Bioimpacts. 2025;15:30792.
doi: 10.34172/bi.30792
Original Article
mtDNA copy number/miR663/AATF axis in invasive ductal carcinoma of the breast
Farzaneh Dahi Conceptualization, Formal analysis, Investigation, Writing – original draft, 1 
Shirin Shahbazi Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Writing – original draft, 1, * 
Loabat Geranpayeh Methodology, Writing – original draft, 2
Author information:
1Department of Medical Genetics, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran
2Department of Surgery, Sina Hospital, Tehran University of Medical Sciences, Tehran, Iran
Abstract
Introduction:
Mitochondrial DNA (mtDNA) copy number variations have been reported in multiple human cancers. Previous studies indicate that mitochondrial retrograde signaling regulates miR663, which plays a key role in tumorigenesis, including regulating apoptosis antagonizing transcription factor (AATF). This study investigates the expression of miR663 and AATF in relation to mtDNA copy number in invasive ductal carcinoma (IDC) of the breast.
Methods:
Paired primary tumors and adjacent non-tumor tissues were analyzed to assess changes in miR663 and AATF expression using fold-change analysis. The mtDNA copy number was quantified using COX1 as the mitochondrial gene and COX4 as the nuclear control gene. To validate the findings, publicly available data from The Cancer Genome Atlas (TCGA) were also analyzed.
Results:
A significant reduction in tumor miR663 expression was observed (fold change=0.139), with a strong correlation between miR663 and AATF expression. A significant Z-score difference was also detected between miR663 and mtDNA copy number. miR663 was predominantly expressed in grade I tumors but significantly downregulated in higher-grade tumors, whereas AATF expression increased with tumor grade. In silico analysis of TCGA data confirmed elevated AATF expression, with notable variations across breast cancer subtypes.
Conclusion:
We observed reduced expression of miR663 and mtDNA copy number in breast tumors, along with variations in AATF levels across subtypes. The decrease in miR663 could be associated with lower mtDNA copy numbers and impaired retrograde signaling, impacting AATF expression and function. Our findings underscore the therapeutic promise of targeting the mtDNA/miR-663/AATF axis, which could lead to advancements in breast cancer treatment.
Keywords: Breast cancer, miRNAs, Tumorigenesis
Copyright and License Information
© 2025 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
There are no founding sources for the present work.
Introduction
Evidence suggests that breast cancer is not solely a genetic disease but also a metabolic disorder.1 Defects in mitochondrial metabolism, particularly the reduction of mitochondrial DNA (mtDNA) content, play a crucial role in breast tumorigenesis.2 Several studies have emphasized the importance of mitochondria in carcinogenesis, including metabolic reprogramming, apoptosis evasion, genomic instability, and tumor metastasis.3,4
Mitochondria primarily generate cellular energy through oxidative phosphorylation(OXPHOS) via the electron transport chain (ETC). Disruptions in this process increase reactiveoxygen species (ROS) production, leading to oxidative stress, a major contributor to DNAdamage and tumorformation.4 Mitochondria contain multiple copies of mtDNA to ensure efficient ETC proteinproduction. Variations in mtDNA copy number contribute to oxidative stress and are implicated in the progression of multiple tumors, including breast cancer.5,6
Studies indicate that somaticmtDNA mutations accumulate in primary breast tumors with age. Low mtDNA levels have also been associated with tumor invasion and chemotherapy resistance.2 A recent study analyzed somatic mutations in 205 genes involved in glycolysis and OXPHOS pathways using The Cancer Genome Atlas (TCGA) dataset to investigate mitochondrial dysfunction in breast cancer. Among 968 patient samples, mutations were identified in 132 genes, with seven genes recognized as potential biomarkers forbreast cancer.7
Mitochondria regulate nuclear gene expression via mitochondrial retrograde signalingpathways.8 This retrograde signaling plays a fundamental role in cellular function, influencing a broad spectrum of genes, including microRNAs (miRNAs). As tissue-specific regulators, miRNAs are integral to tumorigenesis. Specific miRNAs support tumor cells by shifting metabolism from OXPHOS to aerobic glycolysis, a phenomenon known as the Warburg effect.9
The miR663 gene on chromosome 20p11.1 is among the miRNAs associated with mitochondrial function in breast cancer. Carden et al demonstrated that miR663 expression decreased in MCF-7 and MDA-MB-231 breast cancer cells lacking mtDNA. Restoration of mtDNA in these cells reversed this effect, suggesting that miR663 is epigenetically regulated through retrograde signaling. Bisulfite sequencing further revealed that miR663 promoter activity is suppressed via methylation induced by ROS. The study also indicated that miR663 regulates nuclear respiratory chain subunits in ETC complexes I, II, III, and IV.10
One of the primary targets of miR663 is the apoptosis antagonizing transcription factor (AATF), whose degradation is a key step in cellular apoptosis. AATF, predominantly a nuclear protein, exerts pro-proliferative effects by regulating G1/S and G2/M checkpoints in the cell cycle.11 Increasing evidence suggests that AATF protein is also localized in mitochondria, where it plays an active role in the cellular response to oxidativestress.12
Benakanakere et al identified miR663 as an apoptosis-inducing factor by targeting AATF mRNA in oral epithelialcells.13 The AATF (Che-1) gene on chromosome 17q12 encodes a 558-amino acid protein featuring an acidic N-terminal region, a leucine zipper motif, phosphorylation sites for kinases, and three nuclear receptor binding motifs. Given its role in cell cycle arrest, AATF has been investigated in hepatocellular carcinoma and identified as a potential driver of hepatocarcinogenesis.14 A 2024 study proved that TNF-α converting enzyme inhibition through AATF reduction could be a novel targetedtherapy.15 However, analysis of the AATF gene in over 100 breast cancer families failed to identify coding sequence mutations linked to cancer susceptibility, shifting research focus toward miRNA-mediated silencing mechanisms.16
Based on these findings, this study investigates the functional axis of mitochondria/miR663/AATF in breast cancer. To minimize confounding variables, invasive ductal carcinoma (IDC) was selected as the study focus, as it represents 80% of all breast cancer cases.
Materials and Methods
Patients and samples
This study was approved by the Ethics Committee at Tarbiat Modares University, Tehran, Iran (IR.MODARES.REC.1399.131), and all patients provided informed consent. A total of 22 fresh frozen breast tumor tissues and paired adjacent non-tumor tissues were collected from Sina and Farmaniyeh hospitals in Tehran. Demographic data, including tumor grade, lymphatic invasion, family history, and hormone receptor expression, are presented in Table 1.
Table 1.
