Bioimpacts. 10(4):259-268.
doi: 10.34172/bi.2020.33
Original Research
Designing a new generation of expression toolkits for engineering of green microalgae; robust production of human interleukin-2
Jaber Dehghani 1, 2  , Khosro Adibkia 3, *
, Khosro Adibkia 3, *  , Ali Movafeghi 2, Mohammad M. Pourseif 1, Yadollah Omidi 1, 3, 4, *
, Ali Movafeghi 2, Mohammad M. Pourseif 1, Yadollah Omidi 1, 3, 4, * 
Author information:
1Research Center for Pharmaceutical Nanotechnology, Biomedicine Institute, Tabriz University of Medical Sciences, Tabriz, Iran
2Department of Plant Biology, Faculty of Natural Sciences, University of Tabriz, Tabriz, Iran
3Department of Pharmaceutics, Faculty of Pharmacy, Tabriz University of Medical Sciences, Tabriz, Iran
4Department of Pharmaceutical Sciences, College of Pharmacy, Nova Southeastern University, Fort Lauderdale, Florida 33328, USA
Abstract
Introduction:
Attributable to some critical features especially the similarity of the protein synthesis machinery between humans and microalgae, these microorganisms can be utilized for the expression of many recombinant proteins. However, low and unstable gene expression levels prevent the further development of microalgae biotechnology towards protein production.
Methods:
Here, we designed a novel "Gained Agrobacterium -2A plasmid for microalgae expression" (named GAME plasmid) for the production of the human interleukin-2 using three model microalgae, including Chlamydomonas reinhardtii, Chlorella vulgaris , and Dunaliella salina . The GAME plasmid harbors a native chimeric hsp70/Int-1/rbcS2 promoter, the microalgae specific Kozak sequence, a novel hybrid 2A peptide, and Int-1 and Int-3 of the rbcS2 gene in its expression cassette.
Results:
The obtained data confirmed that the GAME plasmid can transform the microalgae with high transformation frequency. Molecular and proteomic analyses revealed the stable and robust production of the hIL-2 by the GAME plasmid in the microalgae. According to the densimetric analysis, the microalgae can accumulate the produced protein about 0.94% of the total soluble protein content. The ELISA data confirmed that the produced hIL-2 possesses the same conformation pattern with the acceptable biological activity found naturally in humans.
Conclusion:
Most therapeutic proteins need post-translational modifications for their correct conformation, biological function, and half-life. Accordingly, microalgae could be considered as a cost-effective and more powerful platform for the production of a wide range of recombinant proteins such as antibodies, enzymes, hormones, and vaccines.
Keywords: 2A peptide, Agrobacterium tumefaciens,
Chlamydomonas reinhardtii
,
Chlorella vulgaris
,
Dunaliella salina
, Interleukin-2
Copyright and License Information
© 2020 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
In recent years, much attention has been paid to microalgae in terms of their applications in food, cosmetics, biofuel, and pharmaceutics industries.
1-3
Microalgae are considered as enriched resources of vital compounds such as proteins, antioxidants, unsaturated lipids, and vitamins. As a result, they can be exploited for different biomedical applications even the treatment of various diseases such as cancer.
4
Moreover, the eyespot and channelrhodopsins in the microalgae are located in the center of optogenetic and neuroscience for illuminating the fundamentals of eyes and brain functions.
5-7
Although the genetic manipulation of microalgae is lower than that of the other expression hosts, their key characteristics make them an interesting expression platform for the production of recombinant proteins.
8,9
They can be used as promising biofactory for the production of a wide range of proteins, in large part because of being “generally regarded as safe” (GRAS) for human usages, the possibility for the scale-up in closed/controlled bioreactors, and the capability to assemble of multi-subunit proteins.
10-12
Nowadays, the potentials of microalgae species such as Chlamydomonas reinhardtii , Phaeodactylum tricornutum , Dunaliella sp. and Chlorella vulgaris , have been evaluated for the expression of different type of proteins such as enzymes, hormones, monoclonal antibodies (mAbs) and the other therapeutic biologics.
8,9,13-15
The transformable compartments of microalga include mitochondria, chloroplast, and nucleus. Of these, the transformations of the two latter organelles have gained more attention for the production of recombinant proteins,
16
while the lack of gene silencing and the possibility of simultaneous insertion of multiple genes are the most important advantages of the chloroplast transformation.
17,18
The chloroplast engineering, however, may face some drawbacks, including the absence of N-glycosylation machinery, intense codon usage bias, and difficulties of the transformation procedures.
19
Altogether, the nuclear transformation seems to provide some benefits such as the capability to perform post-translational modifications such as N-glycosylation and disulfide bridges. Further, it provides a great possibility for the targeting of the expressed proteins into the subcellular compartments or secreting them into the culture media.
8,10
However, the gene expression by the nuclear transformation is low and unstable, in large part because of the use of poor promoters, position effects, transgene silencing, and bias in codon usage.
20
The gene silencing is deemed to happen at both transcriptional and translational levels due to the functional presence of nuclease and proteases in the microalgae cytosol.
4
In addition, the insertion of the genes of interest (GOIs) into the nuclear genome might occur randomly, causing the truncate of the important host genes or even cell death.
10
Recent researches identified codon optimization using native and chimeric strong promoters and showed that the untranslated regions (UTRs) might be involved in the gene expression of microalgae.
21
Further studies affirmed that the introns (Ints) can impose positive effects on the gene expression levels in eukaryotic cells through the control of mRNA formation and their stabilities and also exporting the transcripts into the nuclear.
22
However, the number, position, and orientation of the inserted Ints in the expression cassette might influence the gene expression efficiency.
23
The 2A peptide is recently utilized to improve the gene expression levels in the different eukaryotic hosts. This peptide consists of ~20 amino acids and involves in the self-cleavage reaction. In fact, during the elongation step of protein translation, a peptide bond could not be formed in the two last amino acids, resulting in the cleavage of the 2A peptide.
24,25
Such peptides were first identified in the foot and mouth disease virus (the so-called P2A), which was then shown to be able to increase the transcription and translation efficiency of the genes.
26
Afterward, several 2A-like peptides (e.g., T2A, E2A, BmCPV 2A and, BmIFV 2A) have been identified in some viruses and named based on their original hosts.
27,28
Further, owing to the transferable nature of the T-DNA of the pTi plasmid, the use of Agrobacterium -based plasmids is another approach for the stable transformation of microalgae and also a wide range of hosts such as plants, yeasts, fungi, and even human cells.
4,10,29,30
Herein, we constructed a novel Agrobacterium -2A-based (the so-called GMAE plasmid) system, containing hsp70/Int-1/rbcs2 chimeric promoter, microalgae specific Kozak sequence, a chimeric 2A peptide, Int-1 and -3 of the rbcS2 gene and UTRs optimized for the robust and stable expression of hIL-2 gene in the nuclear of the C. reinhardtii . The GMAE plasmid is also successfully utilized for the efficient transformation of C. vulgaris and D. salina cells with the hIL-2 gene. Molecular and proteomic analyses showed that the GMAE plasmid can stably transform the microalgal cells and robustly produce the hIL-2 protein while the ELISA data confirmed the produced hIL-2 has acceptable biological activity.
Materials and Methods
Materials
The antibiotics (kanamycin, rifampicin, cefotaxime, and chloramphenicol) and also the Ni-Sepharose™ 6 Fast Flow resin were purchased from the Sigma-Aldrich Co. (Missouri, United States). The Taq DNA Polymerase Master Mix for the PCR experiments was obtained from the Ampliqon Co. (Odense, Denmark). The reagents and enzymes used for RNA extraction, cDNA synthesis, and qPCR experiments were purchased from Thermo Fisher Scientific Co. (Waltham, MA, USA). The primary antibodies (i.e., anti-His-tag, anti-hIL-2, and anti-CmR) and also the goat anti-rabbit IgG (alkaline phosphatase conjugate) antibodies, for western blotting, were respectively purchased from the Santa Cruz Biotechnology Co. (Heidelberg, Germany) and Abcam Co. (Cambridge, MA, USA). The reagents for the ELISA assay were obtained from the Bio-Techne Co. (Minnesota, USA).
Microalgae cultivation
The C. reinhardtii , D. salina , and C. vulgaris cells were respectively grown in tris-acetate–phosphate (TAP), modified Johnson’s, and BG-11 media. The microalgae were cultured under continuous illumination by white florescent lamps (80 μmol photon m-2s-1 irradiance) at 25°C temperature, and 16:8 (light: dark) photoperiod.
