Bioimpacts. 2025;15:31211.
doi: 10.34172/bi.31211
Review
Biomarkers for colorectal cancer detection: An insight into colorectal cancer and FDA-approved biomarkers
Mohammad Yasin Zamanian Conceptualization, Project administration, Supervision, Writing – original draft, Writing – review & editing, 1, 2, * 
Darmadi Darmadi Validation, Writing – review & editing, 3
Razieh Darabi Validation, Writing – review & editing, 4
Raed Fanoukh Aboqader Al-Aouadi Data curation, Resources, 5
Mehraveh Sadeghi Ivraghi Data curation, Resources, 6
Esra Küpeli Akkol Conceptualization, Project administration, Supervision, Writing – original draft, Writing – review & editing, 7, * 
Author information:
1Department of Physiology, School of Medicine, Hamadan University of Medical Sciences, Hamadan 6718773654, Iran
2Department of Pharmacology and Toxicology, School of Pharmacy, Hamadan University of Medical Sciences, Hamadan 6718773654, Iran
3Department of Internal Medicine, Faculty of Medicine, Universitas Sumatera Utara, Medan, Indonesia
4Department of Internal Medicine, School of Medicine, Hamadan University of Medical Sciences, Hamadan 6718773654, Iran
5College of Medicine, Al-Ayen Iraqi University, AUIQ, An Nasiriyah, Iraq
6School of Medicine, Qazvin University of Medical Sciences, Qazvin, Iran
7Department of Pharmacognosy, Faculty of Pharmacy, Gazi University, Ankara 06330, Türkiye
Abstract
Colorectal cancer (CRC) constitutes a significant global health challenge, accounting for a considerable proportion of cancer cases and associated mortality. Projections indicate a potential increase in new cases by 2040, attributed to demographic factors such as aging and population growth. Although advancements in the understanding of CRC pathophysiology have broadened treatment options, challenges such as drug resistance and adverse effects persist, highlighting the necessity for enhanced diagnostic methodologies. Timely detection markedly improves survival rates; however, colonoscopy, regarded as the gold standard for CRC screening, is constrained by its invasiveness and reliance on practitioner expertise. Consequently, the development of novel diagnostic approaches is imperative. Cancer biomarkers, which serve as indicators of cancer progression, show significant promise for improving diagnosis, prognosis, and treatment strategies. This study investigates molecular and cellular biomarkers, including proteins, DNA mutations, methylation markers, and microRNAs, that are pivotal in precision medicine and the monitoring of CRC progression. Additionally, emerging biomarkers such as circular RNAs (circRNAs) and long non-coding RNAs (lncRNAs) present opportunities for early detection. Current Food and Drug Administration (FDA)-approved CRC biomarkers reflect a shift towards personalized medicine, enhancing patient compliance and clinical outcomes. Nevertheless, further research is essential for the discovery of novel biomarkers and for deepening the understanding of CRC etiology, thereby advancing personalized care. Addressing standardization challenges will be crucial for ensuring global patient access to biomarker-based strategies.
Keywords: Colorectal cancer, Biomarkers, FDA-approved, Epigenetic, Liquid biopsy
Copyright and License Information
© 2025 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Funding Statement
None.
Introduction
In recent years, colorectal cancer (CRC) has become a major concern and a leading cause of cancer-related deaths worldwide.1 In 2020, CRC accounted for 10% of global cancer cases and 9.4% of cancer-related fatalities.2 Experts project a substantial increase in the incidence of new CRC cases worldwide by the year 2040. This forecast considers various factors, including demographic aging, population growth, and advancements in human development.2,3 Significant advancements have been made in the understanding of CRC pathophysiology, leading to an expanded array of treatment options that include endoscopic and surgical excision, radiotherapy, immunotherapy, and targeted therapy.4,5 Although these treatments have effectively halted the progression of cancer, they are associated with limitations such as drug resistance and undesirable side effects in the context of CRC.1,6 However, early detection significantly increases the five-year survival rate to approximately 90%.7 Colonoscopy is widely regarded as the most reliable method for CRC screening. However, this procedure may be invasive and uncomfortable for patients, and its accuracy is significantly affected by the operator's expertise and experience.8 In light of these clinical challenges, it is imperative to identify innovative approaches that enhance the rapid diagnosis and monitoring of patient prognosis.
A cancer biomarker is a quantifiable characteristic that indicates the potential development, incidence, or outcome of cancer.9 These characteristics can be classified into molecular, cellular, physiological, or imaging-based categories.9 The primary objective of this study is to investigate cancer biomarkers at the molecular and cellular levels. One aspect of biomarker testing in oncology involves analyzing tumors or bodily fluids to identify alterations in DNA, RNA, proteins, or other biomolecules. These changes can provide valuable insights for cancer diagnosis, prognosis, precision medicine, guiding treatment strategies, predicting drug responses, and monitoring cancer progression.10,11 Currently, researchers have explored various categories of colorectal biomarkers, including proteins, mutations, methylation markers, microRNAs, volatile organic compounds, and alterations in gut microbiome composition.12,13
This review aims to provide a comprehensive overview of the prognostic and predictive biomarkers currently used in clinical settings while also examining emerging biomarkers that may have significant implications for the monitoring and treatment of CRC patients. Furthermore, this review will address the major challenges encountered in biomarker research and their application in clinical practice.
Search strategy
A comprehensive literature search was conducted to identify relevant studies for this review. The following electronic databases were searched: PubMed, Google Scholar, Scopus, and Web of Science. The search terms included a combination of keywords and Medical Subject Headings (MeSH) such as “liquid biopsy,” “colorectal cancer,” “biomarker,” “cancer biomarkers,” “clinical implementation,” and “FDA.” Boolean operators (AND, OR) were used to combine terms as appropriate.
The inclusion criteria were (1) original research articles published in English; (2) studies focusing on the use of biomarkers in cancer diagnosis, prognosis, or treatment monitoring; and (3) studies involving human subjects. Exclusion criteria included (1) case reports, editorials, or conference abstracts; (2) studies not directly related to the clinical application of liquid biopsy; and (3) articles without available full text.
Titles and abstracts were screened for relevance, followed by full-text review of potentially eligible articles. Reference lists of included studies were also examined to identify additional relevant publications. Discrepancies in study selection were resolved through discussion among the authors.
Overview of biomarkers
There are specific germline or hereditary variations that increase the risk of cancer.14 Based on their frequency and ability to induce disease, germline variations can be categorized into three categories: uncommon variants exhibiting significant penetration, relatively common variants with moderate penetration, and common variants with limited penetration.15 The first ones have a significant correlation with cancer-predisposing syndromes and hereditary malignancies, making them excellent candidates for use as biomarkers for cancer risk assessment. Different cancer types might be more or less susceptible to germline variations. For instance, Lynch syndrome-associated mutations in the genes epithelial cell adhesion molecule(EPCAM), MutL protein 1 (MLH1), MutS homolog 2 (MSH2), mutS homolog 6 (MSH6), and postmeiotic segregation increased 2 (PMS2) are more strongly associated with CRC than pancreatic cancer (PC).15,16 In addition to helping determine a person's vulnerability to cancer, germline genetic markers are significant prognostic and predictive indicators for targeted treatments.17 Many cancers can develop due to the accumulation of somatic mutations, some unique to a specific type of cancer and others common among different types of cancer. Chromosomal abnormalities encompass both numerical and structural variations, serving as significant biomarkers.18-20 The translocation of chromosomes 22 and 9, resulting in the fusion of breakpoint cluster region-Abelson murine leukemia viral oncogene homolog (BCR-ABL), led to the discovery of the Philadelphia chromosome as the initial chromosomal abnormality in cancer.21 Fig. 1 illustrates the translocation between chromosome 22 and chromosome 9.
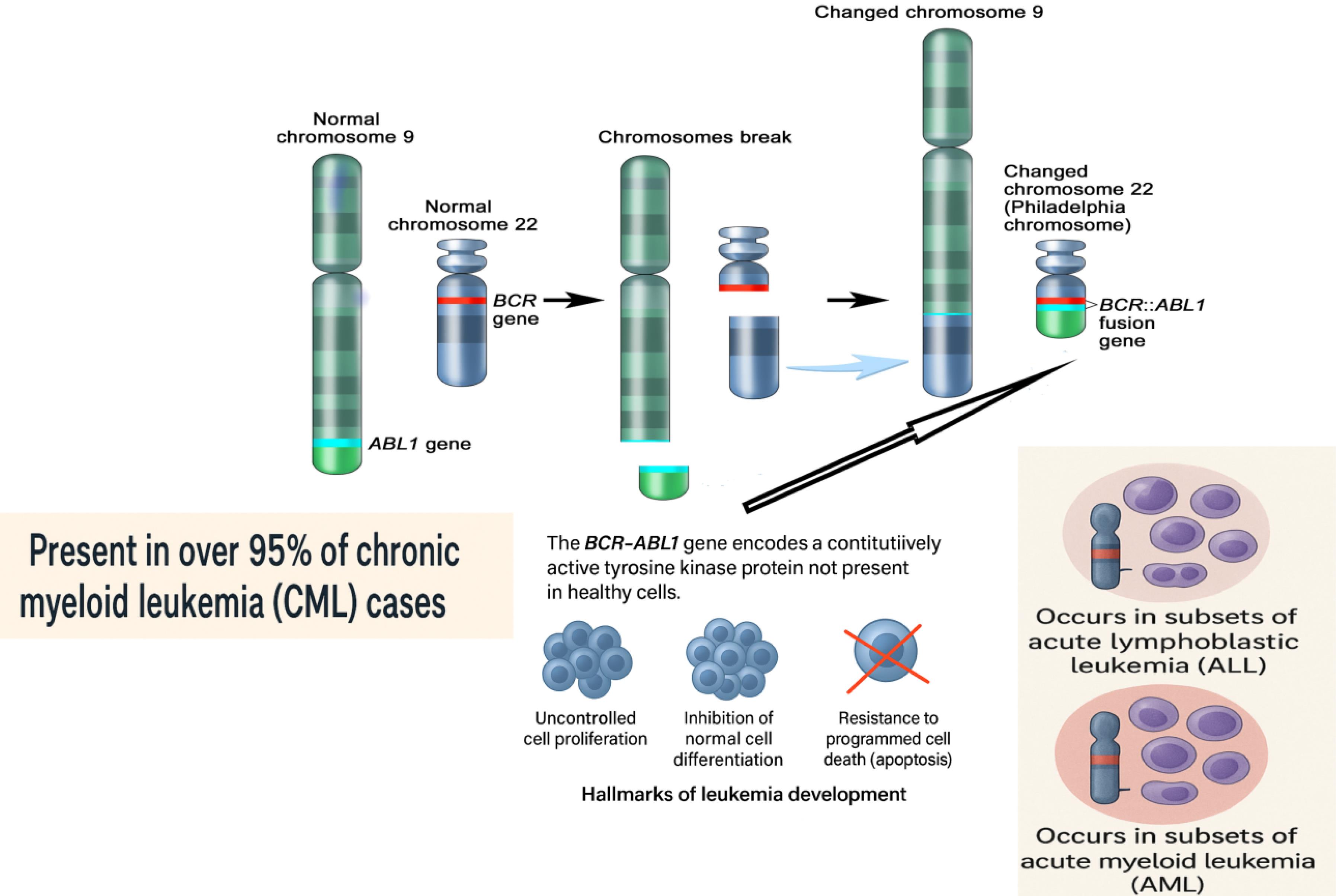

Fig. 1.
The Philadelphia chromosome arises from a reciprocal translocation between the long arms of chromosome 9 and chromosome 22, cytogenetically designated as t(9;22)(q34;q11). During this chromosomal rearrangement, a segment of the ABL1 gene from chromosome 9 is translocated to chromosome 22, where it fuses with the BCR gene. This event produces a shortened, abnormal chromosome 22 known as the Philadelphia chromosome and a derivative, elongated chromosome 9. The fusion of the BCR and ABL1 genes on the Philadelphia chromosome results in the formation of the BCR-ABL1 fusion gene, which encodes a constitutively active tyrosine kinase protein. Unlike normal tyrosine kinases, this fusion protein is always active, leading to uncontrolled cell proliferation, inhibition of normal cell differentiation, and resistance to apoptosis. These effects are central to the development of leukemia. The Philadelphia chromosome is present in over 95% of chronic myeloid leukemia (CML) cases and is also found in subsets of acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML). Different breakpoints within the BCR gene result in various BCR-ABL1 fusion transcripts, including p210, which is typical in classical CML and some Philadelphia chromosome-positive ALL cases; p190, which is most common in Philadelphia chromosome-positive ALL and rare in CML; and p230, which is associated with chronic neutrophilic leukemia and rare CML variants. The presence of the Philadelphia chromosome is a defining diagnostic marker for CML and plays a critical role in guiding prognosis and treatment, particularly with the use of targeted tyrosine kinase inhibitors.
.
The Philadelphia chromosome arises from a reciprocal translocation between the long arms of chromosome 9 and chromosome 22, cytogenetically designated as t(9;22)(q34;q11). During this chromosomal rearrangement, a segment of the ABL1 gene from chromosome 9 is translocated to chromosome 22, where it fuses with the BCR gene. This event produces a shortened, abnormal chromosome 22 known as the Philadelphia chromosome and a derivative, elongated chromosome 9. The fusion of the BCR and ABL1 genes on the Philadelphia chromosome results in the formation of the BCR-ABL1 fusion gene, which encodes a constitutively active tyrosine kinase protein. Unlike normal tyrosine kinases, this fusion protein is always active, leading to uncontrolled cell proliferation, inhibition of normal cell differentiation, and resistance to apoptosis. These effects are central to the development of leukemia. The Philadelphia chromosome is present in over 95% of chronic myeloid leukemia (CML) cases and is also found in subsets of acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML). Different breakpoints within the BCR gene result in various BCR-ABL1 fusion transcripts, including p210, which is typical in classical CML and some Philadelphia chromosome-positive ALL cases; p190, which is most common in Philadelphia chromosome-positive ALL and rare in CML; and p230, which is associated with chronic neutrophilic leukemia and rare CML variants. The presence of the Philadelphia chromosome is a defining diagnostic marker for CML and plays a critical role in guiding prognosis and treatment, particularly with the use of targeted tyrosine kinase inhibitors.
Genetic alterations frequently utilized as cancer biomarkers include mutations that impact a single nucleotide or a limited number of nucleotides.22 Cell-free DNA, originating from the lysis of cancer cells or active secretion, is termed circulating tumor DNA (ctDNA) and holds promise as a biomarker for cancer.23,24 Detecting ctDNA presents significant challenges due to its low concentration, necessitating the employment of highly sensitive methodologies.25 To develop effective cancer biomarkers, it is essential to consider various factors, including tumor DNA content, sample type, and detection techniques. DNA sequencing and polymerase chain reaction (PCR)-based methods are frequently utilized to investigate small DNA alterations, whereas fluorescence in situ hybridization (FISH) and array comparative genomic hybridization are employed for the analysis of larger DNA fragments.26
Protein biomarkers in cancer encompass a diverse array of proteins reflective of specific cancer characteristics. These proteins can be identified through tumor tissue analysis.27 Immunohistochemistry (IHC) is typically employed to assess proteins within tumor tissues, while the enzyme-linked immunosorbent assay (ELISA) is a widely used method for analyzing protein biomarkers in bodily fluids.28 Also, various techniques such as chromatography, Western blotting, laser capture microdissection (LCM), protein microarrays, and gel-based approaches are available for measuring proteins in small sample sizes.29 A comprehensive proteomics map study involving 949 human cell lines has developed a wide range of 189 biomarkers for drug sensitivity prediction, significantly contributing to cancer research and drug application.30 The Food and Drug Administration (FDA) has approved the programmed death-1/programmed death-ligand 1 (PD-1/PD-L1) antibody for the initial or second course of treatment for several types of cancer.31 Regarding gastroesophageal malignancies, a comprehensive evaluation is conducted to determine the most suitable immunotherapy treatment. This evaluation includes assessing microsatellite instability (MSI) or DNA mismatch repair (MMR), tumor mutational burden, and PD-L1 expression.32
Epigenetic variations lead to alterations in DNA methylation and modifications of histone proteins, all without affecting the underlying DNA coding sequence. However, these changes significantly influence the structure and stability of DNA, playing a pivotal role in the pathogenesis of cancer.33,34 MLH1 methylation serves as a widely recognized biomarker in clinical settings for the assessment of Lynch syndrome, a hereditary condition linked to an elevated risk of cancer.34 Moreover, DNA methylation biomarkers hold considerable potential in forecasting the efficacy of cancer therapies.35 One of the notable advantages of these diagnostic tests is their capacity to accurately identify cancer in its early stages and to detect any residual disease. The FDA has sanctioned several biomarkers for methylation-based testing. These include septin 9 (SEPT9), which can be detected in plasma utilizing the Epi proColon test, as well as a combination of NDRG family member 4 (NDRG4) and bone morphogenetic protein 3 (BMP3), which can be found in stool samples for screening purposes.34
It is noteworthy that less than 2% of transcripts are protein-coding genes that generate translational messenger RNA (mRNA) transcripts.36 Microarrays and RNA sequencing are advanced techniques used to analyze the transcriptome.37 Micro RNAs (miRNAs) are approximately 22 nucleotides long and have a crucial role in regulating various cellular processes.38,39 They control protein expression by modulating mRNA expression, thereby influencing cell growth, cell cycle, cell differentiation, apoptosis, invasion, and metastasis.39-41 When it comes to diagnosing CRC, several studies have found that the expression of certain miRNAs in patients significantly differs from that in the general population. These variations can serve as biomarkers for the early detection of CRC and hold significant importance in the early diagnosis and prognosis.39 CircRNA is a recently discovered form of single-stranded RNA. These circRNAs have the potential to serve as biomarkers for early cancer detection.42-44 The potential of circRNAs as biomarkers for CRC diagnosis was noted in a study by Jiang W. et al. In addition, circRNAs can interact with other proteins or RNAs to enhance diagnostic accuracy for CRC.45,46 However, additional research is necessary to fully comprehend the mechanisms of cancer.47
Long non-coding RNAs (lncRNAs), which are non-translated RNA transcripts, can play a crucial role in suppressing tumors by interacting with the p53 tumor suppressor gene.48
The lncRNA activator of enhancer domains is upregulated by p53, while decreased expression is linked to CRC, breast cancer, and androgen-sensitive prostate cancer.49,50 Researchers have found that elevated levels of long intergenic non-coding RNA (lincRNA)-p21 in CRC enhance the response to radiotherapy, facilitating apoptosis. Conversely, reduced levels of lincRNA-p21 are associated with a higher risk of disease progression.51 The changes in lncRNA expression levels can serve as indicators and potential biomarkers for the early detection of CRC.
