Bioimpacts. 14(4):29957.
doi: 10.34172/bi.2023.29957
Review
Cancer treatment comes to age: from one-size-fits-all to next-generation sequencing (NGS) technologies
Sepideh Parvizpour Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Validation, Writing – original draft, 1, 2, * 
Hanieh Beyrampour-Basmenj Visualization, Writing – original draft, 2
Jafar Razmara Software, Validation, Writing – review & editing, 3, * 
Farhad Farhadi Resources, 4
Mohd Shahir Shamsir Investigation, 5
Author information:
1Research Center for Pharmaceutical Nanotechnology, Biomedicine Institute, Tabriz University of Medical Sciences, Tabriz, Iran
2Department of Medical Biotechnology, School of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
3Department of Computer Science, Faculty of Mathematics, Statistics and Computer Science, University of Tabriz, Tabriz, Iran
4Food and Drug Administration, Tabriz University of Medical Sciences, Tabriz, Iran
5Bioinformatics Research Group, Faculty of Science, Universiti Teknologi Malaysia, Johor Bahru, Malaysia
Abstract
Cancer is one of the leading causes of death worldwide and one of the greatest challenges in extending life expectancy. The paradigm of one-size-fits-all medicine has already given way to the stratification of patients by disease subtypes, clinical characteristics, and biomarkers (stratified medicine). The introduction of next-generation sequencing (NGS) in clinical oncology has made it possible to tailor cancer patient therapy to their molecular profiles. NGS is expected to lead the transition to precision medicine (PM), where the right therapeutic approach is chosen for each patient based on their characteristics and mutations. Here, we highlight how the NGS technology facilitates cancer treatment. In this regard, first, precision medicine and NGS technology are reviewed, and then, the NGS revolution in precision medicine is described. In the sequel, the role of NGS in oncology and the existing limitations are discussed. The available databases and bioinformatics tools and online servers used in NGS data analysis are also reviewed. The review ends with concluding remarks.
Keywords: Cancer, Next-generation sequencing, One-size-fits-all medicine, Precision medicine, Personalized medicine, Stratified medicine
Copyright and License Information
© 2024 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Globally, cancer continues to threaten human lives and is still a major cause of death for millions of people despite continuous advances in its diagnosis and treatment.1-4 According to statistics, the number of cancer cases will increase by 47% from 19.3 million to 28.4 million by 2040.5 Several factors, including economic growth and lifestyle changes, may contribute to this increasing rate. The disease is heterogeneous, affecting different organs of the human body and characterized by several subtypes. The development of cancer is the result of uncontrolled cell division caused by mutations in specific genes, including proto-oncogenes, cell cycle regulators, and tumor suppressor genes.6
One-size-fits-all medicine pertains to a treatment strategy that is standardized and uniformly implemented among all patients, disregarding individual variations in genetic composition, disease attributes, or response to treatment. Historically, this has been the conventional approach in medicine, entailing the development of a single treatment protocol based on the average reaction observed within a population. Nonetheless, this approach presents certain limitations, especially in the field of oncology, given the highly diverse nature of cancer as a disease with varied molecular profiles and treatment outcomes.7
The variation in therapeutic platforms among patients is influenced by genetic variations, tumor heterogeneity, and individual patient characteristics. This heterogeneity creates difficulties in achieving optimal treatment outcomes for all patients using a standardized approach. Conversely, individualized or precision medicine seeks to customize treatment strategies based on the distinct characteristics of each patient, such as their genetic profile, tumor molecular features, and other relevant factors. This personalized approach enables targeted therapies that are more likely to be effective and reduce unnecessary treatments or adverse effects.8
In the context of cancer, the significance of universal medicine in therapeutic applications is its potential to enhance treatment outcomes and patient survival rates. Healthcare providers can improve treatment effectiveness by considering individual patient characteristics, such as unique genetic mutations or biomarkers, and selecting appropriate treatments while avoiding ineffective ones. This approach can result in more accurate and focused therapies, potentially increasing treatment response rates and decreasing the likelihood of treatment-related toxicities.9
Thanks to recent advances in high-throughput genomics and proteomics, scientists can now gain insight into the role of these genetic mutations and the signaling pathways involved in cancer cells.10 However, most cancers are genetically complex and are best described by pathway activation rather than specific mutations. Following the success of the Human Genome Project, similar projects were launched to understand how the genome contributes to different types of cancer. As a result of this success and the increased accessibility and reliability of sequencing, genomic research has now entered clinical practice. NGS has introduced a new approach to cancer treatment called precision medicine (PM).11
Before the advent of NGS, Sanger sequencing and PCR-based techniques were used to determine the mutational status of cancer; NGS panels, on the other hand, allow the identification of alterations even in sparse biopsy tissue and allow a comprehensive test to examine several genes at once. NGS can be used to obtain genetic information with a simple blood sample, especially when it is unsafe to collect tissue for molecular testing, such as in brain, lung, and peritoneal lesions.12 According to this concept, medical therapies are tailored to the individual characteristics and conditions of each patient.13 Despite widespread use in a variety of healthcare settings, NGS is most advanced in oncology, where physicians use NGS to analyze tumors and match them to therapy that targets the genetic alterations that drive tumor growth.14 Roy Chowdhury's study was the first to use NGS in personalized oncology.15 By using these techniques, he was able to demonstrate that patient health care could be improved and genetic information could be used most effectively.
NGS can predict risk factors, diagnose disease through sequencing and medical imaging, identify biomarkers, identify therapeutic targets for new drugs, and accurately predict prognosis.16,17 Cancer has numerous genomic aberrations that make it a multifaceted disease. However, by using NGS, it is possible to detect these mutations and aberrations in cancer-causing genes. Through NGS, genomic sequencing is becoming more accessible for clinical use, and genomic profiling is a promising approach for precision oncology in the future. Cancer genomics has benefited the most from NGS, as it allows for the reanalysis, comparison, and sequencing of normal and tumor genomes from the same patient. Because of its ability to detect a large number of variants related to complicated mechanisms of oncogenesis and tumor heterogeneity, this technology has been used in many cancer studies.18
This review provides an overview of the role of NGS technology in cancer treatment based on the precision medicine strategy. The original research and review papers with the keywords of Next-Generation sequencing (NGS), precision medicine (PM), one-size-fits-all medicine, personalized medicine, and bioinformatics algorithms and databases in NGS were reviewed and described in this paper. In the sequel, the existing limitations and challenges of the technology and experts’ opinion are discussed. We also describe the available databases and bioinformatics tools and online servers used in NGS for analyzing variants from NGS data generated for the analysis of cancer data.
Precision medicine
The development of prevention and treatment methods does not take into account individual differences between people. Over the past 30 years, medicine has shown that effective treatments can also be ineffective and have side effects.19 Precision medicine promises to deliver the right treatment to the right patient at the right time. Several terms associated with personalized medicine are often used interchangeably, including stratified medicine, individualized medicine, and precision medicine.
