Bioimpacts. 2025;15:30520.
doi: 10.34172/bi.30520
Original Article
Recombinant receptor-binding motif of spike COVID-19 vaccine candidate induces SARS-CoV-2 neutralizing antibody response
Hossein Samiei-Abianeh Data curation, Formal analysis, Methodology, Validation, Writing – original draft, 1, 2 
Shahram Nazarian Conceptualization, Methodology, Validation, Writing – review & editing, 2, * 
Emad Kordbacheh Methodology, 2
Alireza Felegary Formal analysis, Methodology, Validation, 2
Author information:
1Department of Medical Biotechnology and Nanotechnology, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran
2Department of Biology, Faculty of Basic Sciences, Imam Hossein University, Tehran, Iran
Abstract
Introduction:
The SARS-CoV-2 pandemic necessitates effective therapeutic solutions. The receptor-binding motif (RBM) is a subdomain of the spike protein's receptor-binding domain (RBD) and is critical for facilitating the binding of SARS-CoV-2 to the human ACE2 receptor. This study investigates the use of the receptor-binding motif (RBM) domain as an immunogen to produce potent neutralizing antibodies against SARS-CoV-2.
Methods:
The RBM gene was codon-optimized and cloned into the pET17b vector for expression in E. coli BL21 (DE3) cells, induced with 1 mM IPTG. The recombinant RBM protein was purified using Ni-NTA affinity chromatography. After validating the recombinant RBM by Western blotting with anti-His tag antibodies, BALB/c mice were immunized with 20 µg of the purified RBM protein. Anti-RBM IgG was subsequently purified using protein G resin, and its neutralizing capacity was assessed using the Pishtaz Teb Zaman Neutralization Assay Kit.
Results:
The recombinant RBM protein, with a molecular weight of 10 kDa, was expressed as inclusion bodies. the typical yield of purification was 27 mg/L of bacterial culture. The neutralization test demonstrated a concentration of 36 µg/mL of neutralizing antibodies in the immunized serum, preventing the spike protein from binding to ACE2.
Conclusion:
Our study demonstrated that anti-RBM antibodies exhibited neutralization effects on SARS-CoV-2. These findings provide evidence for the development of a vaccine candidate through the induction of antibodies against the RBM, necessitating further studies with adjuvants suitable for human use to evaluate its potential for human vaccination.
Keywords: COVID-19, Spike glycoprotein, Receptor-binding domain, Receptor-binding motif, Recombinant protein, Subunit vaccine
Copyright and License Information
© 2025 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Introduction
The COVID-19 pandemic, a worldwide health crisis, originated from the novel coronavirus SARS-CoV-2. This virus attaches to human cells by binding to the angiotensin-converting enzyme 2 (ACE2) receptor through its spike (S) glycoprotein, a mechanism first observed during the outbreak in China.1 The S protein of SARS-CoV-2 consists of two parts: S1, which is responsible for receptor binding, and S2, which enables the virus to merge with the host cell membrane.2
The receptor-binding motif (RBM), a crucial subdomain within the receptor-binding domain (RBD) of the spike protein, is essential for the attachment of SARS-CoV-2 to the human ACE2 receptor. The RBM contains a total of 72 amino acids and is intimately connected to the ACE2 receptor, specifically between amino acids 437 and 508.3 Within the RBM, six key amino acid residues - L455, F486, Q493, S494, N501, and Y505 - are responsible for enabling the virus to bind to ACE2.4 RBM is an important target for therapeutic interventions against SARS-CoV-2 because mAbs targeting this site compete with ACE2 binding.5
The global emergence of COVID-19 urgently necessitated effective therapeutic strategies against SARS-CoV-2. Initially, the drug repurposing approach, encompassing corticosteroids and antivirals, emerged as a cost-effective and timely strategy for novel pathogens. For instance, Remdesivir, initially developed for Ebola, received FDA authorization in October 2020.6 Another example is Dexamethasone, which has been shown to reduce 28-day mortality in hospitalized COVID-19 patients who are receiving mechanical ventilation or oxygen support.7 WHO strongly recommends nirmatrelvir/ritonavir, known as 'Paxlovid,' for high-risk individuals, especially those who are immunosuppressed.8 Paxlovid, an oral antiviral pill, demonstrated an 89% reduction in the risk of hospitalization and death among unvaccinated individuals in clinical trials.9 Between December 31, 2020, and April 30, 2021, WHO issued emergency use authorizations for COVID-19 vaccines developed by Pfizer, AstraZeneca/Oxford, Johnson & Johnson, and Moderna. These vaccines have been created using different technological platforms by various companies viz., mRNA-based nucleic acid vaccine (Pfizer and Moderna), adenoviral vector-based vaccines (Oxford-AstraZeneca- Serum Institute and Johnson and Johnson), inactivated vaccine (Bharat Biotech), and recombinant subunit protein vaccine (Novavax).
Recombinant vaccines offer distinct advantages in the landscape of COVID-19 prevention, particularly in terms of safety profiles and efficacy. Compared to inactivated vaccines and adenovirus-based vectors, recombinant vaccines demonstrate heightened safety profiles with minimal side effects. Park et al demonstrated that priming with an mRNA vaccine provides better protection against influenza virus compared to a protein vaccine. However, in their study, protein vaccines have shown to be more effective as boosters.10 Consequently, it is hypothesized that in scenarios requiring ongoing vaccination due to emerging coronavirus variants, protein vaccines might provide a safety benefit over mRNA vaccines. Furthermore, certain recombinant vaccines display temperature stability, which is crucial for broadening access to COVID-19 vaccines, especially in low- and middle-income nations. Notably, several recombinant vaccines have demonstrated good prevention efficacy, as seen with Novavax, which showed approximately 31 percent effectiveness in preventing infection.11
Most recombinant COVID-19 vaccines leverage either the full-length S protein or the RBD as an immunogen to prompt the elicitation of binding and neutralizing antibodies (NAbs). RBD-based vaccines have consistently demonstrated high efficacy, with over 90% of SARS-CoV-2 NAbs in convalescent sera or vaccine recipients targeting the RBD.12-14 Positive clinical effectiveness has been observed in trials of certain RBD-based vaccines against COVID-19.15-18 Moreover, recent studies have indicated that RBD possesses five glycosylation sites, with two glycosylation sites (T323 and S325) potentially playing pivotal roles in mediating spike and ACE2 receptor interactions.19,20 The absence of glycosylation in RBM may enhance its structural integrity and activity when expressed in bacterial systems such as E. coli, compared to expressing the RBD which contains glycosylation sites.
Nazarian et al have previously expressed recombinant proteins of the RBD and Nucleocapsid (N) in a prokaryotic host, and their immune response has been evaluated.21,22 Based on previous studies of RBM-ACE-2 interactions, we have designed our study on an rRBM protein immunogen capable of neutralizing SARS-CoV-2, thereby preventing its attachment to ACE-2 on host cells.
Materials and Methods
Sequence analysis and vector design
We obtained the sequence for the RBM protein of the SARS-CoV-2 strain WuhanHu-1 (NCBI Reference Sequence: YP_009724390.1), which codes for residues N437 to Y508 of spike. To optimize this nucleotide sequence for expression in E. coli, we employed the Genscript Optimization GeneTM algorithm server. The GenscriptTM Codon Optimization algorithm utilizes a comprehensive approach, assessing and adjusting for over 200 factors that influence gene expression. These factors include GC content, codon usage, codon content index, RNase splicing sites, and cis-acting mRNA destabilizing motifs. Following optimization, the optimized sequence underwent analysis using the GenRCA Rare Codon Analysis Tool (https://www.genscript.com/tools/rare-codon-analysis). The optimized RBM gene was then inserted into the expression vector pET17b with a 6xHis-tag and a stop codon at the 3' end for purification purposes. Restriction enzyme sites (NdeI and BamHI) were presented at the ends of the sequence following the restriction site identification using the NEBcutter online tool.23 Before conducting laboratory techniques, we used online tools to analyze the physicochemical characteristics of the designed recombinant protein and predict its 3D structure. The physicochemical characteristics of the designed recombinant protein were analyzed using the ProtParam server.24 The 3D structure of the protein was predicted using the I-TASSER ab initio online software and visualized using Accelrys Discovery Studio 2.5.25 The resulting structure was validated using ProSA-web 26 and the quality of the resulting stereochemistry of the structure was analyzed using the Ramachandran plot in PROCHECK software.27 Finally, Shinegene (China)was tasked with synthesizing the recombinant pET17b-RBM.
