Bioimpacts. 2025;15:30232.
doi: 10.34172/bi.30232
Original Article
Performance of protein N linear epitopes in serodiagnosis of COVID-19 infection
Sahar Farajnia Conceptualization, Data curation, Investigation, Methodology, Resources, 1, 2 
Nazli Khajenasiri Writing – review & editing, 1
Safar Farajnia Funding acquisition, Project administration, Supervision, 2, 3, * 
Farzin Seyrafi Formal analysis, Writing – original draft, 3
Nasim Bakhtiyari Visualization, 3, 4
Author information:
1Department of Biology, University of Rabe Rashidi, Tabriz, Iran
2Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
3Biotechnology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
4Student Research Committee, Tabriz University of Medical Sciences, Tabriz, Iran
Abstract
Introduction:
Despite the efforts to contain the spread of COVID-19, the virus remains in circulation, posing a considerable risk to populations across the globe. Hence, rapid and early detection of this infection is essential for effective disease control. The nucleocapsid (N) protein of the virus serves as a primary target for antibody response during CoV2 infections, making it a potential candidate for COVID-19 detection. This study aims to prepare and evaluate the linear epitopes of the N protein for serodiagnosis of COVID-19 infection.
Methods:
The linear epitope of the N protein gene was identified using ABCpred, BCpred, and IEDB. These epitopes were subsequently amplified by RT-PCR, cloned, and expressed in soluble form in the E. coli BL21 strain. The recombinant protein was purified using the Ni-NTA column. The reactivity of purified N protein with sera from SARS-CoV-2 patients was analyzed using an ELISA assay.
Results:
Sequencing analysis demonstrated the successful cloning of the linear epitopes of the N protein into the PET-28a vector, along with an n-terminal His-tag fusion. The recombinant protein was produced in E. coli BL21 and purified with a Ni-NTA column. The analysis demonstrated that the N protein linear epitopes were expressed in a soluble form and appeared as a 50 kDa band in the SDS-PAGE. Examination for the reactivity of the purified N protein with the COVID-19 patient’s sera by ELISA revealed that the N protein recognizes the infection with high sensitivity and specificity.
Conclusion:
The results of this study indicated that linear epitopes of the SARS-CoV-2 N protein are highly immunogenic and could be exploited for serodiagnosis of infection in patients suspected of COVID-19 infection.
Keywords: SARS-CoV-2, N protein, Linear epitopes, Serodiagnosis, COVID-19
Copyright and License Information
© 2025 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
The support provided by the Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran (grant no. 69285).
Introduction
The coronavirus infection (COVID-19) was discovered in China in December 2019 and rapidly disseminated worldwide. Then, the epidemic was classified as a global pandemic on March 12, 2020. However, its rapid expansion poses a significant threat to the worldwide population, giving rise to a wide range of illnesses, ranging from asymptomatic infections to severe pneumonia and even fatalities.1,2 The SARS-CoV-2 30 kb positive-sense RNA genome contains 14 ORFs encoding 27 different proteins consisting of orf1ab and orf1a open reading frames that encode 15 non-structural proteins (NSPs), NSP1-NSP10, and NSP12-NSP16. The 3′end of the genome encrypts four viral structural proteins and a large number of auxiliary proteins labeled ORF3-ORF10.3,4 Proteins such as membrane (M), spike (S), envelope (E), and nucleocapsid (N) serve as essential structural elements.4 The N protein possesses diverse functions, playing essential roles in various stages of the viral life cycle, including viral budding, viral assembly, regulation of the host cell cycle, and viral mRNA replication.5,6 Given the virus's high transmission rate, early disease detection has gained significant importance. Serological diagnosis, a noninvasive technique, offers numerous advantages in diagnosing the disease. However the development of a proper serological diagnosis method requires the identification of epitopes that are the target of the antibody response against the virus.7,8 Hence, various antigens have been attempted and introduced as a candidates for serodiagnosis of COVID-19, including the nucleocapsid (N) protein, the S protein, the S1 subunit, and theRBD.
To date, the development of serology-based diagnostic tests has primarily focused on anti-S and anti-N antibodies in SARS-CoV-2. These proteins possess high immunogenicity and elicit a robust antibody response during the initial stages of infection. Moreover, these proteins play a crucial role as the specific targets for neutralizing antibodies.3,9-11 It has been shown that the SARS-CoV-2 nucleocapsid protein is the main target of antibody responses upon SARS-CoV-2 infections, and their antibodies can identify early infection more reliably than spike protein antibodies.12,13 The production of sensitive and reliable serological assays necessitate the recombinant production of the SARS-CoV-2 N antigen.14,15 Although various expression systems have been tried to produce recombinant proteins, including mammalian, yeast, insect, and bacterial expression systems, the expression in E. coli displays several advantages. It has been shown that protein production in prokaryotic systems like E. coli provides an inexpensive tool for rapidly producing large quantities of recombinant protein. The main disadvantage of this system is its inability to express conformationally complex epitopes. In the present study, the linear epitopes of the SARS-CoV-2 N protein were identified and expressed in the E. coli expression system in soluble form. The recombinant protein was then purified, and its performance for serodiagnosis of SARS-CoV-2 was evaluated using the ELISA method.
Materials and Methods
Linear epitope prediction and assessment
Credible B cell epitope prediction software holds significant importance in numerous clinical and biotechnological applications, including vaccine development and the production of therapeutic antibodies.16
Bepipred 2.0 and the IEBD (the Immune Epitope Database) web server interface were used to predict linear B-cell epitopes in the SARS-CoV-2 N-protein, which could potentially elicit an antibody response.17 The N-protein sequence was retrieved from the NCBI protein database. The Bepipred 2.0 tool was employed to assess whether an amino acid within the protein could potentially be part of an epitope.18 All predictions exceeding the designated threshold (defaulted to 0.5) are denoted as 'E' in the 'Epitopes' line located above the sequence.19
After analyzing the epitopes with the highest scores, the B-cell epitope-enriched regions were chosen as a template for amplification, cloning, and subsequent production.20
RNA extraction and PCR amplification
Samples from the initial outbreak of SARS-CoV-2 were used for RNA isolation and PCR amplification. The total RNA extraction was done using the RNX-plus solution following the manufacturer's instructions. Briefly, one milliliter of RNX-PLUS solution was added to the homogenized sample, vortexed, and incubated at 25 °C for 5 minutes. After adding chloroform, the tube contents were well mixed and incubated on ice. After centrifuging at 12 000 rpm for 15 minutes, the upper phase was transferred to a new tube, combined with isopropanol, and incubated on ice for 15 minutes. The pellet was then washed with 1 ml of 75% ethanol, dried at RT for 15 minutes, and dissolved in 50 μl of DEPC-treated water.21,22
PCR amplification and cloning
First-strand cDNA was synthesized by the Sina Clon first-strand cDNA synthesis kit using viral RNA as a template, according to the protocol recommended by the producer. Two microliters of cDNA were entered into the PCR and amplified using primers designed for the SARS-CoV-2 protein N linear epitope-enriched region (Table 1). The PCR product was purified, ligated into the pET28a vector after digestion with Sal1 and Nhe1, and transformed into the E. coli DH5 strain. 23,24 The integrity and sequence of the cloned gene were confirmed by PCR and sequencing.25 The sequence of Linear epitopes of SARS-CoV-2 was deposited in GENE BANK under accession number PP275143.
