BioImpacts. 8(3):201-209.
doi: 10.15171/bi.2018.23
Original Research
In silico identification of albendazole as a quorum sensing inhibitor and its in vitro verification using CviR and LasB receptors based assay systems
Shaminder Singh 1, 2, *, Sonam Bhatia 3
Author information:
1Biochemical Engineering Research and Process Development Centre (BERPDC), Institute of Microbial Technology (IMTECH), Chandigarh – 160036, India (Previous address)
2Department of Industrial Microbiology, Jacob Institute of Biotechnology and Bioengineering, Sam Higginbottom University of Agriculture, Technology and Sciences (SHUATS), Allahabad, Uttar-Pradesh-211007, India (Present address)
3Department of Pharmaceutical Sciences, Shalom Institute of Health and Allied Sciences (SIHAS), Sam Higginbottom University of Agriculture, Technology and Sciences (SHUATS), Allahabad, Uttar-Pradesh-211007, India
Abstract
Introduction:
Quorum sensing inhibition (QSI) is one of the vital tools to overcome emerging virulence of pathogenic bacteria which aims at curbing bacterial resistance. Targeting QS (quorum sensing) as chemotherapy is less likely to generate resistance among pathogens as it targets only the adaptation and not the survival mechanism of the pathogen. Several QS inhibitors were developed in the recent past but none of them managed to have clinical application due to known toxic effects for human consumption. A rapid development of QS inhibitor drugs could be achieved by verification of the QSI activity of drugs which are already in clinical use with known pharmacology. Recently, a known FDA approved clinical drug niclosamide belonging to an anthelmintic class is found to exhibit QSI activity.
Methods:
We have focused our study on Albendazole, another FDA approved clinical drug belonging to the same class for its potential to act as QSI. The structure-based molecular docking is used for finding putative interactions made by this drug with the CviR and LasB receptor protein of Chromobacterium violaceum and Pseudomonas aeruginosa , respectively. Further, the in vitro activity of this drug has been evaluated by employing CviR and LasB receptor-based bioassay. The efficacy of this drug alone and in combination with antibiotic Tobramycin to inhibit P. aeruginosa based biofilms was also analyzed by developing the biofilms on chambered glass slides and performing anti-biofilm assay.
Results:
Further, this drug found to inhibit purple pigment violacein production in C. violaceum , which is under the control of C6-AHL-CviR mediated QS in this human pathogen. The in vivo bioassays results suggested that albendazole has great potential to act as a QS inhibitor as found inhibiting violacein production in C. violaceum and biofilm formation in P. aeruginosa , respectively.
Conclusion:
It is that structure-based molecular docking guided bioassay evaluation is an efficient tool for finding the new therapeutic use of old drugs which could have more chances to come easily in clinical application for their newly identified therapeutic uses.
Keywords: Albendazole, CviR, Docking, LasR, Quorum sensing inhibitors
Copyright and License Information
© 2018 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Researchers all around the world are engaged in finding remedies for the problems that are associated with chemotherapy against Pseudomonas aeruginosa and Acinetobacter sp. in the immune-compromised persons. These organisms have emerged to pose deadly threat for hospitalized patients.
1,2
Mostly, the patients, who have gone through the organ transplant or undergoing cystic fibrosis treatment often caught infections from either of these pathogens.
3-5
Moreover, these organisms have developed multiple drug resistance along with their behavior to dwell in complex biofilms which further rescue them from the attack of antibiotics. The main driving force for bacterial biofilm formation and virulence gene expression comes from a well-known process of quorum sensing (QS), which is cell density-dependent mode of communication used by these bacteria.
6-8
Both the Gram-positive and Gram-negative bacteria share the same basic principles of QS-regulated gene expression but the signaling and molecular mechanism operating in the QS circuit get deferred. The interruption of QS is the way to fight against these pathogenic bacteria. The discovery of halogenated furanones from the marine alga Delisea pulchra has generated interest among the scientific community to screen several compounds of natural and synthetic origin against QS signaling.
9-12
Various antibiotics like azithromycin, ciprofloxacin and ceftazidime are known to inhibit bacterial QS at the concentrations below their respective minimum inhibitory concentrations (MICs).
13,14
In several other reports, conjugates of antibiotics (e.g., ciprofloxacin, gentamicin, tobramycin, clarithromycin, piperacillin, etc) and N-Acyl homoserine lactones (AHL) analogues (antibiotic-AHL) have been administered to the biological systems.
15
It was found that these conjugates enhance the uptake of antibiotics and hence shown better outcomes in terms of bacterial clearance load. Previously, aminoglycoside antibiotic tobramycin in the sub-MICs, found to disrupt biofilm formation in P. aeruginosa as well as affecting the swarming motility and protease production. However, this organism has shown resistance against several marketed antibiotic formulations.
13,16
Several QS inhibitors have been developed with a great potential of QS inhibition activity, but unfortunately, none of them so far is available for the clinical use due to the pharmacokinetics and toxic implications. Here, we have tried to use the principle of evaluating “old drugs for new use” to curb the QS-mediated virulence in P. aeruginosa.
17
The screening of known drugs for the other activities by employing the SOSA (selective optimization of side activities of drug molecules) approach had been already implied by Imperi et al and thereby niclosamide was found that possess a QS inhibition (QSI) activity.
18
This approach is highly advantageous as the resultant hits can be directly tested under the clinical studies or can be used as a lead on drug discovery platform. This process can yield safe drug like compounds at extremely reduced cost than what generally associated with drug discovery pipeline.
Our study is focused on albendazole (Fig. 1), a benzimidazole derivative that is given orally for the treatment of parasitic worms.
19
We are working in the area of QS and screening several secondary metabolites produced by microorganisms subsequent to their QSI activity.
20,21
We have standardized various bioassays by testing several known inhibitors of QS against violacein pigment production in Chromobacterium violaceum and also green fluorescence protein production in P. aeruginosa lasB-gfp strain. As mentioned earlier, both of the assayed traits are under the control of QS.
20,21
Interestingly, we found that albendazole has anti-QS activities in sub-MICs in a dose-dependent manner. Therefore, it can inhibit violacein production in C. violaceum. Albendazole also interferes with virulence production and biofilm formation in Pseudomonas aeruginosa lasB-gfp strain. Further, the key interactions and binding pose of albendazole inside the active site pocket of CviR and LasR receptor proteins were evaluated by performing molecular docking analysis. To the best of our knowledge, this is the first report on (i) QS inhibitory property of albendazole, and (ii) demonstrating a reduction in virulence gene expression and biofilm formation at sub-MIC concentration.
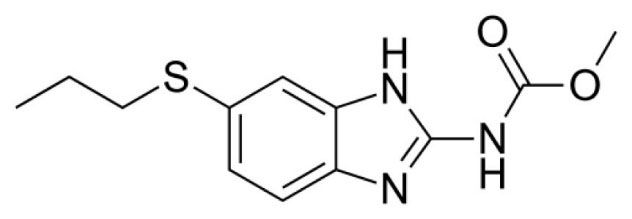
Fig. 1.
