BioImpacts. 9(4):219-225.
doi: 10.15171/bi.2019.27
Original Research
Association of MS4A6A, CD33, and TREM2 gene polymorphisms with the late-onset Alzheimer’s disease
Elham Mehdizadeh 1, Mohammad Khalaj-Kondori 2, Zeinab Shaghaghi-Tarakdari 3, Saeed Sadigh-Eteghad 1, Mahnaz Talebi 1, *, Sasan Andalib 4, 5, 6, 7, 8, 9, *
Author information:
1Neurosciences Research Center (NSRC), Tabriz University of Medical Sciences, Tabriz, Iran
2Department of Biology, Faculty of Natural Sciences, University of Tabriz, Tabriz, Iran
3Department of Genetics, Animal Biology Group, Faculty of Natural Sciences, University of Tabriz, Tabriz, Iran
4Neuroscience Research Center, Poursina Hospital, Guilan University of Medical Sciences, Rasht, Iran
5Department of Neurosurgery, Poursina Hospital, School of Medicine, Guilan University of Medical Sciences, Rasht, Iran
6Department of Nuclear Medicine, Odense University Hospital, Odense, Denmark
7Center for Applied Neuroscience, Brain Research - Interdisciplinary Guided Excellence, BRIDGE, Department of Clinical Research, Faculty of Health Sciences, University of Southern Denmark, Odense, Denmark
8Department of Clinical Research, Faculty of Health Sciences, University of Southern Denmark, Odense, Denmark
9Department of Psychiatry, Psychiatry in the Region of Southern Denmark, Odense, Denmark
Abstract
Introduction:
Alzheimer’s disease (AD), which is a progressive neurodegenerative disorder, causes structural and functional brain disruption. MS4A6A, TREM2, and CD33 gene polymorphisms loci have been found to be associated with the pathobiology of late-onset AD (LOAD). In the present study, we tested the hypothesis of association of LOAD with rs983392, rs75932628, and rs3865444 polymorphisms in MS4A6A, TREM2, CD33 genes, respectively.
Methods:
In the present study, 113 LOAD patients and 100 healthy unrelated age- and gender-matched controls were selected. DNA was extracted from blood samples by the salting-out method and the genotyping was performed by RFLP-PCR. Electrophoresis was carried out on agarose gel. Sequencing was thereafter utilized for the confirmation of the results.
Results:
Only CD33 rs3865444 polymorphism revealed a significant difference in the genotypic frequencies of GG (P = 0.001) and GT (P = 0.001), and allelic frequencies of G (P = 0.033) and T (P = 0.03) between LOAD patients and controls.
Conclusion:
The evidence from the present study suggests that T allele of CD33 rs3865444 polymorphism is associated with LOAD in the studied Iranian population.
Keywords: Late onset Alzheimer’s disease, LOAD, MS4A6A, CD33, TREM2, Polymorphisms
Copyright and License Information
© 2019 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Alzheimer’s disease (AD), which is an age-related neurodegenerative disorder, is the most prevalent form of dementia in the elderly. Hallmarks of AD include intraneuronal neurofibrillary tangles (NFTs) of hyperphosphorylated tau protein and cerebral senile plaques with extracellular deposits of β-amyloid peptide (Aβ). The sporadic AD is categorized into early- and late-onset with a prevalence of 3–5% and 95–97%, respectively.
1-4
Genetics has been found to be a contributing factor in neurodegenerative diseases such as Parkinson’s disease,
5
multiple sclerosis,
6
Leber’s Hereditary Optic Neuropathy,
7
and AD.
8
Amyloid precursor protein (APP)
9,10
and presenilin-1-2 (PSEN1-2) mutations
11,12
have been found in less than 5% of the AD patients.
13,14
Moreover, allelic variation of the Apolipoprotein E (Apo E) locus on chromosome 19 has been considered as a risk factor of late-onset AD (LOAD).
15
Potential involvement of genes on chromosomes 6, 9, 10, 11, 12, 14, 18, 19, and X in LOAD has been demonstrated.
16,17
It has been shown that mutations and differential expression of microglial receptors are associated with an increased risk of developing AD.
18
TREM2, which stands for triggering receptor expressed on myeloid cells 2, is an innate immune receptor on chromosome 6q21.1. It is expressed on the cell surface of macrophages, osteoclasts, microglia, and immature dendritic cells. It facilitates phagocytosis and downregulation of inflammation in the central nervous system.
19
TREM2 strongly increases the risk of developing AD, indicating microglial role in the pathogenesis of AD.
20
R47H (rs75932628) missense mutation within TREM2 gene has been reported to be associated with an increased risk of AD (OR = 2.92) in Icelandic population.
19
Such association has also been found in American, German, Dutch, and Norwegian populations.
21,22
Elsewhere, there was an association of rare R47H and R62H variants of TREM2 gene and increased risk of LOAD.
