Bioimpacts. 2025;15:30160.
doi: 10.34172/bi.30160
Review
Molecular dynamics simulation in tissue engineering
Ali Rahmani Conceptualization, Visualization, Writing – original draft, 1, 2 
Rahim Jafari Project administration, Supervision, Writing – original draft, Writing – review & editing, 2, * 
Samad Nadri Project administration, 3, 4, 2, * 
Author information:
1Student Research Committee, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran
2Department of Medical Nanotechnology, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran
3Zanjan Pharmaceutical Nanotechnology Research Center, Zanjan University of Medical Sciences, Zanjan, Iran
4Zanjan Metabolic Diseases Research Center, Zanjan University of Medical Sciences, Zanjan, Iran
Abstract
Introduction:
In tissue engineering, the interaction among three primary elements, namely cells, material scaffolds, and stimuli, plays a pivotal role in determining the fate of cells and the formation of new tissue. Understanding the characteristics of these components and their interplay through various methodologies can significantly enhance the efficiency of the designed tissue engineering system. In silico methods, such as molecular dynamics (MD) simulation, use mathematical calculations to investigate molecular properties and can overcome the limitations of laboratory methods in delivering adequate molecular-level information.
Methods:
The studies that used molecular dynamics simulation, either alone or in combination with other techniques, have been reviewed in this paper.
Results:
The review explores the use of molecular dynamics simulations in studying substrate formation mechanism and its optimization. It highlights MD simulations' role in predicting biomolecule binding strength, understanding substrate properties' impact on biological activity, and factors influencing cell attachment and proliferation. Despite limited studies, MD simulations are considered a reliable tool for identifying ideal substrates for cell proliferation. The review also touches on MD simulations' contribution to cell differentiation studies, emphasizing their role in designing engineered extracellular matrix for desired cell fates.
Conclusion:
Molecular dynamics simulation as a non-laboratory tool has many capabilities in providing basic and practical information about the behavior of the molecular components of the cell as well as the interaction of the cell and its components with the surrounding environment. Using this information along with other information obtained from laboratory tools can ultimately lead to the advancement of tissue engineering through the development of more appropriate and efficient methods.
Keywords: Tissue engineering, Molecular Dynamics, Simulation, Modeling
Copyright and License Information
© 2025 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Introduction
Despite tremendous advances in biology and medicine, many diseases and physical disabilities remain unresolved. These conditions include nervous system disorders such as spinal cord injuries,1 various metabolic disorders such as diabetes,2 blindness, or low vision caused by damage to a specific part of the eye, such as the retina,3 etc. Conventional therapies have been incapable of overcoming these conditions and are used more in a managerial aspect. Therefore, researchers have taken various approaches to provide effective treatments, including stem cell therapy,4 gene therapy,5 and tissue engineering.6
Tissue engineering aims to create functional constructs that restore, maintain, or improve damaged tissues or whole organs.7 Tissue engineering studies, which utilize biomaterials for mechanical support or as systems for stem cell or drug delivery, have demonstrated significant potential in the management of diseases. For example, in the case of spinal cord injuries, tissue engineering can provide scaffolds that support the regeneration of neural tissue and guide the growth of axons across the lesion site.8 Moreover, tissue engineering can incorporate stem cells, growth factors, and biomimetic materials to enhance the recovery of neurological function. Similarly, for metabolic disorders such as type 1 diabetes the goal is to use biomaterials that can hold cells, such as islets or β-cells, and provide them with a suitable environment outside and inside the body to keep them alive and functioning.9
The flexibility of tissue engineering distinguishes it from other methods since various types of cells, chemical, biological and physical stimuli, and different substrates such as hydrogels and nanofiber scaffolds can be used according to the intended purpose.
Here, a comprehensive molecular-level understanding of the tissue engineering components in terms of physico-chemical properties, formation mechanisms, and their interaction with living materials will greatly help the design of more advanced systems. As a result, several techniques are routinely utilized in the study of interactions between cells, proteins, and DNA with substrates and nanostructures, offering valuable information from various perspectives. Of these techniques, UV-vis, fluorescence spectroscopy, dynamic light scattering, and atomic force microscopy for the study of binding and its strength; circular dichroism, Fourier transform infrared spectroscopy, Raman spectrometry, X-ray crystallography, and nuclear magnetic resonance to study conformational changes; mass spectroscopy and N-terminal microsequencing for identification; and finally, quartz crystal microbalance and surface plasmon resonance methods to study the interaction kinetics can be mentioned. For example, using Raman tweezers spectroscopy, Barkur et al have revealed the deoxygenation of red blood cells in the presence of gold and silver nanoparticles (NPs) due to the adhesion of the NPs to cell surfaces.10 Although these laboratory techniques provide excellent details about the interactions and the fabricated structure as a whole, there is still a knowledge gap regarding what happens at the molecular level. The main drawbacks of these techniques are their poor spatial and temporal resolution, which limits them for providing in-depth descriptions on the molecular events that occur very quickly.11 Researchers, therefore, use computer simulation techniques, especially molecular dynamics (MD) simulation, to overcome these challenges.
MD simulations allow researchers to study molecular interactions and events at an atomic level, providing a detailed view of the behavior of individual atoms and molecules. This high resolution allows researchers to study the molecular mechanisms underlying biological processes in greater detail than experimental methods can offer. Also, due to the use of very short time steps, usually 1-2 femtoseconds, MD simulations can provide detailed information about fast events such as protein conformational changes or self-assembly of molecules which typically occur within the range of a few nanoseconds to microseconds.12
This review article begins with a brief overview of MD simulations and tissue engineering. Then it covers the use of MD simulations in the study of production of biomaterials and their structural optimization. We discuss self-assembly as a highly useful technique in the production of biomaterials. Self-assembly is the process by which molecules come together to create multi-component structures through precise interactions without the need for external forces and with the help of environmental factors. Because natural and synthetic polymer substrates used in tissue engineering are typically formed by self-assembly, understanding their molecular dynamics can help create more efficient substrates.13,14 Following that, the adsorption and desorption of molecules on tissue engineering substrates is discussed. The surface of cell culture substrates can be functionalized by attaching various substances for a variety of reasons, such as enhancing hydrophilicity, boosting cell adhesion and proliferation, and inducing cell differentiation. Here, MD simulations play a key role in investigation of the interaction between these molecules/moieties and the components of the culture substrate. The use of MD simulations in drug delivery systems, with a focus on tissue engineering, has also been reviewed. Finally, we explored further aspects of tissue engineering from a molecular perspective, including cell proliferation and differentiation, using MD simulations. A summary of the reviewed articles is presented in Table 1.
Table 1.
A brief review of MD simulation studies related to tissue engineering
|
|
Authors
|
Aim
|
Findings
|
| Optimization of substrate |
Hao et al 15 |
Use of experimental and MD simulation methods to investigate the adsorption and bioactivity of fibronectin (Fn) on surfaces with varying chemistries with the ultimate goal of providing insights for the rational design of fibronectin-activating biomaterials. |
Surfaces with -CH3 head groups have strong Fn adsorption but poor bioactivity, those with -NH2 and -COOH have efficient Fn adsorption and excellent bioactivity, and that with -OH groups has poor Fn adsorption but non-negligible bioactivity. This difference has been attributed to the amount of exposure of RGD and PHSRN motifs and the deformation of the protein on different surfaces. |
| Huang et al 16 |
To clarify the adsorption and desorption dynamics of bone morphogenetic protein-2 (BMP-2) on various nano-textured hydroxyapatite (HAP) surfaces at the atomic level, hoping to offer important insights for creating BMP-2-based tissue engineering implants/scaffolds |
The adsorption strength of BMP-2 on HAP surfaces increases with the increase of surface roughness. The adsorption orientation and conformation of BMP-2 on HAP surfaces are dependent on the surface nano-texture type. |
| Raffaini et al17 |
To study how fibronectin type I module adsorbs on hydrophobic graphite surface and how it changes its conformation and function |
The fibronectin module strongly adsorbs on the graphite surface due to hydrophobic interactions, losing its native structure as it spreads on the surface of the graphite |
| Biswas et al18 |
To investigate how two types of cell-adhesion peptides, RGD and YIGSR, interact with two types of biomaterial surfaces, hydroxyapatite and TiO2, which are used for bone and dental implants. The study can help understand how the peptides can improve the biocompatibility and functionality of the implant |
Both peptides bind to the titanium surface more strongly than to the hydroxyapatite surface. RGD maintained its “hairpin”-like structure during adsorption on a HA surface, and a slightly “relaxed hairpin” structure on TiO2 surface. |
| Self-assembly |
Tekin 19 |
To understand the structural properties of peptide amphiphile (PA)-based cylindrical nanofibers and the factors that play a role in the self-assembly process, which can provide insights for designing novel PA-based nanomaterials |
The secondary structure that peptides adopt in the nanofiber are mainly random coil and beta sheet. The self-assembly of the nanofiber is due to the hydrophobic interactions between VVAG–VVAG parts and electrostatic attraction of D–Na+ and E–R |
| Sun et al 20 |
Developing a bioinspired supramolecular nanofiber hydrogel through self-assembly of biphenyl-tripeptide for tissue engineering. MD simulations were used to investigate the self-assembly behavior of biphenyl-tripeptide in water and to explore the thermodynamic mechanism involved in the process |
The sequences with FF motifs were able to form single aggregates faster and more stable than the others. The FAF and FGF motifs had more hydrogen bonds within the molecules, which hindered the hydrogen bonding between the molecules, making them more compact and likely to precipitate. |
| Lee, et al 21 |
Investigation of the self-assembly of a peptide amphiphile into cylindrical nanofibers and understanding its structure and dynamics. This information can be used to better design new nanofiber structures with improved properties. |
Formation of a stable nanofiber in a 40 ns simulation. Epitope sequence IKVAV was located on the surface of the nanofiber which is important for promoting neurite sprouting and cell adhesion |
| Cell proliferation |
Shamloo et al22 |
To investigate the adhesive characteristics of different polymer-protein systems using MD simulation and to explore their relation to cell adhesion and proliferation. This information can be used to develop novel polymer-protein systems with optimized adhesive characteristics |
Hydrophobic surface of PCL generally led to stronger protein adhesion and better cell proliferation compared to PVA, with the exception of albumin subdomain. The findings also demonstrated a direct correlation between stronger adhesion of ECM proteins to the surface and more desirable cell adhesion and proliferation |
| Cell differentiation |
Mehralitabar et al22 |
To investigate how the combination of bioactive and nonbioactive alkyl-peptides forms a more stable nanofiber structure for differentiating neural stem cells. |
Comparison between nanofibers made of the combination of bioactive and nonbioactive alkyl-peptides and that composed of bioactive alkyl-peptides only, it has been shown that the former is more stable structure and have a more favorable surface environment for neural cell adhesion and proliferation. This findings would contribute to the development of more efficient and effective scaffolds for neural tissue engineering |
| Bock et al23 |
Evaluating the mouse mesenchymal stem cells behavior in response to terahertz radiation. The MD simulation used to explore the molecular mechanisms that are involved in the terahertz radiation-induced cellular reprogramming |
Local breathing dynamics, that is the dynamics of transient opening and closing of the DNA double helix of in the PPARG promoter region DNA occur simultaneously with the gene-specific response to the THz radiation |
| Drug delivery |
Hasani-Sadrabadi et al24 |
To understand the interaction between microfluidics synthesized chitosan nanoparticles and dexamethasone (as modulator of osteoblast) at the molecular level with the aim of optimizing the design of the nanoparticles for sustained intracellular delivery of the drug. |
Strong interactions between dexamethasone (Dex) molecules and chitosan (CS) chains, in which significant role played by van der Waals interactions in the CS-Dex system; Also CS functional groups are able to have hydrogen bonding interactions with Dex drugs |
MD simulation and its applications in biomedical study
The underlying basis of MD simulation is rooted in Newton’s equations of motion. MD simulation, as a computational methodology, utilizes these equations to generate configurations of a molecular system over time. The simulation begins with an initial configuration of atoms which can be generated randomly or based on experimental data. The forces acting on each atom are calculated based on the interactions between atoms, which can be modeled using force fields. The simulation then proceeds by calculating the velocities of atoms and updating their positions. This process is repeated many times, allowing the simulation to explore the behavior of the system over time. The trajectory of a system is the sequence of atomic positions and velocities over time. The trajectory can be used to study the behavior of a system in detail, such as studying the conformational changes of proteins due to folding and unfolding, the diffusion of drug molecules out of their carriers, and the structural changes of biomolecules upon adsorption on material surfaces, etc.25,26
There are numerous MD software available to run such simulations and typically consist of three main components: Force fields, main MD program and auxiliary programs.
A force field is a set of mathematical expressions and parameters (constants) that correlate the configurations of a system of atoms/particles to its energies. Some force fields use parameters obtained by ab initio quantum mechanics calculation, while in empirical force fields the parameters are mostly derived from fitting calculated observables to experimental data in physics and chemistry. Despite the diversity in force fields, they typically consist of two main parts: equations for calculating bonded energies based on bond lengths, bond angles and dihedral angles, and those for calculating non-bonded energies due to electrostatics and van der Waals interactions27 (equation 1).
