Bioimpacts. 11(1):53-57.
doi: 10.34172/bi.2021.07
Original Research
Considerable increase in Poly(3-hydroxybutyrate) production via phbC gene overexpression in Ralstonia eutropha PTCC 1615
Farzaneh Barati 1  , Ezat Asgarani 1, *
, Ezat Asgarani 1, *  , Sara Gharavi 1, Mohammad Reza Soudi 2
, Sara Gharavi 1, Mohammad Reza Soudi 2
Author information:
1Department of Biotechnology, Faculty of Biological Sciences, Alzahra University, Tehran, Iran
22 Department of Microbiology, Faculty of Biological Sciences, Alzahra University, Tehran, Iran
Abstract
Introduction:
Poly(3-hydroxybutyrate) (PHB) is a well-known biodegradable polymer produced by some microorganisms and can be a suitable alternative for petrochemical plastics. PHB synthase encoded by phb C gene is the main enzyme in PHB biosynthesis pathway in Ralstonia eutropha. The aim of current study was the transformation of R. eutropha PTCC 1615 with its own phb C gene and evaluation of the overexpression effect on PHB accumulation.
Methods:
DNA fragment including phbC gene and its promoter and terminator regions, was isolated from R. eutropha PTCC 1615, inserted into pET28a(+) vector, and transferred to the competent bacteria using calcium chloride and heat shock method. The effect of the cloned gene expression on PHB production was investigated with absorption of crotonic acid produced through PHB dehydration. Statistical analyses were carried out by SPSS software.
Results:
PHB content of cells of the engineered strain was 1.4 times more than that of the native bacteria. This significant difference can be an important finding for improvement of biopolymer production.
Conclusion:
Overexpression of phb C, the critical gene in PHB biosynthesis pathway, in R. eutropha PTCC 1615 had considerable effect on PHB accumulation.
Keywords: Ralstonia eutropha, Poly(3-hydroxybutyrate), PHB synthase, Transformation
Copyright and License Information
© 2021 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/
). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Today, there is a wide range of applications for synthetic plastics. However due to their resistance against natural destruction, nonrenewable sources, and toxic substances generated during their long run degradation in nature, they have become a great environmental problem.
1
Much attention has been paid to bioplastics which have sustainable sources and environmentally friendly elimination.
2
Bioplastics are polymers which are produced as storage materials by different organisms. An important group of biopolymers is called polyhydroxyalkanoate (PHA) that accumulates in some microorganisms as an energy reservoir. High molecular weight of the natural polyesters in range of 50-1000 kDa has led to properties like chemically synthesized plastics and so resulted in many uses.
3
Poly(3-hydroxybutyrate) (PHB) is a well-known member of PHA family. Ralstoniaeutropha (also Alcaligenes eutrophus or Cupriavidus necator) is a model organism for PHB production.
4
Three enzymes conduct the polymer biosynthesis pathway in this bacterium. First step of the pathway is catalyzed by β-ketothiolase which combines two acetyl-CoA molecules to form acetoacetyl-CoA. Reduction of acetoacetyl-CoA to (R)-3-hydroxybutyryl-CoA is the next step which is carried out by acetoacetyl-CoA reductase. Finally, polymerization of (R)-3-hydroxybutyryl-CoA monomers as the last step occurs in the presence of PHB synthase. These enzymes are encoded by phbA, phbB and phbC genes, respectively.
5,6
Some PHAs such as PHB are produced commercially, but because of high production costs, they cannot yet compete with synthetic plastics. To decrease expenses, various strategies have been employed such as DNA manipulation or utilization of cheap carbon sources.
7
For instance, in some studies, overexpression of phbC gene and PHB synthase engineering, have led to significant increase in PHB production. These efforts have showed that PHB synthase is the most critical enzyme in the polymer biosynthesis.
8-10
In the present work, the effect of phbC gene overexpression on PHB production was investigated in R. eutropha PTCC 1615. For this purpose, precise and cost-effective methods were employed.
Materials and Methods
Bacterial strains, culture condition, and media
Ralstoniaeutropha PTCC 1615 and Escherichia coli DH5α were utilized as host strain and source of pET28a (+) vector, respectively. These strains were provided by Iranian Research Organization for Science and Technology. LB medium was, used for cultivation of these bacteria, contained 10 g.L-1, bactopeptone; 5 g.L-1, yeast extract and 5 g.L-1, NaCl and pH was adjusted at 7.
8
LB medium was supplemented with 50 μg.mL-1 kanamycin for E. coli DH5α. Incubation temperature was 30˚C for R. eutropha PTCC 1615 and 37˚C for E. coli DH5α, and liquid cultures placed in a shaker incubator at 150 rpm for both.
6,8
Isolation of target DNA fragment
Primer design
Cloning primers were designed with Gene runner and Oligo7 software, using genome sequence of R. eutropha H16 (accession number: NC_008313.1) available in nucleotide database of NCBI after which they were evaluated by NCBI Primer Blast. Primers were designed to include upstream and downstream transcriptional regulatory sequences of phbC gene in the amplified DNA fragment. Regions upstream of phbC gene in the form of GG-10N-GC, are promoter sequences which are recognized by a protein that is homologous of σ54 in E. coli.