22
The MS4A, which refers to membrane-spanning 4-domains subfamily A gene, is located on chromosome 11q12.2. It serves an important role in immunity. In addition, MS4A families have a significant effect on tau phosphorylation, Aβ generation, and apoptosis through the regulation of calcium homeostasis. MS4A gene is highly expressed in monocytes and myeloid cells.
23
Recently LOAD genome-wide association studies (GWASs) have detected single nucleotide polymorphisms (SNPs) of MS4A6A gene such as rs983392 in association with increased risk of LOAD.
24,25
MS4A6A expression level is associated with an increased generation of plaque and tangle in AD patients.
26
It is suggested that AD specific brain structures are influenced by MS4A6A genotypes, indicating a possible role of the polymorphism in AD-related neuroimaging phenotype.
27
CD33 or Siglec-3 is a type I transmembrane protein belonging to the sialic acid-binding immunoglobulin-like lectins expressed on the cells of myeloid lineage. It plays an important role in mediating clathrin-independent endocytosis, cell growth and survival regulation by the induction of apoptosis, cell–cell interaction, and immune cells inhibition.
28,29
The association of CD33 gene polymorphisms with the LOAD has been demonstrated in several GWASs.
30,31
In addition, it was shown that CD33 rs3865444 polymorphism was associated to CD33 overexpression and amyloid plaque intensity by the impairment of Aβ microglia-mediated clearance in the brain of AD patients.
32
Little attention has been paid to the association of these polymorphisms with AD in the Iranian population. In the present study, our aim was to test the hypothesis of association of the three polymorphisms, rs983392,rs75932628, and rs3865444of MS4A6A, TREM2, and CD33 genes, respectively, in LOAD patients in an Iranian population in the north west of Iran.
Materials and methods
Participants
In this case–control study, 113 LOAD patients and 100 unrelated healthy controls were recruited. All the subjects were selected from neurologic wards of hospitals of Tabriz University of Medical Sciences, Tabriz, Azerbaijan Province, Iran. Inclusion criteria were LOAD diagnosis according to its diagnostic protocol.
33
Exclusion criteria were history of other neurodegetrative diseases and inherited diseases. Unrelated healthy control subjects were selected by advertisement from the general population and controlled by a neurologist for not suffering from LOAD. Age and gender matching was carried out in the two groups to control their possible confounding effects. Peripheral blood samples were obtained from LOAD patients and elderly control group and written informed consent was obtained from all of the subjects. Additionally, information on demographic characteristics such as gender, age, education, and MMSE score was collected.
Genotyping
DNA was extracted from blood samples (5 mL) by the salting-out of the cellular proteins. The salting-out was carried out by dehydration and precipitation using a saturated NACL solution.
34
By using Primer 3 software, forward and reverse primers were designed for each DNA fragment (Table 1). DNA was amplified by polymerase chain reaction (PCR) (Table 2). Restriction fragment length polymorphism (RFLP) was performed by certain restriction enzymes (Thermo Fisher Scientific, USA) (Table 1). Restriction products were separated by electrophoresis in 3% agarose gel with the aid of DNA safe stain and visualized in a UV transilluminator. The gels were thereafter evaluated in a UV transilluminator and images were captured. In addition, sequencing of 10% of PCR products were carried out by Sanger method to confirm the results of RFLP analysis.
Table 1.
Forward and reverse primers and restriction enzymes used for electrophoresis of the investigated polymorhisyms
|
Gene
|
SNP
|
Forward primer
|
Reverse primer
|
Restriction enzyme
|
| MS4A6A |
rs983392 (A/G) |
5’-GCCCAGAATATGTCAGCAAAAAC-3’ |
3’-AAGAAGAGCTAGCATGCACAGA-5’ |
NlaIII |
| TREM2 |
rs75932628 (C/T) |
5’-GTTGTAGATTCCGCAGCG-3’ |
3’-AAGACCAAGTGCCTCCAGA-5’ |
Hinp1I |
| CD33 |
rs3865444 (G/T) |
5’-ACAACTGTTTACACCAGGGC-3’ |
3’-AGTGTTTCTCCGAGATGACG-5’ |
N1aIII |
Table 2.
PCR reaction condition for MS4A6A, TREM2, and CD33 gene polymorphisms
|
Reaction component
|
MS4A6A gene polymorphism
|
TREM2 gene polymorphism
|
CD33 gene polymorphism
|
| WATER, Nuclease-free |
17 µL |
18 µL |
17 µL |
| 10x FastDigest or 10x FastDigest Green Buffer |
2 µL |
2 µL |
2 µL |
| PCR reaction mixture |
10 µL (0.2 µg DNA) |
10 µL(0.1-0.5 µg DNA) |
10 µL(0.2µg DNA) |
| FastDigest enzyme |
1 µL (NlaIII) |
1-2 µL (HinP1I) |
1 µL (NlaIII) |
| Expexted size of digested products |
67+174 bp |
164+369 bp |
282+366 bp |
Data analysis was performed by SPSS version 16.0 by chi-square test. Hardy–Weinberg equilibrium was assessed using Fisher exact test (P > 0.05). Furthermore, the odds ratios (OR) with 95% confidence interval (CI) were estimated for data. A P ≤ 0.05 was considered as statistically significant.
