Bioimpacts. 12(6):567-588.
doi: 10.34172/bi.2022.23616
Review
Recent advances in electrochemical strategies for bacteria detection
Alireza Khoshroo 1, *  , Maryamosadat Mavaei 2, Masoume Rostami 3, Bahare Valinezhad-Saghezi 3, Ali Fattahi 2, 4, *
, Maryamosadat Mavaei 2, Masoume Rostami 3, Bahare Valinezhad-Saghezi 3, Ali Fattahi 2, 4, * 
Author information:
1Nutrition Health Research center, Hamadan University of Medical Sciences, Hamadan, Iran
2Pharmaceutical Sciences Research Center, Health Technology Institute, Kermanshah University of Medical Sciences, Kermanshah, Iran
3Student Research Committe, Kermanshah University of Medical Sciences, Kermanshah, Iran
4Medical Biology Research Center, Health Institute, Kermanshah University of Medical Sciences, Kermanshah, Iran
Abstract
Introduction:
Bacterial infections have always been a major threat to public health and humans' life, and fast detection of bacteria in various samples is significant to provide early and effective treatments. Cell-culture protocols, as well-established methods, involve labor-intensive and complicated preparation steps. For overcoming this drawback, electrochemical methods may provide promising alternative tools for fast and reliable detection of bacterial infections.
Methods:
Therefore, this review study was done to present an overview of different electrochemical strategy based on recognition elements for detection of bacteria in the studies published during 2015-2020. For this purpose, many references in the field were reviewed, and the review covered several issues, including (a) enzymes, (b) receptors, (c) antimicrobial peptides, (d) lectins, (e) redox-active metabolites, (f) aptamer, (g) bacteriophage, (h) antibody, and (i) molecularly imprinted polymers.
Results:
Different analytical methods have developed are used to bacteria detection. However, most of these methods are highly time, and cost consuming, requiring trained personnel to perform the analysis. Among of these methods, electrochemical based methods are well accepted powerful tools for the detection of various analytes due to the inherent properties. Electrochemical sensors with different recognition elements can be used to design diagnostic system for bacterial infections. Recent studies have shown that electrochemical assay can provide promising reliable method for detection of bacteria.
Conclusion:
In general, the field of bacterial detection by electrochemical sensors is continuously growing. It is believed that this field will focus on portable devices for detection of bacteria based on electrochemical methods. Development of these devices requires close collaboration of various disciplines, such as biology, electrochemistry, and biomaterial engineering.
Keywords: Bacterial infection, Electrochemical sensor, Rapid detection of infection, Bioreceptor, Bacteriophage
Copyright and License Information
© 2022 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
The presence of pathogenic bacteria in water, food, and environment around human beings endangers their health, and the infections caused by them pose a significant threat of morbidity and mortality worldwide. Excessive and inappropriate use of antibiotics creates resistance and difficulty in treatment.
1
This is while progress in development of new antibiotics does not match the rise in risk of increasing resistance, and fewer options remain for physicians and health care specialists for treatment of certain infections.
2,3
It is estimated that the annual mortality due to antimicrobial resistance will rise to 10 million cases worldwide by 2050. This problem has been addressed globally, where several countries are facing with emergence of bacteria that are entirely resistant to available antibiotics. Therefore, certain country-specific action plans have been prepared based on the worlds҆ action plan of the World Health Organization (WHO) to deal with antimicrobial resistance.
4
The ability of most pathogenic bacteria to evolve quickly allows them to be adapted and grow in various pH, temperature, and pressure conditions as well as a wide range of salt concentrations, which makes combating with them difficult.
4,5
Mycobacterium tuberculosis, Acinetobacter baumannii, Bacillus cereus, Brucella, and E. coli are only some of challenging bacterial species affecting humans҆ life. M. tuberculosis, causing development of tuberculosis, is a common airborne infectious microorganism leading to millions of deaths each year worldwide with the highest mortality rate of 1.8 million deaths.
5,6
Acinetobacter baumannii is a non-motile gram-negative member of the Gammaproteobacteria. These microorganisms are responsible for some infections, i.e., bloodstream infections, wound infections, ventilator-acquired pneumonia, and urinary tract infections.
7,8
Bacillus cereus is a gram-positive bacillus that causes food-borne diseases, i.e., emetic and diarrheal syndromes.
9
Brucella bacteria are encapsulated gram-negative coccobacilli known to affect animals, such as bovines, camels, sheep, and goats. B. abortus, B. melitensis, B. canis, and B. suis are the primary Brucella bacteria capable of causing diseases in human beings.
10
The symptoms of these diseases include undulant fever, headache, chills, myalgia, and arthralgia. E. coli O157: H7 via foodborne poisoning and water pollution causes hemolytic uremic syndrome, colitis bleeding, and diarrhea.
11
Therefore, there is a need for a simple method to detect bacteria so as to improve quality of life and humans҆ health. Early detection of bacterial infections is essential for health care; it can prevent propagation of infections and antibiotic resistance and help to choose right treatment for patients.
12-16
To this end, rapid, easy, cost-effective, and accurate detection of infections and correct distinguishing between the infected and uninfected people have an enormous effect on controlling drug resistance. Conventional bacterial diagnosis techniques, such as cell culture, colony counting, polymerase chain reaction (PCR), enzyme-linked immunosorbent assay (ELISA), and fluorescence-based technique, are inappropriate in this case because, they are labor-intensive and time-consuming and require sample preparation before analysis.
5,7
Furthermore, low concentration of bacteria, limit of detection (LOD) in the range of 10410-5 CFU/mL, is a challenge for these methods.
4
In the recent years, new methods have been applied based on electrochemical approaches to improve bacterial detection.
17-22
These methods have received a great deal of attention due to their sensitivity, high selectivity, acceptable precision (5-7%), and fast analysis time (5-10 minutes). There is also no complicated pretreatment or high skill required for analysis.
21-23
The electrochemical sensor used in this regard is made up of three parts, including a recognition element on the sensing platform, an effective transducer, and a digital signal processor (Fig. 1). The receptors used in construction of electrochemical sensors are illustrated in Fig. 1.
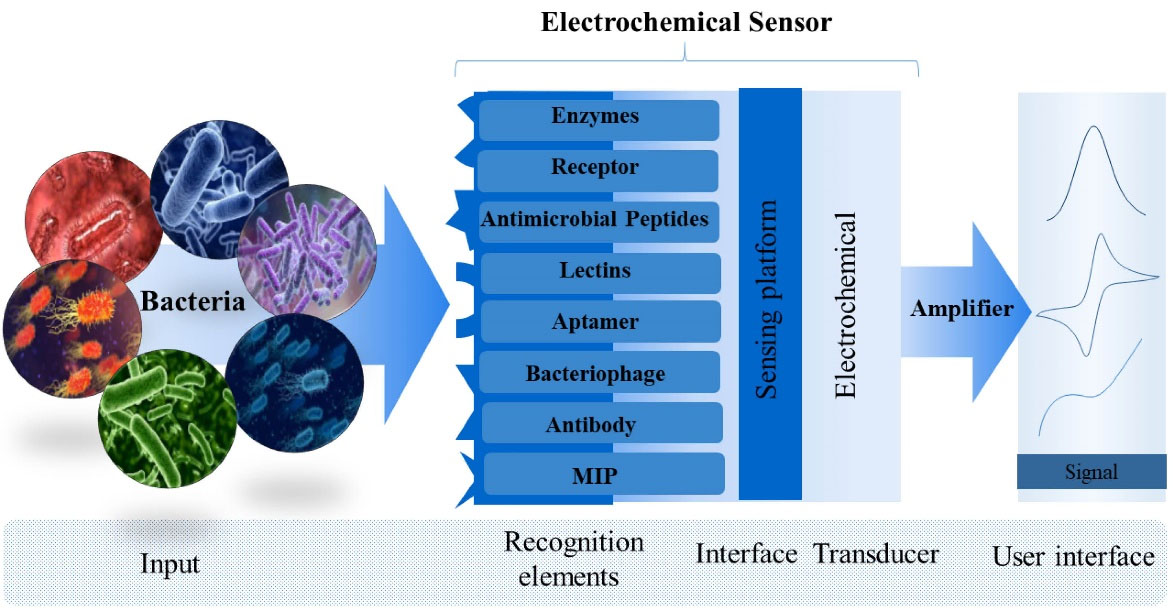
Fig. 1.
Structure of an electrochemical sensor showing recognition element, interface, transducer and output signal.
.
Structure of an electrochemical sensor showing recognition element, interface, transducer and output signal.
The present study is a review study on strategies used for building sensors developed during 2015-2020. In this respect, there are references, including the studies by Majdinasab et al
24
who presented various aptasensors for detection of pathogenic bacteria in food samples, Richter et al
25
who reviewed different bacteriophage-based methods for bacterial detection, Amiri et al
26
who published a comprehensive overview of the current electrochemical methods used for detection of pathogens, and Kuss et al
27
who reported the recent advancements and challenges in electrochemical detection of pathogenic bacteria. This review study focuses on designing bacteria-detecting sensors based on recognition elements that have not been reviewed so far. For this purpose, many references in the field were reviewed, and the review covered several issues, including (a) enzymes, (b) receptors, (c) antimicrobial peptides, (d) lectins, (e) redox-active metabolites, (f) aptamer, (g) bacteriophage, (h) antibody, and (i) molecularly imprinted polymers. Explanation is provided for detection method in each case in the following.
Enzymes
Enzymes, as catalytic proteins, are common examples of biomarkers used in enzymatic electrochemical biosensors for recognition of bacteria.
28
Enzymatic biosensors work on the basis of reaction of an enzyme with its substrate so that, substrate is metabolized by enzyme or inhibits enzymatic reaction.
29
Despite the advantages offered by enzyme-based sensors, several restrictions are associated with their use. Cost and instability of enzymes are a part of challenges in wide application of enzyme-modified assays.
28
For overcoming this problem, Mubarok et al,
30
developed a convenient, simple, and rapid electrochemical assay to monitor neuraminidase activity in saliva, human blood, urine, and nasal swab samples. The assay was based on a facile synthesized probe N-acetyl-2-O-(4-aminophenyl)-neuraminic acid (AP-Neu5Ac) as a non-protein substrate on sensing platform to determine neuraminidase activities. Thus, an electrochemical sensor was developed based on cleavage of the AP-Neu5Ac. Release of p-aminophenol, resulting from the neuraminidase-catalyzed hydrolysis of AP-Neu5Ac was followed using a differential pulse voltammetry (DPV) technique. The developed sensor showed good sensitivity with a LOD of 5.6 ng/mL. The main advantage of the proposed method based on AP-Neu5Ac is its applicability in real samples.
30
More recently, enzymatic biosensors have been used to identify foodborne pathogenic bacteria. The amperometric enzymatic biosensor developed by Huang et al,
31
can detect Listeria monocytogenes and determine bacterial count and somatic cell count (SCC) in raw milk. Bacterial infection was evaluated through bacterial count on the basis of changes in electroactive metabolites of microorganisms. The signals resulting from lactate dehydrogenase (LDH) activity were used to determine SCC. LDH, as an enzyme biomarker, can catalyze conversion of lactate acid and NAD+ as substrates into pyruvic and NADH as electrochemical active substances. Detection range was found to be from 102 to 108 CFU/mL within detection time of 1–10 hours by amperometric technique. SCC detection could be performed from 350 to 780 thousand SCC/mL in about 60 seconds.
31
As a biomarker and endogen enzyme, β-galactosidase (β-gal) can be applied for detection of microorganisms by creating a clear signal. In their study, Tarditto et al
32
reported an electrochemical magneto immunosensor to detect E. coli in swine feces. This immunosensor worked based on activity of β-gal to generate p-aminophenol (p-AP) from p-aminophenyl-β-D-galactopyranoside (p-APG). For this purpose, magnetic beads conjugated with enterotoxigenic E. coli polyclonal antibody (anti-ETEC F4) were used, and LOD of 33 CFU/mL was obtained in less than 2 hours by square wave voltammetry (SWV) technique.
Recently, an electrochemical sensor has been presented to detect of coliform bacteria on the food samples. This voltammetric sensor, reported by Badalyan et al,
33
is based on indirect sensing of β-gal activity which product by coliform bacteria. Detection range was found to be from 1.6 log10−6.6 log10 CFU/mL
1
for coliform bacteria. The screen-printed electrode (SPE) was modified with graphene and polyacrylamide gel (GR/PAAGC). In this study, 6-chloro-3-indoxyl-β-D-galactopyranoside (6-CIGP) was applied as a novel substrate to monitor electroactive 6,6′-dichloro-Indigo (6-DI) generated by enzymatic reaction of 6-CIGP with ß-gal. β-gal product by coliform bacteria via lacZ gene. The sensor proved to have a detection limit of 0.1 log10 CFU/mL in 30 minutes by cyclic voltammetric (CV) technique.
Lightness, biocompatibility, user friendliness, and low cost of paper-based sensors have made them a top priority to monitor bacteria through colorimetric analysis and electrochemical sensing.
34,35
Adkins et al
36
evaluated enzyme activity in water and food samples containing Enterococcus ssp. and E. coli using the printed carbon electrodes on transparency films. Production of β-glucuronidase and β- galactosidase by E. coli and expression of β-glucosidase by Enterococcus ssp. were tracked calorimetrically using a smartphone and a cardboard box (Fig. 2). Alternatively, these bacterial enzymes could be directly detected electrochemically. Limit of quantitation (LOQ) was as low as 10 CFU/mL for E. coli and 100 CFU/mL for E. faecalis and E. faecium by SWV technique. The electrochemical method did not decrease analysis time, but a comparison of that method with the colorimetric method in terms of detection limits demonstrated superiority of the electrochemical method.
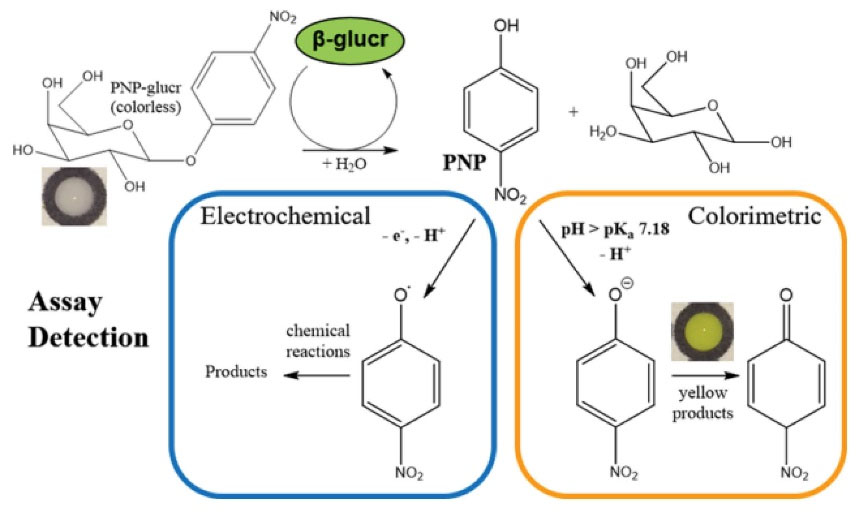
Fig. 2.