Common sample types used for cancer biomarker analysis include blood, urine, stool, and other body fluids such as exhaled breath, saliva/buccal swabs, cerebrospinal fluid, and sputum.34,35 For many years, the field of cancer diagnosis and monitoring has extensively utilized protein biomarkers, particularly from plasma samples. Urine is frequently used for biomarker testing in cases of bladder or prostate cancer. The Alere NMP22 Bladder Check Test is a reliable protein-based diagnostic and monitoring tool for bladder cancer.25 An FDA-approved screening test called Cologuard can identify DNA alterations associated with CRC in stool samples, achieving 92% sensitivity.25 Stool samples are also valuable for microbiota profiling, which can help predict therapy responses. For instance, researchers have found that the ratio of Fusobacterium nucleatum to Bifidobacterium probiotic (Fn/Bb) demonstrates high sensitivity (84.6%) and specificity (92.3%) in detecting CRC. The combined ratio of Fn/Bb and F. nucleatum to Faecalibacterium prausnitzii (Fn/Fp) increases sensitivity in detecting stage I CRC to 90%.25,52
Table 1 provides an overview of biomarker types and detection methods used in CRC research and diagnostics.
Table 1.
Overview of biomarker types and detection methods
|
Biomarker category
|
Key points
|
References
|
| Germline variants |
Variants linked to hereditary cancers, like Lynch syndrome, are useful for risk assessment. |
14-17
|
| Somatic mutations |
Chromosomal abnormalities (e.g., BCR-ABL) serve as key cancer biomarkers. |
18-21
|
| Circulating tumor DNA |
Low levels require sensitive detection; potential as cancer biomarkers. |
9,23,24
|
| Protein biomarkers |
Detected by IHC, ELISA; FDA-approved PD1/PD-L1 for cancer treatment. |
26-30
|
| Epigenetic markers |
DNA methylation (e.g., MLH1) used for early detection and treatment prediction. |
31-33
|
| Transcriptome biomarkers |
miRNAs, circRNAs, and lncRNAs are key for early cancer detection, prognosis, and therapy response. |
34-49
|
| Sample types |
Blood, urine, stool, saliva, etc.; stool DNA testing (e.g., Cologuard) for CRC detection. |
9,33-50
|
FDA-approved genetic biomarkers
APC: The adenomatous polyposis coli (APC) gene, located at the 5q22.2 locus, produces a protein that acts as a growth suppressor. This protein regulates cell proliferation and adhesion to adjacent tissues.53 The alteration of the APC gene is responsible for approximately 85% of CRC patients.7 Given the prevalence of APC mutations among patients and their close association with Wnt/β-catenin signaling, targeting this pathway could potentially benefit individuals with APC mutations.54 Despite the emergence of various methods and drugs for treating patients with APC mutations, the significant side effects of traditional medications have proven to be a major challenge in effectively inhibiting CRC caused by these mutations. Therefore, identifying novel target genes for managing CRC in patients with APC mutations is essential.
TP53: The gene TP53, also known as p53, is essential for suppressing the cell cycle, cell division, and proliferation via regulating intercellular communication.55 It is important to highlight that the incidence of P53 gene mutations is higher in distal bowel and rectal cancers compared to proximal tumors.55 This suggests a potential correlation between the p53 gene mutation and the occurrence of distant metastasis as well as vascular infiltration in CRC.56 The nucleoside reverse transcriptase inhibitor 3TC demonstrated significant therapeutic activity in p53-mutated CRC.57 Phase II clinical trials revealed promising results, offering a novel therapeutic approach for the treatment.58
PIK3CA: The PIK3CA oncogene is a crucial component of the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) pathway, which has a significant impact on numerous biological processes such as cell replication, death, and motility. PIK3CA is commonly found in CRC, with a mutation frequency of around 10-15%.59-61 Research has indicated that aspirin, also known as acetylsalicylic acid, can have a beneficial impact on CRCs caused by PIK3CA gene mutations.62 Compounds like Selaginella doederleinii Hieron ethyl acetate (SDEA) have been found to effectively induce cell death in CRC by inhibiting the PI3K signaling pathway.63 However, most PI3K inhibitors have demonstrated toxic or undesirable side effects during in vitro testing. Therefore, it is essential to continue developing PI3K signaling pathway inhibitors that are both safer and more effective.7
SMAD Family Member 4 (SMAD4): SMAD4 is essential for the TGF-β signaling pathway, facilitating the transmission of signals from the extracellular environment to the nucleus.64 The SMAD4 mutation is found in approximately 10-15% of individuals diagnosed with CRC.65 The absence of the SMAD4 gene does not directly cause the formation of tumors, but it does play a role in the spread of tumors, which has implications for the prognosis and rates of survival of individuals diagnosed with stage III CRC.64,66-68 Possible biomarkers for predicting a poor outcome for cetuximab-based treatment in Chinese patients with metastatic CRC may include mutations of SMAD4.69 Given the impact of SMAD4 mutations on the TGF-β signaling pathway, it is worthwhile to consider TGF-β signaling pathway inhibitors as a potential treatment for CRC.70
KRAS: The mutation of the RAS gene is prevalent in a significant majority of patients diagnosed with CRC. Specifically, the KRAS gene mutation accounts for approximately 85% of these alterations, while mutations in the NRAS and HRAS genes collectively constitute the remaining 15%.71,72 The proto-oncogene KRAS encodes the P21 Ras protein, which plays a pivotal role in various signal transduction pathways, angiogenesis, and tumorigenesis. In CRC patients, mutations in the KRAS gene frequently occur at the 35th nucleotide within the 12th codon. This particular mutation is significantly associated with tumor infiltration and lymphatic metastasis.73,74 Approximately 85% of CRC patients exhibit a KRAS mutation. The KRAS gene is integral to numerous essential cellular processes, including the epidermal growth factor receptor (EGFR) and RAS signaling pathways, as well as cellular growth and differentiation. Therefore, the KRAS gene is of considerable significance in the diagnosis and treatment of CRC.73,75,76 Cetuximab and panitumumab are commonly used treatments for patients with metastatic CRC who have wild-type KRAS and NRAS. Nevertheless, additional resistance mechanisms have been observed, including acquired mutations of the EGFR and RAS genes and amplification of ERBB2, RAS, or MET.77,78 The effectiveness of rechallenge therapy is shown by a favorable outcome from prior anti-EGFR treatment and significant gaps between previous treatment plans.79 Developing effective therapeutics for other KRAS mutations is the next research frontier.80
BRAF: The BRAF gene plays a critical role in cell proliferation, differentiation, and programmed cell death in normal physiological conditions.81 Mutations in the BRAF gene result in the persistent activation of signaling pathways, which can lead to uncontrolled cellular proliferation and the subsequent development of cancer. A prior study indicated that approximately 10% of patients with CRC possess a mutation in the BRAF gene, with the majority of these cases (90%) characterized by the BRAFV600E mutation.82 The BRAFV600E mutation is linked to certain clinicopathological characteristics, including being female, being older, having a tumor in the proximal colon, being at an advanced TNM stage, having poor differentiation, and having mucinous histology.83 The BRAF mutant status proved to be a dependable predictor of survival in metastatic CRC. Patients with BRAF-mutant CRC experienced a decline in disease-free survival (DFS) and overall survival (OS) following recurrence subsequent to postoperative adjuvant therapy.84 Additionally, the BRAF gene is strongly linked to the RAS-RAF-MAPK signaling pathway. As a result, it is not feasible to treat CRC by suppressing the BRAF gene mutation using a single method. Utilizing a combination of EGFR, BRAF, and MEK inhibition has emerged as a promising clinical approach.85-87 The (Binimetinib, Encorafenib, and Cetuximab Combined to Treat BRAF-Mutant CRC) BEACON CRC trial examined the potential of combining encorafenib, binimetinib, and cetuximab to treat patients. The compelling results of combining sorafenib with cetuximab, with or without binimetinib, highlight the significant advantages of using BEACON combinations.88 Above all, the KRAS and BRAF genes are widely recognized as critical biomarkers for early detection.89 Analyzing the KRAS and BRAF genes can diagnose patients with Stage III CRC. It is noteworthy that this specific type of cancer exhibits a greater propensity for hepatic metastasis compared to cancers characterized by a single gene mutation.89-91
MSI Status: Microsatellites are short tandem repeats of DNA sequences, and MSI status is a consequence of a compromised MMR system, typically resulting from the inactivation of four crucial MMR genes (MSH2, MLH1, MSH6, and PMS2).92 An inadequate MMR system hampers the ability to correct errors that occur during DNA replication, especially those involving the addition or deletion of repeating units.93 This ultimately leads to a hypermutable phenotype, characterized by instability at two or more loci (MSI-high). There are two different methods to determine MSI status: IHC or PCR.94 IHC can determine the expression of the MLH1, MSH2, MSH6, and PMS2 genes, which helps identify tumors as MSI (deficient MMR, dMMR) rather than MSS (proficient MMR, pMMR).88 Approximately 15% of patients diagnosed with CRC exhibit microsatellite instability (MSI) tumors, with a small subset of these cases attributable to Lynch syndrome—a hereditary cancer syndrome that significantly increases the risk of CRC.95,96 This condition is also referred to as Hereditary Non-Polyposis CRC (HNPCC). The first application of MSI was as a screening method for Lynch syndrome detection.97 It is important to note that the occurrence of MSI varies depending on the stage of the disease. Approximately 15% of stage II/III CRCs exhibit dMMR, whereas the prevalence of dMMR in stage IV CRCs is only around 4%-5%.98 Tumors characterized by MSI exhibit distinct clinical and pathological features, occurring more frequently in the right colon and being predominantly observed in younger patient populations. These tumors also display poor differentiation and are characterized by a robust lymphocyte infiltrate. Generally, patients with MSI-high have a more favorable prognosis when compared to patients with MSI-low (MSS).97,99 In the field of onco-immunology, the success of checkpoint inhibitors in various tumor types has sparked interest among researchers and clinicians.100 In 2017, pembrolizumab, a monoclonal anti-PD1 antibody, received FDA approval for use in MSI-high patients, regardless of their specific cancer type.101 Additionally, the FDA has approved Nivolumab and Ipilimumab for patients with refractory stage IV MSI-high.102 It is worth noting that MSI status could potentially serve as a predictive marker for patients with stage III MSI.88 However, it is important to note that not all patients with metastatic CRC experience a positive response to immunotherapy, particularly those with MSI-high. In patients receiving immunotherapy, the expression of PD-L1 in tumors did not correlate with improved survival outcomes, raising questions about the reliability of PD-L1 as a prognostic indicator for checkpoint inhibition treatment in metastatic CRC.103 There is a need for further studies to identify biomarkers in this rapidly evolving field of clinical research.104 Fig. 2 illustrates key genetic mutations involved in CRC pathways.
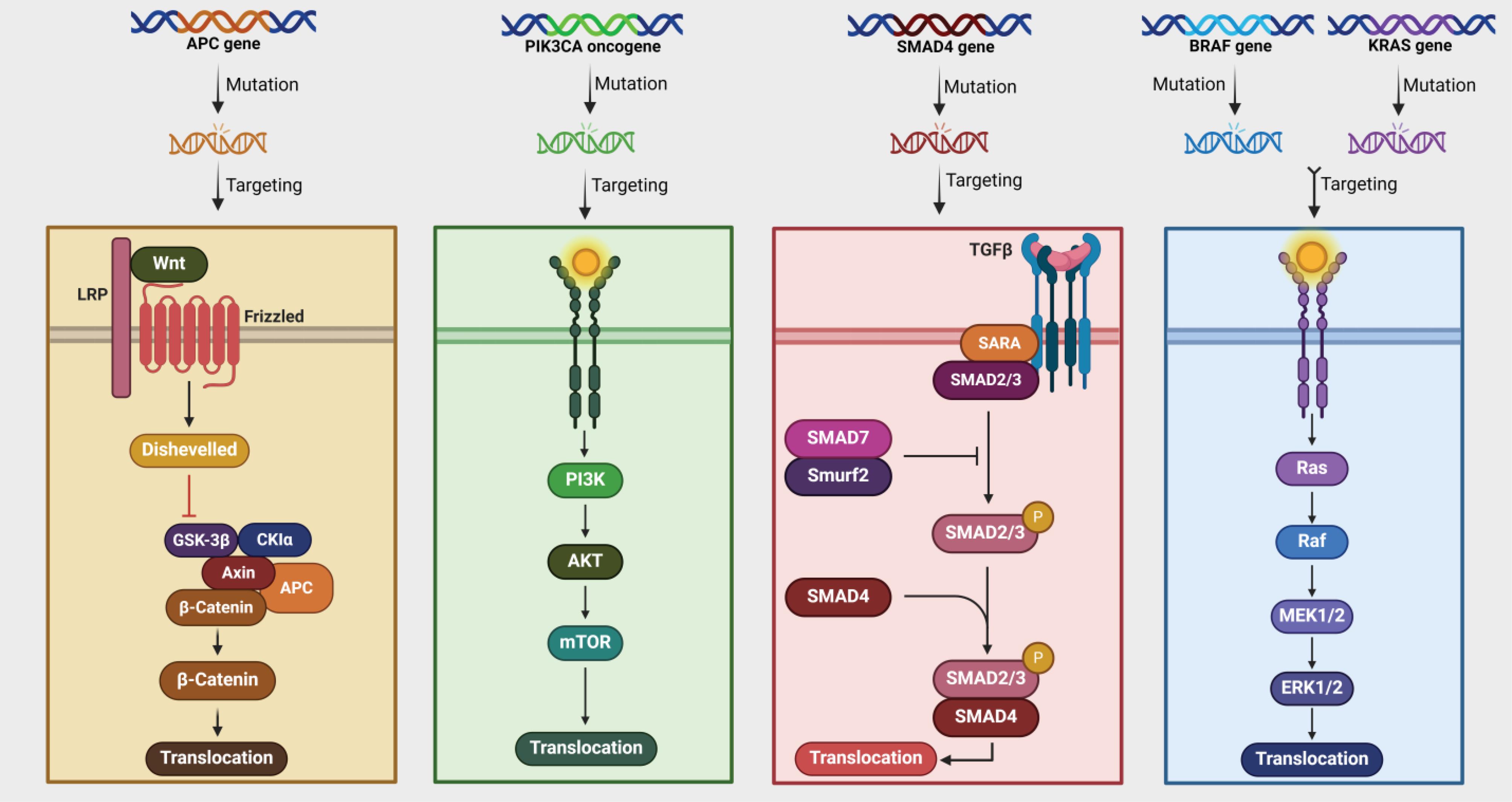

Fig. 2.
