BioImpacts. 8(4):253-261.
doi: 10.15171/bi.2018.28
Original Research
Characterization and partial purification of an antibacterial agent from halophilic actinomycetes Kocuria sp. strain rsk4
Ravi Ranjan Kumar 1, *, Vasantba J Jadeja 2
Author information:
1Department of Biotechnology, Shree M. & N. Virani Science College, Kalawad Road, Rajkot 360005, Gujarat, India
2Department of Microbiology, Shree M. & N. Virani Science College, Kalawad Road, Rajkot 360005, Gujarat, India
Abstract
Introduction
: The inevitable rise of antibiotic-resistant bacteria is a global health problem. These pathogens erode the utility of available antibiotics. Staphylococcus aureus is one of the major causes of community-acquired infections. The aim of work was to evaluate the marine actinomycetes for production of the antibacterial agent against pathogens.
Methods
: Halophilic actinomycetes were isolated, characterized and screened for production of antibacterial agent against pathogenic bacteria. The antibacterial compounds were extracted by solvent extraction and separated by TLC based bioautography. Antibacterial compound was further purified by flash chromatography followed by high-performance liquid chromatography (HPLC) techniques. The active fraction was characterized by spectroscopy techniques. The minimum inhibitory concentration of antibiotic was determined against pathogens.
Results
: A new halophilic actinomycetes strain rsk4 was isolated from marine water. It was designated as Kocuria sp. based on the physiological, biochemical and 16S rDNA sequencebased characters. It was able to produce broad-spectrum antibacterial compound and exhibited significant inhibitory activities against antibiotic-resistant S. aureus. The antibacterial compound was secreted optimally at 5% NaCl and neutral pH in the starch casein medium during stationary phase. The crude ethyl acetate extract was separated by chloroform-methanol, 24:1, v/v having Rf value 0.45. Bioassay of HPLC fractions confirms the presence of antibiotics picks at retention time: 3.24 minutes. The UV-Visible and mass spectra of the compound revealed that the active compound was different from other known antibiotics. The lowest minimum inhibitory concentration was recorded against S. aureus (30 µg/mL).
Conclusion
: The result suggests that a broad-spectrum antibacterial compound obtained from halophilic actinomycetes is effective against pathogenic bacteria. This compound may be a good alternative treatment against antibiotic-resistant pathogen S. aureus.
Keywords: Antibacterial activity, Halophiles, Kocuria, Minimum inhibitory concentration
Copyright and License Information
© 2018 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Disease-causing bacteria resistance to commercially available antibiotics has become increasingly widespread in the recent decades.
1
Therefore, investigation of antibiotic producer against antibiotic-resistant pathogens is continued to be an important sector of research. Among microorganisms, actinomycetes are the most reliable groups from which a large number of economically and industrially important antimicrobial compounds have been obtained.
2
Actinomycetes are widespread in most ecological niches. They are accountable for the production of more than 70% of the natural antibiotics available in the market.
3
Most of the valuable antibiotics have been derived from terrestrial actinomycetes. Intensive exploitation diminishes the efforts for novel investigation of terrestrial actinomycetes. Exploring members of actinomycetes from other habitats have been continued to thrive; hence, researchers switched over to new environments in search of the novel antimicrobial structure against human and plant pathogen.
4
Marine environments are extreme habitats, where actinomycetes populations are mostly unexplored for the discovery of novel bioactive secondary metabolites. Diverse actinomycetes have been isolated from the marine environment. Phylogenetic diversity of marine actinomycetes was reported from an Indian coastal solar saltern.
6
Bacterial strains isolated from marine habitats were thought to be a function of terrestrial contaminants. Recent research data unveiled that several actinomycetes taxa are natural or well-adopted inhabitants of the marine environment.
Marine environment has been shown to be a crucial source for the discovery of novel antibiotics producing microorganisms.
5
Marine derived actinomycetes shown great potential in the synthesis of diverse novel bioactive secondary metabolites such as antibiotics and other chemotherapeutic agents with unusual structures and properties.
7
Kocuria species is one of the poorly studied microorganisms in terms of its industrial applications. The genus Kocuria was named after Slovakian microbiologist Miroslav Kocur, to accommodate phylogenetically distinct actinobacteria. Kocuria species belongs to the family Micrococcaceae, suborder Micrococcineae, order Actinomycetales and class Actinobacteria.
8
In this study, isolation, separation and purification of an antibacterial compound from a novel halophilic actinomycetes Kocuria sp. strain rsk4 has been reported.
Materials and Methods
Sample collection
The halophilic actinomycetes strain rsk4 was isolated from a marine water sample collected from Porbandar region (21°37'48"N 69°36'0"E) at Gujarat, Western India. The sample was collected using a sterile container and instantly brought to the laboratory for further processing. The physicochemical parameters such as pH, temperature, and salinity were measured during sample collection.
Isolation of halophilic actinomycetes
Halophilic actinomycetes were isolated from marine water sample by serial dilution method in three different media: Actinomycetes Isolation Agar (Sodium caseinate, 2.0 g/L; Asparagine, 0.1 g/L; Sodium propionate, 4.0 g/L; Dipotassium phosphate, 0.5 g/L; Magnesium sulphate, 0.1 g/L; Ferrous sulphate, 0.001 g/L; Agar 20.0 g/L; pH 7.0), Starch casein agar (starch, 10 g/L; casein, 10 g/L; peptone, 5 g/L; yeast extract, 5 g/L; agar, 30 g/L; pH 7.2) and Seghal and Gibbons agar (Yeast extract, 10 g/L; Casamino acids, 7.5 g/L; Na citrate, 3 g/L; MgSO4·7 H2O, 2 g/L; KCl, 2 g/L; FeCl2, 0.023 g/L; agar, 30 g/L; pH 7.2) supplemented with 5%-15% NaCl for salinity. To minimize the bacterial and fungal growth, each media was supplemented with Nalidixic acid (20 µg/mL) and Nystatin (25 µg/mL) respectively and incubated at 30˚C for 2-4 days. Morphologically distinct colonies were picked and further re-streaked to ensure the purity of colony onto Starch casein agar with respective salinities. All isolates were preserved at 4˚C on slants and as glycerol stocks at -20˚C.
Antibacterial potential of strain rsk4
Strain was grown for 8 days and cell free supernatant was check for its antibacterial activity using agar well diffusion technique.
9
Samples were withdrawn at 24 hour intervals periods and cell biomass were separated by filtration. Dry weight of bacterial growth was measured as per ml of the sample. Bacterial pathogens were obtained from Microbial type culture collection (MTCC) and Gene Bank of the Institute of Microbial Technology (IMTECH), Chandigarh, India. The bacterial pathogens used were: Staphylococcus aureus (Clinical isolates), Enterococcus faecalis MTCC 439, Bacillus megaterium MTCC 2444, Bacillus subtilis MTCC 441, Mycobacterium smegmatis MTCC 6, Escherichia coli (Clinical isolates), Klebsiella pneumoniae MTCC 109, Salmonella typhimurium MTCC 1251, Proteus vulgaris MTCC 1771 and Pseudomonas aeruginosa MTCC 2453. Bacterial pathogens were poured into Mueller Hinton agar (MHA) (Himedia), 6 mm size well has been prepared and 30 µl cell free supernatant was added into the well. 30 µL starch casein medium was added into one of the well as a negative control. Zones of inhibition were measured after 24 hours incubation at 37˚C using Hi-Antibiotic ZoneScale (HiMedia). Antibacterial assays were repeated in three replicates to confirm the activity and consistent production of antibacterial metabolites. Staphylococcus aureus was pathogenic antibiotic resistant bacteria and it was found to be comparatively more sensitive to the antibacterial agent; therefore, further studies was carried out with it.