Etotal = (Ebond length + Ebond angle + Edihedral angle) + (Eelectrostatic + Evan der Waals)
Various force fields may use different functional form for each of the above energy contributions. For example, energy changes due to covalent bond stretching can be modeled using either harmonic potential (Hooke’s law) or Morse potential, while van der Waals energies can be calculated using either Lennard-Jones or Buckingham potentials. Another difference between force fields lies in their parameter values, such as partial charge of a certain atom type or bond strength for a certain covalent bond.28
Force fields are developed to capture different aspects of molecular behavior and vary in their suitability for specific types of molecules or systems.
For example, COMPASS (Condensed-phase Optimized Molecular Potentials for Atomistic Simulation Studies) is a general ab initio-derived force field for MD simulations of common organic molecules, inorganic small molecules, and polymers.29 In the case of biomacromolecules, there is a group of specialized empirical force fields that are more accurate in simulating structural and dynamic features compared to general force fields. These force field families which include OPLS,30 CHARMM,31-33 GROMOS34 and AMBER35 are parametrized to simulate proteins, carbohydrates, lipids, and nucleic acid molecules. They are optimized for systems that solely contain biomacromolecules. However, for heterogeneous systems containing both biological molecules and non-biological compounds such as metals, polymers, and mineral structures, there are derivatives of the mentioned force fields that can be used for the simulation of the interactions between different components of the system. For instance, the INTERFACE force field comprises a set of parameters well-suited for the MD simulation of adsorption process of biomolecules on various surface types.36 Additionally, GAFF (General Amber Force Field) and CGenFF (CHARMM General Force Field) are two other force fields that are extensions to the standard AMBER and CHARMM force fields covering various kinds of small organic molecules and drugs.
Validation of force fields is an essential step in their development to ensure their accuracy and reliability. This includes evaluating whether the simulation results match the behavior and properties of molecules and systems in the real world under different conditions. For this purpose, data on molecular structure, energy and thermodynamic properties obtained by MD simulations are compared against those from experimental methods such as X-ray crystallography, spectroscopy (e.g., NMR) and calorimetry.37-39 A thoroughly validated force field that can reproduce laboratory findings with a suitable accuracy can provide confidence that in similar conditions its simulation results on different systems are reliable. It is very important to note that the outcome of MD simulations is dependent on the initial state of the system, that is, the starting positions and velocities of its constituent particles.
As a result, to enhance the reliability of simulation outcomes, one common practice is to conduct multiple MD simulations with varying initial conditions. If the results of the simulations do not differ much from each other, it suggests that the simulation results are robust and reliable, and that any observed trends or patterns are not due to random fluctuations or initial conditions.40
There are many different software programs available for running MD simulations such as GROMACS, NAMD, CHARMM, AMBER, LAMMPS, Desmond, Materials Studio, SAMSON, YASARA, etc., some of which are commercial, and some are free. These software are different from each other in several ways. 1- Some have a graphical interface and others have a command line interface. 2- In terms of the variety of tools needed to analyze the results as well as other auxiliary tools 3- Supported simulation methods 4- Types of supported force fields and accordingly the types of molecules and materials. Among the available software, the first four mentioned software are more popular in simulating systems containing biomolecules, which is due to built-in force fields and specialized analysis tools. Although these software are primarily developed to simulate biomolecules, they can also be used for systems containing non-biological components (e.g., polymers, metals, minerals, etc.). In such cases, certain tools can be used to make topology and force field parameters for the non-biological components of the systems. A notable example is CHARMM-GUI (https://www.charmm-gui.org/)41,42 which can generate input files compatible with CHARMM force field and suitable for user-selected MD program(s).
Nowadays, high-tech computers and their capacity to run complicated problems make it possible to use MD simulation as a routine tool in computational chemistry and biomedical studies.43
MD simulations surmount experimental constraints through their capability to access microsecond and nanometer time and length scales. The full atomistic representation and dynamic behavior inherent in MD simulations, for instance, enable the identification of specific molecular interactions and the comprehension of how proteins undergo structural changes induced by binding.11 Hence, MD simulations can be used to provide predictions of biological responses to various modifications, including mutation, phosphorylation, protonation, or interaction with other molecules/materials.44
Biological systems exhibit inherent complexity due to the presence of various molecules and components such as proteins, lipids, small organic molecules, etc., that interact with each other in a complex network. Environmental factors and unknown variables further increase this complexity. MD simulation software can be utilized to simulate the effects of factors such as temperature, pressure, pH, type of solvent, and ions on various biological phenomena. However, it is crucial to acknowledge that MD simulations are merely a representation of reality that employ certain assumptions and are not perfect. Moreover, MD simulations are limited to studying small parts of the system and over relatively short time frames, rather than encompassing the entire system. In spite of the limitations associated with MD simulation methods, they can still offer valuable insights that cannot be obtained through traditional laboratory methods.
Given the recent advancements in tissue engineering and the ongoing studies in this field, comprehending the intricate molecular aspects of the involved components from both structural and biological viewpoints is of paramount significance.
Tissue engineering
Tissue engineering is an interdisciplinary research field. The ultimate goal of research in tissue engineering is to achieve a dynamic, temporary replacement system for damaged tissues that can mimic tissue function and provide requirements for restoring normal tissue function and homeostasis. There are many functional components in tissue engineering, however, cells, stimulating factors, and suitable substrates are generally referred to as the three main pillars. According to studies, the precise engineering of components in tissue engineering can determine the cellular fate and guide it in the desired direction.45-48 Increasing our understanding at the molecular level of how biomolecules interact with each other as well as with non-biological materials such as substrates, and also understanding the underlying mechanisms of stimuli, ultimately gives us the ability to have greater control over the factors influencing cell behavior such as such as adhesion, proliferation, and differentiation. This enhanced understanding ultimately enables us to maximize the efficiency of designed tissue engineering systems.
A variety of cells have been used in tissue engineering, but stem cells have been studied the most. Stem cells, due to their unique properties, including the ability to self-reproduce, multiply and differentiate into other cell lines, have presented exciting opportunities to tissue engineering researchers. The functions of stem cells, including their proliferation and differentiation, can be adjusted by external chemical, biological and physical stimuli.49-51
Another component of tissue engineering is the cell substrate. According to prior researches, polymers have demonstrated significant potential as substrates in tissue engineering investigations owing to their ability to mimic the extracellular matrix (ECM) of cells in a compelling manner. Since cells are restrained in their natural niche in a protein scaffold called ECM, a structure that can imitate this natural niche the most is at the center of attention of many tissue engineering studies. On the other hand, different features of the scaffold, such as stiffness and rigidity, porosity, hydrophilicity and hydrophobicity, and many other properties are involved in determining cell fate. In tissue engineering, various types of stimuli are employed to prompt cells to behave in specific ways. Notably, growth factors and physical stimuli are commonly utilized, and these can exert a powerful influence on cellular behavior and fate.52-55
The precise engineering of these components is essential to guide cellular fate in the desired direction. For cells, it is important to understand their intrinsic properties, such as differentiation potential, self-renewal capacity, and epigenetic regulation. These properties can be influenced by factors such as culture conditions, growth factors, and gene transfer. By manipulating these factors, researchers can induce or inhibit specific cellular fates and functions.56 For stimulating factors, it is important to select the appropriate type, concentration, timing, and delivery method for each tissue type and cell type. Different factors have different effects on cell proliferation, differentiation, migration, survival, and apoptosis. By optimizing these parameters, researchers can enhance or suppress specific cellular responses. Lastly, for substrates, it is important to design the material properties that match the mechanical requirements of each tissue type and cell type. Different substrates have different stiffnesses, deformabilities, adhesivities, biodegradabilities, and biocompatibilities. By modifying these properties, researchers can create or modify specific microenvironments that support or inhibit specific cellular behaviors.57
Therefore, considering the complexity of tissue engineering, fundamental studies in the molecular level hold significance for the detailed investigation of tissue engineering structures and their interactions in determining cell fate. For example, understanding the interaction of cells with scaffolds and growth factors or drugs with cells and various cell components are essential in the optimal design of tissue engineering systems with high level of precision.52,58 However, achieving this level of precision is not without challenges. One major challenge is the variability of cell behavior in response to different stimuli. Cells can respond differently to the same stimulus depending on their phenotype, culture conditions, and environmental cues. An additional challenge is the complexity of cell-substrate interactions. Cells adhere differently to various substrates as a result of differences in surface chemistry, surface topography, and stiffness, among other factors. Interactions at molecular level are characterized using a variety of approaches, such as spectroscopy and, more recently, molecular dynamics simulation methods, which is a promising technology.44
Fabrication of biomaterials and optimization of structures
In tissue engineering, substrates have been used to provide the desired space for tissue regeneration during their temporary operation, promote cell proliferation and/or differentiation, and allow the entrance and exit of nutrients, drugs, and other bioingredients.59 In other words, scaffolds and biomaterials should match the target tissue in terms of shape and be able to temporarily evoke the function of the target tissue. In bone tissue, for instance, the scaffold’s spatial shape should be either extended or wide, depending on the type of bone and the planned location. Also, from a structural point of view, the fabricated structure must have the capability to mimic the characteristics of the bone, including mechanical resistance, stiffness, tension, etc.60,61
The proliferation and differentiation of cells in scaffolds is another aspect that should be considered in biomaterials design. For example, it has been reported that scaffolds with high porosity can promote vascularization.62 Likewise, special attention has recently been paid to electrically conductive biomaterials in the field of nerve tissue engineering.3,63,64 These and similar examples demonstrate how crucial scaffold design and biomaterial optimization are for tissue engineering. In the following, we discuss the application of MD simulations in the self-assembly-based fabrication and optimization of scaffold structures used in tissue engineering.
Polymeric self-assembly
Tissue engineering scaffolds can be fabricated through both top-down and bottom-up approaches. The bottom-up fabrication methods are based on self-assembly processes through which disordered systems with separate molecules spontaneously turn into ordered structures via different local interactions.65,66 This process is the underlying mechanism for forming most of the nanostructures in biological systems, including cell membranes, the helical structure of DNA, and ribosomes. Moreover, several other critical biological functions, such as the interaction of a ligand with its receptor, depend on self-assembly.67 Self-assembly can be used to fabricate a variety of nanostructures, such as nanotubes and nanofibers, using different sorts of components, including amino acids, oligo- and polypeptides, nucleic acids, polymers, etc.68,69 The main forces involved in the self-assembly process are the attractive driving forces, repulsive opposition forces, and directional forces. The balance between attractive and repulsive forces initiates the formation of self-assembled aggregates (Fig. 1).69,70 Given its widespread application in the fabrication of tissue engineering scaffolds, understanding the self-assembly molecular details helps in the rational design of novel and more effective nanostructures. MD simulations have the potential to facilitate the exploration of the physicochemical properties of peptides, as well as their intricate interactions with solvents. It would enable researchers to investigate the stability of peptide molecules and how it affects their self-assembling capability, thereby facilitating the development of functional peptide scaffolds for tissue engineering.20 Here we have reviewed several important MD simulations of self-assembled structures used in tissue engineering.
