BioImpacts. 6(3):125-133.
doi: 10.15171/bi.2016.19
Original Article
Interactions of cephalexin with bovine serum albumin: displacement reaction and molecular docking
Hamed Hamishehkar 1, Soheila Hosseini 2, Abdolhossein Naseri 3, Azam Safarnejad 2, Farzaneh Rasoulzadeh 1, *
Author information:
1Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
2Biotechnology Research Center and School of Pharmacy, Tabriz University of Medical Sciences, Tabriz, Iran
3Department of Analytical Chemistry, Faculty of Chemistry, University of Tabriz, Tabriz, Iran
Abstract
Introduction:
The drug-plasma protein interaction is a fundamental issue in guessing and checking the serious drug side effects related with other drugs. The purpose of this research was to study the interaction of cephalexin with bovine serum albumin (BSA) and displacement reaction using site probes.
Methods:
The interaction mechanism concerning cephalexin (CPL) with BSA was investigated using various spectroscopic methods and molecular modeling method. The binding sites number, n, apparent binding constant, K, and thermodynamic parameters, ΔG0, ΔH0, and ΔS0 were considered at different temperatures. To evaluate the experimental results, molecular docking modeling was calculated.
Results:
The distance, r=1.156 nm between BSA and CPL were found in accordance with the Forster theory of non-radiation energy transfer (FRET) indicating energy transfer occurs between BSA and CPL. According to the binding parameters and ΔG0= negative values and ΔS0= 28.275 j mol-1K-1, a static quenching process is effective in the CPL-BSA interaction spontaneously. ΔG0 for the CPL-BSA complex obtained from the docking simulation is -28.99 kj mol-1, which is close to experimental ΔG of binding, -21.349 kj mol-1 that indicates a good agreement between the results of docking methods and experimental data.
Conclusion:
The outcomes of spectroscopic methods revealed that the conformation of BSA changed during drug-BSA interaction. The results of FRET propose that CPL quenches the fluorescence of BSA by static quenching and FRET. The displacement study showed that phenylbutazon and ketoprofen displaced CPL, indicating that its binding site on albumin is site I and Gentamicin cannot be displaced from the binding site of CPL. All results of molecular docking method agreed with the results of experimental data.
Keywords: Bovine serum albumin, Cephalexin, Circular dichroism, Fluorescence spectroscopy, Gentamicin displacement, Molecular modeling
Copyright and License Information
© 2016 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
The drug–protein interaction may establish a protein–drug complex, which has an important effect on the drug tissue distribution and metabolic rate that aids in understanding the drug pharmacokinetics and pharmacodynamics.
1
Therefore, the information of the interactions among plasma bio-macromolecules and drugs is a crucial issue in biological and the medical sciences. Albumin is the most important protein of plasma for carrying many exogenous and endogenous compounds.
3
It rises the apparent solubility of hydrophobic drugs in the plasma and moderates their transport to cells. Subsequently, the drug protein binding and probable interactions are routinely investigated by this protein.
4
Cephalexin (CPL) (Fig. 1) is a first-generation cephalosporin antibiotic that has a broad spectrum antibiotic and is extensively administered therapeutically for the most common and uncomplicated infections including upper breathing infections, ear infections, skin infections, and urinary organs infections with an acceptable level of side effects. Antibiotics are also a regularly prescribed medication for in-patient use in hospitals, particularly the intensive care units (ICUs).
5
The probability for pharmacokinetic properties changes by β-lactam antibiotics is increased with administration of a combination of multiple medications, particularly in long suffering diagnosed with kidney issues and variable plasma albumin levels. The knowledge of drug-plasma protein interaction is a crucial issue in predicting and preventing serious drug side effects associated with other drugs, which contain a high protein binding.
6-10
One of the medicines that can interact with CPL is gentamicin. These interactions can affect the metabolism of the drugs and cause kidney damage or other things. For this purpose, it is necessary to define the bond connection of CPL to bovine serum albumin (BSA) and displacement interaction of BSA-CPL with other drugs such as gentamicin and study the thermodynamic parameters of CPL- BSA interaction. Fluorescence and UV–Vis absorption spectroscopy techniques, Fourier transform infrared spectroscopy (FT-IR) and circular dichroism (CD) are powerful tools for investigating the reactivity of chemical and biological systems. To access this function, the spectroscopic properties of CPL in the presence of BSA were investigated. The interactions of β-lactam with other antibiotics which affect their protein binding characteristics, have been reported in a number of earlier studies.
11
In this work, we use spectroscopic methods to study the binding of CPL to BSA in aqueous solution under biological conditions. We use also molecular modeling to approve the outcomes of spectroscopic methods. Gaining interaction evidence concerning quenching mechanisms, thermodynamic parameters, binding parameters, probing the binding site, binding modes, effect of metal ions, and intermolecular binding distance were our purposes. Moreover, the possibilities of gentamicin interaction with protein binding characteristics of CPL were investigated.
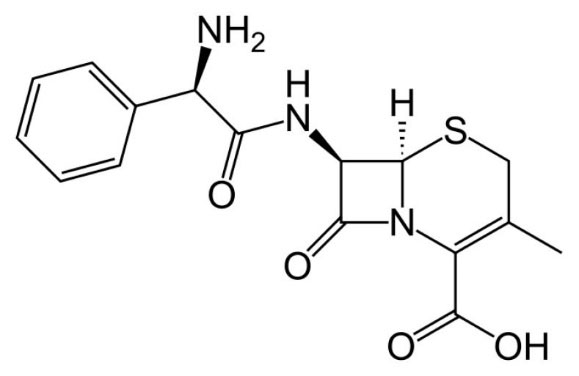
Fig. 1
.
Molecular structure of cephalexin.
.
Molecular structure of cephalexin.
Materials and methods
Materials
Drug and BSA purchased from Sigma-Aldrich Co. (Poole, England) was daily used to make the stock solution of 10-4M. Standard stock solutions of CPL was set by dissolving a suitable amount of the pure drug (98%) in distilled water as required to 10-3M. All solutions in this study were diluted with 0.1M phosphate buffer (pH 7.4).
Apparatus
All fluorescence spectra were studied on a RF-5301 spectrofluorophotometer (Shimadzu, Kyoto Japan) and a 10 mm quartz cuvette. The widths of both the excitation and emission slits were set at 5 nm. The optimum excitation and emission wavelengths for BSA were obtained as 280 and 340 nm, respectively. To estimate and eliminate the inner filter effect (IFE), the correction ways were performed based on the absorbance amounts of solutions at excitation and emission wavelengths of albumin. UV spectra measurements were performed on a Shimadzu 2550 UV– spectrometer using a 10 mm cell at 20 nm intervals. FTIR spectra were acquired on a Shimadzu FTIR–8400 S (Shimadzu, Kyoto, Japan).
Circular Dichroism (CD) studies were made on an Aviv, model 215, spectrophotometer (Aviv Biomedical, Inc., Lakewood, NJ, USA) using 1 mm path length at room temperature with a scan rate of 500 nm/min and a reaction time of 0.5 s. The scans were repeated three times for each spectrum.
Spectroscopic investigations
All solutions must be relaxed at least 10 min before measuring the spectrum. To correct the fluorescence or absorption background, proper blanks corresponding to the buffer were deduced. The following measurable analysis was obtained by using the corrected fluorescence intensities at λem=340 nm at three temperatures.