Results
Demographic characteristics of patients such as gender, age, education, and MMSE score are shown in Table 3. Allelic and genotypic frequencies of MS4A6A (rs983392), TREM2 (rs75932628), and CD33 (rs3865444) gene polymorphisms were assessed in 113 LOAD patients and 100 control individuals. For MS4A6A gene polymorphism (rs983392), G allele produced digestion by NlaIII detecting CATG/ sequence, in contrast to A allele abolishing the restriction site. For the TREM2 (rs75932628), C allele produced digestion by Hinp1I detecting G/CGC sequence, in contrast to T allele abolishing the restriction site. For the CD33 (rs3865444), G allele produced digestion by NlaIII detecting CATG/sequence, in contrast to A allele abolishing the restriction site. Genotypic and allelic frequencies of MS4A6A polymorphism (rs983392) showed no significant difference between the LOAD patients and the control group (Table 4). In TREM2 polymorphism (rs75932628), there was a non-significant difference between the frequency of allele T and the others (Table 5). There was also a non-significant difference between TT genotypic frequency and the others (Table 5). We also found a significant difference in genotypic (GG, P = 0.001; GT, P = 0.001) and allelic (G, P = 0.033; T, P = 0.03) frequencies of CD33 gene polymorphism (rs3865444) between the LOAD patients and the controls (Table 6). Fig. 1 shows RFPL results of MS4A6A, TREM2, and CD33 gene polymorphisms. Fig. 2 illustrates sequencing confirmation of the PCR-RFLP results.
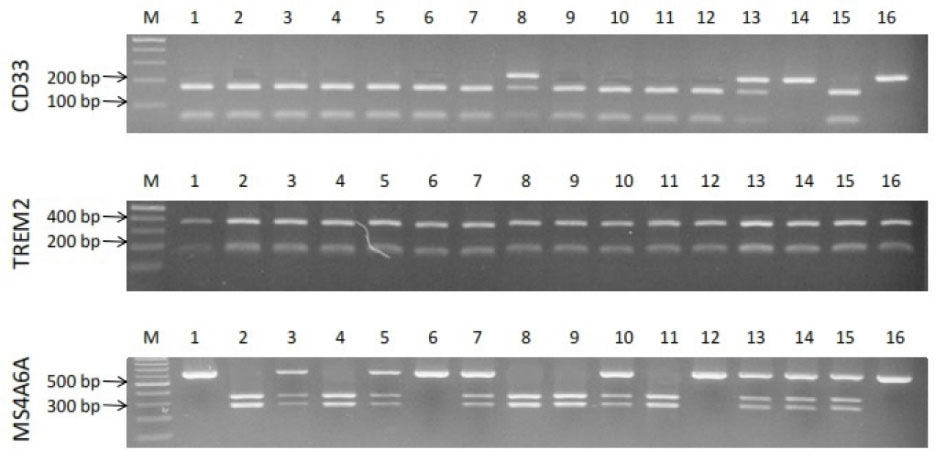
Fig. 1.
Electrophoresis results of MS4A6A, TREM2, and CD33 gene polymorphisms.
.
Electrophoresis results of MS4A6A, TREM2, and CD33 gene polymorphisms.
