Bioimpacts. 11(3):187-197.
doi: 10.34172/bi.2021.09
Original Research
Characterization of isolated compounds from Morus spp. and their biological activity as anticancer molecules
Aditya Rao Rao Shimoga Janakirama 1, 2, *  , Suma Mathad Shivayogi 1, Jamuna Kolkar Satyanarayana 1, Ramesh Chapeyil Kumaran 1
, Suma Mathad Shivayogi 1, Jamuna Kolkar Satyanarayana 1, Ramesh Chapeyil Kumaran 1
Author information:
1Molecular Biomedicine laboratory, PG Department of Studies and Research in Biotechnology, Sahyadri Science College campus, Kuvempu University, Shimoga, Karnataka, India
2Department of Plant Cell Biotechnology, CSIR- Central Food Technological Research Institute, Mysore, Karnataka, India
Abstract
Introduction:
The genus Morus is well known for its medicinal benefits from time immemorial. The present work reported the health-promoting properties of the biologically active molecules present in different species of the genus Morus.
Methods:
Different solvent extracts of the three plant species of Morus were investigated initially for their antioxidant effects, followed by in vitro anticancer studies against MCF7 and 3T3 cell lines along with their bioactive isolates viz. cathafuran-B, moracin-M, and Ursolic acid. Further, in silico docking studies were performed for the isolated compounds to predict their probable mode of interaction with P38Map Kinase.
Results:
The results indicated that all three species under study possessed remarkable antioxidant effects which are supported by a linear and positive correlation between different antioxidant activities. The in vitro cell antiproliferative test indicated that the cell survivability decreased with an increase in the concentration of extracts and compounds. Among the extracts, M. laevigata methanol extract showed 21.57, 6.27% of cell survival against MCF7 and 3T3 cell lines at 800 µg/mL concentration while among the isolated compounds, ursolic acid showed 8.46, 17.58% of cell survival at 200 µg/mL concentration. Among the three compounds docked, ursolic acid showed greater binding affinity towards the target protein in terms of its binding energy (-9.97 kJ/mol) compared to Cathafuran B (-8.35 kJ/mol) and Moracin M (-6.91 kJ/mol).
Conclusion:
The study generated interesting results in terms of health benefits of Morus species by documenting their antioxidant and anticancer activities, thereby validating the folk claims of therapeutic benefits of mulberry.
Keywords: Antioxidants, Phytochemicals, Cytotoxicity, Correlation, In silico docking
Copyright and License Information
© 2021 The Author(s)
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Overproduction of various forms of reactive oxygen species, such as oxygen radicals and non-free radical species is considered to be the main contributor to oxidative stress, which has been linked to several diseases like cancer, atherosclerosis, and tissue damage in rheumatoid arthritis.
1,2
Plants are an exemplary source of medicines and edible plant parts are rich sources of natural antioxidants that possess the ability to protect the body from damage caused by free radicals induced oxidative stress.
3
The preventive mechanisms of natural phytochemicals on tumor promotion range from the inhibition of genotoxic effects to increased antioxidants and anti-inflammatory activity, inhibition of proteases and cell proliferation, protection of intracellular communications to modulate apoptosis and signal transduction pathways.
4-6
Mulberry is medicinally the most important plant which belongs to genera Morus. Indigenous medicinal practitioners, for centuries, have used different parts of the mulberry plant for treating diseases and symptoms of many serious diseases like diabetes mellitus, atherosclerosis, and hypertension.
7
Evidence has shown that large numbers of biomolecules of medicinal interest are present in the genus Morus, but extensive work is being carried out by targeting mainly Morus alba.
1,8
The presence of phytochemicals vary in terms of species, varieties, within the different parts of the plants, and also with the influence by the seasons and different agronomic conditions, a comparative study among the different species could yield encouraging results in terms of their phytopharmacological effects. Hence, the present study aims to compare the antioxidant potentials among the three species belonging to the genus Morus and antiproliferative effects of the solvent extracts along with their bioactive isolates.Further, in silico docking studies were performed to predict the binding modes of the bioactive isolates as inhibitors to P38 MAP kinase, a key protein involved in cancer growth and development.
9
The study also attempts to statistically relate the connection between different phytoconstituents and their association in producing antioxidant effects.
Materials and Methods
Phytochemistry
Extract preparation
The leaf materials of M. alba, M. serrata and M. laevigata were collected from authentic germplasm source and were shade dried, powdered. Two hundred grams of the powder was subjected to hot solvent extraction using a soxhlet extractor utilizing 500 mL of petroleum ether, chloroform, and methanol sequentially. Solvent recovery was done for 3 days using a rotary vacuum evaporator and concentrated extracts (15 g) were preserved in desiccators for further analysis. All the phytochemical activities were performed from chemicals and reagents procured from Merck Life Science Private Limited, Bengaluru, India.
Isolation and characterization of bioactive compounds
The plant extracts were column chromatographed over 60-120 mesh sized silica gel column (Merck, India), using stepwise gradient elution with different solvents based on their polarity. The collected eluent from column chromatography was monitored by thin-layer chromatography (TLC). Compounds on TLC were spotted by exposure to UV light at 350 nm using the UV chamber (spectroline UV viewing cabinet, Sigma-Aldrich, USA). The fractions with similar TLC pattern with a single spot and Rf (retention factor) values were combined and kept for the evaporation of the solvent, and completely condensed compounds were stored using microcentrifuge tubes in a desiccator.
The plant extracts were initially subjected to fractionation using organic solvents with increasing polarity (N-hexane, petroleum ether, chloroform, ethyl acetate, and methanol). Each fraction was chromatographed for the isolation of bioactive molecules and only the compounds obtained in their purest form were described here which were later subjected for spectroscopic analysis. Compound-1 was eluted from the petroleum ether fraction of pet. ether extract of M. laevigata(4.8 g) using a mixture of petroleum ether and ethyl acetate (40:60). The ethyl acetate fraction of the methanol extract of M. alba(4.5 g) was chromatographed and compound-2 was eluted with a mixture of methanol and ethyl acetate (60:40) and further, its petroleum ether fraction eluted with a mixture of ethyl acetate and petroleum ether (70:30) yielded compound-3. The structure of the isolated compounds was determined based on infrared spectroscopy (IR) (Shimadzu Fourier transformed infrared (FT-IR) spectrophotometer), liquid chromatography-mass spectroscopy (LC-MS) (Shimadzu, LCMS 2010A, Japan), and nuclear magnetic resonance (NMR) spectral analyses (Bruker 400 MHz spectrometer).
Evaluation of in vitro antioxidant activities and their statistical significance
The antioxidant ability of any biological mixture depends upon the mode of action of its bioactive constituents. The quantitative assessment of such bioactive constituents can relate to their qualitative effects like radical scavenging, chelating, or reducing ability. The statistical significance of such a relationship can be estimated by calculating the correlation coefficient (r), which gives a measure of the degree of closeness between these variables. With this perspective following antioxidant activities were performed.
Total phenolic content estimation
The total phenolic content (TPC) was estimated using the method prescribed by Makkar.
10
To this end, 100 µg/mL of each extract with the final volume made up to 1 mL with distilled water was used for TPC estimation. The tubes were added with 0.5 mL of 2 M Folin-Ciocalteu reagent (1:1 with water) and 2.5 mL of 1.9 M sodium carbonate solution. The contents were mixed and the tubes were incubated at 90oC for one minute. The absorbance was recorded at 725 nm against the reagent blank. The standard curve constructed using catechol (0-100 µg/mL) was used to calculate the TPC and the results were expressed as catechol equivalent.
Total flavonoid content estimation
Total flavonoid contents of all the extracts were determined by the method of Zhishen
11
and using the standard curve of catechol, the results were expressed as catechol equivalent in µg/mg of extract. 100 µg/mL concentration of extracts and standard solution of catechol (20, 40, 60, 80 and 100 mg/mL) was added with 0.3 ml of 0.725 M sodium nitrite, 0.3 mL of 0.75M aluminium chloride. The mixture was incubated for 5 minutes at room temperature (25 ± 2°C) and then it was added with 2 mL of 1M sodium hydroxide. The final volume was made up to 10 mL using distilled water. The content was mixed well and the absorbance was measured at 510 nm.