Genetic Mutations and Targeted Pathways. This figure illustrates key genetic mutations and their associated pathways in cancer. It highlights mutations in the APC, PIK3CA, SMAD4, BRAF, and KRAS genes, each associated with different signaling pathways. The APC gene mutation affects the Wnt signaling pathway, leading to the activation of β-catenin and its translocation into the nucleus. This pathway involves proteins like Dishevelled, GSK-3β, and CKIα. The PIK3CA oncogene mutation activates the PI3K/AKT/mTOR pathway, which is crucial for cell growth and survival. This pathway is often targeted in cancer therapies due to its role in tumor progression. The SMAD4 gene mutation disrupts the TGF-β signaling pathway, which involves proteins such as SARA, SMAD2/3, and SMAD7. This pathway is essential for regulating cell proliferation and differentiation. The BRAF gene mutation activates the MAPK/ERK pathway, which includes Ras, Raf, MEK1/2, and ERK1/2 proteins. This pathway is critical for cell division and is a common target in cancer treatment strategies. KRAS gene mutations also affect the MAPK/ERK pathway, similar to BRAF mutations. These mutations are prevalent in many cancers and are associated with resistance to certain therapies. Targeting these pathways can help manage cancer progression by inhibiting key proteins involved in cell signaling and growth. Overall, this figure underscores the complexity of cancer biology and the importance of targeted therapies. By understanding these pathways, researchers can develop more effective treatments that specifically address the underlying genetic mutations driving cancer growth.
.
Genetic Mutations and Targeted Pathways. This figure illustrates key genetic mutations and their associated pathways in cancer. It highlights mutations in the APC, PIK3CA, SMAD4, BRAF, and KRAS genes, each associated with different signaling pathways. The APC gene mutation affects the Wnt signaling pathway, leading to the activation of β-catenin and its translocation into the nucleus. This pathway involves proteins like Dishevelled, GSK-3β, and CKIα. The PIK3CA oncogene mutation activates the PI3K/AKT/mTOR pathway, which is crucial for cell growth and survival. This pathway is often targeted in cancer therapies due to its role in tumor progression. The SMAD4 gene mutation disrupts the TGF-β signaling pathway, which involves proteins such as SARA, SMAD2/3, and SMAD7. This pathway is essential for regulating cell proliferation and differentiation. The BRAF gene mutation activates the MAPK/ERK pathway, which includes Ras, Raf, MEK1/2, and ERK1/2 proteins. This pathway is critical for cell division and is a common target in cancer treatment strategies. KRAS gene mutations also affect the MAPK/ERK pathway, similar to BRAF mutations. These mutations are prevalent in many cancers and are associated with resistance to certain therapies. Targeting these pathways can help manage cancer progression by inhibiting key proteins involved in cell signaling and growth. Overall, this figure underscores the complexity of cancer biology and the importance of targeted therapies. By understanding these pathways, researchers can develop more effective treatments that specifically address the underlying genetic mutations driving cancer growth.
Table 2 provides information about FDA-approved genetic biomarkers for CRC.
Table 2.
FDA-approved genetic biomarkers for CRC
|
Biomarker
|
Mutation frequency
|
Clinical relevance
|
Therapeutic approaches
|
Ref.
|
| APC |
85% of CRC patients |
Impacts Wnt/β-catenin signaling. |
Targeting the Wnt/β-catenin pathway |
1,7,52
|
| TP53 (p53) |
Higher in distal bowel/rectal cancers |
Associated with metastasis |
3TC (nucleoside reverse transcriptase inhibitor) |
53-55
|
| PIK3CA |
10-15% of CRC patients |
Affects PI3K/Akt/mTOR pathway |
Aspirin, SDEA, PI3K inhibitors |
56-59
|
| SMAD4 |
~15% of CRC patients |
Tumor progression, poor prognosis |
TGF-β signaling inhibitors |
60-65
|
| KRAS |
85% of RAS mutations |
Tumor infiltration, EGFR/RAS signaling |
Cetuximab, panitumumab, rechallenge therapy |
66-74
|
| BRAF |
~10% of CRC patients (90% BRAFV600E) |
Poor survival in metastatic CRC |
Combination therapies (EGFR, BRAF, MEK inhibitors) |
75-83
|
| MSI status |
~15% of CRC patients |
Better prognosis, linked to Lynch syndrome |
Immunotherapies (pembrolizumab, nivolumab, ipilimumab) |
80-92
|
FDA-approved protein biomarkers
CEA, a tumor marker, is a molecule involved in intracellular adhesion, produced in embryonic gastrointestinal tissue as well as in tumor cells, and plays a role in angiogenesis.105,106 Elevated levels of CEA in the blood are indicative of several cancers including gastrointestinal tract, breast, lung, ovary, and pancreatic cancer.107,108 However, an increase in CEA levels can also occur in various non-malignant conditions, including cigarette smoking, alcoholism, chronic inflammatory bowel disease, diverticulitis, pancreatitis, and liver disease.108 The CEA protein is widely used as a marker for CRC. It is effective in identifying CRC and is beneficial for assessing patient prognosis.109 A possible correlation between CEA, CRC metastasis, and tumor growth was suggested by the study, which found a strong relationship between a positive CEA status before surgery and vascular spreading, nerve infiltration, and the size of the tumor. In order to more successfully guide clinical therapy and prognosis, pretreatment CEA levels in the blood may be utilized to predict the status of CRC, including the stage of cancer and lymph node involvement.110 Additionally, previous findings have shown that the serum CEA level in CRC patients can independently predict both OS and DFS time.111-113 In the world of patients with CRC, CEA is an appropriate choice for further investigation of circulating tumor cells (CTCs) or circulating epithelial cells (CECs). According to a number of research studies, patients with greater blood levels of CEA protein prior to surgery may have a higher risk of metastasis and recurrence.114 However, it is important to note that the levels of CEA in the bloodstream may change considerably as a result of external variables such as liver and lung disorders or daily behaviors, which may in turn impact their correlation with the original malignant tissue.115 As a result, although the blood CEA level has been extensively used in clinical practice to diagnose and predict CRC.116,117 they have demonstrated inadequate sensitivity, specificity, and repeatability for characterizing CRC.118
CA 19-9: CA19-9 was discovered in 1979 and is now frequently used to detect pancreatic cancers early.119 It is a type of monoclonal antibody that binds to E-Selectin.120 Elevated levels of CA19-9 can indicate both malignant and non-malignant conditions. This tumor marker is mainly produced by pancreatic, lung, gastrobiliary tract, and CRCs. However, higher CA19-9 levels can also be found in patients with chronic liver disease, diabetes mellitus, endometriosis, or bronchiectasis.121 guidelines do not support the use of CA19-9 for prognostic evaluation.122-124 However, certain studies have indicated that CA19-9 holds comparable importance to CEA in terms of predicting survival.125 Individuals with CA19-9 levels exceeding 200 U/mL experienced a significantly reduced 5-year survival rate of only 8%. Several other studies have reported comparable findings with slightly varying cut-off values ranging from 31 to 37 U/mL.125-128 In various studies, it has been found that CA19-9, although not highly sensitive on its own, is closely related to CEA and has the potential to enhance the overall sensitivity of CEA.127,129-132 However, a study by Bagaria et al demonstrated that analyzing both tumor markers together did not increase the sensitivity to CEA.133 Currently, it is recommended to use CEA in combination with other screening methods to assess prognosis, monitor surveillance following therapeutic resection, and track treatment progress, such as chemotherapy or radiation. Using CA19-9 by itself is not advised to identify CRC or evaluate ongoing treatment or monitoring because of its low sensitivity.122,123,134
The decrease in serum CEA levels is a common outcome observed in numerous studies following effective treatment with anti-EGFR monoclonal antibodies or KRAS G12C inhibitors.135,136 In a retrospective study of the cases of 215 patients with CRC, it was observed that a KRAS mutation was associated with elevated initial CEA levels. As a result, the researchers suggested that elevated initial CEA levels might serve as an indicator of the presence of a KRAS mutation.137 However, a different retrospective cohort study looked back at the data of 183 patients with CRC and discovered that CEA levels did not significantly impact the identification of KRAS mutations. Other research has also shown that CEA cannot be used as a marker to determine KRAS mutation status or EGFR expression in CRC. Nonetheless, numerous studies have observed decreased CEA levels in the blood following successful treatment with anti-EGFR monoclonal antibodies or KRAS G12C inhibitors.135,136 In a study by Hong Jae Jeon et al, researchers suggested that the CEA-expressing circulating epithelial cells (CCEC) might serve as a supplementary biomarker for blood CEA levels, enabling the prediction of prognosis in patients receiving different treatment modalities.138 CEA and CA19-9 levels in the blood are commonly used as biomarkers to predict patient prognosis in CRC, and numerous studies have verified that both are increased in patients with advanced malignancies.139 A previous analysis examining the predictive significance of serum CA19-9 levels in CRC patients found that a more significant number of patients with higher CA19-9 levels also had elevated CEA levels compared to those with lower CA19-9 levels.140 The two serum markers did not show a significant correlation in certain instances. However, it was discovered that the CCEC count was higher in individuals with serum CA19-9 levels more than 10 U/mL compared to those with lower CA19-9 levels (1.83 ± 1.59 cells/mL vs. 0.72 ± 0.90 cells/mL; P = 0.043).138 The combined CEA result also increased among individuals with elevated serum CA19-9 levels, but these results were not as significant as CCEC counts because of the weak correlation between serum CEA and CA19-9 levels.
These findings suggest that CCEC may offer valuable insights into CRC pathology that serum CEA levels might overlook. To address these limitations, CECs were quantified by measuring CEA expression. The results confirmed that the CCEC count demonstrates excellent diagnostic capability compared to serum CEA levels.
This approach not only aids in diagnosing CRC but also in assessing tumor invasiveness and its potential for metastasis.138 The presence of CCECs was found exclusively in individuals with CEA-positive tumors, which aligns with the fact that CEA is highly expressed in various CRC types.141 Although there is a risk of missing specific cancer-related cell subtypes, the CCEC count remains a promising biomarker for CRC. It is essential to recognize that evaluating multiple factors can enhance the sensitivity of the tests. For example, pairing CEA with CA242 significantly increases sensitivity compared to using either marker alone.142 Additionally, Wang and colleagues showed that the combination of CEA, CA19-9, and CA242 enhanced the precision of prognostic forecasting in surgical patients with CRC.143 The Luo et al research found a notable connection between serum CA19-9 and CA125 levels and both lymph node metastasis and pTNM staging. A significantly higher percentage of stage III + IV patients tested positive for CA19-9 or CA125 compared to stage I + II patients.110 These results are consistent with previous reports,144-146 and suggest that CA19-9 and CA125 could be valuable for differential diagnosis, disease monitoring, and therapeutic evaluation of numerous malignant tumors. Nevertheless, guidelines recommend CEA as a valuable predictor of OS.122,124,134 Table 3 provides information about FDA-approved protein biomarkers for CRC.
Table 3.
FDA-approved protein biomarkers for CRC
|
Biomarker
|
Source
|
Clinical Relevance
|
Limitations
|
Ref.
|
| CEA |
Tumor marker from gastrointestinal tissue and tumor cells |
Indicates various cancers (CRC, breast, lung, ovarian, pancreatic); prognostic marker for CRC |
Sensitive to non-malignant conditions; inadequate specificity and repeatability |
93-107
|
| CA 19-9 |
Monoclonal antibody (E-Selectin) |
Primarily used for pancreatic cancer; correlates with CEA for survival prediction |
Low sensitivity alone; not recommended for CRC diagnosis or treatment monitoring |
108-123
|
| CECs |
Blood-based marker |
Predicts prognosis in CRC; potential to identify invasive tumors |
Limited correlation with serum CEA; may overlook specific cancer cell subtypes |
124-132
|
FDA-approved epigenetic biomarkers
The SEPT9 gene functions as a tumor suppressor and is involved in several essential physiological processes, including cytokinesis, DNA repair, cell movement, and apoptosis.147 Abnormal methylation decreases the transcriptional activity of the SEPT9 gene, resulting in dysregulated gene expression and impaired physical function, which may ultimately contribute to cancer development.148 Toth et al149 found that SEPT9 protein expression in CRC cells is significantly lower than in normal cells, and mRNA expression of SEPT9 decreases during the transformation from adenoma to CRC. Moreover, Wasserkort et al150 also suggested that hypermethylation of SEPT9 may occur later in the progression from adenomas to CRC. This difference in timing could explain why the detection sensitivity of SEPT9 gene methylation is lower in adenomas compared to CRC. A recent meta-analysis found that plasma mSEPT9 has high diagnostic value and is significantly associated with the stage of CRC.151 In addition, survival analysis indicated that there was a negative correlation between SEPT9 methylation levels and DFS after CRC surgery.152 In a study consisting of 1544 CRC samples (stages I-IV), assessment of the SEPT9 trial was retrospectively conducted. The study reported a sensitivity and specificity of 68.2 and 78.8% for CRC and a sensitivity of 21.6% for advanced adenomas (AA). The main performance characteristics of mSEPT9 were demonstrated by a Church TR study in 2014,152 and mSEPT9 was eventually approved by the FDA for CRC screening.153
The methylation of the SEPT9 gene promoter in plasma is one of the most researched noninvasive DNA methylation biomarkers for diagnosing CRC. SEPT9, a GTP-binding protein, plays a role in actin dynamics, cytoskeletal remodeling, vesicle trafficking, and exocytosis. Many studies have examined the accuracy of this biomarker in diagnosing CRC in large groups of patients, and the findings showed a sensitivity ranging from 48.2% to 95.6% and a specificity ranging from 79.1% to 99%.154-156 Epi proColon 2.0 CE, a plasma-based test, detects SEPT9 methylation with a sensitivity of 68.2% to 81.0% and a specificity of 87.4% to 99.0%.157,158 The Epi proColon (SEPT9) test is approved by the FDA.159
SEPT9 and SDC2 aberrant methylation have been examined in the stool and plasma of patients with CRC in various studies.160,161 The ColoDefense test is a blood-based methylation assay designed for early CRC screening, combining the methylation of two biomarkers (SEPT9 and SDC2) in a single reaction to improve the detection rate for early-stage CRC and AA.161 Recent studies demonstrate that ColoDefense is a potent, suitable, and practical approach with high sensitivity and specificity for early screening. The simultaneous identification of mSEPT9 and mSDC2 in blood shows excellent promise for partially non-invasive CRC screening. Additionally, the ColoDefense stool test has demonstrated greater accuracy in detecting AAs and CRC compared to the detection of mSEPT9 or mSDC2 alone.160-162
DNA methylation can be detected using various methods, such as methylation-specific PCR,163 DNA sequencing, MethyLight,164 methylation-specific melting curve analysis,165 pyrosequencing,166 microarray analysis,167 And liquid chromatography (LC). Notably, LC was the first platform used to quantify global DNA methylation, which was represented by the total 5-methyldeoxycytidine content in DNA samples.168 Due to technological progress, the combination of LC and mass spectrometry offers a precise and extremely sensitive approach for comprehensive quantification of DNA methylation.169 Several methylation markers are associated with the initiation and progression of CRC.170-172 Certain markers have the potential to detect CRC early, evaluate prognosis, and even predict the response to treatment. The status of these methylation markers is linked to various factors, including tumor size, grade, and metastasis.
Interest in microbiome science has led to the sequencing and profiling of an unprecedented number of sample types and tissues. This effort has uncovered microbial signals in locations previously thought to be devoid of microbial communities. However, the reliability and significance of these signals are not always evident.173 The influence of certain microorganisms on cancer development is widely recognized. However, with the recent surge in interest in microbiome science, we are on the brink of potentially changing our understanding of cancer. It is estimated that approximately 20% of the overall cancer burden is related to infectious agents, a finding that could revolutionize cancer research.174 F. nucleatum, a bacterium, has become significant in microbiome research on CRC. Its potential role in the progression and development of CRC has attracted considerable interest and is a matter of immediate concern in our field.175 Fusobacteria are Gram-negative anaerobic bacilli with species-specific reservoirs in the human mouth, gastrointestinal tract, and elsewhere.173,176 F. nucleatum has been found in CRC tissues in 10% to 90% of cases, with a higher occurrence in the proximal colon compared to the distal colon. It is frequently linked to advanced disease, resistance to chemotherapy, metastasis, and a grim prognosis.177 Therefore, an association between F. nucleatum and human CRC has emerged across patient populations and disease stages.178 Research has demonstrated that F. nucleatum strengthens the ability of CRC cells to adhere to each other, promotes the attachment of CRC cells to endothelial cells, and aids in the process of moving out of blood vessels and forming metastases by activating the Nuclear factor kappa B (NF-κB) pathway and increasing the expression of Keratin7-antisense (KRT7-AS) and Keratin7 (KRT7) in CRC cells.179,180 The level of F. nucleatum was significantly higher in fecal samples of CRC patients with positive lymph node metastasis179 The fecal samples of patients with carcinoma showed an enrichment of F. nucleatum and other bacteria associated with intestinal inflammation compared to those of healthy subjects.181 In another study, findings showed that the combination of F. nucleatum infection and high tumor mutational burden strongly predicted poor prognosis.182
The association between F. nucleatum and CRC has generated interest in identifying F. nucleatum DNA and cells in stool using a quantitative PCR assay, as well as in intestinal tissues and antibodies against F. nucleatum.173,183,184 These markers could be used to detect tumors and predict outcomes for individuals with CRC. The presence of F. nucleatum in stool samples could aid in non-invasive screening. In addition to stool-based diagnostics, the identification of IgA or IgG antibodies against F. nucleatum in serum shows promise as a diagnostic tool.173 Table 4 provides information about FDA-Approved Epigenetic Biomarkers for CRC.