Fluorescence quenching spectra of BSA
Fluorescence quenching spectra of BSA with CPL were recorded at excitation wavelength (280 nm) and emission wavelength (300–450 nm) at three different temperatures (299, 305 and 311 K) and pH=7.4. Fluorescence spectra of these solutions were investigated upon addition of Ibuprofen, Phenylbutazon, Ketoprofen, Gentamicin, and some metal ions at the same conditions. The synchronous fluorescence spectra were also scanned from 280 to 400 nm at ∆λ=15 and 60 nm, respectively.
UV–Visible absorption spectra
The absorption spectra of BSA in the presence of different concentrations of CPL were determined in the range of 200–400 nm at room temperature. The concentration of BSA was set at 3.33 ×10-6M while that of CPL was changed from 0 to 1.30×10-4M.
FT-IR spectroscopic measurements
The FTIR spectrum of BSA in the absence and presence of CPL in phosphate buffer was found in the range of 4000–500 cm-1 with a small resolution of 2 cm-1 and 100 scans. The concentrations of the CPL in the complex mixtures were 20 ×10-6M with a final BSA concentration of 3.33×10-6M. Spectra were gathered after 10 min of incubation of BSA with CPL solution at room temperature.
Circular dichroism measurements
CD measurements were studied at room temperature in the wavelength range of 200–240 nm. These measurements were carried out by mixing a fixed concentration of BSA (3.33×10-6M) with varying concentration of CPL (0, 3.33, 10)×10-6M. All scans were measured under continuous nitrogen atmosphere.
Molecular docking studies
The three-dimensional (3D) structure of CPL was drawn by ChemBioDraw Ultra and energy optimized using HyperChem software by AM1 semi-empirical method with RMS gradient of 0.001 kcalmol-1. The crystal structure of BSA (pdb id: 4f5s) was downloaded from Protein Data Bank. CPL was docked to 6 subdomains of BSA by Autodock Vina with grid map of (22×22×22) and maps were located on each subdomain to determine the binding site of BSA, then the best docking was further considered by AutoDock. Molecular docking was finished in model1 and model2 by AutoDock Tools. Water molecules were removed and polar hydrogen atoms were added to BSA and then charged using kollman charges. The grid maps to (60×60×60Å) and a grid spacing of 0.375A0. Center of the grid box (the x-, y, z-axes) was set to 0.527 Å, 28.13 Å, and 102.81Å with 100 docking runs. Population size was set to 150 with 2500000 energy evaluations (medium). Conformational searching was made using the Lamarckian genetic algorithm (LGA), analysis made by Autodock tools, and VDW scaling factor was 1.
Results
Analysis of fluorescence quenching data
By using fluorescence spectroscopy, the interaction between these drugs and BSA was investigated to collect the necessary information regarding its quenching mechanism, binding constants and binding sites. Some drug molecules absorb light at the excitation and emission wavelength of BSA that influences the fluorescence intensity. It is known as the IFE. To estimate the impact of IFE in this method, the absorbance of CPL at 280 nm, excitation wavelength of BSA, and 340 nm emission wavelength of BSA were determined. The absorbance of concentrated solutions (CCPL= 1.30×10-4M) was higher than 0.1, approximately about 0.25, at 2800 nm (Fig. 2). In such cases the effect of IFE should be corrected using Eq. 1.
12,13
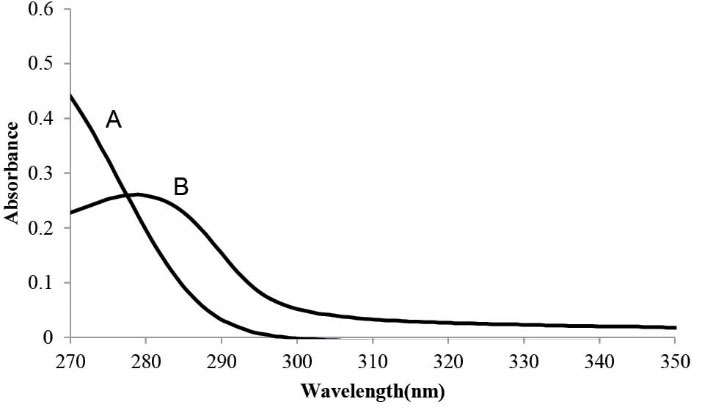
Fig. 2
.
UV spectra of A: CPL (130×10-6M) and B: albumin
(3.33×10-6M).
.
UV spectra of A: CPL (130×10-6M) and B: albumin
(3.33×10-6M).
(Eq. 1)
Where Fcor and Fobs are the corrected and observed fluorescence, respectively. Aex and Aem are the absorbance of the drug at excitation wavelength and emission wavelength, respectively.
Tryptophan residues (Trp) and tyrosine residues (Tyr) are two types of fluorophores in BSA that can be excited at 280 nm. As demonstrated and noted in Fig. 3 with the increasing amounts of CPL, the fluorescence intensities decreased without any shifts in λmax.
14,15
If the conditions of pH, temperature, and ionic strength are fixed, two different mechanisms can occur in the fluorescence quenching, static quenching, and dynamic quenching. Stern Volmer equation can describe both of them. Normally, increasing temperature causes the increase of quenching rate constants for dynamic quenching and the reverse effect was observed for the static quenching. To configure which mechanism has an important role in the interaction, fluorescence quenching data were studied by the stern–Volmer equation (Eq. 2)
16
:
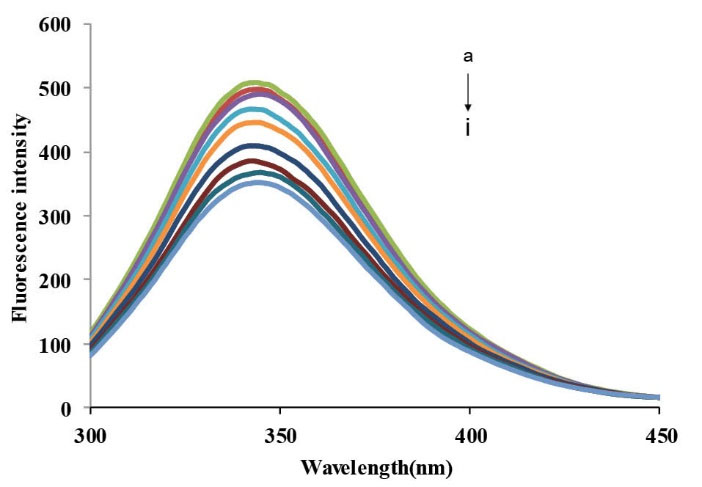
Fig. 3
.
Fluorescence emission spectra of BSA in the presence
of different concentrations of CPL; [BSA] = 3.33×10-6M; [CPL] =
(0,10,30,50,70,90,110,120 ,130)×10-6M (λex = 280 nm, T = 293 K,
pH =7.4).
.
Fluorescence emission spectra of BSA in the presence
of different concentrations of CPL; [BSA] = 3.33×10-6M; [CPL] =
(0,10,30,50,70,90,110,120 ,130)×10-6M (λex = 280 nm, T = 293 K,
pH =7.4).
(Eq. 2)
Where F0 and F are the steady state fluorescence intensities of BSA in the absence and presence of quencher, respectively. KSV and [Q] are the Stern–Volmer quenching constant and the concentration of the quencher, respectively. Kq is the quenching rate constant and τ0 is the average lifetime of the BSA in the absence of any quencher and is generally equal to 10-8 s.
