Bioimpacts. 14(6):30118.
doi: 10.34172/bi.2024.30118
Original Article
Association of tumour mutation burden with prognosis and its clinical significance in stage III gastric cancer
Ya-Lin Han Conceptualization, Data curation, Project administration, Writing – original draft, Writing – review & editing, 1, 2, # 
Li Chen Conceptualization, Data curation, Project administration, Writing – original draft, Writing – review & editing, 3, #
Xu-Ning Wang Conceptualization, Data curation, Investigation, Project administration, Visualization, Writing – original draft, Writing – review & editing, 4
Mao-Lin Xu Conceptualization, Visualization, Writing – original draft, Writing – review & editing, 1
Rui Qin Conceptualization, Writing – original draft, Writing – review & editing, 5
Fang-Ming Gong Investigation, Methodology, Writing – original draft, Writing – review & editing, 1
Peng Sun Methodology, Writing – original draft, Writing – review & editing, 1
Hong-Yi Liu Investigation, Methodology, Writing – original draft, Writing – review & editing, 1
Zhi-Peng Teng Data curation, Investigation, Validation, Writing – original draft, Writing – review & editing, 1
Zhao-Xia Li Formal analysis, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing, 2, *
Guang-Hai Dai Formal analysis, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing, 3, * 
Author information:
1Department of General Surgery, The First Medical Centre, Chinese PLA General Hospital, Beijing 100853, China
2Department of Oncology, PLA Rocket Force Characteristic Medical Centre, Beijing 100088, China
3Department of Oncology, Fifth Medical Center of Chinese PLA General Hospital, Beijing 100071, China
4Department of General Surgery, The Air Force Hospital of Northern Theater PLA, Shenyang 110042, China
5Department of Gastroenterology, The 305 Hospital of PLA, Beijing 100017, China
#These authors contributed equally to this study.
Abstract
Introduction:
To explore the correlation between the tumour mutation burden (TMB) and prognosis and its clinical significance among patients with stage III gastric cancer (GC).
Methods:
Patients with stage III GC were divided into a high TMB and low TMB group in both a study cohort of 38 patients and the Cancer Genome Atlas (TCGA) cohort of 173 patients. In the study cohort, next-generation sequencing was used to detect mutated GC genes and obtain TMB data. In the TCGA cohort, gene set enrichment analysis was performed, and the relationship between TMB, prognosis and clinicopathologic factors was analysed. Western blot and quantitative real-time polymerase chain reaction were used to detect the expression levels of both proteins and genes. Cell viability was measured using methyl thiazolyl tetrazolium and transwell cell assays.
Results:
Patients in the high TMB group had better overall survival (OS) rates than patients in the low TMB group for both cohorts and TMB was associated with age, mutation signature 1 and mutation signature 17. The Cox regression analysis revealed that age, not TMB, was an independent prognosis factor. Furthermore, genes with high-frequency mutations were significantly enriched in the RTK-RAS and Notch signalling pathways. The activation of these pathways was lower in the high TMB compared with the low TMB group, and the proliferation and migration abilities of GC cells showed a similar pattern in both TMB groups.
Conclusion:
Patients in the high TMB group had better OS rates than patients in the low TMB group. Genes with high-frequency mutations were significantly enriched in the RTK-RAS and Notch pathways. Hence, TMB could serve as a prognosis biomarker with potential clinical significance.
Keywords: Mutated genes, Molecular characteristic, Prognosis, Stage III gastric cancer, Tumour mutation burden
Copyright and License Information
© 2024 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
None.
Introduction
Gastric cancer (GC) is one of the predominant malignant tumours with high morbidity and lethality worldwide.1 Its incidence appears to be regional, with high-incidence areas mainly concentrated in eastern Asian countries.2 In 2020, there were 479 000 new cases of GC and 374 000 new deaths related to the disease in China, accounting, respectively, for 43.9% and 48.6% of all cases worldwide, which presents a serious threat to human health.3 According to Chinese Union for Gastrointestinal Tumour Surgery data from 2014–2017, the proportion of advanced GC in China is as high as 80.3%.4 The mainstay of treatment for GC is surgery, both Chinese and Japanese guidelines for the management of GC recommend D2 radical gastrectomy for advanced GC.5,6 Presently, treatment has aided in managing the disease, and the 5-year survival rate for early-stage GC (stage II or lower) after surgery ranges from 60% to 80%. Conversely, patients with stage III tumours after surgery have a 5-year survival rate of 18%–50%.7 The diagnosis of stage III GC includes patients with invasion depth up to the serosa layer and/or with lymph node metastasis, according to the pTNM staging of GC (eighth edition), jointly formulated by the American Joint Committee on Cancer and the Union for International Cancer Control.8 Postoperative adjuvant chemotherapy is recommended by the National Comprehensive Cancer Network guidelines for all patients with stage III GC.9
Gastric cancer is, however, a heterogeneous collection of diseases with differing responses to immunotherapy. Numerous biomarkers have been investigated to identify susceptibility to immunotherapy,10 and tumour mutation burden (TMB) has been identified as an emerging cancer biological marker characterised by microsatellite instability and is considered to be a predictor of response to GC immunotherapy.11 By definition, TMB is the cumulative total number of somatic mutations detected per million bases, as well as the total number of mutations per million bases in the coding region of gene exons in tumour cells, including substitutions, insertions and deletions.12 Highly mutated tumours are thought to have immunogenicity and neoantigen burdens, two factors that are thought to respond to immunotherapy.13 In this study, the authors found several patients in the same pathologic stage and an identical postoperative in-clinic chemotherapy regimen; however, there was still a significant difference in survival rates. To explore the discrepancy of molecular characteristics in patients with stage III GC with different survival prognoses, the present study was initiated. The results are considered to have profound significance for judging therapeutic efficacy and prognosis.
This study included two cohorts of stage III GC: 1) a study cohort of 38 patients; and 2) a TCGA cohort of 173 patients. The mutated genes and TMB data of both cohorts were assessed. We aimed to obtain pathway enrichment and examine the clinical significance of TMB on stage III GC, as well as identify the TMB level and pathway prediction for prognosis significance in stage III GC to provide a foundation for future clinical trials.
Materials and methods
Clinical data
Sample selection
The tumour tissue of 38 patients with stage III GC was assessed using the next-generation sequencing (NGS) molecular biological technique at the First Medical Centre of Chinese PLA General Hospital from January 2009 to December 2014. Patient information including sex, tumour location, surgical resection modality, pathologic type, postoperative adjuvant therapy and the survival period was collected. The individual characteristics are shown in Table 1. A total of 38 cases were divided into the long-term survival group (survival time ≥5 years) and the short-term survival group (survival time <3 years) based on the patient survival rates. All of the above 38 samples were detected using FoundationOne CDx, a comprehensive companion diagnostic test for pan-tumours, approved by the Food and Drug Administration.
Table 1.
