Bioimpacts. 10(4):243-250.
doi: 10.34172/bi.2020.31
Original Research
Promoter methylation and expression pattern of DLX3, ATF4, and FRA1 genes during osteoblastic differentiation of adipose-derived mesenchymal stem cells
Sevda Rahimzadeh 1, 2  , Reza Rahbarghazi 1, Somayeh Aslani 2, Hadi Rajabi 2, Zeinab Latifi 2, Majid Farshdousti Hagh 3, Alireza Nourazarian 2, Hojjatollah Nozad Charoudeh 4, Mohammad Nouri 5, *
, Reza Rahbarghazi 1, Somayeh Aslani 2, Hadi Rajabi 2, Zeinab Latifi 2, Majid Farshdousti Hagh 3, Alireza Nourazarian 2, Hojjatollah Nozad Charoudeh 4, Mohammad Nouri 5, *  , Alireza Abhari 1, 2, *
, Alireza Abhari 1, 2, * 
Author information:
1Stem Cell Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
2Department of Biochemistry and Clinical Laboratories, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
3Department of Hematology, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
4Anatomical Sciences Department, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
5Department of Reproductive Biology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
Abstract
Introduction:
Nowadays, mesenchymal stem cells are touted as suitable cell supply for the restoration of injured bone tissue. The existence of osteogenic differentiation makes these cells capable of replenishing damaged cells in the least possible time. It has been shown that epigenetic modifications, especially DNA methylation, contribute to the regulation of various transcription factors during phenotype acquisition. Hence, we concentrated on the correlation between the promoter methylation and the expression of genes DLX3, ATF4 , and FRA1 during osteoblastic differentiation of adipose-derived mesenchymal stem cells in vitro after 21 days.
Methods:
Adipose-derived mesenchymal stem cells were cultured in osteogenesis differentiation medium supplemented with 0.1 µM dexamethasone, 10 mM β-glycerol phosphate, and 50 µM ascorbate-2-phosphate for 21 days. RNA and DNA extraction was done on days 0, 7, 14, and 21. Promoter methylation and expression levels of genes DLX3 , ATF4 , and FRA1 were analyzed by methylation-specific quantitative PCR and real-time PCR assays, respectively.
Results:
We found an upward expression trend with the increasing time for genes DLX3, ATF4, and FRA1 in stem cells committed to osteoblast-like lineage compared to the control group (P <0.05). On the contrary, methylation-specific quantitative PCR displayed decreased methylation rates of DLX3 and ATF4 genes, but not FRA1 , over time compared to the non-treated control cells (P <0.05). Bright-field images exhibited red-colored calcified deposits around Alizarin Red S-stained cells after 21 days compared to the control group. Statistical analysis showed a strong correlation between the transcription of genes DLX3 and ATF4 and methylation rate (P <0.05).
Conclusion:
In particular, osteoblastic differentiation of adipose-derived mesenchymal stem cells enhances DLX3 and ATF4 transcriptions by reducing methylation rate for 21 days.
Keywords: DNA methylation, Osteoblastic differentiation, Adipose-derived mesenchymal stem cells, DLX3, ATF4, FRA1
Copyright and License Information
© 2020 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Bone tissue is composed of a discrete calcified matrix with collagen fibers and different cell types such as osteoblasts, osteocytes, osteogenic cells, and osteoclasts.
1
Although bone tissue has the potency to restore and repair small-sized defects, critical large-sized bone defects require interventional approaches.
2
Grafting and metal implants are the gold standard methods for bone defects, but these modalities have numerous disadvantages such as implant rejection, tissue dysfunction, and microbial infections.
3
From the past to present, regenerative medicine and stem cell-based therapies have shown promising preclinical and clinical outcomes to accelerate and reconstitute injured tissues and organs.
3,4
Recently, mesenchymal stem cells (MSCs) account for a small fraction of mononuclear cells in different tissues and have been touted as a promising supply for cell-based therapies.
5-8
Due to the existence of multipotentiality and self-renewal bioactivity, it potentiates MSCs to give rise to various lineages such as osteoblasts, adipocytes, and chondrocytes.
9
It has been shown that lineage-specific differentiation potential of stem cells correlates with the expression of special transcription factors which are per se regulated by diverse extracellular stimuli such as hormones, cytokines and etc.
10,11
To induce osteogenic differentiation of MSCs, the critical role of several transcription factors has been previously determined.
12-14
For instance, Runx2 and Osterix are two osteoblastic specific transcription factors that regulate the differentiation of MSCs to osteoblasts.
14
Later on, other bone markers such distal‐less homeobox 3, 5, and 6 (DLX3, 5 , and6 ), activating transcription factor 4 (ATF4) , FRA1 , MSX1 , MSX2, and Twist participate in osteoblastic differentiation of MSCs.
15
Accumulating data support a notion that ATF4 is activated in the late stages of osteoblastic differentiation leads to the expression of osteogenic specific genes.
16
An orchestrated interactions of ATF4 and Runx2 with OSE2s (Runx2-binding sites) and OSE1 (ATF4-binding site) promotes the transcription of gene osteocalcin (OCN ).
15,16
The phosphorylation of DLX3 at serine 10 via protein kinase A (PKA) activity enhances BMP2-induced osteoblastic differentiation.
17
A highly orchestrated osteogenic differentiation follows by the activation of Fos-related antigen 1 (Fra-1) belongs to the activator protein-1 (AP-1) family.
18
RUNX2 can induce osteoblastic differentiation of C2C12 myogenic progenitor cells by positively targeting FRA-1.