Structure of the drug candidate albendazole used in this study.
.
Structure of the drug candidate albendazole used in this study.
Materials and Methods
Molecular docking studies
Molecular docking is one of the most frequently used methods in the structure-based drug design strategy, where small ligands are docked into the active site of target receptor proteins. These ligands are then ranked on the basis of their binding affinity and orientation in the active site. This technique is highly useful for understanding the ligand-binding interactions into the active site, and the ligand-receptor intermolecular forces were analyzed based on the docking score of reference ligand.
22
For performing molecular docking analysis, the following three steps have to be carried out: (1) ligand and protein preparation, (2) grid generation, and (3) ligand docking.
Protein and ligand preparation
The structure of CviR (PDB ID: 3QP5) and LasR (PDB ID: 3IX3) protein co-crystallized with chlorolactone (HLC) MF: (C14H16ClNO4) and 3-Oxo-N-[(3S)-tetrahydro-2-oxo-3-furanyl]-dodecanamide (OHN) MF: (C16H27NO4) ligands were taken from the depository PDB (protein data bank).
23,24
The protein structures were prepared using the Protein Preparation Wizard incorporated in Maestro.
25
This tool helps in assigning right bond orders to amino acids as well it adds hydrogen atoms to the protein. Finally, the minimization was done using Impref and up to the described state, where the average root mean square deviation (RMSD) of all the atoms comes down to 0.3 Å. Further, for preparation of ligands, Lig-prep module of Maestro was utilized, where low energy ionized, tautomeric and stereoisomeric states of the ligands within a pH range of 7.0±2.0 were generated using the OPLS 2005 force field.
Receptor grid generation
Receptor Grid generation module of GLIDE (grid-based ligand docking with energetics) software
26
was used to generate the grid for molecular docking purpose. The co-crystallized ligand HLC and OHN was used as a reference for the grid generation. The inner grid box of 10 Å was defined around the centroid of bound ligands (HLC and OHN), whereas outer box was extended up to 20 Å.
Ligand docking
For the validation of docking protocol, bound ligand was extracted and then was re-docked to generate the same docking poses as found in their co-crystallized forms. Finally, a set of optimized ligands were docked using Ligand Docking module of GLIDE, and they were analyzed on the basis of their GLIDE docking score and intermolecular interactions.
Visualization software
For the purpose of molecular visualization and binding site analysis, the academic version of PyMOL (https://pymol.org/academic) was used.
Bacterial strains, growth conditions and chemicals used
In this study C. violaceum (CV12472) wild-type strain and P. aeruginosa plasB-gfp (ASV) strain was used. In the wild type strain CV12472, production of purple violacein pigment was under the control of QS.
27
In strain plasB-gfp (ASV), gene expression of green fluorescent protein (GFP) that results in green fluorescence was under the control of QS.
28
The assay plates for the C. violaceum were made using the Luria Bertani (LB) (tryptone 1%, yeast extract 0.5%, NaCl 1% and 1% agar). The plasB-gfp strain was grown using the AT media as described previously.
29
The known QS inhibitor, 4-nitropyridine-N-oxide (4-NPO), was purchased from Sigma-Aldrich and was used at 1 mM concentration as a positive control. Propidium iodide (PI) dye was used for staining biofilms and in 2.5 mg/mL concentration. Antibiotic tobramycin sulfate was purchased from the local vendor of make Tobraneg-80 (equivalent to tobramycin IP 40 mg in 2 mL) and concentration of 2 µg/mL.
Violacein inhibition assay
The production of characteristic purple pigment violacein is under the control of the QS system.
27
In this wild-type strain, violacein is inducible by the auto-inducer C6-AHL.
30
Hence, the presence of anti-QS compounds may compete for the binding in the active site of the receptor and thereby results in inhibition of violacein production. The QSI activity of albendazole was checked on C. violaceum using the agar diffusion method. To determine the MIC of albendazole for QS inhibition activity, agar well diffusion method was performed in which a stock solution of albendazole (1 mg/mL) and subsequent dilutions (0.1, 0.2, 0.4, 0.6 & 0.8 mg/mL) were made and added to the wells of seeded CV12472 plates and incubated overnight at 30°C.
lasB-gfp assay
Followed by preliminary assay on C. violaceum, further analysis of QSI potential of albendazole was carried out using plasB-gfp (ASV) bioassay. We have performed this assay as previously described in one of our work.
20
In test tubes, 10 mL of LB broth was inoculated with plasB-gfp (ASV) strain and was kept in incubation (overnight (O/N), 37 °C). An O/N growth culture was diluted 1:40 (about 125 µL of O/N culture was added to 4.875 mL of LB broth). In each row, 170 µL of the LB broth was added to the first well and 100 µL to the rest of the wells in 96-well microtiter plate. To this, 30 µL of the albendazole (1 mM stock) were added to the first well in each row and 100 µL was serially transferred to the next well up to the fifth well. The last well in each row was added with DMSO as a negative control. Finally, 100 µL of 1:40 diluted O/N culture was added to all the wells resulting in 1:80 dilutions of the culture with concentration doses ranging (75–4.6 mg/mL) of albendazole. The effect of albendazole on the growth and GFP expression was measured by taking OD at 450 nm and fluorescence under the excitation of 485 nm and emission at 535 nm.
Biofilm inhibition assay
The biofilms of plasB-gfp (ASV) were established on microscopic coverslips. 10 µL of an overnight culture was added to the chambers having 200 µL of the LB broth and such 5 chambers assembly was kept for incubation at 37°C. After every six hours, the media was replaced with fresh LB broth. About after 24 hours of incubation, the chambers were added with 10 µL of albendazole (1 mM) and then kept for incubation (for 24 hours).
20
The 3-day old mature biofilms stained with PI, washed twice with phosphate buffer saline (PBS) to remove the unattached cells and subsequently the stained biofilms were fixed using fixative. The biofilms on coverslips were visualized on Nikon Confocal microscope and analyzed using NIS-elements software.
Effect of albendazole on biofilms
The biofilms of P. aeruginosa were developed on microscopic cover-slips. The plasB-gfp (ASV) strain was inoculated in 10 mL AT media and incubated overnight at 37°C with 180 rpm shaking. The O/N culture then was diluted to OD 0.2 and 150 µL of this added to the microfuge-coverslip assembly and kept for incubation for 48 hours at 37°C. A water reservoir has been kept in the incubator to prevent drying of the biofilms. The two days old biofilms then treated with sub-MIC concentration (0.16 µg/mL) of albendazole alone and along with tobramycin (100 µg/mL). Since the GFP expression of this strain is under the control of QS, hence green fluorescence served as an indicator of live cells and propidium iodide (PI, 2.5 mg/mL) was added for staining dead cells. The biofilms developed on microscopic coverslips and given O/N treatments of albendazole. The coverslips were then removed from the microfuge tube assembly and fixed on glass slides. The biofilms were visualized using Nikon A1 Confocal Laser Microscope system. Firstly, the dense area of the biofilm was focused using DIA parameter and then biofilms were exposed with Kr/Ar lasers, where GFP excited with eGFP channel at an excitation of 485 nm and emission at 535 nm and PI excited with the live/dead channel at an excitation of 561 nm and emission of 640 nm. The analysis was done using Nikon NIS-C image analysis software. The captured images were further analyzed using the COMSTAT software equipped with ImageJ package to analyze images and a loci tools.