Clinical and pathological characteristics, and treatment features of 38 postoperative patients with stage III gastric cancer
|
Item
|
Group
|
Number
|
Composition ratio (%)
|
| Gender |
Male |
34 |
89.5 |
| Female |
4 |
10.5 |
| Age |
<60 years |
20 |
52.6 |
| ≥60 years |
18 |
47.4 |
| Tumor location |
Upper |
10 |
26.3 |
| Middle |
8 |
21.1 |
| Lower |
20 |
52.6 |
| Total gastrectomy |
No |
34 |
89.5 |
| Yes |
4 |
10.5 |
| Tumor size |
<5 cm |
22 |
57.9 |
| ≥5 cm |
16 |
42.1 |
| Depth of invasion |
T3 |
22 |
57.9 |
| T4 |
16 |
42.1 |
| Lymph node metastasis |
0 |
7 |
18.4 |
| 1-2 |
14 |
36.9 |
| 3-6 |
16 |
42.1 |
| ≥7 |
1 |
2.6 |
| Borrmann's classification |
type I |
1 |
2.6 |
| type II + III |
33 |
86.9 |
| type IV |
4 |
10.5 |
| WHO histological classification |
Adenocarcinoma |
30 |
78.9 |
| Signet ring cell carcinoma |
8 |
21.1 |
| Lauren's classification |
Intestinal type |
18 |
47.4 |
| Diffuse type |
13 |
34.2 |
| Mixed type |
7 |
18.4 |
| Histological differentiation |
Low/moderately Low differentiation |
32 |
84.2 |
| Moderately high/moderate/high differentiation |
6 |
15.8 |
| Cancer emboli |
Yes |
16 |
42.1 |
| No |
22 |
57.9 |
| Nerve involvement |
Yes |
8 |
21.1 |
| No |
30 |
78.9 |
| Cancer nodules |
Present |
6 |
15.8 |
| Absent |
32 |
84.2 |
| TNM staging |
ⅢA |
22 |
57.9 |
| ⅢB |
12 |
31.6 |
| ⅢC |
4 |
10.5 |
| Treatment plan group |
Containing oxaliplatin regimen |
26 |
68.4 |
| Containing docetaxel regimen |
12 |
31.6 |
| Treatment cycle group |
2-5 Cycles |
11 |
28.9 |
| ≥6 Cycles |
27 |
71.1 |
Relationship between the TMB threshold level and prognosis
Patients with stage III GC were divided into two groups (high and low TMB) using the R package’s ‘surviminer’ tool with the surv_cutpoint function, which determines the optimal cutpoint for one continuous variable with the maximally selected rank statistics from R package 'maxstat' function.14 The survival curve of the differential TMB threshold level was analysed using the Kaplan–Meier method.
The Cancer Genome Atlas data
Data acquisition
All data of 173 patients with stage III GC from the Cancer Genome Atlas database (TCGA) were downloaded from XENA (https://xena.ucsc.edu/),15 and the TMB was calculated using mutation files (‘maf’ files).
Relationship of the TMB threshold level with prognosis and clinicopathology
The general linear model was used to explore the relationship between the TMB and clinical factors. In addition, we compared the prognosis of patients with high and low TMB. The KM survival analysis was used to compare the survival time of patients in the high and low TMB groups. Subsequently, the association between TMB and prognosis was analysed using the multivariate Cox regression model. All the cells of the body could mutate, and mutation signatures related to GC were selected as clinical factors. Mutation signatures 1 and 2 are found in many types of cancers. Mutation signatures 15 and 17 are also found in several cancers, including stomach cancer.
Cell culture
Primary cells were collected using patient tumour tissue samples. Briefly, the tumour tissue samples were washed with phosphate-buffered saline (PBS) 3 times, then soaked in 10 000 U penicillin and streptomycin for 20 minutes and cut into pieces of 3-4 mm2. A fibroblast medium (ScienCell Research Laboratories, San Diego, CA) containing 20% foetal bovine serum (FBS) was added to these small samples for culture to ensure that fibroblasts around the samples were confluent to achieve contact inhibition.16 These small tissue blocks were then digested by trypsin and thereafter passaged. The primary gastric cancer cells of samples passaged 5–8 times were used in this study.16 All cell genotypes were consistent with those of the gastric cancer tissue of origin. Finally, GC cells (2 × 105 cells/well) were plated in triplicate in a six-well plate and incubated. Cells harvested by trypsin were counted using an automated cell counter. Cell lines were cultured in RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 10% FBS and 100 mg/mL of penicillin/streptomycin/glutamine (Gibco) in humidified incubators with a 5% CO2 at 37 ℃.
Western blotting
Proteins were extracted using a RIPA buffer (Solarbio, Beijing, China) and separated by conducting sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were incubated with primary antibodies overnight at 4 °C and then incubated with horseradish peroxidase-conjugated secondary antibodies at room temperature of 15 ℃ to 30 ℃ for 1 hour.17 The primary antibodies used were as follows: Notch-1 rabbit mAb (1: 1000; ab52627, Abcam) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) rabbit mAb (1:1000; 10494-1-AP, Proteintech), AKT rabbit mAb (1:1000; 4691, Cell Signalling Technology), phospho-Akt (Thr308) rabbit mAb (1:1000; 13038, Cell Signalling Technology), phospho-Akt (Ser473) rabbit mAb (1:1000; 4060, Cell Signalling Technology); all secondary antibodies (7074, Cell Signalling Technology) were used at a 1:5000 dilution. The results were quantified using the ImageJ Analysis System (BioRad Laboratories Inc., Hercules, CA, USA) software.
Cell transwell assay
Tumour cells (2,000 cells/well) were seeded into the upper chamber, and the complete medium (containing 20% FBS) was placed into the lower compartment. Later, cells that passed through the upper chamber were fixed with 4% paraformaldehyde (Solarbio) for 10 minutes, washed with PBS 3 times and stained with crystal violet regent (Solarbio) for 5 minutes.18 The cell counts were determined under a microscope (Nikon).
Reverse transcription and quantitative real-time PCR (qPCR)
Total RNAs were extracted from the cells using the UNlQ-10 Column TRIzolTM Total RNA Isolation Kit (Sango Biotech), and cDNA was synthesised by the MonScriptTM RTIII All-in-One Mix with dsDNase (Monad) according to the manufacturer’s instructions. Quantitative RT-PCR for RNA and PCR for genomic DNA was performed using the MonAmpTM ChemoHS qPCR Mix (Monad). The reaction mixtures were incubated at 50°C for 15 minutes, followed by 95 °C for 5 minutes. Next, 35 PCR cycles were performed using the following cycling conditions: 95 °C for 15 seconds, 60 °C for 30 seconds and 72 °C for 1 minute. The gene expression values were normalised to those of GAPDH. All reactions were repeated at least three times and the 2−ΔΔCq method was used to analyze the relative expression.19 The primers are shown in Table 2.
Table 2.
RT-PCR primer sequences
|
Primer
|
Forward
|
Reverse
|
| Hes1 |
TCAACACGACACCGGATAAAC |
GCCGCGAGCTATCTTTCTTCA |
| Hey1 |
GTTCGGCTCTAGGTTCCATGT |
CGTCGGCGCTTCTCAATTATTC |
| Hey2 |
AAGGCGTCGGGATCGGATAA |
AGAGCGTGTGCGTCAAAGTAG |
| CDKN1A |
TGTCCGTCAGAACCCATGC |
AAAGTCGAAGTTCCATCGCTC |
| FASN |
CCGAGACACTCGTGGGCTA |
CTTCAGCAGGACATTGATGCC |
| CDKN1B |
AACGTGCGAGTGTCTAACGG |
CCCTCTAGGGGTTTGTGATTCT |
| ACLY |
ATCGGTTCAAGTATGCTCGGG |
GACCAAGTTTTCCACGACGTT |
| CTSD |
ATTCAGGGCGAGTACATGATCC |
CGACACCTTGAGCGTGTAG |
| GAPDH |
TGACTTCAACAGCGACACCCA |
CACCCTGTTGCTGTAGCCAAA |
Methyl thiazolyl tetrazolium (MTT) assay for cell proliferation
Cells were seeded in triplicate at a density of 1000 cells/well on 96-well plates and incubated for 1–10 days. Then, MTT was added into each well for a final concentration of 0.5 mg/mL and incubated for 4 hours at 37 °C. After the incubation, all of the medium was removed and 100 µL of DMSO was added to each well.20 Absorbance was then assayed using the Biotek Synergy H1 microplate reader at OD490, and the growth curve was drawn according to OD490 values by days.
