Bioimpacts. 11(3):173-179.
doi: 10.34172/bi.2021.23
Original Research
A sensitive homogeneous enzyme assay for euchromatic histone-lysine-N-methyltransferase 2 (G9a) based on terbium-to-quantum dot time-resolved FRET
Mohammad Amjadi 1, *  , Tooba Hallaj 1, 2, Niko Hildebrandt 3, 4
, Tooba Hallaj 1, 2, Niko Hildebrandt 3, 4
Author information:
1Department of Analytical Chemistry, Faculty of Chemistry, University of Tabriz, Tabriz 5166616471, Iran
2Cellular and Molecular Research Center, Cellular and Molecular Medicine Institute, Urmia University of Medical Sciences, Urmia, Iran
3NanoBioPhotonics (nanofret.com), Institute for Integrative Biology of the Cell (I2BC), Université Paris-Saclay, Université Paris-Sud, CNRS, CEA, Orsay, France
4Laboratoire Chimie Organique, Bioorganique, Réactivité et Analyse (COBRA), Université de Rouen Normandie, CNRS, INSA, 76821 Mont-Saint-Aignan, France
Abstract
Introduction:
Histone modifying enzymes include several classes of enzymes that are responsible for various post-translational modifications of histones such as methylation and acetylation. They are important epigenetic factors, which may involve several diseases and so their assay, as well as screening of their inhibitors, are of great importance. Herein, a bioassay based on terbium-to-quantum dot (Tb-to-QD) time-resolved Förster resonance energy transfer (TR-FRET) was developed for monitoring the activity of G9a, the euchromatic histone-lysine N-methyltransferase 2. Overexpression of G9a has been reported in some cancers such as ovarian carcinoma, lung cancer, multiple myeloma and brain cancer. Thus, inhibition of this enzyme is important for therapeutic purposes.
Methods:
In this assay, a biotinylated peptide was used as a G9a substrate in conjugation with streptavidin-coated ZnS/CdSe QD as FRET acceptor, and an anti-mark antibody labeled with Tb as a donor. Time-resolved fluorescence was used for measuring FRET ratios.
Results:
We examined three QDs, with emission wavelengths of 605, 655 and 705 nm, as FRET acceptors and investigated FRET efficiency between the Tb complex and each of them. Since the maximum FRET efficiency was obtained for Tb to QD705 (more than 50%), this pair was exploited for designing the enzyme assay. We showed that the method has excellent sensitivity and selectivity for the determination of G9a at concentrations as low as 20 pM. Furthermore, the designed assay was applied for screening of an enzyme inhibitor, S-(5’-Adenosyl)-L-homocysteine (SAH).
Conclusion:
It was shown that Tb-to-QD FRET is an outstanding platform for developing a homogenous assay for the G9a enzyme and its inhibitors. The obtained results confirmed that this assay was quite sensitive and could be used in the field of inhibitor screening.
Keywords: Enzyme assay, Histone-modifying enzymes, Epigenetic enzymes, Time-resolved FRET, Inhibitor screening
Copyright and License Information
© 2021 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Epigenetics is the study of changes in the regulation of gene activity and expression, independent of gene sequences. An important epigenetic factor is post-translational modifications of histones, such as methylation, acetylation and phosphorylation, which are controlled by various histone-modifying enzymes (HMEs) such as histone methyltransferases, acetyltransferases and phosphoryltransferases.
1
One of the histone methyltransferases (HMT) is euchromatic histone-lysine-N-methyltransferase 2 (EHMT2), also known as G9a, which catalyzes histone H3 lysine 9 mono- and dimethylation, usually associated with transcriptional gene silencing.
2
It has been found that the initiation and growth of cancer not only depend on genetic mutations but also on epigenetic misregulations. For example, it has been reported that G9a is overexpressed in a number of cancers such as ovarian carcinoma, lung cancer, multiple myeloma, and brain cancer.
1-3
This can be attributed to the inhibition effect of G9a on tumor suppressor genes, which arises from enhanced methylation levels in H3 tails as a result of elevated G9a levels.It has also been reported a decrease in the activity of G9a could inhibit cell proliferation in several cancer cell lines. This fact has been attributed to autophagic death through an imbalance in the serine–glycine biosynthetic pathway.
3
So, improvement of methods for HME assays and investigation of small molecules as their inhibitors have attracted great attention.
4,5
Up to now, various compounds have been studied and introduced as G9a inhibitors. They are classified into H3 peptide substrate-competitive inhibitors and S-adenosylmethionine (SAM) cofactor-competitive inhibitors.
6
Study on the ways to inhibit and control the activity of such enzymes and development of new inhibitors, selective toward subsets of HMTs or even a single HMT are an important and active field of research that can open new horizons for therapeutic interventions.
7,8
Various methods including mass spectrometry,
9,10
radioisotope-based methods,
11,12
enzyme-linked immunosorbent assays (ELISA),
13
fluorescence,
14,15
and colorimetric
16
methods (based on measurement of enzymatic reaction by-products) have been reported to study histone methyltransferases as well as their inhibitors.
Förster resonance energy transfer (FRET) is a nonradiative energy transfer process in which the excited donor transfers its energy to a ground state acceptor through a dipole-dipole interaction.
17
Due to the unique properties of FRET-based assays and the possibility of removing interfering background luminescence in the time-resolved (TR) fluorescence methods, TR-FRET has been known as a sensitive fluorescence method in various bioassays. One of the best donors for TR-FRET biosensing is luminescent terbium complexes (LTCs) owing to their sharp and distinct emission bands, stability, and very long excited-state lifetimes (in the millisecond range).
18
Hildebrandt and coworkers
19
applied LTCs as a donor with quantum dots (QDs) as an acceptor in TR-FRET. They indicated that QDs are suitable acceptors for lanthanide ions because of their strong absorption that has a very large spectral overlap with several emission bands of lanthanides.
Even though TR-FRET-based epigenetic enzyme assay using Tb-to-green fluorescent protein (GFP) and Eu-to-XL665 pairs
20,21
have been already reported, to the best of our knowledge, terbium-to-quantum dot (Tb-to-QD) conjugate has not yet been used for this kind of assay. A steady-state FRET-based bioassay was also reported for the detection of the histone acetyltransferase activity using QDs as donors with dye as an acceptor.