17
Fig. 4 shows the Stern–Volmer graphs at three different temperatures.
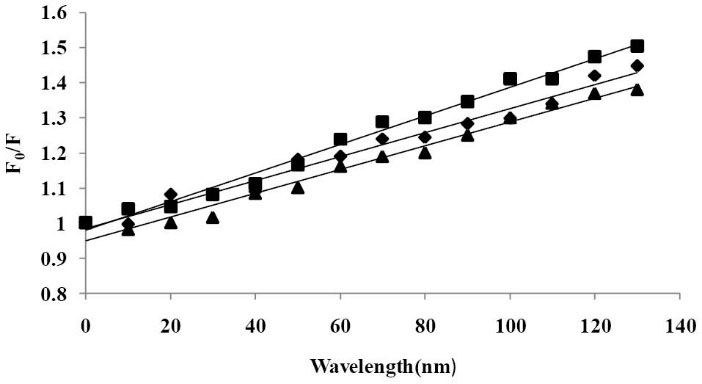
Fig. 4
.
The Stern–Volmer plots for the quenching of BSA by CPL
at different temperatures, 299 K (■), 305 K (♦) and 311 K (▲) λex
= 280 nm, λem = 340 nm and pH= 7.40.
.
The Stern–Volmer plots for the quenching of BSA by CPL
at different temperatures, 299 K (■), 305 K (♦) and 311 K (▲) λex
= 280 nm, λem = 340 nm and pH= 7.40.
It demonstrated the plots are linear for three temperatures and the slopes decrease with increasing temperature. The values of KSV for the interaction of CPL with BSA at three temperatures are shown in Table 1.
Table 1
.
Stern–Volmer quenching constants of CPL –BSA at different temperatures
|
pH
|
T(K)
|
K
sv×
(10
-3
)
|
n
|
R
|
|
7.4
|
299 |
5.450 |
1.246 |
0.984 |
| 305 |
4.793 |
1.795 |
0.976 |
| 311 |
4.484 |
1.525 |
0.979 |
This study found that the Ksv values are related reversely to temperature indicating a static quenching mechanism for the interaction of BSA with CPL.
18
Determination of the binding constant and the number of binding sites
There are n equivalent binding sites on the BSA and there are many methods and equations for the investigation of the binding constant, K, and the number of binding sites, n, for the reaction, P + nD→DnP. But, only in Eq. 3, the total drug concentrations can be used
19
:
(Eq. 3)
Where, F0 and F are the fluorescence intensities in the absence and presence of the quencher, [Dt] and [Pt] are the total quencher concentration and the total protein concentration, respectively. Therefore, the number of binding sites, n, and binding constant, K, can be estimated by the plots of log [(F0−F)/F] versus log (1/([Dt]−(F0−F)[Pt]/F0)), from the slopes and intercepts appropriately (Table 2). The correlation coefficients (larger than 0.98) indicates a good agreement in the interaction of CPL and BSA with Eq. 3. There are about two classes of binding site for CPL at the boundary of BSA. As demonstrated in Table 2, the K values between BSA and CPL decreased with increasing the temperature that shows the complex formation between CPL and BSA supporting the fluorescence quenching mechanism is a static process.
Table 2
.
Modified Stern–Volmer association constant Ka and relative thermodynamic parameters of CPL–BSA
|
T
|
K
a×
(10
-3
)
|
R
|
∆H(Kj mol
-1
)
|
∆G(Kj mol
-1
)
|
∆S(j mol
-1
K
-1
)
|
| 299 |
5.451 |
0.991 |
-12.895 |
-21.349 |
28.275 |
| 305 |
4.793 |
0.981 |
- |
-21.519 |
- |
| 311 |
4.484 |
0.983 |
- |
-21.688 |
- |
UV–Vis absorption spectra
The study of UV–Vis absorption spectra is a helpful method and appropriate to discover the structural change and know the formation of the complex.
20
For validation of the fluorescence quenching mechanism of BSA by CPL, UV–Vis absorption spectra of BSA were recorded before and after adding CPL (Fig. 5). As shown in Fig. 5, not only the absorbance intensity increased by the addition of CPL, but also the absorption spectra maximally shifted to a shorter wavelength region (279→263 nm).
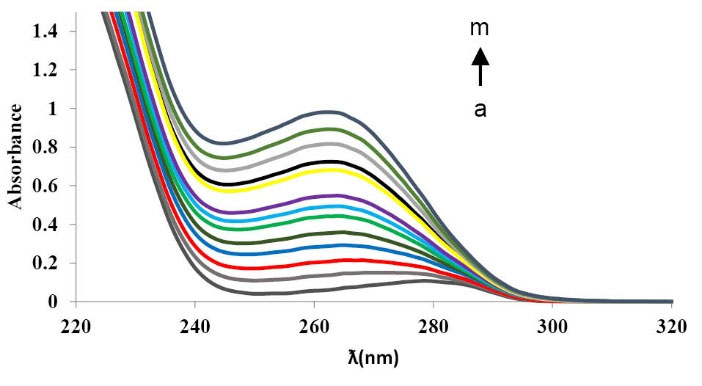
Fig. 5
.
Absorption spectra of BSA in the presence of CPL [BSA]
= 3.33×10-6M; [CPL] = (0,10, 20, 30, 40, 50, 60, 70, 80, 90, 100,
110)×10-6M, from cure a to l), the absorption spectrum of CPL
only(m).
.
Absorption spectra of BSA in the presence of CPL [BSA]
= 3.33×10-6M; [CPL] = (0,10, 20, 30, 40, 50, 60, 70, 80, 90, 100,
110)×10-6M, from cure a to l), the absorption spectrum of CPL
only(m).
FT-IR spectroscopy
FTIR spectroscopy is a simple instrument for providing structural and conformational changes of proteins. IR spectra of proteins display a number of amide bands representing diverse vibrations of the peptide moiety. Both the amide I (ranging from 1600 to 1700 cm−1) and amide II (around 1548 cm−1) bands of the protein are related with secondary structure of protein.
However, the change of protein secondary structure affects the amide I band more than amide II.
21
Fig. 6 shows the FT-IR spectra of free BSA and CPL–BSA in phosphate buffer. As can be noted, there is a shift in the peak position of amide I of BSA from 1652 to 1656 cm−1 after addition of CPL demonstrating a slight change in the secondary structure of BSA.
22
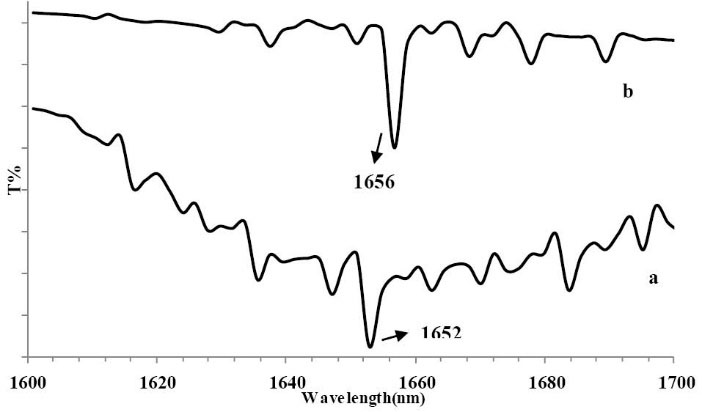
Fig. 6
.