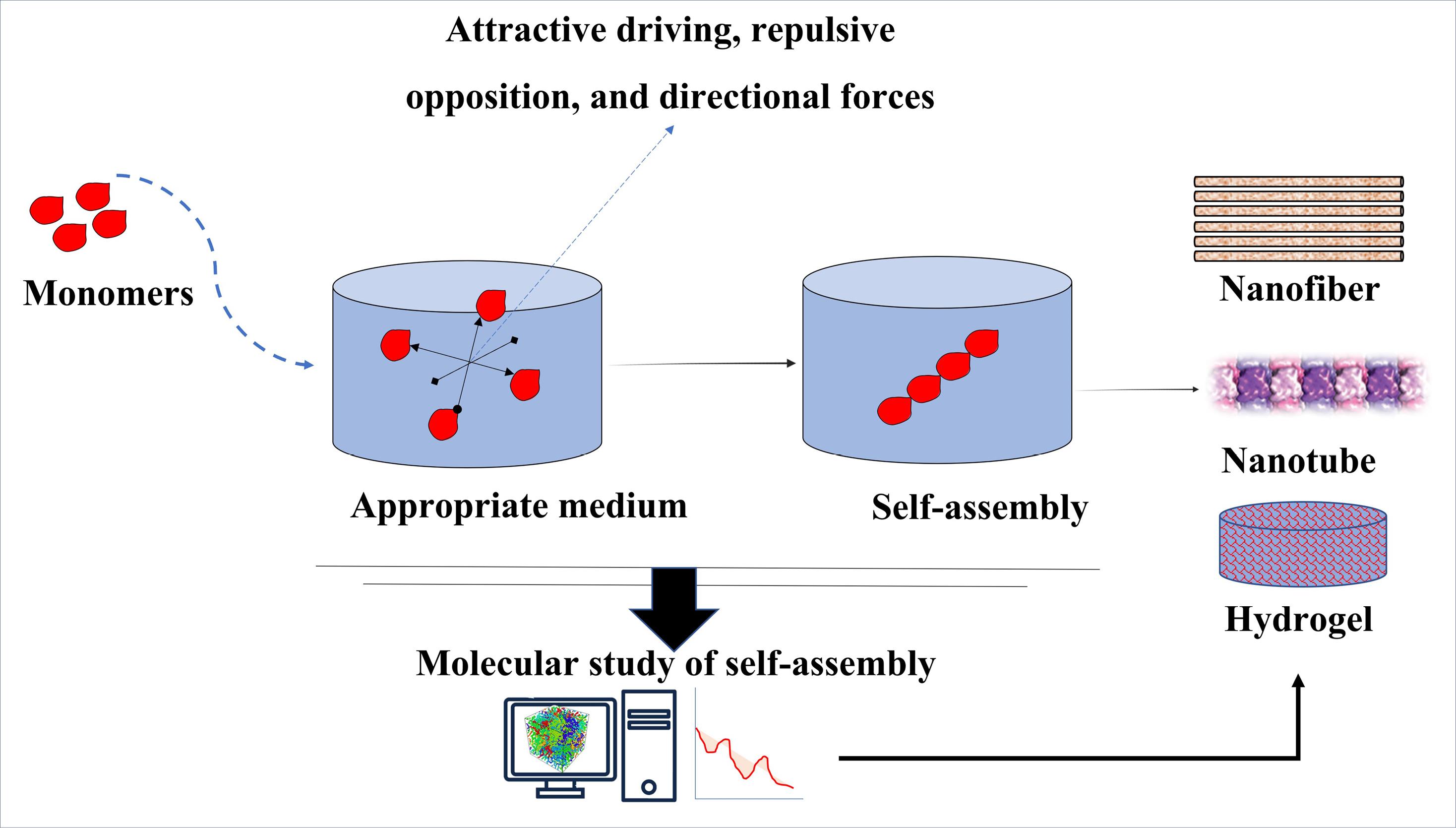
Fig. 1.
Application of MD simulations in studying the mechanism or predicting the outcome of self-assembly process for the fabrication of various nanostructures in tissue engineering. In the MD simulation, the important forces involved in the self-assembly process are investigated, the most important of these forces are attractive driving, repulsive opposition and directional forces. These forces are crucial in the final formation of the structure, which can be in the form of hydrogel, nanotube, nanofiber, etc.
.
Application of MD simulations in studying the mechanism or predicting the outcome of self-assembly process for the fabrication of various nanostructures in tissue engineering. In the MD simulation, the important forces involved in the self-assembly process are investigated, the most important of these forces are attractive driving, repulsive opposition and directional forces. These forces are crucial in the final formation of the structure, which can be in the form of hydrogel, nanotube, nanofiber, etc.
Peptide amphiphiles (PAs) are a group of materials of interest for fabricating functional structures through self-assembly.71 Researchers have used these self-assembling PAs to fabricate nanofiber structures that can be employed in different areas of tissue engineering, including forming blood vessels, wound healing, bone and cartilage regeneration, and axon regeneration.72
Through atomistic MD simulations, Lee et al investigated the self-assembly of an IKVAV-bearing peptide amphiphile into cylindrical nanofibers.21 They used the CHARMM force field to relax a bunch of PAs with cylindrical configurations to observe the formation of a stable nanofiber in their simulation. The initial structure consisted of 16 layers, each containing 9 PA molecules arranged radially, with the alkyl chains oriented toward the interior of the fiber. Each layer was rotated by 20 degrees in relation to the layer before it, and there was a 5-angstrom gap between successive layers. Simulation was performed in explicit water in physiologic concentration of ions and under periodic boundary condition (PBC). The system was simulated for 40 ns using the NPT ensemble and Langevin dynamics at a temperature of 310 K and pressure of 1 atm. At the end of the simulation, the resulting nanofiber demonstrated close alignment with the laboratory findings regarding both its diameter and the peptide’s secondary structure. From structural point of view, they revealed that despite the PAs' similar amino acid sequences, each PA molecule had its own secondary structure, including α-helix, β-sheet, turn, and/or coil. Moreover, they have reported that the epitope sequence IKVAV is located on the surface of the nanofiber. Considering that the IKVAV epitope fragment promotes neurite sprouting and is responsible for cell adhesion, a crucial process in tissue engineering,73,74 their exposure at the surface of the fibers is consistent with their intended design to stimulate neurite growth.
Tekin et al19 used united atom MD simulations to investigate the structural properties of various PA-based cylindrical nanofibers with different arrangements of PA. The peptide amphiphile (PA) molecule studied consisted of a hydrophobic alkyl chain (C12) attached to the N-terminus of peptide sequence VVAGERGD. To form the nanofibers, layers of PA molecules were stacked, with each layer featuring PA molecules arranged radially, positioning the peptide portion outward and the alkyl portion inward. Various starting structures of nanofibers were constructed by using different number of PA molecules per layer to determine the most stable configuration. The simulations were conducted in explicit solvent with PBC at 300 K and 1 bar. For peptide and alkyl segments of PA molecules the GROMOS 53a6 force field and Berger lipid parameters were used respectively. GROMACS software version 4.5.6 was used to run 30 ns or 50 ns simulations in NPT ensemble. They have reported that among various initial configurations, the 19-layered nanofiber containing 12 PAs per layer was the most stable, consistent with experimental findings. Further investigation showed that random coils and β-sheets were the most dominant secondary structures formed in the self-assembled PA molecules, respectively. Moreover, hydrophobic interactions between the VVAG–VVAG moieties of the PA molecules and electrostatic interactions between aspartate and Na+, as well as between glutamate and arginine, were demonstrated to be the leading forces of fiber self-assembly. This study elucidated the forces existing between the components of self-assembled structure, which can guide the design and development of more efficient frameworks for specific tissue engineering purposes.
Hydrogels are another important class of self-assembling structures in tissue engineering due to their high similarity to natural extracellular matrix (ECM).75,76 As a result, many studies have focused on the development of nanofibrous hydrogels to create novel structures that mimic the natural ECM.77,78 Synthetic polypeptide nanofibers can support various critical aspects of tissue engineering, including promoting cell adhesion and regulating cell behaviors.20 It is also possible to enhance the biological properties of hydrogels by blending them with functional polypeptides.79 The design of such biomimetic polypeptide structures requires a better understanding of fiber formation mechanisms.
Sun et al20 investigated the self-assembly of biphenyl-tripeptides for the fabrication of nanofiber hydrogels for tissue engineering. The researchers synthesized six tripeptides of sequence FFG, FFA, FGF, FAF, GFF and AFF, all conjugated with biphenylacetic acid (BPAA), to examine their potential to form nanofibers through self-assembly. The experimental findings revealed that peptides containing 'FF' blocks were capable of forming transparent hydrogels, whereas the two other peptides consisting of BPAA-tripeptide sequences resulted in precipitation. To better understand the process of self-assembly in short peptides, MD simulations were utilized to examine their structural characteristics and conformational dynamics by simulating the aggregation of multiple peptides in a single system. For each BPAA-tripeptide, eight molecular models with random positions were solvated in simulation box containing physiological concentration ions. MD simulations were run for 100 ns in NVT ensemble at 310 K using the NAMD program, with the merged CHARMM36 force field applied to describe the potential functions of BPAA-tripeptides. The progress of self-assembling processes was measured by counting the number of peptides in each peptide aggregate over time. According to their MD simulation study, the “FF” brick (phenylalanine-phenylalanine) interactions were the key drivers for the self-assembly of the nanofibers. However, excessive intramolecular hydrogen bonds in some of the peptide sequences (BPAA-FAF and BPAA-FGF) hindered the formation of intermolecular hydrogen bonds, leading to compact aggregates and precipitations.
These example studies demonstrate how powerful a tool MD simulation can be for the detailed investigation of structural features, such as the position of functional moieties, as well as the forces governing the self-assembly process of fibers. This information, which is difficult to obtain due to the inherent limitations of laboratory studies, can be used to design more efficient self-assembling fibers with predefined applications in tissue engineering.
Adsorption and desorption of molecules on substrates
With the introduction of new scientific and technological disciplines, such as novel drug delivery and tissue engineering techniques, the use of more complex systems involving biomolecules and other materials has become more common. The interaction of biomolecules with material surfaces can significantly impact the physico-chemical and biological properties of the materials, which can then be used to direct a variety of processes in tissue engineering. Binding of proteins, lipids, DNA components, etc., to the substrate can also regulate adhesion, migration, proliferation, and differentiation of cells.
In the case of NPs, the adsorption of proteins on their surfaces forms complex structures known as nanoparticle-protein corona (NP-PC), which significantly influence the biological reactivity of NPs.80,81 Previous studies have shown that the structural arrangement of biomolecules on NPs can change the biological activity of cells with which they interact (Fig. 2). Here, the contact surface characteristics of NPs play an important role in changing the biological activity of biomolecules.82,83 Hence, scientists focus on NP surface design for controlled adsorption of specific biomolecules for various applications, including bioimaging and biosensing. Another primary application of modifying NP surface is to limit the adsorption of biomolecules, a common strategy to prevent them from being recognized by the immune system, as has been shown, for example, with gold NPs (AuNPs).84-87
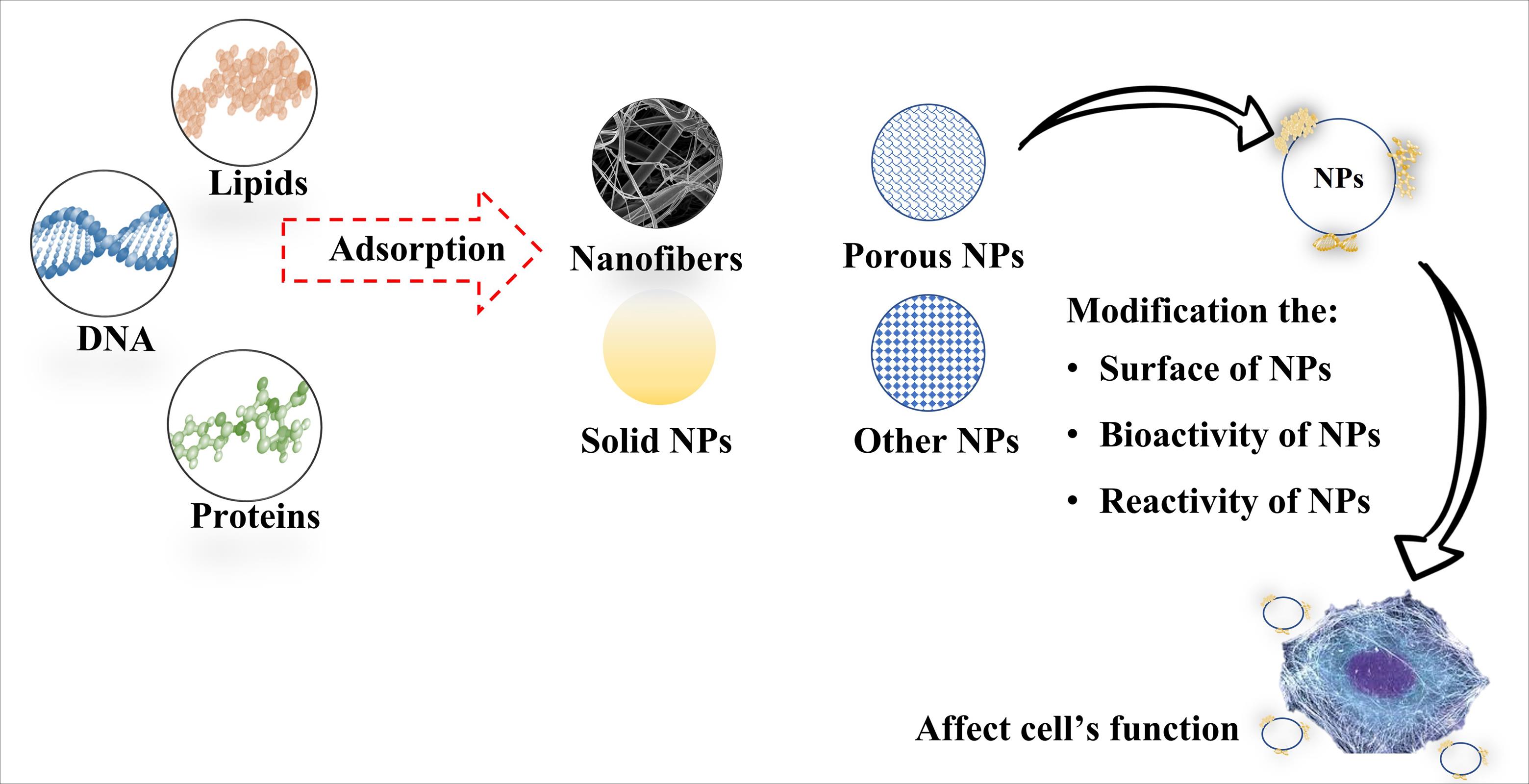
Fig. 2.