Immunohistochemistry (IHC)
The paraffin-embedded tissue sections were processed according to standard procedures for dewaxing and dehydration. The primary antibody was incubated with the sections overnight at 4 °C. A secondary antibody was then applied to the sections and incubated at 37 °C for 30 minutes, and the presence of antigens was revealed using diaminobenzadine tetrachloride (DAB; Dako) and counter-stained with nuclear red or haematoxylin (blue). The relative quantities of IHC reaction were accessed by Image-Pro Plus 6.0. Briefly, scores were applied to rate staining intensity in the cancer cells (no staining: 0; weak: 1; moderate: 2; strong: 3) and the percentage of stained cells (<5%: 0; 5–25%: 1; >25–50%: 2; >50–75%:3; >75%:4). The final score was equal to the intensity multiplied by the percentage. The staining was stratified into low levels (scores 1–3) or high levels of expression (score ≥ 4).20
Statistical analysis
The Cox regression was applied to analyse the prognosis factors, and the general linear model was employed to analyse the TMB-related factors. The forest plot was drawn by the ‘forestplot’ function of the R package, the survival curve was plotted using the Kaplan–Meier method and the waterfall plot of the mutation landscape was plotted using Maftools. The TMB cut-off in the low or high group was determined by the ‘surviminer’ tool in the R package, with maximally selected rank statistics determined by the 'maxstat' function in the R package.
The mutation gene enrichment was conducted by Maftools, and a P value of <0.05 was considered statistically significant. The results are shown as the mean ± SD, and a two-way ANOVA was performed for statistical analysis.
Results
The mutated genes in the study cohort and the TCGA data with stage III GC
The gene mutation profiles of 38 patients in the short- and long-term survival groups were drawn by the GenvisR R package (Fig. 1A). A total of 63 tumour-associated mutated genes were found; the top 5 most frequently mutated genes were TP53 (26/38), CCNE1 (9/38), ARID1A (9/38), KRAS (5/38) and PTEN (4/38). The mutation profile of the TCGA cohort showed that the top 5 mutated genes were TTN, TP53, MUC16, LRP18 and CSMD3 (Figs. 1B, 1C).
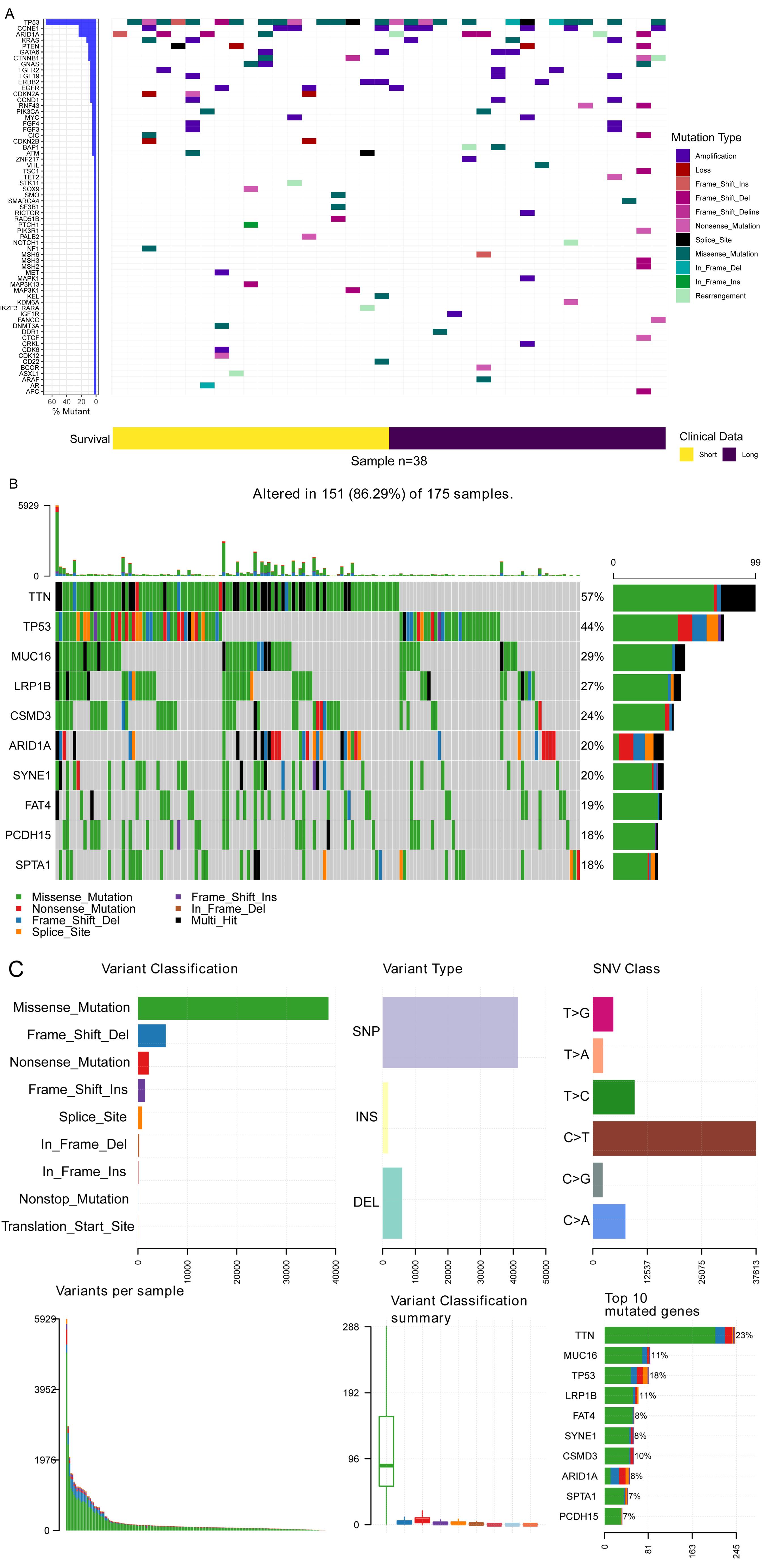
Fig. 1.
Mutation pattern of Stage III gastric cancer. (A) Waterfall plot of mutation in our dataset. (B) Waterfall plot of mutation data of TCGA cohort of Stage III gastric cancer. (C) Mutation summary of Stage III gastric cancer.
.
Mutation pattern of Stage III gastric cancer. (A) Waterfall plot of mutation in our dataset. (B) Waterfall plot of mutation data of TCGA cohort of Stage III gastric cancer. (C) Mutation summary of Stage III gastric cancer.
Clinical significance of TMB in the study cohort and the TCGA data
Patients were divided into groups by TMB both in the study cohort and the TCGA cohort. In the study cohort of 38 patients with stage III GC, the TMB level was 0–37 mutations per megabase unit (Muts/Mb). There were 7 patients in the high TMB group (TMB ≥ 10 Muts/Mb), 26 patients in the low TMB group (TMB < 10 Muts/Mb) and 5 patients with uncertain TMB values. In the TCGA cohort of 173 patients with stage III GC, there were 87 patients in the high TMB group and 86 patients in the low TMB group, with a 114:59 male-to-female ratio.
