Bioimpacts. 14(6):29912.
doi: 10.34172/bi.2024.29912
Original Article
Identification and characterization of antibacterial peptides produced by Lactobacillus plantarum 1407
Silpa Sajan Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Writing – original draft,
Rupachandra Saravanan Supervision, Visualization, Writing – review & editing, , * 
Author information:
Department of Biotechnology, School of Bioengineering, Faculty of Engineering & Technology, SRM Institute of Science and Technology, Kattankulathur, Chennai - 603 203, India
Abstract
Introduction:
Peptides from lactic acid bacteria provide health benefits and can inhibit the growth of pathogenic organisms. The present work aimed to isolate and characterize peptides with antibacterial activity from Lactobacillus plantarum 1407.
Methods:
Peptides were isolated and purified from L. plantarum 1407. The effect of various physiological parameters on the antibacterial activity of the isolated peptides was analyzed. The mode of action of the peptides on indicator organisms was observed using transmission microscopy analysis and flow cytometry analysis.
Results:
Antibacterial activity and mode of action of peptides isolated from L. plantarum 1407 against gram-positive and gram-negative bacteria have been studied. L. plantarum culture exhibited maximum antibacterial activity at 40 °C, pH 8, and 0.7% salt concentration. The cell-free supernatant (CFS) was concentrated using a 3 kDa ultrafiltration membrane and the peptide fractions (<3 kDa) were further fractionated using Sephadex G-25 gel filtration chromatography. The antibacterial activity of the eluted fractions (F1 to F4) was evaluated using flow cytometry and transmission electron microscopy. F3 fraction exhibited increased antibacterial activity than F1, F2, and F4 fractions against the indicator organisms. Cell membrane damage and leakage of cytoplasmic content of the bacterial cells treated with the antibacterial F3 fraction peptides were observed.
Conclusion:
The active peptides from L. plantarum 1407 can be potentially used for the treatment of bacterial infections.
Keywords: Flow cytometry, Gel filtration chromatography, Lactobacillus plantarum, Peptides, Ultrafiltration
Copyright and License Information
© 2024 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Introduction
Lactic acid bacteria (LAB) synthesize various compounds that positively impact the technical and nutritional qualities of fermented food products, in addition to producing lactic acid as the primary by-product of their anaerobic metabolism.1 Bacteriocins are antimicrobial peptides produced by bacteria that can kill or inhibit other bacterial strains.2 The majority of research on the biotechnological applications of various bacteriocins has been on their usage as food preservatives, with nisin serving as the prototype and having been effectively implemented in food technology.3
The ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanii, Pseudomonas aeruginosa, and Enterobacter species) represents a group of bacteria that are resistant to almost all conventional antibiotics and are the causative agents of various skin infections. About 50% of skin and soft tissue infections (SSTIs) are caused by Staphylococcus aureus and Gram-negative pathogens like P. aeruginosa and K. pneumoniae whichcontribute to infections leading to chronic wounds. Increased resistance is reported against topical antibiotics such as vancomycin, fusidic acid, and mupirocin.4 Antimicrobial peptides from LAB offer extraordinary potential as therapeutic agents either by themselves or in conjunction with traditional antimicrobial drugs.5 Bacteriocin-producing Lactobacillus acidophilus strains JCM1132 and L. acidophilus CCFM720 can prevent various bacterial infections.6
Several studies have reported the antibacterial activities of bacteriocins from different LAB species.7 BAS-48 was effective against acute infection in a murine model along with a significant effect on skin pathogens.8 Topical formulations of garvicin KS had a significant antibacterial effect without any toxicity.9 TSU4 from Lactobacillus animalis TSU4 is tested effective against chronic infections caused by prominent pathogens such as S. aureus, Shigella flexneri, and P. aeruginosa. 10 Novel enterocins DD28 and DD93 exhibit anti-staphylococcal activity in multi-drug resistant Staphylococcus aureus (MRSA).11 Lacticin 3147 from Lactococcus lactic subsp. cremoris MG1363 reduced S. aureus Xen 29 infections.12 Peocin produced by Paenibacillus ehimensis NPUST1 significantly decreased clinical and antibiotic-resistant pathogens.13 Gassericin E produced by Lactobacillus gasseri IEV1461 prevented colonization of pathogens such as Mobiluncus curtisii, Prevotella bivia, Eggerthella spp, Sneathia Ispp, andMegasphaera type I spp.14
The antibacterial activity of peptides was found to be related to molecular mass, charge of the sequence, and conformation.15 According to reports, peptides may disrupt the membrane’s structural integrity by interacting with its lipid molecules.16 Nisin repolarizes cell membrane through its high affinity for the cell wall precursor lipid II which represents the central cell wall building block of peptidoglycan synthesis.15 Lactocin 160 from Lacticaseibacillus rhamnosus targets the cytoplasmic membrane of pathogens such as Gardnerella vaginalis and Prevotella bivia causing an outflow of ATP molecules and dispersion of the electrochemical proton gradient.17
This study aimed to investigate the antibacterial activity of peptides isolated from Lactobacillus plantarum MTCC1407. The peptide fractions were isolated, purified, and assessed for antibacterial activity against Gram-positive and Gram-negative pathogens. The membrane interactions between the F3 fraction peptides and bacterial strains like Staphylococcus aureus, Pseudomonas aeruginosa, Enterococcus faecalis, and Klebsiella pneumoniae were further investigated by examining its effect on the bacterial membrane integrity for production of a new effective antibacterial therapeutic agents.
Materials and Methods
Materials
Agar Agar (Cat No: GRM666-500G) was purchased from HiMedia. De Man, Rogosa, and Sharpe (MRS) broth (Cat No:49190) and Nutrient Broth (Cat No:55427) were purchased from Siscon Research Laboratories Pvt. Ltd.
Microorganisms and culture conditions
Lactobacillus plantarum 1407 was acquired from Microbial Type Culture Collection (MTCC). The indicator organisms used in this study were: Staphylococcus aureus ATCC 2593, Enterococcus faecalis ATCC 29212, Klebsiella pneumoniae ATCC 700603 (obtained from American Type Culture Collection (ATCC)), and Pseudomonas aeruginosa MTCC 15159 (obtained from MTCC). These cultures are subcultured in Nutrient Broth (NB) at 37 ºC for 24 hours.
Screening of L. plantarum 1407 for antibacterial activity
Lactobacillus plantarum was inoculated into MRS broth and incubated at 37 ºC for 12-14 hours. The CFS was collected by centrifugation (4000 rpm, 20 minutes, 4 °C), and neutralized to a pH of 6.5–7.0 with 10 M NaOH.18 The L. plantarum MTCC 1407strain was tested for its antibacterial activity using well diffusion assay.19 The indicator organisms (S. aureus, E. faecalis, K. pneumoniae, and P. aeruginosa) were spread evenly over a sterile nutrient agar petri plate using a sterile L rod. A well (8 mm diameter) was punched using a sterile cork borer and 100 µL Cell Free Supernatant (CFS) was loaded in each well. The plates are incubated at 37 °C for 24 hours. The antibacterial activity was estimated by measuring the inhibition zone after incubation.20
Effect of temperature, pH, and salt concentration on bacterial growth
Samples of L. plantarum (2% v/v of 18 hours culture)were cultivated in MRS broth with pH adjusted to 5, 6, 8, 9, 10 and the culture was incubated at 37 °C for 24-48 hours. The samples were inoculated in MRS broth and incubated without aeration at various temperatures (20 °C, 40 °C, 50 °C, 60 °C, 70 °C) for 24-48 hours for determination of the influence of temperature on the growth of L. plantarum. Different concentrations of sodium chloride (NaCl) (0.5%, 0.6%, 0.7%, 0.8%, 0.9%, 1%) were added to MRS broth inoculated with 2% v/v of 18 hours L. plantarum cultureand incubated at 37 °C for 24-48 hours. The OD was measured for every 1 hour at 600 nm and the growth curves were plotted.19 The antibacterial activity was expressed as killing activity using the well diffusion method. For quantitative analysis, 2-fold CFS dilutions were prepared using phosphate buffered saline (PBS), and the arbitrary units (AU/mL) were calculated as ab × 100, where “a” is the dilution factor and “b” is the last dilution that demonstrates the inhibition zone of at least 2mm diameter.21
Sensitivity of L. plantarum CFS to pH and heat
The CFS of L. plantarum 1407 was collected by centrifugation at 4000 rpm for 20 minutes at 4 °C. The pH of the CFS was adjusted to 5, 6, 8, 9 and 10. The CFS was heated to 40 °C, 50 °C, 60 °C and 100 °C for 20 minutes. The untreated CFS was used as the positive control. Well diffusion assay was performed to evaluate the loss of antibacterial activity of the treated CFS. A significant decrease in the zone of inhibition indicates the sensitivity of the antibacterial activity to pH and heat treatment.18
Fractionation and antibacterial activity of purified peptides
Lactobacillus plantarum 1407 was used as an inoculum (2% v/v) and incubated for 24 hours at 37 °C in 100 mL of MRS broth. Ammonium sulfate precipitation, followed by ultrafiltration and freeze drying, was performed to concentrate the CFS. 10 mL of the CFS was mixed with various concentrations (20%, 40%, 60%, and 80%) of ammonium sulfate, and left for 24 hours to precipitate. The pellet was collected after centrifugation at 5000 rpm for 10 minutes and resuspended in 25 mL of 0.05M potassium phosphate buffer. The suspension was agitated for 24 hours at 4 °C and partially purified using a 3 kDa ultrafiltration membrane (Amicon Ultra-0.5 Centrifugal Filter unit UFC5003). The partially purified CFS was centrifuged at 3510 rpm for 15 minutes and the fractions containing peptides below 3 kDa were diluted in 2 mL of deionized water and fractionated using a Sephadex G-25 size exclusion chromatography. The column (1.6 × 50 cm) was packed with Sephadex-G25 and equilibrated with 0.2 M phosphate buffer (pH 7). The eluents (5 mL) were collected at a flow rate of 1 mL/min for all eluted peaks at an absorption wavelength of 280 nm. The concentration of peptides in the fractions was determined by Lowry Assay.22,23
The minimal inhibitory concentrations (MICs) of the peptide fractions were determined.24 50 μL of bacterial culture was diluted with PBS and added to 96 well plates. The peptide fractions (50 μL) diluted by a 2-fold dilution method, were added to the plates and incubated at 37 °C for 24 hours. Bacterial suspension mixed with phosphate buffer was used as a negative control. After incubation, the absorbance at 600 nm was determined using a microplate reader. The overnight culture of bacteria was washed with PBS (pH 7.4), and exposed to the peptide fractions at 37 °C. At fixed time intervals, 200 µL of the aliquots were collected and the absorbance was measured at 600 nm using a microplate reader.25
The vulnerability of all peptide fractions to proteinase K was also tested and evaluated for subsequent antibacterial activity. 500 µL of peptide fractions was treated with equal volume of proteinase K with 1 mg/mL final concentration and a control without treatment was prepared. All preparations were incubated at 37 ℃, followed by antibacterial assay using the indicator organisms.18
Mass spectrometry analysis
The identification was performed using a Nexera X2 UHPLC connected with Impact HD qTOF (Bruker, Germany). The antibacterial peptide fraction was treated with trypsin (0.01 mg/mL) and chromatographic analysis was carried out on a Zorbax Eclipse Plus C18 column (2.6 × 150 mm, 3 µm particle size). The separation was carried out using formic acid (0.1%) and acetonitrile as mobile phase with a flow rate of 0.3 mL/min. The injection volume was set at 10µL and gradient elution was used to run the solvent. Instrumental source parameters were as follows: Dry gas, 8.0l/min; Drying gas temperature, 220 °C; spray voltage, 2000 V and nebulization, 2.0 Bar. The scan range was set at 50-3000 m/z with a total run time of 60 minutes. The mass spectra were obtained using the positive mode of LC- ESI-MS.26
Fourier transform-infrared spectroscopy analysis
Fourier transform-infrared spectroscopy (FT-IR) was used between a range of 4000 to 400 cm-1 to determine the functional groups. The sample was pelletized by mixing with KBr powder. The analysis was characterized by Shimadzu IRTracer-100 (Tokyo, Japan), FTIR spectrophotometer.