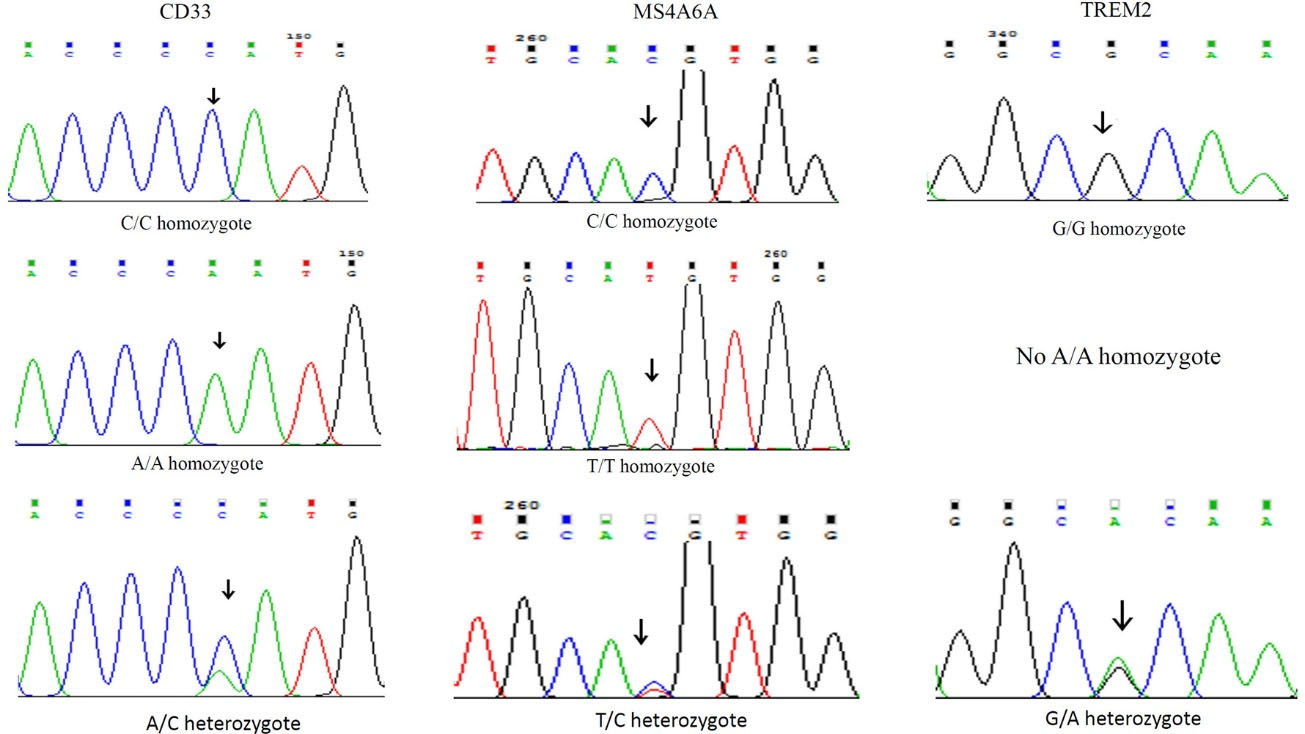
Fig. 2.
Sequencing confirmation of MS4A6A, TREM2, and CD33 gene polymorphisms.
.
Sequencing confirmation of MS4A6A, TREM2, and CD33 gene polymorphisms.
Table 3.
Demographic information of studied healthy controls and AD cases
|
Demographic information
|
Controls No. (%)
|
AD cases
No. (%)
|
| Gender |
Male |
32 (35.95) |
42 (42) |
| Female |
57 (64.04) |
58 (58) |
| Age group (y) |
65-69 |
46 (52.27) |
21 (21.64) |
| 70-74 |
13 (14.77) |
13 (13.4) |
| 75-79 |
18 (20.45) |
27 (27.83) |
| ≥80 |
11 (12.5) |
36 (37.11) |
| Education (y) |
Illiterate |
44 (57.14) |
43 (50) |
| 1-4 years |
28 (36.36) |
28 (32.55) |
| 5-8 years |
7 (9) |
10 (11.62) |
| ≥9 years |
3 (3.89) |
5 (5.81) |
| MMSE score |
<18 group |
0 (0) |
37 (36) |
| ≥18 group |
93 (99) |
66 (63) |
Table 4.
Genotype and allele distribution of MS4A6A gene polymorphism (rs983392) in control and case groups
|
Genotypes and alleles of MS4A6A polymorphism (rs983392)
|
LOAD (n=113)
|
Control (n=88)
|
Odds ratio (95 % CI)
|
P
value
|
|
n
|
F
|
n
|
F
|
| A |
122 |
0.539 |
89 |
0.505 |
1.146 (0.63-2.07) |
0.315 |
| G |
104 |
0.460 |
87 |
0.494 |
0.873 (0.48-1.58) |
0.367 |
| AA |
31 |
0.274 |
23 |
0.261 |
1.069 (0.54-2.09) |
0.405 |
| AG |
60 |
0.530 |
43 |
0.488 |
1.183 (0.65-2.14) |
0.249 |
| GG |
22 |
0.194 |
22 |
0.250 |
0.722 (0.35-1.48) |
0.179 |
Table 5.
Genotype and allele distribution of TREM2 gene polymorphism (rs75932628) in control and case groups
|
Genotypes and alleles of TREM2 poly morphisms (rs75932628)
|
LOAD (n=102)
|
Control (n=86)
|
Odds ratio (95 % CI)
|
P
value
|
|
n
|
F
|
n
|
F
|
| C |
203 |
0.995 |
171 |
0.994 |
1.2 (0.002-1166261.4) |
0.467 |
| T |
1 |
0.004 |
1 |
0.006 |
0.665 (0.00-405.997) |
0.5 |
| CC |
101 |
0.99 |
85 |
0.988 |
1.2 (0.046-41.1) |
0.287 |
| CT |
1 |
0.009 |
1 |
0.012 |
0.755 (0.036-1125592.1) |
0.493 |
| TT |
0 |
0 |
0 |
0 |
- |
- |
Table 6.