Table 4.
FDA-approved epigenetic biomarkers for crc
|
Biomarker
|
Source
|
Clinical Relevance
|
Limitations
|
Ref.
|
| SEPT9 |
Plasma, stool |
Tumor suppressor; abnormal methylation linked to CRC; FDA-approved for screening with high diagnostic value |
Lower sensitivity in adenomas; variable detection sensitivity based on cancer stage |
136-142
|
| ColoDefense |
Blood |
Combines SEPT9 and SDC2 methylation for improved detection of early-stage CRC and AA |
Variable performance data; further validation needed in diverse populations |
149-151
|
| Fusobacterium nucleatum |
Stool, tissue |
Associated with CRC progression; higher levels linked to advanced disease; potential for non-invasive detection |
Detection methods vary; correlation with inflammation requires further investigation |
162-173
|
Multi-target stool DNA test (Cologuard)
The Cologuard is a non-invasive screening test for CRC and precancerous lesions, which received approval from the FDA in 2014. This test targets several genetic mutations and alterations that are commonly associated with CRC. It combines stool DNA analysis with immunochemical detection of hemoglobin to improve sensitivity. The immunochemical component detects hemoglobin in the stool, which may indicate the presence of cancer or polyps causing bleeding. This aspect is particularly valuable for detecting cancers that do not release adequate DNA into the stool but still cause bleeding.185 Cologuard provides offers a broader detection spectrum than traditional fecal immunochemical test (FIT) or guaiac-based fecal occult blood test (FOBT) by capturing both genetic evidence of cancer and functional evidence of bleeding.186 Cologuard detects specific mutations in KRAS, APC, BRAF V600E, NDRG4, BMP3, and methylation in SEPT9 and MLH1.185 Patients only need to collect a stool sample along with a preservative solution using a home collection kit, which enhances patient compliance compared to invasive procedures. The sample is sent to a laboratory. Extraction involves isolating DNA from stool cells and any free-floating DNA fragments. The process is designed to ensure the recovery of both human and cancer-derived DNA.185 PCR is used to amplify the target DNA sequences associated with KRAS and APC mutations. To detect the methylation of the NDRG4 and BMP3 genes, methylation-specific polymerase chain reaction (PCR) techniques are utilized. These methodologies effectively differentiate between methylated and unmethylated DNA, thereby elucidating the presence of cancer-associated epigenetic alterations.187 The results obtained from both genetic/epigenetic assessments and FIT are integrated using proprietary algorithms developed by Exact Sciences to evaluate mutations, methylation, and hemoglobin levels to generate an overall risk. The final report indicates whether further diagnostic procedures, such as a colonoscopy, are recommended.185
Cologuard has been evaluated in multiple studies to determine its clinical effectiveness in comparison to traditional screening methods. The pivotal study leading to FDA approval was a large, multicenter trial that included more than 10,000 participants. The study showed that Cologuard was effective in detecting CRC with a high degree of sensitivity and reasonable specificity. The FDA recognized Cologuard as a suitable alternative to traditional screening methods for individuals at average risk.185 Its approval came with labeling that specifies Cologuard is for use in average-risk adults aged 45 and older who are asymptomatic. It is a screening test and not a diagnostic tool, meaning that positive results should be followed up with a colonoscopy for a definitive diagnosis.185 The results showed that it had a sensitivity of 92% for detecting CRC and 42% for detecting advanced precancerous lesions, compared to 73% for FOBT.185 It has a higher false positive rate than some other screening tests, which may lead to unnecessary follow-up procedures. The test also has a lower sensitivity for detecting AAs compared to CRC, meaning that some precancerous lesions may go undetected. It is more expensive than traditional stool-based tests, which may limit its accessibility. Additionally, it is recommended every three years, as opposed to annual FIT tests. A study by Pickhardt et al. found that, while Cologuard was more sensitive for detecting CRC, CT colonography had a better detection rate for AAs. These findings highlight the complementary role of different screening methods in CRC surveillance.188 Cologuard has demonstrated high sensitivity for detecting CRC. Specificity of approximately 87% for CRC, which means there is a 13% rate of false positives. This is lower compared to FIT, which has a specificity of about 95%. While the lower specificity of Cologuard results in a higher rate of false positives, its higher sensitivity can be advantageous in detecting cancers early.185 The recommendation to use it every 3 years is based on balancing the test's ability to detect CRC with its limitations in detecting all types of precancerous lesions.189 The age range (45 and older) aligns with current guidelines for screening.190 It is particularly beneficial for patients who prefer a non-invasive screening method over a colonoscopy. A positive Cologuard result requires follow-up with a diagnostic colonoscopy for confirmation. The high sensitivity helps to ensure that potential cases are identified, but a colonoscopy remains necessary for further investigation.191
When compared to FIT, Cologuard offers superior sensitivity for CRC detection, but this comes at the cost of lower specificity. A study by Pickhardt et al. found that while Cologuard is more effective at detecting CRC than FIT, it also has a higher rate of false positives. This trade-off is important when considering patient management and follow-up procedures.188 Cologuard's sensitivity for CRC is comparable to that of colonoscopy; however, colonoscopy remains superior in detecting AAs, with Cologuard showing a lower sensitivity for these lesions. A study by Levin et al. found that while Cologuard had a sensitivity of 42% for detecting AAs, colonoscopy’s sensitivity is higher, making it more effective for polyp detection and surveillance.189 Cologuard’s high sensitivity for CRC means that it can effectively identify cancers at an early stage, potentially improving patient outcomes and survival rates.192 Early detection is crucial, as it can significantly reduce mortality.193 Ease of use has been shown to improve screening adherence. A study by Zhu et al. found that patients offered Cologuard were more likely to participate in screening than those offered colonoscopy.194 Long-term studies are ongoing to evaluate the sustained effectiveness of Cologuard in reducing incidence and mortality.192 A negative result suggests a lower likelihood of CRC or AAs but does not completely rule out the possibility of these conditions. Regular screening according to recommended guidelines is still important, as Cologuard does not prevent CRC or guarantee the detection of all lesions.195
Traditional FOBT, such as guaiac-based tests, detects occult blood in the stool, which can be indicative of CRC or large polyps. However, these tests are less sensitive and specific compared to newer methods. FOBT typically has a sensitivity for CRC of around 50-70% and a specificity of about 95%. In contrast, Cologuard shows higher sensitivity at 92% and slightly lower specificity at 87%.196 While FOBT has lower sensitivity, it maintains a high specificity and a long track record of use in screening programs. Additionally, it is relatively inexpensive and widely available.197 FIT is also less affected by dietary and medication-related factors compared to FOBT. FIT is simpler and less costly compared to stool DNA tests. It is effective in detecting CRC, though it lacks the broader detection capabilities provided by stool DNA tests.186
Colonoscopy detects CRC and adenomatous polyps (AAs) with high sensitivity—90-95% for CRC and 70-80% for AAs—and very high specificity.198 In contrast, Cologuard shows a high sensitivity for CRC at 92%, but it is less effective at detecting AAs, with a sensitivity of only 42%.199 Additionally, Cologuard has a lower specificity of 87%, resulting in a higher rate of false positives, which may lead to unnecessary follow-up procedures.200 Colonoscopy provides direct visualization of the colon, allowing for both detecting and immediate removal of polyps, thereby preventing CRC. Cologuard is often used as an initial screening tool, with colonoscopy remaining the definitive diagnostic and treatment procedure. It provides a valuable addition to screening options compared to traditional fecal tests. However, consider its limitations, including lower specificity and reduced sensitivity for AAs, when compared to colonoscopy. Balancing these factors with patient preferences and clinical guidelines is crucial for selecting the most appropriate strategy.
Emerging Biomarkers and Future Directions
The search for novel biomarkers is ongoing, driven by the need to enhance early detection and improve sensitivity and specificity. Identifying specific APC mutations can aid in genetic screening to identify individuals at higher risk, particularly those with familial adenomatous polyposis (FAP) and other hereditary syndromes.201 PIK3CA encodes a subunit of the PI3K enzyme, which is involved in cell growth and survival. PIK3CA encodes a subunit of the PI3K enzyme, which is involved in cell growth and survival. Mutations in PIK3CA are frequently found in CRC and are associated with poor prognosis. Current investigations are focusing on their potential as biomarkers for targeted treatments and for monitoring disease progression.202 Studies are exploring the use of NDRG4 methylation as a biomarker for detection, particularly in stool and plasma samples.203 Research is also focusing on the role of SOX17 methylation in CRC detection and its potential use in combination with other biomarkers.204 Histone H3 lysine 27 trimethylation (H3K27me3), a repressive histone modification associated with gene silencing, indicates tumor presence and progression in CRC.205 Global DNA methylation patterns reflect overall changes in DNA methylation status across the genome and may serve as biomarkers for early detection and monitoring.206 The detection of abnormal Mucin 1, a transmembrane glycoprotein that plays a role in cell signaling and protection, is being investigated for its diagnostic and prognostic potential.207 Changes in Mucin 2, a protein involved in forming the protective mucus layer of the gastrointestinal tract, can be detected in samples. This protein may serve as a marker for detection and could complement existing biomarkers, improving the sensitivity of stool-based tests.208 CEA has primarily been used as a monitoring tool rather than a screening test. However, combining CEA with genetic and epigenetic biomarkers may enhance both screening and monitoring capabilities.209 Elevated levels of Secreted Frizzled-Related Protein 2 (SFRP2), a protein involved in the Wnt signaling pathway and frequently dysregulated in CRC, have been found in the serum of patients.210 Reduced levels of short-chain fatty acids (SCFAs) such as butyrate, propionate, and acetate, along with SCFA profiling, particularly in combination with other fecal-based test could serve as biomarkers for screening.211 Research is concentrating on identifying specific amino acid profiles, including glutamine and tyrosine, within metabolomics studies, as well as examining alterations in lipid metabolites, such as phospholipids and sphingolipids, in patients. These investigations have the potential to enhance diagnostic accuracy.212 Lipid profiles could help diagnose and better understand pathogenesis.213 Proteomics markers in blood or stool, particularly those involved in inflammation or tumorigenesis, may offer additional diagnostic information.212 All of the markers mentioned above offer new avenues for enhancing diagnostic accuracy, monitoring disease progression, and personalizing treatment.
Advancements in detection technologies are focused on enhancing the sensitivity, specificity, and convenience of screening methods, facilitating early diagnosis, improving diagnostic accuracy, optimizing patient outcomes, and offering personalized treatment options. Ongoing research and development efforts are expected to further refine these technologies and broaden their clinical applications. The integration of artificial intelligence (AI) and machine learning is increasingly prevalent in the analysis of extensive datasets, enabling the identification of novel biomarkers. AI algorithms are capable of processing complex data derived from genomics, proteomics, and other fields to unveil potential biomarkers and enhance the formulation of personalized screening strategies.214
Next-generation sequencing (NGS) has become a transformative tool offering detailed insights into genetic alterations, enabling comprehensive profiling, and supporting precision medicine. It is advancing the detection of low-abundance ctDNA and other biomarkers.195 Despite challenges associated with data interpretation, cost, and clinical integration, ongoing advancements in NGS technology and bioinformatics are facilitating enhanced detection and management of CRC. Precise interpretation necessitates the utilization of advanced bioinformatics tools and expertise to differentiate between clinically relevant and benign genetic variants.195 Fig. 3 illustrates the process and key considerations involved in integrating liquid biopsy technologies into clinical practice.
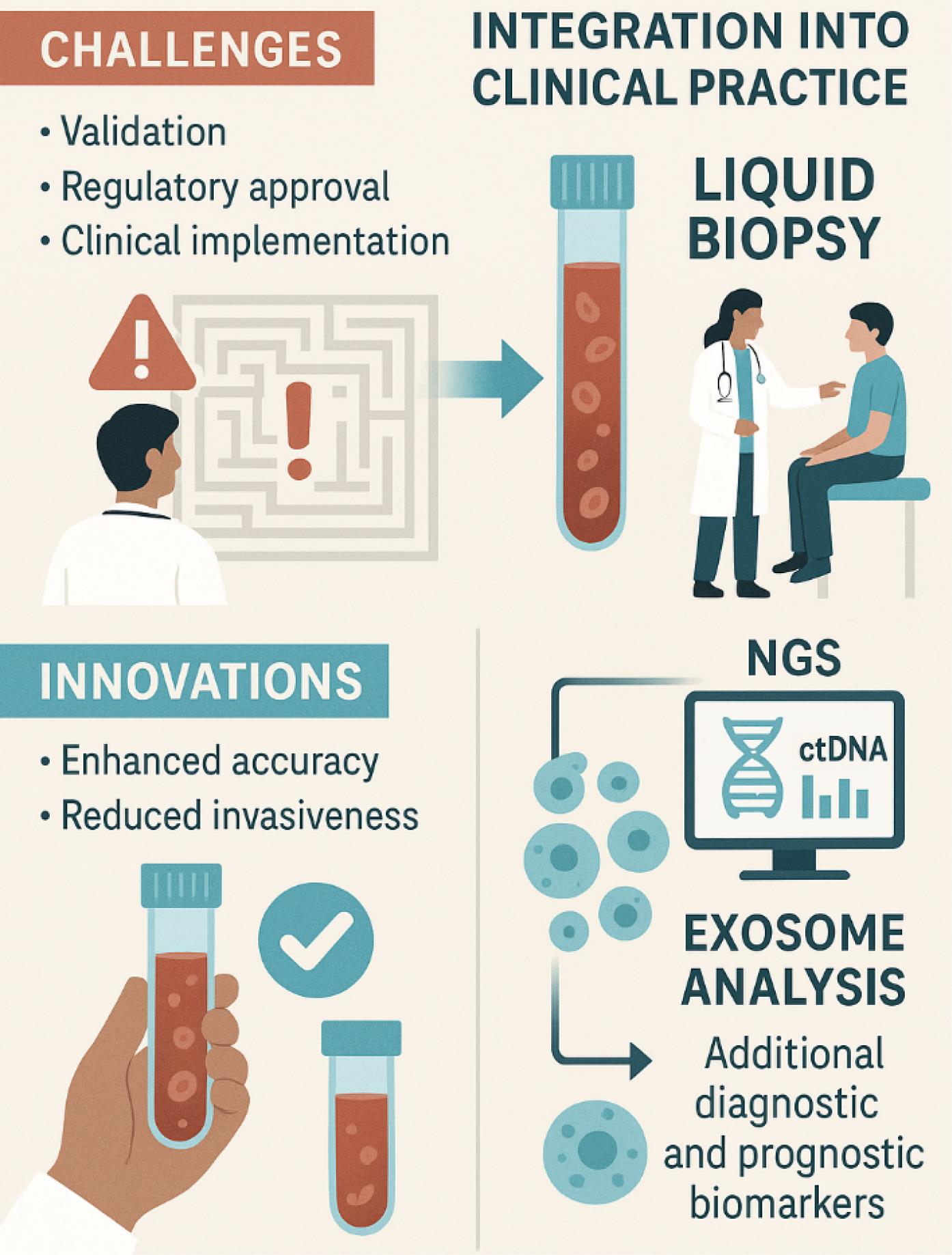
Fig. 3.
Integration of liquid biopsy technologies into clinical practice: challenges, innovations, and opportunities. This figure highlights several challenges, including the need for validation, regulatory approval, and effective clinical implementation. The figure also emphasizes recent innovations in the field, such as enhanced accuracy and reduced invasiveness of liquid biopsy techniques. Furthermore, it showcases the role of NGS for analyzing ctDNA and the use of exosome analysis, which provides additional diagnostic and prognostic biomarkers. Overall, the figure underscores both the opportunities and hurdles associated with adopting liquid biopsy methods in routine patient care.
.
Integration of liquid biopsy technologies into clinical practice: challenges, innovations, and opportunities. This figure highlights several challenges, including the need for validation, regulatory approval, and effective clinical implementation. The figure also emphasizes recent innovations in the field, such as enhanced accuracy and reduced invasiveness of liquid biopsy techniques. Furthermore, it showcases the role of NGS for analyzing ctDNA and the use of exosome analysis, which provides additional diagnostic and prognostic biomarkers. Overall, the figure underscores both the opportunities and hurdles associated with adopting liquid biopsy methods in routine patient care.
