Bioimpacts. 2025;15:30047.
doi: 10.34172/bi.2024.30047
Original Article
An update on technical method of cartilage decellularization: A physical-based protocol
Hengameh Dortaj Conceptualization, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing, 1, 2 
Ahmad Vaez Conceptualization, Supervision, Writing – review & editing, 1 
Ashraf Hassanpour-Dehnavie Formal analysis, Supervision, Visualization, Writing – review & editing, 3 
Ali Akbar Alizadeh Conceptualization, Project administration, Supervision, Writing – review & editing, 1, * 
Author information:
1Department of Tissue Engineering and Applied Cell Sciences, School of Advance Medical Sciences and Technologies, Shiraz University of Medical Sciences, Shiraz, Iran
2Tissue Engineering Research Group (TERG), Department of Anatomy and Cell Biology, School of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran
3Tissue Engineering Lab, Department of Anatomical Sciences, School of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran
Abstract
Introduction:
Despite advances in orthopedic surgery, the lack of effective conventional treatment for cartilage defects has led to research in cartilage tissue engineering. One of the interesting topics is the use of decellularized extracellular matrix (ECM) as a suitable natural scaffold that supports the growth and function of cells cultured in it. A concern with decellularization protocols, especially those with high detergent concentrations, is the disruption of native ECM, which has deleterious effects on subsequent scaffold recellularization. Therefore, this study focused on optimizing cartilage decellularization by physical methods without the use of ionic detergents.
Methods:
The bovine tracheal cartilage fragments were decellularized by a combination of 8 cycles of freeze-thaw and ultrasound techniques. Then, the tissues were immersed and shaken in 0.25% trypsin for 24 hours. Efficient cell removal and preservation of ECM were confirmed by histological and cytocompatibility assessments. The in-vivo studies were performed to evaluate the biocompatibility and bioactivity of the scaffold.
Results:
The histological assessments indicated the appropriate cytocompatibility and the fibroblast cell culture study demonstrated that cells were able to proliferate and migrate on the decellularized cartilage. In-vivo evaluation also showed a reduced adverse immune response, including leukocyte infiltration into the ECM.
Conclusion:
These results suggest that a cartilage scaffold created using a physical decellularization protocol that efficiently removes cells while preserving the native ECM can be a suitable scaffold for cartilage reconstruction. The main advantage of this protocol is the absence of potentially toxic chemicals in the tissues.
Keywords: Physical decellularization, Cartilage, Extracellular matrix, Tissue engineering
Copyright and License Information
© 2025 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
This work was supported by a grant offer from the Vice-Chancellor of Research deputy of Shiraz University of Medical Sciences [grant No. 23498].
Introduction
Despite advances in orthopedic surgery, the lack of effective conventional treatment for cartilage defects has led to research in cartilage tissue engineering.
Tissue engineering is one of the new approaches to medical science that uses the principles of engineering (materials, mechanics, physics, and chemistry) and biological sciences to produce biological substitutes for the repairing and regenerating of damaged tissues and maintaining, or improving their function.1 Cells, scaffolds, and growth-stimulating factors are considered the pillars of tissue engineering, which provide structural support for cell attachment and enable further tissue remodeling.2
Extracellular matrix (ECM) components and cell-matrix dynamics of the cartilage tissue, and cell-matrix dynamics play an important role in the mechanical strength of the tissue framework and the organization of cellular processes. ECM is a natural endogenous scaffold with a complex network of proteins and saccharides such as collagen, elastin, glycoproteins, growth factors, glycosaminoglycans (GAGs), proteoglycans, cytokines, and a variety of enzymes that produce signals.3 Due to the microenvironment content of the mature ECM specific to each tissue, the application of the natural ECM as a scaffold provides a bed for the ideal growth of cells in injured tissues.4,5 Therefore, one of the interesting topics in cartilage tissue engineering is the use of decellularized ECM as a suitable natural scaffold that supports the growth and function of cells cultured in it by maintaining the original components and ultrastructures.
The tissue decellularization (DC) approach has formed a parallel research line to the approaches that use synthetic polymer scaffolds and natural derivative polymers.6,7 Studies show that the scaffolds obtained from DC tissues can be a suitable substrate for cells similar to the original tissue by maintaining the original components.8
During decellularization, maintaining the integrity of the structure and mechanics of the tissue, as well as the active molecules and proteins in the ECM structure, is very important.9 An ideal decellularization protocol effectively removes all cells to prevent toxic effects while minimizing any damage to the structural integrity, composition, and biological activity of the remaining ECM.10 Such scaffolds are highly regarded for their appropriate immunological responses, mild antigenic properties, and ability to improve cell adhesion, and homeostasis.11-13 However, studies have shown that the efficiency of a decellularization method largely depends on the characteristics of the target tissue. According to these results, it is difficult to balance the complete elimination of cells (any cell debris triggers an immune system response) and maintain the biomechanical properties of the tissue.14 Therefore, researchers usually have to find a balance between removing cell contents and maintaining optimal structural, mechanical, and biochemical properties. Recent research has focused on optimal decellularization methods, which generally include physical methods such as freezing, mechanical stress, and mechanical excitation, as well as chemical methods such as anionic, ionic, and hypotonic cleaners like Triton (Triton X-100) and sodium dodecyl sulfate (SDS), and enzymatic methods such as trypsin and nucleases with different concentrations and times.15,16 Most of these methods affect the surface morphology and mechanical properties of the scaffold to some extent. The decellularization process can alter the pattern and distribution of matrix fibers and glycosaminoglycans, which is more evident in the biomechanical function of the matrix. In addition, chemical methods reduce growth factors and GAG content, increase ECM extensibility or stiffness, loosen the collagen network, and leave residual nuclear cages. Thus, a concern with decellularization protocols, especially those with high detergent concentrations, is the disruption of native ECM composition and architecture, which has deleterious effects on subsequent scaffold recellularization.16-18 Therefore, this study focused on optimizing cartilage decellularization by physical methods without the use of ionic detergents through a combination of freeze-thaw and ultrasound techniques. Generally, physical methods are used as the main step in decellularization. The most common physical methods for stimulating tissues are freeze-thaw cycles and mechanical stimulation, or sonication.19 The absence of potentially toxic chemicals in physical methods is considered an important advantage compared to chemical methods. Applying successive cycles of freezing and thawing can reduce the penetration of leukocytes into the ECM and thus limit the immune responses of the host tissue.20 Another method is physical decellularization by ultrasound. This method also causes cell lysis in the ECM structure.21 In this method, the cell wall is destroyed by mechanical processes, and the contents of the cell are fixed. In the next step, the cell contents can be removed from the ECM.22,23 So far, chemical and enzymatic methods have been used in most studies based on the construction of desalinated natural scaffolds, and no comparison and study of physical methods, as well as the study of the remaining ECM structure after the application of these methods, has been reported. There is no precise characterization in the literature of the non-use of ionic detergents for tissue decellularization. This study aimed to optimize and evaluate non-chemical methods of decellularizing cartilage tissue.
Materials and Methods
Materials
All chemical materials and reagents for cellular study, including Dulbecco's Modified Eagle Medium (DMEM) supplemented and Trypsin, were purchased from Shellmax Company USA. Fetal Bovine Serum (FBS) was purchased from Gibco and penicillin/streptomycin was prepared from Sigma Aldrich, Germany. Also, a DNA extraction mini kit was purchased from Qiagen, Germany. Glutaraldehyde 25% and Phosphate Buffer Saline (PBS) prepared from Merck, Germany. MTT kit purchased from Bioidea, Iran.
Tissue collection
The fresh bovine trachea was harvested immediately after slaughter. Adipose tissue and perichondrium were removed from the trachea using sharp scissors. The samples were kept in a 10% penicillin-streptomycin solution on dry ice during the whole transportation time from the slaughterhouse to the laboratory and during the stages of cleaning and preparation of cartilage.
Decellularization process
Fresh cartilage samples were cut into approximately 1 mm2 pieces. The cartilage fragments were decellularized by placing them in a hypotonic Tris-HCL solution, followed by eight cycles of freeze-thawing. To prepare Tris HCL buffer, 2 grams of Tris was dissolved in 750 cc of distilled water. The pH of the solution was adjusted to 8 using hydrochloric acid, and then the volume of the solution was adjusted to 1 liter with distilled water. This solution was used as a buffer during the physical step. The samples were kept for 15 minutes in a nitrogen tank for freezing and then kept at 60 °C for 15 minutes.
For ultrasound, cartilage samples were placed into the container tube with PBS. BANDELIN homogenizer SONOPULS HD 2070 was used for ultrasound. The amount of applied power was equal to 70%, which was applied as a pulse system with a wavelength of 20 kHz for 45 minutes. Then, the tissues were immersed in 0.25% trypsin (Sigma-Aldrich) for 24 hours in a shaker incubator with high agitation. The solution was changed every 8 hours. To remove debris, the samples were washed with PBS. The decellularized tissues were then freeze-dried and stored at -20 °C.
Characterization of decellularized cartilage
To confirm cell removal, the samples were stained with Hematoxylin and Eosin (H&E) and Alcian blue and then, examined by a light microscope. The extracellular matrix (ECM) structures were qualitatively validated with periodic acid Schiff (PAS) staining for glycosaminoglycans (GAG) and Alcian blue staining for collagen fibers. To assess the effectiveness of cell removal, the native and decellularized cartilage sections were incubated in a Hoechst solution for 10 minutes before imaging on a fluorescent microscope. In Hoechst staining, this dye can bind to regions rich in adenine and thymine (A=T) in the DNA structure, so this dye is seen as bright spots in the native cartilage structure containing chondrocyte nuclei using a fluorescent microscope.