Reaction scheme showing the dual electrochemical and colorimetric detection of formed PNP from reacting bacterially produced β-glucr with PNPglucr. Reprinted with permission from Adkins et al.36 Copyright 2022 American Chemical Society.
.
Reaction scheme showing the dual electrochemical and colorimetric detection of formed PNP from reacting bacterially produced β-glucr with PNPglucr. Reprinted with permission from Adkins et al.36 Copyright 2022 American Chemical Society.
Noh et al
37
designed an enzymatic biosensor to detect E. coli. This system was established through redox cycling of isopropyl-β-d-thiogalactopyranoside (IPTG) and permeabilization treatment simultaneously. The expression level of ß-d-galactosidase (gal) was increased by IPTG treatment, and enzymatic reaction of gal substrate was facilitated by permeabilization treatment. The electrochemical detection strategy had a high sensitivity with a LOD of 1.0 CFU/mLby chronocoulograms without a need for DNA amplification.
37
As mentioned before, enzyme-based biosensors have been developed to detect pathogenic bacteria, but some limitations exist in wide application of those biosensors, such as instability of enzymes especially after immobilization on a surface and high cost of protein production.
38,39
Receptor
Regarding the research on sensing, receptor proteins are alluring owing to their generic “receiving functions” as well as “sending functions”.
40
They are also widely used for biosensing because of their high specificity and sensitivity.
28
Toll-like receptors (TLRs) are activated by a broad range of microbial components called as pathogen-associated molecular patterns (PAMPs). These components include substances on outer membranes of bacterial walls (e.g., lipopolysaccharides (LPS), peptidoglycans, and lipopeptides), bacterial flagellin, bacterial DNA, and viral RNA.
34,41,42
They are applied to detection of bacteria because, they possess natural detection capabilities.
43
Unlike other pathogen recognition elements, TLR protein has the specific potential to carry out broad-spectrum detection of endotoxins. Utilizing a wide range of available detection strategies can be a step to develop clinical assays for early warning diagnosis and quick treatment.
44
In an attempt, She et al
44
designed an electrochemical response sensor based on the interaction between TLR5 and bacterial flagellins to monitor endotoxins of both gram-negative (S. typhimurium) and gram-positive (B. subtilis) bacterial strains with LOD of 0.1 μM for each flagellin. Lipoic acid n-hydroxysuccinimide ester (LPA) was firstly modified on an Au electrode. Then, TLR5 was immobilized on sensing surface via an amine coupling reaction. The interaction of TLR5 with endotoxin could be evaluated by electrochemical impedance spectroscopy (EIS) and voltammetry.
In another study for detection of endotoxin, Amini et al
45
used LPS as a recognition element. A complex of TLR4 and myeloid differentiation-2 (MD-2) immobilized on a sensing platform was used to detect LPS from E. coli and Salmonella. Modification of the electrode with TLR4/(MD-2) complex through Lip-NHS as a link allowed detection of LPS from E. coli with a LOD of 1.3×10-4 EU/mL and detection of LPS from Salmonella with a LOD of 1.5×10 -4 EU/mL by EIS tecnique.
45
Mayall et al
46
successfully designed a biosensor using the TLR4 protein to recognize macromolecular LPS and thus, to trace gram-negative bacteria. LOD was only 1 ng/mL. For the receptor biosensor, a self-assembled thiolated monolayer containing carboxylic acid and 11- mercaptoundecanoic acid (MUA) was employed. The COOH groups were activated via 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC)/ N-hydroxysuccinimide (NHS)/ 2-morpholinoethanesulfonic acid (MES), leading to formation of amide bonds. This allowed tethering of nitrilotriacetic acid (NTA) to surface of the modified Au microelectrode. Then, the NTA groups could be coordinated with Ni2+ ions. TLR-4 proteins were also tethered to Ni-NTA functional groups. When the target LPS was added to the solution, electrode impedance was rapidly increased due to dimerization of the TLR-4 proteins around macromolecular LPS.
46
The same authors reported a TLR-4 receptor sensor fabricated with various types of thiol sites that had different terminal functional groups immobilized on a gold surface (Fig. 3). In their study, Mayall et al sought to increase and improve signal magnitude and decrease detection time sensing of the sensor by employing a redox-active functional group (ferrocenyl group). TLR-4 was immobilized on the electrode via interactions with a poly-histidine tag. In the presence of LPS (from lysed Gram-negative bacteria), the TLR-4 dimerized and blocked access of ferrocyanide in the solution to ferrocenyl-terminated thiols, thus increasing probe resistance. It was shown that the ferrocene-based biosensor would not respond selectively to gram-positive bacteria or viruses. Linear range of this sensor was 100-105 cells/mL based on impedance measurements for gram-negative bacteria.
47
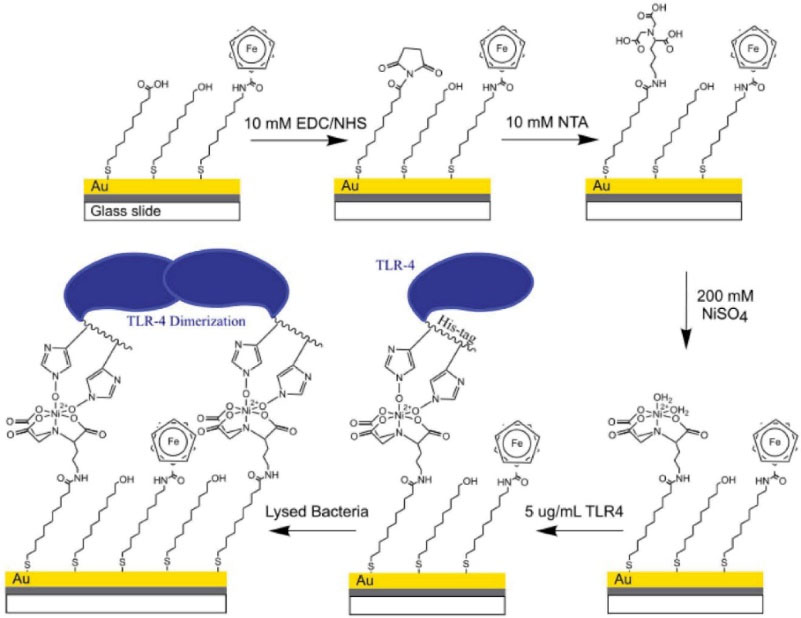
Fig. 3.
Schematic representation of the proposed SAM design containing ferrocenylterminated thiols mixe with similar chain length carboxylic acid and hydroxyl-terminated thiols. Reprinted with permission from Mayall et al.
47
Copyright 2018 American Chemical Society.
.
Schematic representation of the proposed SAM design containing ferrocenylterminated thiols mixe with similar chain length carboxylic acid and hydroxyl-terminated thiols. Reprinted with permission from Mayall et al.
47
Copyright 2018 American Chemical Society.
Antimicrobial peptides
Antimicrobial peptides (AMPs) form another class of recognition components with a positive charge and are composed of 12–50 amino acids with molecular weight of 1-5 kDa. Due to their positive charge, AMPs are extremely specific for pathogenic bacteria, allowing electrostatic interactions with oppositely charged components on membranes of bacteria.
26
The first application of AMPs in electrochemical sensors was reported by Manoor et al,
42
who used magainin I-immobilized interdigitated gold electrodes to detect E. coli O157:H7 and S. typhimurium. AMPs, as synthetic biomolecules, are advantageous for bacterial sensing due to their ease of synthesis, selective binding to targets, and intrinsic stability. However, it is not possible to use AMPs for detection of bacterial strains in real samples. Therefore, a great challenge with this bioreceptor is achieving sensitivity and especially, selectivity within different bacterial strains.
48-50
For AMP-modified biosensors, sensitivity can be finely improved and tuned by various strategies. One of strategies for impedimetric sensors is fabrication of peptidic modulators. Liu et al
51
developed a multi-domain peptide with the sequence of WK3 (QL) 6 K2 G3C for sensitive bacterial detection. In detail, the peptide was immobilized on an Au platform via G3C to selectively detect S. epidermidis, E. coli, Staphylococcus aureus, and P. aeruginosa with a LOD of 102 CFU/mL by EIS technique. This platform had the potential for differentiation and detection of living and dead bacteria. The excellent ability of AMP-modified sensors to detect bacteria was used in the study by Eissa and Zourob
52
to construct a dual colorimetric and electrochemical sensor of S. aureus protease. In this strategy, a gold sensing platform was modified with AMP magnetic nanoparticles, which produced black color of the electrode. When, S. aureus protease was added onto the sensing platform, peptide sequence cleavage and yellow color of the gold electrode appeared. In addition, current changed after peptide cleavage that was proportional to bacteria. LOD of 3.0 CFU/mLwas obtained by SWV technique after a minute.
Another strategy is the use of three-dimensional interdigitated electrode arrays (3D-IDEA) as platforms to improve sensitivity of transducers. This is made possible through separation of electrode digits with insulating barriers. Gil et al reported a similar technique implemented on 3D-IDEA platformsby impedance measurements. Hoyos-Nogués et al
53
focused on N-terminal domain of the human antibacterial protein lactoferrin (LF), or hLF1-11 peptide. Via its N-terminus, the synthetic peptide hlf1-11 was immobilized on surface of 3D-IDEA modified with epoxy silane, and its capacity to rapidly bind onto pathogenic Streptococcus sanguinis was evaluated by EIS. LODs of 10 CFU/mL and 100 CFU/mL were obtained for S. sanguinis in KCl and saliva by EIS technique, respectively. The proposed device presents a promising tool for periodontal implants in monitoring of infections.
Another strategy to improve sensitivity is signal amplification. There are different signal amplifiers, such as carbon nanotubes, organometallic compounds (e.g., ferrocene), metal/metal oxide nanoparticles, and magnetic nanoparticles (MNPs).
54-56
In this regard, Wilson et al
57
used MNPs bioconjugated with melittin (MLT) for bacterial separation. MLT, as a cationic amphipathic molecule, shows antimicrobial properties against different microorganisms. They reported a sensitive method to distinguish gram-positive from gram-negative bacteria, such as E. coli, Salmonella typhimurium, and S. aureus in food samples. The proposed biosensors detected E. coli (LOD: 1 CFU/mL) by EIS technique within 25 minutes very efficiently. Furthermore, Andrade et al
58
introduced a detection assay composed of carbon nanotubes and clavanin A (ClavA) to identify Enterococcus faecalis, Klebsiella pneumoniae, Bacillus subtilis, and E. coli based on EIS measurements. This nanostructured sensor showed a wide range of bacterial counts (102–106 CFU/mL). Also, LOD of the biosensor was equal to 102 CFU/mLfor Klebsiella pneumonia and E. coli while, B. subtilis and Enterococcus faecalis had a LOD of 103 CFU/mL. The results showed that the proposed sensor can distinguish non-pathogenic bacteria from pathogenic ones as well as gram-negative bacteria from gram-positive ones.
36
A few years later, the research group made a change in detection system of E. coli, E. faecalis, P. aeruginosa, S. typhimurium, and S. aureus using the same ClavA but, this time, it was conjugated to Au nanoparticles. This allowed a linear range of detection from 101 to 104 CFU/mL. LOD of the AMP-based impedimetric sensor was equal to 103 CFU/mL.
59
The same authors modified electrode surface with cysteine (Cys)/AuNPs/4-mercaptobenzoic acid (MBA) and then, bound it to ClavA. LOD of the heat-killed bacteria was equal to 103 CFU/mL by EIS technique.
60
Li et al reported a rapid impedimetric biosensor for detection of E. coli.
61
In this study, a combination of ferrocene and magainin I was immobilized on an Au electrode. This modification led to LOD of 103 CFU/mL. Zhang et al
62
also developed an electrochemical M. tuberculosis sensor based on two-dimensional Ti3C2 MXenes as a signal-amplifying material. The target biomarker and capture probe were 16S rDNA of bacteria and peptide nucleic acid, respectively. LOD was equal to 20 CFU/mL in 2 hours.
One of the methods for detection of multiplexed bacteria is using microfluidic systems. Lillehoj et al
63
reported the use of a microfluidic chip modified and interdigitated with two species-specific AMPs, including G10KHc and C16G2cys to detect multiple types of bacteria (e.g., Streptococcus mutans and Pseudomonas aeruginosa). LOD of 105 CFU/mLwas calculated by EIS technique within 25 minutes for both strains of bacteria.
Lectins
In the category of biomarkers, lectins are the proteins that can be exploited to specifically bind to bacterial carbohydrate chains. This biomolecule is used as a bimolecular glue to immobilize proteins and as a molecular recognizer in biosensors.
45
Lectin molecules and carbohydrates in structure of bacteria can recognize each other. The main motivations for using lectin as a sensing platform are its intrinsic stability, high sensitivity, good grafting, small size, and cheap assembly process.
17
In this regard, for clinical applications, Estrela et al
64
reported a metal oxide-based field-effect transistor (FET) sensor developed using an extended- gate FET with an interdigitated metal oxide semiconductor (MOS) for detection of the mannose-specific type 1 fimbriae of E. coli. The authors proposed an extended-gate FET system immobilized with α-D-mannose to detect a label-free target E. coli strain (i.e., uropathogenic E. coli called as UPEC). Using this device, LOQ was found to be 2 × 105 CFU/mLby EIS technique within about only 2 seconds. The results showed that biologically-sensitive FET is capable of the initial discrimination between pathogenic and non-pathogenic UPEC strains and also multiplexed screening.
Di Lorenzo et al
65
used a paper-based electrochemical device to detect bacteria up to 1.9 × 103 CFU/mLin a water sample by EIS technique. The paper-based electrode was constructed with carbon conductive ink on hydrophobic paper. Then, carbon sensing platform was oxidized electrochemically to generate COOH groups on carbon surface. These groups were activated with NHS/EDC to allow immobilization of lectin Con A as a carbohydrate-binding protein and biorecognition element. Con A was chosen due to its potential to selectively link with mono- or oligo-saccharides on bacterial cells.
In 2016, Yang et al
66
designed a label-free Con A-based biosensor platform for rapid identification of E. coli strains by EIS technique. Con A tended towards the target (E. coli) and differentiated gram-negative bacteria from gram-positive ones. For the biosensor, a binary monolayer was made with a combination of 11-mercaptoundecanoic acid and dithiothreitol on surface of an Au electrode and then, Con A was covalently immobilized. E. coli binding on the electrode was monitored with LOD of 75 CFU/mLin a linear range of 1 × 102-1 × 105 CFU/mL.
