Bioimpacts. 2025;15:30424.
doi: 10.34172/bi.30424
Original Article
Targeting hypoxia-inducible factor 1 alpha augments synergistic effects of chemo/immunotherapy via modulating tumor microenvironment in a breast cancer mouse model
Mohsen Rashid Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft, 1 
Mina Ramezani Investigation, Methodology, 2
Ommoleila Molavi Conceptualization, Investigation, Project administration, Supervision, Validation, Writing – review & editing, 3
Zeinab Ghesmati Investigation, Methodology, Software, 4
Behzad Baradaran Project administration, Supervision, 5
Mehdi Sabzichi Writing – review & editing, 6
Fatemeh Ramezani Conceptualization, Data curation, Formal analysis, Investigation, Project administration, Supervision, Validation, Writing – review & editing, 1, * 
Author information:
1Department of Molecular Medicine, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
2Department of Anatomical Sciences, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran
3Biotechnology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
4Department of Medical Biotechnology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
5Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
6School of Pharmacy and Biomedical Sciences, Faculty of Science and Health, University of Portsmouth, Portsmouth, United Kingdom
Abstract
Introduction:
The immunosuppressive context of the tumor microenvironment (TME) is a significant hurdle in breast cancer (BC) treatment. Combinational therapies targeting cancer core signaling pathways involved in the induction of TME immunosuppressive milieu have emerged as a potent strategy to overcome immunosuppression in TME and enhance patient therapeutic outcomes. This study presents compelling evidence that targeting hypoxia-inducible-factor-1 alpha (Hif-1α) alongside chemotherapy and immune-inducing factors leads to substantial anticancer effects through modulation of TME.
Methods:
Chitosan (Cs)/Hif-1alpha siRNA nano-complex was synthesized by siRNA adsorption methods. Nanoparticles were fully characterized using dynamic light scattering and scanning electron microscope. Cs/Hif-1α siRNA cytotoxicity was measured by MTT assay. The anticancer effects of the combinational therapy were assessed in BALB/c bearing 4T1 tumors. qPCR and western blotting were applied to assess the expression of some key genes and proteins involved in the induction of immunosuppression in TME.
Results:
Hif-1α siRNA was successfully loaded in chitosan nanoparticles. Hif-1α siRNA nanocomplexes significantly inhibited the expression of Hif-1α. Triple combination therapy (Paclitaxel (Ptx) + Imiquimod (Imq) + Cs/Hif-1α siRNA) inhibited tumor growth and downregulated cancer progression genes while upregulating cellular-immune-related cytokines. Mice without Cs/Hif-1α siRNA treatments revealed fewer cancer inhibitory effects and more TME immunosuppressive factors. These results suggest that the inhibition of Hif-1α effects synergize with Ptx and Imq to inhibit cancer progression more significantly than other combinational treatments.
Conclusion:
Combining Hif-1α siRNA with Ptx and Imq is promising as a multimodality treatment. It has the potential to attenuate TME inhibitory effects and significantly enhance the immune system's ability to combat tumor cell growth, offering an inspiration of hope in the fight against BC.
Keywords: TME, Combinational therapy, HIF-1α, Imiquimod, Paclitaxel
Copyright and License Information
© 2025 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
This work was financially supported by a grant from the immunology research center at Tabriz University of Medical Science, Tabriz, Iran.
Introduction
Breast cancer (BC) is currently the most prevalent cancer worldwide, based on the GLOBOCAN 2020 Report.1 Due to the high mortality rates from breast cancer (685 000 deaths in 2020), it has become the leading cause of cancer death among women. Chemoresistance is one of the main challenges in BC treatment. Chemoresistance develops in BC through multiple mechanisms that are not completely elucidated. Mutation of the multidrug resistance gene (MDR) upregulation, signaling pathway modification, cancer stem cells’ immortality, hypoxia-inducible factor 1α (HIF-1α) elevation, and tumor microenvironment (TME) are some of the important mechanisms known for the development of chemoresistance in BC.2-5 Conventional chemotherapy methods usually have multiple severe side effects in patients, such as cytotoxicity, which arises from the non-selective targeting of actively proliferating normal cells in the body, leading to malfunction of normal tissues. One of the vital organs affected by the side effects of chemotherapy is bone marrow, which is responsible for making immune cells and components of the body’s immune system.6,7
Paclitaxel (Ptx) is a first-line chemotherapy agent widely used for chemotherapy of metastatic BC.8 Ptx inhibits tumor progression by inducing programmed cell death and immunogenic cell death (ICD) by affecting microtubule stability.9,10 Ptx has been shown to significantly induce pro-apoptotic Bax and Bad gene expression in MCF-7 cells.11,12_ENREF_14 ICD is a subtype of apoptosis in which dying cells release some Th1-type inflammatory factors, which in turn activate dendritic cells, leading to the induction of cell-mediated anticancer immune responses.13-15
Tumor microenvironment (TME) is an environment in which cancer cells and cancer stem cells (CSCs) reside and interact with normal cells, mainly fibroblast and immune cells.16,17 CSCs in TME are also found to be important in tumorigenesis and progression of cancer due to their self-renewing potential.16,18,19 Therefore, manipulation of TME by targeting key signaling pathways involved in the establishment of TME cellular networks is considered a promising approach for the successful treatment of cancer. HIF-1α is known to be induced in many types of human malignancies and plays a key role in the formation of cancer TME.20-22
HIF-1α, a central controller for sensing and adapting to cellular oxygen levels, transcriptionally activates genes modulating oxygen homeostasis and metabolic activation.23 In the hypoxic condition of TME, HIF-1α regulates the transcription of hundreds of genes that are involved in angiogenesis, metastasis, and proliferation of cancer cells.19,24,25 It was shown that inhibition of HIF-1α significantly suppressed tumor growth in the BALB/c mice bearing prostate cancer by modulating the immunosuppressive microenvironment.26,27 Another promising strategy for modulating cancer TME is using immunomodulatory factors known to induce Th1-type inflammatory responses. An important group of immunomodulatory agents that target toll-like receptors (TLR) on dendritic cells (DCs) has been shown to modulate cancer TME and induce potent anticancer immune responses. TLR ligands have been widely studied for activating DCs and anticancer immune responses. Recent studies have revealed that members of the imidazoquinoline family are recognized independently by TLR-7 and TLR-8, activating the signal transduction cascade downstream of these receptors.28,29 There is ample evidence that Imq treatments increased NK1.1 + cells in the TME by upregulating IL-12/IL-6.30-32
Combination therapy (CT), as a multimodality treatment that uses two or more therapeutic agents or methods, has emerged as a promising approach for curing cancer. The synergic efficiency of anticancer agents improves the effectiveness of drugs compared with mono-therapy methods as they target central genes and signaling pathways in a synergistic mode. CT has been shown to reduce drug resistance and boost anti-cancer drug potential, leading to effective inhibition of tumor cell proliferation and angiogenesis, lowering CSCs proliferation and self-renewal, and induction apoptosis.33-36 It has been reported that CT by Ptx and Imq induces activation of DCs and secretion of IL-12 and TNFα within the suppressed TME in C57BL/6 melanoma bearing mice.37 HIF-1α inhibitors and gemcitabine (Gem) were used in the Panc02 cancer model of immune-competent C57BL/6 and nude mice.38 This study reports that while immunotherapy with Gem or PX-478 (HIF-1α inhibitor) significantly suppresses tumor growth, CT with Gem and PX-478 resulted in significantly stronger inhibitory effects on cancer growth in immune-competent and incompetent mice.38
In this study, we investigated the synergistic antitumor effects of a new CT using Hif-1α siRNA alongside chemoimmunotherapy (Ptx + Imq) in a mouse model of breast cancer. This multifaceted approach elicited promising levels of anticancer immune responses within the TME. Our hypothesis posited that by reshaping the TME through Hif-1α inhibition, we could boost the therapeutic effectiveness of Ptx and Imq, ultimately inhibiting tumor growth in the BC mouse model.
Materials and Methods
Materials
The low molecular weight of Cs (99 % Pure, 114 kDa, 75% deacetylation, Sigma-Aldrich, Cas: 9012-76-4), sodium tripolyphosphate (Sigma-Aldrich, CAS: 7758-29-4), Acetic Acid (Merck, CAS: 64-19-7), three types of predesigned 22-mer HIF-1α-specific siRNA duplex that supplied by Bioneer Corporation(Korea, ID: 15251-1, 15251-2, 15251-3) were used for Cs/Hif-1α siRNA nano-complex and cell/tumor treatments (Table 1).Imq powder was purchased from InvivoGen (San Diego, United States). Antibodies for western blot were purchased from Santa Cruz Biotechnology (β-actin sc-47778, HIF-1α sc-13515, STAT3 sc-8019 Santa Cruz, Bax sc-7480, Bcl2 sc-7382, Inf-gamma sc-8423, IL12 sc-74147, IL10 sc-8438, PDL1 sc-518027). Cell culture Flask (APA company, IRAN), RPMI medium (Sigma-Aldrich, Gibco®, MDL: MFCD00217342), Fetal Bovine Serum (FBS, Biosera), 0.25% trypsin-EDTA (1X) (BIO-IDEA, BI-1602), MTT formazan powder (Sigma-Aldrich, Cas: 57360-69-7), DMSO (Sigma-Aldrich, Cas: 67-68-5) was used for cell culture and viability test. The Chemo drug, Ptx (Sigma-Aldrich, CAS: 33069-62-4), was applied for in vivo treatment. TRIzol (Sigma-Aldrich, CAS: 15596026) for RNA extraction, complementary DNA (cDNA) synthesis kit (YTA, Cat: YT4500) for cDNA synthesis, and Real-time master mix SYBR green (Amplicon, no-ROX) was used for real-time PCR method. 4T1 cell line (CRL-2539) was purchased from the Pasteur Institute, Tehran, Iran. BALB/c mice were purchased from Amol Pasteur Institute, Mazandaran, Iran.
Table 1.
Sequences of mice predesigned HIF-1α siRNA
|
ID
|
Type
|
Sequences (5'→3')
|
| 15251-1 |
Sense |
GUGGUUGGGUCUAACACUA |
| Anti-sense |
UAGUGUUAGACCCAACCAC |
| 15251-2 |
Sense |
CUGAUUGCAUCUCCAUCUU |
| Anti-sense |
AAGAUGGAGAUGCAAUCAG |
| 15251-3 |
Sense |
CAGUUACGAUUGUGAAGUU |
| Anti-sense |
AACUUCACAAUCGUAACUG |
Chitosan synthesis based on ionic gelation
Firstly, 1 mg of tripolyphosphate (TPP) dissolved in 1.2 mL of aqueous solution (0.84 mg/mL), and 6 mg of Cs dissolved in 3 mL of 0.1 N acetic acid solution (1 mg/mL). TPP was added to Cs solution dropwise under constant magnetic stirring for 30 minutes at room temperature (chitosan to TPP weight ratio was 6:1). The nanoparticles were incubated for 30 minutes at room temperature before supplementary analysis. Nanoparticles were collected by centrifugation (Labnet Prism R refrigerated centrifuge c2500-R) at 12 000 × g for 15 minutes. The supernatants were discarded, and nanoparticles were resuspended in filtered water.39
siRNA adsorption onto chitosan-TPP nanoparticles
Pre-prepared Cs nanoparticles prepared by ionic gelation as described above were dispersed in distilled water to yield a different Cs concentration based on Cs(µg) =
Formula. N/P ratio (the molar ratio of Cs amino groups/RNA phosphate groups) was prepared, ranging from 100 to 350. To adsorb siRNA onto the surface of Cs nanoparticles, 10 µL (0.026 µg/ µL) of siRNA solution was added to the calculated amount of Cs suspension and quickly mixed by inverting the interaction tube up and down. Then, the particles were incubated for two hours at room temperature before further analysis.39,40
Characterization of the chitosan nanoparticles size and charge
The mean particle diameter (Z-average) was analyzed by scanning electron microscopy (SEM) and dynamic light scattering (DLS) Zetasizer (Malvern instrument, UK), and the zeta potential of the nanoparticles was determined by dynamic light scattering (DLS). The particle size and Zeta potential measurements were made at 25 °C in N/P 350; no further dilution was performed for these particles. Each batch was analyzed in triplicate.