DNA quantification assay was performed using a QIAamp® DNA Blood and Tissue Mini Kit (Qiagen GmbH, Hilden, Germany). To determine the amount of dsDNA, 25 mg of lyophilized sample FATG1 buffer and proteinase K were added to the samples following vortexing and incubation at 60 °C for 2 hours. After that, FATG2 buffer was added to the sample mixture and incubated at 70 °C for 10 minutes, followed by incubation with elution buffer (PH 7.5- 9).24 The samples were then washed and dehydrated using cold ethanol, and then their absorbance was measured in triplicate at 260 nm using the NanoDrop® ND-1000 (Nanodrop Technologies Inc., Wilmington, United States of America) Nanodrops according to the following equation:
DNA concentration (mg/mL-1) ¼ (A260-A320) ×50 × (10 mm per 0.51 mm).25
The qualitative analyses of DNA content were applied by running an electrophoresis for intact and DC tissue. DNA fragments must be less than 200 bp long to confirm decellularization
The GAG content of the tissue was compared in both study groups using the dimethyl methylene blue (DMMB) staining method. For this purpose, first, a dye solution containing 16 mg of DMMB, 1.6 g of NaCl, 3.04 g of glycine, and 95 ml of 0.1 M acetic acid per liter of DI water was prepared so that the pH was set at ~3. For GAG quantification, about 100 mg of each tissue was incubated with 0.5 mg/ml proteinase K at 56 °C overnight in a shaking incubator to completely digest. Twenty μL of the digested samples were added to 200 μL of DMMB in a 48-well plate. After pipetting, the absorbance was measured at 656 nm using a microplate reader.26
For ultrastructure assessment lyophilized and intact samples were prepared for scanning electron microscopy (SEM). The intact samples were fixed using 2.5% glutaraldehyde (Sigma-Aldrich, USA) in 0.1 M PBS and 4% formaldehyde at 4 °C overnight, they were gradually dehydrated via an increasing graded series of ethanol (50,70,90 and 100%) and kept overnight to be air-dried in a fume hood. Pore size and porosity were measured using the ImageJ software.
The size of DC cartilage particles and decellularized extracellular matrix (DC-ECM) microparticles that were dispersed in an aqueous solution was defined using dynamic light scattering (DLS). This is a physical method that is used in the determination of particle size distribution. The light scattered by the particles is related to the particle diameter.27
Sonicated samples were dispersed in PBS (1 mg/mL), and the average hydrodynamic particle size and polydispersity index (PDI) were measured using a particle size analyzer at room temperature.
The cell viability was evaluated on days 1, 3, and 5 using 3T3 fibroblasts. The concentration of 5×103 cells were exposed to 2D conventional cultures and were treated with 1 mg/mL 3-(4, 5-dimethyl thiazolyl-2)-2, 5-diphenyltetrazolium bromide (MTT, M5655; Sigma–Aldrich) for 3 h at 37 °C and 5% CO2. Dimethyl sulfoxide (Sigma-Aldrich) was added to the Formazan crystals, which formed purple precipitates, and placed on a shaker incubator at a temperature of 37 °C for 20 min. The optical density of the eluted MTT was measured by a spectrophotometer at 590 nm.28
To perform the migration test for cell invasion evaluation, fibroblast cells were used at a density of 103 in a volume of 1 mL cultured on 24-well plates on samples from different study groups. The chamber was loaded with DMEM supplemented with 10% FBS. The plates were incubated at 37 °C in 5% CO2 for 10 h. A sterile cotton swab was gently drawn over the surface of the plate to detach non-migrated cells and washed with PBS. Then a groove was made in the middle of each well using a 200 μL pipette tip after the cells reached a density of 80%. The cells were imaged after 24 and 48 hours, by an inverted microscope, and the groove filling was examined.29 A conventional 2D monolayer culture system on a polystyrene 24-well dish was used at the same cell density as the controls. To obtain the same field during the imaging, the scratch was marked on the plate as a reference.
Lyophilized DC and intact tissues were placed in 1% trypsin in PBS (pH 7.2) at 37 °C for 3 weeks; each sample was done in triplicate. The samples were dried and weighed every day for one week, and then every week for 21 days. The weight loss was calculated by the following formula30:
Weight loss (%) = (Wo − Wt)/Wo × 100
Where Wo is the initial dried weight and Wt is the dried weight of the degraded sample.
For the evaluation of the decellularized tissue, Raman spectra were obtained using an excitation laser wavelength of 851 nm, a laser power level of 50 mW, a temperature of -40 °C, and a frequency of 2 Hz. Raman images were generated by scanning 200 points from the selected area after background subtraction. The Raman spectra were analyzed in the range of 300 to 1500 cm-1, with a resolution of 4 cm-1, in this study.31
The surface topography and roughness (Sq.) of the DC and native cartilage samples were obtained using atomic force microscopy (AFM) (Ara Research Company, Iran). The images of the surface topography of the cartilage were obtained. The scan size was 5 × 5 μm. Three samples per group were used.
Xenotransplantation of DC cartilage
Five male rats (Wistar, 65-80 g) were subjected to subcutaneous implantation of DC cartilage samples. Under aseptic conditions, the dorsum of the rats was shaved, 5 mm transverse incisions were made, and a subcutaneous pocket was created. 6-10 pieces of DC samples were inserted in each pocket. After 21 days, animals were sacrificed, and obtained samples were used for histopathological analysis.
Statistical analysis
One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test was used for DNA and GAG quantification in this study. The data were analyzed using SPSS software version 21 and were expressed as the mean value ± standard error of the mean (SEM). A P value of < 0.05 was considered significant.
Results
Characterization of decellularized cartilage
Both H&E and Hoechst staining showed that nuclei were removed from the buffet. In addition, PAS and Alcian blue (PH 2.5) staining showed that GAGs and collagen fibers were well preserved after the decellularization process. As seen in Fig. 1, the absence of bright spots in Hoechst's staining indicates decellularization in the samples.
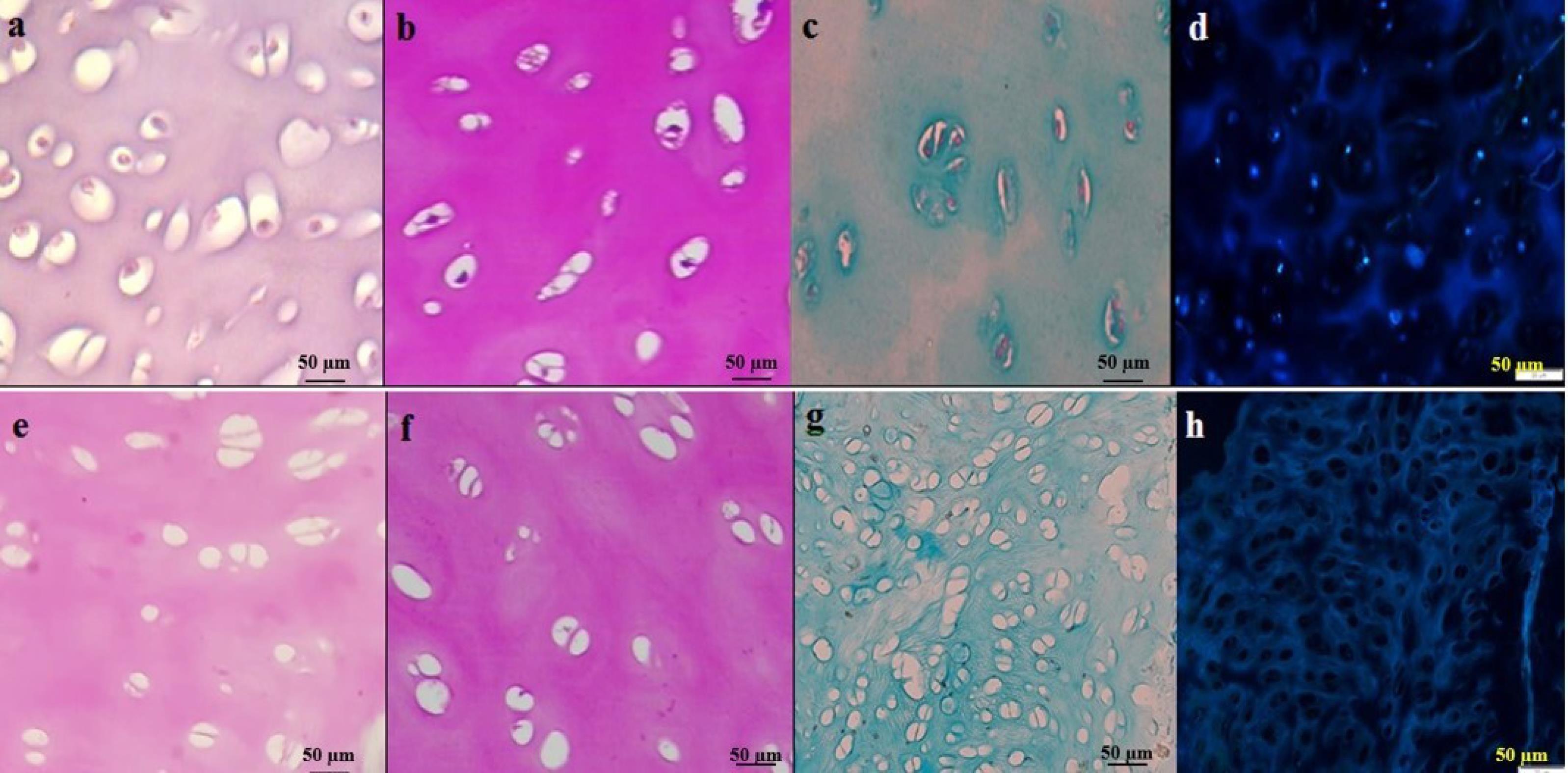
Fig. 1.
Histology was used to visualize the structure and composition of cartilage tissue and to assess the preservation of the extracellular matrix (ECM). Figs. a-d show intact cartilage tissue, while Figs e-h show decellularized (DC) cartilage. Hematoxylin and Eosin (H&E) staining (Figs. a and e) was used to visualize the general structure of the tissue. Periodic acid Schiff (PAS) staining (Figs. b and f) was used to detect glycosaminoglycans (GAGs) in the ECM, while Alcian blue staining (Figs. c and g) was used to detect the presence of collagen fibers. Hoechst staining (Figs. d and h) was used to visualize the nuclei of the cells. Scale bars: 50 μm.
