BioImpacts. 10(1):27-36.
doi: 10.15171/bi.2020.04
Original Research
PEGylated gold nanoparticles-ribonuclease induced oxidative stress and apoptosis in colorectal cancer cells
Mostafa Akbarzadeh Khiavi 1, 2, †  , Azam Safary 2, 3, †
, Azam Safary 2, 3, †  , Jaleh Barar 2, 4
, Jaleh Barar 2, 4  , Hamed Farzi-Khajeh 5, Abolfazl Barzegari 2, Rahimeh Mousavi 2, Mohammad Hossein Somi 1, *
, Hamed Farzi-Khajeh 5, Abolfazl Barzegari 2, Rahimeh Mousavi 2, Mohammad Hossein Somi 1, *  , Yadollah Omidi 2, 4, *
, Yadollah Omidi 2, 4, * 
Author information:
1Liver and Gastrointestinal Diseases Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
2Research Center for Pharmaceutical Nanotechnology, Biomedicine Institute, Tabriz University of Medical Sciences, Tabriz, 51656-65811, Iran
3Connective Tissue Diseases Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
4Department of Pharmaceutics, Faculty of Pharmacy, Tabriz University of Medical Sciences, Tabriz, Iran
5Organosilicon Research Laboratory, Faculty of Chemistry, University of Tabriz, Tabriz, Iran
† These authors have an equal contribution as the joint first authors.
Abstract
Introduction:
Currently, drug-induced reactive oxygen species (ROS) mediating apoptosis pathway have extensively been investigated in designing effective strategies for colorectal cancer (CRC) chemotherapy. Bovine pancreatic ribonuclease A (RNase A) represents a new class of cytotoxic and non-mutagenic enzymes, and has gained more attention as a potential anticancer modality; however, the cytosolic ribonuclease inhibitors (RIs) restrict the clinical application of this enzyme. Nowadays, nanotechnology-based diagnostic and therapeutic systems have provided potential solutions for cancer treatments.
Methods:
In this study, the gold nanoparticles (AuNPs) were synthesized, stabilized by polyethylene glycol (PEG), functionalized, and covalently conjugated with RNase A. The physicochemical properties of engineered nanobiomedicine (AuNPs-PEG-RNase A) were characterized by scanning electron microscope (SEM), dynamic light scattering (DLS), and UV-vis spectrum. Then, its biological impacts including cell viability, apoptosis, and ROS production were evaluated in the SW-480 cells.
Results:
The engineered nanobiomedicine, AuNPs-PEG-RNase A, was found to effectively induce apoptosis in SW-480 cells and result in a significant reduction in cancer cell viability. Besides, the maximum production of ROS was obtained after the treatment of cells with an IC50 dose of AuNPs-PEG-RNase A.
Conclusion:
Based on the efficient ROS-responsiveness and the anticancer activity of RNase A of the engineered nanomedicine, this nanoscaled biologics may be considered as a potential candidate for the treatment of CRC.
Keywords: Bovine pancreatic ribonuclease, Colorectal cancer, Gold nanoparticles, Nanomedicine, PEGylation, Reactive oxygen species
Copyright and License Information
© 2020 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Colorectal cancer (CRC) is considered as one of the most common, death-related cancers, which is mainly attributed to metastasis cancers.
1
From a molecular point of view, cellular reduction/oxidation (redox) system plays a crucial role in the regulation of various aspects of cell function.
2
There are several indications that under different levels of oxidative stress, the expression of antioxidant enzymes such as superoxide dismutase, catalase, and peroxiredoxins and non-enzymatic antioxidants (e.g., vitamins E and A) is upregulated in breast, colon, pancreatic, prostate, and other cancers.
3
Reactive oxygen species (ROS) can be toxic to cells at high levels and lead to DNA damage and mutation.
4
Besides genetic instability, ROS can also stimulate the proliferation of cancer cells, subsequent angiogenesis, invasion, and metastasis.
5
In addition to increasing cell growth, ROS are also involved in apoptosis induction or autophagy and act as an inhibitor for cancer cell proliferation by the oxidation of cellular components such as DNA, lipid, and proteins. To date, researchers have extensively investigated drug-induced ROS mediating apoptosis pathway to design effective strategies for CRC chemotherapy.
6,7
Despite the valuable advantages and therapeutic effects, currently used chemotherapy agents (e.g., irinotecan hydrochloride, capecitabine, 5-fluorouracil, leucovorin calcium, and ziv-aflibercept) often encounter some limitations such as undesired side effects and drug-resistance phenomena.
8,9
To tackle such shortcomings, there is an emerging need for developing novel and effective treatment strategies for CRC.
During the past years, nanotechnology-based diagnostic and therapeutic systems have provided potential solutions for cancer treatments.
10-13
Multifunctional nanosystems (NSs) in conjugation with various imaging probes and targeting agents such as antibodies (Abs)/aptamers (Aps), and enzymes have been recently developed for cancer therapy.
14-16
Among different types of NSs, the greater affinity, specificity, and functionality of enzymes to selectively target the desired cells/tissues make them prominent treatment modalities for a variety of diseases.
17-20
Bovine pancreatic ribonuclease A (RNase A, EC:3.1.27.5) is one of the best characterized small proteins (13.7 kDa), which was investigated for therapeutical purposes as a potential anticancer modality in 1972.
21
RNases represent a new class of cytotoxic and non-mutagenic enzymes, that can inhibit the proliferation of cancer cells through the catalytic cleavage of phosphodiester bonds in various types of single-stranded RNA (e.g., tRNA, rRNA, mRNA, and non-coding RNA), and hence, influence several signaling pathways, as well as protein biosynthesis at transcription and translation stages.
22,23
However, the presence of cytosolic ribonuclease inhibitors (RIs) is one of the most important obstacles against the clinical application of RNases.
24
In this regard, various strategies including conjugation with antibodies (Abs), peptide/proteins, and nanoparticles (NPs) have been employed to resolve RIs inhibitory effects.
24,25
It has been shown that the conjugation of the enzymes with nanoparticles (NPs) has improved the stability and activity of therapeutic enzymes.
26
Among all nanomaterials, gold nanoparticles (AuNPs) are an attractive candidate for delivery of various anticancer agents due to their individual properties including biocompatibility, easy conjugation to biomolecules such as antibody and enzyme, and the unique optical properties, which increase the binding rate to amine and thiol groups, allowing surface modification.
27-30
In our previous study, it was shown that a novel nanobiomedicine composed of AuNPs conjugated with Bovine RNase A (RnaGNPs) was able to overcome the intracellular RIs, induce apoptosis, and inhibit the cellular invasion and metastasis.
10
In the current study, to retrieve and improve biological and therapeutical properties of RnaGNPs, AuNPs were synthesized, stabilized by polyethylene glycol (PEG), and covalently conjugated with RNase A. The physicochemical properties of engineered nanobiomedicine (AuNPs-PEG-RNase A) were characterized by scanning electron microscope (SEM), dynamic light scattering (DLS), and UV-vis spectrum. Then its biological impacts (e.g., cell viability, apoptosis, and oxidative stress induction) were evaluated in the SW-480 cells.
Materials and Methods
Reagents
All chemicals, buffers, and solvents were purchased from commercial sources and used as received. Bovine pancreatic ribonuclease A (≥70 Kunitz units/mg protein), gold (III) chloride trihydrate (HAuCl4•3H2O), 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA), sodium citrate, N-hydroxyl succinimide (NHS), 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC), 3-mercaptopropionic acid (3MPA), ethanol 99%, penicillin-streptomycin 1% (10 000 U/mL penicillin, 10 mg/mL streptomycin), and phosphate-buffered saline (PBS) were received from Sigma-Aldrich company (Munich, Germany). The human CRC SW-480 cell line was purchased from the National Cell Bank of Iran, Pasteur Institute (Tehran, Iran). RPMI 1640 medium, fetal bovine serum (FBS), and trypsin-EDTA (0.02–0.05%) were obtained from Gibco (Paisley, UK). Gene JET RNA purification kit and Maxima SYBER Green/ROX qPCR master mix were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Reverse transcriptase reagent kit and FITC-labeled annexin V- apoptosis detection kit were respectively obtained from TAKARA Co. (Tokyo, Japan) and Applied Biosystems (Foster City, CA, USA). Polyethylene glycol 2000 (PEG 2000), triethylamine (TEA, 99%), 3-Mercaptopropionic acid (MPA, 99%), and succinic anhydride (99%) were purchased from Sigma-Aldrich Co.