Transformation of the recombinant vector
The pET17b-RBM recombinant plasmid was employed for transforming competent E. coli strain BL21 (DE3) cells, prepared using the calcium chloride method. Transformation of the recombinant plasmid into E. coli was facilitated through the heat shock metho.28 Initially, transformed cells were cultivated on LB agar plates supplemented with 100 µg/mL ampicillin at 37°C. These plates were then incubated for 12 to 16 hours at 37 °C. The success of the transformation was confirmed through double digestion of the plasmid with NdeI and BamHI enzymes.29
Expression and purification of the recombinant RBM
A single colony was selected and cultured overnight in LB broth containing 100 µg/mL ampicillin at 37 °C. After the overnight culture, 1 mL of this culture was inoculated into 50 mL of fresh LB broth with 50 µg/mL ampicillin. Upon reaching an optical density of 0.7-1.0 at 600 nm (OD600 nm), IPTG was introduced at a concentration of 0.5 mM to enhance the expression of the RBM protein. Induction was carried out for 5 hours at 37 °C. After induction, E. coli cells were harvested by centrifugation at 5000 rpm at 4 °C for 6 minutes, and the resulting pellets were resuspended in extraction buffer. The cells were then disrupted using ultrasonic treatment with an ultrasonic processor set to an intensity of 20 kHz, performing 10-second pulses with 30-second intervals for a total of three cycles and subsequently centrifuged at 10 000 rpm at 4 °C for 20 minutes. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to distinguish the induced and uninduced samples.30
To evaluate the solubility of the recombinant protein, bacterial pellets were subjected to various extraction buffers. One set of cells was resuspended in PBS buffer, while another set was resuspended in an inclusion body (IB) solubilization solution (8 M urea, 20 mM Tris-HCl, and 100 mM NaCl, pH = 8). Subsequently, both mixtures underwent ultrasonic treatment to disrupt the cells and were then incubated for 1 hour at 37 °C in a shaker incubator. Following centrifugation for 20 minutes at 13 000 rpm, the resulting supernatants were collected and subjected to analysis using SDS-PAGE with a discontinuous buffer system, following the Laemmli method.31
Affinity chromatography using a Ni-NTA column served as the method for protein purification.32 Prior to purification, bacterial lysis containing RBM inclusion bodies underwent refolding by dialyzing with a urea gradient (6, 4, 2, 1, 0.5, and 0 M) in 20 mM Tris-HCl buffer with 500 mM NaCl at pH 8.0. The refolded solution was then applied to the Ni-NTA column for purification. Following the removal of nonspecific bindings through washing with a buffer (50 mM sodium phosphate buffer, pH 8.0, 500 mM sodium chloride, 0.5% Triton X-100, 10% glycerol, 20 mM imidazole), The recombinant RBM protein was eluted using an elution buffer (20 mM sodium phosphate, 0.5 M NaCl, 500 mM imidazole, pH 7.4).33 subsequent purification steps and the purity of the recombinant RBM were assessed using SDS-PAGE.
Antigen characterization by ELISA and western blotting
To prepare the protein samples, they were boiled in 5X reducing dye buffer (500 mM DTT, 250 mM Tris-HCl, 10% (w/v) SDS, 50% (v/v) glycerol, 0.1% bromophenol blue, pH 6.8) for 10 minutes. Subsequently, the proteins were separated on a 12% denaturing polyacrylamide gel and stained with Coomassie blue. For Western blot analysis, the proteins were transferred from a polyacrylamide gel (unstained) to a polyvinylidene difluoride (PVDF) membrane using a Biorad electrophoretic transfer device at 500 mA for 90 minutes. Next, the membrane was blocked overnight in blocking buffer (consisting of 5% (w/v) fat-free dried milk dissolved in PBS + 0.05% (v/v) Tween 20). Following the washing step, the membrane was incubated with mouse anti His-tag antibodies (from Sigma-Aldrich, USA; catalog No.SAB2702219) at a 1:10 000 dilution, targeting the RBM 6xHis tag, at 37 °C for 45 minutes. Subsequently, the substrate (3,3′-diaminobenzidine (DAB) in Tris 50mM, pH = 7.8) was applied to the membrane after washing to expose the band(s), and the process was halted by removing the substrate and adding water to the paper.34
For the enzyme-linked immunosorbent assay (ELISA), Maxisorb plates (Nunc) were coated with 5 µg of recombinant protein and blocked with 5% skim milk in PBST (PBS plus 0.05% Tween 20). The plates were washed and incubated with serially diluted SARS-CoV-2 convalescent human sera (from previous study22) at 37 °C for 30 minutes. After washing with PBST four times, 100 µl of 1 in 2000 dilution of Horseradish Peroxidase (HRP)-conjugated anti-Human IgG were added to each well plate. The plate was then incubated at 37 °C for 30 minutes and washed four times with PBST. Then, 100 µL of the substrate solution containing 3 mg O-Phenylene Diamine (OPD) were added to each well plate, and the reaction was stopped with 2.5 M H2SO4. Finally, the absorbance was measured at 495 nm on a microplate reader.35
Immunization of mice
The animals were maintained in clean standard conditions in the Animal Care Facility of Imam Hossein University. All the animal tests were performed in accordance with Care and Use of Laboratory Animals guidelines.36 Ten BALB/c mice were randomly assigned to each experimental group. Subcutaneous (s.c.) injections were administered as the initial dose and booster immunizations on days 21 and 35. The control groups received 100 μL of antigen diluent alone (PBS) via s.c injection. Meanwhile, one test group received 20 μg of protein in 100 μL PBS for each administration without adjuvant (rRBM free), while another group received antigen with an equal volume (100 μL) of complete Freund's adjuvant for the first administration and incomplete Freund's adjuvant for the booster dose (rRBM + Adj).37 Blood samples were collected from the venous sinus (Retro-orbital) of all mouse groups using a capillary tube/pipette on days 14, 28, and 42. After clot formation, the samples were centrifuged at 3500 g for 10 minutes. The resulting supernatant sera from each group of mice were stored at -20 °C.
At day 42, a total of 100 mg of feces from the test and control groups was collected and resuspended in 1 mL of ice-cold PBS (supplemented with 0.1% sodium azide and 1 mM PMSF). Following homogenization, the samples were centrifuged for 15 minutes at 7000 rpm and 4 °C. Supernatants were transferred to a fresh tube and stored at 4 °C for quick ELISA analysis.38
Serum IgG and feces IgA measurement
Indirect ELISA was employed to detect IgG or IgA antibodies against recombinant RBM protein in serum or fecal samples. Briefly, 96-well polystyrene plates were coated with 5 µg/mL of RBM protein in coating buffer (15 mM Na2CO3, 35 mM NaHCO3, pH 9.6) and incubated overnight at 4 °C. Following a washing step, the plates were blocked with 5% fat-free dry milk in PBS for 1 hour at 37 °C. Dilutions of sera (1:100-1:25 600) or fecal extracts (1:1-1:4) in PBS were then added and incubated overnight at 4 °C. Subsequently, goat horseradish peroxidase-conjugated anti-mouse IgG or IgA (diluted 1:10 000 in PBS) were used as secondary antibodies. After adding a substrate solution (6 mg OPD + 0.1 M citric acid + 1 mM H2O2, pH 4.35), optical density (OD) values at 495 nm were measured using a microplate reader (Bio Tek, USA). All experiments were performed in triplicate, and the results were expressed as mean and standard deviation.
Neutralizing antibody detection
The measure of anti-RBM neutralizing antibody was conducted according to the manufacturer's instructions (Pishtaz Teb Zaman Diagnostics, Tehran, Iran). The SARS-CoV-2 Neutralizing Antibody ELISA Kit uses a competitive method based on quantitative inhibition of RBD and ACE2, capable of identifying all classes of neutralizing antibodies against SARS-CoV-2 within 45 minutes. Initially, a human neutralizing antibody is used as a standard, followed by the addition of the test serum according to the kit's instructions. The neutralizing antibody concentration values, reported in micrograms per milliliter (µg/mL), are derived from the standard curve. The cut-off value for the kit, established based on the mean and three standard deviations of anti-SARS-CoV-2 neutralizing antibody concentration in normal serum specimens, is 2.5 µg/mL, as specified by the manufacturer.
Statistical analysis
Statistical analyses were conducted using Statistica version 13.0. Comparisons among multiple groups were made using a two-way ANOVA with Duncan comparison post hoc test. P values less than 0.01 were considered statistically significant.
Results
In silico analysis of recombinant RBM
A synthetic sequence encoding the RBM of the Spike gene was engineered, tailored to the codon usage preferences of E. coli. Negative cis-acting elements and repetitive sequences were avoided to enhance the synthetic gene's efficiency. The analysis of codon bias and GC content was conducted for both non-optimized and optimized sequences. Post-optimization, the GC content was increased from 35.29% to 53.11%, thereby enhancing mRNA stability. The optimized mRNA's 2D structure exhibited a ∆G of -61.9 kcal/mol, indicating a stable configuration without the formation of stable hairpins or pseudoknots at the 5' end (Fig. 1). Finally, the restriction sites of NdeI (CATATG) and BamHI (GGATCC) were appended to the 5' and 3' ends of the recombinant RBM sequence, respectively (Fig. 2) and sent to ShineGene bio-company (China) for production.