Demographic information of the patients
|
Characteristics
|
Sub-groups
|
Frequency (%)
|
| Histology grade |
Grade I (low-well differentiated) |
4 (18.2) |
| Grade II (intermediate-moderately differentiated) |
12 (54.5) |
| Grade III (high-poor differentiated) |
5 (22.7) |
| Missing |
1 (4.5) |
| Total |
22 (100) |
| Site of primary tumor |
Left breast |
9 (40.9) |
| Right breast |
12 (54.5) |
| Bilateral |
1 (4.5) |
| Total |
22 (100) |
| Lymphatic invasion |
Positive |
13 (59.1) |
| Negative |
8 (36.4) |
| Missing |
1 (4.5) |
| Total |
22 (100) |
| Estrogen receptor |
Positive |
18 (81.8) |
| Negative |
3 (13.6) |
| Missing |
1 (4.5) |
| Total |
22 (100) |
| Progesterone receptor |
Positive |
15 (68.2) |
| Negative |
6 (27.3) |
| Missing |
1 (4.5) |
| Total |
22 (100) |
| Her-2 |
Positive (1-2) |
4 (18.2) |
| Positive (3 + ) |
3 (13.6) |
| Negative |
14 (63.6) |
| Missing |
1 (4.5) |
| Total |
22 (100) |
| Family history |
Positive |
8 (36.3) |
| Negative |
14 (63.6) |
| Total |
22 (100) |
All samples were histologically confirmed as IDC of the breast. Patients had no prior history of chemotherapy, radiotherapy, or hormone therapy. Clinical and pathological data were documented, including tumor size, grade, and the status of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2/neu).
Total DNA extraction and determination of mtDNA content
Total DNA was extracted from tissue samples using the DNrich Tissue Kit (Azma Elixir Pajooh, Iran). The quality and quantity of the extracted DNA were assessed via electrophoresis and a NanoPhotometer (NP80, Germany).
Real-time PCR was performed on a StepOne apparatus (Applied Biosystems), and cycle threshold (Ct) values were recorded. Each tumor mtDNA copy number was compared with its paired adjacent non-tumor tissue.
To quantify the mtDNA copy number, the following formula was applied:
2 [Ct(nucleargene)-Ct(mitochondrialgene)]
As previously described.17 The relative amplification of the nuclear cytochrome c oxidase subunit IV (COX4) gene was compared to that of the mitochondrial cytochrome c oxidase subunit I (MT-COXI) gene.18 Standard curves were generated for each primer, and downstream assays were performed at a 10 ng/μL DNA concentration.
Total RNA extraction and cDNA synthesis
Total RNA was extracted from tissue samples using TRIzol reagent (Yektatajhiz, Iran). RNA precipitation was performed using chloroform and isopropyl alcohol, followed by solubilization in DEPC-treated water and storage at −80 °C until use. The quality and quantity of extracted RNA were assessed, and samples with an A260/A280 ratio of 1.8–2.0 were selected for downstream analyses.
cDNA synthesis was performed using the cDNA Synthesis Kit (Yektatajhiz, Iran). For miR663, cDNA synthesis was conducted using a stem-loop primer (Table 2). A 1000 ng/μL concentration of RNA was the favored concentration for cDNA synthesis.
Table 2.
Primers and stem-loop sequences
|
Primer
|
Sequence (5'-3')
|
|
MT-COX1
|
Forward: TGATCTGCTGCAGTGCTCTGA
Reverse: TCAGGCCACCTACGGTGAA |
|
COX4
|
Forward: GAAAGTGTTGTGAAGAGCGAAGAC
Reverse: GTGGTCACGCCGATCCAT |
|
AATF
|
Forward: TCGGTTTCATGTCCTTAGCAAGC
Reverse: GGAGGTGGGCGATGTCAATC |
|
PUM1
|
Forward: TCGGAAGTAGCAGTTCTCTCG
Reverse: CTGTGTCCAATGGGGGTCAA |
|
miR663
|
Forward: AGGCGGGGTGCTGCGGGA |
|
u47
|
Forward: CCACTGCTGTAATGATTCTGC |
| Universal reverse |
Reverse: CCAGTGCAGGGTCCGAGGTA |
|
miR663 Stem-loop |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCGGTC |
|
U47 Stem-loop |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACCTCAG |
Note: The MT-COX1 and COX4 genes were selected as the target and reference genes to quantify the mtDNA copy number. For expression analysis of AATF and miR663, the PUM1 and U47 genes were used as reference genes. A stem-loop primer was applied for miR663 cDNA synthesis.
Quantitative real-time PCR (qRT-PCR) assay
The PUM1 and U47 genes were selected as internal reference genes (Table 2). According to prior research, these are among the most stable housekeeping genes in normal and tumor breast tissues. PUM1 may play a greater role in regulating cellular functions under hypoxia than other reference genes.19
The efficiency of qRT-PCR primers was evaluated using LinRegPCR software (Academic Medical Center, Amsterdam, the Netherlands). qRT-PCR reactions were performed using SYBR Green Master Mix High ROX (Ampliqon) under the following conditions: initial denaturation at 95°C for 15 minutes, followed by 40 cycles of 95 °C for 15 seconds and 60 °C for 45 seconds. All experiments were duplicated, and a non-template control (NTC) was included in each reaction.
TCGA analysis
The expression of AATF was analyzed across various stages and subtypes of breast cancer using TCGA data. A total of 114 normal samples, 183 stage I samples, 615 stage II samples, 247 stage III samples, and 20 stage IV samples were examined. For breast cancer subtype analysis, 566 samples were from the luminal subgroup, 37 were from the HER2 + subgroup, and 116 were from the triple-negative breast cancer (TNBC) subgroup. All subtypes were compared with 114 normal samples.
Statistical analysis
Real-time PCR data were analyzed using GraphPad Prism 8.0.2 and REST 2009 software (Qiagen, Technical University Munich). Fold-change calculations were based on the 2-ΔΔCt method. The correlation between miR663 expression mtDNA copy number and AATF expression was assessed using IBM SPSS (version 16.0).
Due to the limited sample size and non-normal data distribution, nonparametric tests were employed, including Wilcoxon, Mann-Whitney U, and Kruskal-Wallis tests. Correlations between gene expression levels were evaluated using Spearman's Rank Correlation Coefficient. A P value < 0.05 was considered statistically significant in all analyses.
Results
Relative expression analysis
The melt curves of AATF and miR663, along with those of the internal control genes, are presented in Fig. 1. According to LinRegPCR software, the PCR efficiency was 1.93 for AATF and 1.81 for miR663.