Taxonomic and biochemical characterization
Colony characteristics of strain rsk4 were studied on International Streptomyces Project (ISP) media yeast extract-malt extract agar (ISP-2), inorganic salt-starch agar (ISP-4), glycerol-asparagine agar (ISP-5), tyrosine agar (ISP-7), nutrient agar, tryptone soya agar and starch casein agar. Physiological characteristics were studied at various salt (0-15%, w/v) and pH (5-10) concentration in starch casein agar. Biochemical characteristics including MR, VP, nitrate reduction, indole production, citrate utilization, urease, catalase, oxidase test, H2S production, melanin pigment production carbon utilization, nitrogen utilization, extracellular enzyme production and antibiotic resistance tests were carried for identification of organism up to genus level.
16S rDNA regions-based characterization
The strain was inoculated into starch casein medium and cultivated at 30˚C for 3 days. The culture was harvested by centrifugation and the resulting precipitates were suspended in solution (glucose, 50 mM pH 8.0; Tris-HCl, 25 mM, pH 8.0; EDTA 10 mM). Lysis buffer (200 mL of 5% lysozyme, 20 mL of 1% pronase and 4 mL of 1% RNase) was used to lysed the cells. Genomic DNA was extracted with 250 mL of extraction buffer (Tris-HCl, 20 mM; EDTA, 100 mM; sodium dodecyl sulfate, 1%; proteinase K, 0.01%). The DNA solution was extracted 2 times with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1). DNA was precipitated with two-fold volume ethanol, washed with 70% ethanol and finally resuspended in 20 μL TE buffer (Tris-HCl, 10 mM; EDTA, 1 mM; pH 8.0). DNA was was quantified by spectrophotometer and preserved at -20°C. The GC content of the DNA was determined by the thermal denaturation method.
10
The 16S rDNA sequencing was carried out in thermal reactor thermocycler (GeneAmp® PCR System 9700, Applied Biosystems) using the universal bacterial primers Fd1 5′-TTCCGGTTGATCCTGCC-3′ (forward); and Rd1, 5′-AAGGAGGTGATCCAGCC-3′ (reverse). PCR condition was: 94°C for 3 minutes in the first cycle; denaturing at 94°C for 30 seconds, 30 second annealing at 55°C, 2 minutes elongation at 72°C and a final extension step at 72°C for 10 minutes. The amplified product was purified by 1.2% agarose gel using the GeNeiTM Quick PCR Purification Kit.
DNA sequencing was carried out according to the dideoxy chain termination method by M/s Macrogen Inc., Seoul, Korea. The sequence was screened using BioEdit software. 16S rDNA Sequence was subjected to BLAST search to obtain similarity between sequences.11 The sequences were aligned using the ClustalW program and submitted to the GenBank database. The 16S rDNA sequence based phylogenetic tree was constructed using MEGA6 software by distance based Neighbour-joining method with bootstrap testing.
12
Effect of media, NaCl, pH and incubation period on growth and antibacterial agent
The effect of several media on antibacterial activity of rsk4 was evaluated against S. aureus by agar well diffusion method. The media used were: glucose yeast extract malt extract broth (ISP2), inorganic salts starch broth (ISP4), glycerol asparagines broth (ISP5), starch casein medium (SCM), glucose yeast extract broth (GYEA) and Bennett medium (BM).
13
The effect of the salt on growth and production of the antibacterial compound was studied on starch casein media at varying salt concentrations (0-15% w/v) at pH 7 for 10 days. Similarly, the effect of pH on growth and antibiotic production was studied in the range of pH 5-10 on starch casein media having 5% w/v NaCl for 10 days. The effect of incubation period on the production of the antibacterial agent was also studied by incubating organism for 1-10 days at temperature 30˚C and pH 7.
Fermentation and extraction of active compounds
The isolated strain was grown in 4 L starch casein media seeded with 5% inoculums of active culture and maintained 180 rpm at 30°C for 10 days. After incubation, fermentation broth was filtered through Whatman No.1 filter paper. Filtrate was further centrifuged at 10 000 rpm (10 280 g) at 4°C for 15 minutes. The extraction process was scaled up for extraction of the antibacterial compounds with organic solvents on the basis of the partition coefficient. The cell-free supernatant was adjusted at different pH and extracted thrice with equal volume of culture filtrate and ethyl acetate. The ethyl acetate extract was dried using vacuum rotary evaporator and recuperated in methanol. Extract was bioassayed for the antibacterial activity against S. aureus as an indicator microorganism.
TLC guided bioautography for separation of the antibacterial metabolites
The residual crude active extract was separated by ascending analytical thin layer chromatography (TLC) in a running solvent system. The solvents tested for separation of the antibacterial metabolites included; solvent system 1 (Butanol-Acetic acid-Water, 30:20:70 v/v), solvent system 2 (Chloroform-Methanol, 24:1 v/v), solvent system 3 (Ethyl acetate-Methanol, 6:4 v/v) and solvent system 4 (Hexane-Ethyl acetate, 1:9 v/v). TLC was carried out on commercially available aluminum TLC sheets (20 × 20 cm with 0.2 mm thickness, silica gel GF254, Merck, Darmstadt, Germany). The solvent fronts were marked, Rf values have been measured and the TLC sheet was aseptically processed for bioautography. Agar overlay bioautographic evaluation was conducted to localize the separated antibacterial compound on TLC chromatogram.
14
Molten nutrient agar containing actively grown S. aureus was aseptically sprayed over different developed TLC chromatogram. Sheets were incubated at 37˚C in 100% relative humidity for 24 hours. Clear inhibition zone around the separated active components was recorded.
Purification of the antibacterial metabolites
The enormous quantity of antibacterial compounds has been separated on preparative TLC plate with 1 mm thick silica gel 60 (Merck Ltd.). TLC bands that exhibited antibacterial activity on analytical TLC were scratched off carefully. It was dissolved in methanol and centrifuged at 10 000 rpm for 5 minutes to eliminate silica. Then, supernatant was filtered using 0.22 µm filter and bioassayed against test organism S. aureus for confirmation. This process was repeated multiple times; all the supernatants were merged and concentrated to 30 mg/mL using a rotary evaporator. Antibacterial metabolites were further purified by an efficient method of flash chromatography. Light yellow colored TLC extract (30 mg/mL) was mixed with 1 g Silica gel 60. Methanol was evaporated at 37˚C temperature. Silica gel mixture was packed in the column (Silica Gel 30 µm) and flash chromatography (Yamazen) was performed.