Different NPs are used in tissue engineering. Adsorption of different biomolecules, including proteins, DNA, and lipids can lead to changes in structure, reactivity, bioactivity, bioavailability, biocompatibility, and biodistribution of NPs, thereby modifying cells’ function. MD simulation can provide a deep insight into the interaction between molecules and nanostructures. This information can be used in preparing and optimizing structures in tissue engineering based on hydrophilicity and hydrophobicity and cell adhesion, which can ultimately affect cell fate.
.
Different NPs are used in tissue engineering. Adsorption of different biomolecules, including proteins, DNA, and lipids can lead to changes in structure, reactivity, bioactivity, bioavailability, biocompatibility, and biodistribution of NPs, thereby modifying cells’ function. MD simulation can provide a deep insight into the interaction between molecules and nanostructures. This information can be used in preparing and optimizing structures in tissue engineering based on hydrophilicity and hydrophobicity and cell adhesion, which can ultimately affect cell fate.
Scientists can achieve precise surface design of materials to control the interaction with biomolecules by employing various strategies. For instance, surface modification techniques involve functionalizing the surfaces of materials with specialized chemical groups and/or biomolecules to make customized properties for biomolecular interactions. Besides, adjusting surface topography down to the micron and nanometer scales will also determine biomolecular behavior including cell attachment and proliferation. Finally, using a bio-mimetic approach that leads to recreating the extracellular matrix environment of cells on the surfaces of materials will allow for control or directing a controlled attachment and proliferation of specific types of cells.88-90
There are different methodologies that are being explored to tailor biomolecules-materials interactions for specific tissue engineering applications, given the diversity of biomolecules and material surface characteristics. Coating materials with peptides or proteins is a common strategy for modifying material surfaces to improve their biological performance. For example, the RGD (arginine-glycine-aspartic acid) peptide sequence is commonly used to promote cell adhesion and spreading on material surfaces due to its high affinity for integrin receptors on cells.91 As another example, fibronectin coating on implants promotes osteoblast adhesion and differentiation, which enhances osseointegration.92 As mentioned above, creating nanostructures or patterns on the material surface to control the cell-material interaction is another strategy. For example, nanotopographical features on a substrate can promote cell attachment and differentiation of human osteoblast-like cells compared to a flat surface.93
Proper binding of proteins, such as growth factors, to NP can be critical to their activity. Bone morphogenetic protein-2 (BMP-2) is a promising osteogenetic protein in bone tissue engineering that was approved by FDA in 2002 for bone defect reconstruction and spinal fusion.94-96 One of the main challenges in delivering growth factors appropriately to the target site is to preserve their biological activity. Huang et al evaluated the adsorption and desorption of bone morphogenetic protein-2 on textured hydroxyapatite surfaces (HAP) by conventional and steered MD simulation using GROMACS with the OPLS-AA force field.16 According to their results, compared to the flat model, the HAP-1:1 model (with ridges and grooves at a ratio of 1:1) showed more robust adsorption stability, lower deformation of the BMP-2 molecule upon interaction with the HAP-1:1 model, and higher stability of cysteine-knots of the BMP-2 dimer on the HAP-1:1 model, resulting in higher biological activity of this protein on the HAP-1:1 surface. These findings highlighted the importance of optimizing the adsorption of biomolecules on NP for tissue engineering.
The adsorbed biomolecules play a critical role in the effective implantation and operation of nanosystems in tissue engineering. Among the biomolecules that are used in tissue engineering, fibronectin has unique properties in that it can modulate cell fate and binds to different biomolecules, e.g., collagen,97 fibrin,98 and a variety of growth factors,99 leading to fibrillogenesis and influencing essential cellular processes.100 Hao et al have provided mechanistic insights into the adsorption and bioactivity of fibronectin on surfaces with varying chemistries.15 They used self-assembled monolayers (SAMs) on gold with methyl (-CH3), amino (-NH2), carboxyl (-COOH), and hydroxyl (-OH) groups to replicate various chemical groups found on the surfaces of biomaterials. Fibronectin was subsequently adsorbed onto the surfaces of SAMs. Their results indicated that electrostatic interactions accounted for a considerable portion of the interactions. In SAMs-NH2, the polar interactions led to tight binding of Fn in the “side-on” orientation. On the other hand, they found that the Fn adsorbed on SAMs-CH3 had a poor ability to promote cell proliferation due to the low solvent accessible surface area (SASA) of the RGD and PHSRN motifs as well as the deformation of the protein. However, efficient adsorption of Fn on SAMs-NH2 and exposure of its bioactive sites (RGD and PHSRN) were reported to be the cause of its enhanced bioactivity for cell proliferation, integrin β1 expression, and osteogenic differentiation.15
In another study, Raffaini and Ganazzoli investigated Fn adsorption on graphite through MD simulation to elucidate adsorption mechanism of proteins on hydrophobic surfaces. They demonstrated that the protein adsorption is quantitatively independent of the initial orientation of the domain approaching the surface. Adsorption in its early stages is accompanied by local rearrangement and possibly the loss of some of the secondary structures, especially α-helix structures, in the regions near the surface. After this stage, the protein starts to unfold and increase its contact with the surface as much as possible.17
One of the most important aspects of tissue engineering is cell adhesion to the substrate, which is a vital process in cell viability, proliferation, and differentiation. Biswas et al have investigated the hydration and interaction of cell-adhesion peptides, specifically RGD and YIGSR, with the hydroxyapatite surface and TiO2 surface through MD simulation.18 They found that the initial peptide orientation significantly affects the adsorption energy. Their results showed that YIGSR adsorption on HA-(001) surfaces was more robust than that of RGD. However, the RGD structure has been demonstrated to maintain its “hairpin"-like structure following adsorption on a flat HA-(001) surface but adopts a slightly "relaxed hairpin" structure on the TiO2 (110) surface. They came to the conclusion that titanium oxide is a good candidate for tissue engineering in situations where tissue regeneration occurs through cell signaling because RGD on titanium oxide has a more favorable adsorption energy than HA does.
As evidenced by the studies cited above and related investigations, the interaction of biomolecules with various material surfaces has a significant effect on their biological properties. Furthermore, various aspects of these interactions, such as binding affinity, biomolecule conformational changes, and so on, depend on the type of biomolecule, the material surface chemistry, and topological features. For tissue engineering applications, the precise surface design of materials allows for greater control over their interaction with biomolecules (e.g., growth factors and adhesion proteins) and, as a result, better regulation of cell attachment and proliferation.
Drug delivery in tissue engineering
One of the challenges in tissue engineering is delivering growth factors and other signaling molecules to stem cells at the site of tissue damage. These molecules are important for promoting the proliferation and differentiation of stem cells into specific cell types. Nanoparticles can be engineered to carry drugs or other molecules to specific locations in the body. One advantage of using nanoparticles for drug delivery is that they can be engineered to release their cargo over a longer period of time at a given site, which can improve the overall success of tissue engineering treatments.
Delivering an adequate dosage of a drug to the intended tissue while minimizing any accompanying side effects constitutes a major hurdle in drug delivery systems, necessitating interdisciplinary collaboration to overcome it. In drug delivery, different factors must be considered, including the conditions of the target tissue, such as its anatomical location, the environmental pH of the target site, the type of the treatment, such as immunotherapy, cancer therapy etc., and the DDS targeting strategy.101-104 Here, molecular investigation of DDs from various aspects can advance our knowledge to develop more efficient systems.
Hasani-Sadrabadi et al conducted a study to evaluate the effectiveness of microfluidic synthesized chitosan NPs containing dexamethasone (CS-Dex) on the osteogenic differentiation of mesenchymal stem cells (MSCs). They investigated the fabricated drug delivery systems by MD simulation in order to study the interactions of Dex with the CS NPs. According to their results, Dex molecules moved toward CS chains at the end of the simulation, indicating an affinity between Dex molecules and Cs chains. Energy analysis of intermolecular interactions (van der Waals and electrostatic), hydrogen bonds, and radial distribution function (RDF), all indicated favorable interactions between chitosan strands and Dex molecules. The authors believed that the strong interaction between the drug and chitosan NPs was the cause of the high loading of Dex in the chitosan NPs and its prolonged release.24
Despite the importance of using MD simulation in studying drug delivery systems used in tissue engineering, there are unfortunately very few examples of this type of research available at present. However, due to the ability of MD simulation to help us understand the factors that affect the drug's molecular affinity for the carrier, as well as various factors involved in drug release, it is expected that we will see more and more applications of this technique in optimizing the delivery of drugs and differentiation factors in the future.
Cell proliferation
Cell proliferation is a process that multicellular organisms use to increase their cell numbers and replace dead cells.105 Cell proliferation occurs through a highly organized process called the cell cycle, leading to the copying of older cells into newer ones.106,107 The whole system's survival is the primary objective of cell proliferation. Depending on the requirements of the system, certain groups of cells proliferate independently from other cells. Of course, some cells that are already fully differentiated lose their ability to divide and are unable to participate in this process. These tissues, such as neural tissue are referred to as non-dividing tissues. But, the majority of tissues routinely use cell proliferation to renew themselves, including skin and bone marrow.105
In tissue engineering, cell attachment to the substrate surface is very important for cell proliferation.108 An ideal 3D microenvironment for cell attachment and proliferation can be created by selecting a scaffold or substrate with the right combination of surface properties such as charge, roughness, chemical functionalities, and hydrophobicity/hydrophilicity.109,110 For example, in bone tissue engineering, weak cell adhesion leads to poor proliferation on the surface of the engineered scaffold, causing poor incorporation of the implant, infections, inflammations, and even complete implant failure.111 The molecular mechanisms of cell adhesion on artificial materials can be divided into “direct non-receptor-mediated cell-material binding” occurs through weak forces, such as hydrogen bonding, electrostatic, polar, or ionic interactions between various molecules on the cell membrane and functional chemical groups on the polymers while “receptor-mediated binding” occurs through ECM molecules or their parts, such as fibronectin, vitronectin, collagen, or laminin (Fig. 3).112 Given the importance of scaffold surface characteristics in determining the quality of cell attachment and proliferation, understanding the cell-surface interaction on a molecular scale will greatly help design suitable scaffolds for tissue engineering.
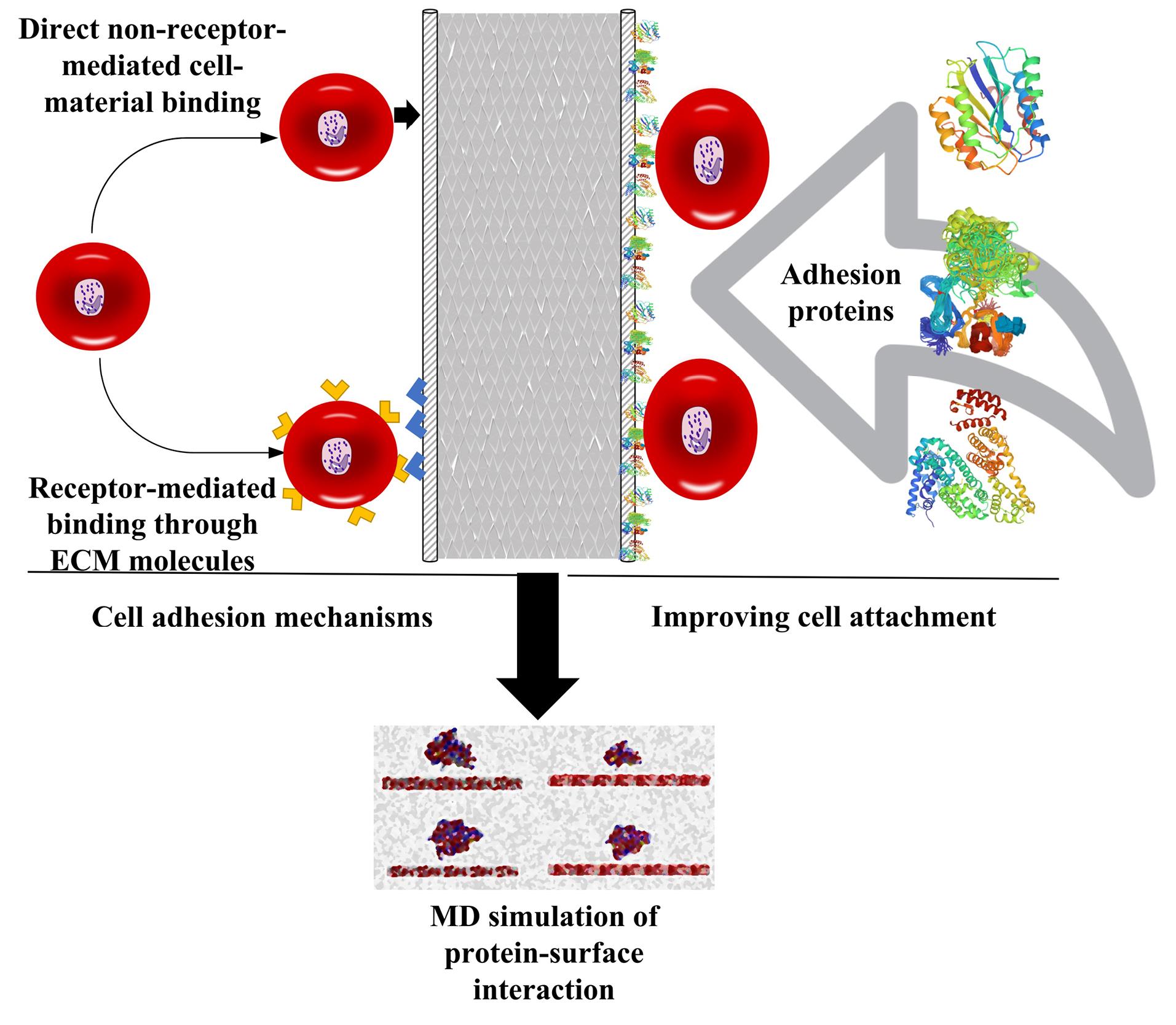
Fig. 3.