19
Based on these findings, it seems that ATF4, DLX3, and FRA-1 genes are essential transcription factors during bone tissue development and regeneration. To our knowledge, few experiments have explored the dynamics ofATF4, DLX3, and FRA-1 genes in different stem cells committed to multiple lineages except osteoblast-like lineage. The activity of ATF4, DLX3, and FRA-1 genes during osteoblastic differentiation of MSCs is not fully addressed before. An intriguing question in the current experiment correlates with the determination of ATF4, DLX3, and FRA-1 genes promoter methylation and expression levels during osteoblastic differentiation of adipose-derived MSCs (ADSCs) after 21 days.
Materials and Methods
Cell expansion
Human ADSCs were obtained from Royan Institute (Tehran, Iran). ADSCs were re-suspended in low glucose content Dulbecco's Modified Eagle Medium (DMEM/LG; Gibco; Ireland) containing 10% fetal bovine serum (FBS; Gibco; Ireland) and 1% Pen-Strep (Gibco). Cells were maintained at 37°C with a humidified atmosphere and 5% CO2. To replenish the exhausted medium, cell supernatants were changed every 3-4 days. Cells at a confluence of 80% were detached by using 0.25% Trypsin- (1mM) EDTA solution (Gibco; Ireland). ADSCs at passages 3-6 were used in different analyses.
Osteoblast-like differentiation of ADSCs
For this propose, ADSCs were transferred into 6-well culture plates (SPL) and allowed to adhere and expand for 48 h. Thereafter, ADSCs were incubated with osteogenic induction medium containing10 mM β-glycerol phosphate (Cat no: G9891; Sigma-Aldrich, Germany), 50 µM ascorbate-2-phosphate (Sigma-Aldrich, Germany), and 0.1 µM dexamethasone (Sigma-Aldrich, Germany) for 21 days. The exhausted medium was exchanged every 3-4 days until the experimental procedure finished.
Alizarin Red S staining
To confirm the osteoblast differentiation of ADSCs, we performed Alizarin Red S (Sigma-Aldrich, Germany) staining to detect calcium deposition. For this propose, ADSCs were seeded at an initial density of 1 × 106 per well of 6-well plates and exposed to the osteogenic culture medium supplemented with 1-2% FBS. After the completion of the differentiation protocol, we stained the cells on days 0, 7, 14, and 21 with Alizarin Red S (2 g Alizarin Red S in 100 mL of distilled water; pH = 4.1-4.3). To stain the cells, the osteogenic medium was discarded and cells were washed with 1.5 ml DPBS and fixed with pre-cooled formalin solution (10%) for 20 minutes at room temperature. After twice washing with PBS, Alizarin Red S solution was poured to each well and kept for 30 minutes at room temperature. Finally, Alizarin Red S solution was discarded and the wells washed with PBS (2 times; each for 5 minutes).
Real-time PCR analysis
The expression of osteopontin (OPN ) and ATF4 , DLX3, and FRA1 was studied at time points 0, 7, 14, and 21 days to confirm the osteoblastic differentiation of ADSCs. For this propose, the RNA was extracted from differentiating ADSCs using the Trizol® Reagent (Cat no: T9424; Sigma-Aldrich) according to the manufacturer’s instruction. Isolated RNAs were qualified by 1% agarose electrophoresis and concentrations measured by using Nanodrop® (Model: ND-1000; Thermo Scientific). Then, RNAs were reverse-transcribed to cDNAs by using cDNA synthesis kit (Cat no: YT4500; Yekta Tajhiz Azma, Tehran, Iran). Real-time PCR was performed using the SYBR Green solution (batch no: 17D2701; Amplicon) and Mic qPCR Real-Time Cycle. The 2-ΔΔCT method was used to calculated relative gene expression. Primer was designated by the Primer Quest tool (version 2.2.3) and the sequence outlined in Table 1.
Table 1.
Primer list used for real-time PCR analysis
|
Gene
|
|
Primer sequences
|
TM (°C)
|
Product size
|
|
ATF4
|
F |
TTCTCCAGCGACAAGGCTAAGG |
61.5 |
122 |
| R |
CTCCAACATCCAATCTGTCCCG |
60.5 |
122 |
|
DLX3
|
F |
AAGCCCAAGAAGGTCCGAAA |
60.80 |
107 |
| R |
TTTCACCTGTGTCTGCGTGA |
60.90 |
107 |
|
FRA1
|
F |
GGAGGAAGGAACTGACCGACTT |
59 |
127 |
| R |
CTCTAGGCGGTCCTTCTGCTTC |
60 |
127 |
|
OPN
|
F |
CGAGGTGATAGTGTGGTTTATGG |
60 |
120 |
| R |
CACCATTCAACTCCTCGCTTTC |
60 |
120 |
DNA extraction and sodium bisulfite treatment
On days 0, 7, 14, and 21, nuclear DNA was extracted from ADSCs. Cells were collected and incubated with white blood cells lysis buffer (0.5 M EDTA, 1 M Tris-base, and 75 mM NaCl) at 56˚C for 1 hour followed by treatment with proteinase K at 56˚C for 30 minutes. After the addition of chloroform, the samples were centrifuged at 12000 rpm for 20 minutes and supernatants transferred into tubes containing EtOH (Merck). Then, the samples were mixed gently and stored at -20˚C for 15 minutes. Finally, samples were air-dried and double distilled water added to each tube. The integrity of isolated DNA was evaluated by a NanoDrop® system (Model: ND-1000; Thermo Scientific). To measure methylation rate, unmethylated cytosine residues of DNA were converted to uracil residues with sodium bisulfite by using EZ DNA Methylation-Gold Kit–Zymo Research (Cat no: D5006). To provide positive control, we used peripheral blood DNA samples and methylated by CpG-specific methylase from Spiroplasma namely SSS1 methylase (Biolabs; USA) as previously described. For negative control, we used EpiTect Control DNA (Cat no: 59655; Qiagen).