11
Terminator sequence of phbC gene was found from WebGeSTer, the largest transcription terminator database (http://pallab.serc.iisc.ernet.in/gester).
12
BamHI and HindIII restriction enzyme recognition sites were added to 5’ end of forward and reverse cloning primers, respectively. Moreover, a toehold with a length of 4 bp was placed at the 5’ end of each cloning primer just behind restriction sites in order to optimize enzyme functions. Hence, the resulting amplified fragment included phbC gene, as well as 308 bp upstream and 92 bp downstream of the gene. To obtain complete sequence of the purpose segment, internal primers were designed and evaluated using software and databases similar to cloning primers. Table 1 displays cloning and internal primer sequences.
Table 1.
The sequences of cloning and internal primers
|
Primers
|
|
Sequence (5’-3’)
|
| Cloning primers |
Forward |
TAAT
G
▼
GATCC TCAAATGTCTCGGAATCG
|
| Reverse |
ATAT
A
▼
AGCTT TCAGTCATTGTGTAGTCC
|
| Internal primers |
Forward |
GCGCTACATGAAGGACTTCTC |
| Reverse |
TTCCAGAACAGCAGGTCGAAC |
Amplification of the target segment
Chromosomal DNA was extracted from R. eutropha PTCC 1615
13
and used as template. PCR mixture (50 μL final volume) with cloning primers, contained 10X PCR buffer, 5 μL; 20 mM MgCl2, 5 μL; 10 mM dNTP mix, 1 μL; 10 μM primer, 1 μL each; template DNA, 100 ng.μL-1, 2 μL; Exprime TaqTM DNA polymerase (GeNet Bio, Daejeon, Korea, 5 U.μL-1), 1.25 μL; and sterile distilled water, 33.75 μL. PCR program included 3 minutes preheating at 95˚C then 5 minutes at 98˚C followed by 30 cycles of 98˚C for 15 seconds, 58.8˚C for 1 minute, and 72˚C for 2.5 minutes, with the final extension at 72˚C for 10 minutes before cooling to 4˚C (Thermocycler: PEQLAB Ltd, Fareham, United Kingdom). PCR tubes containing all components of reaction mixture except the polymerase were preheated to 95˚C in thermocycler, following which the polymerase was added. PCR with internal primers was also performed using the same protocol for components and program except 60˚C annealing temperature. To ensure the correct amplification of target DNA fragment, sequencing of PCR products was carried out by Bioneer Corporation of South Korea. Subsequently, complete sequence of the desired fragment was prepared in Bioedit software, aligned in Nucleotide BLAST server of NCBI database and analyzed. In all steps, 2190 bp DNA piece resulting from PCR reaction with cloning primers was utilized.
Construction of recombinant vector
Vector pET28a (+) was isolated from E. coli DH5α using alkaline lysis method.
14
PCR product gel extraction was performed via GF-1 gel DNA recovery kit (Vivantis, Subang Jaya, Malaysia). Isolated vector and extracted PCR product were digested with BamHI and HindIII restriction enzymes (Sinaclon, Tehran, Iran). To check for the functionality of the enzymes, control reaction for each one was included. Double digested PCR product and plasmid were purified from components of digestion reaction with EZ-10 Spin Column PCR Product Purification Kit (BioBasic, Toronto, Canada) following which the digested PCR product was recombined into the digested vector by T4 DNA ligase (TaKaRa, Kusatsu, Japan) at 16˚C for 4 hours. The digestion and ligation processes were carried out according to the protocol proposed by the companies.
Transformation of R. eutropha PTCC 1615 and determination of colonies harboring recombinant vector
Competent R. eutropha PTCC 1615 was prepared using cold CaCl2 followed by the transfer of recombinant vector into the bacterium using heat shock method.
15
Transformants were selected on LB-agar plate containing kanamycin (200 μg.mL-1).
16
Plasmid DNAs were isolated (with alkaline lysis method) from some colonies and for identifying transformants harboring recombinant plasmids, PCRs were carried out using the extracted plasmids as DNA templates. The cloned gene was sequenced by Bioneer Corporation of South Korea and analyzed as mentioned previously.
Cultivation and PHB quantification
Pre-cultures of the native and engineered strains with optical density of 0.5 were inoculated to flasks containing LB medium and flask cultures were shaken at 150 rpm and 30˚C (Incubator shaker: Vision Scientific, Yuseong-Gu, Korea). This part was carried out three times. Pre-culture and culture of the recombinant strain included kanamycin (200 μg.mL-1). At 48 hours of growth, cells were harvested by centrifugation (Eppendorf centrifuge, Hamburg, Germany) at 9302 ×g and 4˚C for 15 minutes followed by twice washing the pellet with distilled water and an overnight drying at 105˚C. PHB was extracted from dried pellet using chloroform-sodium hypochlorite method.
17
PHB content of the native and recombinant strains was then determined according to Law and Slepecky procedure.
18,19
In this case, PHB in reaction with concentrated sulfuric acid in the vicinity of 100˚C water bath, was converted to crotonic acid through dehydration.