22
Particular advantages of QDs such as the broad absorption spectra with large extinction coefficients, sharp and size-tunable fluorescence spectra, and chemically accessible surface area make them excellent choices as acceptors for FRET-based assays.
17
Considering these facts, herein, we developed a rapid and sensitive homogenous enzyme assay for G9a on the basis of Tb-to-QD TR-FRET. The designed assay was applied for G9a inhibitor screening.
Materials and Methods
Materials
Human G9a with N-terminal GST tag expressed in Baculovirus infected Sf9 cell expression system was obtained from BPS Bioscience (San Diego, USA). Its concentration was 0.1 mg/mL and its MW was 74.6 kDa. It was aliquoted and stored at -80 °C. The working solution of the enzyme was prepared freshly by dilution with assay buffer. Anti-H3K9me2 monoclonal antibody (raised in Mouse against histone H3, dimethylated at lysine 9) with a concentration of 50 µg/50 µL was purchased from BPS Bioscience. Biotinylated Histone H3 (1-21) peptide with the sequence of H-ARTKQTARKSTGGKAPRKQLAGGK (Biotin)-NH2, was obtained from AnaSpec Inc (Fremont, USA). The stock solution (500 µM) was prepared in ultrapure water, aliquoted and stored at -20°C. Working solution (1.0 µM) was prepared daily by diluting 1.0 µL of stock solution to 500 µL with assay buffer. S-(5’-adenosyl)-L-methionine chloride (SAM) as cofactor was obtained from Sigma (USA). Its stock solution (30 mM) was made in 5 mM H2SO4/10%v ethanol, aliquoted and stored at -80°C. The working solution (300 µM) was prepared just before use by mixing 1.0 µL of stock solution with 99 µL of assay buffer. S-(5’-adenosyl)-L-homocysteine (SAH) was also obtained from Sigma and a 25-mM stock solution was prepared in DMSO, aliquoted and stored at -20°C. Poly-L-lysine (0.01%) was obtained from Sigma and used for stopping the enzymatic reaction. Streptavidin (sAv)-coated QDs (with emission wavelengths of 605, 655 and 705) with a concentration of 2.0 µM were obtained from Life Technologies (Thermofisher Scientific). Tris-HCl buffer (pH 8.5) containing 0.1% BSA was used as assay buffer in all experiments.
Instruments and equipment
Absorption spectra were recorded by a Lambda 35 UV/Vis System (PerkinElmer). PL decay curves were obtained by an EI fluorescence plate reader (Edinburgh Instruments) by the use of 4000 detection bins with 2 μs integration time and excitation at 337.1 nm with a nitrogen laser (20 Hz, VSL 337 ND, Spectra-Physics). Optical bandpass filters (Semrock) were (494 ± 10) nm for the Tb detection, (660 ± 7), (607 ± 5) and (707 ± 13) nm for the QD650, QD605 and QD705 detection channels, respectively.
Terbium bioconjugation
The conjugation of antibody (AB) with a terbium complex (Tb) was done after the exchange of AB buffer to carbonate (100 mM, pH 9.0) using Zeba desalting column (7K MWCO, 0.5 mL). Subsequently, 1.04 µL of Tb-NHS (N-Hydroxysuccinimide) (8 mM in DMF) was added to the obtained AB in a centrifuge tube. It should be noted that the Tb/AB ratio was kept at 50. After briefly vortexing, the tube was wrapped in a foil and incubated in an Intelli mixer at 30 rpm for 4 hours. The obtained Tb-AB was purified with Zeba spin desalting columns (7K MWCO), by washing with 100 mM Tris buffer (pH 7.4). These columns contain a high-performance resin, which effectively retains small molecules and passes only larger compounds. To ensure all excess Tb is separated from Tb-AB conjugate, two rounds of purification was performed. The final volume of labeled AB was made to 150 µL with Tris buffer and its absorption spectrum was recorded by Lamda 35 Spectrophotometer.
General procedure for G9a enzyme assay
The mixture of 10 µL of assay buffer,10 µL of G9a with various concentrations (the final concentrations were 0.02, 0.04, 0.08, 0.15 and 0.3 nM), 10 µL of peptide (400 µM) and 10 µL of SAM (100 µM) was incubated in a 96-well black microplate for 40 min. Afterward, 10 µL of detection mixture (containing 0.0005% poly-l-lysine, 0.01 µM Tb-AB and 2.5 nM sAv-QD705) was added into each well and incubated the microplate for more 1 hour. Lastly, PL decay curves of Tb donor and QD acceptor were measured in a time window from 0-8 ms. The time window from 100 to 900 µs was used for the calculation of TR-FRET ratio by the following equation: FRET-Ratio= I (ChA)/I (ChD).
Procedure for inhibitor screening
The inhibitor screening procedure was almost the same as the previous procedure except that 10 µL of inhibitor with various concentrations in assay buffer (containing 2% DMSO (was first pre-incubated with 10 µL of G9a (0.6 nM) for 10 minutes. Final concentrations of inhibitor was 0.01, 0.1, 1.0, 10.0 and 100 µM. Afterward, the procedure was continued by adding peptide, SAM, then detection mixture according to the prior section.
Results
Principle of the bioassay
In this study, a rapid and sensitive homogenous enzyme assay was developed for G9a on the basis of Tb-to-QD time-resolved FRET. This enzyme acts on histone H3 and methylates its lysine 9 location. The amount of methylation is controlled by the activity of the enzyme. The overall strategy for assay was to use a Tb-conjugated antibody and biotinylated peptide together with streptavidin-coated QDs as a FRET donor and acceptor, respectively. We used biotinylated Histone H3 (1-21) peptide as a substrate. After enzymatic reaction with G9a and cofactor (SAM), which leads to the dimethylation of lysine 9 location on the substrate, the Tb-AB conjugate and sAv-QDs were added and FRET ratio was measured, which was proportional to dimethylated peptide concentration and so to G9a activity. The illustration of the principles of this assay is shown in Scheme 1.