Determination of siRNA loading efficiency
Adsorbed (%) siRNA loading efficiency on chitosan nanoparticles was calculated from the concentration of free siRNA in the supernatant recovered after nanoparticles centrifugation (12 000 × g, 15 minutes) by Nanodrop spectrometer (NanoDrop 2000C, USA). Unloaded Cs–TPP nanoparticles supernatant (without siRNA) was used as a blank. siRNA loading efficiency (%) was the percentage of adsorbed or entrapped siRNA that came from the total added siRNA in nanoparticle preparation mines detected free siRNA in the supernatant gathered from nanoparticle-siRNA centrifugation.39
Gel retardation assay
The siRNA binding with Cs was determined by gel retardation assay using 4% agarose gel electrophoresis. Different N/P ratios of Cs to siRNA were loaded (100 to 350). 1:5 dilution of loading dye was added to each well, and electrophoresis was carried out at a constant voltage of 70 V for 1 hour in TAE buffer (0.04 M Tris, 0.001 M EDTA, 0.02 M Acetic Acid, pH 8.3). The siRNA bands were then visualized under a UV transilluminator at a wavelength of 365 nm.39,41
Cell culture and chitosan toxicity assay on 4T1 cell line
The MTT assay was carried out to study the cytotoxicity of the Cs nanoparticles. 4T1 cells were cultured in RPMI medium containing 10% Fetal bovine serum, 100 U/mL penicillin, and 100 µg/mL streptomycin and incubated at 37 °C with 5% CO2 atmosphere and 95% humidity. The cells with 90% confluency were collected from a culture flask with 0.25% trypsin-EDTA solution. The 4T1 cells were seeded at a density of 10 × 103 cells/well in a 96-well plate (200 µL medium/well). After reaching 80% confluence, cells were exposed to varying final concentrations of Cs nanoparticles and Cs/HIF-1α siRNA ranging from 0.062 nmol to 0.683 nmol for 48 h. 4T1 cell lines without nanoparticle treatments were used as a control group.
Subsequently, 100 µL (0.5 mg/mL) MTT solution in Phosphate-buffer saline (PBS) was added per well, and the plate was incubated at 37 °C for four hours in the dark.42 Finally, a DMSO reagent replaced the media and the relative absorbance was measured at 570 nm using an ELISA reader (Mikura Ltd., Horsham, UK).
HIF-1α siRNA treatment and in vitro gene expression of Hif-1α
To study the ability of Cs/Hif-1α siRNA nano-complex in silencing Hif-1a gene expression, BC cells (200 × 103) were seeded on 6-well plates in RPMI media (2000 µL/well containing 10% FBS and 1% penicillin/streptomycin) and incubated at 37 °C in 5% CO2 for Cs/HIF-1α siRNA treatment. After 48 hours, the medium was replaced with RPMI-1640, FBS-free, antibiotic-free. Then, cells were treated with 0.070 nmol (N/P = 350) final concentration of Cs containing 0.26 µg siRNA (mixed of three types). After 5 hours, the medium was replaced with RPMI-1640 medium with 10% FBS and 0.05% antibiotics. 44 hours later, RNA extraction was done for gene expression assay.43 Total RNA isolated by TRIzol reagent from Cs and Cs/Hif-1α siRNA treated and untreated cell lines. Then, isolated RNA was transcribed to cDNA by a cDNA synthases kit according to company protocol (YTA). Quantitative real-time PCR (qPCR) was done to measure HIF-1α and beta-actin gene expression in these cell lines by Amplicon cyber green master mix by using the Roche Life science light cycler 96 system. These primers TTTGGACACTGGTGGCTCAG (F), GAGCTGTGAATGTGCTGTGATC (R) (5'→3') as Hif-1α primers and GGCGGACTGTTACTGAGCTG (F), CGCCTTCACCGTTCCAGTT (R) (5'→3') as Beta actin primers were used to real-time PCR.
Animal studies
Establishment of 4T1 syngeneic mice model
For an in vivo study of Cs/Hif-1α siRNA nano-complex, Cs, and Ptx effects. Five- to eight-week-old female inbreed BALB/c mice (body weight = 18g) were purchased from Amol Pasteur Institute, Mazandaran, Iran, and were maintained in an animal facility in Tabriz Pharmaceutical Biotechnology Center. All maintenance and treatment procedures were performed based on approved protocols and recommendations of the Tabriz Pharmaceutical Biotechnology Center. To create a tumor in mice, 106 cells (in 100 µL medium) were inoculated into the right flank of mice subcutaneously. 12-16 days after implantation of cells, injected cells became palpable as a tumor, with size variety between 50-100 mm3, then mice were randomly allocated to five treatment groups of five.
In-vivo combinational therapies
Based on Table 2, 25 mice were divided into five groups, and each group received defined therapies for eight days. Therapies include (1) PBS, (2) Ptx + Cs/Hif-1α siRNA (Double Combinational Therapy (DCT)), (3) Ptx + Imq (DCT) (4) Imq + Cs/Hif-1α siRNA (DCT), and (5) Ptx + Imq + Cs/Hif-1α siRNA (Triple Combinational Therapy (TCT)). The PBS was injected intratumorally (i.t.) at a dose of 0.1 ml per mouse daily. Ptx was administered intraperitoneally at a dose of 2 mg/kg, Cs/siRNA was injected intratumorally at a concentration of 0.070 nmol (N/P = 350), and Imq was delivered intratumorally at a dose of 1 mg/kg.44,45 Tumors were harvested by tumor-bearing mice sacrificing. Then, Tumors were dissected, photographed, taken, and stored at -70 °C for further analysis.
Table 2.
Twenty-five mice were divided into five groups of five
|
Group
|
Therapy
|
Description
|
| 1 |
PBS |
This group received PBS every day (Control group). |
| 2 |
Ptx + Cs/Hif-1α siRNA |
This group received Ptx on days 1, 3, 5, and 7 and Cs/Hif-1α siRNA on days 2, 4, 6, and 8. |
| 3 |
Ptx + Imq |
This group received Ptx on days 1, 3, 5, and 7 and Imq on days 2, 4, 6, and 8. |
| 4 |
Imq + Cs/Hif-1α siRNA |
This group received Imq on days 1, 3, 5, and 7 and Cs/Hif-1α siRNA on days 2, 4, 6, and 8. |
| 5 |
Ptx + Imq + Cs/Hif-1α siRNA |
This group received Ptx on days 1, 3, 5, and 7 and Imq + Cs/Hif-1α siRNA on days 2, 4, 6, and 8.
|
Each group received the defined therapies.
RNA extraction and quantitative real-time PCR
To calculate and compare gene expression in treated tumors, total RNA was isolated from the treated cell lines and also tumors by TRIzol reagent and qualified by Nanodrop spectrometer (NanoDrop 2000C, USA). 1-5 ng of isolated RNA was transcribed to cDNA by reverse transcriptase PCR kit according to company protocol (YTA).
Table 3 shows targeted gene primer sequences (5' → 3') that participate in the cancer progression pathway, apoptosis, and cytokines involved in the modulation of the immune system. The qPCR reaction was performed using SYBR Green I master mix (no-ROX) in the qPCR thermal cycler (Roche Life Science Light Cycler 96 system, Germany) to evaluate the relative expression level of interested genes. All real-time data was collected by comparative gene expression analysis Delta-Delta Ct methods.46 The Gapdh gene was used to normalize the cycle threshold (Ct) values of the target genes.
Table 3.
Primer sequences (5' → 3') of all interested genes
|
|
Primers
|
Forward
|
Reverse
|
Gene ID
|
| 1 |
Bax
|
AGGGTTTCATCCAGGATCGAG |
TCCACGTCAGCAATCATCCTC |
12028 |
| 2 |
Bcl-2
|
GCCTCTTCACCTTTCAGCATTG |
TTCCCCGTTGGCATGAGATG |
12043 |
| 3 |
Hif-1 alpha
|
TTTGGACACTGGTGGCTCAG |
GAGCTGTGAATGTGCTGTGATC |
15251 |
| 4 |
Il-10
|
CCTGGGTGAGAAGCTGAAGAC |
ATGGCCTTGTAGACACCTTGG |
16153 |
| 5 |
Il-12
|
CTATGGTCAGCGTTCCAACAG |
AGGTGGTTTAGGAGGGCAAG |
16159 |
| 6 |
Inf-gamma
|
ACAATGAACGCTACACACTGC |
CATCCTTTTGCCAGTTCCTCC |
15978 |
| 7 |
Pdl1
|
ATTGTAGTGTCCACGGTCCTC |
TCGACGATCAGAGGGTTCAAC |
NM_021893.3 |
| 8 |
Stat-3
|
AAACCCTCAAGAGCCAAGGAG |
ACGTACTCCATTGCTGACAAG |
20848 |
| 9 |
Βeta-actin
|
GGCGGACTGTTACTGAGCTG |
CGCCTTCACCGTTCCAGTT |
NM_007393.5 |
Western blot analysis
To approve real-time results, the protein expression level of some genes was analyzed. Tumors were surgically removed and lysed by lysis buffer (Tris-HCL 500 µL, PH = 8, EDTA 0.003 g, NaCl 0.08 g, Sodium Deoxycholate 0.025 g, SDS 0.01 g, Protease inhibitor cocktail one tablet, Triton (NP40 (1%) 10 µL). Consequently, the samples were centrifuged (12 000 rpm, 10 minutes, 4 °C), and according to Bradford method protocol (Bio-Rad Laboratories, USA), the protein content in the supernatant was detected in samples by spectrophotometers. The target proteins were separated by using SDS-PAGE gel electrophoresis and then moved to the PVDF membrane (polyvinylidene difluoride membrane) and were blocked with 2% w/v of skim milk and 0.1% v/v of Tween 20 in tris buffered saline (TBS) for masking of unspecific bands. Specific primary antibodies (β-actin sc-47778, HIF-1α sc-13515, STAT3 sc-8019 Santa Cruz, Bax sc-7480, Bcl2 sc-7382, Inf-gamma sc-8423, IL12 sc-74147, IL10 sc-8438, PDL1 sc-518027) were added to the blocked PVDF membranes that contained the target proteins and were incubated overnight at 4 °C. After washing with TBS-T, the membranes were incubated with secondary antibodies (IgG-HRP sc-2357, m-IgGκ BP-HRP: sc-516102 Santa Cruz) for one hour at room temperature. The bands related to the target proteins were visualized using an enhanced chemiluminescence detection kit (ECL advanced reagents composed of non-fat milk and reagent A, B) and were measured with Amersham Imager. Lastly, after normalizing the western blot bands using β-actin expression as a control, the stains were calculated using ImageJ software, version 1.52v.
Statistical analysis
All data in this research were presented as mean ± standard deviation and analyzed using GraphPad Prism v.9 statistical software. Student’s t-test and one-way ANOVA tests were used to evaluate the statistical significance of differences between groups. Significant data: *** P value <0.0001 high significance, **P value<0.001 middle significance, *P value<0.01 low significance.
Results
Fabrication and characterization of Cs
The mean diameter of synthesis Cs was reported to be 220 nm, and the zeta potential was 32 m.v by DLS. siRNA adsorption onto the surface of the Cs/TPP nanostructure reduced the zeta potential to 15 m.v (Fig. 1a). The SEM image reveals the proper morphology and size of Cs, measuring approximately 50-200 nm (Fig. 1b). To find the appropriate ratio of the N/P reaching the maximum encapsulation, the gel retardation assay with Cs/Hif-1α siRNA nano-complex at the different ratios of N/P (100 to 350) was performed. The results indicated that complete complexation was verified on an N/P ratio of 350, corresponding to the Cs-to-siRNA ratio of 84 (Fig. 1c). Approximately 60% loading efficiency was obtained as measured by Nanodrop for adsorbed siRNA at a weight ratio of Cs/siRNA 84:1 (Fig. 1d).