15
Components were separated by isocratic elution with Hexane/ethyl acetate. Flow rate was maintained at 3 ml/min. Forty tubes with 7 mL fraction volume were collected. Each fraction was analyzed for the presence of active components by analytical TLC. After visualization with ultraviolet light, components having the same retention factor (Rf) were pooled, concentrated and bioassayed for confirmation of antibacterial compound. The antibacterial compound was finally purified by analytical high-performance liquid chromatography using a reverse phase C18 column (Sunfire, 4.6 mm × 100 mm, 3.5 µm) at a constant temperature of 40°C.
16
Elution was carried out at flow rate of 0.55 mL/min and the eluate was detected using a UV detector. Fractions were collected using fraction collector. It was concentrated (20 µg/mL) and 30 µL extract were bioassayed. Penicillin (20 µg/mL) was also used to confirm the antibiotic resistance capacity of S. aureus. Streptomycin (20 µg/mL) was considered as a standard antibiotic for comparisons of potency against S. aureus. HPLC was repeated numerous times to get adequate amount of antibacterial compound for further analysis. According to the found antibacterial activity, the active fractions were merged and further characterized.
Characterization of purified compound
The solubility, thermal stability and pH stability test of antibacterial compound was carried out. The UV absorption spectrum of the antibacterial compound was recorded in methanol solution ranging 190 nm to 600 nm by Shimadzu UV 1800 spectrophotometer.
17
The Electro spray ionization mass spectra (ESI-MS) of active compound of strain rsk4 was recorded with a liquid chromatography-ion trap mass spectrometer (Shimadzu LCMS-8030 Triple Quadrupole Liquid Chromatograph Mass Spectrometer) at negative ion detection mode.
18
Minimum inhibitory concentrations
Minimum inhibitory concentration (MIC) of pure antibacterial metabolite was accomplished using conventional agar dilution method against 10 test microorganisms cited above.
19
Test microorganisms were inoculated onto the Mueller-Hinton medium containing different concentrations of the active metabolite (10-100 µg/mL). After an incubation period of 24 hours at 37˚C, the plates were examined for growth. The lowest antibiotic concentration that inhibited the growth of one or more organism was noted. The Mueller-Hinton media without test pathogenic microorganisms and active compounds was used as a control. MIC of broad-spectrum antibiotics streptomycin was also carried out as a standard for comparison.
Results
Isolation of halophilic actinomycetes
Halophilic actinomycetes strain rsk4 was isolated from Porbandar region, Western India. It was gram-positive, coccoid shaped, non-encapsulated, reddish orange colored, aerobic, catalase-positive bacteria which do not produce endospore.
Antibacterial potential of strain rsk4
Culture filtrate of strain rsk4 exhibited a broad spectrum antibacterial activity against both Gram-positive and Gram-negative bacteria. Among the all test pathogenic organisms, Staphylococcus aureus was fund to be most sensitive to the antibacterial agent.
Taxonomic characterization of the organism
The results of physiological characteristics illustrate that rsk4 was halophile and able to grow up to 15% w/v NaCl. Optimum NaCl tolerance was in the range of 5%-10% and optimum requirement of pH was found to be ⁓7. Biochemical characters suggest that it utilizes arabinose, galactose, rhamnose, glucose, lactose and sucrose as sources of carbon along with acid production. Nitrate reduction and alkaline phosphatase tests showed positive results, whereas indole production, methyl red, voges proskauer, coagulase, oxidase and urease showed negative results. It was positive for starch, casein and gelatin hydrolysis but negative for β-galactoside and lipid hydrolysis. Strain rsk4 could also be distinguished from other strain by showing resistance to ampicillin, gentamicin, ofloxacin, penicillin-G and vancomycin (Table 1).
Table 1.
Physiological and biochemical characteristics of strain rsk4
|
S. No.
|
Characteristic
|
Strain rsk4
|
| 1 |
Gram reaction |
+ |
| 2 |
Sporulation |
- |
| 3 |
Melanin pigmentation |
+ |
| 4 |
Anaerobic growth |
- |
| 5 |
Indole production |
+ |
| 6 |
Methyl red |
- |
| 7 |
Voges Proskauer |
- |
| 8 |
Nitrate reduction |
+ |
| 9 |
H2S production
|
- |
| 10 |
Oxidase |
- |
| 11 |
Urease |
- |
| 12 |
β-Galactosidase |
- |
| 13 |
Catalase |
+ |
| 14 |
Coagulase |
- |
| 15 |
Starch hydrolysis |
+ |
| 16 |
Gelatin hydrolysis |
+ |
| 17 |
Lipid hydrolysis |
- |
| 18 |
Casein hydrolysis |
+ |
|
|
Carbon source utilization
|
|
| 19 |
Arabinose |
+ |
| 20 |
Galactose |
+ |
| 21 |
Rhamnose |
+ |
| 22 |
Glucose |
+ |
| 23 |
lactose |
+ |
| 24 |
Sucrose |
+ |
| 25 |
Fructose |
- |
|
|
Amino acid utilization
|
|
| 26 |
Arginine |
- |
| 27 |
Lysine |
- |
| 28 |
Histidine |
- |
| 29 |
Proline |
+ |
“+” positive, “-” negative
The 16S rDNA sequence (1361 bp) of the strain rsk4 was submitted to GenBank under the accession number HQ258887. The sequence was analyzed by BLAST search. It was further aligned with reference species existing in GenBank database for confirmation and identification of the strain rsk4 at genus level. Fig. 1 shows phylogenetic tree generated on the basis of 16S rDNA sequence of rsk4 and nucleotide sequences of closely related strains using distance based neighbor-joining method with bootstrap value.
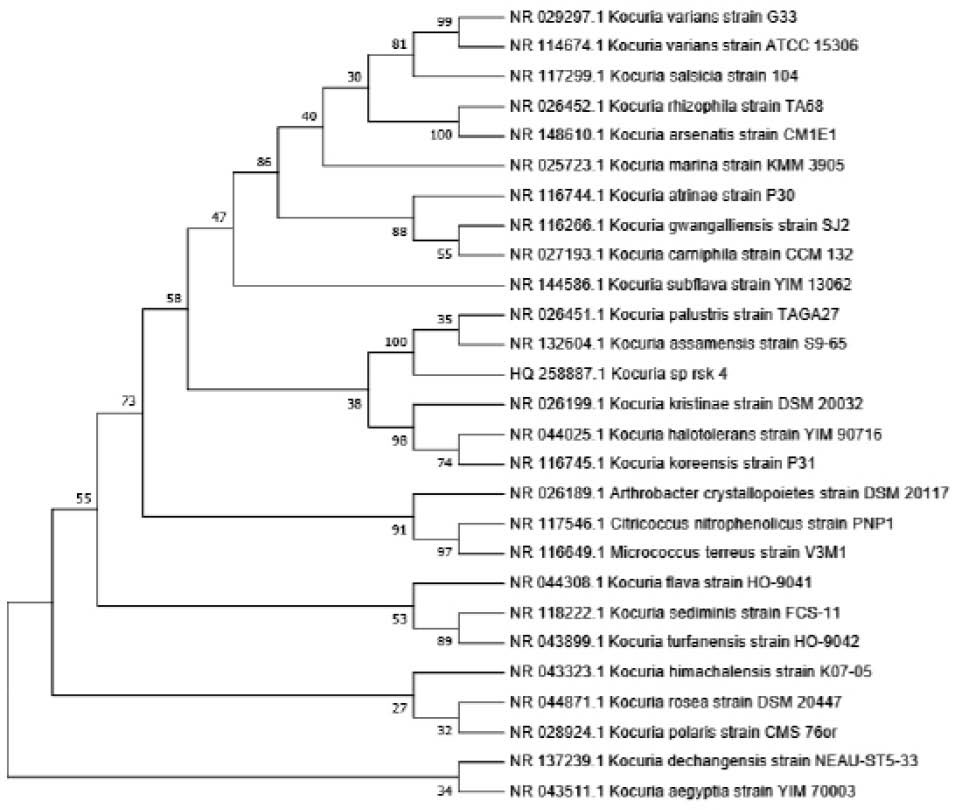
Fig. 1.