Genotype and allele distribution of CD33 gene polymorphism (rs3865444) in control and case groups
|
Genotypes and alleles of CD33 polymorphism (rs3865444)
|
LOAD (n=105)
|
Control (n=91)
|
Odds ratio (95 % CI)
|
P
value
|
|
n
|
F
|
n
|
F
|
| C |
173 |
0.824 |
130 |
0.714 |
1.875 (0.91-3.88) |
0.03 |
| T |
37 |
0.176 |
52 |
0.286 |
0.533 (0.25-1.09) |
0.03 |
| CC |
73 |
0.695 |
43 |
0.472 |
2.549 (1.37-4.74) |
0.001 |
| CT |
27 |
0.257 |
44 |
0.483 |
0.370 (0.19-0.7) |
0.001 |
| TT |
5 |
0.047 |
4 |
0.043 |
1.098 (0.24-4.95) |
0.50 |
Discussion
Recently, several GWASs have identified polymorphisms in MS4A6A, TREM2, and CD33 gene loci in LOAD patients. However, little is known about the abovementioned polymorphisms in Iranian population in Azerbaijan province. One study in an Iranian Azeri population showed that bridging integrator 1 (BIN1), estrogen receptor 1 (ESR1), toll-like receptor 2 (TLR2), chemokine receptor type 2 (CCR2), tumor necrosis factor alpha (TNF α), APOE, and phosphatidylinositol-binding clathrin assembly protein (PICALM) are loci for susceptibility of LOAD.
35
In this population, variants of TNF α, ESR1, CCR2, and APOE showed to be associated in 3 different genetic models. With adjustment for APOE, the genotypic and allelic association with BIN1, CCR2, and ESRα (PvuII) was found only in patients with APOE ε4; nevertheless, without Bonferroni correction, the association with CCR5, was seen only in cases with APOE ε4 allele.
TREM2 is an innate immune receptor and is encoded on the cell surface of microglia, immature dendritic cells, osteoclasts, and macrophages. For the first time, Jonsson et al
19
reported the rs75932628 polymorphism in TREM2 gene associated with an increased AD risk in an Icelandic population. An association of the TREM2 p.R47H substitution with Parkinson’s disease (OR=2.67; P = 0.026) and frontotemporal dementia (OR = 5.06; P = 0.001) were demonstrated.
36
In addition, an association of this SNP with the increased risk of LOAD has been reported.
22
In the present study, no T allele and TT genotype of TREM2 rs75932628 (R47H) polymorphism was seen. The TREM2rs75932628 SNP, identified by Jonsson et al,
19
failed to pass quality control due to a low minor allele frequency (0.0009). The frequency of minor allele varies usually across populations with a reported frequency from the Exome Variant Server of 0.26% among 4300 European Americans and 0.02% among 2203 African Americans.
37
Nevertheless, Mehrjoo et al
38
showed that there were-T one homozygous and 2 heterozygous AD patients and one heterozygous healthy control of TREM2 exon 2 rs75932628.
The MS4A gene serves an important role in immunity, Aβ generation, tau phosphorylation, and apoptosis.
26
MS4A genes are highly expressed in myeloid cells and monocytes.
23
Recent LOAD GWASs have demonstrated several SNPs such as rs983392 polymorphism in MS4A6A associated with LOAD susceptibility.
25,31,39
In spite of the fact that MS4A6E mRNA expression and rs670139 variant are associated with increased generation of tangle and plaque in brain tissues of AD patients,
26
the International Genomics of Alzheimer's Project (IGAP) meta-analysis showed rs983392 variant of MS4A6A gene was associated with reduced LOAD risk. This polymorphism affects active transcription sites and enhancers in human primary monocytes and is linked to chromatin marks presenting low transcription in neurons and brain tissues.
40
Moreover, Qiong et al mentioned that in 261 controls, 47 AD patients and, 456 patients with mild cognitive impairment, rs983392 SNP in MS4A6A gene was associated with an increase in the volume of left inferior temporal regions. Furthermore, this SNP was associated with the volume of right middle temporal at baseline.
41
However, we found no significant association between the rs983392 SNP of MS4A6A gene and susceptibility to LOAD.
CD33 or Siglec-3 is a type I transmembrane protein belonging to the sialic acid-binding immunoglobulin-like lectins and is encoded on the cells of myeloid lineage. It serves an important role in mediating clathrin-independent endocytosis, cell growth and survival regulation by induction of apoptosis, and cell–cell interaction and immune cell functions inhibition. With respect to CD33 rs3865444 polymorphism, our findings revealed an association between GG (P = 0.001) and GT (P = 0.001) genotypes and LOAD. LOAD was also associated with G (P = 0.033) and T (P = 0.03) alleles. It is also worth mentioning that T allele and GT genotype showed a lower odds of AD. The association of CD33 gene polymorphisms with the LOAD has been evaluated by recent GWASs
25,30,31,39
with contradictory findings. The rs3865444 SNP can be seen in upstream of the 5’ UTR of the CD33 gene.
25,31
In addition, CD33 rs3865444 polymorphism was associated with increased CD33 gene expression and amyloid plaque intensity by impairing microglia-mediated clearance of Aβ in the brain of AD patients.