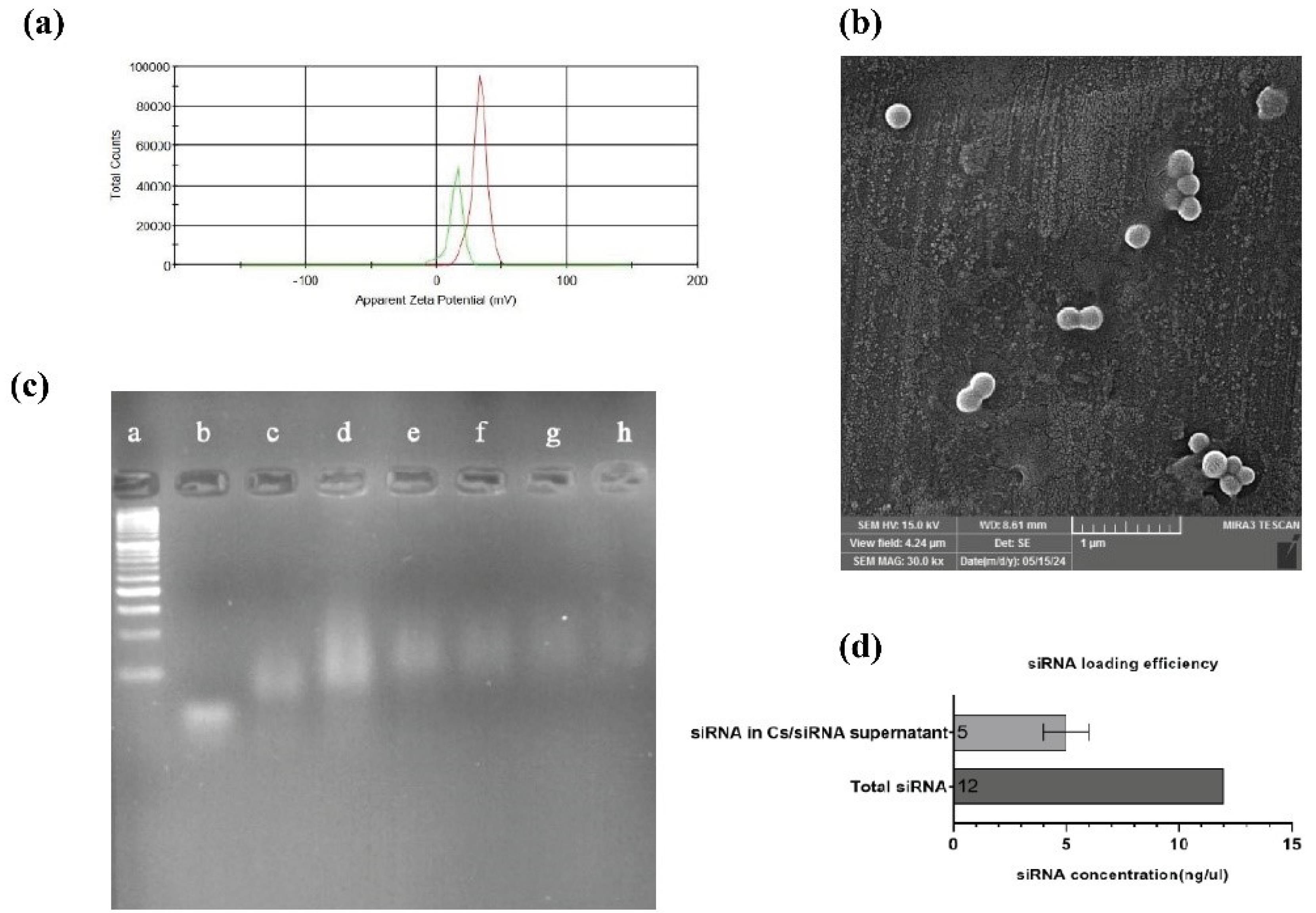
Fig. 1.
Characterization of Nanoparticles size, charge, and siRNA loading potential. (a) Zeta potential graph. The red line describes the zeta potential of chitosan/TPP, and the green line shows the Chitosan/TPP/Hif-1α siRNA charge. (b) Scanning electron microscopy image of chitosan. (c) Gel Retardation assay. a) 50bp ladder. b) free siRNA. c) N/P = 100. d) N/P = 150. e) N/P = 200. f) N/P = 250. g) N/P = 300. h) N/P = 350. (d) siRNA loading efficiency. (All data are presented as the mean ± SD. *P < 0.01, **P < 0.001 and ***P < 0.0001. One-way ANOVA was used to compare the Tests and the control group).
.
Characterization of Nanoparticles size, charge, and siRNA loading potential. (a) Zeta potential graph. The red line describes the zeta potential of chitosan/TPP, and the green line shows the Chitosan/TPP/Hif-1α siRNA charge. (b) Scanning electron microscopy image of chitosan. (c) Gel Retardation assay. a) 50bp ladder. b) free siRNA. c) N/P = 100. d) N/P = 150. e) N/P = 200. f) N/P = 250. g) N/P = 300. h) N/P = 350. (d) siRNA loading efficiency. (All data are presented as the mean ± SD. *P < 0.01, **P < 0.001 and ***P < 0.0001. One-way ANOVA was used to compare the Tests and the control group).
Cytotoxicity study of the chitosan nanoparticles and gene silencing effects of chitosan/Hif-1α siRNA
The MTT assay was done to evaluate the cytotoxicity of the Cs nanoparticle and Cs/siRNA complex against the 4T1 cell line. The results revealed that Cs exhibited minimal toxicity at low doses ranging from 0.062 nmol to 0.186 nmol. However, at doses exceeding 0.1995 nmol, cells demonstrated an intolerance to Cs concentration, resulting in cell death within 48 hours. Based on Graph Pad Prism v.9 analysis, the inhibitory concentration (IC50) of Cs (red line) was about 0.1995 nmol, resulting in a 50% reduction in 4T1 cell viability. The inhibitory concentration of Cs/Hif-1α siRNA was found to be about 0.2586 nmol (P< 0.0001), totally significant compared with Cs. Thus, 0.070 nmol was selected for HIF-1α siRNA loading and cells/tumor treatments, which was a safe concentration of Cs (Fig. 2a).
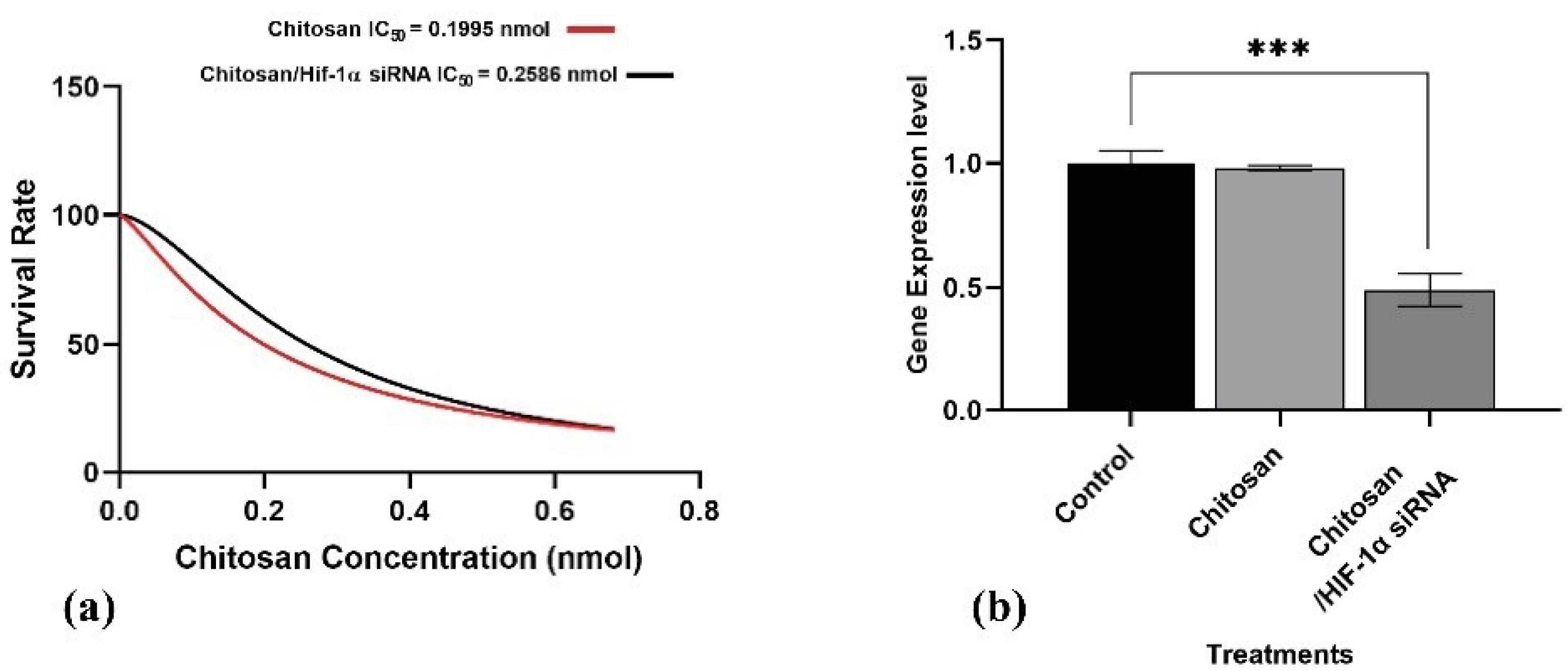
Fig. 2.
a) Survival Rate of 4T1 cell line under chitosan (Red line) and chitosan/Hif-1α siRNA (Black line) treatment. 4T1 cell line treated with 0.062, 0.124, 0.186, 0.310, 0.434, 0.559 and 0.689 nmol of chitosan and chitosan/Hif-1α siRNA for 48 hours. b) Hif-1α expression in the treated cell line. Cs and Cs/Hif-1α siRNA effects on Hif-1α gene expression on 4T1 cell line (All data are presented as the mean ± SD. *P < 0.01, **P < 0.001 and ***P < 0.0001. One-way ANOVA was used to make comparisons between the Tests and the control group).
.
a) Survival Rate of 4T1 cell line under chitosan (Red line) and chitosan/Hif-1α siRNA (Black line) treatment. 4T1 cell line treated with 0.062, 0.124, 0.186, 0.310, 0.434, 0.559 and 0.689 nmol of chitosan and chitosan/Hif-1α siRNA for 48 hours. b) Hif-1α expression in the treated cell line. Cs and Cs/Hif-1α siRNA effects on Hif-1α gene expression on 4T1 cell line (All data are presented as the mean ± SD. *P < 0.01, **P < 0.001 and ***P < 0.0001. One-way ANOVA was used to make comparisons between the Tests and the control group).
Gene silencing efficacy of Cs/Hif-1α siRNA nano-complex was first evaluated in the cell culture. The 4T1 cells were incubated for 48 h by Cs/Hif-1α siRNA nano-complex 0.070 nmol of Cs concentration. Next, gene expression was analyzed by qPCR. It was found that Cs/Hif-1α siRNA nano-complex has an inhibitory effect on Hif-1α gene expression in the 4T1 cell line and reduced Hif-1α expression by 50% compared to the control group (P < 0.0001) (Fig. 2b). To confirm the silencing effects of the Hif-1α siRNA on the Hif-1α gene, the cell line was treated with Cs nanostructure alone. The results indicated no significant differences in Hif-1α gene expression between the control and Cs-treated cell lines (Fig. 2b).