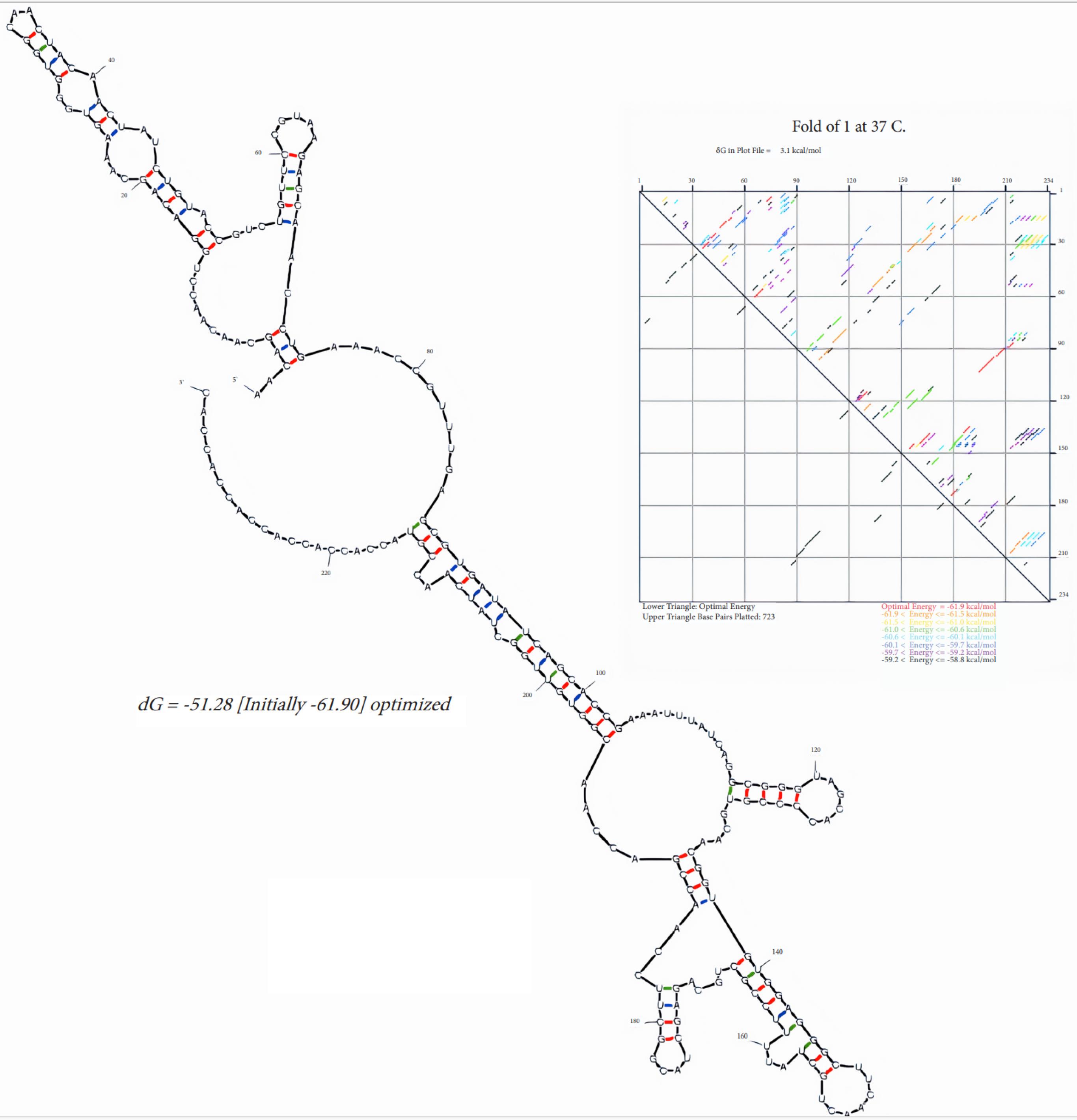
Fig. 1.
Linear RNA folding and energy dot plot predicted by Mfold. The energy dot plot uses different colors to represent varying levels of suboptimality. The optimal base pairs, shown in red, indicate the most stable configurations with the lowest free energy. Blue represents base pairs that are less likely to form, indicating higher energy states. The red color highlights the base pairs in the optimal folding configuration.
.
Linear RNA folding and energy dot plot predicted by Mfold. The energy dot plot uses different colors to represent varying levels of suboptimality. The optimal base pairs, shown in red, indicate the most stable configurations with the lowest free energy. Blue represents base pairs that are less likely to form, indicating higher energy states. The red color highlights the base pairs in the optimal folding configuration.

Fig. 2.
Schematic illustration of optimized nucleotide sequence of RBM for cloning in expression vector
.
Schematic illustration of optimized nucleotide sequence of RBM for cloning in expression vector
Table 1 shows physicochemical characteristics for the recombinant RBM. The 3D structure of the rRBM was created using I-TASSER software (Fig. 3). The confidence score (C-score) from I-TASSER, which assesses the quality of predicted structure, was 0.14, indicating moderate confidence. It is important to note that a higher C-score, which ranges from -5 to 2, typically signifies a model with greater reliability. The model's estimated TM-score was 0.73 ± 0.11, suggesting a likelihood of high structural similarity to known protein structures, while the expected root mean square deviation (RMSD) was 3.2 ± 2.3Å, indicating the average distance between the atoms of the model and the actual structure they represent. The Ramachandran plot generated by the RAMPAGE server showed that approximately 71.4% of residues are in favored regions, while 28.5% are in allowed regions. Additionally, the ProSA tool was used to assess the model by comparing it to known X-ray structures, yielding a Z-score of -1.02 (Fig. 3).
Table 1.
The physicochemical characteristics predicted by ProtParam tool for the recombinant RBM protein
|
Protein sequence
|
MGSSHHHHHHSSGLVPRGSHMNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYHHHHH |
|
Number of aa
|
98 |
Molecular weight (Da)
|
10184.25 |
Total number of atoms
|
1504 |
|
Formula
|
C494H712N150O114S4 |
negatively charged residues
|
5 |
positively charged residues
|
7 |
|
Instability index
|
24.02 (stable) |
Aliphatic index
|
44.69 |
Theoretical pI
|
8.63 |
| pET28a initial fusion tag, Terminal His-Tag |
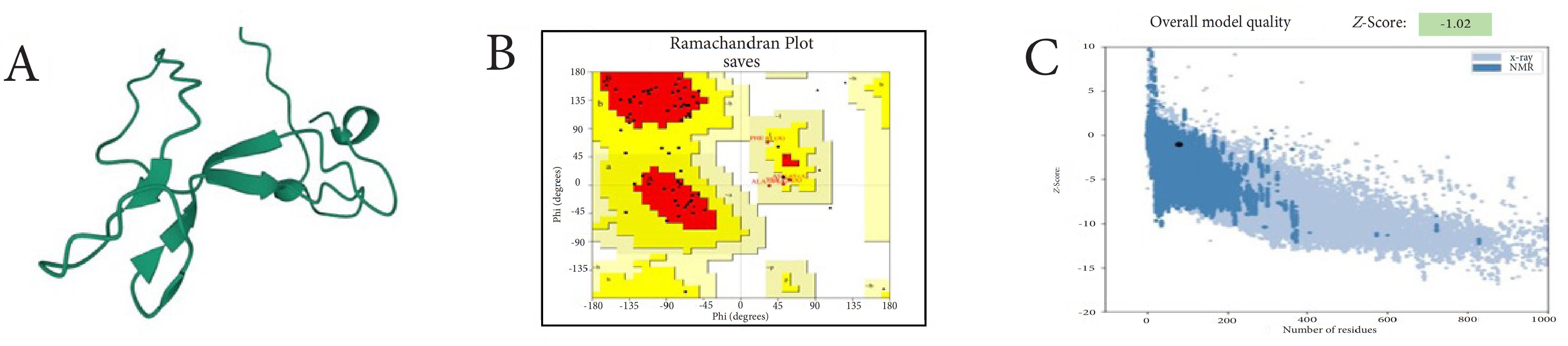
Fig. 3.
(a) 3D representation of the recombinant RBM protein predicted using I-TASSER software, (b) Evaluation of model stability through a Ramachandran plot, and (c) ProSA analysis.
.
(a) 3D representation of the recombinant RBM protein predicted using I-TASSER software, (b) Evaluation of model stability through a Ramachandran plot, and (c) ProSA analysis.
Production of recombinant RBM as inclusion bodies
The rRBM was found as inclusion bodies in the bacterial lysate after induction (Fig. 4A) with a molecular weight of approximately 10 kD. various temperatures (18, 20, 25, and 30°C) and media with different IPTG concentrations to avoid inclusion body formation was tested but observed no expression at temperatures lower than 37 °C. This measurement aligns with the predicted size of the RBM from the in silico analysis. After affinity purification (IMAC) using Ni-NTA column, the recombinant RBM was analyzed via SDS-PAGE (Fig. 4B), and the typical yield of one-step purification was 27 mg/L of bacterial culture. The purity of the purified recombinant RBM band (Lane 6 of Fig. 4B) was calculated to be 97% using GelAnalyzer 23.1 software.