10
Agrobacterium-2A-based plasmid construction
The GMAE plasmid was built on the pCAMBIA1304 backbone. An enhanced hsp70 and rbcS2 regulatory elements which are linked through the Int-1 of the rbcS2 gene were synthesized as a promoter. A novel 5'-UTR, start codon, and microalgae-specific Kozak sequence was placed after the chimeric promoter. Subsequently, the chloramphenicol acetyltransferase (CmR ) gene, which was used for the selection of the transformed microalgae cells and also the coding sequence of the hIL-2, was placed in the construct in the same open reading frame (ORF). Two latter genes were fused together with a novel chimeric 2A peptide. Moreover, two copies of Int-1 were inserted between the hIL-2 exons. Then, six histidines (i.e., 6x His-tag) were placed before the stop codon, utilizing the hIL-2 purification. It should be noted that the recognizing sequence of the XhoI and BamHI restriction enzymes were respectively inserted before and after the IL-2 coding sequence for the easy cloning of the other GOIs. Further, one copy of the Int-3 was located after the hIL-2 coding sequence. The designed construct was synthesized by GenScript Co. and cloned between the SacII and BstEII restriction sites in the pCAMBIA1304 backbone (Figs. S1-S3). The final scheme of the plasmid was achieved by the SnapGene software.
Microalgae transformation procedures
About 200 µL of the microalgae cultures were harvested in the log phase of growth (OD660: 0.4-0.5), cultured on the solidified media and incubated for about five days to form the algal lawns. Then,A. tumefaciens strain EHA101, which harbored the GAME plasmid, was incubated at 28°C and 220 rpm overnight in the 2x LB medium containing 20 mg/L rifampicin and 50 mg/L kanamycin. Further, about 350 μL of the A. tumefaciens cells were inoculated with 25 mL of the 2x LB medium and incubated at 28°C for about four hours, by which the OD660 of the bacterial cells were reached to almost 0.6. Then, the bacterial cells were harvested by centrifugation at 5000 ×g for 5 minutes. The resultant pellets were resuspended in the appropriate liquid media based on the microalgae type. Afterward, about 300 µL of the Agrobacterium cultures were spread on the microalgae lawns and co-cultivated at 23°C for 48 hours in the presence of white light. The cocultured cells were harvested and washed three times with the fresh liquid media containing 500 mg/L cefotaxime to eliminate the Agrobacterium cells. Finally, the washed microalgae cells were cultured on the solidified media containing the appropriated concentration of the chloramphenicol for the selection of the transformants (Fig. S1).
4,10
DNA analysis
Genomic DNAs were extracted at the late log phase of growth from the transformed and untransformed microalgae by using the following buffer (2% CTAB, 100 mM Tris-HCl, 1.4 M NaCl, 1% PVP, 20 mM Na2EDTA, 0.2% LiCl, PH: 8).
31
The presence of the hIL-2 (i.e., the Exs-3-4) and CmR genes was evaluated by polymerase chain reaction (PCR) reactions and the specific primer sets (Table 1).
10
The utilized primer sets were designed by Oligo software version 7 and synthesized by GenScript Company (Nanjing, China). The PCR thermal cycle conditions were initial denaturation at 94˚C for 5 minutes, annealing at 52-62˚C for 1 minute and extension at 72˚C for 1 minute, which was performed for 32 successive cycles followed by a final extension step at 72˚C for 5 minutes (Table 1). The PCR products were analyzed using 1 and 2% agarose gel electrophoresis.
Table 1.
The list of used primers
|
Primers
|
Forward sequence
|
Reverse sequence
|
T
m
(C°)
|
| IL-2 |
5'-ATGGTAGATCTGACTAGTAAAGG-3' |
5'-TCATCCATGCCATGTGTAATC-3' |
52 |
|
CmR
|
5'-ATGAAAAAGCCTGAACTCACCG-3' |
5'-CTATTTCTTTGCCCTCGGACG-3' |
56 |
| hsp70 |
5'-GCAAATTACATATGTCTGCGTG-3' |
5'-GTTTACTGACTCTTAAGCTAGCTG-3' |
52.5 |
| rbcS2 |
5'-TCCCATCAAGCTTGCATGCC-3' |
5'-AAAGAACATAGGCCCCCTGG-3' |
57 |
| Int-1 |
5'-GTGAGTCGACGAGCAAGCCC-3' |
5'- CTGCAAATGGAAACGGCGACG-3' |
60 |
| Int-3 |
5'-GTAAGTCTGGCGAGAGCCCGACG-3' |
5'- CTGCGGGCGCACGGGAAATG-3' |
62 |
| α-tubulin |
5'-CCGCCTGGACCACAAGTTC-3' |
5'-TAGTACTCCTCACCCTCGCC-3' |
58.5 |
RNA analysis
Total RNAs were extracted from the transformed and untransformed microalgae cells using the TRIzol® reagent according to the manufacture instruction. The quality and quantity of the RNAs were evaluated with a NanoDrop ND®-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and electrophoresis on 2% agarose gel. The complementary DNA (cDNA) from the extracted RNAs were synthesized by the RevertAid First Strand cDNA Synthesis Kit based on the manufacture instruction. The PCR reactions with cDNAs as a template and the specific primers were performed in the same manner mentioned about the DNA analysis. The qPCR reactions were performed to measure the expression level of the hIL-2 (i.e., the Exs-3-4) gene using a power SYBRTM Green master mix on a Bio-Rad IQ5 real-time PCR detection system (Marnes- La Coquette, France). The α-tubulin gene from the untransformed C. reinhardtii is used as a housekeeping control (Table 1).
Western blotting assay
The TSPs were extracted from the transformed and untransformed microalgae cells using the CelLytic™ P Cell Lysis Reagent. The concentration and purity of the extracted proteins were determined by the absorbance at 280 nm in the NanoDrop® ND-1000 spectrophotometer. Further, about 20 mg of the TSPs were subjected to 15% SDS-PAGE gel. After the separation, the proteins were electrophoretically transferred to a nitrocellulose membrane using a semi-dry blotting system (Bio-Rad, Hercules, CA, USA) and blocked with nonfat skim milk 5% for 24 hours. Furthermore, the membranes were incubated with the primary anti-His-tag, anti-hIL-2, and anti-CmR antibodies (1:1000), and then, the proteins were detected by goat anti-rabbit IgG-alkaline phosphatase conjugate (1:1000) on the basis of the manufacture procedures.
Protein purification and ELISA assay
The TSPs were extracted from 150 mL microalgae cultures to purify the produced hIL-2 protein. Subsequently, the whole extracted proteins were dialyzed through the dialysis tubes with 10 kDa cut-off (Sigma-Aldrich, USA). Further, protein purification was performed by affinity chromatography in which the proteins were incubated with the Ni-Sepharose™ 6 Fast Flow resin at 4°C for 1 hour. After removing the flow-through, the resin was washed two times by binding buffer (20 mM NaH2PO4, 0.5 M NaCl and 5mM Imidazole) and subsequently, the 6x His-tagged proteins were collected by elution buffer (20 mM NaH2PO4, 0.5 M NaCl and 500 mM Imidazole). The quantity of the purified hIL-2 was evaluated by NanoDrop ND®-1000 spectrophotometer and subsequently, western blotting was performed against the purified hIL-2 by an anti-His-tag antibody.
The ELISA assay was performed for the evaluation of the correct conformation of the microalgae produced hIL-2 compared to that of in human cells. The whole proteins were extracted from the transformed and untransformed C. reinhardtii cells using phosphate-buffered saline (PBS) buffer (10 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCl, and 137 mM NaCl) and sonication and then three concentrations of the extracted proteins (i.e., 10, 20 and 30 µL) were subjected into the ELISA sandwich. Moreover, 10 µL of the total protein from the untransformed microalga was used as a negative control. These approaches were performed three times for each concentration (Table S2). The ELISA analysis was performed by the DuoSet® Human IL-2 kit based on the company instruction.
Statistical analysis
Data were analyzed as the mean values with standard deviation (SD) from three independent experiments. The statistical analyses were utilized by Student t -test or ANOVA. A P value of less than 0.05 was considered statistically significant.
Results
Designing the GMAE plasmid
To be highly effective on the expression level of genes in the C. reinhardtii , the GMAE plasmid was designed as an Agrobacterium -2A-based construct consisting of different elements, including hsp70/rbcS2 promoter, Int-1and Int-3. The hsp70 (300 bp), rbcS2 (200 bp) regions and the Int-1 (145 bp) and Int-3 (238 bp) of the rbcS2 gene were amplified from the C. reinhardtii through the specific primers, and subsequently, their sequences were determined (Fig. 1 and Figs. S2-S4). The coding sequences of the CmR and hIL-2 were extracted from the NCBI database and then optimized based on the C. reinhardtii codon bias. Furthermore, the sequences of the P2A-T2A peptide, in which the coding sequences of GSG amino acids were fused to the C-termini of the peptide. The chimeric promoter (hsp70/Int-1/rbcS2) and expression cassettes (CmR -2A peptide-hIL-2) and regulatory region (Int-3) were placed continuously in the same ORF. Finally, the 25 kb synthetic construct was cloned into the pCAMBIA1304 backbone through SacII and BStEII restriction enzymes and the GMAE plasmid with 8843 bp size was built (Figs. 1 and 2 and Figs. S1-S3).