In precision medicine, genetic, environmental, and lifestyle variables are considered in the treatment and prevention of disease (Fig. 1). Using laboratory and genetic tests, individuals can predict when a disease will begin, whether it will progress or worsen, the likelihood of a particular outcome, and which treatment will be most effective. From disease prevention to health promotion to population-level and public health interventions, precision medicine approaches can be used in all areas of health. Outside of cancer pharmacotherapy, there have been few successful applications of personalized drug therapy. For example, 53 medicines were considered personalized medicine in Germany in 2017, of which 41 (77%) were for cancer treatment and 12 (23%) were for other purposes.13 The purpose of PM in oncology is to identify mutations associated with cancer progression or resistance to treatment. Since NGS has been used and large genetic databases are available, this approach has evolved considerably. Taking into account cancer mutations and a variety of patient characteristics, NGS can initiate the transition to PM and enable physicians to choose the best therapeutic approach for cancer patients.13
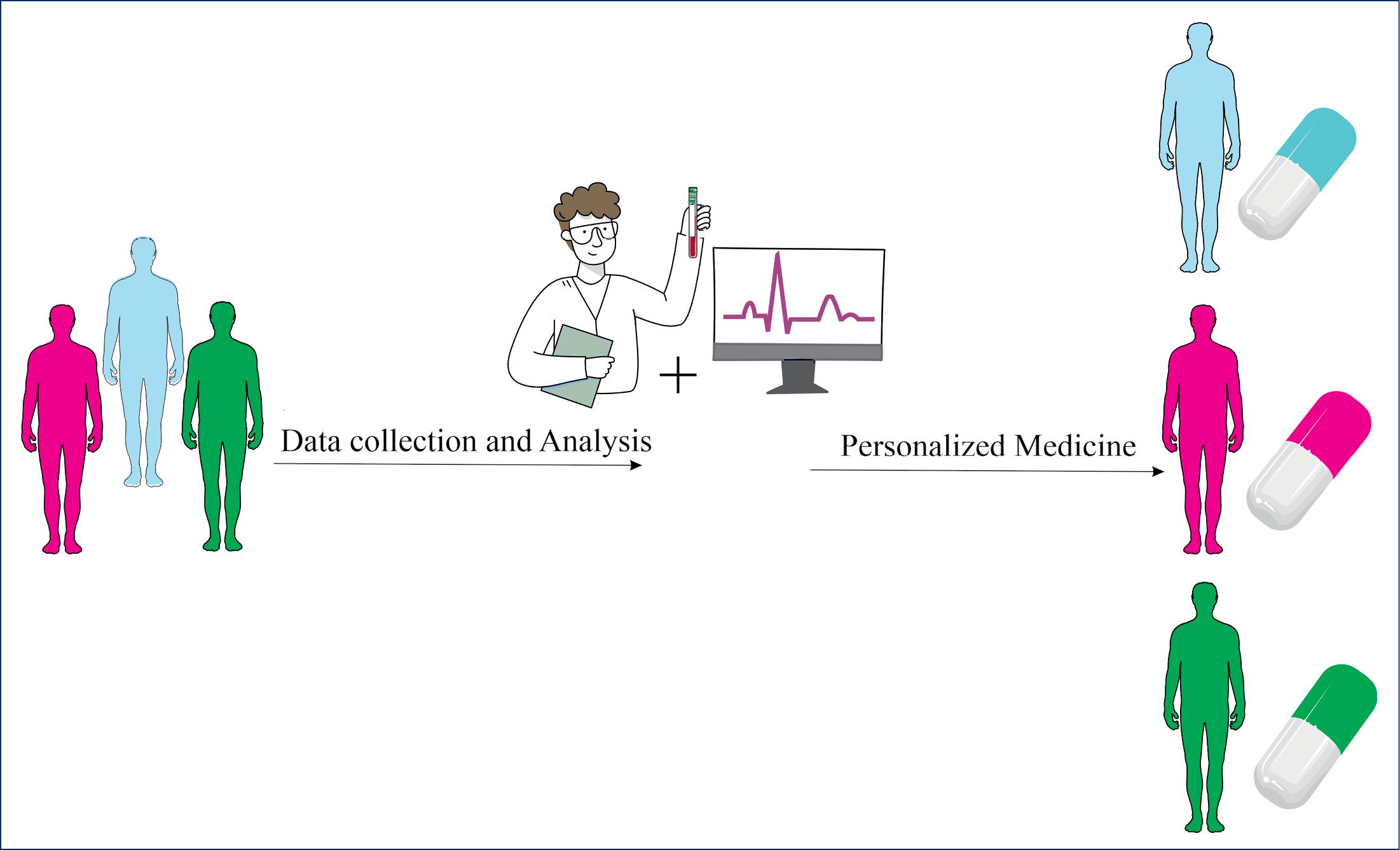
Fig. 1.
Personalized medicine considering genetic, environmental, and lifestyle variables in the treatment and prevention of disease
.
Personalized medicine considering genetic, environmental, and lifestyle variables in the treatment and prevention of disease
NGS technology
With the completion of the Human Genome Project in 2004, researchers were able to identify cancer genes in silico and define their sequences using computational tools. Cancer gene exons are amplified by PCR, and their sequences are determined by fluorescent Sanger sequencing. Because only a limited amount of DNA was available in a given cancer sample and this approach was not scalable, the first efforts of this type were limited to the analysis of a small number of genes.20 After Sanger sequencing, NGS was the first sequencing technology to sequence the human cancer genome. This project contributed to the identification of new subtypes, the development of biomarkers, and the identification of new therapeutic targets for cancer, culminating in the development of the Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov).21 (Fig. 2). Other NGS efforts include the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) for pediatric cancers and the International Cancer Genomics Consortium for adult cancers (ICGC; https://dcc.icgc.org). There are ∼25000 tumors analyzed by TCGA and ICGC for whole-genome sequencing (WGS). It is possible to retrieve curated information on somatic mutations in more than one million tumor samples through a variety of databases, including the Catalog of Somatic Mutations in Cancer (COSMIC) (Table 1).