Table 1.
The list of primers employed for PCR amplification of the N protein linear epitopes
|
Primer type
|
|
Sequence
|
| Vector primers |
T7F |
TAATACGACTCACTATAGGG |
| T7R |
GCTAGTTATTGCTCAGCGG |
| N protein-specific primers |
N F |
TAGCTAGCATGTCTGATAATGGACCCC |
| N R |
ATAGTCGACGGCCTGAGTTGAGTCAGCA |
Recombinant expression and purification
For recombinant expression, the pET28a-SARS-CoV-2 N protein construct was transferred into the E. coli BL21 strain and plated on a kanamycin-containing LB agar. Selected bacterial colonies were cultivated in LB broth containing kanamycin (50 mcg/ml). The cultures were transferred to 200 ml of fresh liquid LB media containing kanamycin. At an OD600 of 0.6, the culture was induced with 0.3 mM IPTG, and incubation was continued at 20 °C for protein expression. The culture was centrifuged at 9000 rpm for 10 minutes to harvest the bacteria. The cells were suspended in the lysis buffer (300 mM NaCl and 100 mM NaH2PO4 at pH 8) and lysed by sonication for 20 cycles (30 s pulse on and 30 s pulse off). The lysate was subjected to centrifugation at 9000 rpm, and the soluble and insoluble fractions were analyzed using 12% SDS-polyacrylamide gel electrophoresis.23,25 For purification, the soluble fraction of the lysate was passed over the Ni-NTA column after equilibration with buffer A (300 mM NaCl, 50 mM NaH2PO4, 10 mM imidazole, pH 8). The column was washed with buffer B (NaH2PO4, NaCl, pH 8, containing 20 mM imidazole). After that, the protein was eluted with buffer A containing imidazole (250 mM), and the protein purity was analyzed using SDS-PAGE. The purified protein was dialyzed against PBS (pH 7.2), and its concentration was determined at 280 nm using a nanodrop spectrophotometer.23,26
ELISA assay
The enzyme-linked immunosorbent assay (ELISA) was utilized to determine the diagnostic efficacy of the SARS-CoV-2 N protein. The purified N protein was diluted in PBS buffer (pH 7.2) and coated in a 96-well microtiter ELISA plate20,27 overnight at 4 °C. The following day, the wells were washed with PBS containing 0.05% Twin 20 (PBS-T), blocked with 0.1% Twin 20, and incubated for one hour at 37 °C. The patient sera were diluted with PBS buffer, added to the wells, and incubated at 37 °C for one hour. The wells were washed with PBS-T and incubated with Horse Reddish Peroxidase-conjugated goat anti-human IgG for 1 hour at RT. Following washing, TMB substrate was added to the plate and incubated for 20 minutes. The level of reactivity was then assessed by measuring the optical densities at 450 nm after the reaction was stopped with 0.1 M HCL.27,28
Statistical analysis
The accuracy of the assay was assessed by analyzing the results of one hundred serum samples obtained during the COVID-19 epidemic. Statistical analysis using GraphPad software was employed to determine the sensitivity and specificity of the ELISA results. The sensitivity and specificity were determined according to the following equations:
(1)
(2)
(3)
(4)
TN represents true negative, TP is true positive, FN is false negative, and FP is false positive.29
Results
Patients’ information
Serum samples were obtained from individuals who underwent RT-PCR testing for COVID-19 at the Central Laboratory of East Azerbaijan Province. Among the 100 patients, 36 were male with an average age of 40 years, while 64 were female with an average age of 37 years. Among the 50 positive cases, 15 were male and 35 were female. Additionally, out of the 50 negative cases, 29 were female and 21 were male.
Predicting linear epitopes on protein N
Bepipred 2.0, BC pred, ABC pred, and the IEBD web server were utilized to predict the linear B-cell epitopes (Tables 2-4). An analysis was conducted on the results obtained from three software programs, and their findings were compared. The selection of epitope regions was based on the examination of scores and the consideration of physicochemical properties, including hydrophilicity ( > 0), flexibility, surface accessibility ( > 0), and antigenicity ( > 0). Fig. 1 illustrates the N protein sequences and its linear epitope enriched regions.
Table 2.
Epitopes predicted by BCpred
|
Position
|
Epitope
|
Score
|
| 361 |
KTFPPTEPKKDKKKKADETQ |
1 |
| 18 |
GGPSDSTGSNQNGERSGARS |
1 |
| 139 |
LNTPKDHIGTRNPANNAAIV |
0.997 |
| 114 |
GTGPEAGLPYGANKDGIIWV |
0.997 |
| 198 |
TPGSSRGTSPARMAGNGGDA |
0.995 |
| 67 |
PRGQGVPINTNSSPDDQIGY |
0.994 |
| 276 |
RRGPEQTQGNFGDQELIRQG |
0.993 |
| 89 |
RATRRIRGGDGKMKDLSPRW |
0.965 |
| 232 |
SKMSGKGQQQQGQTVTKKSA |
0.958 |
| 39 |
QRRPQGLPNNTASWFTALTQ |
0.953 |
| 399 |
DLDDFSKQLQQSMSSADSTQ |
0.891 |
Table 3.
Epitopes predicted by ABCpred
|
Rank |
Sequence
|
Start position
|
Score
|
| 1 |
TRRIRGGDGKMKDLSP |
91 |
0.94 |
| 2 |
KSAAEASKKPRQKRTA |
249 |
0.93 |
| 2 |
EGALNTPKDHIGTRNP |
136 |
0.93 |
| 3 |
NKHIDAYKTFPPTEPK |
354 |
0.91 |
| 3 |
TGSNQNGERSGARSKQ |
24 |
0.91 |
| 3 |
KDGIIWVATEGALNTP |
127 |
0.91 |
| 4 |
SGTWLTYTGAIKLDDK |
327 |
0.88 |
| 5 |
HGKEDLKFPRGQGVPI |
59 |
0.87 |
| 5 |
ASSRSSSRSRNSSRNS |
182 |
0.87 |
| 6 |
ADETQALPQRQKKQQT |
376 |
0.86 |
| 6 |
APRITFGGPSDSTGSN |
12 |
0.86 |
| 7 |
NSSPDDQIGYYRRATR |
77 |
0.85 |
| 7 |
TFPPTEPKKDKKKKAD |
362 |
0.85 |
| 7 |
QELIRQGTDYKHWPQI |
289 |
0.85 |
Table 4.
Epitopes predicted by IEDB
|
No. |
Start
|
End
|
Peptide
|
Length
|
Mean
|
| 1 |
4 |
15 |
NGPQNQRNAPRI |
12 |
0.603 |
| 2 |
17 |
48 |
FGGPSDSTGSNQNGERSGARSKQRRPQGLPNN |
32 |
0.675 |
| 3 |
59 |
105 |
HGKEDLKFPRGQGVPINTNSSPDDQIGYYRRATRRIRGGDGKMKDLS |
47 |
0.557 |
| 4 |
119 |
127 |
AGLPYGANK |
9 |
0.539 |
| 5 |
137 |
163 |
GALNTPKDHIGTRNPANNAAIVLQLPQ |
27 |
0.606 |
| 6 |
165 |
216 |
TTLPKGFYAEGSRGGSQASSRSSSRSRNSSRNSTPGSSRGTSPARMAGNGGD |
52 |
0.661 |
| 7 |
226 |
267 |
RLNQLESKMSGKGQQQQGQTVTKKSAAEASKKPRQKRTATKA |
42 |
0.598 |
| 8 |
276 |
299 |
RRGPEQTQGNFGDQELIRQGTDYK |
24 |
0.555 |
| 9 |
343 |
348 |
DPNFKD |
6 |
0.529 |
| 10 |
358 |
402 |
DAYKTFPPTEPKKDKKKKADETQALPQRQKKQQTVTLLPAADLDD |
45 |
0.634 |

Fig. 1.