.
Histology was used to visualize the structure and composition of cartilage tissue and to assess the preservation of the extracellular matrix (ECM). Figs. a-d show intact cartilage tissue, while Figs e-h show decellularized (DC) cartilage. Hematoxylin and Eosin (H&E) staining (Figs. a and e) was used to visualize the general structure of the tissue. Periodic acid Schiff (PAS) staining (Figs. b and f) was used to detect glycosaminoglycans (GAGs) in the ECM, while Alcian blue staining (Figs. c and g) was used to detect the presence of collagen fibers. Hoechst staining (Figs. d and h) was used to visualize the nuclei of the cells. Scale bars: 50 μm.
The DNA quantification assay revealed a significant reduction in the DNA content following decellularization. Specifically, the amount of DNA was found to be 9.84 ng/mg dry tissue weight in DC samples, compared to 55.35 ng/mg dry tissue weight in intact samples (N=3 per group, P=0.000) as shown in Fig. 2A and 2B.
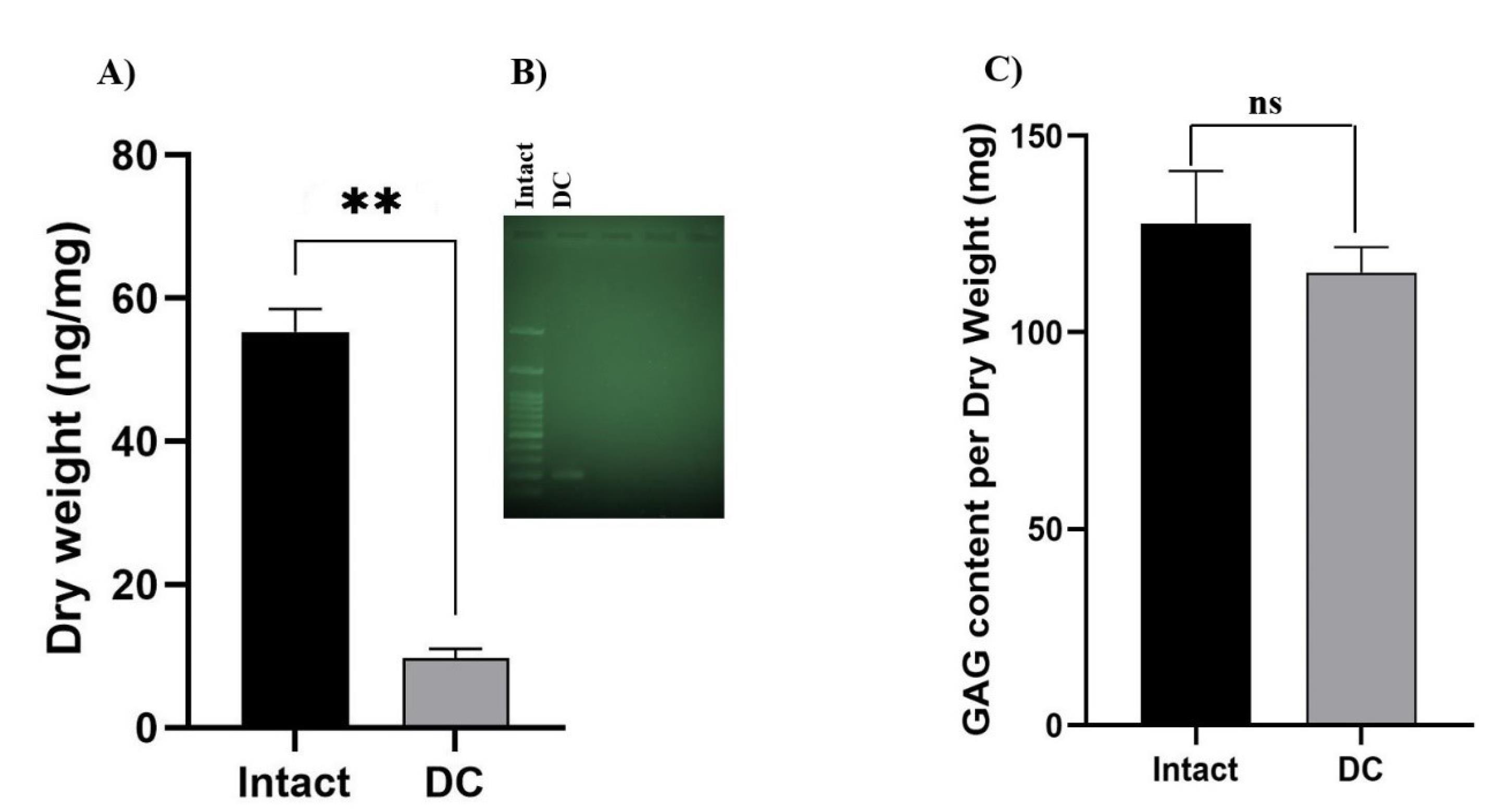
Fig. 2.
(A) Decrease in DNA content after decellularization. Data are expressed as the mean ± standard error of the mean, N = 3 per group, **Indicates significant difference, P ≤ 0.0001. B) The qualitative analyses of DNA content were applied by running electrophoresis for intact and DC tissue and added to the main file. DC tissue showed one band (near to 200 bp) compared to the impact. C) The decrease in GAG content after decellularization was not significant compared to the intact sample. (Data are expressed as the mean ± standard error of the mean, N = 3 per group). Ns: not significant.
.
(A) Decrease in DNA content after decellularization. Data are expressed as the mean ± standard error of the mean, N = 3 per group, **Indicates significant difference, P ≤ 0.0001. B) The qualitative analyses of DNA content were applied by running electrophoresis for intact and DC tissue and added to the main file. DC tissue showed one band (near to 200 bp) compared to the impact. C) The decrease in GAG content after decellularization was not significant compared to the intact sample. (Data are expressed as the mean ± standard error of the mean, N = 3 per group). Ns: not significant.
The preservation of GAG molecules in decellularized tissues is desirable; studies have shown that the amount of GAG is typically lower in DC samples compared to intact cartilage. However, this difference was not statistically significant as shown in Fig. 2C.
SEM assessment showed microarchitecture integrity and efficiency devoid of cells after decellularization as shown in Fig. 3. SEM images showed that, in comparison with intact tissues, DC samples had a cell-less appearance, due to the removal of cellular debris or various ingredients from the cartilage tissue. In SEM analysis, the porosity of the DC cartilage was determined to be 78% ± 0.65%.
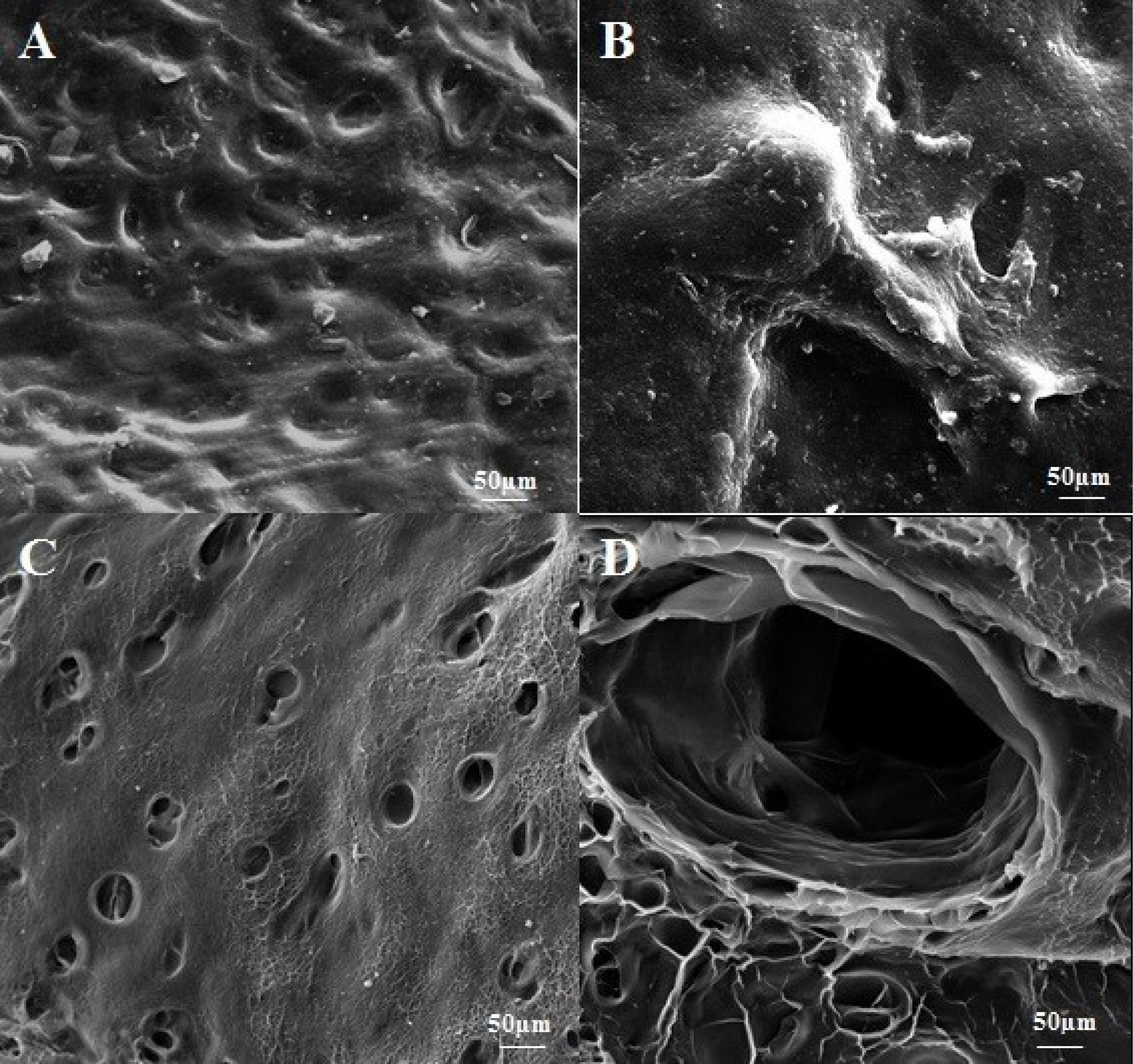
Fig. 3.