20
Absorbance of the resultant molecule was measured at 235 235 nm wavelength in spectrophotometer (Cecil BioAquarius, Cambridge, United Kingdom) against sulfuric acid blank, and quantified pursuant to the PHB standard curve. This experiment was also repeated three times for each strain.
All statistical analyses were done by the SPSS software version 13.0. Between groups (engineered and native) statistical analysis was performed using independent samples t test. P value less than 0.05 was considered significant.
Results
Isolation of the target DNA segment
The 2190 bp and 1059 bp DNA fragments were amplified from R. eutropha PTCC 1615 in PCR reaction using cloning and internal primers, respectively. The size of PCR products was confirmed by 1% (w/v) agarose gel electrophoresis (Fig. 1). Furthermore, the results of the 2190 bp sequence alignment with Nucleotide collection of NCBI database indicated that the cloned DNA fragment shared 99% identity with the same region in R. eutropha H16 genome. Sequences of the proposed DNA segment and the enzyme, PHB synthase, it encodes are shown in Supplementary file 1. The σ54 homologous protein binds to GG-10N-GC promoter sequences in nitrogen limitation condition similar to σ54, and transcription begins. Moreover, PHB production occurs under limited nitrogen availability.
21
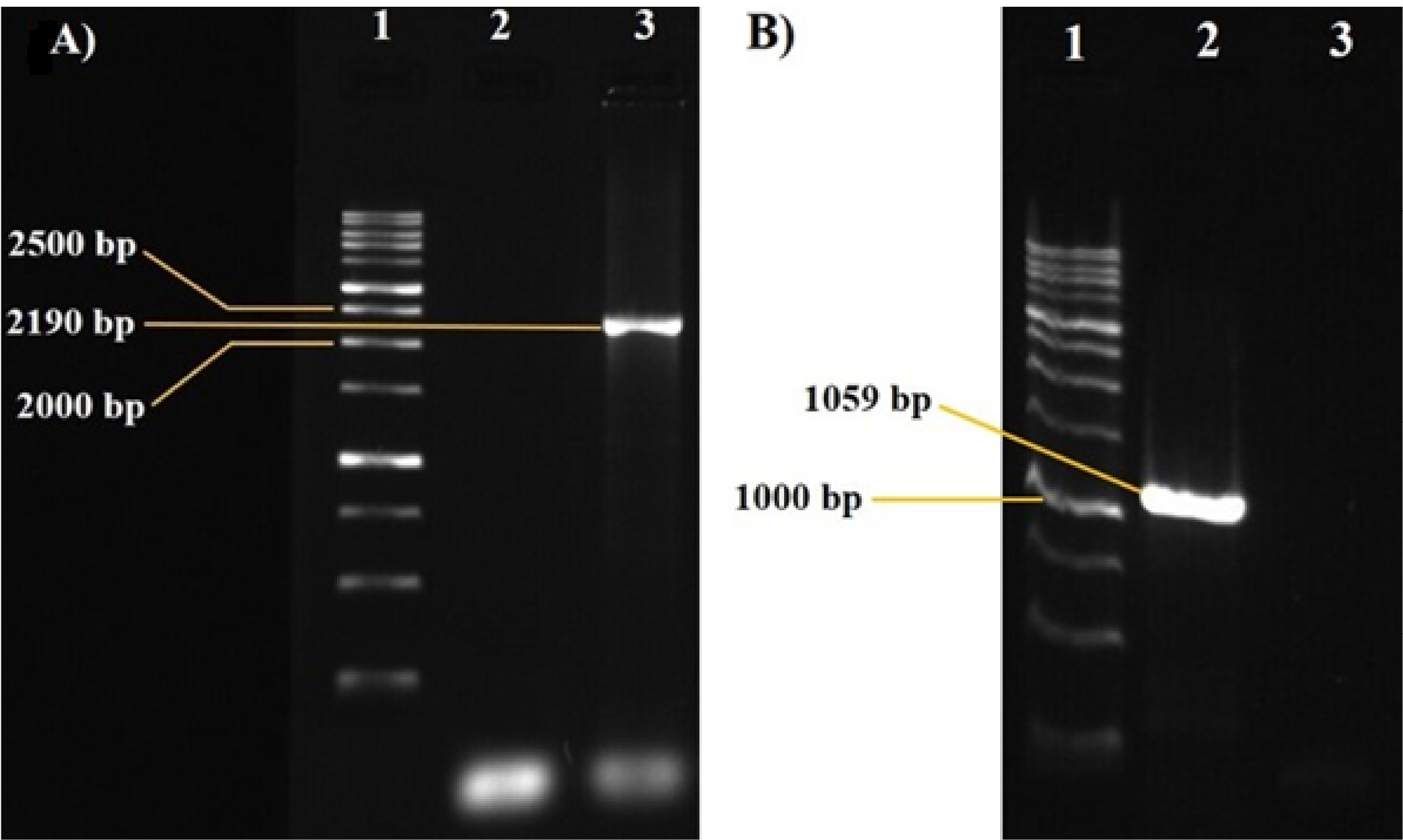
Fig. 1.