The phylogenetic tree of 16S rDNA gene sequences of the strain rsk4.
.
The phylogenetic tree of 16S rDNA gene sequences of the strain rsk4.
Effect of media, NaCl, pH and incubation period on growth and antibiotic production
Among several media studied, starch casein medium was found to be most appropriate growth supportive as well as the synthesis of antibacterial agents by rsk4 (Fig. 2). Strain rsk4 grew at the range of 5-10% (w/v) NaCl and secreted antibiotic optimally at 5% NaCl (Fig. 3). Fig. 4 shows that rsk4 secreted the antibacterial metabolites in the wide range of pH 5-9, but optimum antibacterial activity observed at neutral pH. Strain rsk4 secreted the antibiotic in SCM after 5 days but produces optimally on the seventh day at pH ⁓7 and temperature 30˚C (Fig. 5).
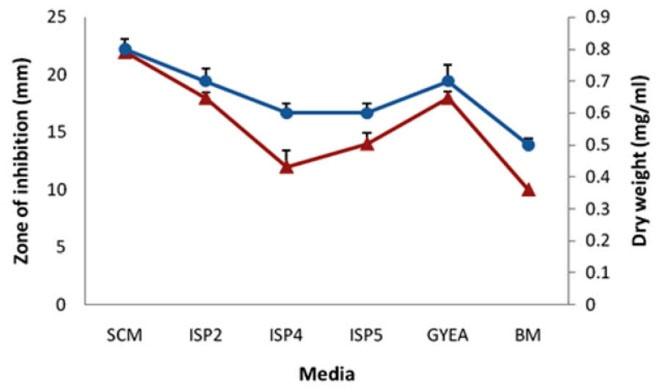
Fig. 2.
Effect of different media on the growth (circles) and the antibiotic production (triangles).
.
Effect of different media on the growth (circles) and the antibiotic production (triangles).
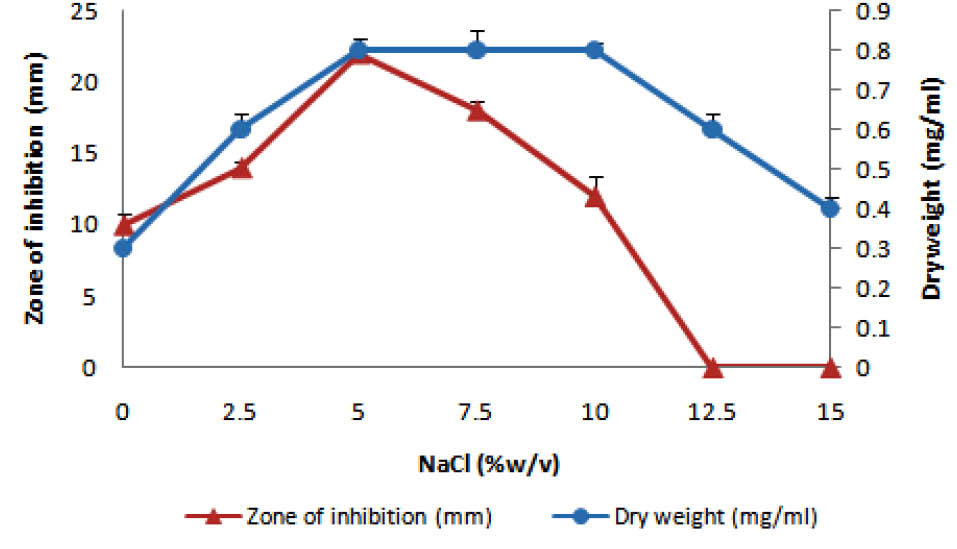
Fig. 3.
Effect of NaCl on the growth (circles) and the antibiotic production (triangles).
.
Effect of NaCl on the growth (circles) and the antibiotic production (triangles).
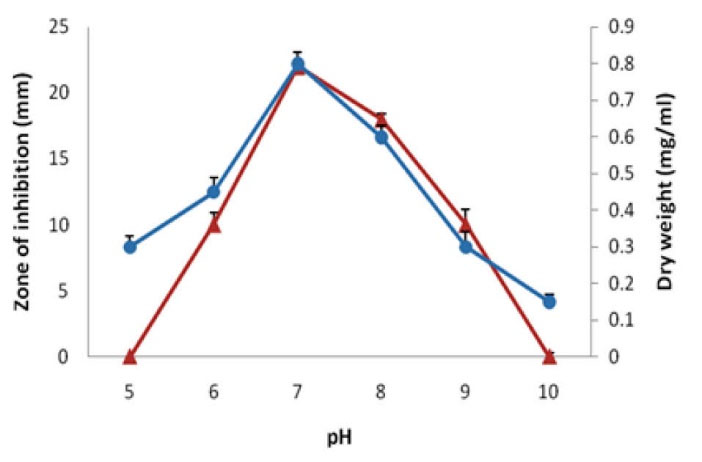
Fig. 4.
Effect of pH on the growth (circles) and the antibiotic production (triangles)
.
Effect of pH on the growth (circles) and the antibiotic production (triangles)
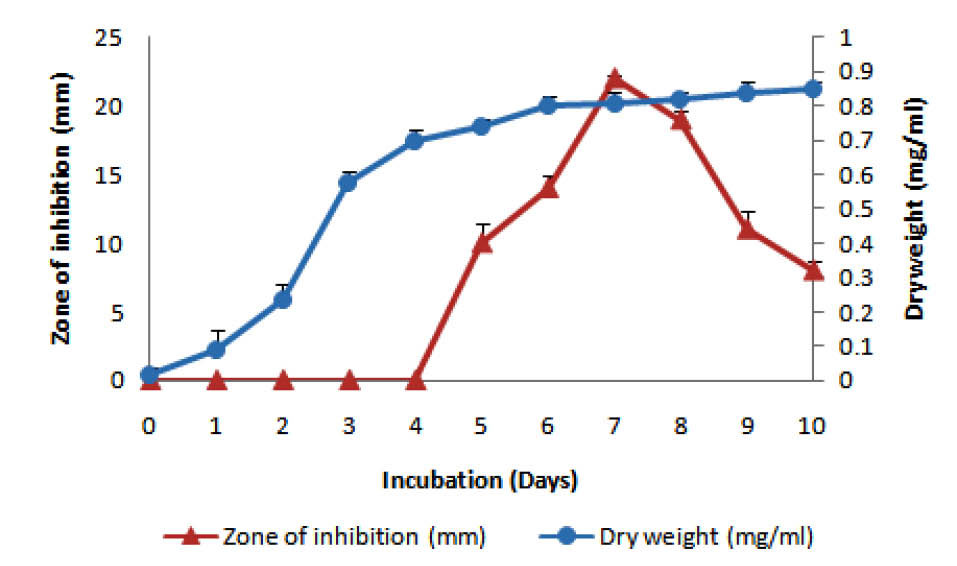
Fig. 5.