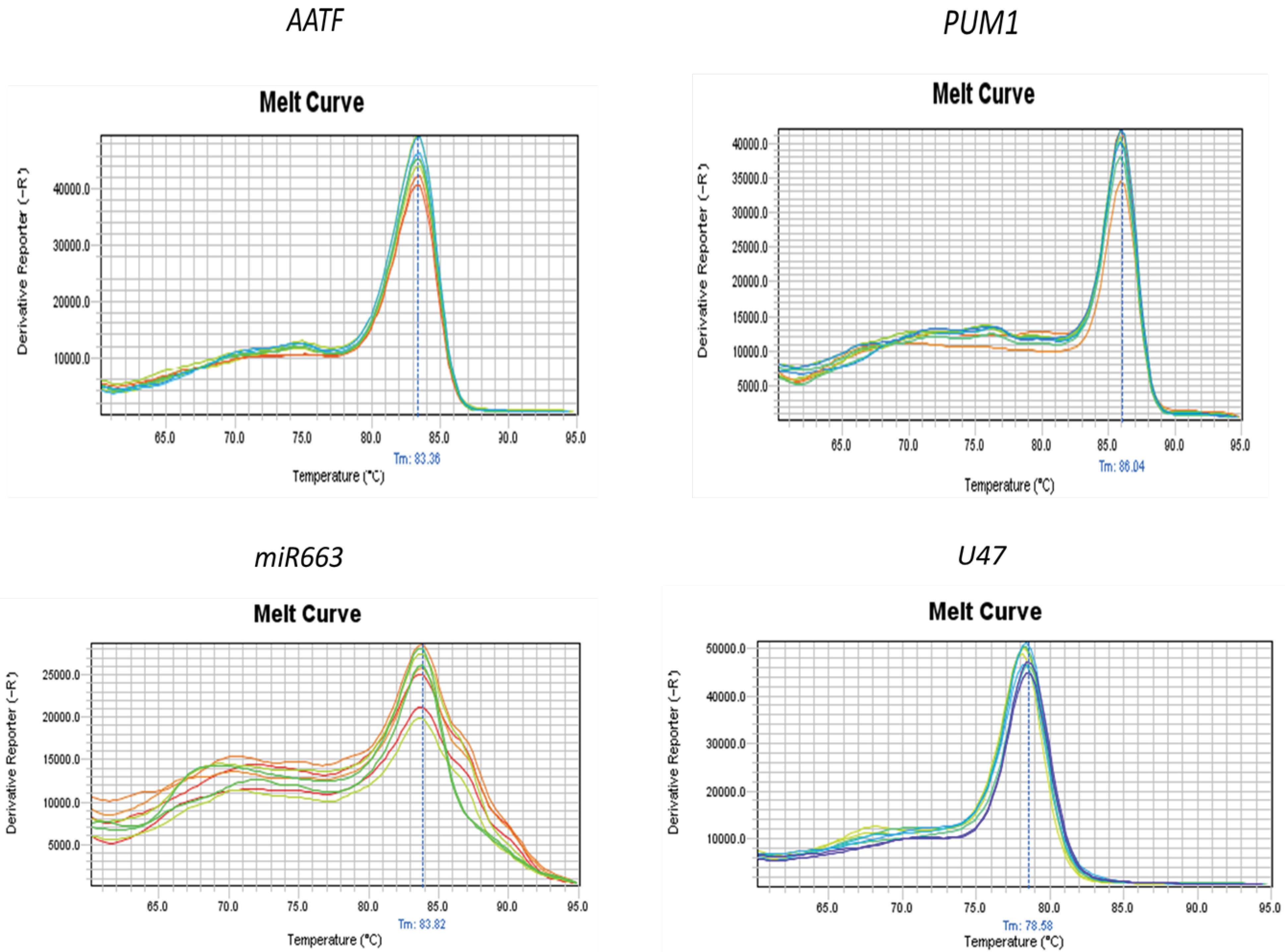
Fig. 1.
Melt curve analysis. Top: AATF and its internal control PUM1. Bottom: miR663 and the U47 control gene. The lack of sharpness in the melt curve of miR663 results from unavoidable primer-dimer interference. No amplification was detected in the NTCs.
.
Melt curve analysis. Top: AATF and its internal control PUM1. Bottom: miR663 and the U47 control gene. The lack of sharpness in the melt curve of miR663 results from unavoidable primer-dimer interference. No amplification was detected in the NTCs.
Using GraphPad Prism and REST software, the relative gene expression of miR663 was calculated after normalization to U47. The results demonstrated a significant decrease in miR663 expression in breast tumors compared to paired adjacent non-tumor tissues (fold change = 0.139, P = 0.000), as illustrated in Fig. 2A.
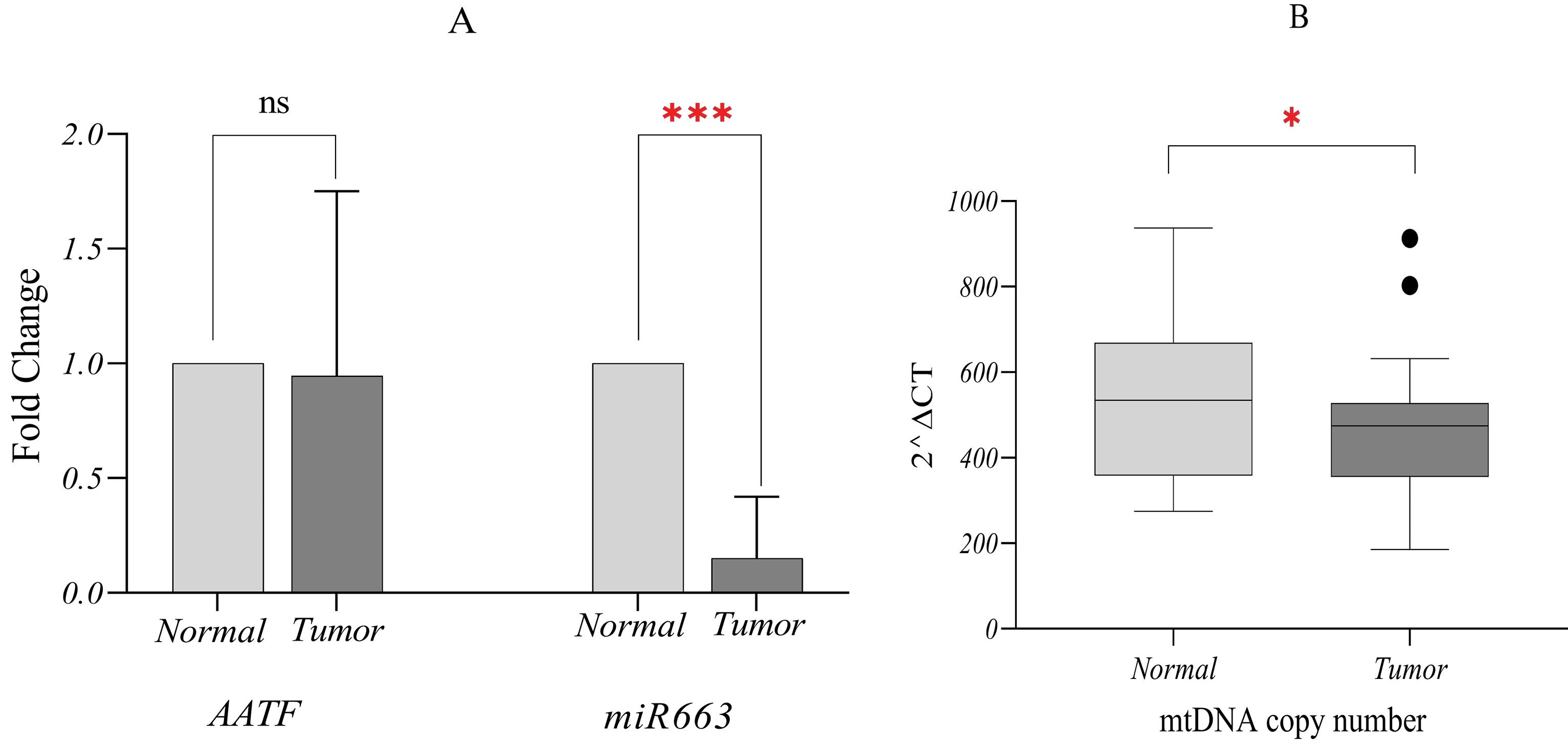
Fig. 2.
