Bioimpacts. 2025;15:30568.
doi: 10.34172/bi.30568
Original Article
Uncovering the anticancer effects of Bhallataka Taila in lung cancer: A metabolomic and network pharmacology approach
G. P Suchitha Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, 1 
Shubham S. Upadhyay Formal analysis, Methodology, Visualization, 1
Ravishankar Pervaje Conceptualization, Resources, 2
T. S Keshava Prasad Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing, 1, * 
Shobha Dagamajalu Conceptualization, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing, 1, * 
Author information:
1Center for Systems Biology and Molecular Medicine, Yenepoya Research Centre, Yenepoya (Deemed to be University), Mangalore 575018, India
2Sushrutha Ayurveda Hospital, Puttur 574201, Karnataka, India
Abstract
Introduction:
Bhallataka (Semecarpus anacardium Linn.) is used in traditional medicine to treat various ailments. The nut extract of Bhallataka, known as Bhallataka taila, has anticancer properties. Although several studies have explored to verify and evaluate its anticancer properties and efficacy against various cancers, the specific target proteins, mode of action, and associated metabolites have not yet been identified. This study aimed to elucidate the biological mechanisms of Bhallataka taila using an integrated metabolomics and systems pharmacology approach with in vitro validation.
Methods:
Untargeted metabolomics using liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed to evaluate the metabolites in Bhallataka taila, identify key protein targets and link them to cellular pathways through bioinformatics-based network pharmacology. Protein targets were mapped using BindingDB, and pathway enrichment was analyzed using STRINGdb. An in vitro study of A549 cells assessed the impact of Bhallataka taila on cellular viability (MTT assay), apoptosis (AO-EB staining), reactive oxygen species (ROS) production (fluorescent spectroscopy and DCFDA staining), and marker validation (immunoblotting and qRT-PCR). The integration of metabolomics, network pharmacology, and in vitro experiments offers a significant understanding of the anticancer mechanisms and pathways influenced by Bhallataka taila in non-small cell lung cancer (NSCLC) cells. Statistical analysis was performed using GraphPad Prism using one-way ANOVA.
Results:
Metabolomics combined with network pharmacology detected 2023 unique metabolites at the MS1 level and 216 metabolites at the MS2 level. Bhallataka taila metabolites were found to interact with 180 human target proteins identified through BindingDB analysis. These target proteins were mapped to key cancer regulatory signaling pathways, along with TNF-related apoptosis-inducing ligand (TRAIL), protease-activated receptor-1 (PAR1)-mediated thrombin signaling, Syndecan-1 and Glypican pathways, and vascular endothelial growth factor receptor (VEGFR)1/2 pathways. In vitro validation demonstrated that Bhallataka taila significantly regulated apoptosis (57%) and ROS production (56%) in A549 cells compared to control while modulating other cancer-related regulatory pathways.
Conclusion:
This data-driven study can help researchers identify promising cancer treatment candidates and validate their efficacy. This approach integrates traditional knowledge with modern scientific techniques to reinforce the anticancer potential of Bhallataka taila and its mechanisms.
Keywords: Bhallataka taila, Metabolites, Protein targets, Pathways, Cancer, Traditional medicine
Copyright and License Information
© 2025 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
This study was supported by the Department of Biotechnology, Government of India, under the "Skill Development in Mass Spectrometry-Based Metabolomics Technology BIC" (BT/PR40202/BTIS/137/53/2023).
Introduction
The Indian traditional medicine systems, including Ayurveda and Siddha, have long used plant-derived products to manage various diseases. Scientific evidence supporting the efficacy of Ayurvedic medicines is steadily expanding, with many plants used in Ayurveda being validated for their anticancer properties.1 Semecarpus anacardium Linn. (Bhallataka) is one such plant, and its nut extract, known as Bhallataka taila, plays a key role in both Ayurveda and Siddha for treating various ailments, including cancer. Known for its cytotoxic, anti-inflammatory, and antioxidant properties,2 Bhallataka taila has emerged as a potent anticancer agent. Studies have shown that flavonoids extracted from Bhallataka taila exhibit cytoprotective effects, protecting normal lung cells against hydrogen peroxide-induced toxicity, reactive oxygen species (ROS) generation, membrane damage, and DNA damage.3 The nut extract contains several bioactive phytoconstituents, including flavonoids, phenolics, bhilawanols, sterols, anacardic acid, and glycosides.4
Plant omics is an evolving field in systems biology that provides a comprehensive strategy for unraveling intricate biological frameworks, including metabolic pathways, regulatory networks, and interactions among genes, proteins, and metabolites.4
Metabolites are biochemical compounds produced through intrinsic metabolic pathways that play a crucial role in delivering protective benefits. In addition to being mere byproducts of metabolism, metabolites also bind to proteins, influencing biological processes. Numerous plant-derived compounds, such as paclitaxel, curcumin, strigolactones, geniposide, etoposide, and camptothecin, have already been approved as drugs for cancer treatment by modulating protein regulation. The growing interest in small molecules as targets of proteins is fueling the research into their potential for cancer management.5-7
LC-MS and Gas chromatography-mass spectrometry (GC-MS) are effective techniques for metabolomics research. These methods have greatly advanced our understanding of metabolite composition and have uncovered many previously unknown metabolites.8 The integration of MS-driven metabolomics, network analysis, and target protein profiling offers a more profound understanding of the therapeutic impact of Bhallataka taila in lung cancer treatment, providing a pathway for its clinical validation and optimization.
In this study, we employed combined metabolomics and systems pharmacology methods to uncover the biochemical mechanisms of Bhallataka taila in a metabolic framework with in vitro validation. This study lays the foundation for further clinical research, not only in cancer therapeutics, but also in exploring the broader potential of traditional treatments. This integrative approach can reveal new perspectives on the therapeutic benefits of Bhallataka taila and other conventional treatments.
Reagents and Methods
Material procurement
This study was approved by the Scientific Review Board (YRC-SRB/126/2021). Bhallataka taila (Lot number 85) was acquired from SDP Remedies and Research Centre, Puttur, a GMP-accredited company. The drug was prepared and maintained at the center and the product was stored under the code SDP/BT/005-2017.
The preparation method involved boiling 200 g Bhallataka nuts in 500 mL of cow milk, which was repeated three times. The nut-milk mixture was then subjected to shodhana detoxification by combining it with 1.5 kg of ghee and boiling at 100 °C until the drug was completely dehydrated. Shodhana is a traditional method used to neutralize potentially harmful or semi-poisonous compounds, enhancing the drug's safety and efficacy. In this case, it was specifically used to eliminate urushiol, a toxic compound found in raw Bhallataka nuts. The final product was filtered and preserved.
Solvents, chemicals, and antibodies
Mass spectrometry analysis and extraction were performed using LC-MS-grade solvents; chromatographic-methanol, formic acid, acetonitrile, and chloroform were sourced from Merck. Dulbecco's modified Eagle medium (DMEM), antibiotic/antimycotic, and fetal bovine serum (FBS) were obtained from Gibco (Thermo Fisher Scientific). Cisplatin was purchased from Celon Labs. The MTT assay reagent was obtained from Himedia, whereas Hoechst-33342, acridine orange (AO), ethidium bromide (EB), and 2′,7′-dichlorofluorescein diacetate (DCFDA) were obtained from Sigma-Aldrich. The bicinchoninic acid (BCA) estimation kit was obtained from Thermo Fisher Scientific. Antibodies against total and phospho-extracellular signal-regulated kinase (pERK) (T202/Y204), beta-actin, hypoxia-inducible factor 1 (HIF-1) α, and p53 were obtained from Cell Signaling Technology. Ki-67 was obtained from Thermo Fisher Scientific (Invitrogen). Clarity ECL and nitrocellulose membrane substrates were obtained from BioRad (USA), and X-ray films were purchased from Carestream (USA).
Metabolite extraction
Metabolite extraction was performed as described by Karthikkeyan et al.4 Briefly, 25 µL of Bhallataka taila was mixed with 1 mL of triple solvent (acetonitrile, methanol, and water) in a 2:2:1 ratio. The solution was vortexed using a Spinix vortex shaker (Tarsons) at room temperature for 5 min, sonicated in an ultrasonic water bath (PCI Analytics) for 10 min at room temperature, and spun down for 15 min at 12 000 g. The upper phase was gently decanted into a fresh vial and dried under vacuum using SpeedVac (Savant SDP1010, Thermo Scientific) without temperature control. The dried supernatants were kept at -20 °C for LC-MS/MS approach.
Untargeted metabolomics profiling using LC-MS/MS
Untargeted metabolomics data acquisition was performed using a QTRAP-6500 mass spectrometer (AB SCIEX, USA) operated with Analyst software (v 1.6.3). The samples were separated on an Agilent Infinity II 1290 chromatography (Agilent Technologies, USA) with a reverse-phase ZORBAX Eclipse Plus C18 RRHD column (2.1 × 150 mm, 1.8 μm). Solvent-A consisted of 0.1% formic acid, while Solvent-B consisted of 0.1% formic acid in 90% acetonitrile, with gradients program. A 10 µL sample was injected and separated at a flow rate of 0.3 mL/min.
Information-dependent acquisition was used, employing enhanced mass spectra and enhanced product ion scans to track the top five most intense ions in the 50–1000 Da range, scanned at 10 000 Da/s. Electrospray ionization (ESI) was applied with a probe temperature of 450 °C, voltage of 4500 V (or -4,500 V), curtain gas pressure of 30 psi, ion source gas pressure of 30 psi, and declustering potential of 100 V (or -100 V). Collision-induced dissociation (CID) was used to fragment precursor ions, with a collision energy of 40 V (or -40 V), a spread of ± 25 V, and entrance potential of 10 V (or -10 V). This broad range ensures the optimal fragmentation of diverse metabolites and detailed structural data capture. Metabolomic data for Bhallataka taila were obtained in both positive and negative ionization modes, each in triplicate, to improve detection sensitivity and capture a broader spectrum of metabolites.8
Data processing
Raw data files from the QTRAP-6500 were reformatted.mzML format through MSConvertGUI and ProteoWizard.9 Data processing was performed using the open-source MZmine 2.53 platform.10 Initially, baseline correction was applied, with the smoothing set to 500 units and an asymmetric factor of 0.001. Mass identification was performed in centroid mode with a noise level cut-off of 1.0E5. Chromatogram restoration required a minimum period retention time of 0.2 min and a peak height threshold of 1.0E4, with a m/z tolerance of 0.05 Da. The isotopic peak grouper algorithm was used to identify local minima and deisotope peaks.