Cell attachment mechanism on scaffolds and improving it through protein adhesion which leads to enhanced cell proliferation. One of the most important factors that affect cell proliferation is the surface of cell culture substrates. Different studies have shown that by surface modification, cell proliferation can be optimized in tissue engineering based on a defined goal. To improve cell adhesion and subsequently promote cell proliferation, substrates modified with proteins can be used, which cause better cell attachment and adhesion to the surface of the structure.
.
Cell attachment mechanism on scaffolds and improving it through protein adhesion which leads to enhanced cell proliferation. One of the most important factors that affect cell proliferation is the surface of cell culture substrates. Different studies have shown that by surface modification, cell proliferation can be optimized in tissue engineering based on a defined goal. To improve cell adhesion and subsequently promote cell proliferation, substrates modified with proteins can be used, which cause better cell attachment and adhesion to the surface of the structure.
Shamloo and Sarmadi evaluated the adhesive characteristic of polymer-protein systems and their correlation with cell adhesion and proliferation through molecular dynamics simulation.113 They calculated the work of adhesion and peeling force of various biomaterial-protein systems using MD simulation. These systems included polycaprolactone (PCL) and polyvinyl alcohol (PVA) polymers, as well as different ECM protein fragments like collagen type-I and fibronectin, and two subdomains of human serum albumin (HSA). In the experimental part of their study, they evaluated cell proliferation and bonding using bone marrow cells on PCL/PVA electrospun scaffolds. They compared the MD simulation results with the experimental findings to reveal any possible correlation between them. Their MD findings showed that proteins adhered strongly to the PCL surface due to its high surface hydrophobicity, but only one albumin subdomain adhered properly to the PVA surface. Consistent with these findings, the cell proliferation assay revealed a higher cell attachment and proliferation rate on PCL compared to the PVA scaffold. They therefor concluded that there is a direct link between stronger adhesion of the aforementioned proteins to the scaffold surface and cell proliferation.113 In another study, they examined the various ratios of PCL and PVA in greater detail and found that samples with more than 50% of PCL exhibited stronger protein adsorption, which was consistent with their simulation results.114
Few studies have been carried out regarding investigating optimal conditions for cell proliferation using MD simulation. But it is undeniable that promoting cell adherence to the tissue engineering substrates is a crucial element in cell proliferation. Further research is needed to confirm the efficacy of MD simulations in all systems, but the aforementioned studies show that it is possible to use MD simulation to successfully determine the optimal conditions for cell adhesion to the scaffold and cell proliferation.
Cell differentiation
The primary purpose of tissue engineering is to regenerate the damaged or destroyed tissue and return it to its normal function. This means that, components of tissue engineering need to be optimized so that they can best stimulate cultured cells to restore function and renew the host tissue. During this process, stem and stromal cells are transformed into a specific type of mature cells through a biological process called cell differentiation. Biological, chemical, and environmental growth factors are just a few of the many elements that play a role in this process. Along with the mentioned factors, ECM also plays a crucial role in deciding cell fate. In the differentiation process, extracellular signals are required. Certain factors are secreted during tissue repair and make their way into the (ECM) and affect injured sites for tissue formation.115,116 Given this, it is crucial to investigate, at the molecular level, how growth factors interact with the intracellular signaling pathways involved in cell differentiation. Recent studies have shown that mechanical and physical stimuli such as substrate pattern, surface hardness, mechanical properties, and electromagnetic current induction can effectively determine cell fate and affect differentiation (Fig. 4). Therefore, it is important to study these stimuli in great detail in order to fully understand their role in the differentiation of cells.3,63,115 In this part of our review, we have summarized the studies that used MD simulations to investigate factors affecting cell differentiation.
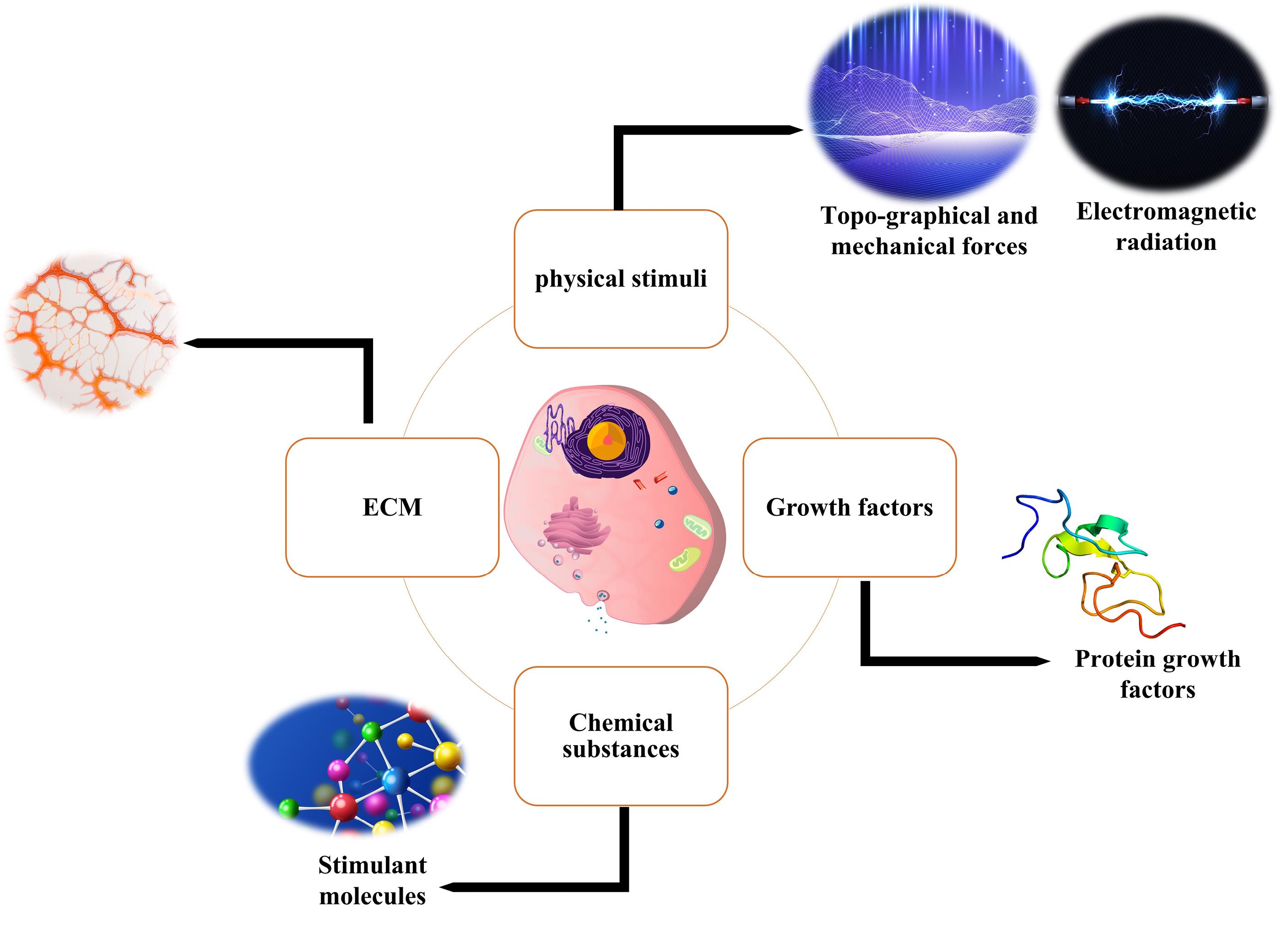
Fig. 4.
Different approaches for inducing cell differentiation. Cell differentiation is a very complex process in which different signaling pathways are involved. Among the most dominant approaches used for cell differentiation, we can mention the use of growth factors or smaller molecules, which under controlled conditions leads to cell differentiation into a specific type. Also, the cells’ ECM, which as an organic structure keeps cells in their natural niche, has emerged as an important factor in the process of cell differentiation; Therefore, today in tissue engineering, most of the researches are directed towards 3D substrates that provide a 3D space similar to a cell niche for cell differentiation. Also, the use of physical stimulation, including electrical stimulation, has been used in research to differentiate cells, although their mechanism is not exactly known, but it is mostly attributed to ionic changes inside the cell.
.
Different approaches for inducing cell differentiation. Cell differentiation is a very complex process in which different signaling pathways are involved. Among the most dominant approaches used for cell differentiation, we can mention the use of growth factors or smaller molecules, which under controlled conditions leads to cell differentiation into a specific type. Also, the cells’ ECM, which as an organic structure keeps cells in their natural niche, has emerged as an important factor in the process of cell differentiation; Therefore, today in tissue engineering, most of the researches are directed towards 3D substrates that provide a 3D space similar to a cell niche for cell differentiation. Also, the use of physical stimulation, including electrical stimulation, has been used in research to differentiate cells, although their mechanism is not exactly known, but it is mostly attributed to ionic changes inside the cell.
The distribution and density of the functional epitopes on a nanofiber's surface is one of the main determinants of how well it can promote cell differentiation, albeit this parameter may also have an impact on the fiber stability. Mehralitabar et al have shown the effect of this factor through MD simulation of a fiber made of the alkyl-peptides containing the epitope (FAQRVPP) that is effective in differentiating neural stem cells.22 They have compared the stability of two fiber structures in the forms of “all functionalized” nanofiber (containing only bioactive alkyl-peptides) and “distributed functionalized” nanofiber (a combination of non-bioactive and bioactive alkyl-peptides with a ratio 2:1). According to their results, the fully functionalized fiber had an unstable structure that broke up into micelle-like structures to reduce the steric hindrance between functional epitopes. In contrast, the nanofiber with distributed functional epitopes exhibited an integrated, stable structure containing a greater number of beta sheets that were neatly ordered and oriented around the hydrophobic core. Furthermore, the involvement of hydrophobic contacts in the formation of the alkyl-chain core, as well as that of hydrogen bonds and electrostatic interactions in the stability of the fiber structure, were also demonstrated in this simulation. The authors emphasized the application of MD simulation in the development of more effective nanofiber scaffolds for tissue engineering with various purposes.22
One type of physical stimulus that can be used to accelerate the process of cell differentiation is electromagnetic radiation. Terahertz radiation (THz) is present in the environment as part of the solar spectrum. Its energy level is within the range of hydrogen bonds, van der Waals interactions, and charge-transfer reactions associated with the molecular motions of biological molecules.117,118 Bock et al23 have evaluated the effect of terahertz radiation on stem cell reprogramming. According to this study's experimental section, the terahertz irradiation promoted stem cell differentiation toward an adipose phenotype. The underlying mechanisms have been attributed to the activation the transcription factor peroxisome proliferator-activated receptor gamma (PPARG). PPARG encodes a nuclear receptor protein belonging to the peroxisome proliferator-activated receptor (Ppar) family, a ligand-activated transcription factor that regulates adipocyte differentiation.119 They used MD simulation to investigate the possibility of a connection between terahertz radiation and PPARG. Their results showed that the local breathing dynamics, that is, the dynamics of the transient opening and closing of the DNA double helix in the PPARG promoter region, occur simultaneously with the gene-specific response to the THz radiation. These findings suggest that the terahertz radiation can modulate the DNA structure and accessibility, and thus affect the gene transcription. The authors propose that this mechanism may explain how terahertz radiation can induce cellular reprogramming.23
The mechanical and phase transition of the scaffold can affect cellular fate, including proliferation and differentiation.120 Developing a new ECM-like scaffold with favorable mechanical properties can be essential for tissue engineering. To the best of our knowledge, there is only one MD study that has evaluated the phase transition effect of ECM on cell differentiation. James et al used MD simulation to investigate a designed laminin-mimetic, elastin-like fusion protein as an artificial ECM. They specifically analyzed the temperature dependent structural/physical behavior of a candidate chimeric protein that contained a laminin globular-like (LG) domain joined to an elastin-like polypeptide (ELP). They reported that the fabricated ELP region constructed using the repeat sequence (VPGXG) tends to be completely flexible and can turn into β-rich secondary structures at temperatures in the physiological ranges (310-315 K). It is well known that the secondary structural features of a peptide correlate with the mechanical properties (density, stiffness, etc.) of peptide-based hydrogels. Also, due to the tendency of the ELP region to adopt β-rich secondary structure which affects its differential solvation, it has been proposed that altering its sequence can be used to systematically change the phase transition characteristics, and hence the overall functioning of this fusion protein.121 ELPs are involved in the proliferation and differentiation of various types of cells and can be used as cross-linked gel fibers or injectable scaffolds for tissue engineering.122 Given the effect of ELPs on cellular fate, MD simulation’s ability to predict some required and expected features of scaffolds can help improve tissue engineering outcomes.