Relative expression analysis normalized to internal controls. (A) Significant decrease in the median expression of miR663 in tumor samples compared to adjacent non-tumor tissues. No statistically significant difference was observed in AATF median expression between tumor and adjacent non-tumor tissues. (B) Differences in median mtDNA copy number between tumor and adjacent non-tumor tissues. The median mtDNA copy number in tumor tissues was approximately 474.6 copies per cell, showing a reduction compared to adjacent non-tumor tissues, which had 534.9 copies per cell. ٭0.01 < P ≤ 0.05, ٭٭٭ 0.0001 < P ≤ 0.001.
.
Relative expression analysis normalized to internal controls. (A) Significant decrease in the median expression of miR663 in tumor samples compared to adjacent non-tumor tissues. No statistically significant difference was observed in AATF median expression between tumor and adjacent non-tumor tissues. (B) Differences in median mtDNA copy number between tumor and adjacent non-tumor tissues. The median mtDNA copy number in tumor tissues was approximately 474.6 copies per cell, showing a reduction compared to adjacent non-tumor tissues, which had 534.9 copies per cell. ٭0.01 < P ≤ 0.05, ٭٭٭ 0.0001 < P ≤ 0.001.
For AATF mRNA quantification, PUM1 was used as the internal control. The analysis showed no significant change in AATF expression (fold change = 0.886, P = 0.668) (Fig. 2A). Although REST software did not indicate a significant increase, mean fold-change calculations suggested AATF overexpression. To further investigate AATF mRNA levels in breast cancer, TCGA data were analyzed, and the findings are presented in the following sections.
Investigation of mtDNA copy number variations
The standard curves for DNA concentrations and the melt curves for COX1 and COX4 amplification are presented in Fig. 3. COX1 exhibited a higher amplification rate than the COX4 nuclear control gene.
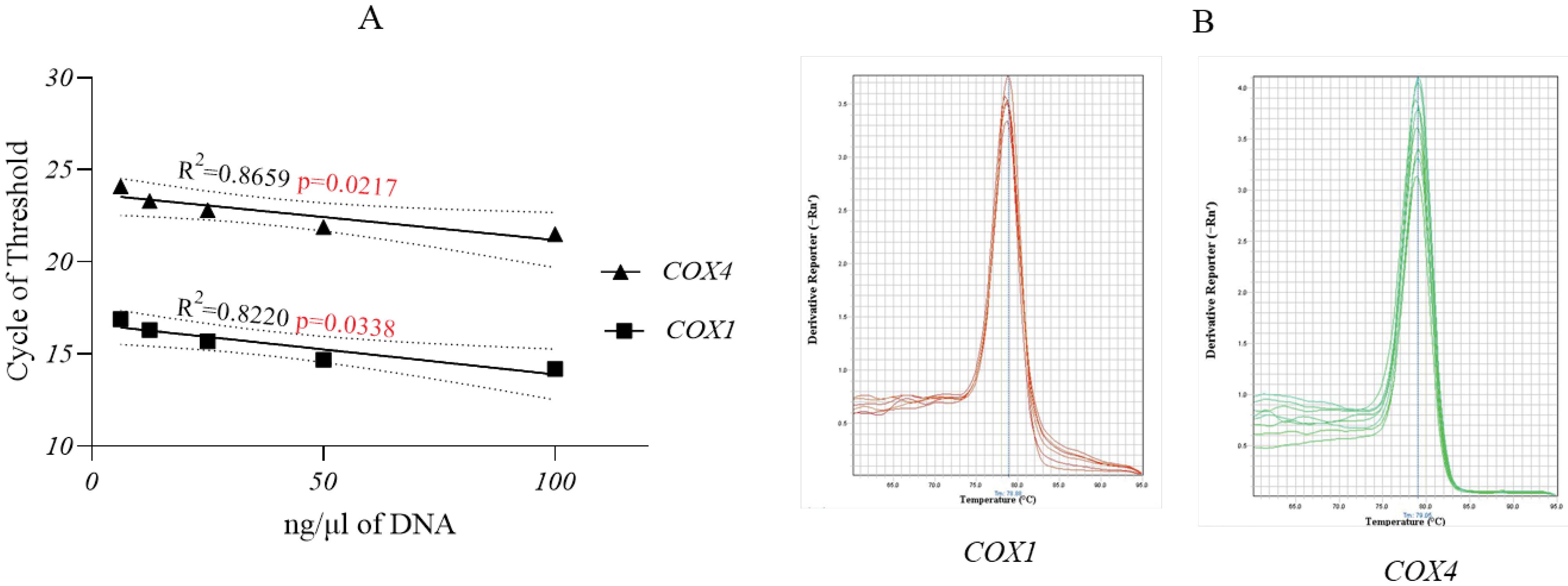
Fig. 3.
mtDNA copy number analysis. (A) Standard curves of five DNA serial dilutions tested for the COX1 and COX4 genes. The P-value was 0.021 for COX4 and 0.033 for COX1 indicating a strong link between the DNA concentration and amplification rate (B) Melt curves of COX1 and COX4 gene amplifications.
.
mtDNA copy number analysis. (A) Standard curves of five DNA serial dilutions tested for the COX1 and COX4 genes. The P-value was 0.021 for COX4 and 0.033 for COX1 indicating a strong link between the DNA concentration and amplification rate (B) Melt curves of COX1 and COX4 gene amplifications.
Using the 2∆Ct method, the median mtDNA copy number in tumor tissues was 474.6 copies per cell, compared to 534.9 copies per cell in adjacent non-tumor tissues. The median mtDNA copy number in tumor samples was analyzed using GraphPad Prism software, revealing a significant difference when outlier points were excluded. These findings align with previous studies reporting a reduction in mtDNA copy number in breast tumor tissues (Fig. 2B).
