Bioimpacts. 14(4):30150.
doi: 10.34172/bi.2024.30150
Original Article
Interaction of toll-like receptors and ACE-2 with different variants of SARS-CoV-2: A computational analysis
Azadeh Zahmatkesh Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing – original draft, 1, * 
Elham Salmasi Formal analysis, Resources, Software, Visualization, Writing – review & editing, 2
Reza Gholizadeh Data curation, Formal analysis, Methodology, Resources, Visualization, Writing – review & editing, 3
Author information:
1Department of Anaerobic Bacterial Vaccines Research and Production, Razi Vaccine and Serum Research Institute, Agricultural Research, Education and Extension Organization, Karaj, Iran
2Department of Biomedical Engineering, School of Medicine, Tsinghua University, Beijing, PR China
3Department of Catalysis and Chemical Reaction Engineering, National Institute of Chemistry, Hajdrihova 19, SI-1001 Ljubljana, Slovenia
Abstract
Introduction:
Computational studies were performed to investigate the unknown status of endosomal and cell surface receptors in SARS-CoV-2 infection. The interactions between Toll-like receptors (TLRs)- 4/7/8/9 or ACE2 receptor and different SARS-CoV-2 variants were investigated.
Methods:
The RNA motifs for TLR7, TLR8 and a CpG motif for TLR9 were analyzed in different variants. Molecular docking and molecular dynamics (MD) simulations were performed to investigate receptor-ligand interactions.
Results:
The number of motifs recognized by TLR7/8/9 in the Alpha, Delta and Iranian variants was lower than in the wild type (WT). Docking analysis revealed that the Alpha, Delta and some Iranian spike variants had a higher affinity for ACE2 and TLR4 than the WT, which may account for their higher transmission rate. The MD simulation also showed differences in stability and structure size between the variants and the WT, indicating potential variations in viral load.
Conclusion:
It appears that Alpha and some Iranian isolates are the variants of concern due to their higher transmissibility and rapid spread. The Delta mutant is also a variant of concern, not only because of its closer interaction with ACE2, but also with TLR4. Our results emphasize the importance of ACE2 and TLR4, rather than endosomal TLRs, in mediating the effects of different viral mutations and suggest their potential therapeutic applications.
Keywords: TLR4, TLR7/8/9, Motif, Alpha variant, Delta variant, Transmission
Copyright and License Information
© 2024 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Toll-like receptors (TLRs) are a class of pathogen recognition receptors (PRRs) that detect pathogen-associated molecular patterns (PAMPs) and are essential for the activation of innate immunity and the initiation of adaptive immunity.1 TLRs have been associated with the infection of various respiratory diseases such as influenza, Middle East Respiratory Syndrome (MERS) and severe acute respiratory syndrome (SARS).2-4 However, there is limited information on the association of TLRs with the recent SARS-CoV-2.5,6
Endosomal TLRs (TLR3, 7, 8 and 9) can detect viral nucleic acids and play a crucial role in triggering immune responses against viruses.7 TLR3 binds double-stranded RNA (dsRNA) and TLR7/8 bind single-stranded RNA (ssRNA) of the viral genome.8 TLR9 is an intracellular TLR that recognizes bacterial and viral DNA molecules. TLR9 detects CpG signaling motifs (GTCGTT) in bacterial and viral DNA.9,10 A study has shown that TLR7 has specific ssRNA sequence preferences. It has complete or moderate affinity for binding consecutive uridine-containing ssRNAs and low affinity for single uridine-containing ssRNAs.11 Although TLR9 is known to detect DNA, it is frequently expressed in peripheral blood mononuclear cells (PBMCs) after infection with SARS-CoV, a single-stranded RNA, which may be through viral CpG motifs. The viral genomic sequence of SARS-CoV has a higher number of TLR9 signaling motifs than some other coronaviruses or respiratory viruses.12 On the other hand, TLR4 recognizes lipopolysaccharide (LPS), the cell wall component of bacteria that can induce cellular responses.13 In an in silico study, a strong association was found between SARS‐CoV and SARS‐CoV‐2 spike protein and its receptor ACE2 as well as TLR4.14
One of the first mutations in SARS-CoV-2 detected in COVID-19 patients in the United Kingdom (2020/12/01) is known as lineage B.1.1.7 or Alpha variant.15 Another mutation that has emerged in India is lineage B.1.617 or the Delta variant. Initial studies indicated that the Alpha and Delta variants are associated with a higher viral load and faster transmission.16,17 Little is known about the interaction between these variants and host receptors. In the present research, we will conduct an in silico study on the interaction of the Wuhan reference sequence, Alpha, Delta and some Iranian sequences of SARS-CoV-2 with the corresponding TLRs or ACE2 and make a comparison to find out whether different mutations are related to the changes in binding affinity to these receptors. This research will shed light on the unexplored status of endosomal and cell-surface receptors in viral infection and make an important contribution to focusing on a specific receptor and finding future potential therapeutics.
Materials and Methods
Genomic sequences and 3D protein structures
The genomic reference sequence of Wuhan SARS-CoV-2 was obtained from the NCBI database (accession number: NC_045512.2). The genomic sequence of SARS-CoV-2 Alpha (UK lineage B.1.1.7) (GISAID: EPI_ISL_1275784) and Delta (lineage B.1.617.2) (EPI_ISL_3221054) variants were retrieved from the GISAID database.18 The PDB files of Wuhan SARS-CoV-2 spike protein (PDB ID: 6M0J), Alpha spike protein (7LWU), human ACE2 (PDB ID: 1R42) and TLR4 (PDB ID: 2Z63) were retrieved from the RCSB protein database (www.rcsb.org). The mutant spike proteins of the Delta sequence and some of the Iran sequences were modeled using the SWISS-MODEL server.
A total of 115 protein sequences for the spike protein of Iranian SARS-CoV-2 variants belonging to different collection dates were retrieved from the NCBI database together with the spike protein sequence of Wuhan (wild type: WT) for multiple sequence alignment. The genomic spike sequences of the Alpha and Delta variants were translated into the protein sequences.
TLR7/8/9 interactions with genomic RNA
The Sequence Searcher software (https://4virology.net/virology-ca-tools/sequence-searcher/) was used to search single-stranded RNA motifs6,19 for TLR7 and TLR8, and a CpG motif for TLR920 in the entire genomic sequences of SARS-CoV-2.
Multiple sequence alignment
The spike protein sequences of the WT, Alpha, Delta and Iranian isolates were aligned using ClustalW and BioEdit software and the most and least variable sequences were determined. In all sequences, mutations in residue 614 and the receptor binding domain (RBD) involved in spike ACE2 receptor binding were analyzed according to Yi et al,21 and Yurkovetskiy et al, 2020.22 Then, the most variable sequence and the sequences with the RBD mutations were further analyzed for docking analysis.
TLR4 and ACE2 interactions with mutated spike proteins
To investigate and compare the binding affinity of different variants of the SARS-CoV-2 spike protein (WT, Alpha, Delta and Iranian isolates with the RBD mutations) and its receptors in the host, the 3D crystal structures of human TLR4 and ACE2 as well as the spike RBDs of WT and Alpha SARS-CoV-2 were retrieved from the RCSB Protein Data Bank. The 3D structure of the RBD molecules of the other variants was modeled using SWISS-MODEL. Co-crystallized ligands and water molecules were removed from the structures using ViewerLite software (version 4.2). For protein-protein docking, the web servers HDOCK (http://hdock.phys.hust.edu.cn/) and Patchdock/Firedock (https://bioinfo3d.cs.tau.ac.il/PatchDock/php.php) were used for RBD-ACE2 and RBD-TLR4, respectively.