31,32
Results
Molecular Modeling studies
CviR and LasR protein
The x-ray crystal structure of full-length CviR (PDB ID: 3QP5) bound with antagonist chlorolactone (HLC) in a resolution of 3.25 Å was reported by Chen et al.
23
It is only known structure for CviR so far, which is co-crystallized with its antagonist. CviR is a homodimer with two binding domains (Ligand binding domain [LBD] and DNA-binding domain [DBD]) that placed in a ‘‘crossed-domain’’ conformation. In this closed conformation of the protein, 2 DBD units are apart which have reduced affinity to bind with DNA as shown in Fig. 2. The antagonist HLC stabilizes this closed conformation of CviR as compare to the agonist and thus prevents the QS activity.
Similarly, the structure of LasR protein, which directs the synthesis of autoinducer OHN [N-(3-oxododecanoyl)-L-homoserine lactone], is provided in Fig. 2. The bound autoinducer forms the hydrogen bonding interaction with key amino acid residues viz. Ser129, Asp73, Trp60. In addition, the hydrophobic interactions played the significant role in stabilizing the protein-ligand complex. This finding is also noticed by us in the previous study where N,N-disubstituted biguanides with aryl groups have shown a significant correlation between π stacking ability and observed biological activity.
21
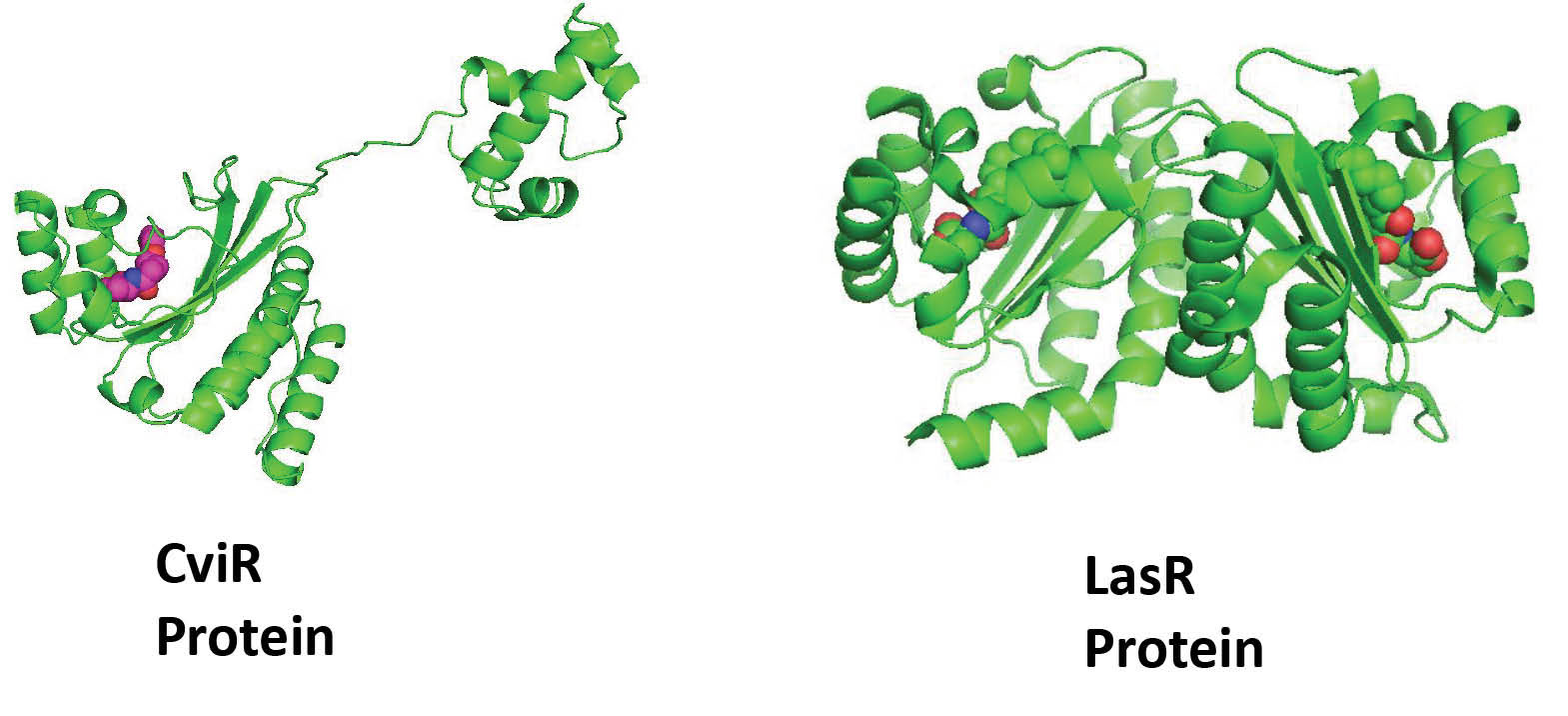
Fig. 2.
Three-dimensional ribbon representation of CviR and LasR receptor proteins. The co-crystallized ligand (HLC) in CviR and (OHN) in LasR receptor is represented by CPK model.
.
Three-dimensional ribbon representation of CviR and LasR receptor proteins. The co-crystallized ligand (HLC) in CviR and (OHN) in LasR receptor is represented by CPK model.
Molecular docking studies were performed to investigate the binding potential of albendazole towards CviR and LasR protein. The docking protocol was standardized by performing the re-docking of bound ligands in order to get the correct binding pose of HLC and OHN as found in their co-crystallized states. HLC was able to get dock inside the active site of protein with almost identical binding pose (GLIDE score of -8.06) as compared to its co-crystallized x-ray structure. The best scored docked conformer of HLC with its crystallized protein showed superimposition with the RMSD of 0.03 (Fig. 3). This protocol was acceptable and was further utilized to dock other sets of ligands. Similarly, the OHN was re-docked (GLIDE Score = -7.52) in the active site of LasR receptor for validating the docking protocol and found suitable to use the protocol for docking albendazole inside its active site pocket.