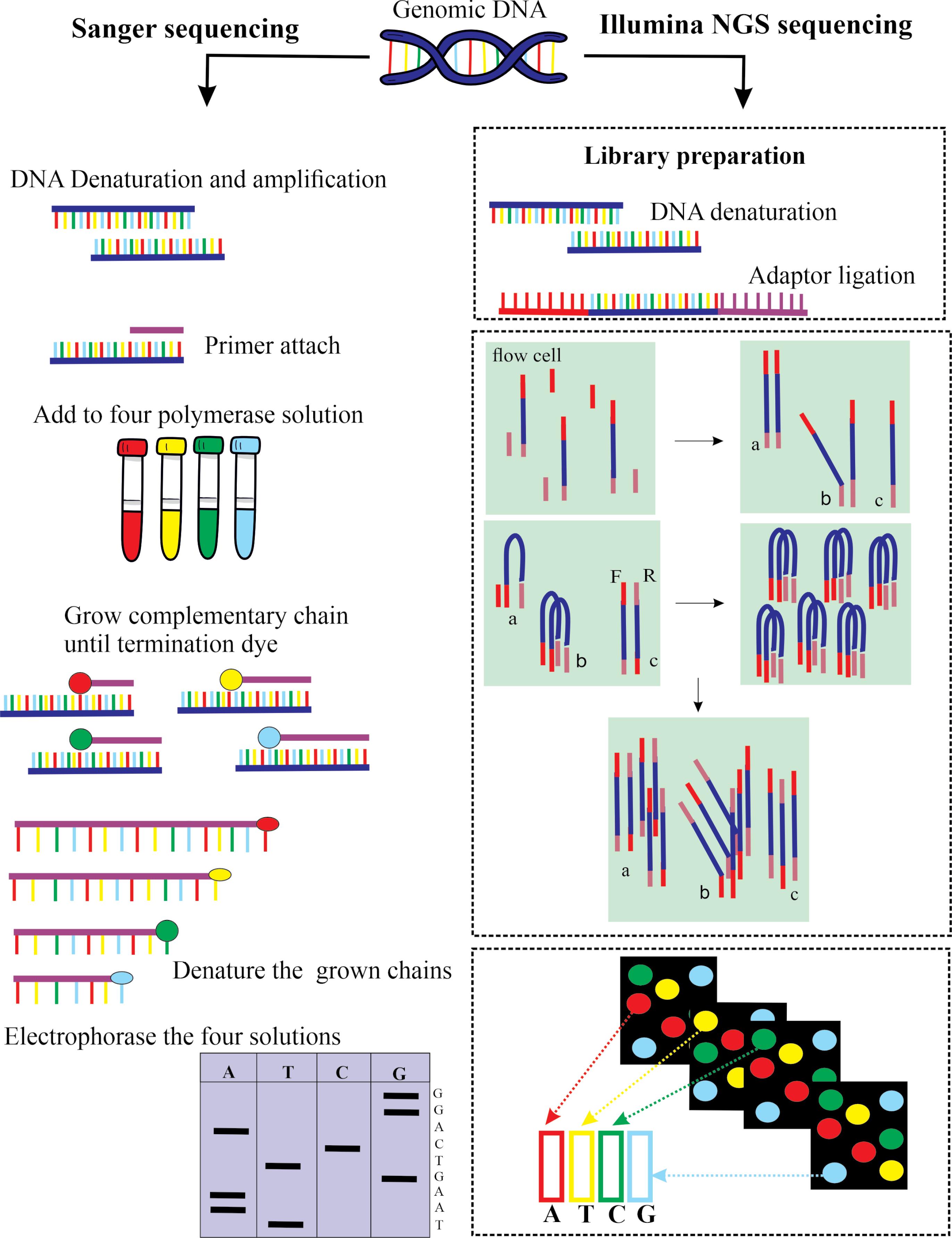
Fig. 2.
Next-generation sequencing (NGS) compared to Sanger sequencing. NGS platforms are able to concurrently extract the sequence of millions of DNA fragments producing vast volumes of data, while Sanger sequencing can identify the DNA fragment in a reaction, up to a maximum of *700 bases.
.
Next-generation sequencing (NGS) compared to Sanger sequencing. NGS platforms are able to concurrently extract the sequence of millions of DNA fragments producing vast volumes of data, while Sanger sequencing can identify the DNA fragment in a reaction, up to a maximum of *700 bases.
Table 1.
A brief overview of databases used for genetic variant characterization
Numerous recurrent genomic aberrations were discovered by the major research consortia. In addition, these studies found a 'long tail' of rare mutations that were often actionable. Molecular subtyping of breast cancer and the discovery of novel genetic alterations in lung cancer have both been aided by genome and transcriptome sequencing in clinical practice. Our understanding of other cancers is also influenced by these findings.
Several NGS platforms are available for high-throughput sequencing. The quality, quantity, and application of sequencing are influenced by different sequencing approaches. NGS technology is broadly divided into short-read and long-read sequencing. The difference between the two is based on the length of the reads, which ranges from 100 to 600 bp in the first technique, while it can reach up to 900 kb in the second. Short-read sequencing is widely used because of its cost and accuracy; however, short-read sequencing has difficulty resolving heterozygous or repetitive sequences, whereas long-read sequencing is most effective. The major platforms using short-read technology are Illumina, Ion Torrent, 454 Life Science, and SOLiD. The first three are based on "by synthesis" sequencing, while the SOLiD platform is based on "by ligation" sequencing. A nanopore-based system, the MinION system, and the PacBio system, which uses a Single Molecule, Real-Time (SMRT) approach, are the main long-read technologies available on the market today.22 A typical NGS run involves genomic DNA extraction from samples, library preparation, adaptor ligation, adaptor sequencing, sample enrichment, and finally sequencing.23 Illumina,24 Ion Torrent,25 and Complete Genomics Technology26 are three of the most popular NGS platforms currently available.27
Regardless of platform, all NGS processes go through the same three phases: library preparation, sequencing, and data analysis.28 Among NGS platforms, Illumina is perhaps the most popular because it offers a wide range of scalable options to meet the needs of different study designs, sequencing costs, and intended uses. An Illumina sequencing platform comparison tool helps researchers select the best sequencing platform for their needs.24 The Ion Torrent sequencing platform offers similar sequencing efficiency to Illumina in terms of volume and speed. Unlike Illumina, which uses fluorescent labels to detect synthesized nucleotides, Ion Torrent uses semiconductors. During DNA polymerization, hydrogen ions are released and measured using solid-state pH meters to detect newly synthesized nucleotides.
Despite shorter sequencing run times than Illumina for similar sequence data, sequencing error rates are a problem, especially for long sequence homopolymers. The Ion Torrent platform supports WGS, whole-exome sequencing (WES), and panel gene sequencing (PGS), as well as molecular clinical applications.25 The Complete Genomics technology was developed by MGI Tech Co. Ltd (MGI), a subsidiary of Beijing Genomics Institute (BGI). The NGS platforms developed by BGI/MGI support a range of sequencing applications, including WGS, PGS, WES, epigenetics, microbial, and clinical trials. Compared with other NGS sequencers, including Illumina, BGI/MGI sequencers offer comparable performance in terms of sequencing time, sequencing quality, and throughput.29
NGS revolution in precision medicine
The advent of NGS technology in the last decade has revolutionized our understanding of cancer biology. After genome sequencing of thousands of tumors from all major cancer types, many genetic and epigenetic alterations were identified and found to potentially contribute to tumorigenesis. Sanger sequencing was the basis of traditional molecular testing methods, now referred to as 'first-generation sequencing'.30 In 1977, Frederick Sanger and colleagues developed the first widely used sequencing method. Applied Biosystems first commercialized the Sanger method in 1986. It provides high-quality sequences for relatively long DNA fragments (up to 900 base pairs).