Future developments will likely enhance the utility of NGS, making it a cornerstone of personalized cancer care. Genomics profiling by NGS helps identify actionable mutations and guide targeted therapies, such as those targeting EGFR or VEGF. Profiling can uncover mechanisms of resistance to therapy and improve it.195
Liquid biopsy integration into clinical practice is fraught with challenges related to validation, regulatory approval, and clinical implementation.215 Innovations in liquid biopsy technologies provide enhanced accuracy and reduced invasiveness in diagnostic tools, as well as a more comprehensive understanding of tumor biology. The integration of liquid biopsy with NGS is becoming increasingly prevalent in the analysis of ctDNA for the detection of genetic alterations.195 Exosome analysis can provide additional diagnostic and prognostic information and is being explored for identifying biomarkers. The utilization of nanoparticles for the isolation and analysis of exosomes has improved sensitivity and specificity.216 Improvements in mass spectrometry can Identify specific metabolites and provide profiling with high sensitivity, thereby discovering new biomarkers. Advances in proteomics techniques enable the development of multi-protein panels and the introduction of protein biomarkers, which can improve diagnostic accuracy and stratify patients.
The integration of NGS with proteomics, metabolomics, and other omics technologies can yield a more holistic understanding of CRC. Such integrative methodologies are poised to enhance biomarker discovery, facilitate treatment stratification, and improve monitoring strategies.195 Advances in bioinformatics and analytical tools are expected to enhance the interpretation of NGS data. Developing more sophisticated algorithms and software will improve variant interpretation and clinical decision-making.195 Establishing standard guidelines will facilitate the widespread adoption of NGS for CRC and ensure high-quality results.213 The pathway to regulatory approval for liquid biopsy tests involves rigorous scrutiny by the FDA. Extensive clinical trials and comprehensive data are required to demonstrate the safety and efficacy of the test. Collaborative efforts among international regulatory bodies can facilitate its global adoption. Effective integration necessitates that healthcare providers receive thorough training in interpreting results and understanding their implications for patient management. Guidelines can offer recommendations, thereby assisting clinicians in making informed decisions.217 Implementing educational programs for healthcare professionals can enhance the acceptance of liquid biopsy technologies. Conducting health economic studies to demonstrate the value of liquid biopsy in terms of cost and improved outcomes can support its integration into healthcare systems.
Table S1 provides a concise comparison of the main FDA-approved biomarkers used in the detection, monitoring, and management of CRC, focusing on their sensitivity, specificity, and clinical indications.
Table S2 classifies key CRC biomarkers by their clinical role—diagnostic, prognostic, or predictive—and indicates whether they are FDA-approved for clinical use. Diagnostic biomarkers, such as CEA and mSEPT9, are primarily used for detecting or confirming the presence of disease, while prognostic biomarkers like BRAF and MSI/dMMR provide information about likely disease outcomes regardless of treatment. Predictive biomarkers, including KRAS/NRAS mutations and MSI/dMMR, help guide therapy by indicating which patients are more likely to benefit from specific treatments such as anti-EGFR antibodies or immunotherapy. Only a subset of these biomarkers, such as CEA, KRAS/NRAS, BRAF, MSI/dMMR, mSEPT9, and Cologuard, are currently FDA-approved, while others like ctDNA, miRNAs, and microbiome-based markers remain investigational and are not yet integrated into standard clinical practice.
Table S3 provides a comprehensive comparison between FDA-approved and investigational biomarkers for CRC. It highlights which biomarkers are currently validated and integrated into clinical practice, such as CEA, KRAS, and MSI, and distinguishes them from emerging biomarkers like certain microRNAs, circRNAs, and novel protein panels that are still under investigation. This clear distinction supports clinicians and researchers in understanding the current landscape of biomarker utility and the future directions of colorectal cancer diagnostics and monitoring.
In summary, recent studies have identified several novel biomarkers that are improving the early detection and management of CRC. Non-coding RNAs, such as circRNAs and lncRNAs, show promise for non-invasive screening and prognosis. DNA methylation markers like SEPT9 and SDC2 have led to FDA-approved blood tests with high sensitivity for CRC detection. Multi-target stool DNA tests, such as Cologuard, combine genetic and methylation markers to enhance detection rates over traditional methods. The presence of Fusobacterium nucleatum DNA in stool is emerging as a marker for CRC progression and prognosis. Additionally, liquid biopsy techniques and proteomic/metabolomic panels are providing new, minimally invasive ways to monitor disease and personalize treatment (Fig. 4).
Fig. 4.
Summary of recent studies on novel biomarkers in CRC.
.
Summary of recent studies on novel biomarkers in CRC.
Conclusion
The current landscape of FDA-approved biomarkers indicates a paradigm shift towards more personalized and effective cancer management. Tools such as Cologuard provide a non-invasive alternative to colonoscopy, significantly enhancing patient adherence to screening protocols due to their high sensitivity and specificity. Similarly, KRAS mutation testing plays a pivotal role in informing treatment decisions, particularly in predicting responses to anti-EGFR therapies. Although not intended for screening purposes, KRAS testing is essential for the customization of targeted treatments. CEA is predominantly utilized for monitoring treatment efficacy and disease recurrence, with sensitivity notably increasing in more advanced disease stages. MSI assays have also become integral in identifying patients eligible for immune checkpoint inhibitor therapies, thereby improving outcomes for individuals with MSI-high tumors. Additionally, methylated DNA markers, such as SEPT9, identify epigenetic alterations and contribute significantly to early detection efforts.
The development of effective and user-friendly biomarker-based tools not only reduces mortality but also facilitates early diagnosis and timely intervention, ultimately enhancing patient survival and quality of life. The future of cancer management appears promising, with advancements in multi-omics, liquid biopsy, artificial intelligence, and next-generation sequencing technologies, which have the potential to improve diagnostic precision, reduce costs, and integrate these biomarkers into standard clinical practice. Despite substantial progress, ongoing research remains essential to discover novel biomarkers and deepen our understanding of cancer biology. Addressing challenges related to standardization and accessibility will be crucial to ensuring the global implementation of these technologies in patient care.
Review Highlights
What is the current knowledge?
-
Colorectal cancer (CRC) is a major global health burden with increasing incidence projected by 2040.
-
Colonoscopy is effective but invasive, limiting compliance and widespread use for population-level screening.
-
FDA-approved biomarkers like CEA, KRAS, and MSI guide treatment or monitoring, but not all support early detection.
What is new here?
-
We highlight emerging biomarkers (e.g., circRNAs, methylated DNA, lncRNAs) with early detection and prognostic potential.
-
The review emphasizes the diagnostic role of multi-omics, AI, and liquid biopsy in future CRC management.
-
Integration of new molecular tools promises more accessible, precise, and personalized strategies for CRC diagnosis and care.
Competing Interests
The authors declare that they have no conflicts of interest.
Ethical Approval
Not applicable.
Supplementary files
Supplementary file 1 contains Tables S1-S3.
(pdf)
References
- Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol 2021; 14:101174. doi: 10.1016/j.tranon.2021.101174 [Crossref] [ Google Scholar]
- Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol 2019; 16:713-32. doi: 10.1038/s41575-019-0189-8 [Crossref] [ Google Scholar]
- Murphy N, Moreno V, Hughes DJ, Vodicka L, Vodicka P, Aglago EK. Lifestyle and dietary environmental factors in colorectal cancer susceptibility. Mol Aspects Med 2019; 69:2-9. doi: 10.1016/j.mam.2019.06.005 [Crossref] [ Google Scholar]
- Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet 2019; 394:1467-80. doi: 10.1016/s0140-6736(19)32319-0 [Crossref] [ Google Scholar]
- Guren MG. The global challenge of colorectal cancer. Lancet Gastroenterol Hepatol 2019; 4:894-5. doi: 10.1016/s2468-1253(19)30329-2 [Crossref] [ Google Scholar]
- Zamanian MY, Golmohammadi M, Abdullaev B, García MO, Alazbjee AA, Kumar A. A narrative review on therapeutic potential of naringenin in colorectal cancer: focusing on molecular and biochemical processes. Cell Biochem Funct 2024; 42:e4011. doi: 10.1002/cbf.4011 [Crossref] [ Google Scholar]
- Wen G, Jia Z, Wang Y, Kang Q, Hu D, Wang Z. Exploring the effect of carbon nanoparticle tracing technique on five-year overall survival and disease-free survival in patients undergoing radical surgery for colorectal cancer: a retrospective study. Front Oncol 2024; 14:1514175. doi: 10.3389/fonc.2024.1514175 [Crossref] [ Google Scholar]
- Garborg K, Holme Ø, Løberg M, Kalager M, Adami HO, Bretthauer M. Current status of screening for colorectal cancer. Ann Oncol 2013; 24:1963-72. doi: 10.1093/annonc/mdt157 [Crossref] [ Google Scholar]
- Mäbert K, Cojoc M, Peitzsch C, Kurth I, Souchelnytskyi S, Dubrovska A. Cancer biomarker discovery: current status and future perspectives. Int J Radiat Biol 2014; 90:659-77. doi: 10.3109/09553002.2014.892229 [Crossref] [ Google Scholar]
- Committee on the Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials, Board on Health Care Services, Board on Health Sciences Policy, Institute of Medicine. In: Micheel CM, Nass SJ, Omenn GS, eds. Evolution of Translational Omics: Lessons Learned and the Path Forward. Washington, DC: National Academies Press (US). 2012. doi: 10.17226/13297.
- Broza YY, Zhou X, Yuan M, Qu D, Zheng Y, Vishinkin R. Disease detection with molecular biomarkers: from chemistry of body fluids to nature-inspired chemical sensors. Chem Rev 2019; 119:11761-817. doi: 10.1021/acs.chemrev.9b00437 [Crossref] [ Google Scholar]
- Loktionov A. Biomarkers for detecting colorectal cancer non-invasively: DNA, RNA or proteins?. World J Gastrointest Oncol 2020; 12:124-48. doi: 10.4251/wjgo.v12.i2.124 [Crossref] [ Google Scholar]
- Binefa G, Rodríguez-Moranta F, Teule A, Medina-Hayas M. Colorectal cancer: from prevention to personalized medicine. World J Gastroenterol 2014; 20:6786-808. doi: 10.3748/wjg.v20.i22.6786 [Crossref] [ Google Scholar]
- Kuiper RP, Ligtenberg MJ, Hoogerbrugge N, Geurts van Kessel A. Germline copy number variation and cancer risk. Curr Opin Genet Dev 2010; 20:282-9. doi: 10.1016/j.gde.2010.03.005 [Crossref] [ Google Scholar]
- Li X, Liu G, Wu W. Recent advances in Lynch syndrome. Exp Hematol Oncol 2021; 10:37. doi: 10.1186/s40164-021-00231-4 [Crossref] [ Google Scholar]
- Iau PT, Macmillan RD, Blamey RW. Germ line mutations associated with breast cancer susceptibility. Eur J Cancer 2001; 37:300-21. doi: 10.1016/s0959-8049(00)00378-6 [Crossref] [ Google Scholar]
- Slade D. PARP and PARG inhibitors in cancer treatment. Genes Dev 2020; 34:360-94. doi: 10.1101/gad.334516.119 [Crossref] [ Google Scholar]
- Cortés-Ciriano I, Lee JJ, Xi R, Jain D, Jung YL, Yang L. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat Genet 2020; 52:331-41. doi: 10.1038/s41588-019-0576-7 [Crossref] [ Google Scholar]
- Schneider BL, Kulesz-Martin M. Destructive cycles: the role of genomic instability and adaptation in carcinogenesis. Carcinogenesis 2004; 25:2033-44. doi: 10.1093/carcin/bgh204 [Crossref] [ Google Scholar]
- Vargas-Rondón N, Villegas VE, Rondón-Lagos M. The role of chromosomal instability in cancer and therapeutic responses. Cancers (Basel) 2017; 10:4. doi: 10.3390/cancers10010004 [Crossref] [ Google Scholar]
- Laurent E, Talpaz M, Kantarjian H, Kurzrock R. The BCR gene and Philadelphia chromosome-positive leukemogenesis. Cancer Res 2001; 61:2343-55. [ Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C. Mutational landscape and significance across 12 major cancer types. Nature 2013; 502:333-9. doi: 10.1038/nature12634 [Crossref] [ Google Scholar]
- Youssef O, Sarhadi VK, Armengol G, Piirilä P, Knuuttila A, Knuutila S. Exhaled breath condensate as a source of biomarkers for lung carcinomas A focus on genetic and epigenetic markers-a mini-review. Genes Chromosomes Cancer 2016; 55:905-14. doi: 10.1002/gcc.22399 [Crossref] [ Google Scholar]
- Sarhadi V, Lahti L, Saberi F, Youssef O, Kokkola A, Karla T. Gut microbiota and host gene mutations in colorectal cancer patients and controls of Iranian and Finnish origin. Anticancer Res 2020; 40:1325-34. doi: 10.21873/anticanres.14074 [Crossref] [ Google Scholar]
- Sarhadi VK, Armengol G. Molecular biomarkers in cancer. Biomolecules 2022; 12:1021. doi: 10.3390/biom12081021 [Crossref] [ Google Scholar]
- Tuononen K, Sarhadi VK, Wirtanen A, Rönty M, Salmenkivi K, Knuuttila A. Targeted resequencing reveals ALK fusions in non-small cell lung carcinomas detected by FISH, immunohistochemistry, and real-time RT-PCR: a comparison of four methods. Biomed Res Int 2013; 2013:757490. doi: 10.1155/2013/757490 [Crossref] [ Google Scholar]
- Stowell SR, Ju T, Cummings RD. Protein glycosylation in cancer. Annu Rev Pathol 2015. 10: 473-510. doi: 10.1146/annurev-pathol-012414-040438.
- Ding Z, Wang N, Ji N, Chen ZS. Proteomics technologies for cancer liquid biopsies. Mol Cancer 2022; 21:53. doi: 10.1186/s12943-022-01526-8 [Crossref] [ Google Scholar]
- Alnabulsi A, Murray GI. Proteomics for early detection of colorectal cancer: recent updates. Expert Rev Proteomics 2018; 15:55-63. doi: 10.1080/14789450.2018.1396893 [Crossref] [ Google Scholar]
- Gonçalves E, Poulos RC, Cai Z, Barthorpe S, Manda SS, Lucas N, et al. Pan-cancer proteomic map of 949 human cell lines. Cancer Cell 2022; 40: 835-49.e8. doi: 10.1016/j.ccell.2022.06.010.
- Lu J, Lee-Gabel L, Nadeau MC, Ferencz TM, Soefje SA. Clinical evaluation of compounds targeting PD-1/PD-L1 pathway for cancer immunotherapy. J Oncol Pharm Pract 2015; 21:451-67. doi: 10.1177/1078155214538087 [Crossref] [ Google Scholar]
- Park R, Da Silva LL, Saeed A. Immunotherapy predictive molecular markers in advanced gastroesophageal cancer: MSI and beyond. Cancers (Basel) 2021; 13:1715. doi: 10.3390/cancers13071715 [Crossref] [ Google Scholar]
- Cheung P, Lau P. Epigenetic regulation by histone methylation and histone variants. Mol Endocrinol 2005; 19:563-73. doi: 10.1210/me.2004-0496 [Crossref] [ Google Scholar]
- Müller D, Győrffy B. DNA methylation-based diagnostic, prognostic, and predictive biomarkers in colorectal cancer. Biochim Biophys Acta Rev Cancer 2022; 1877:188722. doi: 10.1016/j.bbcan.2022.188722 [Crossref] [ Google Scholar]
- Poon MTC, Keni S, Vimalan V, Ip C, Smith C, Erridge S. Extent of MGMT promoter methylation modifies the effect of temozolomide on overall survival in patients with glioblastoma: a regional cohort study. Neurooncol Adv 2021; 3:vdab171. doi: 10.1093/noajnl/vdab171 [Crossref] [ Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004; 431:931-45. doi: 10.1038/nature03001 [Crossref] [ Google Scholar]
- Hrdlickova R, Toloue M, Tian B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip Rev RNA 2017; 8: 10.1002/wrna.1364. doi: 10.1002/wrna.1364.