Effect of incubation period on growth (circles) and antibiotic production (triangles).
.
Effect of incubation period on growth (circles) and antibiotic production (triangles).
Fermentation and extraction of active compounds
The cell-free supernatant was concentrated and antibacterial metabolite was extracted thrice by n-hexane, benzene, chloroform, dichloromethane, ethyl acetate and butanol. Ethyl acetate was found to be an appropriate solvent for extraction. No antibacterial activity was observed with ethyl acetate as a negative control. The organic extract was concentrated to get yellow colored crude extract which was re-dissolved in methanol for bioassay.
Separation and TLC bioautography of the antibacterial metabolites
The results of the bioautography suggest that Rf values were different for diverse solvents but clear separated bands with a sharp zone of inhibition were observed in solvent system chloroform-methanol, 24:1 v/v. A single sharp zone against S. aureus at Rf value 0.45 was marked in bioautography indicated the presence of a separated antibacterial compound (Fig. 6).
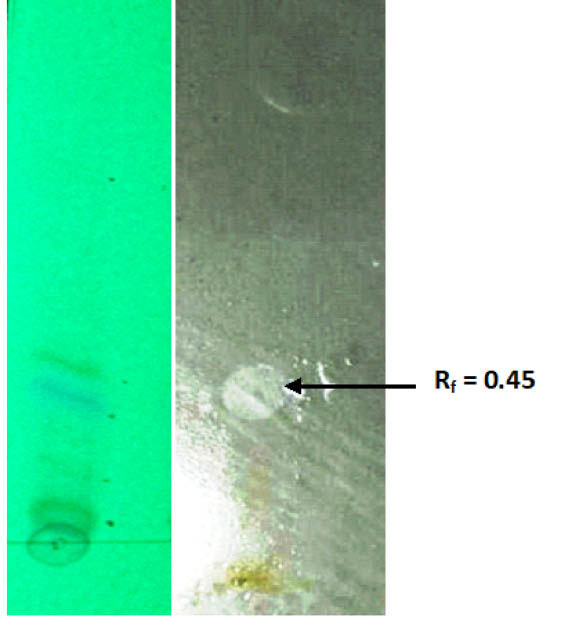
Fig. 6.
TLC-based bioautography exhibit antibacterial activity at the Rf value of 0.45.
.
TLC-based bioautography exhibit antibacterial activity at the Rf value of 0.45.
Purification and characterization of the antibacterial agent
The active band of preparative TLC was reconfirmed by bioassay. The preparative TLC extract was further separated by flash chromatography with isocratic elution using Hexane/ethyl acetate gradient (100% Hexane /0% ethyl acetate to 0% Hexane /100% ethyl acetate) solvent system. Bioassay of fractions confirmed the presence of antibacterial components in the fraction 21 to 23. Antibiotic purification was finally carried out by HPLC using 0.01% glacial acetic Acid (pH 3.7)-acetonitrile; 10:90, v/v. Active component was detected by UV detector at 230 nm. Chromatogram of the separated antibacterial compound obtained two peaks; one merged and one sharp isolated peak (Fig. 7). Bioassay of each peak fraction revealed that only fraction 2 (retention time: 3.24 minutes) possess potent antibacterial activity against S. aureus (Fig. 8). The characterization of antibacterial compound suggests that it was readily soluble in a semi-polar solvent mainly methanol, ethyl acetate and dichloromethane. It was stable at pH 6.8-7.2 but degraded more rapidly below pH 6. It was stable between temperature ranges of 20°C-40ºC and degraded rapidly at more than 55°C. The spectral analysis of the antibacterial compound produced by strain rsk4 revealed the ultraviolet (UV) absorption maxima recorded at 233 nm (Fig. 9). The obtained ESI-MS spectra showed molecular ion peaks at m/z 472.4 [M-H] which leads to a molecular weight of 473 g/mol (Fig. 10).
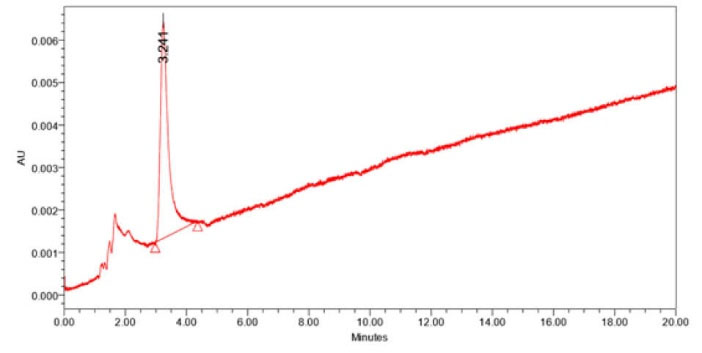
Fig. 7.
Chromatogram of the separated antibacterial compound by HPLC (retention time = 3.24 min).
.
Chromatogram of the separated antibacterial compound by HPLC (retention time = 3.24 min).
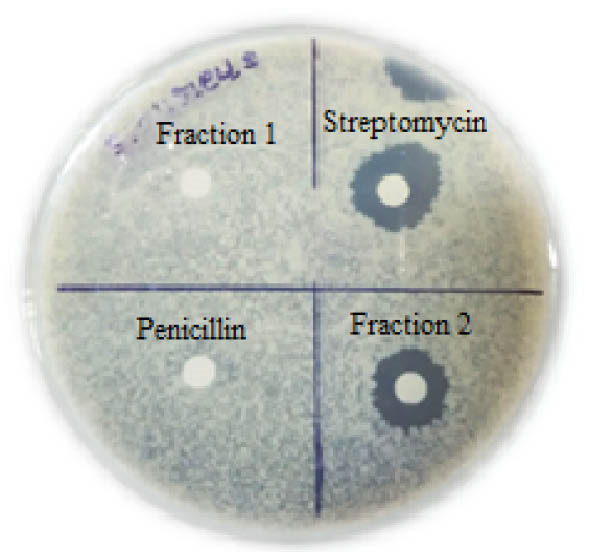
Fig. 8.
Bioassay of the active fraction of HPLC.
.
Bioassay of the active fraction of HPLC.