Despite the efficiency of the Sanger method in sequencing a few short DNA segments, it is cumbersome and ineffective in sequencing long sequences. In contrast, NGS is much faster, cheaper, uses less DNA, and is more accurate than Sanger sequencing.23 For each base sequenced in Sanger, a large number of DNA templates are required (e.g., 100 bp sequences require hundreds of copies, and 1000 bp sequences require thousands) because a complete sequence requires strands that terminate at each base. When using NGS, it is possible to obtain a sequence from a single strand. Sequence validation and contig construction are performed with multiple staggered copies in both types of sequencing. Two things make NGS more efficient than Sanger sequencing. The first difference is that in some versions of NGS, the chemical reaction is combined with signal detection, whereas in Sanger sequencing they are separate. With Sanger sequencing, only one read of 1kb or less is possible at a time, whereas with NGS, 300Gb of DNA can be read in a single run. The first sequencing of the human genome cost around £300 million, due to the labor, less time, and reagents involved with NGS. A human genome would still cost £6 million for Sanger sequencing using the known sequence, but Illumina sequencing would cost just over £1000 today.31
The NGS method relies on many short, overlapping reads, so each DNA or RNA segment is sequenced multiple times because each read is amplified before sequencing. NGS is also more effective and faster because it can perform more repeats than Sanger sequencing. More repeats mean more coverage, resulting in more accurate and reliable sequences, even if the individual reads are less accurate. With Sanger sequencing, sequences can be read for much longer. The parallel nature of NGS allows more contiguous short reads to be assembled into longer reads. For smaller projects and validation of next-gen results, the Sanger method continues to be widely used. It can generate DNA sequencing reads of more than 500 nucleotides, making it a better technology than sequencing technologies with short reads (such as Illumina).
Precision medicine and NGS in oncology
Among healthcare settings where NGS has been tested, oncologists use it most often to match tumors to treatments that target genes that drive cancer growth. These therapies are called sequencing-targeted therapies. In several studies, NGS has been shown to identify cancer mutations with clinical significance. An international data-sharing consortium, the Genomics Evidence Neoplasia Information Exchange (GENIE), estimates a 30% rate of ability for several cancer types.32 According to the GENIE consortium, 30% of tumors with mutations could be treated with existing targeted therapies. Matching cancer therapies to patients' genomes has been shown to improve treatment outcomes. It has been demonstrated that patients with advanced cancer who received sequencing-matched therapy had a higher overall response rate (27% versus 5%), a shorter time to treatment failure (5.2 versus 2.2 months), and longer survival (13.4 versus 9.0 months) than patients who did not receive sequencing-matched therapy.33 These measures assess changes in tumor size; time from initiation of treatment to patient withdrawal from the study due to disease progression, toxicity, or death; or time from initiation of treatment to end of follow-up. Similar results were reported by Radovich et al,34 who found that patients who received matched treatment had longer progression-free survival than patients who did not receive matched treatment (86 vs. 49 days).
Progression-free survival is another statistic used to evaluate oncology, which measures how long it takes for cancer to progress after treatment is started.Sequencing-matched therapy has also been shown to improve overall survival,35,36 progression-free survival,37 and patient tumor response compared with non-matched therapy.37,38 In addition, the development of drugs targeting tumor-driving mutations identified by NGS has made significant progress. A study by Le et al39 found that mismatch repair mutations were effective with PD -1 loss of function in 12 tumor types. As a result of this study, pembrolizumab was first approved by the FDA in 2017 because it is administered solely based on mutations and not tumor type. Drilon et al40 reported that LOXO-195 (selitrectinib) is also histology-independent and can be used in different tumor types depending on gene fusion.
The European Society for Medical Oncology (ESMO) has provided recommendations regarding the use of NGS in the field of oncology. ESMO proposes three levels of recommendations for NGS utilization in different cancer types. According to this recommendation, NGS should be routinely employed on tumor samples in advanced non-squamous non-small-cell lung cancer, prostate cancers, ovarian cancers, and cholangiocarcinoma based on current evidence. In these specific tumors, large multigene panels may be utilized if they offer significant value compared to smaller panels despite higher costs. For colon cancers, NGS could serve as an alternative to PCR. ESMO further suggests that research centers specializing in clinical trials should incorporate multigene sequencing as a screening tool for eligible patients and expedite drug development. Additionally, these centers should collect prospective data to enhance the optimization of NGS utilization. In addition, ESMO advises the assessment of tumor mutational burden (TMB) in specific types of cancer, such as cervical, well and moderately differentiated neuroendocrine, salivary, thyroid, and vulvar cancers, as TMB-high indicates a potential positive response to pembrolizumab in these cases. However, ESMO recognizes that utilizing extensive gene panels may result in limited numbers of patients who clinically benefit. Therefore, ESMO suggests that the administration of off-label drugs tailored to genomics should only occur if a national or regional access program and decision-making procedure have been established. The guidelines from ESMO offer direction on the application of NGS in the field of oncology and emphasize the significance of integrating genomic information into clinical decision-making. NGS enables clinicians to acquire detailed genomic profiles of tumors, aiding in the identification of possible therapeutic targets and informing treatment choices. The objective of these recommendations is to enhance the utilization of NGS across various cancer types and encourage its incorporation into regular clinical practice.41
The limitations of NGS
It appears that NGS may be useful in cancer treatment, but no randomized controlled trials have been conducted.14,42 Because NGS is capable of identifying so many diagnostic subtypes, it is extremely difficult to recruit sufficient numbers of patients to support randomized controlled trials for each subtype of cancer. Pembroluzimab, the first precision medicine therapy approved by the FDA without randomized controlled trials, is an example of how the FDA has understood these limitations.14 The only randomized controlled trial of precision medicine to date showed no significant improvement in patient outcomes when patients were treated with targeted treatments using NGS, regardless of their cancer type. In this phase II trial, 195 patients with advanced cancer were randomly assigned to treatment based on their molecular profile or to the treatment of their physician's choice, with no difference in progression-free survival between the control and test groups.14 One of the reasons for this is the difficulty of treating advanced cancer patients whose tumors are genetically heterogeneous, meaning that different mutations may be present in different cells within a tumor. Although the sequencing results of this study were considered a valid basis for using drugs outside their recommended settings, it raised some important questions about their clinical value.
