Bioimpacts. 2025;15:30222.
doi: 10.34172/bi.30222
Original Article
Identification of key pathways and molecular players potentially involved in endometrial cancer metastasis through integrated bioinformatics analyses
Maryam Rezazadeh Data curation, Investigation, Resources, Visualization, Writing – original draft, Writing – review & editing, 1 
Shahla Danaei-Mehrabad Investigation, Resources, Writing – original draft, Writing – review & editing, 2
Nahideh Afshar Zakariya Investigation, Resources, Writing – original draft, Writing – review & editing, 3
Fatemeh Kazemi Investigation, Resources, Writing – original draft, Writing – review & editing, 3
Marziyeh Sadat Moslehian Data curation, Investigation, Resources, Visualization, Writing – original draft, Writing – review & editing, 4
Amin Tamadon Data curation, Investigation, Resources, Visualization, Writing – original draft, Writing – review & editing, 5, 6, 7
Reza Shirazi Investigation, Resources, Writing – original draft, Writing – review & editing, 8
Mahdi Mahdipour Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Supervision, Validation, 4, 9, * 
Parvin Hakimi Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Supervision, Validation, 3, * 
Author information:
1Department of Medical Genetics, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
2Department of Gynecology, Eastern Azerbaijan ACECR ART Center, Eastern Azerbaijan Branch of ACECR, Tabriz, Iran
3Women's Reproductive Health Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
4Stem Cell Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
5Department of Natural Sciences, West Kazakhstan Marat Ospanov Medical University, Aktobe, Kazakhstan
6Stem Cells Technology Research Center, Shiraz University of Medical Sciences, Shiraz, Iran
7Department of Research and Development, PerciaVista R&D Co., Shiraz, Iran
8Department of Anatomy, School of Biomedical Sciences, Medicine & Health, UNSW Sydney, Sydney, Australia
9Department of Reproductive Biology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
Abstract
Introduction:
Endometrial cancer (EC) is a particularly frequent gynecological cancer, and metastasis is the leading cause of death in patients with EC. Using publicly accessible gene expression data, a bioinformatics study was carried out to increase our knowledge and reveal treatment targets for EC metastasis. This study aimed to identify new important molecular actors and clarify the molecular processes and pathways underlying EC metastasis.
Methods:
The GEOexplorer and R programming languages were used to analyze and visualize gene expression data from EC metastatic gene expression datasets, and differentially expressed genes (DEGs) and differentially expressed lncRNAs (DElncRNAs) were identified using bioinformatics with P-value thresholds of < 0.05, and |log2FC| > 1.5. KEGG pathway enrichment analysis and gene ontology enrichment was used to enrich the observed DEGs, protein-protein interactions were established, and hub genes were identified.
Results:
The findings revealed that DEGs were considerably enriched in a number of pathways, including the "Pathways in cancer", "Breast cancer", and "Rap1 signaling pathway." DEGs were also found to be involved in a number of biological processes, cellular components, and molecular activities. The PPI network included the hub genes CTNNB1, FGFR3, ESR1, and SRSF3 as well as a number of DElncRNAs, such as LINC01541, SNHG17, LINC00520, BHLHE22-AS1, LOC100509445, H19, and HOTAIRM1.
Conclusion:
This study contributes to our understanding of the molecular processes driving EC metastasis, which may result in the development of new treatment targets and indicators for the early identification of EC metastasis. More studies are needed to validate these findings and to understand the functional roles of these key factors in EC metastasis.
Keywords: Endometrial cancer, Metastasis, Bioinformatics analysis, lncRNAs, Pathway enrichment, Hub genes
Copyright and License Information
© 2025 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Introduction
Endometrial carcinoma (EC) is a prevalent form of malignancy that affects women’s health worldwide. According to current data, approximately 65,620 instances of cancer will be detected in the United States by 2020, representing 7% of all newly diagnosed female cancers. From 2007 to 2016, the rate of cancer increased by 1.3% every year, while death rates increased by the same proportion.1 Patients with recurring or advanced illnesses, as well as those with a potentially severe histologic classification, such as high-grade endometrioid, papillary serous, or clear cell histology, have a much poorer clinical prognosis.2,3 This is true despite the fact that, in its preliminary stages, EC has a 5-year survival rate of up to 90%. Previous studies have shown that high-grade carcinomas have a lower incidence rate of new diagnoses, but a higher fatality rate. This demonstrates that high-grade tumors are more inclined to recur and arise at an advanced stage.4 Nonetheless, people with comparable ECs may have varied outcomes, particularly in patients with high-grade EC patients.5 The current findings underscore the need for more complete knowledge of EC molecular genetics, particularly in the context of late-stage and metastatic ECs. The ability of malignant cells to diffuse and colonize distant tissues or organs in the body is referred to as metastasis. The majority of cancers have the capacity to metastasize.6 Metastasis is largely assisted by the circulatory and lymphatic systems, both of which are required for this process.7 In this context, molecular mechanisms are also changed in favor of cancer cell metastasis.8 Thus, identifying the molecular pattern of cancer metastasis can help in understanding the process of metastasis, and bioinformatics analysis can play a vital role in this regard.
Bioinformatic analysis has the potential to play a critical role in identifying the molecular signature of EC metastasis. Research, for example, examined mutational patterns in both primary and metastatic ECs and discovered that the mutational mechanisms underlying ECs differ even among tumors of the identical TCGA molecular classification and in the development from basic to metastatic ECs.9 A recent investigation has unveiled a novel, long-noncoding RNA (lncRNA) paradigm for prognosticating patients diagnosed with EC. This investigation identified lncRNAs that were differentially expressed in association with EC from The Cancer Genome Atlas (TCGA). The findings discovered that a 3-lncRNA profile (CTD-2377D24.6, RP4-616B8.5, and RP11-389G6.3) was significantly correlated with histological classification, developed clinical stage and clinical rating in EC patients.10 LncRNAs are molecules of RNA with more than 200 nucleotides and lack the ability to encode proteins.11 LncRNAs are currently shown to have both oncogenic and tumor-suppressive properties and, therefore, play an important role in the advancement of tumors and metastasis.12 LncRNAs have the ability to modulate gene expression through various mechanisms such as transcriptional regulation, epigenetic modification, RNA or protein storage, and enhancing regulation by interfering with other molecules.13
In this study, we aimed to understand the pathways and molecular factors involved in metastasis, such as lncRNAs and genes, by analyzing data related to EC metastasis. By identifying the molecular signature of EC metastasis and understanding the function of lncRNAs and other genes in this process, we hope to gain a more inclusive understanding of the molecular genetics of EC and improve clinical outcomes in individuals with recurring or severe disease.
Materials and Methods
The current investigation involved a comprehensive examination of the NCBI Gene Expression Omnibus database (GEO, https://www.ncbi.nlm.nih.gov/geo/) using relevant keywords, such as "endometrial cancer," "endometrial carcinoma," "endometrial neoplasm," "endometrial adenocarcinoma," "endometrial hyperplasia," and "metastasis." The following inclusion and exclusion criteria were applied during the dataset selection process to minimize the risk of selection bias and ensure data quality consistency.
Inclusion criteria
-
Studies primarily focus on endometrial cancer and related conditions, such as endometrial carcinoma, neoplasm, adenocarcinoma, hyperplasia, and metastasis.
-
Microarray datasets with comprehensive gene expression profiles.
-
The datasets were sourced from human subjects with documented cases of metastatic and non-metastatic endometrial cancer.
-
Datasets containing information on both Differentially Expressed Genes (DEGs) and long non-coding RNAs (DElncRNAs).
-
Studies conducted on relevant tissues or samples specifically targeted endometrial cancer and metastatic conditions.
Exclusion criteria
-
Studies lacking clear documentation of endometrial cancer cases or relevant clinical information.
-
Datasets with incomplete or ambiguous gene expression profiles.
-
Non-human studies or studies conducted on animal models.
-
Studies lacking explicit information on metastatic status or exclusively focusing on non-metastatic conditions.
-
Datasets with inadequate sample sizes or insufficient statistical power.
-
Studies with inconsistent or unclear methodologies for gene expression analysis.
-
Datasets with substantial missing data or poor data quality, as indicated by incomplete annotations or unreliable experimental procedures.
Of the datasets specified, only two were chosen for additional analysis based on research questions GSE120490 and GSE29436. A systems biology approach utilizing online databases and R software was employed to extract data from the two microarray datasets of metastatic EC obtained from individuals with metastatic and non-metastatic EC. Our objective was to identify differentially expressed genes (DEGs) and lncRNAs (DElncRNAs) using predetermined criteria. The methods employed were executed in adherence to pertinent guidelines and regulations."
The collection of gene expression profile data and Identification of DEGs and DElncRNAs
Both datasets were analyzed using the GEOexplorer14 based on GEO2R15 and were based on a GPL570 chip-based platform (HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array. GEOexplorer, accessible via https://geoexplorer.rosalind.kcl.ac.uk, offers a method for conducting interactive and replicable analyses of microarray and RNA-seq gene expression data.14 The GSE120490 comprised 100 non-metastatic EC samples and 45 metastatic EC samples.16 The GSE29436 contained 8 EC samples, of which 4 were metastatic EC samples, and 4 were non-metastatic EC samples. The criteria utilized to select significant DEGs and DElncRNAs were a p-value less than 0.05 and a |log2FC| > 1.5.