Transmittance of the buffer solution from the spectrum of
the (a) BSA solution, (b) the difference spectra of BSA (subtracting
the absorption of the CPL–freeform from that of CPL–BSA
boundform).
.
Transmittance of the buffer solution from the spectrum of
the (a) BSA solution, (b) the difference spectra of BSA (subtracting
the absorption of the CPL–freeform from that of CPL–BSA
boundform).
Fluorescence resonance energy transfer from BSA to CPL
Fluorescence resonance energy transfer (FRET) is a powerful model that helps to evaluate donor-acceptor interactions by calculating the distance between two fluorophores, donor and acceptor.
23
The distance between two fluorophores should be shorter than 8 nm.
24
The energy transfer efficiency, E, is well-defined by the following Eq. (4), where r0 is the distance from the drug to the tryptophan residue of BSA, and R0 is the Forster critical distance, at which 50% of the excitation energy is transferred to the drug.
12
R0 can be estimated from donor emission, BSA, and acceptor absorption spectra, drug, using the Forster equation (Eq. 5):
(Eq. 4)
(Eq. 5)
(Eq. 6)
In Eq. 5, K2 is the random orientation factor associated to the geometry of the donor and acceptor and K2 = 2/3 for fluid solution; N the average refractive index of medium in the wavelength range where spectral overlap is significant; Φ the fluorescence quantum yield of the donor; j the effect of the spectral overlap between the emission spectrum of the donor and the absorption spectrum of the acceptor, which could be determined by Eq. 6. Here, F(λ) is the corrected fluorescence intensity of the donor in the wavelength range λ to λ+∆λ; ε (λ) is the extinction coefficient of the acceptor at λ. FRET is an important method for studying the diversity of biological systems including energy transfer processes.
The spectral overlap between UV-Vis absorption spectrum of CPL and the fluorescence emission spectrum of free BSA is illustrated in Fig. 7. Because the fluorescence emission of BSA was affected by the excitation light around 288 nm, the spectrum ranging from 300 to 400 nm was selected to analyze the overlapping integral.
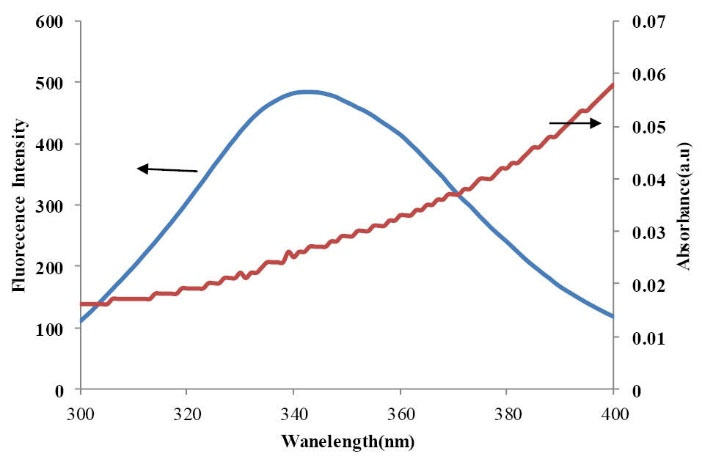
Fig. 7
.
The spectral overlap between UV/vis absorption spectrum
of CPL (10×10-6M) and the fluorescence emission spectrum
of free BSA (10×10-6M).
.
The spectral overlap between UV/vis absorption spectrum
of CPL (10×10-6M) and the fluorescence emission spectrum
of free BSA (10×10-6M).
In this study, N= 1.36 and Φ= 0.15 according to Eqs. 4, 5 and 6. It was calculated that J = 1.97×10-15 cm3 L mol−1, E = 0.0462, R0 = 0.698 nm, and r = 1.156 nm. The average distances between BSA and CPL is less than 8 nm suggesting that energy transfer happens between BSA and CPL. Furthermore, since r is larger than R0, it recommends that CPL quench the fluorescence of BSA by non-radiative energy transferring and static quenching.
Synchronous fluorescence spectra analysis
Synchronous fluorescence spectroscopy is a method to find evidence about the molecular environment close the chromosphere molecules. Since tryptophan (Trp) and tyrosine (Tyr) residues of BSA show the fluorescence property, we can find the specific evidence of tyrosine and tryptophan residues in BSA by setting ∆λ at 15 nm and 60 nm, respectively and determining the probable shift in wavelength emission maximum λmax. There is no significant shift (299-301) in synchronous fluorescence spectra of BSA by addition of CPL enhancement at λ=15 nm demonstrating that CPL has slight effect on the environment of the Tyr residues in BSA. By contrast, when ∆λ=60 nm (Fig. 8), the emission maximum wavelength shows a red shift (from 342 to 335 nm) representing that the environment of the tryptophan residues were altered and hydrophobicity near this residue decreased resulting the interaction between BSA and CPL.
25-27
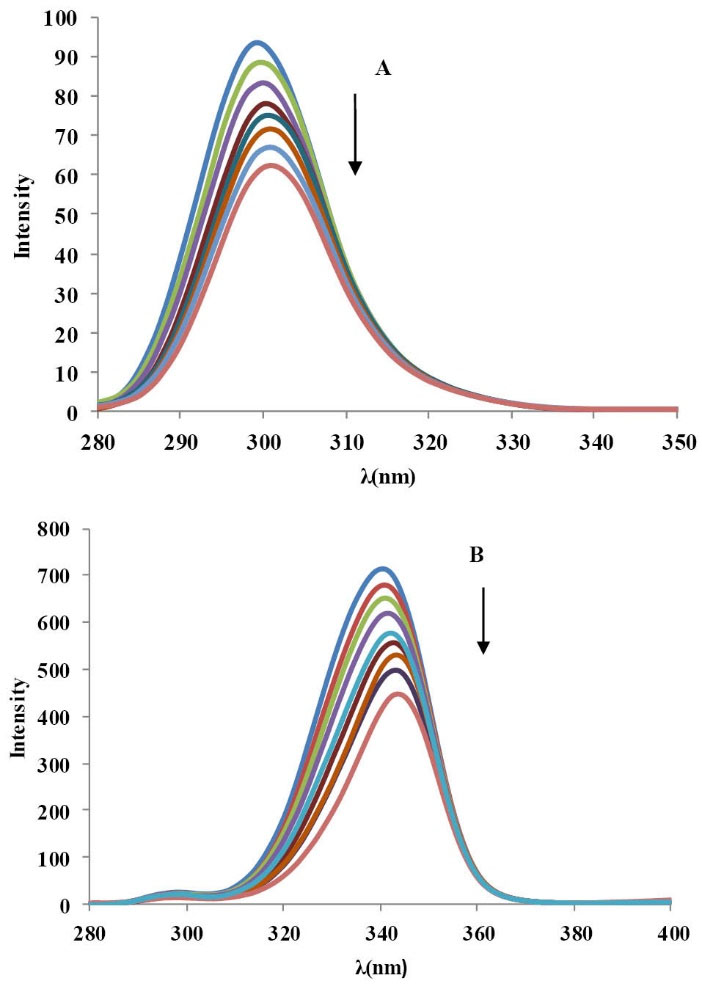
Fig. 8
.
