of oligos into Jurkat cells using TurboFectTM reagent
Jurkat cells were transfected with 50 nM of FAM-labeled Scramble using TurboFectTM transfection reagent (Thermo Fisher, USA). The effectiveness of the transfection was assessed by examining the cells under a fluorescent microscope. The results showed that the majority of cells exhibited a luminous color, indicating successful entry of Scramble into the cells (Fig. 4A). To further confirm the transfection efficiency, flow cytometry was performed to determine the exact number of FAM-labeled (transfected) cells. The data revealed that more than 75% of Jurkat cells were successfully transfected with FAM-labeled Scramble (Fig. 4B). It should be noted that transfection of suspension cells can be a challenging and inefficient process; fortunately, our optimized transfection procedure was able to achieve a notably high transfection rate.
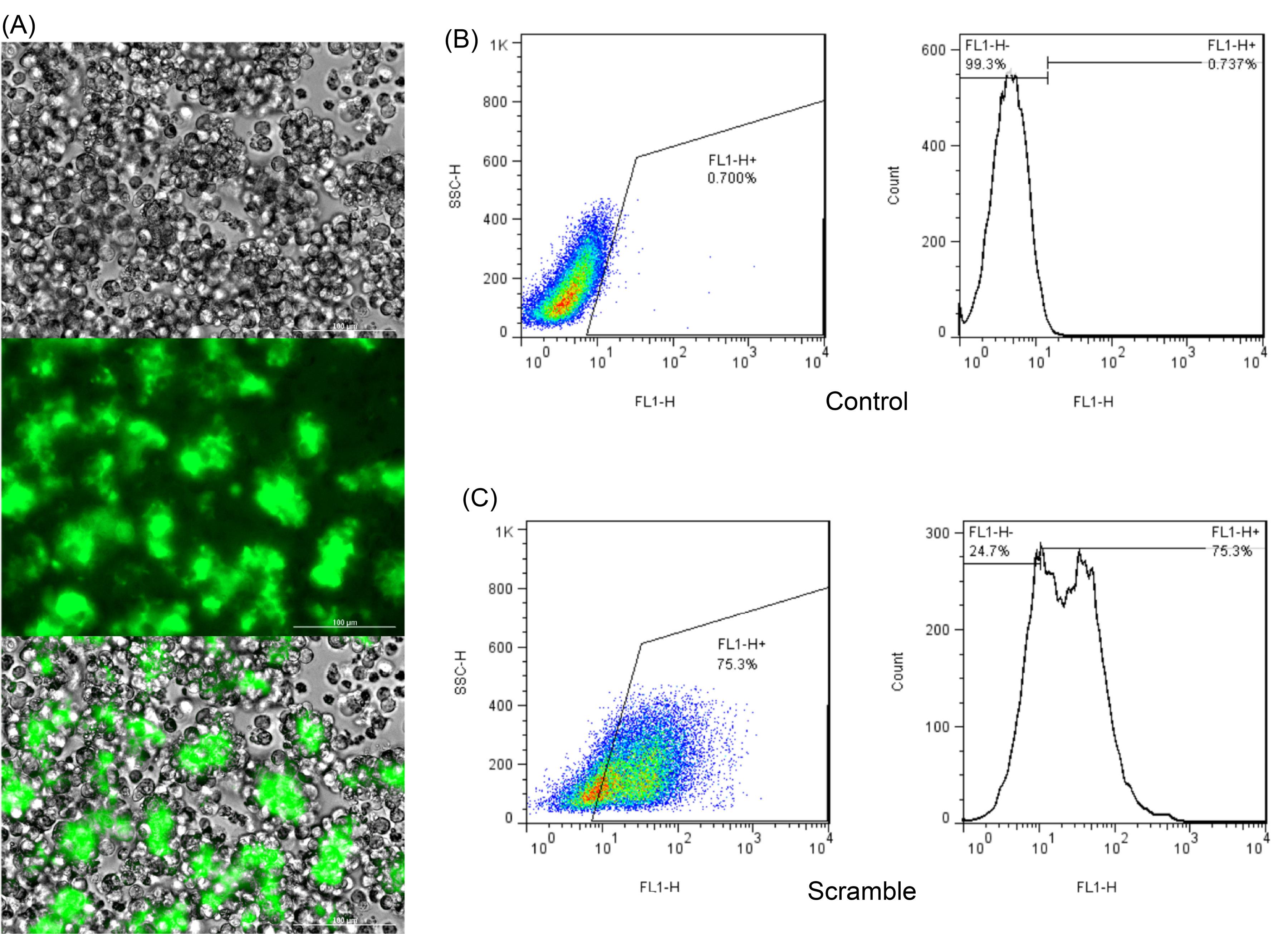
Fig. 4.
(A) Establishment of transient transfection into the Jurkat cell line. To validate the transfection efficacy, cells were transfected with FAM-labeled Scramble and examined under a fluorescent microscope 48 hours post transfection applying TurboFectTM. According to the observations, high percentage of Jurkat cells were FAM-labeled 48 hours after treatment. (B) Counting the FAM-labeled cells (FL1-H+: transfected cells) using flow cytometer in un-transfected cells (negative control, the number is below 1 that shows no transfection); (C) Counting the FAM-labeled cells (FL1-H+: transfected cells) using flow cytometer showed that more than 75% of Jurkat cells were transfected with FAM-labeled Scramble.
.
(A) Establishment of transient transfection into the Jurkat cell line. To validate the transfection efficacy, cells were transfected with FAM-labeled Scramble and examined under a fluorescent microscope 48 hours post transfection applying TurboFectTM. According to the observations, high percentage of Jurkat cells were FAM-labeled 48 hours after treatment. (B) Counting the FAM-labeled cells (FL1-H+: transfected cells) using flow cytometer in un-transfected cells (negative control, the number is below 1 that shows no transfection); (C) Counting the FAM-labeled cells (FL1-H+: transfected cells) using flow cytometer showed that more than 75% of Jurkat cells were transfected with FAM-labeled Scramble.