Currently, NGS strategies for precision medicine are also limited by differences between sequenced and unsequenced groups within a single study and by differences between studies and populations. For example, certain cancers have a higher prevalence of actionable mutations than others, such as prostate cancer because of the high prevalence of BRAF mutations in melanoma.43 However, the question arises whether patients with targeted mutations may have less aggressive cancers or cancers that are inherently easier to treat. Although study populations differ, many studies have shown clinical benefits in using sequencing results to treat patients. In addition, sequencing-targeted therapies only work for a small percentage of cancer patients with "actionable mutations" identified by NGS. Although practical obstacles prevent sequencing-matched therapy, this phenomenon raises questions about the clinical utility of metrics for "actionable mutations".38,44,45 The term "actionable mutation" is not uniformly defined, so recent studies may define it much more broadly to include mutations that have prognostic value or are indicative of an inherited cancer syndrome.46 In these cases, many patients with triggerable mutations may not be matched to cancer therapy based on their sequencing information.47
To get the most benefit from genetic studies, the method they use to evaluate mutations must be transparent and precise, and that clear distinctions are made between different categories of mutations and their potential effects. For example, in the GENIE study, actionable mutations were clearly defined based on a scale from level 1 gene mutations, indicating standard treatment of same cancer, to level 3B gene mutations, indicating promising research therapies.32 In addition, the increased financial burden of NGS should be considered. The cost of technical and human resources for specialized clinicians has increased substantially in recent years. It is becoming increasingly difficult to spend time on post-sequencing tasks, including filtering variants, comparing data, interpreting, and writing reports, as the number of genes required for comprehensive diagnosis and treatment decisions continues to increase. In addition, NGS procedures in hematology use extensive gene/hotspot panels for different hematologic subtypes, which requires in-depth knowledge and experience.48
Expert opinion on next-generation sequencing
Traditional ethical frameworks to protect patients and research participants are being challenged by the use of NGS in research and clinical projects. There are many types of clinical relevance associated with NGS data and results. Appropriate procedures must be put in place to effectively protect, manage, and communicate the data. Informed consent, privacy, and disclosure of results are three of the most important ethical considerations in NGS.49 The current research landscape encourages data sharing and the use of big data techniques, which has implications for the ethical framework of NGS research. Collaboration and data sharing are essential to advance scientific knowledge. In addition, increasing access to online datasets and advances in analytical techniques have made genomic information easier to re-identify.50
Genetic information can have devastating consequences for individuals, including effects on their insurance, social status, or relationships. NGS will raise additional privacy concerns as it is increasingly used for forensic purposes. The vast amounts of data generated by NGS must be secured and stored to appropriate standards by institutions using the technology. Genomic data should be stored in a way that minimizes the risk of re-identification, according to some privacy recommendations. Rather than making it impossible to identify individuals, it might be more appropriate to focus on methods that protect individuals from potential harm and misuse of their genomic data.49
Ethical, regulatory, and analytical challenges in NGS
Interpretation of NGS data remains a challenge despite its rapidly growing popularity. Despite available pipelines, it is still difficult to regulate NGS data, especially when these data are intended for clinical management. Due to the lack of uniformity in data processing strategies, data results are not comparable or reproducible.23,51,52 Several efforts have been made to establish standardized methods of bioinformatics analysis, including the development of shareable workflows.53,54 There is no standardized clinical interpretation for identified variants in all diseases. Standardized interpretations, reporting guidelines, and analyses are being developed to address this problem.55-57
The use of incomparable results in clinical applications should be regulated because of the enormous impact they bring.58 Obtaining WGS or other genetic data raises a number of ethical issues. Before genetic data are collected from patients, genetic counseling is essential to consider the implications of potential unintended analyzes. In addition, written informed consent must be provided. The cost of testing must be accurately estimated, and it must be verified that it can be approved by the insurance because, without insurance, the facility or individual must pay for the cost. Before testing, the patient must be informed of any incidental findings. For this problem, accredited laboratory guidelines such as those recommended by the ACMG (American College of Medical Genetics and Genomics)59 or those provided by reliable organizations should be followed.60
The handling of genomic patient data is a sensitive topic that is the subject of much debate. Regulations protecting the sharing of genomic data for research purposes are of particular concern to many people. Governments worldwide have enacted legislation to protect their citizens' genomic data, such as the South African Personal Data Protection Act (POPIA).61 Although these regulations have increased public confidence, there are still ambiguities that need to be addressed, particularly when it comes to international collaborations involving the sharing of personal genomic data. An article published in Science on October 30, 2019, described a scandal involving DNA collected from Africans and misused by scientists at the Wellcome Sanger Institute in the United Kingdom.62 A dispute arose over claims that Sanger scientists had developed a commercial chip based on shared DNA, which Stellenbosch College and the College of Kwa-Zulu Natal said was not part of the material transfer agreements (MTAs). As a result of this scandal, future genomics collaborations with Africa may be at risk as compliance with MTAs is questioned. It could also undermine public trust, limiting access to personal genomic data.
Bioinformatics algorithms and NGS
By screening an individual's NGS profile, scientists and physicians will be able to develop highly effective cancer prevention and treatment strategies. To achieve this goal, computational methods must be used to analyze biomedical data. To this end, machine learning is the best choice to create tasks that learn from the data and improve performance based on experience. In these techniques, a set of covariates is mapped to a single dependent outcome using a large set of training data. A model can be developed to diagnose a disease or predict its outcome based on a patient's clinical and genetic profile, but its success depends directly on the availability of large training data sets.
In cancer research, NGS technology has enabled the sequencing of individual genomes. In the last decade, machine learning techniques have been widely used to diagnose and treat various types of diseases, especially malignant cancers, using clinical and genetic profiles. Two types of techniques are used: supervised and unsupervised. In supervised techniques, training data are labeled and a model is created. A model is then created to predict an unknown input, while unsupervised techniques use unlabeled data to explore and analyze the relationships between them. Classification tasks are usually solved by supervised techniques, while clustering tasks are usually solved by unsupervised techniques.
To gain insights from studies of tumor microenvironments and other complex genomic data, researchers need easy-to-use bioinformatics applications. To deal with genetically heterogeneous diseases, further improvements in bioinformatics algorithms are needed despite advances in NGS technology and bioinformatics. We believe that machine learning algorithms, especially neural networks and support vector machines, as well as new developments in artificial intelligence, will play an important role in improving NGS platforms and software in the future. By improving clinical diagnostics and opening new avenues for the development of novel therapies, scientists and clinicians will be able to solve complex biological challenges.54 See Table 2 for an overview of algorithms for validating and interpreting genetic variants.
Table 2.