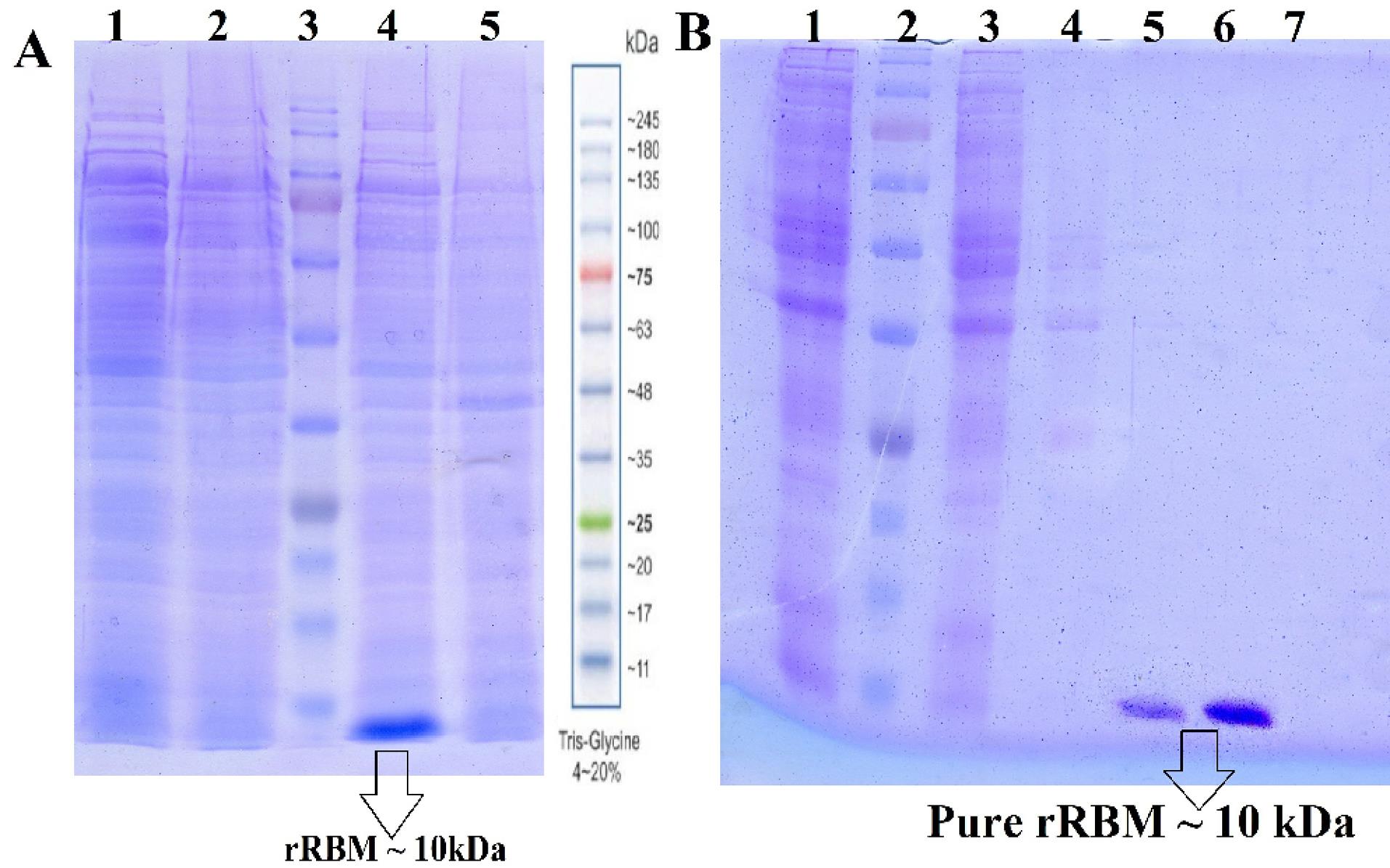
Fig. 4.
A) 12%SDS-PAGE Analysis of rRBM Expression, Soluble fractions of rRBM post-induction (lane 1 and 2), protein marker (lane 3), insoluble fractions containing inclusion bodies (lane 4) and non-induced (lane5) fractions. B) Purification analysis of induced bacterial lysis. solution after soup loaded (Lane 1), protein marker (lane2), washing the non-specific bonds by washing buffer (lanes 3 and 4), elution fraction by imidazole buffer (lanes 5 and 6).
.
A) 12%SDS-PAGE Analysis of rRBM Expression, Soluble fractions of rRBM post-induction (lane 1 and 2), protein marker (lane 3), insoluble fractions containing inclusion bodies (lane 4) and non-induced (lane5) fractions. B) Purification analysis of induced bacterial lysis. solution after soup loaded (Lane 1), protein marker (lane2), washing the non-specific bonds by washing buffer (lanes 3 and 4), elution fraction by imidazole buffer (lanes 5 and 6).
Validation of rRBM antigen
To validate the recombinant RBM protein produced from E. coli, an anti-His antibody Western blot was conducted. This method confirms the presence and integrity of the His-tagged RBM protein. The results confirmed the identification and antigenic activity of the rRBM sequence, as the immunoreactive protein displayed the expected molecular weight of RBM (Fig. 5). Furthermore, ELISA analysis demonstrated the capability of accurately detecting the recombinant protein in sera from convalescent COVID-19 patients, which contained anti-SARS-CoV-2 antibodies (Fig. 6).
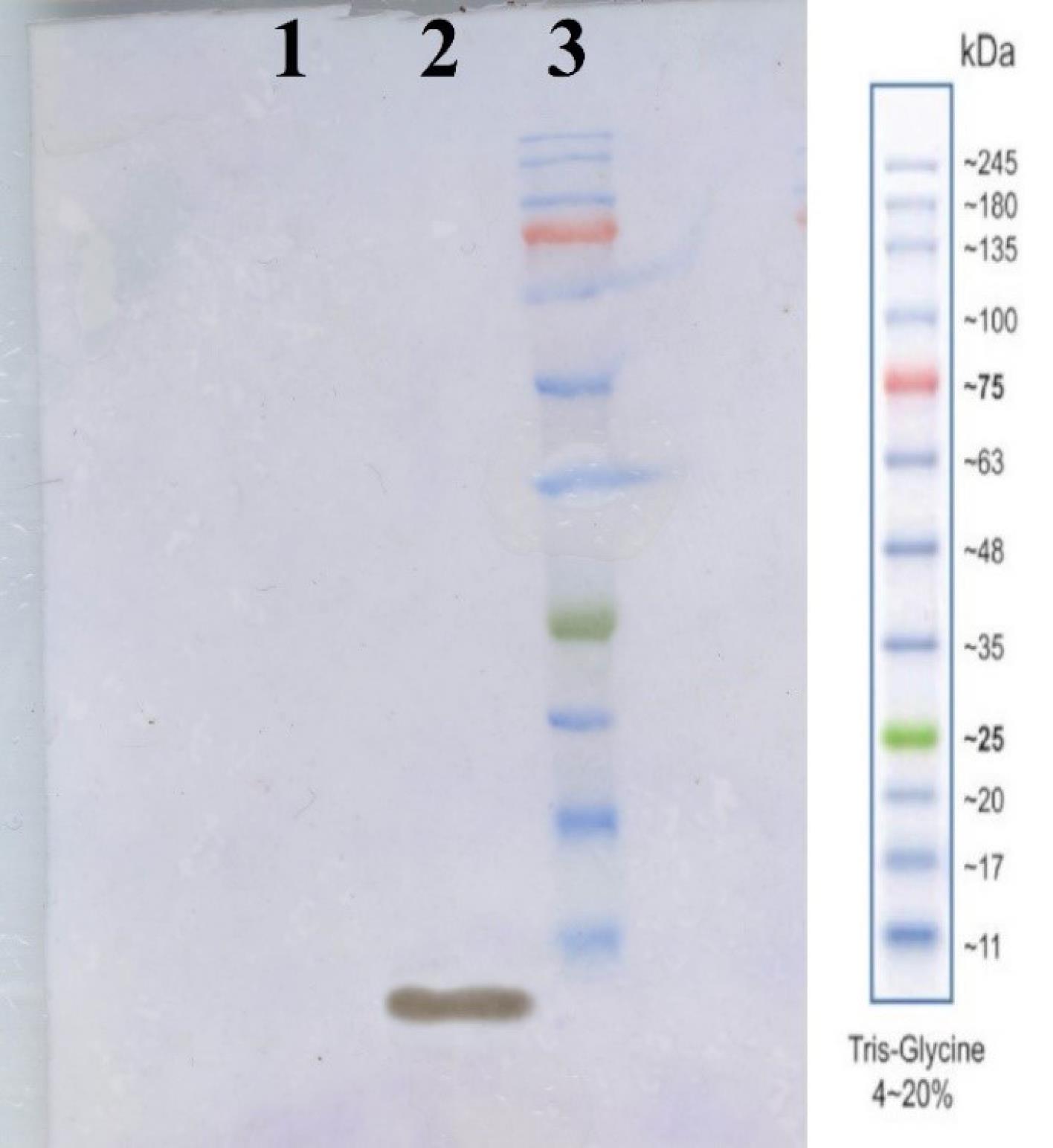
Fig. 5.