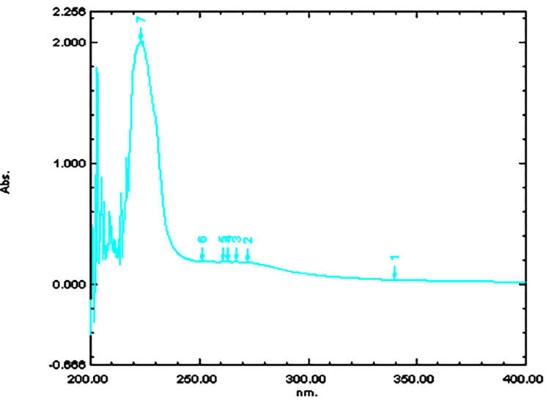
Fig. 9.
Absorption maxima at 233 nm.
.
Absorption maxima at 233 nm.
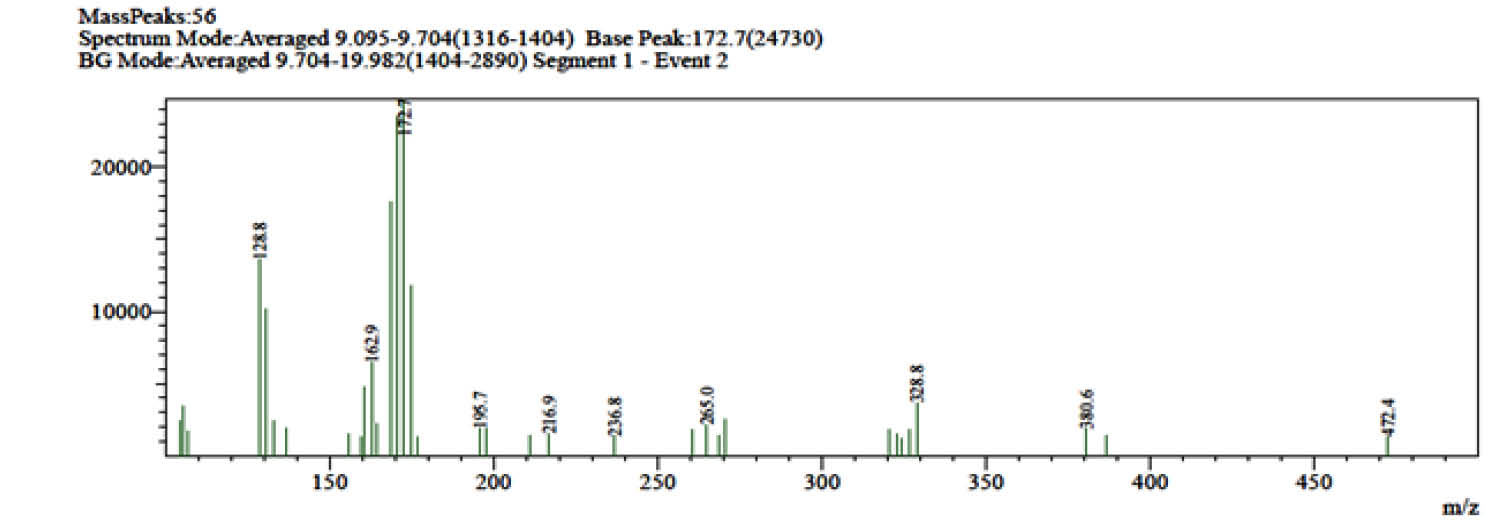
Fig. 10
.
Mass spectra of the purified compound from Kocuria sp. strain rsk4.
.
Mass spectra of the purified compound from Kocuria sp. strain rsk4.
Minimum inhibitory concentrations
MICs of broad-spectrum antimicrobial metabolite are summarized in Table 2. The MIC values of antibacterial compound was comprised between 30-60 µg/mL. The lowest MIC was recorded against S. aureus (30 µg/mL) and highest against Klebsiella pneumoniae (60 µg/mL).
Table 2.
Minimum inhibitory concentrations (MICs) of the antibacterial compounds produced by the strain rsk4 against several target microorganisms
|
Target microorganisms
|
MIC (µg/mL)
|
|
Rsk4
|
Streptomycin
|
|
Staphylococcus aureus MTCC 96
|
30 |
20 |
|
Mycobacterium smegmatis MTCC 6
|
50 |
20 |
|
Bacillus subtilis MTCC 441
|
30 |
20 |
|
Bacillus megaterium MTCC 2444
|
40 |
20 |
|
Enterococcus faecalis MTCC 439
|
40 |
20 |
|
Klebsiella pneumoniae MTCC 109
|
60 |
10 |
|
Proteus vulgaris MTCC 1771
|
50 |
10 |
|
Salmonella typhimurium MTCC 1251
|
40 |
10 |
|
Pseudomonas aeruginosa MTCC 2453
|
60 |
10 |
|
Escherichia coli MTCC 739
|
40 |
10 |
Discussion
Actinomycetes are always recognized as potential microorganism for the production of antibiotics. Discovery of novel antibiotics from unexplored extremophilic bacteria is still a thrust of many researchers. The higher possibilities of production of antimicrobial metabolites by bacteria from the halophilic environment may be associated with the defensive or aggressive role of the organism for maintaining their ecological niche. The thriving of organisms in such extreme and competitive environment assumes that their metabolic compatibility is strongly influenced by natural selection. Recent discoveries suggested that halophilic actinomycetes habitats are rich with bioactive antibiotics.
20
With this perspective; halophilic actinomycetes rsk4 was isolated from the marine water of Porbandar region, Western India. This coastal area is still untouched for the exploration of antibiotic-producing microorganism. Actinomycetes strain rsk4 isolated from marine water exhibited good growth in starch casein agar. Strain rsk4 exhibited antibacterial activity against both Gram-positive and Gram-negative bacteria including multiple antibiotic resistant S. aureus. S. aureus is one of the major human pathogens and multiple drug resistance Gram-positive bacteria which can cause various infections like dermal, pulmonary and blood infections. These infections are common on pediatric patients and healing options are also limited.
21
The result is being consistent with the report of Manimaran et al.
22
Strain rsk4 was characterized for their taxonomic classification. The morphological and biochemical properties were matched with Bergey’s manual of determinative bacteriology.
23
The 16S rRNA gene sequence of the rsk4 had only 94% homology with other Kocuria species which suggest that rsk4 is novel species. On the basis of physiological, morphological, biochemical and 16S rDNA sequence based character, rsk4 was designated as Kocuria sp. strain rsk4. The DNA G+C content is derived as 55.89% but other known Kocuria sp. are ranged from 60.0 to 75.3 mol% which illustrate the uniqueness of our species.
The study on antimicrobial production requires a search of a suitable culture medium. The antibacterial activity of rsk4 was found to be optimum on “starch casein medium”. The differential influence of antibacterial activity may be due to specific nitrogen source present in “starch casein medium”. The result shows similarity with the previous reports of optimum antibiotic production using organic nitrogen sources for better yield.
24
Effect of salt on antibiotic production reveals that it shows maximum antibacterial activity at higher salt concentrations (10% NaCl). A similar result was reported by Kekar which secret antibacterial metabolites from halophilic Kocuria sp. SBSK-120 against S. aureus at high salt concentrations.