Synthesis of PEG ligands SH-PEG-COOH
In the first step, a 100 mL flask was charged with poly (ethylene glycol) MW 2000 (1.5 mM), 3-mercaptopropionic acid (1.5 mM), and dry toluene. To this reaction mixture, concentrated sulfuric acid was added. The reaction flask was equipped with an azeotropic distillation apparatus and the mixture was refluxed at 130°C under N2 for 4 hours. Toluene was removed under reduced pressure. The recovered polymer was recrystallized from tetrahydrofuran (THF) or a mixture of methanol and diethyl ether. In the second step, the obtained polymer from the previous step was dissolved in dry toluene, after the addition of succinic anhydride (1 mM) and TEA, the reaction mixture started to boiling at 150°C under N2 for 5 hours with an azeotropic distillation apparatus. Toluene was removed under reduced pressure. Then the white solid precipitation was collected and washed with tetrahydrofuran and diethyl ether/methanol to obtain 0.8 g AAOh (yield: 80%).
31
The FTIR spectra of PEG ligands SH-PEG-COOH was recorded on the dried pellet obtained after centrifugation and removal of water from the samples. IR (KBr): 3500; 2878; 2775; 2377; 1745; 1490; 1280; 1100 cM-1.
Synthesis of gold nanoparticles (AuNPs)
First, AuNPs were synthesized from HAuCl4 by wet chemical methods using trisodium citrate as a reducing agent.
32
Briefly, water solutions of HAuCl4 and sodium citrate (1% w/v) were mixed under heavy boiling, resulting in gold particles with a net negative charge from the adsorption of citrate ions, which stabilized the particles.
Preparation of AuNPs-PEG-RNase A conjugates
Carboxylate-functionalized AuNPs-PEG was accomplished using carbamide and succinimide (EDC-NHS) chemistry.
31
Briefly, EDC (0.4 mmol) and NHS (0.4 mmol) were added to the AuNPs (0.33 µmol) solution. After 2 hours, 15 µL of RNase A (5mg/mL) was added to the reaction mixture and incubated under shaking with constant mixing at 4°C for 3 hours. The AuNPs-PEG-RNase A was collected after centrifugation at 10 000 rpm and washed (×3) with PBS.
Determination of loading and unloading of RNase A to AuNPs-PEG
The amount of conjugated RNase A was determined by the bicinchoninic acid (BCA) protein assay kit for the quantitation of total protein in a supernatant.
33
The Enzyme loading (%) was calculated using the following equation:
Where unloaded protein and protein in nanoparticles are given as mg/mL.
Physical characterization of the enzyme-loaded nanostructure
Particle size and zeta potential of AuNPs and AuNPs-PEG-RNase A were measured by dynamic light scattering using a Nanotrac Wave™ (Microtrac, San Diego, CA, USA).
34
For shape and morphological analysis, the AuNPs-PEG-RNase A was deposited onto a clean gold substrate and dried at room temperature and evaluated by a scanning electron microscope (SEM) (TEScan Mira-3, Brno, Czech). Absorbance measurements were performed on 96-well plate cultures containing 100 µL of compounds per well by Cytation 5 Cell Imaging Multi-Mode Reader (Cytation 5, BioTek, Winooski, USA) in the range of 200–800 nm.
Confirmation of AuNPs-PEG-RNase A conjugates and enzyme activity assay
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis was further performed to confirm the AuNPs-PEG-RNase A conjugation.
35
For this purpose, free RNase A and AuNPs-PEG-RNase A conjugates were loaded on 12% polyacrylamide gel and then protein bands were visualized by staining with coomassie brilliant blue G-250. Besides, the RNase A activity in free and conjugated form was determined by measuring the RNA degradation using the method of Kalnitsky et al.
36
Briefly, the solution of 1 mg/mL of the enzyme in 0.10 M sodium acetate was prepared (pH 5.0). Afterward, 1 mL of enzyme solution was added to different concentrations of 1% RNA (0.5, 1, 2, 5 and 10 μg/mL) and then all samples were incubated at 37°C for 5 minutes. Before analysis, the reaction of samples was stopped by the addition of 1 mL uranyl acetate-perchloric acid solution. Then, the samples were transferred to the ice bath and cooled for 5 minutes. The degradation of RNA was determined by a UV-visible spectrophotometer (λmax=260 nm). The sodium acetate buffer (0.10 M, pH 5.0) with 1% RNA was considered as a blank.
Cell culture and cell viability assay
The experimental study was performed on the SW-480 cell line belonging to human CRC. SW-480 cell line was purchased from the cell bank of Pasture Institute (Iran, Tehran), and maintained in RPMI-1640 medium supplemented with 10% FBS, 1% antibiotics (100 unit/mL penicillin and 100 μg/mL streptomycin). Cells were cultured at 37ºC and 5% Co2. The cells were passaged every 3 days following detaching with 0.25% trypsin/EDTA and the experiment was performed during the logarithmic phase of cell growth. The cell viability was analyzed using the MTT assay.
37
The SW-480 cells were seeded in 96-well microplates at a density of 1.0×104 cells/well/200 μL. The cells were then incubated in 5% CO2 humidified atmosphere at 37°C. After 24 hours of incubation, the cells were treated with a serial concentration (50,100,150,200, and 250 µg/mL) of free AuNPs-PEG, RNase A, and AuNPs-PEG-RNase A incubated for 24, 48, and 72 hours. After the mentioned time intervals, the medium of each well was replaced with 200 μL of fresh MTT reagent (2 mg/mL) for 4 hours. Then, the MTT reagent was removed and 200 μL DMSO and 25 μL Sorensen’s buffer (0.133 M, pH 7.2) were mixed and added to stop the reaction and then the cells were incubated at 37°C for additional 15 minutes. The optical density was measured using BioTeck Elisa reader (Winooski, VT) at 570 nm wavelength. The viability of cells was evaluated relative to theoretical absorbance. All experiments were performed in triplicate.
Apoptosis assay by DAPI
The effect of AuNPs-PEG-RNase A on apoptosis induction was evaluated by DAPI staining assay.
38
Approximately 2×103 cells were seeded in 96-well cell culture plate. After incubation at 37°C for 24 hours, the cells were washed 3 times with PBS (NaCl, KCl, KH2PO4, and Na2HPO4, pH 7.4). Then ice-cold paraformaldehyde (PFA) 4% was added to each well for fixation of cells and then was incubated for 20 minutes at 37°C. Afterward, the wells were washed with PBS and then Triton-X-100 solution (0.1%) was added to each well and incubated for 5 minutes at 37°C for permeabilization. After being rewashed with PBS, the DAPI solution (0.1%) was added to each well and incubated for 7 minutes. Then, the cells were washed (×3) with 0.1% Triton X-100 in PBS, and the cells with condensed and fragmented chromatin were analyzed with a live imaging system (Cytation 5, Biotek, Winooski, USA).
Apoptosis assay by Annexin V
To detect the extent of apoptosis induced by free RNase A, AuNPs-PEG, and AuNPs-PEG-RNase A in intact SW-480 cells, the flow cytometry analysis was used via staining the cells with annexin V-FITC.
39
Briefly, SW-480 cells (1.0 × 105 cells/mL) were exposed to RNase A, AuNPs-PEG, and AuNPs-PEG-RNase A for 48 hours. After the incubation, the cells were collected and re-suspended in 200 μL of annexin V-binding buffer and 5 𝜇L of propidium iodide (PI) and incubated for 10 minutes at room temperature in the dark. The cells were finally analyzed by the flow cytometry (BD FACS Calibur) with an emission filter of 600 nm for PI (red) and 515–545 nm for FITC (green).
2,7-DCF oxidation assay
The DCF assay was performed to determine the formation of ROS in SW-480 cells.
40
Briefly, 2×105 the SW-480 cells were seeded in a 6-well plate and treated with different formulations of AuNPs-PEG, RNase A, and AuNPs-PEG-RNase A for 48 hours. Then cells were washed with PBS and incubated with 0.5 mL of medium containing 5 μM of 2′,7′ dichlorodihydrofluorescein diacetate (DCFH-DA) for 30 minutes at 37°C and fluorescence was detected using a live imaging system (Cytation 5, Biotek, Winooski, USA).
Flow cytometry analysis of ROS production
The intracellular levels of ROS were evaluated using DCFH-DA.
41
The 2×105 SW-480 cells were seeded in a 6-well plate and treated with different formulations of AuNPs-PEG, RNase A, and AuNPs-PEG-RNase A for 48 hours. The cells were harvested by trypsin and washed with PBS. Then, the cells were incubated with 0.5 mL of medium containing 5 μM of the ROS-sensitive probe DCFH-DA at 37°C for 20 minutes. The cells were subsequently gathered and washed (×3) with cell culture medium without FBS. All samples were analyzed by the flow cytometry.
Statistical analysis
Data were performed to determine the statistical differences by one-way ANOVA and Student’s t-test. The statistical analysis software was SPSS Version 16.0 (IBM Corporation, NY, USA) and P < 0.05 was considered statistically significant. All the data were expressed as mean values ± standard deviation (SD).