Methylation-specific quantitative PCR (MS-qPCR)
DNA methylation of ATF4 , DLX3, and FRA1 was analyzed using MS-qPCR on days 0, 7, 14, and 21. Two sets of primers for the detection of methylated and unmethylated genes ATF4 , DLX3, and FRA1 are designated using Gene Runner software and MethPrimer website (Table 2). For MS-qPCR analysis, 7.5 µl SYBR Green reagent (Amplicon Master Mix) was mixed with 2 μL of each primer, 4.5 μL of DDW MS-qPCR and 1 μL DNA samples template. The percentage of ATF4 , DLX3 and FRA1 methylation was estimated using the formula as follows: 100/1 + (efficiency MetCt(Met))/(efficiency UnmetCt(unmet)). We considered index 2 for efficiency Met and Unmet.
Table 2.
Primer list used for MS-qPCR assay
|
Gene
|
|
|
Primer sequence
|
TM ( °C)
|
Product size
|
|
ATF4
|
Methylated |
F |
5’-ATCGGGAAAGCGTAGTCG-3’ |
60.6 |
203 |
| R |
5’-CAAATACGACCAAAACGACCG-3’ |
61.6 |
203 |
| Unmethylated |
F |
5’-GATTGGGAAAGTGTAGTTGGG-3’ |
60.1 |
206 |
| R |
5’-TCCAAATACAACCAAAACAACCA-3’ |
60.8 |
206 |
|
DLX3
|
Methylated |
F |
5’-GTAATGGTGTAAGCGTTTTTCG-3’ |
60 |
177 |
| R |
5’-ACCACTCATCCTAACGAACG-3’ |
60.9 |
177 |
| Unmethylated |
F |
5’-GGTAATGGTGTAAGTGTTTTTTGG-3’ |
60.2 |
179 |
| R |
5’-AACCACTCATCCTAACAAACACT-3’ |
61.4 |
179 |
|
FRA1
|
Methylated |
F |
5’-TTGGTAGGTGCGTTAGTTCG-3’ |
61.5 |
111 |
| R |
5’-CCCGAAAACTACGAACCCG-3’ |
62.2 |
111 |
| Unmethylated |
F |
5’-TTTTGGTAGGTGTGTTAGTTTGTAG-3’ |
60.7 |
114 |
| R |
5’-CCCCAAAAACTACAAACCCAC-3’ |
61.1 |
114 |
Statistical analysis
Data are presented as mean ± SD. One-way ANOVA analysis with Tukey post hoc test was used to find statistical differences between groups by using Graph Pad Prism software version 6.07. P value < 0.05 was considered statistically significant in this study.
Results
Confirmation of osteoblastic differentiation
ADSCs were incubated with the osteogenic differentiation medium for 0, 7, 14, and 21 days. We performed Alizarin Red S staining to detect calcium mineral deposition in extracellular space (Fig. 1A). According to our results, red-color calcium deposits were observed in differentiating ADSCs 7 days after incubation with osteogenic medium and these features reached the maximum levels after 21 days, confirming successful osteoblast-like differentiation of ADSCs while no calcium mineral deposition was found in the control group (Fig. 1A). We also monitored the transcription of the osteoblast-specific marker, OPN to assess the osteogenic potential of ADSCs (Fig. 1B). Based on the data from real-time PCR analysis, OPN was up-regulated in ADSCs committed to osteoblast-like cells. According to our data, the expression of OPN was increased by time and reached at day 21. Compared to cells from various time points, the control ADSCs did not express OPN expanded in normal culture medium (Fig. 1B). These data demonstrated that ADSCs had the potential to give rise to osteoblast-like cells after being exposed to osteogenic medium over 21 days.
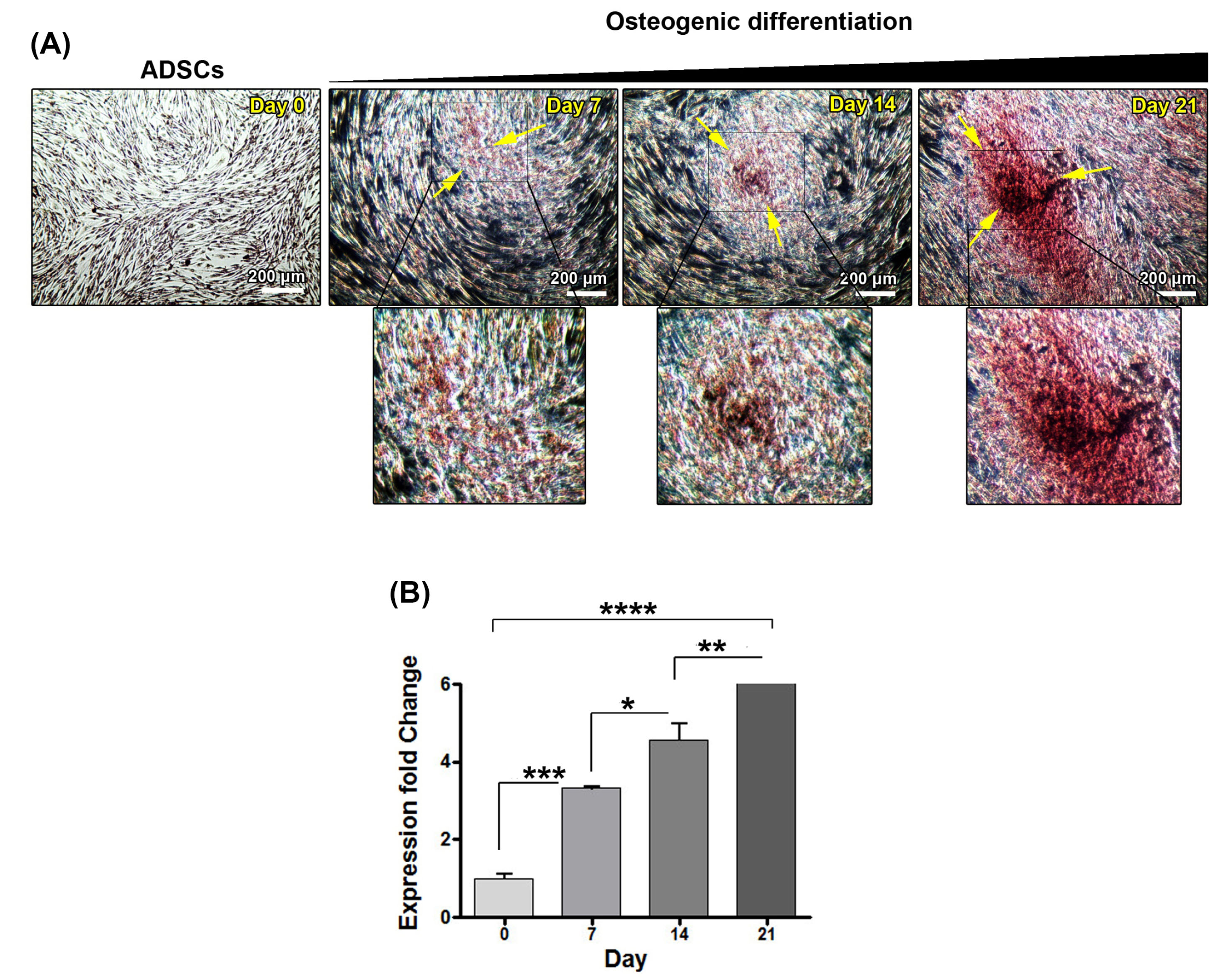
Fig. 1.