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6:857-66. doi: 10.1038/nrc1997 [Crossref] [ Google Scholar]
- Rapado-González Ó, Majem B, Álvarez-Castro A, Díaz-Peña R, Abalo A, Suárez-Cabrera L. A novel saliva-based miRNA signature for colorectal cancer diagnosis. J Clin Med 2019; 8:2029. doi: 10.3390/jcm8122029 [Crossref] [ Google Scholar]
- Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 2010; 127:118-26. doi: 10.1002/ijc.25007 [Crossref] [ Google Scholar]
- Gurer T, Aytekin A, Caki E, Gezici S. [miR-485-3p and miR-4728-5p as tumor suppressors in pathogenesis of colorectal cancer]. Mol Biol (Mosk) 2022; 56:516-20. doi: 10.31857/s0026898422030077.[Russian] [Crossref] [ Google Scholar]
- Han Y, Zhang H, Bian C, Chen C, Tu S, Guo J. Circular RNA expression: its potential regulation and function in abdominal aortic aneurysms. Oxid Med Cell Longev 2021; 2021:9934951. doi: 10.1155/2021/9934951 [Crossref] [ Google Scholar]
- Zheng R, Zhang K, Tan S, Gao F, Zhang Y, Xu W. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Mol Cancer 2022; 21:49. doi: 10.1186/s12943-021-01471-y [Crossref] [ Google Scholar]
- Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015; 25:981-4. doi: 10.1038/cr.2015.82 [Crossref] [ Google Scholar]
- Jiang W, Zhang X, Chu Q, Lu S, Zhou L, Lu X. The circular RNA profiles of colorectal tumor metastatic cells. Front Genet 2018; 9:34. doi: 10.3389/fgene.2018.00034 [Crossref] [ Google Scholar]
- Qi L, Pan Y, Tang M, Chen X. Circulating cell-free circRNA panel predicted tumorigenesis and development of colorectal cancer. J Clin Lab Anal 2022; 36:e24431. doi: 10.1002/jcla.24431 [Crossref] [ Google Scholar]
- Radanova M, Mihaylova G, Nazifova-Tasinova N, Levkova M, Tasinov O, Ivanova D. Oncogenic functions and clinical significance of circular RNAs in colorectal cancer. Cancers (Basel) 2021; 13:3395. doi: 10.3390/cancers13143395 [Crossref] [ Google Scholar]
- Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci 2018; 109:2093-100. doi: 10.1111/cas.13642 [Crossref] [ Google Scholar]
- Léveillé N, Melo CA, Rooijers K, Díaz-Lagares A, Melo SA, Korkmaz G. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun 2015; 6:6520. doi: 10.1038/ncomms7520 [Crossref] [ Google Scholar]
- Zhao H, Kim Y, Wang P, Lapointe J, Tibshirani R, Pollack JR. Genome-wide characterization of gene expression variations and DNA copy number changes in prostate cancer cell lines. Prostate 2005; 63:187-97. doi: 10.1002/pros.20158 [Crossref] [ Google Scholar]
- Wang G, Li Z, Zhao Q, Zhu Y, Zhao C, Li X. LincRNA-p21 enhances the sensitivity of radiotherapy for human colorectal cancer by targeting the Wnt/β-catenin signaling pathway. Oncol Rep 2014; 31:1839-45. doi: 10.3892/or.2014.3047 [Crossref] [ Google Scholar]
- Guo S, Li L, Xu B, Li M, Zeng Q, Xiao H. A simple and novel fecal biomarker for colorectal cancer: ratio of Fusobacterium nucleatum to probiotics populations, based on their antagonistic effect. Clin Chem 2018; 64:1327-37. doi: 10.1373/clinchem.2018.289728 [Crossref] [ Google Scholar]
- Li Y, Song Y, Huang J, Wang K. APC mutations as a predictive marker of endometrial cancer immunotherapy: a retrospective cohort study. Lancet Oncol 2022; 23:S9. doi: 10.1016/s1470-2045(22)00408-9 [Crossref] [ Google Scholar]
- Zhang W, Kong L, Zhu H, Sun D, Han Q, Yan B. Retinoic acid-induced 2 (RAI2) is a novel antagonist of Wnt/β-catenin signaling pathway and potential biomarker of chemosensitivity in colorectal cancer. Front Oncol 2022; 12:805290. doi: 10.3389/fonc.2022.805290 [Crossref] [ Google Scholar]
- Liebl MC, Hofmann TG. The role of p53 signaling in colorectal cancer. Cancers (Basel) 2021; 13:2125. doi: 10.3390/cancers13092125 [Crossref] [ Google Scholar]
- Russo A, Bazan V, Iacopetta B, Kerr D, Soussi T, Gebbia N. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol 2005; 23:7518-28. doi: 10.1200/jco.2005.00.471 [Crossref] [ Google Scholar]
- Zhang Y, Wang Y, Zhang B, Li P, Zhao Y. Methods and biomarkers for early detection, prediction, and diagnosis of colorectal cancer. Biomed Pharmacother 2023; 163:114786. doi: 10.1016/j.biopha.2023.114786 [Crossref] [ Google Scholar]
- Rajurkar M, Parikh AR, Solovyov A, You E, Kulkarni AS, Chu C. Reverse transcriptase inhibition disrupts repeat element life cycle in colorectal cancer. Cancer Discov 2022; 12:1462-81. doi: 10.1158/2159-8290.Cd-21-1117 [Crossref] [ Google Scholar]
- Rosty C, Young JP, Walsh MD, Clendenning M, Sanderson K, Walters RJ. PIK3CA activating mutation in colorectal carcinoma: associations with molecular features and survival. PLoS One 2013; 8:e65479. doi: 10.1371/journal.pone.0065479 [Crossref] [ Google Scholar]
- Zhang M, Jang H, Nussinov R. The structural basis for Ras activation of PI3Kα lipid kinase. Phys Chem Chem Phys 2019; 21:12021-8. doi: 10.1039/c9cp00101h [Crossref] [ Google Scholar]
- Brown KK, Toker A. The phosphoinositide 3-kinase pathway and therapy resistance in cancer. F1000Prime Rep 2015; 7:13. doi: 10.12703/p7-13 [Crossref] [ Google Scholar]
- Hall DC, Benndorf RA. Aspirin sensitivity of PIK3CA-mutated colorectal cancer: potential mechanisms revisited. Cell Mol Life Sci 2022; 79:393. doi: 10.1007/s00018-022-04430-y [Crossref] [ Google Scholar]
- Stefani C, Miricescu D, Stanescu-Spinu II, Nica RI, Greabu M, Totan AR. Growth factors, PI3K/AKT/mTOR and MAPK signaling pathways in colorectal cancer pathogenesis: where are we now?. Int J Mol Sci 2021; 22:12260. doi: 10.3390/ijms221910260 [Crossref] [ Google Scholar]
- Mehrvarz Sarshekeh A, Advani S, Overman MJ, Manyam G, Kee BK, Fogelman DR. Association of SMAD4 mutation with patient demographics, tumor characteristics, and clinical outcomes in colorectal cancer. PLoS One 2017; 12:e0173345. doi: 10.1371/journal.pone.0173345 [Crossref] [ Google Scholar]
- Wismayer R, Matthews R, Whalley C, Kiwanuka J, Kakembo FE, Thorn S. Determination of the frequency and distribution of APC, PIK3CA, and SMAD4 gene mutations in Ugandan patients with colorectal cancer. BMC Cancer 2024; 24:1212. doi: 10.1186/s12885-024-12967-3 [Crossref] [ Google Scholar]
- Zhao M, Mishra L, Deng CX. The role of TGF-β/SMAD4 signaling in cancer. Int J Biol Sci 2018; 14:111-23. doi: 10.7150/ijbs.23230 [Crossref] [ Google Scholar]
- Pfohl U, Loskutov J, Bashir S, Kühn R, Herter P, Templin M. A RAS-independent biomarker panel to reliably predict response to MEK inhibition in colorectal cancer. Cancers (Basel) 2022; 14:3252. doi: 10.3390/cancers14133252 [Crossref] [ Google Scholar]
- Wang C, Sandhu J, Tsao A, Fakih M. Presence of concurrent TP53 mutations is necessary to predict poor outcomes within the SMAD4 mutated subgroup of metastatic colorectal cancer. Cancers (Basel) 2022; 14:3644. doi: 10.3390/cancers14153644 [Crossref] [ Google Scholar]
- Mei Z, Shao YW, Lin P, Cai X, Wang B, Ding Y. SMAD4 and NF1 mutations as potential biomarkers for poor prognosis to cetuximab-based therapy in Chinese metastatic colorectal cancer patients. BMC Cancer 2018; 18:479. doi: 10.1186/s12885-018-4298-5 [Crossref] [ Google Scholar]
- Kang YH, Wang JH, Lee JS, Lee NH, Son CG. Coptidis Rhizoma suppresses metastatic behavior by inhibiting TGF-β-mediated epithelial-mesenchymal transition in 5-FU-resistant HCT116 cells. Front Pharmacol 2022; 13:909331. doi: 10.3389/fphar.2022.909331 [Crossref] [ Google Scholar]
- Afolabi H, Md Salleh S, Zakaria Z, Seng CE, Mohd Nafil SN, Abdul Aziz AA. A systematic review and meta-analysis on the occurrence of biomarker mutation in colorectal cancer among the Asian population. Biomed Res Int 2022; 2022:5824183. doi: 10.1155/2022/5824183 [Crossref] [ Google Scholar]
- Damit D, Patnaik R, Chaw LL, Lu SK, Telisinghe PU, Lu ZH. KRAS mutation: characterization and its impact on survival outcome of patients with metastatic colorectal cancer. Front Biosci (Landmark Ed) 2022; 27:213. doi: 10.31083/j.fbl2707213 [Crossref] [ Google Scholar]
- Sui X, Chen Y, Liu B, Li L, Huang X, Wang M. The relationship between KRAS gene mutation and intestinal flora in tumor tissues of colorectal cancer patients. Ann Transl Med 2020; 8:1085. doi: 10.21037/atm-20-5622 [Crossref] [ Google Scholar]
- Sammoud S, Khiari M, Semeh A, Amine L, Ines C, Amira A. Relationship between expression of ras p21 oncoprotein and mutation status of the K-ras gene in sporadic colorectal cancer patients in Tunisia. Appl Immunohistochem Mol Morphol 2012; 20:146-52. doi: 10.1097/PAI.0b013e3182240de1 [Crossref] [ Google Scholar]
- Vitiello PP, Cardone C, Martini G, Ciardiello D, Belli V, Matrone N. Receptor tyrosine kinase-dependent PI3K activation is an escape mechanism to vertical suppression of the EGFR/RAS/MAPK pathway in KRAS-mutated human colorectal cancer cell lines. J Exp Clin Cancer Res 2019; 38:41. doi: 10.1186/s13046-019-1035-0 [Crossref] [ Google Scholar]
- Tosi F, Magni E, Amatu A, Mauri G, Bencardino K, Truini M. Effect of KRAS and BRAF mutations on survival of metastatic colorectal cancer after liver resection: a systematic review and meta-analysis. Clin Colorectal Cancer 2017; 16:e153-63. doi: 10.1016/j.clcc.2017.01.004 [Crossref] [ Google Scholar]
- Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov 2014; 4:1269-80. doi: 10.1158/2159-8290.Cd-14-0462 [Crossref] [ Google Scholar]
- Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015; 21:827. doi: 10.1038/nm0715-827b [Crossref] [ Google Scholar]
- Chong LC, Hardingham JE, Townsend AR, Piantadosi C, Rico GT, Karapetis C. Rechallenge with anti-EGFR therapy in metastatic colorectal cancer (mCRC): results from South Australia mCRC registry. Target Oncol 2020; 15:751-7. doi: 10.1007/s11523-020-00760-8 [Crossref] [ Google Scholar]
- Bando H, Ohtsu A, Yoshino T. Therapeutic landscape and future direction of metastatic colorectal cancer. Nat Rev Gastroenterol Hepatol 2023; 20:306-22. doi: 10.1038/s41575-022-00736-1 [Crossref] [ Google Scholar]
- Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell 2008; 132:363-74. doi: 10.1016/j.cell.2007.12.032 [Crossref] [ Google Scholar]
- Beeram M, Patnaik A, Rowinsky EK. Raf: a strategic target for therapeutic development against cancer. J Clin Oncol 2005; 23:6771-90. doi: 10.1200/jco.2005.08.036 [Crossref] [ Google Scholar]
- Chen D, Huang JF, Liu K, Zhang LQ, Yang Z, Chuai ZR. BRAFV600E mutation and its association with clinicopathological features of colorectal cancer: a systematic review and meta-analysis. PLoS One 2014; 9:e90607. doi: 10.1371/journal.pone.0090607 [Crossref] [ Google Scholar]
- Margonis GA, Buettner S, Andreatos N, Kim Y, Wagner D, Sasaki K. Association of BRAF mutations with survival and recurrence in surgically treated patients with metastatic colorectal liver cancer. JAMA Surg 2018; 153:e180996. doi: 10.1001/jamasurg.2018.0996 [Crossref] [ Google Scholar]
- Barras D, Missiaglia E, Wirapati P, Sieber OM, Jorissen RN, Love C. BRAF V600E mutant colorectal cancer subtypes based on gene expression. Clin Cancer Res 2017; 23:104-15. doi: 10.1158/1078-0432.Ccr-16-0140 [Crossref] [ Google Scholar]
- Tak E, An HI, Lee AS, Han K, Choi J, Kim HD. Antitumor effects of immunotherapy combined with BRAF and MEK inhibitors in BRAF V600E metastatic colorectal cancer. Cancer Immunol Immunother 2025; 74:154. doi: 10.1007/s00262-025-04005-3 [Crossref] [ Google Scholar]
- Ambrosini M, Tougeron D, Modest D, Guimbaud R, Kopetz S, Decraecker M. BRAF + EGFR + /- MEK inhibitors after immune checkpoint inhibitors in BRAF V600E mutated and deficient mismatch repair or microsatellite instability high metastatic colorectal cancer. Eur J Cancer 2024; 210:114290. doi: 10.1016/j.ejca.2024.114290 [Crossref] [ Google Scholar]
- Koncina E, Haan S, Rauh S, Letellier E. Prognostic and predictive molecular biomarkers for colorectal cancer: updates and challenges. Cancers (Basel) 2020; 12:319. doi: 10.3390/cancers12020319 [Crossref] [ Google Scholar]
- Li Y, Xiao J, Zhang T, Zheng Y, Jin H. Analysis of KRAS, NRAS, and BRAF mutations, microsatellite instability, and relevant prognosis effects in patients with early colorectal cancer: a cohort study in East Asia. Front Oncol 2022; 12:897548. doi: 10.3389/fonc.2022.897548 [Crossref] [ Google Scholar]
- Wang C, Pan D. Mutation patterns and prognostic analysis of BRAF/KRAS/PIK3CA in colorectal cancer. J Clin Lab Anal 2022; 36:e24444. doi: 10.1002/jcla.24444 [Crossref] [ Google Scholar]
- Formica V, Sera F, Cremolini C, Riondino S, Morelli C, Arkenau HT. KRAS and BRAF mutations in stage II and III colon cancer: a systematic review and meta-analysis. J Natl Cancer Inst 2022; 114:517-27. doi: 10.1093/jnci/djab190 [Crossref] [ Google Scholar]
- Ismael NE, El Sheikh SA, Talaat SM, Salem EM. Mismatch repair proteins and microsatellite instability in colorectal carcinoma (MLH1, MSH2, MSH6 and PMS2): histopathological and immunohistochemical study. Open Access Maced J Med Sci 2017; 5:9-13. doi: 10.3889/oamjms.2017.003 [Crossref] [ Google Scholar]
- Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev 2008; 129:391-407. doi: 10.1016/j.mad.2008.02.012 [Crossref] [ Google Scholar]
- Chen J, Yan Q, Sun J, Wang Q, Tao Y, Xiao D. Microsatellite status detection of colorectal cancer: evaluation of inconsistency between PCR and IHC. J Cancer 2023; 14:1132-40. doi: 10.7150/jca.81675 [Crossref] [ Google Scholar]
- Gelsomino F, Barbolini M, Spallanzani A, Pugliese G, Cascinu S. The evolving role of microsatellite instability in colorectal cancer: a review. Cancer Treat Rev 2016; 51:19-26. doi: 10.1016/j.ctrv.2016.10.005 [Crossref] [ Google Scholar]
- Sun BL. Current microsatellite instability testing in management of colorectal cancer. Clin Colorectal Cancer 2021; 20:e12-20. doi: 10.1016/j.clcc.2020.08.001 [Crossref] [ Google Scholar]
- Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med 2003; 348:919-32. doi: 10.1056/NEJMra012242 [Crossref] [ Google Scholar]
- Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med 2000; 342:69-77. doi: 10.1056/nejm200001133420201 [Crossref] [ Google Scholar]
- Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 2005; 23:609-18. doi: 10.1200/jco.2005.01.086 [Crossref] [ Google Scholar]
- Basile D, Garattini SK, Bonotto M, Ongaro E, Casagrande M, Cattaneo M. Immunotherapy for colorectal cancer: where are we heading?. Expert Opin Biol Ther 2017; 17:709-21. doi: 10.1080/14712598.2017.1315405 [Crossref] [ Google Scholar]
- Marcus L, Lemery SJ, Keegan P, Pazdur R. FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin Cancer Res 2019; 25:3753-8. doi: 10.1158/1078-0432.Ccr-18-4070 [Crossref] [ Google Scholar]
- Bilgin B, Sendur MA, Bülent Akıncı M, Şener Dede D, Yalçın B. Targeting the PD-1 pathway: a new hope for gastrointestinal cancers. Curr Med Res Opin 2017; 33:749-59. doi: 10.1080/03007995.2017.1279132 [Crossref] [ Google Scholar]
- Sahin IH, Akce M, Alese O, Shaib W, Lesinski GB, El-Rayes B. Immune checkpoint inhibitors for the treatment of MSI-H/MMR-D colorectal cancer and a perspective on resistance mechanisms. Br J Cancer 2019; 121:809-18. doi: 10.1038/s41416-019-0599-y [Crossref] [ Google Scholar]
- Morse MA, Hochster H, Benson A. Perspectives on treatment of metastatic colorectal cancer with immune checkpoint inhibitor therapy. Oncologist 2020; 25:33-45. doi: 10.1634/theoncologist.2019-0176 [Crossref] [ Google Scholar]
- Yakabe T, Nakafusa Y, Sumi K, Miyoshi A, Kitajima Y, Sato S. Clinical significance of CEA and CA19-9 in postoperative follow-up of colorectal cancer. Ann Surg Oncol 2010; 17:2349-56. doi: 10.1245/s10434-010-1004-5 [Crossref] [ Google Scholar]
- Johnson B, Mahadevan D. Emerging role and targeting of carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) in human malignancies. Clin Cancer Drugs 2015; 2:100-11. doi: 10.2174/2212697x02666150602215823 [Crossref] [ Google Scholar]
- Sisik A, Kaya M, Bas G, Basak F, Alimoglu O. CEA and CA 19-9 are still valuable markers for the prognosis of colorectal and gastric cancer patients. Asian Pac J Cancer Prev 2013; 14:4289-94. doi: 10.7314/apjcp.2013.14.7.4289 [Crossref] [ Google Scholar]
- Hao C, Zhang G, Zhang L. Serum CEA levels in 49 different types of cancer and noncancer diseases. Prog Mol Biol Transl Sci 2019; 162:213-27. doi: 10.1016/bs.pmbts.2018.12.011 [Crossref] [ Google Scholar]
- Fakih MG, Padmanabhan A. CEA monitoring in colorectal cancer What you should know. Oncology (Williston Park) 2006; 20:579-87. [ Google Scholar]
- Luo H, Shen K, Li B, Li R, Wang Z, Xie Z. Clinical significance and diagnostic value of serum NSE, CEA, CA19-9, CA125 and CA242 levels in colorectal cancer. Oncol Lett 2020; 20:742-50. doi: 10.3892/ol.2020.11633 [Crossref] [ Google Scholar]
- Takagawa R, Fujii S, Ohta M, Nagano Y, Kunisaki C, Yamagishi S. Preoperative serum carcinoembryonic antigen level as a predictive factor of recurrence after curative resection of colorectal cancer. Ann Surg Oncol 2008; 15:3433-9. doi: 10.1245/s10434-008-0168-8 [Crossref] [ Google Scholar]
- Wang WS, Lin JK, Chiou TJ, Liu JH, Fan FS, Yen CC. Preoperative carcinoembryonic antigen level as an independent prognostic factor in colorectal cancer: Taiwan experience. Jpn J Clin Oncol 2000; 30:12-6. doi: 10.1093/jjco/hyd003 [Crossref] [ Google Scholar]
- Wiratkapun S, Kraemer M, Seow-Choen F, Ho YH, Eu KW. High preoperative serum carcinoembryonic antigen predicts metastatic recurrence in potentially curative colonic cancer: results of a five-year study. Dis Colon Rectum 2001; 44:231-5. doi: 10.1007/bf02234298 [Crossref] [ Google Scholar]
- Yang KM, Park IJ, Kim CW, Roh SA, Cho DH, Kim JC. The prognostic significance and treatment modality for elevated pre- and postoperative serum CEA in colorectal cancer patients. Ann Surg Treat Res 2016; 91:165-71. doi: 10.4174/astr.2016.91.4.165 [Crossref] [ Google Scholar]
- Duffy MJ, McDermott EW, Crown J. Blood-based biomarkers in breast cancer: from proteins to circulating tumor cells to circulating tumor DNA. Tumour Biol 2018; 40:1010428318776169. doi: 10.1177/1010428318776169 [Crossref] [ Google Scholar]
- Mego M, Mani SA, Cristofanilli M. Molecular mechanisms of metastasis in breast cancer--clinical applications. Nat Rev Clin Oncol 2010; 7:693-701. doi: 10.1038/nrclinonc.2010.171 [Crossref] [ Google Scholar]
- Okada M, Nishio W, Sakamoto T, Uchino K, Yuki T, Nakagawa A. Effect of histologic type and smoking status on interpretation of serum carcinoembryonic antigen value in non-small cell lung carcinoma. Ann Thorac Surg 2004; 78:1004-9. doi: 10.1016/j.athoracsur.2004.03.019 [Crossref] [ Google Scholar]
- Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 2008; 83:584-94. doi: 10.4065/83.5.584 [Crossref] [ Google Scholar]
- Koprowski H, Steplewski Z, Mitchell K, Herlyn M, Herlyn D, Fuhrer P. Colorectal carcinoma antigens detected by hybridoma antibodies. Somatic Cell Genet 1979; 5:957-71. doi: 10.1007/bf01542654 [Crossref] [ Google Scholar]
- Nakayama T, Watanabe M, Teramoto T, Kitajima M. Slope analysis of CA19-9 and CEA for predicting recurrence in colorectal cancer patients. Anticancer Res 1997; 17:1379-82. [ Google Scholar]
- Kim S, Park BK, Seo JH, Choi J, Choi JW, Lee CK. Carbohydrate antigen 19-9 elevation without evidence of malignant or pancreatobiliary diseases. Sci Rep 2020; 10:8820. doi: 10.1038/s41598-020-65720-8 [Crossref] [ Google Scholar]
- Duffy MJ, van Dalen A, Haglund C, Hansson L, Holinski-Feder E, Klapdor R. Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur J Cancer 2007; 43:1348-60. doi: 10.1016/j.ejca.2007.03.021 [Crossref] [ Google Scholar]
- Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol 2006; 24:5313-27. doi: 10.1200/jco.2006.08.2644 [Crossref] [ Google Scholar]
- Bolocan A, Ion D, Ciocan DN, Paduraru DN. Prognostic and predictive factors in colorectal cancer. Chirurgia (Bucur) 2012; 107:555-63. [ Google Scholar]
- Lakemeyer L, Sander S, Wittau M, Henne-Bruns D, Kornmann M, Lemke J. Diagnostic and prognostic value of CEA and CA19-9 in colorectal cancer. Diseases 2021; 9:21. doi: 10.3390/diseases9010021 [Crossref] [ Google Scholar]
- Ueda T, Shimada E, Urakawa T. The clinicopathologic features of serum CA 19-9-positive colorectal cancers. Surg Today 1994; 24:518-25. doi: 10.1007/bf01884571 [Crossref] [ Google Scholar]
- Shin JK, Kim HC, Lee WY, Yun SH, Cho YB, Huh JW. High preoperative serum CA 19-9 levels can predict poor oncologic outcomes in colorectal cancer patients on propensity score analysis. Ann Surg Treat Res 2019; 96:107-15. doi: 10.4174/astr.2019.96.3.107 [Crossref] [ Google Scholar]
- Zhang LN, OuYang PY, Xiao WW, Yu X, You KY, Zeng ZF. Elevated CA19-9 as the most significant prognostic factor in locally advanced rectal cancer following neoadjuvant chemoradiotherapy. Medicine (Baltimore) 2015; 94:e1793. doi: 10.1097/md.0000000000001793 [Crossref] [ Google Scholar]
- Lu Z, Peng J, Wang Z, Pan Z, Yuan Y, Wan D. High preoperative serum CA19-9 level is predictive of poor prognosis for patients with colorectal liver oligometastases undergoing hepatic resection. Med Oncol 2016; 33:121. doi: 10.1007/s12032-016-0838-5 [Crossref] [ Google Scholar]
- Zhang SY, Lin M, Zhang HB. Diagnostic value of carcinoembryonic antigen and carcinoma antigen 19-9 for colorectal carcinoma. Int J Clin Exp Pathol 2015; 8:9404-9. [ Google Scholar]
- Filella X, Molina R, Grau JJ, Piqué JM, Garcia-Valdecasas JC, Astudillo E. Prognostic value of CA 199 levels in colorectal cancer. Ann Surg 1992; 216:55-9. doi: 10.1097/00000658-199207000-00008 [Crossref] [ Google Scholar]
- Ozawa T, Ishihara S, Kawai K, Nozawa H, Yamaguchi H, Kitayama J. Prognostic significance of preoperative serum carbohydrate antigen 19-9 in patients with stage IV colorectal cancer. Clin Colorectal Cancer 2016; 15:e157-63. doi: 10.1016/j.clcc.2016.04.012 [Crossref] [ Google Scholar]
- Bagaria B, Sood S, Sharma R, Lalwani S. Comparative study of CEA and CA19-9 in esophageal, gastric and colon cancers individually and in combination (ROC curve analysis). Cancer Biol Med 2013; 10:148-57. doi: 10.7497/j.issn.2095-3941.2013.03.005 [Crossref] [ Google Scholar]
- Duffy MJ, Lamerz R, Haglund C, Nicolini A, Kalousová M, Holubec L. Tumor markers in colorectal cancer, gastric cancer and gastrointestinal stromal cancers: European group on tumor markers 2014 guidelines update. Int J Cancer 2014; 134:2513-22. doi: 10.1002/ijc.28384 [Crossref] [ Google Scholar]
- Jia J, Zhang P, Gou M, Yang F, Qian N, Dai G. The role of serum CEA and CA19-9 in efficacy evaluations and progression-free survival predictions for patients treated with cetuximab combined with FOLFOX4 or FOLFIRI as a first-line treatment for advanced colorectal cancer. Dis Markers 2019; 2019:6812045. doi: 10.1155/2019/6812045 [Crossref] [ Google Scholar]
- Alese OB, Wu C, Chapin WJ, Ulanja MB, Zheng-Lin B, Amankwah M. Update on emerging therapies for advanced colorectal cancer. Am Soc Clin Oncol Educ Book 2023; 43:e389574. doi: 10.1200/edbk_389574 [Crossref] [ Google Scholar]
- Selcukbiricik F, Bilici A, Tural D, Erdamar S, Soyluk O, Buyukunal E. Are high initial CEA and CA 19-9 levels associated with the presence of K-ras mutation in patients with metastatic colorectal cancer?. Tumour Biol 2013; 34:2233-9. doi: 10.1007/s13277-013-0763-6 [Crossref] [ Google Scholar]
- Jeon HJ, Seo Jh, Jeong E, Son CY, Rawding PA, Hwang Y. Carcinoembryonic antigen-positive circulating epithelial cells as a biomarker for the diagnosis and prognosis of colorectal cancer. Biotechnol Bioprocess Eng 2024; 29:877-89. doi: 10.1007/s12257-024-00115-4 [Crossref] [ Google Scholar]
- Berretta M, Alessandrini L, De Divitiis C, Nasti G, Lleshi A, Di Francia R. Serum and tissue markers in colorectal cancer: state of art. Crit Rev Oncol Hematol 2017; 111:103-16. doi: 10.1016/j.critrevonc.2017.01.007 [Crossref] [ Google Scholar]
- Yu H, Son GM, Joh YG. The clinical significance of preoperative serum levels of carbohydrate antigen 19-9 in colorectal cancer. J Korean Surg Soc 2013; 84:231-7. doi: 10.4174/jkss.2013.84.4.231 [Crossref] [ Google Scholar]
- Kim Y, Bu J, Cho YH, Son IT, Kang SB. A viable circulating tumor cell isolation device with high retrieval efficiency using a reversibly deformable membrane barrier. J Micromech Microeng 2017; 27:025015. doi: 10.1088/1361-6439/aa53ff [Crossref] [ Google Scholar]
- Dong D, Zhang L, Jia L, Ji W, Wang Z, Ren L. Identification of serum periostin as a potential diagnostic and prognostic marker for colorectal cancer. Clin Lab 2018; 64:973-81. doi: 10.7754/Clin.Lab.2018.171225 [Crossref] [ Google Scholar]
- Wang J, Wang X, Yu F, Chen J, Zhao S, Zhang D. Combined detection of preoperative serum CEA, CA19-9 and CA242 improve prognostic prediction of surgically treated colorectal cancer patients. Int J Clin Exp Pathol 2015; 8:14853-63. [ Google Scholar]
- Ning S, Wei W, Li J, Hou B, Zhong J, Xie Y. Clinical significance and diagnostic capacity of serum TK1, CEA, CA 19-9 and CA 72-4 levels in gastric and colorectal cancer patients. J Cancer 2018; 9:494-501. doi: 10.7150/jca.21562 [Crossref] [ Google Scholar]
- Haque A, Ray SK, Cox A, Banik NL. Neuron specific enolase: a promising therapeutic target in acute spinal cord injury. Metab Brain Dis 2016; 31:487-95. doi: 10.1007/s11011-016-9801-6 [Crossref] [ Google Scholar]
- Baqar AR, Wilkins S, Staples M, Angus Lee CH, Oliva K, McMurrick P. The role of preoperative CEA in the management of colorectal cancer: a cohort study from two cancer centres. Int J Surg 2019; 64:10-5. doi: 10.1016/j.ijsu.2019.02.014 [Crossref] [ Google Scholar]
- Wang Y, Chen PM, Liu RB. Advance in plasma SEPT9 gene methylation assay for colorectal cancer early detection. World J Gastrointest Oncol 2018; 10:15-22. doi: 10.4251/wjgo.v10.i1.15 [Crossref] [ Google Scholar]
- Sun J, Zheng MY, Li YW, Zhang SW. Structure and function of septin 9 and its role in human malignant tumors. World J Gastrointest Oncol 2020; 12:619-31. doi: 10.4251/wjgo.v12.i6.619 [Crossref] [ Google Scholar]
- Tóth K, Galamb O, Spisák S, Wichmann B, Sipos F, Valcz G. The influence of methylated septin 9 gene on RNA and protein level in colorectal cancer. Pathol Oncol Res 2011; 17:503-9. doi: 10.1007/s12253-010-9338-7 [Crossref] [ Google Scholar]
- Wasserkort R, Kalmar A, Valcz G, Spisak S, Krispin M, Toth K. Aberrant septin 9 DNA methylation in colorectal cancer is restricted to a single CpG island. BMC Cancer 2013; 13:398. doi: 10.1186/1471-2407-13-398 [Crossref] [ Google Scholar]
- Hu J, Hu B, Gui YC, Tan ZB, Xu JW. Diagnostic value and clinical significance of methylated SEPT9 for colorectal cancer: a meta-analysis. Med Sci Monit 2019; 25:5813-22. doi: 10.12659/msm.915472 [Crossref] [ Google Scholar]
- Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014; 63:317-25. doi: 10.1136/gutjnl-2012-304149 [Crossref] [ Google Scholar]
- Peterse EF, Meester RG, de Jonge L, Omidvari AH, Alarid-Escudero F, Knudsen AB. Comparing the cost-effectiveness of innovative colorectal cancer screening tests. J Natl Cancer Inst 2021; 113:154-61. doi: 10.1093/jnci/djaa103 [Crossref] [ Google Scholar]
- Fu B, Yan P, Zhang S, Lu Y, Pan L, Tang W. Cell-free circulating methylated SEPT9 for noninvasive diagnosis and monitoring of colorectal cancer. Dis Markers 2018; 2018:6437104. doi: 10.1155/2018/6437104 [Crossref] [ Google Scholar]
- Song L, Jia J, Peng X, Xiao W, Li Y. The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: a meta-analysis. Sci Rep 2017; 7:3032. doi: 10.1038/s41598-017-03321-8 [Crossref] [ Google Scholar]
- Wu D, Zhou G, Jin P, Zhu J, Li S, Wu Q. Detection of colorectal cancer using a simplified SEPT9 gene methylation assay is a reliable method for opportunistic screening. J Mol Diagn 2016; 18:535-45. doi: 10.1016/j.jmoldx.2016.02.005 [Crossref] [ Google Scholar]
- Lamb YN, Dhillon S. Epi proColon® 20 CE: a blood-based screening test for colorectal cancer. Mol Diagn Ther 2017; 21:225-32. doi: 10.1007/s40291-017-0259-y [Crossref] [ Google Scholar]