Data alignment was performed using the Random Sample Consensus (RANSAC) algorithm, followed by gap filling using the peak finder algorithm. Metabolite detection at the MS1 level was conducted using the MS2Compound tool,11 with a m/z error tolerance of 0.05 Da. The considered adducts included [M + H], [M + 2H], [2M + H], [M-H], [M-2H], and [2M-H] for both positive and negative ionization modes. The identified metabolites were further analyzed by MS/MS using the MS2Compound tool, which searched the relative intensity results files in.mgf format from MZmine 2.53. The search parameters were 0.05 Da for precursors and 0.5 Da for fragment ions.
To minimize false discoveries, data processing involved stringent quality controls using precise m/z tolerance levels, the RANSAC algorithm for alignment, and filtering of rank 1 metabolites. These steps ensured accurate metabolite identification and minimized false positives.
Bioinformatic assessment
Metabolites identified from the PlantCyc and KEGG databases were employed to assess pathway enrichment, and their chemical classification was performed using MetaboAnalyst 5.0.12 The SMILES IDs for the metabolites were retrieved via the PubChem Identifier Exchange Service. The protein targets of these metabolites were assessed using BindingDB (2024), a database of experimentally validated protein-small molecule interactions.13
Gene ID conversion was performed using the DAVID platform (version 2024.2.0), transforming UniProt IDs of protein targets into gene identifiers, which were then categorized by source organism.14 Gene ontology classification, pathway enrichment, and protein interaction mapping were performed using gprofiler (version 0.2.3), FunRich (version 3.1.4), and STRINGdb (version 12.0), with a p-value cut-off of ≤ 0.05 for statistical relevance.15-17 To enhance the reliability of the results, a 1% false discovery rate (FDR) was applied. Pathway enrichment analysis based on KEGG pathway annotations (version 110.1) revealed several cancer-related signaling pathways for genes linked to Homo sapiens.18
Cell culture
A549 cells were procured from the National Center for Cell Science, Pune, India. The cells were maintained in DMEM containing 10% FBS and 1X antibiotic/antimycotic solution and incubated at 37°C with 5% CO2 in a humidified incubator.
Evaluation of cytotoxicity by MTT assay
A549 cells were grown in 96-well plates at a density of 5000 cells/well. Cell viability was assessed after 48 h of treatment with varying concentrations of Bhallataka taila (0.78–7800 µg/mL) and the positive control cisplatin (2–10 µg/mL) using the MTT assay. Following treatment, the cells were exposed to MTT dye for 3-4h, and the formed formazan crystals were solubilized in a 1:1 mixture of ethanol and dimethyl sulfoxide. The absorbance was measured at 570 nm, and the viability of cells was quantified as a percentage compared with the control. To minimize variability, treatments were standardized by ensuring uniform cell density and growth conditions. All experiments were performed in triplicate, with positive and negative controls were included to further reduce experimental variability.
Detection of ROS using fluorescence spectroscopy and DCFDA staining
Fluorescence spectroscopy was used to assess the ability of Bhallataka taila and cisplatin to induce ROS formation in A549 cells. Cells were cultured at a population of 1 × 105 per well in a six-well plate and administered them with the IC50 concentrations of Bhallataka taila and cisplatin. Upon 48 h of treatment, one set of cells was trypsinized and centrifuged. The pellet obtained was reconstituted in cell lysis buffer and incubated for 30 min, with disruption occurring every 5 min. After incubation, the cells were spun down at 1200 rpm for 20 min at 4ºC and the upper phase was collected. Next, 25µM DCFDA stain was added to the supernatant and maintained for 30 min at 37 ºC in the dark. DCFDA is converted into the fluorescent compound DCF in the existence of ROS. The ROS intensity was measured using fluorescence spectroscopy at 485/600 nm.19 In a separate group of cells, a combination of 5 μg/mL Hoechst and 25µM DCFDA was used for 15 min of staining in the dark, and the images were acquired using a fluorescence imager (ZOE, BioRad).
Apoptosis assessment by AO-EB staining
Apoptosis in A549 cells after Bhallataka taila and cisplatin treatment was assessed using AO-EB assay.Cells were maintained in a six-well plate at a population of 1 × 105 per well and administered with IC50 values of Bhallataka taila and cisplatin. After 48 h of treatment, the cells were stained with a 2 µg/mL solution of AO: EB (1:1) for 15 minutes at 37ºC, shielded from light.20 Following staining, cells were gently rinsed with 1X phosphate-buffered saline (PBS) to clear excess dye. Apoptotic cells were then imaged using a fluorescence imager (ZOE, BioRad) under both green and red fluorescence fields.
Immunoblotting
Proteins were selected based on their involvement in key cancer-related pathways identified through BindingDB analysis, pathway enrichment, ROS, and apoptosis experiments, ensuring their relevance to the study objectives. A549 cells were cultured at a population of 1 × 105 cells per well and exposed to IC50 doses of Bhallataka taila and cisplatin for 48 h. After treatment, cells were trypsinized, rinsed with 1X PBS, and proteins were extracted using 2% cell lysis buffer. The cell extract was sonicated, incubated at 95 °C for 10 min, and then centrifuged. The protein concentration was assessed using a BCA protein quantification kit.
The protein expression levels of total and phospho-ERK1/2 (T202/Y204), HIF-1α, Ki-67, p53, and beta-actin were assessed by immunoblotting. Approximately 10 μg of protein per condition was loaded into each well and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Following electrophoresis, proteins were transferred to a nitrocellulose membrane. The membranes were blocked for 1 h at room temperature with 5% skim milk powder and then subjected to primary antibodies (1:1000 dilution) overnight at 4°C. Anti-rabbit IgG was employed as the secondary antibody and incubated at room temperature for 1 h. Protein bands were detected using an ECL substrate and imaged with X-ray films. Densitometry analysis was conducted using ImageJ software.4
Quantitative real time-polymerase chain reaction (qRT -PCR)
Genes were selected based on their regulatory roles in cancer pathways identified through pathway enrichment analysis, ROS, and apoptosis experiments to complement the protein expression findings. A549 cells were cultured at a population of 1 × 105 cells/well and exposed to IC50 doses of Bhallataka taila and cisplatin for 48 h. Upon treatment, cells were extracted and RNA was isolated using the Trizol method. The RNA quantity was measured using a microvolume spectrometer (Colibri, Germany) and then processed to cDNA using a cDNA synthesis kit (Takara). The levels of genes comprising Bcl-2-associated X protein (BAX), B cell lymphoma-2 (Bcl2), HIF-1α, p53, matrix metalloproteinases (MMP)-2, and MMP-9 were measured using qRT-PCR (BioRad).20
Statistical analysis
Cell culture assays, AO-EB staining, ROS detection, immunoblotting, and qRT-PCR were performed in biological triplicate, with results depicted as the standard error of the mean. Data from the MTT assay, AO-EB staining, and immunoblotting were statistically assessed using one-way ANOVA with GraphPad Prism. For both bioinformatics and in vitro experiments, a p-value ≤ 0.05 was considered statistically significant.
Results
Detection of Bhallataka taila metabolites
An untargeted metabolomics approach was used to investigate and compare molecular signals using the PlantCyc, KEGG, and Human Metabolome Database (HMDB). The total ion chromatograms of Bhallataka taila in both ionization modes are shown in Supplementary file 1. Metabolite detection at the precursor ion level revealed 2023 non-redundant metabolites at MS1, with 1814 and 217 identified in positive and negative modes, respectively. The annotated metabolite profiles, including their intensities and polarities, are provided in Supplementary files 2 and 3. At the MS/MS level, 216 metabolites were identified from both polarities using the MS2Compound tool.8 A list of these metabolites, including their names and corresponding MS/MS level data, is provided in Supplementary file 4. Despite the use of multiple databases, a significant portion of the data from Bhallataka taila remained unassigned (Supplementary files 5 and 6).
Identification of previously identified metabolites in Bhallataka taila
We searched for previously identified Bhallataka taila metabolites in the literature at the precursor and fragment levels and found very few available studies. However, we successfully mapped the dodecanoic acid at the fragment level, as identified by Shebriya et al in their GC-MS analysis of Bhallataka nut extract.21 We also detected undecane at the precursor level, which was previously identified by Ruhila et al (Table 1).22 The spectra of the identified metabolites are shown in Fig. 1A–B. The detection of the previously described metabolites helped validate our data.
Table 1.