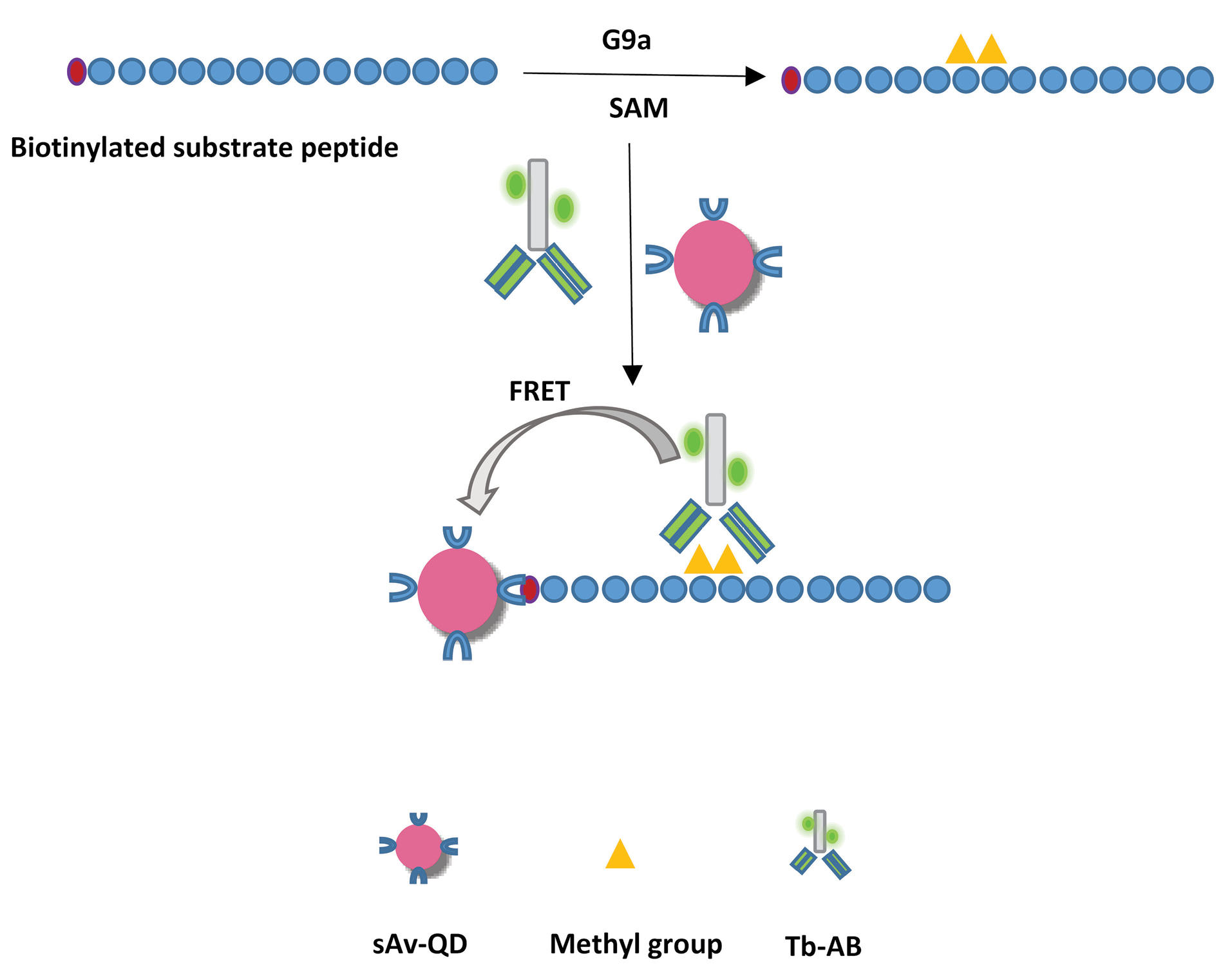
Scheme 1.
Principles of the G9a assay
.
Principles of the G9a assay
Conjugation of Tb to antibody
The most common technique for AB labeling involves the use of a chemical group that reacts with –NH2. Due to the especially nucleophilic property of primary amines, they are suitable targets for conjugation with reactive groups including NHS Esters. Here a macrocyclic ligand-based Tb complex [23] activated by NHS was used for random labeling of AB. A high excess of Tb-NHS was used with respect to AB (50 molar excess). The absorption spectrum of purified Tb-AB conjugate is shown in Fig. S1 (Electronic Supplementary Material, ESM). The peak at 340 nm is due to Tb and the one at 280 nm is ascribed to AB. This confirms the binding of Tb to AB.
FRET pairs
In order to achieve suitable sensitivity in the enzyme assay, three different acceptors including QD605, QD655 and QD705 with the same Tb donor were tested. A macrocyclic ligand-based Tb complex
23
with high brightness was applied as a FRET donor. Each FRET pair was used for enzyme assay according to the general procedure. The obtained PL decay curves are shown in Fig. 1. The sensitization can be seen in the decay curves of the QD detection channels; the long-lifetime parts of these decay curves remarkably increase by adding G9a and SAM. These results show that FRET occurs with all pairs and so, Tb-to-QD FRET can be applied for the G9a assay. Control experiments with only Tb-AB and only QD-Peptide showed the background of Tb PL (spectral crosstalk in the QD detection channels) and directly excited QD PL. Furthermore, FRET efficiencies for the three mentioned FRET pairs were calculated using both Tb donor quenching and QD acceptor sensitization. All fitting was performed by FAST software (Edinburgh Instruments). The results are given in Table 1. According to the obtained results, QD705 produced higher FRET efficiency and FRET ratio than the other QDs, so this QD was used for subsequent experiments.