As is shown in Figs. 2A–2B, the patients with high TMB had better overall survival (OS) rates than patients with low TMB in both cohorts. The present study also explored the different mutated genes between the high and low TMB groups in the TCGA cohort, and the top 10 mutated genes were identified (P < 0.05, Fig. 2C). Furthermore, the immunohistochemistry analysis of tumour-infiltrating CD8+ T-cells in the study cohort provided evidence supporting the role of anti-tumour immunity in the improved prognosis of patients with GC who have a high TMB (Fig. 2D, please refer to Fig. S1 of Supplementary file 1 for specific quantitative results of CD8+ cells).
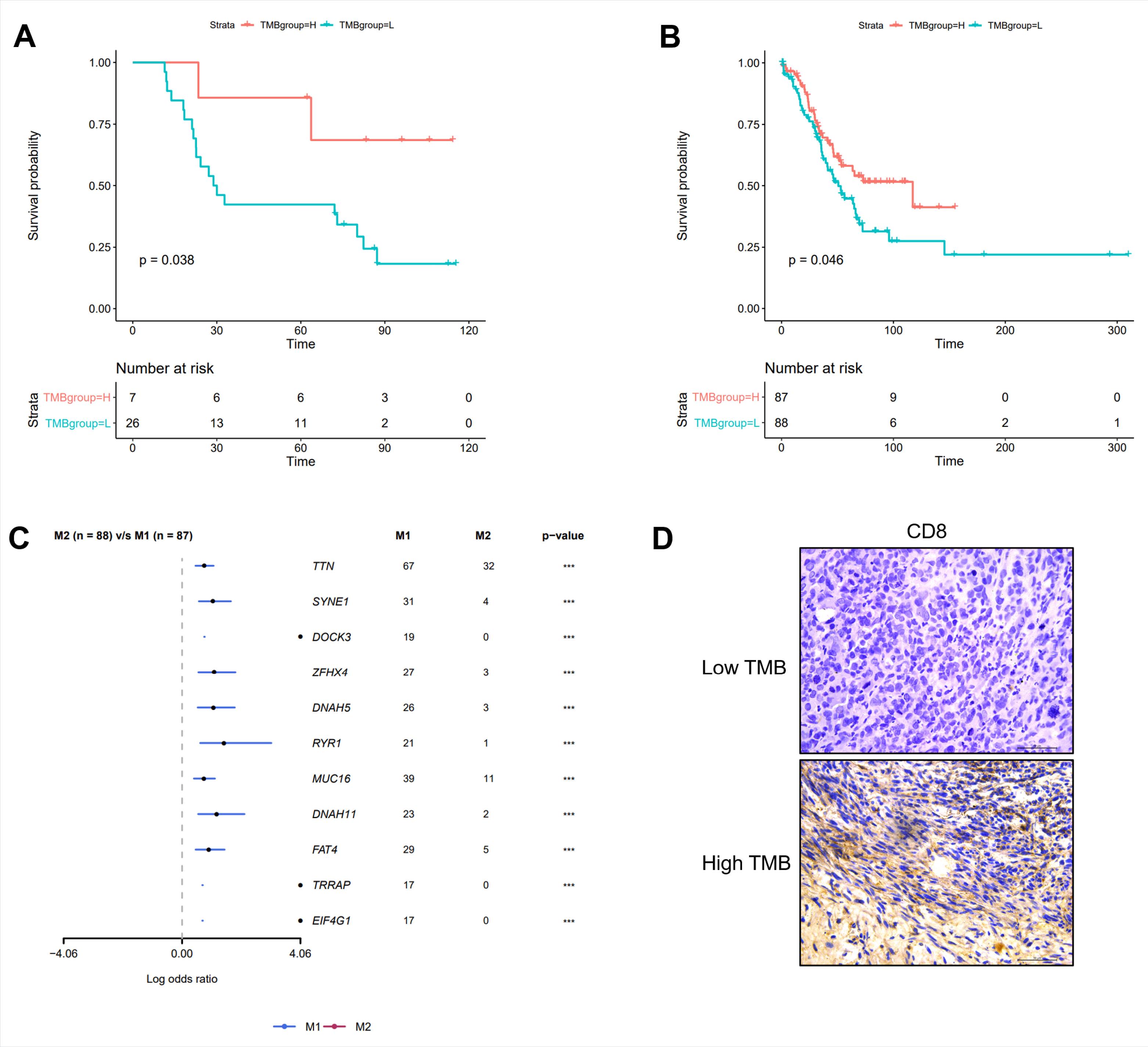
Fig. 2.
TMB is a prognosis biomarker. Patients with high TMB had better survival than those patients with low TMB both in our dataset (A) and TCGA dataset (B). (C) Different mutation genes between low TMB group and high TMB group. (D) Immunohistochemistry analysis of tumor-infiltrating CD8+ T cells in low TMB group and high TMB group. M1 represents the high TMB (Tumor Mutation Burden) group, while M2 represents the low TMB group.
.
TMB is a prognosis biomarker. Patients with high TMB had better survival than those patients with low TMB both in our dataset (A) and TCGA dataset (B). (C) Different mutation genes between low TMB group and high TMB group. (D) Immunohistochemistry analysis of tumor-infiltrating CD8+ T cells in low TMB group and high TMB group. M1 represents the high TMB (Tumor Mutation Burden) group, while M2 represents the low TMB group.
In the TCGA cohort, TMB levels were significantly associated with age (P = 0.008), signature 1 (P < 0.001) and signature 17 (P = 0.045) but not with the T-stage, N-stage, mutation signature 2 or mutation signature 15 (Fig. 3A).
The multivariate Cox regression analysis revealed that age (P = 0.01) but not TMB (P = 0.09) was an independent prognosis factor (Fig. 3B).