Scanning electron micrographs of decellularized (DC) cartilage were obtained to assess the effectiveness of the decellularization process. The micrographs revealed that the cells had been efficiently removed and that the 3D structures and integrity of the tissue were well-preserved. Figs. A and B show intact cartilage tissue, with Fig. A displaying the surface structure of the intact tissue and Fig. B presenting chondrocytes in the intact tissue. The place of removed cells is clear in Figs C and D of DC cartilage.
.
Scanning electron micrographs of decellularized (DC) cartilage were obtained to assess the effectiveness of the decellularization process. The micrographs revealed that the cells had been efficiently removed and that the 3D structures and integrity of the tissue were well-preserved. Figs. A and B show intact cartilage tissue, with Fig. A displaying the surface structure of the intact tissue and Fig. B presenting chondrocytes in the intact tissue. The place of removed cells is clear in Figs C and D of DC cartilage.
The DLS measurements showed that the particle distribution was homogeneous, with a polydispersity index of less than 0.3. Additionally, the particle sizes were found to be less than 1 nm as shown in Supplementary file 1.
Examination of the culture of fibroblast cells revealed that the cells were capable of proliferating and migrating on DC cartilage. Cell invasion was assessed at 24 and 48 hours using an inverted microscope. After 24 hours, the scratch gap was observed to be 70% smaller than the control, indicating that the cells had migrated towards the scratch area. After 48 hours, the scratch was filled, indicating that cell proliferation had occurred as shown in Fig. 4.
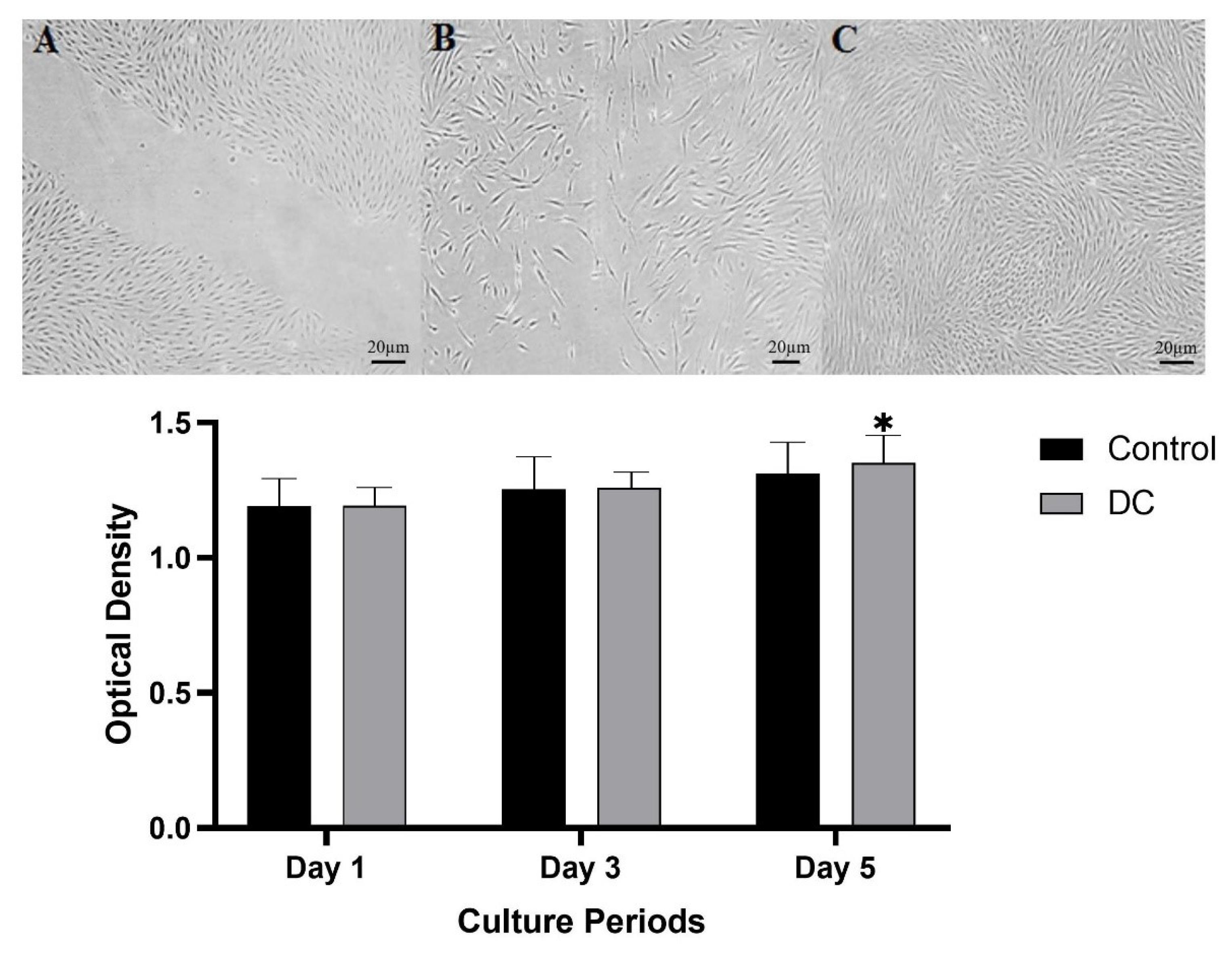
Fig. 4.
Analysis of fibroblast cell migration by in vitro assay. Photographs were taken of the gaps observed under the microscope at 0 h (A), 24 h cell migration shown in the space (B). And 48 h cells reach 80% of confluence (C). Cytocompatibility assessment of DC tissue by MTT test showed that the cell proliferation on the DC cartilage was parallel with the control culture up to day 3, whereas, at longer culture intervals, a higher optical density value was detected in the seeded DC tissue than in the control group. Data are expressed as the mean ± standard error.
.
Analysis of fibroblast cell migration by in vitro assay. Photographs were taken of the gaps observed under the microscope at 0 h (A), 24 h cell migration shown in the space (B). And 48 h cells reach 80% of confluence (C). Cytocompatibility assessment of DC tissue by MTT test showed that the cell proliferation on the DC cartilage was parallel with the control culture up to day 3, whereas, at longer culture intervals, a higher optical density value was detected in the seeded DC tissue than in the control group. Data are expressed as the mean ± standard error.
The MTT test results demonstrated that cells cultured on DC tissue were able to survive and proliferate over time. The cells showed an increase in optical density (OD) values on consecutive days, indicating cell growth. On the first day of culture, the level of cell proliferation on DC tissue was comparable to that of the control culture. However, over longer culture periods, the OD values of cells on decellularized tissue increased, even when compared to the control group. Although this difference was not significant, it suggests that the decellularized tissue was able to support cell growth and survival over time as shown in Fig. 4.
After 21 days, the in vitro degradation rate of lyophilized DC cartilage was found to be 45.03%. However, this difference was not statistically significant when compared to intact samples, where the degradation rate was measured to be 42.58% as shown in Fig. 5.
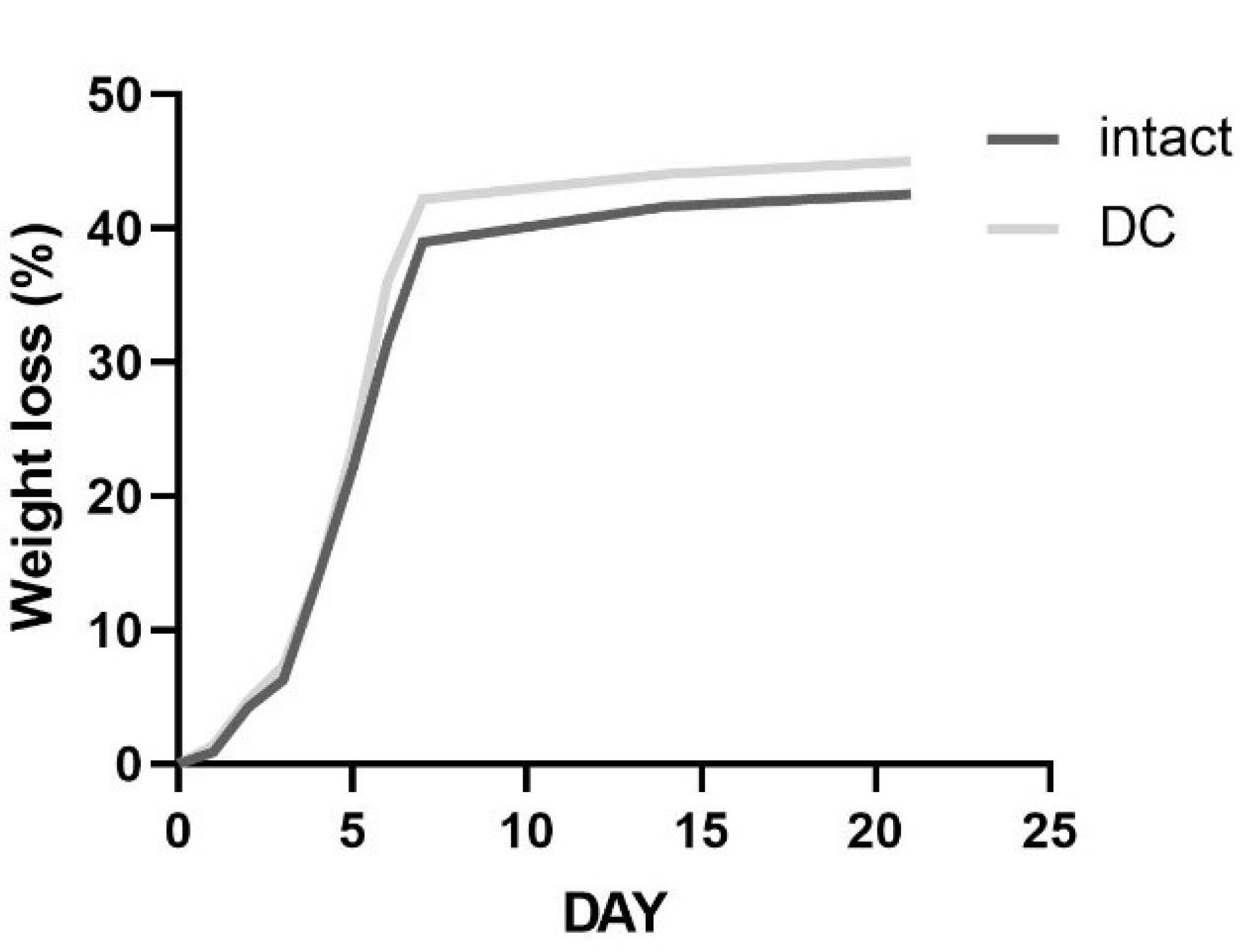
Fig. 5.