Flow cytometry analysis
The membrane integrity was determined using flow cytometry. The bacterial suspension was centrifuged at 2500 rpm for 10 minutes, and the pellet was washed thrice with PBS (pH: 7.4). The cells were exposed to F3 peptide fraction for different time intervals at 37 °C. The treated cells were washed with PBS, and incubated in the propidium iodide (PI) solution for 10 min at 37 °C. The intensity of fluorescence was measured using a BD FACSCaliburTM Flow Cytometer (US) after removing the unbound dye. 10 000 events were collected for each sample and detected at an excitation wavelength of 488 nm and an emission wavelength of 525 nm. The setup of the flow cytometer was as follows: FL1=600; Gate Side Scatter (SSC)= 350, FL2=799, flow rate=low. The logarithmic mode was used to collect the readings. The presentation of data as FL2/SSC density plots allowed the differentiation between stained bacteria from other microbial cells and background noise. PBS buffer blanks were analyzed identically to facilitate noise correction of bacterial counts.27,28
Preparation of bacterial cells for transmission electron microscopy (TEM)
F3 peptide fraction (0.25 mg/mL) was added to 1 mL of bacterial suspensions. Deionized water (1 mL) was used as the negative control. The samples were incubated for 15 and 30 minutes, at 37 ºC, respectively. The sample solution (4 µL) was added to a TEM grid followed by 4 µL of 1% phosphotungstic acid solution and stained for a min.29 The samples were observed under a JEM-210 plus high-resolution transmission electron microscope (JEOL, Japan).
Cell viability assay
L929, a mouse fibroblast cell line was acquired from National Centre for Cell Science (NCCS), Pune. The cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; HiMedia -AL007S) with fetal bovine serum (10%) (HiMedia- RM-9955) and 1% Antibiotic-Antimycotic Solution (HiMedia-A002) at 37 °C in 5% CO2.
MTT assay was used to explore the effect of purified F3 peptide fraction on the viability of L929 fibroblast cells with modification.30 Fibroblast cells (1 × 105 cell/mL) seeded in 96-well plates were treated with various concentrations of purified F3 peptide fraction (50-250 µg/mL) and incubated for 24 hours at 37 °C. After removing the treatment mixture from each well, 50 µL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) solution (1mg/mL in DMEM; SRL-33611) was added to each well and incubated for 4 h in the dark. 100 µL of dimethylsulfoxide (DMSO; HiMedia- TC185) solution was added to dissolve the formazan crystals. The Optical density (OD) was measured at 570 nm using a microplate reader.31 The experiment was carried out in triplicates. The cell viability percentage was calculated as:
Percentage of cell viability = (OD of test/OD of untreated control) × 100
Statistical analysis
Tukey’s multiple comparison tests were used for examining differences between various parameters in GraphPad Prism version 8. All data were presented as mean ± standard deviation (SD) of three independent experiments. P < 0.05 was considered significant.
Results
Antibacterial activity of Lactobacillus plantarum 1407
The CFS from L. plantarum 1407 exhibited strong antibacterial activity against all four strains of pathogens. The diameter of the zone of inhibition against S. aureus was 13.4 mm, followed by K. pneumoniae (11.8 mm), E. faecalis (10.3 mm),and Pseudomonas aeruginosa (6 mm) (Fig. 1).
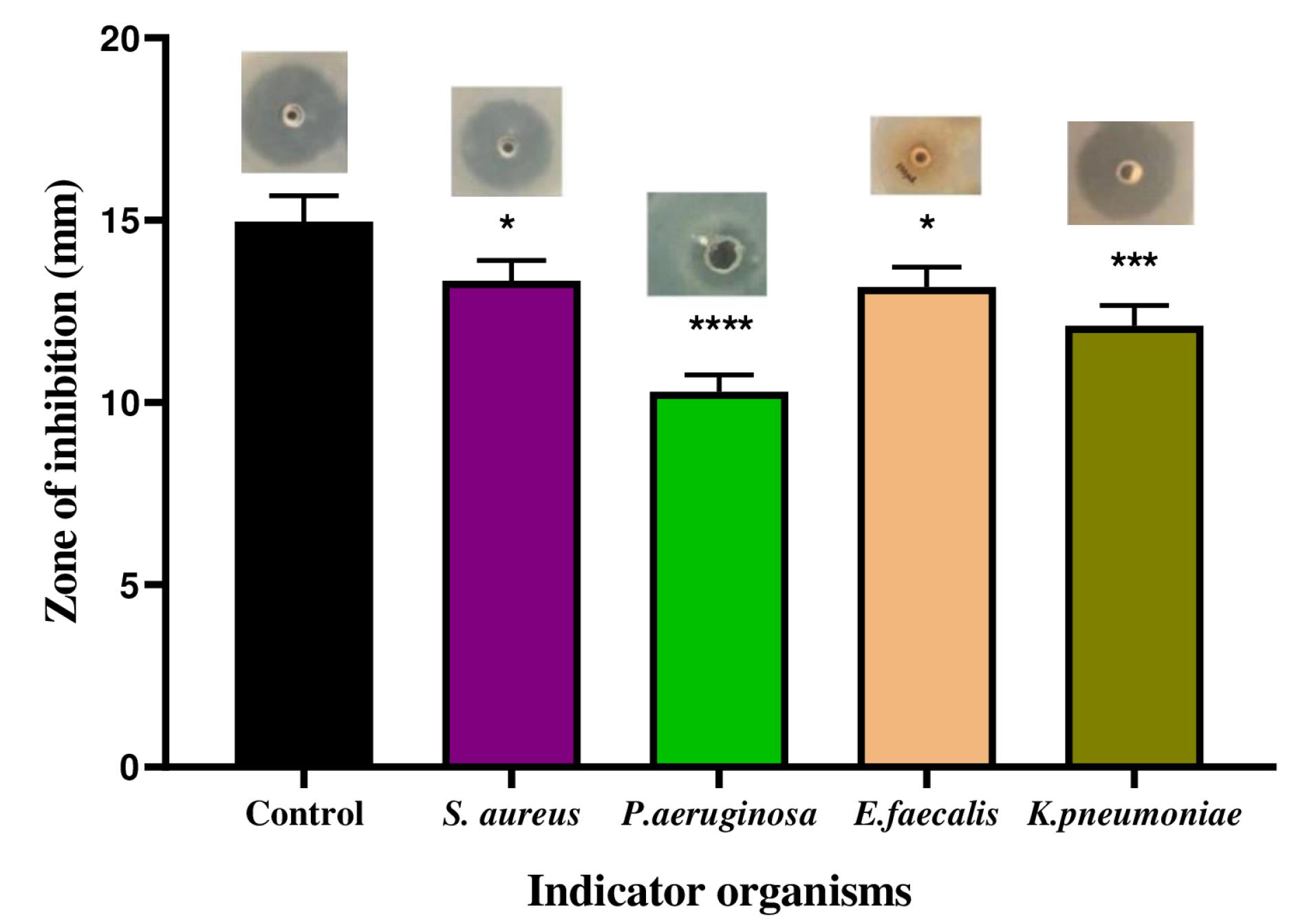
Fig. 1.
Antibacterial activity of the L plantarum 1407 CFS against indicator organisms. Data are representative of three independent experiments and are plotted as mean ± SD (n=3); *P < 0.05; ***P <0.001; ****P < 0.0001
.
Antibacterial activity of the L plantarum 1407 CFS against indicator organisms. Data are representative of three independent experiments and are plotted as mean ± SD (n=3); *P < 0.05; ***P <0.001; ****P < 0.0001
Influence of pH, temperature, and salt concentration on the bacterial growth
Lactobacillus plantarum 1407 exhibited increased antibacterial activity at pH 8. The antibacterial activity was less at pH 5 and 10. The maximum antibacterial activity was observed at a temperature of 40 °C. Regarding various salinity (NaCl%) tested from 0.5%-1% NaCl, increased antibacterial activity was observed at 0.7%. Decreased antibacterial activity was observed at NaCl concentrations beyond 0.7% (Supplementary file 1, Figs. S1-S4).
Effect of pH and temperature on antibacterial activity of CFS
Peptides differ greatly in their pH sensitivity. The CFS remained stable after 3 hours of incubation at a pH range of 5-8, but its antibacterial activity was reduced at pH levels higher than 8. L. plantarum 1407 CFS was active after 20 minutes of exposure to temperatures ranging from 40 to 100 °C (Fig. S5). After 15-20 minutes of heat treatment at 100 ºC, the antibacterial activity has decreased. After heat treatment, approximately 60%-70% of the bacterial inhibitory activity was retained. The results demonstrate that the CFS is stable in increased temperature conditions.
Purification of the peptides
The peptides from CFS were purified through ammonium sulfate precipitation. Well diffusion agar assay with serial dilution was performed after each purification step to monitor the activity of the peptides. The initial activity was calculated as 700 (AU/mL) against S. aureus. The use of an ultrafiltration system with a 3 kDa cut-off membrane increased the inhibitory activity from 700 (AU/mL) to 19200 (AU/mL). The gel filtration chromatography on G-25 fine Sephadex fractionated the peptide fractions obtained from ultrafiltration (Fig. 2). About 4 peptide fractions were obtained named F1, F2, F3, and F4 fractions at the absorbance of 280 nm. The eluted fractions were further analyzed for antibacterial activity. Out of the 4 fractions, the F3 fraction exhibited increased inhibitory activity. The data presented in Table S1 summarises the purification details of the peptide fractions.