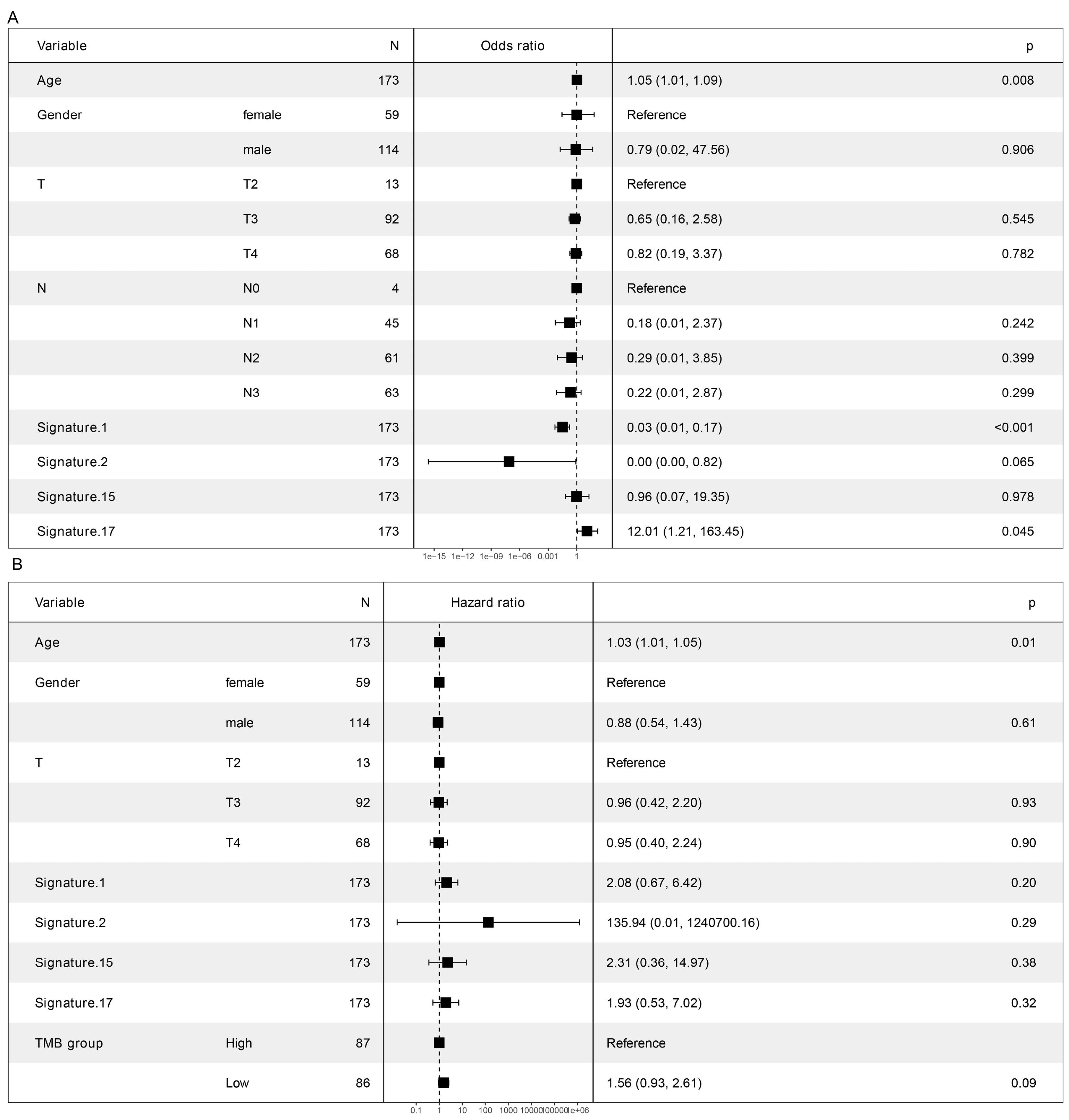
Fig. 3.
Association of clinical factors and TMB. (A) Association of TMB and clinical factors by general liner model. (B) Prognosis related factors in Stage III gastric cancer.
.
Association of clinical factors and TMB. (A) Association of TMB and clinical factors by general liner model. (B) Prognosis related factors in Stage III gastric cancer.
Oncogenic signalling pathway enrichment
For further analysis of the molecular character of stage III GC, the authors of the present study plotted the driver genes, including FERMT2 and WDR55, using Maftools to detect cancer driver genes according to positional clustering21 (Fig. 4A). Several oncogenic signalling pathways were enriched (Fig. 4B), including RTK-RAS, Notch and WNT. A detailed account of the RTK-RAS signalling pathways’ enriched genes is shown in Fig. 4C.
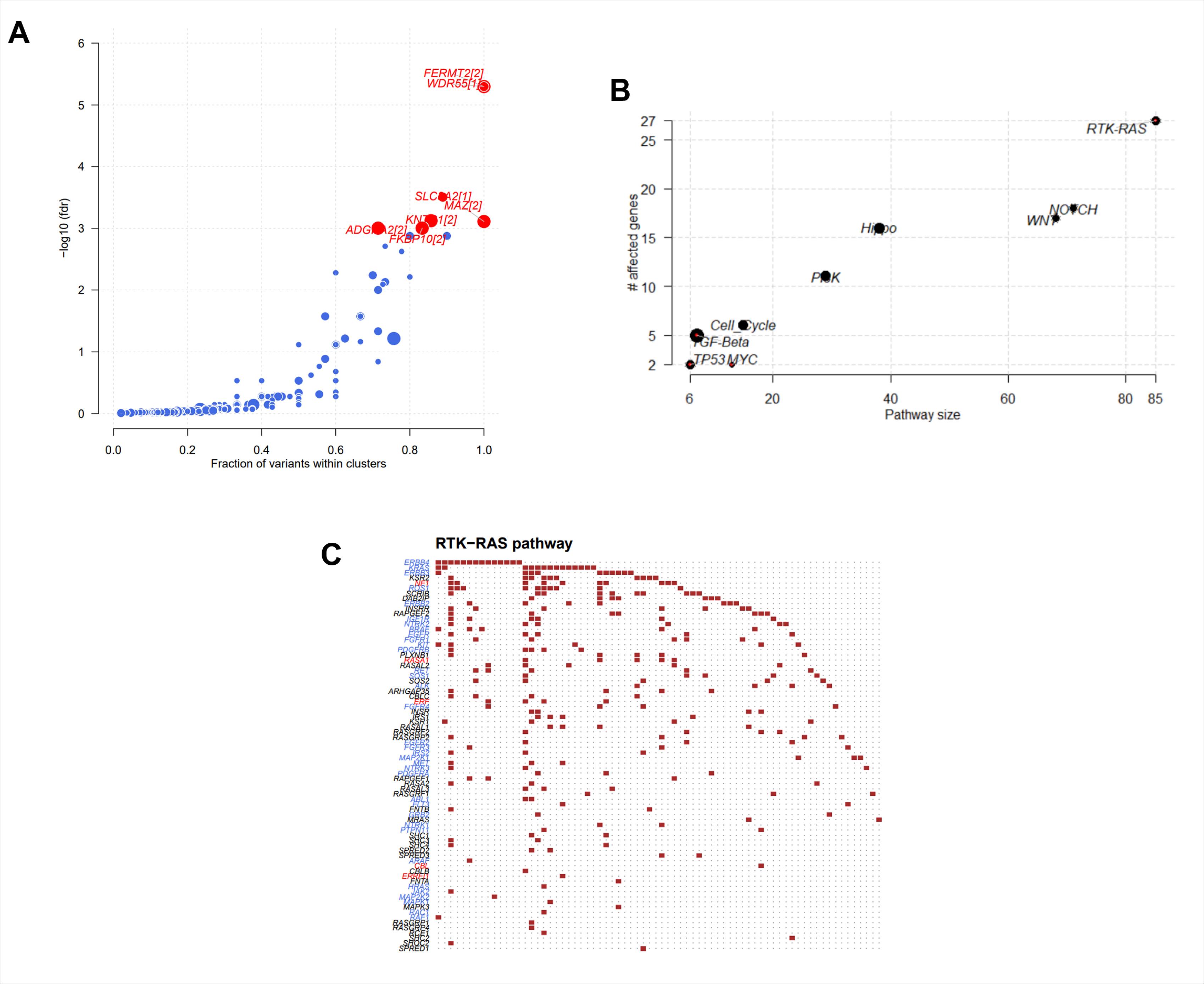
Fig. 4.
Enrichment analysis of tumor signaling pathways in Stage III gastric cancer. A. Scatter plot of driver genes in Stage III gastric cancer. B. The main pathways enriched in Stage III gastric cancer. The presentation reveals 9 signaling pathways predominantly enriched with mutated genes in Stage III gastric cancer. C. Detailed mutation genes in the top enriched oncogenic pathway of RTK-RAS.
.
Enrichment analysis of tumor signaling pathways in Stage III gastric cancer. A. Scatter plot of driver genes in Stage III gastric cancer. B. The main pathways enriched in Stage III gastric cancer. The presentation reveals 9 signaling pathways predominantly enriched with mutated genes in Stage III gastric cancer. C. Detailed mutation genes in the top enriched oncogenic pathway of RTK-RAS.