KEGG and GO enrichment analyses of DEGs
To examine the potential biological processes (BP), molecular functions (MF), and cellular components (CC) linked to the DEGs, we employed the EnrichR database for functional annotation and pathway enrichment analysis. This analysis encompassed the utilization of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.17-20 The utilization of GO as a prominent bioinformatics instrument facilitates the annotation of genes and the analysis of biological processes associated with these genes. The KEGG database is a useful resource for understanding the overarching functionality of biological systems resulting from large molecular datasets generated by large-scale experimental approaches. Statistical significance was set at P < 0.05.
Protein-protein interaction (PPI) network of DEGs and hub genes
A web-based tool for evaluating PPI data is the Search Tool for the Retrieval of Interacting Genes database (STRING, https://string-db.org/).21 Assessing the functional interactions of proteins may provide insights into the processes of disease creation or progression. DEGs discovered in the two datasets were submitted to the STRING database to investigate their possible interactions. The rationale behind the selection of interactions was based on the cumulative score, which was set greater than 0.4. This score threshold was chosen because it represents a balance between sensitivity and specificity, with the aim of capturing meaningful interactions while minimizing false positives. The interactions meeting this significance criterion were retrieved and utilized for PPI network construction through Cytoscape software, a freely accessible bioinformatics tool for visualizing biological interaction networks.22 Furthermore, we determined hub genes that are potentially important in EC metastatic development were determined by assessing the Maximal Clique Centrality (MCC) of each protein node in Cytoscape using the CytoHubba plugin.23 To conclude, the top 10 genes in terms of substantial MCC in EC metastasis were identified as hub genes.
Validation of hub genes
In order to validate the dependability of hub genes identified through our detection, we conducted an analysis of their prognostic and expression characteristics in EC utilizing Gene Expression Profiling Interactive Analysis (GEPIA). The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression datasets were used to compile a massive collection of 8,587 normal samples and 9,736 tumor samples that made up the interactive online application tool known as GEPIA.23 Subsequently, the survival curve and box plot were used to depict the associations. The reference gene GAPDH was used to standardize the data obtained from the shared genes.
Results
Identification of DEGs and DElncRNAs in EC metastasis
DEGs retrieved from two microarray datasets, GSE12490 and GSE29436, were evaluated using the R program and limma package, respectively. The evaluation criteria for inclusion were |log2FC| ≥ 1.5 and P values < 0.05. As illustrated in Fig. 1, the "Volcano plot" for the datasets was constructed by plotting the DEGs against each dataset's corresponding p-values and fold changes in the R software using the EnhancedVolcano package. DEGs that lacked annotations in the final outcome of the analysis were eliminated from subsequent investigations. Based on the assessment of the datasets according to the given criteria, GSE12490 and GSE29436 included 79 and 56 DEGs, respectively. DElncRNAs in EC metastasis are presented in detail in Table 1.
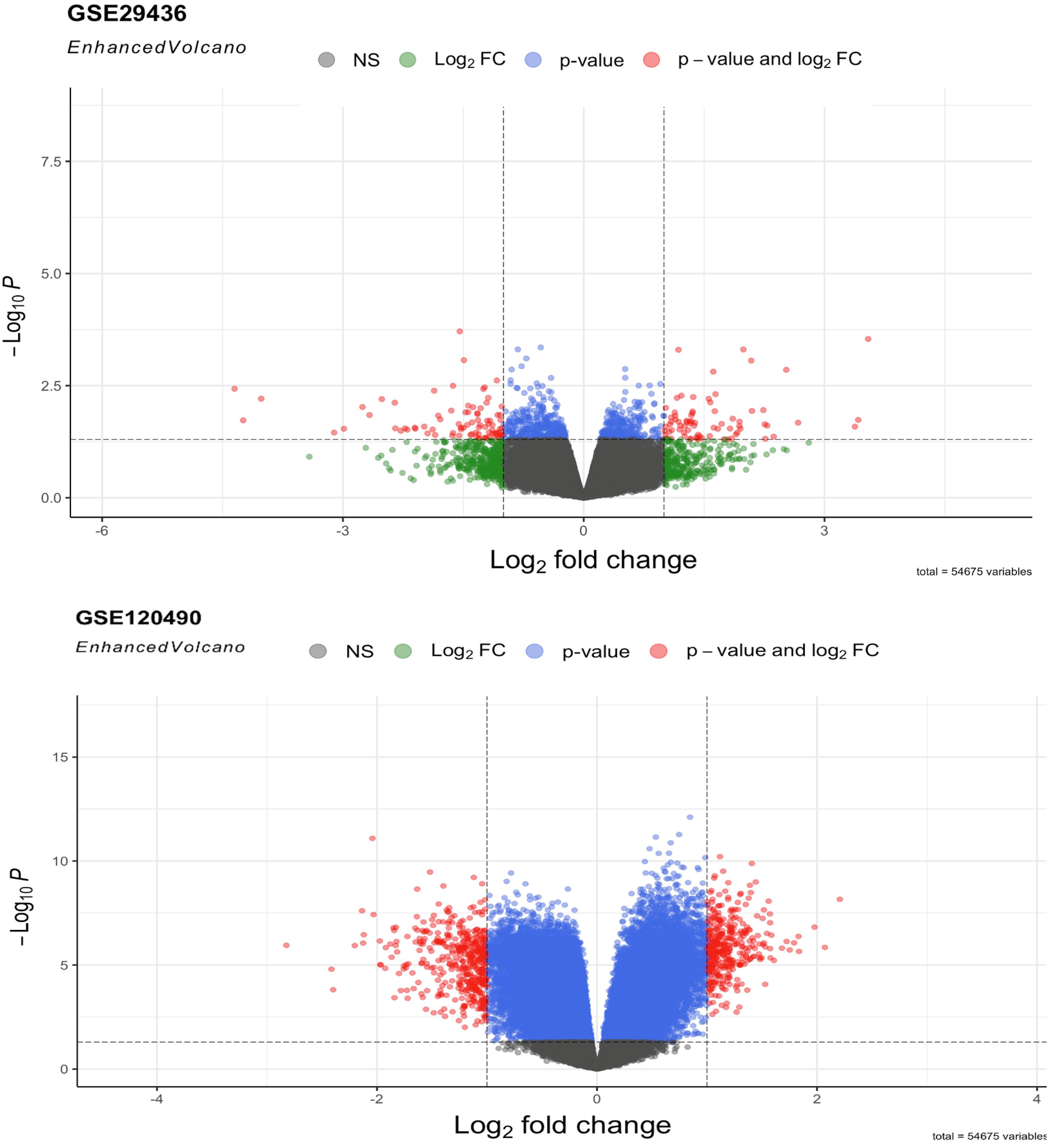
Fig. 1.
Volcano plot of DEGs related to EC metastasis, including GSE129480 and GSE29436 datasets. Screening for DEGs was performed using a P value < 0.05 and |log2FC| ≥ 1.5.
.
Volcano plot of DEGs related to EC metastasis, including GSE129480 and GSE29436 datasets. Screening for DEGs was performed using a P value < 0.05 and |log2FC| ≥ 1.5.
Table 1.
DElncRNAs in EC metastasis
|
LncRNAs
|
Log2FC
|
Adjusted
P
value
|
P
value
|
|
LINC01541
|
-2.3982562 |
0.00065329 |
0.00015417 |
|
SNHG17
|
-2.2020088 |
3.16E-05 |
1.16E-06 |
|
LINC00520
|
-1.8791358 |
2.95E-05 |
9.78E-07 |
|
LOC401463
|
-1.6075514 |
2.71E-05 |
7.52E-07 |
|
LOC100509445
|
-2.7563043 |
0.99992982 |
0.0094775 |
|
H19
|
2.26218638 |
0.99992982 |
0.04827739 |
|
HOTAIRM1
|
1.63256877 |
0.99992982 |
0.01177712 |
Functional enrichment analysis
The KEGG pathway analysis revealed that "Pathways in cancer," "Breast cancer," "Rap1 signaling pathway," "PI3K-Akt signaling pathway," and "Adherens junction" were the top five enriched pathways associated with the DEGs. The top five enriched biological processes were "Positive Regulation of Glycogen Biosynthetic Process (GO:0045725)," "Positive Regulation of Glycogen Metabolic Process (GO:0070875)," "Pulmonary Valve Morphogenesis (GO:0003184)," "Pulmonary Valve Development (GO:0003177)," and "Retinal Ganglion Cell Axon Guidance (GO:0031290). Furthermore, "Anaphase-Promoting Complex (GO:0005680)," "Catenin Complex (GO:0016342)," "P-body (GO:0000932)," "Adherens Junction (GO:0005912)," and "Apicolateral Plasma Membrane (GO:0016327)" were the top five enriched cellular components identified. The top five enriched molecular functions were, in order, "N6-methyladenosine-containing RNA Binding (GO:1990247)," "Transcription Corepressor Binding (GO:0001222)," "Insulin-Like Growth Factor Receptor Binding (GO:0005159)," and "mRNA 3'-UTR Binding (GO:0003730)." The functional enrichment results are shown in more detail in Fig. 2.