Confirmation of osteoblastic differentiation evaluating Alizarin Red S staining (A) and osteopontin gene (B) expressions Adipose-derived mesenchymal stem cells were imaged during osteoblastic differentiation on days 0, 7, 14 and 21 by an inverted microscope. The cells were stained at respective time points by Alizarin Red S solution. By the time the level of red-colored deposition increased, indicating the calcium deposits at extracellular space. These changes were evident on day 21 (A). Osteopontin (OPN ) gene expression levels have been shown on days 0, 7, 14, and 21 during osteoblastic differentiation(B). The expression of this gene was increased by time and reached a maximum level on day 21. *P <0.05; **P <0.01; ***P <0.001; ****P <0.0001.
.
Confirmation of osteoblastic differentiation evaluating Alizarin Red S staining (A) and osteopontin gene (B) expressions Adipose-derived mesenchymal stem cells were imaged during osteoblastic differentiation on days 0, 7, 14 and 21 by an inverted microscope. The cells were stained at respective time points by Alizarin Red S solution. By the time the level of red-colored deposition increased, indicating the calcium deposits at extracellular space. These changes were evident on day 21 (A). Osteopontin (OPN ) gene expression levels have been shown on days 0, 7, 14, and 21 during osteoblastic differentiation(B). The expression of this gene was increased by time and reached a maximum level on day 21. *P <0.05; **P <0.01; ***P <0.001; ****P <0.0001.
Methylation of ATF4, DLX3, but not FRA1, genes was decreased during osteogenesis
Considering the role of epigenetics in osteogenic differentiation of ADSCs, we examined whether promoter methylation of ATF4 , DLX3, and FRA1 could be varied during differentiation. Based on our results, the rate of methylation in the promoter region of ATF4 and DLX3 was increased by time in ADSCs cultured in osteogenic medium (Fig. 2). However, we found an irregular pattern related to methylation in the promoter region of FRA1 in ADSCs exposed to osteogenic differentiation medium (Fig. 2). These data added a notion that during osteogenic differentiation of ADSCs, the methylation in the promoter region of some genes such as ATF4 and DLX3 was modulated in ADSCs after being-exposed to osteogenic medium over 21 days.
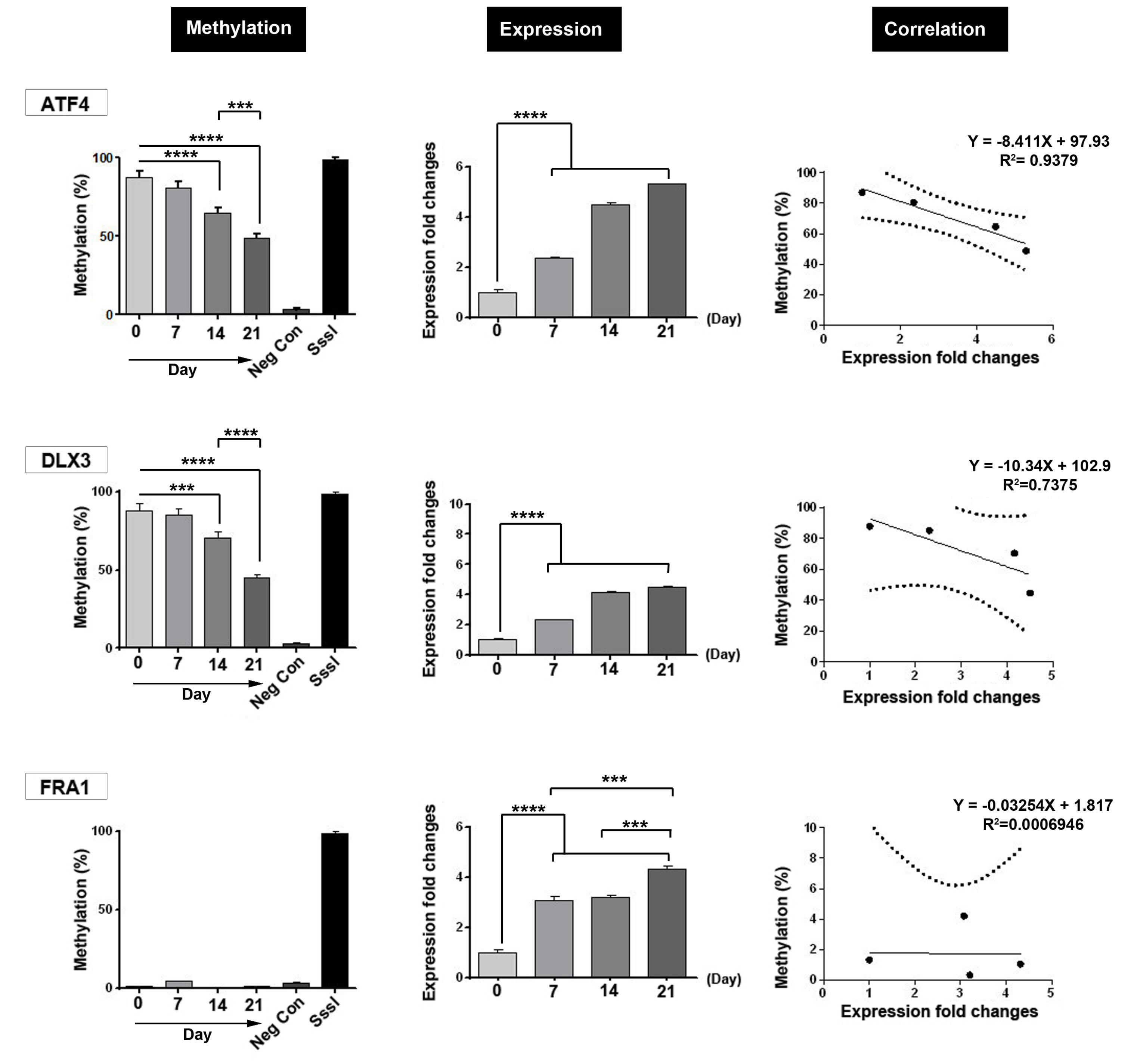
Fig. 2.
Representative image of DNA methylation, mRNA level and the correlation between methylation and expression for ATF4 , DLX3 and FRA1 genes during osteoblastic differentiation of ADSCs on 0, 7, 14 and 21 days. A significant increase in the mRNA level of all three genes was observed during MSCs differentiation. The amount of DNA methylation of both genes ATF4 and DLX3 was significantly reduced, but no significant changes were observed in the DNA methylation level of FRA1 . Also, correlation analysis revealed a strong relationship between ATF4 and DLX3 gene expression and DNA methylation. SssI: CpG-specific methylase from Spiroplasma ***P <0.001; ****P <0.0001.
.