Amplified fragments using cloning A) and internal B) primers. A) Lane 1, Molecular weight marker 250 bp; lane 2, negative control of PCR reaction; and lane 3, 2190 bp PCR product. B) Lane 1, Molecular weight marker 250 bp; lane 2, 1059 bp PCR product; and lane 3, negative control.
.
Amplified fragments using cloning A) and internal B) primers. A) Lane 1, Molecular weight marker 250 bp; lane 2, negative control of PCR reaction; and lane 3, 2190 bp PCR product. B) Lane 1, Molecular weight marker 250 bp; lane 2, 1059 bp PCR product; and lane 3, negative control.
Transformation and selection
The recombinant vector with about 7.5 kbp size was transferred into R. eutropha PTCC 1615 and transformants grew on an LB-agar plate containing kanamycin. As Fig. 2 indicates, six of seven colonies received the plasmid carrying the 2190 bp DNA segment following which sequencing data analysis was carried out showing that the DNA fragment has 99% identity with the same area in R. eutropha H16 genome.
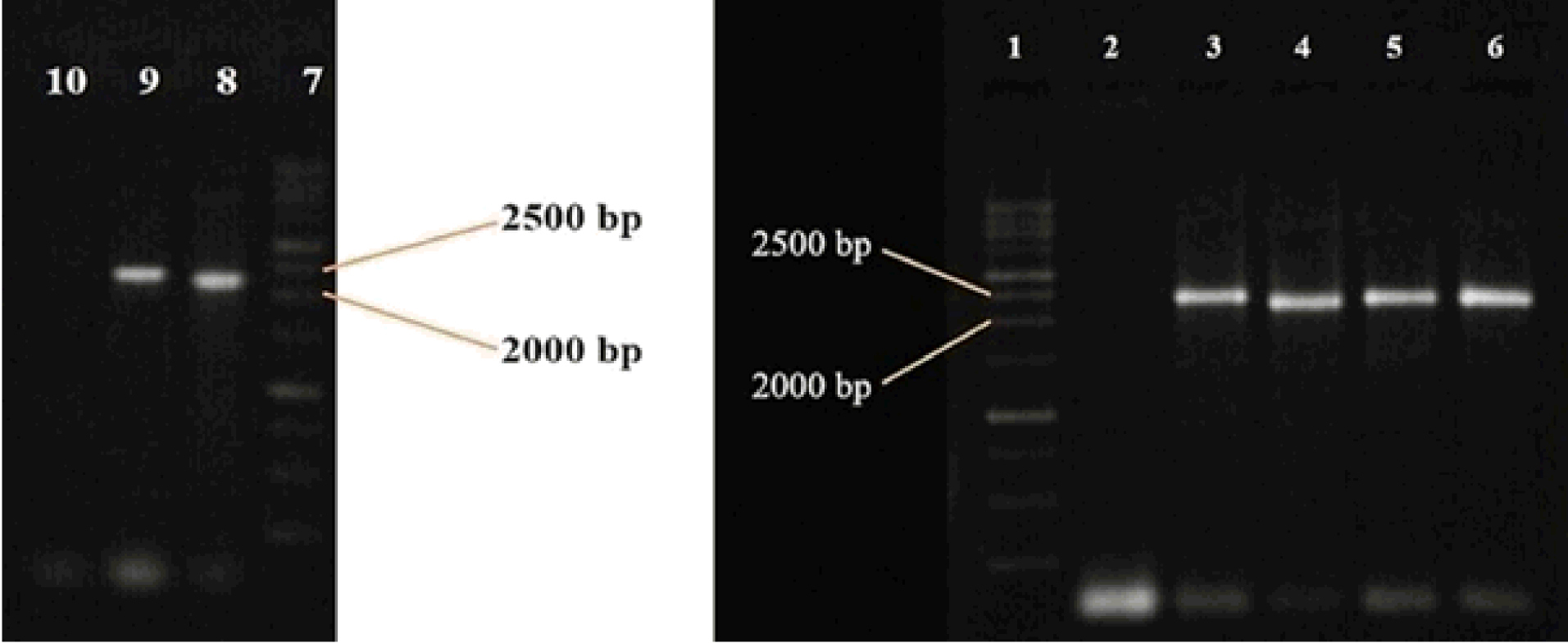
Fig. 2.
Identification of transformants harboring recombinant plasmids. PCR products were loaded on agarose gel. Lanes 1 and 7, Molecular weight marker 250 bp; lane 2, colony without recombinant plasmid; lanes 3-6 and 8-9, colonies containing recombinant plasmid; and lane 10, negative control of PCR reaction.
.
Identification of transformants harboring recombinant plasmids. PCR products were loaded on agarose gel. Lanes 1 and 7, Molecular weight marker 250 bp; lane 2, colony without recombinant plasmid; lanes 3-6 and 8-9, colonies containing recombinant plasmid; and lane 10, negative control of PCR reaction.