Results
Synthesis and characterization of AuNPs-PEG-RNase A
The AuNPs were prepared by the citrate-reduction method. Fig. 1A represents the FT-IR spectra of the SH-PEG-COOH. Absorption at 1700 cm-1 and 2250 cm-1 confirms the successful coupling of PEG with COOH and SH, respectively. The functionalized NSs were analyzed by SEM (Fig. 1B), as well as DLS. As shown in Fig. 1 (C and D) , the size of pegylated AuNPs (AuNPs-PEG) and AuNPs-PEG-RNase A measured ∼24 and ∼36 nm, respectively. AuNPs-PEG and AuNPs-PEG-RNase A displayed zeta potential value of –10 and –23.4 mV.
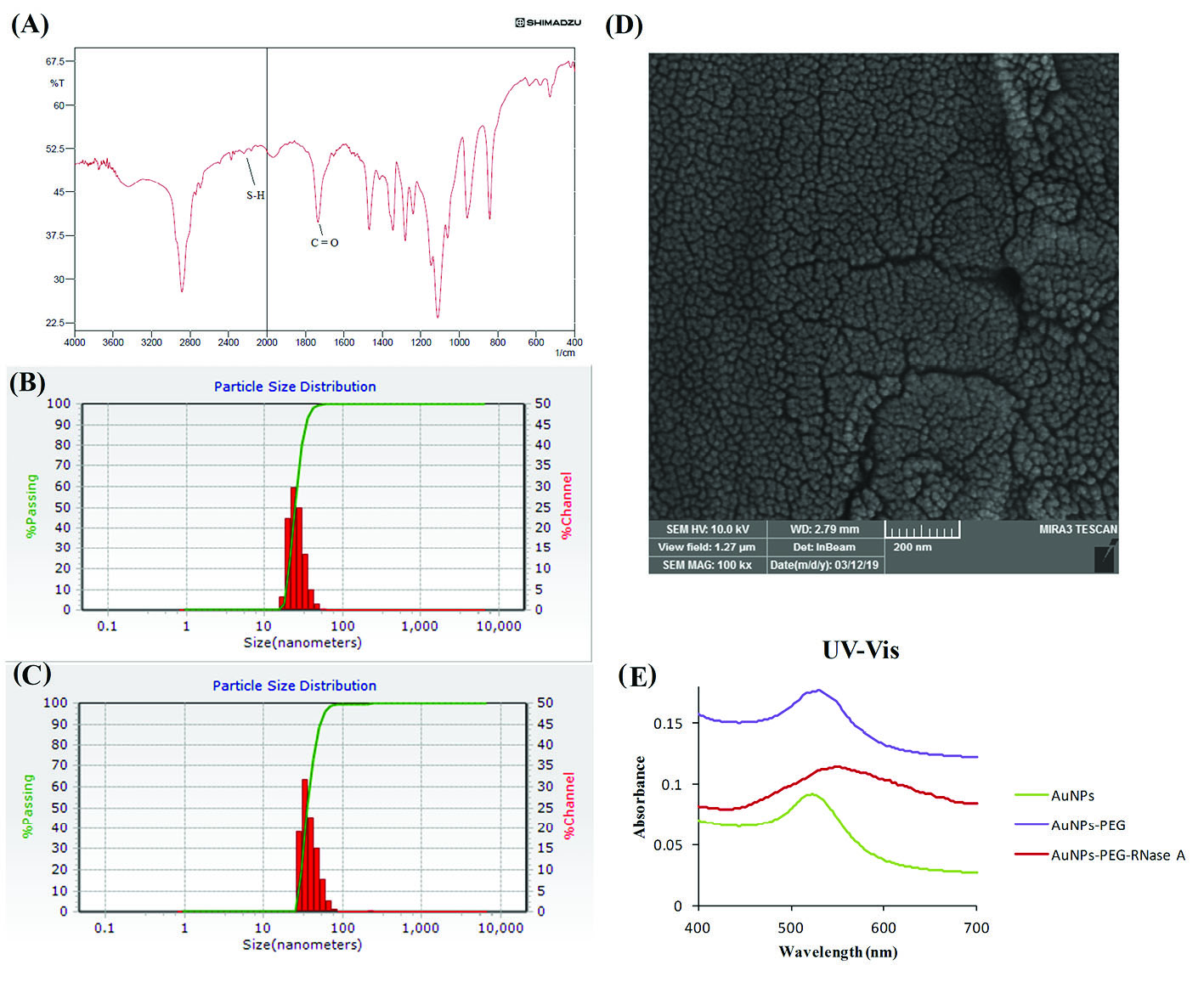
Fig. 1.
Characterization of the engineered AuNPs-PEG-RNase A nanobiosystem. (A) FT-IR spectra of SH-PEG-COOH. (B) DLS measurement of AuNPs-
PEG, and (C) AuNPs-PEG-RNase A nanobiosystem. (D) SEM image of AuNPs-PEG-RNase A. (E) UV absorption spectra of AuNPs, AuNPs-PEG, and
AuNPs-PEG-RNase A nanobiosystem.
.
Characterization of the engineered AuNPs-PEG-RNase A nanobiosystem. (A) FT-IR spectra of SH-PEG-COOH. (B) DLS measurement of AuNPs-
PEG, and (C) AuNPs-PEG-RNase A nanobiosystem. (D) SEM image of AuNPs-PEG-RNase A. (E) UV absorption spectra of AuNPs, AuNPs-PEG, and
AuNPs-PEG-RNase A nanobiosystem.
UV–vis absorption spectra of AuNPs, Au-PEG, and AuNPs-PEG-RNase A are presented in Fig. 1E. The unmodified NPs illustrated an absorption peak at ∼522 nm, while Au-PEG nanoparticles showed an absorption peak at ∼528 nm. However, it was demonstrated that the maximum absorption peak was increased and shifted to ∼548 nm in the engineered AuNPs-PEG-RNase A nanobiomedicine.
Confirmation of AuNPs-PEG-RNase A conjugates and enzyme activity
SDS-PAGE analysis was used for monitoring and evaluating the AuNPs-PEG-RNase A conjugates. Using Coomassie brilliant blue staining, RNase A was significantly indicated by the appearance of a strong band (∼13 kDa) in all samples including free RNase A (lane 1) and AuNPs-PEG-RNase A conjugates (lane 2, 3), as shown in Fig. 2A. Based on the SDS-PAGE results, conjugation of RNase A to AuNPs-PEG was performed successfully. Besides, the activity of conjugated RNase A was also confirmed by analyzing the degradation rate of different concentrations of RNA. The preliminary activity assay of RNase A showed that the conjugated enzyme to AuNPs-PEG was completely active (Fig. 2B).
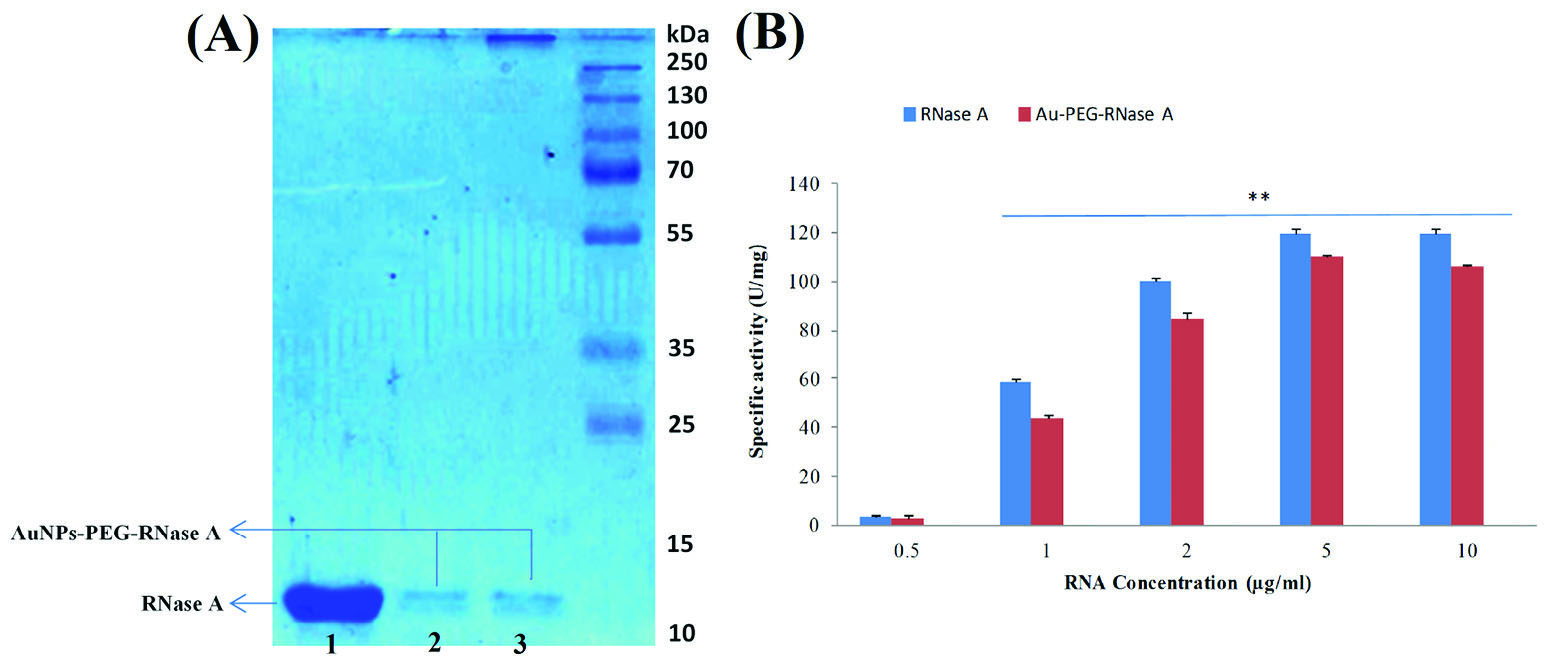
Fig. 2.