The resulting docked structures were then analyzed by Molecular Dynamics (MD) simulations on the bound state (complex). The MD simulations were performed using the Forcite module of Biovia's Materials Studio package on a high-performance computing (HPC) cluster. The final systems with about 7419 atoms were first minimized for 10 ps and then equilibrated for 1000 ps in the NPT ensemble (P=10‒4 GPa bar and T=250 K). For proteins, the universal force field was considered.23 For water and ions, the TIP3P model24,25 and the standard force field26 were used, respectively. The periodic boundary conditions were applied for all three dimensions. The long-range Coulomb interactions were calculated using particle-mesh Ewald full electrostatics with a grid size of about 1 Å in each dimension. A smooth (10–12 Å) cut-off was used to calculate the van der Waals energies between atoms. The temperature T was kept at 250 K by applying the Nose method27 and the pressure was kept constant at 10‒4 GPa using the Andersen method.28 The simulation time step was set to 1 fs for bonded and non-bonded interactions (van der Waals, improper, dihedral, and angle) using the SETTLE algorithm29 with all bonds held rigid. Electrical interactions were also calculated every 4 fs using the algorithms with multiple time steps.30 After performing 1000 ps of molecular dynamics simulations, the mean square deviation (MSD) of the trajectory was calculated to check whether the systems had reached stability. In addition, the radius of gyration was analyzed to determine the compactness of the mutant structures compared to WT.
Accession numbers
Wuhan SARS-CoV-2: (GenBank: NC_045512.2); SARS-CoV-2 Alpha: (GISAID: EPI_ISL_1275784); Delta: (GISAID: EPI_ISL_3221054).
Results
Motif analysis
The data analysis showed that the total number of genomic sequence motifs detected by TLR7/8/9 in the SARS-CoV-2 Alpha and Delta variants and the Iranian sequences was lower than in the SARS-CoV-2 WT sequence (Table 1). The detectable motifs in the Iranian sequences showed different variability compared to the reference sequence (30 to 54 numbers lower for MT994849.1 and MT889692.1, respectively).
Table 1.
Comparison of SARS-CoV-2 sequences based on the number of TLR motifs
|
|
WT (Wuhan reference sequence NC_045512.2)
|
Alpha
|
Delta
|
Iran SARS-CoV-2 Sequences
|
|
MT994286.1
|
MT994881.1
|
MT994849.1
|
MT994632.1
|
MT889692.1
|
MT447177.1
|
MT320891.2
|
MT240479.1
|
|
No. of nucleotides
|
29903 |
29782 |
29823 |
29794 |
29822 |
29819 |
29822 |
29800 |
29793 |
29822 |
29836 |
|
Collection date
|
2019-12 |
2020-12-01 |
2021-06-30 |
2020-05-01 |
2020-05-01 |
2020-05-01 |
2020-05-01 |
2020-05-01 |
2020-03-26 |
2020-03-09 |
2020-03-04 |
|
TLR7 motifs
|
|
|
|
|
|
|
|
|
|
|
|
| UUU |
654 |
642 |
643 |
641 |
643 |
642 |
641 |
639 |
642 |
642 |
642 |
| (UUU)2 |
7 |
2 |
2 |
2 |
3 |
2 |
2 |
2 |
2 |
2 |
2 |
| UUC |
535 |
534 |
536 |
534 |
535 |
535 |
533 |
532 |
534 |
536 |
535 |
| (UUC)2 |
23 |
23 |
23 |
23 |
23 |
23 |
22 |
22 |
23 |
23 |
23 |
| (UUC)3 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
| UUG |
703 |
697 |
701 |
699 |
699 |
700 |
702 |
698 |
699 |
699 |
701 |
| (UUG)2 |
28 |
28 |
28 |
28 |
28 |
28 |
28 |
28 |
28 |
28 |
28 |
| (UUG)3 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
| UUA |
719 |
720 |
718 |
716 |
717 |
718 |
719 |
719 |
716 |
717 |
717 |
| (UUA)2 |
20 |
20 |
20 |
20 |
20 |
20 |
20 |
21 |
20 |
20 |
20 |
| (UUA)3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
TLR8 motifs
|
|
|
|
|
|
|
|
|
|
|
|
| UG |
2084 |
2074 |
2075 |
2078 |
2079 |
2079 |
2080 |
2077 |
2078 |
2078 |
2081 |
| (UG)2 |
148 |
147 |
147 |
148 |
148 |
148 |
148 |
147 |
148 |
148 |
148 |
| (UG)3 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
| (UG)4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| (UG)5 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
TLR7/8 motifs
|
|
|
|
|
|
|
|
|
|
|
|
| GU |
2023 |
2016 |
2011 |
2015 |
2018 |
2019 |
2016 |
2015 |
2016 |
2017 |
2018 |
| UUGU |
276 |
273 |
273 |
274 |
274 |
276 |
275 |
273 |
274 |
274 |
275 |
| GUUC |
95 |
95 |
96 |
93 |
96 |
96 |
98 |
95 |
94 |
96 |
95 |
| GUUU |
191 |
191 |
189 |
189 |
191 |
190 |
188 |
189 |
190 |
190 |
190 |
| UUUC |
166 |
166 |
166 |
167 |
167 |
167 |
165 |
164 |
167 |
167 |
167 |
| UGUU |
248 |
246 |
247 |
247 |
247 |
248 |
248 |
245 |
247 |
247 |
247 |
| UCUC |
77 |
76 |
77 |
77 |
76 |
76 |
77 |
76 |
76 |
77 |
77 |
| AUGU |
195 |
194 |
195 |
195 |
195 |
196 |
195 |
194 |
195 |
195 |
195 |
| UAUA |
130 |
130 |
130 |
129 |
130 |
128 |
129 |
129 |
129 |
130 |
129 |
| AUAU |
103 |
105 |
104 |
103 |
103 |
102 |
103 |
104 |
103 |
103 |
103 |
| AUAC |
94 |
96 |
95 |
94 |
93 |
94 |
94 |
94 |
94 |
93 |
93 |
| UAUU |
139 |
139 |
139 |
140 |
140 |
141 |
140 |
139 |
139 |
140 |
140 |
| UUAU |
155 |
156 |
156 |
155 |
155 |
157 |
155 |
156 |
155 |
155 |
155 |
| CUAC |
116 |
113 |
113 |
114 |
115 |
115 |
114 |
116 |
115 |
115 |
115 |
| GUAC |
131 |
130 |
130 |
131 |
131 |
130 |
131 |
132 |
131 |
131 |
131 |
| UAUC |
81 |
81 |
81 |
81 |
81 |
81 |
81 |
81 |
81 |
81 |
81 |
|
TLR9 motif
|
|
|
|
|
|
|
|
|
|
|
|
| GTCGTT |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
|
Total
|
9153 |
9106 |
9107 |
9105 |
9119 |
9123 |
9116 |
9099 |
9108 |
9116 |
9120 |
Multiple sequence alignment
Alignment of the Alpha, Delta and the Iranian spike protein sequences showed a variable number of mutations compared to the WT spike sequence, regardless of the time of their collection. The most variable sequence was Iran MW040527 (QOC89639.1) (collection date: 2020-04-22), with 41 mutations in the spike protein, none of which were compatible with the mutations in the Alpha spike sequence. The least variable sequence belonged to Iran MW039533 (QOC77761.1) (collection date: 2020-03-20), which had no mutation. The D614G mutation was detected in 41 of 115 Iran spike sequences, including the most variable sequence of MW040527 (Table 2). Other mutations in RBD-related positions were detected in eight sequences, half of which had different amino acid substitutions (at the same locus) than the previously analyzed amino acid changes (Table 2).
Table 2.
Comparison of the different amino acid substitutions in SARS-CoV-2 variants and the previously investigated amino acid changes
|
RBD mutations in previous studies
|
Observed mutations in similar positions
|
Iran spike protein sequences
(Gene accession number)
|
| P499T |
P499T |
MW090849 (QOJ75919.1) |
| Q493N |
Q493P |
MW055425 (QOE84221.1) |
| F486L |
F486L |
MW055425 (QOE84221.1)
MW090920 (QOJ75940.1) |
|
|
N501Y |
Alpha |
| N501T |
N501K
N501R |
MW055425 (QOE84221.1)
MW093140 (QOJ86685.1) |
| Q498Y |
Q498P
Q498Y |
MW045452 (QOD59279.1)
MW090849 (QOJ75919.1) |
| T470N |
T470N |
MW045459 (QOD59282.1) |
|
|
L450R |
Delta |
| L452K |
L452P |
MW136446 (QOL79333.1) |
| D614G |
D614G |
MW045462, MW040527, MW040525, MW040523, MW040514, MW040511, MW055435, MW055256, MW055255, MW045471, MW045470, MW090877, MW090872, MW090871, MW090866, MW090867, MW090863, MW090854, MW090851, MW063481, MW090921, MW090920, MW090904, MW090900, MW090879, MW136261, MW136260, MW135333, MW165496, MW165494, MW165491, MW136352, MW136351, MW136350, MW136262, MW548636, MW548609, MW548595, MW548639, MW548638, MW548637 |
Molecular docking
Fig. 1a and 1b show the docked structures for WT RBD-TLR4 and WT RBD-ACE2, respectively. The docking results of the human ACE2 receptor and the mutant spike RBDs are shown in Table 3. The Alpha variant showed the highest affinity with ACE2 (with RBD mutation of N501Y), followed by MW090849 (with RBD mutations of Q498Y, P499T), Delta (with RBD mutations of L450R and T476K), MW045459 (with RBD mutations of I402N, N448K, R457S, F464L, D467K, S469Q, T470N and I472N), MW136446 (with RBD mutation of L452P), WT and MW040527 (with non-RBD mutation of D614G), MW093140 (with the RBD mutations of P463L, T500I, N501R, R509K, V511E, S514P, H519I, P521Q, A522P, T523P and P527T), MW045452 (with the RBD mutations of N487K and Q498P), MW090920 (with the RBD mutations of N440H and F486L and the non-RBD mutation of D614G) and MW055425 (with the RBD mutations of F486L, N487I, Q493P, N501K and V510E), respectively. The Alpha variant had a higher docking score than the Delta variant for ACE2 binding, and both had higher scores than WT.