List of previously reportedmetabolites representing as signature metabolites for Bhallataka taila
|
Metabolite name
|
Molecular formula
|
Monoisotopic mass
|
MS1/MS2
|
| Dodecanoic acid |
C12H24O2 |
200.17 |
MS2 |
| Undecane |
C11H24 |
156.18 |
MS1 |
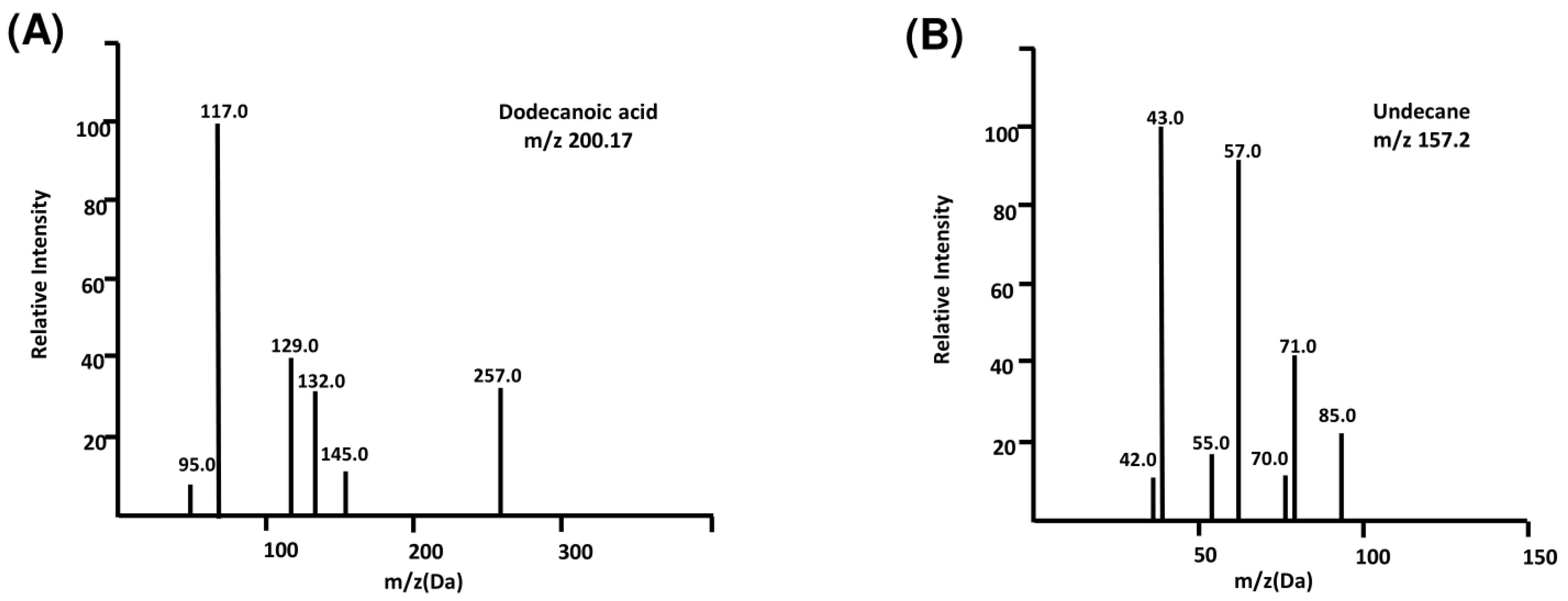
Fig. 1.
MS detection of previously reported metabolites. (A) Identification of dodecanoic acid metabolite in Bhallataka taila with 2M + H adduct (B) Identification of undecane metabolite in Bhallataka taila with M + H adduct.
.
MS detection of previously reported metabolites. (A) Identification of dodecanoic acid metabolite in Bhallataka taila with 2M + H adduct (B) Identification of undecane metabolite in Bhallataka taila with M + H adduct.
Chemical classification and pathway enrichment
The metabolites detected in Bhallataka taila were enriched in specific chemical classes (Fig. 2A). These included pheniramines, hydroxyindoles, alkanes, hydrocarbons, eicosanoids, and thiazoles (Supplementary file 7). Pathway analysis identified these metabolites as predominantly associated with key metabolic pathways, such as arachidonic acid metabolism, tyrosine/pyrimidine/purine metabolism, and cytochrome P450-mediated metabolism of xenobiotics, which contribute significantly to cancer regulation (Fig. 2B) (Supplementary file 8).
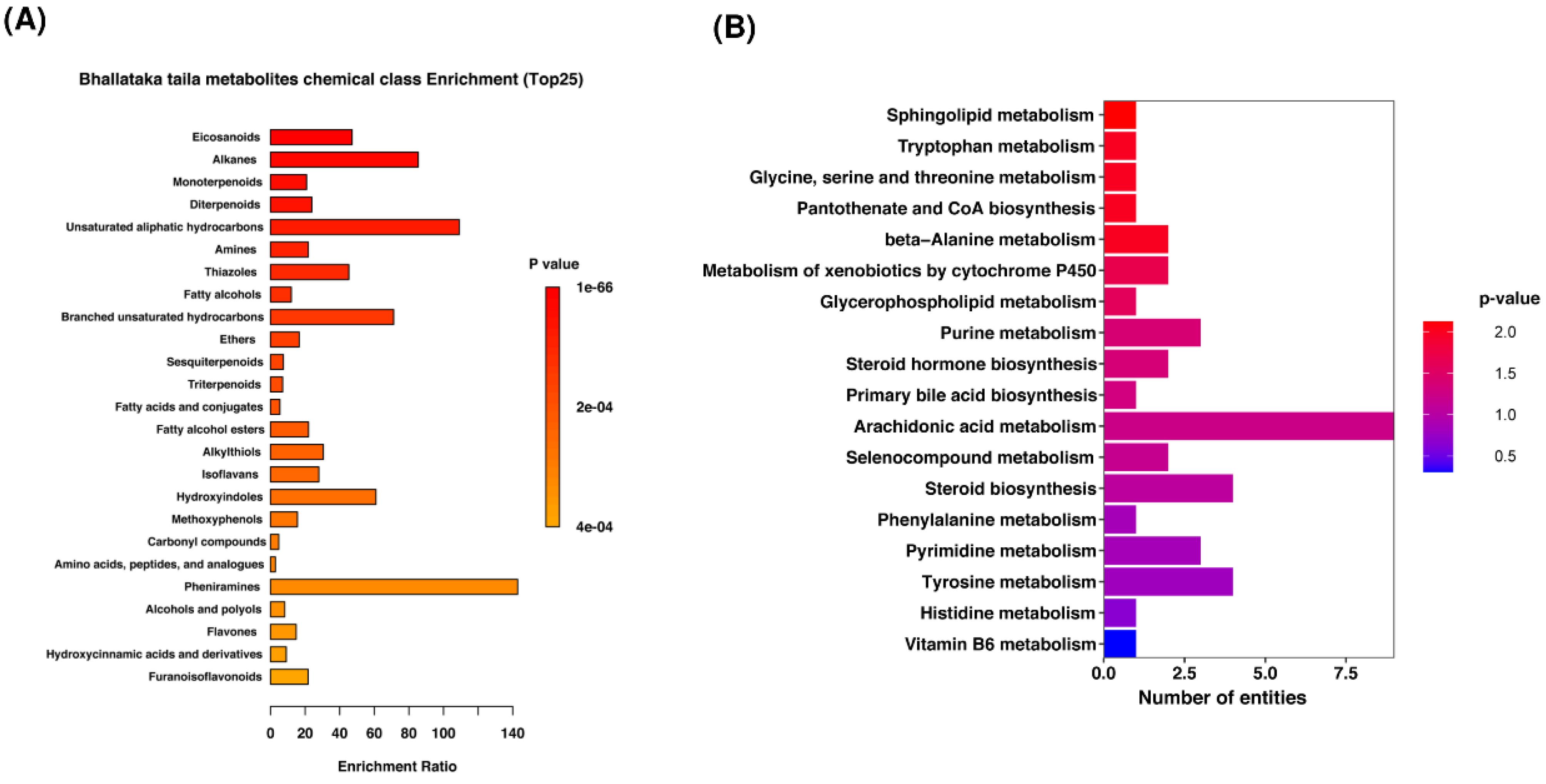
Fig. 2.
Metabolite categorization. (A) Overall visualization of Bhallataka taila metabolites, showing the top 25 enriched chemical classes (B) Enriched pathways of Bhallataka taila metabolites.
.
Metabolite categorization. (A) Overall visualization of Bhallataka taila metabolites, showing the top 25 enriched chemical classes (B) Enriched pathways of Bhallataka taila metabolites.
Target proteins of Bhallataka taila metabolites
Metabolites interact with various proteins involved in biochemical pathways, allowing them to play active roles in these pathways beyond merely serving as indicators of metabolic processes. To identify potential molecular targets of Bhallataka taila, we mapped its metabolites to their protein targets. These protein interactions provide valuable insights into the molecular targets of cancer. Using the online tool BindingDB, which provides information on protein-metabolite interactions based on the metabolite's SMILES ID, we identified 180 human protein targets for Bhallataka taila metabolites (Supplementary file 9).
These proteins were categorized according to their biological processes, subcellular localization, and molecular functions (Fig. 3A). Pathway enrichment analysis showed that these proteins were highly enriched in pathways involved in cancer regulation, including TRAIL signaling, PAR1-mediated thrombin signaling, ErbB receptor signaling, Glypican pathway, Syndecan-1 signaling, PDGF receptor signaling, and VEGFR1/2 signaling (Fig. 3B, Supplementary file 10). To further explore the protein-interacting partners of these targets, we generated a protein-protein interaction network using STRINGdb. The network analysis revealed that Bhallataka taila target proteins were clustered into three major groups (Fig. 3C).