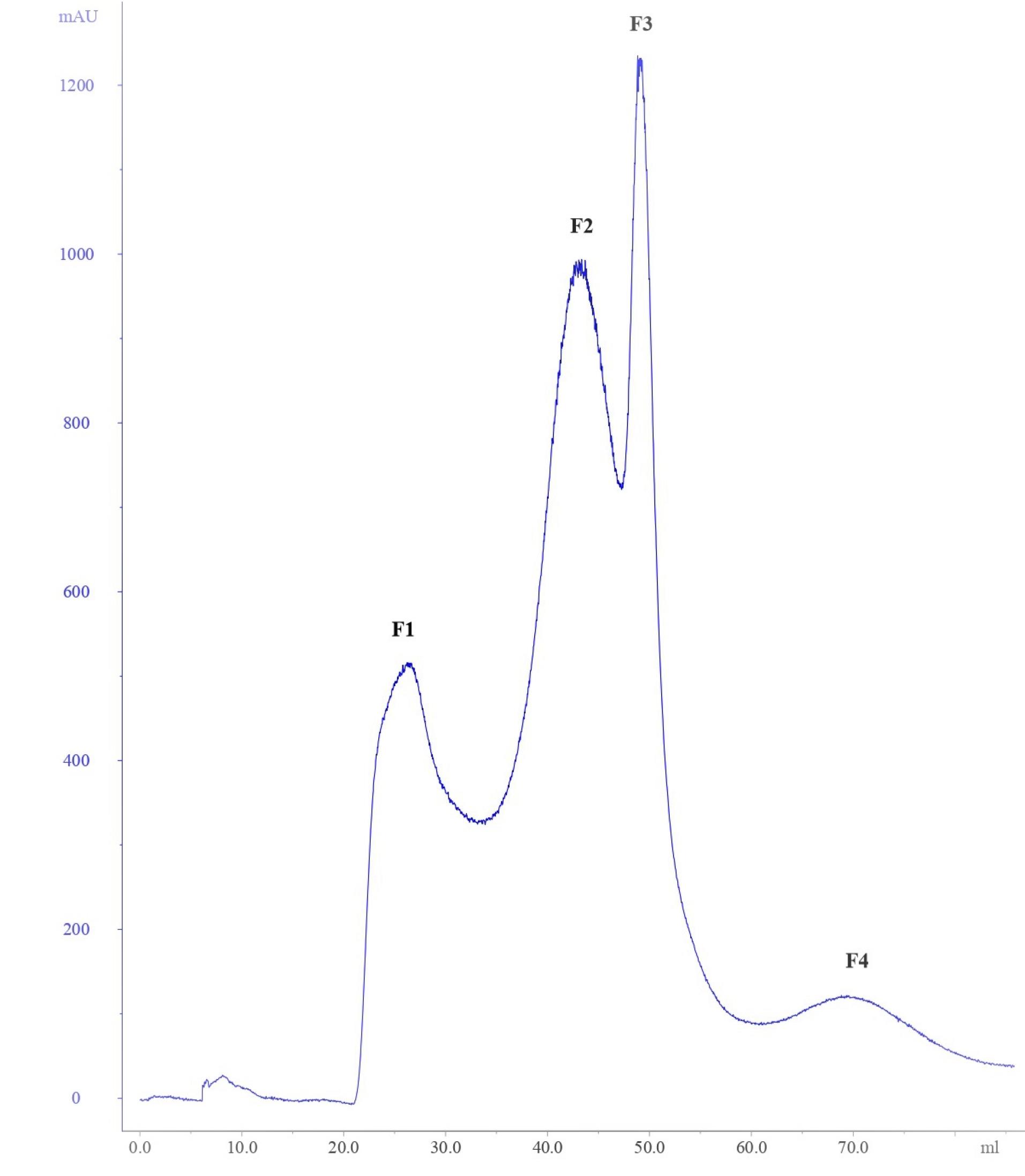
Fig. 2.
Purification of peptide fractions from ultrafiltration by Sephadex gel filtration (G-25) chromatography.
.
Purification of peptide fractions from ultrafiltration by Sephadex gel filtration (G-25) chromatography.
Antibacterial Activity of purified peptide fractions
The MIC of the F1-F4 fractions obtained by gel filtration chromatography was evaluated (Fig. 3). All the fractions exhibited antibacterial activity against the indicator organisms such as S. aureus, P. aeruginosa, E. faecalis, and K. pneumoniae. F3 fraction exhibited the highest MIC than the other fractions (F1, F2, F4) and was used for further studies. The growth curve analysis of the peptide fractions against all the indicator organisms depicted similar results (Fig. S6). A decrease in growth was observed for indicator organisms treated with F3 fraction and the growth was completely inhibited within 180 minutes. These results demonstrated that the F3 fraction peptides exhibited rapid inhibitory activity against gram-positive (S. aureus, E. faecalis) and gram-negative (P. aeruginosa, K. pneumoniae).
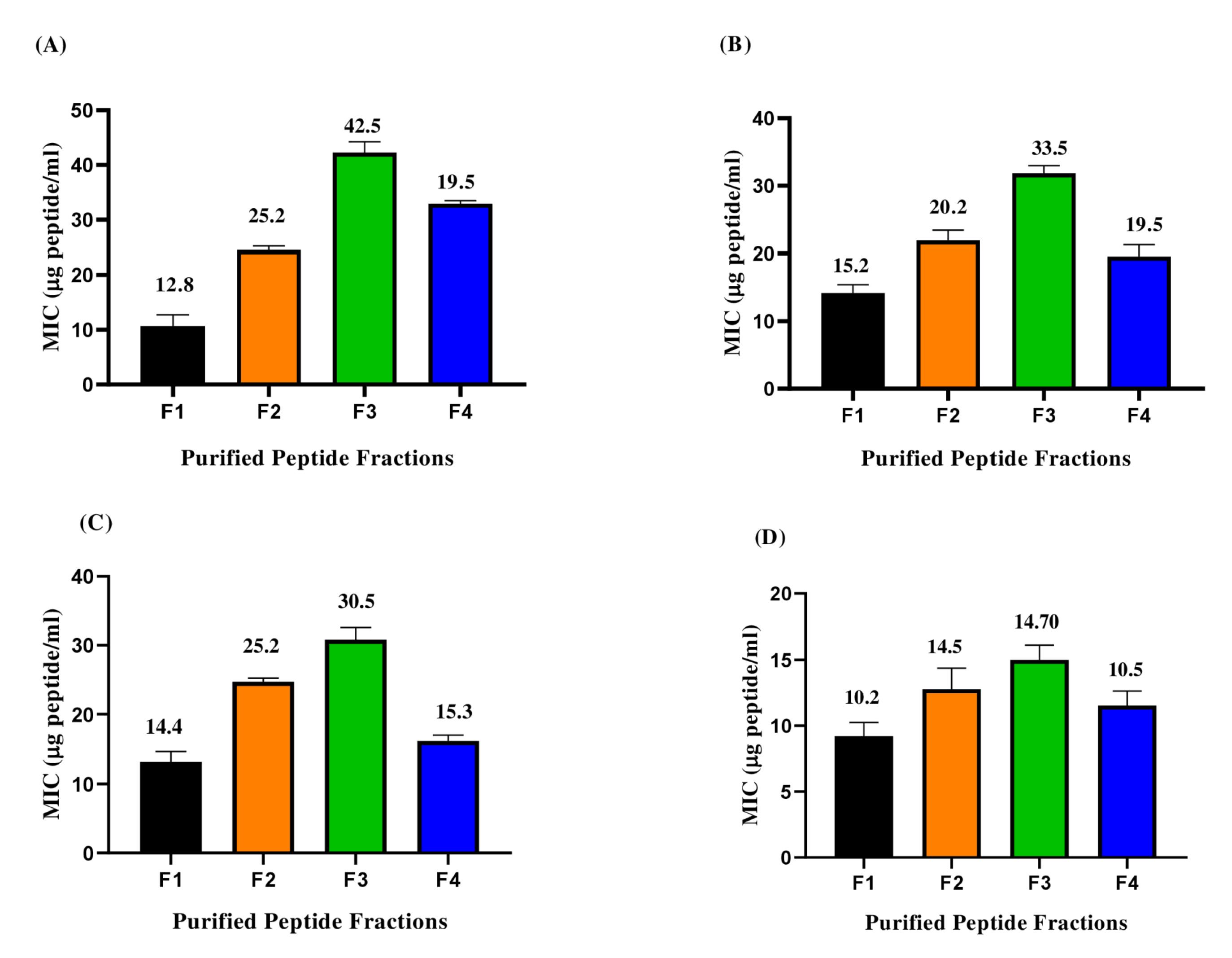
Fig. 3.
Minimum inhibitory concentration of purified peptide fractions (F1-F4) against indicator organisms: (A) S. aureus (B) P. aeruginosa (C) E. faecalis (D) K. pneumoniae.
.
Minimum inhibitory concentration of purified peptide fractions (F1-F4) against indicator organisms: (A) S. aureus (B) P. aeruginosa (C) E. faecalis (D) K. pneumoniae.
Enzyme stability analysis of peptide fractions
The enzyme stability of all the peptide fractions was evaluated and only F3 fraction peptides were stable after the treatment with proteinase K. A loss of antibacterial activity was observed for F1, F2 and F4 fractions after the proteinase K treatment. About 50% antibacterial activity was retained by F3 fraction peptides. The results of enzyme stability test is summarized in Table S2 (See Supplementary file 1).
Identification of molecular weight of F3 fraction peptides
The peptide sequences in fraction 3 which exhibited potent antibacterial activity were identified using LC-MS/MS (Fig. 4) and a total of 10 peptides were identified with molecular weight ranging between 300–800 Da (Table 1). The amino acid sequences of the peptides were different.