Total antioxidant capacity
The spectrophotometric method prescribed by Prieto et al
12
was employed to estimate the total antioxidant capacity. The extracts were prepared with a concentration of 100 µg/mL and mixed with 1 mL of reagent solution consisting of 0.6M Sulfuric acid, 28mM sodium phosphate, 4mM ammonium molybdate. After incubating the tubes for 90 minutes at 95oC, they were cooled to room temperature and the optical density was read at 695 nm against a blank. The standard curve of ascorbic acid (0-100 µg/mL) was constructed to express the results in terms of its equivalents.
Reducing power assay
The method prescribed by Oyaizu
13
was employed for the assessment of the reducing power of the extracts and ascorbic acid was used as the standard. The extracts with concentration ranging between 0-1000 µg/mL were taken in test tubes and were mixed with 2.5 mL of 0.2M phosphate buffer (pH 6.6) and 2.5 mL of 1% potassium ferricyanide. The tubes were incubated at 50oC for 20 minutes and later were added with 2.5 mL of 10% Trichloroacetic acid. The tubes were centrifuged for 10 minutes at 3000 rpm and 2.5 mL of the upper layer was collected and mixed with an equal volume of distilled water followed by 0.5 mL of 0.1% Ferric chloride. The optical density was read at 700 nm. An increase in absorbance indicated increased reducing power and the EC50 (µg of extract/mL), the effective concentration at which the absorbance was 0.5 for reducing power was calculated.
14
DPPH (2,2-diphenyl-1-picrylhydrazyl) radical scavenging activity
DPPH free radical scavenging activity was measured by employing the method of Wong et al
15
using ascorbic acid as standard. The different concentrations of each of the extracts ranging between 0-1000 µg/mL were dissolved in methanol and were added to 3 mL of 0.1mM DPPH dissolved in methanol. The contents were mixed well and incubated for 30 minutes at room temperature in the dark. Absorbance was read after incubation at 517 nm. The inhibition percentage was calculated using the following formula and the final results were expressed as an effective concentration (EC50), which is the amount of antioxidants necessary to decrease the initial concentration by 50%.
% of Inhibition= (A0-A1)/A0 x 100,
where A0 is the absorbance of the control (without test samples) and A1 is the absorbance of test samples.
Nitric oxide radical scavenging activity
For the assessment of nitric oxide scavenging activity, the method reported by Garrat
16
was employed and ascorbic acid was used as standard. 10 mM of sodium nitroprusside prepared in phosphate buffer saline (PBS) was mixed with the extracts prepared in different concentrations (0-1000 µg/mL the amount of antioxidants necessary) and incubated at 25oC for 180 minutes. To this mixture, equal volume of freshly prepared Griess reagent consisting of 0.058M sulphanilamide, 0.004M naphthylethylenediamine dichloride, and 0.30M phosphoric acid were added and the absorbance was read at 546 nm. The percentage of inhibition was calculated using the formula prescribed previously and the results are expressed as EC50.
Hydroxyl radical scavenging activity
The method described by Halliwell et al
17
was followed to estimate the hydroxyl radical scavenging activity of all the extracts. The reaction mixture consisting of 60 µL of 1.0 mM FeCl3, 90 µL of 1 mM 1,10-phenanthroline, 2.4 mL of 0.2M phosphate buffer (pH 7.8), 150 µL of 0.17M H2O2 was added to different concentrations of the extracts (0-1000 µg/mL). The contents were thoroughly mixed and the tubes were kept for incubation at room temperature for 5 minutes. The optical density was read at 560 nm. The hydroxyl radicals scavenging activity was calculated using the formula prescribed before and the results are expressed as EC50.
Fe2+chelating activity
The chelation of ferrous ions by extracts was estimated by the method of Dinis.
18
To 0.5 mL of different concentrations of the extracts (0-1000 µg/mL), 1.6 mL of de-ionized water and 0.05 mL of 2mM FeCl2 was added. After 30 seconds, 0.1 mL of 5 mM ferrozine was added. Ferrozine (3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine-p,p′-disulfonic acid monosodium salt hydrate) reacted with divalent iron to form stable magenta complex species that were very soluble in water. After 10 minutes at room temperature, the absorbance of the Fe2+- Ferrozine complex was measured at 562 nm. Disodium EDTA (ethylenediaminetetraacetic acid) was used as a standard metal-chelating agent.
19
The chelating activity of the extract for Fe2+ was calculated using the formula prescribed earlier and the results are expressed as EC50.
Correlation analysis
For bivariate analysis, Pearson correlation coefficient (r) was calculated using dot plot method
20
for different qualitative estimations, viz. reducing power assay, DPPH radical scavenging activity, hydroxyl radical scavenging activity, nitric oxide radical scavenging activity, and iron chelating activity versus different quantitative estimations viz total phenolic estimation, total flavonoid estimation, and total antioxidant estimations.
21
Statistical analyses
All the experiments were carried out in triplicates. Difference between the group’s mean was assessed by one-way analysis of variance (ANOVA). The results obtained were compared with the control group. P value <0.01 was considered statistically significant. The % inhibition values from each trial were used to generate the regression line and EC50 was calculated. The results were pooled and mean and standard deviation (SD) was calculated. All the statistical analysis was done using the SPSS software version 20 and the graphs were plotted using GraphPad Prism v.7.0.
In vitro cell antiproliferative test
Cytotoxicity of the sample on tumor cells was measured by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assay.
22
Since superior anticancer effects were observed in in vivo models,
23
along with antioxidant effects by the methanolic extracts of the three species, they were also selected to assess in vitroanticancer activity along with the isolated bioactive compounds.
Maintenance of cell culture
The MCF7 and 3T3 cell lines were cultured using Dulbecco's Modified Eagle Medium (DMEM, Sigma–Aldrich) media with 10% heat-inactivated fetal bovine serum (FBS) (Thermo Scientific, North American origin) and antibiotics penicillin-streptomycin powder (Hi-Media) 0.4 µg/mL final concentration. Once the cells in the culture dish (T25 flask) reach confluence, the monolayer cells were detached and single-cell suspensions were made using trypsin-ethylene diamine tetraacetic acid (EDTA). Trypan blue was added to check the viability of the cells and a hemocytometer was used to do the cell counting. 10 × 103 cells/well/100 μL were seeded in a 96-well plate. Cells were allowed to attach to the well plate for 24 hours at 37°C, 5% CO2, 95% relative humidity.
The cells were treated with different concentrations of the test samples after 24 hours of incubation. The required final drug concentrations of 50, 100, 200, 400, 800 µg/mL for extracts and 1.8, 3.6, 7.25, 12.5, 25, 50, 100, and 200 µg/mL for isolated compounds were obtained from the stock of 1mg/ml by serial dilution and the final volume was made up to 200 μL/well using media. After the addition of the drug, the plates were incubated for an additional 48 hours at 37oC. The plates were maintained in triplicate for all concentrations.
Cytotoxicity assay
After 48 hours incubation, cell viability was determined by adding tetrazolium salt (Sigma) as a cytotoxicity indicator. 20 μL from MTT stock of 5mg/ml (PBS was used to make the stock) was mixed with 80 μL of media to give a final concentration of 1 mg/mL was added to each well and incubated at 37oC for 4 hours. The medium with MTT was discarded and 100 μL of DMSO per well was added to solubilize the formazan crystals. The absorbance was measured using a microplate reader at 570 nm. Tetrazolium salts are cleaved to formazan dye by cellular enzyme mitochondrial succinate dehydrogenase present only in the viable cells. The wells containing cells that received no treatment were considered to represent 100% viability. The % of cell survival was determined using the following formula.