Association between miR663, AATF expression changes, and mtDNA copy number
Spearman's correlation analysis revealed a significant association between miR663 and AATF expression (P= 0.012). Additionally, the nonparametric Wilcoxon test indicated a statistically significant difference between miR663 and AATF expression (P= 0.007, Z = −2.711). A significant difference was also observed between miR663 expression and mtDNA copy number (P =0.017, Z = −2.386).
However, based on Spearman's correlation and Wilcoxon tests, no statistically significant correlation was detected between mtDNA content and AATF mRNA expression. Although the two components at the beginning and end of the proposed mtDNA/miR663/AATF axis did not exhibit a direct statistical correlation, it is plausible that while the decrease in miR663 did not lead to a significant increase in AATF mRNA, it may have contributed to an increase in AATFprotein levels. Further research should investigate this potential regulatory mechanism at the AATF protein level.
Correlation between tumor grades and miR663, AATF expression
Clinical data indicated that most tumors were classified as grade II. The correlation between tumor grade and the expression of miR663 and AATF was assessed using the Kruskal-Wallis test in SPSS software. The results demonstrated a progressive decrease in miR663 mean fold-change levels and a corresponding increase in AATF mean fold-change with increasing tumor grade (Fig. 4).
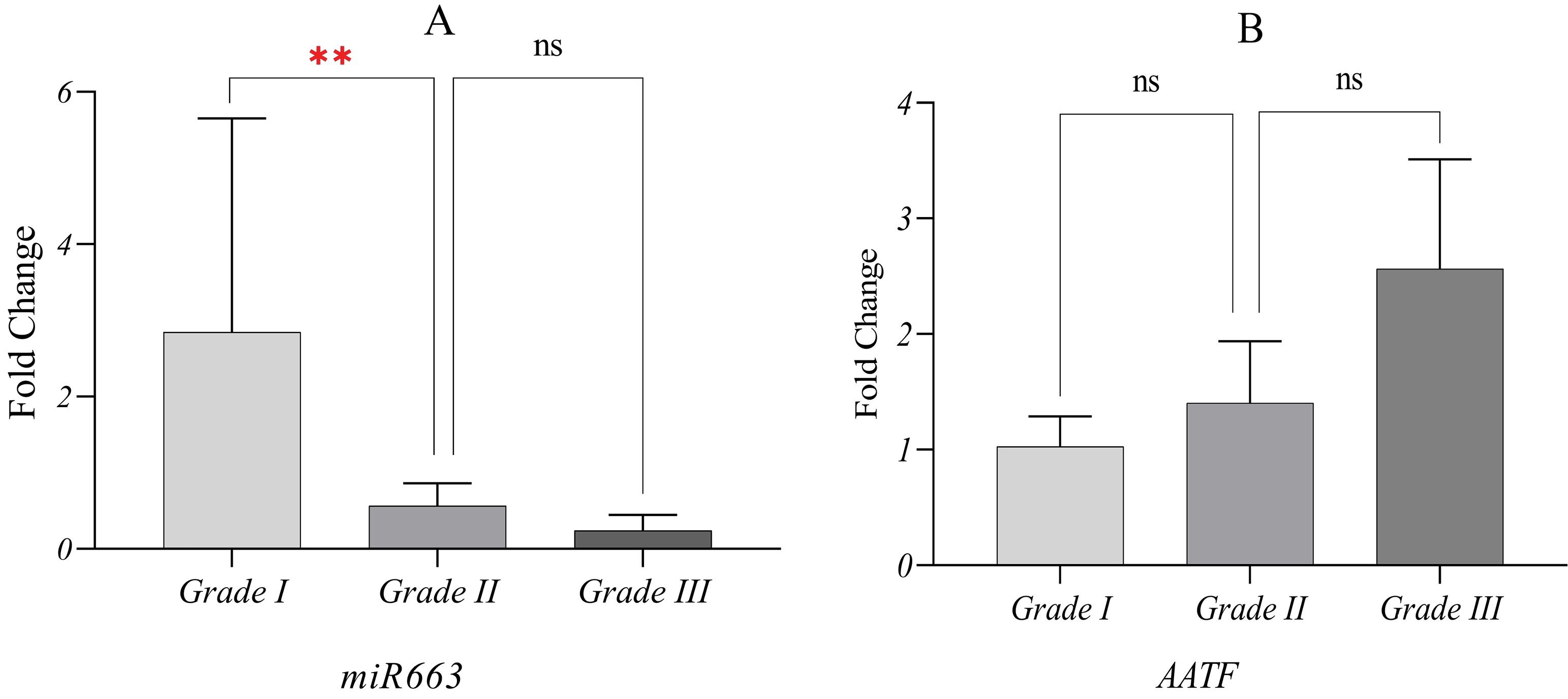
Fig. 4.
Mean fold-change of miR663 and AATF based on tumor pathological grade. The significance of group differences was analyzed using the Kruskal-Wallis test, and standard errors of the mean (SEM) are presented. (A) With decreased tumor cell differentiation, miR663 expression declined. The P-value was 0.00 for the comparison between Grade I and Grade II. The difference between Grade I and Grade III also yielded the same rate with a P-value of 0.00 (not indicated). The difference between grade II and grade III was not statistically significant (P = 0.42) (B) No statistically approved link between AATF expression and tumor grade was observed. However, it was found that as tumor cell differentiation decreased, AATF was overexpressed. ٭٭ 0.001 < P ≤ 0.01, ns; not significant P > 0.05.
.
Mean fold-change of miR663 and AATF based on tumor pathological grade. The significance of group differences was analyzed using the Kruskal-Wallis test, and standard errors of the mean (SEM) are presented. (A) With decreased tumor cell differentiation, miR663 expression declined. The P-value was 0.00 for the comparison between Grade I and Grade II. The difference between Grade I and Grade III also yielded the same rate with a P-value of 0.00 (not indicated). The difference between grade II and grade III was not statistically significant (P = 0.42) (B) No statistically approved link between AATF expression and tumor grade was observed. However, it was found that as tumor cell differentiation decreased, AATF was overexpressed. ٭٭ 0.001 < P ≤ 0.01, ns; not significant P > 0.05.
Consistent with overall findings, miR663 mean fold-change was significantly reduced in grade II and grade III tumors, whereas grade I tumors increased miR663 expression, reaching levels comparable to non-tumor tissues or even higher.
Based on available evidence, as tumor cells progress to higher grades and lose differentiation, the inhibition of miR663 intensifies, weakening its tumor-suppressive effects. This reduction in miR663 removes its inhibitory influence on AATF, allowing the anti-apoptotic properties of tumor cells to develop.
Analysis of TCGA data
The analysis of TCGA data revealed that AATF expression was significantly higher in tumors than in normal tissues, slightly increasing as the disease stage progressed. The median AATF expression in normal samples was 38.5 transcripts per million (TPM). However, expression levels increased to 45.6, 49.3, 49.1, and 50.0 TPM in stages I, II, III, and IV, respectively (Fig. 5A).