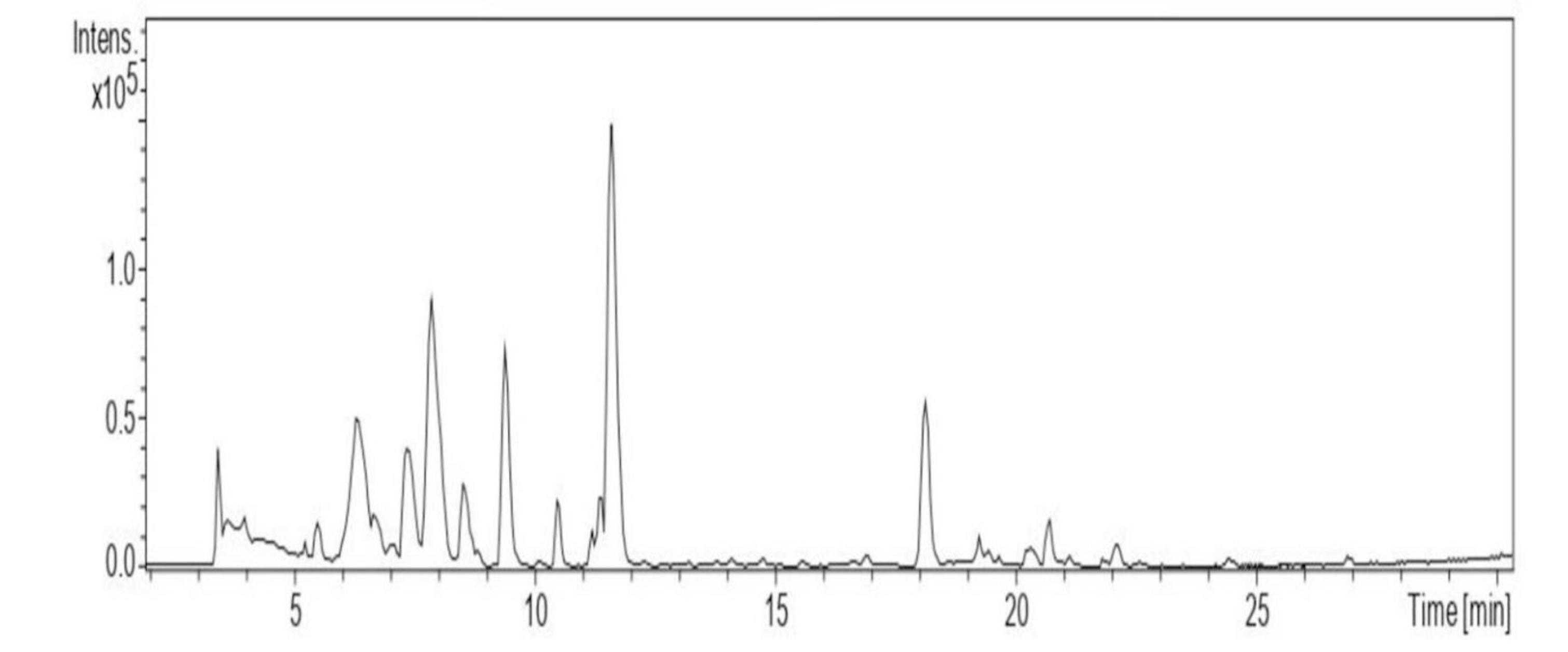
Fig. 4.
LC-MS chromatogram of the peptides identified from F3 fraction.
.
LC-MS chromatogram of the peptides identified from F3 fraction.
Table 1.
Peptide sequences identified in F3 fraction from L. plantarum obtained using Sephadex G-25 gel filtration chromatography
|
Retention time (min)
|
Molecular weight (Da)
|
Peptide Sequence
|
Amino acid sequence
|
| 3.5 |
742.277 |
GGGLWPGV |
Glycine-Glycine-Glycine-Leucine-Tryptophan-Proline-Glycine-Valine |
| 5.4 |
320.904 |
GGCC |
Glycine-Glycine-Cysteine- Cysteine |
| 6.1 |
215.134 |
SGA |
Serine-Glycine-Alanine |
| 7.3 |
270.150 |
HGG |
Histidine-Glycine-Glycine |
| 18 |
372.179 |
AAGGP |
Alanine-Alanine-Glycine-Glycine- Proline |
| 19.1 |
596.328 |
SGKAYA |
Serine-Glycine-Lysine-Alanine-Tyrosine-Alanine |
| 20 |
479.296 |
DGGGGN |
Aspartic acid--Glycine-Glycine-Glycine-Glycine-Asparagine |
| 21.9 |
564.347 |
FGRNA |
Phenylalanine-Glycine-Arginine-Asparagine-Alanine |
| 24.3 |
518.312 |
LGVDD |
Leucine-Glycine-Valine-Aspartic acid-Aspartic acid |
| 27 |
679.497 |
AFAGAGW |
Alanine-Phenylalanine-Alanine-Glycine-Alanine-Glycine-Tryptophan |
Characterization of the F3 fraction peptides
FTIR revealed the spectral range of the functional group of peptide mixture between 400-4500 cm-1 (Fig. 5). Peak signals were observed at 2374.41, 2741.85, 3439.17, 3522.07 and 3674.12 cm-1. The peak at 2374.41 cm-1 represents strong O=C-O stretching. The presence of an aldehyde group is indicated by a peak at 2741.85 cm-1. The peak at 3439.17 cm-1 is an indication of the presence of a primary amine group (N-H stretching). Finally, the peaks at 3522.07 (strong broad OH-stretching) and 3674 cm-1 represent the presence of alcohol.
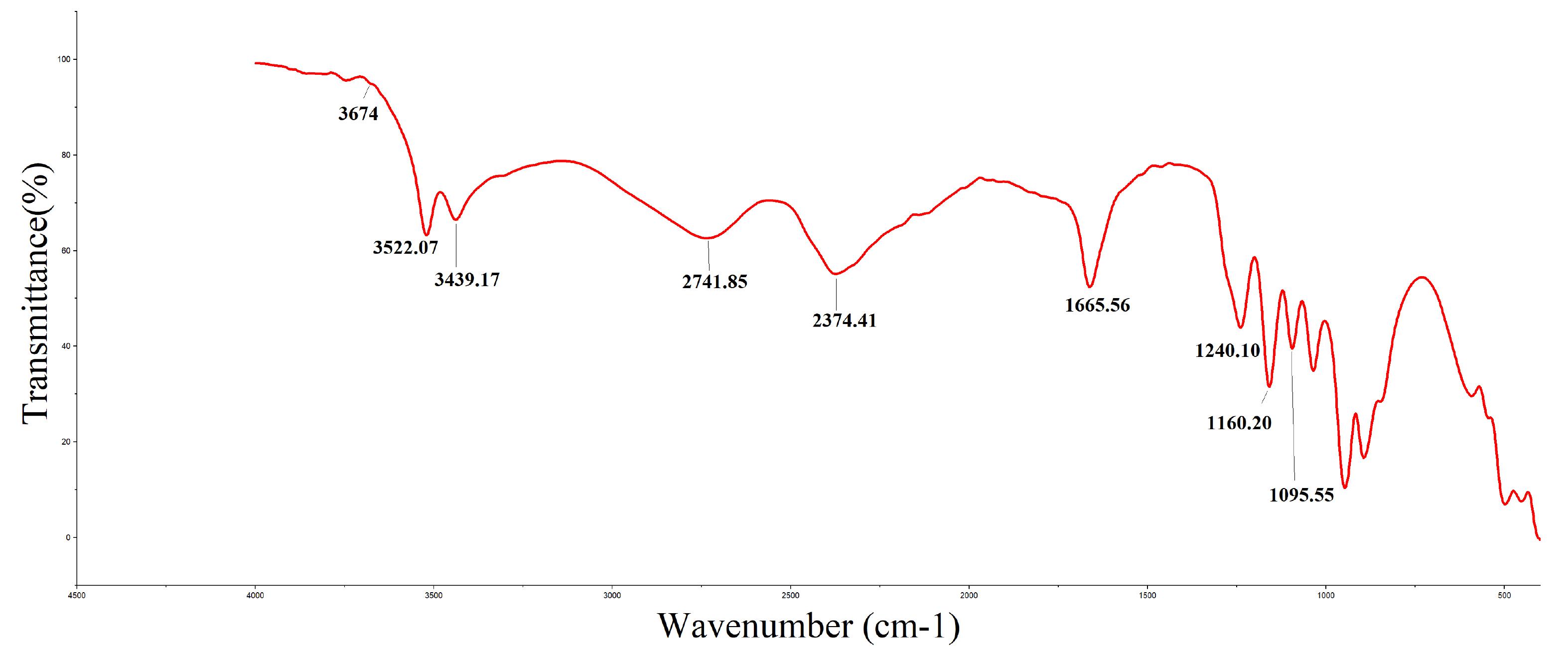
Fig. 5.
FTIR spectra of F3 fraction peptides.
.
FTIR spectra of F3 fraction peptides.
Analysis of membrane integrity of bacterial cells treated with F3 fraction peptides
The membrane integrity of bacterial cells was assessed using PI, which can only penetrate damaged or dead cells and fluoresce by binding to nucleic acids within the cell, but not live cells with intact membranes.32
A gradual decrease in the percentage of live cells was observed after treatment with F3 fraction peptides (Fig. 6). The subsequent increase in the percentage of dead cells suggests that the cell membrane integrity was persistently disrupted in the presence of peptide.33 Viable cells with intact membranes are stained pink whereas cells with compromised membranes appear blue following membrane integrity impairment. The F3 fraction peptides permeabilize the cell membrane in a concentration independent manner but this effect was time-dependent and maximum permeabilization occurred after 120 min (Fig. 6). After 20 minutes of exposure around 57.8%, 8.84%,19.14% and 55.18% cells showed membrane permeabilization for S. aureus, P. aeruginosa, E. faecalis and K. pneumoniae respectively. After 60 minutes exposure the percentage of membrane permeabilized cells increased to 58.1%, 46.7%, 64.7% and 56.76% for S. aureus, P. aeruginosa, E. faecalis and K. pneumoniae respectively. Finally maximum permeabilization was observed on exposure to peptides for 120 minutes as indicated by the increase in percentage of PI stained cells (78.16%, 52.5%, 88.4% and 61.9% for S. aureus, P. aeruginosa, E. faecalis and K. pneumoniae respectively) (Fig. 6; Supplementary Figs. S7-S10).