Combination of miR-3143, miR-3182 and rapamycin significantly promotes Jurkat cell apoptosis
In this study, the induction of apoptosis was investigated using flow cytometric analysis of Annexin V-fluorescein-isothiocyanate-stained cells (Fig. 5A-5G). Following treatment with rapamycin at a concentration of 13 μM for 48 hours, there was a significant increase in the proportion of late apoptotic cells as observed in the upper right quadrant of the flow cytometry plot (Fig. 5D). These findings suggest that rapamycin has the ability to trigger cellular apoptosis. Moreover, when miR-3143 and miR-3182 were combined with rapamycin and transfected into Jurkat cells, the same increase in apoptosis was observed compared to other study groups and the percentage of apoptotic cells reached approximately 60%, which is notably higher than the control (**P<0.0001) (Fig. 5G). Overall, these data suggest that the combination of miR-3143, miR-3182, and rapamycin can effectively induce apoptosis in Jurkat cells.
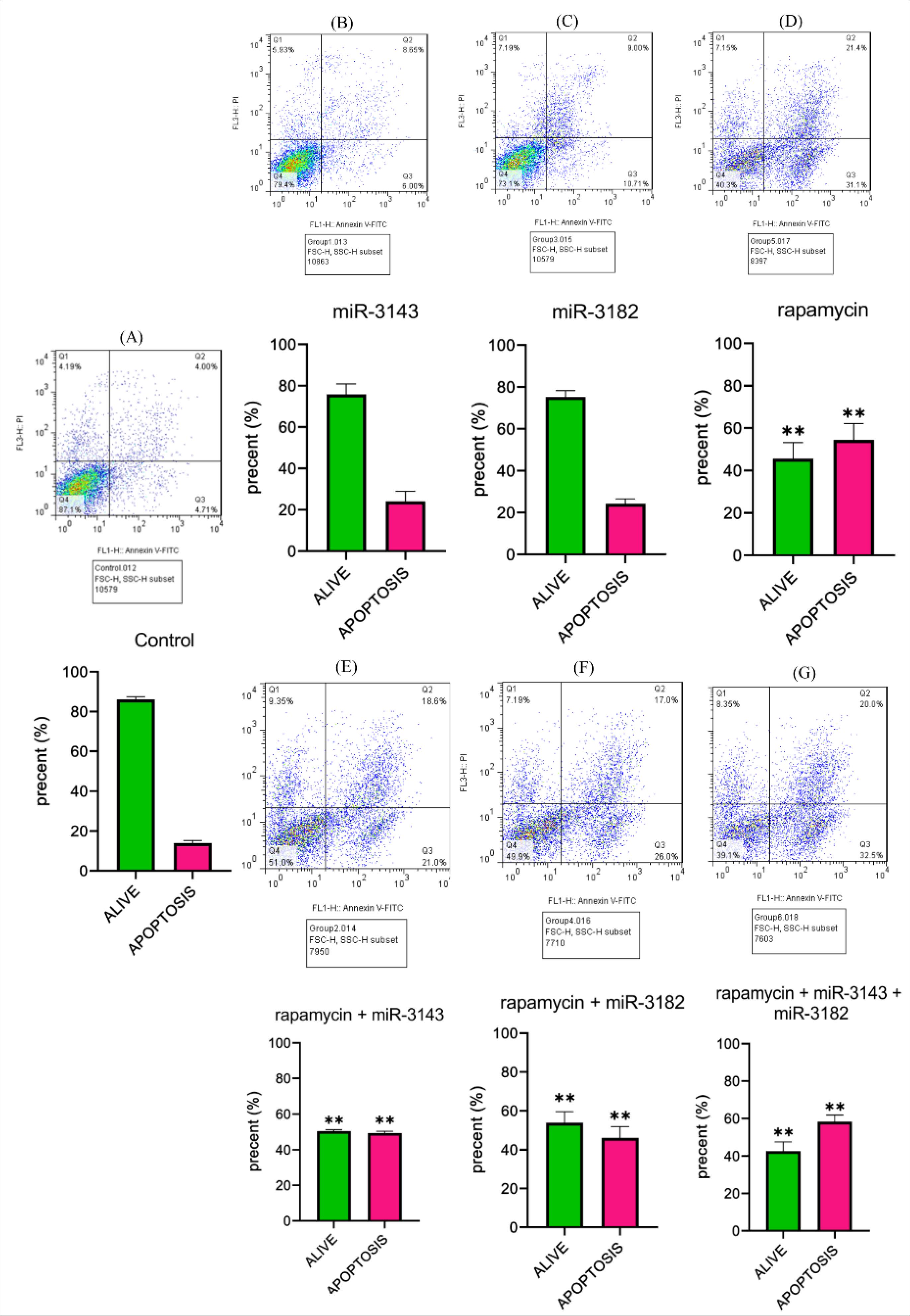
Fig. 5.
Apoptosis assay in seven different study groups: (A) Non-transfected Jurkat cells, (B) Jurkat cells transfected with miR-3143 mimic, (C) Jurkat cells transfected with miR-3182, (D) Jurkat cells transfected with rapamycin(IC50=13µM), (E) Jurkat cells transfected with miR-3143 mimic plus rapamycin, (F) Jurkat cells transfected with miR-3182 mimic plus rapamycin, and (G) Jurkat cells transfected with miR-3143 and miR-3182 plus rapamycin. Our results indicated that the group treated with rapamycin showed a significant increase in apoptosis in the Jurkat cell line (**P<0.0001). This pattern was also observed in the groups treated with miR-3143 plus rapamycin, miR-3182 plus rapamycin, and miR-3143 and miR-3182 plus rapamycin (**P<0.0001). The statistical data presented in this graph represent the findings from two independent experiments(n=2).
.