Animal studies
Synergic effects of combination therapy on inhibition of tumor growth
The potential of combinational therapy was studied by administering Ptx, Imq, and Cs/Hif-1α siRNA according to the therapy protocol (Table 2) to syngeneic 4T1 tumor-bearing BALB/c mice. The eight-day therapy of Ptx, Imq, and Cs/Hif-1α siRNA at designated doses according to the scheduled treatment was well tolerated by the mice under examination. To assess the effect of CTs on tumor growth, initially, we surveyed their effectiveness on phenotypes of tumors. Figs. 3a and 3b display the inhibitory potential of combination treatments on tumor volume and size. While the tumor size and volume were significantly increased in the PBS (control) group, The TCT and DCT groups demonstrated substantial inhibition of tumor growth and development (P < 0.0001). By administering Ptx, Imq, and Cs/Hif-1α siRNA simultaneously, tumor growth was gradually inhibited from day 1 to 8 when compared with the control group. TCT, as a complete treatment, effectively inhibited syngeneic 4T1 tumors on BALB/c (Fig. 3a) (P < 0.0001). However, complete tumor regression was not attained during these therapies. Based on our observation, tumor growth was gradually suppressed from the first day of the experiment until its conclusion and the tumor volume was reduced significantly (Fig. 3b). Overall, TCT demonstrated more effective inhibition of tumor growth and a more significant reduction in volume compared to DCT (P < 0.0001).
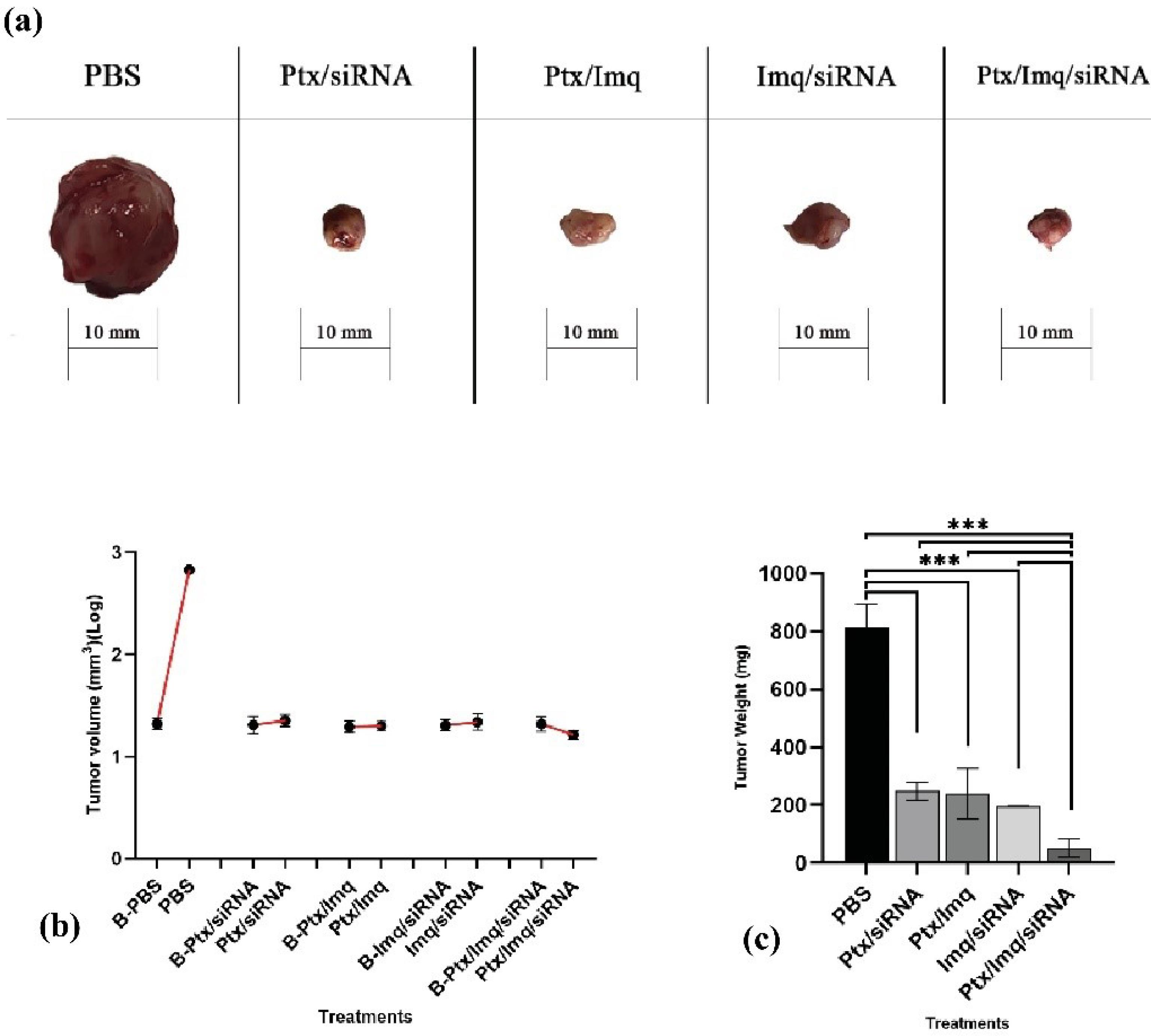
Fig. 3.
Tumor image, size, and weight after combinational therapy. (a) Tumor’s image. After eight days of therapy, all mice were sacrificed under anesthesia. Tumors were removed carefully, and the photo was taken immediately. (b) Effects of defined therapies on tumor volume. The first spot (●) of each treatment, which is indicated by B-, means tumor volume before treatments, and the second spot (●), means tumor volume after treatments. (c) Tumor weight analysis in breast cancer mouse model. Tumors were removed after eight days of therapy and were weighted by precise scale (All data are presented as the mean ± SD. *P < 0.01, **P < 0.001 and ***P < 0.0001. One-way ANOVA was used to make comparisons between the Tests and the PBS (control) group).
.
Tumor image, size, and weight after combinational therapy. (a) Tumor’s image. After eight days of therapy, all mice were sacrificed under anesthesia. Tumors were removed carefully, and the photo was taken immediately. (b) Effects of defined therapies on tumor volume. The first spot (●) of each treatment, which is indicated by B-, means tumor volume before treatments, and the second spot (●), means tumor volume after treatments. (c) Tumor weight analysis in breast cancer mouse model. Tumors were removed after eight days of therapy and were weighted by precise scale (All data are presented as the mean ± SD. *P < 0.01, **P < 0.001 and ***P < 0.0001. One-way ANOVA was used to make comparisons between the Tests and the PBS (control) group).
As shown in Fig. 3c, TCT and DCT reduced the weight of the tumor in all examined groups. Notably, all combinational therapies, such as TCT and DCT, significantly decreased tumor weight compared with the PBS group (P < 0.0001). In summary, the triple combination therapy (Ptx + Imq + Cs/Hif-1α siRNA) significantly reduced tumor weight compared to the PBS group (P < 0.0001) and all DCT groups (P < 0.0001).
Effects of combinational therapies on cancer apoptosis and progression-related genes
To reveal the molecular mechanisms behind observed anticancer effects of proposed combinational treatments, we analyzed tumor samples for the expression level of critical genes involved in anticancer immune responses and cancer progression, and immune-inducing and immune-inhibiting cytokines, tumor samples were analyzed by qPCR. The efficacy of CT was investigated on cell proliferation and cancer progression-related genes. Hif-1α and signal transducer and activation of Stat3 plus vein endothelial growth factor (VEGF) genes play a crucial role in the hypoxic TME. They are involved in numerous aspects of tumor progression, such as metastasis, angiogenesis, and immune evasion.47,48 Hif-1α and Stat3 genes were selected as target genes for cancer therapy. In this study, Hif-1α as a cancer progression essential gene was inhibited by TCT and all DCT treatments significantly (P < 0.0001) (Fig. 4a). Further, Stat3 was downregulated by Ptx + Imq + Cs/Hif-1α siRNA more significantly than DCT (P < 0.0001) (Fig. 4b).
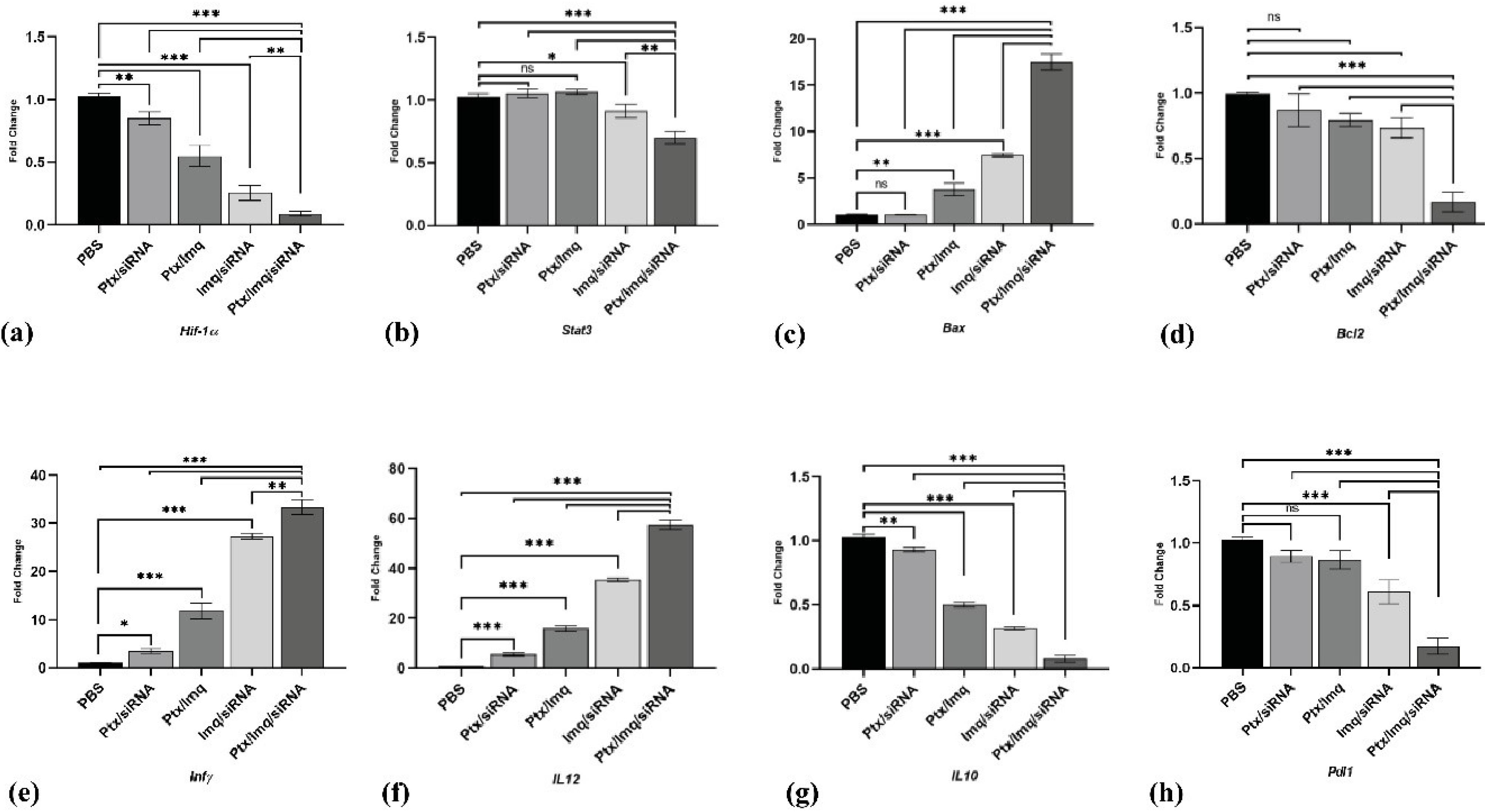
Fig. 4.