32
CD33 has been identified as a modifier of Aβ pathology in vivo. The protective T allele of rs3865444 SNP is associated with reduced levels of insoluble Aβ42, CD33 microglial expression, and plaque burden in brains of AD patients. CD33 microglial is overexpressed in AD but the expression reduced in patients with T allele of the CD33 SNP rs3865444 because of the lower numbers of CD33-positive microglial cells. Therefore, CD33 may serve an important role in regulating Aβ microglial clearance and consequently may be a treating and preventive target of AD.
42
Recently, therapies targeting CD33 have been developed in acute myeloid leukemia (AML).
28,43,44
Walkeret al
45
investigated the rs3865444 polymorphism effects on the AD development and the expression of CD33 mRNA and protein in 96 controls and 97 AD patients and showed that the overexpression of CD33 mRNA was seen to be associated with the pathology of AD in temporal cortex samples and also identified that homozygous individuals with A/A (or T/T) alleles resulted in reduced levels of CD33 protein in temporal cortex.
45
A meta-analysis of 2634 LOAD patients and 4201 controls comprising six case-control studies from the USA and Europe also showed a significant association of CD33 rs3865444 SNP and AD (OR = 0.92, P = 0.049).
46
Investigation of fifty-eight SNPs in 229 LOAD cases and 318 control individuals from mainland China showed that rs6656401-rs3865444 (CR1-CD33) pairs were associated with the reduction of LOAD risk.
47
Conclusion
MS4A6A, TREM2, and CD33 gene polymorphisms loci have been found to be associated with the susceptibility to LOAD. In the present study, we tested the hypothesis of association of LOAD with rs983392, rs75932628, and rs3865444 polymorphisms in MS4A6A, TREM2, CD33 genes, respectively.The findings from the present study suggest that T allele of CD33 rs3865444 polymorphism is associated with LOAD in the studied Iranian population. Further investigations at mRNA and protein level may provide insights on the role of these mutation in susceptibility to LOAD.
Acknowledgement
We thank Neurosciences Research Center of Tabriz University of Medical Sciences for providing the support.
Funding sources
Funding was provided by Neurosciences Research Center of Tabriz University of Medical Sciences.
Conflicts of interest
The authors declare no conflicts of interest.
Ethical statement
The present study was approved by the Ethics Committee of Tabriz University of Medical Sciences.
Authors’ contribution
EM contributed to data handling and draft preparation. MKK contributed to conceptualism, experiment design, data analysis, and draft preparation. ZST contributed to data handling and draft preparation. SSE contributed to study consultation and draft preparation. MT contributed to conceptualism, experiment design, supervision, data handling, draft preparation, and writing and reviewing. SA contributed to consultation, data analysis, draft preparation, and writing and reviewing.
Research Highlights
What is the current knowledge?
simple
-
√ MS4A6A, CD33, and TREM2 gene polymorphisms are said
to be associated with LOAD.
What is new here?
simple
-
√ AD and control subjects were studied for carrying MS4A6A,
CD33, and TREM2 gene polymorphisms.
-
√ T allele of CD33 rs3865444 polymorphism was associated
with LOAD.
References
- Chiang K, Koo EH. Emerging therapeutics for Alzheimer's disease. Annu Rev Pharmacol Toxicol 2014; 54:381-405. doi: 10.1146/annurev-pharmtox-011613-135932 [Crossref] [ Google Scholar]
- Francis PT, Nordberg A, Arnold SE. A preclinical view of cholinesterase inhibitors in neuroprotection: do they provide more than symptomatic benefits in Alzheimer's disease?. Trends Pharmacol Sci 2005; 26:104-11. doi: 10.1016/j.tips.2004.12.010 [Crossref] [ Google Scholar]
- Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell 2012; 148:1204-22. doi: 10.1016/j.cell.2012.02.040 [Crossref] [ Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 2002; 297:353-6. doi: 10.1126/science.1072994 [Crossref] [ Google Scholar]
- Andalib S, Vafaee MS, Gjedde A. Parkinson's disease and mitochondrial gene variations: a review. J Neurol Sci 2014; 346:11-9. doi: 10.1016/j.jns.2014.07.067 [Crossref] [ Google Scholar]