A robust method of detecting bacterial endotoxin on a 3D-IDEA platform was reported by pursuit of charge changes in surface of the platform. Changes in charge were induced by interaction of the immobilized Con A and LPS as a ubiquitous marker in test solution. The bacterial LPS was detected within 20 minutes. Screening LPS of E. coli binding by EIS changes yielded a low LOD of 2 µg/mL. Brosel-Oliu et al
67
employed polyethylenimine (PEI) polycation as an initial anchoring layer negatively charged at neutral pH. Then, Con A was immobilized through deposition on the 3D-IDEA surface. For evaluation of capturing LPS with the PEI interface, several blocking strategies were tried until a selective sensor response was gained between LPS and Con A.
67
A brief summary indicating some of the characteristics of representative the sensors covered in the review is presented in Table 1.
Table 1.
Overview of sensors proposed for bacteria detection divided by the type of recognition elements
|
Analyte
|
Recognition elements
|
Detectionmethod
|
Linear range / CFU/mL
|
LOD CFU/mL
|
Ref.
|
| Listeria monocytogenes |
LDH |
Amperometry |
102 to 108
|
- |
30
|
|
E. coli
|
β-gal |
SWV |
5 × 101 to 5 × 103
|
33 |
32
|
| Coliform |
β-gal |
CV |
1.6 log10−6.6 log10
|
0.1 log10
|
33
|
|
Enterococcus ssp.
|
β-glucosidase |
SWV |
- |
100 |
|
|
E. coli
|
Glucuronidase |
SWV |
- |
10 |
36
|
|
E. coli
|
β-gal |
Chronocoulometry |
1 to 106
|
1 |
32
|
| Gram-negative bacteria |
TLR-4 |
EIS |
102 to 105
|
- |
47
|
|
E. coli
|
Peptides |
EIS |
104 to 107
|
- |
42
|
|
S. epidermidis
|
Peptides |
EIS |
102 to 106
|
100 |
51
|
|
E. coli
|
Peptides |
EIS |
102 to 106
|
100 |
51
|
|
Staphylococcus aureus
|
Peptides |
EIS |
102 to 106
|
100 |
51
|
|
P. aeruginosa
|
Peptides |
EIS |
102 to 106
|
100 |
51
|
|
S. aureus
|
Peptide |
SWV |
10 to 108
|
3 |
52
|
|
S. sanguinis
|
Peptide |
EIS |
102 to 100
|
10 |
53
|
|
E. coli
|
Melittin |
EIS |
1 to 106
|
1 |
57
|
|
S. aureus
|
Melittin |
EIS |
1 to 106
|
10 |
57
|
|
S. typhi
|
melittin |
EIS |
10 to 104
|
10 |
57
|
|
Enterococcus faecalis
|
Clavanin A |
EIS |
102 to 106
|
100 |
58
|
|
Klebsiella pneumoniae
|
Clavanin A |
EIS |
102 to 106
|
100 |
58
|
|
E. coli
|
Clavanin A |
EIS |
101 to 104
|
103
|
59
|
|
E. coli
|
Peptide |
EIS |
103 to 107
|
103
|
61
|
|
M. tuberculosis
|
rDNA fragments |
EIS |
102 to 108
|
20 |
62
|
|
Streptococcus mutans
|
Peptide |
EIS |
104 to 107
|
103
|
63
|
|
Pseudomonas aeruginosa
|
Peptide |
EIS |
104 to 107
|
103
|
63
|
|
E. coli
|
Aminoethyl glycosides |
EIS |
- |
2 × 105
|
64
|
| Bacteria |
Con A |
EIS |
103 to 106
|
1.9 × 103 |
65
|
|
E. coli
|
Con A |
EIS |
102 to 105
|
75 |
66
|
Redox-active bacterial metabolites
Another set of agents in the category of biomarkers are secondary metabolites as redox-active compounds. Bacterial species depend on a broad range of factors, such as signaling molecules, signal detection methods, and signal-transduction mechanisms, which can coordinate gene regulation.
68
Various pigments as secondary metabolites released by bacteria play vital roles in pathogenesis of bacteria. These diagnostic bio-molecules and redox-active biomarkers include integral components of bacterial quorum sensing (QS), small secondary metabolites, and virulence factors associated with bacterial functions.
69
QS molecules in cell-to-cell communication process are the molecules that are involved in production and detection of diffusible signaling molecules.
In this regard, 2-heptyl-4-hydroxyquinoline (HHQ), producing Pseudomonas quinolone signals (PQSs), can act as a potential diagnostic biomarker for specific detection of P. aeruginosa (PA) in early-stage infections. It is also likely to prevent biofilm formation with well-timed medical treatment. One class of QS secondary metabolites and controlled extracellular factors is set for phenazine compounds, which are present in pyocyanin (PYO), phenazine-1-carboxylic acid (PCA), 5-methylphenazine-1-carboxylic acid (5-MCA), and 1-phenazine-1-carboxamide (PCN).
70
PYO, as a redox-active material, is both a quorum sensing agent and an extracellular virulence factor for PA.
71
Generation of several virulence factors by PA and other species is controlled by cell-to-cell signaling mediated by QS molecules. PA also secretes secondary metabolites as well as small molecules (2’-aminoacetophenone, 2AA) in a culture supernatant.
72
Indeed, barakacin as a signaling molecule and pyoverdine (PVD) as a virulence factor serve as biomarkers generated by PA.
73
Clinical diagnostic methods for positive detection and determination of PA involve a time-consuming and expensive culture growth process. Therefore, certain standard laboratory redox-active biomarkers have recently been developed. Electrochemical detection of PYO through transparent carbon ultramicroelectrode arrays (T-CUAs) was proposed by Stevenson et al.
74
Electro-active nature of PYO led to electrochemical conversion of PYO at T-CUA using the voltammetric technique. Also, through SWV, the sensor could detect PYO with a LOD of 1-1.6 µM in a linear dynamic range of 1-250 µM.
In another report,
75
the same researchers detected some PYO secreted on T-CUAs in a biological growth medium. In their previous study, they had conducted tests in a buffer solution without growth media. Recognition of cellular PYO agents in biological growth media indicated that bacterial species were exposed to a plethora of compounds (e.g., sugars and proteins); when these compounds were adsorbed onto sensing platform, they probably hindered signal of PYO agents. The authors concluded that their proposed technique was a proper one to apply in studies of biotoxins through T-CUAs.
Inspired by the in-situ use of electrocatalytic current for bio-sensing, Yong et al
76
pioneered in introduction of a whole-cell electrochemical biosensor based on S. oneidensis MR-1 cell as an electroactive type of bacteria and a proof and redox cycling module for detection of PYO agents with obviously magnified detection signals. This system could achieve ultrahigh sensitivity (1.3 μA/nM) and low LOD (47 pM) by CV technique. Redox reactivation/cycling systems are applied as efficient tools to enhance electrochemical signals in sensitive detection of pathogens for recognition of PYO agents at the point of care (POC). Also, electrochemical signal output can be significantly amplified by integration of biochemical redox cycling systems to continuously regenerating target analytes. Although the CV technique is not readily adapted to a POC device, the speed and ease with which CV curves can be obtained make this detection method ideal for ensuring quality control when carrying out each step in the sensor surface modification procedure. Under these premises, Yuan et al
77
reported an in-vivo two-way redox cycling system based on whole-cell bidirectional electron transfer for simultaneous recognition of two types of warfare toxins. This system provided the simultaneous use of lactate (as an electron donor) for reductive cycling reactions and fumarate (as an electron acceptor) for oxidative cycling reactions. Using 1-hydroxyphenazine (OHP), PYO, and voltammetry as a recognition method, ultrasensitive detection was done in the range of 304 ± 4 pM for PYO and 1.5 ± 0.2 nM for OHP. The system showed a good linear concentration range of 0.5 - 50 nM for PYO and OHP by CV technique. Also, good rates of sensitivity were achieved for PYO (1.85 μA/nM) and OHP (4.0 μA/nM).
Electrochemical sensors for detection of PYO have been also reported by other researchers. For instance, Sismaet et al
78
studied production of a PYO target in the presence of various amino acids (as regulatory molecules). In this trial, SWV response was improved due to the presence of amino acids. They also performed SWV scanning of the PYO target using commercially available carbon-based electrodes linked to an Ag/AgCl reference. They proved able to recognize differences in PYO production rates among the clinical strains obtained from various hospital settings.
72
Bentley et al
79
proposed a novel bio-based redox capacitor film for detection of PYO. Chronocoulometry signals of PYO molecules were amplified by a catechol-grafted chitosan film, and LOD was lowered. The researchers designed a miniaturized electrochemical system and stabilized it inside a microdevice enclosed in a chip holder.
Cristea et al
80
for the first time, integrated chemical finger-based printed sensors onto a glove to simultaneously detect PVD and PYO derived from PA through the voltammetric method within 4 minutes (Fig. 4). The sensors displayed linearity in the range of 5-50 µM for PVD and 0.01−0.1 µM for PYO, sensitivity of 1.09 nA/µM for PVD and 2.51 µA/µM for PYO (R2 = 0.995 and 0.990, respectively), and LOD of 1.66 µM for PVD and 3.33 nM for PYO were also obtained by SWV technique.
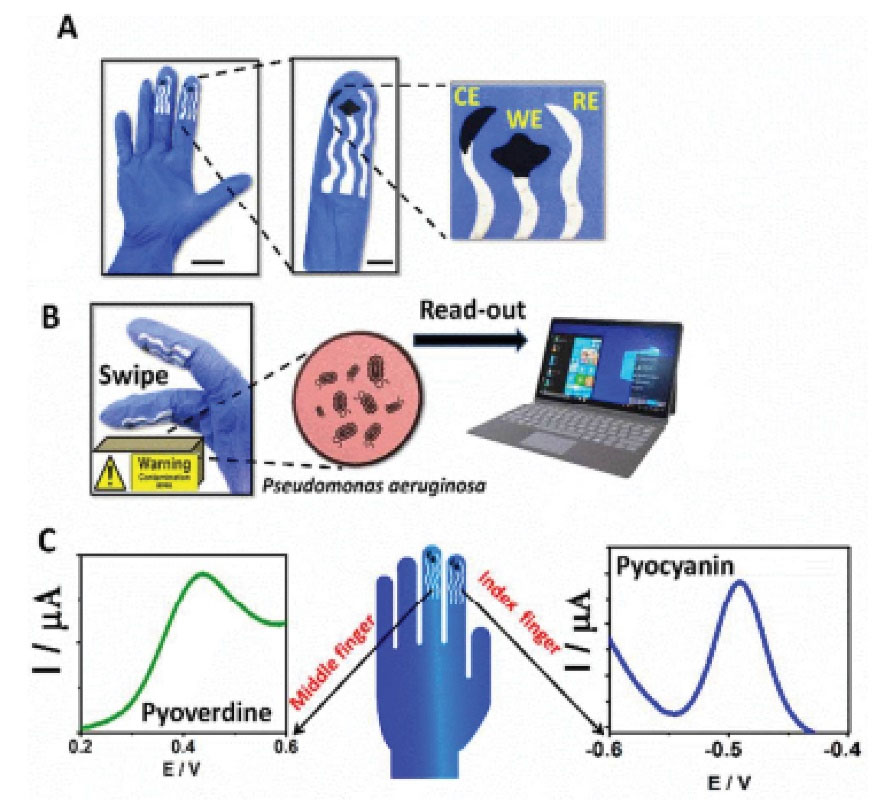
Fig. 4.
Finger-based printed sensors onto a glove (A) Screen-printed sensing glove: image of the real printed glove (A left, scale bar 3 cm) and the details of its electrodes design (A right, scale bar 1 cm). (B) Images of the on-glove swiping approach for sampling P. aeruginosa residues from furniture surfaces (B, left) and transmitting the data to a laptop (B, right). (C) SWV data recorded by the middle-finger, detecting PyoV (C, left) and by the index finger, identifying PyoC (C, right). Reprinted with permission from Ciui et al.
80
Copyright 2018 American Chemical Society.
.
Finger-based printed sensors onto a glove (A) Screen-printed sensing glove: image of the real printed glove (A left, scale bar 3 cm) and the details of its electrodes design (A right, scale bar 1 cm). (B) Images of the on-glove swiping approach for sampling P. aeruginosa residues from furniture surfaces (B, left) and transmitting the data to a laptop (B, right). (C) SWV data recorded by the middle-finger, detecting PyoV (C, left) and by the index finger, identifying PyoC (C, right). Reprinted with permission from Ciui et al.
80
Copyright 2018 American Chemical Society.
There are different papers on simultaneous detection of PYO and PQS. In this case, a sensor was applied with a conductive polymer film to increase electroactive surface area.
81
In a later study, PA strains were grown on surface of the electrode to concentrate electrochemical signals of PQS and PYO.
82
Table 2 presents some electrochemical sensors developed for detection of redox-active compounds.
Table 2.
electrochemical pathogen sensors based on the redox-active bacterial metabolites
|
Analyte
|
Detection method
|
Sensitivity
|
LOD
|
Linear range
|
Ref.
|
| PYO |
SWV |
19.1-267.0 µM |
0.13-1.81 µM |
1-100 µM |
71
|
| PYO |
Amperometry |
-
|
125 nM |
125 nM–100 μM |
83
|
| PYO |
CV |
- |
2µM |
2–100 µM |
84
|
PYO
HHQ
PQS
|
DPV |
- |
50 nM
250 nM
250 nM
|
2–100μM
2–75
2–100
|
68
|
PYO
HHQ
PQS
|
DPV |
- |
2.06 µM
3.61 µM
4.85 µM
|
5 - 50 µM
|
85
|
| Barakacin |
DPV |
- |
5, 100, 125, and 10 nM in different pH |
1–10 μM |
70
|
| IQS |
CV |
- |
46, 20 and 12 nM in different pH |
14, 12 and 15 μM |
86
|
| 2-AA |
CV |
|
4.86 mM |
10 - 60 mM |
87
|
| PCA |
CV |
- |
50 mM |
1.52 - 50.0 mM |
88
|
| PVD |
DPV |
0.14 µA/µM |
66.90 nM |
0.5 - 100 µM |
73
|
| PVD |
ESI, DPV, CV |
0.076 µM |
0.33 μM |
1 - 100 μM |
89
|
| PVD |
DPV |
0.134 µM |
0.33 μM |
1–100 μM |
90
|
| PYO |
DPV |
- |
- |
5-50 μM |
91
|
PYO
HHQ
PQS
2-AA
|
CV, SWV |
- |
- |
- |
69
|
| PYO |
SWV |
0.0234, 0.0841 and
0.0588 μA/μM respectively
|
0.17 μM, 0.15 μM and 0.09 μM in HS, BRB and SWF respectively |
0.183–20 μM, 0.336–10 μM, and 0.336–20 μM respectively |
92
|
PYO
PCA
PCN
5-MCA
|
SWV |
- |
2.6 mM |
- |
93
|
PYO, Pyocyanin; HHQ, 2-Heptyl-4(1H)-quinolone; PQS, Pseudomonas Quinolone Signal IQS, 2-(2-hydroxyphenyl)-thiazole-4-carbaldehyde; 2-AA, 2-aminoacetophenone; PCA,Phenazine-1-carboxylic acid; PVD, Pyoverdine; HS, human serum; BRB, Britton-Robinson buffer; SWF, simulated wound fluid; PCA, phenazine-1-carboxylic acid; PCN, phenazine-1-carboxamide; 5-MCA, 5-methylphenazine-1-carboxylic acid.