Gene expression analysis by real-time PCR. (a, b)Hif-1α and Stat3 as a progression pathway genes expression level. (c)Bax as a pro-apoptotic gene expression level. (d) Bcl-2 as an anti-apoptosis gene expression level. (e, f)Inf-γ and IL12 as an immune-inducing gene expression level. (g, h)IL10 and Pdl1 as an immune inhibiting genes expression level. (All data are presented as the mean ± SD of the gene expression levels. *P < 0.01, **P < 0.001 and ***P < 0.0001. One-way ANOVA was used to compare the Tests and the PBS (control) group).
.
Gene expression analysis by real-time PCR. (a, b)Hif-1α and Stat3 as a progression pathway genes expression level. (c)Bax as a pro-apoptotic gene expression level. (d) Bcl-2 as an anti-apoptosis gene expression level. (e, f)Inf-γ and IL12 as an immune-inducing gene expression level. (g, h)IL10 and Pdl1 as an immune inhibiting genes expression level. (All data are presented as the mean ± SD of the gene expression levels. *P < 0.01, **P < 0.001 and ***P < 0.0001. One-way ANOVA was used to compare the Tests and the PBS (control) group).
In apoptosis-related genes, the pro-apoptotic gene Bax showed a significant increase with DCT like Ptx + Imq, Imq + Cs/Hif-1α siRNA (P < 0.001) and, significantly higher increases with the triple combinational therapy Ptx + Imq + Cs/Hif-1α siRNA (P < 0.0001). Interestingly, Bcl2, an anti-apoptotic gene, exhibited downregulation with Ptx + Imq + Cs/Hif-1α siRNA (P < 0.0001), while DCT showed no significant effects on Bcl2. TCT demonstrated effective upregulation of pro-apoptotic genes and inhibition of the anti-apoptotic gene. This aligns with findings from Yonghong et al, who reported that HIF-1α siRNA could induce apoptosis in the BC cell line MDA-MB-231 and GC-2 cells by blocking the HIF-1α (Fig. 4c and Fig. 4d).49,50
The impact of combinational therapies on antigen-presenting cells (APC) function and antitumor immunity
TLR-7 agonists have been considered in cancer therapy based on antitumoral activity; TLR-7/8 activation up-regulated central transcription factor nuclear factor-κβ and IL-12, which leads to activation of DCs and production of Th1-type proinflammatory cytokines.28 It was reported that IL-12 produced by APC, such as DC, inhibited tumor development and increased survival of BC patients significantly.51 We discovered a significant increase in IL-12 levels in the tumors of mice treated with TCT and DCT, demonstrating a marked upregulation compared to the control group (P < 0.0001) (Fig. 4f). INF-γ, known for inducing CD80/CD86 expression and promoting immune response, was also upregulated significantly by TCT and DCT therapies (P < 0.0001) (Fig. 4e).52,53
Inhibition of Th2 type cytokines following combination therapies
IL-10, as an immune-inhibiting cytokine, affects the function of APC by inhibiting the MHC and co-stimulatory molecules and inducing T-regulatory cells.54,55 Malignant cells release IL-10 to generate and support tumor progression and development.56 PD-L1 is expressed by tumor cells as an “adaptive immune mechanism” to escape anti-tumor responses.57,58 Based on the results of this research, the expression levels of IL-10 and PDL-1 were significantly suppressed in treated mice, particularly with Ptx + Imq + Cs/Hif-1α siRNA. (P < 0.0001) (Fig. 4g, 4h).
Western blotting analysis
To confirm qPCR results, western blotting was done on HIF-1α, STAT3, BAX, BCL2, IL12, INF-γ, IL10, and PDL1. HIF-1α protein expression level significantly downregulated by DCT and TCT (P < 0.0001) (Fig. 5b). STAT3 protein expression level diminished by TCT (P < 0.001), and all DCT had no significant effects on STAT3 protein level (Fig. 5c). BAX protein as a pro-apoptotic element increases by Ptx + Imq, Imq + siRNA, and Ptx + Imq + Cs/Hif-1α siRNA significantly (P < 0.0001) (Fig. 5d). Also, BCL2 as an anti-apoptotic element diminished by Ptx + Imq, Imq + siRNA, and Ptx + Imq + Cs/Hif-1α siRNA significantly (P < 0.0001) (Fig. 5e). IL12 and INF-γ as an immune inducing factor significantly up-regulated by DCT and TCT except Ptx + Cs/Hif-1α siRNA (P < 0.0001) (Fig. 5f, 5g). IL10 and PDL1 as an immune inhibiting element down-regulated by DCT and TCT except Ptx + Cs/Hif-1α siRNA (P < 0.0001) (Fig. 5h, 5i).
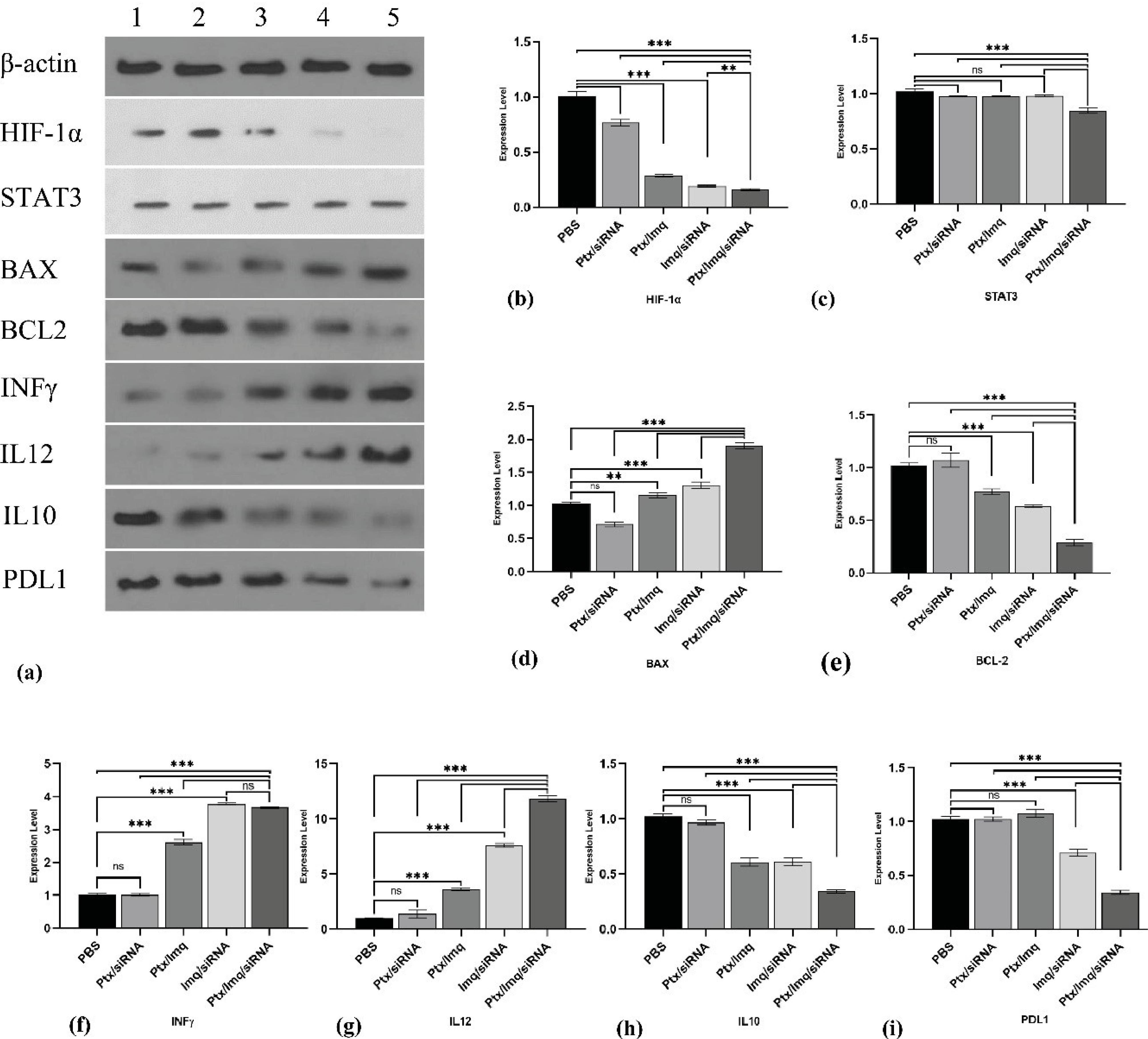
Fig. 5.
Western blotting analysis after combinational therapy. (a) western blotting gel β-actin 43kDa as a housekeeping, HIF-1α 130kDa, STAT3 97.86kDa, Bax 23kDa, Bcl2 26kDa, Inf-γ 20.25kDa, IL12 70kDa, IL10 20kDa, Pdl1 33kDa. 1) PBS treatments 2) Ptx + Cs/Hif-1α siRNA 3) Ptx + Imq 4) Imq + Cs/Hif-1α siRNA 5) Ptx + Imq + Cs/Hif-1α siRNA (b) HIF-1α protein expression level. (c) STAT3 protein expression level (d) Bax protein expression level (e) Bcl2 expression level (f) Inf-γ protein expression level (g) IL12 protein expression level (h) IL10 protein expression level (i) PDl1 protein expression level. (All data are presented as the mean ± SD of the protein expression levels. *P < 0.01, **P < 0.001, and ***P < 0.0001. One-way ANOVA was used to make comparisons between the Tests and the PBS (control) group).
.
Western blotting analysis after combinational therapy. (a) western blotting gel β-actin 43kDa as a housekeeping, HIF-1α 130kDa, STAT3 97.86kDa, Bax 23kDa, Bcl2 26kDa, Inf-γ 20.25kDa, IL12 70kDa, IL10 20kDa, Pdl1 33kDa. 1) PBS treatments 2) Ptx + Cs/Hif-1α siRNA 3) Ptx + Imq 4) Imq + Cs/Hif-1α siRNA 5) Ptx + Imq + Cs/Hif-1α siRNA (b) HIF-1α protein expression level. (c) STAT3 protein expression level (d) Bax protein expression level (e) Bcl2 expression level (f) Inf-γ protein expression level (g) IL12 protein expression level (h) IL10 protein expression level (i) PDl1 protein expression level. (All data are presented as the mean ± SD of the protein expression levels. *P < 0.01, **P < 0.001, and ***P < 0.0001. One-way ANOVA was used to make comparisons between the Tests and the PBS (control) group).
Discussion
Cancer-induced immunosuppression in TME and multiple drug resistance are two important barriers to cancer treatment. The paper provides valuable insights into overcoming chemoresistance and cancer induced immunosuppression in a BC model by targeting Hif-1α alongside an ICD inducing chemotherapeutic agent (i.e., Ptx) and an immunomodulatory agent (i.e., TLR7) which is known as to induce Th1 type inflammatory immune response.59 Numerous studies have reported robust outcomes for chemoimmunotherapy compared to traditional chemotherapy for cancer; however, these advancements have not been translated into improved therapeutic outcomes for cancer patients. It has been substantiated that the immunosuppressive milieu within the TME constitutes a primary factor contributing to the failure of immunotherapy. Therefore, modifying the TME holds promise for enhancing the response rates of chemoimmunotherapy.60,61
In this study, Cs nanoparticles were effectively employed for delivering Hif-1α siRNA as an integral component of our designed combination therapy. In line with our studies several other studies have reported the effective delivery of siRNA to cancer cells using Cs nanoparticles. Haliza Katas et al. have reported that Cs nanoparticles absorbing siRNA (entrapped siRNA between TPP and Cs method) could effectively deliver siRNA to target cells.62 Also, Justein Malmo et al. blocked genes in the H1299 cell line (human lung carcinoma) using Cs nanoparticles/siRNA strongly efficiently and without toxicity.63 In some other studies, PDGF-D and PDFGR-β genes were effectively blocked using Cs/siRNA nanoplexes in breast cancer mouse model.64 Shaymaa et al. reported that the oncogenic microRNA, miRNA-21, was effectively blocked using GE11-peptide conjugated small interfering RNA-loaded chitosan nanoparticles(GE11-siRNA-CsNPs) in EGFR overexpressed triple-negative breast cancer (TNBC).65 In agreement with previous research and Cs potentials to siRNA delivery and low toxicity, we successfully blocked the HIF-1α gene expression using Cs/HIF-1α siRNA in vitro and in vivo. Consistent with prior research highlighting Cs's potential for siRNA delivery and its low toxicity profile, we achieved successful inhibition of HIF-1α gene expression both in vitro and in vivo using Cs/HIF-1α siRNA.