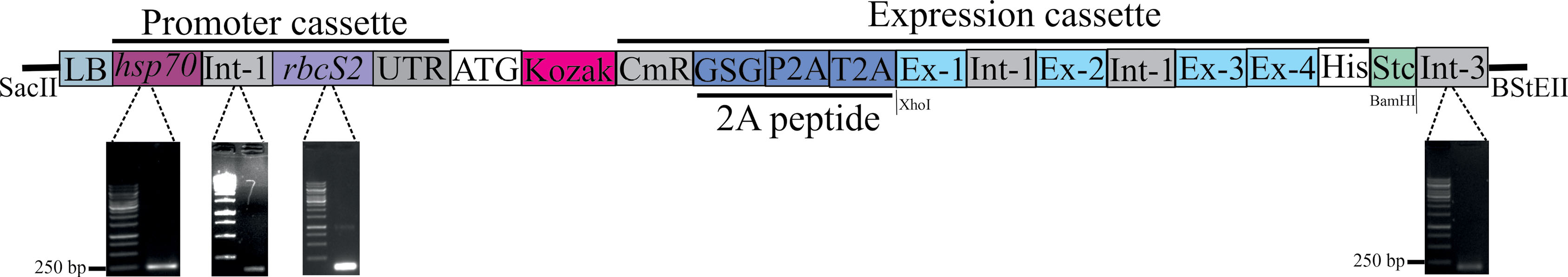
Fig. 1.
The schematic image of the deigned construct containing a promoter and expression cassettes. The hsp70, Int-1, rbcS2, and Int-3 elements were isolated and sequenced from the C. reinhardtii genome. LB: Left border from the A. tumefaciens , Ints: Intron 1 of the rbcS2 gene, CmR : Chloramphenicol acetyltransferase, Stc: Stop codon (i.e., TAA). The synthetic elements were cloned into the pCAMBIA1304 backbone through the SacII and BStEII restriction enzymes.
.
The schematic image of the deigned construct containing a promoter and expression cassettes. The hsp70, Int-1, rbcS2, and Int-3 elements were isolated and sequenced from the C. reinhardtii genome. LB: Left border from the A. tumefaciens , Ints: Intron 1 of the rbcS2 gene, CmR : Chloramphenicol acetyltransferase, Stc: Stop codon (i.e., TAA). The synthetic elements were cloned into the pCAMBIA1304 backbone through the SacII and BStEII restriction enzymes.
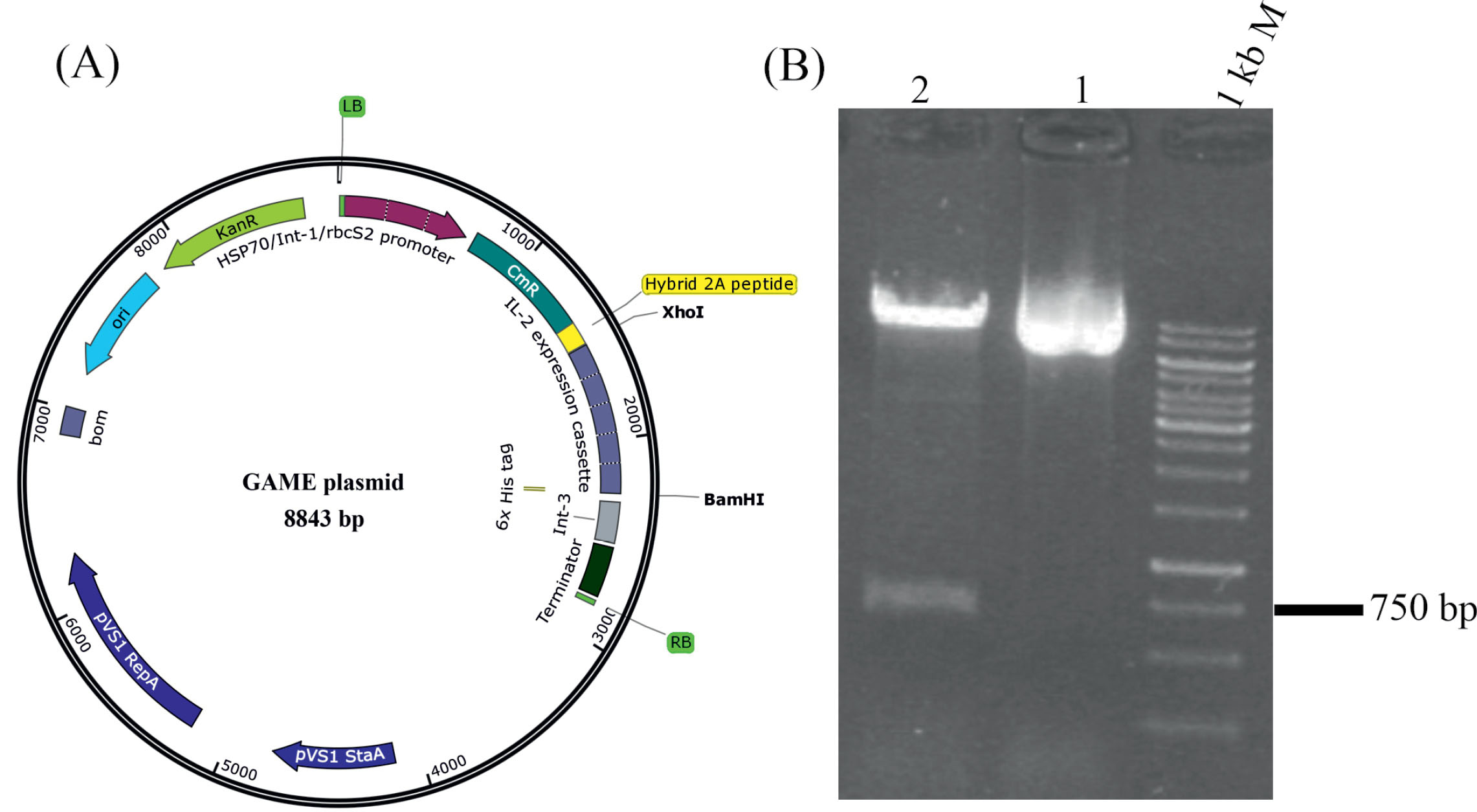
Fig. 2.
The structure of the GMAE plasmid and its electrophoresis. (A) The plasmid is an Agrobacterium -2A-mediated construct optimized based on the C. reinhardtii codon bias. (B) The size of the GMAE plasmid is 8843 bp (Lan 1) according to electrophoresis analysis. For more validation about the construction of GMAE plasmid, the vector was double digested with BamHI and XhoI restriction enzymes with restriction sites at the first and end of the hIL-2 expression cassette. As expected, two bands with ~760 bp and ~8 kb were determined after electrophoresis (Lan 2).
.
The structure of the GMAE plasmid and its electrophoresis. (A) The plasmid is an Agrobacterium -2A-mediated construct optimized based on the C. reinhardtii codon bias. (B) The size of the GMAE plasmid is 8843 bp (Lan 1) according to electrophoresis analysis. For more validation about the construction of GMAE plasmid, the vector was double digested with BamHI and XhoI restriction enzymes with restriction sites at the first and end of the hIL-2 expression cassette. As expected, two bands with ~760 bp and ~8 kb were determined after electrophoresis (Lan 2).
Microalgae transformation and its efficiency
Although the GMAE plasmid is optimized for the
C.
reinhardtii
, its potential was evaluated for the nuclear
transformation of C. vulgaris and D. salina as well. The
chloramphenicol sensitivity assays revealed that 60 and 130
mg/L of the antibiotic were suitable for the transformation
of D. salina and C. reinhardtii or C. vulgaris, respectively
(Fig. S1). Microalgae transformations were performed via
the cocultivation of the C. reinhardtii, C. vulgaris, and
D.
salina
(OD660: 0.5) and EHA101 strain of A. tumefaciens
which held the GAME plasmid. Further, the microalgae
are transferred into the solidified media containing the
appropriate concentration of chloramphenicol and after
about 10 days the transformed cells appeared in the media
(Fig. 3). Our data, based on the number of survived cells
on the plates containing chloramphenicol from a count of
106 cfu, showed that the GAME plasmid could transform
the mentioned microalgae with high frequency. The data revealed the transformation efficiency is relatively higher
in the C. reinhardtii (157 ± 0.8 cfu per 106 cells) compared
to the C. vulgaris (157 ± 0.8 cfu per 106 cells), and D. salina
(157 ± 0.8 cfu per 106 cells), as shown in Fig. 3 and Table 2.