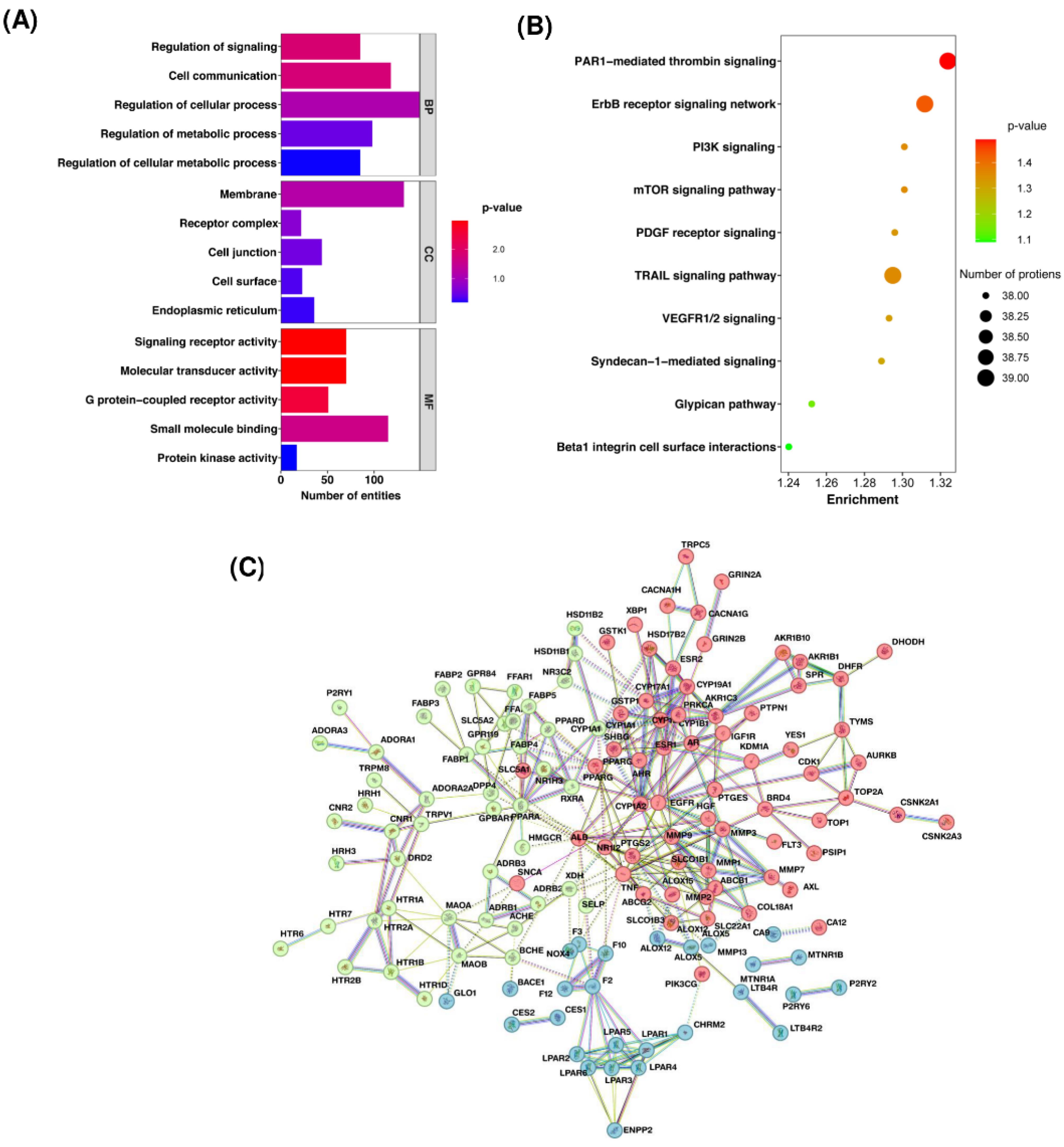
Fig. 3.
Analysis of protein targets of Bhallataka metabolites. (A) Bar graph illustrating the categorization of target proteins according to biological processes, molecular functions, and cellular locations. (B) Bubble plot depicting the enriched pathways of Bhallataka metabolite-protein targets. (C) Protein-protein interaction network of Bhallataka taila protein targets. The three different colors in k-means clustering represent various protein clusters based on interaction or functional similarity patterns.
.
Analysis of protein targets of Bhallataka metabolites. (A) Bar graph illustrating the categorization of target proteins according to biological processes, molecular functions, and cellular locations. (B) Bubble plot depicting the enriched pathways of Bhallataka metabolite-protein targets. (C) Protein-protein interaction network of Bhallataka taila protein targets. The three different colors in k-means clustering represent various protein clusters based on interaction or functional similarity patterns.
Regulation of cancer signaling pathways
Pathways in cancer
Protein target enrichment analysis using KEGG revealed that several key cancer-related pathways are regulated by Bhallataka taila. The enriched protein targets were associated with cellular functions including cytokine-cytokine receptor interaction, extracellular matrix (ECM)-receptor interaction, Wnt signaling pathway, HIF-1 signaling pathway, peroxisome proliferator-activated receptor (PPAR) signaling pathway, and VEGF signaling pathway. Key proteins associated with these pathways include coagulation factor II (F2), G protein-coupled receptors (GPCR), Rho guanine nucleotide exchange factor (RhoGEF), androgen receptor (AR), epidermal growth factor receptor (EGFR), FLT3LG, Fms-like tyrosine kinase 3 (FLT3), hepatocyte growth factor (HGF), insulin-like growth factor receptor (IGFR), protein kinase C (PKC), rapidly accelerated fibrosarcoma (Raf), cyclooxygenase (COX2), HIF-1β, VEGF, Pim-1 proto-oncogene (PIM1/2), MMPs, cyclin-dependent kinases (CDK4/6), retinoic X receptor (RXR), estrogen receptor (ER), Fas cell surface death receptor (Fas), Fas ligand (FasL), and telomerase reverse transcriptase (TERT), all of which are modulated by Bhallataka taila in cancer pathways (Fig. 4A).
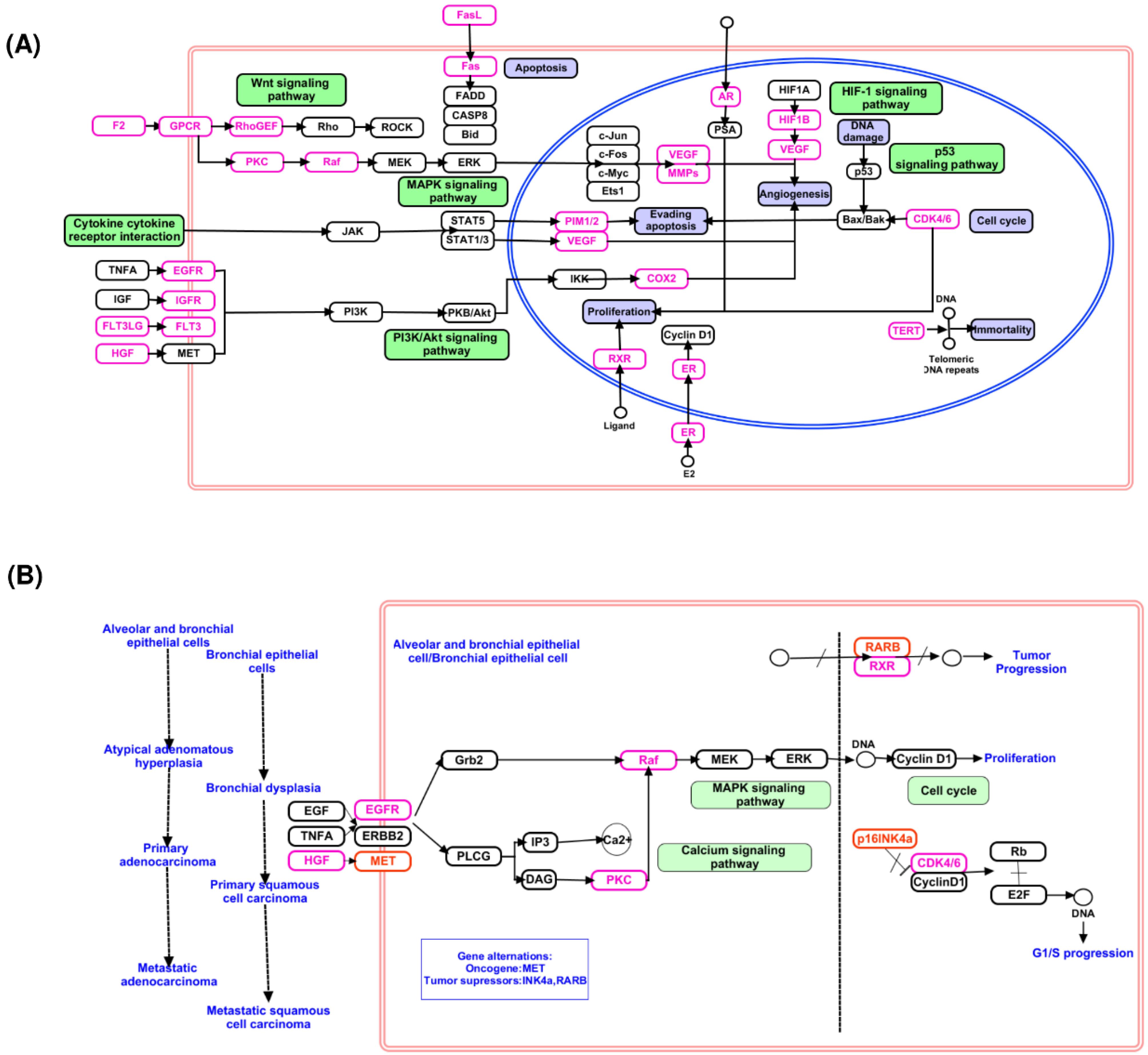
Fig. 4.
The proteins involved in the cancer signaling pathway are modulated by Bhallataka taila metabolite target proteins (A) Cancer pathway, (B) NSCLC.
.
The proteins involved in the cancer signaling pathway are modulated by Bhallataka taila metabolite target proteins (A) Cancer pathway, (B) NSCLC.
In addition, several of these enriched protein targets, including EGFR, HGF, PKC, Raf, CDK4/6, and RXR, are known to regulate key signaling pathways involved in NSCLC, including the mitogen-activated protein kinases (MAPK) signaling pathway, apoptosis, proliferation, phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) pathway, and cell cycle pathways (Fig. 4B).
In vitro validation of the anticancer activity of Bhallataka taila
Lung adenocarcinoma A549 cells were used to confirm the predictive value of Bhallataka taila protein targets and to investigate their anticancer activity.
Cell cytotoxicity of Bhallataka taila in A549 cells
The anticancer mechanisms of Bhallataka taila were assessed in A549 cells using MTT assay. Both Bhallataka taila and cisplatin reduced the cell viability in a dose-responsive manner (Fig. 5A and 5B). IC50 values for Bhallataka taila and cisplatin were 78 µg/mL and 6 μg/mL, respectively, 48 h post-treatment, with untreated cells serving as the control. The IC50 values were used for subsequent investigations (* P < 0.05, ** P < 0.01, *** P < 0.001 relative to control).