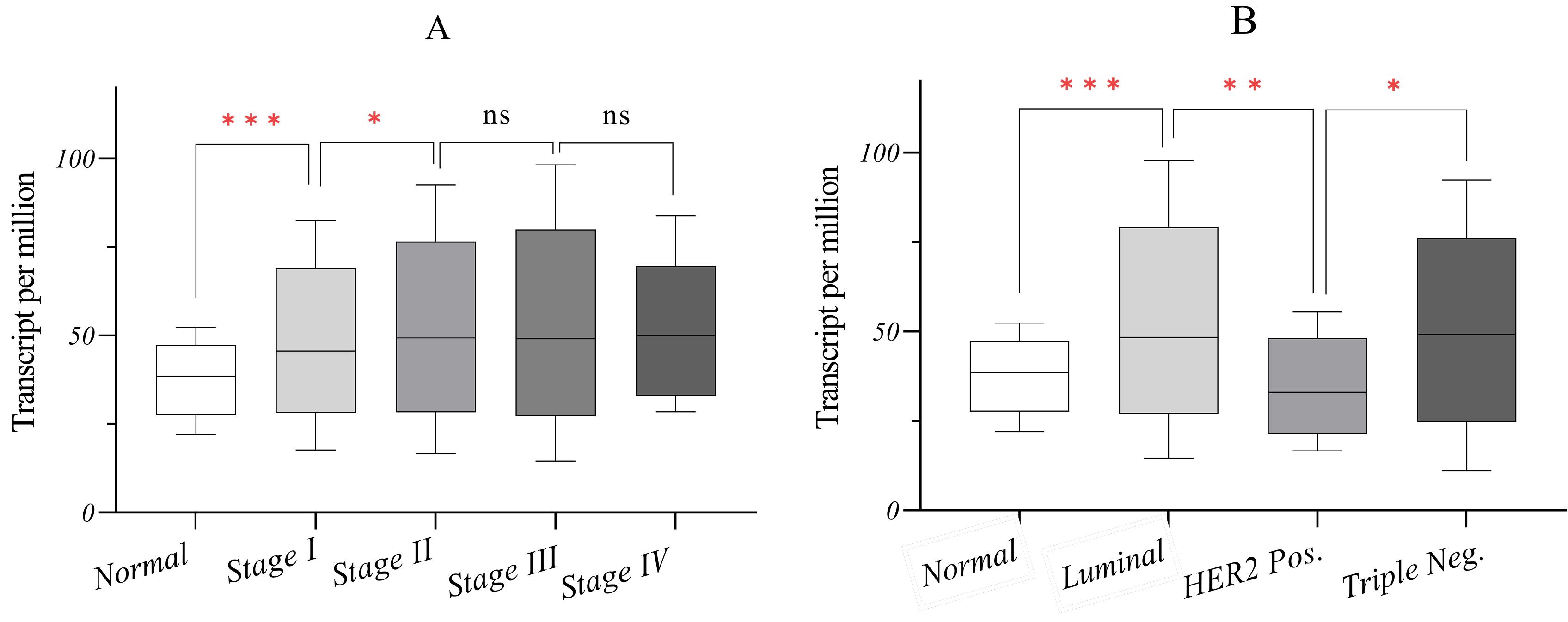
Fig. 5.
In silico analysis of AATF expression in breast cancer based on TCGA data. (A) AATF expression significantly increases in tumors compared to normal breast tissues. (B) AATF expression is reduced in HER2 + tumors, whereas luminal and TNBC tumors exhibit elevated expression levels. ٭0.01 < P ≤ 0.05, ٭٭ 0.001 < P ≤ 0.01, ٭٭٭ 0.0001 < P ≤ 0.001, ns; not significant P > 0.05.
.
In silico analysis of AATF expression in breast cancer based on TCGA data. (A) AATF expression significantly increases in tumors compared to normal breast tissues. (B) AATF expression is reduced in HER2 + tumors, whereas luminal and TNBC tumors exhibit elevated expression levels. ٭0.01 < P ≤ 0.05, ٭٭ 0.001 < P ≤ 0.01, ٭٭٭ 0.0001 < P ≤ 0.001, ns; not significant P > 0.05.
In addition, AATF expression varied across breast cancer subtypes. While AATF levels were reduced in HER2 + tumors, they were elevated in luminal and TNBC tumors compared to normal tissues. The median AATF expression was 48.3 TPM in luminal tumors, 33.07 TPM in HER2 + tumors, and 49.1 TPM in TNBC tumors (Fig. 5B).
These findings are noteworthy, as when all subtypes are analyzed collectively, the data appear normalized, reflecting a relative increase in AATF expression. However, when analyzed separately, a significant increase in luminal and TNBC subtypes becomes evident despite their distinct pathogenesis. Conversely, AATF expression in HER2 + tumors is markedly reduced, reaching levels comparable to or even lower than those in normal tissues.
Discussion
Identifying miR663 as a target of mitochondrial retrograde signaling in breast tumor cells has introduced new insights into mitochondrial–nuclear crosstalk.10 This study demonstrated the downregulation of mtDNA/miR663 in primary breast cancer tissues. The reduction in miR663 expression was substantial, nearly diminishing in tumors with higher pathological grades. These findings, consistent with previous studies, suggest that mitotherapy and miRNA-based therapies could be potential strategies for breast cancer treatment.10,20
A comprehensive study by Carden et al established miR663 activity in retrograde signaling and revealed clinically relevant findings. In silico analysis of TCGA data showed a significant reduction in miR663 expression in metastatic tumors compared to primary tumors. Furthermore, an increase in miR663 expression was observed in stage II tumors compared to stage I, followed by a significant decline in stages III and IV. Kaplan-Meier survival analysis indicated that higher miR663 expression was associated with more prolonged patient survival.10
A 2021 study by Wang et al further demonstrated that reduced miR663a expression was significantly associated with lymph node metastasis and poor survival outcomes in breast cancer patients. The researchers proposed miR663a as an independent prognostic factor for breast cancer.20
Experimental evidence has established a correlation between mtDNA copy number variations and breast cancer; however, findings across blood and tumor tissue studies remain inconsistent. Several studies in breast cancer patients have reported increased mtDNA copy numbers in blood samples compared to matched controls.21-23 A comparative study analyzing breast cancer patient samples found that mtDNA copy numbers in buffy coat and tumor tissues were lower than those in serum and adjacent non-tumor tissue.24 A recent study identified an inverse correlation between leukocyte mtDNA copy number and breast cancer patient survival.25 Rai et al conducted a simultaneous analysis of blood and tumor tissue samples from patients with metastatic breast cancer, confirming that mtDNA copy number is regulated independently in blood and tumor tissues. Their findings indicated a decrease in mtDNA copy number in blood but an increase in tumor tissue.26 A study on Mexican breast cancer patients reported a reduction in tumor mtDNA copy number, along with three deletions at A249del, A290del, and A291del, as well as C16327T mutations in mtDNA.27
Our investigation of AATF, a key target of miR663, did not reveal any significant changes in mRNA expression. However, TCGA breast cancer data analysis showed that AATF expression varied across different subtypes. Specifically, AATF expression was markedly reduced in HER2 + tumors while elevated in luminal and TNBC subtypes compared to normal breast tissues.