Percentage of weight loss of intact and DC cartilage in 21 days. DC samples did not have any significant reduction in weight loss when compared to intact cartilage. Data were expressed as the mean ± standard error.
.
Percentage of weight loss of intact and DC cartilage in 21 days. DC samples did not have any significant reduction in weight loss when compared to intact cartilage. Data were expressed as the mean ± standard error.
The Raman spectra of the DC and intact tissue showed differences in the intensity of certain peaks, which provided information about the ECM content. Specifically, peaks at 509 cm-1, 856 cm-1, 940 cm-1, and 1003 cm-1 were observed and assigned to different molecules. The peak at 509 cm-1 was assigned to collagen, which is a major component of the ECM. The peak at 856 cm-1 was assigned to hydroxyproline, an amino acid that is found almost exclusively in collagen. The peak at 940 cm-1 was assigned to cysteine, which is often present in proteins that contain disulfide bonds. Finally, the peak at 1003 cm-1 was assigned to phenylalanine, an amino acid that is a component of many proteins.32,33 These peaks were demonstrated in both groups of tissues. A peak at 548 cm-1 was sharper in intact samples compared to DC samples, which were candidates for cholesterol.33,34 The Raman spectra showed that there were two distinct vibrational peaks at 1120 cm-1 and 1333 cm-1, which were identified as representing a strong C-O bond of ribose in RNA and guanine, respectively. However, these peaks were not observed in the decellularized (DC) samples.35 Furthermore, a bond observed at 1450 cm-1 was attributed to other carbohydrates that are present in the cell membrane36 as shown in Fig. 6.
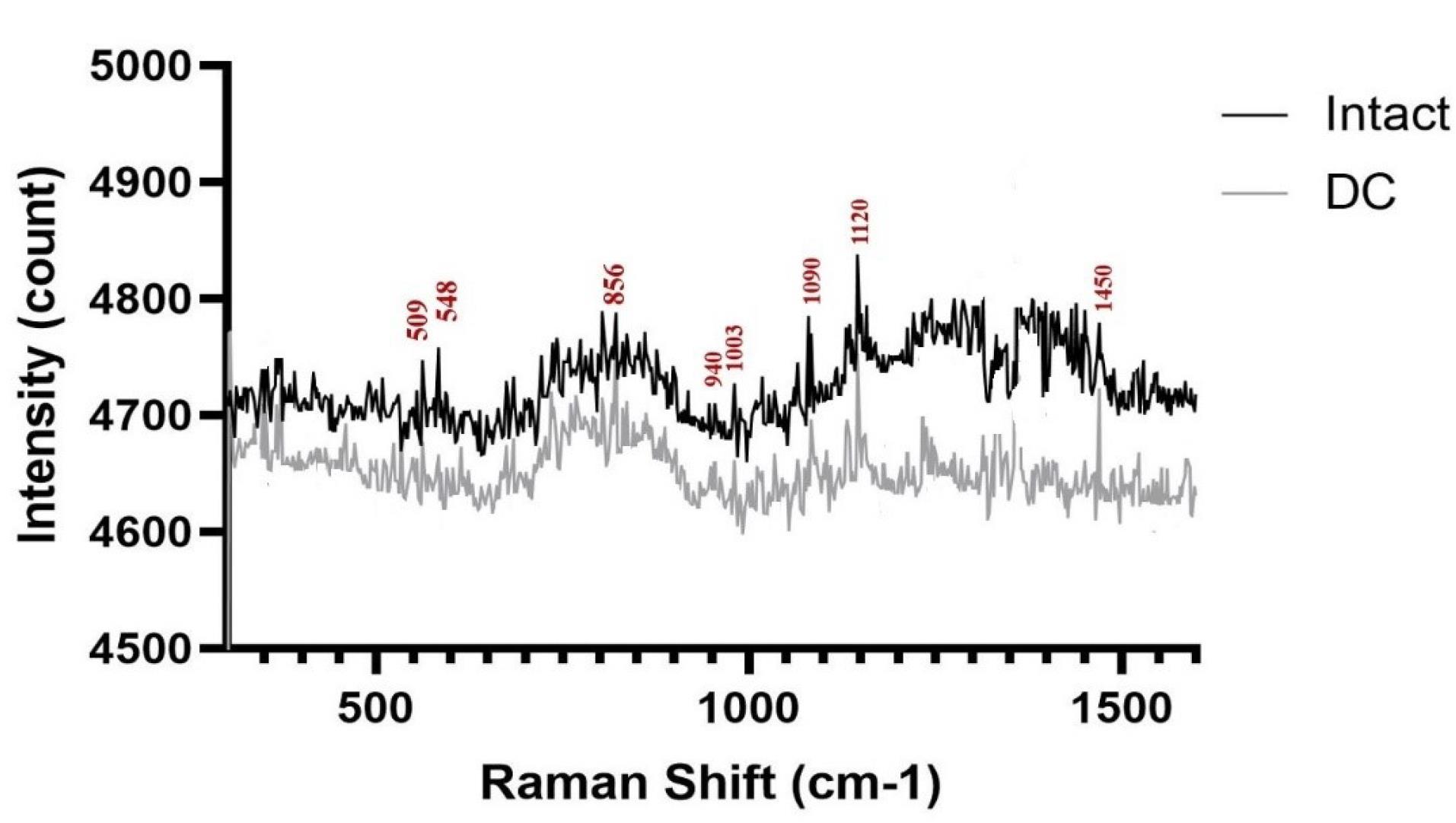
Fig. 6.
After normalization, the Raman spectra of intact and DC cartilage were found to be nearly identical. Both spectra exhibited the characteristic bands of collagen and RNA. This indicates that the overall composition of the ECM was preserved in the DC cartilage, despite the removal of cellular components.
.
After normalization, the Raman spectra of intact and DC cartilage were found to be nearly identical. Both spectra exhibited the characteristic bands of collagen and RNA. This indicates that the overall composition of the ECM was preserved in the DC cartilage, despite the removal of cellular components.
The topographic changes and roughness of the intact and DC cartilage as well as their 3D representations were analyzed by AFM. The mean roughness value (Ra) was calculated using AFM images shown in Table 1 and Fig. 7.
Table 1.
Mean value surface roughness of intact and decellularized cartilage tissue
|
Sample
|
Average surface roughness (Ra) (nm)
|
| Intact cartilage |
6.872±029 |
| Decellularized cartilage |
5.824±0.63 |
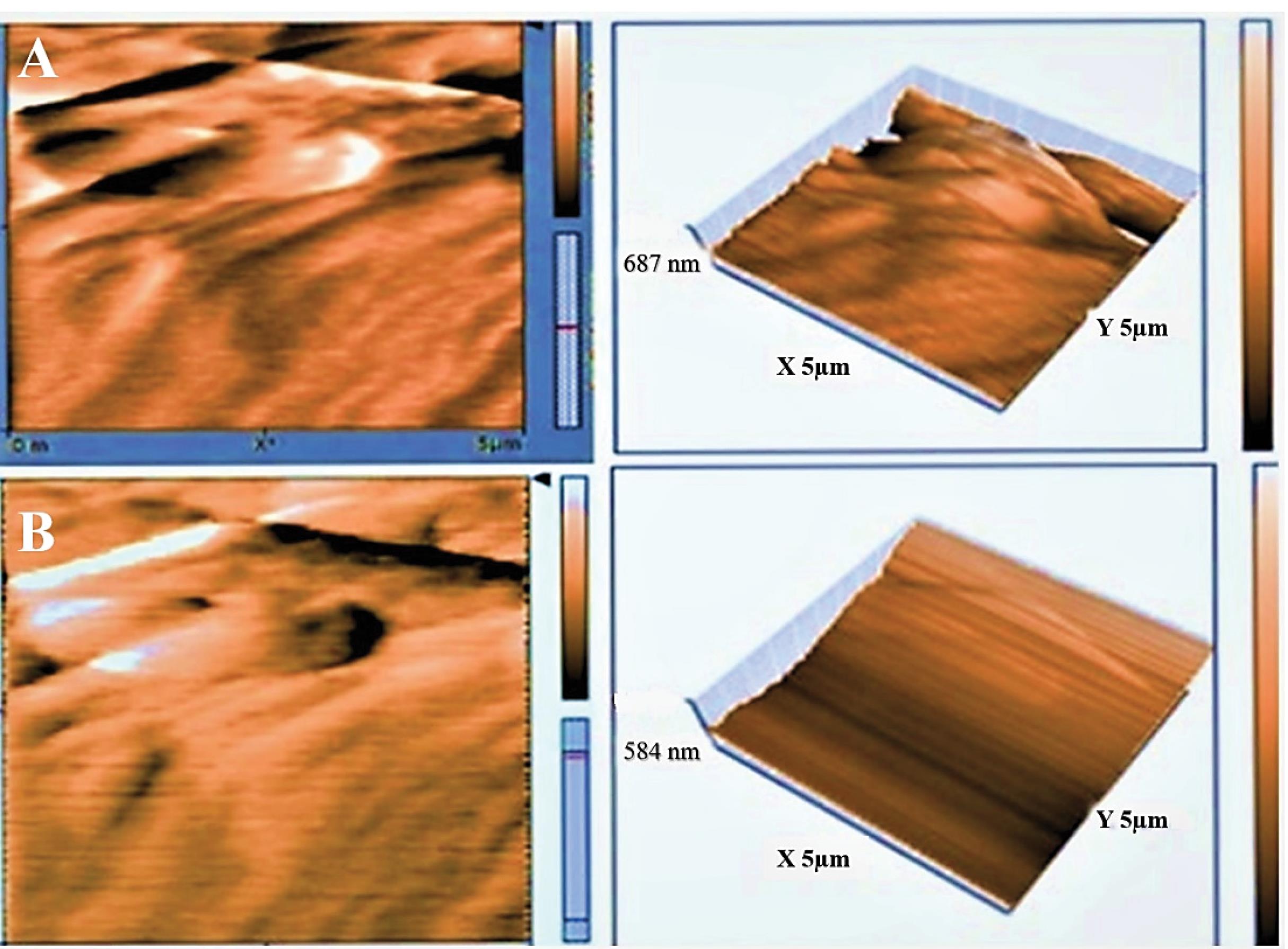
Fig. 7.