Growth and migration ability of GC cells in the different groups
The expression of the Notch-1 and RTK-RAS pathway-related proteins was detected using WB. The results demonstrated that the expression of the Notch-1 pathway-related proteins (Notch-1, Hes1 and Hey1) was lower in the high (P1-3) than in the low (P4-6) TMB group; this finding was confirmed via immunohistochemistry (Fig. 5A). Moreover, RTK-RAS pathway activation was also lower in the high compared to the low TMB group (Fig. 5B). The relevant target genes’ expressions of Notch-1 and RTK-RAS pathways were also similar to the expression of the Notch-1 and RTK-RAS pathway-related proteins (Figs. 5C, D). Proteins such as CDKN1A and CDKN1B are involved in the regulation of the cell cycle and may be associated with the dysregulation of cancer cell proliferation and apoptosis.
Both the proliferation rate of GC cells (Fig. 5E) and their migration ability were lower in the high TMB group compared with the low TMB group (Fig. 5F). The above results revealed that GC cell damage was lower in the high than in the low TMB group. Nevertheless, the present data already indicates that interfering with the expression of Notch-1 and the KRAS pathway can affect various phenotypes in TMB-low cells. However, further evidence is required to fully understand the exact mechanisms involved.
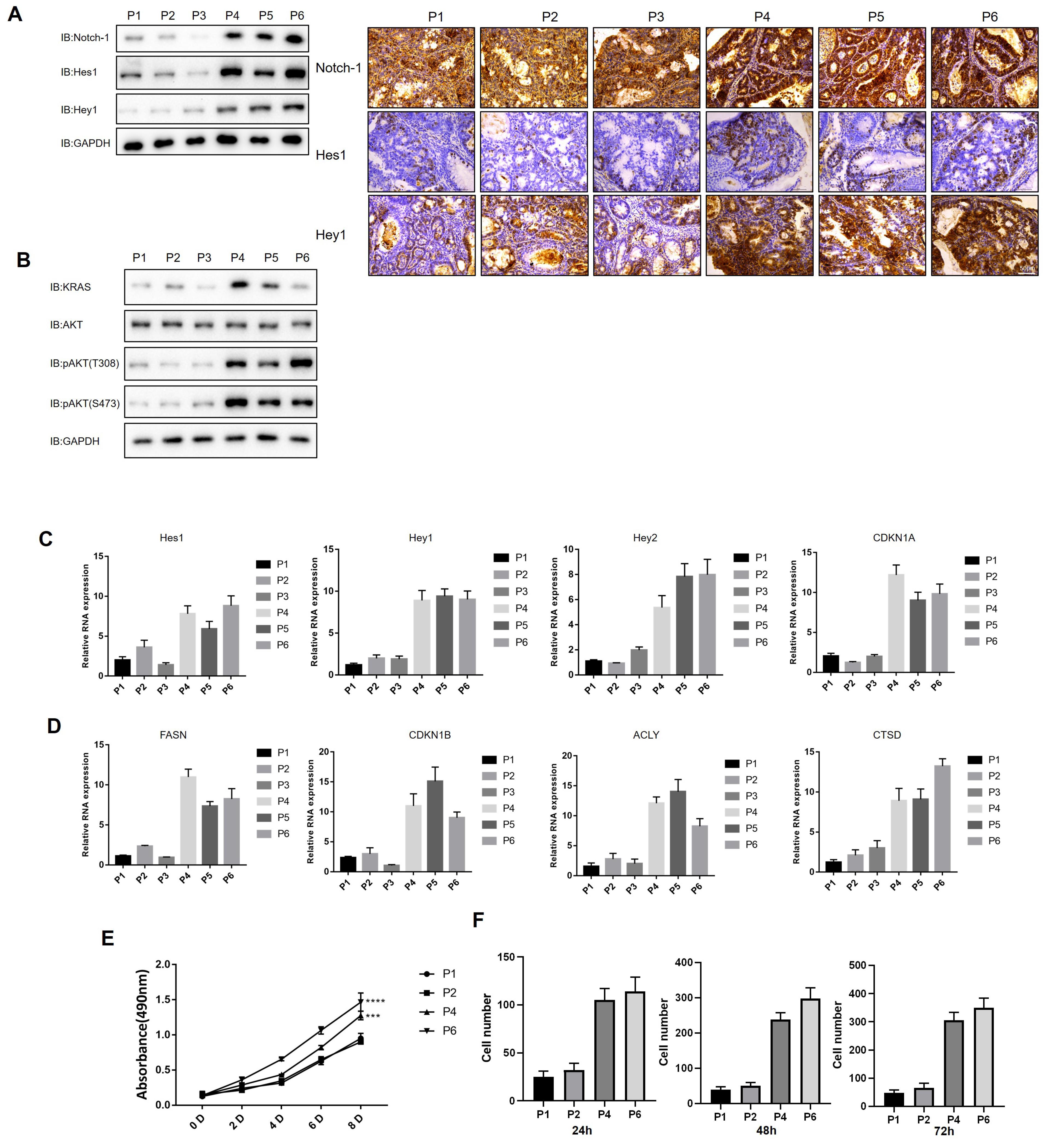
Fig. 5.
Validation of RTK-RAS and Notch-1 pathway in patients with gastric cancer. A. Expression of Notch-1 pathway-related proteins in gastric cancer samples. B. Expression of RTK-RAS pathway-related proteins in gastric cancer samples. C. Expression of Notch-1 pathway-related protein mRNA in gastric cancer samples. CDKN1A: Cyclin-Dependent Kinase Inhibitor 1A; FASN: Fatty Acid Synthase; CDKN1B: Cyclin-Dependent Kinase Inhibitor 1B; ACLY: ATP Citrate Lyase; CTSD: Cathepsin D. D. Expression of RTK-RAS pathway-related protein mRNA in gastric cancer samples. E. Growth curves of different gastric cancer cell lines. F. Transwell results of different gastric cancer cell. All data expressed by mean±SD, by two-way ANOVA analysis, * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
.
Validation of RTK-RAS and Notch-1 pathway in patients with gastric cancer. A. Expression of Notch-1 pathway-related proteins in gastric cancer samples. B. Expression of RTK-RAS pathway-related proteins in gastric cancer samples. C. Expression of Notch-1 pathway-related protein mRNA in gastric cancer samples. CDKN1A: Cyclin-Dependent Kinase Inhibitor 1A; FASN: Fatty Acid Synthase; CDKN1B: Cyclin-Dependent Kinase Inhibitor 1B; ACLY: ATP Citrate Lyase; CTSD: Cathepsin D. D. Expression of RTK-RAS pathway-related protein mRNA in gastric cancer samples. E. Growth curves of different gastric cancer cell lines. F. Transwell results of different gastric cancer cell. All data expressed by mean±SD, by two-way ANOVA analysis, * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Discussion
Stage III GC differs from early-stage disease; in patients in the early stage of GC, emphasis is placed on tumour resection rather than on systemic chemotherapy.22 Given that most cases are diagnosed at a late stage, it is important to identify the detailed molecular characteristics of stage III GC, and TMB is currently being investigated as a GC biomarker.10 In this study, the mutation landscape was plotted both in the study dataset and the TCGA set. The authors explored the clinical significance of TMB in stage III GC. The study demonstrated that TMB could serve as a prognostic biomarker, although it was not an independent prognostic factor.