Confirmation of AuNPs-PEG-RNase A conjugates and enzyme activity. (A) SDS-PAGE analysis of AuNPs-PEG-RNase A conjugates. (B) RNase A
activity assay in the presence of different concentrations of RNA in free and conjugated forms using Uv-vis. Ez: RNase A.
.
Confirmation of AuNPs-PEG-RNase A conjugates and enzyme activity. (A) SDS-PAGE analysis of AuNPs-PEG-RNase A conjugates. (B) RNase A
activity assay in the presence of different concentrations of RNA in free and conjugated forms using Uv-vis. Ez: RNase A.
Cell viability analysis
The cytotoxicity effects of AuNPs-PEG, RNase A, and AuNPs-PEG-RNase A were determined by MTT assays. The viability of the SW-480 cancer cells was analyzed in the presence of different concentrations of AuNPs-PEG, RNase A, and AuNPs-PEG-RNase A for 24, 48, and 72 hours time points. As revealed in Fig. 3, there was not a significant change in various amounts of AuNPs-PEG and RNase A in comparison to the untreated control group (P> 0.05). Meanwhile, after the treatment with different concentrations of engineered AuNPs-PEG-RNase A (50-250 μg/mL), the viability of the SW-480 cells was significantly decreased. The IC50 values for AuNPs-PEG-RNase A after 24, 48, and 72 hours were obtained 167 μg/mL, 138 μg/mL, and 97 μg/mL, respectively.
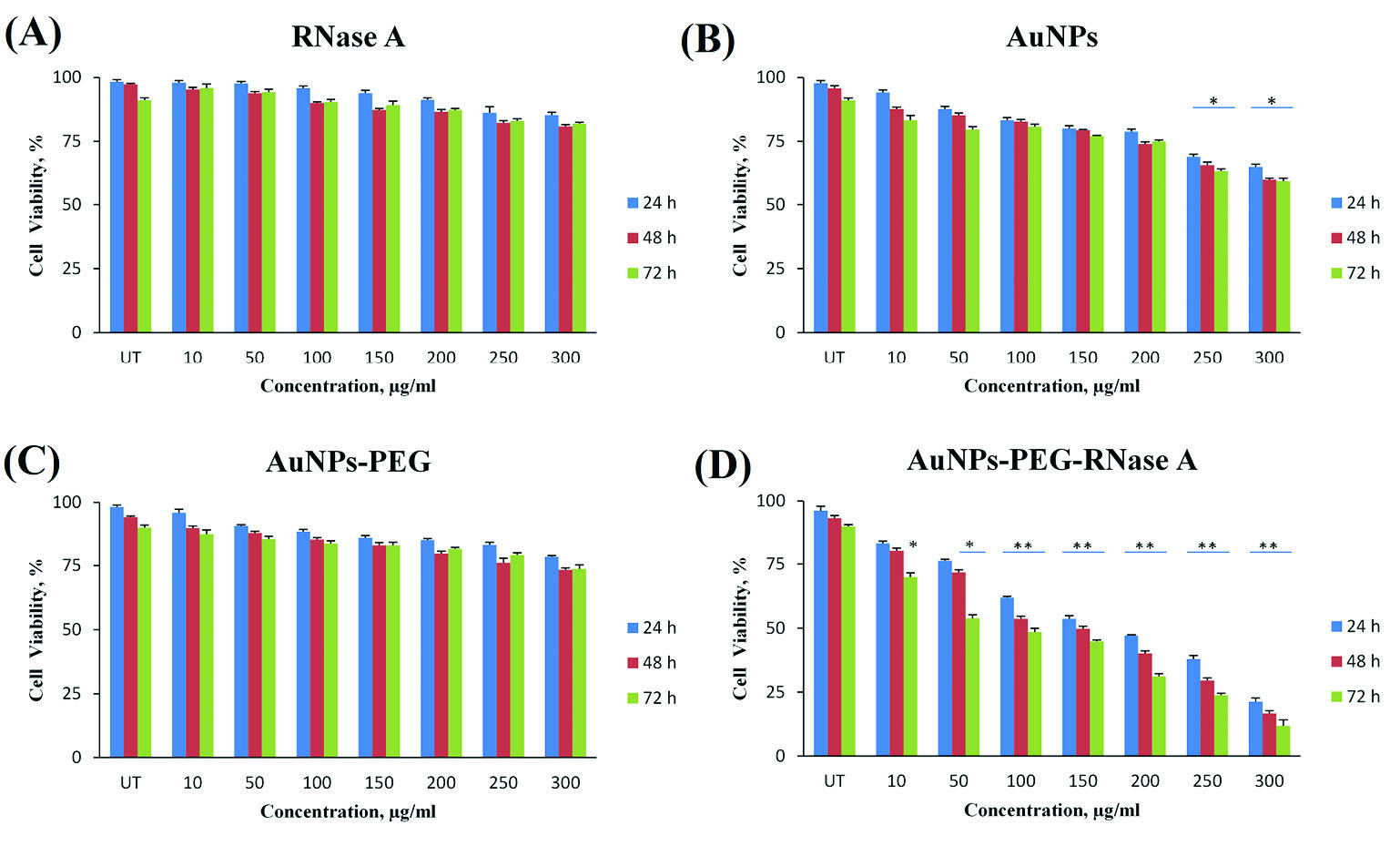
Fig. 3.
Cell viability assay of the SW-480 cell line in different concentrations (0-300 μg/mL) of (A) RNase A, (B) AuNPs, (C) AuNPs-PEG, and (D) AuNPs-
PEG-RNase A nanobiosystem after 24, 48, and 72 hours. The data present means ±SD of three separate experiments. Quantification is given from three
independent experiments. UT: untreated groups. *P< 0.05, and **P < 0.01.
.
Cell viability assay of the SW-480 cell line in different concentrations (0-300 μg/mL) of (A) RNase A, (B) AuNPs, (C) AuNPs-PEG, and (D) AuNPs-
PEG-RNase A nanobiosystem after 24, 48, and 72 hours. The data present means ±SD of three separate experiments. Quantification is given from three
independent experiments. UT: untreated groups. *P< 0.05, and **P < 0.01.
Apoptosis assay by DAPI staining
Chromatin condensation/remodeling and nuclear fragmentation that are typical markers of apoptosis were evaluated in the SW-480 cells treated with the AuNPs-PEG, free RNase A, and AuNPs-PEG-RNase A using DAPI staining assay. As shown in Fig. 4, the morphological changes of the nucleus as a biochemical characteristic of apoptosis in the SW-480 cells treated with AuNPs-PEG-RNase A was more detectable than the cells with AuNPs and RNase A alone, that was shown confirming the occurrence of higher cytotoxicity effects of the AuNPs-PEG-RNase A.