The synchronous fluorescence spectra of BSA in the
presence of CPL (T=293K, pH=7.4). [BSA]=3.33×10-6M; [CPL]=
(0, 10, 30, 50, 70, 90, 110, 120,130) × 10-6M. (A) Δλ=15 nm, (B)
Δλ = 60 nm.
.
The synchronous fluorescence spectra of BSA in the
presence of CPL (T=293K, pH=7.4). [BSA]=3.33×10-6M; [CPL]=
(0, 10, 30, 50, 70, 90, 110, 120,130) × 10-6M. (A) Δλ=15 nm, (B)
Δλ = 60 nm.
Circular dichroism studies
CD spectroscopy investigates the spectrum of a protein for secondary structure with highly reliability.
28
To study the structural change of BSA by the addition of CPL, the CD spectra of BSA was measured before and after the addition of CPL (Fig. 9). There are two negative bands in the far-UV region at ~208 and ~222 nm which are characteristic of the helical structure of proteins. As estimated, the CD spectrum displays a strong negative ellipticity at 208 nm and 222 nm.
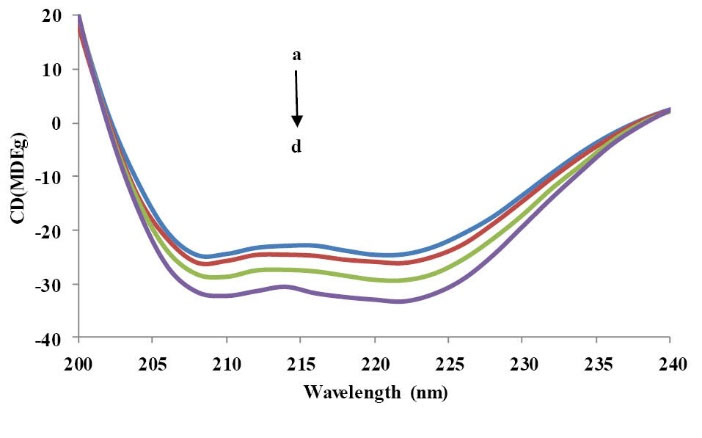
Fig. 9
.
Circular dichroic spectra in the 200–240 nm range; (a)
BSA, 3.33×10-6M; (b) BSA: CPL = 1:1; (c) 1:3; (d)1:4.
.
Circular dichroic spectra in the 200–240 nm range; (a)
BSA, 3.33×10-6M; (b) BSA: CPL = 1:1; (c) 1:3; (d)1:4.
The negative ellipticity in the region of far-UV CD was increased by the addition of the CPL to BSA (1:1, 1:3, and 1:4), without any substantial shift of peaks. This showed that the binding of CPL to BSA made an intense increase in the content of α-helical structure of the BSA clarifying the stabilization of the BSA, secondary structure as a consequence of the BSA–CPL interaction in the binary and ternary system.
29
This was supposed to be the result of the complex formation of BSA-CPL. However, the similarity between the shapes of the CD spectra relating to BSA before and after addition of the CPL in all relating systems proposed that the structure of BSA was still mainly α –helical.
30
Discussion
Thermodynamic analysis for binding mode between CPL and BSA
Small particles are bound to macromolecule typically by four acting forces containing hydrogen bond, Vander Waals force, electrostatic force, and hydrophobic interaction force.
There are thermodynamic parameters dependent to temperature, enthalpy change (ΔH), entropy change (ΔS), and free energy change (ΔG) of the reaction that are significant for characterizing the binding type. Therefore, the thermodynamic parameters were investigated to describe the acting forces between CPL and BSA.
The enthalpy change (∆H°) did not differ meaningfully with the temperature range considered, and the thermodynamic factor of ∆H°, entropy change (∆S°), and free energy (∆G°) were calculated by the Van 't Hoff equation:
Where K is the binding constant and R is the gas constant. The values of H° and ∆S° were evaluated using Eq. 7 by the plot of ln K versus 1/T.
10
The value of ∆G° was calculated using the following equation
31
:
The calculated thermodynamic parameters for CPL- BSA interaction are illustrated in Table 2. The values of ΔG are negative indicating that the binding procedure is spontaneous. The enthalpy (ΔH) and entropy (ΔS) of the interaction of CPL and BSA are negative and positive, respectively. According to the report of Ross and Subramanian,
32
the positive ΔH and ΔS value shows that the binding is mainly entropy-driven and a hydrophobic effect contributes in the interaction between CPL and BSA. The negative ΔH and ΔS values are related with hydrogen bonding and Van der Waals force. As a final point, low positive or negative ΔH and positive ΔS values are categorized by electrostatic interactions. Consequently, the interaction of CPL with BSA might contain the electrostatic interaction.
Displacement experiments using site probes and Gentamicin
BSA consists of amino acid chains making a single polypeptide, which has three homologous-helices in domains (I–III). Each domain is divided into anti-parallel six helices and four sub-domains (A and B). A cluster of two sub-domains with their grooves to each other forms a domain, and three of such domains form an albumin molecule. Sites I and II are two main definite drug-binding sites in serum albumin, which are situated in particular holes in sub-domains IIA and IIIA, respectively.
33
The majority of small molecules, which are identified to combine with BSA, form a complex at site I, and only a few at site II. Though, it is challenging to find the real site from the structure of the small molecule involved. Therefore, it is proposed that site I of serum albumin demonstrated an attraction for Ketoprofen and Phenylbutazone (PB), and site II for Ibuprofen (IB) and others. To recognize the position of the CPL binding site on BSA, the displacement experiments were performed by the site probes Ibuprofen, ketoprofen, and phenylbutazon.
The percentage of fluorescence probe displaced by the drug was calculated by determining the variations in fluorescence intensity according to the method suggested by Sudlow et al.
34
Relative fluorescence (RF) can be used to display the alterations in fluorescence intensity in the presence of probes as Eq. 9:
Where, F0 and F represent the fluorescence of CPL plus BSA in the absence and presence of the probe, respectively. According to the spectral data determined in the displacement studies, the plots of F/F0 against site probe concentration were found and are shown in Fig. 10.
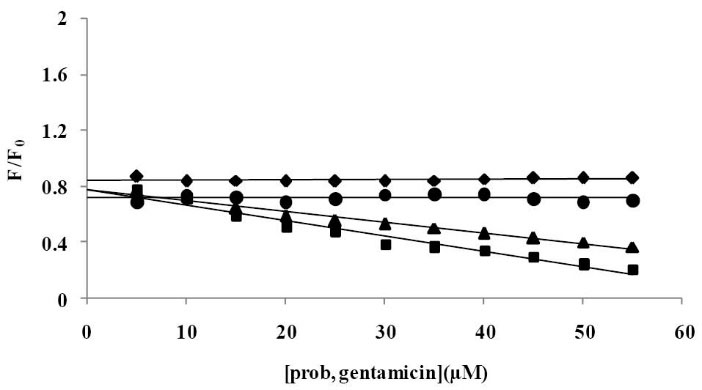
Fig. 10
.
Effect of site marker probes (Ketoprofen ■,
Phenylbutazone▲, Ibuprofen ♦) and Gentamicin (●) on the
fluorescence of CPL–BSA; [BSA] = 3.33×10-6M; [CPL]=60×10-6M;
[prob, Gentamicin]=(0, 5, 10, 15, 20, 25, 30, 35, 40, 45, 50) ×
10-6M.
.