Overview of algorithms used for genetic variant validation and interpretation
NGS is a high-throughput sequencing technology that has revolutionized genomics research. There are several computational tools that are widely used in NGS-generated data analysis. Table 3 represents some of these computational tools. In addition, several algorithms have been developed analyzing NGS data, each serving a specific purpose. Some commonly used algorithms are introduced in Table 4.
Table 3.
Some computational tools for analyzing NGS generated data
Table 4.
Some commonly used algorithms developed for NGS data analysis
|
Application
|
Description
|
Popular algorithms
|
| Base calling |
Converts raw sequencing signals into DNA base calls |
Phred, Solexa, and Illumina's Real-Time Analysis (RTA) |
| Read mapping |
Reads mapping algorithms align short reads generated by NGS to a reference genome or transcriptome |
Bowtie, BWA, and HISAT |
| Variant calling |
Identifies genetic variations (e.g., single nucleotide variants, insertions, deletions) from aligned reads |
GATK (Genome Analysis Toolkit), Samtools, and FreeBayes |
| De novo assembly |
Reconstructs genomes or transcriptomes without relying on a reference genome |
Velvet, SPAdes, and Trinity |
| Transcript quantification |
Estimates the abundance of RNA transcripts from RNA-seq data |
Cufflinks, StringTie, and Salmon |
| ChIP-seq peak calling |
Identifies regions of the genome enriched with protein-DNA interactions |
MACS2, SICER, and HOMER |
| Pathway analysis |
Analyzes gene expression data to identify enriched biological pathways or functional categories. |
DAVID, Enrichr, and Gene Set Enrichment Analysis (GSEA) |
Success paradigms in the NGS marketing
During the forecast period, the growing population and increasing use of personalized medicine will drive the growth of the NGS industry. In North America, many of the major companies are striving to develop innovative products to reduce the cost of genome sequencing. This, in turn, is expected to drive regional market expansion. From 2019 to 2027, the NGS market is expected to reach USD 11,776 million, growing at a CAGR of 19.1%. Using first-generation Sanger sequencing techniques, the human genome was sequenced nearly 15 years ago, requiring the collaboration of hundreds of laboratories around the world and costing approximately $100 million.63 Since 2005, NGS has overcome many of the earlier problems by allowing multiple samples to be sequenced in parallel at a fraction of the cost ($1 million) and time (two months). The technology and sequencing platforms continue to evolve. A genome sequence is increasingly used by physicians to guide patient treatment.
Life Science launched the first NGS platform in 2005, enabling cost-effective and time-efficient high-throughput sequencing.64 The Centers for Medicare and Medicaid Services (CMS) has taken steps to adopt NGS testing for patients with germline ovarian or breast cancer approved by the FDA. In recent years, CMS has actively followed the rapid innovation of NGS testing and the evolution of cancer diagnostics. Because of their ability to detect multiple types of genetic mutations simultaneously, NGS tests are among the most comprehensive genetic tests available for cancer patients. Patients with advanced cancer who meet a set of criteria were covered by Medicare for laboratory diagnostic testing with NGS for the first time in March 2018. This decision will allow more Medicare patients to access NGS to treat other types of hereditary cancers and reduce mortality.
Concluding remarks
Effective action is needed to address both the health and economic burdens of cancer. In cancer research as well as in all other areas of human genetics, NGS is proving to be a cornerstone of modern research. Compared to Sanger sequencing, NGS offers several advantages, such as high throughput, higher speed, and lower cost. NGS provides high-quality data at an affordable price, with improved data processing capabilities, a faster computing system, and a more efficient process. Clinical diagnostics and genetic medicine have greatly benefited from bioinformatics tools that facilitate NGS-based genetic analysis strategies. The size of the global next-generation sequencing market is expected to increase significantly in the coming years.
Review Highlights
What is the current knowledge?
√ Next-generation sequencing has introduced a new approach to cancer treatment called precision medicine, in which medical therapies are tailored to each patient's characteristics and condition.
√ Genome sequencing is becoming more accessible for clinical use, and genomic profiling is a promising approach for precision oncology in the future.
√ Clinical diagnostics and genetic medicine have benefited greatly from bioinformatics tools that facilitate NGS-based genetic analysis strategies.
What is new here?
√ The available databases and bioinformatics tools and online servers used in NGS data analysis have been reviewed.
√ The way that NGS technology facilitate cancer treatment have been highlighted.
√ The NGS revolution in precision medicine have been described.
Acknowledgments
The authors like to acknowledge the Research Center for Pharmaceutical Nanotechnology at Tabriz University of Medical Sciences for their financial support (#72560) and the Department of Computer Science, Faculty of Mathematical Sciences, University of Tabriz for their technical support.
Competing Interests
None to be declared.
Ethical Statement
Not applicable.
Funding
This study was supported by the Research Center for Pharmaceutical Nanotechnology, Tabriz University of Medical Sciences (#72560).
References
- Krzyszczyk P, Acevedo A, Davidoff EJ, Timmins LM, Marrero-Berrios I, Patel M. The growing role of precision and personalized medicine for cancer treatment. Technology (Singap World Sci) 2018; 6:79-100. doi: 10.1142/S2339547818300020 [Crossref] [ Google Scholar]
- Parvizpour S, Razmara J, Omidi Y. Breast cancer vaccination comes to age: impacts of bioinformatics. Bioimpacts 2018; 8:223-35. doi: 10.15171/bi.2018.25 [Crossref] [ Google Scholar]
- Salehi M, Razmara J, Lotfi S. Development of an Ensemble Multi-stage Machine for Prediction of Breast Cancer Survivability. Journal of AI and Data Mining 2020; 8:371-8. doi: 10.22044/jadm.2020.8406.1978 [Crossref] [ Google Scholar]
- Parvizpour S, Razmara J, Pourseif MM, Omidi Y. In silico design of a triple-negative breast cancer vaccine by targeting cancer testis antigens. Bioimpacts 2019; 9:45-56. doi: 10.15171/bi.2019.06 [Crossref] [ Google Scholar]
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021. 10.3322/caac.21660.
- Parvizpour S, Masoudi-Sobhanzadeh Y, Pourseif MM, Barzegari A, Razmara J, Omidi Y. Pharmacoinformatics-based phytochemical screening for anticancer impacts of yellow sweet clover, Melilotus officinalis (Linn.) Pall. Comput Biol Med 2021; 138:104921. doi: 10.1016/j.compbiomed.2021.104921 [Crossref] [ Google Scholar]
- Kohler S. Precision medicine–moving away from one-size-fits-all. Quest 2018; 14:12-5. [ Google Scholar]
- Kuchuk I, Clemons M, Addison C. Time to put an end to the “one size fits all” approach to bisphosphonate use in patients with metastatic breast cancer? Multidisciplinary Digital Publishing Institute; 2012. p. 303-4.