Early studies on AATF function in breast cancer demonstrated its role as a stimulatory factor for ER expression.28 The immunoblotting analysis further confirmed AATF overexpression in the MCF-7 breast cancer cell line compared to the non-tumorigenic MCF-10A cell line.29 Beyond breast cancer, research in multiple myeloma has identified new roles for AATF, including reducing genomic instability and mitigating DNA damage by clearing R-loops (triple-stranded RNA: DNA hybrids).30 A separate study in non-mammary cell lines showed that AATF depletion led to histone deacetylation and reduced cell proliferation.31
A direct link between mitochondrial function and AATF was established in cisplatin-treated bladder cancer cell lines, where AATF inhibited drug-induced apoptosis by reducing cytochrome c levels, a key component of OXPHOS.32 A recent and interesting study disclosed new localization and function of AATF protein in mitochondria in interaction with HCLS1-associated protein X-1 (HAX1). HAX1 regulates cell migration, a key process in carcinogenesis and metastasis. Furthermore, it supports the anti-apoptotic activity exerted by AATF. They revealed that the HAX1/AATF complex was located in the mitochondria of the MCF-7 cell line, acting in cellular response to oxidative stress.12
Due to the limited sample size, this study had insufficient statistical power to fully elucidate the functional role of the mtDNA/miR663/AATF axis in breast cancer pathogenesis. Future studies focusing on AATF protein expression, tumor grade, and breast cancer subtypes are necessary to clarify further the impact of this axis on breast cancer progression.
Conclusion
We identified a significant reduction in miR663 and mtDNA copy number in breast tumors. A strong correlation was also observed between miR663 and AATF expression. During the experimental phase of our study we noticed that miR663 was significantly reduced in Grade II and III tumors compared to Grade I. In the bioinformatics analysis we found a significant increase in AATF in luminal and TNBC subtypes and a reduction in AATF in HER2 + tumors.
These results, combined with emerging evidence on the functional coordination of the mtDNA/miR663/AATF axis, suggest that mitotherapy and miRNA-based therapies could serve as potential strategic approaches for breast cancer treatment. This may contribute to the advancement of personalized management strategies for specific breast cancer subtypes.
Research Highlights
What is the current knowledge?
-
mtDNA copy number variation has been reported in various human cancers.
-
Mitochondrial retrograde signaling regulates miR663.
-
Increased miR663 is associated with improved breast cancer patient survival.
-
The AATF gene is a target of miR663.
-
AATF is primarily a nuclear protein but relocates to mitochondria in response to oxidative stress.
What is new here?
-
A significant reduction in miR663 expression and mtDNA copy number was identified in breast tumor tissues.
-
A strong correlation was observed between miR663 and AATF expression in breast tumors.
-
miR663 mean fold change was significantly reduced in Grade II and III tumors compared to Grade I.
-
AATF expression was reduced in HER2 + tumors but elevated in luminal and TNBC subtypes.
Competing Interests
The authors declare that there is no conflict of interests.
Ethical Approval
This study was approved by the Ethics Committee at Tarbiat Modares University in Tehran, Iran (IR.MODARES.REC.1399.131) and the patients signed the informed consent.
Acknowledgements
The authors would like to thank the patients who participated in this survey. Additionally, Graphical abstract image provided by Servier Medical Art (https://smart.servier.com/), used under a Creative Commons BY 4.0 license.
References
- Wang L, Zhang S, Wang X. The metabolic mechanisms of breast cancer metastasis. Front Oncol 2020; 10:602416. doi: 10.3389/fonc.2020.602416 [Crossref] [ Google Scholar]
- Weerts MJ, Sleijfer S, Martens JW. The role of mitochondrial DNA in breast tumors. Drug Discov Today 2019; 24:1202-8. doi: 10.1016/j.drudis.2019.03.019 [Crossref] [ Google Scholar]
- Giampazolias E, Tait SW. Mitochondria and the hallmarks of cancer. FEBS J 2016; 283:803-14. doi: 10.1111/febs.13603 [Crossref] [ Google Scholar]
- Dahi F, Mortezanejad S, Geranpayeh L, Shahbazi S. A review on the role of mitochondrial DNA mutations in cancer. Arch Med Lab Sci 2021; 7:1-12. doi: 10.22037/amls.v7.34736 [Crossref] [ Google Scholar]
- Hu L, Yao X, Shen Y. Altered mitochondrial DNA copy number contributes to human cancer risk: evidence from an updated meta-analysis. Sci Rep 2016; 6:35859. doi: 10.1038/srep35859 [Crossref] [ Google Scholar]
- Abd Radzak SM, Mohd Khair SZ, Ahmad F, Patar A, Idris Z, Mohamed Yusoff AA. Insights regarding mitochondrial DNA copy number alterations in human cancer (review). Int J Mol Med 2022; 50:104. doi: 10.3892/ijmm.2022.5160 [Crossref] [ Google Scholar]
- de Oliveira RC, Cavalcante GC, Soares-Souza GB. Exploring aerobic energy metabolism in breast cancer: a mutational profile of glycolysis and oxidative phosphorylation. Int J Mol Sci 2024; 25:12585. doi: 10.3390/ijms252312585 [Crossref] [ Google Scholar]
- Yang D, Kim J. Mitochondrial retrograde signalling and metabolic alterations in the tumour microenvironment. Cells 2019; 8:275. doi: 10.3390/cells8030275 [Crossref] [ Google Scholar]
- Suriya Muthukumaran N, Velusamy P, Akino Mercy CS, Langford D, Natarajaseenivasan K, Shanmughapriya S. MicroRNAs as regulators of cancer cell energy metabolism. J Pers Med 2022; 12:1329. doi: 10.3390/jpm12081329 [Crossref] [ Google Scholar]
- Carden T, Singh B, Mooga V, Bajpai P, Singh KK. Epigenetic modification of miR-663 controls mitochondria-to-nucleus retrograde signaling and tumor progression. J Biol Chem 2017; 292:20694-706. doi: 10.1074/jbc.M117.797001 [Crossref] [ Google Scholar]