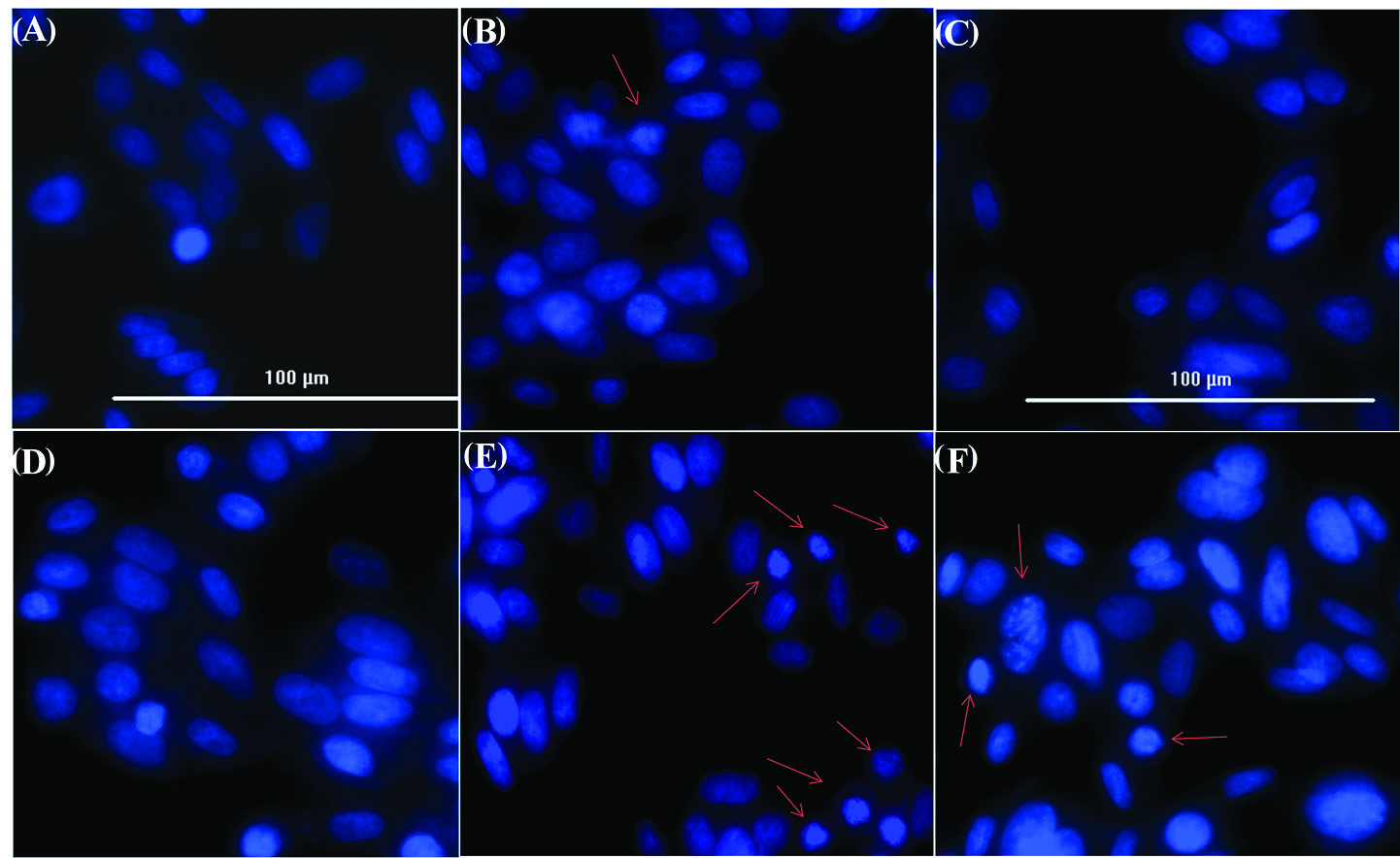
Fig. 4.
Apoptosis assay by DAPI staining of SW-480 nucleus. (A) control group, (B) treated with AuNPs, (C) AuNPs-PEG, (D), free RNase A, and (E, F)
AuNPs-PEG-RNase A nanobiosystem. Red arrows depict chromatin condensation and fragmented nuclei.
.
Apoptosis assay by DAPI staining of SW-480 nucleus. (A) control group, (B) treated with AuNPs, (C) AuNPs-PEG, (D), free RNase A, and (E, F)
AuNPs-PEG-RNase A nanobiosystem. Red arrows depict chromatin condensation and fragmented nuclei.
Apoptosis assay by Annexin V
The FITC-labeled Annexin V/PI flow cytometry analysis was performed to quantify the occurrence of apoptosis upon treatment with AuNPs-PEG, free RNase A, and AuNPs-PEG-RNase A of the SW-480 cells. As shown in Fig. 5, the AuNPs-PEG-RNase A could significantly induce 26.58% of early and 22.63% of late apoptosis in the treated SW-480 cells. Meanwhile, the apoptosis (both early and late) rates in the treated cells with free RNase A and AuNPs-PEG were found around 5.0% and 5.6%, respectively.
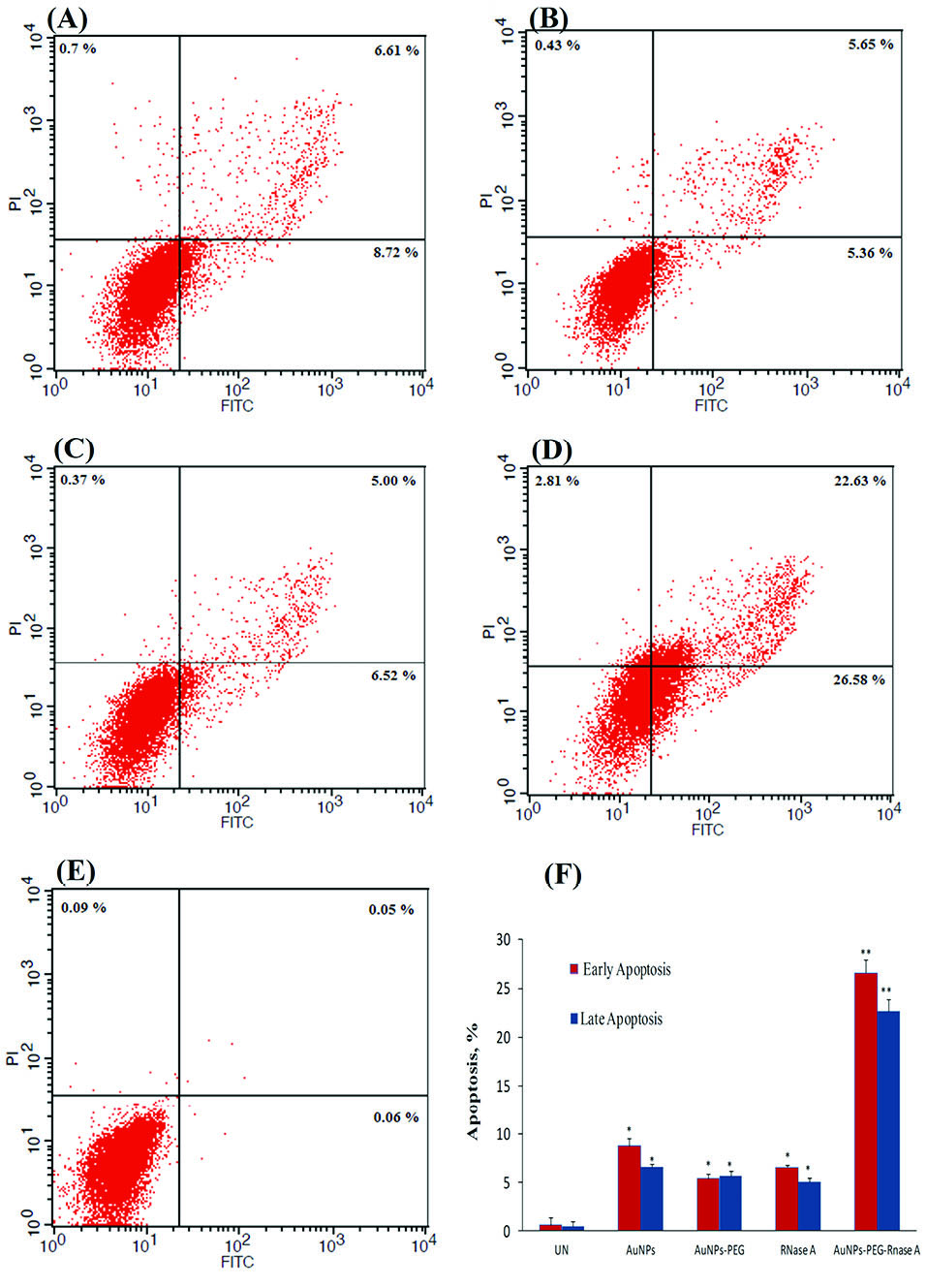
Fig. 5.
Apoptosis assay using Annexin-V flow cytometry analysis on SW-
480 cancer cells. (A) treated with AuNPs, (B) AuNPs-PEG, (C) free RNase
A, (D) AuNPs-PEG-RNase A nanobiosystem, and (E) control group. (F)
The rate of apoptosis (early and late) in the treated cells with AuNPs,
AuNPs-PEG, free RNase A, and AuNPs-PEG-RNase A. UT:untreated
control. Quantification is given from three independent experiments. *P <
0.05, and **P < 0.01.
.
Apoptosis assay using Annexin-V flow cytometry analysis on SW-
480 cancer cells. (A) treated with AuNPs, (B) AuNPs-PEG, (C) free RNase
A, (D) AuNPs-PEG-RNase A nanobiosystem, and (E) control group. (F)
The rate of apoptosis (early and late) in the treated cells with AuNPs,
AuNPs-PEG, free RNase A, and AuNPs-PEG-RNase A. UT:untreated
control. Quantification is given from three independent experiments. *P <
0.05, and **P < 0.01.