Apoptosis assay in seven different study groups: (A) Non-transfected Jurkat cells, (B) Jurkat cells transfected with miR-3143 mimic, (C) Jurkat cells transfected with miR-3182, (D) Jurkat cells transfected with rapamycin(IC50=13µM), (E) Jurkat cells transfected with miR-3143 mimic plus rapamycin, (F) Jurkat cells transfected with miR-3182 mimic plus rapamycin, and (G) Jurkat cells transfected with miR-3143 and miR-3182 plus rapamycin. Our results indicated that the group treated with rapamycin showed a significant increase in apoptosis in the Jurkat cell line (**P<0.0001). This pattern was also observed in the groups treated with miR-3143 plus rapamycin, miR-3182 plus rapamycin, and miR-3143 and miR-3182 plus rapamycin (**P<0.0001). The statistical data presented in this graph represent the findings from two independent experiments(n=2).
Combination of miR-3143, miR-3182, and rapamycin induces cell cycle arrest in Jurkat cells
The present study utilized flow cytometry to evaluate cell cycle indices and establish the precise mechanism by which rapamycin and miRNAs inhibit cell viability. The obtained results, as shown in Fig. 6A, indicate that rapamycin therapy induced cell cycle arrest at the G0/G1 phase and prevented entry into the S phase when compared to the control group. Similarly, miR-3143, miR-3182, and rapamycin group significantly induced cell cycle arrest at the G0/G1 phase (*P < 0.05). These findings suggest that the combination of miR-3143, miR-3182, and rapamycin may cause cell cycle arrest by regulating the expression of key proteins involved in the regulation of G0/G1 phase in cells. As illustrated in Fig. 6B, relevant statistical comparisons are provided to support these observations.
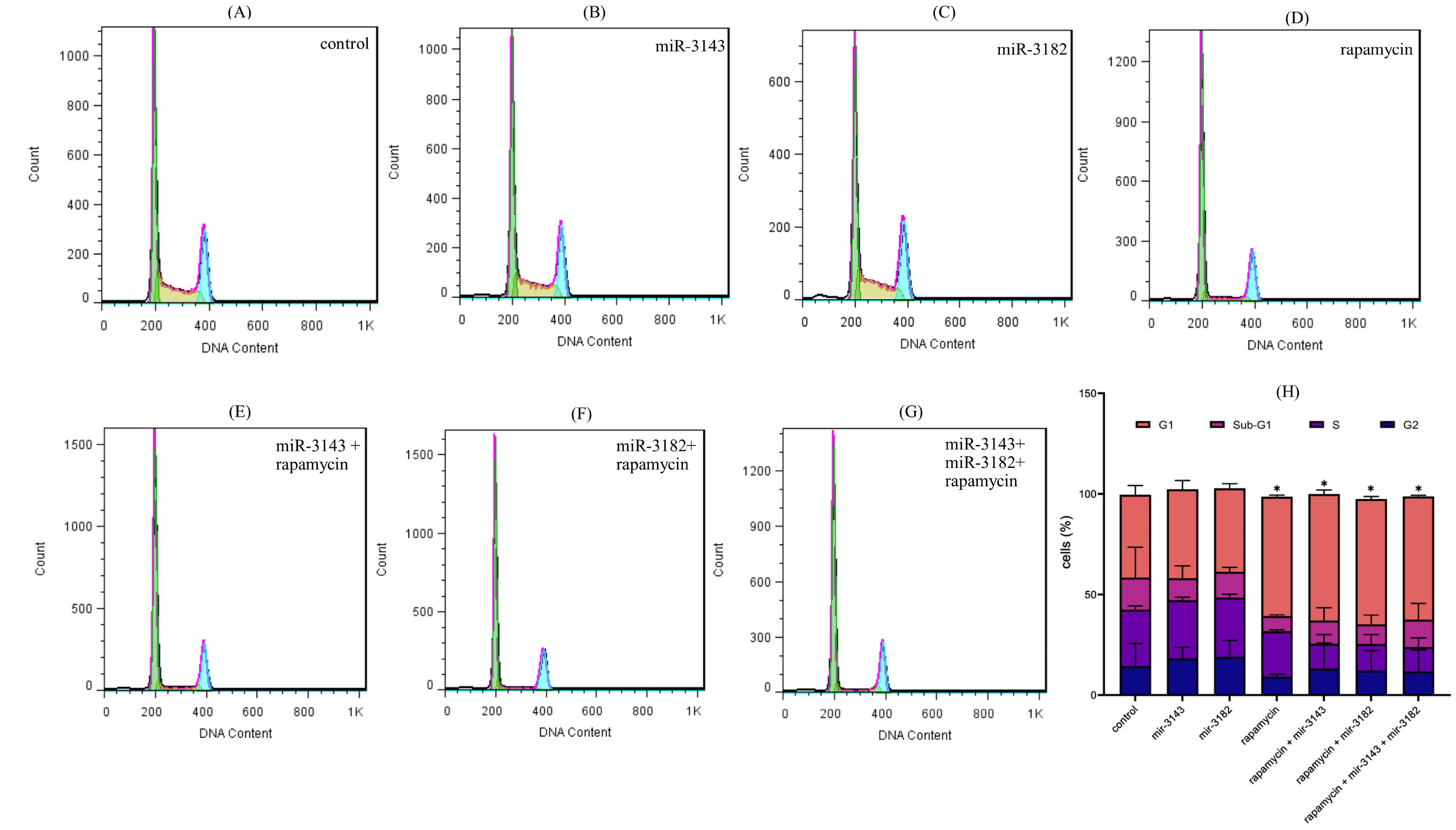
Fig. 6.
A) Cell cycle assay in seven different study groups: (A) Non-transfected Jurkat cells, (B) Jurkat cells transfected with miR-3143 mimic, (C) Jurkat cells transfected with miR-3182, (D) Jurkat cells transfected with rapamycin (IC50=13µM), (E) Jurkat cells transfected with miR-3143 mimic plus rapamycin, (F) Jurkat cells transfected with miR-3182 mimic plus rapamycin, and (G) Jurkat cells transfected with miR-3143 and miR-3182 plus rapamycin. (H) The statistical data from the cell cycle assay which indicated that compared with the control group, rapamycin group significantly induced G1/sub G1 arrest and reduced S phase entry. Exactly the same pattern was observed for miR-3143 plus rapamycin and miR-3143 plus rapamycin. Furthermore, it was found that miR-3143, miR-3182, and rapamycin significantly induced G1/sub G1 arrest (* P<0.05). The statistical data presented in this graph represent the findings from two independent experiments (n=2).