Effect of site marker probes (Ketoprofen ■,
Phenylbutazone▲, Ibuprofen ♦) and Gentamicin (●) on the
fluorescence of CPL–BSA; [BSA] = 3.33×10-6M; [CPL]=60×10-6M;
[prob, Gentamicin]=(0, 5, 10, 15, 20, 25, 30, 35, 40, 45, 50) ×
10-6M.
The comparative fluorescence intensity meaningfully reduced after adding the phenylbutazon and ketoprofen, whereas the addition of Ibuprofen produced no observed changes, which shows that phenylbutazon and ketoprofen can displace the CPL, however, Ibuprofen has little effect on the binding of CPL to BSA. In all displacement tests, the implication was that CPL fixes to the site I in sub-domain IIA of BSA. These findings are in agreement with the results reported previously.
35,36
The reasonable procedure was examined for gentamicin, the importance of these studies is that gentamicin is classified to the aminoglycozide and are drugs typically prescribed in combination with CPL (illustrated in Fig. 10). The fluorescence demonstrated no significant change with the addition of gentamicin to the same solution, which indicates that gentamicin cannot be displaced from the binding site of CPL. Then it can be decided that gentamicin was bound to site II of BSA.
The effect of common metal ions on the binding constant
If there are metal ions present in the solution, the binding properties between CPL and BSA may be affected. A number of trace metal ions exist within the human body. Therefore, this aspect is particularly important and necessary in this research.
The effect of common ions such as Fe3+, Zn2+, K+ and Na+on the CPL– BSA binding was studied at 293 K by investigating the fluorescence intensity of CPL–BSA compound in the presence of each ion, distinctly in the range of 300–500 nm with the excitation wavelength at 280 nm. The studied cations in the phosphate buffer have no effect on the CPL–BSA interaction under the experimental conditions. The effect of properties of such cations on the interaction between a drug and BSA has been described in the former works.
37
The fluorescence emission spectrum of CPL in the presence of common ions (Table 3) shows no interaction between the ions and CPL. However, there is a binding reaction between the common ion and protein and thus the presence of common ion directly affects the binding between CPL and BSA.
Table 3
.
The binding constants (L mol−1) between CPL and BSA at 293 K in the presence of common ions
|
Ions
|
Association constant(K)
|
| 0 |
3650.32 |
|
Fe3+
|
3850.72 |
|
K+
|
4007.52 |
|
Na+
|
4237.43 |
|
Zn2+
|
4251.78 |
The CPL–BSA binding constant was increased in the presence of studied ions. Therefore, the binding force between BSA and CPL was improved which extended the serum-level of CPL in the blood plasma and improved the extreme efficiency of CPL.
Molecular docking results
The lowest energy in AutoDock Vina outcome (Table 4) specified that CPL was bound to subdomain IIA better than other sites. The results of further analysis proposed that CPL interacts with BSA of the hydrophobic cavity of subdomain IIA of BSA (Fig. 11 A). CPL was surrounded by the hydrophobic and negatively charged residues such as Arg194, Arg198, Arg256, Ala290, His241, His287, and Leu237 through hydrogen bond between CPL and Arg198 (3.03A0, 2.92A0) and Arg256 (2.86A0) (Fig. 11 D). The existence of the hydrogen bond makes a reduction in the hydrophilicity and a rise in the hydrophobicity which becomes stable in the CPL-BSA complex.
38
It shows that CPL-BSA binding is mainly hydrophobic in nature. Distance between Trp213 residue and CPL was 15.9 Ǻ and free energy for the CPL-BSA complex found from the docking simulation was -6.93 kcal mol-1 (-28.99 kj mol-1) (Table 5), which is close to experimental ΔG of binding (-21.349 kj mol-1) (Table 2).
Table 4
.
ΔG0 of binding CPL-BSA (kcal mol-1) for docking positions
|
Complex
|
Subdomain
|
|
IA
|
IIA
|
IIIA
|
IB
|
IIB
|
IIIB
|
| CPL-BSA |
-5.2 |
-8.9 |
-7.5 |
-7.6 |
-5.2 |
-6.0 |
Table 5
.
Amino acid residues involved in CPL-BSA intraction with ΔG0 and Hydrogen bond distance
|
Grid size
|
Subdomain
|
Inhibition Constant, Ki
(μM)
|
ΔG
0a
kcal mol-1
|
ΔE
1
b
kcal mol-1
|
ΔE
2
c
kcal mol-1
|
| 60×60×60 |
IIA |
8.33 |
-6.93 |
-8.66 |
+0.54 |
ΔG0a is estimated Free Energy of Binding in the binding process. ΔE1b is Vander Waals energy. ΔE2c is electrostatic energy.
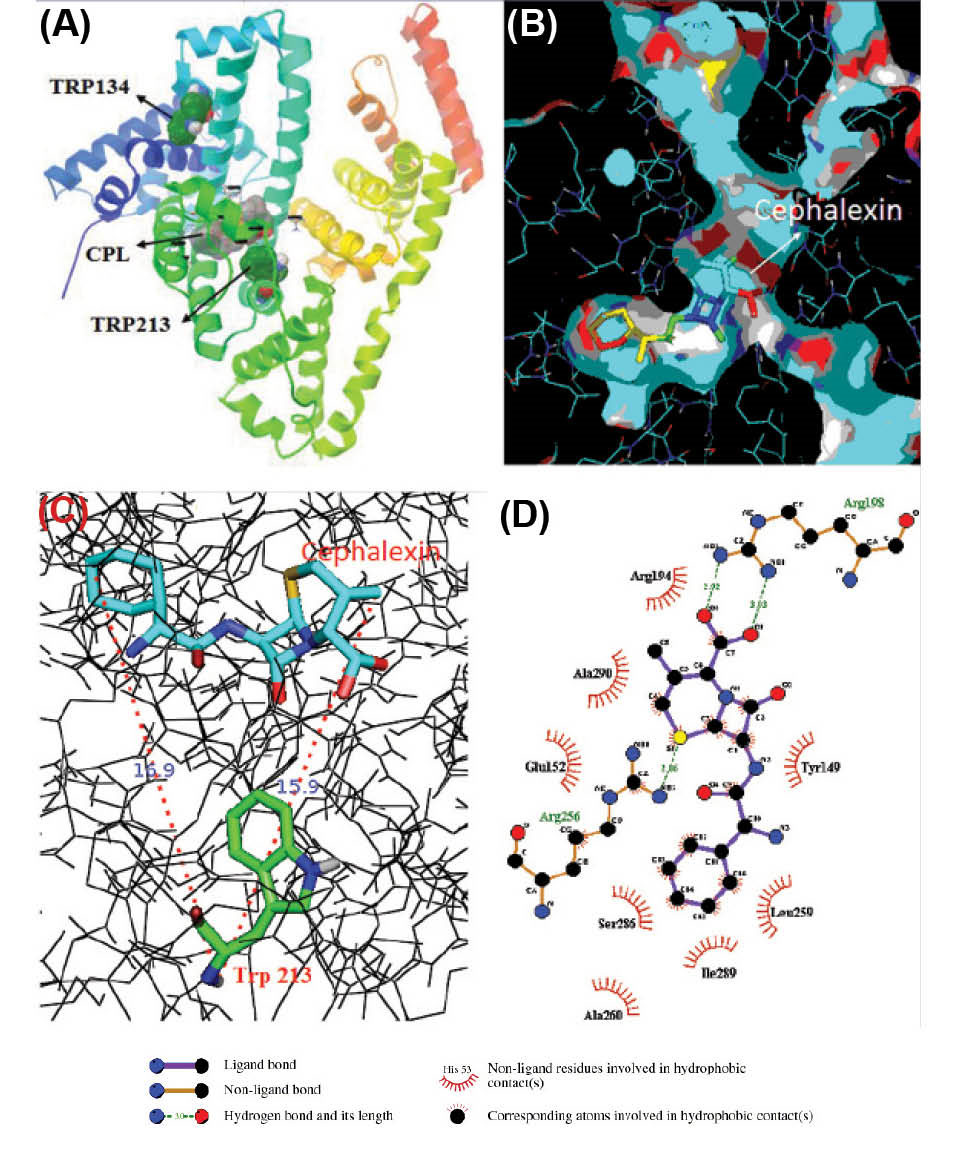
Fig. 11
.