SARS-CoV-2 N protein sequences and its predicted linear epitopes.
.
SARS-CoV-2 N protein sequences and its predicted linear epitopes.
Extraction of RNA and synthesis of cDNA
RNA extracted using the RNX-plus solution showed high quality in the nanodrop evaluation. cDNA synthesized using the extracted viral RNA as a template was amplified by PCR using primers specific to the N protein sequence linear epitopes. Analysis by agarose gel electrophoresis revealed a PCR product of about 1260 bp that was in a predicted size (Fig. 2). The PCR product was cloned into the pET 28a expression vector and confirmed by PCR using target and pET vector specific primers (Fig. 3).
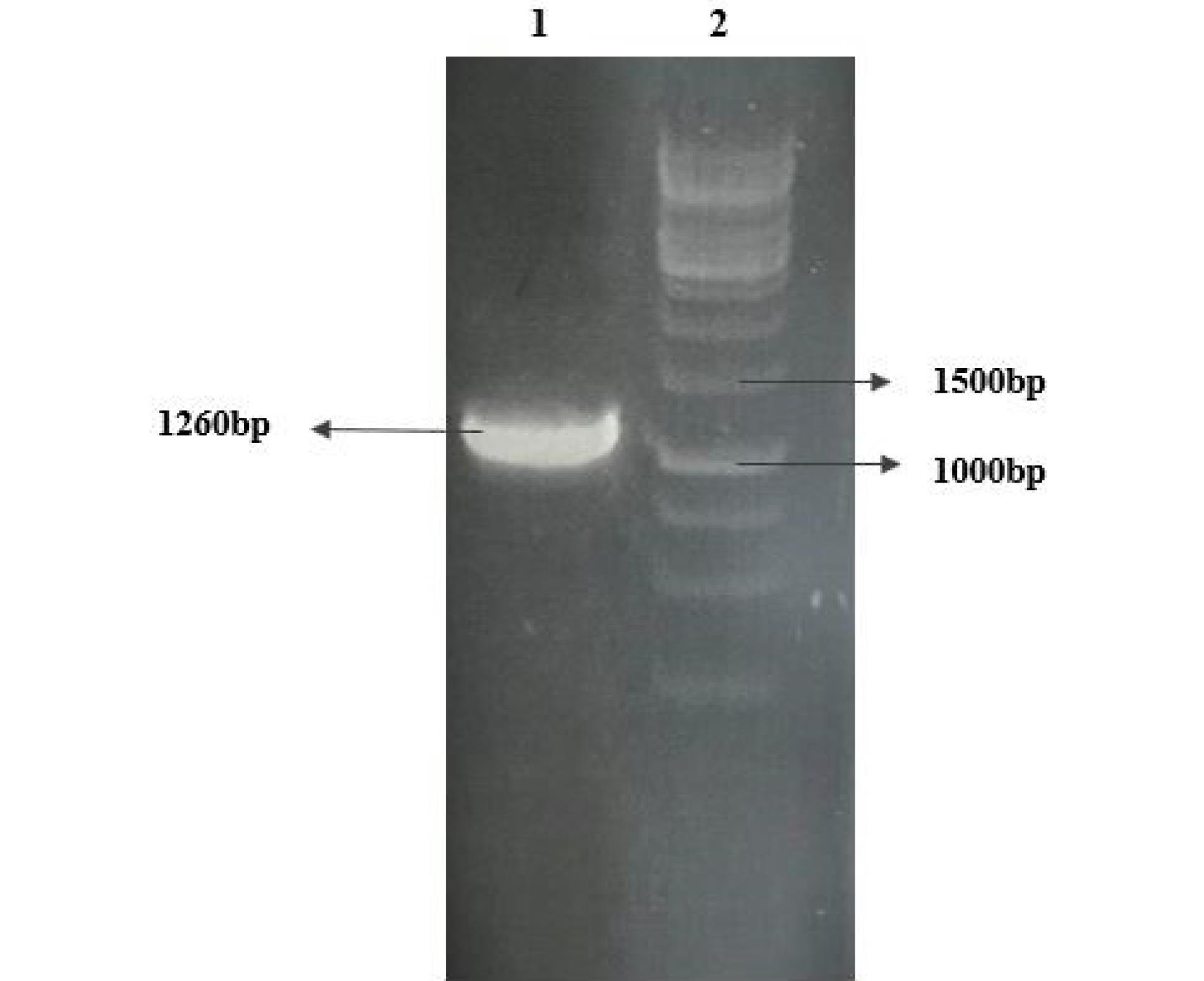
Fig. 2.
PCR amplification of the sequence encoding for the N protein linear epitope by RT-PCR.Lane1: PCR product and Lane2: DNA size marker.
.
PCR amplification of the sequence encoding for the N protein linear epitope by RT-PCR.Lane1: PCR product and Lane2: DNA size marker.