PHB production comparison between the native and engineered strain
As shown in Fig. 3, results indicated that the mean PHB content of dry cell weight (DCW) of the engineered was significantly higher than the native bacteria (7.74 ± 0.64 versus 5.51 ± 0.43 percent, respectively; P= 0.045; mean difference = 2.23). This finding indicated that the manipulated strain was 40% more productive than the wild type. Similar results were obtained regarding biomass (g.L-1) and PHB (g.L-1) production (Fig. 4). The mean biomass production by the engineered bacteria was 1.32 ± 0.004 and by the native strain was 1.26 ± 0.011 g.L-1 (P= 0.007). Moreover, PHB (g.L-1) production by the engineered and native bacteria were 0.102 ± 0.008 and 0.069 ± 0.005, respectively (P= 0.032).
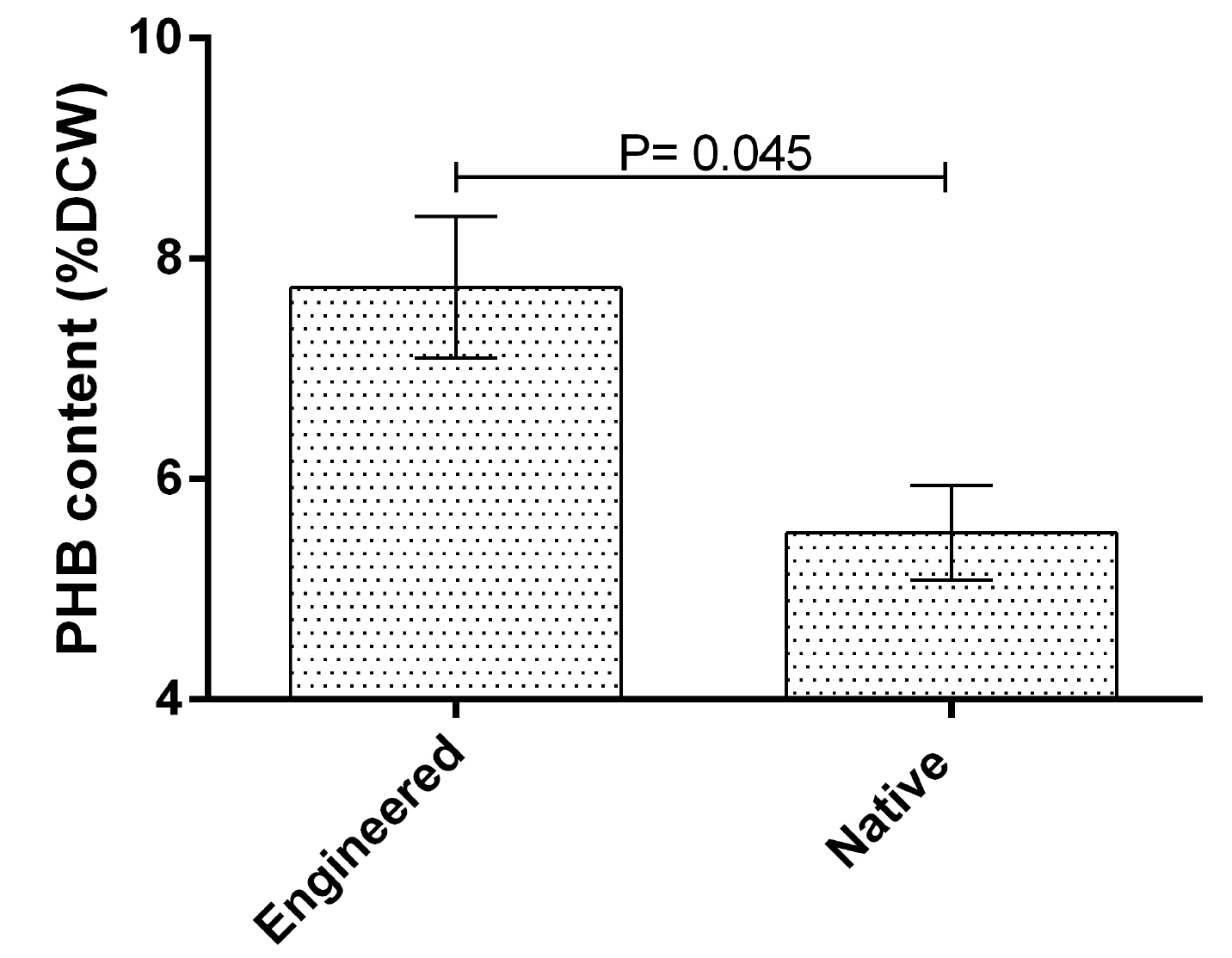
Fig. 3.
PHB content (%DCW) by the engineered and native bacteria. X-axis represents the study groups. Y-axis represents PHB content. Statistical analysis was done by the independent sample t test. A P value less than 0.05 was considered significant
.