- Jin P, Kang Q, Wang X, Yang L, Yu Y, Li N. Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol 2015; 30:830-3. doi: 10.1111/jgh.12855 [Crossref] [ Google Scholar]
- Koch A, Joosten SC, Feng Z, de Ruijter TC, Draht MX, Melotte V. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol 2018; 15:459-66. doi: 10.1038/s41571-018-0004-4 [Crossref] [ Google Scholar]
- Chen Y, Wang Z, Zhao G, Sun C, Ma Y, Zhang L. Performance of a novel blood-based early colorectal cancer screening assay in remaining serum after the blood biochemical test. Dis Markers 2019; 2019:5232780. doi: 10.1155/2019/5232780 [Crossref] [ Google Scholar]
- Zhao G, Li H, Yang Z, Wang Z, Xu M, Xiong S. Multiplex methylated DNA testing in plasma with high sensitivity and specificity for colorectal cancer screening. Cancer Med 2019; 8:5619-28. doi: 10.1002/cam4.2475 [Crossref] [ Google Scholar]
- Zhao G, Liu X, Liu Y, Li H, Ma Y, Li S. Aberrant DNA methylation of SEPT9 and SDC2 in stool specimens as an integrated biomarker for colorectal cancer early detection. Front Genet 2020; 11:643. doi: 10.3389/fgene.2020.00643 [Crossref] [ Google Scholar]
- Iyer GR, Hasan Q. Alteration of methylation status in archival DNA samples: a qualitative assessment by methylation specific polymerase chain reaction. Environ Mol Mutagen 2020; 61:837-42. doi: 10.1002/em.22398 [Crossref] [ Google Scholar]
- Fu T, Sharmab A, Xie F, Liu Y, Li K, Wan W. Methylation of MGMT is associated with poor prognosis in patients with stage III duodenal adenocarcinoma. PLoS One 2016; 11:e0162929. doi: 10.1371/journal.pone.0162929 [Crossref] [ Google Scholar]
- Azuara D, Aussó S, Rodriguez-Moranta F, Guardiola J, Sanjuan X, Lobaton T. New methylation biomarker panel for early diagnosis of dysplasia or cancer in high-risk inflammatory bowel disease patients. Inflamm Bowel Dis 2018; 24:2555-64. doi: 10.1093/ibd/izy255 [Crossref] [ Google Scholar]
- Pichon F, Shen Y, Busato F, S PJ, Jacquelin B, Grand RL. Analysis and annotation of DNA methylation in two nonhuman primate species using the Infinium Human Methylation 450K and EPIC BeadChips. Epigenomics 2021; 13:169-86. doi: 10.2217/epi-2020-0200 [Crossref] [ Google Scholar]
- Li D, Guo J, Wang S, Zhu L, Shen Z. Identification of novel methylated targets in colorectal cancer by microarray analysis and construction of co-expression network. Oncol Lett 2017; 14:2643-8. doi: 10.3892/ol.2017.6506 [Crossref] [ Google Scholar]
- Magaña AA, Wrobel K, Caudillo YA, Zaina S, Lund G, Wrobel K. High-performance liquid chromatography determination of 5-methyl-2'-deoxycytidine, 2'-deoxycytidine, and other deoxynucleosides and nucleosides in DNA digests. Anal Biochem 2008; 374:378-85. doi: 10.1016/j.ab.2007.11.026 [Crossref] [ Google Scholar]
- Li S, Tollefsbol TO. DNA methylation methods: global DNA methylation and methylomic analyses. Methods 2021; 187:28-43. doi: 10.1016/j.ymeth.2020.10.002 [Crossref] [ Google Scholar]
- Wang C, Liu Y, Guo W, Zhu X, Ahuja N, Fu T. MAPT promoter CpG island hypermethylation is associated with poor prognosis in patients with stage II colorectal cancer. Cancer Manag Res 2019; 11:7337-43. doi: 10.2147/cmar.S206731 [Crossref] [ Google Scholar]
- Jones PA, Ohtani H, Chakravarthy A, De Carvalho DD. Epigenetic therapy in immune-oncology. Nat Rev Cancer 2019; 19:151-61. doi: 10.1038/s41568-019-0109-9 [Crossref] [ Google Scholar]
- Kang B, Lee HS, Jeon SW, Park SY, Choi GS, Lee WK. Progressive alteration of DNA methylation of Alu, MGMT, MINT2, and TFPI2 genes in colonic mucosa during colorectal cancer development. Cancer Biomark 2021; 32:231-6. doi: 10.3233/cbm-203259 [Crossref] [ Google Scholar]
- Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol 2019; 17:156-66. doi: 10.1038/s41579-018-0129-6 [Crossref] [ Google Scholar]
- Hashemi Goradel N, Heidarzadeh S, Jahangiri S, Farhood B, Mortezaee K, Khanlarkhani N. Fusobacterium nucleatum and colorectal cancer: a mechanistic overview. J Cell Physiol 2019; 234:2337-44. doi: 10.1002/jcp.27250 [Crossref] [ Google Scholar]
- Li R, Shen J, Xu Y. Fusobacterium nucleatum and colorectal cancer. Infect Drug Resist 2022; 15:1115-20. doi: 10.2147/idr.S357922 [Crossref] [ Google Scholar]
- Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol 2015; 23:141-7. doi: 10.1016/j.mib.2014.11.013 [Crossref] [ Google Scholar]
- Rubinstein MR, Baik JE, Lagana SM, Han RP, Raab WJ, Sahoo D, et al. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/β-catenin modulator Annexin A1. EMBO Rep 2019; 20. doi: 10.15252/embr.201847638.
- Li J, Chen Z, Wang Q, Du L, Yang Y, Guo F. Microbial and metabolic profiles unveil mutualistic microbe-microbe interaction in obesity-related colorectal cancer. Cell Rep Med 2024; 5:101429. doi: 10.1016/j.xcrm.2024.101429 [Crossref] [ Google Scholar]
- Zhang Y, Zhang L, Zheng S, Li M, Xu C, Jia D. Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF-κB/ICAM1 axis. Gut Microbes 2022; 14:2038852. doi: 10.1080/19490976.2022.2038852 [Crossref] [ Google Scholar]
- Chen S, Su T, Zhang Y, Lee A, He J, Ge Q. Fusobacterium nucleatum promotes colorectal cancer metastasis by modulating KRT7-AS/KRT7. Gut Microbes 2020; 11:511-25. doi: 10.1080/19490976.2019.1695494 [Crossref] [ Google Scholar]
- Narayanan V, Peppelenbosch MP, Konstantinov SR. Human fecal microbiome-based biomarkers for colorectal cancer. Cancer Prev Res (Phila) 2014; 7:1108-11. doi: 10.1158/1940-6207.Capr-14-0273 [Crossref] [ Google Scholar]
- Hsieh YY, Kuo WL, Hsu WT, Tung SY, Li C. Fusobacterium nucleatum-induced tumor mutation burden predicts poor survival of gastric cancer patients. Cancers (Basel) 2022; 15:269. doi: 10.3390/cancers15010269 [Crossref] [ Google Scholar]
- Fan JQ, Zhao WF, Lu QW, Zha FR, Lv LB, Ye GL. Fecal microbial biomarkers combined with multi-target stool DNA test improve diagnostic accuracy for colorectal cancer. World J Gastrointest Oncol 2023; 15:1424-35. doi: 10.4251/wjgo.v15.i8.1424 [Crossref] [ Google Scholar]
- Tunsjø HS, Gundersen G, Rangnes F, Noone JC, Endres A, Bemanian V. Detection of Fusobacterium nucleatum in stool and colonic tissues from Norwegian colorectal cancer patients. Eur J Clin Microbiol Infect Dis 2019; 38:1367-76. doi: 10.1007/s10096-019-03562-7 [Crossref] [ Google Scholar]
- Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014; 370:1287-97. doi: 10.1056/NEJMoa1311194 [Crossref] [ Google Scholar]
- Jin P, You P, Fang J, Kang Q, Gu F, Cai Y. Comparison of performance of two stool DNA tests and a fecal immunochemical test in detecting colorectal neoplasm: a multicenter diagnostic study. Cancer Epidemiol Biomarkers Prev 2022; 31:654-61. doi: 10.1158/1055-9965.Epi-21-0991 [Crossref] [ Google Scholar]
- Raut JR, Guan Z, Schrotz-King P, Brenner H. Fecal DNA methylation markers for detecting stages of colorectal cancer and its precursors: a systematic review. Clin Epigenetics 2020; 12:122. doi: 10.1186/s13148-020-00904-7 [Crossref] [ Google Scholar]
- Pickhardt PJ, Graffy PM, Weigman B, Deiss-Yehiely N, Hassan C, Weiss JM. Diagnostic performance of multitarget stool DNA and CT colonography for noninvasive colorectal cancer screening. Radiology 2020; 297:120-9. doi: 10.1148/radiol.2020201018 [Crossref] [ Google Scholar]
- Levin B, Lieberman DA, McFarland B, Andrews KS, Brooks D, Bond J. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology 2008; 134:1570-95. doi: 10.1053/j.gastro.2008.02.002 [Crossref] [ Google Scholar]
- Jayasinghe M, Prathiraja O, Caldera D, Jena R, Coffie-Pierre JA, Silva MS. Colon cancer screening methods: 2023 update. Cureus 2023; 15:e37509. doi: 10.7759/cureus.37509 [Crossref] [ Google Scholar]
- Rex DK, Johnson DA, Anderson JC, Schoenfeld PS, Burke CA, Inadomi JM. American College of Gastroenterology guidelines for colorectal cancer screening 2009 [corrected]. Am J Gastroenterol 2009; 104:739-50. doi: 10.1038/ajg.2009.104 [Crossref] [ Google Scholar]
- Fisher DA, Princic N, Miller-Wilson LA, Wilson K, Fendrick AM, Limburg P. Utilization of a colorectal cancer screening test among individuals with average risk. JAMA Netw Open 2021; 4:e2122269. doi: 10.1001/jamanetworkopen.2021.22269 [Crossref] [ Google Scholar]
- Li J, Li Z-P, Ruan W-J, Wang W. Colorectal cancer screening: The value of early detection and modern challenges. World J Gastroenterol 2024; 30:2726. doi: 10.3748/wjg.v30.i20.2726 [Crossref] [ Google Scholar]
- Zhu X, Parks PD, Weiser E, Fischer K, Griffin JM, Limburg PJ. National survey of patient factors associated with colorectal cancer screening preferences. Cancer Prev Res (Phila) 2021; 14:603-14. doi: 10.1158/1940-6207.Capr-20-0524 [Crossref] [ Google Scholar]
- Imperiale TF, Porter K, Zella J, Gagrat ZD, Olson MC, Statz S. Next-generation multitarget stool DNA test for colorectal cancer screening. N Engl J Med 2024; 390:984-93. doi: 10.1056/NEJMoa2310336 [Crossref] [ Google Scholar]
- Grobbee EJ, Wisse PHA, Schreuders EH, van Roon A, van Dam L, Zauber AG. Guaiac-based faecal occult blood tests versus faecal immunochemical tests for colorectal cancer screening in average-risk individuals. Cochrane Database Syst Rev 2022; 6:CD009276. doi: 10.1002/14651858.CD009276.pub2 [Crossref] [ Google Scholar]
- Gómez-Molina R, Suárez M, Martínez R, Chilet M, Bauça JM, Mateo J. Utility of stool-based tests for colorectal cancer detection: a comprehensive review. Healthcare (Basel) 2024; 12:1645. doi: 10.3390/healthcare12161645 [Crossref] [ Google Scholar]
- Rex DK, Boland CR, Dominitz JA, Giardiello FM, Johnson DA, Kaltenbach T. Colorectal cancer screening: recommendations for physicians and patients from the US Multi-Society Task Force on Colorectal Cancer. Am J Gastroenterol 2017; 112:1016-30. doi: 10.1038/ajg.2017.174 [Crossref] [ Google Scholar]
- Mannucci A, Goel A. Stool and blood biomarkers for colorectal cancer management: an update on screening and disease monitoring. Mol Cancer 2024; 23:259. doi: 10.1186/s12943-024-02174-w [Crossref] [ Google Scholar]
- McDaniels R. The Effectiveness of the FIT and FIT-DNA Screening Tests to Aid in the Detection of Abnormalities Within the Colorectal Epithelium [dissertation]. Grambling State University; 2024.
- Kanth P, Inadomi JM. Screening and prevention of colorectal cancer. BMJ 2021; 374:n1855. doi: 10.1136/bmj.n1855 [Crossref] [ Google Scholar]
- Tan ES, Fan W, Knepper TC, Schell MJ, Sahin IH, Fleming JB. Prognostic and predictive value of PIK3CA mutations in metastatic colorectal cancer. Target Oncol 2022; 17:483-92. doi: 10.1007/s11523-022-00898-7 [Crossref] [ Google Scholar]
- Ji X, Sha J, Qian H, Zhang G, He T, Dang Y. Highly sensitive fecal DNA testing of NDRG4 12b methylation is a promising marker for detection of colorectal precancerosis. J BUON 2021; 26:1239-45. [ Google Scholar]
- Vagionas A, Balgkouranidou I, Koukaki T, Biziota E, Amarantidis K, Kakolyris S. Prognostic significance of SOX17 and WNT5a promoter methylation status in circulating cell-free DNA metastatic colorectal cancer patients. Hippokratia 2023; 27:7-11. [ Google Scholar]
- Chen Y, Ren B, Yang J, Wang H, Yang G, Xu R. The role of histone methylation in the development of digestive cancers: a potential direction for cancer management. Signal Transduct Target Ther 2020; 5:143. doi: 10.1038/s41392-020-00252-1 [Crossref] [ Google Scholar]
- Luo H, Zhao Q, Wei W, Zheng L, Yi S, Li G. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med 2020; 12:eaax7533. doi: 10.1126/scitranslmed.aax7533 [Crossref] [ Google Scholar]
- Cox KE, Liu S, Lwin TM, Hoffman RM, Batra SK, Bouvet M. The mucin family of proteins: candidates as potential biomarkers for colon cancer. Cancers (Basel) 2023; 15:1491. doi: 10.3390/cancers15051491 [Crossref] [ Google Scholar]
- Li C, Zuo D, Yin L, Lin Y, Li C, Liu T. Prognostic value of MUC2 expression in colorectal cancer: a systematic review and meta-analysis. Gastroenterol Res Pract 2018; 2018:6986870. doi: 10.1155/2018/6986870 [Crossref] [ Google Scholar]
- Takeda Y, Nakano T, Yanagaki M, Takada N, Kumamoto T, Furukawa K. The time-dependent changes in serum carcinoembryonic antigen impact on posthepatectomy outcomes of colorectal liver metastasis. Surgery 2022; 172:625-32. doi: 10.1016/j.surg.2022.03.039 [Crossref] [ Google Scholar]
- Zhang W, Zhang K, Ma Y, Song Y, Qi T, Xiong G. Secreted frizzled-related proteins: A promising therapeutic target for cancer therapy through Wnt signaling inhibition. Biomed Pharmacother 2023; 166:115344. doi: 10.1016/j.biopha.2023.115344 [Crossref] [ Google Scholar]
- Genua F, Mirković B, Mullee A, Levy M, Gallagher WM, Vodicka P. Association of circulating short chain fatty acid levels with colorectal adenomas and colorectal cancer. Clin Nutr ESPEN 2021; 46:297-304. doi: 10.1016/j.clnesp.2021.09.740 [Crossref] [ Google Scholar]
- Kang JS. Dietary restriction of amino acids for cancer therapy. Nutr Metab (Lond) 2020; 17:20. doi: 10.1186/s12986-020-00439-x [Crossref] [ Google Scholar]
- Abbes S, Baldi S, Sellami H, Amedei A, Keskes L. Molecular methods for colorectal cancer screening: progress with next-generation sequencing evolution. World J Gastrointest Oncol 2023; 15:425-42. doi: 10.4251/wjgo.v15.i3.425 [Crossref] [ Google Scholar]
- Sun J, Zhao J, Jiang F, Wang L, Xiao Q, Han F. Identification of novel protein biomarkers and drug targets for colorectal cancer by integrating human plasma proteome with genome. Genome Med 2023; 15:75. doi: 10.1186/s13073-023-01229-9 [Crossref] [ Google Scholar]
- Xiao Y, Wang X, Weng H, Ding Z, Qian K, Jin W. Ultrasensitive tumour-agnostic non-invasive detection of colorectal cancer recurrence using ctDNA methylation. Clin Transl Med 2022; 12:e1015. doi: 10.1002/ctm2.1015 [Crossref] [ Google Scholar]
- Wang X, Tian L, Lu J, Ng IO. Exosomes and cancer - diagnostic and prognostic biomarkers and therapeutic vehicle. Oncogenesis 2022; 11:54. doi: 10.1038/s41389-022-00431-5 [Crossref] [ Google Scholar]
- Berkley-Brown MJ. Developing a Clinical Practice Guideline for Colorectal Cancer Screening in Primary Care [dissertation]. Walden University; 2021.