- Andalib S, Talebi M, Sakhinia E, Farhoudi M, Sadeghi-Bazargani H, Motavallian A. Multiple sclerosis and mitochondrial gene variations: a review. J Neurol Sci 2013; 330:10-5. doi: 10.1016/j.jns.2013.04.018 [Crossref] [ Google Scholar]
- Andalib S, Talebi M, Sakhinia E, Farhoudi M, Sadeghi-Bazargani H, Masoudian N. No evidence of association between optic neuritis and secondary LHON mtDNA mutations in patients with multiple sclerosis. Mitochondrion 2017; 36:182-5. doi: 10.1016/j.mito.2017.08.005 [Crossref] [ Google Scholar]
- Chouraki V, Seshadri S. Genetics of Alzheimer’s disease. Adv Genet 2014; 87:245-94. doi: 10.1016/B978-0-12-800149-3.00005-6 [Crossref] [ Google Scholar]
- Chartier-Harlin M-C, Crawford F. Early-Onset Alzheimer's Disease Caused by Mutations at Codon 717 of the (Beta)-Amyloid Precursor Protein Gene. Nature 1991; 353:844. doi: 10.1038/353844a0 [Crossref] [ Google Scholar]
- Goate A, Chartier-Harlin M-C. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature 1991; 349:704. doi: 10.1038/349704a0 [Crossref] [ Google Scholar]
- Levy-Lahad E, Wasco W, Poorkaj P, Romano DM. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science 1995; 269:973. doi: 10.1126/science.7638622 [Crossref] [ Google Scholar]
- Sherrington R, Rogaev E, Liang Ya, Rogaeva E. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature 1995; 375:754. doi: 10.1038/375754a0 [Crossref] [ Google Scholar]
- Grazina M, Pratas J, Silva F, Oliveira S, Santana I, Oliveira C. Genetic basis of Alzheimer's dementia: role of mtDNA mutations. Genes, Brain and Behavior 2006; 5:92-107. doi: 10.1111/j.1601-183X.2006.00225.x [Crossref] [ Google Scholar]
- Hoenicka J. Genes in Alzheimer's disease. Revistadeneurologia 2006; 42:302-5. [ Google Scholar]
- Strittmatter WJ, Weisgraber KH, Huang DY, Dong L-M, Salvesen GS, Pericak-Vance M. Binding of human apolipoprotein E to synthetic amyloid beta peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proceedings of the National Academy of Sciences 1993; 90:8098-102. [ Google Scholar]
- Burns LC, Minster RL, Demirci FY, Barmada MM, Ganguli M, Lopez OL. Replication study of genome-wide associated SNPs with late-onset Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet 2011; 156B:507-12. doi: 10.1002/ajmg.b.31194 [Crossref] [ Google Scholar]
- Sillen A, Forsell C, Lilius L, Axelman K, Bjork BF, Onkamo P. Genome scan on Swedish Alzheimer's disease families. Mol Psychiatry 2006; 11:182-6. doi: 10.1038/sj.mp.4001772 [Crossref] [ Google Scholar]
- Zhao L. CD33 in Alzheimer’s Disease – Biology, Pathogenesis, and Therapeutics: A Mini-Review. Gerontology 2018. doi: 10.1159/000492596 [Crossref]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J. Variant of TREM2 associated with the risk of Alzheimer's disease. N Engl J Med 2013; 368:107-16. doi: 10.1056/NEJMoa1211103 [Crossref] [ Google Scholar]
- Gratuze M, Leyns CEG, Holtzman DM. New insights into the role of TREM2 in Alzheimer's disease. Mol Neurodegener 2018; 13:66. doi: 10.1186/s13024-018-0298-9 [Crossref] [ Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E. TREM2 variants in Alzheimer's disease. N Engl J Med 2013; 368:117-27. doi: 10.1056/NEJMoa1211851 [Crossref] [ Google Scholar]
- Jin SC, Benitez BA, Karch CM, Cooper B, Skorupa T, Carrell D. Coding variants in TREM2 increase risk for Alzheimer's disease. Hum Mol Genet 2014; 23:5838-46. doi: 10.1093/hmg/ddu277 [Crossref] [ Google Scholar]
- Zuccolo J, Deng L, Unruh TL, Sanyal R, Bau JA, Storek J. Expression of MS4A and TMEM176 Genes in Human B Lymphocytes. Front Immunol 2013; 4:195. doi: 10.3389/fimmu.2013.00195 [Crossref] [ Google Scholar]
- Antúnez C, Boada M, González-Pérez A, Gayán J, Ramírez-Lorca R, Marín J. The membrane-spanning 4-domains, subfamily A (MS4A) gene cluster contains a common variant associated with Alzheimer's disease. Genome medicine 2011; 3:33. doi: 10.1186/gm249 [Crossref] [ Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat gen 2011; 43:436-41. doi: 10.1038/ng.801 [Crossref] [ Google Scholar]
- Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PloS One 2012; 7:e50976. doi: 10.1371/journal.pone.0050976 [Crossref] [ Google Scholar]