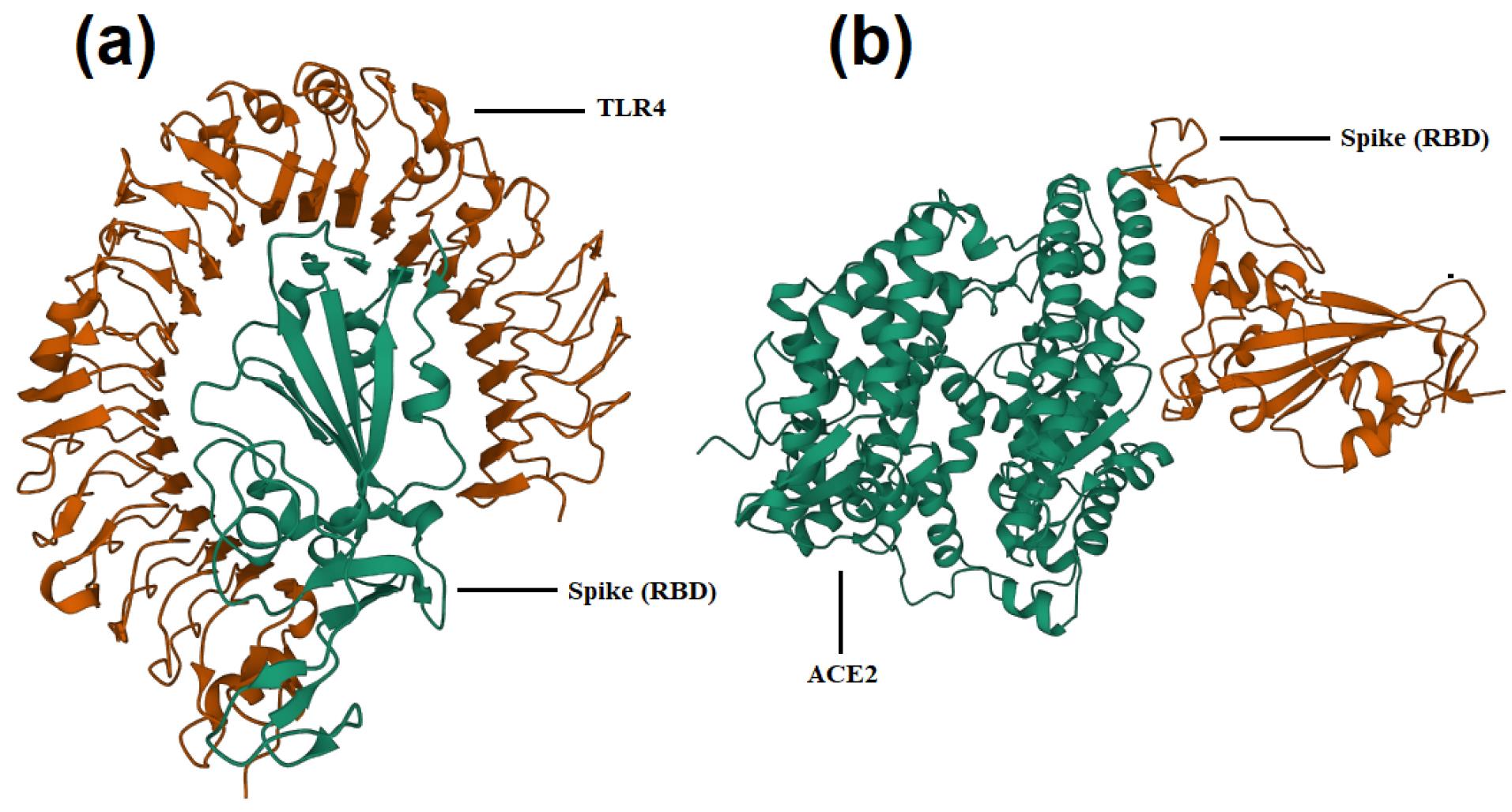
Fig. 1.
Interaction of WT spike RBD and TLR4 (a) or ACE2 (b) according to docking analysis
.
Interaction of WT spike RBD and TLR4 (a) or ACE2 (b) according to docking analysis
Table 3.
Docking score for ACE2-Spike and docking global energy for TLR4-Spike
|
Variants/Isolates
|
ACE2
|
TLR4
|
|
Score
|
RMSD
|
Global Energy
|
RMSD
|
| Wild type |
-345.14 |
0.60 |
-34.13 |
2 |
| Alpha |
-362.53 |
0.36 |
-43.24 |
2 |
| Delta |
-352.16 |
0.46 |
-36.78 |
2 |
| MW045452 |
-336.78 |
0.47 |
-49.68 |
2 |
| MW055425 |
-324.12 |
0.94 |
-46.23 |
2 |
| MW136446 |
-347.03 |
0.45 |
-30.4 |
2 |
| MW090849 |
-361.06 |
0.37 |
-42.37 |
2 |
| MW040527 |
-345.14 |
0.60 |
-48.63 |
2 |
| MW045459 |
-348.80 |
0.64 |
-49.76 |
2 |
| MW093140 |
-343.72 |
0.46 |
-40.51 |
2 |
| MW090920 |
-325.54 |
0.48 |
-27.7 |
2 |
Docking analysis of human TLR4 and mutated spike RBDs showed a higher affinity for TLR4 and all spike variants except MW090920 and MW136446, compared to the WT spike (Table 3). According to global energies, the highest affinity for TLR4 belongs to MW045459 and MW045452, followed by MW040527, MW055425, Alpha variant, MW090849, MW093140, Delta variant, WT, MW136446 and MW090920, respectively. The Alpha variant had a higher global energy than the Delta variant for binding to TLR4 and both had a higher energy than WT.
Molecular dynamics simulation
In MD simulation studies, we analyzed the mean square deviation (MSD) and radius of gyration (Rg) data for all receptor-ligand complex structures. The dynamic behavior of the mutants showed different profiles compared to WT. The results of MSD and Rg versus time for all ACE2 spike structures are shown in the plots in Fig. 2a and b, respectively. The A plot shows that the stability of the ACE2-Delta, -Alpha, -MW055425, -MW093140 and -MW090849 structures was lower than the ACE2-WT structure due to the higher MSD. Other structures showed higher stability compared to the WT structure. The average MSD values for all complexes were as follows; WT: 375.933 Å2, Alpha: 379.170 Å2, Delta: 430.581 Å2, MW055425: 379.206 Å2, MW090849: 395.596 Å2, MW090920: 346.941 Å2, MW040527: 370.113 Å2, MW045452: 365.858 Å2, MW045459: 354.233 Å2, MW093140: 385.805 Å2, MW136446: 368.681 Å2. The order of the MSD values was MW090920 < MW045459 < MW045452 < MW136446 < MW040527 < WT < Alpha < MW055425 < MW093140 < MW090849 < Delta.