Representative image of DNA methylation, mRNA level and the correlation between methylation and expression for ATF4 , DLX3 and FRA1 genes during osteoblastic differentiation of ADSCs on 0, 7, 14 and 21 days. A significant increase in the mRNA level of all three genes was observed during MSCs differentiation. The amount of DNA methylation of both genes ATF4 and DLX3 was significantly reduced, but no significant changes were observed in the DNA methylation level of FRA1 . Also, correlation analysis revealed a strong relationship between ATF4 and DLX3 gene expression and DNA methylation. SssI: CpG-specific methylase from Spiroplasma ***P <0.001; ****P <0.0001.
Expression of ATF4, DLX3, and FRA1 was induced in ADSCs during osteogenesis
Levels of ATF4 , DLX3, and FRA1 mRNA were examined on days 0, 7, 14, and 21. During osteoblasts differentiation of MSCs, the transcription levels of ATF4 , DLX3, and FRA1 were semi-quantified by real-time PCR assay. Based on data from this panel, a significant increase in mRNA expression of ATF4 , DLX3, and FRA1 was shown by time and these values reached a maximum level at the end of the experiment. We found a significant difference in the transcription level of three genes on days 14 and 21 compared to the initial time point (day 0; P < 0.05). These results showed a possible critical role of ATF4 , DLX3, and FRA1 during osteoblast differentiation of ADSCs. Based on the data from this panel, we found a strong correlation between transcription level and methylation in the promoter region of genes ATF4 (P < 0.05; R2= 0.9379) and DLX3 (R2= 0.7375, P > 0.05). We found a non-significant relationship between methylation in the promoter region and the expression of FRA1 (P > 0.05; R2 ≃ 0).
Discussion
Technologies for the restoration of bone diseases in the clinical setting would be a basic requirement in human medicine. The osteogenic potential of various stem cell types, especially ADSCs, offer hope for healing of injured bone tissue.
15
Here, we aimed to monitor the expression and promoter methylation rate of ATF4 , DLX3, and FRA1 during ADSCs differentiation into osteoblast-like cells. Alizarin Red S in vitro staining confirmed the efficiency of the current protocol to induce of ADSCs differentiation into osteoblast-like cells after 21 days. By applying the osteogenic differentiation medium, we noted the novel phenotype acquisition coincided with the formation of red-colored calcium mineral deposits required for bone matrix construction. Along with these changes, the expression of OPN was simultaneously induced and reached a maximum on day 21. Although as yet not well understood, the osteogenic potential of stem cells coincides with the changes in the expression of genes such as OCN , OPN , OSN, and formation of calcium hydroxyapatite.
20,21
Our data confirmed that ADSCs' exposure to osteogenic differentiation medium led to the up-regulation of genes ATF4 , DLX3, and FRA1 .
22-24
It has been shown that ATF4 participates in osteoblast survival and proliferation at both early and late phases of osteogenesis by modulating specific gene transcription via RSK2-dependent activity and promotion of DDR2-mediated p38 MAPK signaling pathway.
16,25
In support of ATF4 role on bone function, Yang and colleagues found that the suppression of ATF4 reduced bone formation capacity by the inhibition of OCN in mice model. All these changes contributed to decreased bone mineral density and deposition of the extracellular matrix.
16
Interestingly, it is well-established that the expression of ATF4 could promote osteogenic capacity in non-osteoblastic lineages.