The surface topography and 3D structure of intact and decellularized (DC) cartilage were analyzed using atomic force microscopy (AFM). Fig. A shows the surface topography of intact cartilage, while Fig. B shows the surface topography of DC cartilage.
.
The surface topography and 3D structure of intact and decellularized (DC) cartilage were analyzed using atomic force microscopy (AFM). Fig. A shows the surface topography of intact cartilage, while Fig. B shows the surface topography of DC cartilage.
In vivo assessment
In vivo studies were conducted to assess the biocompatibility and bioactivity of DC cartilage after transplantation. The results indicated that none of the rats died during the transplant experiment, and no significant complications or infections were observed during the 2-week follow-up period after surgery. Macroscopic observations also revealed no signs of graft rejection. However, histological examination using H&E staining revealed the presence of a few number of host cells, such as lymphocytes and fibroblasts. Additionally, a few pieces of cartilage were observed after 2 weeks, which had integrated into the skin of the animal as shown in Fig. 8.
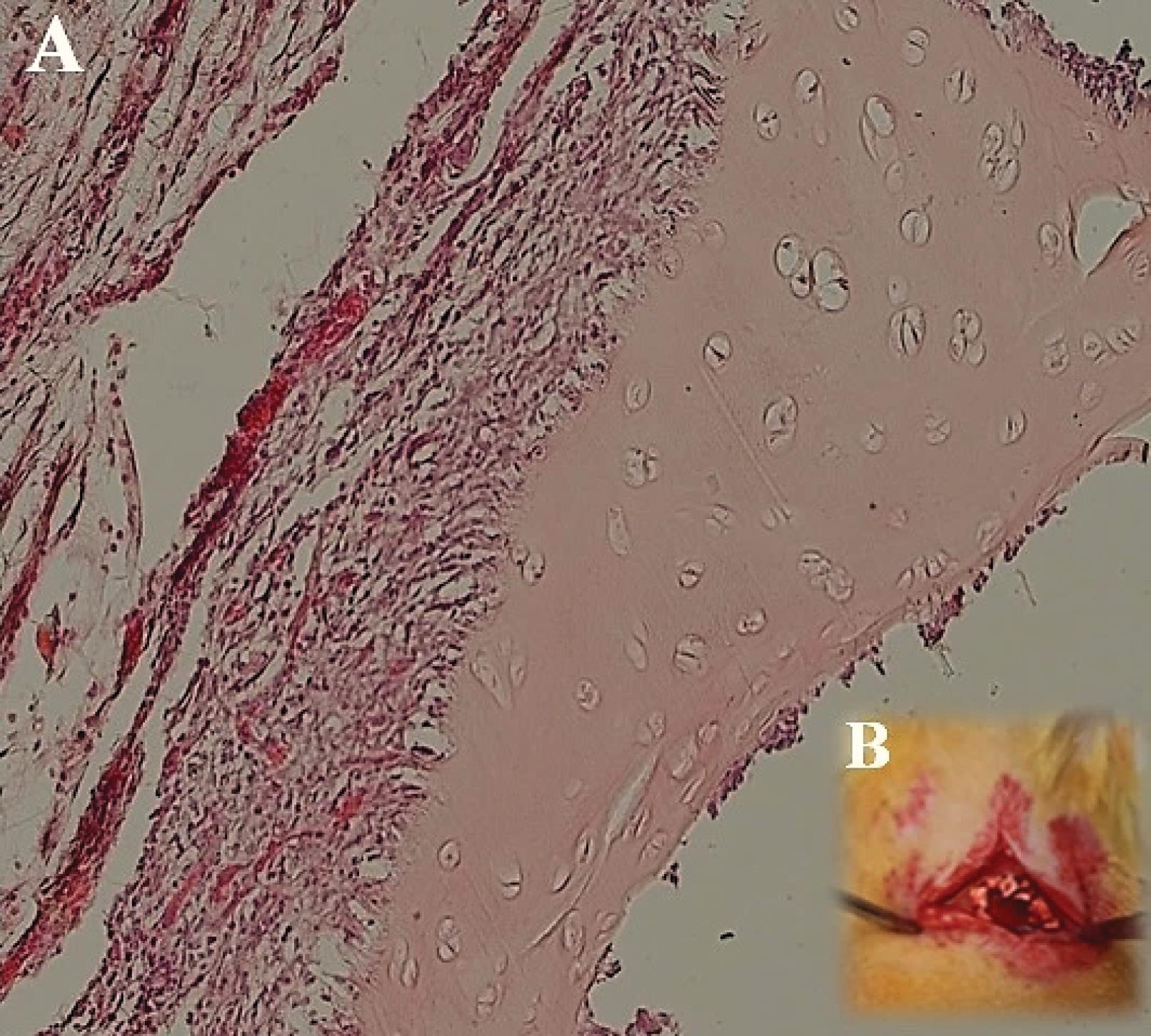
Fig. 8.
After implantation of decellularized (DC) cartilage under the skin of rats, no signs of rejection were observed 2 weeks after surgery. The cartilage had integrated into the skin layer within this time frame. Hematoxylin and Eosin (H&E) staining at 40x magnification showed the presence of host cells, such as lymphocytes and fibroblasts. Fig. A shows the DC cartilage implanted in the skin after 2 weeks, while Fig. B depicts the surgical procedure using DC cartilage under the skin of a rat.
.
After implantation of decellularized (DC) cartilage under the skin of rats, no signs of rejection were observed 2 weeks after surgery. The cartilage had integrated into the skin layer within this time frame. Hematoxylin and Eosin (H&E) staining at 40x magnification showed the presence of host cells, such as lymphocytes and fibroblasts. Fig. A shows the DC cartilage implanted in the skin after 2 weeks, while Fig. B depicts the surgical procedure using DC cartilage under the skin of a rat.
Discussion
In our study, we used physical and sectioning methods to achieve optimal enzyme penetration in bovine tracheal cartilage for decellularization purposes. The results of our study were encouraging, and our technique proved to be practical, economical, and non-destructive. This method could potentially be applied to decellularize other tissues as well. Previous studies have used different detergents and methods for decellularization. For instance, some studies have reported using SDS, a chemical detergent, to remove liver ECM.16,18,37 SDS acts as a protein degrading agent that has great activity to interfere with the structure of proteins by removing non-bonding compounds.38 Damaging the ECM and basement membrane can have severe consequences. To address this issue, DC scaffolds have been utilized in many tissue engineering applications to regenerate various tissues and organs. In particular, DC cartilage is an ideal substrate for investigating cartilage-matrix interactions and can serve as a valuable tool for studying arthritis and other rheumatic diseases.39 In previous studies, most decellularization protocols are based on the application of enzymatic and chemical substances.40 In a study by Zang et al, rat tracheas were decellularized using a modified enzyme-detergent treatment. Tracheal tissue treated with five detergent-enzymatic treatment cycles demonstrated good preservation of the extracellular matrix structure.41 The duration and number of decellularization cycles are crucial factors that influence tissue preservation. However, several studies have demonstrated that all decellularization methods can eventually lead to some degree of impairment to the original tissue architecture, texture, and composition. The potential for structural and compositional damage must be considered during the decellularization process.42,43 The results of the study showed that the modified approach successfully decellularized the cartilage tissue, producing non-cytotoxic DC tissues. The use of a combination of physical methods was found to enhance the quality of decellularization. The study's findings are significant because they represent a step towards using decellularized human cadaveric donor tracheae as scaffolds for airway and joint bioengineering. The expedited preparation broadens the clinical scenarios in which decellularization can be utilized. DLS analysis revealed that no intact cells remained in the samples after the decellularization process. H&E staining confirmed the effectiveness of the decellularization process in reducing the number of cellular remnants, which was supported by the DNA content analysis results. Overall, the study's findings suggest that the modified approach for decellularizing cartilage is a promising method for producing high-quality DC tissues that can be used in tissue engineering applications.44,45 Exposure to detergents leads to the leaching of macromolecules, such as GAGs and other important molecules.46 The commonly used enzymes in related works are nucleases that digest DNA and RNA. These enzymes are generally observed to degrade rapidly and are also reported to possess certain cytotoxic properties.47 Several studies have reported a decrease in GAGs in the heart, articular cartilage, and liver decellularized with Triton X-100 and SDS.48,49 After decellularization, the amount of dsDNA remaining per mg of ECM should be less than 50 ng/mg.50 Also, the length of the remaining DNA fragments should be less than 200 bp (considered standard).51 When using DC tissues as scaffolds, GAGs such as chondroitin sulfate are important for chondrogenic differentiation and the establishment of a physiological microenvironment.52 Collagen regulates cell phenotype, signal evolution, and conduction between cells and scaffolds.53 Therefore, Collagen is a crucial protein in the ECM of many tissues, as it provides strength and mechanical consistency, and influences cell migration, proliferation, and differentiation. Therefore, the preservation of collagen during the decellularization process is essential for successful tissue regeneration after transplantation. In this study, non-chemical detergents were used to decellularize the tissue, resulting in the preservation of collagen. This was confirmed by staining and Raman spectrophotometry. The success of the optimal decellularization protocol was also demonstrated by the loss of DNA, maximal preservation of glycosaminoglycans, and precise tissue architecture.54 To further characterize the scaffolds, Raman and Atomic Force Microscopy spectra were obtained and reported. The results of confocal spectrometry and SEM showed that the decellularization protocol used in this study effectively protected collagen fibers. This approach can alleviate concerns about the potential adverse effects of residual enzymes and detergents in the decellularized tissue. The use of non-chemical protocols for the decellularization of tissues represents a novel approach in the field of tissue engineering and regenerative medicine. Traditionally, chemical detergents have been used for decellularization, but they can have adverse effects on the ECM components of the tissue, including degradation of collagen and GAGs, which can negatively affect the structural and functional properties of the tissue. The novelty and importance of non-chemical protocols for decellularization lie in their ability to effectively remove cells while preserving the ECM components of the tissue. These techniques can improve the quality and functionality of the resulting decellularized tissue, making them more suitable for use in tissue engineering and regenerative medicine applications. Non-chemical protocols for decellularization can also reduce the risk of immune rejection when used in transplantation, as they eliminate the need for harsh chemical agents that can cause tissue damage and inflammation. Additionally, non-chemical protocols can be more cost-effective and environmentally friendly than chemical protocols, as they may require fewer reagents and generate less hazardous waste.