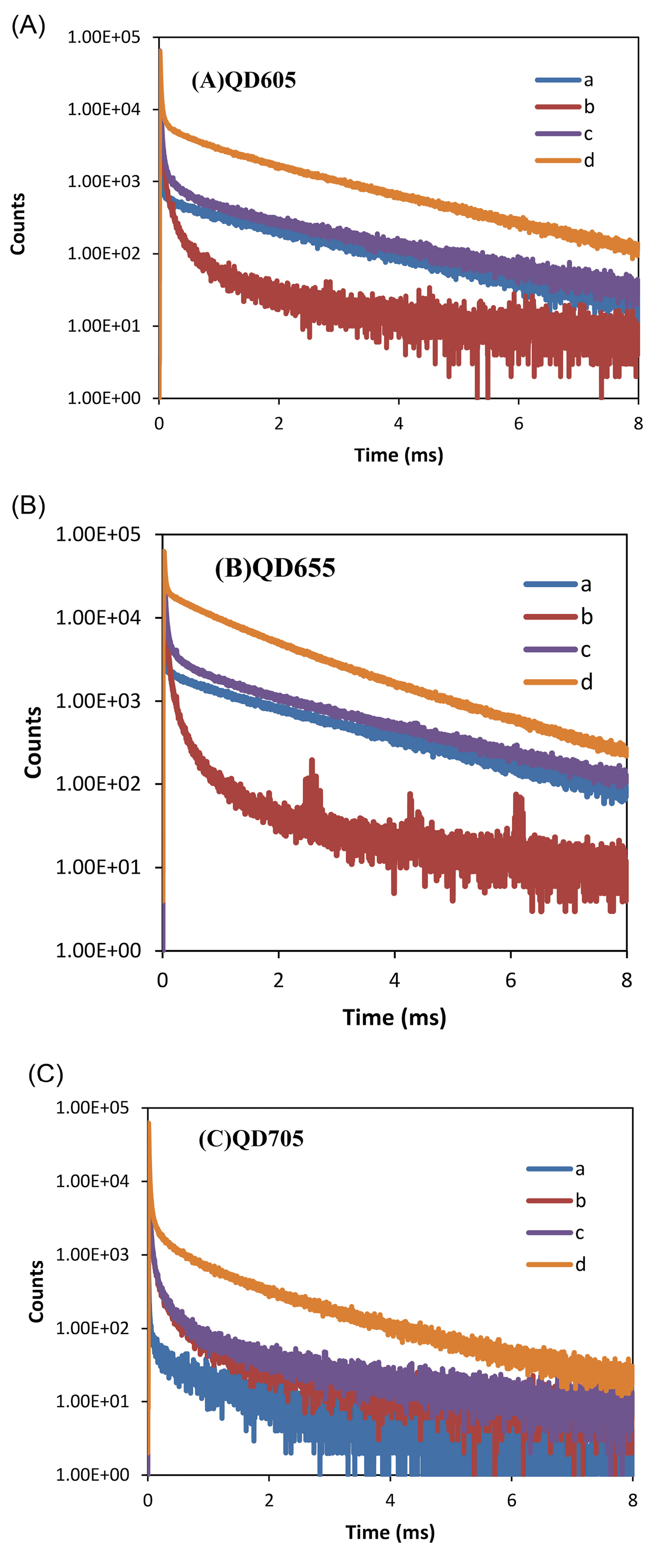
Fig. 1.
PL decay curves of G9a assay obtained from acceptor channel for (A) QD605, (B) QD655 and (C) QD705. In each graph, (a) QD + peptide; (b); Tb-AB (c) Tb-AB + QD + peptide; (d) Tb-AB + QD + peptide + G9a + SAM.
.
PL decay curves of G9a assay obtained from acceptor channel for (A) QD605, (B) QD655 and (C) QD705. In each graph, (a) QD + peptide; (b); Tb-AB (c) Tb-AB + QD + peptide; (d) Tb-AB + QD + peptide + G9a + SAM.
Table 1.
The FRET efficiencies using donor quenching and acceptor sensitization
|
QD
|
ηFRET (D)%
|
ηFRET (A)%
|
| 605 |
55 |
40 |
| 655 |
50 |
43 |
| 705 |
54 |
62 |
G9a assay
The effects of several parameters, such as peptide and SAM concentration and reaction time, were examined to obtain the best condition for assaying the activity of G9a. It is known that the G9a enzymatic reaction takes place at an alkaline solution.
24
We chose pH 8.5 which is reasonably close to physiological pH, and at the same time, the enzymatic reaction is fast enough at this condition. The amount of H3(1-21) peptide on the enzymatic reaction was studied at a constant amount of G9a and saturating amount of SAM. The results are graphically shown in Fig. 2A. As can be seen, the methylation saturation is obtained above 100 nM of the peptide substrate. So this concentration of peptide was chosen for enzyme assay in the next experiments. The other factor that should be taken into account in the G9a enzymatic reaction is the concentration of cofactor, SAM, which provides the required methyl group for methylation reaction. As can be seen in Fig. 2B, above 20 µM, the reaction and so the FRET signal reaches a plateau. Based on this result, the amount of SAM was adjusted in 20 µM for subsequent works.
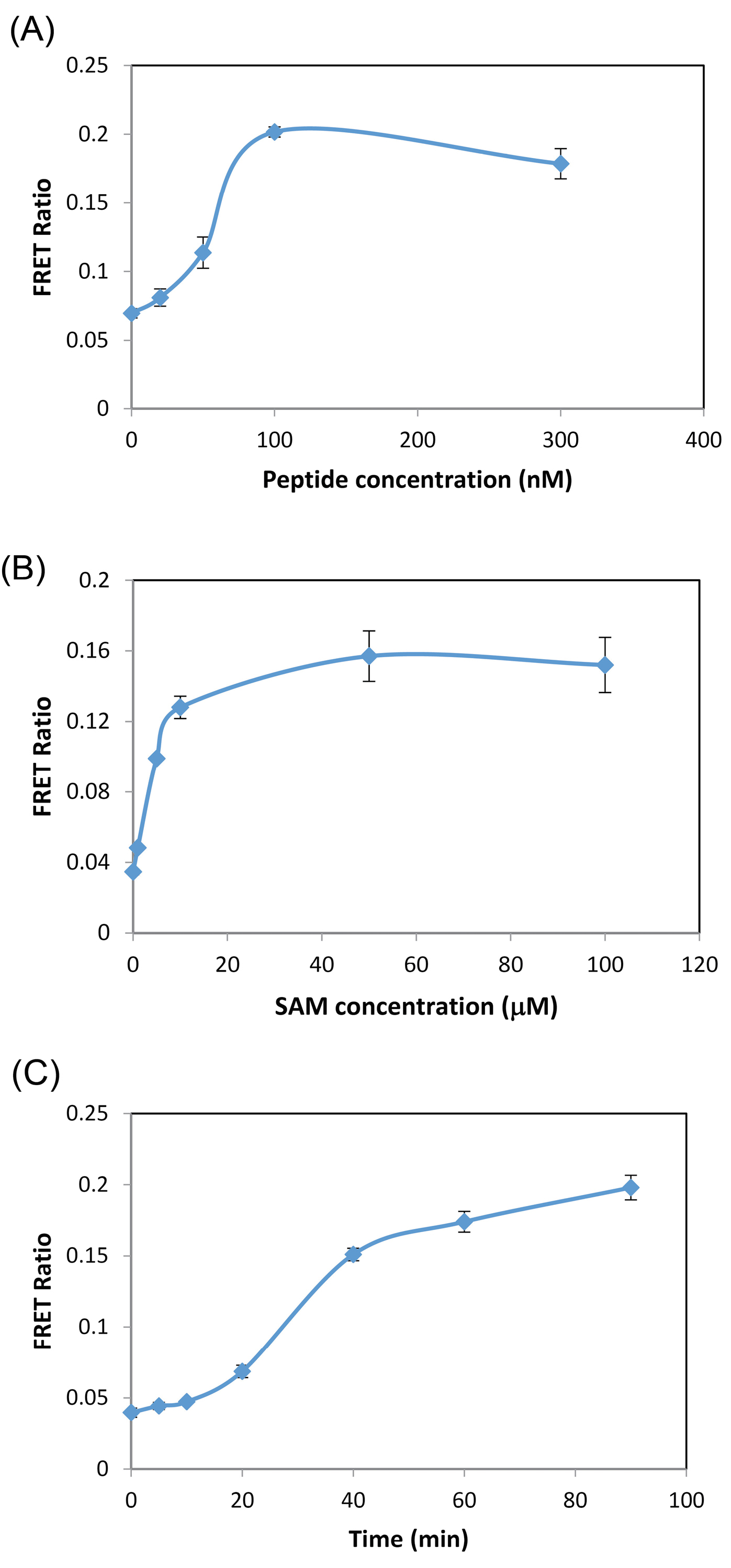
Fig. 2.