Aptamer
A single strand in a sequence of constitutive units of DNA or RNA is called as an aptamer. Favored for its selectivity and a low detection limit, it is immobilized on electroactive materials, such as nanoparticles, magnetic particles, carbon-based nanomaterials, and nanofibers and is used for biorecognition of elements consisting of a DNA sequence.
94-96
Biosensors with aptamers, as a recognition element, are being developed for various infectious diseases, due to their rapidity, small dimensions, low cost, and compatibility with minimization assay scales.
97-99
Aptamers are easily modified due to the existence of abundant amine functional groups in their structures. Also, because of their unique spatial structure, they attach to targets, such as bacteria, drug molecules, surface proteins of cells, and viruses.
100
An aptamer is considered advantageous for its high specificity and easy synthesis. Regarding immunoassay development, production of antibodies requires the use of animals, and a further disadvantage is that antibodies have a short lifetime in a working environment or a storage position. For this reason, aptasensors are used to prepare sensors in order to study elements, such as bacteria, viruses, and biomolecules.
101-104
A lot of studies have been done on aptasensors to detect bacteria. In 2018, Shahrokhian et al
105
reported an electrochemical aptasensor based on Au nanoparticles/carbon nanoparticles/cellulose nanofibers nanocomposite for impedimetric detection of S. aureus as a photogenic bacterium. In that study, cellulose nanofibers were modified with carbon nanofibers to be used in modification of electrodes, due to their properties, such as porosity, high surface area, and biocompatibility. Finally, Au nanoparticles were used to increase surface area and electrical conductivity of electrodes. Strong binding of Au to a thiol group made it favorable for sensor assembly. The aptasensor displayed linearity within the range of 1.2 × 101 -1.2 × 108 CFU/mL and LOD of 1 CFU/mL for S. aureus by EIS technique. Furthermore, the proposed sensor could detect Staphylococcus aureus in humans҆ blood serum as a clinical sample with a complex matrix.
In another study by this group, an electrochemical aptasensor was reported for detection of Salmonella typhimurium based on nanoporous gold. This aptasensor showed some significant advantages, such as the ability to distinguish between living and dead bacterial cells and detect S. typhimurium in linear range of 6.5 × 102- 6.5 × 108 CFU/mL with LOQ of 6.5 × 101 CFU/mL and LOD of 1 CFU/mL by EIS technique.
106
Au-based materials have been widely used for immobilization of aptamers via a thiol group to efficiently amplify electrochemical signals while maintaining biological activity. In this regard, Lai et al
100
used Au nanoholes on indium tin oxide (ITO) to identify S. aureus 16 sRNA. The Au nanoholes were formed through spin-coating of an AuNP solution on ITO covered with a monolayer of polystyrene to create a nanosphere lithography (NSL) patterned-ITO substrate. Then, a single strand of a thiolated oligonucleotide probe (ssHS probe for targeting S. aureus 16S rRNA) was immobilized on the electrode. Performance of the aptamer on the Au nanohole-modified ITO was studied to detect Staphylococcus aureus through CV techniques. The Au-nanohole arrays could enhance electrochemical signals, compared to non-template AuNP structures, which improved DNA hybridization tracing by 23% with low LOD of 10 pM. Moreover, in the presence of non-complementary sequences, the proposed sensor showed high selectivity for distinguishing S. aureus 16 sRNA from E. coli and PA.
McLamore et al
107
presented the functionalized graphene-nitrocellulose paper with platinum nano-cauliflower to immobilize a thiolated 64-mer RNA aptamer for detection of E. coli O157:H7. This aptasensor showed LOD of ≈4 CFU/mL for E. coli O157:H7 by EIS technique. Abbaspour et al
108
developed a sensitive dual-aptamer-based sandwich immunosensor for detection of S. aureus. The sandwich aptasensor was prepared from a primary biotinylated anti-S. aureus aptamer (on streptavidin-coated magnetic beads) and a secondary aptamer (on silver nanoparticles). S. aureus was captured in a particular interaction with the aptamer. Then, the sandwich system was completed by addition of secondary anti-S. aureus aptamer-conjugated AgNPs. Finally, the sandwich separated via a magnet was analyzed by differential pulse stripping voltammetric signals resulting from silver particles, which was proportional to the S. aureus. This electrochemical sensor showed wide dynamic range of 10 to 106 CFU/mL with LOD of 1.0 CFU/mL.
Since, pathogen bacteria are menacing to humans҆ health, detection and treatment of bacterial infections is important. Antibiotic resistance is increased if diagnosis and treatment of infectious diseases is prolonged. In this respect, Yoo et al
109
prepared an aptamer-functionalized capacitance sensor array to follow real-time bacterial growth and antibiotic susceptibility. They cultured S. aureus and E. coli and observed the increased capacity in proliferation of bacteria. When the bacteria were exposed to certain MIC (the minimum inhibitory concentration) of antibiotic, capacity in the capacitance sensor was decreased because the antibiotic prevented bacterial growth.
109
LOD was equal to 10 CFU/mL within an hour. Ozkan-Ariksoysal et al
110
reported a genosensor based on DNA-wrapped multi-walled carbon nanotubes for detection of E. coli from PCR-amplified real samples. The capture probe was immobilized on carbon nanotubes and twisted on carbon nanotubes by π-stacking interactions. Then, chitosan was used to stabilize the designed sensor via covalent bonds between amine group of chitosan and carboxyl groups on carbon nanotubes. This sensor had LOD of 17 nM with a linear section in the range of 7-12 µg/mL by DPV technique. Sheikh-Zeinoddin et al reported an electrochemical DNA sensor without PCR amplification of samples.
9
It was used to detect B. cereus with LOD of 9.4 × 10-12 mol L -1 by EIS technique, which was better than those reported for antigen-antibody and cell-based sensors. The sensing platform consisted of PGE-gold nanoparticles self-assembled with single-stranded DNA of the nheA gene. An increase in charge transfer resistance in EIS technique due to hybridization of the ssDNA with the target DNA served as an electrochemical signal. In the same year, Gonzalez-Rodriguez et al
111
reported a sensitive electrochemical DNA biosensor for detection of pathogenic Bacillus anthracis. Mercaptohexanol was used to backfill the electrode after DNA immobilization, thus regulating the probes҆ surface density. The sensor could detect DNA samples with LODs as low as 10 pM by CV technique and identify DNA of Bacillus anthracis. Heli et al
112
reported a sensitive biosensor based on gold nanoribbons covered by gold nanoblooms, which emerged to be a dense p-ssDNA layer (Fig. 5). MB could interact with both ssDNA and dsDNA through different modes,including electrostatic attractions and intercalation. Changes were recorded by the DPV method to produce signals before and after hybridization of the DNA and ssDNA probes. The biosensor showed a linear range from 10 zmol dm-3 to 10 pmol dm-3 and LOD of 1.71 zmol dm-3 for Brucella genome. In 2016, the same authors published another paper,
10
entitled “An Ultrasensitive Electrochemical Genosensor for Brucella Based on Palladium Nanoparticles”. The genosensor could detect complementary sequences and had a high sensitivity of 0.02 µA dm3 mol-1 to detect brucellosis in real samples even without using PCR amplification. Linear concentration of the genosensor was from 1.0×10-12 to1.0×10-19 mol dm -3 with LOD of 2.7×10 -20 mol dm-3 by the DPV method.
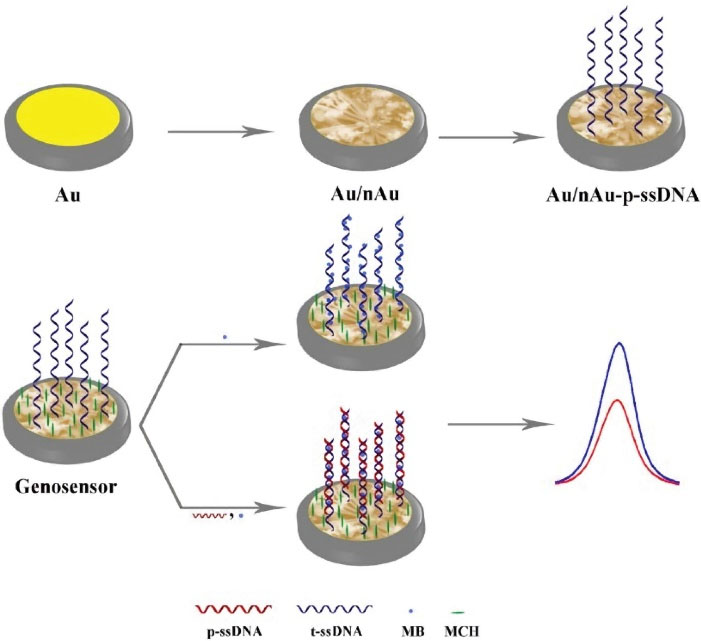
Fig. 5.
Fabrication protocol of the genosensor and detection of t-ssDNA.
112
(Creative Commons CC BY license).
.
Fabrication protocol of the genosensor and detection of t-ssDNA.
112
(Creative Commons CC BY license).
In 2016, Bachmann et al
113
conducted the first label-free and amplification-free EIS-based detection of 16S ribosomal RNA (E. coli 16S rRNA). The assay was a screen-printed dual gold electrode modified with thiol-peptide nucleic acid. LODs of the employed fluorescence-based microarray and the EIS assay were equal to 20 pM and 50 pM, respectively. However, a kinetic EIS assay format allowed specific detection of E. coli 16S rRNA within 10 minutes.
Hamidi-Asl et al
114
developed an aptasensor based on core-shell Ag@Au nanoparticles for detection of E. coli cells. The nanoparticles were fixed on the electrode via cysteamine and then, a thiolated aptamer was immobilized on the nanoparticles. This structure provided chemical stability and biocompatibility to stabilize the aptamer. The sensor also showed LOD of 90 CFU/mLwhile the LOD reported for this aptamer in the literature is equal to 370 CFU/mLby EIS method.
Pividori et al
115
designed an electrochemical genosensor based on silica magnetic particles to detect Salmonella, Listeria, and E. coli. For the first time, they used silica MPs as a platform for DNA immobilization and obtained the tagged amplified DNA by choosing gene targets, including the invA gene (tagged with fluorescein) for S. enterica, the prfA gene (tagged with biotin) for L. monocytogenes, and the eaeA gene (tagged with digoxigenin) for E. coli. The sensor could detect and distinguish 0.04, 0.13, and 0.05 ng/mL of S. enterica, L. monocytogenes, and E. coli, respectively within 3 hours. In 2018, Tamand et al
116
developed an electrochemical DNA sensor based on core-shell cerium oxide nanorod@polypyrrole composite to detect Salmonella by EIS method. This nanocomposite was synthesized by polymerization of pyrrole monomer on cerium oxide nanorods. Then, an ssDNA sequence was immobilized on it. This DNA-based sensor had a linear range of 0.01–0.4 nM with low LOD of 0.084 nM. Luo et al
117
used E. coli O157:H7 aptamer (apt-E) to construct an aptasensor based on aptamer-induced catalyzed hairpin assembly. In this method, signals were amplified using apt-E, hairpin H1, and H2. Concentration of E. coli O157:H7 was proportional to amount of apt-E, and apt-E, in turn, was proportional to amount of H1/H2 complexes. Thus, concentration of bacteria could be determined by detection of H1/H2 complexes with the fabricated aptasensor. LOD was equal to75 CFU/mLfor this aptasensor by electrophoretic method.
Bacteriophage
Bacteriophages were first investigated by Twort in 1915 and were studied later by Deherelle.
111
They are viruses attacking bacteria but do not affect eukaryotes. Each virus has its bacteriophage. After this discovery, it was immediately used to eliminate bacterial cells as a therapeutic agent against bacterial infections. Although, Deherelle used phages for treatment of dysentery in 1919, they were re-introduced 2 years later by Bruynaghe Maising to treat staphylococci.
112
In the 1940s, bacteriophages were mostly replaced by antibiotics, and then phage studies continued. Nowadays, they are used as suitable tools for phage display vaccines in the field of biotechnology of biosensors and control factors. Phages are encapsulated in DNA- or RNA -encapsulated proteins in various shapes.
In the recent years, bacteriophages have emerged in electrochemistry as scaffolds and molds for new electrode materials. They gain a natural ability to connect selectively to specific and adaptive agents and to be incorporated into certain chemical applications through genetic modification. This has introduced various attractive topics to the field of electrochemical research. Due to their durability and ease of preparation, phages are widely used as diagnostic elements in biosensors and scaffolds. The use of bacteriophages for sensing of bacteria is very important because, they act in a specific way.
118
In 2013, Shabani et al
119
performed some magnetic manipulations to improve detection of bacteria by EIS method. In this method, bacteriophage T4 coated with beads was used specifically to detect and capture E. coli k12 cells. The biosensor was tested in real milk samples. For capturing a high amount of bacteria in the sample, 2% milk was mixed with 108 CFU/mLofE. coli k12 elicited from milk. Surface modification of magnetic beads by phages and bacterial attachment were confirmed in a fluorescence analysis. The fluorescein isothiocyanate (FITC) and fluorescence labels serve to ensure binding of phages T4 to magnetic beads.
Also, flow cytometry was conducted as a conventional method to check magnetic beads and the phages coated on them. In another study, Tlili et al
120
designed a biosensor based on T4 phages for identification and measurement of E. coli count from living cells of bacteria by EIS method (Fig. 6). The sensor was used in two ways. The first one involved survival assays and tests and the use of phages as diagnostic elements through free-labeled electrochemical impedance. The second way was loop-mediated isothermal amplification (LAMP) to reinforce selectivity of the E. coli’s Tuf gene. As the impedance results indicated, the E. coli bacteria could be detected rapidly and accurately with LOD of 800 CFU/mLwhile well isolated from other types of E. coli bacteria even the dead ones. Overall, the aim of this study was screening in the shortest possible time. Integration of LAMP to modify the biosensor so as to sense phages in less than 1 hour helped to achieve this goal.