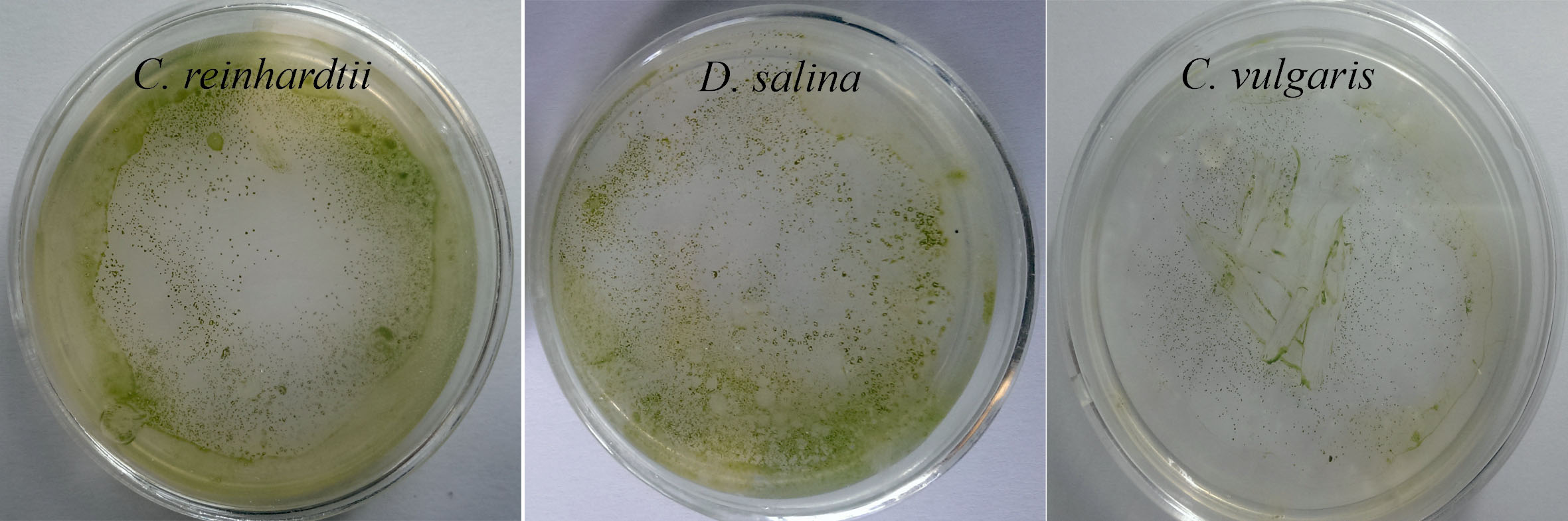
Fig. 3.
Appearing the chloramphenicol resistance microalgae cells in the solid media containing the appropriate concentration of the antibiotic.The transformation frequency in the C. reinhardtii is relatively higher than the D. salina and C. vulgaris .
.
Appearing the chloramphenicol resistance microalgae cells in the solid media containing the appropriate concentration of the antibiotic.The transformation frequency in the C. reinhardtii is relatively higher than the D. salina and C. vulgaris .
Table 2.
Transformation frequency by GMAE plasmid
|
Microalgae
|
No. of cells used for transformation
|
No. of transformants
|
Transformation frequency ( cfu /10
6
cells)
|
|
C. reinhardtii
|
4.6 × 106
|
815 |
177 ± 1.7 |
|
D. salina
|
5.1 × 106
|
758 |
148 ± 6.2 |
|
C. vulgaris
|
4.8 × 106
|
563 |
117 ± 2.9 |
T-DNA integration into the microalgae genomes
The PCR experiments from the genomic DNA of the transformed C. reinhardtii , C. vulgaris , and D. salina with the specific primers (i.e., ~654 bp and ~252 bp amplicons as shown in Table 1) confirmed the successful integration of the CmR and hIL-2 (i.e., the Exs-3-4) genes into the microalgae genomes respectively (Figs. 4A and 4C). Further, the PCR reactions by the primers (Table 1) and the cDNAs from the transformed and untransformed microalgae as template validated the same results, indicating the successful transcription of the genes (Figs. 4B and 4D). Furthermore, the detection of the hIL-2 was reconfirmed by the qPCR, in which the results showed the expression level of the hIL-2 is stronger in the C. reinhardtii in comparison with those of the C. vulgaris , and D. salina (Fig. 5C).
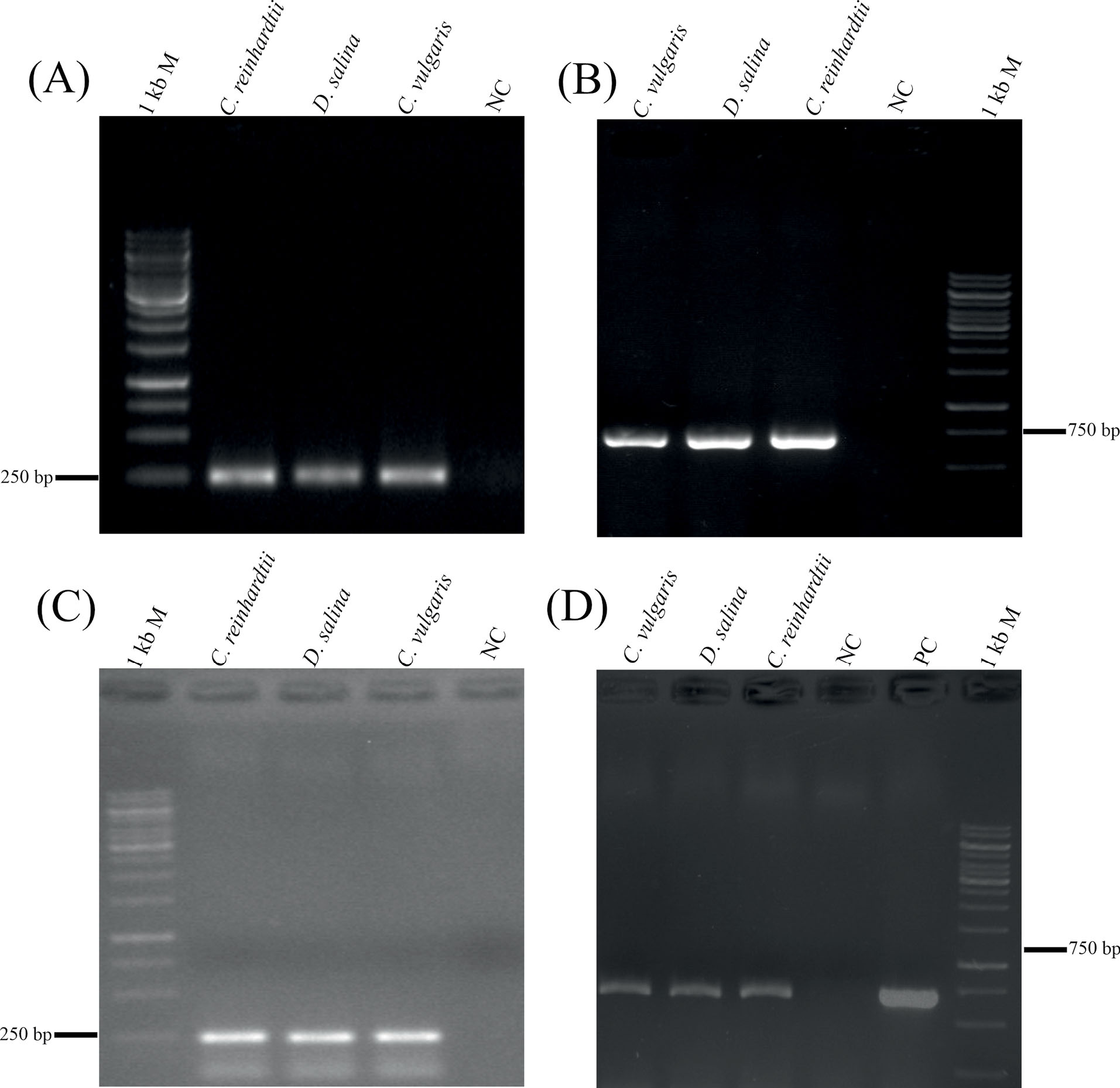
Fig. 4.
Successful integration and transcription of the IL-2 (panels A and C) and CmR (panels B and D) genes into the genome and transcriptome of the C. reinhardtii , D. salina , and C. vulgaris . NC: negative control (i.e., untransformed C. reinhardtii ), PC: Positive control (GMAE plasmid).
.
Successful integration and transcription of the IL-2 (panels A and C) and CmR (panels B and D) genes into the genome and transcriptome of the C. reinhardtii , D. salina , and C. vulgaris . NC: negative control (i.e., untransformed C. reinhardtii ), PC: Positive control (GMAE plasmid).