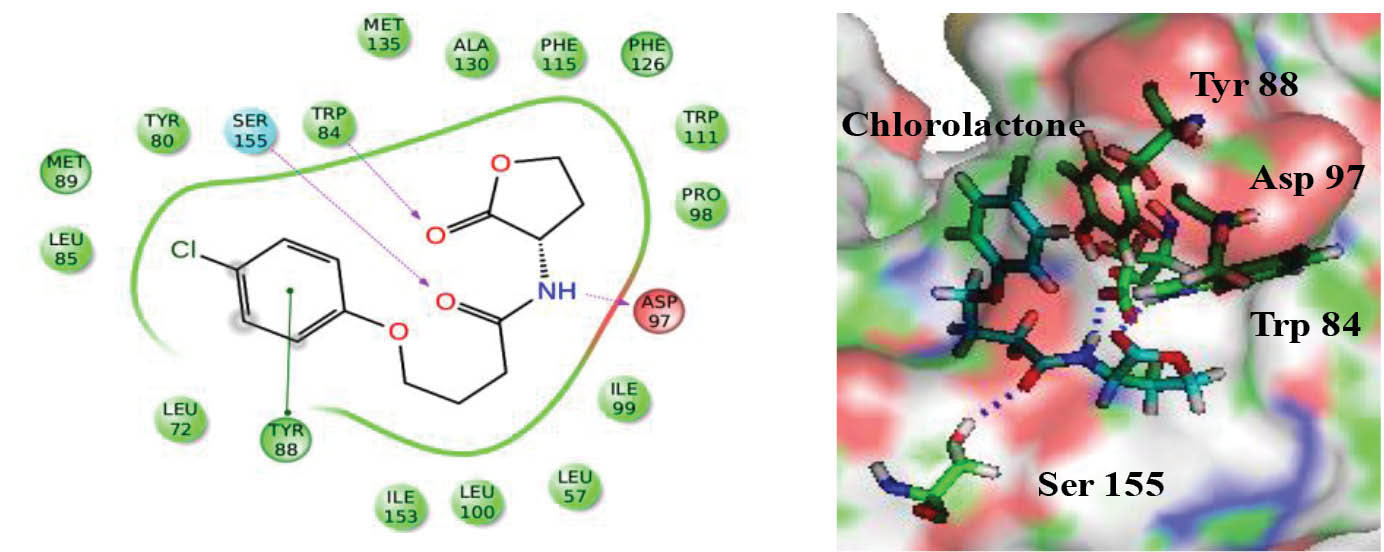
Fig. 3.
Docking pose of positive control chlorolactone (HLC) in the active site of ligand CviR.
.
Docking pose of positive control chlorolactone (HLC) in the active site of ligand CviR.
The 3D structure of albendazole in SDF format was taken from the PubMed library. Further, the structure was prepared for docking by using Ligprep module of Maestro interface, which was optimized by using the OPLS 2005 force field. The optimized structure was docked inside the active site, where 10 different docked conformers of albendazole were generated. The best-docked conformer of albendazole showed a GLIDE score of -6.89. Fig. 4 gives the binding model of albendazole inside the active site of CviR protein along with the key interactions. Although, the amide hydrogen (2.07 Å) and ring hydrogen (N-H, 2.49 Å) of albendazole were engaged in the formation of a bifurcated hydrogen bond with Asp97 (1.89 Å), however, in HLC the amide hydrogen involved in the hydrogen bond formation. Secondly, imidazole ring nitrogen also forms hydrogen bonding interaction with Trp84 (2.35 Å), thus it plays the function of lactone moiety of HLC (Fig. 4). The benzene ring that is attached to imidazole ring, showed a hydrophobic interaction with Tyr80 residue and involved in π-π stacking. Similarly, in the active site pocket of LasR receptor, albendazole showed hydrogen bonding interaction with Tyr56 (2.06 Å) and Ser129 (2.41 Å). Both these amino acids act as hydrogen bonding donors as shown in Fig. 4. The presence of albendazole in the hydrophobic pocket of CviR and LasR receptors demonstrated the presence of hydrophobic interactions that were formed by benzimidazole framework and hydrophobic amino acid residues. Thus, the molecular docking studies have established the potential of the structural framework of albendazole to quench the mechanism of QS. Table 1 provides the values of docking scores of different compounds against CviR and LasR receptors.
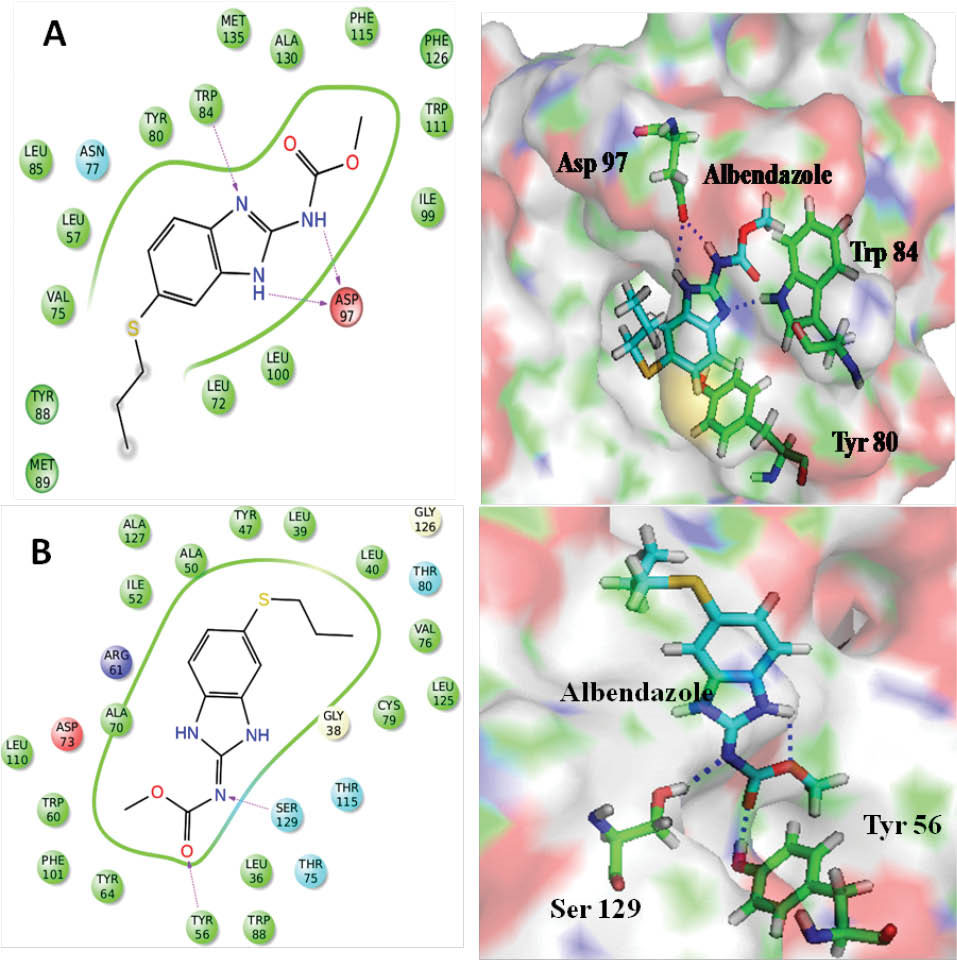
Fig. 4.