Some research has consistently indicated that combinational therapy involving Hif-1α targeting can reduce drug resistance and improve drug efficacy, thereby mitigating tumor cell proliferation and angiogenesis. This approach also diminishes cancer stem cell proliferation and self-renewal while inducing apoptosis.33-36 It was shown that inhibiting HIF-1α by IDF-11774 (HIF-1α inhibitor) in K1 and BCPAP thyroid cancer cell line significantly decreases tumor cell proliferation and migration.66 Also, inhibition of HIF-1α suppressed angiogenesis and tumor growth in HCT116 cells and colon cancer mouse models.67 Inhibition of Hif-1α strongly repressed tumor progression in 4T1 xenograft tumor, which was related to the phosphorylationof eIF4E regulated by both MAPK and mTOR signaling pathways.68 As our data shows, inhibition of HIF-1α in combination with Imq and Ptx significantly reduces tumor growth by inhibiting HIF-1α (P < 0.0001) and STAT3 gene expression levels(P < 0.0001). HIF-1α was knocked down by adenovirus-expressing shRNA in the HCC cell line by Sung Hoon Chei et al. under hypoxic conditions (1% O2, 24h). Interestingly, inhibition of HIF-1α up-regulated the expression of apoptotic factors while downregulating anti-apoptotic factors simultaneously.69 Targeting Hif-1α has been reported to attenuate PD-L1 expression on tumor cells and tumor-infiltrating myeloid cells, which causes tumor rejection.70
In this study, we demonstrate that combinational therapy of Ptx with Cs/HIF-1α substantially inhibited tumor growth by suppressing the expression of HIF-1α protein in the 4T1 breast cancer mouse model. Ptx is known as a first-line chemotherapeutic drug in BC treatment and is commonly used as an immunogenic cell death inducer in the clinical treatments of advanced BC.9 Despite the notable therapeutic effects of Ptx, the systemic cytotoxicity has restricted its application.71 Thus, combining Ptx with other therapies has the potential to amplify its therapeutic efficacy while mitigating associated side effects. HIF-1α plays a crucial role in promoting cancer stem cell specification and maintenance under hypoxic conditions. Furthermore, it contributes to multidrug resistance in breast cancer, particularly against Ptx, through various mechanisms. Hence, targeting hypoxia represents a critical preparatory step prior to Ptx administration.72-75 Previous studies also indicate that inhibiting Hif-1α in conjunction with chemodrug treatment leads to synergistic anticancer effects. Additionally, research has demonstrated that combining a Hif-1α inhibitor with gemcitabine, a first-line chemodrug for pancreatic cancer, induces immunogenic cell death (ICD) and augments the infiltration of tumor-infiltrating lymphocytes.38,76 It has been demonstrated that HIF-1α renders MDA-MB-231 cells resistant to Ptx-induced apoptosis. Consequently, combining Ptx with HIF-1α siRNA therapy initiates the apoptosis pathway in MDA-MB-231 cells by modulating the expression of genes involved in both pro- and anti-apoptotic mechanisms in breast cancer cell lines.77
We observed that incorporating Imq alongside TLR7, Ptx, and Cs/HIF-1ɑ yielded more potent inhibitory effects on cancer growth compared to combination therapies involving only Ptx with Cs or Cs/HIF-1ɑ in the 4T1 mouse breast cancer model. The therapeutic effects of Imq as TLR7 ligand alone or in combination with standard conventional therapy have been widely studied in cancer therapy. Imq as a TLR7 agonist increases the apoptosis index in basal cell carcinoma patients by decreasing the Bcl2 gene level. TLR7 agonists were shown to stimulate up-regulation of IL12 and INF-γ in TME, resulting in strong NK cell activation.78-80 Studies have shown that combining Ptx with the TLR agonist Imq induces DC proliferation by approximately 250%. Moreover, co-treatment with paclitaxel and imiquimod enhances the secretion of pro-inflammatory and Th1 cytokines in DCs.37 In a study by Wenlu Yan et al., it was reported that combining Irinotecan, a classic clinical chemotherapy agent, with Imq in a CT-26 colorectal cancer mouse model promoted dendritic cell (DC) maturation and the secretion of pro-inflammatory cytokines.81 Also, Imq combination with GEM, induces immune cells and establishes strong anticancer activity against 4T1 breast tumors. Also, it showed that infiltration of Th1 monocytes increased at the site of cancer cells treated with Imq/GEM.82 There is ample evidence that Imq treatments increased NK1.1+ cells in the TME by upregulation of IL-12/IL-6.30,31 Imq treatment in the mouse tumor model led to theupregulationofIL-12, which would enhance CD8+T cell immune responses, antitumor effects, and prolonged survival in treated mice. Based on Shi-Xing Yang et al., IL-12 upregulation enhanced CD4+/CD8+T cells, INF-γ in TME, while also mediating tumor suppressive effects by CD8+T cells and producing INF-γ by CD8 + cells causing apoptosis in cells.83 In line with previous studies we found that, combinational therapy of Ptx and Imq significantly increases inflammatory cytokines IL-12 and INFγ and down-regulated immune inhibitory cytokines. Imq combination therapy with doxorubicin using pH-sensitive micelle nanoparticles inhibits 4T1 cells based xenograft model, showed that IL-12 and INFγ production by Imq treatments induces apoptosis in tumor cells and suppresses tumor growth.83,84_ENREF_78It was shownthat Imq treatments would upregulate Caspase 3, 7, 9, and Bad/Bax and downregulate BCL-2 to accelerate apoptosis in SCl-1, SCC-13, HaCat, and TRAMP-C2 cells.85-87 It was confirmed that Imq increases apoptosis in BCC/KMC1, AGS, Hela, and B16 cell lines after 24 hours. Imq remarkably increased the level of CRT on the cell surface and the secretion of extracellular ATP.88 In line with previous reports,our findings show that Imq combinational treatments significantly increase apoptosis by up-regulating BAX (P < 0.001) and down-regulation BCL2 (P < 0.0001) in the 4T1 breast cancer mouse model.
In agreement with previous research, Within the experimental groups, the most robust effects on inhibiting tumor growth and modifying the expression levels of genes associated with cancer progression were noted in mice with cancer who underwent treatment with TCT (Ptx + Imq + Cs/Hif-1α siRNA)(P < 0.0001). Our findings suggest that TCT exerted its effects by downregulating cancer progression and proliferation genes, including Hif-1α and Stat-3, while simultaneously inducing apoptosis. This was achieved through the upregulation of pro-apoptosis genes and the downregulation of Bcl2. Moreover, the TCT regulates the TME by augmenting the release of immune-inducing cytokines, specifically INF-γ and IL12, while concurrently suppressing immune-inhibiting cytokines. Our observations indicate that TCT exhibits more pronounced effects compared to DCT within the study groups. The elevated levels of Th1 inflammatory cytokines observed in tumors treated with TCT create a favorable microenvironment for the activation and functionality of anticancer immune responses. This results in a more robust inhibition of tumors in the TCT group compared to other experimental groups, including DCT. Furthermore, the downregulation of Hif-1α by Cs/HIF-1α siRNA can sensitize cancer cells to both direct cytotoxic effects of Ptx and indirect killing by anticancer immune responses activated by TCT.
Conclusion
In summary, the findings of this study suggest that a triple combination therapy (TCT) involving an ICD-inducing chemotherapeutic agent (Ptx), a TLR7 ligand (Imq), and a Hif-1α blocking agent (i.e., Cs/Hif-1α siRNA) is a promising approach to modify TME which is a major barrier to cancer treatment. The immunosuppressive milieu of TME is identified as a primary factor contributing to the failure of immunotherapy, emphasizing the need for TME alteration to enhance chemoimmunotherapy response rates. The synergistic application of Cs/Hif-1α siRNA nanoparticles in combination with Ptx + Imq demonstrated a significantly higher level of cytokines, indicative of enhanced immune responses. Moreover, the mice treated with the TCT exhibited significantly stronger inhibition of tumor growth compared to those receiving monotherapy. The study's findings emphasize the significant impact of TCT on the TME, suppressing tumor growth, modulating the expression of critical genes involved in immunosuppression and cancer progression, and promoting antitumor immune responses. The study provides valuable insights that underscore the significance of combination therapies in the quest for more effective and tailored cancer treatments.
An important limitation of our study lies in our assessment of the anticancer effects of the triple combination therapy in a mouse cancer model. Given the variances in the components and functions of the immune system between mice and humans, the responses to combinational therapy may vary between these two species. Therefore, we recommend evaluating the therapeutic efficacy of the designed combination therapy in non-human primates. These primates offer exceptional suitability for biomedical research due to their genetic and physiological resemblances to humans.
Research Highlights
What is the current knowledge?
HIF-1α, as an oxygen-sensing factor in tumor hypoxia, was shown to be up-regulated in many cancers and increases the expression of tumor progressive genes as well.
What is new here?
HIF-1α gene blocking using combinational therapy decreases the expression of tumor progressive genes and modifies immunomodulatory cytokines in TME.
Competing Interests
None declared.
Ethics Statement
Ethical approval for this study was obtained from Tabriz University of Medical Science (IR.TBZMED.VCR.REC.1399.162).
Acknowledgements
The authors thank the Advanced Medical Sciences College and Immunology Research Center of Tabriz for supporting the facilities.