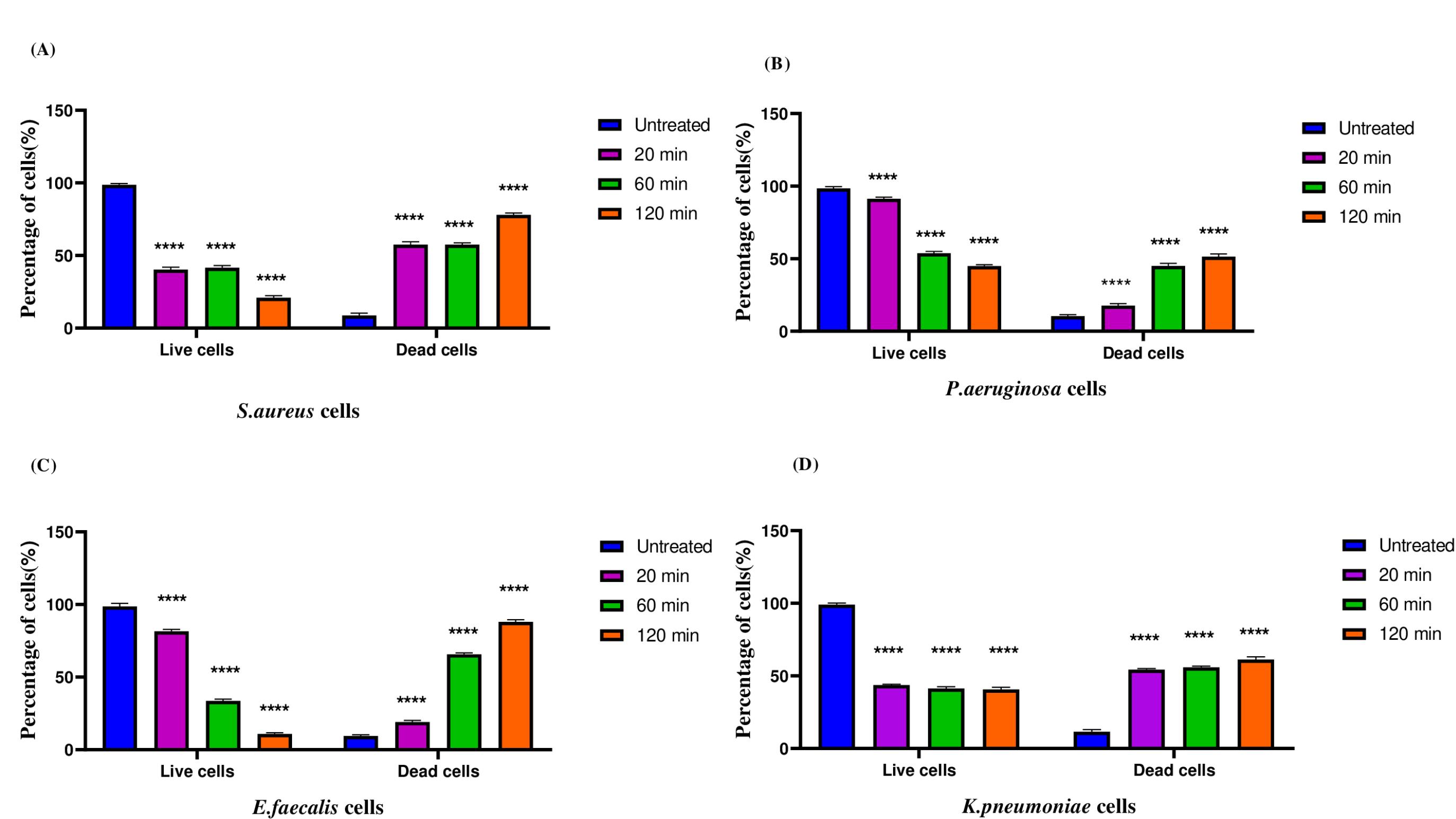
Fig. 6.
Effect of F3 fraction peptides on the membrane integrity of (A) S. aureus (B) P. aeruginosa (C) E. faecalis (D) K. pneumoniae. Data are representative of three independent experiments and are plotted as mean ± SD (n=3); ****P < 0.0001.
.
Effect of F3 fraction peptides on the membrane integrity of (A) S. aureus (B) P. aeruginosa (C) E. faecalis (D) K. pneumoniae. Data are representative of three independent experiments and are plotted as mean ± SD (n=3); ****P < 0.0001.
Analysis of the intracellular organization of bacterial cells treated with F3 fraction peptides using TEM
The indicator organisms treated with F3 fraction peptides were observed under the TEM. The untreated samples were used as control. The intracellular organization of untreated S. aureus samples (Fig. 7A) indicated intact cell membrane. The samples treated with F3 fraction peptides show blebbing of the S. aureus cells, disrupted cell membrane and pore formation as indicated by the arrows (Fig. 7B). The typical rod shaped structure was observed for untreated K. pneumoniae samples (Fig. 7C). An electron-dense transparent zone between cytoplasm was observed in K. pneumoniae cells treated with F3 fraction peptides which indicates change in cytoplasmic density. Disintegration of cell membrane and formation of pores were also observed (Fig. 7D). An intact membrane was observed for untreated E. faecalis samples (Fig. 7E). The time dependent treatment of E. faecalis cells with F3 fraction peptides resulted in pore formation and gradual efflux of cytoplasmic contents. The distorted E. faecalis cells with blurred boundaries is an indication of alteration of cell morphology (Fig. 7F). The typical Gram-negative structure with uniform cytoplasm and distinct cell integrity was seen in untreated P. aeruginosa samples (Fig. 7G). An irregular cell profile and cell wall debris in the P. aeruginosa cells treated with F3 fraction peptides indicate cell wall damage. The leaked cytoplasmic material of P. aeruginosa can be found around the cell as a result of possible membrane permeability transformation (Fig. 7H). The density of the cytoplasm is reduced and the cytoplasm disappears in K. pneumoniae and P. aeruginosa cells treated with F3 fraction peptides (Fig. 7D and Fig.7H).
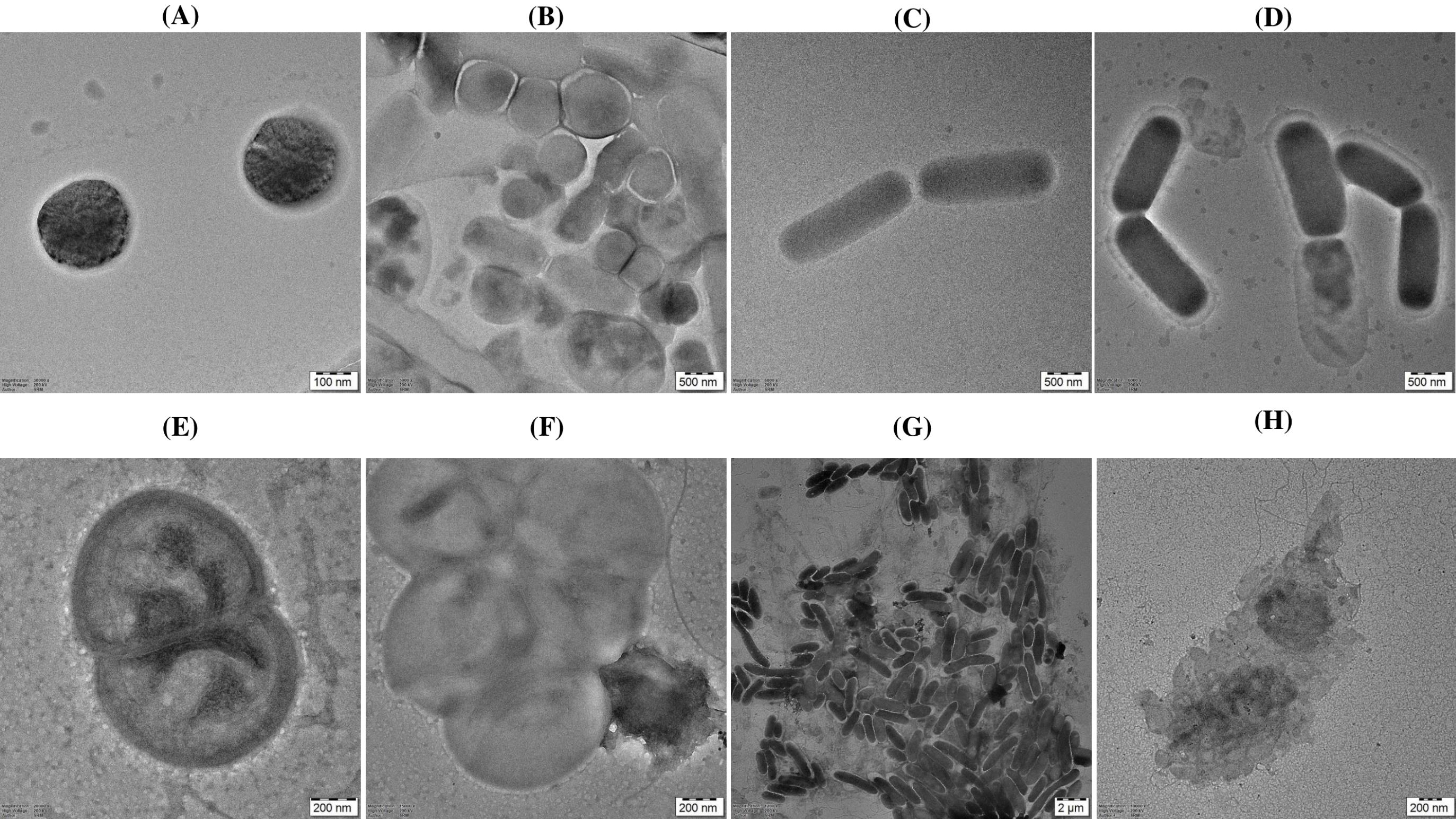
Fig. 7.
TEM photomicrograph of (A) S. aureus control (B) S. aureus treated with F3 fraction peptides for 120 min (C) K. pneumoniae control (D) K. pneumoniae treated with F3 fraction peptides for 120 min (E) E. faecalis control (F) E. faecalis treated with F3 fraction peptides for 120 min (G) P. aeruginosa, control (H) P. aeruginosa treated with F3 fraction peptides for 120 min.
.
TEM photomicrograph of (A) S. aureus control (B) S. aureus treated with F3 fraction peptides for 120 min (C) K. pneumoniae control (D) K. pneumoniae treated with F3 fraction peptides for 120 min (E) E. faecalis control (F) E. faecalis treated with F3 fraction peptides for 120 min (G) P. aeruginosa, control (H) P. aeruginosa treated with F3 fraction peptides for 120 min.
Cell viability assay
The cytotoxicity of the F3 fraction peptides on L929 fibroblast cell lines was evaluated by MTT Assay. No significant cytotoxicity was observed in the concentration range between 50-250 µg/mL (Fig. 8). The MTT results indicate that the F3 fraction peptides are nontoxic to cells.