26
These data show the unique role of ATF4 in ADSCs undergone a phenotypic trans-differentiation to osteoblast-like phenotype. However, it is yet vague that what extent ATF4 participates in the direct orientation of ADSCs toward osteoblast lineage.
DLX3 plays a crucial role in the osteogenic orientation of progenitor cells during the development of the embryo.
27
The activity of DLX3 has been shown in the placenta, periosteum, osteoblasts, and chondrocytes and cells differentiating toward osteoblasts.
27,28
Remarkably, DLX3 transcription factor controls the co-expression of OCN and RUNX2 with a particular activity on the promoter regions.
29,30
Further molecular assessments revealed that the suppression of DLX3 contributed to mouse pup death even prior to bone development.
31
Similar to finding described by Choi and co-workers, they found that DLX3 ablation accelerated differentiation of multipotent mesenchymal C2C12 mouse myoblasts to osteoblast lineage at late stages.
32
However, this group did not describe an underlying mechanism related to the activity of DLX3 on osteoblastic differentiation. This hypocrisy may be explained by the fact that DLX3 activity is solely associated with the stem cell type undergo differentiation into osteoblast-like cells at different time points.
Fos-related protein (Fra-1) is a member of the AP-1 transcription factor family. Previous experiments revealed thatFra-1 expressing transgenic mice developed osteosclerosis characterized by high bone mass coincided with enhanced osteoblastic differentiation and bone formation.
33
In support of this claim,Fra-1 knockout (fra1-) mice developed osteopenia, a disorder with low bone mass. Also, it was observed that the expression of bone matrix components including osteocalcin and matrix Gla protein decreased in these mice.
34
Over-expression of Fra-1 in adipose-derived stromal cells inhibited osteoarthritis in mice.
35
Similar to the expression of the above-mentioned genes, we found an increased expression of FRA1 . These data demonstrated that the expression of all genes including ATF4 , DLX3, and FRA1 was increased during the osteogenic differentiation of ADSCs. As above-mentioned, changes in the expression of these factors are integral to the trans-differentiation of MSCs. The methylation of DNA, changes in nucleosome histone tails and the remodeling of a chromosome are epigenetic changes, which are crucial in cellular genomic profile expression during differentiation to different lineages. Recent data in the field of embryonic stem cell differentiation revealed an inevitable role for dynamic epigenetic regulation.
36
In this regard, Aranda et al stated that the differentiation potential of SCs correlated with epigenetic status changes.
37
Among these modulations, the changes related to DNA methylation is the most dominant procedure. DNA methylation includes the addition of a methyl group to the 5 cytosines in a CpG dinucleotide, altering genomic integrity, and modulating the various genes. These changes may relate to gene silencing.
38
A recently published data demonstrated that embryonic stem cells differentiating to neuronal lineage showed the CpG methylation at a distinct locus.
39,40
In MSCs, DNA methylation of Trip10 promoter had the potential to accelerate the neuronal and osteoblast differentiation of MSCs.
41
The analysis of promoter methylation revealed different patterns for FRA1 , DLX3, and ATF4 genes. FRA1 promoter remained in the hypomethylated form pre- and post-osteoblastic differentiation procedure. On the other hand, ATF4 andDLX3 promoter region showed the decrease of DNA methylation by time which reached a minimum level on day 21. Correlation of DNA methylation and gene expression changes of ATF4 andDLX3 genes demonstrated a strong relationship. In a better word, osteogenic differentiation promoted the expression of these genes while decreased the methylation rate. Previous works showed that hypermethylation could decrease the transcription and bioactivity of a distinct gene.
Conclusion
Our data added a notion that continuous incubation of ADSCs with osteogenesis medium decreased the net promoter methylation rate of ATF4 andDLX3 genes which could possibly control the expression of these genes. These data showed that monitoring the methylation and expression of specific genes during stem cell differentiation into target cell types could enable us to efficiently control differentiation and orientation rates.
Acknowledgments
Authors thank the personnel of Stem Cell Research Center and Department of Biochemistry and Clinical laboratories affiliated to Tabriz University of Medical Sciences.
Funding sources
This study was supported by a grant (IR.TBZMED.REC.1395.1344) from Tabriz University of Medical Sciences.
Ethical statement
Not applicable.
Competing interests
There is no conflict of interests.
Authors’ contribution
Conceptualization: AB, MFH; Experiments design: RR, MN; Data analysis: SR, ZL; Provision of study materials and equipment: AB, AN, HNC; Study validation: RR; Supervision: AB, MFH; Data presentation: SR, SA; Draft preparation: HR; Manuscript writing and edition: SR, RR.
Research Highlights
What is the current knowledge?
simple
-
√ Mesenchymal stem cells have great potential in the
restoration of bone-related injuries.
-
√ Promoter methylation and expression of specific genes
promote lineage-specific differentiation of stem cells.
What is new here?
simple
-
√ Promoter methylation of special transcription factors
(DLX3, ATF4, and FRA1 genes) induces osteoblastic
differentiation of adipose-derived MSCs.