(A) Minimum energy docking conformation obtained from
docking in subdomain (IIA) of BSA, Trp-213, Trp 134 and CPL have
been identified. (B) the surface representation of BSA showing
the binding pocket of CPL by PyMOL. (C) The distance between
Trp-213 and CPL from docking simulation in minimum energy is
shown by dotted line.(D) Representation of CPL interaction with
its binding pocket in 2- dimensional view, generated by LIGPLOT
.
(A) Minimum energy docking conformation obtained from
docking in subdomain (IIA) of BSA, Trp-213, Trp 134 and CPL have
been identified. (B) the surface representation of BSA showing
the binding pocket of CPL by PyMOL. (C) The distance between
Trp-213 and CPL from docking simulation in minimum energy is
shown by dotted line.(D) Representation of CPL interaction with
its binding pocket in 2- dimensional view, generated by LIGPLOT
Conclusion
In conclusion, this investigative research on the interaction of CPL and BSA was studied by spectroscopic techniques with fluorescence and UV–Vis absorption spectroscopy. The experimental outcomes confirmed that the mechanism of interaction between BSA and CPL is static.
Based on the synchronous fluorescence technique, the secondary construction of BSA was reformed in the presence of CPL. It was also demonstrated that Trp residue participates more than Tyr in the interaction between CPL and BSA. The results of the current research revealed the presence of a single binding site and hydrophobic interaction in the CPL - BSA complex. Gentamicin changed the binding constants of the CPL–BSA complex. Therefore, the variation of the kinetic and dynamic properties of CPL by gentamicin through alteration of the binding capacity of CPL to BSA cannot be negligible. Furthermore, the displacement investigation demonstrated that the location of CPL is in site I of BSA. In addition, the molecular docking studies confirmed the results of experimental studies.
Ethical approval
There is no ethical issue to be considered.
Competing interests
We wish to confirm that there are no known conflicts of interests associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Acknowledgments
The authors acknowledge financial support from the Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran.
Research Highlights
What is current knowledge?
simple
-
√ Participation of both Tyr and particularly Trp residues in
the interaction between CPL and BSA.
What is new here?
simple
-
√ Docking simulation for CPL-BSA and comparing to the
results of experimental methods.
References
- Hegde AH, Sandhya B, Seetharamappa J. Evaluation of binding and thermodynamic characteristics of interactions between a citrus flavonoid hesperitin with protein and effects of metal ions on binding. Mol Biol Rep 2011; 38:4921-9. doi: 10.1007/s11033-010-0634-9 [Crossref] [ Google Scholar]
- He XM, Carter DC. Atomic structure and chemistry of human serum albumin. Nature 1992; 358:209-15. [ Google Scholar]
- Ran D, Wu X, Zheng J, Yang J, Zhou H, Zhang M. Study on the Interaction between Florasulam and Bovine Serum Albumin. J Fluoresc 2007; 17:721-6. doi: 10.1007/s10895-007-0226-9 [Crossref] [ Google Scholar]
- Kamat BP. Study of the interaction between fluoroquinolones and bovine serum albumin. J Pharm Biomed Anal 2005; 39:1046-50. doi: 10.1016/j.jpba.2005.05.013 [Crossref] [ Google Scholar]
-
Goljan EF. Rapid review pathology: with student consult online access: Elsevier Health Sciences; 2013.
- Nerli B, Romanini D, Picó G. Structural specificity requirements in the binding of beta lactam antibiotics to human serum albumin. Chem Biol Interact 1997; 104:179-202. doi: 10.1016/S0009-2797(97)00024-0 [Crossref] [ Google Scholar]
- Phillips GO, Power DM, Robinson C, Davies JV. Interactions of bovine serum albumin with penicillins and cephalosporins studied by pulse radiolysis. Biochim Biophys Acta 1973; 295:8-17. doi: 10.1016/0005-2795(73)90068-8 [Crossref] [ Google Scholar]
- Wang Z, Song Z, Chen D. Study on the binding behavior of bovine serum albumin with cephalosporin analogues by chemiluminescence method. Talanta 2010; 83:312-9. doi: 10.1016/j.talanta.2010.09.029 [Crossref] [ Google Scholar]
- Naseri A, Hosseini S, Rasoulzadeh F, Rashidi MR, Zakery M, Khayamian T. Interaction of norfloxacin with bovine serum albumin studied by different spectrometric methods; Displacement studies, molecular modeling and chemometrics approaches. J Lumin 2015; 157:104-12. doi: 10.1016/j.jlumin.2014.08.031 [Crossref] [ Google Scholar]
- Rasoulzadeh F, Asgari D, Naseri A, Rashidi MR. Spectroscopic studies on the interaction between erlotinib hydrochloride and bovine serum albumin. DARU 2010; 18:179. doi: 10.1016/j.chemosphere.2012.12.069 [Crossref] [ Google Scholar]
- Frimodt-Møller N, Thomsen VF. Interaction between beta-lactam antibiotics and gentamicin against Streptococcus pneumoniae in vitro and in vivo. Acta Pathol Microbiol Immunol Scand B 1987; 95B:269-75. doi: 10.1111/j.1699-0463.1987.tb03124.x [Crossref] [ Google Scholar]
- Ehteshami M, Rasoulzadeh F, Mahboob S, Rashidi MR. Characterization of 6-mercaptopurine binding to bovine serum albumin and its displacement from the binding sites by quercetin and rutin. J Lumin 2013; 135:164-9. doi: 10.1016/j.jlumin.2012.10.044 [Crossref] [ Google Scholar]
- Anbazhagan V, Renganathan R. Study on the binding of 2,3-diazabicyclo[222]oct-2-ene with bovine serum albumin by fluorescence spectroscopy. J Lumin 2008; 128:1454-8. doi: 10.1016/j.jlumin.2008.02.004 [Crossref] [ Google Scholar]
- Sułkowska A, Bojko B, Rownicka J, Pentak D, Sułkowski W. Effect of urea on serum albumin complex with antithyroid drugs: fluorescence study. J Mol Struct 2003; 651:237-43. doi: 10.1016/S0022-2860(02)00635-X [Crossref] [ Google Scholar]
- Bi S, Sun Y, Qiao C, Zhang H, Liu C. Binding of several anti-tumor drugs to bovine serum albumin: Fluorescence study. J Lumin 2009; 129:541-7. doi: 10.1016/j.jlumin.2008.12.010 [Crossref] [ Google Scholar]
- He Y, Wang Y, Tang L, Liu H, Chen W, Zheng Z. Binding of Puerarin to Human Serum Albumin: A Spectroscopic Analysis and Molecular Docking. J Fluoresc 2008; 18:433-42. doi: 10.1007/s10895-007-0283-0 [Crossref] [ Google Scholar]
-
Chen G-Z, Huang X-Z, Xu J-G, Zheng Z, Wang Z. The methods of fluorescence analysis. Beijing: Science Press; 1990. p. 112.