PHB content (%DCW) by the engineered and native bacteria. X-axis represents the study groups. Y-axis represents PHB content. Statistical analysis was done by the independent sample t test. A P value less than 0.05 was considered significant
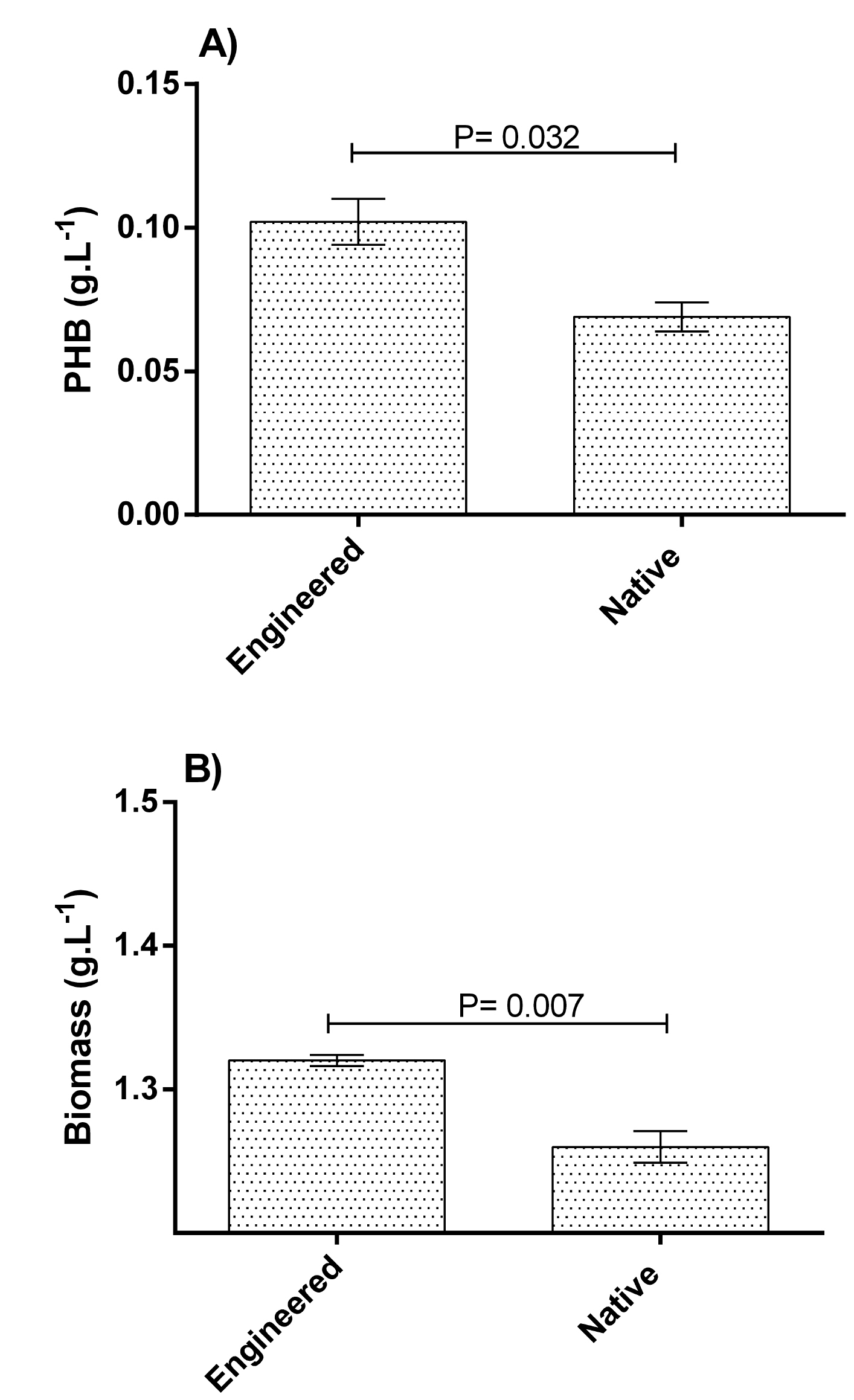
Fig. 4.
A) PHB (g.L-1) and B) biomass (g.L-1) production by the engineered and native bacteria. X-axis represents the study groups. Y-axis represents PHB and biomass production. Statistical analysis was done by the independent sample t test. A P value less than 0.05 was considered significant.
.
A) PHB (g.L-1) and B) biomass (g.L-1) production by the engineered and native bacteria. X-axis represents the study groups. Y-axis represents PHB and biomass production. Statistical analysis was done by the independent sample t test. A P value less than 0.05 was considered significant.
Discussion
Some previous studies have indicated that phbC is a critical gene so that its overexpression or modification has a significant effect on PHB production compared with other PHB biosynthesis genes.
8,10,16
In this study the main gene was cloned in R. eutropha PTCC 1615, a strain from Persian Type Culture Collection, with differences in methods than similar studies.
In previous studies, phbC gene has been cloned in R. eutropha H16,
8
as well as A. latus.
9
In both of these efforts, pHB52 vector harboring phbCAB operon has been used and for construction of the vector, DNA fragment containing phbCAB operon has been isolated by colony hybridization with synthetic oligodeoxyribonucleotides as probes.
22
On the other hand, in the mentioned studies, to obtain the plasmid comprising phbC gene (instead of PHB synthesis operon) and kanamycin resistance marker, pHB52 vector has been constructed using time-consuming and cost-intensive procedures.