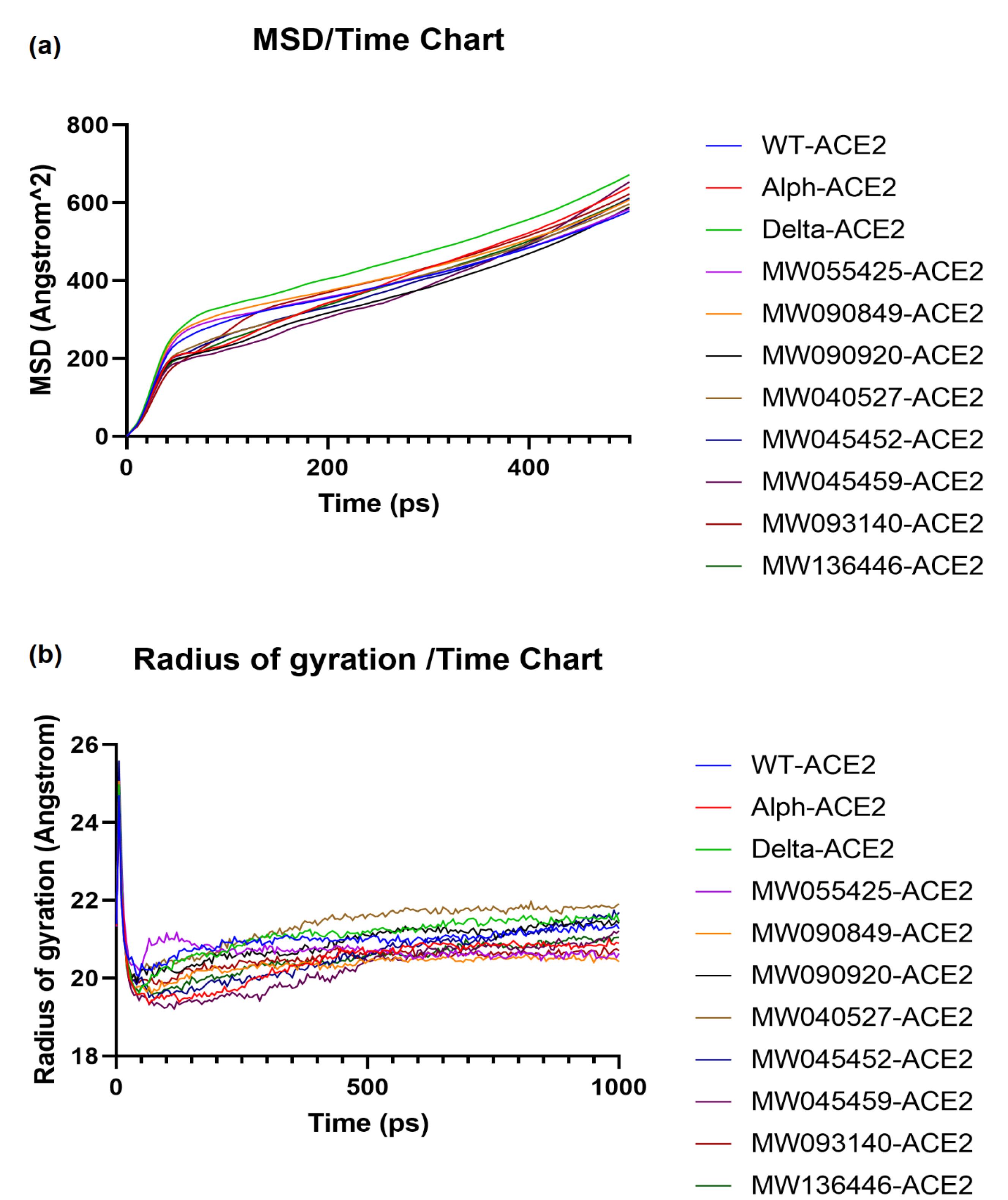
Fig. 2.
Molecular dynamics simulation study of the mutations of spike RBD-ACE2 complex on different parameters for different variants, (a) MSD (b) Rg score.
.
Molecular dynamics simulation study of the mutations of spike RBD-ACE2 complex on different parameters for different variants, (a) MSD (b) Rg score.
By quantifying compactness using Rg measurements in the complexes, we were able to relate the structural differences caused by mutations in the SARS-CoV-2 variants to the experimentally observed variations in binding affinity and stability parameters. The results of Rg versus time (B plot) showed that after ~ 450 ps almost all structures had reached a stable conformation. The Rg values for ACE2-Delta and ACE2-MW040527 were slightly lower than the WT values before 250 ps, but then increased and were higher than the WT values, indicating a slightly lower compactness. The Rg value for ACE2-MW055425 was slightly higher than that of the WT for the first 200 ps, but then gradually decreased to a lower level. Other structures, including ACE2–Alpha, showed relatively higher compactness than the WT. The average Rg values for all complexes were as follows; WT: 20.997 Å, Alpha: 20.459 Å, Delta: 21.125 Å, MW055425: 20.722 Å, MW090849: 20.389 Å, MW090920: 20.953 Å, MW040527: 21.381 Å, MW045452: 20.634 Å, MW045459: 20.290 Å, MW093140: 20.531 Å, MW136446: 20.529 Å. The order of Rg values was MW045459 < MW090849 < Alpha < MW093140 < MW136446 < MW045452 < MW055425 < MW090920 < WT < Delta < MW040527. Since only minor deviations were observed in mutated complexes compared to the WT complex, it appears that the structures were kept compact and stable despite the mutations.
Fig. 3a shows that all structures had an almost higher MSD compared to the WT structure, except TLR4-MW136446 and TLR4-Delta, which had a lower value and showed an increase after 350 ps. The average MSD values for all complexes were as follows; WT: 237.150 Å2, Alpha: 263.201 Å2, Delta: 225.129 Å2, MW040527: 251.516 Å2, MW045452: 250.288 Å2, MW045459: 272.140 Å2, MW055425: 264.102 Å2, MW090849: 321.587 Å2, MW090920: 280.955 Å2, MW093140: 318.752 Å2, MW136446: 203.840 Å2. The MSD order was MW136446 < Delta < WT < MW045452 < MW040527 < Alpha < MW055425 < MW045459 < MW090920 < MW093140 < MW090849.
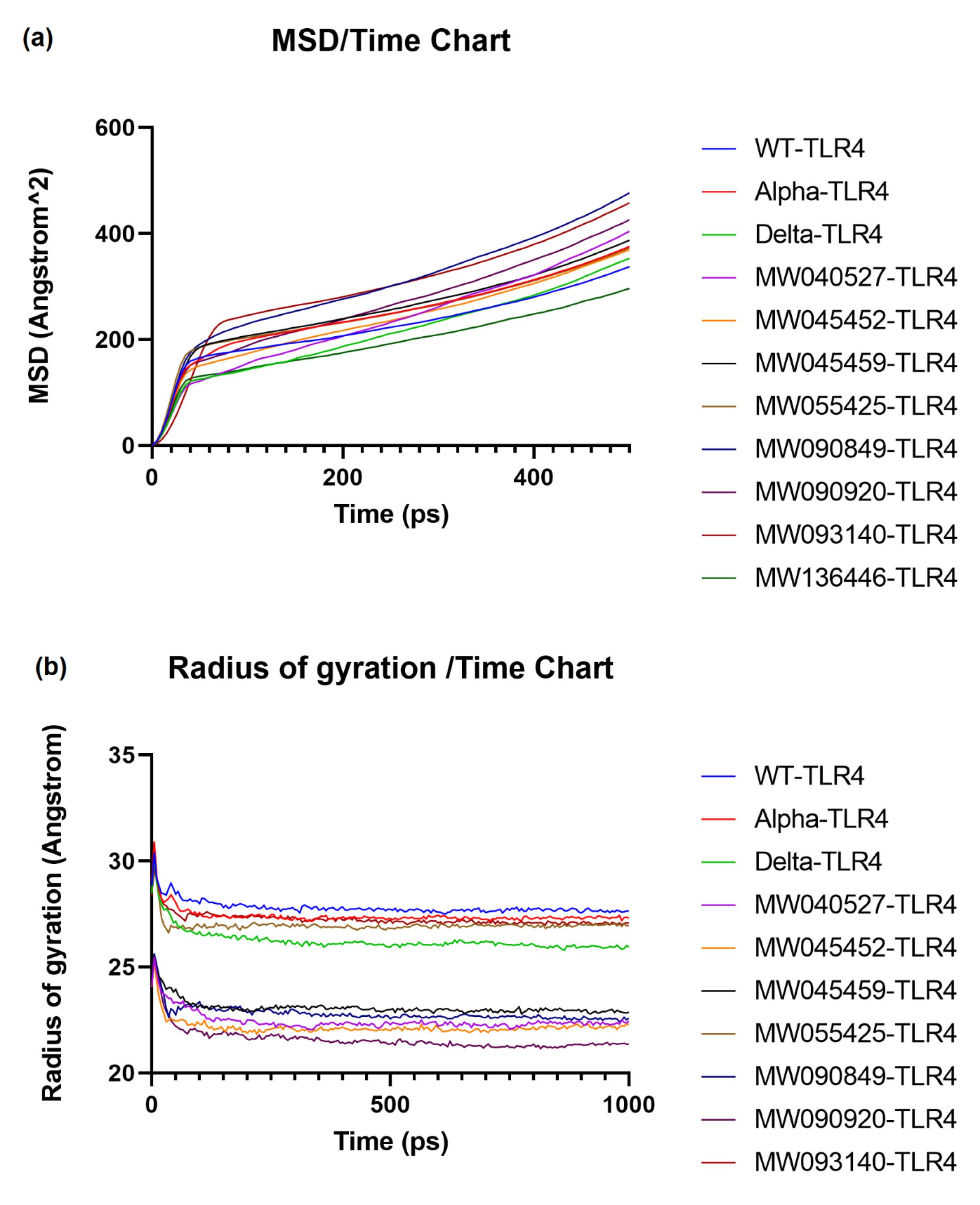
Fig. 3.