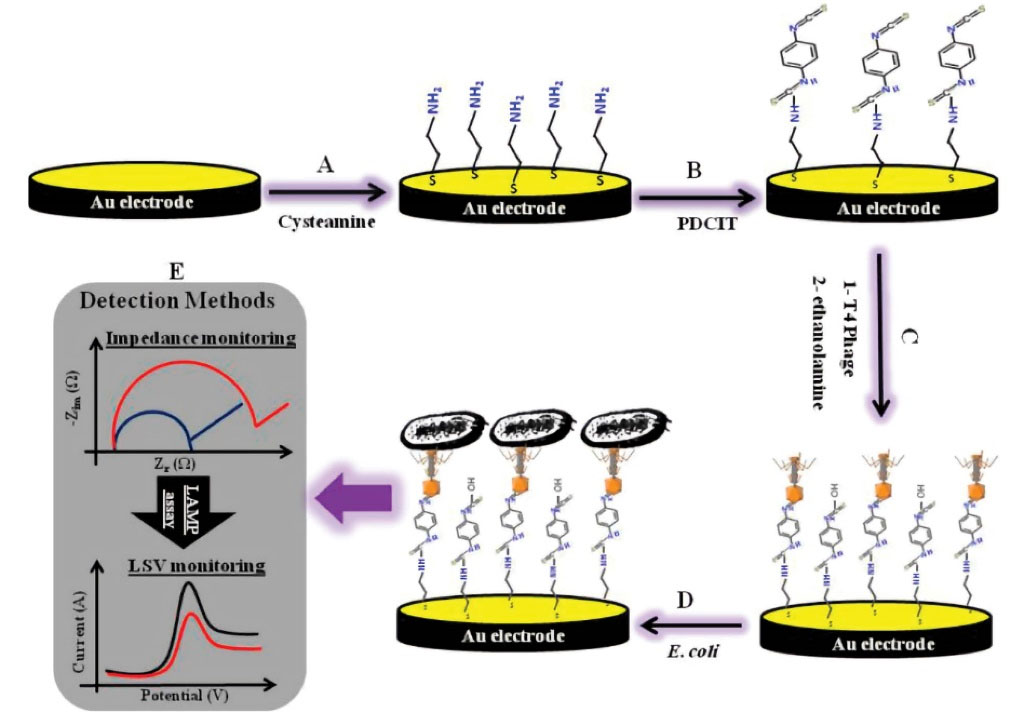
Fig. 6.
Schematic representation of the T4-bacteriophage biosensors. (A) Cysteamine-assembly, (B) Activation with 1,4-dithiocyanate, (C) Immobilization of the T4 phage, (D) Capturing of the E. coli cells. (E) Detection method for E. coli based on the impedimetric/LAMP dual-response. Reprinted with permission from Tlili et al.
120
Copyright 2013 American Chemical Society.
.
Schematic representation of the T4-bacteriophage biosensors. (A) Cysteamine-assembly, (B) Activation with 1,4-dithiocyanate, (C) Immobilization of the T4 phage, (D) Capturing of the E. coli cells. (E) Detection method for E. coli based on the impedimetric/LAMP dual-response. Reprinted with permission from Tlili et al.
120
Copyright 2013 American Chemical Society.
In 2016, Moghtader et al
121
effectively diagnosed pathogenic bacteria using bacteriophages and graphite electrodes decorated with gold nanoparticles. In this study, EIS was used for rapid, inexpensive, and selective detection of pathogenic bacteria. The bacterium type studied was T4 phage. Indeed, it served as a diagnostic probe. Also, E. coli was studied as a target bacterium for T4 phage. The phages were absorbed onto the electrode by simple incubation at room temperature, and rate of absorption was increased with an increase in charge-transfer resistance value (Rct). The increase in Rct value was due to the effect of absorbing layers in the bacteria. In case of non-target bacteria, conductivity was reduced significantly. Impedance measurements showed that the electrode employed as an electrochemical device was fast, direct, and low-cost enough for detection of bacteria. In 2016, Wang et al
122
published a paper entitled "Macrophage-Based Electrochemical Sensors", in which they evaluated toxicity of LPS in pathogens. They investigated LPS and showed that dose-dependent toxicity exists in mouse macrophages. Electrochemical signals changed as concentration of [Ca+2] ions changed. In addition, a sensor was developed in a simple mouse macrophage cell to detect LPS rapidly and investigate the effect of its toxicity. Properties of MNPs facilitated reuse of the sensor. The results showed a significant reduction in amount of LPS at a dose-dependent electrochemical impedance rate in the range of 1-5 µg/mL. Value of impedance at various concentrations of LPS was between 1-50 µg/mL with a LOD of 0.15 µg/mL. Impedance was correlated with concentration of calcium in cells, indicating that calcium production occurs in cells after incubation, and its presence on LPS induces an electrical signal. Another study was conducted in the same year by Neha Bhardwaj et al, who investigated the immobilized-free graphene in bacteriophages for susceptibility to Staphylococcus. The aim of study was designing a sensor for the family of coagulase-negative Staphylococcus. This biosensor had a linear range of 2-2.2 ×106 CFU/mL and a low LOD of 2 CFU/mL by EIS method. Response time was about 2 seconds. The sensor also proved to have advantages, such as long half-life of about 3 months, fine dimensions, good coating, as well as being portable, disposable, and environmentally friendly. Owing to its satisfactory test results with real samples, such as apple juice, application of the sensor can be extended to detect other pathogens. In 2017, Yan et al,
123
conducted a study on natural origin of S. aureus-specific lytic bacteriophage P-S. aureus-9. Bacteriophage of a water sample was used to perform specific palaeomagnetic assembly and capture S. aureus in the sample. S. aureus cells were identified by the horseradish peroxidase (HRP) label that interacted with protein A and the Fc region of immunoglobulin (IgG) in a mouse.
The phage was mixed with magnetic beads, and S. aureus was linked to the phage after the HRP-labeled goat anti-mouse IgG was added to the S. aureus-bacteriophage. Finally, absorbance value was found to be 450 nm. This device was successfully used in real samples, such as phosphate-buffered saline (PBS) and apple juice. In another study by Zhang et al,
11
phages were mixed with magnetic beads and were used for rapid detection of E. coli O157:H7. Advantage of this study was application of natural phages for preparation of phagomagnetic beads. Using this system in food samples was useful to detect contamination. The purpose of working with a simple bacteriophage, such as O157-IOV-4 is removing (E. coli) O157:H1 contamination from water and purifying it. Finally, once the magnetic beads were produced in the presence of (E. coli) O157:H1, they had the highest absorbance of 450 nm, compared to other bacteria. Zhou et al
124
developed a new type of carbon nanotube-based biosensor for rapid identification of living bacterial cells. In this regard, T2 Phage-based biosensors were used to diagnose E. coli. The T2 Phages acted as biosensing elements covalently bonded to the functionalized carbon nanotubes on the electrode surface. As fluorescence microscopy indicated, the immobilized T2 phage particles were highly capable of capturing bacteria. In this method, an electric field-induced (EFI) charge was used to immobilize the bacteriophage onto surface of the modified electrode to detect E. coli. This sensor had a LOD of 103 CFU/mL by EIS method.
In 2020, Lee et al
125
developed a wild-type T4 bacteriophage sensor for detection of viable pathogenic bacteria. Capturing efficiency of the phages immobilized on sensing platform reached a maximum when the Debye length was comparable to the phage size. A low LOD of 14 ± 5 CFU/mLand a wide dynamic range of 1.9 × 101-1.9 × 108 CFU/ mL were obtained by DPV method. Furthermore, the electrochemical sensor modified with T4 bacteriophage proved to have the ability to distinguish viable and dead bacterial cells.
Antibody
Immunosensors are the devices working based on the interactions between antibodies and antigens on a converter surface.
126-129
Either an antibody or an antigen can be considered as the species immobilized on a sensing platform to detect an antigen or an antibody, respectively.
130
Antibodies are glycoproteins, referred to as a heavy chain of the same Doppler peptides, and two smaller identical polypeptides, called as a light chain. Protein chains are connected by disulfide bonds to form heavy chains. Amino-terminal pairs of heavy chains and light chains provide bonds to antigens. Antibodies are a group of functional glycoproteins produced in response to a foreign body in human and animal serums. Immobilized antibodies create immune responses through certain steps; so that, they bind to antigens, prevent them from binding to target cells,and cover them so that immune system can identify and destroy them.
Viswanathana et al
131
reported an electrochemical immunosensor based on a metal sulfide and multi-walled carbon nanotube poly(allylamine) to simultaneously detect pathogens in foods. This electrochemical immunosensor consisted of three antibodies (i.e., E. coli-CdS, Campylobacter-PbS, and Salmonella-CuS) immobilized on a specific nanocrystal with releasable metal ions. Corresponding non-overlapping curves were obtained by electrochemical measurements. Multi-walled carbon nanotubes improve electrochemical performance of substrates in reactions and increase sensitivity and detection of multiple bacteria. This type of sensors is used to control quality of food, although valid laboratory tests are required to determine its sensitivity and diagnostic properties.
131
Over the last few years, various nanoparticles have been used to enhance performance of electrochemical sensors.
132-135
Xiang et al
136
used bio-compatible nanocomposites to detect Salmonella. In this study, chitosan and GNPs were mixed up to improve performance of a film of gold composite. As a result, a linear range of 10-105 CFU/mL and a LOD of 5 CFU/mL were achieved by DPV method. Also, the immunosensor provided satisfactory results in detection of Salmonella in real-life samples. In 2016, Marion et al
137
used silica nanoparticles to connect antibodies and a poly-electrolyte layer. They manufactured a miniaturized electrochemical biosensor (lab-on-a-chip device) to detect and degrade E. coli. Detection of E. coli cells will finally be estimated through CV and quartz crystal microbalance (QCM) measurements. Farka et al
138
used a specific antibody on a screen-printed electrodes activated with a cysteamine monolayer. Microbial cells were treated by sonication and heat, and the effect of treatment was studied by atomic force microscopy (AFM). Wide linear response was obtained in the range between 103-108 CFU/mL by EIS method. Around the same time, Altintas et al
139
reported a microfluidic-based electrochemical sensor for detection of pathogenic bacteria. Integration and electrical connections of microfluidic materials are a great challenge in the field of designing and manufacturing sensors. Concentration range of detection was between 0.99 × 104 - 3.98 × 109 CFU/mL. This sensor was used to determine E. coli in water through functionalization of its AuNPs. In 2015, Xinai et al
140
studied an amplified immunoassay for E. coli in dairy products using the functionalized gold nanorod-based labels. In this diagnostic method, rapid response time and high sensitivity was achieved by DPV method. In another study, Zhong et al
141
described application of cadmium sulfide quantum dots (CdS QDs)-encapsulated metal-organic frameworks as signal-amplifying tags for ultrasensitive electrochemical detection of E. coli O157:H7. In optimal conditions, they achieved a linear range of 108 CFU/mL and a LOD of 3 CFU/mLfor E. coli by DPV method. This method has not been investigated for other immune sensors yet. In 2016, Krithiga et al
142
reported application of an immunosensor to detect PA in water. Monoclonal antibody immobilized on surface of the sensor was modified with CCLP (calcium cross -linked pectin) and gold nanoparticles. Detection range of 101-107 CFU/mLand LOD of 9 CFU/mLwere calculated by linear sweep voltammetry (LSV) method.
In the series of studies on biosensors, the principle of G-EFT (graphene FETs) was first established by Mao et al.
143
On the basis of this principle, bonding of probes and target proteins can significantly change electrical conductivity of sensors (Fig. 7). For detection of E. coli with such devices vertically-oriented graphene (VG) sheets are directly grown on sensing platforms through the plasma-enhanced chemical vapor deposition (PECVD) method. These sheets function as a sensing channel. Sensitivity of this type of biosensors was found to be 2 ng/mL.
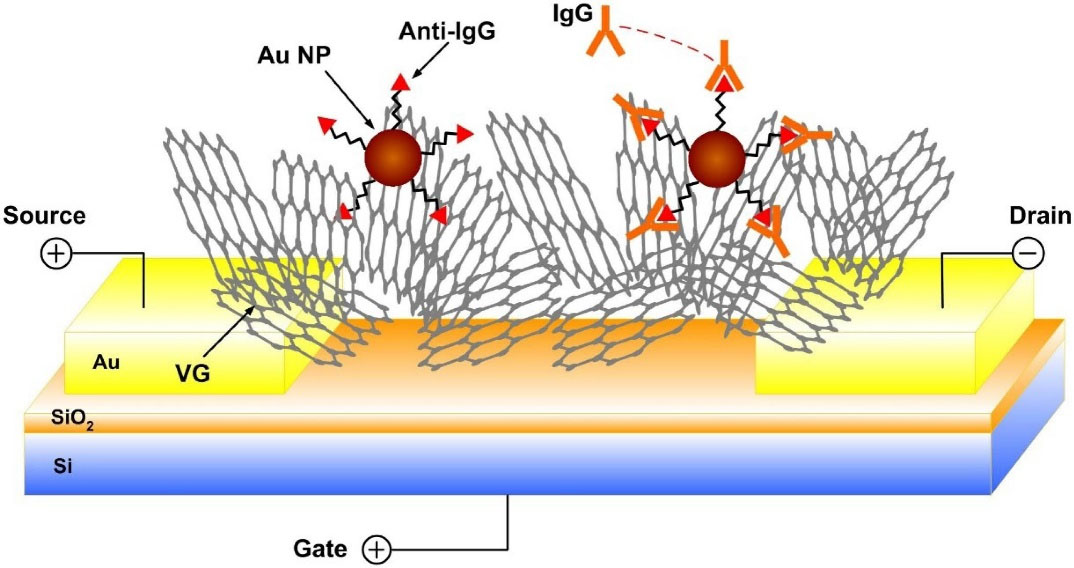
Fig. 7.
Schematic of the VG FET sensor by direct growth of VG between the drain and the source electrodes. Probe antibody is labeled to the VG surface through Au NPs.
.
Schematic of the VG FET sensor by direct growth of VG between the drain and the source electrodes. Probe antibody is labeled to the VG surface through Au NPs.
Following this method, in 2016, Wu et al
144
reported the use of the G-EFT principle for detection of E. coli. Maria et al
145
used an electrochemical magnetic microbeads-based sensing platform for POC diagnosis of brucellosis in human beings and animals. An antigen-coated magnetic microsphere was used as a solid support phase for detection of biomolecules, such as antibodies, peptides, and proteins. On this platform, there were supermagnetic micro-toads coated with antigens, which were then used with HRP-conjugated antibodies. Also, electrochemical scans were performed on 8-channel cell-electrode cartridges with potentiometric acetate. This system allows detection of many pathogenic microorganisms like parasitic protozoa, bacteria, and viruses in the shortest time and lowest cost. In 2016, Wang et al
146
reported an electrochemical sensor for detection of Salmonella using a redox cycling-based electrochemical method. Antibodies were immobilized on magnetic beads and were used to separate Salmonella from real samples. LOD was equal to 7.6 × 102 CFU/mLand 10 CFU/mL,which was calculated by choronocoulograms. In another study,
147
magnetic beads modified with anti-Salmonella antibodies were used to separate Salmonella from a saline buffer and agricultural water. The sensing platform was a screen-printed carbon electrode modified with Au nanoparticle-RGO and magnetite beads. First S. pullorum antibody was added to magnetic beads coated with silica and then, reaction was done by reduction of chloroauric acid and graphene oxide. LOD of 89 CFU/mLwas calculated by DPV method.