Western blot analysis for rRBM using anti-His antibody, lane1 BSA as a negative control, lane2 rRBM pure protein, lane3 protein ladder.
.
Western blot analysis for rRBM using anti-His antibody, lane1 BSA as a negative control, lane2 rRBM pure protein, lane3 protein ladder.
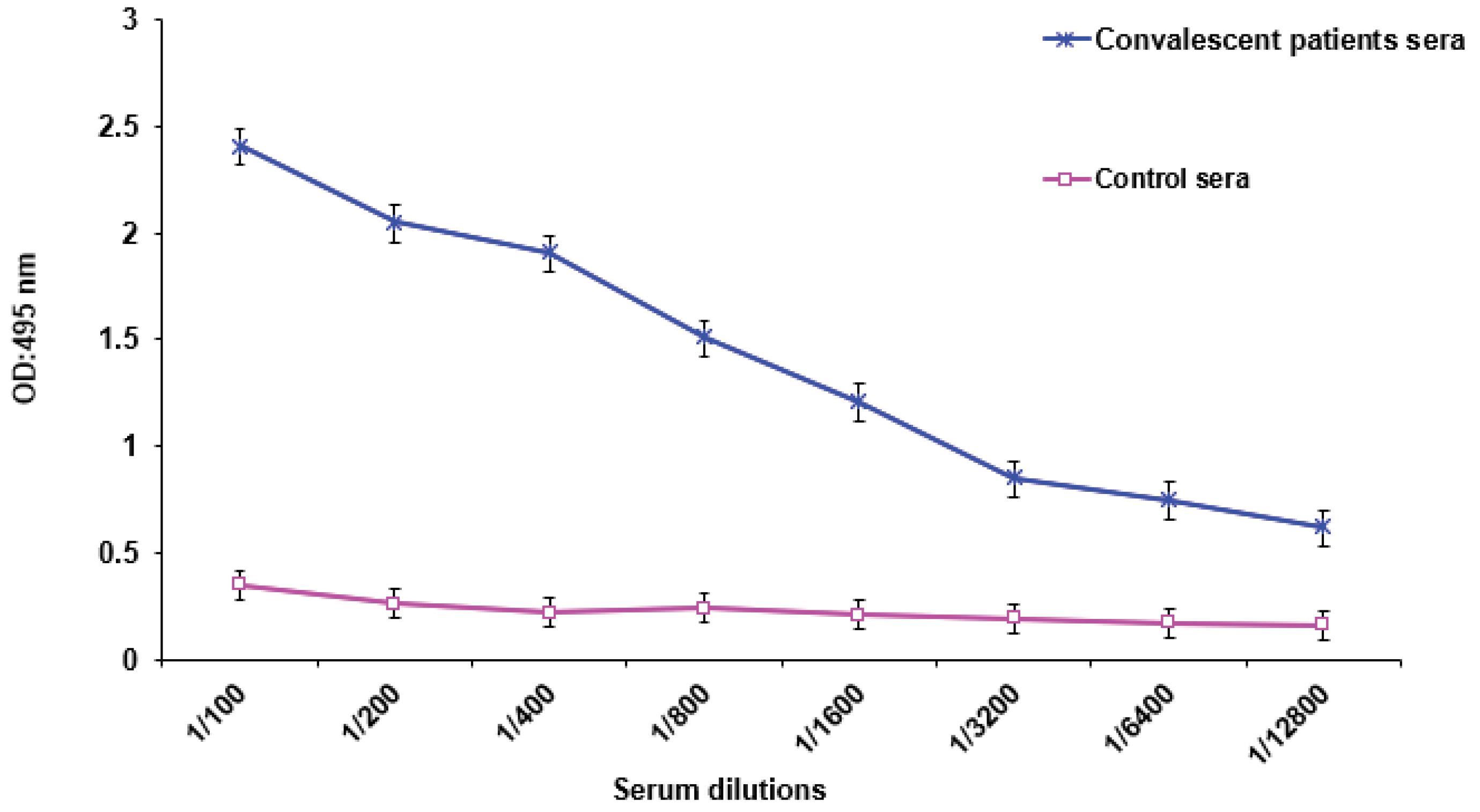
Fig. 6.
Serum Reactivity using convalescent sera obtained from known COVID-19 patients with rRBM protein antigen.
.
Serum Reactivity using convalescent sera obtained from known COVID-19 patients with rRBM protein antigen.
Humoral response to RBM and post-vaccination antibody response
To investigate the humoral immune responses elicited by recombinant RBM, we employed an ELISA. Sera obtained on day 14 after the initial vaccine dose, from both the adjuvanted and unadjuvanted groups, exhibited robust IgG responses against recombinant RBM. Pre-immunization sera and control animals treated with phosphate-buffered saline (PBS) displayed minimal antibody responses, indicating background levels. Notably, the administration of recombinant RBM alone successfully induced specific antibody production. However, the inclusion of Freund's adjuvant significantly enhanced antibody induction, resulting in higher levels of specific antibodies by day 14 and further elevated levels by day 28 (Fig. 7A and B). Sera collected on day 48, seven days after the third vaccination, demonstrated a significant antibody response in mice. The IgG serum titers were 1:12 800 and 1:3200 for the rRBM + Adj and rRBM groups, respectively (Fig. 7C).
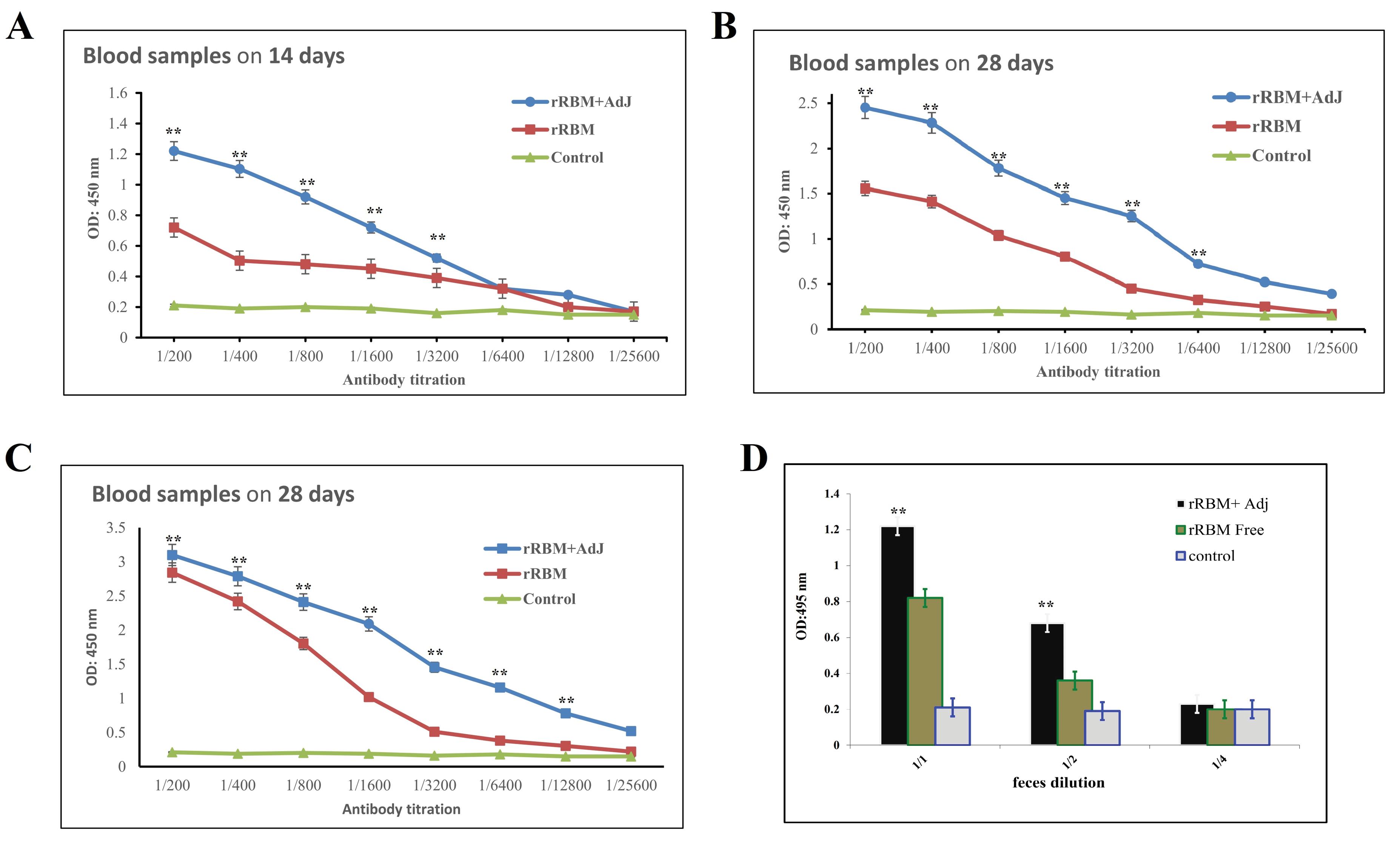
Fig. 7.