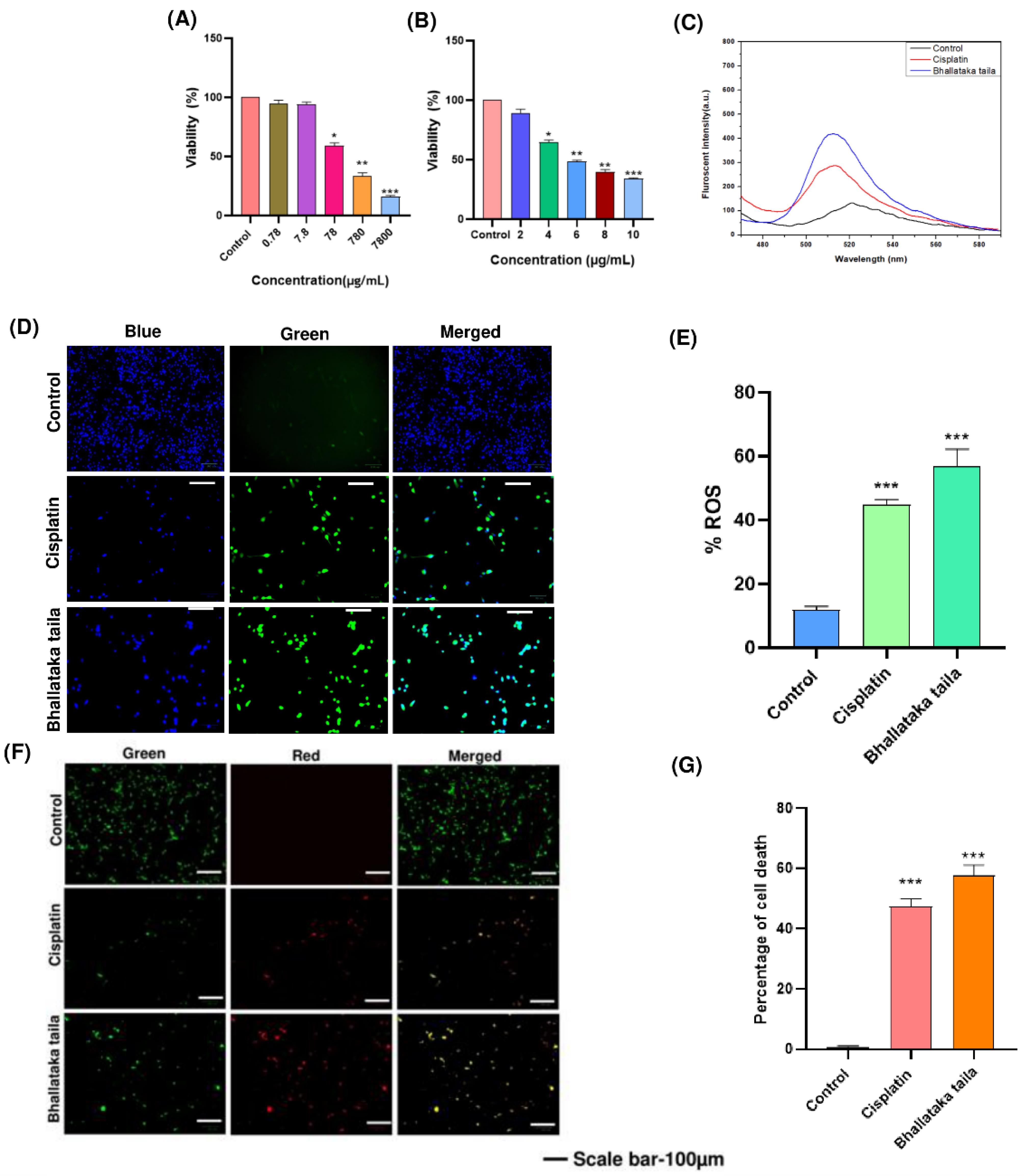
Fig. 5.
Cytotoxic effects on lung adenocarcinoma A549 cells were assessed using MTT assay after 48 h of treatment with (A) Bhallataka taila and (B) cisplatin. (C) Fluorescence spectroscopy and (D) fluorescent microscopic images revealed elevated ROS levels in the A549 cells following treatment with Bhallataka taila and cisplatin. (E) Graphical representation of the percentage of ROS after treatment compared with the control. (F) AO-EB dual staining indicated cell death. The control exhibited live cells with unaltered nuclei displaying green fluorescence, and treated cells displayed early apoptotic cells with yellow fluorescence, signifying apoptosis triggered by Bhallataka taila and cisplatin. (G) Graphical depiction of cell death percentage after treatment in contrast to the control. (* P < 0.05, ** P < 0.01, *** P < 0.001, relative to control).
.
Cytotoxic effects on lung adenocarcinoma A549 cells were assessed using MTT assay after 48 h of treatment with (A) Bhallataka taila and (B) cisplatin. (C) Fluorescence spectroscopy and (D) fluorescent microscopic images revealed elevated ROS levels in the A549 cells following treatment with Bhallataka taila and cisplatin. (E) Graphical representation of the percentage of ROS after treatment compared with the control. (F) AO-EB dual staining indicated cell death. The control exhibited live cells with unaltered nuclei displaying green fluorescence, and treated cells displayed early apoptotic cells with yellow fluorescence, signifying apoptosis triggered by Bhallataka taila and cisplatin. (G) Graphical depiction of cell death percentage after treatment in contrast to the control. (* P < 0.05, ** P < 0.01, *** P < 0.001, relative to control).
ROS assessment of Bhallataka taila
Increased levels of ROS within the intracellular region can significantly impact cellular activity, often leading to cellular apoptosis. In the current study, Bhallataka taila-treated A549 cells exhibited a marked elevation in intracellular ROS levels compared with the cisplatin-treated and control groups (Fig. 5C-5E).
Apoptosis exerted by Bhallataka taila
A549 cells were treated for 48 h with Bhallataka taila and cisplatin, followed by AO-EB dual nuclear staining to assess apoptosis. Treatment with Bhallataka taila led to an increase in late-phase apoptotic and necrotic cells, as indicated by the orange fluorescence from EB uptake, a pattern similar to that observed with cisplatin induction (Fig. 5F and 5G).
Impact of Bhallataka taila on the cancer-regulatory pathway in A549 cells
The mechanisms responsible for Bhallataka taila-mediated regulation of the ERK1/2 pathway and its anticancer activity were investigated by assessing the expression of total and phosphorylated ERK1/2, HIF-1α, Ki-67, and p53 using immunoblotting. The results revealed that treatment with Bhallataka taila and cisplatin reduced the expression of key pathway regulatory proteins, including phosphorylated ERK1/2, HIF-1α, and Ki-67, compared with the control. Additionally, both Bhallataka taila and cisplatin enhanced the expression of p53, a critical tumor suppressor protein (Fig. 6) (* P < 0.05, ** P < 0.01, *** P < 0.001, relative to control, ## P < 0.01, ### P < 0.001, relative to cisplatin).
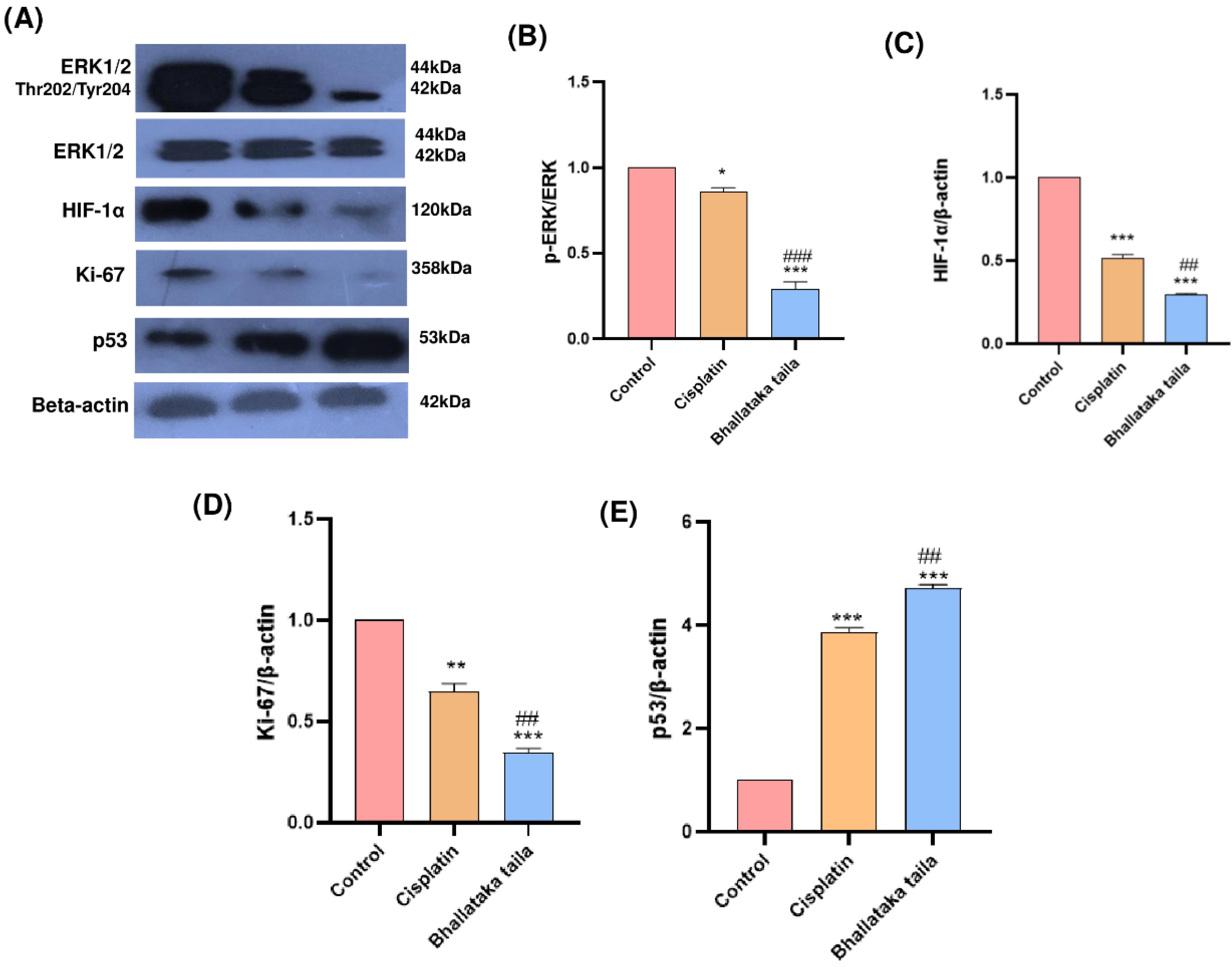
Fig. 6.