.
A) Cell cycle assay in seven different study groups: (A) Non-transfected Jurkat cells, (B) Jurkat cells transfected with miR-3143 mimic, (C) Jurkat cells transfected with miR-3182, (D) Jurkat cells transfected with rapamycin (IC50=13µM), (E) Jurkat cells transfected with miR-3143 mimic plus rapamycin, (F) Jurkat cells transfected with miR-3182 mimic plus rapamycin, and (G) Jurkat cells transfected with miR-3143 and miR-3182 plus rapamycin. (H) The statistical data from the cell cycle assay which indicated that compared with the control group, rapamycin group significantly induced G1/sub G1 arrest and reduced S phase entry. Exactly the same pattern was observed for miR-3143 plus rapamycin and miR-3143 plus rapamycin. Furthermore, it was found that miR-3143, miR-3182, and rapamycin significantly induced G1/sub G1 arrest (* P<0.05). The statistical data presented in this graph represent the findings from two independent experiments (n=2).
PI3K, AKT, mTOR, S6KB1 gene expression significantly decreased in Jurkat cells transfected with miR-3143, miR-3182 mimic and rapamycin
The study groups, including Jurkat cells transfected with miR-3143 mimic, Jurkat cells transfected with miR-3182, Jurkat cells transfected with rapamycin, Jurkat cells transfected with miR-3143 mimic plus rapamycin, Jurkat cells transfected with miR-3182 mimic, and Jurkat cells transfected with miR-3143 and miR-3182 plus rapamycin underwent qRT-PCR analysis of PI3K, AKT, mTOR, and S6KB1. TurboFectTM reagent was utilized to transfect miR-3143, miR-3182, and their respective combinations into Jurkat cells. The results indicated that the miR-3143 mimic-transfected group showed a significant reduction in the expression levels of PI3K, AKT, mTOR, and S6KB1 (****P<0.0001) compared to non-transfected Jurkat cells (Fig. 7A). In contrast, the miR-3182 mimic group only exhibited decreased expression levels of mTOR and S6KB1 compared to non-transfected Jurkat cells (***P<0.001) (Fig. 7B). Rapamycin was applied to regulate the expression levels of the mTOR gene and its downstream signaling pathway in Jurkat cells. The results showed a considerable decrease in mTOR expression (****P <0.0001) and a significant increase in PI3K and S6KB1 gene expression (**P <0.01) in the rapamycin-treated group (Fig. 7C). The results also revealed a significant decrease in these genes' expression levels in the group that received both rapamycin and miR-3143 (P<0.001, P<0.0001) compared to the control group (Fig. 7D). Similar findings were observed in the group that received miR-3182 plus rapamycin (Fig. 7E). It is noteworthy that the combined treatment with miR-3143, miR-3182, and rapamycin resulted in a greater reduction in the expression levels of the target genes compared to the groups that received rapamycin or any of the miRNAs alone (P<0.0001) (Fig. 7F).
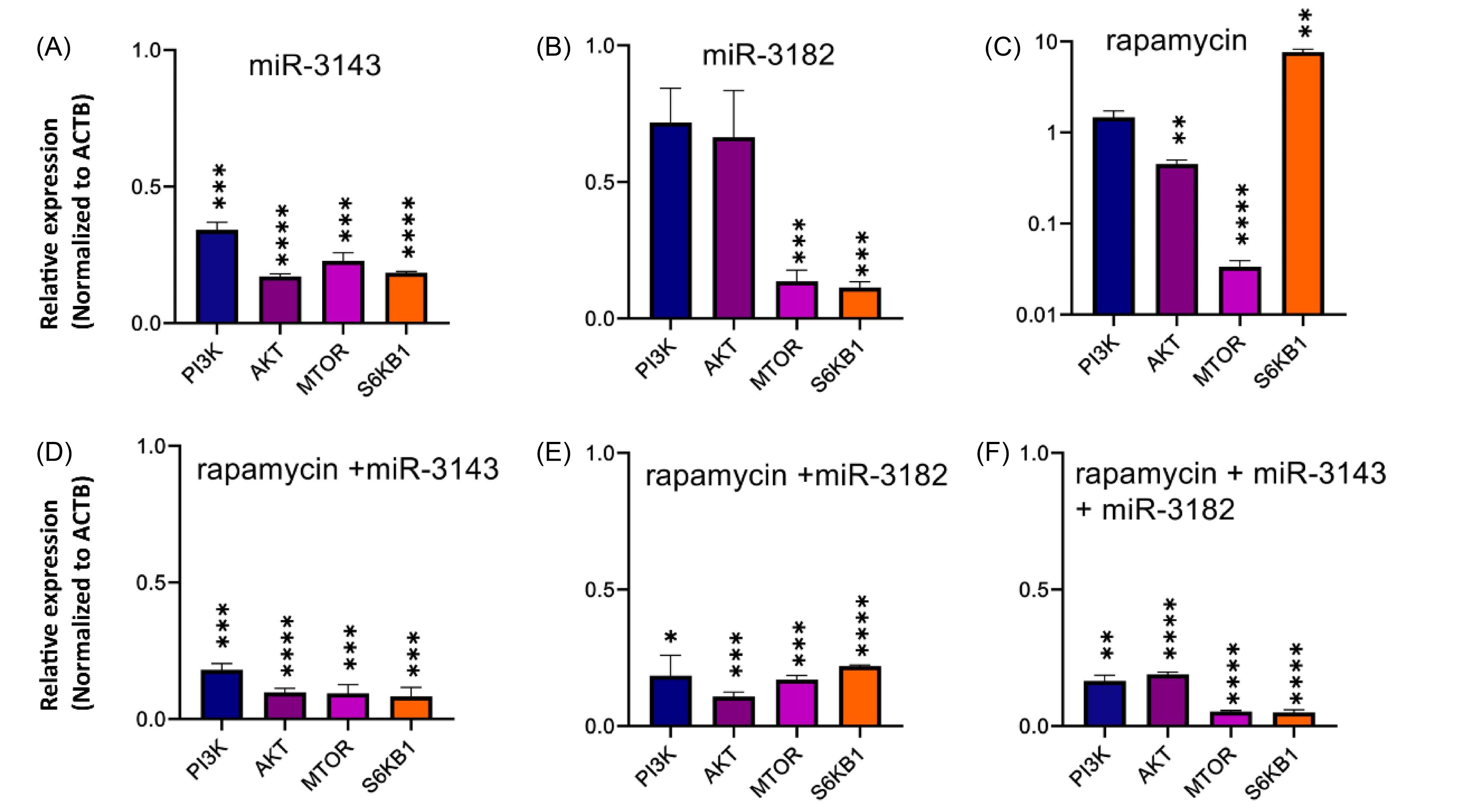

Fig. 7.