%of cell survival = Absorbance of sample/ Absorbance of control x 100
In silico binding studies on isolated compounds with P38 MAP Kinase
Tools and servers used
The structure of the ligand molecules was drawn using ChemDraw Ultra 6.0and their 3-D geometrical optimization was done using Chemsketch V.12.01. Open babel, a standalone tool was used to generate 3D coordinates for all the ligands. Energy minimization was done using Discovery studio V.2.5 and CHARMm force field was added. All the in-silico studies were performed on Lenovo G50-80 machine (Intel Core i3-5005U Processor 2.0 GHz, 4 GB memory with Microsoft Windows 10 operating system).
The P38 MAP kinase inhibitors are found to be effective against several disease models, including inflammation, arthritis, and other joint diseases, septic shock, and myocardial injury. Treatment with P38 MAP kinase inhibitors attenuated both P38 activation and disease severity.
9
The protein structure file (PDB ID: 1OUK) was retrieved from the protein data bank and was edited to remove the heteroatoms. Later it was added with C-terminal oxygen, polar hydrogen, CHARMm force field, and Gasteiger charges.
24
The binding site information is obtained from ligplot and the residues forming the pocket were identified. Protein-ligand interactive visualization and analysis were carried out in Python Molecular Viewer V.1.5.6 and Discovery Studio V.2.5.
25
Molecular docking
Automated docking was used to study the interactions of ligand molecules to the binding pocket of the macromolecule.
26
A genetic algorithm method implemented in the AutoDock V.4.2 was employed to study appropriate binding modes of the ligand in different conformations.
27
For the ligand molecules, all the torsions were allowed to rotate during docking. The grid box was set around the residues forming the active pocket.
28,29
Grid log file was generated using AutoGrid program and Lamarckian genetic algorithm and the pseudo-Solis and Wets methods were applied for energy minimization using default parameters to generate dock parameter file. Docking energies were calculated using the following equation.
30
Docking energy = Intermolecular energy + Total internal energy + Torsional free energy + Unbound energy
The results were validated by comparing the binding interactions generated by Autodock program with LibDock program of Discovery studio V.2.5. Hydrogen bonds were added and energy was minimized using CHARMm force field. The ligands were docked into the binding site using Libdock program which was highlighted using a binding sphere. Absolute energy, CHARMm energy, and Libdock score were obtained from the study.
Results
Phytochemistry
Isolation and characterization of bioactive compounds
The molecular formula C24H26O4 of Compound 1 was obtained from the TOF-MS m/z379 [M + H]+ with the combination of IR(KBr) cm-1: 3441 (OH), 2918 (alkyl) and1H NMR, 13C NMR data (Supplementary file 1). The compound 1 was confirmed as cathafuran B, by spectroscopic analysis, and comparison with its literature data.
31
The molecular formula C30H48O3 of Compound 2 was obtained from the TOF-MS m/z457 [M + H]+ with the combination of IR(KBr) cm-1: 3362 (OH), 2924 (alkyl), 1658 (-C=O) and 1H NMR, 13C NMR data (Supplementary file 1). Compound 2 was confirmed as ursolic acid, by spectroscopic analysis, and comparison with its literature data.
32
The molecular formula C14H10O4 of Compound 3 was obtained from the TOF-MS m/z241 [M + H]- with the combination of IR(KBr) cm-1: 3332 (OH) and 1H NMR, 13C NMR data (Supplementary file 1). Compound 3 was confirmed as Moracin M, by spectroscopic analysis, and comparison with its literature data.
33
The structures determined by IR, 1H NMR 13C NMR and LC-MS analyses were compared with the literature data which are in agreement with predicted structures (Supplementary file 1). The structures of the compounds 1-3 are presented in Fig. 1.
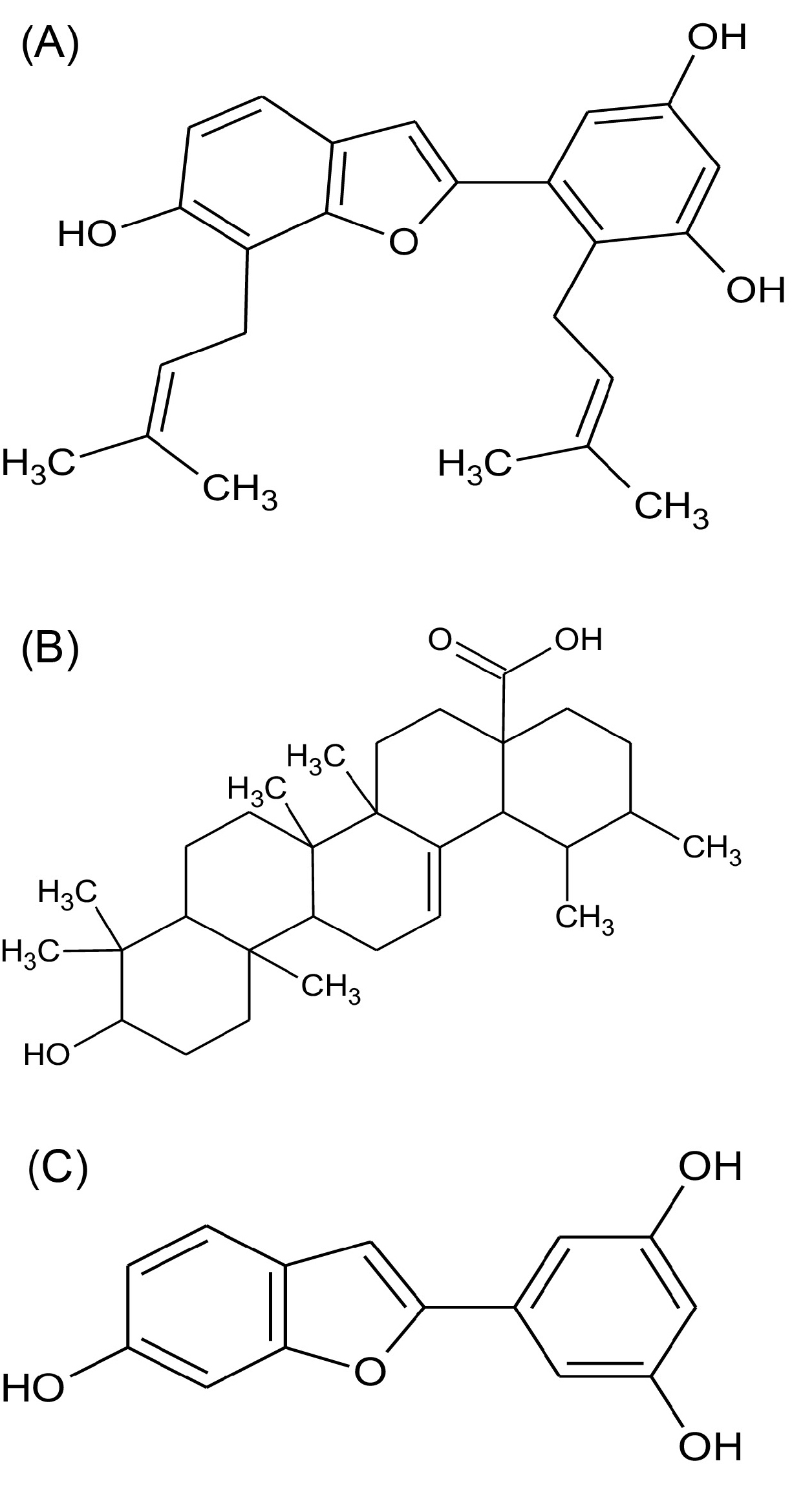
Fig. 1.
Molecular structure of Cathafuran B (A), Ursolic acid (B) and Moracin M (C).
.
Molecular structure of Cathafuran B (A), Ursolic acid (B) and Moracin M (C).