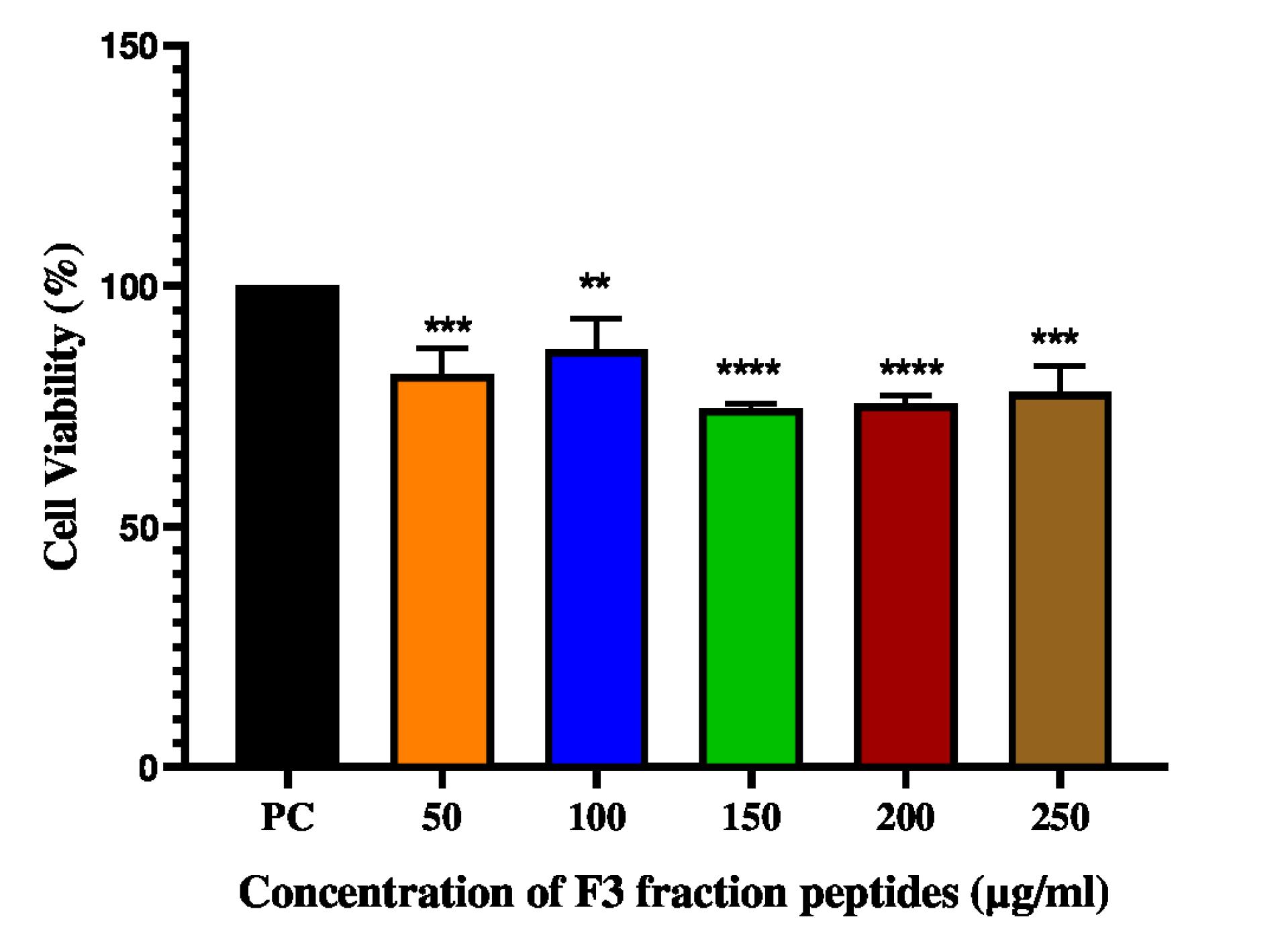
Fig. 8.
Cell viability Assay of F3 fraction peptides on L929 fibroblast cell lines. Data are representative of three independent experiments and are plotted as mean ± SD (n=3); ****P < 0.0001; ***P<0.001; **P<0.01
.
Cell viability Assay of F3 fraction peptides on L929 fibroblast cell lines. Data are representative of three independent experiments and are plotted as mean ± SD (n=3); ****P < 0.0001; ***P<0.001; **P<0.01
Discussion
LAB is recognized as a healthy bacteria in dairy products and processed vegetables, playing a crucial role in preservation. The present investigation focuses on the isolation, characterization, and antibacterial activity of peptides from L. plantarum 1407. The strain was tested for its antibacterial activity against four pathogens and the highest inhibitory activity was found against S. aureus (Fig. 1). Previous studies34 have reported the inhibition of S. aureus by CFS of LAB. Few studies have mentioned that the lyophilized CFS of L. casei can inhibit the growth of E. coli, Salmonella typhi, P. aeruginosa, S. aureus and methicillin-resistant Staphylococcus aureus (MRSA).35
The growth and antibacterial activity of L. plantarum 1407 was affected by initial pH, incubation temperature, and salt concentration. The bacteria exhibited antibacterial activity even at high pH and temperature and at a salt concentration of 0.7% (Supplementary file 1, Figs. S2-S4). In previous literature works the maximum growth of Leuconostoc mesenteroides L124 and L. curvatus L442 was reported at a temperature of 30°C and pH between 6-8.23 L. lactis showed maximum growth at a salt concentration of 0.8%.36 Similar results were revealed by Soltani et al33 that Pediococcus exhibited antibacterial activity at a salt concentration between 0.7%-0.8%. Enhanced nisin and pediocin production was observed at higher pH drop gradients.37 The CFS retained its antibacterial activity under varying conditions of pH and temperature. The results are consistent with previous reports wherein maximum antibacterial activity was observed between pH 6 and 8.38 Similar results were observed for peptide microcin J25 that exhibited increased antibacterial activity at pH 8 and temperature 37 °C.39 The CFS was tested for its sensitivity to extreme physiological conditions and it was observed that even at high temperature and pH, the CFS retained its antibacterial activity. The CFS retained >80% of its inhibitory activity after heat treatment at 60 °C or 80 °C for 10-20 minutes and was stable at a pH between 5-10. The antibacterial activity nearly decreased at pH >10. There have been reports on the stability of cell free supernatant of lactic acid bacteria after treatment for 30 min at 90 °C and 121 °C.40 Similarly, CFS from L. plantarum 27, Lactobacillus acidophilus, and Enterococcus durans were reported stable at 100 ºC for 60 minutes.21 Heat stable antibacterial peptides can be used in various sectors such as Food & Process Engineering industry, Cosmeceutical industry and pharmaceutical industry. The major applications of heat stable peptides are given in Table S3.
The isolated peptides were fractionated using size exclusion gel chromatography (Sephadex G-25) and among the four fractions obtained (Fig. 2), Fraction 3 (F3) exhibited increased antibacterial activity and was further analyzed. The minimum inhibitory concentration (MIC) of the F1-F4 fraction peptides showed that the F3 fraction exhibited the highest MIC (Fig. 3) and was in accordance with the MIC of other antimicrobial peptides from LAB.41 The MIC of nisin was 32 µg/mL4 which indicates that nisin and the F3 fraction peptides have antibacterial activity within a similar range (33 µg/mL). It was discovered that plantaricin 827 had a time and dose-dependent action on S. aureus cells.42 The antimicrobial activity of a newly discovered bacteriocin plantaricin GZ1-27 against MRSA was reported in a recent study.7 The MIC values and the growth curve analysis in the present study indicate that the F3 fraction peptides showed similar activity to plantaricin 827 and plantaricin GZ-127. The peptide fractions were further tested for their sensitivity to proteinase K. F3 fraction peptides retained about 60% of the antibacterial activity whereas F1, F2 and F4 fractions lost their antibacterial activity after treatment with proteinase K. Similar results were reported for plantaricin K25 and peptide ST8KF.43 Salivaricin from Ligilactobacillus salivarius also exhibited high residual activity after enzyme treatment.44 As F3 fraction peptides retained their antibacterial activity after treatment with proteinase K, MS/MS analysis has been performed to identify the molecular weight and amino acid sequences.
The MS/MS analysis of amino acids of F3 fraction peptides (Fig. 4) revealed the presence of arginine, lysine, histidine, glycine, and phenylalanine, all of which contributes to the effective binding ability of peptides with bacterial membranes. The presence of basic amino acids like arginine and lysine in F3 fraction peptides impart a highly positive charge to the peptides attracting them to the negatively charged membrane surface.45 Arginine in the F3 fraction peptides facilitates their interaction with negatively charged surfaces such as lipopolysaccharides (LPS) as reported previously.32 The presence of histidine can cause changes in membrane permeability.46 The hydrophobicity of phenylalanine influences the antimicrobial activity of peptides by neutralizing LPS on the bacterial membrane.47
FTIR revealed the spectral range of the functional group of F3 fraction between 400-4500 cm-1 (Fig. 5). The prominent groups were the Carbonyl group ( O=C-O stretching) at 2374.41 cm-1, aldehyde group at 2741.85 cm-1 and primary amine (N-H stretching) at 3439.17 cm-1. The peak at 1665.56 cm-1 (C=O stretching)attributes to the amide I of the peptides.
The membrane integrity analysis of the bacterial cells treated with peptides was monitored by flow cytometry. Flow cytometry analysis indicated that treatment of indicator organisms with F3 fraction peptides enhanced uptake of PI suggesting the loss of membrane integrity of the bacterial membrane. More than 50% cells were stained with PI after exposure to F3 fraction peptides for about 60 minutes. The difference in percentage rates of P. aeruginosa and K. pneumoniae suggest the different rates of membrane permeabilization for gram-negative bacteria. The sequence of steps occurring at the membrane appears to begin with depolarization followed by more significant membrane disruption. The percentage of dead cells indicates the ability of F3 fraction peptides to penetrate the bacterial cell membrane (Fig. 6; Figure S7-S10). The interactions of F3 fraction peptides with bacterial cells may also be affected by strain, concentration, and exposure time, which is supported by previous works.48 Propidium iodide cannot pass through intact cell membranes but can bind to cellular DNA. Blue signals were intensified at 20, 60 and 120 minutes indicating membrane integrity loss in the majority of the cells treated with F3 fraction peptides. The accumulation of dead cells strongly suggests that the inhibitory activity of the F3 fraction peptides is mediated by membrane disruption gradually leading to cell death. The F3 fraction peptides destroyed cell membrane integrity, causing the outflow of intracellular contents and, ultimately, cell death. This is in agreement with the activity of antimicrobial peptide tachyplesin that caused damage to the cell wall and cell membrane.49 As per the previous reports, the recognition of LPS on the outer membrane of Gram-negative bacteria and gradual membrane integrity loss by the F3 fraction peptides can be ascribed to the presence of alanine and arginine in the peptides.50
The TEM results suggested that the mechanism of action of F3 fraction peptides includes the formation of cytoplasmic membrane pores. The inhibition of pore formation in the membrane and cell wall synthesis in Gram-positive bacteria contributes to the loss of cellular structure after treatment with F3 fraction peptides.51,52 The progressive damage induced by the F3 fraction peptides to the cell membrane of bacteria leads to lysis of the indicator organisms (Fig. 7A-7H). The results are comparable to those obtained with antimicrobial peptides such as lactocin XN8-A, NK-2, L10, lactocin MXJ, and plantaricin GZ1-27.7,50 The presence of arginine in F3 fraction attracts negative outer membrane components such as LPS, lipoteichoic acid (LTA), phospholipids, and peptidoglycan precursor lipid II via electrostatic interaction which can be the initiating the antibacterial action.50 The presence of arginine in the F3 fraction peptides can potentially cause cytoplasmic internalization via the endocytic and direct translocation mechanisms.53 Arginine facilitates F3 fraction adsorption onto the outer membrane surface through electrostatic interactions between peptides and LPS. The peptides generate hydrogen bonds with the phosphate groups after adsorption, destroying salt bridges between phosphate and divalent cations and destabilizing the outer membrane.54 The hydrophobic moieties of peptides interact with the lipid tails of LPS molecules, further destabilizing the outer membrane. The peptides diffuse into the periplasmic region and adsorbs onto the surface of the cytoplasmic membrane once the outer membrane is disrupted.54 When peptides reach the cytoplasmic membrane, they cause disturbances that culminate in the loss of transmembrane potential and bacterial cell death. Gram-positive bacteria contain a layer of crosslinked peptidoglycans with teichoic acid encircling the cytoplasmic membrane, which is responsible for the bacterial cell's stiffness.54 The F3 fraction peptides diffuse across this peptidoglycan and act on the cytoplasmic membrane. The TEM images show that the favourable interactions between teichoic acid and cationic amino acids in peptides facilitate the accumulation of peptides on the surface of the cytoplasmic membrane, leading to its rupture.54
Hydrogen donors include basic amino acids such as arginine and lysine. Hydrophobic residues in the F3 fraction peptides permeate and disorganize the lipid tail region of the bacterial membrane. Increased accumulation of peptides caused membrane thinning, leading to lateral expansion of the membrane, which impacts the mechanical properties of the membrane.55 The membrane expansion lowers the packing of the lipid molecules, resulting in the creation of cavities and a reduction in the free energy of water molecules translocating across the lipid tail area. As a result, the membrane becomes perforated due to the collapse of the transmembrane potential followed by additional membrane malfunctions like inhibition of ATP generation and loss of proton motive force gradually resulting in the bacterial death.56 Nisin from L. lactis exhibited a similar mode of antibacterial action by creating cell membrane pores and cell wall biosynthesis interference through a specific lipid II interaction.57 As indicated by the morphology and cellular lysis, the primary antibacterial mechanism of F3 fraction peptides is surface binding of the bacterial membrane leading to membrane disruption.