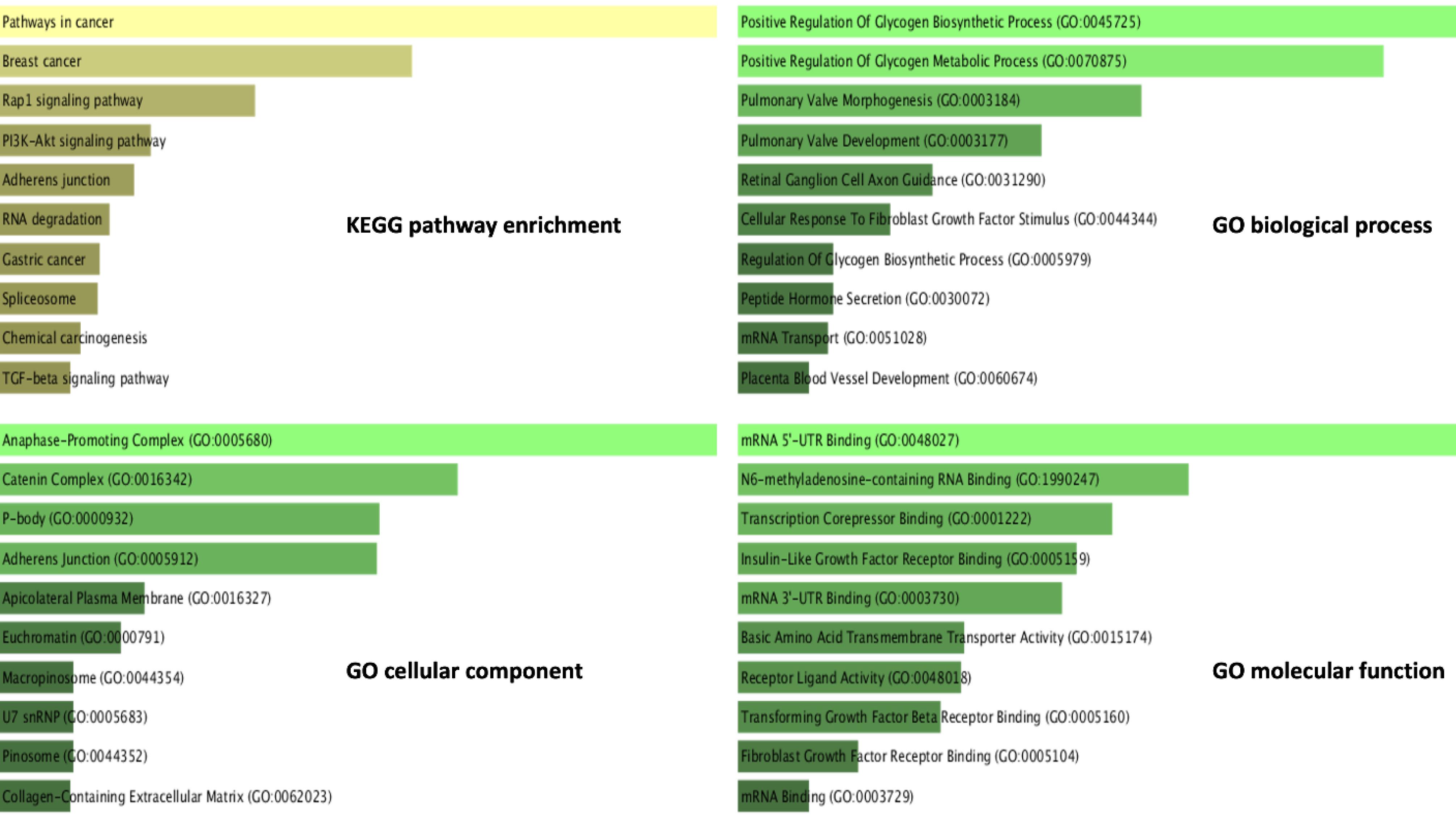
Fig. 2.
Functional enrichment of the EC metastasis DEGs.Functional enrichment, including KEGG pathway enrichment and GO analysis, was conducted using the EnrichR database. The length of each bar corresponds to the level of significance within the respective category, arranged by the p-value. It should be noted that a higher degree of correlation exists between a given category and the corresponding bar when the color intensity of the latter is lower.
.
Functional enrichment of the EC metastasis DEGs.Functional enrichment, including KEGG pathway enrichment and GO analysis, was conducted using the EnrichR database. The length of each bar corresponds to the level of significance within the respective category, arranged by the p-value. It should be noted that a higher degree of correlation exists between a given category and the corresponding bar when the color intensity of the latter is lower.
Identification of hub genes and network analysis
The PPI network connected to the DEGs related to EC metastasis was studied using the Cytoscape STRING plugin. Using the Cytoscape Cytohubba plugin, the top 10 hub genes were identified according to MCC. The genes that have been classified are CTNNB1, IRS1, FGFR3, FGF9, FGF20, ESR1, ALYREF, ARHGEF7, SRSF3, and SNRPE (Fig. 3). Table 2 presents comprehensive information regarding hub genes associated with metastasis in EC.
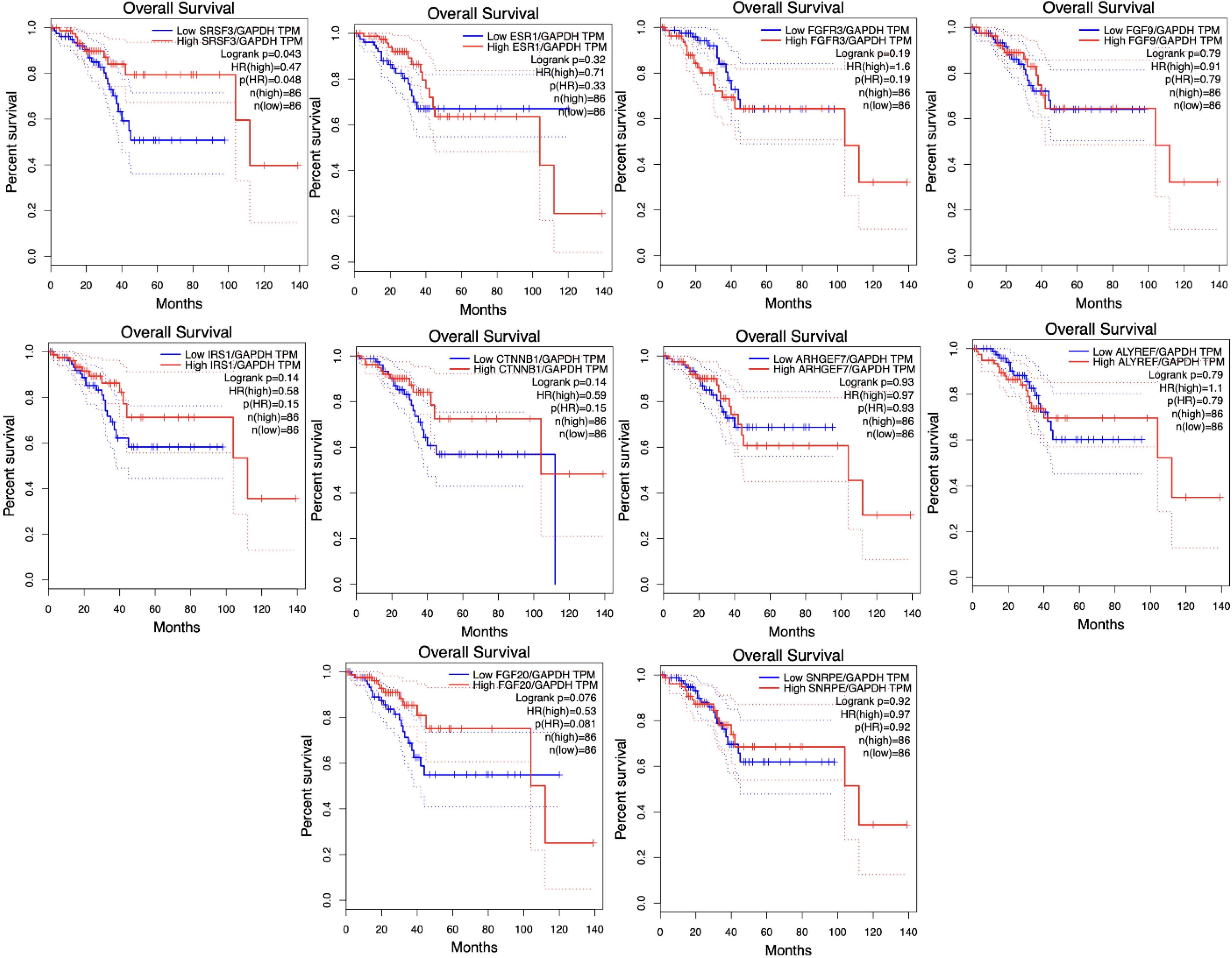
Fig. 3.