Molecular dynamics simulation study of the mutations of spike RBD-TLR4 complex on different parameters for different variants, (a) MSD (b) Rg score.
.
Molecular dynamics simulation study of the mutations of spike RBD-TLR4 complex on different parameters for different variants, (a) MSD (b) Rg score.
The Rg score of all protein structures was lower than that of WT, indicating more compact complexes (Fig. 3b). The most compact systems were TLR4-MW045459, TLR4-MW090849, TLR4-MW040527, TLR4-MW045452 and TLR4-MW090920. The average Rg values for all complexes are as follows; WT: 27.771 Å, Alpha: 27.355 Å, Delta: 26.167 Å, MW055425: 26.934 Å, MW090849: 22.747 Å, MW090920: 21.503 Å, MW040527: 22.386 Å, MW045452: 22.108 Å, MW045459: 23.027 Å, MW093140: 27.207 Å, MW136446: Å. The order of the Rg values were MW090920 < MW045452 < MW040527 < MW090849 < MW045459 < Delta < MW055425 < MW093140 <Alpha < WT.
Discussion
During the COVID-19 pandemic, TLRs have attracted the attention of researchers as their agonists or antagonists have been associated to vaccines or drug strategies against the virus.31 TLR7 is predominantly expressed in human plasmacytoid dendritic cells (DC) and B lymphocytes, and TLR8 is expressed in monocytes and neutrophils.32 Cells expressing TLR733 and TLR834,35 have been shown to be particularly abundant in lung tissue. Several studies have shown the stimulation of TLR7/8 by various viruses. Influenza virus can induce TLR7-positive plasmacytoid DC to produce high levels of type I interferon.36 GU-rich ssRNA oligonucleotides derived from human immunodeficiency virus-1 (HIV-1) can be recognized by murine TLR7 and human TLR8.34 Adverse side effects of TLR7 and/or 8 on immune responses have also been reported. There is some evidence of a cytokine storm leading to upregulation of inflammatory cytokines such as IL-6 and TNF-α in respiratory infections or diseases such as SARS-CoV-1,37 SARS-CoV-2,38 and asthma.39
The lower number of genomic sequence motifs detected by TLR7/8/9 in the Alpha and Delta variants compared to the WT sequence suggests that these motifs may not play a critical role in the detection rate by host TLR7/8/9 and the higher transmission ability of these variants. The lower number of motifs in Iranian isolates compared to the WT sequence suggests that the genomic sequences of Iranian variants may have a lower binding affinity to endosomal TLRs. An immunoinformatic study on the SARS-CoV-2 genome showed that it had more ssRNA motif sequences that could be recognized by TLR7/8 than the SARS-CoV genome, suggesting a greater possibility of interaction with TLR7/8. This has been linked to the potential ability of SARS-CoV-2 to stimulate proinflammatory responses via TLR7/8, leading to severe lung injury and death.6
The sequences of UUU, GU and UG were the most variable motifs compared to the WT sequence. The number of all these motifs was lower in Alpha, Delta and all Iranian isolates compared to the Wuhan sequence. GU-rich oligoribonucleic acids (ORNs containing at least two Us in combination with G or C) were found to stimulate signaling in TLR7/8-expressing cells and induce high IFN-α and TNF-α production. In contrast, AU-rich sequences (ORNs containing A in combination with U) induce the highest TNF-α production in TLR8-expressing cells, but no IFN-α induction.19 The lower number of GU-rich motifs (1042 vs. 1053) in the Iranian isolate MT889692.1 may be associated with lower proinflammatory cytokine production, although there is no information on the immunologic differences of this isolate versus WT. Although a lower number of GU motifs was detected in the Alpha and Delta variants, the number of GU-rich sequences was not significantly different from WT (1047 and 1048 vs. 1053, respectively). No significant difference was found in the number of AU-rich motifs between the studied sequences and the WT sequence.
Mutations in the SARS-CoV-2 spike protein that affect its binding activity to hACE2 have been investigated. Two types of amino acid substitutions were identified; one type included P499T, Q493N, F486L, A475P, and L455Y, which reduced receptor binding, and the other included N501T, Q498Y, E484P, T470N, L452K, and N439R, which enhanced receptor binding activity.21 In our study with Iranian isolates, mutations were detected at most of the above positions (with the exception of A475P, L455Y, E484P and N439R). The Alpha variant had the N501Y mutation, which, as expected, resulted in a higher affinity for hACE2 than the WT variant. The Alpha variant is reported to be susceptible to neutralization by antibodies and is not a major problem for the available vaccines.40 The Delta variant had the mutations of L450R (corresponding to WT amino acid 452) and T476K (corresponding to WT amino acid 478) in the RBD region and had a higher docking score compared to the WT. The L452R mutation in the spike protein of the Delta variant may be responsible for its higher transmissibility than the Alpha variant.41 The Delta variant has been described as less susceptible to neutralizing antibodies mobilized by vaccines than other SARS-CoV-2 variants of concern.42 These capabilities have been attributed to the structural variations due to the L452R and T478K mutations in the RBD.43,44 Higher docking scores show that Alpha and Delta variants can enter the host cell more efficiently than the WT.
The MW090849 isolate showed both reducing (P499T) and enhancing (Q498Y) mutations and had a higher docking score than the WT sequence. However, the Q498Y + P499T mutations resulted in non-lethal virulence in BALB/c mice.45 The MW055425 isolate carried one reducing mutation (F486L) and two mutations at residues 493 (reducing) and 501 (enhancing), which differed in the nature of the amino acid changes from the previously studied mutations. Together, these mutations resulted in a lower binding affinity to ACE2 compared to the WT spike sequence. In a study of an RBD generated with both the Q493N and F486L mutations, weaker binding to ACE2 than the WT protein and an RBD with Q493N was reported.46 For MW090920, the docking score was lower than that of the WT isolate. This result appears to be due to the reducing mutation of F486L, although the D614G mutation was present. The MW093140 isolate had a mutation at residue 501, but with a different amino acid substitution than in the previous studies (N501R versus the enhancing mutation N501Y). Its binding affinity to hACE2 was slightly lower than that of the WT. The MW045452 isolate also had a different mutation at residue 498 (Q498P) based on previous studies (with the enhancing mutation Q498Y) and had a lower docking score compared to WT. The MW045459 isolate had one enhancing mutation (T470N) and its docking score with ACE2 was slightly higher than that of WT. The MW136446 isolate had the L452P mutation with a different amino acid substitution from the previous studies (the enhancing mutation L452K); however, this mutation also resulted in a higher affinity. Another substitution at this position (L452R) has been associated with low sensitivity to neutralizing antibodies.43 Different amino acid substitutions in the isolates of MW055425, MW093140, MW045452 and MW136446 compared to previous studies make it difficult to predict the outcome of the mutations.