25
Neutral pH was found to be most suitable for optimum product formation. This result is also comparable with halophilic Actinobacter Q70, which produces broad-spectrum antibiotic at pH 7.
26
Study on the growth curve and product formation confirms that antibacterial compound was produced during stationary phase; therefore, the antibacterial compound is the secondary metabolite. The nature of antibacterial compound produced by strain rsk4 must be considered as antibiotics. The bulk production of antibiotic was proceeded by submerging the culture. Presence of antibiotic in broth was confirmed by bioassay. It was stated that most of the antibacterial secondary metabolites were secreted in the extracellular medium by actinomycetes.
27
Ethyl acetate was found to be most suitable for solvent extraction. Ethyl acetate was previously reported as an extraction solvent for crude extracts of actinomycetes.
28
Antibacterial bioassay of the crude extract against S. aureus confirms that metabolites are not degraded during solvent extraction.
The most appropriate solvent system for TLC separation was found to be chloroform-methanol, 24:1 v/v. The result is being consistent with Ravi and Atta who separated antibiotics by TLC using the similar solvent system.
29,30
Localization of antibiotic on developed TLC chromatogram was confirmed by bioautography. Recently, TLC guided bioautography was performed by Tomas for visualization of an active band of antibacterial molecule 13-deoxytetrodecamycin against S. aureus.
31
Preparative TLC was performed using the same solvent system to get an enormous quantity of antibacterial compound. Flash chromatography was further used to separate bulk quantity in a single run. This technique is less familiar for antibiotic separation but effective technique for purification of compound. Presence of active antibiotic compound in flash chromatographic fractions was confirmed by bioassay. The bioassay-guided fractionation using flash chromatography was recently used by Pan for the purification of spirotetronate antibiotics; Lobophorins.
32
Antibiotics was finally purified by HPLC.Optimization of mobile phase for HPLC separation shows that 0.01% glacial acetic acid (pH 3.7)-acetonitrile; 10:90, v/v was suitable for the purification of compound of interest. Acetonitrile - acetic acid mobile phase was also used by several researchers for separation of ciprofloxacin and sulfonamide antibiotics.
33
Analytical HPLC was repeated several times to get enormous fractions. The active fraction was pulled and further characterized. The antibiotic substance was readily soluble in the semi-polar solvent, which suggests its nature as the semi-polar compound. The molecular weight of antibacterial compound (473 g/mol) does not match with any known antibacterial antibiotics which advocate the uniqueness of this molecule. MIC value was obtained by the agar dilution method and compared with standard antibiotic streptomycin. Similar method of MIC was done by Aouiche against several test microorganisms using broad-spectrum antibiotics saquayamycins.
34
The MIC of the antibacterial compound (30 µg/mL) is comparable with standard antibiotics streptomycin (20 µg/mL). The present report is the first attempt to purify antibacterial metabolites from Kocuria sp. Antibiotics isolated from strain rsk4 could be an important therapeutic alternative, especially against antibiotic-resistant pathogen S. aureus.
Conclusion
Screening of antibacterial metabolites from taxonomically unique populations of extremophilic actinomycetes has been considered to a promising option. Actinomycetes from saline environments have potent biotechnological applications, including production of antibacterial agents. In this study rsk4 isolated from marine water has been identified to be Kocuria sp. on the basis of physiological, biochemical and 16S rDNA sequence analysis. Strain rsk4 shows broad-spectrum activity against bacteria including antibiotic-resistant bacterial pathogen S. aureus. Purification followed by characterization of antibiotic reveals that it is unique from other known antibiotics. Further structural elucidation and investigations of this compound may solve the problem in treating infections caused by antibiotic-resistant bacteria.
Acknowledgments
The authors are thankful to the management of Shree M. & N. Virani Science College, Rajkot (India) for providing necessary research facilities.
Funding sources
This work was supported by University Grant Commission, Pune, India (grant No. 47-566/13).
Ethical statement
The authors declare no ethical issues.
Competing interests
The authors declared no competing interests.
Authors contribution
RRK: conceptualization, data handling, experiments design, data analysis, study validation, data presentation, draft preparation, draft writing, draftreviewing and project administration. VJJ: supervision, study consultation, provision of study materials and equipment.
Supplementary materials
Supplementary data contains Figs. S1-S3 and tables S1-S3.
(pdf)
Research Highlights
What is the current knowledge?
simple
-
√ Staphylococcus aureus is one of the antibiotic resistant
pathogen, causes community acquired infections.
-
√ Marine actinomycetes are relatively unexplored.
-
√ Marine bacteria are considered as potential hub of novel
bioactive metabolites.
What is new here?
simple
-
√ Halophilic Kocuria sp. is able produces antibiotics against
S. aureus.
-
√ Purified antibacterial compound is unique.
References
- Jones RJ. Key considerations in the treatment of complicated staphylococcal infections. Clin Microbiol Infect 2008; 14:3-9. doi: 10.1111/j.1469-0691.2008.01923.x [Crossref] [ Google Scholar]
- Mahajan GB, Balachandran L. Antibacterial agents from Actinomycetes - a review. Front Biosci (Elite Ed) 2012; 4:240-253. doi: 10.2741/373 [Crossref] [ Google Scholar]
- Takahashi Y, Omura S. Isolation of new actinomycete strains for the screening of new bioactive compounds. J Gen Appl Microbiol 2003; 49:141-54. doi: 10.2323/jgam.49.141 [Crossref] [ Google Scholar]
- Ramesh S, William A. Marine actinomycetes: An ongoing source of novel bioactive metabolites. Microbiol Res 2012; 167:571-80. doi: 10.1016/j.micres.2012.06.005 [Crossref] [ Google Scholar]
- Jose PA, Jebakumar SR. Phylogenetic diversity of actinomycetes cultured from coastal multipond solar saltern in Tuticorin, India. Aquat Biosyst 2012; 8:23. doi: 10.1186/2046-9063-8-23 [Crossref] [ Google Scholar]