Construction of the PPI networks. A. The PPI network was established by utilizing the STRING database. B. The PPI network was constructed by applying a threshold of combined score > 0.4 and subsequently identifying the hub genes. Rectangular shapes are used to symbolize genes, whereas lines are employed to depict the interaction of proteins between genes. The hub genes are distinguished in a network with a wide range of red to yellow colors.
.
Construction of the PPI networks. A. The PPI network was established by utilizing the STRING database. B. The PPI network was constructed by applying a threshold of combined score > 0.4 and subsequently identifying the hub genes. Rectangular shapes are used to symbolize genes, whereas lines are employed to depict the interaction of proteins between genes. The hub genes are distinguished in a network with a wide range of red to yellow colors.
Table 2.
Hub genes in EC metastasis
|
Gene symbol
|
LogFC
|
P
value
|
Adjusted
P
value
|
|
CTNNB1
|
1.836041664 |
2.21E-06 |
4.31E-05 |
|
IRS1
|
1.556989876 |
2.43E-07 |
1.67E-05 |
|
FGFR3
|
2.5234038 |
0.00140257 |
0.99992982 |
|
FGF9
|
1.6782271 |
2.40E-05 |
0.00016559 |
|
FGF20
|
-3.1093853 |
0.03528495 |
0.99992982 |
|
ESR1
|
1.504688762 |
1.38E-05 |
0.00011473 |
|
ALYREF
|
1.680948574 |
1.55E-06 |
3.63E-05 |
|
ARHGEF7
|
-1.586372361 |
2.19E-07 |
1.58E-05 |
|
SRSF3
|
-1.5772856 |
8.52E-08 |
9.99E-06 |
|
SNRPE
|
1.55954423 |
8.50E-09 |
3.52E-06 |
Survival analysis
The effects of altered CTNNB1, IRS1, FGFR3, FGF9, FGF20, ESR1, ALYREF, ARHGEF7, SRSF3, and SNRPE expression on the median survival time and overall survival rate of patients with EC metastases were examined in this study. The results showed that SRSF3 (log-rank P = 0.043) was significantly associated with the prognosis and overall survival of EC metastasis, whereas the expression levels of CTNNB1, IRS1, FGFR3, FGF9, FGF20, ESR1, ALYREF, ARHGEF7, and SNRPE were not (P = 0.05). Fig. 4 depicts the survival analysis curve of hub genes, revealing an advantageous relationship between increased SRSF3 levels and improved survival rates. Bioinformatics analysis showed a substantial decrease in SRSF3 expression in EC (log2FC = -1.577, p-value = 8.52E-08). GEPIA has the potential to perform predictive analyses of gene survival rates using RNA-seq data acquired from the TCGA database.
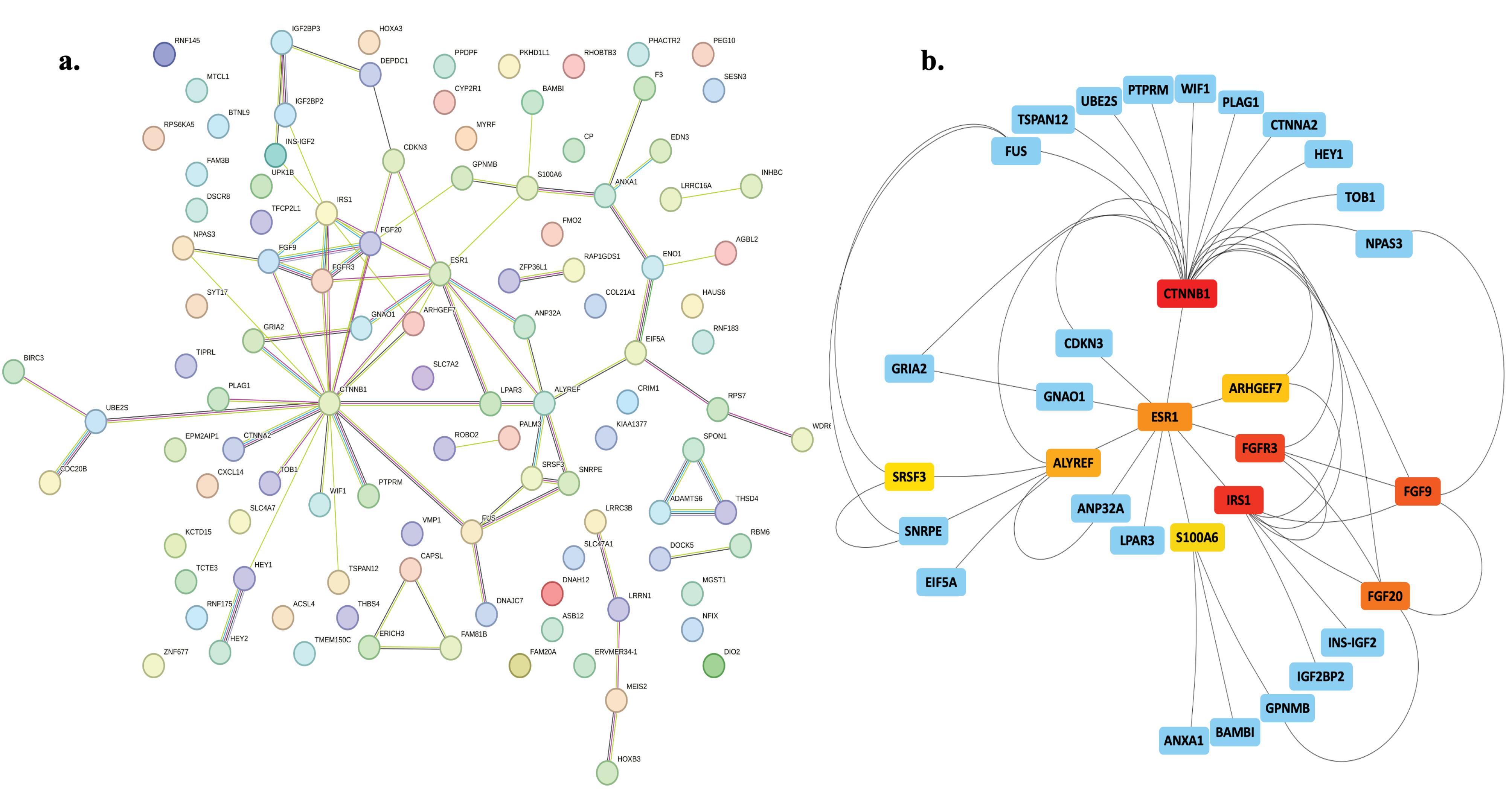
Fig. 4.
Relationships between hub gene expression and clinical outcomes in EC patients. Elevated SRSF3 levels are associated with increased survival rates. The prognostic significance of CTNNB1, IRS1, FGFR3, FGF9, FGF20, ESR1, ALYREF, ARHGEF7, and SNRPE expression levels was found to be insignificant in relation to the outcome of patients with EC. GAPDH was used as a reference gene to standardize the data obtained from the hub genes.
.
Relationships between hub gene expression and clinical outcomes in EC patients. Elevated SRSF3 levels are associated with increased survival rates. The prognostic significance of CTNNB1, IRS1, FGFR3, FGF9, FGF20, ESR1, ALYREF, ARHGEF7, and SNRPE expression levels was found to be insignificant in relation to the outcome of patients with EC. GAPDH was used as a reference gene to standardize the data obtained from the hub genes.
Discussion
The incidence and mortality rates of EC, A Few common type of cancer that affects women, have increased globally.24 A few instances of distant metastasis have been reported, which has prevented a thorough study of EC epidemiology. Approximately 3% of EC patients who received therapy between 1999 and 2001 were diagnosed with stage IV disease, according to the twenty-sixth annual statement of the International Federation of Gynecology and Obstetrics (FIGO).25 Similar to the poorly characterized EC metastasis epidemiology, the molecular mechanisms and pathways of EC metastasis need to be assessed further. Regarding this matter, a certain study has revealed that the exhaustive genomic examination of EC by TCGA has resulted in the identification of four unique molecular subtypes, which possess significant prognostic implications.26 A recent investigation revealed that Berberine (BBR) exhibited the ability to impede the proliferation, motility, infiltration, and metastasis of EC cells in vitro and in vivo.27 Moreover, a particular study discovered that events resembling epithelial-mesenchymal transition (EMT) serve a central function in the advancement of tumors and the development of malignancy, granting the nascent cancerous cell invasive and metastatic characteristics.28 The advancement, invasion, and metastasis of EC have been linked to EMT. EMT includes a number of cellular alterations, such as the breakdown of cell adhesion, the activation of mesenchymal marker expression, and the restructuring of actin filaments.29 In this regard, the molecular mechanisms and pathways of EC metastasis require further assessment.