Conclusion
Preservation of the natural elements of the ECM can serve as a viable platform to explore cell behavior. Furthermore, the decellularized scaffolds can be employed as bioinks in 3D bioprinting or hydrogel fabrication. However, the success of this strategy depends on various factors, including tissue density and thickness, and the effectiveness of the decellularization procedure. Here, we developed a streamlined decellularization protocol for cartilage by using a combination of freezing-thawing and sonication without any chemical detergent. The resulting decellularized constructs have the potential to serve as effective graft alternatives for the regeneration of cartilage and the treatment of osteoarthritis or cartilage damage. Furthermore, a detergent-free technique can be employed to utilize cartilage fragments as scaffolds for cell sheet technology, which presents an invaluable asset for exploring regenerative medicine and tissue engineering.
Research Highlights
What is the current knowledge?
√ The decellularized scaffolds with original components can be a suitable substrate for cells similar to the original tissue.
√ Recent research has focused on optimal decellularization methods, which generally include physical and chemical methods.
√ A concern with decellularization chemical protocols, especially those with high detergent concentrations, is the disruption of native ECM
What is new here?
√ This study focused on optimizing cartilage decellularization by physical methods without the use of ionic detergents.
√ The advantage of this protocol is the absence of potentially toxic chemicals in the tissue and proper native ECM preservation
√ According to this result, a cartilage scaffold based on a physical decellularization protocol can be an ideal scaffold for cartilage reconstruction.
Competing Interests
The authors declare that they have no competing interests.
Ethical Statement
This study was approved by the Ethics Committee of Shiraz University of Medical Sciences (Registration number: IR.SUMS.REC.1401.580).
Supplementary files
Supplementary file 1 contains Fig. S1.
(pdf)
Acknowledgements
The authors would like to thank Shiraz University of Medical Sciences and also pathology department of Nemazee Hospital. The authors would like to thank Shiraz University of Medical Sciences, Shiraz, Iran, and also the Center for Development of Clinical Research of Nemazee Hospital and Dr. Nasrin Shokrpour for editorial assistance.
References
- Mantha S, Pillai S, Khayambashi P, Upadhyay A, Zhang Y, Tao O. Smart Hydrogels in Tissue Engineering and Regenerative Medicine. Materials (Basel) 2019; 12:3323. doi: 10.3390/ma12203323 [Crossref] [ Google Scholar]
- Ding X, Zhao H, Li Y, Lee AL, Li Z, Fu M. Synthetic peptide hydrogels as 3D scaffolds for tissue engineering. Advanced Drug Delivery Reviews 2020; 160:78-104. doi: 10.1016/j.addr.2020.10.005 [Crossref] [ Google Scholar]
- Theocharis AD, Skandalis SS, Gialeli C, Karamanos NK. Extracellular matrix structure. Adv Drug Deliv Rev 2016; 97:4-27. doi: 10.1016/j.addr.2015.11.001 [Crossref] [ Google Scholar]
- Yu Y, Zhang W, Liu X, Wang H, Shen J, Xiao H. Extracellular matrix scaffold-immune microenvironment modulates tissue regeneration. Composites Part B: Engineering 2022; 230:2022; 230. doi: 10.1016/j.compositesb.2021.109524 [Crossref] [ Google Scholar]
- Gattazzo F, Urciuolo A, Bonaldo P. Extracellular matrix: a dynamic microenvironment for stem cell niche. BiochimBiophys Acta 2014; 1840:2506-19. doi: 10.1016/j.bbagen.2014.01.010 [Crossref] [ Google Scholar]
- Stratton S, Shelke NB, Hoshino K, Rudraiah S, Kumbar SG. Bioactive polymeric scaffolds for tissue engineering. Bioact Mater 2016; 1:93-108. doi: 10.1016/j.bioactmat.2016.11.001 [Crossref] [ Google Scholar]
- Yao S, Liang Z, Lee YW, Yung PSH, Lui PPY. Bioactive Decellularized Tendon-Derived Stem Cell Sheet for Promoting Graft Healing After Anterior Cruciate Ligament Reconstruction. Am J Sports Med 2023; 51:66-80. doi: 10.1177/03635465221135770 [Crossref] [ Google Scholar]
- Badylak SF, Taylor D, Uygun K. Whole-organ tissue engineering: decellularization and recellularization of three-dimensional matrix scaffolds. Annu Rev Biomed Eng 2011; 13:27-53. doi: 10.1146/annurev-bioeng-071910-124743 [Crossref] [ Google Scholar]
- Yi S, Ding F, Gong L, Gu X. Extracellular Matrix Scaffolds for Tissue Engineering and Regenerative Medicine. Curr Stem Cell Res Ther 2017; 12:233-46. doi: 10.2174/1574888X11666160905092513 [Crossref] [ Google Scholar]
- Gilbert TW, Sellaro TL, Badylak SF. Decellularization of tissues and organs. Biomaterials 2006; 27:3675-83. doi: 10.1016/j.biomaterials.2006.02.014 [Crossref] [ Google Scholar]
- Xing H, Lee H, Luo L, Kyriakides TR. Extracellular matrix-derived biomaterials in engineering cell function. Biotechnol Adv 2020; 42:107421. doi: 10.1016/j.biotechadv.2019.107421 [Crossref] [ Google Scholar]
- Hussey GS, Dziki JL, Badylak SF. Extracellular matrix-based materials for regenerative medicine. Nature Reviews Materials 2018; 3:159-73. doi: 10.1038/s41578-018-0023-x [Crossref] [ Google Scholar]
- Tonti OR, Larson H, Lipp SN, Luetkemeyer CM, Makam M, Vargas D. Tissue-specific parameters for the design of ECM-mimetic biomaterials. Acta Biomater 2021; 132:83-102. doi: 10.1016/j.actbio.2021.04.017 [Crossref] [ Google Scholar]
- Mendibil U, Ruiz-Hernandez R, Retegi-Carrion S, Garcia-Urquia N, Olalde-Graells B, Abarrategi A. Tissue-Specific Decellularization Methods: Rationale and Strategies to Achieve Regenerative Compounds. Int J Mol Sci 2020; 21:5447. doi: 10.3390/ijms21155447 [Crossref] [ Google Scholar]
- Das P, Singh YP, Mandal BB, Nandi SK. Tissue-derived decellularized extracellular matrices toward cartilage repair and regeneration. Methods Cell Biol 2020: 157: 185-221.
- Gilpin A, Yang Y. Decellularization Strategies for Regenerative Medicine: From Processing Techniques to Applications. Biomed Res Int 2017; 2017:9831534. doi: 10.1155/2017/9831534 [Crossref] [ Google Scholar]
- Kawecki M, Łabuś W, Klama‐Baryla A, Kitala D, Kraut M, Glik J. A review of decellularization decellularization methods caused by an urgent need for quality control of cell‐free extracellular matrix'scaffolds and their role in regenerative medicine. Journal of Biomedical Materials Research Part B: Applied Biomaterials 2018; 106:2018; 106. doi: 10.1002/jbm.b.33865 [Crossref] [ Google Scholar]
- Keane TJ, Swinehart IT, Badylak SF. Methods of tissue decellularization used for the preparation of biologic scaffolds and in vivo relevance. Methods 2015; 84:25-34. doi: 10.1016/j.ymeth.2015.03.005 [Crossref] [ Google Scholar]
- Rabbani M, Zakian N, Alimoradi N. Contribution of Physical Methods in Decellularization of Animal Tissues. J Med Signals Sens 2021; 11:1-11. doi: 10.4103/jmss.JMSS_2_20 [Crossref] [ Google Scholar]
- Parmaksiz M, Dogan A, Odabas S, Elcin AE, Elcin YM. Clinical applications of decellularized extracellular matrices for tissue engineering and regenerative medicine. Biomed Mater 2016; 11:022003. doi: 10.1088/1748-6041/11/2/022003 [Crossref] [ Google Scholar]
- Sevastianov VI, Basok YB, Grigoriev AM, Nemets EA, Kirillova AD, Kirsanova LA. Decellularization of cartilage microparticles: Effects of temperature, supercritical carbon dioxide and ultrasound on biochemical, mechanical, and biological properties. J Biomed Mater Res A 2023; 111:543-55. doi: 10.1002/jbm.a.37474 [Crossref] [ Google Scholar]
- Malcova T, Nacu V, Rojnoveanu G, Andree B, Hilfiker A, editors. Evaluation of ultrasound application for the decellularization of small caliber vessels. International Conference on Nanotechnologies and Biomedical Engineering. Springer; 2021.