8,9,14
PHB extraction was carried out with the chloroform-sodium hypochlorite method, which obtains purer PHB than other methods.
23
According to a research on optimization of PHB production in R. eutropha PTCC 1615, this strain can produce PHB in a range of 2.9%-90.1% DCW depending on components of medium and physical culture conditions.
24
In this study, overexpression of phbC gene resulted in 40% DCW increase in PHB production. While, in similar studies mentioned earlier, 10 and 20% DCW increasing has been obtained in R. eutropha H16 and A. latus, respectively.
8,9
Conclusion
In this study, the effect of the cloned phbC gene on PHB production in R. eutropha PTCC 1615 was investigated. The results indicated that PHB content increased by 40% DCW in engineered strain compared to that of the native bacteria. This finding shows a higher PHB production compared to similar efforts. Factors including amplification using special primers and Taq DNA polymerase with proofreading activity which results in the desired DNA fragment without errors, and also the existence of transcriptional regulatory sequences in cloned DNA fragment and concurrent transcription of the transgene with the host gene, are potential factors accountable for this finding. Moreover, it should be noted that the expression of the recombinant gene without induction and procedures used for vector construction and transformation were easy and cost-effective actions compared to similar efforts. On the other hand, the results revealed that the last part of PHB biosynthesis pathway which is catalyzed by PHB synthase enzyme, is likely to be the rate-limiting step, but further studies are required to verify this finding.
Acknowledgments
This manuscript was part of the thesis written by Farzaneh Barati, MSc student of Microbial Biotechnology. We would like to thank Research Chancellor of Alzahra University.
Funding sources
This work was financially supported by Alzahra University.
Ethical statement
None declared.
Competing interests
None declared.
Authors’ contribution
FB: Formal analysis, investigation, data curation, manuscript preparation; EA: Supervision, conceptualization, methodology and manuscript editing; SG: Project administration, validation, and manuscript editing; MRS: Methodology, validation, and manuscript editing.
Supplementary Materials
Supplementary file 1 contains phbC gene and its upstream and downstream sequences in R. eutropha H16.
(pdf)
Research Highlights
What is the current knowledge?
simple
-
√ PHB is an important biodegradable polymer with various applications.
-
√ Industry is attempting to find cost-effective methods for PHB production.
What is new here?
simple
-
√ Accurate isolation and a flawless DNA fragment resulted from precise primer design and using a polymerase with proofreading activity.
-
√ High transformation rate because of the suitable size of the recombinant vector.
-
√ Reliable and cost-effective employed methods for recombinant vector construction and transformation.
-
√ The 40% increasing of PHB content in the engineered strain compared with the native.
References
- Emadian SM, Onay TT, Demirel B. Biodegradation of bioplastics in natural environments. Waste Manag 2017; 59:526-36. doi: 10.1016/j.wasman.2016.10.006 [Crossref] [ Google Scholar]
- Balaji S, Gopi K, Muthuvelan B. A review on production of poly β hydroxybutyrates from cyanobacteria for the production of bio plastics. Algal Research 2013; 2:278-85. doi: 10.1016/j.algal.2013.03.002 [Crossref] [ Google Scholar]
- Keshavarz T, Roy I. Polyhydroxyalkanoates: bioplastics with a green agenda. CurrOpin Microbiol 2010; 13:321-6. doi: 10.1016/j.mib.2010.02.006 [Crossref] [ Google Scholar]
- Reinecke F, Steinbuchel A. Ralstonia eutropha strain H16 as model organism for PHA metabolism and for biotechnological production of technically interesting biopolymers. J Mol Microbiol Biotechnol 2009; 16:91-108. doi: 10.1159/000142897 [Crossref] [ Google Scholar]
- Suriyamongkol P, Weselake R, Narine S, Moloney M, Shah S. Biotechnological approaches for the production of polyhydroxyalkanoates in microorganisms and plants - a review. Biotechnol Adv 2007; 25:148-75. doi: 10.1016/j.biotechadv.2006.11.007 [Crossref] [ Google Scholar]
- Tran H-D, Huynh V-H, Pham C-H, Chung A-D, Tran Q-D. Cloning phbCAB operon of Alcaligenes eutrophus H16 into Escherichia coli DH5 [alpha] to manufacture Poly 3-Hydroxybutyrate using molasses as carbon source. Journal of Chemical, Biological and Physical Sciences (JCBPS) 2016; 6:1017. [ Google Scholar]
-
Chen G-Q. Industrial Production of PHA. In: Chen GG-Q, ed. Plastics from Bacteria: Natural Functions and Applications. Berlin, Heidelberg: Springer Berlin Heidelberg; 2010:121-32.