In vitro antioxidant activities
Total phenolic content estimation
The TPC of the three leaf extracts was plotted on the standard curve of catechol. From the results, it was found that among the three plants, M. albaexhibited the highest TPC followed by M. serrata and M. laevigatain their respective solvent extracts.Among the three solvents used, methanol extract showed prominent total phenolic activity (76, 68, and 54 µg of catechol/mg of extract) when compared with that of chloroform (57, 55, and 49 µg of catechol/mg of extract) and petroleum ether (26, 21, and 19 µg of catechol/mg of extract) in the above-said species (Fig. 2 and Table S1).
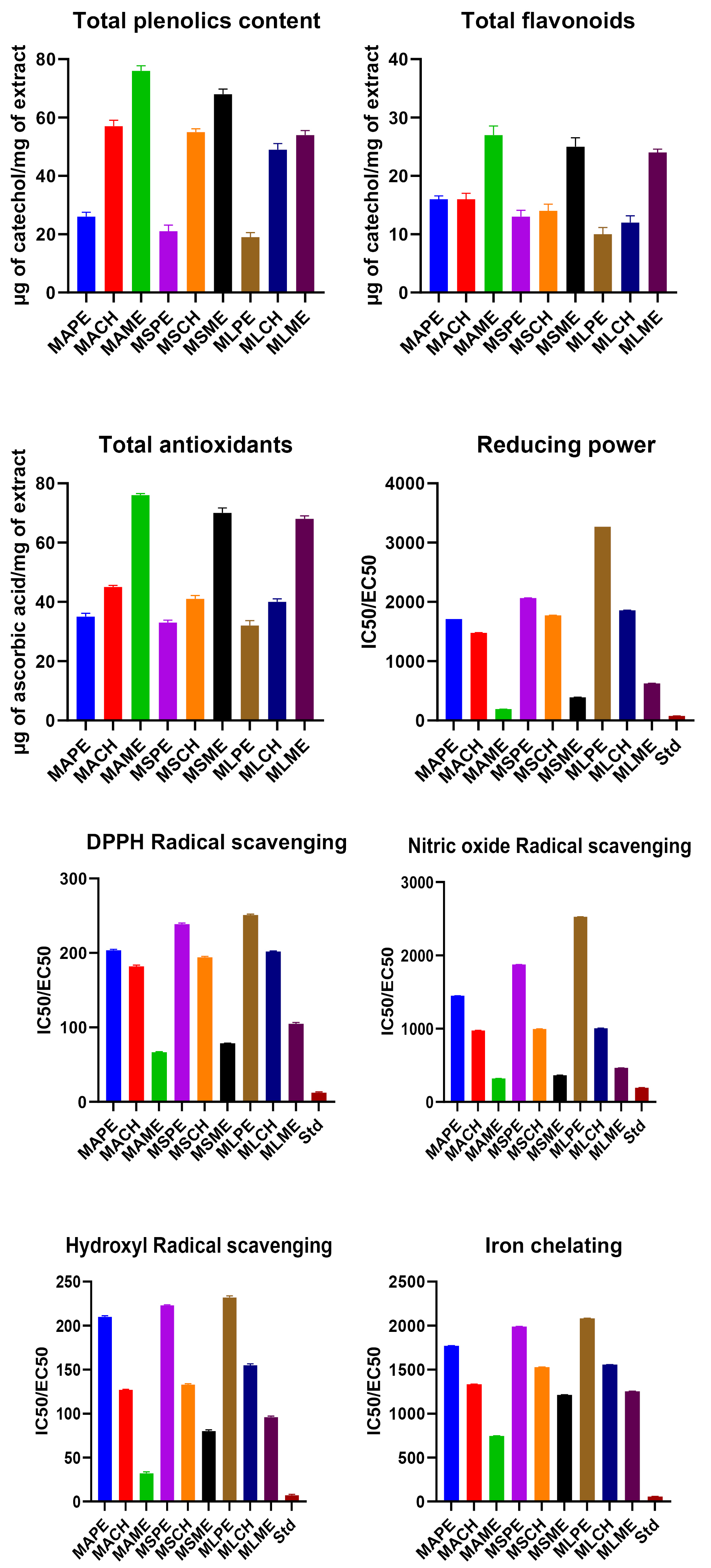
Fig. 2.
IC50/EC50 values of antioxidant activities of different solvent extracts of three species of genus Morus. (Where: MAPE: Morus alba petroleum ether extract, MACH: Morus alba chloroform extract, MAME: Morus alba methanol extract, MSPE: Morus serrata petroleum ether extract, MSCH: Morus serrata chloroform extract, MSME: Morus serrata methanol extract, MLPE: Morus laevigata petroleum ether extract, MLCH: Morus laevigata chloroform extract, MLME: Morus laevigata methanol extract).
.
IC50/EC50 values of antioxidant activities of different solvent extracts of three species of genus Morus. (Where: MAPE: Morus alba petroleum ether extract, MACH: Morus alba chloroform extract, MAME: Morus alba methanol extract, MSPE: Morus serrata petroleum ether extract, MSCH: Morus serrata chloroform extract, MSME: Morus serrata methanol extract, MLPE: Morus laevigata petroleum ether extract, MLCH: Morus laevigata chloroform extract, MLME: Morus laevigata methanol extract).
Total flavonoid content estimation
The results of total flavonoid content estimation were similar to that of TPCs. The methanol extract of M. alba found to have maximum flavonoid content of 27 µg of catechol/mg of extract than M. serrata and M. laevigatawhich was found to be 25 µg and 24 µg of catechol/mg respectively.In the chloroform extracts, it was 16 µg, 14 µg, and 13 µg of catechol/mg while, in petroleum ether extracts it was 16 µg, 13 µg, and 10 µg of catechol/mg of extract respectively in the above-said species (Fig. 2 and Table S1).
Total antioxidant capacity
The results of total antioxidant capacity revealed that all the species of Morus under study have prominent antioxidant potentials. Among the three species, M. alba had higher total antioxidant capacity compared to M. serrataand M. laevigata. Among the three solvent extracts, methanol extract has shown higher activity (76, 70, and 68 μg/mg of extract) than chloroform (45, 41 and 40 μg/mg of extract) and petroleum ether extracts (35, 33, and 32 μg/mg of extract) among the above-said species (Fig. 2 and Table S1).
Reducing power assay
All the plants showed prominent reducing power in a dose-dependent manner. M.alba methanol extract had the highest reducing power with an effective concentration (EC50) of 190.47 µg/mL compared to the chloroform extract (1479.29 µg/mL) and the petroleum ether extracts (1712.32 µg/mL). Methanol, chloroform, and petroleum ether extracts of M. Serrata had EC50 values of 392.15, 1773.05 and 2066.11 µg/mL respectively. Methanol, chloroform, and petroleum ether extracts of M. laevigata showed 626.30, 1858.73, and 3267.97 µg/mL respectively. Standard ascorbic acid showed an EC50 value of 78.43 µg/mL (Fig. 2 and Table S1).
DPPH radical scavenging activity
The results of DPPH radical scavenging activity were found to be highest in methanol extracts (66.71, 78.74, and 104.86 µg/mL) followed by chloroform (181.81, 194.12, and 201.97 µg/mL) and pet. Ether extracts (203.74, 238.72, and 250.81 µg/mL) of M. alba, M. serrataand M. laevigata respectively. The results were compared with the standard, ascorbic acid with an EC50 value of 12.32 µg/mL (Fig. 2 and Table S1).
Nitric oxide radical scavenging activity
The results of nitric oxide radical scavenging activity revealed that M. albapossessed better scavenging activity than M. serrata and M. laevigata irrespective of the solvent. Among the three solvents, methanol (320, 365, and 465 μg/mL) had better scavenging ability than chloroform (975.90, 997.25, and 1007.04 µg/mL) and petroleum ether (1449.27, 1876.34, and 2526.52 μg/mL). Ascorbic acid recorded 194.44 µg/mL of EC50 (Fig. 2 and Table S1).