References
- Rahimzadeh S, Burczynska B, Ahmadvand A, Sheidaei A, Khademioureh S, Pazhuheian F. Geographical and socioeconomic inequalities in female breast cancer incidence and mortality in Iran: a Bayesian spatial analysis of registry data. PLoS One 2021; 16:e0248723. doi: 10.1371/journal.pone.0248723 [Crossref] [ Google Scholar]
- Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N. Drug resistance in cancer: an overview. Cancers (Basel) 2014; 6:1769-92. doi: 10.3390/cancers6031769 [Crossref] [ Google Scholar]
- Dong X, Bai X, Ni J, Zhang H, Duan W, Graham P. Exosomes and breast cancer drug resistance. Cell Death Dis 2020; 11:987. doi: 10.1038/s41419-020-03189-z [Crossref] [ Google Scholar]
- Ji X, Lu Y, Tian H, Meng X, Wei M, Cho WC. Chemoresistance mechanisms of breast cancer and their countermeasures. Biomed Pharmacother 2019; 114:108800. doi: 10.1016/j.biopha.2019.108800 [Crossref] [ Google Scholar]
- Shivhare S, Das A. Cell density modulates chemoresistance in breast cancer cells through differential expression of ABC transporters. Mol Biol Rep 2023; 50:215-25. doi: 10.1007/s11033-022-08028-2 [Crossref] [ Google Scholar]
- Khdair A, Chen D, Patil Y, Ma L, Dou QP, Shekhar MP. Nanoparticle-mediated combination chemotherapy and photodynamic therapy overcomes tumor drug resistance. J Control Release 2010; 141:137-44. doi: 10.1016/j.jconrel.2009.09.004 [Crossref] [ Google Scholar]
- Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature 2019; 575:299-309. doi: 10.1038/s41586-019-1730-1 [Crossref] [ Google Scholar]
- Paridaens R, Biganzoli L, Bruning P, Klijn JG, Gamucci T, Houston S. Paclitaxel versus doxorubicin as first-line single-agent chemotherapy for metastatic breast cancer: a European Organization for Research and Treatment of Cancer Randomized Study with cross-over. J Clin Oncol 2000; 18:724-33. doi: 10.1200/jco.2000.18.4.724 [Crossref] [ Google Scholar]
- Hajjar A, Ergun MA, Alagoz O, Rampurwala M. Cost-effectiveness of adjuvant paclitaxel and trastuzumab for early-stage node-negative, HER2-positive breast cancer. PLoS One 2019; 14:e0217778. doi: 10.1371/journal.pone.0217778 [Crossref] [ Google Scholar]
- Ghiringhelli F, Rébé C. Using immunogenic cell death to improve anticancer efficacy of immune checkpoint inhibitors: from basic science to clinical application. Immunol Rev 2024; 321:335-49. doi: 10.1111/imr.13263 [Crossref] [ Google Scholar]
- Zhao S, Tang Y, Wang R, Najafi M. Mechanisms of cancer cell death induction by paclitaxel: an updated review. Apoptosis 2022; 27:647-67. doi: 10.1007/s10495-022-01750-z [Crossref] [ Google Scholar]
- Behroozaghdam M, Hashemi M, Javadi G, Mahdian R, Soleimani M. Expression of Bax and Bcl2 genes in MDMA-induced hepatotoxicity on rat liver using quantitative real-time PCR method through triggering programmed cell death. Iran Red Crescent Med J 2015; 17:e24609. doi: 10.5812/ircmj.24609 [Crossref] [ Google Scholar]
- Kroemer G, Galluzzi L, Kepp O, Zitvogel L. Immunogenic cell death in cancer therapy. Annu Rev Immunol 2013; 31:51-72. doi: 10.1146/annurev-immunol-032712-100008 [Crossref] [ Google Scholar]
- Ladoire S, Enot D, Andre F, Zitvogel L, Kroemer G. Immunogenic cell death-related biomarkers: impact on the survival of breast cancer patients after adjuvant chemotherapy. Oncoimmunology 2016; 5:e1082706. doi: 10.1080/2162402x.2015.1082706 [Crossref] [ Google Scholar]
- Liu P, Zhao L, Zitvogel L, Kepp O, Kroemer G. Immunogenic cell death (ICD) enhancers-drugs that enhance the perception of ICD by dendritic cells. Immunol Rev 2024; 321:7-19. doi: 10.1111/imr.13269 [Crossref] [ Google Scholar]
- Giraldo NA, Sanchez-Salas R, Peske JD, Vano Y, Becht E, Petitprez F. The clinical role of the TME in solid cancer. Br J Cancer 2019; 120:45-53. doi: 10.1038/s41416-018-0327-z [Crossref] [ Google Scholar]
- Aponte PM, Caicedo A. Stemness in cancer: stem cells, cancer stem cells, and their microenvironment. Stem Cells Int 2017; 2017:5619472. doi: 10.1155/2017/5619472 [Crossref] [ Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74. doi: 10.1016/j.cell.2011.02.013 [Crossref] [ Google Scholar]
- Zarychta E, Ruszkowska-Ciastek B. Cooperation between angiogenesis, vasculogenesis, chemotaxis, and coagulation in breast cancer metastases development: pathophysiological point of view. Biomedicines 2022; 10:300. doi: 10.3390/biomedicines10020300 [Crossref] [ Google Scholar]
- Kheshtchin N, Hadjati J. Targeting hypoxia and hypoxia-inducible factor-1 in the tumor microenvironment for optimal cancer immunotherapy. J Cell Physiol 2022; 237:1285-98. doi: 10.1002/jcp.30643 [Crossref] [ Google Scholar]
- Jing X, Yang F, Shao C, Wei K, Xie M, Shen H. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol Cancer 2019; 18:157. doi: 10.1186/s12943-019-1089-9 [Crossref] [ Google Scholar]
- Huang R, Zhou PK. HIF-1 signaling: a key orchestrator of cancer radioresistance. Radiat Med Prot 2020; 1:7-14. doi: 10.1016/j.radmp.2020.01.006 [Crossref] [ Google Scholar]
- Choudhry H, Harris AL. Advances in hypoxia-inducible factor biology. Cell Metab 2018. 27: 281-98. 10.1016/j.cmet.2017.10.005.
- Ke Q, Costa M. Hypoxia-inducible factor-1 (HIF-1). Mol Pharmacol 2006; 70:1469-80. doi: 10.1124/mol.106.027029 [Crossref] [ Google Scholar]
- Semenza GL. Expression of hypoxia-inducible factor 1: mechanisms and consequences. Biochem Pharmacol 2000; 59:47-53. doi: 10.1016/s0006-2952(99)00292-0 [Crossref] [ Google Scholar]
- Duechler M, Wilczynski JR. Hypoxia inducible factor-1 in cancer immune suppression. Curr Immunol Rev 2010; 6:260-71. doi: 10.2174/157339510791823853 [Crossref] [ Google Scholar]
- Shen Z, Pei Q, Zhang H, Yang C, Cui H, Li B. Hypoxia-inducible factor-1α inhibition augments efficacy of programmed cell death 1 antibody in murine prostatic cancer models. Anticancer Drugs 2022; 33:587-94. doi: 10.1097/cad.0000000000001294 [Crossref] [ Google Scholar]
- Schön MP, Schön M. TLR7 and TLR8 as targets in cancer therapy. Oncogene 2008; 27:190-9. doi: 10.1038/sj.onc.1210913 [Crossref] [ Google Scholar]
- Schön MP, Schön M, Klotz KN. The small antitumoral immune response modifier imiquimod interacts with adenosine receptor signaling in a TLR7- and TLR8-independent fashion. J Invest Dermatol 2006; 126:1338-47. doi: 10.1038/sj.jid.5700286 [Crossref] [ Google Scholar]
- Chuang CM, Monie A, Hung CF, Wu TC. Treatment with imiquimod enhances antitumor immunity induced by therapeutic HPV DNA vaccination. J Biomed Sci 2010; 17:32. doi: 10.1186/1423-0127-17-32 [Crossref] [ Google Scholar]
- Craft N, Bruhn KW, Nguyen BD, Prins R, Lin JW, Liau LM. The TLR7 agonist imiquimod enhances the anti-melanoma effects of a recombinant Listeria monocytogenes vaccine. J Immunol 2005; 175:1983-90. doi: 10.4049/jimmunol.175.3.1983 [Crossref] [ Google Scholar]
- de Mello Palma V, Frank LA, Balinha DM, Rados PV, Pohlmann AR, Guterres SS. Is imiquimod a promising drug to treat oral mucosa diseases? A scoping review and new perspectives. Br J Clin Pharmacol 2024; 90:427-39. doi: 10.1111/bcp.15923 [Crossref] [ Google Scholar]
- Bayat Mokhtari R, Homayouni T, Baluch N, Morgatskaya E, Kumar S, Das B. Combination therapy in combating cancer. Oncotarget 2017; 8:38022-43. doi: 10.18632/oncotarget.16723 [Crossref] [ Google Scholar]
- Eriksson E, Lövgren T, Loskog AS. Targeting tumor-induced immunosuppression using conventional cancer therapeutics. Med Res Arch 2021; 9:1-10. doi: 10.18103/mra.v9i12.2606 [Crossref] [ Google Scholar]
- Kerbel RS, Yu J, Tran J, Man S, Viloria-Petit A, Klement G. Possible mechanisms of acquired resistance to anti-angiogenic drugs: implications for the use of combination therapy approaches. Cancer Metastasis Rev 2001; 20:79-86. doi: 10.1023/a:1013172910858 [Crossref] [ Google Scholar]
- Liang P, Ballou B, Lv X, Si W, Bruchez MP, Huang W. Monotherapy and combination therapy using anti-angiogenic nanoagents to fight cancer. Adv Mater 2021; 33:e2005155. doi: 10.1002/adma.202005155 [Crossref] [ Google Scholar]
- Seth A, Heo MB, Lim YT. Poly (γ-glutamic acid) based combination of water-insoluble paclitaxel and TLR7 agonist for chemo-immunotherapy. Biomaterials 2014; 35:7992-8001. doi: 10.1016/j.biomaterials.2014.05.076 [Crossref] [ Google Scholar]
- Zhao T, Ren H, Jia L, Chen J, Xin W, Yan F. Inhibition of HIF-1α by PX-478 enhances the anti-tumor effect of gemcitabine by inducing immunogenic cell death in pancreatic ductal adenocarcinoma. Oncotarget 2015; 6:2250-62. doi: 10.18632/oncotarget.2948 [Crossref] [ Google Scholar]
- Katas H, Alpar HO. Development and characterisation of chitosan nanoparticles for siRNA delivery. J Control Release 2006; 115:216-25. doi: 10.1016/j.jconrel.2006.07.021 [Crossref] [ Google Scholar]
- Mao S, Sun W, Kissel T. Chitosan-based formulations for delivery of DNA and siRNA. Adv Drug Deliv Rev 2010; 62:12-27. doi: 10.1016/j.addr.2009.08.004 [Crossref] [ Google Scholar]
- Lee D, Zhang W, Shirley SA, Kong X, Hellermann GR, Lockey RF. Thiolated chitosan/DNA nanocomplexes exhibit enhanced and sustained gene delivery. Pharm Res 2007; 24:157-67. doi: 10.1007/s11095-006-9136-9 [Crossref] [ Google Scholar]
- Kumar P, Nagarajan A, Uchil PD. Analysis of cell viability by the MTT assay. Cold Spring Harb Protoc 2018; 2018:pdb-rot095505. doi: 10.1101/pdb.prot095505 [Crossref] [ Google Scholar]
- Park S, Jeong EJ, Lee J, Rhim T, Lee SK, Lee KY. Preparation and characterization of nonaarginine-modified chitosan nanoparticles for siRNA delivery. Carbohydr Polym 2013; 92:57-62. doi: 10.1016/j.carbpol.2012.08.116 [Crossref] [ Google Scholar]
- Wei J, Long Y, Guo R, Liu X, Tang X, Rao J. Multifunctional polymeric micelle-based chemo-immunotherapy with immune checkpoint blockade for efficient treatment of orthotopic and metastatic breast cancer. Acta Pharm Sin B 2019; 9:819-31. doi: 10.1016/j.apsb.2019.01.018 [Crossref] [ Google Scholar]
- Franco MS, Roque MC, de Barros ALB, de Oliveira Silva J, Cassali GD, Oliveira MC. Investigation of the antitumor activity and toxicity of long-circulating and fusogenic liposomes co-encapsulating paclitaxel and doxorubicin in a murine breast cancer animal model. Biomed Pharmacother 2019; 109:1728-39. doi: 10.1016/j.biopha.2018.11.011 [Crossref] [ Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta CT) Method. Methods 2001; 25:402-8. doi: 10.1006/meth.2001.1262 [Crossref] [ Google Scholar]
- You L, Wu W, Wang X, Fang L, Adam V, Nepovimova E. The role of hypoxia-inducible factor 1 in tumor immune evasion. Med Res Rev 2021; 41:1622-43. doi: 10.1002/med.21771 [Crossref] [ Google Scholar]
- Rashid M, Rostami Zadeh L, Baradaran B, Molavi O, Ghesmati Z, Sabzichi M. Up-down regulation of HIF-1α in cancer progression. Gene 2021; 798:145796. doi: 10.1016/j.gene.2021.145796 [Crossref] [ Google Scholar]
- Shi Y, Chang M, Wang F, Ouyang X, Jia Y, Du H. Role and mechanism of hypoxia-inducible factor-1 in cell growth and apoptosis of breast cancer cell line MDA-MB-231. Oncol Lett 2010; 1:657-62. doi: 10.3892/ol_00000115 [Crossref] [ Google Scholar]
- Wang Z, Zhu C, Song Y, Chen X, Zheng J, He L. SIRT3 inhibition suppresses hypoxia-inducible factor 1α signaling and alleviates hypoxia-induced apoptosis of type B spermatogonia GC-2 cells. FEBS Open Bio 2023; 13:154-63. doi: 10.1002/2211-5463.13523 [Crossref] [ Google Scholar]
- Oka N, Markova T, Tsuzuki K, Li W, El-Darawish Y, Pencheva-Demireva M. IL-12 regulates the expansion, phenotype, and function of murine NK cells activated by IL-15 and IL-18. Cancer Immunol Immunother 2020; 69:1699-712. doi: 10.1007/s00262-020-02553-4 [Crossref] [ Google Scholar]
- Subauste CS, de Waal Malefyt R, Fuh F. Role of CD80 (B71) and CD86 (B72) in the immune response to an intracellular pathogen. J Immunol 1998; 160:1831-40. [ Google Scholar]
- Almeida FS, Vanderley SE, Comberlang FC, de Andrade AG, Cavalcante-Silva LH, Dos Santos Silva E. Leishmaniasis: immune cells crosstalk in macrophage polarization. Trop Med Infect Dis 2023; 8:276. doi: 10.3390/tropicalmed8050276 [Crossref] [ Google Scholar]
- Sato T, Terai M, Tamura Y, Alexeev V, Mastrangelo MJ, Selvan SR. Interleukin 10 in the tumor microenvironment: a target for anticancer immunotherapy. Immunol Res 2011; 51:170-82. doi: 10.1007/s12026-011-8262-6 [Crossref] [ Google Scholar]
- Adeegbe DO, Nishikawa H. Regulatory T cells in cancer; can they be controlled?. Immunotherapy 2015; 7:843-6. doi: 10.2217/imt.15.52 [Crossref] [ Google Scholar]
- Mantovani A, Barajon I, Garlanda C. IL-1 and IL-1 regulatory pathways in cancer progression and therapy. Immunol Rev 2018; 281:57-61. doi: 10.1111/imr.12614 [Crossref] [ Google Scholar]
- Han Y, Liu D, Li L. PD-1/PD-L1 pathway: current researches in cancer. Am J Cancer Res 2020; 10:727-42. [ Google Scholar]
- Wajant H. The role of TNF in cancer. In: Kalthoff H, ed. Death Receptors and Cognate Ligands in Cancer. Berlin, Heidelberg: Springer. 2009. p. 1-15. 10.1007/400_2008_26.