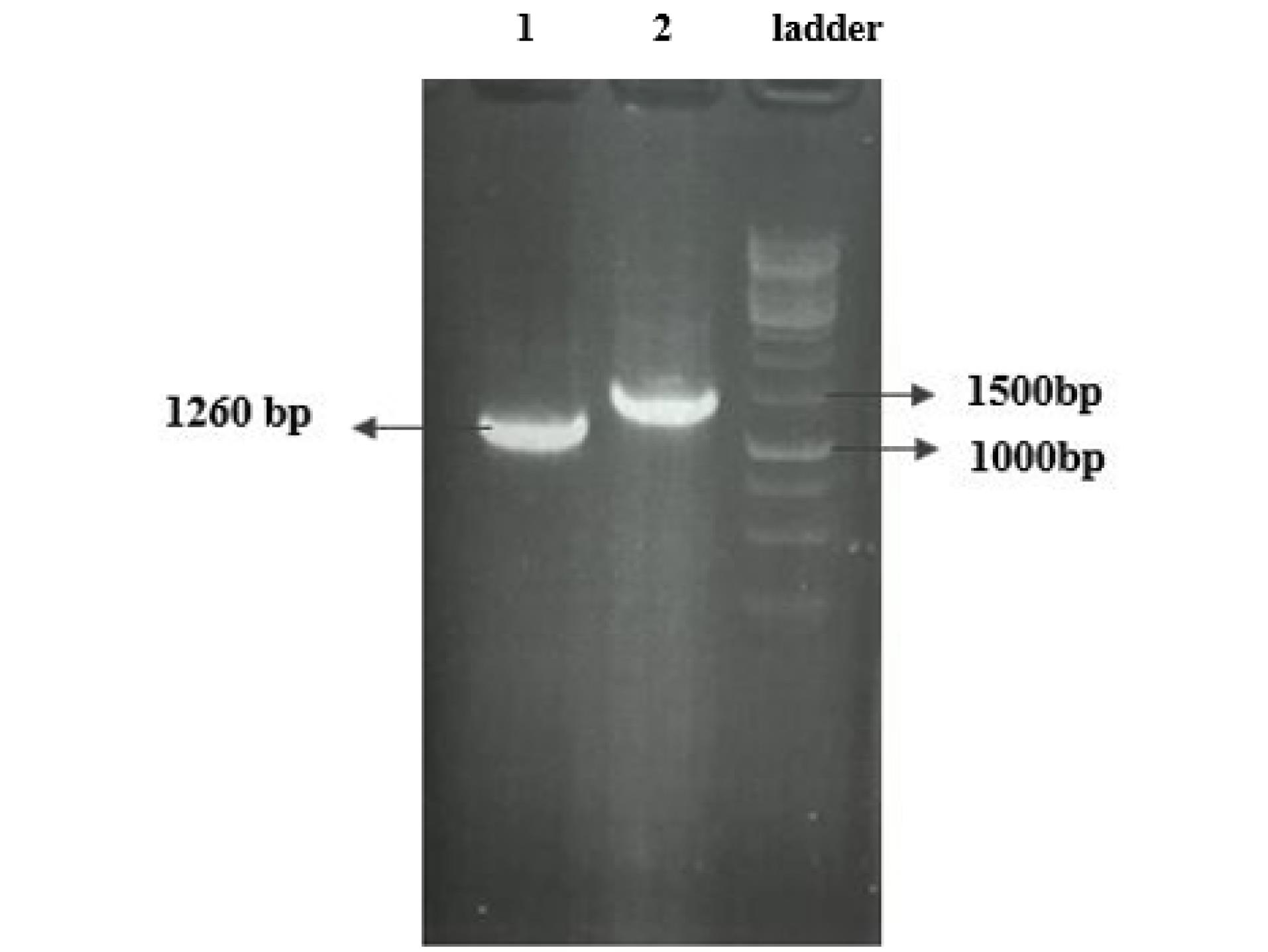
Fig. 3.
PCR amplification of the target gene cloned in pET 28a expression vector: lane 1, using specific primers for the target gene; lane 2, using universal primers for the PET vector and lane 3, DNA size marker.
.
PCR amplification of the target gene cloned in pET 28a expression vector: lane 1, using specific primers for the target gene; lane 2, using universal primers for the PET vector and lane 3, DNA size marker.
Recombinant expression of Sars-cov2 N protein linear epitopes
For high-level expression of recombinant N protein, the BL21 cells containing the N protein expression vector were cultured in LB-kanamycin (100 mcg/ml) broth media. When the cell density reached around OD 600, the culture was induced by adding 1 mM IPTG for 3, 6, 8, and 24 hours at a temperature of 20 °C. SDS-PAGE analysis revealed the successful expression of the recombinant N protein in E. coli, with a yield of about 20% of total cell protein, as evidenced by the presence of a 50kDa band in SDS-PAGE. It was discovered that cells of the BL21 strain of E. coli could be transformed with a gene construct encoding a complete N protein. The expression of the N protein was observed on a 12% SDS-PAGE gel following induction with 1 mM IPTG (Fig. 4).
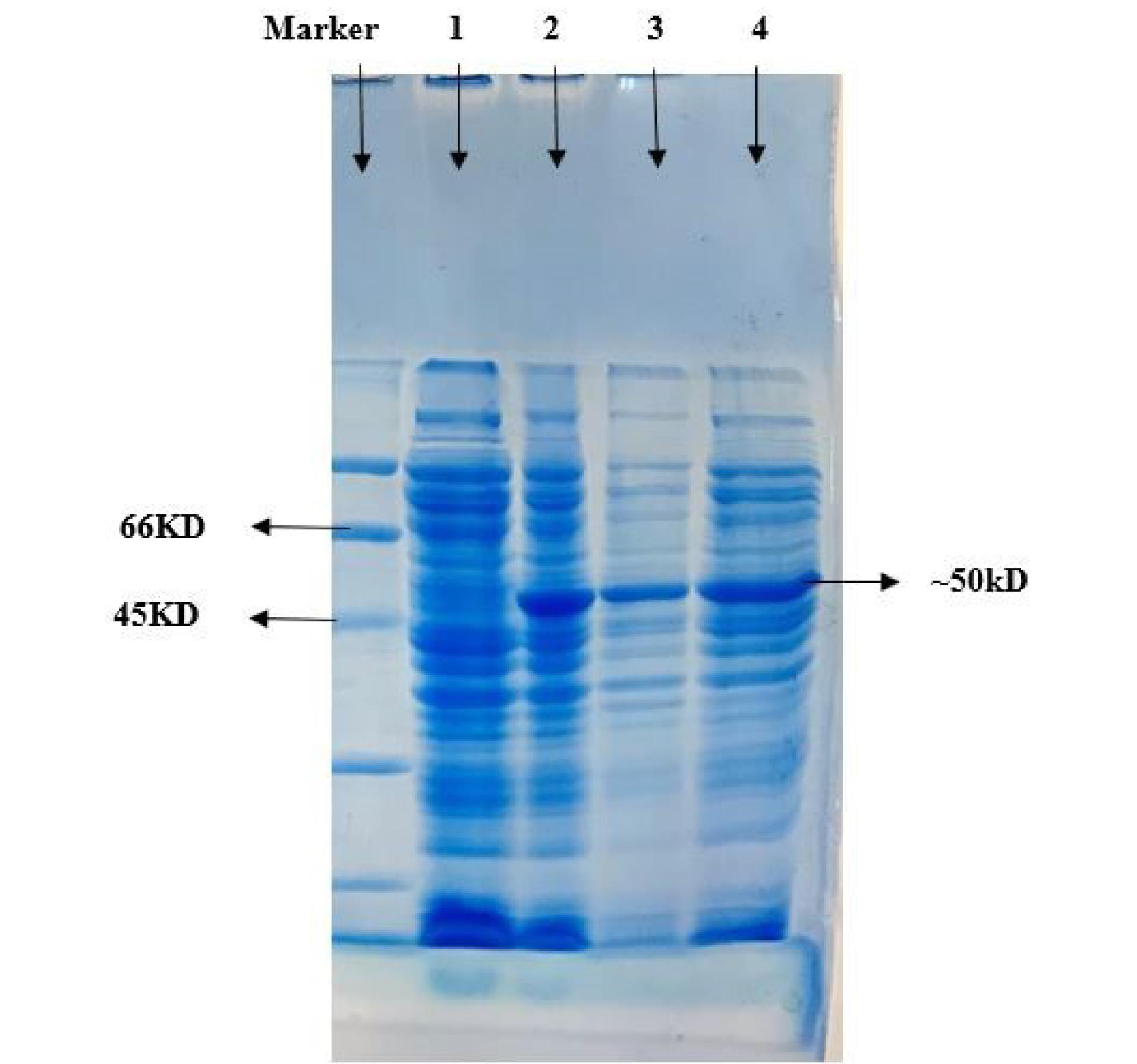
Fig. 4.
The expression of recombinant N protein was analyzed using SDS-PAGE before and after induction with 1 mM IPTG for 3h. In Lane 1: cell lysate before induction; Lane 2: cell lysate 3h after induction; Lane 3: insoluble fraction after sonication; Lane 4: soluble fraction after sonication.