Hydroxyl radical scavenging activity
The hydroxyl radical scavenging activity of ascorbic acid was found to be 6.58µg/mL and that of extracts was in the following order. M. alba methanol extract (32 µg/mL), M. laevigata methanol extract (80 µg/mL), M. serrata methanol (96 µg/mL), Chloroform extract of M. laevigata(127 μg/mL), M. alba(133 µg/mL), M. serrata(155 µg/mL), and pet. ether extract of M. alba (210 µg/mL), M. laevigata (223 µg/mL), M. serrata(232 µg/mL) (Fig. 2 and Table S1).
Fe2+ chelating activity
Among the three plants under study, M. alba recorded the highest chelating activity followed by M. laevigataand M. serrata. Among the three solvents, methanol extracts (745.34, 1212.61, and 1252.34 µg/mL) had higher activity than chloroform (1333.33, 1528.27, and 1556.84 µg/mL) and petroleum ether (1772, 1989.24 and 2081.88 µg/mL). The results were expressed in terms of EC50 values (Fig. 2 and Table S1).
Correlation analyses
Between the different antioxidant activities performed for different solvent extracts of all the three species of genus Morus, a simple and positive correlation was observed. Among the three quantitative activities, the highest average value of correlation coefficient (r) of 0.915, 0.911, and 0.798 are observed for total antioxidant, total phenolics, and total flavonoids respectively against all the qualitative estimations. Among the qualitative activities, the highest average value of ‘r’ was shown by DPPH (0.925) followed by reducing power (0.868), hydroxyl radical (0.867), nitric oxide radical (0.826), and iron-chelating (0.872) against all the quantitative estimations. Correlation coefficients (r) are shown in Table 1.
Table 1.
Correlation coefficient (r) of different antioxidant activities vs. total phenolic, total flavonoid and total antioxidant activities
|
Quantitative estimation
|
Correlation coefficient (r)
|
|
DPPH
|
NO
2-
|
•OH
|
Fe
2+
|
Reducing Power
|
Average
|
| Total phenolic estimation |
0.8648 |
0.9230 |
0.9386 |
0.9423 |
0.8902 |
0.9117 |
| Total flavonoid estimation |
0.9252 |
0.70 |
0.7409 |
0.7713 |
0.8573 |
0.7989 |
| Total antioxidant estimation |
0.9874 |
0.8549 |
0.9214 |
0.9033 |
0.9088 |
0.9151 |
| Average |
0.9258 |
0.8260 |
0.8670 |
0.8723 |
0.8688 |
|
In vitro cell antiproliferative test
The in vitro cell antiproliferative studies were performed against human cancer cell lines; MCF7 (breast cancer cell lines) and 3T3 (non-cancer immortal cell lines). MTT assay was used to analyze the cell growth inhibition and cell viability (Fig. 3 and Table S2). Among the bioactive isolates tested; the results were observed in a dose-dependent manner. The wells treated with ursolic acid showed the minimum cell survival of 8.46 and 17.58% against MCF7 and 3T3 cell lines respectively at the highest treatment concentration of 200 µg/mL, whereas Cathafuran B treatment showed 8.89% and 20.58% cell survival and Moracin M treatment showed 16.09% and 21.6% cell survival. Among the wells treated with plant extracts at their highest concentration of 800 µg/mL, M. laevigatamethanol extract showed the minimum cell survival of 21.57% and 6.27% against MCF7 and 3T3 cell lines, whereas, the treatment with methanolic extract of M. serrata showed 27.96% and 14.84% and M. alba treated wells showed 34.78% and 15.86% respectively.
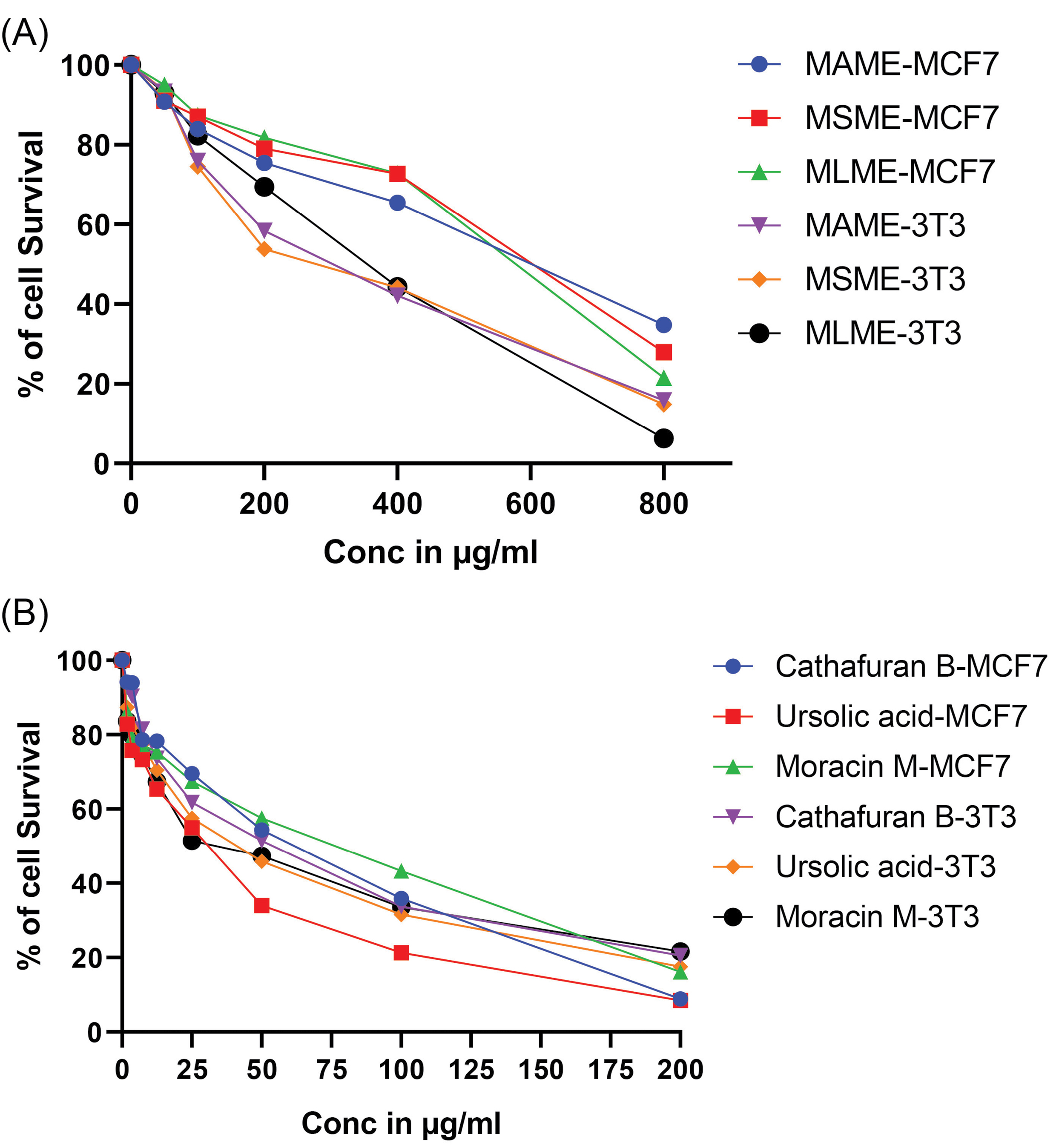
Fig. 3.
Cytotoxic effects of methanol extracts of three species of genus Morus (A), along with the bioactive isolates Cathafuran B, Ursolic acid, and Moracin M (B) against 3T3 and MCF7 cell lines.
.
Cytotoxic effects of methanol extracts of three species of genus Morus (A), along with the bioactive isolates Cathafuran B, Ursolic acid, and Moracin M (B) against 3T3 and MCF7 cell lines.