- Starnecker F, Konig F, Hagl C, Thierfelder N. Tissue-engineering acellular scaffolds-The significant influence of physical and procedural decellularization factors. J Biomed Mater Res B Appl Biomater 2018; 106:153-62. doi: 10.1002/jbm.b.33816 [Crossref] [ Google Scholar]
- Obata T, Tsuchiya T, Akita S, Kawahara T, Matsumoto K, Miyazaki T. Utilization of Natural Detergent Potassium Laurate for Decellularization in Lung Bioengineering. Tissue Eng Part C Methods 2019; 25:459-71. doi: 10.1089/ten.TEC.2019.0016 [Crossref] [ Google Scholar]
- Gilbert TW, Freund JM, Badylak SF. Quantification of DNA in biological scaffold materials. J Surg Res 2009; 152:135-9. doi: 10.1016/j.jss.2008.02.013 [Crossref] [ Google Scholar]
- Sun Y, Tsui YK, Yu M, Lyu M, Cheung K, Kao R. Integration of a miniaturized DMMB assay with high-throughput screening for identifying regulators of proteoglycan metabolism. Sci Rep 2022; 12:1083. doi: 10.1038/s41598-022-04805-y [Crossref] [ Google Scholar]
- Stetefeld J, McKenna SA, Patel TR. Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophys Rev 2016; 8:409-27. doi: 10.1007/s12551-016-0218-6 [Crossref] [ Google Scholar]
- Tolosa L, Donato MT, Gómez-Lechón MJ. General cytotoxicity assessment by means of the MTT assay. Protocols in in vitro hepatocyte research: Springer; 2015. p. 333-48. 10.1007/978-1-4939-2074-7_26.
- Pijuan J, Barcelo C, Moreno DF, Maiques O, Siso P, Marti RM. In vitro Cell Migration, Invasion, and Adhesion Assays: From Cell Imaging to Data Analysis. Front Cell Dev Biol 2019; 7:107. doi: 10.3389/fcell.2019.00107 [Crossref] [ Google Scholar]
- Sharma C, Dinda AK, Potdar PD, Chou CF, Mishra NC. Fabrication and characterization of novel nano-biocomposite scaffold of chitosan-gelatin-alginate-hydroxyapatite for bone tissue engineering. Mater Sci Eng C Mater Biol Appl 2016; 64:416-27. doi: 10.1016/j.msec.2016.03.060 [Crossref] [ Google Scholar]
- Power LJ, Fasolato C, Barbero A, Wendt DJ, Wixmerten A, Martin I. Sensing engineered engineered engineered engineered engineered engineered engineered tissue-engineered cartilage quality with Raman spectroscopy and statistical learning for the development of advanced characterization assays. BiosensBioelectron 2020; 166:112467. doi: 10.1016/j.bios.2020.112467 [Crossref] [ Google Scholar]
- Barthold JE, Martin BM, Sridhar SL, Vernerey F, Schneider SE, Wacquez A. Recellularization and Integration of Dense Extracellular Matrix by Percolation of Tissue Microparticles. Adv Funct Mater 2021; 31:2103355. doi: 10.1002/adfm.202103355 [Crossref] [ Google Scholar]
- Bergholt MS, Serio A, Albro MB. Raman Spectroscopy: Guiding Light for the Extracellular Matrix. Front BioengBiotechnol 2019; 7:303. doi: 10.3389/fbioe.2019.00303 [Crossref] [ Google Scholar]
- Albro MB, Bergholt MS, St-Pierre JP, Vinals Guitart A, Zlotnick HM, Evita EG. Raman spectroscopic imaging for quantification of depth-dependent and local heterogeneities in native and engineered cartilage. NPJ Regen Med 2018; 3:3. doi: 10.1038/s41536-018-0042-7 [Crossref] [ Google Scholar]
- Kumar R, Gronhaug KM, Afseth NK, Isaksen V, de Lange Davies C, Drogset JO. Optical investigation of osteoarthritic human cartilage (ICRS grade) by confocal Raman spectroscopy: a pilot study. Anal Bioanal Chem 2015; 407:8067-77. doi: 10.1007/s00216-015-8979-5 [Crossref] [ Google Scholar]
- Hanifi A, Palukuru U, McGovernC McGovernC, Shockley M, Frank E, Grodzinsky A. Near-infrared spectroscopic assessment of developing engineered tissues: correlations with compositional and mechanical properties. Analyst 2017; 142:1320-32. doi: 10.1039/c6an02167k [Crossref] [ Google Scholar]
- Pan MX, Hu PY, Cheng Y, Cai LQ, Rao XH, Wang Y. An efficient method for decellularization of the rat liver. J Formos Med Assoc 2014; 113:680-7. doi: 10.1016/j.jfma.2013.05.003 [Crossref] [ Google Scholar]
- Luo Z, Bian Y, Su W, Shi L, Li S, Song Y. Comparison of various reagents for preparing a decellularized porcine cartilage scaffold. American Journal of Translational Research 2019; 11:1417. [ Google Scholar]
- Shen W, Berning K, Tang SW, Lam YW. Rapid and Detergent-Free Decellularization of Cartilage. Tissue Eng Part C Methods 2020; 26:201-6. doi: 10.1089/ten.TEC.2020.0008 [Crossref] [ Google Scholar]
- Sutherland AJ, Beck EC, Dennis SC, Converse GL, Hopkins RA, Berkland CJ. Decellularized cartilage may be a chondroinductive material for osteochondral tissue engineering. PloS one 2015; 10:e0121966. doi: 10.1371/journal.pone.0121966 [Crossref] [ Google Scholar]
- Zang M, Zhang Q, Chang EI, Mathur AB, Yu P. Decellularized tracheal matrix scaffold for tracheal tissue engineering: in vivo host response. PlastReconstr Surg 2013; 132:549e-59e. doi: 10.1097/PRS.0b013e3182a013fc [Crossref] [ Google Scholar]
- Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials 2011; 32:3233-43. doi: 10.1016/j.biomaterials.2011.01.057 [Crossref] [ Google Scholar]
- Dimou Z, Michalopoulos E, Katsimpoulas M, Dimitroulis D, Kouraklis G, Stavropoulos-Giokas C. Evaluation of a Decellularization Protocol for the Development of a Decellularized Tracheal Scaffold. Anticancer Res 2019; 39:145-50. doi: 10.21873/anticanres.13090 [Crossref] [ Google Scholar]
- Butler CR, Hynds RE, Crowley C, Gowers KH, Partington L, Hamilton NJ. Vacuum-assisted decellularization: an accelerated protocol to generate tissue-engineered human tracheal scaffolds. Biomaterials 2017; 124:95-105. doi: 10.1016/j.biomaterials.2017.02.001 [Crossref] [ Google Scholar]
- Lange P, Greco K, Partington L, Carvalho C, Oliani S, Birchall MA. A pilot study of a novel vacuum-assisted method for decellularization of tracheae for clinical tissue engineering applications. J Tissue Eng Regen Med 2017; 11:800-11. doi: 10.1002/term.1979 [Crossref] [ Google Scholar]
- Rana D, Zreiqat H, Benkirane-Jessel N, Ramakrishna S, Ramalingam M. Development of decellularized scaffolds for stem cell-driven tissue engineering. J Tissue Eng Regen Med 2017; 11:942-65. doi: 10.1002/term.2061 [Crossref] [ Google Scholar]
- Kim BS, Das S, Jang J, Cho DW. Decellularized Extracellular Matrix-based Bioinks for Engineering Tissue- and Organ-specific Microenvironments. Chem Rev 2020; 120:10608-61. doi: 10.1021/acs.chemrev.9b00808 [Crossref] [ Google Scholar]
- Bruyneel AAN, Carr CA. Ambiguity in the Presentation of Decellularized Tissue Composition: The Need for Standardized Approaches. Artif Organs 2017; 41:778-84. doi: 10.1111/aor.12838 [Crossref] [ Google Scholar]
- Elder BD, Athanasiou KA. Systematic assessment of growth factor treatment on biochemical and biomechanical properties of engineered articular cartilage constructs. Osteoarthritis Cartilage 2009; 17:114-23. doi: 10.1016/j.joca.2008.05.006 [Crossref] [ Google Scholar]
- Taylor DA, Sampaio LC, Ferdous Z, Gobin AS, Taite LJ. Decellularized matrices in regenerative medicine. Acta Biomater 2018; 74:74-89. doi: 10.1016/j.actbio.2018.04.044 [Crossref] [ Google Scholar]
- Arenas-Herrera JE, Ko IK, Atala A, Yoo JJ. Decellularization for whole organ bioengineering. Biomed Mater 2013; 8:014106. doi: 10.1088/1748-6041/8/1/014106 [Crossref] [ Google Scholar]
- Wang Z, Li Z, Li Z, Wu B, Liu Y, Wu W. Cartilaginous extracellular matrix derived from decellularized chondrocyte sheets for the reconstruction of osteochondral defects in rabbits. Acta Biomater 2018; 81:129-45. doi: 10.1016/j.actbio.2018.10.005 [Crossref] [ Google Scholar]
- Hortensius RA, Harley BA. The use of bioinspired alterations in the glycosaminoglycan content of collagen-GAG scaffolds to regulate cell activity. Biomaterials 2013; 34:7645-52. doi: 10.1016/j.biomaterials.2013.06.056 [Crossref] [ Google Scholar]
- Owen SC, Shoichet MS. Design of three-dimensional biomimetic scaffolds. J Biomed Mater Res A 2010; 94:1321-31. doi: 10.1002/jbm.a.32834 [Crossref] [ Google Scholar]