.
The expression of recombinant N protein was analyzed using SDS-PAGE before and after induction with 1 mM IPTG for 3h. In Lane 1: cell lysate before induction; Lane 2: cell lysate 3h after induction; Lane 3: insoluble fraction after sonication; Lane 4: soluble fraction after sonication.
Purification of recombinant N protein
After the cells were harvested and lysed by sonication, the soluble fraction was obtained through centrifugation at 10 000 g for 10 minutes. The recombinant protein was then purified from the supernatant of the E. coli cell lysate using an affinity chromatography method (Fig. 5).
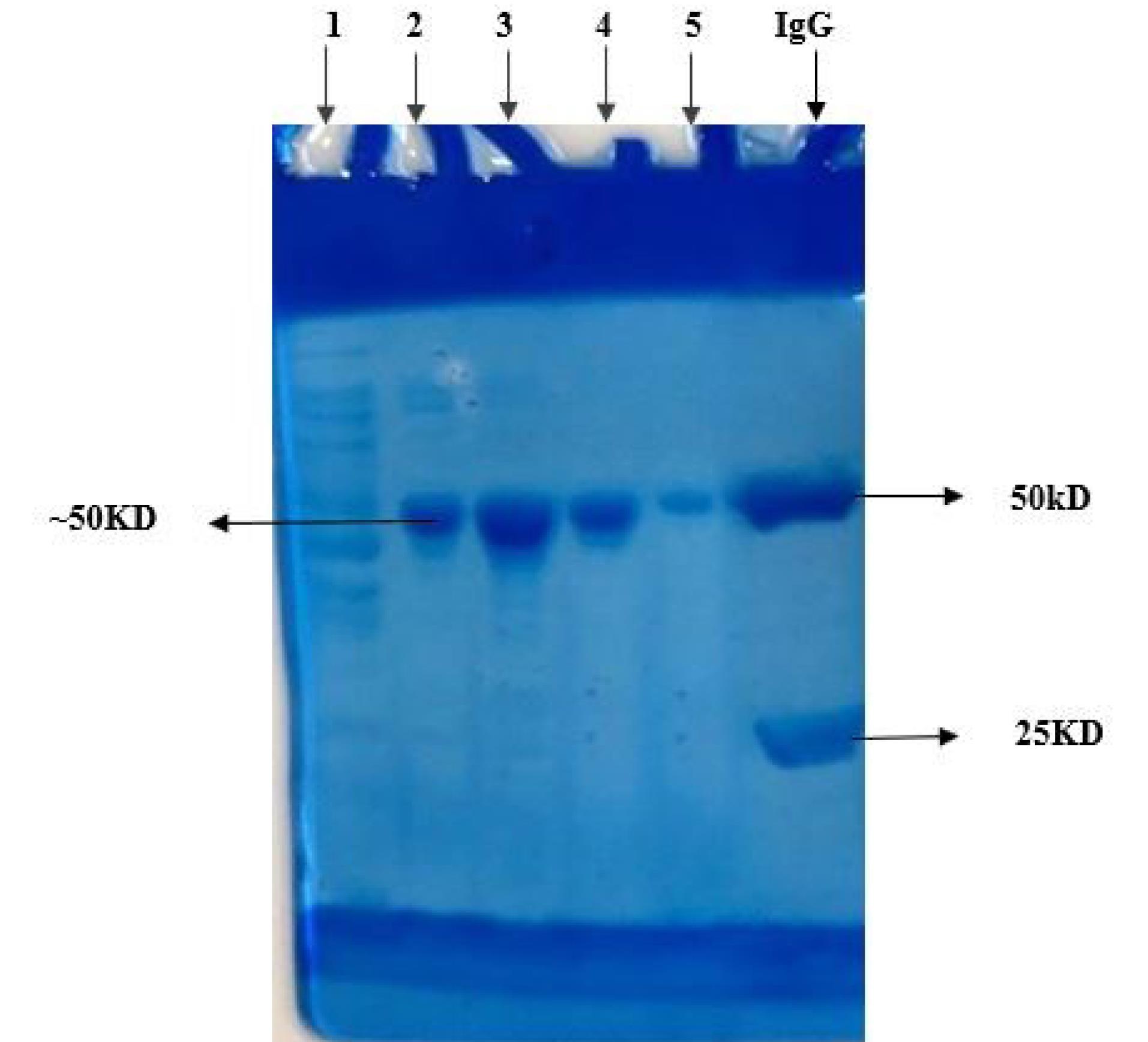
Fig. 5.
The recombinant N protein expressed in BL21 was purified by affinity chromatography and subjected to SDS-PAGE analysis. Lane 1: the flow through; Lane 2-5: purified recombinant N protein (elute fraction1-4); and Lane 6: IgG as a marker
.
The recombinant N protein expressed in BL21 was purified by affinity chromatography and subjected to SDS-PAGE analysis. Lane 1: the flow through; Lane 2-5: purified recombinant N protein (elute fraction1-4); and Lane 6: IgG as a marker
Reactivity of recombinant N protein with sera from COVID-19 patients
The ELISA technique was employed to assess the diagnostic efficacy of purified SARS-CoV-2 recombinant N protein linear epitopes on a collection of one hundred serum samples obtained during the COVID-19 outbreak. Out of the total 50 positive samples, 47 samples yielded positive results, while 48 out of the 50 negative samples were correctly identified as negative (Table 5). To evaluate the sensitivity and specificity of the ELISA findings, a statistical analysis was performed using GraphPad software (Figs. 6 and 7).
Table 5.