In silico binding studies on isolated compounds with P38 MAP Kinase
The results of in silico binding studies were compared with adenosine triphosphate (ATP) in terms of binding (-4.91 kJ/mol) and docking energies (-3.64 kJ/mol). Among the three compounds studied, ursolic acid showed better binding concerning its binding (-9.97 kJ/mol) and docking energy (-9.35 kJ/mol). Interactions with cathafuran B and moracin M showed the binding energies of -8.35 kJ/mol and -6.91kJ/mol and docking energies of -10.5kJ/mol and -7.41 kJ/mol respectively. The binding interactions were validated using LibDock program. The results were in agreement with autodock with a LibDock score of 111.06 for ATP molecule, 83.02 for cathafuran B, 109.3 for ursolic acid and 100.80 for moracin M respectively (Fig. 4, Table 2).
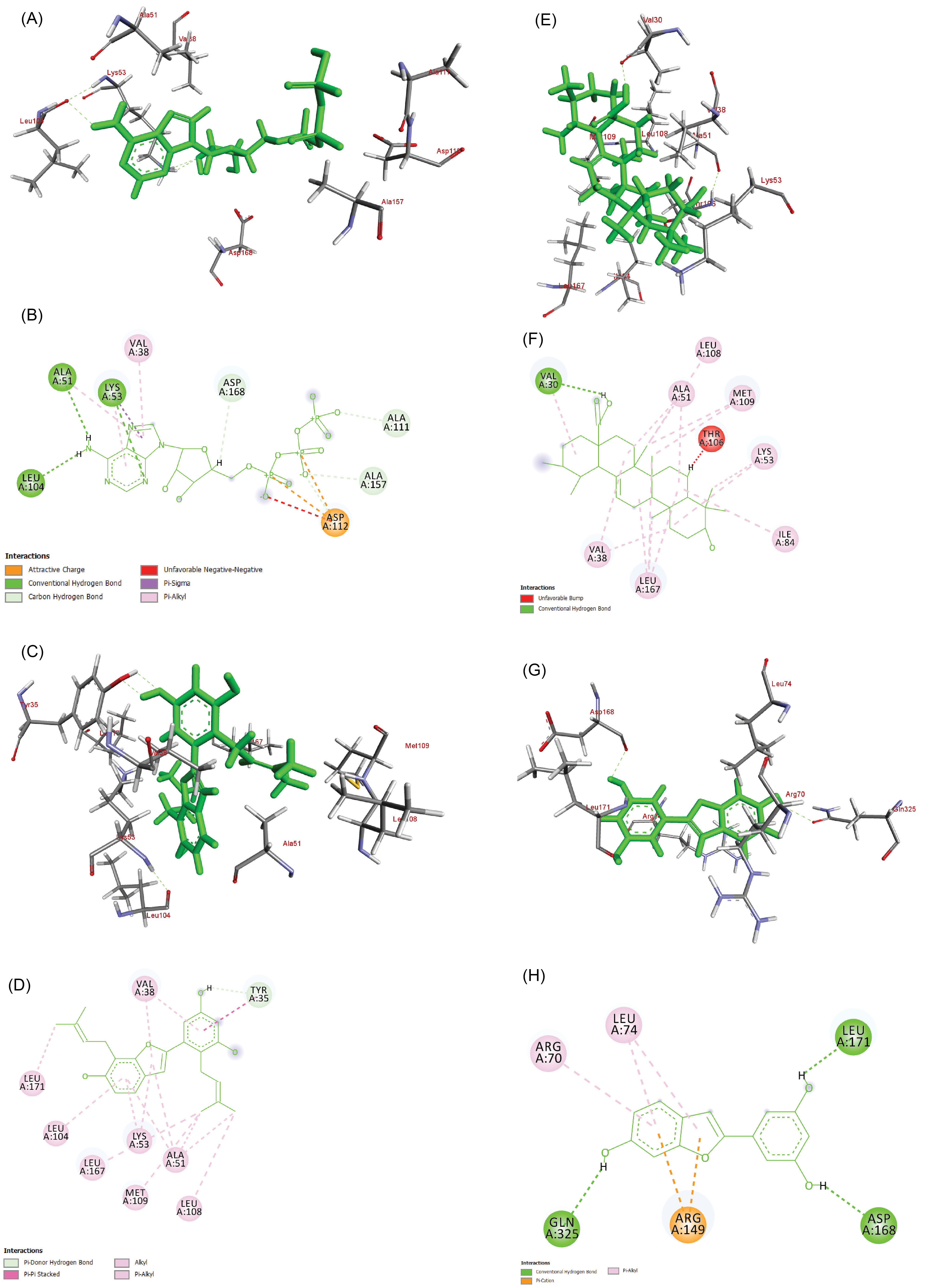
Fig. 4.
3D and 2D Interactions showing the binding of ATP (A, B), Cathafuran B (C, D), Ursolic acid (E, F), and Moracin M (G, H) with 1OUK respectively.
.
3D and 2D Interactions showing the binding of ATP (A, B), Cathafuran B (C, D), Ursolic acid (E, F), and Moracin M (G, H) with 1OUK respectively.
Table 2.
binding studies of isolated compounds with P38 Mitogen-activated protein kinases (P38 MAPk)
|
Compound
|
Docking energy
a
|
Binding energy
a
|
Intermol energy
a
|
Total internal energy
|
Torsional energy
a
|
Unbound energy
|
Ligand efficiency
|
RMS
|
H-bonds
|
Bonding
|
Absolute Energy
|
CHARMm Energy
|
LibDock Score
|
| ATP |
-3.64 |
-4.91 |
-9.39 |
0.64 |
4.47 |
0.64 |
-0.16 |
0 |
2 |
ASP168, PHE169 |
56.09 |
40.084 |
111.06 |
| Cathafuran B |
-10.5 |
-8.35 |
-9.55 |
-1.07 |
1.19 |
-1.07 |
-0.3 |
0 |
1 |
LYS53 |
117.19 |
47.41 |
83.02 |
| Ursolic acid |
-9.35 |
-9.97 |
-10.27 |
0.31 |
0.3 |
0.31 |
-0.3 |
0 |
0 |
- |
60.24 |
77.25 |
109.03 |
| Moracin M |
-7.41 |
-6.91 |
-7.21 |
-0.25 |
0.3 |
-0.25 |
-0.38 |
0 |
2 |
MET109, ASP168 |
99.14 |
25.56 |
100.80 |
Discussion
Antioxidant research continues to grow and emerge as a new beneficial component of plants based on drug discovery. Several antioxidants have been found pharmacologically active as therapeutic agents for many diseases.
34
In the present study, the methanolic extracts of Morus exhibited a significant antioxidant capacity for scavenging and chelating, with a reducing power when compared to chloroform and petroleum ether extracts. Further, it was supported by quantitative estimation namely total phenolic, flavonoid, and total antioxidants. The antioxidant characters of the plant derivatives can be attributed to their phenolic contents.
35
Majority of the antioxidant activities are most probably from phenolic compounds but other than phenolics, small molecules such as ascorbic acid, R-tocopherol, β-carotene, and reduced glutathione also play a crucial role.
36,37
The degree of correlation ranging between 0.7 and 0.98 suggests a simple, positive, and high degree of correlation existing between the variables tested. Several reports have indicated that there is a direct correlation between antioxidant activity and the presence of phenolics, flavonoids and total antioxidants in plants.
38,39
Thus, being a rich source of phytochemicals,
40
the present study supports the therapeutic benefits of Mulberry leaves which have been used in China for hundreds of years to treat hyperglycemia, inflammation, cough, hypertension, cancer, and fever.
8
Computer-aided drug discovery has gained enormous importance in the modern drug discovery pipeline. Its role is essential in reducing the virtual chemical space of drug molecules in synthesizing, modifying, or isolating in their purest form and to screen against a specific disease target into a manageable number of chemicals, leading towards a potential drug candidate.
41-43
The modality of cell death can be studied based on necrosis, apoptosis, necroptosis, autophagic cell death, etc while, the measure of mitochondrial respiration indirectly proposed as a test for cell toxicity, by the inhibition of P38 MAPk which is likely to affect cell proliferation.