The F3 fractions peptides did not exert any cytotoxic effects on L929 fibroblast cell lines. The MTT assay showed >70% viability at all concentrations of the F3 fraction peptides (Fig. 8). Peptides have been reported to show about 91% cell viability on HEK293 cell lines.31 The differences in the effect of peptides on cell lines can be attributed to the difference in their preparation, exposure time and incubation conditions.31
Conclusion
The low molecular weight F3 fraction peptides from L. plantarum 1407 inhibited the growth of pathogens like S. aureus, P. aeruginosa, E. faecalis, and K. pneumoniae. The four peptide fractions obtained from Sephadex (G-25) Chromatography demonstrated a broad-spectrum antibacterial activity at low concentrations. Fraction 3 exhibited maximum antibacterial activity against the indicator organisms. A total of 10 peptides were identified from the F3 fraction through LC-MS/MS analysis. The sensitivity of the F3 fraction peptides to different temperature and pH changes have been evaluated and the F3 fraction peptides retained their antibacterial activity even at high temperature and pH. The incubation of the F3 fraction peptides with the above-mentioned bacterial strains resulted in cell membrane damage and outflow of contents of cytoplasm. These low molecular weight antibacterial peptides from L. plantarum 1407 could potentially serve as therapeutic agents to control bacterial infections in humans with further studies to address their in vivo effects.
Research Highlights
What is the current knowledge?
√ Peptides from LAB are powerful antimicrobial agents with great potential in the prevention and treatment of various bacterial infections.
√ The activity of the peptides is rapid, and they show a lower chance to develop resistance than conventional antibiotics.
What is new here?
√ Novel peptides from L. plantarum 1407 with antibacterial activity have been identified and their role as potential candidates for treating infections has been explored.
Acknowledgment
The authors of this study gratefully acknowledge SRM Central Instrumentation Facility (SCIF), Nanotechnology Research Center (NRC), Department of Translational Medicine and Research (TMR), Interdisciplinary Institute of Indian System of Medicine (IIISM), Research facility (I, II, III) and SRM Institute of Science and Technology (SRMIST) for providing the instrument facilities for various analysis.
Competing Interests
The authors declare there is no conflict of interest.
Ethical Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Supplementary files
Supplementary file 1 contains Figs. S1-S10 and Tables S1-S3.
(pdf)
References
- Ruiz Rodríguez LG, Mohamed F, Bleckwedel J, Medina R, De Vuyst L, Hebert EM. Diversity and Functional Properties of Lactic Acid Bacteria Isolated From Wild Fruits and Flowers Present in Northern Argentina. Front Microbiol 2019; 10:1091. doi: 10.3389/fmicb.2019.01091 [Crossref] [ Google Scholar]
- Yang SC, Lin CH, Sung CT, Fang JY. Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front Microbiol 2014; 5:241. doi: 10.3389/fmicb.2014.00241 [Crossref] [ Google Scholar]
- Gupta V, Datta P. Next-generation strategy for treating drug resistant bacteria: Antibiotic hybrids. Indian J Med Res 2019; 149:97-106. doi: 10.4103/ijmr.IJMR_755_18 [Crossref] [ Google Scholar]
- Pfalzgraff A, Brandenburg K, Weindl G. Antimicrobial Peptides and Their Therapeutic Potential for Bacterial Skin Infections and Wounds. Front Pharmacol 2018; 9:281. doi: 10.3389/fphar.2018.00281 [Crossref] [ Google Scholar]
- Zimina M, Babich O, Prosekov A, Sukhikh S, Ivanova S, Shevchenko M. Overview of Global Trends in Classification, Methods of Preparation and Application of Bacteriocins. Antibiotics (Basel) 2020; 9:553. doi: 10.3390/antibiotics9090553 [Crossref] [ Google Scholar]
- Wongsen S WD, Tumwasorn S. Lactobacillus plantarum B7 attenuates Salmonella typhimurium infection in mice: preclinical study in vitro and in vivo. Asian Biomed 2019; 12:211-8. doi: 10.1515/abm-2019-0022 [Crossref] [ Google Scholar]
- Du H, Chi H, Yao H, Lu Z, Bie X, Zhang C. The antibacterial activity of plantaricin GZ1-27 against MRSA and its bio-preservative effect on chilled pork in combination with chitosan. Int J Food Microbiol 2022; 365:109539. doi: 10.1016/j.ijfoodmicro.2022.109539 [Crossref] [ Google Scholar]
- Ovchinnikov KV, Kranjec C, Thorstensen T, Carlsen H, Diep DB. Successful Development of Bacteriocins into Therapeutic Formulation for Treatment of MRSA Skin Infection in a Murine Model. Antimicrob Agents Chemother 2020; 64:e00829-20. doi: 10.1128/aac.00829-20 [Crossref] [ Google Scholar]
- Al Atya AK, Belguesmia Y, Chataigne G, Ravallec R, Vachée A, Szunerits S. Anti-MRSA Activities of Enterocins DD28 and DD93 and Evidences on Their Role in the Inhibition of Biofilm Formation. Front Microbiol 2016; 7:817. doi: 10.3389/fmicb.2016.00817 [Crossref] [ Google Scholar]
- Piper C, Casey PG, Hill C, Cotter PD, Ross RP. The Lantibiotic Lacticin 3147 Prevents Systemic Spread of Staphylococcus aureus in a Murine Infection Model. Int J Microbiol 2012; 2012:806230. doi: 10.1155/2012/806230 [Crossref] [ Google Scholar]
- Hanny ELL, Mustopa AZ, Budiarti S, Darusman HS, Ningrum RA, Fatimah Fatimah. Efficacy, toxicity study and antioxidant properties of plantaricin E and F recombinants against enteropathogenic Escherichia coli K11 (EPEC K11). Mol Biol Rep 2019; 46:6501-12. doi: 10.1007/s11033-019-05096-9 [Crossref] [ Google Scholar]
- Tseng CC, Murni L, Han TW, Arfiati D, Shih HT, Hu SY. Molecular Characterization and Heterologous Production of the Bacteriocin Peocin, a DNA Starvation/Stationary Phase Protection Protein, from Paenibacillusehimensis NPUST1. Molecules 2019; 24:2516. doi: 10.3390/molecules24132516 [Crossref] [ Google Scholar]
- Phumisantiphong U, Siripanichgon K, Reamtong O, Diraphat P. A novel bacteriocin from Enterococcus faecalis 478 exhibits a potent activity against vancomycin-resistant enterococci. PLoS One 2017; 12:e0186415. doi: 10.1371/journal.pone.0186415 [Crossref] [ Google Scholar]
- Epand RF, Maloy WL, Ramamoorthy A, Epand RM. Probing the "charge cluster mechanism" in amphipathic helical cationic antimicrobial peptides. Biochemistry 2010; 49:4076-84. doi: 10.1021/bi100378m [Crossref] [ Google Scholar]
- Galván Márquez IJ, McKay B, Wong A, Cheetham JJ, Bean C, Golshani A. Mode of action of nisin on Escherichia coli. Can J Microbiol 2020; 66:161-8. doi: 10.1139/cjm-2019-0315 [Crossref] [ Google Scholar]
- Luz C SF, Luciano FB, Mañes J, Meca G. In vitro antifungal activity of bioactive peptides produced by Lactobacillus plantarum against Aspergillus parasiticus and Penicillium expansum. LWT- Food Sci Technol 2017; 81:128-35. doi: 10.1016/j.lwt.2017.03.053 [Crossref] [ Google Scholar]
- Turovskiy Y, Ludescher RD, Aroutcheva AA, Faro S, Chikindas ML. Lactocin 160, a Bacteriocin Produced by Vaginal Lactobacillus rhamnosus, Targets Cytoplasmic Membranes of the Vaginal Pathogen, Gardnerella vaginalis. Probiotics Antimicrob Proteins 2009; 1:67-74. doi: 10.1007/s12602-008-9003-6 [Crossref] [ Google Scholar]
- Golneshin A, Gor MC, Williamson N, Vezina B, Van TTH, May BK. Smith AT Discovery and characterisation of circular bacteriocin
plantacyclin B21AG from Lactiplantibacillus plantarum B21. Heliyon 2020; 6:e04715. doi: 10.1016/j.heliyon.2020.e04715 [Crossref] [ Google Scholar]
- Karthikeyan V SS. Isolation and partial characterization of bacteriocin produced from Lactobacillus plantarum. Afr J Microbiol Res 2009; 3:233-9. [ Google Scholar]
- Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: A review. J Pharm Anal 2016; 6:71-9. doi: 10.1016/j.jpha.2015.11.005 [Crossref] [ Google Scholar]
- Todorov SD. Bacteriocin production by Lactobacillus plantarum AMA-K isolated from Amasi, a Zimbabwean fermented milk product and study of the adsorption of bacteriocin AMA-K TO Listeria sp. Braz J Microbiol 2008; 39:178-87. doi: 10.1590/s1517-838220080001000035 [Crossref] [ Google Scholar]
- Muhialdin BJ, Algboory HL, Kadum H, Mohammed NK, Saari N, Hassan Z. Antifungal activity determination for the peptides generated by Lactobacillus plantarum TE10 against Aspergillus flavus in maize seeds. Food Control 2020; 109:106898. [ Google Scholar]
- Mataragas M, Drosinos EH, Tsakalidou E, Metaxopoulos J. Influence of nutrients on growth and bacteriocin production by Leuconostoc mesenteroides L124 and Lactobacillus curvatus L442. Antonie Van Leeuwenhoek 2004; 85:191-8. doi: 10.1023/B:ANTO.0000020291.01957.a2 [Crossref] [ Google Scholar]