Along the same lines, an electrochemical sensor was used to quickly detect E. coli O157:H7 in pure culture media and food samples based on bifunctional glucose oxidase-polydopamine nanocomposites and Prussian blue. LODs within an hour were obtained as 52 and 190 CFU/mL by amperometric detection in the culture media and food samples, respectively. This sensor needed just a short detection time with a low LOD value owing to efficient amplification with the bifunctional composite.
148
In 2014, this bacterium was again diagnosed with another modified electrode. An attempt
149
was made for sensitive detection of bacteria using an electrode modified with sulfonated graphene poly-(3,4-ethylenedioxythiophene) gold nanoparticles (SG-PEDOT-AuNPs). This antibody has immunodeficiency properties, which increase connectivity and the ability to select biosensors. With a LOD of 3.4× 10 CFU/mL, the tests were conducted in real samples, such as milk and spring water by DPV method. The electrode proved to have a good selectivity for E. coli 0157:H7.
In 2018, Tufa et al
150
reported a biosensor for detection of TB with Fe3O4 nanoparticle and AgNO3. A core-shell structure with graphene quantum dot (GQD) was used to enhance performance of the biosensor. Gold nanoparticles were conjugated to the CPE antibody to amplify signal. These layers had three different roles; AgNO3 was used to increase conductivity, Fe3O4 NPs were used to increase surface-to-volume ratio, and GQD was used to load CFP-10 antibody. Jasim et al
151
used microfluidic-based biosensors to detect low levels of Salmonella in poultry and fresh products. This method actually intended to simultaneously detect multiple Salmonella serotypes with high sensitivity and determine sensitivity of the antibody-antigen binding process; this bond is an indication of change in impedance and the presence of bacteria. The microfluidic device consisted of three microchannels, each of which contained a site for a high concentration of Salmonella cells and was used for positive electrophoresis. The biosensor could also eliminate false-positive results. The sensing spot of bacteria contained an electrode array of 10 finger pairs.
Molecularly imprinted polymers
A rather different approach for monitoring pathogenic bacteria is entrapment of bacteria itself or bacterial recognition ligands in sol-gel and polymer phases or matrixes referred to as molecularly imprinted polymers (MIPs). Indeed, this system is nothing but an artificial receptor ligand.
26
To date, MIPs, as recognition elements, have been resistant to degradation and inactivation. Also, as artificial receptors, they have attracted enormous interest owing to significant advantages, such as predictability of structure, recognition specificity, and application universality.
26,152
Some researchers have used MIPs for electrochemical monitoring of pathogens.
153-157
Microorganism imprinting offers certain advantages for pathogen detection, including simplicity, rapidity, excellent stability, high selectivity, eco-friendliness,and low cost.
158
The first attempt for microorganism imprinting was made by Dickert’s team in 2001
159
using yeast as a template molecule via surface imprinting of polyurethane, which was considered as a salient application of MIPs.
Khan et al
160
offered an approach for detecting protein A (PA) from S. aureus by EIS method with a LOD and a recovery factor of 16.83 nM and 91.1 ± 6.6%, respectively. The imprinted polymers were assembled on a film of single-walled carbon nanotubes (SWCNTs)-SPEs. Synthesis of MIP materials took place via 3-aminophenol electropolymerized with the trapped target molecules and a protein template (PA) on surface of SWCNTs-SPEs using CV. The PA biomolecules were entrapped in polymeric layers and were digested by proteolytic activity of proteinase K. They were subsequently removed from surface polymer to create vacant places.
Some bacteria, e.g., Proteus mirabilis, have flagellar fragments as markers located on their outer surface. Khan et al
161
employed SWCNTs-SPEs with a homemade carbon-printed electrode. The electrode printing was done by coating of a filter paper with hydrophobic paraffin wax and manual printing of the formed electrodes with carbon ink. Some artificial receptors were formed on this platform using polyphenol mixed with flagellar fragments as template molecules. LOD of the platform was equal to 0.7 ng/mL and 0.9 ng/mL, which was calculated through the EIS and SWV methods, respectively. There was also a negligible interference from flagellar filaments or globular proteins of other bacteria.
Some bacteria, such as Bacillus anthracis are able to survive in harsh conditions and generate endospores when they undergo environmental stress. On this basis, Lahcen et al
162
produced a sensing platform on a carbon paste electrode to discover Bacillus cereus spores. This system was based on the use of MIPs, polypyrrole as a conducting polymer and a certain amount of B. cereus spores as a template. The platform showed concentration range of 102-105 CFU/mL by CV method. In 2016, an electrochemical method was used to detect protein levels of bacteria. In this study, an MIP was used to indirectly detect bacteria by targeting their outer membrane protein. For this purpose, the protein A was placed on outer surface of S. aureus directly on SWCNTs. The nanotubes were also laid on a film pressed on a printed electrode plate. Efficiency of the electrode was investigated through EIS. This biosensor had a low LOD (16.83 nM) and a low cost.
160
Jiang et al
163
fabricated an economical sensing platform for synthesis of magnetic MIP so as to quantify the Gram-negative bacterial quorum signaling molecule N-acyl homoserine-lactones (AHLs). The assay was formed on an amino group-functionalized Fe3O4@SiO2 surface, and AHLs were absorbed into cavities through connecting sites of MIPs. The platform could detect oxidative current of AHLs via DPV in the range of 2.5 -100 nM and with a LOD lower than 0.8 nM.
Golabi et al
164
utilizing cell-imprinted polymers (CIPs) as recognition receptors successfully fabricated an aminophenylboronic acid (3-APBA)-based CIP for detection of bacterial cells. The boronic acids҆ functional groups in the 3-APBA monomers, with their ability to reversibly and specifically attach and interact with cis-diol-containing molecules, allowed creation of a polymeric matrix that had both chemical and morphological recognition abilities. This matrix could facilitate release of the entrapped bacterial cell templates and then regenerate the CIPs. The researchers used S. epidermidis as a target and the EIS method for measurements. They gained detection range of 103-107 CFU/mL.
Mugo et al
165
also reported an imprinted polymer electrochemical sensor for detection of E. coli. The sensing platform was based on layer-by-layer assembly of multi-walled carbon nanotubes, or nanocellulose films, integrated with poly (aniline)-doped phenylboronic acid. A pathogen-imprinted polymer layer was coated on the prepared films. The proposed sensor exhibited a low LOD for detection of E. coli (8.7 ± 0.5 CFU/mL) and a rapid response of ≤ 5 minutes by CV technique.
Idil et al
166
designed and employed a type of sensor chips to monitor E. coli cells. The chips were based on a combination of capacitive biosensing approach and micro-contact imprinting methodology. An imprinted substrate was prepared using an amino acid-based biorecognition site and a mixture of monomers and cross-linkers under UV-polymerization. The real-time monitoring of E. coli was conducted within the range of 102-107 CFU/mL. Also, a LOD of 70 CFU/mL was obtained in spiked apple juice and river water samples.
Chen et al
167
proposed a novel electrochemiluminescence (ECL) biosensor to monitor E. coli O157:H7. Dopamine was electropolymerized to produce an N-GQDs/polydopamine film on surface of an electrode and then, immobilization was performed with the E. coli O157:H7 cells as a template on polydopamine film in electropolymerization step. Finally, a surface-imprinted polymer was extracted. The captured target molecules, i.e., E. coli O157:H7 cells, were labeled with N-GQDs antibody, and LOD of 8 CFU/mLwas achieved for the E. coli O157:H7 cells. There was also a linear relationship between the E. coli O157:H7 cells and ECL intensity of GQDs from 10 to 107 CFU/mL. In another recent work, Wu et al
168
selected E. coli O157:H7 as target bacteria and transferred them onto a polypyrrole layer on surface of an electrode (Fig. 8). Since, the layer was conductive; they monitored binding of the target bacteria through EIS. The impedimetric sensor measured the target in drinking water, milk, and apple juice samples. The sensor displayed a recovery range from 96 to 107.9%, with relative standard derivations (RSDs) less than 4% and a LOD of about 103 CFU/mL within an hour.
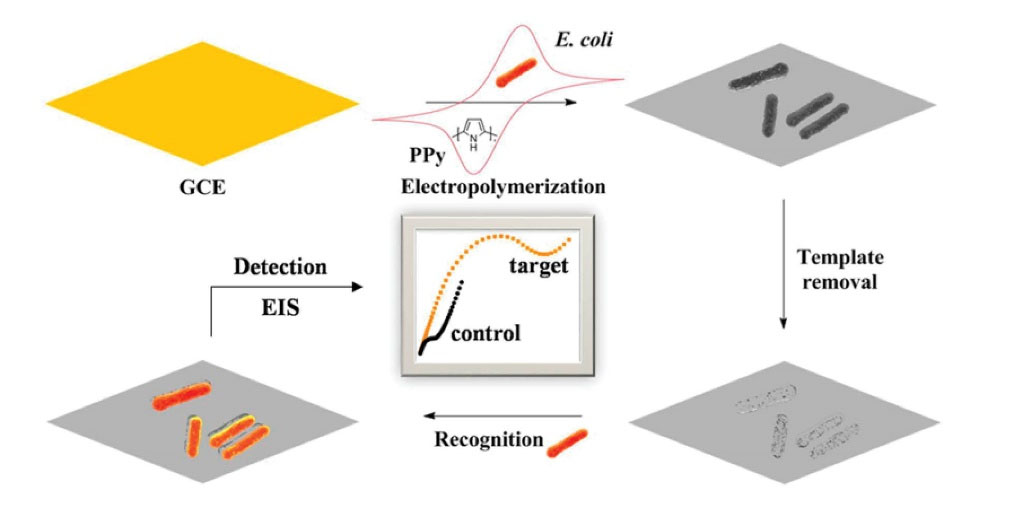
Fig. 8.
Schematic illustration showing the construction of BIP film based sensor for E. coli O157:H7 detection. Reprinted with permission from Wu et al.
168
Copyright 2018 American Chemical Society.
.
Schematic illustration showing the construction of BIP film based sensor for E. coli O157:H7 detection. Reprinted with permission from Wu et al.
168
Copyright 2018 American Chemical Society.
Jafari et al
169
reported an impedimetric sensor to recognize E. coli cells (E. coli UTI89) using artificial receptors. In that study, imprinting of the cells as templates with a rod-shaped morphology was done using thin sol-gel films of organically modified inorganic silica coated on gold electrodes. These imprinted sites had complementary cavities to identify the target bacterial species; thus, high binding affinity was achieved. The biosensor showed a linear range of 1×101- 1×104 CFU/mL and a LOD below than 1 CFU/mLby EIS method. A brief summary indicating some of the characteristics of representative the sensors covered in the aptamer, bacteriophage, antibody and MIP sections are presented in Table 3.
Table 3.
Overview of sensors proposed for bacteria detection divided by the type of recognition elements
|
Analyte
|
Recognition elements
|
Detection method
|
Linear range/CFU/mL
|
LOD CFU/mL
|
Ref.
|
|
S. aureus
|
Aptamer |
EIS |
12 to 1.2 × 108
|
1 |
105
|
|
S. typhimurium
|
Aptamer |
EIS |
6.5 × 102to 6.5 × 108
|
1 |
106
|
|
E. coli
|
RNA aptamer |
EIS |
4 to 105
|
4 |
107
|
|
S. aureus
|
Aptamer |
DPV |
10 to 106
|
1 |
108
|
|
E. coli
|
Aptamer |
EIS |
100 to 105
|
90 |
114
|
|
E. coli
|
Phage T4 |
EIS |
103 to 108
|
103
|
119
|
|
E. coli
|
Phage T4 |
EIS |
104 to 107
|
800 |
120
|
|
E. coli
|
Phage T4 |
EIS |
102 to 106
|
100 |
121
|
|
Staphylococcus
|
Bacteriophage |
EIS |
2 to 2.2 ×106
|
2 |
170
|
|
E. coli
|
T2 phage |
EIS |
103 to 107
|
103
|
124
|
|
E. coli
|
Phage T4 |
DPV |
19 to 1.9 × 108
|
14 |
125
|
|
Salmonella
|
Antibody |
DPV |
10 to 105
|
5 |
136
|
|
Salmonella
|
Antibody |
EIS |
103 to 108
|
103
|
138
|
|
E. coli
|
Antibody |
CV |
0.99 × 104 to 3.98 × 109
|
50 |
139
|
|
E. coli
|
Antibody |
DPV |
3 to 108
|
3 |
141
|
|
P. aeruginosa
|
Antibody |
LSV |
101 to 107
|
9 |
142
|
|
Salmonella
|
Antibody |
Choronocoulometry |
0.4 × 102 to 4 × 105
|
7.6 × 102
|
146
|
|
Salmonella
|
Antibody |
DPV |
102 to 106
|
89 |
147
|
|
E. coli
|
Antibody |
Amperometry |
102 to 106
|
52 |
148
|
|
E. coli
|
Antibody |
DPV |
78 to 7.8 × 106
|
3.4× 10 |
149
|
|
Salmonella
|
Antibody |
EIS |
- |
7 |
151
|
|
B. cereus
|
MIP |
CV |
102 to 105
|
- |
162
|
|
E. coli
|
MIP |
CV |
- |
8.7 |
165
|
|
E. coli
|
MIP |
CV |
102 to 107
|
70 |
166
|
|
E. coli
|
MIP |
EIS |
103 to 108
|
103
|
168
|
|
E. coli
|
MIP |
EIS |
10 to 104
|
1 |
169
|
Review Highlights
What is the current knowledge?
√ Previous reviews on this topic covered modification and strategies to develop electrochemical sensor for bacteria.
What is new here?
√ This review presents an overview of different recognition elements on an electrochemical diagnostic for bacteria.
Concluding Remarks
Bacterial infections pose important threats to public health and humans҆ life. Ttraditional techniques for detection of bacteria are labor-intensive and time-consuming. For overcoming these limitations, electrochemical methods can serve as appropriate ways of detecting bacteria rapidly and sensitively. The great versatility presented by such methods is based on wide range of biosensor designs and their various applications. However, several issues, including reproducibility and working stability remain to be solved. Addressing these technical problems will allow electrochemical sensors to become reliable tools for rapid diagnosis of bacteria in various samples. Detection limits at the level of 1 to 103 CFU/mL for various bacteriawere achieved by electrochemical sensors, which, in general, are below the maximum accepted level by the WHO. In general, the field of bacterial detection by electrochemical sensors is continuously growing. It is believed that this field will focus on portable devices for detection of bacteria based on electrochemical methods. Development of these devices requires close collaboration of various disciplines, such as biology, electrochemistry, and biomaterial engineering. Furthermore, the emerging technologies of lab-on-chip devices offer opportunities for the development of commercial sensors. Commercial analytical devices have, small size and simple construction that made them perfect for point-of-care sensing. How to simply and successfully decrease size without affecting their performance, has become a critical step.
Acknowledgments
The authors gratefully acknowledge the Research Council of Kermanshah University of Medical Sciences for financial support.
Funding sources
No special funding received for the study.
Ethical statement
The present article presents no study with human participants or animals performed by any of the authors.