The expression level of PI3K, AKT, mTOR, and S6KB1 genes in study groups. (A) Jurkat cells transfected with miR-3143, the expression level of PI3K, AKT, mTOR and S6KB1 genes indicated a significant reduction compared to the control (***P<0.001, **** P<0.0001). (B) The expression level of mTOR and S6Kb1 genes in Jurkat cells transfected with miR-3182 indicated a considerable decrease compared to non-transfected cells ( *** P<0.001). (C) In Jurkat cells treated with rapamycin, the expression of mTOR genes was significantly reduced ( **** P<0.0001), while the expression of S6KB1 and AKT was significantly increased compared to the control ( ** P<0.01). (D) In Jurkat cells transfected with miR-3143 and treated with rapamycin, the mRNA expression of PI3K, AKT, mTOR, and S6KB1 genes was significantly decreased ( *** P<0.001, **** P<0.0001). (E) In the Jurkat cells transfected with miR-3182 and treated with rapamycin, the expression of PI3K, AKT, mTOR, and S6KB1 was significantly reduced (*P<0.05, ***P<0.001, ****P<0.0001). (F) In the Jurkat cells that were simultaneously transfected with rapamycin, miR-3143 and miR-3182, the expression level of all target genes (PI3K, AKT, mTOR, and S6KB1) especially mTOR and S6KB1 showed a significant decrease compared with control and single/double treatment ( ****P<0.0001). All the results were compared to the control untreated Jurkat cells.
.
The expression level of PI3K, AKT, mTOR, and S6KB1 genes in study groups. (A) Jurkat cells transfected with miR-3143, the expression level of PI3K, AKT, mTOR and S6KB1 genes indicated a significant reduction compared to the control (***P<0.001, **** P<0.0001). (B) The expression level of mTOR and S6Kb1 genes in Jurkat cells transfected with miR-3182 indicated a considerable decrease compared to non-transfected cells ( *** P<0.001). (C) In Jurkat cells treated with rapamycin, the expression of mTOR genes was significantly reduced ( **** P<0.0001), while the expression of S6KB1 and AKT was significantly increased compared to the control ( ** P<0.01). (D) In Jurkat cells transfected with miR-3143 and treated with rapamycin, the mRNA expression of PI3K, AKT, mTOR, and S6KB1 genes was significantly decreased ( *** P<0.001, **** P<0.0001). (E) In the Jurkat cells transfected with miR-3182 and treated with rapamycin, the expression of PI3K, AKT, mTOR, and S6KB1 was significantly reduced (*P<0.05, ***P<0.001, ****P<0.0001). (F) In the Jurkat cells that were simultaneously transfected with rapamycin, miR-3143 and miR-3182, the expression level of all target genes (PI3K, AKT, mTOR, and S6KB1) especially mTOR and S6KB1 showed a significant decrease compared with control and single/double treatment ( ****P<0.0001). All the results were compared to the control untreated Jurkat cells.
Discussion
The PI3K/Akt/mTOR signaling pathway has been found to play a crucial role in the pathogenesis of T-ALL by promoting cell proliferation, survival, and resistance to chemotherapy. Rapamycin is a potent mTOR inhibitor that has been extensively studied for its effects on T-ALL cell lines. Studies have shown that rapamycin can induce cell cycle arrest and apoptosis in T-ALL cells by inhibiting downstream effectors of the PI3K/Akt/mTOR pathway, including p70S6 kinase and 4E-BP1.30 Other studies have reported that rapamycin can sensitize T-ALL cells to conventional chemotherapeutic agents by enhancing their cytotoxic effects. Several clinical trials have employed this approach, with rapamycin undergone a clinical trial for patients with advanced acute leukemia (NCT00874562).30,31 MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression at the post-transcriptional level. Several miRNAs have been identified as regulators of the PI3K/Akt/mTOR pathway in T-ALL.32-34 Combination therapy using miRNAs and rapamycin has been explored as a potential approach to enhance the therapeutic efficacy of mTOR inhibitors in T-ALL. For example, a study, investigating the use of doxorubicin and miR-34a as a combination therapy for T-ALL, showed that the combination of doxorubicin with miR-34a led to a significant decrease in cell viability compared to either agent alone.35 By targeting multiple components of the PI3K/Akt/mTOR pathway, combination therapy may be more effective and overcome drug resistance. The present study aimed to investigate the potential cytotoxic effects of rapamycin and PI3K/AKT/mTOR inhibitory miRNAs, alone or in combination, against NOTCH1-driven T-ALL. Through the use of bioinformatic tools, we identified six miRNAs that target the PI3K/AKT/mTOR pathway, including miR-3143, miR-3182, miR-99a/100, miR-155, miR-576-5p, and miR-501-3. Further analysis revealed that miR-3143 and miR-3182 were down-regulated in both clinical samples and the Jurkat human T-cell leukemia cell line. Using TurboFectTM reagent, miR-3143 and miR-3182 were over-expressed in Jurkat cells. To our knowledge, the functions and effects of these miRNAs have not been previously investigated in hematological malignancies. We found that PI3K, AKT, mTOR, and S6KB1 could be direct downstream targets of miR-3143 and miR-3182, and there is a negative correlation between these miRNAs and PI3K/AKT/mTOR genes. Interestingly, treatment with rapamycin led to a significant increase in the expression of PI3K/S6KB1 genes, while markedly decreasing mTOR gene expression may be attributed to a compensatory mechanism triggered by mTOR downregulation and linked to T-ALL recurrence. In contrast, transfection of Jurkat cells with miR-3143 or miR-3182 and subsequent treatment with rapamycin resulted in a significant decrease in the mRNA expression of PI3K, AKT, mTOR, and S6KB1 genes. Furthermore, this downregulation was more pronounced in cells treated with both