- Malik JA, Kaur G, Agrewala JN. Revolutionizing medicine with toll-like receptors: a path to strengthening cellular immunity. Int J Biol Macromol 2023; 253:127252. doi: 10.1016/j.ijbiomac.2023.127252 [Crossref] [ Google Scholar]
- Sasse AD, Sasse EC, Clark LG, Ulloa L, Clark OA. Chemoimmunotherapy versus chemotherapy for metastatic malignant melanoma. Cochrane Database Syst Rev 2007; CD005413. 10.1002/14651858.CD005413.pub2.
- Rabinovich GA, Gabrilovich D, Sotomayor EM. Immunosuppressive strategies that are mediated by tumor cells. Annu Rev Immunol 2007; 25:267-96. doi: 10.1146/annurev.immunol.25.022106.141609 [Crossref] [ Google Scholar]
- Katas H, Alpar HO. Development and characterisation of chitosan nanoparticles for siRNA delivery. J Control Release 2006; 115:216-25. doi: 10.1016/j.jconrel.2006.07.021 [Crossref] [ Google Scholar]
- Malmo J, Sørgård H, Vårum KM, Strand SP. siRNA delivery with chitosan nanoparticles: Molecular properties favoring efficient gene silencing. J Control Release 2012; 158:261-8. doi: 10.1016/j.jconrel.2011.11.012 [Crossref] [ Google Scholar]
- Şalva E, Özbaş S, Alan S, Özkan N, Ekentok-Atıcı C, Kabasakal L. Combination therapy with chitosan/siRNA nanoplexes targeting PDGF-D and PDGFR-β reveals anticancer effect in breast cancer. J Gene Med 2023; 25:e3465. doi: 10.1002/jgm.3465 [Crossref] [ Google Scholar]
- Abdulmalek SA, Saleh AM, Shahin YR, El Azab EF. Functionalized siRNA-chitosan nanoformulations promote triple-negative breast cancer cell death via blocking the miRNA-21/AKT/ERK signaling axis: in-silico and in vitro studies. Naunyn Schmiedebergs Arch Pharmacol 2024. 10.1007/s00210-024-03068-w.
- Kim MH, Lee TH, Lee JS, Lim DJ, Lee PC. Hif-1α inhibitors could successfully inhibit the progression of differentiated thyroid cancer in vitro. Pharmaceuticals (Basel) 2020; 13:208. doi: 10.3390/ph13090208 [Crossref] [ Google Scholar]
- Ban HS, Kim BK, Lee H, Kim HM, Harmalkar D, Nam M. The novel hypoxia-inducible factor-1α inhibitor IDF-11774 regulates cancer metabolism, thereby suppressing tumor growth. Cell Death Dis 2017; 8:e2843. doi: 10.1038/cddis.2017.235 [Crossref] [ Google Scholar]
- Zheng Z, Xu T, Liu Z, Tian W, Jiang ZH, Zhu GY. Cryptolepine suppresses breast adenocarcinoma via inhibition of HIF-1 mediated glycolysis. Biomed Pharmacother 2022; 153:113319. doi: 10.1016/j.biopha.2022.113319 [Crossref] [ Google Scholar]
- Choi SH, Park JY, Kang W, Kim SU, Kim DY, Ahn SH. Knockdown of HIF-1α and IL-8 induced apoptosis of hepatocellular carcinoma triggers apoptosis of vascular endothelial cells. Apoptosis 2016; 21:85-95. doi: 10.1007/s10495-015-1185-2 [Crossref] [ Google Scholar]
- Bailey CM, Liu Y, Liu M, Du X, Devenport M, Zheng P. Targeting HIF-1α abrogates PD-L1-mediated immune evasion in tumor microenvironment but promotes tolerance in normal tissues. J Clin Invest 2022; 132:e150846. doi: 10.1172/jci150846 [Crossref] [ Google Scholar]
- Liebmann JE, Cook JA, Lipschultz C, Teague D, Fisher J, Mitchell JB. Cytotoxic studies of paclitaxel (Taxol®) in human tumour cell lines. Br J Cancer 1993; 68:1104-9. doi: 10.1038/bjc.1993.488 [Crossref] [ Google Scholar]
- Tian T, Han J, Huang J, Li S, Pang H. Hypoxia-induced intracellular and extracellular heat shock protein gp96 increases paclitaxel-resistance and facilitates immune evasion in breast cancer. Front Oncol 2021; 11:784777. doi: 10.3389/fonc.2021.784777 [Crossref] [ Google Scholar]
- Xiang L, Semenza GL. Hypoxia-inducible factors promote breast cancer stem cell specification and maintenance in response to hypoxia or cytotoxic chemotherapy. Adv Cancer Res 2019; 141:175-212. doi: 10.1016/bs.acr.2018.11.001 [Crossref] [ Google Scholar]
- Yong L, Tang S, Yu H, Zhang H, Zhang Y, Wan Y. The role of hypoxia-inducible factor-1 alpha in multidrug-resistant breast cancer. Front Oncol 2022; 12:964934. doi: 10.3389/fonc.2022.964934 [Crossref] [ Google Scholar]
- Feng L, Shen F, Zhou J, Li Y, Jiang R, Chen Y. Hypoxia-induced up-regulation of miR-27a promotes paclitaxel resistance in ovarian cancer. Biosci Rep 2020; 40:BSR20192457. doi: 10.1042/bsr20192457 [Crossref] [ Google Scholar]
- Sun X, Kanwar JR, Leung E, Lehnert K, Wang D, Krissansen GW. Gene transfer of antisense hypoxia inducible factor-1 alpha enhances the therapeutic efficacy of cancer immunotherapy. Gene Ther 2001; 8:638-45. doi: 10.1038/sj.gt.3301388 [Crossref] [ Google Scholar]
- Flamant L, Notte A, Ninane N, Raes M, Michiels C. Anti-apoptotic role of HIF-1 and AP-1 in paclitaxel exposed breast cancer cells under hypoxia. Mol Cancer 2010; 9:191. doi: 10.1186/1476-4598-9-191 [Crossref] [ Google Scholar]
- Hart OM, Athie-Morales V, O'Connor GM, Gardiner CM. TLR7/8-mediated activation of human NK cells results in accessory cell-dependent IFN-gamma production. J Immunol 2005; 175:1636-42. doi: 10.4049/jimmunol.175.3.1636 [Crossref] [ Google Scholar]
- Salerno F, Freen-van Heeren JJ, Guislain A, Nicolet BP, Wolkers MC. Costimulation through TLR2 drives polyfunctional CD8 + T cell responses. J Immunol 2019; 202:714-23. doi: 10.4049/jimmunol.1801026 [Crossref] [ Google Scholar]
- Smits EL, Cools N, Lion E, Van Camp K, Ponsaerts P, Berneman ZN. The toll-like receptor 7/8 agonist resiquimod greatly increases the immunostimulatory capacity of human acute myeloid leukemia cells. Cancer Immunol Immunother 2010; 59:35-46. doi: 10.1007/s00262-009-0721-8 [Crossref] [ Google Scholar]
- Yan W, Li Y, Zou Y, Zhu R, Wu T, Yuan W. Co-delivering irinotecan and imiquimod by pH-responsive micelle amplifies anti-tumor immunity against colorectal cancer. Int J Pharm 2023; 648:123583. doi: 10.1016/j.ijpharm.2023.123583 [Crossref] [ Google Scholar]
- Singh B, Maharjan S, Pan DC, Zhao Z, Gao Y, Zhang YS. Imiquimod-gemcitabine nanoparticles harness immune cells to suppress breast cancer. Biomaterials 2022; 280:121302. doi: 10.1016/j.biomaterials.2021.121302 [Crossref] [ Google Scholar]
- Yang SX, Wei WS, Ouyan QW, Jiang QH, Zou YF, Qu W. Interleukin-12 activated CD8 + T cells induces apoptosis in breast cancer cells and reduces tumor growth. Biomed Pharmacother 2016; 84:1466-71. doi: 10.1016/j.biopha.2016.10.046 [Crossref] [ Google Scholar]
- Wen YH, Hsieh PI, Chiu HC, Chiang CW, Lo CL, Chiang YT. Precise delivery of doxorubicin and imiquimod through pH-responsive tumor microenvironment-active targeting micelles for chemo- and immunotherapy. Mater Today Bio 2022; 17:100482. doi: 10.1016/j.mtbio.2022.100482 [Crossref] [ Google Scholar]
- Schön M, Bong AB, Drewniok C, Herz J, Geilen CC, Reifenberger J. Tumor-selective induction of apoptosis and the small-molecule immune response modifier imiquimod. J Natl Cancer Inst 2003; 95:1138-49. doi: 10.1093/jnci/djg016 [Crossref] [ Google Scholar]
- Han JH, Lee J, Jeon SJ, Choi ES, Cho SD, Kim BY. In vitro and in vivo growth inhibition of prostate cancer by the small molecule imiquimod. Int J Oncol 2013; 42:2087-93. doi: 10.3892/ijo.2013.1898 [Crossref] [ Google Scholar]
- Sohn KC, Li ZJ, Choi DK, Zhang T, Lim JW, Chang IK. Imiquimod induces apoptosis of squamous cell carcinoma (SCC) cells via regulation of A20. PLoS One 2014; 9:e95337. doi: 10.1371/journal.pone.0095337 [Crossref] [ Google Scholar]
- Huang SW, Wang ST, Chang SH, Chuang KC, Wang HY, Kao JK, et al. Imiquimod exerts antitumor effects by inducing immunogenic cell death and is enhanced by the glycolytic inhibitor 2-deoxyglucose. J Invest Dermatol 2020; 140: 1771-83.e6. 10.1016/j.jid.2019.12.039.