(A) Effect of amount of peptide on the enzymatic reaction. Conditions: G9a, 0.15 nM; SAM, 50 µM; incubation time, 40 min; Tb-AB 2 nM; QDs, 0.5 nM; (B) Effect of SAM concentration on the enzymatic reaction. Conditions: H3(1-21) Peptide, 100 nM, other condition same as (A); (C) Progress curve for G9a reaction. Conditions: same as (B).
.
(A) Effect of amount of peptide on the enzymatic reaction. Conditions: G9a, 0.15 nM; SAM, 50 µM; incubation time, 40 min; Tb-AB 2 nM; QDs, 0.5 nM; (B) Effect of SAM concentration on the enzymatic reaction. Conditions: H3(1-21) Peptide, 100 nM, other condition same as (A); (C) Progress curve for G9a reaction. Conditions: same as (B).
Moreover, a time course of methylation using peptide substrate H3(1-21) was monitored in the presence of SAM. The results are shown in Fig. 2C. As can be seen, the signal increased sharply up to 40 min and after that, the rate of reaction slowed down, possibly because of the formation of SAH.
At last, under the optimum conditions, a set of experiments was performed to investigate the relationship between FRET signal and concentration of G9a. An increasing concentration of recombinant G9a was added to the reaction mixture. The reaction was allowed to proceeds for 40 min and then the FRET ratios were measured. The obtained results are shown in Fig. 3. As can be seen, FRET signal increases proportionally with the concentration of enzyme up to about 0.3 nM. At concentrations higher than this amount, the reaction reaches a steady state. There is a linear relationship between G9a concentration and signal in the range of 20-150 pM (Fig. 3 inset). The performance of the method is comparable with
25
or better than
26
some previously reported methods for G9a assay. Additionally, compared to the radioisotope-based method, which suffers from radioactive wastes and laborious steps, our method is simple and sensitive, which can be also applied for kinetic studies.
12
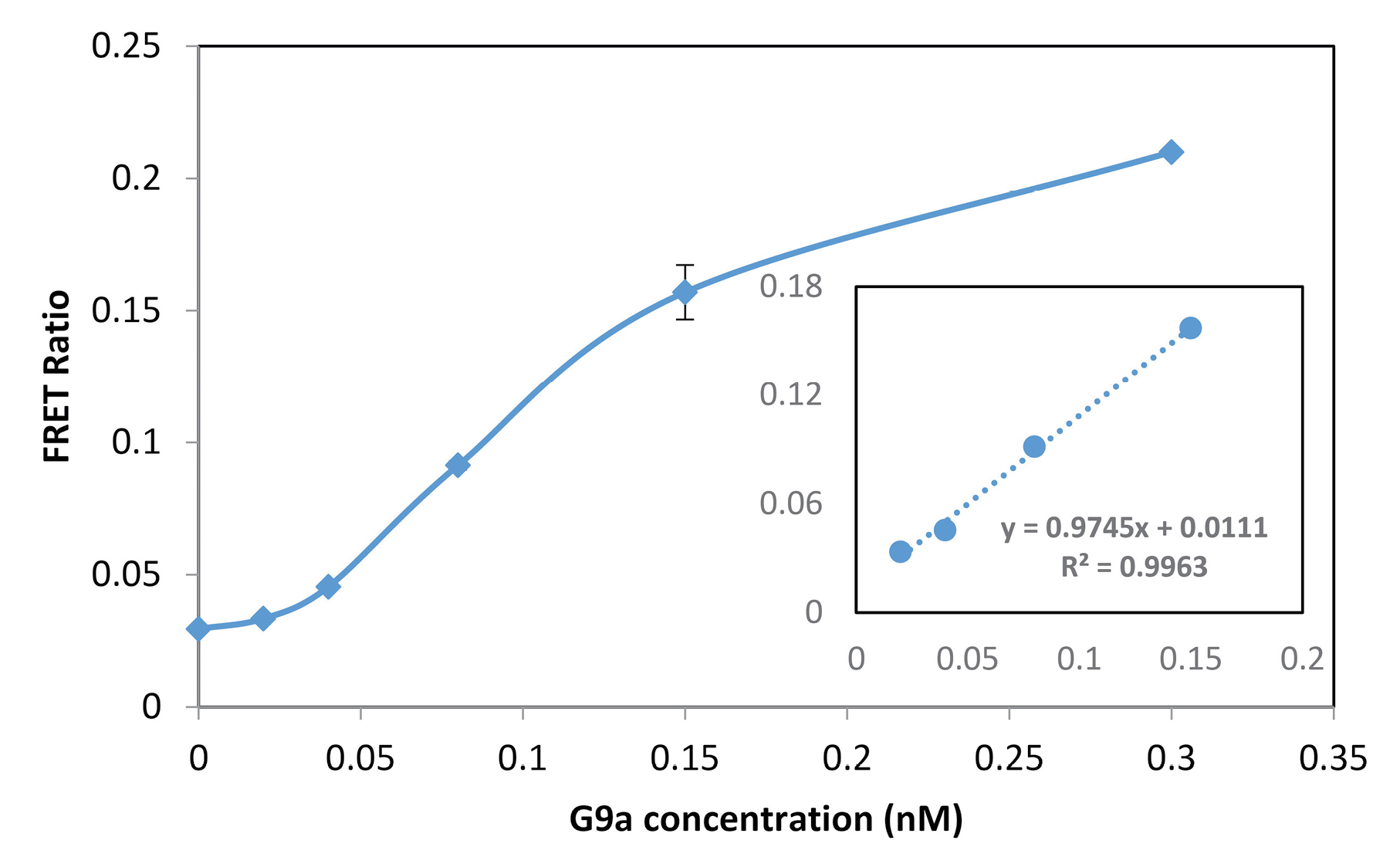
Fig. 3.