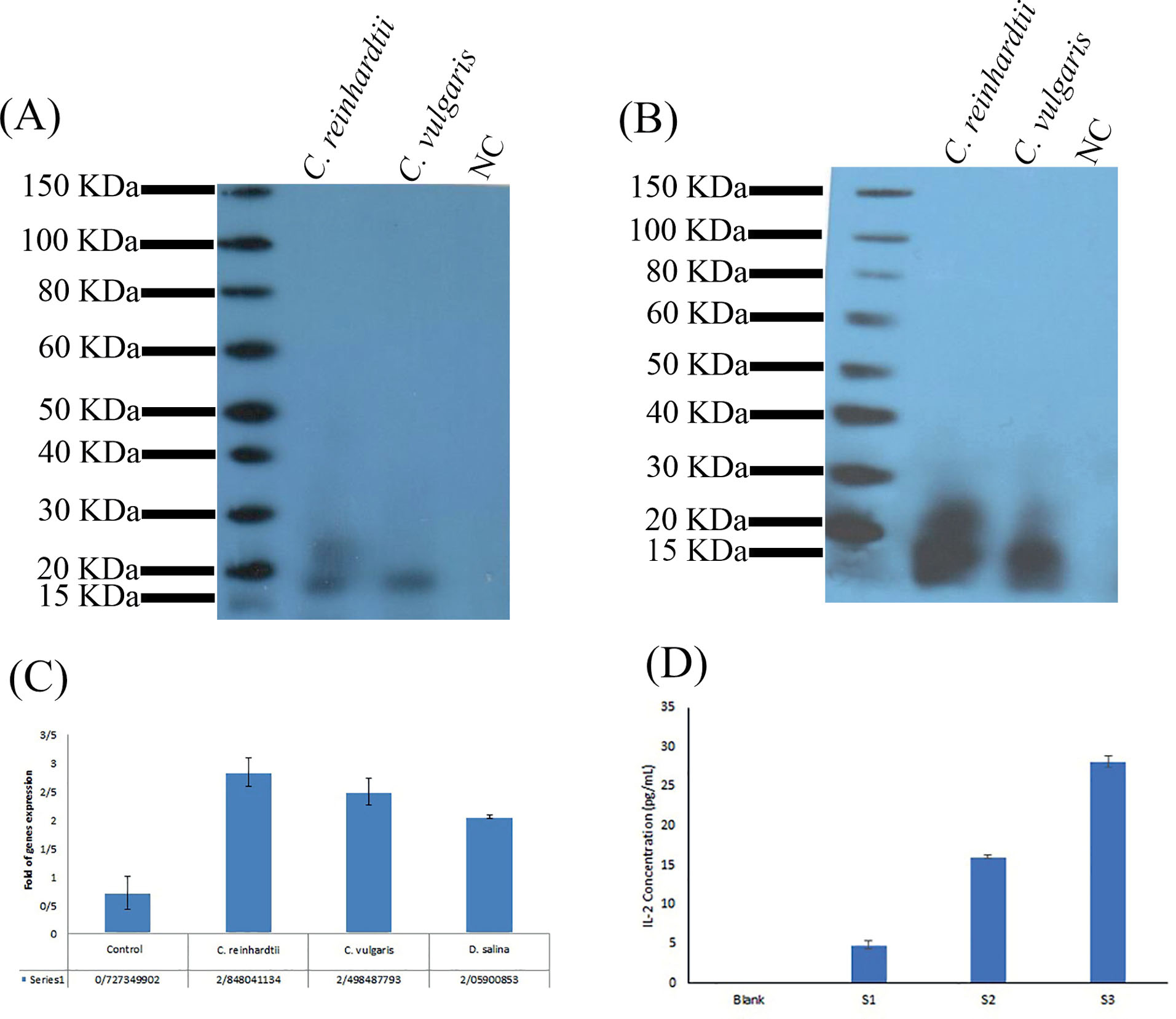

Fig. 5.
Stable hIL-2 expression, production, purification, and biological activity assays. (A) After five months from the transformation, western blotting with the anti-His-tag antibody confirmed the presence of hIL-2 in the transformed C. reinhardtii . (B) Successful purification of hIL-2 from 150 mL culture of the C. reinhardtii cells. Densitometry analysis of western blotting bands by the ImageJ software identified the hIL-2 can accumulate approximately 0.94% and 0.59% of the total soluble proteins of the C. reinhardtii and C. vulgaris respectively. (C) The qPCR data identified the expression level of hIL-2 in the transformed C. reinhardtii is somewhat higher than the C. vulgaris and D. salina cells. (D) The ELISA analysis of the whole extracted proteins from the transformed C. reinhardtii revealed the acceptable biological activity of the expressed hIL-2. Control: α-tubulin gene of the untransformed C. reinhardtii , S1, 2 and 3: Different elution of the whole extracted proteins from the transformedC. reinhardtii , Blank: whole extracted proteins from the untransformedC. reinhardtii .
.
Stable hIL-2 expression, production, purification, and biological activity assays. (A) After five months from the transformation, western blotting with the anti-His-tag antibody confirmed the presence of hIL-2 in the transformed C. reinhardtii . (B) Successful purification of hIL-2 from 150 mL culture of the C. reinhardtii cells. Densitometry analysis of western blotting bands by the ImageJ software identified the hIL-2 can accumulate approximately 0.94% and 0.59% of the total soluble proteins of the C. reinhardtii and C. vulgaris respectively. (C) The qPCR data identified the expression level of hIL-2 in the transformed C. reinhardtii is somewhat higher than the C. vulgaris and D. salina cells. (D) The ELISA analysis of the whole extracted proteins from the transformed C. reinhardtii revealed the acceptable biological activity of the expressed hIL-2. Control: α-tubulin gene of the untransformed C. reinhardtii , S1, 2 and 3: Different elution of the whole extracted proteins from the transformedC. reinhardtii , Blank: whole extracted proteins from the untransformedC. reinhardtii .
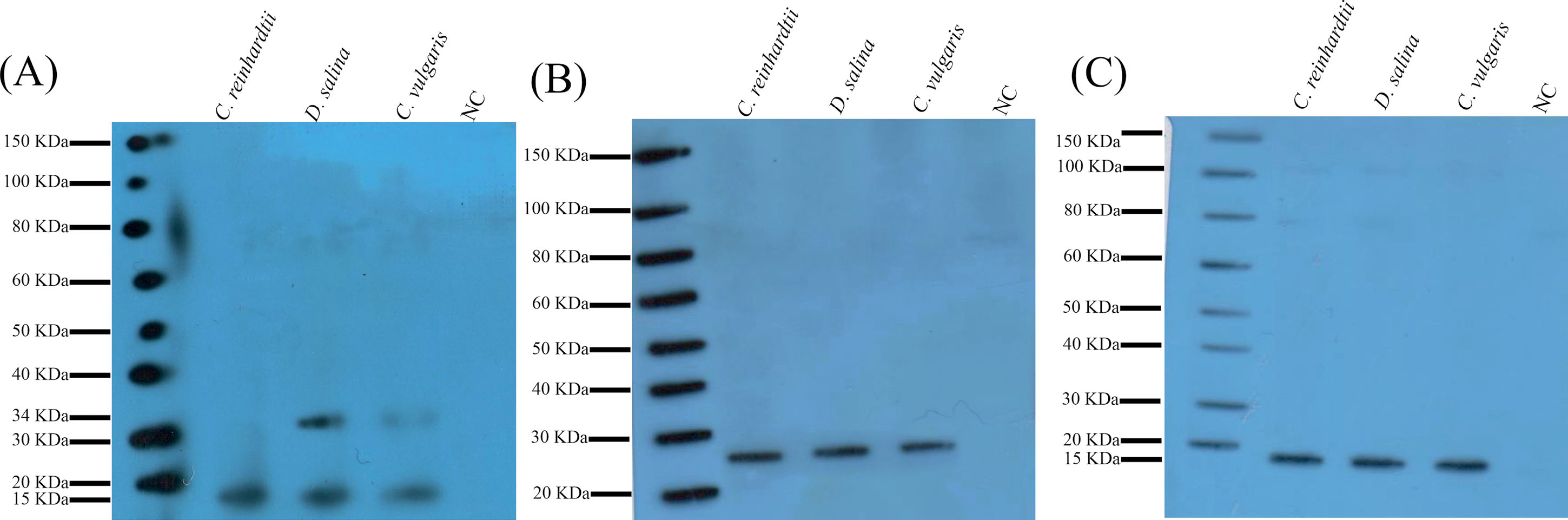
Fig. 6.
The western blotting analyses of the transformed microalgae cells. (A) Detection of hIL-2 with the size of 15 kDa (similar to the human IL-2) in the C. reinhardtii , D. salina , and C. vulgaris . The additional 34 kDa band shows the dimerized IL-2 in the D. salina and C. vulgaris . Panels B and C represent the western blotting analyses with the anti-CmR and anti-His-tag antibodies respectively. These data show that the transformed microalgae can entirely process the GSG-P2A-T2A self-cleaving peptides.
.