References
- Han Y, You X, Xing W, Zhang Z, Zou W. Paracrine and endocrine actions of bone—the functions of secretory proteins from osteoblasts, osteocytes, and osteoclasts. Bone Res 2018; 6:16. doi: 10.1038/s41413-018-0019-6 [Crossref] [ Google Scholar]
- Ozdemir MT, Kir MÇ. Repair of long bone defects with demineralized bone matrix and autogenous bone composite. Indian J Orthop 2011; 45:226-30. doi: 10.4103/0019-5413.80040 [Crossref] [ Google Scholar]
- Fernandez de Grado G, Keller L, Idoux-Gillet Y, Wagner Q, Musset A-M, Benkirane-Jessel N. Bone substitutes: a review of their characteristics, clinical use, and perspectives for large bone defects management. J Tissue Eng 2018; 9:2041731418776819. doi: 10.1177/2041731418776819 [Crossref] [ Google Scholar]
- Gorjipour F, Hosseini-Gohari L, Alizadeh Ghavidel A, Hajimiresmaiel SJ, Naderi N, Darbandi Azar A. Mesenchymal stem cells from human amniotic membrane differentiate into cardiomyocytes and endothelial-like cells without improving cardiac function after surgical administration in rat model of chronic heart failure. J Cardiovasc Thorac Res 2019; 11:35-42. doi: 10.15171/jcvtr.2019.06 [Crossref] [ Google Scholar]
- Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D. Minimal criteria for defining multipotent mesenchymal stromal cells The International Society for Cellular Therapy position statement. Cytotherapy 2006; 8:315-7. doi: 10.1080/14653240600855905 [Crossref] [ Google Scholar]
- Barrett AN, Fong C-Y, Subramanian A, Liu W, Feng Y, Choolani M. Human Wharton's Jelly Mesenchymal Stem Cells Show Unique Gene Expression Compared with Bone Marrow Mesenchymal Stem Cells Using Single-Cell RNA-Sequencing. Stem Cells Dev 2019; 28:196-211. doi: 10.1089/scd.2018.013 [Crossref] [ Google Scholar]
- Keyhanmanesh R, Rahbarghazi R, Aslani MR, Hassanpour M, Ahmadi M. Systemic delivery of mesenchymal stem cells condition media in repeated doses acts as magic bullets in restoring IFN-γ/IL-4 balance in asthmatic rats. Life Sci 2018; 212:30-6. doi: 10.1016/j.lfs.2018.09.049 [Crossref] [ Google Scholar]
- Keyhanmanesh R, Rahbarghazi R, Ahmadi M. Systemic transplantation of mesenchymal stem cells modulates endothelial cell adhesion molecules induced by ovalbumin in rat model of asthma. Inflammation 2018; 41:2236-45. doi: 10.1007/s10753-018-0866-8 [Crossref] [ Google Scholar]
- Rahbarghazi R, Nassiri SM, Ahmadi SH, Mohammadi E, Rabbani S, Araghi A. Dynamic induction of pro-angiogenic milieu after transplantation of marrow-derived mesenchymal stem cells in experimental myocardial infarction. Int J Cardiol 2014; 173:453-66. doi: 10.1016/j.ijcard.2014.03.008 [Crossref] [ Google Scholar]
- Bock C, Beerman I, Lien W-H, Smith ZD, Gu H, Boyle P. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell 2012; 47:633-47. doi: 10.1016/j.molcel.2012.06.019 [Crossref] [ Google Scholar]
- Zamani ARN, Saberianpour S, Geranmayeh MH, Bani F, Haghighi L, Rahbarghazi R. Modulatory effect of photobiomodulation on stem cell epigenetic memory: a highlight on differentiation capacity. Lasers Med Sci 2019; 35(2):299-306. doi: 10.1007/s10103-019-02873-7 [Crossref] [ Google Scholar]
- Zaher W, Harkness L, Jafari A, Kassem M. An update of human mesenchymal stem cell biology and their clinical uses. Arch Toxicol 2014; 88:1069-82. doi: 10.1007/s00204-014-1232-8 [Crossref] [ Google Scholar]
- Cook D, Genever P. Regulation of mesenchymal stem cell differentiation. Adv Exp Med Biol 2013; 786:213-29. doi: 10.1007/978-94-007-6621-1_12 [Crossref] [ Google Scholar]
- Hanna H, Mir LM, Andre FM. In vitro osteoblastic differentiation of mesenchymal stem cells generates cell layers with distinct properties. Stem Cell Res Ther 2018; 9:203. doi: 10.1186/s13287-018-0942-x [Crossref] [ Google Scholar]
- Goto N, Fujimoto K, Fujii S, Ida-Yonemochi H, Ohshima H, Kawamoto T. Role of MSX1 in Osteogenic Differentiation of Human Dental Pulp Stem Cells. Stem Cell Int 2016; 2016:8035759. doi: 10.1155/2016/8035759 [Crossref] [ Google Scholar]
- Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell 2004; 117:387-98. doi: 10.1016/s0092-8674(04)00344-7 [Crossref] [ Google Scholar]
- Li H, Jeong HM, Choi YH, Kim JH, Choi JK, Yeo CY. Protein kinase a phosphorylates Dlx3 and regulates the function of Dlx3 during osteoblast differentiation. J Cell Biochem 2014; 115:2004-11. doi: 10.1002/jcb.24872 [Crossref] [ Google Scholar]
- Viale-Bouroncle S, Klingelhöffer C, Ettl T, Reichert TE, Morsczeck C. A protein kinase A (PKA)/β-catenin pathway sustains the BMP2/DLX3-induced osteogenic differentiation in dental follicle cells (DFCs). Cell Signal 2015; 27:598-605. doi: 10.1016/j.cellsig.2014.12.008 [Crossref] [ Google Scholar]
- Yu S, Geng Q, Sun F, Yu Y, Pan Q, Hong A. Osteogenic differentiation of C2C12 myogenic progenitor cells requires the Fos-related antigen Fra-1 - a novel target of Runx2. Biochem Biophys Res Commun 2013; 430:173-8. doi: 10.1016/j.bbrc.2012.11.033 [Crossref] [ Google Scholar]