G9a titration curve and corresponding calibration curve (inset), Conditions: H3(1-21) peptide, 100 nM; SAM, 50 µM; incubation time, 40 min; Tb-AB 2 nM; QDs, 0.5 nM.
.
G9a titration curve and corresponding calibration curve (inset), Conditions: H3(1-21) peptide, 100 nM; SAM, 50 µM; incubation time, 40 min; Tb-AB 2 nM; QDs, 0.5 nM.
We also investigated the selectivity of the enzyme assay by applying nonspecific peptide substrate (H4 (1-22), H3 (22-44)), co-enzyme (acetyl coenzyme A) and enzyme (GC5, SET7/SET9). As demonstrated in Fig. S2 (ESM), the signals from nonspecific compounds are comparable with that of control (contains no G9a) and so the method has good selectivity for G9a. In summary, the results confirmed that Tb-to-QD time-resolved FRET can be used for bioassay of G9a with satisfactory results.
Enzyme inhibition
In order to evaluate the possibility of application of the assy for inhibitor screening the effect SAH,
27
as an inhibitor, on the enzymatic reaction was examined. For this purpose, G9a was pre-incubated with various concentrations of inhibitor (in DMSO) before initiating reaction by adding peptide and SAM. Then, the designed FRET assay was exploited for the detection of enzymatic reaction products. As can be seen in Fig. 4, the sigmoidal dose-response curve was obtained for the inhibitor and IC50 was calculated to be about 0.5 µM for SAH.
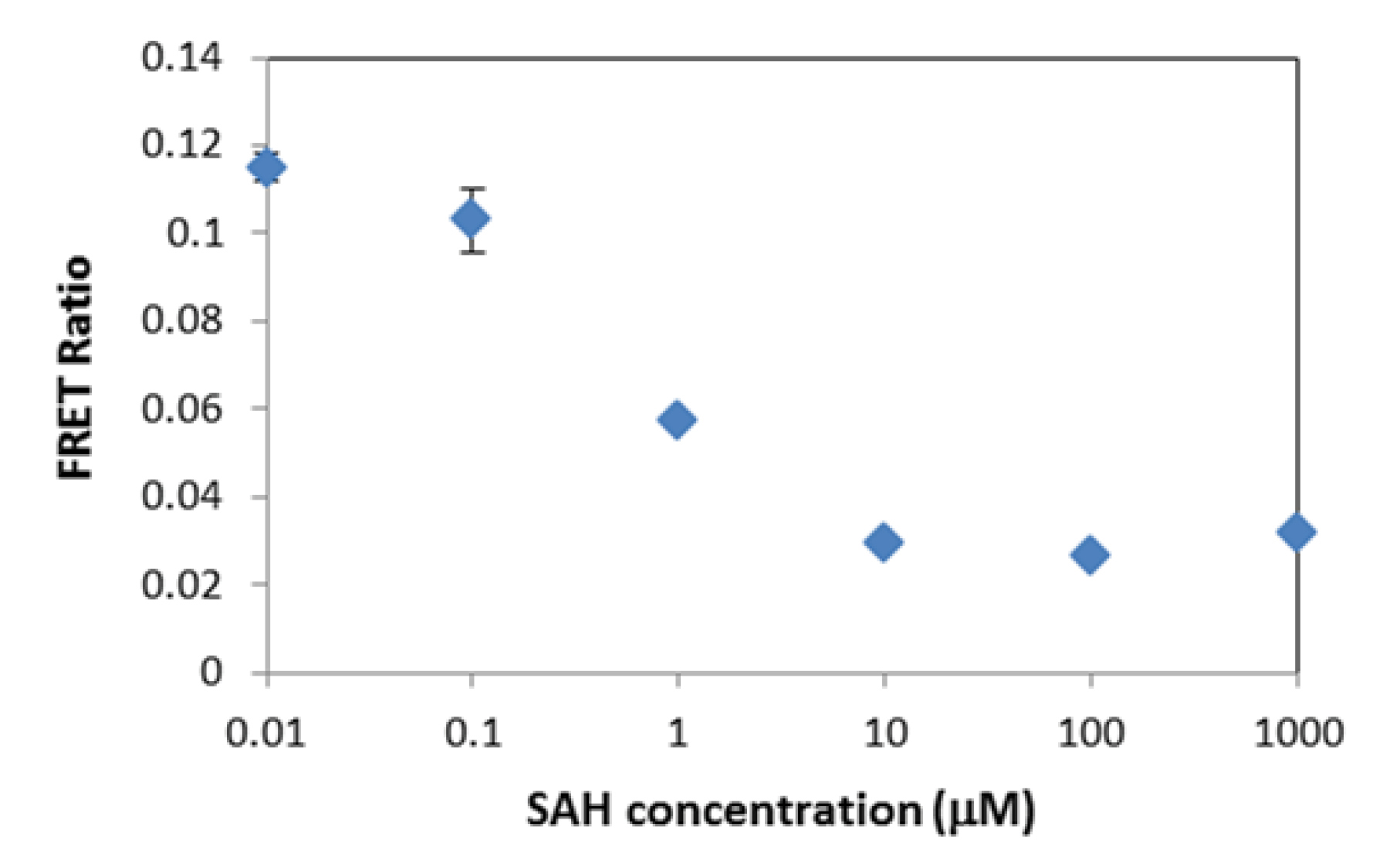
Fig. 4.