- Bonini MG, Gantner BN. The multifaceted activities of AMPK in tumor progression—why the “one size fits all” definition does not fit at all?. IUBMB Life 2013; 65:889-96. [ Google Scholar]
- Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018; 173:321-37 e10. doi: 10.1016/j.cell.2018.03.035 [Crossref] [ Google Scholar]
- Gagan J, Van Allen EM. Next-generation sequencing to guide cancer therapy. Genome Med 2015; 7:80. doi: 10.1186/s13073-015-0203-x [Crossref] [ Google Scholar]
- Ignatiadis M, Dawson SJ. Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality?. Ann Oncol 2014; 25:2304-13. doi: 10.1093/annonc/mdu480 [Crossref] [ Google Scholar]
- Shin SH, Bode AM, Dong Z. Precision medicine: the foundation of future cancer therapeutics. NPJ Precis Oncol 2017; 1:12. doi: 10.1038/s41698-017-0016-z [Crossref] [ Google Scholar]
- Morash M, Mitchell H, Beltran H, Elemento O, Pathak J. The Role of Next-Generation Sequencing in Precision Medicine: A Review of Outcomes in Oncology. J Pers Med 2018; 8:30. doi: 10.3390/jpm8030030 [Crossref] [ Google Scholar]
- Roychowdhury S, Iyer MK, Robinson DR, Lonigro RJ, Wu Y-M, Cao X. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Trans Med 2011; 3:111ra121. [ Google Scholar]
- Dlamini Z, Francies FZ, Hull R, Marima R. Artificial intelligence (AI) and big data in cancer and precision oncology. Comput Struct Biotechnol J 2020; 18:2300-11. doi: 10.1016/j.csbj.2020.08.019 [Crossref] [ Google Scholar]
- Di Resta C, Galbiati S, Carrera P, Ferrari M. Next-generation sequencing approach for the diagnosis of human diseases: open challenges and new opportunities. EJIFCC 2018; 29:4-14. [ Google Scholar]
- Del Vecchio F, Mastroiaco V, Di Marco A, Compagnoni C, Capece D, Zazzeroni F. Next-generation sequencing: recent applications to the analysis of colorectal cancer. J Transl Med 2017; 15:246. doi: 10.1186/s12967-017-1353-y [Crossref] [ Google Scholar]
- Roden DM. Cardiovascular pharmacogenomics: current status and future directions. J Hum Genet 2016; 61:79-85. doi: 10.1038/jhg.2015.78 [Crossref] [ Google Scholar]
- Mardis ER. The Impact of Next-Generation Sequencing on Cancer Genomics: From Discovery to Clinic. Cold Spring Harb Perspect Med 2019; 9:a036269. doi: 10.1101/cshperspect.a036269 [Crossref] [ Google Scholar]
- Qin D. Next-generation sequencing and its clinical application. Cancer Biol Med 2019; 16:4-10. doi: 10.20892/j.issn.2095-3941.2018.0055 [Crossref] [ Google Scholar]
- Morganti S, Tarantino P, Ferraro E, D’Amico P, Viale G, Trapani D, et al. Complexity of genome sequencing and reporting: next generation sequencing (NGS) technologies and implementation of precision medicine in real life. Crit Rev Oncol Hematol 2019. 133: 171-82.
- Kanzi AM, San JE, Chimukangara B, Wilkinson E, Fish M, Ramsuran V. Next Generation Sequencing and Bioinformatics Analysis of Family Genetic Inheritance. Front Genet 2020; 11:544162. doi: 10.3389/fgene.2020.544162 [Crossref] [ Google Scholar]
- Buermans HP, den Dunnen JT. Next generation sequencing technology: Advances and applications. Biochim Biophys Acta 2014; 1842:1932-41. doi: 10.1016/j.bbadis.2014.06.015 [Crossref] [ Google Scholar]
- Heather JM, Chain B. The sequence of sequencers: The history of sequencing DNA. Genomics 2016; 107:1-8. doi: 10.1016/j.ygeno.2015.11.003 [Crossref] [ Google Scholar]
- Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 2016; 17:333-51. doi: 10.1038/nrg.2016.49 [Crossref] [ Google Scholar]
- Levy SE, Myers RM. Advancements in Next-Generation Sequencing. Annu Rev Genomics Hum Genet 2016; 17:95-115. doi: 10.1146/annurev-genom-083115-022413 [Crossref] [ Google Scholar]
- Hess JF, Kohl TA, Kotrova M, Ronsch K, Paprotka T, Mohr V. Library preparation for next generation sequencing: A review of automation strategies. Biotechnol Adv 2020; 41:107537. doi: 10.1016/j.biotechadv.2020.107537 [Crossref] [ Google Scholar]
- Zhu FY, Chen MX, Ye NH, Qiao WM, Gao B, Law WK. Comparative performance of the BGISEQ-500 and Illumina HiSeq4000 sequencing platforms for transcriptome analysis in plants. Plant Methods 2018; 14:69. doi: 10.1186/s13007-018-0337-0 [Crossref] [ Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 1977; 74:5463-7. doi: 10.1073/pnas.74.12.5463 [Crossref] [ Google Scholar]
- Calistri A, Palu G. Editorial commentary: Unbiased next-generation sequencing and new pathogen discovery: undeniable advantages and still-existing drawbacks. Clin Infect Dis 2015; 60:889-91. doi: 10.1093/cid/ciu913 [Crossref] [ Google Scholar]
- Consortium APG. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 2017; 7:818-31. doi: 10.1158/2159-8290.CD-17-0151 [Crossref] [ Google Scholar]
- Tsimberidou AM, Iskander NG, Hong DS, Wheler JJ, Falchook GS, Fu S. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res 2012; 18:6373-83. doi: 10.1158/1078-0432.CCR-12-1627 [Crossref] [ Google Scholar]
- Radovich M, Kiel PJ, Nance SM, Niland EE, Parsley ME, Ferguson ME. Clinical benefit of a precision medicine based approach for guiding treatment of refractory cancers. Oncotarget 2016; 7:56491-500. doi: 10.18632/oncotarget.10606 [Crossref] [ Google Scholar]
- Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014; 311:1998-2006. [ Google Scholar]
- Aisner D, Sholl LM, Berry LD, Haura EB, Ramalingam SS, Glisson BS, et al. Effect of expanded genomic testing in lung adenocarcinoma (LUCA) on survival benefit: The Lung Cancer Mutation Consortium II (LCMC II) experience. American Society of Clinical Oncology; 2016.