(A) Expression of indicated proteins in A549 lung adenocarcinoma cells treated with Bhallataka taila and cisplatin. Beta-actin served as the loading control. Densitometry graph showing the levels of (B) p-ERK (C) HIF-1α (D) Ki-67, and (E) p53 in lung cancer cells (* P < 0.05, ** P < 0.01, *** P < 0.001, relative to control, ## P < 0.01, ### P < 0.001, relative to cisplatin).
.
(A) Expression of indicated proteins in A549 lung adenocarcinoma cells treated with Bhallataka taila and cisplatin. Beta-actin served as the loading control. Densitometry graph showing the levels of (B) p-ERK (C) HIF-1α (D) Ki-67, and (E) p53 in lung cancer cells (* P < 0.05, ** P < 0.01, *** P < 0.001, relative to control, ## P < 0.01, ### P < 0.001, relative to cisplatin).
Gene expression studies
Gene expression associated with apoptosis, tumor inhibition, hypoxia, and metastasis were assessed using qRT-PCR. The findings indicated that Bhallataka taila and cisplatin treatment significantly increased the levels of pro-apoptotic genes BAX and p53, while inhibiting the levels of anti-apoptotic and metastasis-related genes, including Bcl2, HIF-1α, MMP-2, and MMP-9, relative to the control (Fig. 7) (* P < 0.05, ** P < 0.01, relative to control, # P < 0.005, relative to cisplatin).
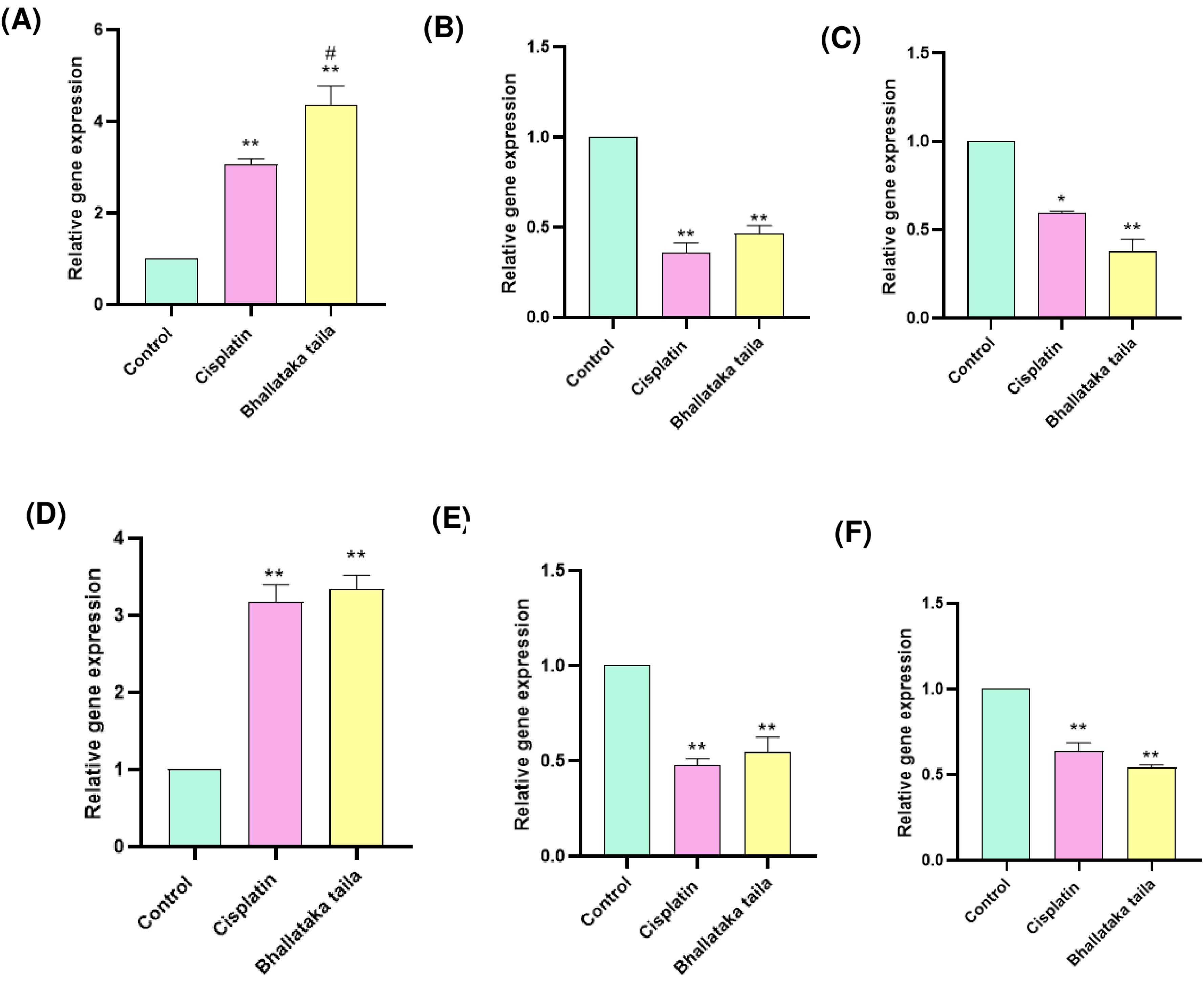
Fig. 7.
Gene expression profiling of (A) BAX (B) Bcl2 (C) HIF-1α (D) p53 (E) MMP-2 (F) MMP-9 in A549 cells exposed to Bhallataka taila and cisplatin as analyzed using qRT-PCR. (* P < 0.05, ** P < 0.01, relative to control, # P < 0.005, relative to cisplatin).
.
Gene expression profiling of (A) BAX (B) Bcl2 (C) HIF-1α (D) p53 (E) MMP-2 (F) MMP-9 in A549 cells exposed to Bhallataka taila and cisplatin as analyzed using qRT-PCR. (* P < 0.05, ** P < 0.01, relative to control, # P < 0.005, relative to cisplatin).
Discussion
Bhallataka taila, derived from S. anacardium Linn., has been used in traditional medicine because of its anti-atherogenic, anti-inflammatory, antimicrobial, hypoglycemic, and anticarcinogenic properties.23 Its toxic effects are minimized and its efficacy is enhanced through purification processes like Shodhana in Ayurveda.24,25 To elucidate the molecular mechanisms responsible for these effects, we employed LC-MS/MS-based global metabolomics and network pharmacology approaches to profile the metabolites of Bhallataka taila and investigate its anticancer potential, particularly in lung adenocarcinoma using A549 cells.
Our analysis identified key molecular signatures of Bhallataka taila, revealing metabolites such as dodecanoic acid and undecane as major constituents. Shibriya et al reported that dodecanoic acid, a lead compounds from Bhallataka nut, has a higher binding affinity to caspase 3, a proteolytic enzyme that is necessary for the execution of apoptosis. As a result, dodecanoic acid appears to be a promising anticancer agent because it inhibits active enzymes and can enhance the efficacy of cancer treatment by reducing the risk of chemoresistance and recurrence.21 Similarly, undecane, identified through GC-MS, has been associated with antioxidant propertiesand anti-inflammatory effects,22,25,26 as demonstrated in studies on keratinocytes and mast cells.27 These findings further support the biological activity of Bhallataka taila.
Bioinformatics analysis predicted that Bhallataka taila metabolites interact with proteins involved in several cancer-related pathways, including the TRAIL signaling, PAR1-mediated thrombin signaling, ErbB receptor signaling network, Syndecan-1 and Glypican pathway, PDGF receptor signaling, VEGFR1/2 signaling, all of which are essential in mediating cancer progression.28-33 Additional pathway enrichment analysis unveiled the involvement of critical cancer pathways, such as cytokine-cytokine receptor interactions, ECM-receptor interactions, Wnt, HIF-1, PPAR, MAPK, apoptosis, proliferation, PI3K/Akt, and cell cycle, suggesting that these pathways may contribute to the anticancer effects observed in NSCLC.34-41
Increased ROS levels significantly trigger the release of proteins that promote apoptosis by disrupting the cellular redox balance and activating pro-apoptotic signals. Higher ROS levels are associated with increased tumor-suppressive signaling in cancer by triggering oxidative stress-mediated tumor cell apoptosis.42 Our study demonstrated that Bhallataka taila induces ROS production in A549 cells, similar to cisplatin.The apoptotic effects of Bhallataka were studied in a myeloid leukemic and breast cancer cell line with a special emphasis on mitochondrial apoptotic effects. This study confirmed the presence of apoptotic morphological changes, providing evidence for the mechanism of action of Bhallataka, and highlighting its broader therapeutic implications.2
Lung cancer is known to be triggered by phosphorylation of the oncogenic signaling proteins AKT and ERK.43 In NSCLC cells, ERK1/2 phosphorylation facilitates proliferation and resistance to chemotherapy.44 Our investigation into the ERK1/2 signaling pathway revealed that Bhallataka taila treatment significantly reduced phospho-ERK 1/2 regulation in A549 cells, inducing a reduction in cell proliferation. Numerous investigations have indicated that increased Ki-67 level is a marker of adverse clinical outcomes among patients suffering from NSCLC.45,46 Our study revealed the decreased Ki-67 protein expression in Bhallataka taila-treated cells, indicating an enhanced survival rate. p53 is crucial for cell cycle regulation, apoptosis, and repair mechanisms.47 HIF-1α activates the glycolysis pathway, leading to increased cellular apoptosis during hypoxia in A549 cells.48 We also observed elevated expression of p53 and HIF-1α in A549 cells exposed with Bhallataka taila at protein and gene expression compared to the control.