44,45
The preventive mechanisms of natural phytochemicals on tumor promotion range from the inhibition of genotoxic effects, increased antioxidants and anti-inflammatory activity, inhibition of proteases and cell proliferation, protection of intracellular communications to modulate apoptosis and signal transduction pathways.
4
The presence of biologically active components such as alkaloids, steroids, triterpenoids, and flavonoids in the leaf extracts of three plants understudy may form the basis for the cell antiproliferative effects as these compounds have proven to exhibit anticancer effects.
46
Further, the presence of specific anticancer compounds in mulberry viz. ursolic acid, oxyresveratrol, moracin and deoxynojirimycin-1,
7
and so on could be contributing significantly to antiproliferative effects.
Conclusion
Despite the various biological activities of mulberry leaf extracts, the bioactive constituents responsible for its antioxidant activities are not clear. In this line of approach, the major groups of phytoconstituents responsible for antioxidant activities in different solvent extracts of three Morus species were evaluated in vitro and further an attempt was made in selecting a suitable antioxidant assay reflecting the phytochemical profile of the plant extract. The results revealed that all the three different extracts of three Morus species possess potential antioxidant activities and the phenolic compounds have a major contribution in their antioxidant ability in chemical assays in vitro. The remarkable antiproliferative effect demonstrated by the methanolic extract of M. laevigataindicates a species-specific distribution of bioactive molecules which depends greatly on the seasons, different agronomic conditions, and other abiotic and biotic factors. This work represents an advance in biomedical science because the role of phenolics as significant contributors to the antioxidant capacity of plants is well established. These findings endorse further investigations on these plants to determine the active principles and their mode of action.
Funding sources
None.
Ethical statement
There is none to be declared.
Competing interests
Authors declare no conflict of interests.
Authors’ contribution
ARSJ and RCK: Conceptualization & experiments design; ARSJ, SMS & JKS: Performed the experiments; ARSJ, RCK & JKS: Writing and reviewing; RCK: Project administration.
Supplementary file 1 contains Tables S1 and S2 as well as FTIR, 1H NMR, 13C NMR and LCMS of Compounds 1-3.
(pdf)
Research Highlights
What is the current knowledge?
simple
-
√ The role and contribution of natural products to human health care is a well-documented fact and its impact on modern drug development is enormous.
-
√ Given that plants are the major natural sources for therapeutics, many plants have been exploited extensively for this purpose.
-
√ It is the need of the hour to explore new plants which can reduce the exploitation load present in some highly utilized plant species.
-
√ Plants like Morus alba which are well known for its antioxidant and anticancer effects require an alternative source to fulfill human needs
What is new here?
simple
-
√ The current study focused on identifying such alternate plant sources belonging to the same genus as they share most of the common characters and hence are expected to produce similar antioxidant and cytotoxic effects as M. alba.
-
√ The study depicted the relationship between the presence of different groups of phytochemicals and their contribution to achieving the antioxidant effect.
-
√ The study also justified the connection between antioxidant molecules being potential anticancer molecules.
-
√ Computational studies predicted the probable mechanism of binding of isolated molecules at the ATP binding site of P38 MAPk protein and inhibiting its activity in cancerous cells.
References
- Butt MS, Nazir A, Sultan MT, Schroën K. Morus alba L nature's functional tonic. Trends Food Sci Technol 2008; 19:505-12. doi: 10.1016/j.tifs.2008.06.002 [Crossref] [ Google Scholar]
- Rohman A, Riyanto S, Yuniarti N, Saputra WR, Utami R, Mulatsih W. Antioxidant activity, total phenolic, and total flavaonoid of extracts and fractions of red fruit (Pandanus conoideus Lam). Int Food Res J 2010; 17:97-106. [ Google Scholar]
- Ozsoy N, Can A, Yanardag R, Akev N. Antioxidant activity of Smilax excelsa L leaf extracts. Food Chem 2008; 110:571-83. doi: 10.1016/j.foodchem.2008.02.037 [Crossref] [ Google Scholar]
- Soobrattee MA, Bahorun T, Aruoma OI. Chemopreventive actions of polyphenolic compounds in cancer. Biofactors 2006; 27:19-35. [ Google Scholar]
- Aditya Rao SJ, Ramesh CK, Basavaraj Padmashali JK. Evaluation of anti-inflammatory and analgesic activity in three Morus species. Res J Pharm Biol Chem Sci 2013; 4:822-30. [ Google Scholar]
- Moghtaderi H, Sepehri H, Delphi L, Attari F. Gallic acid and curcumin induce cytotoxicity and apoptosis in human breast cancer cell MDA-MB-231. BioImpacts 2018; 8:185. doi: 10.15171/bi.2018.21 [Crossref] [ Google Scholar]
- Venkatesh Kumar R, Chauhan S. Mulberry: life enhancer. J Med Plant Res 2008; 2):271-8. [ Google Scholar]
- Chan EW, Lye PY, Wong SK. Phytochemistry, pharmacology, and clinical trials of Morus alba. Chin J Nat Med 2016; 14:17-30. doi: 10.3724/SP.J.1009.2016.00017 [Crossref] [ Google Scholar]
- Lee JC, Kumar S, Griswold DE, Underwood DC, Votta BJ, Adams JL. Inhibition of p38 MAP kinase as a therapeutic strategy. Immunopharmacol 2000; 47):185-201. doi: 10.1016/S0162-3109(00)00206-X [Crossref] [ Google Scholar]
- Makkar HP, Blümmel M, Borowy NK, Becker K. Gravimetric determination of tannins and their correlations with chemical and protein precipitation methods. J Sci Food Agric 1993; 61:161-5. doi: 10.1002/jsfa.2740610205 [Crossref] [ Google Scholar]
- Jia Z, Tang M, Wu J. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem 1999; 64:555-9. [ Google Scholar]
- Prieto P, Pineda M, Aguilar M. Spectrophotometric quantitation of antioxidant capacity through the formation of a phosphomolybdenum complex: specific application to the determination of vitamin E. Anal Biochem 1999; 269:337-41. doi: 10.1006/abio.1999.4019 [Crossref] [ Google Scholar]
- Oyaizu M. Oyaizu MStudies on Products of Browning ReactionAntioxidative activities of products of browning reaction prepared from glucosamine. Japanese J Nutr Diet 1986; 44:307-15. doi: 10.5264/eiyogakuzashi.44.307 [Crossref] [ Google Scholar]
- Jing L, Ma H, Fan P, Gao R, Jia Z. Antioxidant potential, total phenolic and total flavonoid contents of Rhododendron anthopogonoides and its protective effect on hypoxia-induced injury in PC12 cells. BMC Complement Altern Med 2015; 15:287. doi: 10.1186/s12906-015-0820-3 [Crossref] [ Google Scholar]
- Wong SP, Leong LP, Koh JH. Antioxidant activities of aqueous extracts of selected plants. Food Chem 2006; 99:775-83. doi: 10.1016/j.foodchem.2005.07.058 [Crossref] [ Google Scholar]
- Johnson EI. The quantitative analysis of drugs By DC Garratt. J Pharm Pharmacol 1964; 16:772. doi: 10.1111/j.2042-7158.1964.tb07408.x [Crossref] [ Google Scholar]