To identify novel molecular entities and pathways implicated in EC metastasis, we conducted bioinformatics analysis to assess the association of differential genes with these conditions. Bioinformatic analysis was employed to identify differentially expressed genes associated with EC metastasis, with the aim of identifying new functional genes and pathways involved in this process. The spread of cancerous cells from the main tumor to distant organs or tissues is known as metastasis, and it contributes significantly to the morbidity and mortality of cancer.30 The identification of genes and pathways involved in metastasis can provide insight into potential targets for therapeutic intervention and biomarkers for early detection. In this work, the KEGG pathway analysis showed that "Pathways in cancer," "Breast cancer," "Rap1 signaling pathway," "PI3K-Akt signaling pathway," and "Adherens junction" were the topmost five enriched pathways related to DEGs in cancer metastasis. The compilation known as "Pathways in cancer" comprises pivotal signaling pathways implicated in the onset and progression of cancer, including the MAPK31 and PI3K-Akt32 signaling pathways. MAPK and PI3K-Akt pathways have been implicated in cancer metastasis. The MAPK pathway controls cell growth, differentiation, and survival and plays a role in angiogenesis. Mutations or overexpression of components of this pathway can lead to uncontrolled cell growth and increased invasiveness.33 Similarly, the PI3K-Akt pathway controls cell proliferation, survival, metabolism, and angiogenesis. Mutations or overexpression of components of this pathway can lead to increased cell survival, proliferation, invasion, and regulation of gene expression involved in tumor cell metastasis and invasion.32 The Rap1 signaling pathway is a central factor in the processes of cellular adhesion and migration, both of which are fundamental stages in the metastasis of cancer. The signaling pathway of Rap1 is responsible for the regulation of cell adhesion mediated by integrin or cadherin, protease expression levels such as matrix metalloproteinase, and alterations in the cytoskeleton. These changes have been found to be associated with the processes of growth of tumor cells, invasion, and metastasis.34-36 Cell-cell adhesion complexes called adherens junctions are essential for preserving tissue stability and homeostasis.37 Dysregulation of adherent junctions is involved in cancer development and metastasis. The progression of metastatic breast cancer is typified by the demise of tissue integrity, which imitates the developmental EMT mechanism.38 Adherens junctions are essential for maintaining the integrity of interactions between cells and tissues.39
The top five biological processes linked to DEG enrichment in cancer metastasis were identified using GO analysis. The augmentation of glycogen metabolism and biosynthesis has been observed to potentially confer advantages to cancer characteristics, as evidenced by a study that demonstrated an increase in glycogen metabolism in tumor cells in vivo and cancer cells in vitro after exposure to hypoxia.40 The metabolic processes of cancer cells undergo substantial alterations as they acquire metastatic characteristics and adjust to survive in diverse environments that present differing supplies of nutrients, oxygen concentrations, and signals from outside the cell.41 The activation of p38α MAPK in cancer-associated fibroblasts (CAFs) was found to be a decisive factor for the mobilization of glycogen in cancer cells. The ablation of p38α in CAFs and the suppression of glycogen phosphorylase activity in cancer cells led to a reduction in metastatic spread in an in vivo setting. This suggests that cancer cells employ glycogen as a supply of energy to facilitate the proliferation of metastatic tumors.42 The top five enriched cellular components associated with DEGs in cancer metastasis were the "Anaphase-Promoting Complex," "Catenin Complex," P-body, "Adherens Junction," and "Apicolateral Plasma Membrane". The Anaphase-Promoting Complex (APC) is a protein complex composed of multiple subunits that serves a crucial function in the regulation of the cell cycle by facilitating the degradation of cyclins and other proteins involved in cellular regulation.43 Dysregulation of APC activity, either through increased or decreased activity, may aggravate neoplasms.43 The promotion of pancreatic cancer metastasis by human PIWIL1 is facilitated through its co-activation of the anaphase-promoting complex/cyclosome (APC/C).44 The protein complex known as Catenin is of paramount importance in both intercellular adhesion and signaling processes. The E-cadherin–catenin complex is essential for intercellular adhesion and typical and malignant tissue structures.45 The disruption of this intricate system has been associated with the initiation and progression of cancer. The diminished expression of this particular complex in malignant ailments has been linked to the invasion of tumors, metastasis, and an unfavorable prognosis, as evidenced by previous studies.46-48 P-bodies are cytoplasmic granules that participate in the degradation of mRNA and the suppression of translation. These entities are predominantly composed of mRNAs that are translationally repressed and proteins that are associated with mRNA decay. This implies that they are involved in the regulation of post-transcriptional processes.48 The precise function of P-bodies in the process of cancer metastasis remains uncertain. A recent study suggested that these mRNA decay machines may benefit malignant cells by enhancing the expression of oncogenes and decreasing the expression of tumor suppressor genes.49 The P-body-based mRNA metabolic regulators have a significant impact on the advancement and progression of cancer.49 Adherens junctions are membrane-bound structures involved in cell-cell adhesion and signaling. Cadherins and catenins are the primary molecules responsible for cell-cell adhesion, as evidenced by their central role in the formation of these adhesions.50 The implications of cancer development and metastasis have been linked to the dysregulation of adherent junctions. Alterations in the manifestation and operation of cadherins and catenins have been detected in human malignancies and have been linked to clinical consequences for patients.50
Examination of the interplay among DEGs in the context of EC metastasis involved the development of PPI networks. This approach facilitates the identification of hub genes within these networks. Remarkably, CTNNB1 is a well-known gene that is frequently mutated in EC. CTNNB1 mutations are predominantly located at β-catenin phosphorylation sites and are restricted to the endometrial subgroup lacking a specific molecular profile. Mutations in CTNNB1 cause changes in the Wnt/β-catenin signaling pathway, which promotes EC formation and progression by promoting transcription of target genes that regulate the cell cycle.51,52 Notably, recent study revealed that in low-grade endometrial endometrioid carcinomas (EECs), CTNNB1 mutations were significantly associated with local recurrence, while KRAS mutations were linked to distant metastasis/recurrence, indicating a potential genotype-dependent conditioning of these distinct progression types in EC metastasis.53 In addition, in ECs with CTNNB1 exon 3 mutations, increased β-catenin and MMP7 expression, rather than VEGF-A, were associated with bevacizumab responsiveness, suggesting a mechanistic link where overexpressed and secreted MMP7 potentially digests VEGFR-1, releasing VEGF-A and promoting the formation of permeable vessels, contributing to tumor progression and metastasis.54 Likewise, the presence of the CTNNB1p.D32A mutation, resulting in abnormal activation of β-catenin as indicated by immunohistochemistry, suggests a potential association with lung metastasis in a patient initially diagnosed with low-grade early-stage EECs.55
Of these, the remaining genes identified in EC are not known, although their involvement in the metastasis of other cancers has been identified. FGFR3, also referred to as fibroblast growth factor receptor 3, is important for tissue homeostasis, cancer, and metastasis. FGFR3, along with its multiple FGF ligands and signaling partners, is often dysregulated in cancer growth and is one of the contributing factors to therapy resistance.56 FGFR3-altered tumors in urothelial carcinoma correspond with a non-T cell-inflamed phenotype and are therefore thought to be less susceptible to immune checkpoint blockage (ICB).57 However, a study revealed that both FGFR3-mutated and wild-type bladder malignancies respond equally to ICB.57 FGFR3 is significantly expressed in melanoma and is associated with elevated Breslow thickness and lymph node metastases. FGFR3 promotes melanoma development, metastasis, and EMT characteristics, most likely through influencing ERK, AKT, and EGFR phosphorylation levels.58 In the context of metastasis, Zheng et al. indicates that FGFR3 is highly expressed in human lung cancer tissues and is closely associated with lymphatic metastasis, suggesting a potential role for the PRMT5/FGFR3/Akt signaling axis in regulating lung cancer progression and metastasis.59 In addition, FGFR3 plays a crucial role in hepatocellular carcinoma metastasis by promoting angiogenesis, particularly through the upregulation of monocyte chemotactic protein 1 (MCP-1).60
The estrogen receptor alpha (ERα) gene ESR1 encodes, and mutations in ESR1 may result in uncontrolled transcriptional activity and decreased susceptibility to endocrine treatment. These mutations are especially harmful in metastatic breast cancer, where they are seen in up to 36% of patients.61 ESR1 mutations have been shown to preexist in primary tumors and to be enriched during metastasis in clinical and preclinical studies. Furthermore, ESR1 mutations have a distinct transcriptional profile that promotes tumor growth, suggesting that certain ESR1 mutations may impact metastasis.62 ESR1 mutations, particularly the Y537S and D538G variants, demonstrate a role in promoting metastasis in estrogen receptor-positive (ER + ) breast cancer, as evidenced by altered cell adhesion and migration pathways, leading to the formation of larger circulating tumor cell clusters, ultimately suggesting potential therapeutic strategies for targeting ESR1-mutant metastatic breast cancer.63 Notably, Abdel-Hamid et al. revealed that Serum ESR1 expression exhibited a significant positive correlation with preoperative levels of the long non-coding RNA HOTAIR and miR-130a in Egyptian breast cancer patients, suggesting a potential involvement of ESR1 in metastasis and cancer progression.64 In gastric cancer (GC), Shang et al revealed that CAF-derived exosomes enhance GC cell viability, migration, and invasion by upregulating interleukin 32 (IL-32), which interacts with ESR1 and negatively regulates its expression, thereby implicating the IL-32/ESR1 axis as a crucial factor in promoting metastatic behavior in GC cells.65
SRSF3 participates in the growth and progression of cancer. It increases cell proliferation, cell cycle, and metastasis, while inhibiting cell senescence, programmed cell death, and autophagy. SRSF3 knockdown significantly inhibited tumor cell proliferation and metastatic ability.66,67 Notably, based on our results, SRSF3 was the only hub gene associated with the EC patient’s outcome, and its expression was decreased (log2FC = -1.577, P value = 8.52E-08). This pivotal role of SRSF3 in cancer progression aligns with the findings from the study by Zhang et al., where the investigation into cervical cancer cells, particularly SiHa cells, revealed that reducing the SRSF3 level effectively inhibits viability and metastasis by suppressing the PI3K/AKT/mTOR signaling pathway.68 The up-regulating of SRSF3 by LINC01210 in the context of CRC adds another layer to the intricate role of SRSF3 in cancer biology. In CRC cells, LINC01210 has been identified as an accelerator of proliferation and invasion, operating through the epigenetic upregulation of SRSF3.69 This revelation underscores the multifaceted impact of SRSF3 on cancer progression across diverse malignancies.