A key tactic in the fight against cancer is the activation of apoptotic pathways, which constitute an established progressive program of cell death. Pro-apoptotic Bax and anti-apoptotic Bcl-2 contribute to the regulation of apoptosis and carcinogenesis. Studies have reported that Vitexin triggered apoptotic responses and inhibited the Bcl-2/Bax expression ratio in human NSCLC A549 cells.49,50 Samarghandian et al observed that chrysin induces Bax expression while decreasing Bcl-2 expression in A549 cells.51,52 In the current study, treatment with Bhallataka taila induces elevated Bax gene activity and decreases Bcl-2 gene expression in A549 cells compared with the control. MMPs play key roles in facilitating tumor invasion and metastasis by breaking down the ECM and basement membrane.53 MMP-2 is important for tumor development and angiogenesis, so a strong MMP-2 inhibitor is important for managing lung cancer. MMP-9 is strongly involved in the metastatic properties of lung cancer. This suggests that MMP-9 is a potential target for therapy and prognosis in NSCLC.54,55 In our study, Bhallataka taila significantly decreased MMP-2 and MMP-9 gene expression in treated A549 cells, indicating the suppression of the invasion and migration of A549 cells.
These findings highlight the potential of metabolomics for identifying new protein targets and pathways based on identified metabolites. Although some pathways identified in this study were validated in vitro, further investigations into specific metabolite-protein interactions are needed to confirm these targets. Protein identification is heavily based on metabolite input, which, in turn, requires accurate detection. The gaps in metabolite annotation highlight a significant challenge in protein-target detection, highlighting the necessity for a more extensive database for identifying plant-derived metabolites. Despite using plant-specific databases, many metabolites remained unassigned, suggesting that existing resources may be insufficient for detailed metabolomic profiling. In summary, our findings offer a critical understanding of the anticancer mechanisms of Bhallataka taila, emphasizing the importance of improving metabolomics databases to enhance the identification of plant-derived metabolites and their protein targets.
Conclusions
The present study examined the metabolite profile and mode of action of Bhallataka taila in lung cancer cells. We observed that the modulation of the ERK1/2- HIF-1α-p53 signaling axis plays a pivotal role in its anticancer effects. These findings suggest that Bhallataka taila is a promising therapeutic agent in both in vitro and preclinical models of lung cancer and other cancers. The findings not only confirm the anticancer properties of the drug but also highlight new potential targets for therapeutic strategies. Moreover, this study provides a foundation for clinical trials to evaluate the efficacy of Bhallataka taila and other traditional medicines in the treatment of cancer.
Competing Interests
The authors declare no conflict of interest.
Ethical Approval
None to be declared
Supplementary files
Supplementary file 1. The total ion chromatogram of Bhallataka taila from (A) positive and (B) negative modes.
(pdf)
Supplementary file 2. List of identified Bhallataka taila metabolites in positive mode.
(xlsx)
Supplementary file 3. List of identified Bhallataka taila metabolites in negative mode.
(xlsx)
Supplementary file 4. List of Bhallataka taila metabolites at MS2 level.
(xlsx)
Supplementary file 5. List of unassigned metabolite features in Bhallataka taila positive mode.
(xlsx)
Supplementary file 6. List of unassigned metabolite features in Bhallataka taila negative mode.
(xlsx)
Supplementary file 7. List of identified chemical class in Bhallataka taila metabolites.
(xlsx)
Supplementary file 8. List of pathways involved in Bhallataka taila metabolites.
(xlsx)
Supplementary file 9. List of predicted protein targets of Bhallataka taila metabolites.
(xlsx)
Supplementary file 10. List of enriched pathways for the protein targets of Bhallataka taila.
(xlsx)
Acknowledgements
The authors would like to acknowledge Dr. Harkrishna Panaje, Shree Durga Parameshwari (SDP) Remedies, and the Research Centre for sample verification and providing Bhallataka taila and related information. We thank The London School of Hygiene & Tropical Medicine (LSHTM) for the Cardiovascular Biology grant. We also thank the Department of Biotechnology (DBT) National Facility grant for supporting the Center for Systems Biology and Molecular Medicine (CSBMM) at Yenepoya (Deemed to be University).
References
- Poornima P, Efferth T. Ayurveda for cancer treatment. Med Aromat Plants (Los Angel) 2016; 5:e178. doi: 10.4172/2167-0412.1000e178 [Crossref] [ Google Scholar]
- Patil VA, Dubewar AP, Bhonde R, Bhargava C, Kakodkar P. Bhallataka rasayana: a boon in cancer treatment. Int J Ayurvedic Med 2021; 12:763-9. doi: 10.47552/ijam.v12i4.2215 [Crossref] [ Google Scholar]
- Kumar AD, Bevara GB, Kaja LK, Badana AK, Malla RR. Protective effect of 3-O-methyl quercetin and kaempferol from Semecarpus anacardium against H2O2 induced cytotoxicity in lung and liver cells. BMC Complement Altern Med 2016; 16:376. doi: 10.1186/s12906-016-1354-z [Crossref] [ Google Scholar]
- Karthikkeyan G, Pervaje R, Subbannayya Y, Patil AH, Modi PK, Prasad TSK. Plant omics: metabolomics and network pharmacology of liquorice, Indian Ayurvedic Medicine Yashtimadhu. OMICS 2020; 24:743-55. doi: 10.1089/omi.2020.0156 [Crossref] [ Google Scholar]
- Fridlender M, Kapulnik Y, Koltai H. Plant derived substances with anti-cancer activity: from folklore to practice. Front Plant Sci 2015; 6:799. doi: 10.3389/fpls.2015.00799 [Crossref] [ Google Scholar]
- Habtemariam S, Lentini G. Plant-derived anticancer agents: lessons from the pharmacology of geniposide and its aglycone, genipin. Biomedicines 2018; 6:39. doi: 10.3390/biomedicines6020039 [Crossref] [ Google Scholar]
- Gahtori R, Tripathi AH, Kumari A, Negi N, Paliwal A, Tripathi P. Anticancer plant-derivatives: deciphering their oncopreventive and therapeutic potential in molecular terms. Futur J Pharm Sci 2023; 9:14. doi: 10.1186/s43094-023-00465-5 [Crossref] [ Google Scholar]
- Parate SS, Upadhyay SS, Amrutha S, Karthikkeyan G, Pervaje R, Abhinand CS. Comparative metabolomics and network pharmacology analysis reveal shared neuroprotective mechanisms of Bacopa monnieri (L) Wettst and Centella asiatica (L) Urb. Mol Neurobiol 2024; 61:10956-78. doi: 10.1007/s12035-024-04223-3 [Crossref] [ Google Scholar]
- Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 2012; 30:918-20. doi: 10.1038/nbt.2377 [Crossref] [ Google Scholar]
- Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 2010; 11:395. doi: 10.1186/1471-2105-11-395 [Crossref] [ Google Scholar]
- Behera SK, Kasaragod S, Karthikkeyan G, Narayana Kotimoole C, Raju R, Prasad TS. MS2Compound: a user-friendly compound identification tool for LC-MS/MS-based metabolomics data. OMICS 2021; 25:389-99. doi: 10.1089/omi.2021.0051 [Crossref] [ Google Scholar]
- Pang Z, Zhou G, Ewald J, Chang L, Hacariz O, Basu N. Using MetaboAnalyst 50 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat Protoc 2022; 17:1735-61. doi: 10.1038/s41596-022-00710-w [Crossref] [ Google Scholar]
- Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res 2016; 44:D1045-53. doi: 10.1093/nar/gkv1072 [Crossref] [ Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 2003; 4:P3. [ Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler--a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res 2007; 35:W193-200. doi: 10.1093/nar/gkm226 [Crossref] [ Google Scholar]
- Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015; 15:2597-601. doi: 10.1002/pmic.201400515 [Crossref] [ Google Scholar]
- von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res 2005; 33:D433-7. doi: 10.1093/nar/gki005 [Crossref] [ Google Scholar]
- Rahbar Saadat Y, Pourseif MM, Zununi Vahed S, Barzegari A, Omidi Y, Barar J. Modulatory role of vaginal-isolated Lactococcus lactis on the expression of miR-21, miR-200b, and TLR-4 in CAOV-4 cells and in silico revalidation. Probiotics Antimicrob Proteins 2020; 12:1083-96. doi: 10.1007/s12602-019-09596-9 [Crossref] [ Google Scholar]
- Pavan SR, Venkatesan J, Prabhu A. Anticancer activity of silver nanoparticles from the aqueous extract of Dictyotaciliolata on non-small cell lung cancer cells. J Drug Deliv Sci Technol 2022; 74:103525. doi: 10.1016/j.jddst.2022.103525 [Crossref] [ Google Scholar]
- Sajida Sajida, Prabhu A. Anti-angiogenic, apoptotic and matrix metalloproteinase inhibitory activity of Withaniasomnifera (ashwagandha) on lung adenocarcinoma cells. Phytomedicine 2021; 90:153639. doi: 10.1016/j.phymed.2021.153639 [Crossref] [ Google Scholar]
-
Shebriya A, Santhy KS. In silico approach in the identification of chemo therapeutical lead compounds from Semecarpus anacardium L.f. related to apoptotic pathway proteins. Journal of Xi'an Shiyou University (Natural Science Edition) 2021. doi: 10.17605/osf.io/qfb5k.