- Yan J, Wang Q, Pan Q, Rao Z, Su Y, Li H. Assessment of the interaction between fraxinellone and bovine serum albumin by optical spectroscopy and molecular modeling methods. J Lumin 2013; 137:180-5. doi: 10.1016/j.jlumin.2012.12.036 [Crossref] [ Google Scholar]
- Bi S, Song D, Tian Y, Zhou X, Liu Z, Zhang H. Molecular spectroscopic study on the interaction of tetracyclines with serum albumins. Spectrochim Acta A Mol Biomol Spectrosc 2005; 61:629-36. doi: 10.1016/j.saa.2004.05.028 [Crossref] [ Google Scholar]
- Ashoka S, Seetharamappa J, Kandagal PB, Shaikh SMT. Investigation of the interaction between trazodone hydrochloride and bovine serum albumin. J Lumin 2006; 121:179-86. doi: 10.1016/j.jlumin.2005.12.001 [Crossref] [ Google Scholar]
- Hemmateenejad B, Shamsipur M, Samari F, Khayamian T, Ebrahimi M, Rezaei Z. Combined fluorescence spectroscopy and molecular modeling studies on the interaction between harmalol and human serum albumin. J Pharm Biomed Anal 2012; 67–68:201-8. doi: 10.1016/j.jpba.2012.04.012 [Crossref] [ Google Scholar]
- Katrahalli U, Kalanur SS, Seetharamappa J. Interaction of bioactive coomassie brilliant blue g with protein: insights from spectroscopic methods. Sci Pharm 2010; 78:869. doi: 10.3797/scipharm.1008-15 [Crossref] [ Google Scholar]
- Thipperudrappa J, Biradar DS, Hanagodimath SM. Simultaneous presence of static and dynamic component in the fluorescence quenching of Bis-MSB by CCl4 and aniline. J Lumin 2007; 124:45-50. doi: 10.1016/j.jlumin.2006.02.001 [Crossref] [ Google Scholar]
- Rasoulzadeh F, Jabary HN, Naseri A, Rashidi M-R. Fluorescence quenching study of quercetin interaction with bovine milk xanthine oxidase. Spectrochim Acta A Mol Biomol Spectrosc 2009; 72:190-3. doi: 10.1016/j.saa.2008.09.021 [Crossref] [ Google Scholar]
- Zhang L-N, Wu F-Y, Liu A-H. Study of the interaction between 2, 5-di-[2-(4-hydroxy-phenyl) ethylene]-terephthalonitril and bovine serum albumin by fluorescence spectroscopy. Spectrochim Acta A Mol Biomol Spectrosc 2011; 79:97-103. doi: 10.1016/j.saa.2011.02.013 [Crossref] [ Google Scholar]
- Guo M, Lü W-J, Li M-H, Wang W. Study on the binding interaction between carnitine optical isomer and bovine serum albumin. Eur J Med Chem 2008; 43:2140-8. doi: 10.1016/j.ejmech.2007.11.006 [Crossref] [ Google Scholar]
- Guo X-J, Sun X-D, Xu S-K. Spectroscopic investigation of the interaction between riboflavin and bovine serum albumin. J Mol Struct 2009; 931:55-9. doi: 10.1371/journal.pone.0059106 [Crossref] [ Google Scholar]
- Zhang Y, Qi Z, Zheng D, Li C, Liu Y. Interactions of Chromium (III) and Chromium (VI) with Bovine Serum Albumin Studied by UV Spectroscopy, Circular Dichroism, and Fluorimetry. Biol Trace Elem Res 2009; 130:172-84. doi: 10.1007/s12011-009-8322-0 [Crossref] [ Google Scholar]
- Chamani J, Vahedian-Movahed H, Saberi MR. Lomefloxacin promotes the interaction between human serum albumin and transferrin: A mechanistic insight into the emergence of antibiotic’s side effects. J Pharm Biomed Anal 2011; 55:114-24. doi: 10.1016/j.jpba.2010.12.029 [Crossref] [ Google Scholar]
- Zohoorian-Abootorabi T, Sanee H, Iranfar H, Saberi MR, Chamani J. Separate and simultaneous binding effects through a non-cooperative behavior between cyclophosphamide hydrochloride and fluoxymesterone upon interaction with human serum albumin: Multi-spectroscopic and molecular modeling approaches. Spectrochim Acta A Mol Biomol Spectrosc 2012; 88:177-91. doi: 10.1016/j.saa.2011.12.026 [Crossref] [ Google Scholar]
- Li D, Jiang X, Yan Z. Study of interaction between sorbitol and bovine serum albumin by fluorescence spectrometry. Guang Pu Xue Yu Guang Pu Fen Xi 2008; 28:1312-6. doi: 10.1016/j.talanta.2008.03.037 [Crossref] [ Google Scholar]
- Ross PD, Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry 1981; 20:3096-102. doi: 10.1021/bi00514a017 [Crossref] [ Google Scholar]
- Ni Y, Wang S, Kokot S. Spectrometric study of the interaction between Alpinetin and bovine serum albumin using chemometrics approaches. Analytica Chimica Acta 2010; 663:139-46. doi: 10.1016/j.aca.2010.01.053 [Crossref] [ Google Scholar]
- Guo X-j, Jing K, Guo C, Jiang Y-c, Tong J, Han X-w. The investigation of the interaction between oxybutynin hydrochloride and bovine serum albumin by spectroscopic methods. J Lumin 2010; 130:2281-7. doi: 10.1016/j.jlumin.2010.07.005 [Crossref] [ Google Scholar]
- Yamasaki K, Chuang VTG, Maruyama T, Otagiri M. Albumin–drug interaction and its clinical implication. Biochim Biophys Acta 2013; 1830:5435-43. doi: 10.1016/j.bbagen.2013.05.005 [Crossref] [ Google Scholar]
- Xu T, Guo X, Zhang L, Pan F, Lv J, Zhang Y. Multiple spectroscopic studies on the interaction between olaquindox, a feed additive, and bovine serum albumin. Food Chem Toxicol 2012; 50:2540-6. doi: 10.1016/j.fct.2012.04.007 [Crossref] [ Google Scholar]
- Cui F, Kong X, Qin L, Zhang G, Liu Q, Lei B. Specific interaction of 4′-O-(a-l-Cladinosyl) daunorubicin with human serum albumin: The binding site II on HSA molecular using spectroscopy and modeling. J Photochem Photobiol B 2009; 95:162-9. doi: 10.1016/j.jphotobiol.2009.03.001 [Crossref] [ Google Scholar]
- Dhar S, Rana DK, Pal A, Bhattacharya SC. Photobehavior and docking simulations of drug within macromolecules: Binding of an antioxidative isoquinolindione to a serine protease and albumin proteins. J Photochem Photobiol B 2013; 129:69-77. doi: 10.1016/j.jphotobiol.2013.09.007 [Crossref] [ Google Scholar]