The limitations of the present analysis are as follows: (1) a small sample size and (2) extreme heterogeneity among patients with GC, especially in stage III of the disease. Although the present study did not discover any ground-breaking rule introduced by the mutation of a single gene, it was found that genes with high-frequency mutations were significantly enriched in pathways, including the RTK-RAS and Notch signalling pathways. Additionally, the results indicated that inhibiting the RTK-RAS signalling pathway can impede proliferation and induce the apoptosis of cancer cells.23 Previous studies confirmed that the inhibition of upstream regulatory molecules of the Notch signalling pathway can achieve the purpose of impeding GC cell invasion, proliferation and migration.24-26 The present results were consistent with existing studies and showed that most of the mutated genes in stage III GC were enriched in these two signalling pathways, thereby underscoring them as the main signalling pathways involved in the evolutionary deterioration of GC. In the high TMB group, Notch and RTK-RAS pathway activation was lower than in the low TMB group (Figs. 5A–5D). The proliferation and migration ability of GC cells were lower in the high than in the low TMB group (Figs. 5E, 5F). The results suggest that no definite single molecular marker with a significant difference is present at the gene level in patients with stage III GC; however, pathway prediction may be more valuable than a single molecular marker.
In addition, TMB was also found to be associated with survival prognosis. The tumour mutation burden is defined as the total number of somatic mutations after the removal of germline mutations from the tumour genome, which refers to the total number of substitutions, insertions, deletions and mutations per megabase occurring in the exonic coding region of genes assessed in tumour tissue.12,27,28 Tumour cells mutate to produce new proteins that are recognised by the autoimmune system as ‘nonself antigens’ and, as a result, activate T-cells and trigger antitumour immune responses. Therefore, the higher the TMB of tumour cells, the more easily they can be recognised by the immune system and the more strongly the antitumour immune response will be, giving T-cells a greater chance to detect and eliminate cancer cells.29,30 Thus, it follows that high TMB can theoretically serve as a biomarker for antitumour immunotherapy. There is increasing evidence that TMB and clinical benefits are correlated and that a higher TMB is associated with a better prognosis.31-33 A study showed that TMB was more effective than PDL1 in predicting the treatment efficacy of toripalimab in advanced GC, in which the OS was significantly higher in the high (≥12 Muts/Mb) than in the low (<12 Muts/Mb) TMB group (14.9 months vs 4.0 months).34 Another study reported that the median OS rates of patients with metastatic breast cancer were significantly different in the low and high TMB groups (44.9 months vs 85.8 months, respectively).35 The results of the present study were consistent with the results of the above studies indicating that patients in the low TMB group had better OS rates than those in the high TMB group. However, the Cox regression showed that TMB was not an independent prognostic factor. According to previous research,36,37 TMB can be used to predict the prognosis of various types of cancer, but its impact on different cancers is controversial.
A meta-analysis37 found that gastric patients in the high TMB group showed significantly longer OS than in the low TMB group, particularly in Asian patients; however in the non-Asian subgroup, the survival benefit was observed to be not statistically significant. Li et al36 found that the combination of TMB and a positive postoperative tumour marker had significantly better predictive value than the traditional predictive marker; furthermore, TMB can provide information for the prognosis of gastric cancer and different TMB cutoffs, indicating TMB as being a prognostic biomarker. Our research findings indicate that the prognostic value of TMB should be evaluated together with different cutoff values and/or other clinical pathological features or biomarkers, which may be superior to evaluating TMB on its own.
Cho et al examined TMB in 330 patients with GC, 11% of which had high TMB (≥10.5 Muts/Mb), patients with the Lauren intestinal type of GC and early TNM were more likely to have high TMB, however, there was no correlation with HER-2, EGFR, FGFR-2, MET and the expressions of other genes.38 At present, there is no unified standard for establishing a TMB threshold. The standards of different detection platforms vary, and factors such as a lack of support from clinical trial data have resulted in TMB not yet being widely employed to guide clinical treatment. It is believed that as TMB continues to gain momentum as a biomarker of immunotherapy response and standardised assays emerge to enable clinical application, guiding therapy using TMB testing will become increasingly important in precision medicine in the future. However, in stage III GC, a highly heterogeneous tumour, a combination of biomarkers may be more effective than any single such marker. Clinical trials using a combination of TMB and other biomarkers should thus be conducted in the future.
In summary, the TMB level and pathway prediction have directional significance in stage III GC, and relevant research can provide important guiding value for clinical treatment. Tumour mutation burden is closely associated with the prognosis of stage III GC, and further prospective multicentre research is needed for future validation of the prognostic value of TMB in GC.
Conclusion
This single-centre cohort study found that patients with stage III GC in the high TMB group had better OS rates than those in the low TMB group, which indicated the need to conduct prospective multicentre research for the future validation of the prognostic value of TMB in GC. This study also redefined the enriched signalling pathways of stage III GC and found that the activation levels of these pathways varied between the high and low TMB groups. These findings provide new directions for the follow-up study of stage III GC.
Research Highlights
What is the current knowledge?
√ The stage III GC displayed different molecular characteristics.
What is new here?
√ TMB could serve as a potential prognosis biomarker for these patients.
Acknowledgement
We would like to express our gratitude to all those who helped us during the writing of this manuscript.
Competing Interests
The authors declare that they have no competing interests in this work.
Ethical Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Chinese PLA General Hospital (ethical batch number: S2020-326-01, date: 2020.09.24). All participants signed an informed consent form for inclusion in the study.
Supplementary files
Supplementary file 1 contains Fig. S1.