miRNAs and rapamycin, compared to cells treated with either agent alone, indicating the additive effects of these compounds on the expression levels of PI3K/AKT/mTOR genes. Interestingly, co-transfection of miR-3143 and miR-3182 with rapamycin into Jurkat cells led to a significant increase in the percentage of apoptotic cells and cells in the G0/G1 phase, suggesting enhanced cytotoxicity. However, this effect was not statistically different when miR-3143 or miR-3182 were transfected alone. These findings suggest that combination therapy using miRNAs and rapamycin may be a promising approach for enhancing the therapeutic efficacy of mTOR inhibitors in the treatment of T-ALL. Recent studies demonstrated that blocking mTOR kinase with rapamycin leads to the dephosphorylation of critical downstream signaling proteins, p70S6K, which limits cell cycle proliferation and causes G1 growth arrest.15,16 The mTOR blockage causes nuclear localization of the p27 inhibitor in T-ALL cells, according to a second functional study utilizing rapamycin. Gu et al demonstrated that rapamycin induces G0/G1 cell cycle arrest and upregulates p21 and p27 inhibitors while downregulating cyclin D1 in T-ALL cell lines.30 Kong et al revealed that rapamycin arrests the cell cycle in T-ALL cells by lowering MYCN mRNA and protein expression in T-ALL patients and T-ALL cell line.36 The results of the presented study are totally compatible with the aforementioned findings. In a study conducted by Evangelisti et al, the PI3K/AKT/mTOR axis transcription inhibitor ZSTK-474 was utilized to decrease T-ALL proliferation. It was found that this suppression was greatly enhanced when combined with a Wnt/β-catenin pathway inhibitor, suggesting that the inhibition of multiple pathways may be an effective therapeutic strategy for T-ALL treatment.37 Similarly, Shi et al discovered that FAK downregulation significantly increased rapamycin-induced decrease of cell proliferation, G0/G1 cell cycle arrest, and apoptosis, further supporting the idea that combining multiple components could yield more effective treatment outcomes.38 As previously mentioned, several studies have suggested that targeting the PI3K/AKT/mTOR pathway could be a promising therapeutic strategy for T-ALL. In fact, these studies have shown that combining such therapy with other components leads to a significant improvement in efficacy, which is consistent with the findings of our study. Multiple differentially expressed miRNAs were found by Valiollahi et al, including miR-3143, whose dysregulation could alter leukemogenesis and stemness capacities in Ph+ B-ALL. The mentioned study suggested that dysregulation of miR-3143 could play a role in the development and progression of leukemia.39 Muciño-Olmos et al hypothesized that miR-3143 affects carbon metabolism in breast cancer cell lines by targeting hexokinase-2 (MCF7).40 Razaviyan et al reported downregulation of miR-3182 in MCF-7 cells compared to normal mammary epithelial cells (MCF-10A) and discovered a substantial simultaneous decrease in the mTOR and S6KB1 genes in cancer cells.41 Finally, Khazaie-Poul et al utilized human umbilical cord mesenchymal stem cell-loaded exosomes to overexpress miR-3143 and miR-3182 in the MDA-MB-231 cell line, ultimately identifying a substantial reduction in PI3K/AKT/mTOR genes as possible therapeutic targets.42
Conclusion
The PI3K/AKT/mTOR signaling pathway is dysregulated in NOTCH1-overexpressed T-ALL, and inhibiting this pathway may be beneficial in blocking the progression of leukemia. In a cellular model, we investigated the action of rapamycin in combination with miR-3143 and miR-3182 and found that the combination treatment was highly effective, most likely through the downregulation of key genes in the PI3K/AKT/mTOR signaling pathway. Importantly, our findings are consistent with current literature on the effects of antitumor synergy and suggest that the novel combination of miR-3143 and miR-3182 with rapamycin can be proposed as a complementary therapy based on miRNA mimics. However, functional and preclinical studies are necessary to verify these claims.
Research Highlights
What is the current knowledge?
√ Overexpression of PI3K/AKT/mTOR signaling pathway contributes to the pathogenesis of T-cell acute lymphoblastic leukemia (T-ALL), as well as resistance to chemotherapy.
√ Rapamycin, which blocks a crucial component of the PI3K/AKT/mTOR pathway, has been widely used to investigate therapeutic effects on T-ALL.
√ Evidence supports the idea that microRNAs (miRNAs) are crucial in the pathogenesis and therapeutic resistance.
What is new here?
√ The study identifies specific miRNAs that can inhibit the PI3K/AKT/mTOR pathway in T-ALL.
√ The study shows that combining rapamycin with these miRNAs can have a synergistic effect in inducing apoptosis and cell cycle arrest in T-ALL cells.
√ These findings provide new insights into potential treatments for chemotherapy-resistant T-ALL.
Acknowledgements
The authors thank the Cellular and Molecular Biology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran, for providing technical support.
Competing of Interests
The authors declare that they have no conflicts of interests.
Ethical Statement
This study was executed under the supervision of the Ethics Committee of Zanjan University of Medical Sciences, Zanjan, Iran (IR.ZUMS.REC.1399.012).
Funding
This project was funded by Zanjan University of Medical Sciences, Zanjan, Iran (Grant No. A-12-190-28) and Shahid Beheshti University of Medical Sciences, Tehran, Iran (Grant No.27596, ethics code: IR.SBMU.RETECH.REC.1400.112). This manuscript is extracted from Ph.D. research thesis of Fateme Arjmand submitted to Zanjan University of Medical Sciences, Zanjan, Iran.