ROS production assay
To determine the effect of AuNPs-PEG, free RNase A, and AuNPs-PEG-RNase A on intracellular ROS production, DCHF-DA was used as a fluorescent probe using a live imaging system (Cytation 5, Biotek, Winooski, USA) and flow cytometry technique. In this method, DCFH-DA hydrolyzes to DCFH through intracellular esterases and the level of ROS is determined by the oxidation rate of DCFH to DCF. The DCF intensity could reveal the level of ROS production. Fig. 6 shows the increasing fluorescence signal intensity in the treated cells with AuNPs-PEG-RNase A compared to free RNase A and AuNPs-PEG, which can precisely reveal intracellular ROS production. The flow cytometry results indicated that the maximum production of ROS was obtained after the treatment of cells with IC50 dose of AuNPs-PEG-RNase A. In comparison with untreated cells, nearly 4% and 25% ROS were generated in the cells treated with RNase A and AuNPs-PEG-RNase A after 24 hours, respectively. Also, nearly 22% of ROS was generated in the cells treated with H2O2 (0.05%) as a positive control.
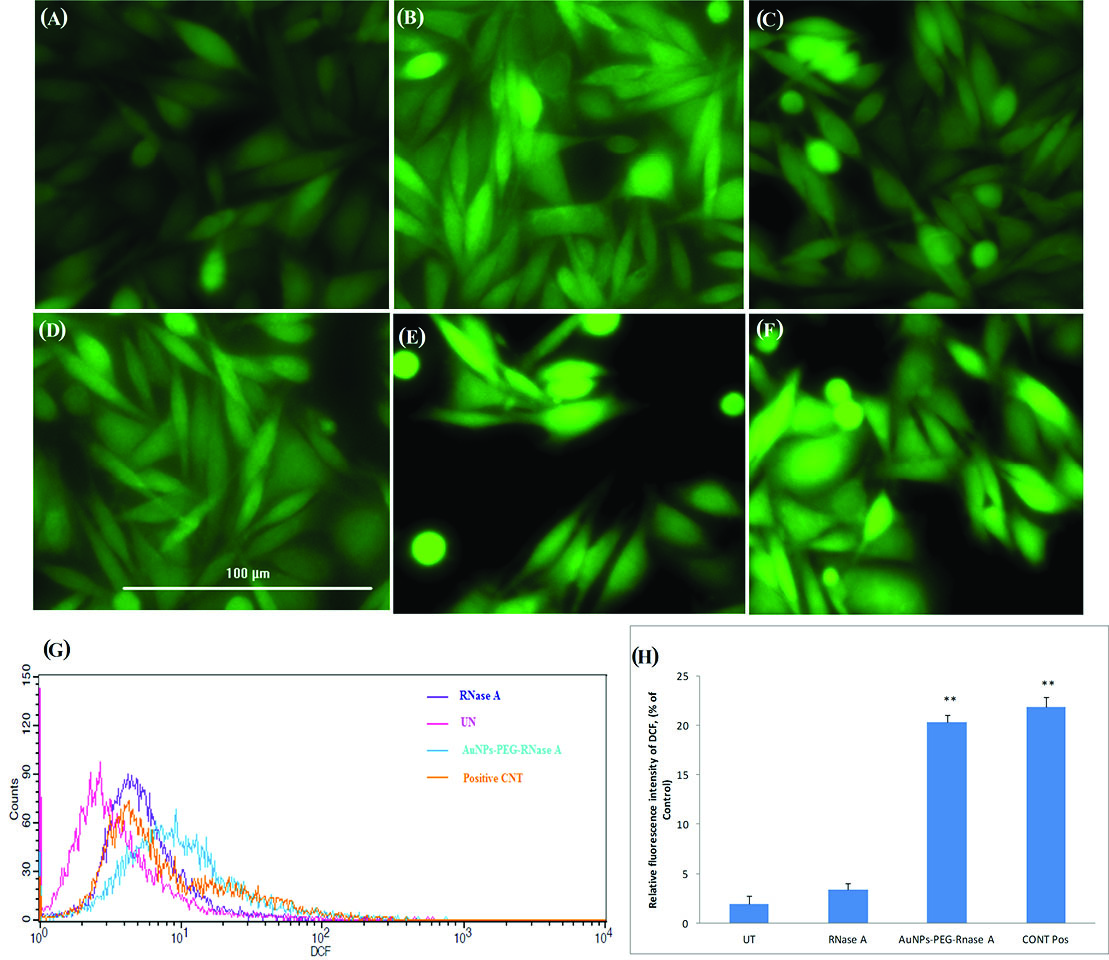
Fig. 6.
ROS production assay on SW-480 cancer cells using DCHF-DA fluorescent dye. Cytation 5 imaging from (A) untreated group, (B) treated cells with
AuNPs, (C) AuNPs-PEG, (D) free RNase A, (E) AuNPs-PEG-RNase A nanobiosystem, and (F) positive control treated with H2O2. (G) The flow cytometry
analysis of ROS production. (H) The rate of DCF intensity in the treated cells with free RNase A, AuNPs-PEG-RNase A, and positive control. UT:untreated
group. Quantification is given from three independent experiments. **P < 0.01.
.
ROS production assay on SW-480 cancer cells using DCHF-DA fluorescent dye. Cytation 5 imaging from (A) untreated group, (B) treated cells with
AuNPs, (C) AuNPs-PEG, (D) free RNase A, (E) AuNPs-PEG-RNase A nanobiosystem, and (F) positive control treated with H2O2. (G) The flow cytometry
analysis of ROS production. (H) The rate of DCF intensity in the treated cells with free RNase A, AuNPs-PEG-RNase A, and positive control. UT:untreated
group. Quantification is given from three independent experiments. **P < 0.01.
Discussion
Ribonucleases present special therapeutic properties, however, the intracellular RIs are one of the most important obstacles for the clinical applications of RNases in cancer therapy. Different strategies have been introduced to increase the therapeutic impacts of RNases, their stability and activity in clinical applications. The main purpose of the current study was to improve the characteristics of designed nanobiosystems (AuNPs-RNase A) in the previous study
10
using pegylated gold nanoparticles for increasing anticancer effects of RNase A and decreasing effective dose against CRC line. Additionally, the role of engineered AuNPs-PEG-RNase A was explored on oxidative stress induction, ROS production, and apoptosis stimulation in SW-480 cancer cells.
Physical characterization of AuNPs-PEG-RNase A nanobiosystem using DLS and SEM (Fig. 1) revealed the size of the engineered NSs approximately ∼24 and ∼36 nm before and after modification. The PEGylated AuNPs and AuNPs-PEG-RNase A showed a surface charge of -10 mV and -24 mV, respectively. The size and zeta potential characteristics of synthesized NSs caused their suitable distribution in solvents without any aggregation. It has been demonstrated that there is a strong correlation between the size of the NPs and their penetration into the cancer cells. Therefore it can be speculated that the PEGylated AuNPs-RNase A with a size~36 nm might be taken up by cells easily, which was confirmed by our preliminary study.
10
The formulation process did not alter the specific activity of the enzyme (Fig. 2).
Based on the cytotoxicity assay results, the AuNPs-PEG-RNase A nanobiosystem showed higher toxicity on the SW-480 cells compared to the untreated group (Fig. 3). Besides, the viability of the SW-480 cells treated with AuNPs was considerably lower than that of the AuNPs-PEG in the same incubation time (P< 0.05). It can be concluded that the pegylation of AuNPs significantly decreased the toxicity of the free AuNPs, which makes it eligible for the in vivo and clinical applications.
The IC50 values of the AuNPs-PEG-RNase A were obtained 167, 138, and 97 μg/mL after 24, 48, and 72 hours treatment, respectively, that was significantly decreased as compared to the non-PEGylated form of conjugates (AuNPs-RNase A), described in our previous study.
10
It seems that PEGylation can probably prevent high loading RNase A on AuNPs surface, lead to a reduced interference between molecules, and subsequently better performance and activity of enzymes.
To evaluate apoptosis induction, DAPI staining was carried out for the assessment of the chromatin condensation and crescent-shaped nuclei within the nuclear membrane of the cells treated with AuNPs and AuNPs-PEG-RNase A (Fig. 4). Furthermore, we assayed the alternation of membrane phospholipids using annexin V flow cytometry, which is based on the existence of phosphatidylserine (Ptd-L-Ser) on the cell plasma membrane as a sign of annexin V affinity for the apoptotic cells.
42
According to the results, the highest frequency of apoptotic cells was found in the AuNPs-PEG-RNase A-treated SW-480 cells (Fig. 5).