Competing interests
The authors declare no conflict of interest.
Authors’ contribution
AK and AF developed the concept and designed the study. All authors contributed to the reviewed the data, extracted the data and writing of the final version of the manuscript. All authors read and approved the final manuscript.
References
- Asadi A, Razavi S, Talebi M, Gholami M. A review on anti-adhesion therapies of bacterial diseases. Infection 2019; 47:13-23. doi: 10.1007/s15010-018-1222-5 [Crossref] [ Google Scholar]
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 2009; 48:1-12. doi: 10.1086/595011 [Crossref] [ Google Scholar]
- Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis 2008; 46:155-164. doi: 10.1086/524891 [Crossref] [ Google Scholar]
- Organization WH. Monitoring and evaluation of the global action plan on antimicrobial resistance: framework and recommended indicators. World Health Organization; 2019.
- Costa MP, Andrade CAS, Montenegro RA, Melo FL, Oliveira MDL. Self-assembled monolayers of mercaptobenzoic acid and magnetite nanoparticles as an efficient support for development of tuberculosis genosensor. J Colloid Interface Sci 2014; 433:141-148. doi: 10.1128/AAC.01464-06 [Crossref] [ Google Scholar]
- Organization WH. Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline, Including Tuberculosis. World Health Organization; 2017.
- Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 2007; 51:3471-3484. doi: 10.1128/AAC.01464-06 [Crossref] [ Google Scholar]
- Bahavarnia F, Mobed A, Hasanzadeh M, Saadati A, Hassanpour S, Mokhtarzadeh A. Bio-assay of Acintobacter baumannii using DNA conjugated with gold nano-star: A new platform for microorganism analysis. Enzyme Microb Technol 2020; 133:109466. doi: 10.1016/j.enzmictec.2019.109466 [Crossref] [ Google Scholar]
-
Izadi Z, Sheikh-Zeinoddin M, Ensafi AA, Soleimanian-Zad S. Fabrication of an electrochemical DNA-based biosensor for Bacillus cereus detection in milk and infant formula. Biosens Bioelectron 2016; 80: 582-589. 10.1016/j.bios.2016.02.032
- Rahi A, Sattarahmady N, Heli H. An ultrasensitive electrochemical genosensor for Brucella based on palladium nanoparticles. Anal Biochem 2016; 510:11-17. doi: 10.1016/j.ab.2016.07.012 [Crossref] [ Google Scholar]
- Zhang Y, Yan C, Yang H, Yu J, Wei H. Rapid and selective detection of E. coli O157:H7 combining phagomagnetic separation with enzymatic colorimetry. Food Chem 2017; 234:332-338. doi: 10.1016/j.foodchem.2017.05.013 [Crossref] [ Google Scholar]
- Eissa S, Zourob M. Ultrasensitive peptide-based multiplexed electrochemical biosensor for the simultaneous detection of Listeria monocytogenes and Staphylococcus aureus. Microchim Acta 2020; 187:1-11. doi: 10.1007/s00604-020-04423-3 [Crossref] [ Google Scholar]
- Luo F, Lu Y, Li Z, Dai G, Chu Z, Zhang J. Probe-lengthening Amplification-assisted Microchip Electrophoresis for Ultrasensitive Bacteria Screening. Sens Actuators B Chem 2020; 325:128784. doi: 10.1016/j.snb.2020.128784 [Crossref] [ Google Scholar]
- Das R, Chaterjee B, Kapil A, Sharma TK. Aptamer-NanoZyme mediated sensing platform for the rapid detection of Escherichia coli in fruit juice. Sens Bio-Sensing Res 2020; 27:100313. doi: 10.1016/j.sbsr.2019.100313 [Crossref] [ Google Scholar]
- Karbelkar AA, Furst AL. Electrochemical Diagnostics for Bacterial Infectious Diseases. ACS Infect Dis 2020; 7:1567-1571. doi: 10.1021/acsinfecdis.0c00342 [Crossref] [ Google Scholar]
- Sohouli E, Ghalkhani M, Zargar T, Joseph Y, Rahimi-Nasrabadi M, Ahmadi F. A new electrochemical aptasensor based on gold/nitrogen-doped carbon nano-onions for the detection of Staphylococcus aureus. Electrochim Acta 2022; 403:139633. [ Google Scholar]
- Templier V, Roux A, Roupioz Y, Livache T. Ligands for label-free detection of whole bacteria on biosensors: A review. TrAC Trends Anal Chem 2016; 79:71-79. doi: 10.1016/j.trac.2015.10.015 [Crossref] [ Google Scholar]
- Khoshroo A, Hosseinzadeh L, Sobhani-Nasab A, Rahimi-Nasrabadi M, Ahmadi F. Silver nanofibers/ionic liquid nanocomposite based electrochemical sensor for detection of clonazepam via electrochemically amplified detection. Microchem J 2019; 145:1185-1190. doi: 10.1016/j.microc.2018.12.049 [Crossref] [ Google Scholar]
- Khoshroo A, Mazloum-Ardakani M, Forat-Yazdi M. Enhanced performance of label-free electrochemical immunosensor for carbohydrate antigen 15-3 based on catalytic activity of cobalt sulfide/graphene nanocomposite. Sens Actuators B Chem 2018; 255:580-587. doi: 10.1016/j.snb.2017.08.114 [Crossref] [ Google Scholar]
- Hosseinzadeh L, Khoshroo A, Adib K, Rahimi-Nasrabadi M, Ahmadi F. Determination of homocysteine using a dopamine-functionalized graphene composite. Microchem J 2021; 165:106124. doi: 10.1016/j.microc.2021.106124 [Crossref] [ Google Scholar]
- Mohammadnia MS, Naghian E, Ghalkhani M, Nosratzehi F, Adib K, Zahedi MM. Fabrication of a new electrochemical sensor based on screen-printed carbon electrode/amine-functionalized graphene oxide-Cu nanoparticles for Rohypnol direct determination in drink sample. J Electroanal Chem 2021; 880:114764. doi: 10.1016/j.jelechem.2020.114764 [Crossref] [ Google Scholar]
- Sohouli E, Ghalkhani M, Shahdost-fard F, Khosrowshahi EM, Rahimi-Nasrabadi M, Ahmadi F. Sensitive sensor based on TiO2 NPs nano-composite for the rapid analysis of Zolpidem, a psychoactive drug with cancer-causing potential. Mater Today Commun 2021; 26:101945. doi: 10.1016/j.mtcomm.2020.101945 [Crossref] [ Google Scholar]
- Zhang Z, Zhou J, Du X. Electrochemical Biosensors for Detection of Foodborne Pathogens. Micromachines 2019; 10:222. doi: 10.3390/mi10040222 [Crossref] [ Google Scholar]
- Majdinasab M, Hayat A, Marty JL. Aptamer-based assays and aptasensors for detection of pathogenic bacteria in food samples. TrAC Trends Anal Chem 2018; 107:60-77. doi: 10.1016/j.trac.2018.07.016 [Crossref] [ Google Scholar]
- Richter Ł, Janczuk-Richter M, Niedziółka-Jönsson J, Paczesny J, Hołyst R. Recent advances in bacteriophage-based methods for bacteria detection. Drug Discov Today 2018; 23:448-455. doi: 10.1016/j.drudis.2017.11.007 [Crossref] [ Google Scholar]
- Amiri M, Bezaatpour A, Jafari H, Boukherroub R, Szunerits S. Electrochemical methodologies for the detection of pathogens. ACS Sens 2018; 3:1069-1086. doi: 10.1021/acssensors.8b00239 [Crossref] [ Google Scholar]
- Kuss S, Amin HMA, Compton RG. Electrochemical detection of pathogenic bacteria—recent strategies, advances and challenges. Chem Asian J 2018; 13:2758-2769. doi: 10.1002/asia.201800798 [Crossref] [ Google Scholar]
- Furst AL, Francis MB. Impedance-based detection of bacteria. Chem Rev 2018; 119:700-726. doi: 10.1021/acs.chemrev.8b00381 [Crossref] [ Google Scholar]
- Hoyos-Nogués M, Gil FJ, Mas-Moruno C. Antimicrobial peptides: powerful biorecognition elements to detect bacteria in biosensing technologies. Molecules 2018; 23:1683. doi: 10.3390/molecules23071683 [Crossref] [ Google Scholar]
- Mubarok AZ, Mani V, Huang C-H, Chang P-C, Huang S-T. Label-free electrochemical detection of neuraminidase activity: A facile whole blood diagnostic probe for infectious diseases. Sens Actuators B Chem 2017; 252:641-648. doi: 10.1016/j.snb.2017.06.061 [Crossref] [ Google Scholar]
- Huang Y-M, Hsu H-Y, Hsu C-L. Development of electrochemical method to detect bacterial count, Listeria monocytogenes, and somatic cell count in raw milk. J Taiwan Inst Chem Eng 2016; 62:39-44. doi: 10.1016/j.jtice.2016.01.030 [Crossref] [ Google Scholar]
- Tarditto LV, Zon MA, Ovando HG, Vettorazzi NR, Arévalo FJ, Fernández H. Electrochemical magneto immunosensor based on endogenous β-galactosidase enzyme to determine enterotoxicogenic Escherichia coli F4 (K88) in swine feces using square wave voltammetry. Talanta 2017; 174:507-513. doi: 10.1016/j.talanta.2017.06.059 [Crossref] [ Google Scholar]
- Badalyan G, Díaz C, Bücking M, Lipski A. Novel sensor platform for rapid detection and quantification of coliforms on food contact surfaces. J Microbiol Methods 2018; 153:74-83. doi: 10.1016/j.mimet.2018.09.009 [Crossref] [ Google Scholar]
- Jiang H, Yang J, Wan K, Jiang D, Jin C. Miniaturized Paper-Supported 3D Cell-Based Electrochemical Sensor for Bacterial Lipopolysaccharide Detection. ACS Sens 2020; 5:1325-1335. doi: 10.1021/acssensors.9b02508 [Crossref] [ Google Scholar]
- Mavaei M, Chahardoli A, Fattahi A, Khoshroo A. A Simple Method for Developing a Hand‐Drawn Paper‐Based Sensor for Mercury; Using Green Synthesized Silver Nanoparticles and Smartphone as a Hand‐Held‐Device for Colorimetric Assay. Glob Challenges 2021; 5:2000099. doi: 10.1002/gch2.202000099 [Crossref] [ Google Scholar]
- Adkins JA, Boehle K, Friend C, Chamberlain B, Bisha B, Henry CS. Colorimetric and electrochemical bacteria detection using printed paper-and transparency-based analytic devices. Anal Chem 2017; 89:3613-3621. doi: 10.1021/acs.analchem.6b05009 [Crossref] [ Google Scholar]
- Noh S, Choe Y, Tamilavan V, Hyun MH, Kang HY, Yang H. Facile electrochemical detection of Escherichia coli using redox cycling of the product generated by the intracellular β-d-galactosidase. Sens Actuators B Chem 2015; 209:951-956. doi: 10.1016/j.snb.2014.12.073 [Crossref] [ Google Scholar]
-
Marques AC, Pinheiro T, Martins GV, Cardoso AR, Martins R, Sales MG, et al. Non-enzymatic lab-on-paper devices for biosensing applications. In: Comprehensive Analytical Chemistry. Vol 89. Elsevier; 2020:189-237. 10.1016/bs.coac.2020.05.001
- Khoshroo A, Sadrjavadi K, Taran M, Fattahi A. Electrochemical system designed on a copper tape platform as a nonenzymatic glucose sensor. Sens Actuators B Chem 2020; 325:128778. doi: 10.1016/j.snb.2020.128778 [Crossref] [ Google Scholar]
- Chambers JP, Arulanandam BP, Matta LL, Weis A, Valdes JJ. Biosensor Recognition Elements. Texas Univ at San Antonio Dept of Biology; 2008.