- Ruhila A, Srivastava A, Yadav P, Galib R, Prajapati PK. Preliminary GC-MS profiling of bhallataka oil. Asian J Res Chem 2021; 14:108-10. doi: 10.5958/0974-4150.2021.00019.5 [Crossref] [ Google Scholar]
- Sundaram R, Muthu K, Shanthi P, Sachdanandam P. Antioxidant and antihyperlipidemic activities of catechol derivatives and biflavonoid isolated from Semecarpus anacardium seeds. Toxicol Mech Methods 2022; 32:123-31. doi: 10.1080/15376516.2021.1973170 [Crossref] [ Google Scholar]
- Sulaiman CT, Deepak M, Praveen TK, Lijini KR, Anandan EM, Salman M. Purification of Bhallathaka (Semecarpus anacardium Lf) enhanced anti-cancer activity. RegulToxicolPharmacol 2021; 122:104898. doi: 10.1016/j.yrtph.2021.104898 [Crossref] [ Google Scholar]
- Maurya SK, Seth A, Laloo D, Singh NK, Gautam DN, Singh AK. Śodhana: an Ayurvedic process for detoxification and modification of therapeutic activities of poisonous medicinal plants. Anc Sci Life 2015; 34:188-97. doi: 10.4103/0257-7941.160862 [Crossref] [ Google Scholar]
- Rai N, Gupta P, Verma A, Tiwari RK, Madhukar P, Kamble SC. Ethyl acetate extract of Colletotrichum gloeosporioides promotes cytotoxicity and apoptosis in human breast cancer cells. ACS Omega 2023; 8:3768-84. doi: 10.1021/acsomega.2c05746 [Crossref] [ Google Scholar]
- Choi D, Kang W, Park T. Anti-allergic and anti-inflammatory effects of undecane on mast cells and keratinocytes. Molecules 2020; 25:1554. doi: 10.3390/molecules25071554 [Crossref] [ Google Scholar]
- Snajdauf M, Havlova K, Vachtenheim J Jr, Ozaniak A, Lischke R, Bartunkova J. The TRAIL in the treatment of human cancer: an update on clinical trials. Front Mol Biosci 2021; 8:628332. doi: 10.3389/fmolb.2021.628332 [Crossref] [ Google Scholar]
- Del Re M, Cucchiara F, Petrini I, Fogli S, Passaro A, Crucitta S. erbB in NSCLC as a molecular target: current evidences and future directions. ESMO Open 2020; 5:e000724. doi: 10.1136/esmoopen-2020-000724 [Crossref] [ Google Scholar]
- Parimon T, Brauer R, Schlesinger SY, Xie T, Jiang D, Ge L. Syndecan-1 controls lung tumorigenesis by regulating miRNAs packaged in exosomes. Am J Pathol 2018; 188:1094-103. doi: 10.1016/j.ajpath.2017.12.009 [Crossref] [ Google Scholar]
- Li N, Spetz MR, Ho M. The role of glypicans in cancer progression and therapy. J HistochemCytochem 2020; 68:841-62. doi: 10.1369/0022155420933709 [Crossref] [ Google Scholar]
- Heldin CH. Targeting the PDGF signaling pathway in tumor treatment. Cell Commun Signal 2013; 11:97. doi: 10.1186/1478-811x-11-97 [Crossref] [ Google Scholar]
- Ceci C, Atzori MG, Lacal PM, Graziani G. Role of VEGFs/VEGFR-1 signaling and its inhibition in modulating tumor invasion: experimental evidence in different metastatic cancer models. Int J Mol Sci 2020; 21:1388. doi: 10.3390/ijms21041388 [Crossref] [ Google Scholar]
- Lee M, Rhee I. Cytokine signaling in tumor progression. Immune Netw 2017; 17:214-27. doi: 10.4110/in.2017.17.4.214 [Crossref] [ Google Scholar]
- Yuan Z, Li Y, Zhang S, Wang X, Dou H, Yu X. Extracellular matrix remodeling in tumor progression and immune escape: from mechanisms to treatments. Mol Cancer 2023; 22:48. doi: 10.1186/s12943-023-01744-8 [Crossref] [ Google Scholar]
- Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene 2017; 36:1461-73. doi: 10.1038/onc.2016.304 [Crossref] [ Google Scholar]
- Huang R, Zhou PK. HIF-1 signaling: A key orchestrator of cancer radioresistance. Radiat Med Prot 2020; 1:7-14. doi: 10.1016/j.radmp.2020.01.006 [Crossref] [ Google Scholar]
- Cheng HS, Yip YS, Lim EK, Wahli W, Tan NS. PPARs and tumor microenvironment: the emerging roles of the metabolic master regulators in tumor stromal-epithelial crosstalk and carcinogenesis. Cancers (Basel) 2021; 13:2153. doi: 10.3390/cancers13092153 [Crossref] [ Google Scholar]
- Alam M, Hasan GM, Eldin SM, Adnan M, Riaz MB, Islam A. Investigating regulated signaling pathways in therapeutic targeting of non-small cell lung carcinoma. Biomed Pharmacother 2023; 161:114452. doi: 10.1016/j.biopha.2023.114452 [Crossref] [ Google Scholar]
- Liu G, Pei F, Yang F, Li L, Amin AD, Liu S. Role of autophagy and apoptosis in non-small-cell lung cancer. Int J Mol Sci 2017; 18:367. doi: 10.3390/ijms18020367 [Crossref] [ Google Scholar]
- Li XQ, Cheng XJ, Wu J, Wu KF, Liu T. Targeted inhibition of the PI3K/AKT/mTOR pathway by ( + )-anthrabenzoxocinone induces cell cycle arrest, apoptosis, and autophagy in non-small cell lung cancer. Cell Mol Biol Lett 2024; 29:58. doi: 10.1186/s11658-024-00578-6 [Crossref] [ Google Scholar]
- Arfin S, Jha NK, Jha SK, Kesari KK, Ruokolainen J, Roychoudhury S. Oxidative stress in cancer cell metabolism. Antioxidants (Basel) 2021; 10:642. doi: 10.3390/antiox10050642 [Crossref] [ Google Scholar]
- Crosbie PA, Crosbie EJ, Aspinall-O'Dea M, Walker M, Harrison R, Pernemalm M. ERK and AKT phosphorylation status in lung cancer and emphysema using nanocapillary isoelectric focusing. BMJ Open Respir Res 2016; 3:e000114. doi: 10.1136/bmjresp-2015-000114 [Crossref] [ Google Scholar]
- Vicent S, López-Picazo JM, Toledo G, Lozano MD, Torre W, Garcia-Corchón C. ERK1/2 is activated in non-small-cell lung cancer and associated with advanced tumours. Br J Cancer 2004; 90:1047-52. doi: 10.1038/sj.bjc.6601644 [Crossref] [ Google Scholar]
- Ishibashi N, Maebayashi T, Aizawa T, Sakaguchi M, Nishimaki H, Masuda S. Correlation between the Ki-67 proliferation index and response to radiation therapy in small cell lung cancer. Radiat Oncol 2017; 12:16. doi: 10.1186/s13014-016-0744-1 [Crossref] [ Google Scholar]
- Wang D, Ye W, Shi Q. Prognostic value of Ki-67 expression in advanced lung squamous cell carcinoma patients treated with chemotherapy. Cancer Manag Res 2021; 13:6429-36. doi: 10.2147/cmar.S326189 [Crossref] [ Google Scholar]
- Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer 2009; 9:738-48. doi: 10.1038/nrc2718 [Crossref] [ Google Scholar]
- Luo F, Liu X, Yan N, Li S, Cao G, Cheng Q. Hypoxia-inducible transcription factor-1alpha promotes hypoxia-induced A549 apoptosis via a mechanism that involves the glycolysis pathway. BMC Cancer 2006; 6:26. doi: 10.1186/1471-2407-6-26 [Crossref] [ Google Scholar]
- Rahman N, Khan H, Zia A, Khan A, Fakhri S, Aschner M. Bcl-2 modulation in p53 signaling pathway by flavonoids: a potential strategy towards the treatment of cancer. Int J Mol Sci 2021; 22:11315. doi: 10.3390/ijms222111315 [Crossref] [ Google Scholar]
- Liu X, Jiang Q, Liu H, Luo S. Vitexin induces apoptosis through mitochondrial pathway and PI3K/Akt/mTOR signaling in human non-small cell lung cancer A549 cells. Biol Res 2019; 52:7. doi: 10.1186/s40659-019-0214-y [Crossref] [ Google Scholar]
- Samarghandian S, Nezhad MA, Mohammadi G. Role of caspases, Bax and Bcl-2 in chrysin-induced apoptosis in the A549 human lung adenocarcinoma epithelial cells. Anticancer Agents Med Chem 2014; 14:901-9. doi: 10.2174/1871520614666140209144042 [Crossref] [ Google Scholar]
- Talebi M, Talebi M, Farkhondeh T, Simal-Gandara J, Kopustinskiene DM, Bernatoniene J. Emerging cellular and molecular mechanisms underlying anticancer indications of chrysin. Cancer Cell Int 2021; 21:214. doi: 10.1186/s12935-021-01906-y [Crossref] [ Google Scholar]
- Cai X, Zhu H, Li Y. PKCζ, MMP-2 and MMP-9 expression in lung adenocarcinoma and association with a metastatic phenotype. Mol Med Rep 2017; 16:8301-6. doi: 10.3892/mmr.2017.7634 [Crossref] [ Google Scholar]
- Merchant N, Nagaraju GP, Rajitha B, Lammata S, Jella KK, Buchwald ZS. Matrix metalloproteinases: their functional role in lung cancer. Carcinogenesis 2017; 38:766-80. doi: 10.1093/carcin/bgx063 [Crossref] [ Google Scholar]
- Winkler K, Kowalczyk A, Bereza P, Regulska K, Kasprzak A, Bamburowicz-Klimkowska M. Levels of active forms of MMP-1, MMP-2, and MMP-9 as independent prognostic factors for differentiating the stage and type of lung cancer (SCLC and NSCLC). Sens Actuators B Chem 2024; 406:135421. doi: 10.1016/j.snb.2024.135421 [Crossref] [ Google Scholar]