Inhibition curve for SAH; Conditions: G9a, 0.15 nM; H3(1-21) Peptide, 100 nM; SAM, 10 µM; DMSO, 2%; incubation time, 40 min; Tb-AB 2 nM; QDs, 0.5 nM.
.
Inhibition curve for SAH; Conditions: G9a, 0.15 nM; H3(1-21) Peptide, 100 nM; SAM, 10 µM; DMSO, 2%; incubation time, 40 min; Tb-AB 2 nM; QDs, 0.5 nM.
Discussion
The presented results show that an efficient FRET occurs between Tb and QDs after their conjugation with dimethylated peptide. QDs with an emission wavelength of 705 nm gives the highest FRET efficiency. This is due to the higher overlap between the absorption spectrum of QD705 and the fluorescence spectrum of Tb. In addition, at this wavelength, the background signal would be lower, which can be considered as another advantage. The use of time-gated detection also leads to a low background signal. Compared to methods like ELISA, our assay is much simpler because it is a homogeneous assay and does not need any separation step. One important advantage of using QDs as FRET acceptors is the possibility of performing multiplex assays. Currently, our group is working in this direction.
Study on the inhibition of epigenetic enzyme activity plays an important role in drug discovery and may open new ways in treatments of cancer.
28,29
Indeed, the ultimate goal of this study was developing a high-throughput screening method for inhibitors. Therefore, in order to show the applicability of the G9a assay for studying the enzyme inhibition, SAH, which is a by-product of HMTs enzymatic reaction, was chosen as a well-known inhibitor of G9a, which acts through competition with SAM in the enzymatic reaction.
30
The results confirm that the method is suitable for inhibitor study.
Conclusion
We here demonstrated that Tb-to-QD TR-FRET can be applied as a sensitive tool for assay of G9a and subsequently for screening of its inhibitors in a simple homogenous format. Terbium complex served as an efficient energy donor and streptavidin-coated ZnS/CdSe core-shell QDs acted as acceptors. The FRET efficiency was more than 50% and enzyme concentrations as low as 20 pM could be detected. We anticipate that this approach can be used as a versatile sensing platform for the development of assays for other epigenetic enzymes by changing the substrate peptide and corresponding antibody. These assays would be suitable for inhibitor screening and drug-discovery applications.
Acknowledgment
The authors would like to thank the Center for International Scientific Studies & Collaborations (CISSC).
Funding sources
This work has been supported by the Center for International Scientific Studies & Collaborations (CISSC).
Ethical statement
None to be declared.
Competing interests
There is no conflict of interests to be reported.
Authors’ contribution
MA: Supervision, experiments design, data analysis, study consultation, writing and reviewing. TH: Experiments design, data analysis, draft preparation. NH: Provision of study materials and equipment, study consultation, writing and reviewing.
Supplementary file 1 contains electronic supplementary material.
(pdf)
Research Highlights
What is the current knowledge?
simple
-
√ Time-resolved FRET is a selective and powerful method for bioassay.
-
√ QDs are bright acceptors in FRET-based sensors.
What is new here?
simple
-
√ Tb-to-QD time-resolved FRET was used for sensitive detection of G9a.
-
√ A fast and homogeneous bioassay was developed for G9a and its inhibitors.
References
- Wei L, Chiu DK, Tsang FH, Law CT, Cheng CL, Au SL. Histone methyltransferase G9a promotes liver cancer development by epigenetic silencing of tumor suppressor gene RARRES3. J Hepatol 2017; 67(4):758-769. doi: 10.1016/j.jhep.2017.05.015 [Crossref] [ Google Scholar]
- Zhang K, Wang J, Yang L, Yuan YC, Tong TR, Wu J. Targeting histone methyltransferase G9a inhibits growth and Wnt signaling pathway by epigenetically regulating HP1α and APC2 gene expression in non-small cell lung cancer. Mol Cancer 2018; 17:153. doi: 10.1186/s12943-018-0896-8 [Crossref] [ Google Scholar]
- Lehnertz B, Pabst C, Su L, Miller M, Liu F, Yi L. The methyltransferase G9a regulates HoxA9-dependent transcription in AML. Genes Dev 2014; 28:317-327. doi: 10.1101/gad.236794.113 [Crossref] [ Google Scholar]
- Wang X, Zhu WG. Advances in histone methyltransferases and histone demethylases. Ai Zheng 2008; 27:1018-1025. [ Google Scholar]
- Martinez NJ, Simeonov A. Cell-based assays to support the profiling of small molecules with histone methyltransferase and demethylase modulatory activity. Drug Discov Today Technol 2015; 18:9-17. doi: 10.1016/j.ddtec.2015.10.004 [Crossref] [ Google Scholar]
- Cao H, Li L, Yang D, Zeng L, Yewei X, Yu B. Recent progress in histone methyltransferase (G9a) inhibitors as anticancer agents. Eur J Med Chem 2019; 179:537-546. doi: 10.1016/j.ejmech.2019.06.072 [Crossref] [ Google Scholar]
- Yokoyama M, Chiba T, Zen Y, Oshima M, Kusakabe Y, Noguchi Y. Histone lysine methyltransferase G9a is a novel epigenetic target for the treatment of hepatocellular carcinoma. Oncotarget 2017; 8:21315-21326. doi: 10.18632/oncotarget.15528 [Crossref] [ Google Scholar]
- Copeland RA, Olhava EJ, Scott MP. Targeting epigenetic enzymes for drug discovery. Curr Opin Chem Biol 2010; 14:505-510. doi: 10.1016/j.cbpa.2010.06.174 [Crossref] [ Google Scholar]