Immuno response induced by rRBM protein in mice. The IgG level in sera collected on days 14, 28, and 42 were evaluated using ELISA at the specified dilutions (A,B,C). IgA levels measured in feces (D). The P values indicate the comparisons between the RBM + Adj group and the PBS or RBM groups. Significance levels were denoted as **P < 0.01. In the case of IgG, a significant distinction was observed between the RBM + Adj group and the RBM alone group at dilutions up to 1:3200, 1:6400, and 1:12800 for days 14, 28, and 42, respectively. The P values for these comparisons were all less than 0.01. For IgA, a significant disparity was found between the RBM + Adj group and the RBM alone group at dilutions of 1:1 and 1:2, with P values of less than 0.01.
.
Immuno response induced by rRBM protein in mice. The IgG level in sera collected on days 14, 28, and 42 were evaluated using ELISA at the specified dilutions (A,B,C). IgA levels measured in feces (D). The P values indicate the comparisons between the RBM + Adj group and the PBS or RBM groups. Significance levels were denoted as **P < 0.01. In the case of IgG, a significant distinction was observed between the RBM + Adj group and the RBM alone group at dilutions up to 1:3200, 1:6400, and 1:12800 for days 14, 28, and 42, respectively. The P values for these comparisons were all less than 0.01. For IgA, a significant disparity was found between the RBM + Adj group and the RBM alone group at dilutions of 1:1 and 1:2, with P values of less than 0.01.
Furthermore, fecal samples were assessed for IgA responses, revealing substantial levels of anti-rRBM IgA. In contrast, the PBS group exhibited a minimal signal (Fig. 7D). These findings indicate that the administered vaccines successfully elicited appropriate humoral immune responses at both the systemic and mucosal levels.
Neutralizing antibody levels in response to rRBM vaccination
The neutralizing capacity of the antibodies induced by the RBM vaccination was evaluated Pishtaz Teb Zaman Neutralization Assay Kit. As shown in Table 2, the average levels of NAbs in the sera of vaccinated and non- vaccinated animals were measured after each dose time points. Mice that received three doses of recombinant rRBM vaccination with adjuvant exhibited a neutralizing antibody level of 31.6 µg/mL, while mice that were vaccinated with recombinant protein alone or treated with the control showed levels of 18.2 µg/mL and 2.2 µg/mL, respectively. These results demonstrated that the recombinant RBM vaccination, particularly when used in combination with the adjuvant, elicited neutralizing antibody response in mice.
Table 2.
Investigation of Nabs obtained from the sera of mice vaccinated with rRBM vaccine
|
|
Time frame for mice bleeding (day)
|
Antibody (µg/mL) in Immunized mice
|
Antibody (µg/mL) in normal animal control |
| rRBM + AdJ |
1st bleeding (14) |
10.1 ± 0.025 |
2.4 ± 0.004 |
| 2nd bleeding (28) |
23.8 ± 0.048 |
1.6 ± 0.022 |
| 3rd bleeding (42) |
31.6 ± 0.028 |
2.5 ± 0.005 |
| rRBM + PBS |
1st bleeding (14) |
4.5 ± 0.024 |
2.4 ± 0.006 |
| 2nd bleeding (28) |
9.7 ± 0.045 |
2.3 ± 0.009 |
| 3rd bleeding (42) |
18.2 ± 0.036 |
1.8 ± 0.018 |
| PBS |
1st bleeding (14) |
2.4 ± 0.018 |
2.4 ± 0.007 |
| 2nd bleeding (28) |
2.1 ± 0.035 |
2.2 ± 0.012 |
| 3rd bleeding (42) |
2.2 ± 0.026 |
2.5 ± 0.004 |
The results are presented as the mean of triplicate tests ± SEM. The cutoff value of the kit instrument is indicated as 2.5 µg/mL.
Discussion
The global pandemic caused by the SARS-CoV-2 virus, which is responsible for COVID-19 respiratory infection, has necessitated effective immunological interventions to control the spread of the disease and induce robust transmission-blocking immunity through vaccination. Initially, a range of vaccines, including inactivated viral vaccines, mRNA vaccines, and vector-based vaccines, were reported in the early stages of the outbreak.39 However, inactivated virus vaccines have limitations in terms of immunogenicity and the strength and duration of immune responses they induce.40 To address these concerns, most current vaccine investigations are focused on highly purified recombinant proteins or subunits of pathogens. Recombinant protein vaccines have the potential to overcome the drawbacks of inactivated virus vaccines and eliminate post-vaccination reactions.41 One drawback of protein vaccines is that the development of new formulations to protect against emerging variants of the coronavirus takes longer compared to mRNA vaccines, which can be more easily adapted. However, research indicates that mRNA vaccines may carry a heightened risk of adverse reactions, including myocarditis or hypersensitivity responses, especially with repeated administrations, in contrast to conventional protein vaccines.42,43 Repeated inoculations with mRNA vaccines have been associated with an increased incidence of adverse effects like myocarditis or hypersensitivity reactions, compared to traditional protein vaccines.44 Therefore, given the ongoing need for recurrent vaccination to address emerging coronavirus variants, protein-based vaccines may offer a safety advantage over mRNA counterparts. Moreover, several investigations have underscored the effectiveness of a heterologous prime-boost vaccination approach, which involves combining different vaccine types, over a homologous prime-boost strategy. Consequently, protein vaccines could serve as a viable option for booster doses subsequent to mRNA vaccination.45,46 The objective of this study was to develop a recombinant vaccine candidate targeting SARS-CoV-2, considering the aforementioned factors and the need for an effective and adaptable immunization approach.
The RBM is a specific region on the S protein of SARS-CoV-2, plays a crucial role in binding to the ACE2 receptor. Antibodies targeting the RBM can disrupt this interaction, effectively inhibiting viral entry into host cells.47 Consequently, the development of a vaccine capable of eliciting Nabs against RBM represents a promising approach in combating the pandemic.48 Recent studies have shown that neutralizing activity is primarily directed against the RBD in COVID-19 patients' sera and human monoclonal antibodies, further supporting the use of RBM as the immunogenic target in our vaccine strategy.49,50 Previous studies have demonstrated that recombinant proteins derived from the RBD and Nucleocapsid of SARS-CoV-2 can induce Nabs.21 Building upon these findings, the RBM of the spike protein was selected and designed to develop a recombinant protein in this research.
To enhance RBM protein expression, the gene's codons were adapted to match E. coli's codon bias. The resulting chimeric gene achieved a codon adaptation index of 0.96, significantly higher than the wild-type gene's score of 0.46 (Fig. 1). These synthetic genes facilitated robust expression in E. coli, yielding 27 mg of purified protein per liter of bacterial culture. This output matched or exceeded those reported in comparable studies,51,52 indicating that the high-level expression of these proteins was largely due to our designed synthetic genes and the pET expression system we used. During the expression process, we observed the RBM protein accumulating as inclusion bodies in the bacteria lysis precipitates. The purified RBM protein exhibited an apparent molecular mass of around 10 kDa, which was consistent with the calculated molecular mass based on the RBM amino acid sequence (approximately 10184 Da as indicated in Table 1). To achieve proper folding and functionality, the RBM antigen, initially in inclusion bodies, underwent purification and refolding steps. Solubilization in urea facilitated purification using Immobilized Metal Affinity Chromatography (IMAC) with Ni2+ ions. Gradual dialysis in urea buffers of decreasing concentration promoted protein refolding. Western blotting with anti-His tag antibodies confirmed the integrity of the recombinant RBM protein.
After three subcutaneous immunizations of mice with purified rRBM antigen (20 µg) along with Freund's adjuvant, significant serum anti-RBM IgG responses were observed (P < 0.01). Additionally, modest anti- RBM IgA responses were detected, indicating the potential of the rRBM antigen to induce neutralizing antibodies against SARS-CoV-2. It is noteworthy that subcutaneous administration of rRBM resulted in both systemic IgG responses and modest IgA responses in mucosal compartments.53 Immunization with adjuvants was found to elicit more effective antibody responses compared to immunization with the antigen alone.54 Importantly, the vaccine candidate induced serum IgA responses, which can neutralize the virus locally on mucosal surfaces without enhancing inflammation via Fc receptors.55 However, subcutaneous immunization resulted in lower mucosal IgA responses, and it remains to be determined whether rRBM immunization can induce mucosal IgA responses in humans. To enhance mucosal immune responses, it is essential to co-administer the target antigen with specific mucosal adjuvants. These adjuvants can help in effectively presenting the antigen to the immune cells present in mucosal tissues, thus stimulating a stronger local immune response. For instance, using saponin-based adjuvants for intranasal spray may increase the mucosal response.56 The inclusion of such mucosal adjuvants in the vaccination strategy could significantly improve the induction of IgA responses at mucosal surfaces, which are the primary entry points for many pathogens including SARS-CoV-2.