It has been established that ROS molecules such as hydrogen peroxide, hydroxyl radical, superoxide anion, and peroxynitrites can be involved in CRC initiation and tumor progression. Oxidative stress condition commonly leads to increasing the ROS level inside the cancer cells, which can contribute to activating the survival pathway upon stress response.
43
Further, high-levels production of ROS through damaging cellular components (e.g., DNA, protein, and lipid) can induce apoptosis in cancer cells. Thus, drug-induced ROS mediated pathway have been investigated for several agents used in cancer chemotherapy.
44
Given the importance of ROS in various physiological phenomena, we evaluated ROS production in human CRC SW-480 cells treated with AuNPs, AuNPs-PEG, RNase A, and AuNPs-PEG-RNase A nanobiosystem using both of flow cytometry and Cytation 5 Cell Imaging techniques (Fig. 6). Our results are in agreement with those of recently published studies that demonstrated the correlation between oxidative stress, the production of ROS, and the induction of apoptosis.
45,46
It has been revealed that the ROS induction pathway can be beneficial for developing therapeutic perspectives of cancer.
46,47
There are a few reports which reported the role of ribonuclease and/or its modified form in the ROS production and apoptosis induction.
23,48
Conclusion
In summary, the current study introduces an efficient ROS-responsive nanobiosystem and its biological impact in CRC therapy. Our previous and present results demonstrated that the conjugation of RNase A to AuNPs and its improvement with PEGylation could retrieve the anticancer activity of the enzyme and overcome the inhibition effects of intracellular RIs. Based on physicochemical properties, AuNPs-PEG-RNase A effectively induced apoptosis in SW-480 cells and resulted in a significant reduction in cancer cell viability. Further, the results of the ROS production assay revealed that the engineered nanomedicine was able to induce ROS production and emphasize the potential of this system in cancer therapy through the ROS-induced mediated pathway. Taken all, we propose the AuNPs-PEG-RNase A nanomedicine as a de novo treatment modality against CRC.
Acknowledgment
This study, as a joint program with the Liver and Gastrointestinal Diseases Research Center (LGDRC) and Research Center for Pharmaceutical Nanotechnology (RCPN). The study was carried out at the RCPN, Biomedicine Institute, Tabriz University of Medical Sciences.
Funding sources
This work is a part of a Ph.D. thesis supported (grant No: 145/261) by the LGDRC, Tabriz University of Medical Sciences and Iran National Science Foundation (INSF) (grant#: 96010102).
Ethical statement
The present study was approved by the Ethics Committee of Tabriz University of Medical Sciences (Ethical No. IR.TBZMED.REC.1395243).
Conflicts of interest
The authors declare no conflicts of interest.
Authors’ contribution
The conception and the design of the study were performed by MAK, AS and HFK. The experiments was carried out by MAK. Data collection and analyses were conducted by MAK. Data interpretation were performed by AS, HFK, JB, AB and MAK. The statistical analysis was performed by AS and RM. The project supervised by YO and MHS. The manuscript was drafted by MAK and revised by AS, JB and YO. All authors participated in manuscript in the critical review process of the manuscript and approval of the final version.
Research Highlights
What is the current knowledge?
simple
-
√ RNase A is one of the best characterized anticancer
enzymes.
-
√ Currently, the cytosolic ribonuclease inhibitors restrict the
clinical application of RNase A.
What is new here?
simple
-
√ An efficient novel nanobiosystem AuNPs-PEG-RNase A
was synthesized and characterized.
-
√ AuNPs-PEG-RNase A effectively induced ROS production
in SW-480 cells.
-
√ AuNPs-PEG-RNase A effectively induced apoptosis in SW-
480 cells.
-
√ AuNPs-PEG-RNase A retrieved and improved the
anticancer activity of RNase A.
References
- Somi MH, Golzari M, Farhang S, Naghashi S, Abdollahi L. Gastrointestinal cancer incidence in East Azerbaijan, Iran: update on 5 year incidence and trends. Asian Pac J Cancer Prev 2014; 15:3945-9. doi: 10.7314/apjcp.2014.15.9.3945 [Crossref] [ Google Scholar]
- Kesarwani P, Murali AK, Al-Khami AA, Mehrotra S. Redox regulation of T-cell function: from molecular mechanisms to significance in human health and disease. Antioxid Redox Signal 2013; 18:1497-534. doi: 10.1089/ars.2011.4073 [Crossref] [ Google Scholar]
- Karihtala P, Soini Y. Reactive oxygen species and antioxidant mechanisms in human tissues and their relation to malignancies. Apmis 2007; 115:81-103. doi: 10.1111/j.1600-0463.2007.apm_514.x [Crossref] [ Google Scholar]
- Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol 2014; 24:R453-62. doi: 10.1016/j.cub.2014.03.034 [Crossref] [ Google Scholar]
- Kumari S, Badana AK, G MM, G S, Malla R. Reactive Oxygen Species: A Key Constituent in Cancer Survival. Biomark Insights 2018; 13:1177271918755391. doi: 10.1177/1177271918755391 [Crossref] [ Google Scholar]
- Chang D, Wang F, Zhao YS, Pan HZ. Evaluation of oxidative stress in colorectal cancer patients. Biomed Environ Sci 2008; 21:286-9. doi: 10.1016/s0895-3988(08)60043-4 [Crossref] [ Google Scholar]
- Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ 2015; 22:377-88. doi: 10.1038/cdd.2014.150 [Crossref] [ Google Scholar]
- Cassidy J, Clarke S, Diaz-Rubio E, Scheithauer W, Figer A, Wong R. XELOX vs FOLFOX-4 as first-line therapy for metastatic colorectal cancer: NO16966 updated results. Br J Cancer 2011; 105:58-64. doi: 10.1038/bjc.2011.201 [Crossref] [ Google Scholar]
- Gressett SM, Stanford BL, Hardwicke F. Management of hand-foot syndrome induced by capecitabine. J Oncol Pharm Pract 2006; 12:131-41. doi: 10.1177/1078155206069242 [Crossref] [ Google Scholar]
- Khiavi MA, Safary A, Aghanejad A, Barar J, Rasta SH, Golchin A. Enzyme conjugated gold nanoparticles for combined enzyme and photothermal therapy of colon cancer cells. Colloids Surf A Physicochem Eng Asp 2019; 572:333-44. doi: 10.1016/j.colsurfa.2019.04.019 [Crossref] [ Google Scholar]
- Zhao CY, Cheng R, Yang Z, Tian ZM. Nanotechnology for Cancer Therapy Based on Chemotherapy. Molecules 2018:23. doi: 10.3390/molecules23040826 [Crossref]
- Calixto G, Bernegossi J, Fonseca-Santos B, Chorilli M. Nanotechnology-based drug delivery systems for treatment of oral cancer: a review. Int J Nanomedicine 2014; 9:3719-35. doi: 10.2147/ijn.s61670 [Crossref] [ Google Scholar]
- Barar J, Kafil V, Majd MH, Barzegari A, Khani S, Johari-Ahar M. Multifunctional mitoxantrone-conjugated magnetic nanosystem for targeted therapy of folate receptor-overexpressing malignant cells. J Nanobiotechnology 2015; 13:26. doi: 10.1186/s12951-015-0083-7 [Crossref] [ Google Scholar]
- Fathi M, Majidi S, Zangabad PS, Barar J, Erfan-Niya H, Omidi Y. Chitosan-based multifunctional nanomedicines and theranostics for targeted therapy of cancer. Med Res Rev 2018; 38:2110-36. doi: 10.1002/med.21506 [Crossref] [ Google Scholar]
- Akbarzadeh Khiavi M, Safary A, Somi MH. Recent advances in targeted therapy of colorectal cancer: Impacts of monoclonal antibodies nanoconjugates. Bioimpacts 2019; 9:139-42. doi: 10.15171/bi.2019.16 [Crossref] [ Google Scholar]
- Omidi Y, Barar J. Induction of human alveolar epithelial cell growth factor receptors by dendrimeric nanostructures. Int J Toxicol 2009; 28:113-22. doi: 10.1177/1091581809335177 [Crossref] [ Google Scholar]