- Henry RA, Kuo YM, Andrews AJ. Differences in specificity and selectivity between CBP and p300 acetylation of histone H3 and H3/H4. Biochemistry 2013; 52:5746-5759. doi: 10.1021/bi400684q [Crossref] [ Google Scholar]
- Li S, Gu XJ, Hao Q, Fan H, Li L, Zhou S, Zhao K. A liquid chromatography/mass spectrometry-based generic detection method for biochemical assay and hit discovery of histone methyltransferases. Anal Biochem 2013; 443:214-221. doi: 10.1016/j.ab.2013.08.029 [Crossref] [ Google Scholar]
- Dhayalan A, Dimitrova E, Rathert P, Jeltsch A. A continuous protein methyltransferase (G9a) assay for enzyme activity measurement and inhibitor screening. J Biomol Screen 2009; 14:1129-1133. doi: 10.1177/1087057109345528 [Crossref] [ Google Scholar]
- Horiuchi KY, Eason MM, Ferry JJ, Planck JL, Walsh CP, Smith RF. Assay development for histone methyltransferases. Assay Drug Dev Technol 2013; 11:227-236. doi: 10.1089/adt.2012.480 [Crossref] [ Google Scholar]
- Kuninger D, Lundblad J, Semirale A, Rotwein P. A non-isotopic in vitro assay for histone acetylation. J Biotechnol 2007; 131:253-260. doi: 10.1016/J.JBIOTEC.2007.07.498 [Crossref] [ Google Scholar]
- Dillon MB, Bachovchin DA, Brown SJ, Finn MG, Rosen H, Cravatt BF. Novel inhibitors for PRMT1 discovered by high-throughput screening using activity-based fluorescence polarization. ACS Chem Biol 2012; 7:1198-1204. doi: 10.1021/cb300024c [Crossref] [ Google Scholar]
- Kumar M, Zielinski T, Lowery RG. Biochemical assay development for histone methyltransferases using a transcreener-based assay for S-Adenosylhomocysteine. Assay Drug Dev Technol 2015; 13:200-209. doi: 10.1089/adt.2014.609 [Crossref] [ Google Scholar]
- Hendricks CL, Ross JR, Pichersky E, Noel JP, Zhou ZS. An enzyme-coupled colorimetric assay for S-adenosylmethionine-dependent methyltransferases. Anal Biochem 2004; 326:100-105. doi: 10.1016/j.ab.2003.11.014 [Crossref] [ Google Scholar]
-
Medintz IL, Hildebrandt N. FRET - Förster Resonance Energy Transfer: From Theory to Applications. Wiley-VCH Verlag GmbH; 2014.
- Hildebrandt N, Wegner KD, Algar WR. Luminescent terbium complexes: Superior Förster resonance energy transfer donors for flexible and sensitive multiplexed biosensing. Coord Chem Rev 2014; 273-274:125-138. doi: 10.1016/J.CCR.2014.01.020 [Crossref] [ Google Scholar]
- Hildebrandt N, Spillmann CM, Algar WR, Pons T, Stewart MH, Oh E. Energy transfer with semiconductor quantum dot bioconjugates: A versatile platform for biosensing, energy harvesting, and other developing applications. Chem Rev 2017; 117:536-711. doi: 10.1021/acs.chemrev.6b00030 [Crossref] [ Google Scholar]
- Machleidt T, Robers MB, Hermanson SB, Dudek JM, Bi K. TR-FRET Cellular assays for interrogating posttranslational modifications of histone H3. J Biomol Screen 2011; 16:1236-1246. doi: 10.1177/1087057111422943 [Crossref] [ Google Scholar]
- Yu V, Fisch T, Long AM, Tang J, Lee JH, Hierl M. High-Throughput TR-FRET Assays for Identifying Inhibitors of LSD1 and JMJD2C Histone Lysine Demethylases. J Biomol Screen 2012; 17:27-38. doi: 10.1177/1087057111418228 [Crossref] [ Google Scholar]
- Ghadiali JE, Lowe SB, Stevens MM. Quantum-dot-based FRET detection of histone acetyltransferase activity. Angew Chem Int Ed Engl 2011; 50:3417-3420. doi: 10.1002/anie.201008263 [Crossref] [ Google Scholar]
- Xu J, Corneillie TM, Moore EG, Law GL, Butlin NG, Raymond KN. Octadentate cages of Tb(III) 2-hydroxyisophthalamides: A new standard for luminescent lanthanide labels. J Am Chem Soc 2011; 133:19900-19910. doi: 10.1021/ja2079898 [Crossref] [ Google Scholar]
- Patnaik D, Hang GC, Estève PO, Benner J, Jacobsen SE, Pradhan S. Substrate specificity and kinetic mechanism of mammalian G9a histone H3 methyltransferase. J Biol Chem 2004; 279:53248-53258. doi: 10.1074/jbc.M409604200 [Crossref] [ Google Scholar]
- Ma F, Liu M, Wang Z, Zhang C. Multiplex detection of histone-modifying enzymes by total internal reflection fluorescence-based single-molecule detection. Chem Commun 2016; 52:1218-1221. doi: 10.1039/C5CC08797J [Crossref] [ Google Scholar]
- Klink TA, Staeben M, Twesten K, Kopp AL, Kumar M, Dunn RS. Development and validation of a generic fluorescent methyltransferase activity assay based on the transcreener AMP/GMP assay. J Biomol Screen 2012; 17:59-70. doi: 10.1177/1087057111421624 [Crossref] [ Google Scholar]
- Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 2007; 25:473-481. doi: 10.1016/j.molcel.2007.01.017 [Crossref] [ Google Scholar]
- Zagni C, Chiacchio U, Rescifina A. Histone methyltransferase inhibitors: novel epigenetic agents for cancer treatment. Curr Med Chem 2012; 20:167-185. doi: 10.2174/0929867311320020002 [Crossref] [ Google Scholar]
- Copeland RA, Olhava EJ, Scott MP. Targeting epigenetic enzymes for drug discovery. Curr Opin Chem Biol 2010; 14:505-510. doi: 10.1016/J.CBPA.2010.06.174 [Crossref] [ Google Scholar]
- Richon VM, Johnston D, Sneeringer CJ, Jin L, Majer CR, Elliston K. Chemogenetic analysis of human protein methyltransferases: chemogenetic analysis of human protein methyltransferases. Chem Biol Drug Des 2011; 78:199-210. doi: 10.1111/j.1747-0285.2011.01135.x [Crossref] [ Google Scholar]