Assessment of IgG enzyme-linked immunosorbent assay (ELISA) performance using the N protein linear epitopes
|
Cut off 0.750
|
True positive
|
True negative
|
PPV
|
NPV
|
Sensitivity
|
Specificity
|
| 47/50 |
48/50 |
96% |
94% |
94% |
96% |
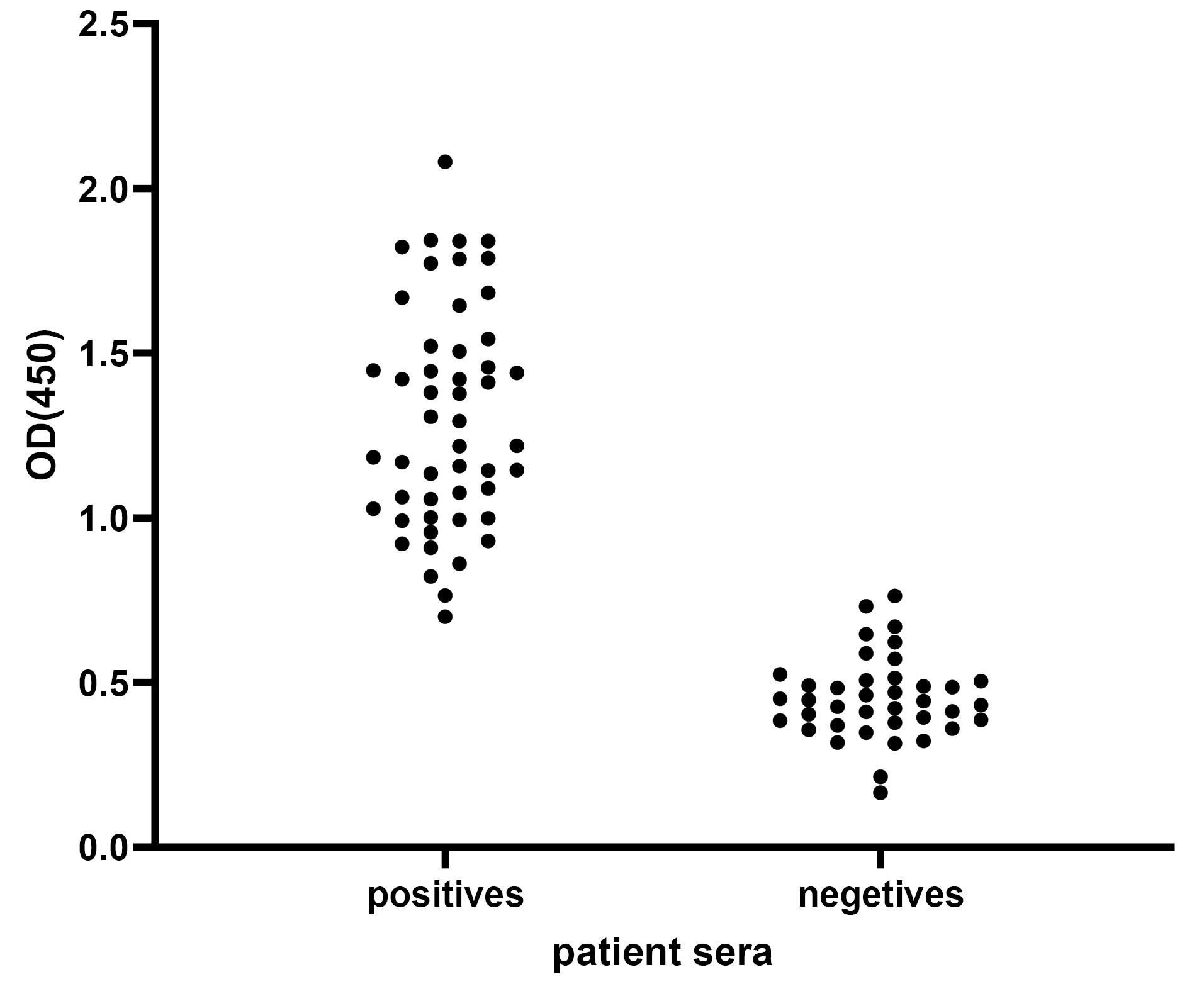
Fig. 6.
To verify the reactivity of recombinant N protein linear epitopes with sera from COVID-19 patients, an ELISA assay was conducted. The purified recombinant protein was immobilized on a 96-well microtiter plate and then exposed to sera from 50 healthy individuals and 50 COVID-19-infected patients. The reactivity was determined by measuring the optical density (OD) at 450nm after the addition of TMB substrate.
.
To verify the reactivity of recombinant N protein linear epitopes with sera from COVID-19 patients, an ELISA assay was conducted. The purified recombinant protein was immobilized on a 96-well microtiter plate and then exposed to sera from 50 healthy individuals and 50 COVID-19-infected patients. The reactivity was determined by measuring the optical density (OD) at 450nm after the addition of TMB substrate.
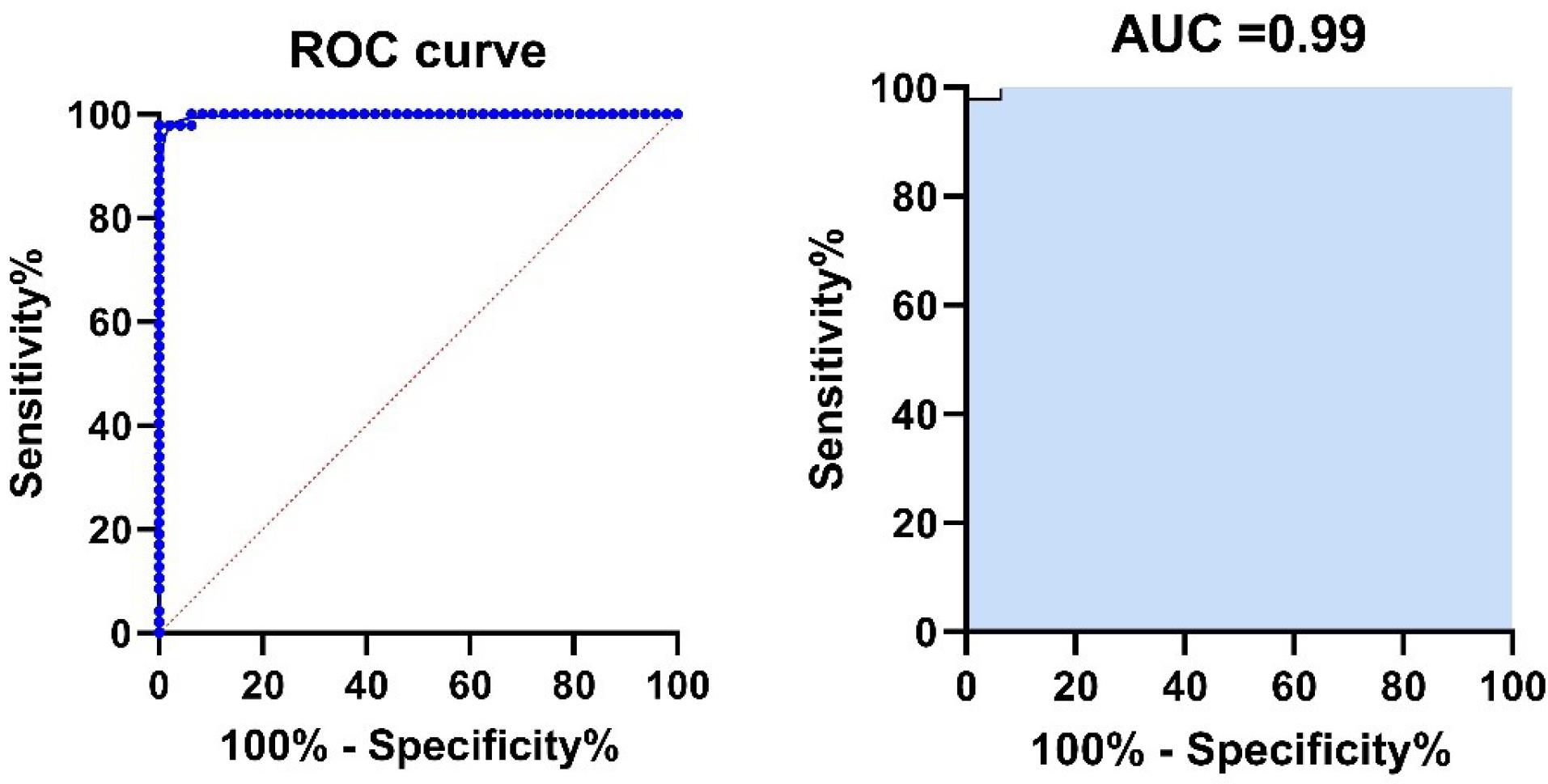
Fig. 7.