- Schwaederle M, Parker BA, Schwab RB, Daniels GA, Piccioni DE, Kesari S. Precision Oncology: The UC San Diego Moores Cancer Center PREDICT Experience. Mol Cancer Ther 2016; 15:743-52. doi: 10.1158/1535-7163.MCT-15-0795 [Crossref] [ Google Scholar]
- Stockley TL, Oza AM, Berman HK, Leighl NB, Knox JJ, Shepherd FA. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret IMPACT/COMPACT trial. Genome Med 2016; 8:109. doi: 10.1186/s13073-016-0364-2 [Crossref] [ Google Scholar]
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017; 357:409-13. doi: 10.1126/science.aan6733 [Crossref] [ Google Scholar]
- Drilon A, Nagasubramanian R, Blake JF, Ku N, Tuch BB, Ebata K. A Next-Generation TRK Kinase Inhibitor Overcomes Acquired Resistance to Prior TRK Kinase Inhibition in Patients with TRK Fusion-Positive Solid Tumors. Cancer Discov 2017; 7:963-72. doi: 10.1158/2159-8290.CD-17-0507 [Crossref] [ Google Scholar]
- Mosele F, Remon J, Mateo J, Westphalen C, Barlesi F, Lolkema M. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol 2020; 31:1491-505. [ Google Scholar]
- Moscow JA, Fojo T, Schilsky RL. The evidence framework for precision cancer medicine. Nat Rev Clin Oncol 2018; 15:183-92. doi: 10.1038/nrclinonc.2017.186 [Crossref] [ Google Scholar]
- Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015; 163:1011-25. doi: 10.1016/j.cell.2015.10.025 [Crossref] [ Google Scholar]
- Meric-Bernstam F, Brusco L, Shaw K, Horombe C, Kopetz S, Davies MA. Feasibility of Large-Scale Genomic Testing to Facilitate Enrollment Onto Genomically Matched Clinical Trials. J Clin Oncol 2015; 33:2753-62. doi: 10.1200/JCO.2014.60.4165 [Crossref] [ Google Scholar]
- Beltran H, Eng K, Mosquera JM, Sigaras A, Romanel A, Rennert H. Whole-Exome Sequencing of Metastatic Cancer and Biomarkers of Treatment Response. JAMA Oncol 2015; 1:466-74. doi: 10.1001/jamaoncol.2015.1313 [Crossref] [ Google Scholar]
- Bryce AH, Egan JB, Borad MJ, Stewart AK, Nowakowski GS, Chanan-Khan A. Experience with precision genomics and tumor board, indicates frequent target identification, but barriers to delivery. Oncotarget 2017; 8:27145-54. doi: 10.18632/oncotarget.16057 [Crossref] [ Google Scholar]
- West HJ. No Solid Evidence, Only Hollow Argument for Universal Tumor Sequencing: Show Me the Data. JAMA Oncol 2016; 2:717-8. doi: 10.1001/jamaoncol.2016.0075 [Crossref] [ Google Scholar]
- Bacher U, Shumilov E, Flach J, Porret N, Joncourt R, Wiedemann G. Challenges in the introduction of next-generation sequencing (NGS) for diagnostics of myeloid malignancies into clinical routine use. Blood Cancer J 2018; 8:113. doi: 10.1038/s41408-018-0148-6 [Crossref] [ Google Scholar]
- Martinez-Martin N, Magnus D. Privacy and ethical challenges in next-generation sequencing. Expert Rev Precis Med Drug Dev 2019; 4:95-104. doi: 10.1080/23808993.2019.1599685 [Crossref] [ Google Scholar]
- Hansson MG, Lochmuller H, Riess O, Schaefer F, Orth M, Rubinstein Y. The risk of re-identification versus the need to identify individuals in rare disease research. Eur J Hum Genet 2016; 24:1553-8. doi: 10.1038/ejhg.2016.52 [Crossref] [ Google Scholar]
- Kulkarni N, Alessandri L, Panero R, Arigoni M, Olivero M, Ferrero G. Reproducible bioinformatics project: a community for reproducible bioinformatics analysis pipelines. BMC Bioinformatics 2018; 19:349. doi: 10.1186/s12859-018-2296-x [Crossref] [ Google Scholar]
- Kanwal S, Khan FZ, Lonie A, Sinnott RO. Investigating reproducibility and tracking provenance - A genomic workflow case study. BMC Bioinformatics 2017; 18:337. doi: 10.1186/s12859-017-1747-0 [Crossref] [ Google Scholar]
- Baichoo S, Souilmi Y, Panji S, Botha G, Meintjes A, Hazelhurst S. Developing reproducible bioinformatics analysis workflows for heterogeneous computing environments to support African genomics. BMC Bioinformatics 2018; 19:457. doi: 10.1186/s12859-018-2446-1 [Crossref] [ Google Scholar]
- Pereira R, Oliveira J, Sousa M. Bioinformatics and Computational Tools for Next-Generation Sequencing Analysis in Clinical Genetics. J Clin Med 2020; 9. 10.3390/jcm9010132.
- Hutchins RJ, Phan KL, Saboor A, Miller JD, Muehlenbachs A, Workgroup CNQ. Practical Guidance to Implementing Quality Management Systems in Public Health Laboratories Performing Next-Generation Sequencing: Personnel, Equipment, and Process Management (Phase 1). J Clin Microbiol 2019; 57:e00261-19. doi: 10.1128/JCM.00261-19 [Crossref] [ Google Scholar]
- Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med 2018; 142:321-46. doi: 10.5858/arpa.2017-0388-CP [Crossref] [ Google Scholar]
- Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 2017; 19:4-23. doi: 10.1016/j.jmoldx.2016.10.002 [Crossref] [ Google Scholar]
- Endrullat C, Glokler J, Franke P, Frohme M. Standardization and quality management in next-generation sequencing. Appl Transl Genom 2016; 10:2-9. doi: 10.1016/j.atg.2016.06.001 [Crossref] [ Google Scholar]
- Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v20): a policy statement of the American College of Medical Genetics and Genomics. Genet Med 2017; 19:249-55. [ Google Scholar]
- Green SM. When do clinical decision rules improve patient care?. Ann Emerg Med 2013; 62:132-5. [ Google Scholar]
- Staunton C, Adams R, Botes M, Dove ES, Horn L, Labuschaigne M. Safeguarding the future of genomic research in South Africa: Broad consent and the Protection of Personal Information Act No4 of 2013. S Afr Med J 2019; 109:468-70. doi: 10.7196/SAMJ.2019.v109i7.14148 [Crossref] [ Google Scholar]
- Stokstad E. Genetics lab accused of misusing African DNA. American Association for the Advancement of Science; 2019.
- Gordon LG, White NM, Elliott TM, Nones K, Beckhouse AG, Rodriguez-Acevedo AJ. Estimating the costs of genomic sequencing in cancer control. BMC Health Serv Res 2020; 20:492. doi: 10.1186/s12913-020-05318-y [Crossref] [ Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005; 437:376-80. doi: 10.1038/nature03959 [Crossref] [ Google Scholar]