- Park J-S, Park H-C, Huh T-L, Lee Y-H. Production of poly-β-hydroxybutyrate by Alcaligenes eutrophus transformants harbouring cloned phbCAB genes. Biotechnol Lett 1995; 17:735-40. doi: 10.1007/bf00130360 [Crossref] [ Google Scholar]
- Seo I-S, Jung Y-M, Lee Y-H. Production of P (3-hydroxybutyrate-3-hydroxyvalerate) and P (3-hydroxybutyrate-4-hydroxybutyrate) using transformant Alcaligenes latus enforcing its own phbC gene. J Microbiol Biotechnol 2001; 11:333-6. [ Google Scholar]
- Kichise T, Taguchi S, Doi Y. Enhanced accumulation and changed monomer composition in polyhydroxyalkanoate (PHA) copolyester by in vitro evolution of Aeromonas caviae PHA synthase. Appl Environ Microbiol 2002; 68:2411-9. doi: 10.1128/aem.68.5.2411-2419.2002 [Crossref] [ Google Scholar]
- Schubert P, Krüger N, Steinbüchel A. Molecular analysis of the Alcaligenes eutrophus poly(3-hydroxybutyrate) biosynthetic operon: identification of the N terminus of poly(3-hydroxybutyrate) synthase and identification of the promoter. Journal of bacteriology 1991; 173:168-75. doi: 10.1128/jb.173.1.168-175.1991 [Crossref] [ Google Scholar]
- Mitra A, Kesarwani AK, Pal D, Nagaraja V. WebGeSTer DB--a transcription terminator database. Nucleic Acids Res 2011; 39:D129-35. doi: 10.1093/nar/gkq971 [Crossref] [ Google Scholar]
- Wright MH, Adelskov J, Greene AC. Bacterial DNA Extraction Using Individual Enzymes and Phenol/Chloroform Separation. J Microbiol Biol Educ 2017; 18. doi: 10.1128/jmbe.v18i2.1348 [Crossref]
- Li H, Bo H, Wang J, Shao H, Huang S. Separation of supercoiled from open circular forms of plasmid DNA, and biological activity detection. Cytotechnology 2011; 63:7-12. doi: 10.1007/s10616-010-9322-9 [Crossref] [ Google Scholar]
- Chan WT, Verma CS, Lane DP, Gan SK. A comparison and optimization of methods and factors affecting the transformation of Escherichia coli. Biosci Rep 2013; 33:e00086. doi: 10.1042/bsr20130098 [Crossref] [ Google Scholar]
- Park H-C, Park J-S, Lee Y-H, Huh T-L. Manipulation of the genes for poly-β-hydroxybutyric acid synthesis in Alcaligenes eutrophus. Biotechnol Lett 1995; 17:729-34. doi: 10.1007/bf00130359 [Crossref] [ Google Scholar]
- Tripathi AD, Srivastava SK. Statistical Optimization of Parameters Affecting Polyhydroxybutyrate (PHB) Recovery by Dispersion Method from Alcaligenes Cells and Its Characterization. J Bioprocess Biotech 2015; 5:7. doi: 10.4172/2155-9821.1000238 [Crossref] [ Google Scholar]
- Law JH, Slepecky RA. Assay of poly-beta-hydroxybutyric acid. J Bacteriol 1961; 82:33-6. doi: 10.1128/JB.82.1.33-36.1961 [Crossref] [ Google Scholar]
- Venkateswar Reddy M, Mawatari Y, Yajima Y, Seki C, Hoshino T, Chang YC. Poly-3-hydroxybutyrate (PHB) production from alkylphenols, mono and poly-aromatic hydrocarbons using Bacillus sp CYR1: A new strategy for wealth from waste. Bioresour Technol 2015; 192:711-7. doi: 10.1016/j.biortech.2015.06.043 [Crossref] [ Google Scholar]
- Quillaguaman J, Munoz M, Mattiasson B, Hatti-Kaul R. Optimizing conditions for poly(beta-hydroxybutyrate) production by Halomonas boliviensis LC1 in batch culture with sucrose as carbon source. Appl Microbiol Biotechnol 2007; 74:981-6. doi: 10.1007/s00253-006-0754-2 [Crossref] [ Google Scholar]
- Lardi M, Aguilar C, Pedrioli A, Omasits U, Suppiger A, Carcamo-Oyarce G. sigma54-Dependent Response to Nitrogen Limitation and Virulence in Burkholderia cenocepacia Strain H111. Appl Environ Microbiol 2015; 81:4077-89. doi: 10.1128/aem.00694-15 [Crossref] [ Google Scholar]
- Kim G-T. Construction of the recombinant phbCAB operon of Alcaligenes eutrophus for accumulation of poly-β-hydroxybutyric acid in Escherichia coli. Kor J Appl Microbiol Biotechnol 1993; 21:221-8. [ Google Scholar]
- Xu A, Lao Y, Zhang Q, Li J, Xia J. Extraction and characterization of PHB from Acidiphilium cryptum DX1-1. Journal of Wuhan University of Technology-Mater Sci Ed 2010; 25:938-43. doi: 10.1007/s11595-010-0124-x [Crossref] [ Google Scholar]
- Aramvash A, Akbari Shahabi Z, Dashti Aghjeh S, Ghafari MD. Statistical physical and nutrient optimization of bioplastic polyhydroxybutyrate production by Cupriavidus necator. Int J Environ Sci Technol (Tehran) 2015; 12:2307-16. doi: 10.1007/s13762-015-0768-3 [Crossref] [ Google Scholar]