The MW040527 isolate, which carried the enhancing D614G mutation, showed no difference in binding affinity to ACE2 compared to the WT mutation. The D614G mutation improves the ability to bind the receptor.22,47 D614G variants have shown higher replication capacity and effective infection in human lung epithelial cells and faster transmission than the wild-type virus.48 Nevertheless, the D614G substitution increases the susceptibility of the virus to neutralization by antibodies and is not expected to be an obstacle for vaccine development.49 In our study, the D614G mutation was detected in 41 Iranian isolates. A previous study has shown that this mutation appeared in Iran in May 2020.50 Of the eight Iranian isolates studied, all have more than one RBD mutation, and the interaction of these mutations could affect binding to the ACE2 receptor. According to the docking results, the effect of the RBD mutations was not consistent with previous studies in some cases. This could be due to the mutations in the spike protein next to or near the RBD region and also due to the fact that we analyzed a structure with multiple mutations and not just a single mutation.
The RBD mutations of N501Y51 and L452R43 with higher binding affinity to ACE2 have shown lower affinity and lower susceptibility to monoclonal antibodies, respectively, leading to antibody escape. However, a different result has been obtained for the D614G mutation, which has a higher binding affinity for both ACE2 and neutralizing antibodies.49 Thus, it appears that each mutation acts in a specific way in interaction with hACE2 or neutralizing antibodies. Also, a combination of mutations in RBD would lead to different results compared to each mutation individually. The information on docking score and affinity will be helpful in determining viral replication and the most transmissible variants. However, the most important issue for vaccine development is the efficacy of the antibodies produced to neutralize new mutations.
The MD simulation evaluates the effects of different mutations on the structure and size of the protein complex and simulates the behavior of the protein in a natural environment, providing information on the stability and degree of compactness.52 Changes in protein stability and flexibility can lead to pathological changes.53 According to the docking analysis, the affinity of spike-ACE2 for Alpha, Delta, MW090849, MW045459 and MW136446 was higher than that of the WT protein; however, the stability followed the same order only for MW045459 and MW136446. The complexes of the Delta, Alpha, MW090849, MW093140 and MW055425 variants had lower stability than the WT during the simulation, while the other variants showed higher stability. Although slight deviations were observed in the Rg plots of the mutant complexes compared to the WT complex, the mobility/structural stability was not lost during the MD simulation. This proves their strong interaction with hACE2. Similar findings were also reported for the Alpha and Delta variants.44
The Alpha-ACE2 complex was formed with a high affinity and compactness, but showed lower stability. Docking and MD simulations showed that the Delta-ACE2 complex was docked with high affinity and good compactness, but had a higher MSD (lower stability) than the WT complex. This was consistent with previous studies. MD simulations of different SARS-CoV-2 variant-ACE2 complexes have shown a higher RMSD (lower stability) of Delta-ACE2 and Alpha-ACE2 than WT-ACE2. Nevertheless, all these studies have revealed that both the Alpha and Delta variants interact more strongly with the host receptor hACE2.54,55 Overall, considering the results of docking to ACE2 and the MD simulation, it appears that the variants of Alpha, Delta, MW090849, MW045459 and MW136446 are the variants of concern for higher transmissibility and rapid spread.
Four 9-mer antigen epitopes have been identified for the SARS-CoV-2 spike protein that bind to the TLR4/MD-2 complex. Two of them bind directly to the TLR4 protein and are located in the RBD.56 Based on this information, the spike RBD region was considered for both ACE2 and TLR4 docking and MD simulation. According to the docking result and MD simulation, the Alpha-TLR4 complex was formed with higher compactness due to higher binding affinity, although it had lower stability (higher MSD) than WT-TLR4. The poor MSD value for the Alpha-TLR4 complex despite good affinity and Rg value suggests that the mutations in the Alpha variant may have a complex influence on the stability and dynamics of the interaction with TLR4. The increased MSD value indicates a stronger fluctuation and lower stability of the complex over time. This could be due to the specific structural changes induced by the mutations in the Alpha variant, leading to a more dynamic interaction despite the stronger binding and compactness. The Delta-TLR4 complex was formed with a higher affinity and stability and only a slight lower compactness than WT-TLR4. Delta mutations would allow a closer association with TLR4 in longer simulations. It appears that the Delta mutant is a variant of concern not only due to its tighter interaction with hACE2, but also with TLR4. Most Iranian sequences (except MW136446 and MW090920) were docked with higher affinity to TLR4 and had lower Rg values than WT, indicating higher compactness. However, most of them had lower stability than WT (except MW136446), as shown by the MSD values. A significant decrease in Rg values was observed for complexes of MW090920-, MW045452-, MW040527-, MW090849- and MW045459-TLR4 compared to WT. Considering their high TLR4 binding affinity compared to WT, these variants may be important for higher transmission rate and rapid spread. A vaccine construct against SARS-CoV-2 has been designed and evaluated by molecular docking and MD simulation. A high binding affinity to the extracellular domains of TLR4 was observed.57 It would be advantageous to include different RBD mutations of SARS-CoV-2 in the design of vaccine constructs to prevent immune escape.
The mutations in the RBDs of the different variants should be tested for their affinity to neutralizing antibodies to further investigate their ability to escape the host immune system. These interactions will be essential for design of therapeutics and development of new vaccines that can overcome immune evading mutants. Therapies that block the binding of spikes to ACE2, either through receptor modifications or treatment with antibodies, could provide inhibition against new strains. Agents that suppress TLR4 signaling could also attenuate the harmful inflammation in COVID-19. However, it is not clear to what extent the RBD will evolve to evade the antibodies. Improving the interaction of SARS-CoV-2 and ACE2 increases the transmissibility of the virus by increasing the number of infected cells and viral load in mucosal secretions and enhancing the ability of the virus to initiate infection in a new host.58 This was the case with mutations of D614G22,59 and N501Y.60 However, mutations that escape the immune system play an important role in maintaining high transmissibility in different populations.61 Changes in pathogenicity were first reported for Alpha and then for Beta, Gamma and Delta, resulting in more hospitalizations and deaths compared to WT.62,63 Alpha and Delta were mainly associated with increased transmissibility and ability to escape the immune system compared to WT.64 However, the Delta variant was replaced by Omicron due to its higher immune escape properties.65 Nevertheless, Omicron lineages were associated with lower pathogenicity.66 This may be partly due to changes in host resistance as well as changes in the virus. When drawing an evolutionary map for different variants, one should keep in mind that all mutations related to receptor affinity (higher transmissibility) and antibody affinity (higher immune escape) are crucial factors in predicting the behavior of a variant.
Conclusion
In this study, we investigated the changes in the interaction between different SARS-CoV-2 variants and TLR4/7/8/9 and ACE2. The results showed that the endosomal TLRs may not play a crucial role in the higher transmissibility of the Alpha and Delta variants. Both the Alpha and Delta spike variants and some of the Iranian isolates had a higher binding affinity to ACE2 and TLR4, which is a possible reason for their higher transmission rate (although higher transmission was not investigated for the Iranian isolates). The Alpha-ACE2 complex was formed with a high affinity and compactness and a lower stability than WT. Delta-ACE2 complex had higher affinity and good compactness but lower stability as reported in other studies. The overall results show that mutations in the Alpha and Delta variants lead to higher transmissibility than the WT variant. Compared to WT-TLR4, higher affinity and compactness as well as lower stability were observed for the Alpha-TLR4 complex. The Delta-TLR4 complex was formed with higher affinity and stability than WT-TLR4 and with good compactness. It appears that the Delta mutant is a variant of concern not only because of its tighter interaction with ACE2 but also with TLR4. Overall, our results demonstrates a strong effect of ACE2 and TLR4 in controlling viral action through different mutations in SARS-CoV-2. These results support the notion that the most critical therapeutics are the factors affecting these two receptors, while the endosomal receptors are of lesser importance.
Research Highlights
What is the current knowledge?
√ TLRs have been associated with the infection of various respiratory diseases such as influenza, MERS and SARS.
√ In an in silico study, a strong association was found between the bat SARS spike protein and its receptor ACE2 and also TLR4.
√ The Alpha and Delta variants of SARS-CoV-2 are associated with higher viral load and faster transmission.
What is new here?