The western blotting analyses of the transformed microalgae cells. (A) Detection of hIL-2 with the size of 15 kDa (similar to the human IL-2) in the C. reinhardtii , D. salina , and C. vulgaris . The additional 34 kDa band shows the dimerized IL-2 in the D. salina and C. vulgaris . Panels B and C represent the western blotting analyses with the anti-CmR and anti-His-tag antibodies respectively. These data show that the transformed microalgae can entirely process the GSG-P2A-T2A self-cleaving peptides.
Microalgae transformation and its efficiency
Although the GMAE plasmid is optimized for the C. reinhardtii , its potential was evaluated for the nuclear transformation of C. vulgaris and D. salina as well. The chloramphenicol sensitivity assays revealed that 60 and 130 mg/L of the antibiotic were suitable for the transformation of D. salina and C. reinhardtii or C. vulgaris , respectively (Fig. S1). Microalgae transformations were performed via the cocultivation of the C. reinhardtii , C. vulgaris , and D. salina (OD660: 0.5) and EHA101 strain of A. tumefaciens which held the GAME plasmid. Further, the microalgae are transferred into the solidified media containing the appropriate concentration of chloramphenicol and after about 10 days the transformed cells appeared in the media (Fig. 3). Our data, based on the number of survived cells on the plates containing chloramphenicol from a count of 106 cfu, showed that the GAME plasmid could transform the mentioned microalgae with high frequency. The data revealed the transformation efficiency is relatively higher in the C. reinhardtii (157 ± 0.8 cfu per 106 cells) compared to the C. vulgaris (157 ± 0.8 cfu per 106 cells), and D. salina (157 ± 0.8 cfu per 106 cells), as shown in Fig. 3 and Table 2.
Production of hIL-2 protein in the microalgae
Western blotting data identified a 15 kDa sharp band in the transformed C. reinhardtii , D. salina , and C. vulgaris (Fig. 6A). The data showed an additional band with a size of ~34 kDa is presented in the transformed C. vulgaris , and D. salina cells, which appeared to be the dimerized hIL-2 proteins (Fig. 6A). Interestingly, semi-quantitative data revealed that the expression levels of hIL-2 displayed no significant differences in the transformants, in large part because of the possible similarity in the codon bias of C. reinhardtii , C. vulgaris , and D. salina .
Functional analysis of the 2A peptide in the microalgae
Regarding the self-cleaving characteristic of the 2A peptides, the correct processing of the expression cassette (i.e., CmR-2A-hIL-2 ) during the translation could make CmR and hIL-2 as the discrete proteins. The screening of chloramphenicol resistance colonies in the C. reinhardtii , D. salina , and C. vulgaris could be the first evidence of the proper processing of the 2A peptide (i.e., GSG-P2A-T2A) in the microalgae (Fig. 3). Moreover, western blotting with anti-His-tag and anti-CmR antibodies showed the mentioned microalgae can completely process the 2A peptide in which ~15 (related to the hIL-2) and ~26 kDa (related to the CmR) distinct bands could determine in the TSPs of the transformants. Moreover, the unprocessed CmR-2A-hIL-2 protein, which has a molecular weight of about 41 kDa, was not identified during the immunoblotting analyses (Figs. 6B and 6C).
Protein expression stability and quantity
The stability and quantity of the expressed hIL-2 are assessed in the extracted proteins of the transformed C. reinhardtii and D. salina after 5 months from the transformation. The western blotting analyses represented that the transformed colonies could stably produce the hIL-2 protein in similar concentration to the freshly transformed microalgae (Fig. 5A).
Protein purification and ELISA assay
The produced hIL-2 in theC. reinhardtii and C. vulgaris was extracted and purified from the 150 mL of the microalgae culture in the late exponential phase of growth through 6x His-tag. Western blotting with anti-hIL-2 antibody confirmed the robust quality and quantity of the produced protein (Fig. 5B). The semi-quantitative analyses from the TSPs and purified hIL-2 revealed that the transformed C. reinhardtii and C. vulgaris could respectively produce about 0.94% and 0.59% of the recombinant protein (Figs. 6A and Fig. 5B). Furthermore, the ELISA analysis confirmed the C. reinhardtii produced IL-2 with proper conformation compared to that of the human cells. Based on these findings, the produced IL-2 showed acceptable biological activity (Fig. 5D, Figs. S5 and Table S1).
Discussion
Most of the currently approved/used biologics are complex proteins, which contain glycosylation sites and disulfide bridges required for their correct conformation, biological functions and pharmacokinetic and pharmacodynamic properties
32
. The hIL-2 protein harbors one glycosylation site in the Thr-3 and two disulfide bridges in the 58 and 105 amino acid residues. As a vital secreted cytokine, hIL-2 plays a fundamental role in the regulation and activation of different immune cascades.
33
This protein is routinely used for the immunotherapy of various malignancies, including kidney, liver, gastric, and colorectal cancers.
34
Besides, hIL-2 could be utilized as a strong adjuvant for the deigning of various human and animal vaccines.
35
Currently, the recombinant hIL-2 is produced commercially in the Escherichia coli cells (Aldesleukin™ or Proleukin™ marketed by Prometheus Laboratories Inc.), which are a non-glycosylated molecule. Of note, the glycosylation is necessary not only for the biological activity of hIL-2 but also for increasing the solubility and in vivo half-life of this protein.
36
Further, the lower solubility of the non-glycosylated hIL-2 causes somewhat hurdles in terms of the production, formulation, and administration. Moreover, the formation of insoluble inclusion bodies during E. coli protein expression is another important problem, upon which hIL-2 protein may aggregate and show immunogenicity to some extent.
37
In this sense, exploring a novel powerful expression system is highly demanded for the efficient production of various biotech drugs.
Microalgae are considered as an alternative expression platform for the production of a wide range of different biologics, in large part because of some critical features such as showing great resilience in the production of glycosylated proteins, and similarity of the protein synthesis machinery to that of the human.
4,10
However, the main bottleneck of the microalgae transformation is the transient and poor expression of GOIs from the nuclear genome which may limit its usefulness for the production of protein-based pharmaceuticals.
8
To tackle these challenges, we described a novel approach for the achievement of stable and high expression levels of recombinant proteins during the nuclear transformation. We previously reported the usefulness of the pCAMBIA1304, as an Agrobacterium -based plasmid and showed the stable expression of the green fluorescent protein and β-glucuronidase as a reporter (gfp:gus ) fused genes in the green microalga Dunaliella pseudosalina .
10
Further, we have implemented the same approach for the stable and efficient expression of the gfp:gus fused genes in the blue-green microalga Spirulina platensis , which contains more than ten resident restriction enzymes.
4
Similarly, the LBA4404 strain of A. tumefaciens is utilized to stably transform the pCAMBIA1304 into the genome of C. reinhardtii.
38
The stable expression of enhanced gfp:gus genes in the Schizochytrium protoplasts has already been reported in A. tumefaciens strain EHA105 and LBA4404, where the latter strain displayed the higher transformation capacity.
39
Of note, the A. tumefaciens strains possess a group of genes (VirD2 , VirE2 , VirE3, and VirF ), whose products are effective on the correct and efficient nuclear transformation. Indeed, VirD2 protein is linked to the 5ʹ end and also the VirE2 is covered the whole length of the GOI, which can protect the GOI from the inadvertent degradation by the host endonucleases.
10
After importing the gene into the host cell, the nuclear localization signal existed in the VirD2 and VirE2 proteins and also VirE2 help the gene to direct into the nucleus. In the nucleus, the VirF protein mediates the uncoating process in which the gene is integrated into the transcriptionally active regions of the host genome.
40
Accordingly, we built our expression cassette into the pCAMBIA1304 backbone (Fig. 1) to achieve the Agrobacterium -based expression system (Fig. 2), which resulted in a specific transformation of the microalgae with high efficiency and stability (Figs. 3, 4 and 5). In this line, we used the hsp70 with an additional 3'-UTRs and rscS2 promoters that fused together through the Int-1 of rscS2 gene. Moreover, an enhanced 5'-UTR and the microalgae specific Kozak sequence were added in the expression cassette, in which the position of start codon is presented for the first time in the microalgae transformation procedures. Because of the regulatory nature of the Int-3 from the rscS2 gene in the transcriptional and post-transcriptional levels, the element was inserted in the downstream of the expression cassette (Figs. S1-S4). Further, the codon usage optimization of the heterologous genes and use of the strong native promoters or chimeric constitutive promoters (e.g., hsp70/rbcS2 ) could elicit a significant increase in the level of expressed genes.
41,42
Besides, consistent with our results, codon-optimized hepatitis B surface antigen (HBsAg) and its corresponding Ab have been expressed in a high expression level in the microalga P. tricornutum using nitrate reductase (NR ) promoter and terminator.