Binding model of albendazole and its hydrogen bonding interactions (blue dashed lines) with (A) CviR (PDB ID: 3QP5) and (B) LasR (PDB ID: 3IX3).
.
Binding model of albendazole and its hydrogen bonding interactions (blue dashed lines) with (A) CviR (PDB ID: 3QP5) and (B) LasR (PDB ID: 3IX3).
Table 1.
Docking scores of different compounds on CviR and LasR receptor
|
Molecular formula
|
Inhibitor/Autoinducer
|
Docking score
|
|
CviR Protein
|
LasR
|
|
(3QP5)
|
(3IX3)
|
|
C14H16ClNO4
|
Chlorolactone (HLC) |
-8.06 |
-- |
|
C10H15NO
|
C6-HSL |
-6.54 |
-- |
|
C16H27NO4
|
(OHN) |
-- |
-7.52 |
|
C5H4N2O3
|
4-nitro pyridine oxide (NPO) |
-5.90 |
-- |
|
C12H15N3O2S
|
Albendazole |
-6.89 |
-7.14 |
Biological studies
Based on the results obtained from the molecular docking analysis, the experimental studies were designed. Albendazole was purchased from Indo-AM and then various in vitro assays were performed to evaluate the presence of QSI activity in this drug.
QSI activity on CviR
The QSI activity was checked using CV12472 bio-monitor strain. Albendazole was found to show the QSI activity as indicated by the zone of inhibition of violacein production without any effect on the growth of the indicator organism (Fig. 5).

Fig. 5.
Dose dependent effect of albendazole on violacein production by CV12472.
.
Dose dependent effect of albendazole on violacein production by CV12472.
QSI activity on LasR
The QSI activity of albendazole on lasR gene expression was checked using bioassay strain and based on P. aeruginosa LasR receptor i.e. plasB-gfp (ASV). The decreasing concentrations of albendazole were increased the GFP expression as indicated by the dose-response assay plot. This finding confirmed the QSI activity of albendazole against LasR without any significant effect on the growth of the microorganism (Figs. 6 and 7).
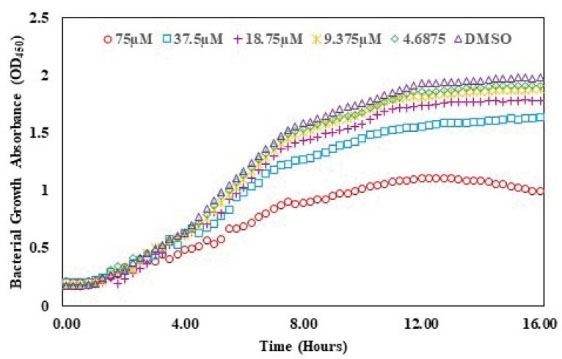
Fig. 6.
Effect of albendazole on the growth of plas-gfp.
.
Effect of albendazole on the growth of plas-gfp.
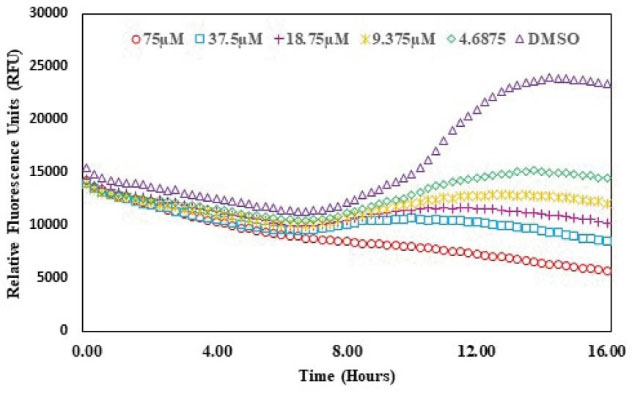
Fig. 7.
Effect of albendazole on the GFP production of plas-gfp
.
Effect of albendazole on the GFP production of plas-gfp
Effect of albendazole on Pseudomonas aeruginosa biofilms
The bacterial cells that were remained on the microscopic coverslips were considered as plasBgfp (ASV) biofilms. And yet the planktonic cells were removed every six hours of growth, whereas the old broth was replaced with fresh broth. On the confocal image analysis of the biofilms, it was found that the control biofilm was formed as a dense mat of green fluorescence bacteria with only 0.01% dead cells (Fig. 8A). The film that was treated with albendazole alone, was found with few partial death of cells. This result implies the biofilm clearance (Fig. 8D-E). The biofilm treated with both albendazole and tobramycin showed an enhanced cell death in the region of biofilm clearance, clearly indicating that at sub-MIC concentration albendazole has acted as QS inhibitor (Fig. 8G-H). It is supported by the fact its sub-MIC concentration is only clearing biofilm with partial cell death. Thus, it is properly permeable for tobramycin to complete cell death in the penetrated area (Fig. 8G-H). Hence, it indicates that albendazole can act as a potent QS inhibitor and can be used with tobramycin in adjunct therapy to curb the pathogenesis of P. aeruginosa.