In this section, we discussed the application of MD simulation to the study of the connection between tissue engineering elements and cell differentiation using some illustrative examples. In this regard, the use of simulations to understand the location and structure of nanofiber functional groups, the molecular basis of physical stimuli, and the prediction of ECM's mechanical characteristics was discussed. Despite the great importance of controlling the cell differentiation process in tissue engineering, few simulation studies have been done on this topic. Since various physical, chemical, and biological processes are involved in cell differentiation, there is a lot of room for MD studies in the field. Among them, we can highlight the study of the interaction of natural or engineered growth factors with the scaffold and cell receptors, as well as the role of the physicochemical characteristics of the substrate in the activation of molecular mechanisms involved in the process of cell differentiation. Also, due to computational and time constraints, MD simulation studies can be combined with in vitro studies to aid the investigation of the processes involved in cell differentiation.
Concluding remarks
The present work was conducted to review the applications of molecular dynamics simulation in tissue engineering studies. To the best of our knowledge, this study is the first of its kind, which is its strength. The authors tried to review the key aspects of tissue engineering that can be studied using MD simulations. A summary of the reviewed articles is shown in Table 1. We began this study by providing an overview of tissue engineering, its main components, and the necessity for developing such novel approaches when dealing with human disease, abnormalities, and defects. Following that, we covered the fabrication and optimization of nanostructures through self-assembly and adsorption processes, respectively. The significance of molecular dynamics simulation for studying the forces regulating the self-assembly process and predicting various aspects of self-assembled structures was explored. In addition to this, we investigated the adsorption of biological components on the substrates from the point of view of tissue engineering and reviewed the research that has been conducted in this area. Collectively, the results of such studies demonstrate the capability of MD simulations to predict and compare the binding strength of biomolecules used in tissue engineering, as well as to investigate the effect of the substrate's physical and chemical properties on the biological activity of these molecules. The use of MD simulation in the investigation of some effective factors in cell attachment to the substrate, an important factor in cell proliferation, was reviewed. There were few studies available regarding cell proliferation but it is clear from the reviewed studies that MD simulations can reliably predict the relationship between protein binding strength and substrate surface hydrophobicity, which is crucial for cell attachment and proliferation. Hence researchers can use MD simulation as a tool to predict ideal substrates to stimulate cell proliferation. Finally, we reviewed the use of MD simulation in cell differentiation studies, looking at topics such as the effect of physical stimuli (such as terahertz waves) on differentiation and the role of simulation in assisting the design of engineered ECM with suitable physical characteristics for cell differentiation. It should be acknowledged that this study had some limitations, the most important of which is the lack of sufficient studies in this field and their dispersion, which makes it challenging to group the studies and assign them to a relevant topic. There are many gaps in our knowledge regarding the basic molecular event in cell interaction with substrates or its microenvironment. In addition, the precise mechanisms by which different surfaces guide cells to a specific fate are still not well understood. This is where MD simulation can be used to answer these questions. Based on this, the MD simulation method’s capabilities can greatly aid in the design of more efficient substrates, better control of differentiation processes, and overall improvement of tissue engineering practices. There are some challenges in the application of MD simulations in tissue engineering. MD simulations require a large number of computational resources and time to simulate complex biological systems with realistic details and accuracy. This problem is more pronounced in the case of simulations related to tissue engineering, because here the systems are larger and have more components. One way to increase the speed of calculations is to use coarse-grained models which simplify the representation of the atoms and molecules in the system by grouping them into larger units or beads. This reduces the number of degrees of freedom and the complexity of the interactions, allowing for longer and larger simulations. Another challenge is the lack of special force fields for such simulations. It is challenging to develop and validate force fields that are suitable for tissue engineering applications, such as modeling biomaterials, bioactive molecules, or cell-biomaterial interactions. To overcome this challenge, researchers are developing more accurate force fields, using machine learning methods or experimental data to optimize the force field parameters. Apart from technical challenges in simulations, our understanding of the intricacies of cellular processes such as adhesion, proliferation, and differentiation is incomplete at present. Without a comprehensive understanding of the interplay between different molecules and molecular pathways that underlie cellular behavior, it is impossible to accurately predict how cells will respond to different environmental factors through simulation. As scientific knowledge expands and we gain a more detailed picture of the molecular-level events that take place within cells, we can expect significant progress in the development of molecular simulations for tissue engineering applications.
Review Highlights
What is the current knowledge?
√ The use of MD simulations to understand time-dependent behavior of systems at the molecular scale
√ The application of MD simulations in drug design, protein engineering, etc.
What is new here?
√ Using MD simulations to predict the interaction between biomolecules and material surfaces
√ Using MD simulations to understand substrate properties' impact on cell attachment and proliferation.
√ Using MD simulations to understand the molecular mechanism of cell differentiation stimuli.
√ Capabilities of MD simulations in designing ideal substrates for tissue engineering.
Competing Interests
The authors have no conflicts of interest to declare.
Ethical Statement
Not applicable.
References
- Kim YH, Ha KY, Kim SI. Spinal Cord Injury and Related Clinical Trials. Clin Orthop Surg 2017; 9:1-9. doi: 10.4055/cios.2017.9.1.1 [Crossref] [ Google Scholar]
- Barati G, Nadri S, Hajian R, Rahmani A, Mostafavi H, Mortazavi Y. Differentiation of microfluidic-encapsulated trabecular meshwork mesenchymal stem cells into insulin producing cells and their impact on diabetic rats. J Cell Physiol 2019; 234:6801-9. doi: 10.1002/jcp.27426 [Crossref] [ Google Scholar]
- Nekouian S, Sojoodi M, Nadri S. Fabrication of conductive fibrous scaffold for photoreceptor differentiation of mesenchymal stem cell. J Cell Physiol 2019; 234:15800-8. doi: 10.1002/jcp.28238 [Crossref] [ Google Scholar]
- Biehl JK, Russell B. Introduction to stem cell therapy. J Cardiovasc Nurs 2009; 24:98-103; quiz 4. doi: 10.1097/JCN.0b013e318197a6a5 [Crossref] [ Google Scholar]
- Dunbar CE, High KA, Joung JK, Kohn DB, Ozawa K, Sadelain M. Gene therapy comes of age. Science 2018; 359. 10.1126/science.aan4672.
- Hasan A, Morshed M, Memic A, Hassan S, Webster TJ, Marei HE. Nanoparticles in tissue engineering: applications, challenges and prospects. Int J Nanomedicine 2018; 13:5637-55. doi: 10.2147/ijn.S153758 [Crossref] [ Google Scholar]
- Mandrycky C, Phong K, Zheng Y. Tissue engineering toward organ-specific regeneration and disease modeling. MRS Commun 2017; 7:332-47. doi: 10.1557/mrc.2017.58 [Crossref] [ Google Scholar]
- Khodabukus A. Tissue-Engineered Skeletal Muscle Models to Study Muscle Function, Plasticity, and Disease. Front Physiol 2021; 12:619710. doi: 10.3389/fphys.2021.619710 [Crossref] [ Google Scholar]
- Amer LD, Mahoney MJ, Bryant SJ. Tissue engineering approaches to cell-based type 1 diabetes therapy. Tissue Eng Part B Rev 2014; 20:455-67. doi: 10.1089/ten.TEB.2013.0462 [Crossref] [ Google Scholar]
- Barkur S, Lukose J, Chidangil S. Probing Nanoparticle-Cell Interaction Using Micro-Raman Spectroscopy: Silver and Gold Nanoparticle-Induced Stress Effects on Optically Trapped Live Red Blood Cells. ACS Omega 2020; 5:1439-47. doi: 10.1021/acsomega.9b02988 [Crossref] [ Google Scholar]
- Casalini T, Limongelli V, Schmutz M, Som C, Jordan O, Wick P. Molecular Modeling for Nanomaterial-Biology Interactions: Opportunities, Challenges, and Perspectives. Front BioengBiotechnol 2019; 7:268. doi: 10.3389/fbioe.2019.00268 [Crossref] [ Google Scholar]
- Jackson Jackson, B M. On the time scale and time course of protein conformational changes. J Chem Phys 1993; 99:7253-9. doi: 10.1063/1.465418 [Crossref] [ Google Scholar]
- Fu IW, Markegard CB, Chu BK, Nguyen HD. The role of electrostatics and temperature on morphological transitions of hydrogel nanostructures self-assembled by peptide amphiphiles via molecular dynamics simulations. Adv Healthc Mater 2013; 2:1388-400. doi: 10.1002/adhm.201200400 [Crossref] [ Google Scholar]
- Haider A, Haider S, Rao Kummara M, Kamal T, Alghyamah A-AA, Jan Iftikhar F. Advances in the scaffolds fabrication techniques using biocompatible polymers and their biomedical application: A technical and statistical review. Journal of Saudi Chemical Society 2020; 24:186-215. doi: 10.1016/j.jscs.2020.01.002 [Crossref] [ Google Scholar]
- Hao L, Li T, Wang L, Shi X, Fan Y, Du C. Mechanistic insights into the adsorption and bioactivity of fibronectin on surfaces with varying chemistries by a combination of experimental strategies and molecular simulations. Bioact Mater 2021; 6:3125-35. doi: 10.1016/j.bioactmat.2021.02.021 [Crossref] [ Google Scholar]
- Huang B, Lou Y, Li T, Lin Z, Sun S, Yuan Y. Molecular dynamics simulations of adsorption and desorption of bone morphogenetic protein-2 on textured hydroxyapatite surfaces. Acta Biomater 2018; 80:121-30. doi: 10.1016/j.actbio.2018.09.019 [Crossref] [ Google Scholar]
- Raffaini G, Ganazzoli F. Molecular dynamics simulation of the adsorption of a fibronectin module on a graphite surface. Langmuir 2004; 20:3371-8. doi: 10.1021/la0357716 [Crossref] [ Google Scholar]
- Biswas S, Becker UJJoB, Nanobiotechnology Nanobiotechnology. Molecular modeling of cell adhesion peptides on hydroxyapatite and TiO 2 surfaces: Implication in biomedical implant devices. J biomaternanobiotechnol 2013; 4:351. doi: 10.4236/jbnb.2013.44044 [Crossref] [ Google Scholar]
- Tekin ED. Molecular dynamics simulations of self-assembled peptide amphiphile based cylindrical nanofibers. RSC Advances 2015; 5:66582-90. doi: 10.1039/C5RA10685K [Crossref] [ Google Scholar]
- Sun Y, Li X, Zhao M, Chen Y, Xu Y, Wang K. Bioinspired supramolecular nanofiber hydrogel through self-assembly of biphenyl-tripeptide for tissue engineering. Bioactive Materials 2022; 8:396-408. doi: 10.1016/j.bioactmat.2021.05.054 [Crossref] [ Google Scholar]
- Lee O-S, Stupp SI, Schatz GC. Atomistic Molecular Dynamics Simulations of Peptide Amphiphile Self-Assembly into Cylindrical Nanofibers. Journal of the American Chemical Society 2011; 133:3677-83. doi: 10.1021/ja110966y [Crossref] [ Google Scholar]
- Mehralitabar H, Taghdir M, Naderi-Manesh H. A combination of bioactive and nonbioactive alkyl-peptides form a more stable nanofiber structure for differentiating neural stem cells: a molecular dynamics simulation survey. J Biomol Struct Dyn 2019; 37:3434-44. doi: 10.1080/07391102.2018.1516571 [Crossref] [ Google Scholar]
- Bock J, Fukuyo Y, Kang S, Phipps ML, Alexandrov LB, Rasmussen K. Mammalian stem cells reprogramming in response to terahertz radiation. PLoS One 2010; 5:e15806. doi: 10.1371/journal.pone.0015806 [Crossref] [ Google Scholar]
- Hasani-Sadrabadi MM, Hajrezaei SP, Emami SH, Bahlakeh G, Daneshmandi L, Dashtimoghadam E. Enhanced osteogenic differentiation of stem cells via microfluidics synthesized nanoparticles. Nanomedicine 2015; 11:1809-19. doi: 10.1016/j.nano.2015.04.005 [Crossref] [ Google Scholar]
- Hansson T, Oostenbrink C, van Gunsteren W. Molecular dynamics simulations. CurrOpin Struct Biol 2002; 12:190-6. doi: 10.1016/s0959-440x(02)00308-1 [Crossref] [ Google Scholar]
- Karplus M, Petsko GA. Molecular dynamics simulations in biology. Nature 1990; 347:631-9. doi: 10.1038/347631a0 [Crossref] [ Google Scholar]
- MacKerell AD. Empirical Force Fields. In: Xu Y, D Xu, J Liang, editors. Computational Methods for Protein Structure Prediction and Modeling: Volume 1: Basic Characterization. New York, NY: Springer New York; 2007. p. 45-69. 10.1007/978-0-387-68372-0_2.