42
Additionally, the existing of UTRs (e.g., 3'-UTRs) on the downstream of heterologous genes is effective on the gene expression efficiency. The 3'-UTRs contain elements, which are involved in the correct termination of the transcripts via regulation translation efficiency, the stability of mRNA, and polyadenylation signals.
43,44
Surprisingly, it seems the first intron of rbcS2 gene in C. reinhardtii and also intron 1 or introns 9 and 10 of NR gene in Volvox carteri could improve the post-transcriptional and post-transitional expression levels of GOI.
45,46
Using the 2A peptides in the expression cassette is another trend to increase the quantity and stability of microalgae expressed genes. It is noteworthy that the linking of the P2A peptide to GFP was shown to significantly increase the fluorescent density during the nuclear expression of the C. reinhardtii .
8
In addition, the peptide is inserted between sh-ble gene and the coding sequence of the xylanase I (Xyn1), as a key industrial enzyme. This method could highly improve the accumulation of the protein (i.e., almost 100 times) compared to unlinked Xyn1 expression.
16
Some previous reports showed that the cleavage efficiency of T2A-P2A was higher overusing solely of the peptides in the eukaryotic organisms.
28
Moreover, it is identified that the presence of the glycine-serine-glycine (GSG) spacer on the N-termini of the 2A peptides could highly increase the cleavage efficiency in the expression hosts.
27
Accordingly, we used a chimeric 2A peptide (i.e., GSG-T2A-P2A), which was inserted between the CmR and coding sequences of hIL-2 genes (Figs. 1 and S3). There is only one report about the gene expression via 2A peptides in microalgae, in which the P2A peptide was specifically used for the expression of GFP and Xyn1 proteins. However, the microalga could not entirely process the P2A peptide, in which the un-cleaved proteins (i.e., ble-P2A-GFP and ble-P2A-Xyl1) were produced in the C. reinhardtii cytosol.
8
Previously, the heavy and light chains of a human monoclonal antibody (i.e., GL4mAb) were expressed in P. tricornutum , with results showing the transformed microalga could robustly produce the recombinant mAb.
42
Moreover, in another study, the stable expression of Xyn1 in the C. reinhardtii revealed the microalga can accumulate the produced protein about 0.25% of the TSP.
8
Our data, however, showed that the used chimeric 2A peptide could entirely proceed in the C. reinhardtii , C. vulgaris , and D. salina and no additional un-cleaved protein (i.e., CmR-chimeric 2A-hIL-2) was found to be exhibited in their cytoplasmic proteins (Fig. 6). Moreover, our results showed that the GMAE plasmid could stably transform the C. reinhardtii , in which the produced IL-2 accumulated about 0.94% of TSP (Fig. 5).
To the best of our knowledge, the GMAE plasmid can reach the highest level of expression in the nuclear transgenes in the microalgae. Collectively, this study is the first successful attempt in the microalgae towards the production of biologics by the proposed strategy using the GAME plasmid.
Conclusion
The major limitations of different expression systems (e.g., bacteria, yeasts, plant, and mammalian cells) and the tempting features of the microalgae (e.g., glycosylation similarity to the human) have inspired many scientists to capitalize on the microalgae for the production of recombinant proteins. However, the transient and low gene expression seems to be the two important bottlenecks during microalgae transformation which limited microalgae-based recombinant protein production. Here, for the first time, we showed that the engineered GAME plasmid system could result in stable and robust production of IL-2 in three microalgae, including C. reinhardtii , C. vulgaris , and D. salina . The GAME plasmid system can indeed pave the way towards the stable microalgae-based production of biomacromolecules such as IL-2 with appropriate post-translational modifications. Despite being able to produce IL-2 in the microalgae (i.e., in C. reinhardtii , C. vulgaris , and D. salina ), we believe that the functionality of the produced protein should be validated in an animal model before its translation into clinical applications. Further, the large-scale production of any biologics in this host demands further optimization for the industrial scale. Taken all, for the first time, we introduce the proposed GAME plasmid system that can be used for the high yield production of biologicals with excellent stability in microalgae.
Acknowledgments
We wish to express our appreciation to the Research Center for Pharmaceutical Nanotechnology (RCPN) at Tabriz University of Medical Sciences, Iran National Science Foundation (INSF) and the University of Tabriz for their financial and technical support.
Funding sources
This work is part of a Postdoc program supported by RCPN at Tabriz University of Medical Sciences (Grant ID: RCPN59560) and INSF (Grant ID: 96015195).
Ethical statement
This study was approved by the ethical committee of Tabriz University of Medical Sciences ( Ethics No. IR.TBZMED.REC.1396.781).
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
All authors conceived and planned the project. JD performed the experiments. JD wrote the draft and KA, and AM finalized the latest version of the manuscript. All authors contributed to discussing the results.
Supplementary Materials
Supplementary file 1 contains Figs. S1-S5 and Tables S1-S2.
(pdf)
Research Highlights
What is the current knowledge?
simple
-
√ Microalgae can consider as a more powerful platform for
the production of different types of therapeutic proteins such
as hIL-2.
-
√ Because of the poor and unstable gene expression in the
microalgae, exploring the new approaches for solving these
limitations have intensely demanded.
What is new here?
simple
-
√ A novel Agrobacterium-2A-based plasmid (so-called
GAME) was constructed for the engineering green
microalgae to robustly producing the human IL-2 (hIL-2).
-
√ The engineered microalgae can stably accumulate the hIL-
2 about 0.94% of the total soluble protein with acceptable
biological activity.
References
- Khan MI, Shin JH, Kim JD. The promising future of microalgae: current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb Cell Fact 2018; 17:36. doi: 10.1186/s12934-018-0879-x [Crossref] [ Google Scholar]
- Chisti Y. Biodiesel from microalgae beats bioethanol. Trends Biotechnol 2008; 26:126-31. doi: 10.1016/j.tibtech.2007.12.002 [Crossref] [ Google Scholar]
- Oey M, Sawyer AL, Ross IL, Hankamer B. Challenges and opportunities for hydrogen production from microalgae. Plant Biotechnol J 2016; 14:1487-99. doi: 10.1111/pbi.12516 [Crossref] [ Google Scholar]
- Dehghani J, Adibkia K, Movafeghi A, Barzegari A, Pourseif MM, Maleki Kakelar H. Stable transformation of Spirulina (Arthrospira) platensis: a promis ing microalga for production of edible vaccines. Appl Microbiol Biotechnol 2018; 102:9267-78. doi: 10.1007/s00253-018-9296-7 [Crossref] [ Google Scholar]
- Stehfest K, Ritter E, Berndt A, Bartl F, Hegemann P. The branched photocycle of the slow-cycling channelrhodopsin-2 mutant C128T. J Mol Biol 2010; 398:690-702. doi: 10.1016/j.jmb.2010.03.031 [Crossref] [ Google Scholar]
- Watanabe HC, Welke K, Sindhikara DJ, Hegemann P, Elstner M. Towards an understanding of channelrhodopsin function: simulations lead to novel insights of the channel mechanism. J Mol Biol 2013; 425:1795-814. doi: 10.1016/j.jmb.2013.01.033 [Crossref] [ Google Scholar]
- Tichy AM, Gerrard EJ, Legrand JMD, Hobbs RM, Janovjak H. Engineering Strategy and Vector Library for the Rapid Generation of Modular Light-Controlled Protein-Protein Interactions. J Mol Biol 2019; 431:3046-55. doi: 10.1016/j.jmb.2019.05.033 [Crossref] [ Google Scholar]
- Rasala BA, Lee PA, Shen Z, Briggs SP, Mendez M, Mayfield SP. Robust expression and secretion of Xylanase1 in Chlamydomonas reinhardtii by fusion to a selection gene and processing with the FMDV 2A peptide. PLoS One 2012; 7:e43349. doi: 10.1371/journal.pone.0043349 [Crossref] [ Google Scholar]