References
- Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest 2012; 122:3398-406. doi: 10.1172/JCI61269 [Crossref] [ Google Scholar]
- Durinck K, Goossens S, Peirs S, Wallaert A, Van Loocke W, Matthijssens F. Novel biological insights in T-cell acute lymphoblastic leukemia. Exp Hematol 2015; 43:625-39. doi: 10.1016/j.exphem.2015.05.017 [Crossref] [ Google Scholar]
- Terwilliger T, Abdul-Hay M. Acute lymphoblastic leukemia: a comprehensive review and 2017 update. Blood Cancer J 2017; 7:e577. doi: 10.1038/bcj.2017.53 [Crossref] [ Google Scholar]
- Teitell MA, Pandolfi PP. Molecular genetics of acute lymphoblastic leukemia. Annu Rev Pathol 2009; 4:175-98. doi: 10.1146/annurev.pathol.4.110807.092227 [Crossref] [ Google Scholar]
- Puckett Y, Chan O. Acute lymphocytic leukemia. StatPearls [Internet]: StatPearls Publishing; 2022.
- Madhusoodhan PP, Carroll WL, Bhatla T. Progress and prospects in pediatric leukemia. CurrProblPediatrAdolesc Health Care 2016; 46:229-41. doi: 10.1016/j.cppeds.2016.04.003 [Crossref] [ Google Scholar]
- Raetz EA, Teachey DT. T-cell acute lymphoblastic leukemia. Hematology 2014, the American Society of Hematology Education Program Book 2016; 2016:580-8. doi: 10.1182/asheducation-2016.1.580 [Crossref] [ Google Scholar]
- Zhao W. Targeted therapy in T-cell malignancies: dysregulation of the cellular signaling pathways. Leukemia 2010; 24:13-21. doi: 10.1038/leu.2009.223 [Crossref] [ Google Scholar]
- Schubbert S, Cardenas A, Chen H, Garcia C, Guo W, Bradner J. Targeting the MYC and PI3K Pathways Eliminates Leukemia-Initiating Cells in T-cell Acute Lymphoblastic LeukemiaTargeting Leukemia-Initiating Cells in T-ALL. Cancer Res 2014; 74:7048-59. doi: 10.1158/0008-5472.CAN-14-1470 [Crossref] [ Google Scholar]
- Belver L, Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer 2016; 16:494-507. doi: 10.1038/nrc.2016.63 [Crossref] [ Google Scholar]
- Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, Belver L, Wendorff AA. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med 2014; 20:1130-7. doi: 10.1038/nm.3665 [Crossref] [ Google Scholar]
- Hales EC, Taub JW, Matherly LH. New insights into Notch1 regulation of the PI3K–AKT–mTOR1 signaling axis: Targeted therapy of γ-secretase inhibitor resistant T-cell acute lymphoblastic leukemia. Cell Signal 2014; 26:149-61. doi: 10.1016/j.cellsig.2013.09.021 [Crossref] [ Google Scholar]
- Khoshamooz H, Kaviani S, Atashi A, Hassankiadeh SHM. Combination effect of Notch1 and PI3K/AKT/mTOR signaling pathways inhibitors on T-ALL cell lines. Int J Hematol Oncol Stem Cell Res 2020; 14:99. doi: 10.18502/ijhoscr.v14i2.2673 [Crossref] [ Google Scholar]
- Batista A, Barata JT, Raderschall E, Sallan SE, Carlesso N, Nadler LM. Targeting of active mTOR inhibits primary leukemia T cells and synergizes with cytotoxic drugs and signaling inhibitors. Exp Hematol 2011; 39:457-72 e3. doi: 10.1016/j.exphem.2011.01.005 [Crossref] [ Google Scholar]
- Zhao YM, Zhou Q, Xu Y, Lai XY, Huang H. Antiproliferative effect of rapamycin on human T-cell leukemia cell line Jurkat by cell cycle arrest and telomerase inhibition. Acta Pharmacol Sin 2008; 29:481-8. doi: 10.1111/j.1745-7254.2008.00767.x [Crossref] [ Google Scholar]
- Jiao G, Yan L, Qiang L, Xia G, Ling G, Gui Z. Resveratrol induces apoptosis and autophagy in T-cell acute lymphoblastic leukemia cells by inhibiting Akt/mTOR and activating p38-MAPK. Biomed Environ Sci 2013; 26:902-11. doi: 10.3967/bes2013.019 [Crossref] [ Google Scholar]
- Ghalavand M, Dorostkar R, Borna H, Mohammadi-Yeganeh S, Hashemi SM. MicroRNA-122 Is More Effective than Rapamycin in Inhibition of Epithelial-mesenchymal Transition and mTOR Signaling Pathway in Triple Negative Breast Cancer. Iran J Allergy Asthma Immunol 2023. 10.18502/ijaai.v22i1.12006.