Finally, the results of this study identified several DElncRNAs that might be involved in EC metastasis, including LINC01541, SNHG17, LINC00520, BHLHE22-AS1, LOC100509445, H19, and HOTAIRM1. In this regard, LINC01541 plays an important role in 17-estradiol (17-E2)-stimulated endometrial stromal cells (ESCs) by functioning as a molecular reservoir to decrease the bioavailability of miR-506-5p.70 SNHG17, in particular, induces colorectal carcinogenesis and metastasis by modulating the Trim23-PES1 axis and the miR-339-5p-FOSL2-SNHG17 positive feedback circuit.71 Furthermore, via activating the miR-125b-5p/EIF5A2 axis, LINC00520 promotes malignant melanoma development and metastasis.72 BHLHE22, on the other hand, is a member of the basic helix loop helix transcription factor family that acts as an inhibitor of transcription and is involved in cellular differentiation.73 A prior investigation of the methylome has revealed that BHLHE22 exhibits hypermethylation in endocervical tissues and can be identified via a Pap-smear specimen.73 The expression of the BHLHE22 protein was observed to be considerably reduced in EC in comparison to the normal endometrium.73 The study found a significant correlation between elevated levels of BHLHE22 and the microsatellite-instable subtype, endometrioid type, grade, and age of the subjects.73 BHLHE22-AS1 lncRNA, as BHLHE22 antisense RNA due to its decreased expression in EC metastatic samples and increased expression of BHLHE22 in EC, can be a new target for further studies on EC. It has been discovered that H19 lncRNA is aberrantly expressed in human aggressive tumors, where it serves as an oncogene.74 H19 has been linked to cell proliferation, spread, migration, EMT, metastasis, and programmed cell death.75 In addition to sequestering microRNAs, H19 facilitates a molecular regulatory mechanism with multiple layers.75 HOTAIRM1 is a lncRNA that is localized to the 5' end of the homeobox A (HOXA) gene cluster. It is also known as HOXA transcript antisense RNA myeloid-specific 1. It is an endogenous antisense transcript of the HOXA1 gene that is expressed in the myeloid lineage76 and activated during neural development.77 The gene HOTAIRM1 is significantly involved in the process of myeloid maturation and exhibits high expression levels in cases of acute myeloid leukemia, thereby exerting a notable influence on the prognosis of affected individuals.78,79 HOTAIRM1 has been identified as having low expression levels in both tissues and plasma of colorectal cancer patients, indicating its potential as a diagnostic biomarker for this disease.80
This study utilized publicly accessible transcriptome datasets with an awareness of potential biases related to sample collection, processing, and analysis. Although the identification of DEGs and DElncRNAs associated with EC metastasis was a focus, the study did not delve into their in vitro or in vivo functions. To address this limitation and enhance robustness, we have incorporated suggestions for improvement by advocating the validation of key biomarkers using quantitative polymerase chain reaction (qPCR). Despite these acknowledged constraints, our work contributes to the understanding of the molecular underpinnings of EC metastasis, offering valuable insights for future research directions and potential treatment options.
Conclusion
Metastasis is a key determinant of poor prognosis and treatment results for EC, which is one of the largest causes of female deaths globally. This study utilized bioinformatic analyses of publicly accessible transcriptome datasets to identify EC metastatic molecular pathways. DEGs associated with EC metastasis were prominent in cancer and cell cycle regulatory pathways in KEGG and GO pathway studies. According to PPI network research, EC metastases may include hub genes CTNNB1, FGFR3, ESR1, and SRSF3. EC metastasis may be linked to several DElncRNAs. Additional experimental studies are needed to validate these results, clarify the precise roles of the identified genes and lncRNAs in EC metastasis, and pave the way for the development of novel diagnostic and therapeutic approaches to EC metastasis. Identifying the major molecular participants and pathways involved in EC metastasis is essential to establish effective diagnostic and treatment methods. Bioinformatics analysis revealed the complicated molecular pathways of EC metastasis and provided new research areas.
Research Highlights
What is the current knowledge?
-
The incidence and mortality rates of endometrial cancer (EC) have increased globally, with limited understanding of distant metastasis.
-
Approximately 3% of patients with EC were diagnosed with stage IV disease between 1999 and 2001.
-
However, the molecular mechanisms and pathways underlying EC metastasis remain poorly characterized.
What is new here?
-
Glycogen metabolism has emerged as a potential contributor to cancer characteristics, impacting the energy supply to tumor cells.
-
Key cellular components, such as the anaphase-promoting complex and adherens junctions, have been implicated in cancer metastasis.
-
Hub genes, including CTNNB1, FGFR3, ESR1, and SRSF3, have been identified in the context of EC metastasis.
-
Several differentially expressed long non-coding RNAs (DElncRNAs) such as LINC01541, SNHG17, and H19 have been identified as potential players in EC metastasis.
Competing Interests
The author Amin Tamadon, was employed by PerciaVista R&D Co. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethical Statement
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Tabriz University of Medical Sciences (IR.TBZMED.VCR.REC.1399.444).
Acknowledgements
We would like to thank Stem Cell Research Center (SCRC) and Women's Reproductive Health Research Center, Tabriz University of Medical Sciences for supporting this research.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020; 70:7-30. doi: 10.3322/caac.21590 [Crossref] [ Google Scholar]
- Dou Y, Kawaler EA, Cui Zhou D, Gritsenko MA, Huang C, Blumenberg L, et al. Proteogenomic characterization of endometrial carcinoma. Cell 2020; 180: 729-48.e26. 10.1016/j.cell.2020.01.026.
- Mitamura T, Watari H, Wang L, Kanno H, Kitagawa M, Hassan MK. microRNA 31 functions as an endometrial cancer oncogene by suppressing Hippo tumor suppressor pathway. Mol Cancer 2014; 13:97. doi: 10.1186/1476-4598-13-97 [Crossref] [ Google Scholar]
- Connor EV, Rose PG. Management strategies for recurrent endometrial cancer. Expert Rev Anticancer Ther 2018. 18: 873-85. 10.1080/14737140.2018.1491311.