- Leach AR. Molecular Modelling: Principles and Applications: Prentice Hall; 2001.
- Sun H. COMPASS: An ab Initio Force-Field Optimized for Condensed-Phase ApplicationsOverview with Details on Alkane and Benzene Compounds. J Phys Chem B 1998; 102:7338-64. doi: 10.1021/jp980939v [Crossref] [ Google Scholar]
- Jorgensen WL, Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc 1988; 110:1657-66. doi: 10.1021/ja00214a001 [Crossref] [ Google Scholar]
- MacKerell AD, Jr. Jr., Bashford D, Bellott M, Dunbrack RL, Jr. Jr., Evanseck JD, Field MJ. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J Phys Chem B 1998; 102:3586-616. doi: 10.1021/jp973084f [Crossref] [ Google Scholar]
- Guvench O, Mallajosyula SS, Raman EP, Hatcher E, Vanommeslaeghe K, Foster TJ. CHARMM Additive All-Atom Force Field for Carbohydrate Derivatives and Its Utility in Polysaccharide and Carbohydrate–Protein Modeling. J Chem Theory Comput 2011; 7:3162-80. doi: 10.1021/ct200328p [Crossref] [ Google Scholar]
- Klauda JB, Venable RM, Freites JA, O'Connor JW, Tobias DJ, Mondragon-Ramirez C. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B 2010; 114:7830-43. doi: 10.1021/jp101759q [Crossref] [ Google Scholar]
- van Gunsteren WF, Billeter S, Eising A, Hünenberger P, Krüger P, Mark A. Biomolecular simulation: the GROMOS96 manual and user guide. Hochschulverlag AG an der ETH Zürich, Zürich 1996; 86:1-1044. [ Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. Journal of the American Chemical Society 1995; 117:5179-97. doi: 10.1021/ja00124a002 [Crossref] [ Google Scholar]
- Heinz H, Lin T-J, Kishore Mishra R, Emami FS. Thermodynamically Consistent Force Fields for the Assembly of Inorganic, Organic, and Biological Nanostructures: The INTERFACE Force Field. Langmuir 2013; 29:1754-65. doi: 10.1021/la3038846 [Crossref] [ Google Scholar]
- Huang J, MacKerell AD, Jr Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J Comput Chem 2013; 34:2135-45. doi: 10.1002/jcc.23354 [Crossref] [ Google Scholar]
- Soares TA, Daura X, Oostenbrink C, Smith LJ, van Gunsteren WF. Validation of the GROMOS force-field parameter set 45A3 against nuclear magnetic resonance data of hen egg lysozyme. Journal of Biomolecular NMR 2004; 30:407-22. doi: 10.1007/s10858-004-5430-1 [Crossref] [ Google Scholar]
- Asche TS, Behrens P, Schneider AM. Validation of the COMPASS force field for complex inorganic–organic hybrid polymers. Journal of Sol-Gel Science and Technology 2017; 81:195-204. doi: 10.1007/s10971-016-4185-y [Crossref] [ Google Scholar]
- Singh S, Singh VK. Molecular Dynamics Simulation: Methods and Application. In: Singh DB, T Tripathi, editors. Frontiers in Protein Structure, Function, and Dynamics. Singapore: Springer Singapore; 2020. p. 213-38. 10.1007/978-981-15-5530-5_9.
- Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 2008; 29:1859-65. doi: 10.1002/jcc.20945 [Crossref] [ Google Scholar]
- Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J Chem Theory Comput 2016; 12:405-13. doi: 10.1021/acs.jctc.5b00935 [Crossref] [ Google Scholar]
- Rácz A, Mihalovits LM, Bajusz D, Héberger K, Miranda-Quintana RA. Molecular Dynamics Simulations and Diversity Selection by Extended Continuous Similarity Indices. J Chem Inf Model 2022; 62:3415-25. doi: 10.1021/acs.jcim.2c00433 [Crossref] [ Google Scholar]
- Hollingsworth SA, Dror RO. Molecular Dynamics Simulation for All. Neuron 2018; 99:1129-43. doi: 10.1016/j.neuron.2018.08.011 [Crossref] [ Google Scholar]
- Vacanti CA. The history of tissue engineering. J Cell Mol Med 2006; 10:569-76. doi: 10.1111/j.1582-4934.2006.tb00421.x [Crossref] [ Google Scholar]
- Vacanti JP, Vacanti CA. Chapter 1 - The History and Scope of Tissue Engineering. In: Lanza R, R Langer, J Vacanti, editors. Principles of Tissue Engineering (Fourth Edition). Boston: Academic Press; 2014. p. 3-8.
- Langer R, Vacanti J. Advances in tissue engineering. J Pediatr Surg 2016; 51:8-12. doi: 10.1016/j.jpedsurg.2015.10.022 [Crossref] [ Google Scholar]
- Courtenay JC, Deneke C, Lanzoni EM, Costa CA, Bae Y, Scott JL. Modulating cell response on cellulose surfaces; tunable attachment and scaffold mechanics. Cellulose (Lond) 2018; 25:925-40. doi: 10.1007/s10570-017-1612-3 [Crossref] [ Google Scholar]
- Bianco P, Robey PG. Stem cells in tissue engineering. Nature 2001; 414:118-21. doi: 10.1038/35102181 [Crossref] [ Google Scholar]
- Polak JM, Bishop AE. Stem cells and tissue engineering: past, present, and future. Ann N Y Acad Sci 2006; 1068:352-66. doi: 10.1196/annals.1346.001 [Crossref] [ Google Scholar]
- Howard D, Buttery LD, Shakesheff KM, Roberts SJ. Tissue engineering: strategies, stem cells and scaffolds. J Anat 2008; 213:66-72. doi: 10.1111/j.1469-7580.2008.00878.x [Crossref] [ Google Scholar]
- Nerem RM, Sambanis A. Tissue engineering: from biology to biological substitutes. Tissue Eng 1995; 1:3-13. doi: 10.1089/ten.1995.1.3 [Crossref] [ Google Scholar]
- Bose S, Roy M, Bandyopadhyay A. Recent advances in bone tissue engineering scaffolds. Trends Biotechnol 2012; 30:546-54. doi: 10.1016/j.tibtech.2012.07.005 [Crossref] [ Google Scholar]
- Ashammakhi N, GhavamiNejad A, Tutar R, Fricker A, Roy I, Chatzistavrou X. Highlights on Advancing Frontiers in Tissue Engineering. Tissue Eng Part B Rev 2022; 28:633-64. doi: 10.1089/ten.TEB.2021.0012 [Crossref] [ Google Scholar]
- Zhang S, Vijayavenkataraman S, Lu WF, Fuh JYH. A review on the use of computational methods to characterize, design, and optimize tissue engineering scaffolds, with a potential in 3D printing fabrication. J Biomed Mater Res B Appl Biomater 2019; 107:1329-51. doi: 10.1002/jbm.b.34226 [Crossref] [ Google Scholar]
- Sachs PC, Mollica PA, Bruno RD. Tissue specific microenvironments: a key tool for tissue engineering and regenerative medicine. J Biol Eng 2017; 11:34. doi: 10.1186/s13036-017-0077-0 [Crossref] [ Google Scholar]
- Xing F, Li L, Zhou C, Long C, Wu L, Lei H. Regulation and Directing Stem Cell Fate by Tissue Engineering Functional Microenvironments: Scaffold Physical and Chemical Cues. Stem Cells Int 2019; 2019:2180925. doi: 10.1155/2019/2180925 [Crossref] [ Google Scholar]
- Pedrero SG, Llamas-Sillero P, Serrano-López J. A Multidisciplinary Journey towards Bone Tissue Engineering. Materials (Basel) 2021; 14. 10.3390/ma14174896.
- Hollister SJ, Maddox RD, Taboas JM. Optimal design and fabrication of scaffolds to mimic tissue properties and satisfy biological constraints. Biomaterials 2002; 23:4095-103. doi: 10.1016/s0142-9612(02)00148-5 [Crossref] [ Google Scholar]
- Hutmacher DW. Scaffold design and fabrication technologies for engineering tissues--state of the art and future perspectives. J Biomater Sci Polym Ed 2001; 12:107-24. doi: 10.1163/156856201744489 [Crossref] [ Google Scholar]
- Hutmacher DW, Schantz T, Zein I, Ng KW, Teoh SH, Tan KC. Mechanical properties and cell cultural response of polycaprolactone scaffolds designed and fabricated via fused deposition modeling. J Biomed Mater Res 2001; 55:203-16. doi: 10.1002/1097-4636(200105)55:2<203::aid-jbm1007>3.0.co;2-7 [Crossref] [ Google Scholar]
- Mikos AG, Sarakinos G, Lyman MD, Ingber DE, Vacanti JP, Langer R. Prevascularization of porous biodegradable polymers. BiotechnolBioeng 1993; 42:716-23. doi: 10.1002/bit.260420606 [Crossref] [ Google Scholar]
- Rahmani A, Nadri S, Kazemi HS, Mortazavi Y, Sojoodi M. Conductive electrospun scaffolds with electrical stimulation for neural differentiation of conjunctiva mesenchymal stem cells. Artif Organs 2019; 43:780-90. doi: 10.1111/aor.13425 [Crossref] [ Google Scholar]
- Zhang J, Qiu K, Sun B, Fang J, Zhang K, Ei-Hamshary H. The aligned core-sheath nanofibers with electrical conductivity for neural tissue engineering. J Mater Chem B 2014; 2:7945-54. doi: 10.1039/c4tb01185f [Crossref] [ Google Scholar]
- Mendes AC, Baran ET, Reis RL, Azevedo HS. Self-assembly in nature: using the principles of nature to create complex nanobiomaterials. Wiley Interdiscip Rev NanomedNanobiotechnol 2013; 5:582-612. doi: 10.1002/wnan.1238 [Crossref] [ Google Scholar]
- Stoffelen C, Huskens J. Soft Supramolecular Nanoparticles by Noncovalent and Host-Guest Interactions. Small 2016; 12:96-119. doi: 10.1002/smll.201501348 [Crossref] [ Google Scholar]
- Cho NH, Guerrero-Martínez A, Ma J, Bals S, Kotov NA, Liz-Marzán LM. Bioinspired chiral inorganic nanomaterials. Nature Reviews Bioengineering 2023; 1:88-106. doi: 10.1038/s44222-022-00014-4 [Crossref] [ Google Scholar]
- Correa NM, Silber JJ, Riter RE, Levinger NE. Nonaqueous polar solvents in reverse micelle systems. Chem Rev 2012; 112:4569-602. doi: 10.1021/cr200254q [Crossref] [ Google Scholar]
- Genix AC, Oberdisse J. Nanoparticle self-assembly: from interactions in suspension to polymer nanocomposites. Soft Matter 2018; 14:5161-79. doi: 10.1039/c8sm00430g [Crossref] [ Google Scholar]
- Yadav S, Sharma AK, Kumar P. Nanoscale Self-Assembly for Therapeutic Delivery. Front BioengBiotechnol 2020; 8:127. doi: 10.3389/fbioe.2020.00127 [Crossref] [ Google Scholar]
- Shimizu T, Masuda M, Minamikawa H. Supramolecular nanotube architectures based on amphiphilic molecules. Chem Rev 2005; 105:1401-43. doi: 10.1021/cr030072j [Crossref] [ Google Scholar]
- Behanna HA, Donners JJ, Gordon AC, Stupp SI. Coassembly of amphiphiles with opposite peptide polarities into nanofibers. J Am Chem Soc 2005; 127:1193-200. doi: 10.1021/ja044863u [Crossref] [ Google Scholar]
- Storrie H, Guler MO, Abu-Amara SN, Volberg T, Rao M, Geiger B. Supramolecular crafting of cell adhesion. Biomaterials 2007; 28:4608-18. doi: 10.1016/j.biomaterials.2007.06.026 [Crossref] [ Google Scholar]
- Lee OS, Cho V, Schatz GC. Modeling the self-assembly of peptide amphiphiles into fibers using coarse-grained molecular dynamics. Nano Lett 2012; 12:4907-13. doi: 10.1021/nl302487m [Crossref] [ Google Scholar]
- Gačanin J, Hedrich J, Sieste S, Glaßer G, Lieberwirth I, Schilling C. Autonomous Ultrafast Self-Healing Hydrogels by pH-Responsive Functional Nanofiber Gelators as Cell Matrices. Adv Mater 2019; 31:e1805044. doi: 10.1002/adma.201805044 [Crossref] [ Google Scholar]
- Wang Y, Zhang W, Gong C, Liu B, Li Y, Wang L. Recent advances in the fabrication, functionalization, and bioapplications of peptide hydrogels. Soft Matter 2020; 16:10029-45. doi: 10.1039/d0sm00966k [Crossref] [ Google Scholar]
- Zhou L, Fan L, Yi X, Zhou Z, Liu C, Fu R. Soft Conducting Polymer Hydrogels Cross-Linked and Doped by Tannic Acid for Spinal Cord Injury Repair. ACS Nano 2018; 12:10957-67. doi: 10.1021/acsnano.8b04609 [Crossref] [ Google Scholar]
- Tang JD, Mura C, Lampe KJ. Stimuli-Responsive, Pentapeptide, Nanofiber Hydrogel for Tissue Engineering. J Am Chem Soc 2019; 141:4886-99. doi: 10.1021/jacs.8b13363 [Crossref] [ Google Scholar]
- Chen Z, Zhang Q, Li H, Wei Q, Zhao X, Chen F. Elastin-like polypeptide modified silk fibroin porous scaffold promotes osteochondral repair. Bioact Mater 2021; 6:589-601. doi: 10.1016/j.bioactmat.2020.09.003 [Crossref] [ Google Scholar]
- Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. Time evolution of the nanoparticle protein corona. ACS Nano 2010; 4:3623-32. doi: 10.1021/nn901372t [Crossref] [ Google Scholar]
- Cedervall T, Lynch I, Foy M, Berggård T, Donnelly SC, Cagney G. Detailed identification of plasma proteins adsorbed on copolymer nanoparticles. Angew Chem Int Ed Engl 2007; 46:5754-6. doi: 10.1002/anie.200700465 [Crossref] [ Google Scholar]
- Verma A, Stellacci F. Effect of surface properties on nanoparticle-cell interactions. Small 2010; 6:12-21. doi: 10.1002/smll.200901158 [Crossref] [ Google Scholar]
- Saptarshi SR, Duschl A, Lopata AL. Interaction of nanoparticles with proteins: relation to bio-reactivity of the nanoparticle. J Nanobiotechnology 2013; 11:26. doi: 10.1186/1477-3155-11-26 [Crossref] [ Google Scholar]
- Perera YR, Xu JX, Amarasekara DL, Hughes AC, Abbood I, Fitzkee NC. Understanding the Adsorption of Peptides and Proteins onto PEGylated Gold Nanoparticles. Molecules 2021; 26. 10.3390/molecules26195788.