In our study, we found that the average concentration of neutralizing antibodies in the sera of immunized mice was 31.6 µg/mL. This result compares favorably to a study by Rezaie et al, who used RBD as a recombinant vaccine in mice and achieved a final concentration of 19.48 µg/mL neutralizing antibodies after the third administration21 Additionally, using the Pishtaz Teb Zaman neutralization kit, 102 patients with COVID-19 were found to have an average of 56.7 µg/mL neutralizing antibodies. Based on these findings, it appears that the concentration of neutralizing antibodies produced after administration with rRBM was appropriate. Considering the dynamic nature of the SARS-CoV-2 virus and the ongoing emergence of new variants, we emphasize the importance of advocating for additional studies. Further investigations are necessary to evaluate the efficacy of the RBM vaccine in neutralizing diverse SARS-CoV-2 variants.
Our study is among the early efforts report the expression of the RBM of the spike glycoprotein without fusion in a prokaryotic host, offering advantages in terms of cost-effectiveness and scalability for vaccine development. However, it has some limitations that need to be considered. Firstly, we did not measure the T-Cells (Th1) response in our study that could be crucial to mount the effective antiviral response against SARS CoV-2. The decision not to assess Th1 response was influenced by the initial focus on humoral responses to evaluate the neutralizing antibody potential of the RBM domain. Additionally, while BALB/c mice are commonly used for immunological studies, their response characteristics can sometimes add complexity to interpreting Th1 responses. Future studies will include a comprehensive assessment of both cellular and humoral immune responses to provide a more complete understanding of the immunogenicity of the RBM domain. Secondly, we applied Freund's adjuvant, which is not used for humans and animals in some countries. Further investigation with other adjuvants, for example, alum, is needed. Thirdly, we did not remove the His-tag from our final recombinant protein before immunization. This decision was based on the low molecular weight of the His-tag (~1.6 kDa). However, it is worth noting that Lin et al demonstrated that the addition of a His-tag to the recombinant vaccine may significantly impair protein immunogenicity against SARS-CoV-2.57 Despite these limitations, our study was successful in inducing receptor antibodies against SARS-CoV-2. However, it is important to note that these results cannot be extrapolated to predict transmission inhibition in vivo. Further studies are needed to evaluate the efficacy of rRBM as a potential vaccine candidate in humans.
Conclusions
In conclusion, this study highlights the importance of targeting the RBM domain of the spike protein in the design of effective vaccines against SARS-CoV-2. Our results provide evidence for the development of an immunogenic vaccine that induces antibodies targeting the RBM domain. Despite the limitations of our study, the induction of receptor antibodies against SARS-CoV-2 with our vaccine candidate is promising and warrants further investigation.
Research Highlights
-
Vaccines targeting the RBD of SARS-CoV-2 have been shown to elicit neutralizing antibodies.
-
The RBM on the RBD domain of SARS-CoV-2 is critical for virus entry into host cells.
-
Recombinant RBM protein (10 kDa) was successfully expressed as an inclusion body in a prokaryotic host.
-
A vaccine targeting the RBM of the S protein of SARS-CoV-2 effectively induces neutralizing antibodies against the RBD.
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethical Statement
This article does not include any studies involving human participants conducted by the authors. All relevant international, national, and institutional guidelines for the care and use of animals were strictly adhered to throughout the research. This study received approval from the Ethics Committee of Imam Hossein Comprehensive University (approval code: 1400.876).
Acknowledgements
We would like to thank the laboratory technicians at Imam Hossein University in Tehran, Iran, for their valuable contributions to the success of this project.
References
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579:270-3. doi: 10.1038/s41586-020-2012-7 [Crossref] [ Google Scholar]
- Rossi GA, Sacco O, Mancino E, Cristiani L, Midulla F. Differences and similarities between SARS-CoV and SARS-CoV-2: spike receptor-binding domain recognition and host cell infection with support of cellular serine proteases. Infection 2020; 48:665-9. doi: 10.1007/s15010-020-01486-5 [Crossref] [ Google Scholar]
- Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol 2020. 94: e00127-20. 10.1128/jvi.00127-20.
- Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med 2020; 26:450-2. doi: 10.1038/s41591-020-0820-9 [Crossref] [ Google Scholar]
- Lopez E, Barthélémy M, Baronti C, Masse S, Falchi A, Durbesson F. Endonuclease-based genotyping of the RBM as a method to track the emergence or evolution of SARS-CoV-2 variants. iScience 2021; 24:103329. doi: 10.1016/j.isci.2021.103329 [Crossref] [ Google Scholar]
- Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 2016; 531:381-5. doi: 10.1038/nature17180 [Crossref] [ Google Scholar]
- Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L. Dexamethasone in hospitalized patients with COVID-19. N Engl J Med 2021; 384:693-704. doi: 10.1056/NEJMoa2021436 [Crossref] [ Google Scholar]
- World Health Organization (WHO). WHO Recommends Against the Use of Remdesivir in COVID-19 Patients. WHO; 2023. Available from: https://www.who.int/news/item/10-11-2023-who-updates-guidelines-on-treatments-for-covid-19.
- Hammond J, Leister-Tebbe H, Gardner A, Abreu P, Bao W, Wisemandle W. Oral nirmatrelvir for high-risk, nonhospitalized adults with COVID-19. N Engl J Med 2022; 386:1397-408. doi: 10.1056/NEJMoa2118542 [Crossref] [ Google Scholar]
- Park HJ, Bang YJ, Kwon SP, Kwak W, Park SI, Roh G. Analyzing immune responses to varied mRNA and protein vaccine sequences. NPJ Vaccines 2023; 8:84. doi: 10.1038/s41541-023-00684-0 [Crossref] [ Google Scholar]
- Mateo-Urdiales A, Sacco C, Petrone D, Bella A, Riccardo F, Del Manso M. Estimated effectiveness of a primary cycle of protein recombinant vaccine NVX-CoV2373 against COVID-19. JAMA Netw Open 2023; 6:e2336854. doi: 10.1001/jamanetworkopen.2023.36854 [Crossref] [ Google Scholar]
- Kleanthous H, Silverman JM, Makar KW, Yoon IK, Jackson N, Vaughn DW. Scientific rationale for developing potent RBD-based vaccines targeting COVID-19. NPJ Vaccines 2021; 6:128. doi: 10.1038/s41541-021-00393-6 [Crossref] [ Google Scholar]
- Feng S, Phillips DJ, White T, Sayal H, Aley PK, Bibi S. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat Med 2021; 27:2032-40. doi: 10.1038/s41591-021-01540-1 [Crossref] [ Google Scholar]
- Premkumar L, Segovia-Chumbez B, Jadi R, Martinez DR, Raut R, Markmann A. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol 2020; 5:eabc8413. doi: 10.1126/sciimmunol.abc8413 [Crossref] [ Google Scholar]
- Yang S, Li Y, Dai L, Wang J, He P, Li C. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect Dis 2021; 21:1107-19. doi: 10.1016/s1473-3099(21)00127-4 [Crossref] [ Google Scholar]
- ai L, Gao L, Tao L, Hadinegoro SR, Erkin M, Ying Z. Efficacy and safety of the RBD-dimer-based COVID-19 vaccine ZF2001 in adults. N Engl J Med 2022; 386:2097-111. doi: 10.1056/NEJMoa2202261 [Crossref] [ Google Scholar]
- Boulton S, Poutou J, Martin NT, Azad T, Singaravelu R, Crupi MJ. Single-dose replicating poxvirus vector-based RBD vaccine drives robust humoral and T cell immune response against SARS-CoV-2 infection. Mol Ther 2022; 30:1885-96. doi: 10.1016/j.ymthe.2021.10.008 [Crossref] [ Google Scholar]
- Salimian J, Ahmadi A, Amani J, Olad G, Halabian R, Saffaei A, et al. Safety and immunogenicity of a recombinant receptor-binding domain-based protein subunit vaccine (Noora vaccineTM) against COVID-19 in adults: a randomized, double-blind, placebo-controlled, Phase 1 trial. J Med Virol 2023; 95. 10.1002/jmv.28097.