- Halliwell B, Gutteridge JM, Aruoma OI. The deoxyribose method: a simple “test-tube” assay for determination of rate constants for reactions of hydroxyl radicals. Anal Biochem 1987; 165:215-9. doi: 10.1016/0003-2697(87)90222-3 [Crossref] [ Google Scholar]
- Dinis TC, Madeira VM, Almeida LM. Action of phenolic derivatives (acetaminophen, salicylate, and 5-aminosalicylate) as inhibitors of membrane lipid peroxidation and as peroxyl radical scavengers. Arch Biochem Biophys 1994; 315:161-9. doi: 10.1006/abbi.1994.1485 [Crossref] [ Google Scholar]
- Zhao L, Pei RS, Ji BP, Luo YC, Zhang D, Xu ZY, Jia XN. Antioxidant activity of aqueous extract fractions of velvet antler (Cervus elaphus Linnaeus). J Food Drug Anal 2010; 18:319-27. [ Google Scholar]
- Li X, Wu X, Huang L. Correlation between antioxidant activities and phenolic contents of radix Angelicae sinensis (Danggui). Molecules 2009; 14(12):5349-61. doi: 10.3390/molecules14125349 [Crossref] [ Google Scholar]
- Mai TT, Fumie N, Van Chuyen N. Antioxidant activities and hypolipidemic effects of an aqueous extract from flower buds of Cleistocalyx operculatus (Roxb) Merr and Perry. J Food Biochem 2009; 33:790-807. doi: 10.1111/j.1745-4514.2009.00251.x [Crossref] [ Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983; 65:55-63. doi: 10.1016/0022-1759(83)90303-4 [Crossref] [ Google Scholar]
- Aditya Rao SJ, Ramesh CK, Mahmood R, Jamuna KS, Prabhakar BT. Antitumor activity of two species of mulberry against EAT cell lines in mice. World J Pharm Res 2015; 4(3):1934-43. [ Google Scholar]
- Gasteiger J, Marsili M. Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 1980; 36:3219-28. doi: 10.1016/0040-4020(80)80168-2 [Crossref] [ Google Scholar]
- Paramesha M, Manivannan S, Rao SA, Srikanth KS, Neelwarne B, Shetty NP. Augmentation of pyrethrins content in callus of Chrysanthemum cinerariaefolium and establishing its insecticidal activity by molecular docking of NavMS Sodium Channel Pore receptor. 3 Biotech 2018; 8:367. doi: 10.1007/s13205-018-1387-8 [Crossref] [ Google Scholar]
- Raghavendra S, Rao SA, Kumar V, Ramesh CK. Multiple ligand simultaneous docking (MLSD): A novel approach to study the effect of inhibitors on substrate binding to PPO. Comput Biol Chem 2015; 59:81-6. doi: 10.1016/j.compbiolchem.2015.09.008 [Crossref] [ Google Scholar]
- Sanner MF. Python: a programming language for software integration and development. J Mol Graph Model 1999; 17(1):57-61. doi: 10.1016/S1093-3263(99)99999-0 [Crossref] [ Google Scholar]
- Fitzgerald CE, Patel SB, Becker JW, Cameron PM, Zaller D, Pikounis VB, O'Keefe SJ, Scapin G. Structural basis for p38α MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity. Nat Struct Mol Biol 2003; 10(9):764-9. doi: 10.1038/nsb949 [Crossref] [ Google Scholar]
- Aditya Rao SJ, Jeevitha B, Smitha R, Ramesh CK, Paramesha M, Jamuna KS. Wound healing activity from the leaf extracts of Morus laevigata and in silico binding studies from its isolates with gsk 3-β Int. J Res Dev Pharm Life Sci 2015; 4(4):1686-96. [ Google Scholar]
- Sanner ME, Olson A, Spehner J. An extension of spherical harmonics to region-based rotationally invariant descriptors for molecular shape description and comparison. Biopolymers 1996; 38(3):305-20. doi: 10.1002/(SICI)1097-0282(199603)38:3<305::AID-BIP4>3.0.CO;2-Y [Crossref] [ Google Scholar]
- Ni G, Zhang QJ, Zheng ZF, Chen RY, Yu DQ. 2-Arylbenzofuran derivatives from Morus cathayana. J Nat Prod 2009; 72(5):966-8. doi: 10.1021/np800789y [Crossref] [ Google Scholar]
- Babalola IT, Shode FO. Ubiquitous ursolic acid: a potential pentacyclic triterpene natural product. J Pharmacogn Phytochem 2013; 2:214-22. [ Google Scholar]
- Yang Y, Yang X, Xu B, Zeng G, Tan J, He X, Hu C, Zhou Y. Chemical constituents of Morus alba L and their inhibitory effect on 3T3-L1 preadipocyte proliferation and differentiation. Fitoterapia 2014; 98:222-7. doi: 10.1016/j.fitote.2014.08.010 [Crossref] [ Google Scholar]
- Bellamakondi PK, Godavarthi A, Ibrahim M. Caralluma umbellata Haw protects liver against paracetamol toxicity and inhibits CYP2E1. BioImpacts 2018; 8:23-30. doi: 10.15171/bi.2018.04 [Crossref] [ Google Scholar]
- Rice-evans CA, Miller NJ, Bolwell PG, Bramley PM, Pridham JB. The relative antioxidant activities of plant-derived polyphenolic flavonoids. Free Radic Res 1995; 22:375-83. doi: 10.3109/10715769509145649 [Crossref] [ Google Scholar]
- Kaur C, Kapoor HC. Anti‐oxidant activity and total phenolic content of some Asian vegetables. Int J Food Sci Technol 2002; 37:153-61. doi: 10.1046/j.1365-2621.2002.00552.x [Crossref] [ Google Scholar]
- Wang H, Cao G, Prior RL. Total antioxidant capacity of fruits. J Agric Food Chem 1996; 44:701-5. doi: 10.1021/jf950579y [Crossref] [ Google Scholar]
- Zhao J, Zhao X, Chao L, Zhang W, You T, Zhang J. Diversity change of microbial communities responding to zinc and arsenic pollution in a river of northeastern China. J Zhejiang Univ Sci B 2014; 15:670-80. doi: 10.1631/jzus.B1000173 [Crossref] [ Google Scholar]
- Ye CL, Liu JW, Wei DZ, Lu YH, Qian F. In vivo antitumor activity by 2′, 4′-dihydroxy-6′-methoxy-3′, 5′-dimethylchalcone in a solid human carcinoma xenograft model. Cancer Chemother Pharmacol 2005; 55:447-52. doi: 10.1007/s00280-004-0975-y [Crossref] [ Google Scholar]
- Aditya Rao SJ, Ramesh CK, Mahmood R, Prabhakar BT. Anthelmintic and antimicrobial activities in some species of mulberry. Int J Pharm Pharm Sci 2012; 4:335-8. [ Google Scholar]
- Reymond JL, Awale M. Exploring chemical space for drug discovery using the chemical universe database. ACS Chem Neurosci 2012; 3:649-57. doi: 10.1021/cn3000422 [Crossref] [ Google Scholar]
- Aditya Rao SJ, Ramesh CK, Raghavendra S, Paramesha M. Dehydroabietylamine, a diterpene from Carthamus tinctorious L showing antibacterial and anthelmintic effects with computational evidence. Curr Comput Aided Drug Des 2020; 16:231-7. doi: 10.2174/1573409915666190301142811 [Crossref] [ Google Scholar]
- Jinadatta P, Rajshekarappa S, Rao KS, Subbaiah SG, Shastri S. In silico, in vitro: antioxidant and antihepatotoxic activity of gnetol from Gnetum ula Brongn. BioImpacts 2019; 9:239. doi: 10.15171/bi.2019.29 [Crossref] [ Google Scholar]
- Maher P. p38 mitogen-activated protein kinase activation is required for fibroblast growth factor-2-stimulated cell proliferation but not differentiation. J Biol Chem 1999; 274:17491-8. doi: 10.1074/jbc.274.25.17491 [Crossref] [ Google Scholar]
- Kavitha MD, Gouda KG, Rao SJ, Shilpa TS, Shetty NP, Sarada R. Atheroprotective effect of novel peptides from Porphyridium purpureum in RAW 2647 macrophage cell line and its molecular docking study. Biotechnol Lett 2019; 41:91-106. doi: 10.1007/s10529-018-2621-5 [Crossref] [ Google Scholar]
- Grbović F, Stanković MS, Ćurčić M, Đorđević N, Šeklić D, Topuzović M, Marković S. In vitro cytotoxic activity of Origanum vulgare L on HCT-116 and MDA-MB-231 cell lines. Plants 2013; 2:371-8. doi: 10.3390/plants2030371 [Crossref] [ Google Scholar]