Receiver operating characteristic (ROC) curves for ELISA for the detection of antibodies against SARS-CoV-2 N protein linear epitopes. The blue line shows the mean area under the curve (AUC) plot, with the AUC value. The sensitivity (94%) and specificity (96%) values correspond to the points in the plots
.
Receiver operating characteristic (ROC) curves for ELISA for the detection of antibodies against SARS-CoV-2 N protein linear epitopes. The blue line shows the mean area under the curve (AUC) plot, with the AUC value. The sensitivity (94%) and specificity (96%) values correspond to the points in the plots
Discussion
The effective control of SARS-CoV-2 pandemics significantly relies on improving the sensitivity of prevailing diagnostic tools, including serological tests. This goal is mainly dependent on the choice of antigens used for serodiagnosis. Among the various antigens of SARS-CoV-2, the N protein has demonstrated both high immunogenicity and conservancy. Its crucial role in viral replication positions it as a promising target for antiviral interventions. Additionally, its distinctive characteristics make it highly suitable as a diagnostic antigen. In this study, linear epitopes of N protein were successfully expressed and purified from the E. coli expression system. An ELISA assay was conducted to determine the sensitivity and specificity of the purified N protein epitopes in detecting COVID-19 infection. Analysis of the results revealed that out of the 50 SARS-CoV-2 positive samples, the N protein ELISA correctly identified 47 samples as positive. Similarly, out of the 50 SARS-CoV-2 negative samples, the N protein ELISA test accurately identified 48 samples as negative. These findings indicate a sensitivity of 94% and a specificity of 96%. Consequently, the N protein linear epitopes exhibit a remarkable ability to detect SARS-CoV-2 infection with high sensitivity and specificity. These results were consistent with previous reports on the performance of N protein in the serodiagnosis of SARS-CoV-2 infection.
Earlier studies have shown that the utilization of SARS-CoV-2 recombinant fragments of the nuclear envelope protein and N protein (rfNP; 58–419 aa) can enable a cost-effective diagnosis with exceptional sensitivity and specificity.23 Batra et al have found that the IgG antibodies targeting the SARS-CoV-2 N protein exhibit a substantial concentration, which can be effectively employed for the detection of COVID-19. Nevertheless, they have also documented that elevated levels of anti-N-protein IgG pose a notable risk for admission to the MICU.30 Hou et al evaluated chemiluminescence immunoassays for the detection of IgM and IgG antibodies in COVID-19 patients for serological diagnosis. They observed a high sensitivity and specificity of the IgG response.31 Ge et al conducted a study on the detection of the SARS-CoV-2 N protein using electronic techniques. They employed the aptamer/antibody sandwich technique for this purpose. However, the findings indicated that the electronic method had a lower limit of quantification for N protein compared to the traditional dual sandwich-based ELISA.32 Another study introduced a swift and efficient method to detect the sars-cov2 nucleocapsid protein (NP) using the fluorescent immunochromatographic (FIC) assay. This approach showed remarkable levels of sensitivity and specificity.33
Spike protein, which is the most important protein in the pathogenesis of SARS-CoV-2, is another antigen that plays a vital role in the identification of the COVID-19. Spike protein is also crucial in the identification of the COVID-19. Also, the findings revealed high sensitivity and specificity of linear epitopes of S protein. Poh et al identified two immunodominant linear B-cell epitopes, S14P5 and S21P2, on the SARS-CoV-2 S glycoprotein, and demonstrated the functional capacity of COVID-19 patient sera antibodies against these regions.34 Additionally, Burbelo et al showed that about 14 days after symptom onset, antibodies against SARS-CoV-2 nucleocapsid protein showed 100% sensitivity and 100% specificity, whereas antibodies to spike protein were detected with 91% sensitivity and 100% specificity. They also reported that the antibody to the nucleocapsid protein of SARS-CoV-2 is more sensitive than the spike protein antibody for detecting early infection.35
Conclusion
The N protein of SARS-CoV-2 has been identified as one of the immunodominant proteins responsible for inducing humoral immune responses. This study demonstrated that the N-protein linear epitopes of SARS-CoV-2 are expressed in a soluble form in E. coli. Through ELISA analysis, it was observed that the protein exhibited a strong reactivity with COVID-19 sera, suggesting its potential application in the serodiagnosis of SARS-CoV-2 infection.
Acknowledgments
The authors would like to thank all patients for participation in this work.
Competing Interests
The authors certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
Ethical Statement
This work was approved by the ethical committee of Tabriz University of Medical Sciences, Tabriz, Iran (IR.TBZMED.REC.03.366).
References
- Kumar V, Doshi KU, Khan WH, Rathore AS. COVID‐19 pandemic: mechanism, diagnosis, and treatment. J Chem Technol Biotechnol 2021; 96:299-308. doi: 10.1002/jctb.6641 [Crossref] [ Google Scholar]
- Gao T, Gao Y, Liu X, Nie Z, Sun H, Lin K. Identification and functional analysis of the SARS-COV-2 nucleocapsid protein. BMC Microbiol 2021; 21:58. doi: 10.1186/s12866-021-02107-3 [Crossref] [ Google Scholar]
- Chang CK, Hou MH, Chang CF, Hsiao CD, Huang TH. The SARS coronavirus nucleocapsid protein--forms and functions. Antiviral Res 2014; 103:39-50. doi: 10.1016/j.antiviral.2013.12.009 [Crossref] [ Google Scholar]
- Di D, Dileepan M, Ahmed S, Liang Y, Ly H. Recombinant SARS-CoV-2 nucleocapsid protein: expression, purification, and its biochemical characterization and utility in serological assay development to assess immunological responses to SARS-CoV-2 infection. Pathogens 2021; 10:1039. doi: 10.3390/pathogens10081039 [Crossref] [ Google Scholar]
- McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses 2014; 6:2991-3018. doi: 10.3390/v6082991 [Crossref] [ Google Scholar]
- de Haan CA, Kuo L, Masters PS, Vennema H, Rottier PJ. Coronavirus particle assembly: primary structure requirements of the membrane protein. J Virol 1998; 72:6838-50. doi: 10.1128/jvi.72.8.6838-6850.1998 [Crossref] [ Google Scholar]
- Jacob CO, Leitner M, Zamir A, Salomon D, Arnon R. Priming immunization against cholera toxin and Ecoli heat-labile toxin by a cholera toxin short peptide-beta-galactosidase hybrid synthesized in Ecoli. EMBO J 1985; 4:3339-43. doi: 10.1002/j.1460-2075.1985.tb04086.x [Crossref] [ Google Scholar]
- Ahmad TA, Eweida AE, Sheweita SA. B-cell epitope mapping for the design of vaccines and effective diagnostics. Trials Vaccinol 2016; 5:71-83. doi: 10.1016/j.trivac.2016.04.003 [Crossref] [ Google Scholar]
- Cubuk J, Alston JJ, Incicco JJ, Singh S, Stuchell-Brereton MD, Ward MD. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat Commun 2021; 12:1936. doi: 10.1038/s41467-021-21953-3 [Crossref] [ Google Scholar]
- Okba NM, Müller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. SARS-CoV-2 specific antibody responses in COVID-19 patients. medRxiv [Preprint]. March 20, 2020. Available from: https://www.medrxiv.org/content/10.1101/2020.03.18.20038059v1.