(pdf)
References
- Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet 2020; 396:635-48. doi: 10.1016/s0140-6736(20)31288-5 [Crossref] [ Google Scholar]
- Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet 2016; 388:2654-64. doi: 10.1016/s0140-6736(16)30354-3 [Crossref] [ Google Scholar]
- Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl) 2021; 134:783-91. doi: 10.1097/cm9.0000000000001474 [Crossref] [ Google Scholar]
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66:115-32. doi: 10.3322/caac.21338 [Crossref] [ Google Scholar]
- Anonymous Anonymous. Japanese gastric cancer treatment guidelines 2018 (5th edition). Gastric Cancer 2021; 24:1-21. doi: 10.1007/s10120-020-01042-y [Crossref] [ Google Scholar]
- Wang FH, Zhang XT, Li YF, Tang L, Qu XJ, Ying JE. The Chinese Society of Clinical Oncology (CSCO): Clinical guidelines for the diagnosis and treatment of gastric cancer, 2021. Cancer Commun (Lond) 2021; 41:747-95. doi: 10.1002/cac2.12193 [Crossref] [ Google Scholar]
- Sexton RE, Al Hallak MN, Diab M, Azmi AS. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev 2020; 39:1179-203. doi: 10.1007/s10555-020-09925-3 [Crossref] [ Google Scholar]
- Mranda GM, Xue Y, Zhou XG, Yu W, Wei T, Xiang ZP. Revisiting the 8th AJCC system for gastric cancer: A review on validations, nomograms, lymph nodes impact, and proposed modifications. Ann Med Surg (Lond) 2022; 75:103411. doi: 10.1016/j.amsu.2022.103411 [Crossref] [ Google Scholar]
- Ajani JA, D'Amico TA, Bentrem DJ, Chao J, Cooke D, Corvera C. Gastric Cancer, Version 22022, NCCN Clinical Practice Guidelines in Oncology. J Natl ComprCancNetw 2022; 20:167-92. doi: 10.6004/jnccn.2022.0008 [Crossref] [ Google Scholar]
- Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin 2021; 71:264-79. doi: 10.3322/caac.21657 [Crossref] [ Google Scholar]
- Scharpf RB, Balan A. Genomic Landscapes and Hallmarks of Mutant RAS in Human Cancers. Cancer Res 2022; 82:4058-78. doi: 10.1158/0008-5472.can-22-1731 [Crossref] [ Google Scholar]
- Yarchoan M, Hopkins A, Jaffee EM. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl J Med 2017; 377:2500-1. doi: 10.1056/NEJMc1713444 [Crossref] [ Google Scholar]
- Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017; 541:321-30. doi: 10.1038/nature21349 [Crossref] [ Google Scholar]
- Mao W, Ding J, Li Y, Huang R, Wang B. Inhibition of cell survival and invasion by Tanshinone IIA via FTH1: A key therapeutic target and biomarker in head and neck squamous cell carcinoma. Exp Ther Med 2022; 24:521. doi: 10.3892/etm.2022.11449 [Crossref] [ Google Scholar]
- Goldman MJ, Craft B. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 2020; 38:675-8. doi: 10.1038/s41587-020-0546-8 [Crossref] [ Google Scholar]
- Xia B, Gu X, Xu T, Yan M, Huang L, Jiang C. Exosomes-mediated transfer of LINC00691 regulates the formation of CAFs and promotes the progression of gastric cancer. BMC Cancer 2023; 23:928. doi: 10.1186/s12885-023-11373-5 [Crossref] [ Google Scholar]
- Boccellato C, Kolbe E, Peters N, Juric V, Fullstone G. Marizomib sensitizes primary glioma cells to apoptosis induced by a latest-generation TRAIL receptor agonist. Cell Death Dis 2021; 12:647. doi: 10.1038/s41419-021-03927-x [Crossref] [ Google Scholar]
- Zhu M, Wei C, Wang H, Han S, Cai L, Li X. SIRT1 mediated gastric cancer progression under glucose deprivation through the FoxO1-Rab7-autophagy axis. Front Oncol 2023; 13:1175151. doi: 10.3389/fonc.2023.1175151 [Crossref] [ Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402-8. doi: 10.1006/meth.2001.1262 [Crossref] [ Google Scholar]
- Wang W, Zhang T, Zhao W, Xu L, Yang Y, Liao Q. A Single Talent Immunogenic Membrane Antigen and Novel Prognostic Predictor: voltage-dependent anion channel 1 (VDAC1) in Pancreatic Cancer. Sci Rep 2016; 6:33648. doi: 10.1038/srep33648 [Crossref] [ Google Scholar]
- Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics 2013; 29:2238-44. doi: 10.1093/bioinformatics/btt395 [Crossref] [ Google Scholar]
- Bollschweiler E, Berlth F, Baltin C, Mönig S, Hölscher AH. Treatment of early gastric cancer in the Western World. World J Gastroenterol 2014; 20:5672-8. doi: 10.3748/wjg.v20.i19.5672 [Crossref] [ Google Scholar]
- Imperial R, Toor OM, Hussain A, Subramanian J, Masood A. Comprehensive pancancer genomic analysis reveals (RTK)-RAS-RAF-MEK as a key dysregulated pathway in cancer: Its clinical implications. Semin Cancer Biol 2019; 54:14-28. doi: 10.1016/j.semcancer.2017.11.016 [Crossref] [ Google Scholar]
- Hibdon ES, Razumilava N, Keeley TM, Wong G, Solanki S, Shah YM. Notch and mTOR Signaling Pathways Promote Human Gastric Cancer Cell Proliferation. Neoplasia 2019; 21:702-12. doi: 10.1016/j.neo.2019.05.002 [Crossref] [ Google Scholar]
- Cui Y, Li Q, Li W, Wang Y, Lv F, Shi X. NOTCH3 is a Prognostic Factor and Is Correlated With Immune Tolerance in Gastric Cancer. Front Oncol 2020; 10:574937. doi: 10.3389/fonc.2020.574937 [Crossref] [ Google Scholar]
- Ma J, Zhao G, Du J, Li J, Lin G, Zhang J. LncRNA FENDRR Inhibits Gastric Cancer Cell Proliferation and Invasion via the miR-421/SIRT3/Notch-1 Axis. Cancer Manag Res 2021; 13:9175-87. doi: 10.2147/cmar.s329419 [Crossref] [ Google Scholar]
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ. Cancer immunologyMutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015; 348:124-8. doi: 10.1126/science.aaa1348 [Crossref] [ Google Scholar]
- Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015; 350:207-11. doi: 10.1126/science.aad0095 [Crossref] [ Google Scholar]
- Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest 2015; 125:3413-21. doi: 10.1172/jci80008 [Crossref] [ Google Scholar]
- Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015; 160:48-61. doi: 10.1016/j.cell.2014.12.033 [Crossref] [ Google Scholar]
- Li X, Wu WK, Xing R, Wong SH, Liu Y, Fang X. Distinct Subtypes of Gastric Cancer Defined by Molecular Characterization Include Novel Mutational Signatures with Prognostic Capability. Cancer Res 2016; 76:1724-32. doi: 10.1158/0008-5472.can-15-2443 [Crossref] [ Google Scholar]
- Kelly RJ. Immunotherapy for Esophageal and Gastric Cancer. Am Soc Clin Oncol Educ Book 2017; 37:292-300. doi: 10.1200/edbk_175231 [Crossref] [ Google Scholar]
- Wei XL, Xu JY, Wang DS. Baseline lesion number as an efficacy predictive and independent prognostic factor and its joint utility with TMB for PD-1 inhibitor treatment in advanced gastric cancer. Ther Adv Med Oncol 2021; 13:1758835921988996. doi: 10.1177/1758835921988996 [Crossref] [ Google Scholar]
- Wang F, Wei XL, Wang FH, Xu N, Shen L, Dai GH. Safety, efficacy and tumor mutational burden as a biomarker of overall survival benefit in chemo-refractory gastric cancer treated with toripalimab, a PD-1 antibody in phase Ib/II clinical trial NCT02915432. Ann Oncol 2019; 30:1479-86. doi: 10.1093/annonc/mdz197 [Crossref] [ Google Scholar]
- Park SE, Park K, Lee E, Kim JY, Ahn JS, Im YH. Clinical implication of tumor mutational burden in patients with HER2-positive refractory metastatic breast cancer. Oncoimmunology 2018; 7:e1466768. doi: 10.1080/2162402x.2018.1466768 [Crossref] [ Google Scholar]
- Li Z, Jia Y, Zhu H, Xing X, Pang F, Shan F. Tumor mutation burden is correlated with response and prognosis in microsatellite-stable (MSS) gastric cancer patients undergoing neoadjuvant. chemotherapy 2021; 24:1342-54. doi: 10.1007/s10120-021-01207-3 [Crossref] [ Google Scholar]
- Ke L, Li S, Huang D. The predictive value of tumor mutation burden on survival of gastric cancer patients treated with immune checkpoint inhibitors: A systematic review and meta-analysis. Int Immunopharmacol 2023; 124:110986. doi: 10.1016/j.intimp.2023.110986 [Crossref] [ Google Scholar]
- Cho J, Ahn S, Son DS, Kim NK, Lee KW, Kim S. Bridging genomics and phenomics of gastric carcinoma. Int J Cancer 2019; 145:2407-17. doi: 10.1002/ijc.32228 [Crossref] [ Google Scholar]