- de Boer SM, Wortman BG, Bosse T, Powell ME, Singh N, Hollema H. Clinical consequences of upfront pathology review in the randomised PORTEC-3 trial for high-risk endometrial cancer. Ann Oncol 2018; 29:424-30. doi: 10.1093/annonc/mdx753 [Crossref] [ Google Scholar]
- Brown D, Smeets D, Székely B, Larsimont D, Szász AM, Adnet PY. Phylogenetic analysis of metastatic progression in breast cancer using somatic mutations and copy number aberrations. Nat Commun 2017; 8:14944. doi: 10.1038/ncomms14944 [Crossref] [ Google Scholar]
- Alitalo A, Detmar M. Interaction of tumor cells and lymphatic vessels in cancer progression. Oncogene 2012; 31:4499-508. doi: 10.1038/onc.2011.602 [Crossref] [ Google Scholar]
- Weber GF. Molecular mechanisms of metastasis. Cancer Lett 2008; 270:181-90. doi: 10.1016/j.canlet.2008.04.030 [Crossref] [ Google Scholar]
- Ashley CW, Da Cruz Paula A, Kumar R, Mandelker D, Pei X, Riaz N. Analysis of mutational signatures in primary and metastatic endometrial cancer reveals distinct patterns of DNA repair defects and shifts during tumor progression. Gynecol Oncol 2019; 152:11-9. doi: 10.1016/j.ygyno.2018.10.032 [Crossref] [ Google Scholar]
- Ding H, Jiang F, Deng L, Wang J, Wang P, Ji M. Prediction of clinical outcome in endometrial carcinoma based on a 3-lncRNA signature. Front Cell Dev Biol 2021; 9:814456. doi: 10.3389/fcell.2021.814456 [Crossref] [ Google Scholar]
- Gao N, Li Y, Li J, Gao Z, Yang Z, Li Y. Long non-coding RNAs: the regulatory mechanisms, research strategies, and future directions in cancers. Front Oncol 2020; 10:598817. doi: 10.3389/fonc.2020.598817 [Crossref] [ Google Scholar]
- Moslehian MS, Shabkhizan R, Asadi MR, Bazmani A, Mahdipour M, Haiaty S. Interaction of lncRNAs with mTOR in colorectal cancer: a systematic review. BMC Cancer 2023; 23:512. doi: 10.1186/s12885-023-11008-9 [Crossref] [ Google Scholar]
- Liu SJ, Dang HX, Lim DA, Feng FY, Maher CA. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer 2021; 21:446-60. doi: 10.1038/s41568-021-00353-1 [Crossref] [ Google Scholar]
- Hunt GP, Grassi L, Henkin R, Smeraldi F, Spargo TP, Kabiljo R. GEOexplorer: a webserver for gene expression analysis and visualisation. Nucleic Acids Res 2022; 50:W367-74. doi: 10.1093/nar/gkac364 [Crossref] [ Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 2013; 41:D991-5. doi: 10.1093/nar/gks1193 [Crossref] [ Google Scholar]
- Casablanca Y, Wang G, Lankes HA, Tian C, Bateman NW, Miller CR. Improving risk assessment for metastatic disease in endometrioid endometrial cancer patients using molecular and clinical features: an NRG Oncology/Gynecologic oncology group study. Cancers (Basel) 2022; 14:4070. doi: 10.3390/cancers14174070 [Crossref] [ Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016; 44:W90-7. doi: 10.1093/nar/gkw377 [Crossref] [ Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-9. doi: 10.1038/75556 [Crossref] [ Google Scholar]
- Gene Ontology Consortium. The gene ontology (GO) project in 2006. Nucleic Acids Res 2006; 34:D322-6. doi: 10.1093/nar/gkj021 [Crossref] [ Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 2016; 44:D457-62. doi: 10.1093/nar/gkv1070 [Crossref] [ Google Scholar]
- Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 2021; 49:D605-d12. doi: 10.1093/nar/gkaa1074 [Crossref] [ Google Scholar]
- Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res 2019; 18:623-32. doi: 10.1021/acs.jproteome.8b00702 [Crossref] [ Google Scholar]
- Li C, Tang Z, Zhang W, Ye Z, Liu F. GEPIA2021: integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res 2021; 49:W242-6. doi: 10.1093/nar/gkab418 [Crossref] [ Google Scholar]
- Sorosky JI. Endometrial cancer. Obstet Gynecol 2012; 120:383-97. doi: 10.1097/AOG.0b013e3182605bf1 [Crossref] [ Google Scholar]
- Creasman WT, Odicino F, Maisonneuve P, Quinn MA, Beller U, Benedet JL. Carcinoma of the corpus uteri FIGO 26th Annual Report on the Results of Treatment in Gynecological Cancer. Int J Gynaecol Obstet 2006; 95 Suppl 1:S105-43. doi: 10.1016/s0020-7292(06)60031-3 [Crossref] [ Google Scholar]
- Devereaux KA, Weiel JJ, Pors J, Steiner DF, Ho C, Charu V. Prospective molecular classification of endometrial carcinomas: institutional implementation, practice, and clinical experience. Mod Pathol 2022; 35:688-96. doi: 10.1038/s41379-021-00963-y [Crossref] [ Google Scholar]
- Wang Y, Zhang S. Berberine suppresses growth and metastasis of endometrial cancer cells via miR-101/COX-2. Biomed Pharmacother 2018; 103:1287-93. doi: 10.1016/j.biopha.2018.04.161 [Crossref] [ Google Scholar]
- Makker A, Goel MM. Tumor progression, metastasis, and modulators of epithelial-mesenchymal transition in endometrioid endometrial carcinoma: an update. Endocr Relat Cancer 2016; 23:R85-r111. doi: 10.1530/erc-15-0218 [Crossref] [ Google Scholar]
- Lax SF, Tamussino KF, Lang PF. [Metastatic mechanisms of uterine malignancies and therapeutic consequences]. Pathologe 2016; 37:549-56. doi: 10.1007/s00292-016-0243-z.[German] [Crossref] [ Google Scholar]
- Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science 2011; 331:1559-64. doi: 10.1126/science.1203543 [Crossref] [ Google Scholar]
- Braicu C, Buse M, Busuioc C, Drula R, Gulei D, Raduly L. A comprehensive review on MAPK: a promising therapeutic target in cancer. Cancers (Basel) 2019; 11:1618. doi: 10.3390/cancers11101618 [Crossref] [ Google Scholar]
- He Y, Sun MM, Zhang GG, Yang J, Chen KS, Xu WW. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct Target Ther 2021; 6:425. doi: 10.1038/s41392-021-00828-5 [Crossref] [ Google Scholar]
- Li J, Wang J, Xie D, Pei Q, Wan X, Xing HR. Characteristics of the PI3K/AKT and MAPK/ERK pathways involved in the maintenance of self-renewal in lung cancer stem-like cells. Int J Biol Sci 2021; 17:1191-202. doi: 10.7150/ijbs.57871 [Crossref] [ Google Scholar]
- Zhang YL, Wang RC, Cheng K, Ring BZ, Su L. Roles of Rap1 signaling in tumor cell migration and invasion. Cancer Biol Med 2017; 14:90-9. doi: 10.20892/j.issn.2095-3941.2016.0086 [Crossref] [ Google Scholar]
- Shah S, Brock EJ, Ji K, Mattingly RR. Ras and Rap1: a tale of two GTPases. Semin Cancer Biol 2019; 54:29-39. doi: 10.1016/j.semcancer.2018.03.005 [Crossref] [ Google Scholar]
- Bailey CL, Kelly P, Casey PJ. Activation of Rap1 promotes prostate cancer metastasis. Cancer Res 2009; 69:4962-8. doi: 10.1158/0008-5472.can-08-4269 [Crossref] [ Google Scholar]
- Kawauchi T. Cell adhesion and its endocytic regulation in cell migration during neural development and cancer metastasis. Int J Mol Sci 2012; 13:4564-90. doi: 10.3390/ijms13044564 [Crossref] [ Google Scholar]
- Bischoff P, Kornhuber M, Dunst S, Zell J, Fauler B, Mielke T. Estrogens determine adherens junction organization and E-cadherin clustering in breast cancer cells via amphiregulin. iScience 2020; 23:101683. doi: 10.1016/j.isci.2020.101683 [Crossref] [ Google Scholar]
- Knights AJ, Funnell AP, Crossley M, Pearson RC. Holding tight: cell junctions and cancer spread. Trends Cancer Res 2012; 8:61-9. [ Google Scholar]
- Favaro E, Bensaad K, Chong MG, Tennant DA, Ferguson DJ, Snell C. Glucose utilization via glycogen phosphorylase sustains proliferation and prevents premature senescence in cancer cells. Cell Metab 2012; 16:751-64. doi: 10.1016/j.cmet.2012.10.017 [Crossref] [ Google Scholar]
- Teoh ST, Lunt SY. Metabolism in cancer metastasis: bioenergetics, biosynthesis, and beyond. Wiley Interdiscip Rev Syst Biol Med 2018; 10:e1406. doi: 10.1002/wsbm.1406 [Crossref] [ Google Scholar]
- Curtis M, Kenny HA, Ashcroft B, Mukherjee A, Johnson A, Zhang Y, et al. Fibroblasts mobilize tumor cell glycogen to promote proliferation and metastasis. Cell Metab 2019; 29: 141-55.e9. 10.1016/j.cmet.2018.08.007.