- Jokerst JV, Lobovkina T, Zare RN, Gambhir SS. Nanoparticle PEGylation for imaging and therapy. Nanomedicine (Lond) 2011; 6:715-28. doi: 10.2217/nnm.11.19 [Crossref] [ Google Scholar]
- Sanchez-Cano C, Carril M. Recent Developments in the Design of Non-Biofouling Coatings for Nanoparticles and Surfaces. Int J Mol Sci 2020; 21. 10.3390/ijms21031007.
- Guerrini L, Alvarez-Puebla RA, Pazos-Perez N. Surface Modifications of Nanoparticles for Stability in Biological Fluids. Materials (Basel) 2018; 11. 10.3390/ma11071154.
- Gui N, Xu W, Myers DE, Shukla R, Tang HP, Qian M. The effect of ordered and partially ordered surface topography on bone cell responses: a review. Biomater Sci 2018; 6:250-64. doi: 10.1039/c7bm01016h [Crossref] [ Google Scholar]
- Kyle DJ, Oikonomou A, Hill E, Vijayaraghavan A, Bayat A. Fabrication and modelling of fractal, biomimetic, micro and nano-topographical surfaces. BioinspirBiomim 2016; 11:046009. doi: 10.1088/1748-3190/11/4/046009 [Crossref] [ Google Scholar]
- Alavi SK, Lotz O, Akhavan B, Yeo G, Walia R, McKenzie DR. Atmospheric Pressure Plasma Jet Treatment of Polymers Enables Reagent-Free Covalent Attachment of Biomolecules for Bioprinting. ACS Appl Mater Interfaces 2020; 12:38730-43. doi: 10.1021/acsami.0c07169 [Crossref] [ Google Scholar]
- Li J, Ding M, Fu Q, Tan H, Xie X, Zhong Y. A novel strategy to graft RGD peptide on biomaterials surfaces for endothelization of small-diamater vascular grafts and tissue engineering blood vessel. J Mater Sci Mater Med 2008; 19:2595-603. doi: 10.1007/s10856-007-3354-5 [Crossref] [ Google Scholar]
- Petrie TA, Reyes CD, Burns KL, García AJ. Simple application of fibronectin-mimetic coating enhances osseointegration of titanium implants. J Cell Mol Med 2009; 13:2602-12. doi: 10.1111/j.1582-4934.2008.00476.x [Crossref] [ Google Scholar]
- Kim J, Bae W-G, Lim K-T, Jang K-J, Oh S, Jang K-J. Density of nanopatterned surfaces for designing bone tissue engineering scaffolds. Materials Letters 2014; 130:227-31. doi: 10.1016/j.matlet.2014.05.107 [Crossref] [ Google Scholar]
- Scheufler C, Sebald W, Hülsmeyer M. Crystal structure of human bone morphogenetic protein-2 at 2.7 A resolution. J Mol Biol 1999; 287:103-15. doi: 10.1006/jmbi.1999.2590 [Crossref] [ Google Scholar]
- Axelrad TW, Einhorn TA. Bone morphogenetic proteins in orthopaedic surgery. Cytokine Growth Factor Rev 2009; 20:481-8. doi: 10.1016/j.cytogfr.2009.10.003 [Crossref] [ Google Scholar]
- Huang B, Yuan Y, Ding S, Li J, Ren J, Feng B. Nanostructured hydroxyapatite surfaces-mediated adsorption alters recognition of BMP receptor IA and bioactivity of bone morphogenetic protein-2. Acta Biomater 2015; 27:275-85. doi: 10.1016/j.actbio.2015.09.007 [Crossref] [ Google Scholar]
- Gold LI, Pearlstein E. Fibronectin-collagen binding and requirement during cellular adhesion. Biochem J 1980; 186:551-9. doi: 10.1042/bj1860551 [Crossref] [ Google Scholar]
- Jara CP, Wang O, Paulino do Prado T, Ismail A, Fabian FM, Li H. Novel fibrin-fibronectin matrix accelerates mice skin wound healing. Bioact Mater 2020; 5:949-62. doi: 10.1016/j.bioactmat.2020.06.015 [Crossref] [ Google Scholar]
- Dayem AA, Won J, Goo HG, Yang GM, Seo DS, Jeon BM. The immobilization of fibronectin- and fibroblast growth factor 2-derived peptides on a culture plate supports the attachment and proliferation of human pluripotent stem cells. Stem Cell Res 2020; 43:101700. doi: 10.1016/j.scr.2020.101700 [Crossref] [ Google Scholar]
- Efthymiou G, Saint A, Ruff M, Rekad Z, Ciais D, Van Obberghen-Schilling E. Shaping Up the Tumor Microenvironment With Cellular Fibronectin. Front Oncol 2020; 10:641. doi: 10.3389/fonc.2020.00641 [Crossref] [ Google Scholar]
- Liu D, Yang F, Xiong F, Gu N. The Smart Drug Delivery System and Its Clinical Potential. Theranostics 2016; 6:1306-23. doi: 10.7150/thno.14858 [Crossref] [ Google Scholar]
- Sadab S, Sahu S, Patel S, Khan R, Khare B, Thakur BS. A Comprehensive Review: Transdermal Drug Delivery System: A Tool For Novel Drug Delivery System. Asian Journal of Dental and Health Sciences 2022; 2:40-7. doi: 10.22270/ajdhs.v2i4.24 [Crossref] [ Google Scholar]
- Patel H, Panchal DR, Patel U, Brahmbhatt T, Suthar M. Matrix type drug delivery system: A review. J Pharm Sci Biosci Res 2011; 1:143-151. [ Google Scholar]
- Ratnaparkhi M, Gupta Jyoti PJT. Sustained release oral drug delivery system-an overview. International Journal of Pharma Research & Review 2013; 3:10-22270. [ Google Scholar]
- Zakrzewski W, Dobrzyński M, Szymonowicz M, Rybak Z. Stem cells: past, present, and future. Stem Cell Res Ther 2019; 10:68. doi: 10.1186/s13287-019-1165-5 [Crossref] [ Google Scholar]
- Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene 2001; 20:2390-400. doi: 10.1038/sj.onc.1204383 [Crossref] [ Google Scholar]
- Liu L, Michowski W, Kolodziejczyk A, Sicinski P. The cell cycle in stem cell proliferation, pluripotency and differentiation. Nat Cell Biol 2019; 21:1060-7. doi: 10.1038/s41556-019-0384-4 [Crossref] [ Google Scholar]
- Thavornyutikarn B, Chantarapanich N, Sitthiseripratip K, Thouas GA, Chen Q. Bone tissue engineering scaffolding: computer-aided scaffolding techniques. Prog Biomater 2014; 3:61-102. doi: 10.1007/s40204-014-0026-7 [Crossref] [ Google Scholar]
- Fröhlich M, Grayson WL, Wan LQ, Marolt D, Drobnic M, Vunjak-Novakovic G. Tissue engineered bone grafts: biological requirements, tissue culture and clinical relevance. Curr Stem Cell Res Ther 2008; 3:254-64. doi: 10.2174/157488808786733962 [Crossref] [ Google Scholar]
- Chang H-Y, Huang C-C, Lin K-Y, Kao W-L, Liao H-Y, You Y-W. Effect of Surface Potential on NIH3T3 Cell Adhesion and Proliferation. The Journal of Physical Chemistry C 2014; 118:14464-70. doi: 10.1021/jp504662c [Crossref] [ Google Scholar]
- Birhanu G, Akbari Javar H, Seyedjafari E, Zandi-Karimi A, Dusti Telgerd M. An improved surface for enhanced stem cell proliferation and osteogenic differentiation using electrospun composite PLLA/P123 scaffold. Artif Cells NanomedBiotechnol 2018; 46:1274-81. doi: 10.1080/21691401.2017.1367928 [Crossref] [ Google Scholar]
- Bacáková L, Filová E, Rypácek F, Svorcík V, Starý V. Cell adhesion on artificial materials for tissue engineering. Physiol Res 2004; 53 Suppl 1:S35-45. [ Google Scholar]
- Shamloo A, Sarmadi M. Investigation of the adhesive characteristics of polymer-protein systems through molecular dynamics simulation and their relation to cell adhesion and proliferation. Integr Biol (Camb) 2016; 8:1276-95. doi: 10.1039/c6ib00159a [Crossref] [ Google Scholar]
- Sarmadi M, Shamloo A, Mohseni M. Utilization of Molecular Dynamics Simulation Coupled with Experimental Assays to Optimize Biocompatibility of an Electrospun PCL/PVA Scaffold. PLoS One 2017; 12:e0169451. doi: 10.1371/journal.pone.0169451 [Crossref] [ Google Scholar]
- Zhao C, Tan A, Pastorin G, Ho HK. Nanomaterial scaffolds for stem cell proliferation and differentiation in tissue engineering. Biotechnol Adv 2013; 31:654-68. doi: 10.1016/j.biotechadv.2012.08.001 [Crossref] [ Google Scholar]
- Dua HS, Joseph A, Shanmuganathan VA, Jones RE. Stem cell differentiation and the effects of deficiency. Eye (Lond) 2003; 17:877-85. doi: 10.1038/sj.eye.6700573 [Crossref] [ Google Scholar]
- Tonouchi M. Cutting-edge terahertz technology. Nature Photonics 2007; 1:97-105. doi: 10.1038/nphoton.2007.3 [Crossref] [ Google Scholar]
- Davies AG, Burnett AD, Fan W, Linfield EH, Cunningham JE. Terahertz spectroscopy of explosives and drugs. Materials Today 2008; 11:18-26. doi: 10.1016/S1369-7021(08)70016-6 [Crossref] [ Google Scholar]
- Porcuna J, Mínguez-Martínez J, Ricote M. The PPARα and PPARγ Epigenetic Landscape in Cancer and Immune and Metabolic Disorders. Int J Mol Sci 2021; 22. 10.3390/ijms221910573.
- Krishna L, Dhamodaran K, Jayadev C, Chatterjee K, Shetty R, Khora SS. Nanostructured scaffold as a determinant of stem cell fate. Stem Cell Res Ther 2016; 7:188. doi: 10.1186/s13287-016-0440-y [Crossref] [ Google Scholar]
- Tang JD, McAnany CE, Mura C, Lampe KJ. Toward a Designable Extracellular Matrix: Molecular Dynamics Simulations of an Engineered Laminin-Mimetic, Elastin-Like Fusion Protein. Biomacromolecules 2016; 17:3222-33. doi: 10.1021/acs.biomac.6b00951 [Crossref] [ Google Scholar]
- Lee S, Lee DS, Jang JH. Recombinant laminin α5 LG1-3 domains support the stemness of human mesenchymal stem cells. Exp Ther Med 2021; 21:166. doi: 10.3892/etm.2020.9597 [Crossref] [ Google Scholar]