- Dehghani J, Adibkia K, Movafeghi A, Maleki-Kakelar H, Saeedi N, Omidi Y. Towards a new avenue for producing therapeutic proteins: Microalgae as a tempting green biofactory. Biotechnol Adv 2019:107499. doi: 10.1016/j.biotechadv.2019.107499 [Crossref]
- Dehghani J, Movafeghi A, Barzegari A, Barar J. Efficient and stable transformation of Dunaliella pseudosalina by 3 strains of Agrobacterium tumefaciens. Bioimpacts 2017; 7:247-54. doi: 10.15171/bi.2017.29 [Crossref] [ Google Scholar]
- Hempel F, Maier UG. Microalgae as Solar-Powered Protein Factories. Adv Exp Med Biol 2016; 896:241-62. doi: 10.1007/978-3-319-27216-0_16 [Crossref] [ Google Scholar]
- Specht E, Miyake-Stoner S, Mayfield S. Micro-algae come of age as a platform for recombinant protein production. Biotechnol Lett 2010; 32:1373-83. doi: 10.1007/s10529-010-0326-5 [Crossref] [ Google Scholar]
- Yan N, Fan C, Chen Y, Hu Z. The Potential for Microalgae as Bioreactors to Produce Pharmaceuticals. Int J Mol Sci 2016:17. doi: 10.3390/ijms17060962 [Crossref]
- Hempel F, Maurer M, Brockmann B, Mayer C, Biedenkopf N, Kelterbaum A. From hybridomas to a robust microalgal-based production platform: molecular design of a diatom secreting monoclonal antibodies directed against the Marburg virus nucleoprotein. Microb Cell Fact 2017; 16:131. doi: 10.1186/s12934-017-0745-2 [Crossref] [ Google Scholar]
- D'Adamo S, Schiano di Visconte G, Lowe G, Szaub-Newton J, Beacham T, Landels A. Engineering the unicellular alga Phaeodactylum tricornutum for high-value plant triterpenoid production. Plant Biotechnol J 2019; 17:75-87. doi: 10.1111/pbi.12948 [Crossref] [ Google Scholar]
- Rasala BA, Chao SS, Pier M, Barrera DJ, Mayfield SP. Enhanced genetic tools for engineering multigene traits into green algae. PLoS One 2014; 9:e94028. doi: 10.1371/journal.pone.0094028 [Crossref] [ Google Scholar]
- Almaraz-Delgado AL, Flores-Uribe J, Perez-Espana VH, Salgado-Manjarrez E, Badillo-Corona JA. Production of therapeutic proteins in the chloroplast of Chlamydomonas reinhardtii. AMB Express 2014; 4:57. doi: 10.1186/s13568-014-0057-4 [Crossref] [ Google Scholar]
- Beligni MV, Yamaguchi K, Mayfield SP. The translational apparatus of Chlamydomonas reinhardtii chloroplast. Photosynth Res 2004; 82:315-25. doi: 10.1007/s11120-004-2440-5 [Crossref] [ Google Scholar]
- Mathieu-Rivet E, Kiefer-Meyer MC, Vanier G, Ovide C, Burel C, Lerouge P. Protein N-glycosylation in eukaryotic microalgae and its impact on the production of nuclear expressed biopharmaceuticals. Front Plant Sci 2014; 5:359. doi: 10.3389/fpls.2014.00359 [Crossref] [ Google Scholar]
- Leon R, Fernandez E. Nuclear transformation of eukaryotic microalgae: historical overview, achievements and problems. Adv Exp Med Biol 2007; 616:1-11. doi: 10.1007/978-0-387-75532-8_1 [Crossref] [ Google Scholar]
- Doron L, Segal N, Shapira M. Transgene Expression in Microalgae-From Tools to Applications. Front Plant Sci 2016; 7:505. doi: 10.3389/fpls.2016.00505 [Crossref] [ Google Scholar]
- Eichler-Stahlberg A, Weisheit W, Ruecker O, Heitzer M. Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta 2009; 229:873-83. doi: 10.1007/s00425-008-0879-x [Crossref] [ Google Scholar]
- Baier T, Wichmann J, Kruse O, Lauersen KJ. Intron-containing algal transgenes mediate efficient recombinant gene expression in the green microalga Chlamydomonas reinhardtii. Nucleic Acids Res 2018; 46:6909-19. doi: 10.1093/nar/gky532 [Crossref] [ Google Scholar]
- Donnelly ML, Hughes LE, Luke G, Mendoza H, ten Dam E, Gani D. The 'cleavage' activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring '2A-like' sequences. J Gen Virol 2001; 82:1027-41. doi: 10.1099/0022-1317-82-5-1027 [Crossref] [ Google Scholar]
- Atkins JF, Wills NM, Loughran G, Wu CY, Parsawar K, Ryan MD. A case for "StopGo": reprogramming translation to augment codon meaning of GGN by promoting unconventional termination (Stop) after addition of glycine and then allowing continued translation (Go). Rna 2007; 13:803-10. doi: 10.1261/rna.487907 [Crossref] [ Google Scholar]
- Ryan MD, King AM, Thomas GP. Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J Gen Virol 1991; 72(Pt 11):2727-32. doi: 10.1099/0022-1317-72-11-2727 [Crossref] [ Google Scholar]
- Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 2011; 6:e18556. doi: 10.1371/journal.pone.0018556 [Crossref] [ Google Scholar]
- Liu Z, Chen O, Wall JBJ, Zheng M, Zhou Y, Wang L. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci Rep 2017; 7:2193. doi: 10.1038/s41598-017-02460-2 [Crossref] [ Google Scholar]
- Hooykaas PJJ, van Heusden GPH, Niu X, Reza Roushan M, Soltani J, Zhang X. Agrobacterium-Mediated Transformation of Yeast and Fungi. Curr Top Microbiol Immunol 2018; 418:349-74. doi: 10.1007/82_2018_90 [Crossref] [ Google Scholar]
- Kunik T, Tzfira T, Kapulnik Y, Gafni Y, Dingwall C, Citovsky V. Genetic transformation of HeLa cells by Agrobacterium. Proc Natl Acad Sci U S A 2001; 98:1871-6. doi: 10.1073/pnas.041327598 [Crossref] [ Google Scholar]
- Atashpaz S, Khani S, Barzegari A, Barar J, Vahed SZ, Azarbaijani R. A robust universal method for extraction of genomic DNA from bacterial species. Mikrobiologiia 2010; 79:562-6. [ Google Scholar]
- Walsh G. Biopharmaceutical benchmarks 2018. Nat Biotechnol 2018; 36:1136-45. doi: 10.1038/nbt.4305 [Crossref] [ Google Scholar]
- Ross SH, Cantrell DA. Signaling and Function of Interleukin-2 in T Lymphocytes. Annu Rev Immunol 2018; 36:411-33. doi: 10.1146/annurev-immunol-042617-053352 [Crossref] [ Google Scholar]
- Mahmoudpour SH, Jankowski M, Valerio L, Becker C, Espinola-Klein C, Konstantinides S. Safety of low-dose subcutaneous recombinant interleukin-2: systematic review and meta-analysis of randomized controlled trials. Sci Rep 2019; 9:7145. doi: 10.1038/s41598-019-43530-x [Crossref] [ Google Scholar]
- Zhang R, Xi X, Wang C, Pan Y, Ge C, Zhang L. Therapeutic effects of recombinant human interleukin 2 as adjunctive immunotherapy against tuberculosis: A systematic review and meta-analysis. PLoS One 2018; 13:e0201025. doi: 10.1371/journal.pone.0201025 [Crossref] [ Google Scholar]
- Kamionka M. Engineering of therapeutic proteins production in Escherichia coli. Curr Pharm Biotechnol 2011; 12:268-74. doi: 10.2174/138920111794295693 [Crossref] [ Google Scholar]
- Rosenberg AS. Effects of protein aggregates: an immunologic perspective. Aaps j 2006; 8:E501-7. doi: 10.1208/aapsj080359 [Crossref] [ Google Scholar]
- Rajam MV, Kumar SV. Green Alga (Chlamydomonas reinhardtii). Methods Mol Biol 2006; 344:421-33. doi: 10.1385/1-59745-131-2:421 [Crossref] [ Google Scholar]
- Cheng R, Ma R, Li K, Rong H, Lin X, Wang Z. Agrobacterium tumefaciens mediated transformation of marine microalgae Schizochytrium. Microbiol Res 2012; 167:179-86. doi: 10.1016/j.micres.2011.05.003 [Crossref] [ Google Scholar]
- Gelvin SB. Traversing the Cell: Agrobacterium T-DNA's Journey to the Host Genome. Front Plant Sci 2012; 3:52. doi: 10.3389/fpls.2012.00052 [Crossref] [ Google Scholar]
- Hempel F, Maier UG. An engineered diatom acting like a plasma cell secreting human IgG antibodies with high efficiency. Microb Cell Fact 2012; 11:126. doi: 10.1186/1475-2859-11-126 [Crossref] [ Google Scholar]
- Hempel F, Lau J, Klingl A, Maier UG. Algae as protein factories: expression of a human antibody and the respective antigen in the diatom Phaeodactylum tricornutum. PLoS One 2011; 6:e28424. doi: 10.1371/journal.pone.0028424 [Crossref] [ Google Scholar]
- Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci 2003; 28:215-20. doi: 10.1016/s0968-0004(03)00052-5 [Crossref] [ Google Scholar]
- Weil JE, Beemon KL. A 3' UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus. RNA 2006; 12:102-10. doi: 10.1261/rna.2129806 [Crossref] [ Google Scholar]
- Lumbreras V, Stevens DR, Purton S. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J 1998; 14:441-7. [ Google Scholar]
- Gruber H, Kirzinger SH, Schmitt R. Expression of the Volvox gene encoding nitrate reductase: mutation-dependent activation of cryptic splice sites and intron-enhanced gene expression from a cDNA. Plant Mol Biol 1996; 31:1-12. [ Google Scholar]