√ Lower number of TLR7/8/9 motifs in Alpha/Delta sequences compared to WT shows no critical role of endosomal TLRs in higher transmission ability of these variants.
√ Higher affinity and good stability/compactness of Delta-ACE2 and Delta-TLR4 complexes may lead to higher transmissibility and viral load compared to WT and Alpha.
√ Higher affinity of Alpha/Delta spikes to ACE2 and TLR4 compared to WT indicates strong effects of these receptors for conducting viral effect through spike various mutations.
Competing interests
The authors declare that they have no competing interest.
Ethical Statement
Not applicable.
Funding
The authors received no funding for this research.
References
- Iwasaki A, Medzhitov R. Regulation of Adaptive Immunity by the Innate Immune System. Science 2010; 327:291-5. doi: 10.1126/science.1183021 [Crossref] [ Google Scholar]
- Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat Rev Immunol 2014; 14:315-28. doi: 10.1038/nri3665 [Crossref] [ Google Scholar]
- Totura AL, Whitmore A, Agnihothram S, Schäfer A, Katze MG, Heise MT. Toll-Like Receptor 3 Signaling via TRIF Contributes to a Protective Innate Immune Response to Severe Acute Respiratory Syndrome Coronavirus Infection. mBio 2015; 6:e00638-15. doi: 10.1128/mBio.00638-15 [Crossref] [ Google Scholar]
- Al-Qahtani AA, Lyroni K, Aznaourova M, Tseliou M, Al-Anazi MR, Al-Ahdal MN, et al. Middle east respiratory syndrome corona virus spike glycoprotein suppresses macrophage responses via DPP4-mediated induction of IRAK-M and PPARγ. Oncotarget 2017; 8. 10.18632/oncotarget.14754.
- Li G, Fan Y, Lai Y, Han T, Li Z, Zhou P. Coronavirus infections and immune responses. J Med Virol 2020; 92:424-32. doi: 10.1002/jmv.25685 [Crossref] [ Google Scholar]
- Moreno-Eutimio MA, López-Macías C, Pastelin-Palacios R. Bioinformatic analysis and identification of single-stranded RNA sequences recognized by TLR7/8 in the SARS-CoV-2, SARS-CoV, and MERS-CoV genomes. Microbes Infect 2020; 22:226-9. doi: 10.1016/j.micinf.2020.04.009 [Crossref] [ Google Scholar]
- Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol 2006; 7:131-7. doi: 10.1038/ni1303 [Crossref] [ Google Scholar]
- Lester SN, Li K. Toll-Like Receptors in Antiviral Innate Immunity. J Mol Biol 2014; 426:1246-64. doi: 10.1016/j.jmb.2013.11.024 [Crossref] [ Google Scholar]
- Akira S, Hemmi H. Recognition of pathogen-associated molecular patterns by TLR family. Immunol Lett 2003; 85:85-95. doi: 10.1016/S0165-2478(02)00228-6 [Crossref] [ Google Scholar]
- Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdörfer B, Giese T. Quantitative expression of toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol 2002; 168:4531-7. doi: 10.4049/jimmunol.168.9.4531 [Crossref] [ Google Scholar]
- Zhang Z, Ohto U, Shibata T, Taoka M, Yamauchi Y, Sato R, et al. Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands. Cell Rep 2018; 25: 3371-81.e5. 10.1016/j.celrep.2018.11.081.
- Ng LFP, Hibberd ML, Ooi E-E, Tang K-F, Neo S-Y, Tan J. A human in vitro model system for investigating genome-wide host responses to SARS coronavirus infection. BMC Infect Dis 2004; 4:34. doi: 10.1186/1471-2334-4-34 [Crossref] [ Google Scholar]
- Naik S, Kelly EJ, Meijer L, Pettersson S, Sanderson IR. Absence of Toll-Like Receptor 4 Explains Endotoxin Hyporesponsiveness in Human Intestinal Epithelium. J Pediatr Gastroenterol Nutr 2001. 32: 449-53. 10.1097/00005176-200104000-00011.
- Choudhury A, Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J Med Virol 2020; 92:2105-13. doi: 10.1002/jmv.25987 [Crossref] [ Google Scholar]
- Rambaut A, Holmes EC, O’Toole Á, Hill V, McCrone JT, Ruis C. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020; 5:1403-7. doi: 10.1038/s41564-020-0770-5 [Crossref] [ Google Scholar]
- Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday J. Estimated transmissibility and impact of SARS-CoV-2 lineage B117 in England. Science 2021; 372:eabg3055. doi: 10.1126/science.abg305517 [Crossref] [ Google Scholar]
- Kang M, Xin H, Yuan J, Ali ST, Liang Z, Zhang J. Transmission dynamics and epidemiological characteristics of Delta variant infections in China. Euro surveill 2022; 27:2100815. [ Google Scholar]
- Khare S, Gurry C, Freitas L, Schultz MB, Bach G, Diallo A. GISAID's Role in Pandemic Response. China CDC Wkly 2021; 3:1049-51. doi: 10.46234/ccdcw2021.255 [Crossref] [ Google Scholar]
- Forsbach A, Nemorin J-G, Montino C, Müller C, Samulowitz U, Vicari AP. Identification of RNA Sequence Motifs Stimulating Sequence-Specific TLR8-Dependent Immune Responses. J Immunol 2008; 180:3729-38. doi: 10.4049/jimmunol.180.6.3729 [Crossref] [ Google Scholar]
- Bauer S, Kirschning CJ, Häcker H, Redecke V, Hausmann S, Akira S. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Nat Acad Sci 2001; 98:9237-42. doi: 10.1073/pnas.161293498 [Crossref] [ Google Scholar]
- Yi C, Sun X, Ye J, Ding L, Liu M, Yang Z. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell Mol Immunol 2020; 17:621-30. doi: 10.1038/s41423-020-0458-z [Crossref] [ Google Scholar]
- Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020; 183: 739-51.e8. 10.1016/j.cell.2020.09.032.
- Rappe AK, Casewit CJ, Colwell KS, Goddard WA, III III, Skiff WM. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J Am Chem Soc 1992; 114:10024-35. doi: 10.1021/ja00051a040 [Crossref] [ Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys 1983; 79:926-35. doi: 10.1063/1.445869 [Crossref] [ Google Scholar]
- Neria E, Fischer S, Karplus M. Simulation of activation free energies in molecular systems. J Chem Phys 1996; 105:1902-21. doi: 10.1063/1.472061 [Crossref] [ Google Scholar]
- Beglov D, Roux B. Finite representation of an infinite bulk system: Solvent boundary potential for computer simulations. J Chem Phys 1994; 100:9050-63. doi: 10.1063/1.466711 [Crossref] [ Google Scholar]
- Rühle V, editor. Berendsen and Nose-Hoover thermostats. 2007.
- Andersen HC. Molecular dynamics simulations at constant pressure and/or temperature. J Chem Phys 1980; 72:2384-93. doi: 10.1063/1.439486 [Crossref] [ Google Scholar]
- Miyamoto S, Kollman PA. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem 1992; 13:952-62. doi: 10.1002/jcc.540130805 [Crossref] [ Google Scholar]
- Tuckerman M, Berne BJ, Martyna GJ. Reversible multiple time scale molecular dynamics. J Chem Phys1992. 97: 1990-2001. 10.1063/1.463137.
- Zahmatkesh A, Bagheri M. Roles and Functions of Toll-Like Receptors in Coronavirus Infections: Forthcoming Vaccine and Therapeutic Strategies for Confronting COVID-19. Vac Res 2021; 8:17-25. doi: 10.52547/vacres.8.2.17 [Crossref] [ Google Scholar]
- Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdörfer B, Giese T. Quantitative Expression of Toll-Like Receptor 1–10 mRNA in Cellular Subsets of Human Peripheral Blood Mononuclear Cells and Sensitivity to CpG Oligodeoxynucleotides. J Immunol 2002; 168:4531-7. doi: 10.4049/jimmunol.168.9.4531 [Crossref] [ Google Scholar]
- Plantinga M, Hammad H, Lambrecht BN. Origin and functional specializations of DC subsets in the lung. Eur J Immunol 2010. 40: 2112-8. 10.1002/eji.201040562.