- Tugaeva KV, Hawkins D, Smith JLR, Bayfield OW, Ker DS, Sysoev AA. The mechanism of SARS-CoV-2 nucleocapsid protein recognition by the human 14-3-3 proteins. J Mol Biol 2021; 433:166875. doi: 10.1016/j.jmb.2021.166875 [Crossref] [ Google Scholar]
- Heffron AS, McIlwain SJ, Amjadi MF, Baker DA, Khullar S, Armbrust T. The landscape of antibody binding in SARS-CoV-2 infection. PLoS Biol 2021; 19:e3001265. doi: 10.1371/journal.pbio.3001265 [Crossref] [ Google Scholar]
- Iyer M, Jayaramayya K, Subramaniam MD, Lee SB, Dayem AA, Cho SG. COVID-19: an update on diagnostic and therapeutic approaches. BMB Rep 2020; 53:191-205. doi: 10.5483/BMBRep.2020.53.4.080 [Crossref] [ Google Scholar]
- Terry JS, Anderson LB, Scherman MS, McAlister CE, Perera R, Schountz T. Development of a SARS-CoV-2 nucleocapsid specific monoclonal antibody. Virology 2021; 558:28-37. doi: 10.1016/j.virol.2021.01.003 [Crossref] [ Google Scholar]
- Khan WH, Hashmi Z, Goel A, Ahmad R, Gupta K, Khan N. COVID-19 pandemic and vaccines update on challenges and resolutions. Front Cell Infect Microbiol 2021; 11:690621. doi: 10.3389/fcimb.2021.690621 [Crossref] [ Google Scholar]
- Amrun SN, Lee CY, Lee B, Fong SW, Young BE, Chee RS. Linear B-cell epitopes in the spike and nucleocapsid proteins as markers of SARS-CoV-2 exposure and disease severity. EBioMedicine 2020; 58:102911. doi: 10.1016/j.ebiom.2020.102911 [Crossref] [ Google Scholar]
- Zhang Q, Wang P, Kim Y, Haste-Andersen P, Beaver J, Bourne PE. Immune epitope database analysis resource (IEDB-AR). Nucleic Acids Res 2008; 36:W513-8. doi: 10.1093/nar/gkn254 [Crossref] [ Google Scholar]
- Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-20: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res 2017; 45:W24-9. doi: 10.1093/nar/gkx346 [Crossref] [ Google Scholar]
- Dhanda SK, Mahajan S, Paul S, Yan Z, Kim H, Jespersen MC. IEDB-AR: immune epitope database-analysis resource in 2019. Nucleic Acids Res 2019; 47:W502-6. doi: 10.1093/nar/gkz452 [Crossref] [ Google Scholar]
- Kwarteng A, Asiedu E, Sakyi SA, Asiedu SO. Targeting the SARS-CoV2 nucleocapsid protein for potential therapeutics using immuno-informatics and structure-based drug discovery techniques. Biomed Pharmacother 2020; 132:110914. doi: 10.1016/j.biopha.2020.110914 [Crossref] [ Google Scholar]
- Zheng X, Deng Y, Xu X, Li S, Zhang Y, Ding J. Comparison of virus concentration methods and RNA extraction methods for SARS-CoV-2 wastewater surveillance. Sci Total Environ 2022; 824:153687. doi: 10.1016/j.scitotenv.2022.153687 [Crossref] [ Google Scholar]
- Wozniak A, Cerda A, Ibarra-Henríquez C, Sebastian V, Armijo G, Lamig L. A simple RNA preparation method for SARS-CoV-2 detection by RT-qPCR. Sci Rep 2020; 10:16608. doi: 10.1038/s41598-020-73616-w [Crossref] [ Google Scholar]
- Djukic T, Mladenovic M, Stanic-Vucinic D, Radosavljevic J, Smiljanic K, Sabljic L. Expression, purification and immunological characterization of recombinant nucleocapsid protein fragment from SARS-CoV-2. Virology 2021; 557:15-22. doi: 10.1016/j.virol.2021.01.004 [Crossref] [ Google Scholar]
- Xie X, Muruato A, Lokugamage KG, Narayanan K, Zhang X, Zou J, et al. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe 2020; 27: 841-8.e3. 10.1016/j.chom.2020.04.004.
- Tanomand A, Farajnia S, Najar Peerayeh S, Majidi J. Cloning, expression and characterization of recombinant exotoxin A-flagellin fusion protein as a new vaccine candidate against Pseudomonas aeruginosa infections. Iran Biomed J 2013; 17:1-7. doi: 10.6091/ibj.22.2012 [Crossref] [ Google Scholar]
- Seyyedhamzeh H, Farajnia S, Kargar M, Kafilzadeh F, Baradaran B. Cloning and expression of UreB-Omp18 recombinant protein from Iranian Hpylori strain. J Microbial World 2022; 15:109-18. doi: 10.30495/jmw.2022.1934280.1988.[Persian] [Crossref] [ Google Scholar]
- Bourassa L, Perchetti GA, Phung Q, Lin MJ, Mills MG, Roychoudhury P. A SARS-CoV-2 nucleocapsid variant that affects antigen test performance. J Clin Virol 2021; 141:104900. doi: 10.1016/j.jcv.2021.104900 [Crossref] [ Google Scholar]
- Timani KA, Ye L, Ye L, Zhu Y, Wu Z, Gong Z. Cloning, sequencing, expression, and purification of SARS-associated coronavirus nucleocapsid protein for serodiagnosis of SARS. J Clin Virol 2004; 30:309-12. doi: 10.1016/j.jcv.2004.01.001 [Crossref] [ Google Scholar]
- Khan WH, Khan N, Mishra A, Gupta S, Bansode V, Mehta D, et al. Dimerization of SARS-CoV-2 nucleocapsid protein affects sensitivity of ELISA based diagnostics of COVID-19. Int J Biol Macromol 2022. 200: 428-37. 10.1016/j.ijbiomac.2022.01.094.
- Batra M, Tian R, Zhang C, Clarence E, Sacher CS, Miranda JN. Role of IgG against N-protein of SARS-CoV2 in COVID19 clinical outcomes. Sci Rep 2021; 11:3455. doi: 10.1038/s41598-021-83108-0 [Crossref] [ Google Scholar]
- Hou H, Wang T, Zhang B, Luo Y, Mao L, Wang F. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin Transl Immunology 2020; 9:e01136. doi: 10.1002/cti2.1136 [Crossref] [ Google Scholar]
- Ge C, Feng J, Zhang J, Hu K, Wang D, Zha L. Aptamer/antibody sandwich method for digital detection of SARS-CoV2 nucleocapsid protein. Talanta 2022; 236:122847. doi: 10.1016/j.talanta.2021.122847 [Crossref] [ Google Scholar]
- Diao B, Wen K, Zhang J, Chen J, Han C, Chen Y, et al. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin Microbiol Infect 2021; 27: 289.e1-289.e4. 10.1016/j.cmi.2020.09.057.
- Poh CM, Carissimo G, Wang B, Amrun SN, Lee CY, Chee RS. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat Commun 2020; 11:2806. doi: 10.1038/s41467-020-16638-2 [Crossref] [ Google Scholar]
- Burbelo PD, Riedo FX, Morishima C, Rawlings S, Smith D, Das S. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J Infect Dis 2020; 222:206-13. doi: 10.1093/infdis/jiaa273 [Crossref] [ Google Scholar]