- Ma J, Zhang W, Tan L, Wang HF, Wan Y, Sun FR. MS4A6A genotypes are associated with the atrophy rates of Alzheimer's disease related brain structures. Oncotarget 2016; 7:58779-88. doi: 10.18632/oncotarget.9563 [Crossref] [ Google Scholar]
- Jandus C, Simon HU, von Gunten S. Targeting siglecs--a novel pharmacological strategy for immuno- and glycotherapy. Biochem Pharmacol 2011; 82:323-32. doi: 10.1016/j.bcp.2011.05.018 [Crossref] [ Google Scholar]
- Tateno H, Li H, Schur MJ, Bovin N, Crocker PR, Wakarchuk WW. Distinct endocytic mechanisms of CD22 (Siglec-2) and Siglec-F reflect roles in cell signaling and innate immunity. Mol Cell Biol 2007; 27:5699-710. doi: 10.1128/MCB.00383-07 [Crossref] [ Google Scholar]
- Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron 2010; 68:270-81. doi: 10.1016/j.neuron.2010.10.013 [Crossref] [ Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet 2011; 43:429-35. doi: 10.1038/ng.803 [Crossref] [ Google Scholar]
- Bradshaw EM, Chibnik LB, Keenan BT, Ottoboni L, Raj T, Tang A. CD33 Alzheimer's disease locus: altered monocyte function and amyloid biology. Nat Neurosci 2013; 16:848-50. doi: 10.1038/nn.3435 [Crossref] [ Google Scholar]
- Dubois B, Feldman HH, Jacova C, DeKosky ST, Barberger-Gateau P, Cummings J. Research criteria for the diagnosis of Alzheimer's disease: revising the NINCDS–ADRDA criteria. The Lancet Neurology 2007; 6:734-46. doi: 10.1016/s1474-4422(07)70178-3 [Crossref] [ Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Research 1988; 16:1215-
. doi: 10.1093/nar/16.3.1215 [Crossref] [ Google Scholar]
- Rezazadeh M, Khorrami A, Yeghaneh T, Talebi M, Kiani SJ, Heshmati Y. Genetic Factors Affecting Late-Onset Alzheimer’s Disease Susceptibility. Neuromolecular Med 2016; 18:37-49. doi: 10.1007/s12017-015-8376-4 [Crossref] [ Google Scholar]
- Rayaprolu S, Mullen B, Baker M, Lynch T, Finger E, Seeley WW. TREM2 in neurodegeneration: evidence for association of the pR47H variant with frontotemporal dementia and Parkinson's disease. Mol Neurodegener 2013; 8:19. doi: 10.1186/1750-1326-8-19 [Crossref] [ Google Scholar]
- Jin SC, Carrasquillo MM, Benitez BA, Skorupa T, Carrell D, Patel D. TREM2 is associated with increased risk for Alzheimer’s disease in African Americans. Mol Neurodegener 2015; 10:19. doi: 10.1186/s13024-015-0016-9 [Crossref] [ Google Scholar]
- Mehrjoo Z, Najmabadi A, Abedini SS, Mohseni M, Kamali K, Najmabadi H. Association Study of the TREM2 Gene and Identification of a Novel Variant in Exon 2 in Iranian Patients with Late-Onset Alzheimer's Disease. Med Princ Pract 2015; 24:351-4. doi: 10.1159/000430842 [Crossref] [ Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 2013; 45:1452-8. doi: 10.1038/ng.2802 [Crossref] [ Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012; 22:1790-7. doi: 10.1101/gr.137323.112 [Crossref] [ Google Scholar]
- Li JQ, Wang HF, Zhu XC, Sun FR, Tan MS, Tan CC. GWAS-Linked Loci and Neuroimaging Measures in Alzheimer's Disease. Mol Neurobiol 2017; 54:146-53. doi: 10.1007/s12035-015-9669-1 [Crossref] [ Google Scholar]
- Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron 2013; 78:631-43. doi: 10.1016/j.neuron.2013.04.014 [Crossref] [ Google Scholar]
- Malik M, Chiles J, Xi HS, Medway C, Simpson J, Potluri S. Genetics of CD33 in Alzheimer's disease and acute myeloid leukemia. Hum Mol Genet 2015; 24:3557-70. doi: 10.1093/hmg/ddv092 [Crossref] [ Google Scholar]
- O'Reilly MK, Paulson JC. Siglecs as targets for therapy in immune-cell-mediated disease. Trends Pharmacol Sci 2009; 30:240-8. doi: 10.1016/j.tips.2009.02.005 [Crossref] [ Google Scholar]
- Walker DG, Whetzel AM, Serrano G, Sue LI, Beach TG, Lue LF. Association of CD33 polymorphism rs3865444 with Alzheimer's disease pathology and CD33 expression in human cerebral cortex. Neurobiol Aging 2015; 36:571-82. doi: 10.1016/j.neurobiolaging.2014.09.023 [Crossref] [ Google Scholar]
- Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F. Replication of EPHA1 and CD33 associations with late-onset Alzheimer's disease: a multi-centre case-control study. Mol Neurodegener 2011; 6:54. doi: 10.1186/1750-1326-6-54 [Crossref] [ Google Scholar]
- Jiao B, Liu X, Zhou L, Wang MH, Zhou Y, Xiao T. Polygenic analysis of late-onset Alzheimer’s disease from mainland China. PloS one 2015; 10:e0144898. doi: 10.1371/journal.pone.0144898 [Crossref] [ Google Scholar]