- Bruno T, De Angelis R, De Nicola F, Barbato C, Di Padova M, Corbi N. Che-1 affects cell growth by interfering with the recruitment of HDAC1 by Rb. Cancer Cell 2002; 2:387-99. doi: 10.1016/s1535-6108(02)00182-4 [Crossref] [ Google Scholar]
- Pisani C, Onori A, Gabanella F, Iezzi S, De Angelis R, Fanciulli M. HAX1 is a novel binding partner of Che-1/AATF Implications in oxidative stress cell response. Biochim Biophys Acta Mol Cell Res 2024; 1871:119587. doi: 10.1016/j.bbamcr.2023.119587 [Crossref] [ Google Scholar]
- Benakanakere MR, Zhao J, Finoti L, Schattner R, Odabas-Yigit M, Kinane DF. MicroRNA-663 antagonizes apoptosis antagonizing transcription factor to induce apoptosis in epithelial cells. Apoptosis 2019; 24:108-18. doi: 10.1007/s10495-018-01513-9 [Crossref] [ Google Scholar]
- Kumar DP, Santhekadur PK, Seneshaw M, Mirshahi F, Uram-Tuculescu C, Sanyal AJ. A regulatory role of apoptosis antagonizing transcription factor in the pathogenesis of nonalcoholic fatty liver disease and hepatocellular carcinoma. Hepatology 2019; 69:1520-34. doi: 10.1002/hep.30346 [Crossref] [ Google Scholar]
- Srinivas AN, Suresh D, Vishwanath PM, Satish S, Santhekadur PK, Koka S. TACE inhibition: a promising therapeutic intervention against AATF-mediated steatohepatitis to hepatocarcinogenesis. Mol Oncol 2024; 18:1940-57. doi: 10.1002/1878-0261.13646 [Crossref] [ Google Scholar]
- Haanpää M, Reiman M, Nikkilä J, Erkko H, Pylkäs K, Winqvist R. Mutation analysis of the AATF gene in breast cancer families. BMC Cancer 2009; 9:457. doi: 10.1186/1471-2407-9-457 [Crossref] [ Google Scholar]
- Jain A, Bakhshi S, Thakkar H, Gerards M, Singh A. Elevated mitochondrial DNA copy numbers in pediatric acute lymphoblastic leukemia: a potential biomarker for predicting inferior survival. Pediatr Blood Cancer 2018; 65. doi: 10.1002/pbc.26874.
- Guha M, Srinivasan S, Raman P, Jiang Y, Kaufman BA, Taylor D. Aggressive triple negative breast cancers have unique molecular signature on the basis of mitochondrial genetic and functional defects. Biochim Biophys Acta Mol Basis Dis 2018; 1864:1060-71. doi: 10.1016/j.bbadis.2018.01.002 [Crossref] [ Google Scholar]
- Mohamadalizadeh-Hanjani Z, Shahbazi S, Geranpayeh L. Investigation of the SPAG5 gene expression and amplification related to the NuMA mRNA levels in breast ductal carcinoma. World J Surg Oncol 2020; 18:225. doi: 10.1186/s12957-020-02001-8 [Crossref] [ Google Scholar]
- Wang G, Chen L, Jian W, Fang L. Low expression of miR-663a indicates poor prognosis and promotes cell proliferation, migration, and invasion in breast cancer. Oncol Res Treat 2021; 44:119-27. doi: 10.1159/000513405 [Crossref] [ Google Scholar]
- Shen J, Platek M, Mahasneh A, Ambrosone CB, Zhao H. Mitochondrial copy number and risk of breast cancer: a pilot study. Mitochondrion 2010; 10:62-8. doi: 10.1016/j.mito.2009.09.004 [Crossref] [ Google Scholar]
- Thyagarajan B, Wang R, Nelson H, Barcelo H, Koh WP, Yuan JM. Mitochondrial DNA copy number is associated with breast cancer risk. PLoS One 2013; 8:e65968. doi: 10.1371/journal.pone.0065968 [Crossref] [ Google Scholar]
- Shen J, Wan J, Song R, Zhao H. Peripheral blood mitochondrial DNA copy number, length heteroplasmy and breast cancer risk: a replication study. Carcinogenesis 2015; 36:1307-13. doi: 10.1093/carcin/bgv130 [Crossref] [ Google Scholar]
- Xia P, Wang HJ, Geng TT, Xun XJ, Zhou WJ, Jin TB, et al. Mitochondrial DNA levels in blood and tissue samples from breast cancer patients of different stages. Asian Pac J Cancer Prev 2014. 15: 1339-44. doi: 10.7314/apjcp.2014.15.3.1339.
- Zhang W, Lin S, Zeng B, Chen X, Chen L, Chen M. High leukocyte mitochondrial DNA copy number contributes to poor prognosis in breast cancer patients. BMC Cancer 2023; 23:377. doi: 10.1186/s12885-023-10838-x [Crossref] [ Google Scholar]
- Rai NK, Panjwani G, Ghosh AK, Haque R, Sharma LK. Analysis of mitochondrial DNA copy number variation in blood and tissue samples of metastatic breast cancer patients (A pilot study). Biochem Biophys Rep 2021; 26:100931. doi: 10.1016/j.bbrep.2021.100931 [Crossref] [ Google Scholar]
- Domínguez-de-la-Cruz E, de Lourdes Muñoz M, Pérez-Muñoz A, García-Hernández N, Moctezuma-Meza C, Hinojosa-Cruz JC. Reduced mitochondrial DNA copy number is associated with the haplogroup, and some clinical features of breast cancer in Mexican patients. Gene 2020; 761:145047. doi: 10.1016/j.gene.2020.145047 [Crossref] [ Google Scholar]
- Sharma M. Apoptosis-antagonizing transcription factor (AATF) gene silencing: role in induction of apoptosis and down-regulation of estrogen receptor in breast cancer cells. Biotechnol Lett 2013; 35:1561-70. doi: 10.1007/s10529-013-1257-8 [Crossref] [ Google Scholar]
- Pan X, Hong X, Li S, Meng P, Xiao F. METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. Exp Mol Med 2021; 53:91-102. doi: 10.1038/s12276-020-00510-w [Crossref] [ Google Scholar]
- Bruno T, Corleone G, Catena V, Cortile C, De Nicola F, Fabretti F. AATF/Che-1 localizes to paraspeckles and suppresses R-loops accumulation and interferon activation in multiple myeloma. EMBO J 2022; 41:e109711. doi: 10.15252/embj.2021109711 [Crossref] [ Google Scholar]
- Catena V, Bruno T, Iezzi S, Matteoni S, Salis A, Sorino C. CK2-mediated phosphorylation of Che-1/AATF is required for its pro-proliferative activity. J Exp Clin Cancer Res 2021; 40:232. doi: 10.1186/s13046-021-02038-x [Crossref] [ Google Scholar]
- Tan S, Fu L, Dong Q. AATF is overexpressed in human bladder cancer and regulates chemo-sensitivity through survivin. Onco Targets Ther 2021; 14:5493-505. doi: 10.2147/ott.S319734 [Crossref] [ Google Scholar]