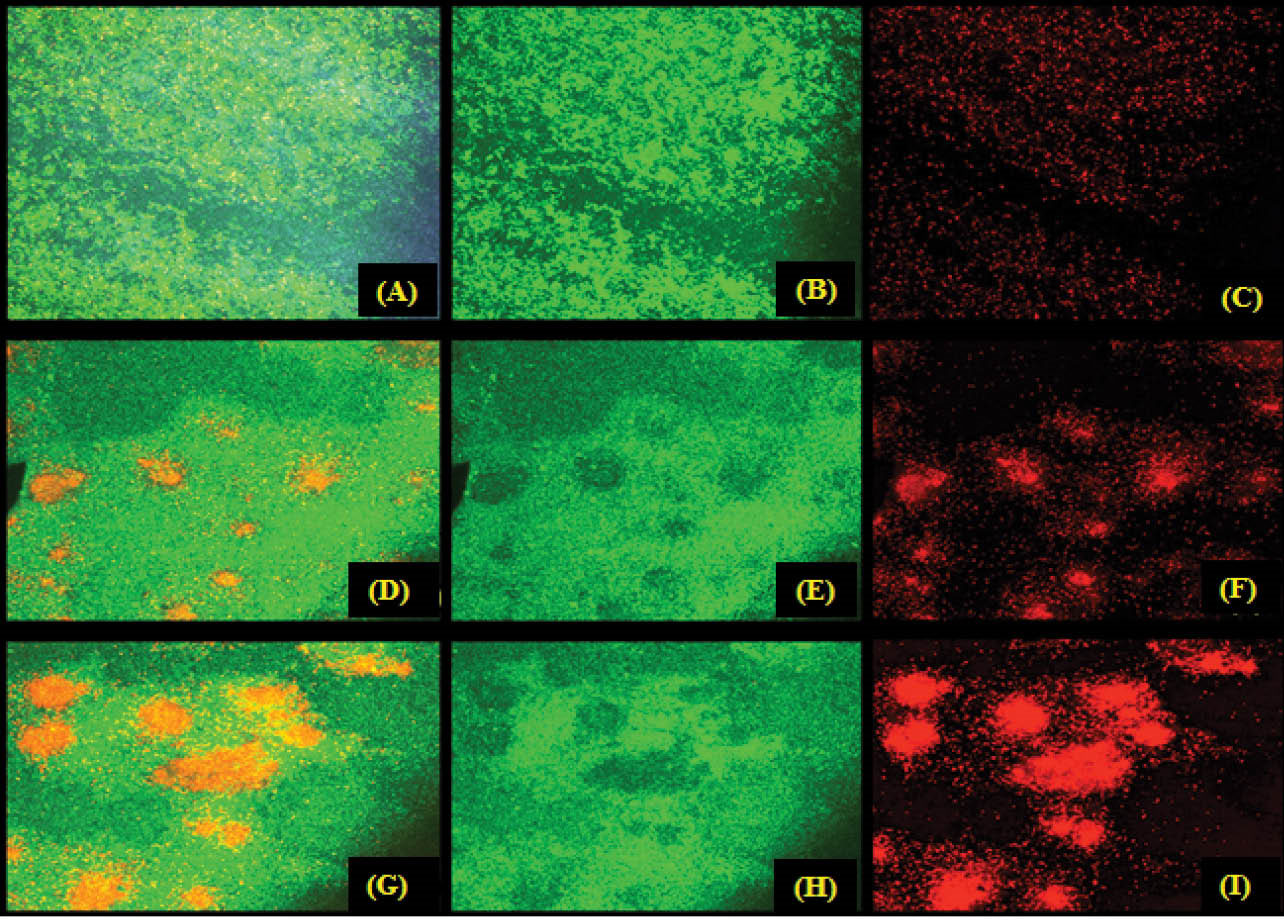
Fig. 8.
Effect of albendazole on the biofilm formation by plas-gfp. The fully matured biofilm controls (A) mixed, (B) GFP alone, (C) PI alone; Biofilms treated with 75 μM albendazole (D) mixed, (E) GFP alone, (F) PI alone and (G) biofilms treated with 75 μM albendazole along with tobramycin (80 mg/mL).
.
Effect of albendazole on the biofilm formation by plas-gfp. The fully matured biofilm controls (A) mixed, (B) GFP alone, (C) PI alone; Biofilms treated with 75 μM albendazole (D) mixed, (E) GFP alone, (F) PI alone and (G) biofilms treated with 75 μM albendazole along with tobramycin (80 mg/mL).
Discussion
The unmet demand for anti-virulence agents with the aim of treating infectious diseases is highly desired. The growing awareness regarding the relationship between QS inhibition and antimicrobial action has led the scientific community to work on the compounds that have the potential for inhibition of the mechanism of QS in pathogenic bacteria.
17,33
The various bio-monitor organisms have been used for screening the QSI activity. Among all the organisms, P. aeruginosa is extensively studied as a model system that can be used for evaluating the QS and biofilm inhibition.
The research on the development of QS inhibitors is largely focused on the identification of structural homologues of autoinducer ligands of QS proteins. Various QS inhibitors belonging to different chemical classes have been discovered by screening libraries of the chemical and natural compounds.
34,35
Use of synthetic methodologies is also adopted as a tool for the development of QS inhibitors.
36
However, some of the compounds prove to be effective in treating P. aeruginosa infection in both in vitro and in vivo studies.
37-39
Despite the fact that multiple leads as QS inhibitors had already been tested, yet the scientific literature supports only three verified clinical trials as listed in the ClinicalTrials.gov database.
40
This situation is mainly due to the toxicity and pharmacokinetic issues that can be associated with QS inhibitors, so far. Garlic extract is the only QS inhibitor which has been tested in humans.
41,42
But the results were not found to be statistically significant, so the improvements were planned. In the following study, ajoene, the active constituent of garlic extract, was synthesized and tested, however, it was found to be less active alone in in vitro models and have shown poor response in in vivo murine model of infection.
43
Thus, despite all the efforts made in the development of QS inhibitors, the clinical application of QS-based therapeutics remains as an imaginative approach yet. The main aim of this study was to find out a potential QS inhibitor as an anti-virulence agent using the alternative strategy. Thus, a new application (anti-QS activity) of the already used drug was explored.
Taking into account the QS potential of anthelmintic drug niclosamide (an amide derivative) and its structural features,
18
another anthelmintic drug albendazole which is an amide ester and contains a benzimidazole framework is selected for investigations. The literature also supports the anti-QS activity of benzimidazole backbone.
44
Hence, evaluation of QS inhibiting potential of albendazole is worth exploring. Initially, the molecular docking studies of albendazole in the active site pocket of CviR and LasR receptors was carried out. It was found that this drug indeed showed interaction with key amino acid residues in the active site of the target proteins. Further, to prove this finding, the biological activity of albendazole was tested using violacein inhibition assay on C. violaceum CV12472 bio-monitor strain. The dose-dependent response of albendazole was also analyzed which confirms QSI activity. Further, the anti-biofilm activity of albendazole on biofilms of P. aeruginosa strain plasB-gfp (ASV) was also estimated. The confocal images of biofilms have been clearly demonstrated the regions of clearance in albendazole treated P. aeruginosa biofilms (Fig. 8). The frequency of resistance in P. aeruginosa isolates to tobramycin, which is an aminoglycoside-based antibiotic is found to be high nearly 30% owing to overexpression of efflux pumps.
45
The biological activity of combined therapeutics (tobramycin with albendazole) was also evaluated and found to be effective more than that of albendazole alone which is also marked by cell death. Hence, it is proven that albendazole show anti-QS activity in sub-MIC concentration and its effectiveness increases to many folds if given in as a complex with an antibiotic.
Conclusion
Development of bacterial-resistant strains has exposed the human population to the large variety of bacterial infections. The QS mechanism in bacteria is involved in the expression of various virulence factors and biofilm formation. Many drug discovery scientists working in the respective field are developing synthetic/natural QS-inhibiting receptor proteins. The compounds that developed so far are associated with the toxicity and pharmacokinetics issues, which have become huge obstacles in their clinical application. Thus, in this study, a contemporary approach was utilized to identify a new perspective for an old drug by keeping in mind its extended advantages.