- Chen Y, Guo Y, Pan Y, Zhao ZJ. Structure analysis of the receptor binding of 2019-nCoV. Biochem Biophys Res Commun 2020; 525:135-40. doi: 10.1016/j.bbrc.2020.02.071 [Crossref] [ Google Scholar]
- Gong Y, Qin S, Dai L, Tian Z. The glycosylation in SARS-CoV-2 and its receptor ACE2. Signal Transduct Target Ther 2021; 6:396. doi: 10.1038/s41392-021-00809-8 [Crossref] [ Google Scholar]
- Rezaei A, Nazarian S, Samiei Abianeh H, Kordbacheh E, Alizadeh Z, Mousavi Gargari SL. Antibodies produced toward recombinant RBD and nucleocapsid neutralize SARS-COV-2. Avicenna J Med Biotechnol 2022; 14:270-7. doi: 10.18502/ajmb.v14i4.10481 [Crossref] [ Google Scholar]
- Nazarian S, Olad G, Abdolhamidi R, Motamedi MJ, Kazemi R, Kordbacheh E. Preclinical study of formulated recombinant nucleocapsid protein, the receptor binding domain of the spike protein, and truncated spike (S1) protein as vaccine candidates against COVID-19 in animal models. Mol Immunol 2022; 149:107-18. doi: 10.1016/j.molimm.2022.06.007 [Crossref] [ Google Scholar]
- Vincze T, Posfai J, Roberts RJ. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res 2003; 31:3688-91. doi: 10.1093/nar/gkg526 [Crossref] [ Google Scholar]
- Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 1999; 112:531-52. doi: 10.1385/1-59259-584-7:531 [Crossref] [ Google Scholar]
- Zhou X, Zheng W, Li Y, Pearce R, Zhang C, Bell EW. I-TASSER-MTD: a deep-learning-based platform for multi-domain protein structure and function prediction. Nat Protoc 2022; 17:2326-53. doi: 10.1038/s41596-022-00728-0 [Crossref] [ Google Scholar]
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 2007; 35:W407-10. doi: 10.1093/nar/gkm290 [Crossref] [ Google Scholar]
- Wlodawer A. Stereochemistry and validation of macromolecular structures. Methods Mol Biol 2017; 1607:595-610. doi: 10.1007/978-1-4939-7000-1_24 [Crossref] [ Google Scholar]
- Froger A, Hall JE. Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp 2007: e253. 10.3791/253.
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual (3 Volume Set). Cold Spring Harbor Laboratory Press; 2001.
- Kielkopf CL, Bauer W, Urbatsch IL. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of proteins. Cold Spring Harb Protoc 2021; 2021:pdb-rot102228. doi: 10.1101/pdb.prot102228 [Crossref] [ Google Scholar]
- Gallagher SR. One-dimensional SDS gel electrophoresis of proteins. Curr Protoc Immunol 2006; Chapter 8: Unit 8.4. 10.1002/0471142735.im0804s75.
- Spriestersbach A, Kubicek J, Schäfer F, Block H, Maertens B. Purification of his-tagged proteins. Methods Enzymol 2015; 559:1-15. doi: 10.1016/bs.mie.2014.11.003 [Crossref] [ Google Scholar]
- Singh SM, Panda AK. Solubilization and refolding of bacterial inclusion body proteins. J Biosci Bioeng 2005; 99:303-10. doi: 10.1263/jbb.99.303 [Crossref] [ Google Scholar]
- Sule R, Rivera G, Gomes AV. Western blotting (immunoblotting): history, theory, uses, protocol and problems. Biotechniques 2023; 75:99-114. doi: 10.2144/btn-2022-0034 [Crossref] [ Google Scholar]
- Tabatabaei MS, Ahmed M. Enzyme-linked immunosorbent assay (ELISA). Methods Mol Biol 2022; 2508:115-34. doi: 10.1007/978-1-0716-2376-3_10 [Crossref] [ Google Scholar]
- National Research Council Committee for the Update of the Guide for the C, Use of Laboratory A. The National Academies Collection: Reports funded by National Institutes of Health. Guide for the Care and Use of Laboratory Animals. Washington (DC): National Academies Press (US); 2011.
- Greenfield EA. Standard immunization of mice, rats, and hamsters. Cold Spring Harb Protoc 2020; 2020:100297. doi: 10.1101/pdb.prot100297 [Crossref] [ Google Scholar]
- Gai W, Zou W, Lei L, Luo J, Tu H, Zhang Y. Effects of different immunization protocols and adjuvant on antibody responses to inactivated SARS-CoV vaccine. Viral Immunol 2008; 21:27-37. doi: 10.1089/vim.2007.0079 [Crossref] [ Google Scholar]
- Gao Q, Bao L, Mao H, Wang L, Xu K, Yang M. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020; 369:77-81. doi: 10.1126/science.abc1932 [Crossref] [ Google Scholar]
- Sanders B, Koldijk M, Schuitemaker H. Inactivated viral vaccines. In: Nunnally BK, Turula VE, Sitrin RD, eds. Vaccine Analysis: Strategies, Principles, and Control. Berlin, Heidelberg: Springer. 2015. p. 45-80. 10.1007/978-3-662-45024-6_2.
- Nascimento IP, Leite LC. Recombinant vaccines and the development of new vaccine strategies. Braz J Med Biol Res 2012; 45:1102-11. doi: 10.1590/s0100-879x2012007500142 [Crossref] [ Google Scholar]
- Oster ME, Shay DK, Su JR, Gee J, Creech CB, Broder KR. Myocarditis cases reported after mRNA-based COVID-19 vaccination in the US from December 2020 to August 2021. JAMA 2022; 327:331-40. doi: 10.1001/jama.2021.24110 [Crossref] [ Google Scholar]
- García-Grimshaw M, Ceballos-Liceaga SE, Hernández-Vanegas LE, Núñez I, Hernández-Valdivia N, Carrillo-García DA. Neurologic adverse events among 704,003 first-dose recipients of the BNT162b2 mRNA COVID-19 vaccine in Mexico: a nationwide descriptive study. Clin Immunol 2021; 229:108786. doi: 10.1016/j.clim.2021.108786 [Crossref] [ Google Scholar]
- Kouhpayeh H, Ansari H. Adverse events following COVID-19 vaccination: a systematic review and meta-analysis. Int Immunopharmacol 2022; 109:108906. doi: 10.1016/j.intimp.2022.108906 [Crossref] [ Google Scholar]
- Rammauro F, Carrión F, Olivero-Deibe N, Fló M, Ferreira A, Pritsch O. Humoral immune response characterization of heterologous prime-boost vaccination with CoronaVac and BNT162b2. Vaccine 2022; 40:5189-96. doi: 10.1016/j.vaccine.2022.07.023 [Crossref] [ Google Scholar]
- Groß R, Zanoni M, Seidel A, Conzelmann C, Gilg A, Krnavek D. Heterologous ChAdOx1 nCoV-19 and BNT162b2 prime-boost vaccination elicits potent neutralizing antibody responses and T cell reactivity against prevalent SARS-CoV-2 variants. EBioMedicine 2022; 75:103761. doi: 10.1016/j.ebiom.2021.103761 [Crossref] [ Google Scholar]
- Grant OC, Montgomery D, Ito K, Woods RJ. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci Rep 2020; 10:14991. doi: 10.1038/s41598-020-71748-7 [Crossref] [ Google Scholar]
- Zhang J, Zeng H, Gu J, Li H, Zheng L, Zou Q. Progress and prospects on vaccine development against SARS-CoV-2. Vaccines (Basel) 2020; 8:153. doi: 10.3390/vaccines8020153 [Crossref] [ Google Scholar]
- Dai L, Zheng T, Xu K, Han Y, Xu L, Huang E, et al. A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell 2020; 182: 722-33.e11. 10.1016/j.cell.2020.06.035.
- Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020; 584:437-42. doi: 10.1038/s41586-020-2456-9 [Crossref] [ Google Scholar]
- He Y, Qi J, Xiao L, Shen L, Yu W, Hu T. Purification and characterization of the receptor-binding domain of SARS-CoV-2 spike protein from Escherichia coli. Eng Life Sci 2021; 21:453-60. doi: 10.1002/elsc.202000106 [Crossref] [ Google Scholar]
- Rahbar Z, Nazarian S, Dorostkar R, Sotoudehnejad Nematalahi F, Amani J. Recombinant expression of SARS-CoV-2 receptor binding domain (RBD) in Escherichia coli and its immunogenicity in mice. Iran J Basic Med Sci 2022; 25:1110-6. doi: 10.22038/ijbms.2022.65045.14333 [Crossref] [ Google Scholar]
- Su F, Patel GB, Hu S, Chen W. Induction of mucosal immunity through systemic immunization: phantom or reality?. Hum Vaccin Immunother 2016; 12:1070-9. doi: 10.1080/21645515.2015.1114195 [Crossref] [ Google Scholar]
- Coffman RL, Sher A, Seder RA. Vaccine adjuvants: putting innate immunity to work. Immunity 2010; 33:492-503. doi: 10.1016/j.immuni.2010.10.002 [Crossref] [ Google Scholar]
- Woof JM, Kerr MA. The function of immunoglobulin A in immunity. J Pathol 2006; 208:270-82. doi: 10.1002/path.1877 [Crossref] [ Google Scholar]
- Chen K, Wang N, Zhang X, Wang M, Liu Y, Shi Y. Potentials of saponins-based adjuvants for nasal vaccines. Front Immunol 2023; 14:1153042. doi: 10.3389/fimmu.2023.1153042 [Crossref] [ Google Scholar]
- Lin TW, Huang PH, Liao BH, Chao TL, Tsai YM, Chang SC. Tag-free SARS-CoV-2 receptor binding domain (RBD), but not C-terminal tagged SARS-CoV-2 RBD, induces a rapid and potent neutralizing antibody response. Vaccines (Basel) 2022; 10:1839. doi: 10.3390/vaccines10111839 [Crossref] [ Google Scholar]