- Safary A, Khiavi MA, Omidi Y, Rafi MA. Targeted enzyme delivery systems in lysosomal disorders: an innovative form of therapy for mucopolysaccharidosis. Cell Mol Life Sci 2019:1-19. doi: 10.1007/s00018-019-03135-z [Crossref]
- Safary A, Khiavi MA, Mousavi R, Barar J, Rafi MA. Enzyme replacement therapies: what is the best option?. BioImpacts: BI 2018; 8:153. doi: 10.15171/bi.2018.17 [Crossref] [ Google Scholar]
- Safary A, Moniri R, Hamzeh-Mivehroud M, Dastmalchi S. Identification and Molecular Characterization of Genes Coding Pharmaceutically Important Enzymes from Halo-Thermo Tolerant Bacillus. Adv Pharm Bull 2016; 6:551-61. doi: 10.15171/apb.2016.069 [Crossref] [ Google Scholar]
- Safary A, Moniri R, Hamzeh-Mivehroud M, Dastmalchi S. Highly efficient novel recombinant L-asparaginase with no glutaminase activity from a new halo-thermotolerant Bacillus strain. Bioimpacts 2019; 9:15-23. doi: 10.15171/bi.2019.03 [Crossref] [ Google Scholar]
- Ardelt W, Ardelt B, Darzynkiewicz Z. Ribonucleases as potential modalities in anticancer therapy. Eur J Pharmacol 2009; 625:181-9. doi: 10.1016/j.ejphar.2009.06.067 [Crossref] [ Google Scholar]
- Mironova N, Patutina O, Brenner E, Kurilshikov A, Vlassov V, Zenkova M. The systemic tumor response to RNase A treatment affects the expression of genes involved in maintaining cell malignancy. Oncotarget 2017; 8:78796-810. doi: 10.18632/oncotarget.20228 [Crossref] [ Google Scholar]
- Ardelt B, Juan G, Burfeind P, Salomon T, Wu JM, Hsieh TC. Onconase, an anti-tumor ribonuclease suppresses intracellular oxidative stress. Int J Oncol 2007; 31:663-9. doi: 10.3892/ijo.31.3.663 [Crossref] [ Google Scholar]
- Attery A, Dey P, Tripathi P, Batra JK. A ribonuclease inhibitor resistant dimer of human pancreatic ribonuclease displays specific antitumor activity. Int J Biol Macromol 2018; 107:1965-70. doi: 10.1016/j.ijbiomac.2017.10.067 [Crossref] [ Google Scholar]
- Vertegel AA, Reukov V, Maximov V. Enzyme–Nanoparticle conjugates for biomedical applications. Methods Mol Biol 2011; 679:165-82. doi: 10.1007/978-1-60761-895-9_14 [Crossref] [ Google Scholar]
- Sirisha VL, Jain A, Jain A. Enzyme Immobilization: An Overview on Methods, Support Material, and Applications of Immobilized Enzymes. Adv Food Nutr Res 2016; 79:179-211. doi: 10.1016/bs.afnr.2016.07.004 [Crossref] [ Google Scholar]
- Qian Y, Qiu M, Wu Q, Tian Y, Zhang Y, Gu N. Enhanced cytotoxic activity of cetuximab in EGFR-positive lung cancer by conjugating with gold nanoparticles. Sci Rep 2014; 4:7490. doi: 10.1038/srep07490 [Crossref] [ Google Scholar]
- Nakhlband A, Barar J, Bidmeshkipour A, Heidari HR, Omidi Y. Bioimpacts of anti epidermal growth factor receptor antisense complexed with polyamidoamine dendrimers in human lung epithelial adenocarcinoma cells. J Biomed Nanotechnol 2010; 6:360-9. doi: 10.1166/jbn.2010.1131 [Crossref] [ Google Scholar]
- Akbarzadeh Khiavi M, Safary A, Barar J, Ajoolabady A, Somi MH, Omidi Y. Multifunctional nanomedicines for targeting epidermal growth factor receptor in colorectal cancer. Cell Mol Life Sci 2019. doi: 10.1007/s00018-019-03305-z [Crossref]
- Samadi Pakchin P, Ghanbari H, Saber R, Omidi Y. Electrochemical immunosensor based on chitosan-gold nanoparticle/carbon nanotube as a platform and lactate oxidase as a label for detection of CA125 oncomarker. Biosens Bioelectron 2018; 122:68-74. doi: 10.1016/j.bios.2018.09.016 [Crossref] [ Google Scholar]
- Du YJ, Brash JL. Synthesis and characterization of thiol‐terminated poly (ethylene oxide) for chemisorption to gold surface. J Appl Polym Sci 2003; 90:594-607. doi: 10.1002/app.12545 [Crossref] [ Google Scholar]
- Kimling J, Maier M, Okenve B, Kotaidis V, Ballot H, Plech A. Turkevich method for gold nanoparticle synthesis revisited. J Phys Chem B 2006; 110:15700-7. doi: 10.1021/jp061667w [Crossref] [ Google Scholar]
- Krieg RC, Dong Y, Schwamborn K, Knuechel R. Protein quantification and its tolerance for different interfering reagents using the BCA-method with regard to 2D SDS PAGE. J Biochem Biophys Methods 2005; 65:13-9. doi: 10.1016/j.jbbm.2005.08.005 [Crossref] [ Google Scholar]
- Kumar D, Meenan BJ, Mutreja I, D'SA R, Dixon D. Controlling the size and size distribution of gold nanoparticles: a design of experiment study. Int J Nanosci 2012; 11:1250023. doi: 10.1142/S0219581X12500226 [Crossref] [ Google Scholar]
- Kim YP, Daniel WL, Xia Z, Xie H, Mirkin CA, Rao J. Bioluminescent nanosensors for protease detection based upon gold nanoparticle-luciferase conjugates. Chem Commun (Camb) 2010; 46:76-8. doi: 10.1039/b915612g [Crossref] [ Google Scholar]
- Kalnitsky G, Hummel JP, Resnick H, Carter J, Barnett LB, Dierks C. The relation of structure to enzymatic activity in ribonuclease. Ann N Y Acad Sci 1959; 81:542-66. doi: 10.1111/j.1749-6632.1959.tb49336.x [Crossref] [ Google Scholar]
-
Kumar P, Nagarajan A, Uchil PD. Analysis of Cell Viability by the MTT Assay. Cold Spring Harb Protoc 2018; 2018: pdb.prot095505. doi:10.1101/pdb.prot095505.
- Mandelkow R, Gumbel D, Ahrend H, Kaul A, Zimmermann U, Burchardt M. Detection and Quantification of Nuclear Morphology Changes in Apoptotic Cells by Fluorescence Microscopy and Subsequent Analysis of Visualized Fluorescent Signals. Anticancer Res 2017; 37:2239-44. doi: 10.21873/anticanres.11560 [Crossref] [ Google Scholar]
- Wlodkowic D, Skommer J, Darzynkiewicz Z. Flow cytometry-based apoptosis detection. Methods Mol Biol 2009; 559:19-32. doi: 10.1007/978-1-60327-017-5_2 [Crossref] [ Google Scholar]
- Karlsson M, Kurz T, Brunk UT, Nilsson SE, Frennesson CI. What does the commonly used DCF test for oxidative stress really show?. Biochem J 2010; 428:183-90. doi: 10.1042/bj20100208 [Crossref] [ Google Scholar]
- Eruslanov E, Kusmartsev S. Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol Biol 2010; 594:57-72. doi: 10.1007/978-1-60761-411-1_4 [Crossref] [ Google Scholar]
- Koopman G, Reutelingsperger C, Kuijten G, Keehnen R, Pals S, Van Oers M. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 1994; 84:1415-20. [ Google Scholar]
- Liu H, Liu X, Zhang C, Zhu H, Xu Q, Bu Y. Redox imbalance in the development of colorectal cancer. J Cancer 2017; 8:1586. doi: 10.7150/jca.18735 [Crossref] [ Google Scholar]
- Sreevalsan S, Safe S. Reactive oxygen species and colorectal cancer. Curr Colorectal Cancer Rep 2013; 9:350-7. doi: 10.1007/s11888-013-0190-5 [Crossref] [ Google Scholar]
- Koul M, Kumar A, Deshidi R, Sharma V, Singh RD, Singh J. Cladosporol A triggers apoptosis sensitivity by ROS-mediated autophagic flux in human breast cancer cells. BMC Cell Biol 2017; 18:26. doi: 10.1186/s12860-017-0141-0 [Crossref] [ Google Scholar]
- Wang M, Sun S, Neufeld CI, Perez‐Ramirez B, Xu Q. Reactive oxygen species‐responsive protein modification and its intracellular delivery for targeted cancer therapy. Angew Chem Int Ed Engl 2014; 53:13444-8. doi: 10.1002/anie.201407234 [Crossref] [ Google Scholar]
- Alexandre J, Hu Y, Lu W, Pelicano H, Huang P. Novel action of paclitaxel against cancer cells: bystander effect mediated by reactive oxygen species. Cancer res 2007; 67:3512-7. doi: 10.1158/0008-5472.CAN-06-3914 [Crossref] [ Google Scholar]
- Wang M, Sun S, Neufeld CI, Perez-Ramirez B, Xu Q. Reactive oxygen species-responsive protein modification and its intracellular delivery for targeted cancer therapy. Angew Chem Int Ed Engl 2014; 53:13444-8. doi: 10.1002/anie.201407234 [Crossref] [ Google Scholar]