- Kazemi-Sefat GE, Keramatipour M, Talebi S, Kavousi K, Sajed R, Kazemi-Sefat NA. The importance of CDC27 in cancer: molecular pathology and clinical aspects. Cancer Cell Int 2021; 21:160. doi: 10.1186/s12935-021-01860-9 [Crossref] [ Google Scholar]
- Yao F, Ma L. piRNA-unbound PIWIL1 promotes metastasis. Nat Cell Biol 2020; 22:359-60. doi: 10.1038/s41556-020-0502-3 [Crossref] [ Google Scholar]
- Tian X, Liu Z, Niu B, Zhang J, Tan TK, Lee SR. E-cadherin/β-catenin complex and the epithelial barrier. J Biomed Biotechnol 2011; 2011:567305. doi: 10.1155/2011/567305 [Crossref] [ Google Scholar]
- Bremnes RM, Veve R, Hirsch FR, Franklin WA. The E-cadherin cell-cell adhesion complex and lung cancer invasion, metastasis, and prognosis. Lung Cancer 2002; 36:115-24. doi: 10.1016/s0169-5002(01)00471-8 [Crossref] [ Google Scholar]
- Kim WK, Kwon Y, Jang M, Park M, Kim J, Cho S. β-catenin activation down-regulates cell-cell junction-related genes and induces epithelial-to-mesenchymal transition in colorectal cancers. Sci Rep 2019; 9:18440. doi: 10.1038/s41598-019-54890-9 [Crossref] [ Google Scholar]
- Luo Y, Na Z, Slavoff SA. P-Bodies: composition, properties, and functions. Biochemistry 2018; 57:2424-31. doi: 10.1021/acs.biochem.7b01162 [Crossref] [ Google Scholar]
- Nsengimana B, Khan FA, Ngowi EE, Zhou X, Jin Y, Jia Y. Processing body (P-body) and its mediators in cancer. Mol Cell Biochem 2022; 477:1217-38. doi: 10.1007/s11010-022-04359-7 [Crossref] [ Google Scholar]
- Vasioukhin V. Adherens junctions and cancer. Subcell Biochem 2012; 60:379-414. doi: 10.1007/978-94-007-4186-7_16 [Crossref] [ Google Scholar]
- Travaglino A, Raffone A, Raimondo D, Reppuccia S, Ruggiero A, Arena A. Prognostic significance of CTNNB1 mutation in early-stage endometrial carcinoma: a systematic review and meta-analysis. Arch Gynecol Obstet 2022; 306:423-31. doi: 10.1007/s00404-021-06385-0 [Crossref] [ Google Scholar]
- Ledinek Ž, Sobočan M, Knez J. The role of CTNNB1 in endometrial cancer. Dis Markers 2022; 2022:1442441. doi: 10.1155/2022/1442441 [Crossref] [ Google Scholar]
- Chibbar R, Foerstner S, Suresh J, Chibbar R, Piche A, Kundapur D. Estrogen/progesterone receptor loss, CTNNB1 and KRAS mutations are associated with local recurrence or distant metastasis in low-grade endometrial endometrioid carcinoma. Appl Immunohistochem Mol Morphol 2023; 31:181-8. doi: 10.1097/pai.0000000000001102 [Crossref] [ Google Scholar]
- Berger AA, Kawaler EA, Dao F, Misirlioglu S, Fernandez EA, Olvera N. The role of CTNNB1 mutations and matrix metalloproteinases (MMPs) in anti-angiogenesis treatment of endometrial carcinoma. Gynecol Oncol 2022; 167:323-33. doi: 10.1016/j.ygyno.2022.09.013 [Crossref] [ Google Scholar]
- Zhong L, Jiang W, RutieYin RutieYin, Liu H, Song L. CTNNB1 pD32A (c95A > C) somatic mutation in stage I grade 1 endometrioid endometrial carcinoma with lung metastasis: a case report. BMC Med Genomics 2023; 16:137. doi: 10.1186/s12920-023-01570-3 [Crossref] [ Google Scholar]
- Krook MA, Reeser JW, Ernst G, Barker H, Wilberding M, Li G. Fibroblast growth factor receptors in cancer: genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance. Br J Cancer 2021; 124:880-92. doi: 10.1038/s41416-020-01157-0 [Crossref] [ Google Scholar]
- Rose TL, Weir WH, Mayhew GM, Shibata Y, Eulitt P, Uronis JM. Fibroblast growth factor receptor 3 alterations and response to immune checkpoint inhibition in metastatic urothelial cancer: a real-world experience. Br J Cancer 2021; 125:1251-60. doi: 10.1038/s41416-021-01488-6 [Crossref] [ Google Scholar]
- Li L, Zhang S, Li H, Chou H. FGFR3 promotes the growth and malignancy of melanoma by influencing EMT and the phosphorylation of ERK, AKT, and EGFR. BMC Cancer 2019; 19:963. doi: 10.1186/s12885-019-6161-8 [Crossref] [ Google Scholar]
- Zheng Y, Lu J, Hu X, Hu X, Gao X, Zhou J. PRMT5/FGFR3/AKT signaling axis facilitates lung cancer cell metastasis. Technol Cancer Res Treat 2023; 22:15330338231161139. doi: 10.1177/15330338231161139 [Crossref] [ Google Scholar]
- Liu X, Jing X, Cheng X, Ma D, Jin Z, Yang W. FGFR3 promotes angiogenesis-dependent metastasis of hepatocellular carcinoma via facilitating MCP-1-mediated vascular formation. Med Oncol 2016; 33:46. doi: 10.1007/s12032-016-0761-9 [Crossref] [ Google Scholar]
- Herzog SK, Fuqua SAW. ESR1 mutations and therapeutic resistance in metastatic breast cancer: progress and remaining challenges. Br J Cancer 2022; 126:174-86. doi: 10.1038/s41416-021-01564-x [Crossref] [ Google Scholar]
- Dustin D, Gu G, Fuqua SA. ESR1 mutations in breast cancer. Cancer 2019; 125:3714-28. doi: 10.1002/cncr.32345 [Crossref] [ Google Scholar]
- Li Z, Wu Y, Yates ME, Tasdemir N, Bahreini A, Chen J. Hotspot ESR1 mutations are multimodal and contextual modulators of breast cancer metastasis. Cancer Res 2022; 82:1321-39. doi: 10.1158/0008-5472.can-21-2576 [Crossref] [ Google Scholar]
- Abdel-Hamid NR, Mohammed EA, Toraih EA, Kamel MM, Abdelhafiz AS, Badr FM. Circulating ESR1, long non-coding RNA HOTAIR and microRNA-130a gene expression as biomarkers for breast cancer stage and metastasis. Sci Rep 2023; 13:22654. doi: 10.1038/s41598-023-50007-5 [Crossref] [ Google Scholar]
- Shang L, Chen X, Zhu T, Chong S, Liu H, Huang W, et al. Cancer-associated fibroblast-secreted exosomes promote gastric cancer cell migration and invasion via the IL-32/ESR1 axis. Appl Biochem Biotechnol 2024. 10.1007/s12010-023-04782-6.
- Xiong J, Chen Y, Wang W, Sun J. Biological function and molecular mechanism of SRSF3 in cancer and beyond. Oncol Lett 2022; 23:21. doi: 10.3892/ol.2021.13139 [Crossref] [ Google Scholar]
- Zhou Z, Gong Q, Lin Z, Wang Y, Li M, Wang L. Emerging roles of SRSF3 as a therapeutic target for cancer. Front Oncol 2020; 10:577636. doi: 10.3389/fonc.2020.577636 [Crossref] [ Google Scholar]
- Zhang L, Li J, Zhang L. SRSF3 restriction eases cervical cancer cell viability and metastasis via adjusting PI3K/AKT/mTOR signaling pathway. Contrast Media Mol Imaging 2022; 2022:8497078. doi: 10.1155/2022/8497078 [Crossref] [ Google Scholar]
- Luo J, Gao K, Chen M, Tian B. LINC01210 promotes malignant phenotypes of colorectal cancer through epigenetically upregulating SRSF3. Pathol Res Pract 2022; 234:153905. doi: 10.1016/j.prp.2022.153905 [Crossref] [ Google Scholar]
- Mai H, Xu H, Lin H, Wei Y, Yin Y, Huang Y. LINC01541 functions as a ceRNA to modulate the Wnt/β-catenin pathway by decoying miR-506-5p in endometriosis. Reprod Sci 2021; 28:665-74. doi: 10.1007/s43032-020-00295-3 [Crossref] [ Google Scholar]
- Qiao D, Qin X, Yang H, Liu X, Liu L, Liu S. Estradiol mediates the interaction of LINC01541 and miR-429 to promote angiogenesis of G1/G2 endometrioid adenocarcinoma in-vitro: a pilot study. Front Oncol 2022; 12:951573. doi: 10.3389/fonc.2022.951573 [Crossref] [ Google Scholar]
- Luan W, Ding Y, Yuan H, Ma S, Ruan H, Wang J. Long non-coding RNA LINC00520 promotes the proliferation and metastasis of malignant melanoma by inducing the miR-125b-5p/EIF5A2 axis. J Exp Clin Cancer Res 2020; 39:96. doi: 10.1186/s13046-020-01599-7 [Crossref] [ Google Scholar]
- Darmawi Darmawi, Chen LY, Su PH, Liew PL, Wang HC, Weng YC. BHLHE22 expression is associated with a proinflammatory immune microenvironment and confers a favorable prognosis in endometrial cancer. Int J Mol Sci 2022; 23:7158. doi: 10.3390/ijms23137158 [Crossref] [ Google Scholar]
- Yang J, Qi M, Fei X, Wang X, Wang K. LncRNA H19: a novel oncogene in multiple cancers. Int J Biol Sci 2021; 17:3188-208. doi: 10.7150/ijbs.62573 [Crossref] [ Google Scholar]
- Garcia-Padilla C, Lozano-Velasco E, Del Mar Muñoz-Gallardo MD, Castillo-Casas JM, Caño-Carrillo S, Martínez-Amaro FJ. LncRNA H19 impairs chemo and radiotherapy in tumorigenesis. Int J Mol Sci 2022; 23:8309. doi: 10.3390/ijms23158309 [Crossref] [ Google Scholar]
- Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 2009; 113:2526-34. doi: 10.1182/blood-2008-06-162164 [Crossref] [ Google Scholar]
- Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS One 2011; 6:e23356. doi: 10.1371/journal.pone.0023356 [Crossref] [ Google Scholar]
- Zhang X, Weissman SM, Newburger PE. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol 2014; 11:777-87. doi: 10.4161/rna.28828 [Crossref] [ Google Scholar]
- Díaz-Beyá M, Brunet S, Nomdedéu J, Pratcorona M, Cordeiro A, Gallardo D. The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget 2015; 6:31613-27. doi: 10.18632/oncotarget.5148 [Crossref] [ Google Scholar]
- Wan L, Kong J, Tang J, Wu Y, Xu E, Lai M. HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J Cell Mol Med 2016; 20:2036-44. doi: 10.1111/jcmm.12892 [Crossref] [ Google Scholar]