- Fattizzo B, Rosa J, Giannotta JA, Baldini L, Fracchiolla NS. The Physiopathology of T- Cell Acute Lymphoblastic Leukemia: Focus on Molecular Aspects. Front Oncol 2020; 10:273. doi: 10.3389/fonc.2020.00273 [Crossref] [ Google Scholar]
- Zhang B-h, Pan X-p. Zhang B-h, Pan X-pCobb GP and Anderson TAmicroRNAs as oncogenes and tumor suppressors. Dev Biol 2007; 302:1-12. doi: 10.1016/j.ydbio.2006.08.028 [Crossref] [ Google Scholar]
- Elliott K, Larsson E. Non-coding driver mutations in human cancer. Nat Rev Cancer 2021; 21:500-9. doi: 10.1038/s41568-021-00371-z [Crossref] [ Google Scholar]
- TR TRPKM. Dinger M Mattick JS Non-coding RNAs: regulators of disease. J Pathol 2010; 2202:126-39. doi: 10.1002/path.2638 [Crossref] [ Google Scholar]
- Ishida M, Selaru FM. miRNA-based therapeutic strategies. CurrPathobiol Rep 2013; 1:63-70. doi: 10.1007/s40139-012-0004-5 [Crossref] [ Google Scholar]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M. DIANA-microT web server v50: service integration into miRNA functional analysis workflows. Nucleic Acids Res 2013; 41:W169-W73. doi: 10.1093/nar/gkt393 [Crossref] [ Google Scholar]
- Agarwal V, Bell G, Nam J, Bartel D. Agarwal V, Bell G, Nam J, Bartel DPredicting effective microRNA target sites in mammalian mRNAsElife. ELife 2015; 4:e05005. doi: 10.7554/eLife.05005 [Crossref] [ Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNAorg resource: targets and expression. Nucleic Acids Res 2008; 36:D149-53. doi: 10.1093/nar/gkm995 [Crossref] [ Google Scholar]
- Dweep H, Gretz N. Dweep H, Gretz NmiRWalk20: a comprehensive atlas of microRNA-target interactions. Nat Methods 2015; 12:697. doi: 10.1038/nmeth.3485 [Crossref] [ Google Scholar]
- Huang H-Y, Lin Y-C-D, Li J, Huang K-Y, Shrestha S, Hong H-C. miRTarBase 2020: updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res 2020; 48:D148-D54. doi: 10.1093/nar/gkz896 [Crossref] [ Google Scholar]
- Mohammadi-Yeganeh S, Paryan M, Mirab Samiee S, Soleimani M, Arefian E, Azadmanesh K. Development of a robust, low cost stem-loop real-time quantification PCR technique for miRNA expression analysis. Mol Biol Rep 2013; 40:3665-74. doi: 10.1007/s11033-012-2442-x [Crossref] [ Google Scholar]
- Horvat L, Antica M, Matulić M. Effect of Notch and PARP pathways’ inhibition in leukemic cells. Cells 2018; 7:58. doi: 10.3390/cells7060058 [Crossref] [ Google Scholar]
- Gu L, Zhou C, Liu H, Gao J, Li Q, Mu D. Rapamycin sensitizes T-ALL cells to dexamethasone-induced apoptosis. J Exp Clin Cancer Res 2010; 29:150. doi: 10.1186/1756-9966-29-150 [Crossref] [ Google Scholar]
- Bressanin D, Evangelisti C, Ricci F, Tabellini G, Chiarini F, Tazzari PL. Harnessing the PI3K/Akt/mTOR pathway in T-cell acute lymphoblastic leukemia: eliminating activity by targeting at different levels. Oncotarget 2012; 3:811. doi: 10.18632/oncotarget.579 [Crossref] [ Google Scholar]
- He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi S. miRNA-based biomarkers, therapies, and resistance in Cancer. Int J Biol Sci 2020; 16:2628-47. doi: 10.7150/ijbs.47203 [Crossref] [ Google Scholar]
- Ratti M, Lampis A, Ghidini M, Salati M, Mirchev MB, Valeri N. MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) as new tools for cancer therapy: first steps from bench to bedside. Target Oncol 2020; 15:261-78. doi: 10.1007/s11523-020-00717-x [Crossref] [ Google Scholar]
- Gandellini P, Profumo V, Folini M, Zaffaroni N. MicroRNAs as new therapeutic targets and tools in cancer. Expert OpinTher Targets 2011; 15:265-79. doi: 10.1517/14728222.2011.550878 [Crossref] [ Google Scholar]
- Najjary S, Mohammadzadeh R, Mansoori B, Vahidian F, Mohammadi A, Doustvandi MA. Combination therapy with miR-34a and doxorubicin synergistically induced apoptosis in T-cell acute lymphoblastic leukemia cell line. Med Oncol 2021; 38:142. doi: 10.1007/s12032-021-01578-8 [Crossref] [ Google Scholar]
- Kong D, Fan S, Sun L, Chen X, Zhao Y, Zhao L. Growth inhibition and suppression of the mTOR and Wnt/β‐catenin pathways in T‐acute lymphoblastic leukemia by rapamycin and MYCN depletion. Hematol Oncol 2021; 39:222-30. doi: 10.1002/hon.2831 [Crossref] [ Google Scholar]
- Evangelisti C, Chiarini F, Cappellini A, Paganelli F, Fini M, Santi S. Acta Pharmacologica Sinica. J Cell Physiol 2020; 235:5413-28. doi: 10.1002/jcp.29429 [Crossref] [ Google Scholar]
- Shi P-J, Xu L-H, Lin K-Y, Weng W-j, Fang J-P. Synergism between the mTOR inhibitor rapamycin and FAK down-regulation in the treatment of acute lymphoblastic leukemia. J Hematol Oncol 2016; 9:1-10. doi: 10.1186/s13045-016-0241-x [Crossref] [ Google Scholar]
- Valiollahi E, Ribera JM, Genesca E, Behravan J. Genome-wide identification of microRNA signatures associated with stem/progenitor cells in Philadelphia chromosome-positive acute lymphoblastic leukemia. Mol Biol Rep 2019; 46:1295-306. doi: 10.1007/s11033-019-04600-5 [Crossref] [ Google Scholar]
- Muciño-Olmos EA, Vázquez-Jiménez A, López-Esparza DE, Maldonado V, Valverde M, Resendis-Antonio O. MicroRNAs regulate metabolic phenotypes during multicellular tumor spheroids progression. Front Oncol 2020; 10:582396. doi: 10.3389/fonc.2020.582396 [Crossref] [ Google Scholar]
- Razaviyan J, Hadavi R, Tavakoli R, Kamani F, Paknejad M, Mohammadi-Yeganeh S. Expression of miRNAs Targeting mTOR and S6K1 Genes of mTOR Signaling Pathway Including miR-96, miR-557, and miR-3182 in Triple-Negative Breast Cancer. Appl BiochemBiotechnol 2018; 186:1074-89. doi: 10.1007/s12010-018-2773-8 [Crossref] [ Google Scholar]
- Khazaei-Poul Y, Mirmotalebisohi SA, Zali H, Molavi Z, Mohammadi-Yeganeh S. Identification of miR-3182 and miR-3143 target genes involved in the cell cycle as a novel approach in TNBC treatment: A systems biology approach. Chem Biol Drug Des 2023; 101:662-77. doi: 10.1111/cbdd.14167 [Crossref] [ Google Scholar]