- Leroy F, De VL. Bacteriocin production by Enterococcus faecium RZS C5 is cell density limited and occurs in the very early growth phase. Int J Food Microbiol 2002; 72:155-64. doi: 10.1016/s0168-1605(01)00635-3 [Crossref] [ Google Scholar]
- Weinstein MP, Lewis JS, 2nd. The Clinical and Laboratory Standards Institute Subcommittee on Antimicrobial Susceptibility Testing: Background, Organization, Functions, and Processes. J Clin Microbiol 2020; 58. 10.1128/jcm.01864-19
- Du H, Yang J, Lu X, Lu Z, Bie X, Zhao H. Purification, Characterization, and Mode of Action of Plantaricin GZ1-27, a Novel Bacteriocin against Bacillus cereus. J Agric Food Chem 2018; 66:4716-24. doi: 10.1021/acs.jafc.8b01124 [Crossref] [ Google Scholar]
- Kong X, Song W, Hua Y, Li X, Chen Y, Zhang C. Insights into the antibacterial activity of cottonseed protein-derived peptide against Escherichia coli. Food Funct 2020; 11:10047-57. doi: 10.1039/d0fo01279c [Crossref] [ Google Scholar]
- Brown MR, Hands CL, Coello-Garcia T, Sani BS, Ott AIG, Smith SJ. A flow cytometry method for bacterial quantification and biomass estimates in activated sludge. J Microbiol Methods 2019; 160:73-83. doi: 10.1016/j.mimet.2019.03.022 [Crossref] [ Google Scholar]
- Li L, Song F, Sun J, Tian X, Xia S, Le G. Membrane damage as first and DNA as the secondary target for anti-candidal activity of antimicrobial peptide P7 derived from cell-penetrating peptide ppTG20 against Candida albicans. J Pept Sci 2016; 22:427-33. doi: 10.1002/psc.2886 [Crossref] [ Google Scholar]
- Khanna S, Bishnoi M, Kondepudi KK, Shukla G. Isolation, characterization and anti-inflammatory mechanism of probiotics in lipopolysaccharide-stimulated RAW 2647 macrophages. World J Microbiol Biotechnol 2020; 36:74. doi: 10.1007/s11274-020-02852-z [Crossref] [ Google Scholar]
- Sornsenee P, Chatatikun M, Mitsuwan W, Kongpol K, Kooltheat N, Sohbenalee S. Lyophilized cell-free supernatants of Lactobacillus isolates exhibited antibiofilm, antioxidant, and reduces nitric oxide activity in lipopolysaccharide-stimulated RAW 2647 cells. PeerJ 2021; 9:e12586. doi: 10.7717/peerj.12586 [Crossref] [ Google Scholar]
- Khalid F, Siddiqi R, Mojgani N. Detection and characterization of a heat stable bacteriocin (Lactocin LC-09) produced by a clinical isolate of lactobacilli. Medical Journal of Islamic Academy of Sciences 1999; 12:67-71. [ Google Scholar]
- Soltani S, Hammami R, Cotter PD, Rebuffat S, Said LB, Gaudreau H. Bacteriocins as a new generation of antimicrobials: toxicity aspects and regulations. FEMS Microbiol Rev 2021; 45:fuaa039. doi: 10.1093/femsre/fuaa039 [Crossref] [ Google Scholar]
- Nowak MG, Skwarecki AS, Milewska MJ. Amino Acid Based Antimicrobial Agents - Synthesis and Properties. ChemMedChem 2021; 16:3513-44. doi: 10.1002/cmdc.202100503 [Crossref] [ Google Scholar]
- Martin-Visscher LA, Gong X, Duszyk M, Vederas JC. The three-dimensional structure of carnocyclin A reveals that many circular bacteriocins share a common structural motif. J Biol Chem 2009; 284:28674-81. doi: 10.1074/jbc.M109.036459 [Crossref] [ Google Scholar]
- Verluyten J, Messens W, De Vuyst L. Sodium chloride reduces production of curvacin A, a bacteriocin produced by Lactobacillus curvatus strain LTH 1174, originating from fermented sausage. Appl Environ Microbiol 2004; 70:2271-8. doi: 10.1128/aem.70.4.2271-2278.2004 [Crossref] [ Google Scholar]
- Guerra NP, Pastrana L. Influence of pH drop on both nisin and pediocin production by Lactococcus lactis and Pediococcus acidilactici. Lett Appl Microbiol 2003; 37:51-5. doi: 10.1046/j.1472-765x.2003.01346.x [Crossref] [ Google Scholar]
- Borrero J, Kelly E, O'Connor PM, Kelleher P, Scully C, Cotter PD. Plantaricyclin A, a Novel Circular Bacteriocin Produced by Lactobacillus plantarum NI326: Purification, Characterization, and Heterologous Production. Appl Environ Microbiol 2018; 84:801-17. doi: 10.1128/aem.01801-17 [Crossref] [ Google Scholar]
- Kapil S, Sharma V. d-Amino acids in antimicrobial peptides: a potential approach to treat and combat antimicrobial resistance. Can J Microbiol 2021; 67:119-37. doi: 10.1139/cjm-2020-0142 [Crossref] [ Google Scholar]
- Iyapparaj P, Maruthiah T, Ramasubburayan R, Prakash S, Kumar C, Immanuel G. Optimization of bacteriocin production by Lactobacillus spMSU3IR against shrimp bacterial pathogens. AquatBiosyst 2013; 9:12. doi: 10.1186/2046-9063-9-12 [Crossref] [ Google Scholar]
- El-Kazzaz SS, Abou El-Khier NT. Effect of the lantibiotic nisin on inhibitory and bactericidal activities of antibiotics used against vancomycin-resistant enterococci. J Glob Antimicrob Resist 2020; 22:263-9. doi: 10.1016/j.jgar.2020.02.031 [Crossref] [ Google Scholar]
- Zhao D, Wang Q, Lu F, Bie X, Zhao H, Lu Z. A novel plantaricin 827 effectively inhibits Staphylococcus aureus and extends shelf life of skim milk. LWT 2022; 154:112849. doi: 10.1016/j.lwt.2021.112849 [Crossref] [ Google Scholar]
- Goh HF, Philip K. Purification and Characterization of Bacteriocin Produced by Weissella confusa A3 of Dairy Origin. PLoS One 2015; 10:e0140434. doi: 10.1371/journal.pone.0140434 [Crossref] [ Google Scholar]
- Wayah SB, Philip K. Purification, characterization, mode of action, and enhanced production of Salivaricin mmaye1, a novel bacteriocin from Lactobacillus salivarius SPW1 of human gut origin. Electronic Journal of Biotechnology 2018; 35:39-47. [ Google Scholar]
- Wang G, Yu Y, Garcia-Gutierrez E, Jin X, He Y, Wang L. Lactobacillus acidophilus JCM 1132 strain and its mutant with different bacteriocin-producing behaviour have various in situ effects on the gut microbiota of healthy mice. Microorganisms 2019; 8:49. doi: 10.3390/microorganisms8010049 [Crossref] [ Google Scholar]
- Kato T, Matsuda T, Ogawa E, Ogawa H, Kato H, Doi U. Plantaricin-149, a bacteriocin produced by Lactobacillus plantarum NRIC 149. Journal of Fermentation and Bioengineering 1994; 77:277-82. doi: 10.1016/0922-338X(94)90234-8 [Crossref] [ Google Scholar]
- Zhu X, Zhao Y, Sun Y, Gu Q. Zhu X, Zhao Y, Sun Y, Gu QPurification and characterisation of plantaricin ZJ008, a novel bacteriocin against Staphylococcus sppfrom Lactobacillus plantarum ZJ008. Food Chem 2014; 165:216-23. doi: 10.1016/j.foodchem.2014.05.034 [Crossref] [ Google Scholar]
- Hong J, Guan W, Jin G, Zhao H, Jiang X, Dai J. Mechanism of tachyplesin I injury to bacterial membranes and intracellular enzymes, determined by laser confocal scanning microscopy and flow cytometry. Microbiol Res 2015; 170:69-77. doi: 10.1016/j.micres.2014.08.012 [Crossref] [ Google Scholar]
- Tang C-H, Ma C-Y. Tang C-H, Ma C-YHeat-induced modifications in the functional and structural properties of vicilin-rich protein isolate from kidney (Phaseolus vulgaris L) bean. Food Chem 2009; 115:859-66. doi: 10.1016/j.foodchem.2008.12.102 [Crossref] [ Google Scholar]
- Wang X, Mishra B, Lushnikova T, Narayana JL, Wang G. Amino Acid Composition Determines Peptide Activity Spectrum and Hot-Spot-Based Design of Merecidin. Adv Biosyst 2018; 2:1700259. doi: 10.1002/adbi.201700259 [Crossref] [ Google Scholar]
- Hasper HE, Kramer NE, Smith JL, Hillman JD, Zachariah C, Kuipers OP. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 2006; 313:1636-7. doi: 10.1126/science.1129818 [Crossref] [ Google Scholar]
- Malanovic N, Lohner K. Gram-positive bacterial cell envelopes: The impact on the activity of antimicrobial peptides. BiochimBiophys Acta 2016; 1858:936-46. [ Google Scholar]
- Nakase I, Takeuchi T, Tanaka G, Futaki S. Methodological and cellular aspects that govern the internalization mechanisms of arginine-rich cell-penetrating peptides. Adv Drug Deliv Rev 2008; 60:598-607. doi: 10.1016/j.addr.2007.10.006 [Crossref] [ Google Scholar]
- Li J, Koh JJ, Liu S, Lakshminarayanan R, Verma CS, Beuerman RW. Membrane Active Antimicrobial Peptides: Translating Mechanistic Insights to Design. Front Neurosci 2017; 11:73. doi: 10.3389/fnins.2017.00073 [Crossref] [ Google Scholar]
- Stevens MJ. Coarse-grained simulations of lipid bilayers. J Chem Phys 2004; 121:11942-8. doi: 10.1063/1.1814058 [Crossref] [ Google Scholar]
- Dimroth P, Kaim G, Matthey U. Crucial role of the membrane potential for ATP synthesis by F(1)F(o) ATP synthases. J Exp Biol 2000; 203:51-9. doi: 10.1242/jeb.203.1.51 [Crossref] [ Google Scholar]
- Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem 2001; 276:1772-9. doi: 10.1074/jbc.M00677020 [Crossref] [ Google Scholar]