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S. Species-Specific Recognition of Single-Stranded RNA via Toll-like Receptor 7 and 8. Science 2004; 303:1526-9. doi: 10.1126/science.1093620 [Crossref] [ Google Scholar]
- Beignon A-S, McKenna K, Skoberne M, Manches O, DaSilva I, Kavanagh DG. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor– viral RNA interactions. J Clin Invest 2005; 115:3265-75. doi: 10.1172/JCI26032 [Crossref] [ Google Scholar]
- Di Domizio J, Blum A, Gallagher-Gambarelli M, Molens JP, Chaperot L, Plumas J. TLR7 stimulation in human plasmacytoid dendritic cells leads to the induction of early IFN-inducible genes in the absence of type I IFN. Blood 2009; 114:1794-802. doi: 10.1182/blood-2009-04-216770 [Crossref] [ Google Scholar]
- Cameron MJ, Bermejo-Martin JF, Danesh A, Muller MP, Kelvin DJ. Human immunopathogenesis of severe acute respiratory syndrome (SARS). Virus Res 2008; 133:13-9. doi: 10.1016/j.virusres.2007.02.014 [Crossref] [ Google Scholar]
- Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis 2020; 71:762-8. doi: 10.1093/cid/ciaa248 [Crossref] [ Google Scholar]
- Tang FSM, Van Ly D, Spann K, Reading PC, Burgess JK, Hartl D. Differential neutrophil activation in viral infections: Enhanced TLR-7/8-mediated CXCL8 release in asthma. Respirol 2016; 21:172-9. doi: 10.1111/resp.12657 [Crossref] [ Google Scholar]
- Wu Y, Wang F. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 2020; 368:1274-8. doi: 10.1126/science.abc2241 [Crossref] [ Google Scholar]
- Farinholt T, Doddapaneni H, Qin X, Menon V, Meng Q, Metcalf G. Transmission event of SARS-CoV-2 Delta variant reveals multiple vaccine breakthrough infections. BMC Med 2021; 19:255. doi: 10.1186/s12916-021-02103-4 [Crossref] [ Google Scholar]
- Planas D, Veyer D, Baidaliuk A. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021; 596:276-80. doi: 10.1038/s41586-021-03777-9 [Crossref] [ Google Scholar]
- Padilla-Sanchez V. Molecular Dynamics of SARS-CoV-2 Delta Variant Receptor Binding Domain in Complex with ACE2 Receptor. ARPHA Preprints; 2021. 10.3897/arphapreprints.e71603.
- Kim S. Differential Interactions between Human ACE2 and Spike RBD of SARS-CoV-2 Variants of Concern. J Chem Theory Comput 2021; 17:7972-9. doi: 10.1021/acs.jctc.1c00965 [Crossref] [ Google Scholar]
- Dinnon KH, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 2020; 586:560-6. doi: 10.1038/s41586-020-2708-8 [Crossref] [ Google Scholar]
- Lima MA, Skidmore M, Khanim F, Richardson A. Development of a nano-luciferase based assay to measure the binding of SARS-CoV-2 spike receptor binding domain to ACE-2. Biochem Biophys Res Commun 2021; 534:485-90. doi: 10.1016/j.bbrc.2020.11.055 [Crossref] [ Google Scholar]
- Mansbach RA, Chakraborty S, Nguyen K, Montefiori DC, Korber B, Gnanakaran S. The SARS-CoV-2 Spike Variant D614G Favors an Open Conformational State. Sci Adv 2021; 7:eabf3671. [ Google Scholar]
- Hou YJ, Chiba S, Halfmann P, Ehre C, Kuroda M, Dinnon KH. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 2020; 370:1464-8. doi: 10.1126/science.abe8499 [Crossref] [ Google Scholar]
- Plante JA, Liu Y, Liu J, Xia H. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021; 592:116-21. doi: 10.1038/s41586-020-2895-3 [Crossref] [ Google Scholar]
- Moradi J, Moghoofei M, Alvandi AH, Abiri R. Variation analysis of SARS-CoV-2 complete sequences from Iran. Future Virol 2022. 10.2217/fvl-2021-0056.
- Chakraborty S. E484K and N501Y SARS-CoV 2 spike mutants Increase ACE2 recognition but reduce affinity for neutralizing antibody. Int Immunopharmacol 2022; 102:108424. doi: 10.1016/j.intimp.2021.108424 [Crossref] [ Google Scholar]
- Krebs BB, De Mesquita JF. Amyotrophic Lateral Sclerosis Type 20 - In Silico Analysis and Molecular Dynamics Simulation of hnRNPA1. PLoS One 2016; 11:e0158939. doi: 10.1371/journal.pone.0158939 [Crossref] [ Google Scholar]
- Khan FI, Wei D-Q, Gu K-R, Hassan MI, Tabrez S. Current updates on computer aided protein modeling and designing. Int J Biol Macromol 2016; 85:48-62. doi: 10.1016/j.ijbiomac.2015.12.072 [Crossref] [ Google Scholar]
- Celik I, Yadav R, Duzgun Z, Albogami S, El-Shehawi AM, Fatimawali Fatimawali. Interactions of the Receptor Binding Domain of SARS-CoV-2 Variants with hACE2: Insights from Molecular Docking Analysis and Molecular Dynamic Simulation. Biology 2021; 10:880. doi: 10.3390/biology10090880 [Crossref] [ Google Scholar]
- Wijayanti NM, Widyananda MH, Muflikhah L, Widodo N. Mutation-Induced Changes in the Stability, B-Cell Epitope, and Antigenicity of the Sars-Cov-2 Variant Spike Protein: A Comparative Computational Stud. Karbala Int J Mod Sci 2023; 9:12. doi: 10.33640/2405-609X.3311 [Crossref] [ Google Scholar]
- Bhattacharya M, Sharma AR, Mallick B, Sharma G, Lee S-S, Chakraborty C. Immunoinformatics approach to understand molecular interaction between multi-epitopic regions of SARS-CoV-2 spike-protein with TLR4/MD-2 complex. Infect Genet Evol 2020; 85:104587. doi: 10.1016/j.meegid.2020.104587 [Crossref] [ Google Scholar]
- Pourseif MM, Parvizpour S, Jafari B, Dehghani J. A domain-based vaccine construct against SARS-CoV-2, the causative agent of COVID-19 pandemic: development of self-amplifying mRNA and peptide vaccines. Bioimpacts 2021; 11:65-84. doi: 10.34172/bi.2021.11 [Crossref] [ Google Scholar]
- Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020; 581:221-4. doi: 10.1038/s41586-020-2179-y [Crossref] [ Google Scholar]
- Ozono S, Zhang Y, Ode H. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat Commun 2021; 12:848. doi: 10.1038/s41467-021-21118-2 [Crossref] [ Google Scholar]
- Liu H, Zhang Q, Wei P, Chen Z, Aviszus K, Yang J. The basis of a more contagious 501YV1 variant of SARS-CoV-2. Cell Res 2021; 31:720-2. doi: 10.1038/s41422-021-00496-8 [Crossref] [ Google Scholar]
- Markov PV, Ghafari M, Beer M, Lythgoe K, Simmonds P, Stilianakis NI. The evolution of SARS-CoV-2. Nat Rev Microbiol 2023; 21:361-79. doi: 10.1038/s41579-023-00878-2 [Crossref] [ Google Scholar]
- Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021; 592:438-43. doi: 10.1038/s41586-021-03402-9 [Crossref] [ Google Scholar]
- Funk T, Pharris A, Spiteri G, Bundle N, Melidou A, Carr M. Characteristics of SARS-CoV-2 variants of concern B117, B1351 or P1: data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Euro Surveill 2021; 26:2100348. doi: 10.2807/1560-7917.es.2021.26.16.2100348 [Crossref] [ Google Scholar]
- Oude Munnink BB, Worp N, Nieuwenhuijse DF. The next phase of SARS-CoV-2 surveillance: real-time molecular epidemiology. Nat Med 2021; 27:1518-24. doi: 10.1038/s41591-021-01472-w [Crossref] [ Google Scholar]
- Viana R, Moyo S, Amoako DG, Tegally H, Scheepers C, Althaus CL. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022; 603:679-86. doi: 10.1038/s41586-022-04411-y [Crossref] [ Google Scholar]
- European Centre for Disease Prevention and Control. Assessment of the further spread and potential impact of the SARS-CoV-2 Omicron variant of concern in the EU/EEA, 19th update - 27 January 2022. ECDC: Stockholm; 2022.