- Bull AT, Ward AC, Goodfellow M. Search and discovery strategies for biotechnology: the paradigm shift. Microbiol Mol Biol Rev 2000; 64:573-606. doi: 10.1128/MMBR.64.3.573-606.2000 [Crossref] [ Google Scholar]
- Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ Microbiol 2005; 7:1039-48. doi: 10.1111/j.1462-2920.2005.00785.x [Crossref] [ Google Scholar]
- Stackebrandt E, Koch C, Gvozdiak O, Schumann P. Taxonomic dissection of the genus Micrococcus: Kocuria gen nov, Nesterenkonia gen nov, Kytococcus gen nov, Dermacoccus gen nov, and Micrococcus Cohn 1872 gen emend. Int J Syst Bacteriol 1995; 45:682-692. doi: 10.1099/00207713-45-4-682 [Crossref] [ Google Scholar]
- Irshad S, Mahmood M, Perveen F. In vitro antibacterial activities of three medicinal plants using agar well diffusion method. Research J Biol 2012; 2:1-8. doi: 10.4172/1948-5948 [Crossref] [ Google Scholar]
- Marmur J, Rownd R, Falkow S, Baron LS, Schildkraut CL, Doty P. The nature of intergeneric episomal infection. Proc Natl Acad Sci USA 1961; 47:972-979. doi: 10.1073/pnas.47.7.972 [Crossref] [ Google Scholar]
- Altschul SF, Thomas LM, Alejandro AS, Zhang J, Zhang Z, Miller W. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25:3389-3402. doi: 10.1093/nar/25.17.3389 [Crossref] [ Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985; 39:783-789. doi: 10.1111/j.1558-5646.1985.tb00420.x [Crossref] [ Google Scholar]
- Jones KL. Fresh isolates of actinomycetes in which the presence of sporogenous aerial mycelium is a fluctuating characteristic. J Bacteriol 1949; 57:141-145. [ Google Scholar]
- Sasidharan S, Chen Y, Saravanan D, Sundram KM, Latha LY. Extraction, isolation and characterization of bioactive compounds from plants’ extracts. Afr J Tradit Complement Altern Med 2011; 8:1-10. doi: 10.4314/ajtcam.v8i1.60483 [Crossref] [ Google Scholar]
- Freyder CP, Zhou W, Doetsch PW, Marzilli LG. Bleomycin A2 and B2 purification by flash chromatography for chemical and biochemical studies. Prep Biochem Biotechnol 1991; 21:257-68. doi: 10.1080/10826069108018577 [Crossref] [ Google Scholar]
- Khurram M, Lawton LA, Edward C, Iriti M, Hamee A, Khan MA. Rapid Bioassay-Guided Isolation of Antibacterial Clerodane Type Diterpenoid from Dodonaea viscosa (L) Jaeq. Int J Mol Sci 2015; 16:20290-307. doi: 10.3390/ijms160920290 [Crossref] [ Google Scholar]
- Maleki H, Mashinchian O. Characterization of Streptomyces isolates with UV, FTIR spectroscopy and HPLC analyses. Bioimpacts 2011; 1:47-52. doi: 10.5681/bi.2011.007 [Crossref] [ Google Scholar]
- Adrielli T, Diogo AS, Daniel S, Caio MM, Gustavo AM, Michele DA. Determination of phenolic profile by HPLC-ESI-MS/MS and antibacterial activity of Eugenia platysma against mollicutes strains. J App Pharm Sci 2017; 7:7-11. doi: 10.7324/JAPS.2017.70502 [Crossref] [ Google Scholar]
- Oki T, Tenmyo O, Tomatsu K, Kamei H. Pradimicins A, B and C: new antifungal antibiotics II In vitro and in vivo biological activities. J Antibiot 1990; 43:763-70. doi: 10.7164/antibiotics.43.763 [Crossref] [ Google Scholar]
- Li F, Maskey RP, Qin S, Sattler I, Fiebig HH, Maier A. Chinikomycins A and B; isolation, structure elucidation, and biological activity of novel antibiotics from a marine Streptomyces sp isolate M045. J Nat Prod 2005; 68:349-353. doi: 10.1021/np030518r [Crossref] [ Google Scholar]
- Pourmand MR, Memariani M, Hoseini M, Yazdchi BS. High prevalence of SEA gene among clinical isolates of Staphylococcus aureus in Tehran. Acta Med Iran 2009; 47:357-361. doi: 10.1016/S0740-0020(02)00180-6 [Crossref] [ Google Scholar]
- Manimaran M, Gopal JV, Kannabiran K. Antibacterial Activity of Streptomyces sp VITMK1 isolated from Mangrove Soil of Pichavaram, Tamil Nadu, India. Proc Natl Acad Sci India Sect B Biol Sci 2015; 87:499-506. doi: 10.1007/s40011-015-0619-5 [Crossref] [ Google Scholar]
-
Buchanan RE, Gibbons NE. Bergey's Manual of Determinative Bacteriology. Baltimore: The Williams and Wilkins Co; 1974.
- Yu J, Liu Q, Liu Q, Liu X, Sun Q, Yan J. Effect of liquid culture requirements on antifungal antibiotic production by Streptomyces rimosus MY02,. Bioresour Technol 2008; 99:2087-2091. doi: 10.1016/j.biortech.2007.03.023 [Crossref] [ Google Scholar]
- Ballav S, Kerkar S, Thomas S, Augustine N. Halophilic and halotolerant actinomycetes from a marine saltern of Goa, India producing anti-bacterial metabolites. J Biosci Bioeng 2015; 119:323-330. doi: 10.1016/j.jbiosc.2014.08.017 [Crossref] [ Google Scholar]
- Hamedi J, Mohammadipanah F, Amoozegar MA, Dehghan S. Isolation of a new moderately halophilic broad-spectrum antibiotic producing actinobacter. Jundishapur J Nat Pharm Prod 2007; 2:94-104. [ Google Scholar]
- Berdy J. Bioactive microbial metabolites: a personal view. J Antibiot 2005; 58:1-26. doi: 10.1038/ja.2005.1 [Crossref] [ Google Scholar]
- Kavitha A, Prabhakar P, Vijayalakshmi M, Venkateswarlu Y. Purification and biological evaluation of the metabolites produced by Streptomyces sp TK-VL_333. Res Microbiol 2010; 161:335-45. doi: 10.1016/j.resmic.2010.03.011 [Crossref] [ Google Scholar]
- Ravi RK, Vasantba J, Bhoomi M, Bonisha T, Bhumika C. Antibacterial Potentials of Actinomycetes Isolated from Gujarat. Int J Pharm Sci Rev Res 2015; 30:78-83. [ Google Scholar]
- Atta HM. Production, purification, physico-chemical characteristics and biological activities of antifungal antibiotic produced by Streptomyces antibioticus AZ-Z710. Am Eur J Sci Res 2010; 5:39-49. [ Google Scholar]
- Tomas G, Michael KH, Sheila P, Geoffrey T, Justin RN. 13-Deoxytetrodecamycin, a new tetronate ring-containing antibiotic that is active against multidrug-resistant Staphylococcus aureus. J Antibiot 2015; 68:698-702. doi: 10.1038/ja.2015.60 [Crossref] [ Google Scholar]
- Pan HQ, Zhang SY, Wang N, Li ZL, Hua HM, Hu JC. New Spirotetronate Antibiotics, Lobophorins H and I, from a South China Sea-Derived Streptomyces sp. 12A35 Mar Drugs 2013; 11:3891-901. doi: 10.3390/md11103891 [Crossref] [ Google Scholar]
- Torres NH, Américo JH, Nazato C, Vilca FZ, Ferreira LF, Maranho LA. Optimization methodology for detection of antimicrobial ciprofloxacin by HPLC FLD. Optimization 2012; 4:59-62. [ Google Scholar]
- Aouiche A, Bijani C, Zitouni A, Mathieu F, Sabaou N. Antimicrobial activity of saquayamycins produced by Streptomyces spp PAL114 isolated from a Saharan soil. J Mycol Med 2014; 24:17-23. doi: 10.1016/j.mycmed.2013.09.001 [Crossref] [ Google Scholar]