To summarize the results, the molecular docking studies of albendazole showed that the active site of CviR and LasR protein are perfectly in close contact with this drug. The binding model revealed that the drug can bind with the similar fashion and form the key interactions as that of bound ligands. Further, we have performed the bioassay of albendazole on CviR and LasR receptor-based systems for evaluating its potential as an anti-QS drug. Among a large number of network of QS proteins, LasR had a vital role in sensing mechanism in P. aeruginosa. The QSI activity was confirmed by performing dose response assay against LasR on basis of the lasB-gfp bioassays. It was also found that this drug can inhibit the virulence gene expression, with restricted exopolysaccharide and biofilm production in P. aeruginosa. Further, it can also be used in the conjugate therapy with tobramycin to show the combined effect of antimicrobial even for drug-resistant strains. In this way, the anti-QS and antibiotic activity of the respective drugs acts in a synergistic manner and thus increased the activity of many folds causing cell death of resistant strain, too. Albendazole is already an FDA approved drug and there will not be a question on the safety profile of this molecule. Hence, it can be interested as a candidate drug in the anti-pathogenic modalities. Here, we highlighted the translational importance of this study in terms of its real-life application. Therefore, albendazole can be utilized as an anti-virulence drug to solve the problem of persisters generation.
Ethical approval
Not applicable.
Competing interests
The authors alone are responsible for the content and writing of the paper.
Acknowledgment
The authors express their great thanks to Prof. Thomas Bjarnsholt for providing plasB-gfp (ASV) strain as well as COMSTAT software. Dr. Shaminder Singh is also thankful to CSIR for providing CSIR-NET scholarship. They are further grateful to BI journal and editorial office in following the process.
Research Highlights
What is current knowledge?
simple
-
√ Amide based anti-helminthic drug Niclosamide have
shown anti-QS activity in P. aeruginosa in in vitro studies.
-
√ Benzimidazole nucleus possess QS inhibiting potential.
-
√ Due to toxic and pharmacokinetic liabilities the clinical
application of active lead compounds is hindered.
What is new here?
simple
-
√ Identification of Albendazole, an amide ester-based drug as
QS inhibitor for C. violaceum and P. aeruginosa.
-
√ New therapeutic target of old FDA-approved drug was
spotted.
-
√ Albendazole was also found to possess anti-biofilm activity
against P. aeruginosa .
-
√ Use of albendazole with tobramycin in adjuvant therapeutic
remedy was also tested and found to be effective against the
anti-biofilm activity, hence new perspective of old drug was
identified.
References
- Burgos F, Torres A, Gonzalez J, de la Bellacasa JP, Rodriguez-Roisin R, Roca J. Bacterial colonization as a potential source of nosocomial respiratory infections in two types of spirometer. Eur Respir J 1996; 9:2612-7. doi: 10.1183/09031936.96.09122612 [Crossref] [ Google Scholar]
- Karlowsky JA, Draghi DC, Jones ME, Thornsberry C, Friedland IR, Sahm DF. Surveillance for antimicrobial susceptibility among clinical isolates of Pseudomonas aeruginosa and Acinetobacter baumannii from hospitalized patients in the United States, 1998 to 2001. Antimicrob Agents Chemother 2003; 47:1681-8. doi: 10.1128/AAC.47.5.1681-1688.2003 [Crossref] [ Google Scholar]
- Botha P, Archer L, Anderson RL, Lordan J, Dark JH, Corris PA. Pseudomonas aeruginosa colonization of the allograft after lung transplantation and the risk of bronchiolitis obliterans syndrome. Transplantation 2008; 85:771-4. doi: 10.1097/TP.0b013e31816651de [Crossref] [ Google Scholar]
- May JR, Herrick NC, Thompson D. Bacterial infection in cystic fibrosis. Arch Dis Child 1972; 47:908-13. doi: 10.1136/adc.47.256.908 [Crossref] [ Google Scholar]
- Corey M, McLaughlin FJ, Williams M, Levison H. A comparison of survival, growth, and pulmonary function in patients with cystic fibrosis in Boston and Toronto. J Clin Epidemiol 1988; 41:583-91. doi: 10.1016/0895-4356(88)90063-7 [Crossref] [ Google Scholar]
- Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet 2001; 35:439-68. doi: 10.1146/annurev.genet.35.102401.090913 [Crossref] [ Google Scholar]
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol 2001; 55:165-99. doi: 10.1146/annurev.micro.55.1.165 [Crossref] [ Google Scholar]
- Whitehead NA, Barnard AML, Slater H, Simpson NJL, Salmond GPC. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev 2001; 25:365-404. doi: 10.1111/j.1574-6976.2001.tb00583.x [Crossref] [ Google Scholar]
- de Nys R, Wright AD, Konig GM, Sticher O. New halogenated furanones from the marine alga Delisea pulchra (cf fimbriata). Tetrahedron 1993; 49:11213-20. doi: 10.1016/S0040-4020(01)81808-1 [Crossref] [ Google Scholar]
- Sarabhai S, Sharma P, Capalash N. Ellagic acid derivatives from Terminalia chebula Retz downregulate the expression of quorum sensing genes to attenuate Pseudomonas aeruginosa PAO1 virulence. PloS One 2013; 8:e53441. doi: 10.1371/journal.pone.0053441 [Crossref] [ Google Scholar]
- Nizalapur S, Kimyon Ãn, Biswas NN, Gardner CR, Griffith R, Rice SA. Design, synthesis and evaluation of N-aryl-glyoxamide derivatives as structurally novel bacterial quorum sensing inhibitors. Org Biomol Chem 2016; 14:680-93. doi: 10.1039/c5ob01973g [Crossref] [ Google Scholar]
- Nizalapur S, Ho KKK, Kimyon Ãn, Yee E, Berry T, Manefield M. Synthesis and biological evaluation of N-naphthoyl-phenylglyoxamide-based small molecular antimicrobial peptide mimics as novel antimicrobial agents and biofilm inhibitors. Org Biomol Chem 2016; 14:3623-37. doi: 10.1039/c6ob00298f [Crossref] [ Google Scholar]
- Skindersoe ME, Alhede M, Phipps R, Yang L, Jensen PO, Rasmussen TB. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2008; 52:3648-63. doi: 10.1128/AAC.01230-07 [Crossref] [ Google Scholar]
- Desai M, Bühler T, Weller PH, Brown MR. Increasing resistance of planktonic and biofilm cultures of Burkholderia cepacia to ciprofloxacin and ceftazidime during exponential growth. J Antimicrob Chemother 1998; 42:153-60. doi: 10.1093/jac/42.2.153 [Crossref] [ Google Scholar]