- Kang ES, Kim DS, Han Y, Son H, Chung YH, Min J. Three-Dimensional Graphene-RGD Peptide Nanoisland Composites That Enhance the Osteogenesis of Human Adipose-Derived Mesenchymal Stem Cells. Int J Mol Sci 2018; 19:669. doi: 10.3390/ijms19030669 [Crossref] [ Google Scholar]
- Otsuki Y, Ii M, Moriwaki K, Okada M, Ueda K, Asahi M. W9 peptide enhanced osteogenic differentiation of human adipose-derived stem cells. Biochem Biophys Res Commun 2018; 495:904-10. doi: 10.1016/j.bbrc.2017.11.056 [Crossref] [ Google Scholar]
- Chen H, Yuan R, Zhang Y, Zhang X, Chen L, Zhou X. ATF4 regulates SREBP1c expression to control fatty acids synthesis in 3T3-L1 adipocytes differentiation. Biochim Biophys Acta 2016; 1859:1459-69. doi: 10.1016/j.bbagrm.2016.07.010 [Crossref] [ Google Scholar]
- Hwang J, Mehrani T, Millar SE, Morasso MI. Dlx3 is a crucial regulator of hair follicle differentiation and cycling. Development 2008; 135:3149-59. doi: 10.1242/dev.022202 [Crossref] [ Google Scholar]
- Matsui M, Tokuhara M, Konuma Y, Nomura N, Ishizaki R. Isolation of human fos-related genes and their expression during monocyte-macrophage differentiation. Oncogene 1990; 5:249-55. [ Google Scholar]
- Lin KL, Chou CH, Hsieh SC, Hwa SY, Lee MT, Wang FF. Transcriptional upregulation of DDR2 by ATF4 facilitates osteoblastic differentiation through p38 MAPK-mediated Runx2 activation. J Bone Miner Res 2010; 25:2489-503. doi: 10.1002/jbmr.159 [Crossref] [ Google Scholar]
- Yang X, Karsenty G. ATF4, the osteoblast accumulation of which is determined post-translationally, can induce osteoblast-specific gene expression in non-osteoblastic cells. J Biol Chem 2004; 279:47109-14. doi: 10.1074/jbc.M410010200 [Crossref] [ Google Scholar]
- Viale-Bouroncle S, Felthaus O, Schmalz G, Brockhoff G, Reichert TE, Morsczeck C. The transcription factor DLX3 regulates the osteogenic differentiation of human dental follicle precursor cells. Stem Cells Dev 2012; 21:1936-47. doi: 10.1089/scd.2011.0422 [Crossref] [ Google Scholar]
- Yang G, Yuan G, Li X, Liu P, Chen Z, Fan M. BMP-2 induction of Dlx3 expression is mediated by p38/Smad5 signaling pathway in osteoblastic MC3T3-E1 cells. J Cell Physiol 2014; 229:943-54. doi: 10.1002/jcp.24525 [Crossref] [ Google Scholar]
- Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol Cell Biol 2004; 24:9248-61. doi: 10.1128/mcb.24.20.9248-9261.2004 [Crossref] [ Google Scholar]
- Shirakabe K, Terasawa K, Miyama K, Shibuya H, Nishida E. Regulation of the activity of the transcription factor Runx2 by two homeobox proteins, Msx2 and Dlx5. Genes Cells 2001; 6:851-6. doi: 10.1046/j.1365-2443.2001.00466.x [Crossref] [ Google Scholar]
- Morasso MI, Grinberg A, Robinson G, Sargent TD, Mahon KA. Placental failure in mice lacking the homeobox gene Dlx3. Proc Natl Acad Sci U S A 1999; 96:162-7. doi: 10.1073/pnas.96.1.162 [Crossref] [ Google Scholar]
- Choi SJ, Song IS, Ryu OH, Choi SW, Hart PS, Wu WW. A 4 bp deletion mutation in DLX3 enhances osteoblastic differentiation and bone formation in vitro. Bone 2008; 42:162-71. doi: 10.1016/j.bone.2007.08.047 [Crossref] [ Google Scholar]
- Jochum W, David JP, Elliott C, Wutz A, Plenk H, Jr Jr, Matsuo K. Increased bone formation and osteosclerosis in mice overexpressing the transcription factor Fra-1. Nat Med 2000; 6:980-4. doi: 10.1038/79676 [Crossref] [ Google Scholar]
- Eferl R, Hoebertz A, Schilling AF, Rath M, Karreth F, Kenner L. The Fos-related antigen Fra-1 is an activator of bone matrix formation. EMBO J 2004; 23:2789-99. doi: 10.1038/sj.emboj.7600282 [Crossref] [ Google Scholar]
- Schwabe K, Garcia M, Ubieta K, Hannemann N, Herbort B, Luther J. Inhibition of Osteoarthritis by Adipose-Derived Stromal Cells Overexpressing Fra-1 in Mice. Arthritis Rheumatol 2016; 68:138-51. doi: 10.1002/art.39425 [Crossref] [ Google Scholar]
- Ozkul Y, Galderisi U. The impact of epigenetics on mesenchymal stem cell biology. J Cell Physiol 2016; 231:2393-401. doi: 10.1002/jcp.25371 [Crossref] [ Google Scholar]
- Aranda P, Agirre X, Ballestar E, Andreu EJ, Roman-Gomez J, Prieto I. Epigenetic signatures associated with different levels of differentiation potential in human stem cells. PLoS One 2009; 4:e7809. doi: 10.1371/journal.pone.0007809 [Crossref] [ Google Scholar]
- Perez-Campo FM, Riancho JA. Epigenetic mechanisms regulating mesenchymal stem cell differentiation. Curr Genomics 2015; 16:368-83. doi: 10.2174/1389202916666150817202559 [Crossref] [ Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008; 454:766-70. doi: 10.1038/nature07107 [Crossref] [ Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 2008; 30:755-66. doi: 10.1016/j.molcel.2008.05.007 [Crossref] [ Google Scholar]
- Hsiao SH, Lee KD, Hsu CC, Tseng MJ, Jin VX, Sun WS. DNA methylation of the Trip10 promoter accelerates mesenchymal stem cell lineage determination. Biochem Biophys Res Commun 2010; 400:305-12. doi: 10.1016/j.bbrc.2010.08.048 [Crossref] [ Google Scholar]