- Kaplan JB. Methods for the treatment and prevention of bacterial biofilms. Expert Opin Ther Pat 2005; 15:955-65. doi: 10.1517/13543776.15.8.955 [Crossref] [ Google Scholar]
- Brackman G, Cos P, Maes L, Nelis HJ, Coenye T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob Agents Chemother 2011; 55:2655-61. doi: 10.1128/AAC.00045-11 [Crossref] [ Google Scholar]
- Rasmussen TB, Givskov M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int J Med Microbiol 2006; 296:149-61. doi: 10.1016/j.ijmm.2006.02.005 [Crossref] [ Google Scholar]
- Imperi F, Massai F, Pillai CR, Longo F, Zennaro E, Rampioni G. New life for an old drug: the anthelmintic drug niclosamide inhibits Pseudomonas aeruginosa quorum sensing. Antimicrob Agents Chemother 2013; 57:996-1005. doi: 10.1128/AAC.01952-12 [Crossref] [ Google Scholar]
- Theodorides VJ, Gyurik RJ, Kingsbury WD, Parish RC. Anthelmintic activity of albendazole against liver flukes, tapeworms, lung and gastrointestinal roundworms. Experientia 1976; 32:702-03. doi: 10.1007/BF01919842 [Crossref] [ Google Scholar]
- Chourasiya SS, Kathuria D, Singh S, Sonawane VC, Chakraborti AK, Bharatam PV. Design, synthesis and biological evaluation of novel unsymmetrical azines as quorum sensing inhibitors. RSC Advs 2015; 5:80027-38. doi: 10.1039/C5RA12925G [Crossref] [ Google Scholar]
- Singh S, Wanjari PJ, Bhatia S, Sonwane VC, Chakraborti AK, Bharatam PV. Design, synthesis, biological evaluation and toxicity studies of N, N-disubstituted biguanides as quorum sensing inhibitors. Med Chem Res 2015; 24:1974-87. doi: 10.1007/s00044-014-1255-y [Crossref] [ Google Scholar]
- Meng X-Y, Zhang H-X, Mezei M, Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 2011; 7:146. doi: 10.2174/157340911795677602 [Crossref] [ Google Scholar]
- Chen G, Swem LR, Swem DL, Stauff DL, O'Loughlin CT, Jeffrey PD. A strategy for antagonizing quorum sensing. Mol Cell 2011; 42:199-209. doi: 10.1016/j.molcel.2011.04.003 [Crossref] [ Google Scholar]
- Zou Y, Nair SK. Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor. Chem Biol 2009; 16:961-70. doi: 10.1016/j.chembiol.2009.09.001 [Crossref] [ Google Scholar]
- Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 2013; 27:221-34. doi: 10.1007/s10822-013-9644-8 [Crossref] [ Google Scholar]
- Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT. Glide: a new approach for rapid, accurate docking and scoring 2 Enrichment factors in database screening. J Med Chem 2004; 47:1750-9. doi: 10.1021/jm030644s [Crossref] [ Google Scholar]
- McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1997; 143:3703-11. doi: 10.1099/00221287-143-12-3703 [Crossref] [ Google Scholar]
- Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, Parsek MR. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 2002; 148:87-102. doi: 10.1099/00221287-148-1-87 [Crossref] [ Google Scholar]
- Bjarnsholt T, van Gennip M, Jakobsen TH, Christensen LD, Jensen PÃ, Givskov M. In vitro screens for quorum sensing inhibitors and in vivo confirmation of their effect. Nat Protoc 2010; 5:282-93. doi: 10.1038/nprot.2009.205 [Crossref] [ Google Scholar]
- Morohoshi T, Kato M, Fukamachi K, Kato N, Ikeda T. N-acylhomoserine lactone regulates violacein production in Chromobacterium violaceum type strain ATCC 12472. FEMS Microbiol Lett 2007; 279:124-30. doi: 10.1111/j.1574-6968.2007.01016.x [Crossref] [ Google Scholar]
- Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Bjarne Kjær Ersbøll. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000; 146:2395-407. doi: 10.1099/00221287-146-10-2395 [Crossref] [ Google Scholar]
-
Vorregaard M. Comstat2-a modern 3D image analysis environment for biofilms: Technical University of Denmark, DTU, DK-2800 Kgs. Lyngby, Denmark; 2008.
- Brackman G, Coenye T. Quorum sensing inhibitors as anti-biofilm agents. Curr Pharm Des 2015; 21:5-11. doi: 10.2174/1381612820666140905114627 [Crossref] [ Google Scholar]
- Geske GD, Wezeman RJ, Siegel AP, Blackwell HE. Small molecule inhibitors of bacterial quorum sensing and biofilm formation. J Am Chem Soc 2005; 127:12762-3. doi: 10.1021/ja0530321 [Crossref] [ Google Scholar]
- Kalia VC. Quorum sensing inhibitors: an overview. Biotechnol Advs 2013; 31:224-45. doi: 10.1016/j.biotechadv.2012.10.004 [Crossref] [ Google Scholar]
- Wu H, Song Z, Hentzer M, Andersen JB, Molin Sr, Givskov M. Synthetic furanones inhibit quorum-sensing and enhance bacterial clearance in Pseudomonas aeruginosa lung infection in mice. Journal of Antimicrobial Chemotherapy 2004; 53:1054-61. [ Google Scholar]
- Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J 2003; 22:3803-15. doi: 10.1093/emboj/cdg366 [Crossref] [ Google Scholar]
- Galloway WRJD, Hodgkinson JT, Bowden S, Welch M, Spring DR. Applications of small molecule activators and inhibitors of quorum sensing in Gram-negative bacteria. Trends Microbiol 2012; 20:449-58. doi: 10.1016/j.tim.2012.06.003 [Crossref] [ Google Scholar]
- Rasmussen TB, Bjarnsholt T, Skindersoe ME, Hentzer M, Kristoffersen P, Kote M. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J Bacteriol 2005; 187:1799-814. doi: 10.1128/JB.187.5.1799-1814.2005 [Crossref] [ Google Scholar]
-
Clinical Trials website. https://clinicaltrials.gov/.
- Wang D, Lin Z, Huo Z, Wang T, Yao Z, Cong Y. Mechanism-based QSAR Models for the Toxicity of Quorum Sensing Inhibitors to Gram-negative and Gram-positive Bacteria. Bull Environ Contam Toxicol 2016; 97:145-50. doi: 10.1007/s00128-016-1801-z [Crossref] [ Google Scholar]
- Wang D, Lin Z, Ding X, Hu J, Liu Y. The Comparison of the Combined Toxicity between Gram-negative and Gram positive Bacteria: a Case Study of Antibiotics and Quorum-sensing Inhibitors. Mol Inform 2016; 35:54-61. doi: 10.1002/minf.201500061 [Crossref] [ Google Scholar]
- Jakobsen TH, van Gennip M, Phipps RK, Shanmugham MS, Christensen LD, Alhede M. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob Agents Chemother 2012; 56:2314-25. doi: 10.1128/AAC.05919-11 [Crossref] [ Google Scholar]
- Frei R, Breitbach AS, Blackwell HE. 2-Aminobenzimidazole Derivatives Strongly Inhibit and Disperse Pseudomonas aeruginosa Biofilms. Angew Chem Int Ed 2012; 51:5226-9. doi: 10.1002/anie.201109258 [Crossref] [ Google Scholar]
- Kiser TH, Obritsch MD, Jung R, McLaren R, Fish DN. Efflux pump contribution to multidrug resistance in clinical isolates of Pseudomonas aeruginosa. Pharmacotherapy 2010; 30:632-8. doi: 10.1592/phco.30.7.632 [Crossref] [ Google Scholar]