- Lin D, Harris KD, Chan NWC, Jemere AB. Nanostructured indium tin oxide electrodes immobilized with toll-like receptor proteins for label-free electrochemical detection of pathogen markers. Sens Actuators B Chem 2018; 257:324-330. doi: 10.1016/j.snb.2017.10.140 [Crossref] [ Google Scholar]
- Li Z, Dai G, Luo F, Lu Y, Zhang J, Chu Z. An electrochemical sensor for bacterial lipopolysaccharide detection based on dual functional Cu2+-modified metal–organic framework nanoparticles. Microchim Acta 2020; 187:1-10. doi: 10.1007/s00604-020-04364-x [Crossref] [ Google Scholar]
- Amini K, Kraatz H-B. Toll-like receptors for pathogen detection in water: challenges and benefits. Int J Environ Anal Chem 2016; 96:836-844. doi: 10.1080/03067319.2016.1209661 [Crossref] [ Google Scholar]
- She Z, Topping K, Shamsi MH, Wang N, Chan NWC, Kraatz H-B. Investigation of the utility of complementary electrochemical detection techniques to examine the in vitro affinity of bacterial flagellins for a toll-like receptor 5 biosensor. Anal Chem 2015; 87:4218-4224. doi: 10.1021/ac5042439 [Crossref] [ Google Scholar]
- Amini K, Ebralidze II, Chan NWC, Kraatz H-B. Characterization of TLR4/MD-2-modified Au sensor surfaces towards the detection of molecular signatures of bacteria. Anal Methods 2016; 8:7623-7631. doi: 10.1039/C6AY01978A [Crossref] [ Google Scholar]
- Mayall RM, Renaud-Young M, Chan NWC, Birss VI. An electrochemical lipopolysaccharide sensor based on an immobilized Toll-Like Receptor-4. Biosens Bioelectron 2017; 87:794-801. doi: 10.1016/j.bios.2016.09.009 [Crossref] [ Google Scholar]
- Mayall RM, Renaud-Young M, Gawron E, Luong S, Creager S, Birss VI. Enhanced Signal Amplification in a Toll-like Receptor-4 Biosensor Utilizing Ferrocene-Terminated Mixed Monolayers. ACS Sens 2018; 4:143-151. doi: 10.1021/acssensors.8b01069 [Crossref] [ Google Scholar]
- Mannoor MS, Zhang S, Link AJ, McAlpine MC. Electrical detection of pathogenic bacteria via immobilized antimicrobial peptides. Proc Natl Acad Sci 2010; 107:19207-19212. doi: 10.1073/pnas.1008768107 [Crossref] [ Google Scholar]
- Sęk S, Vacek J, Dorčák V. Electrochemistry of Peptides. Curr Opin Electrochem 2019; 14:166-172. doi: 10.1016/j.coelec.2019.03.002 [Crossref] [ Google Scholar]
- Malvano F, Pilloton R, Albanese D. A novel impedimetric biosensor based on the antimicrobial activity of the peptide Nisin for the detection of Salmonella spp. Food Chem 2020; 325:126868-126879. doi: 10.1016/j.foodchem.2020.126868 [Crossref] [ Google Scholar]
- Liu X, Marrakchi M, Xu D, Dong H, Andreescu S. Biosensors based on modularly designed synthetic peptides for recognition, detection and live/dead differentiation of pathogenic bacteria. Biosens Bioelectron 2016; 80:9-16. doi: 10.1016/j.bios.2016.01.041 [Crossref] [ Google Scholar]
- Eissa S, Zourob M. A dual electrochemical/colorimetric magnetic nanoparticle/peptide-based platform for the detection of Staphylococcus aureus. Analyst 2020; 145:4606-4614. doi: 10.1039/D0AN00673D [Crossref] [ Google Scholar]
- Hoyos-Nogués M, Brosel-Oliu S, Abramova N, Muñoz FX, Bratov A, Mas-Moruno C. Impedimetric antimicrobial peptide-based sensor for the early detection of periodontopathogenic bacteria. Biosens Bioelectron 2016; 86:377-385. doi: 10.1016/j.bios.2016.06.066 [Crossref] [ Google Scholar]
- Ghanbari MH, Khoshroo A, Sobati H, Ganjali MR, Rahimi-Nasrabadi M, Ahmadi F. An electrochemical sensor based on poly (l-Cysteine)@ AuNPs@ reduced graphene oxide nanocomposite for determination of levofloxacin. Microchem J 2019; 147:198-206. doi: 10.1016/j.microc.2019.03.016 [Crossref] [ Google Scholar]
- Ghanbari MH, Shahdost-Fard F, Salehzadeh H, Ganjali MR, Iman M, Rahimi-Nasrabadi M. A nanocomposite prepared from reduced graphene oxide, gold nanoparticles and poly (2-amino-5-mercapto-1, 3, 4-thiadiazole) for use in an electrochemical sensor for doxorubicin. Microchim Acta 2019; 186:641. doi: 10.1007/s00604-019-3761-6 [Crossref] [ Google Scholar]
- Sohouli E, Keihan AH, Shahdost-fard F, Naghian E, Plonska-Brzezinska ME, Rahimi-Nasrabadi M. A glassy carbon electrode modified with carbon nanoonions for electrochemical determination of fentanyl. Mater Sci Eng C 2020; 110:110684. doi: 10.1016/j.msec.2020.110684 [Crossref] [ Google Scholar]
- Wilson D, Materón EM, Ibáñez-Redín G, Faria RC, Correa DS, Oliveira Jr ON. Electrical detection of pathogenic bacteria in food samples using information visualization methods with a sensor based on magnetic nanoparticles functionalized with antimicrobial peptides. Talanta 2019; 194:611-618. doi: 10.1016/j.talanta.2018.10.089 [Crossref] [ Google Scholar]
- Andrade CAS, Nascimento JM, Oliveira IS, de Oliveira CVJ, de Melo CP, Franco OL. Nanostructured sensor based on carbon nanotubes and clavanin A for bacterial detection. Colloids Surfaces B Biointerfaces 2015; 135:833-839. doi: 10.1016/j.colsurfb.2015.03.037 [Crossref] [ Google Scholar]
- de Miranda JL, Oliveira MDL, Oliveira IS, Frias IAM, Franco OL, Andrade CAS. A simple nanostructured biosensor based on clavanin A antimicrobial peptide for gram-negative bacteria detection. Biochem Eng J 2017; 124:108-114. doi: 10.1016/j.bej.2017.04.013 [Crossref] [ Google Scholar]
- Junior AGS, Oliveira MDL, Oliveira IS, Lima-Neto RG, Sa SR, Franco OL. A simple nanostructured impedimetric biosensor based on clavanin a peptide for bacterial detection. Sensors Actuators B Chem 2018; 255:3267-3274. doi: 10.1016/j.snb.2017.09.153 [Crossref] [ Google Scholar]
- Li Y, Afrasiabi R, Fathi F, Wang N, Xiang C, Love R. Impedance based detection of pathogenic E. coli O157: H7 using a ferrocene-antimicrobial peptide modified biosensor. Biosens Bioelectron 2014; 58:193-199. doi: 10.1016/j.bios.2014.02.045 [Crossref] [ Google Scholar]
- Zhang J, Li Y, Duan S, He F. Highly electrically conductive two-dimensional Ti3C2 Mxenes-based 16S rDNA electrochemical sensor for detecting Mycobacterium tuberculosis. Anal Chim Acta 2020; 1123:9-17. doi: 10.1016/j.aca.2020.05.013 [Crossref] [ Google Scholar]
- Lillehoj PB, Kaplan CW, He J, Shi W, Ho C-M. Rapid, electrical impedance detection of bacterial pathogens using immobilized antimicrobial peptides. J Lab Autom 2014; 19:42-49. doi: 10.1177/2211068213495207 [Crossref] [ Google Scholar]
- Formisano N, Bhalla N, Heeran M, Martinez JR, Sarkar A, Laabei M. Inexpensive and fast pathogenic bacteria screening using field-effect transistors. Biosens Bioelectron 2016; 85:103-109. doi: 10.1016/j.bios.2016.04.063 [Crossref] [ Google Scholar]
- Rengaraj S, Cruz-Izquierdo Á, Scott JL, Di Lorenzo M. Impedimetric paper-based biosensor for the detection of bacterial contamination in water. Sensors Actuators B Chem 2018; 265:50-58. doi: 10.1016/j.snb.2018.03.020 [Crossref] [ Google Scholar]
- Yang H, Zhou H, Hao H, Gong Q, Nie K. Detection of Escherichia coli with a label-free impedimetric biosensor based on lectin functionalized mixed self-assembled monolayer. Sensors Actuators B Chem 2016; 229:297-304. doi: 10.1016/j.snb.2015.08.034 [Crossref] [ Google Scholar]
- Brosel-Oliu S, Galyamin D, Abramova N, Munoz-Pascual F-X, Bratov A. Impedimetric label-free sensor for specific bacteria endotoxin detection by surface charge registration. Electrochim Acta 2017; 243:142-151. doi: 10.1016/j.electacta.2017.05.060 [Crossref] [ Google Scholar]
- Buzid A, Shang F, Reen FJ, Muimhneacháin EÓ, Clarke SL, Zhou L. Molecular signature of Pseudomonas aeruginosa with simultaneous nanomolar detection of quorum sensing signaling molecules at a boron-doped diamond electrode. Sci Rep 2016; 6:30001. doi: 10.1038/srep30001 [Crossref] [ Google Scholar]
- Oziat J, Gougis M, Malliaras GG, Mailley P. Electrochemical characterizations of four main redox–metabolites of Pseudomonas aeruginosa. Electroanalysis 2017; 29:1332-1340. doi: 10.1002/elan.201600799 [Crossref] [ Google Scholar]
- Buzid A, Muimhneacháin EÓ, Reen FJ, Hayes PE, Pardo LM, Shang F. Synthesis and electrochemical detection of a thiazolyl-indole natural product isolated from the nosocomial pathogen Pseudomonas aeruginosa. Anal Bioanal Chem 2016; 408:6361-6367. doi: 10.1007/s00216-016-9749-8 [Crossref] [ Google Scholar]
- Webster TA, Sismaet HJ, Conte JL, Goluch ED. Electrochemical detection of Pseudomonas aeruginosa in human fluid samples via pyocyanin. Biosens Bioelectron 2014; 60:265-270. doi: 10.1016/j.bios.2014.04.028 [Crossref] [ Google Scholar]
- Sismaet HJ, Pinto AJ, Goluch ED. Electrochemical sensors for identifying pyocyanin production in clinical Pseudomonas aeruginosa isolates. Biosens Bioelectron 2017; 97:65-69. doi: 10.1016/j.bios.2017.05.042 [Crossref] [ Google Scholar]
- Gandouzi I, Tertis M, Cernat A, Saidane-Mosbahi D, Ilea A, Cristea C. A Nanocomposite Based on Reduced Graphene and Gold Nanoparticles for Highly Sensitive Electrochemical Detection of Pseudomonas aeruginosa through Its Virulence Factors. Materials (Basel) 2019; 12:1180. doi: 10.3390/ma12071180 [Crossref] [ Google Scholar]
- Elliott J, Simoska O, Karasik S, Shear JB, Stevenson KJ. Transparent carbon ultramicroelectrode arrays for the electrochemical detection of a bacterial warfare toxin, pyocyanin. Anal Chem 2017; 89:6285-6289. doi: 10.1021/acs.analchem.7b00876 [Crossref] [ Google Scholar]
- imoska O, Sans M, Fitzpatrick MD, Crittenden CM, Eberlin LS, Shear JB. Real-Time Electrochemical Detection of Pseudomonas aeruginosa Phenazine Metabolites Using Transparent Carbon Ultramicroelectrode Arrays. ACS Sens 2018; 4:170-179. doi: 10.1021/acssensors.8b01152 [Crossref] [ Google Scholar]
- Yang Y, Yu YY, Wang YZ, Zhang CL, Wang JX, Fang Z. Amplification of electrochemical signal by a whole-cell redox reactivation module for ultrasensitive detection of pyocyanin. Biosens Bioelectron 2017; 98:338-344. doi: 10.1016/j.bios.2017.07.008 [Crossref] [ Google Scholar]
- Yang Y, Yu Y-Y, Shi Y-T, Moradian JM, Yong Y-C. In vivo two-way redox cycling system for independent duplexed electrochemical signal amplification. Anal Chem 2019; 91:4939-4942. doi: 10.1021/acs.analchem.9b00053 [Crossref] [ Google Scholar]
- Sismaet HJ, Webster TA, Goluch ED. Up-regulating pyocyanin production by amino acid addition for early electrochemical identification of Pseudomonas aeruginosa. Analyst 2014; 139:4241-4246. doi: 10.1039/C4AN00756E [Crossref] [ Google Scholar]
- Shang W, Liu Y, Kim E, Tsao C-Y, Payne GF, Bentley WE. Selective assembly and functionalization of miniaturized redox capacitor inside microdevices for microbial toxin and mammalian cell cytotoxicity analyses. Lab Chip 2018; 18:3578-3587. doi: 10.1039/C8LC00583D [Crossref] [ Google Scholar]
- Ciui B, Tertiş M, Cernat A, Săndulescu R, Wang J, Cristea C. Finger-based printed sensors integrated on a glove for on-site screening of Pseudomonas aeruginosa virulence factors. Anal Chem 2018; 90:7761-7768. doi: 10.1021/acs.analchem.8b01915 [Crossref] [ Google Scholar]
- Oziat J, Elsen S, Owens RM, Malliaras GG, Mailley P. Electrochemistry provides a simple way to monitor Pseudomonas aeruginosa metabolites. In: 2015 37th Annual International Conference of the IEEE Engineering inMedicine and Biology Society (EMBC). IEEE. 2015:7522-7525.
- Seviour T, Doyle LE, Lauw SJL, Hinks J, Rice SA, Nesatyy VJ. Voltammetric profiling of redox-active metabolites expressed by Pseudomonas aeruginosa for diagnostic purposes. Chem Commun 2015; 51:3789-3792. doi: 10.1039/C4CC08590F [Crossref] [ Google Scholar]
- Alatraktchi FA, Johansen HK, Molin S, Svendsen WE. Electrochemical sensing of biomarker for diagnostics of bacteria-specific infections. Nanomedicine 2016; 11:2185-2195. doi: 10.2217/nnm-2016-0155 [Crossref] [ Google Scholar]
- Alatraktchi F, Breum Andersen S, Krogh Johansen H, Molin S, Svendsen W. Fast selective detection of pyocyanin using cyclic voltammetry. Sensors 2016; 16:408. doi: 10.3390/s16030408 [Crossref] [ Google Scholar]
- Buzid A, Reen FJ, Langsi VK, Muimhneacháin EÓ, O’Gara F, McGlacken GP. Direct and Rapid Electrochemical Detection of Pseudomonas aeruginosa Quorum Signaling Molecules in Bacterial Cultures and Cystic Fibrosis Sputum Samples through Cationic Surfactant‐Assisted Membrane Disruption. ChemElectroChem 2017; 4:533-541. doi: 10.1002/celc.201600590 [Crossref] [ Google Scholar]
- Shang F, Muimhneacháin EÓ, Reen FJ, Buzid A, O’Gara F, Luong JHT. One step preparation and electrochemical analysis of IQS, a cell–cell communication signal in the nosocomial pathogen Pseudomonas aeruginosa. Bioorg Med Chem Lett 2014; 24:4703-4707. doi: 10.1016/j.bmcl.2014.08.023 [Crossref] [ Google Scholar]
- Metters JP, Kampouris DK, Banks CE. Electrochemistry provides a point-of-care approach for the marker indicative of Pseudomonas aeruginosa infection of cystic fibrosis patients. Analyst 2014; 139:3999-4004. doi: 10.1039/C4AN00675E [Crossref] [ Google Scholar]
- Hameed A, Pi H, Lin S, Lai W, Young L, Liu Y. Direct Electrochemical Sensing of Phenazine‐1‐carboxylic Acid Secreted by Pseudomonas chlororaphis subsp. aureofaciens BCRC 11057T Using Disposable Screen‐printed Carbon Electrode. Electroanalysis 2016; 28:846-853. doi: 10.1002/elan.201500278 [Crossref] [ Google Scholar]
- Gandouzi I, Tertis M, Cernat A, Bakhrouf A, Coros M, Pruneanu S. Sensitive detection of pyoverdine with an electrochemical sensor based on electrochemically generated graphene functionalized with gold nanoparticles. Bioelectrochemistry 2018; 120:94-103. doi: 10.1016/j.bioelechem.2017.11.014 [Crossref] [ Google Scholar]
- Cernat A, Tertis M, Gandouzi I, Bakhrouf A, Suciu M, Cristea C. Electrochemical sensor for the rapid detection of Pseudomonas aeruginosa siderophore based on a nanocomposite platform. Electrochem commun 2018; 88:5-9. doi: 10.1016/j.elecom.2018.01.009 [Crossref] [ Google Scholar]
- Li S, Mou Q, Feng N, Leung PHM. A Selective Medium for Pyocyanin-Dependent Fast Electrochemical Detection of Pseudomonas aeruginosa in Environmental Microbial Samples. Int J Electrochem Sci 2018; 13:3789-3798. doi: 10.20964/2018.04.20 [Crossref] [ Google Scholar]
- Burkitt R, Sharp D. Submicromolar quantification of pyocyanin in complex biological fluids using pad-printed carbon electrodes. Electrochem Commun 2017; 78:43-46. doi: 10.1016/j.elecom.2017.03.021 [Crossref] [ Google Scholar]
- Bellin DL, Sakhtah H, Rosenstein JK, Levine PM, Thimot J, Emmett K. Integrated circuit-based electrochemical sensor for spatially resolved detection of redox-active metabolites in biofilms. Nat Commun 2014; 5:3256. doi: 10.1038/ncomms4256 [Crossref] [ Google Scholar]
- Mazloum-Ardakani M, Hosseinzadeh L. A Sensitive Electrochemical Aptasensor for TNF-α Based on Bimetallic Ag@ Pt Core-Shell Nanoparticle Functionalized Graphene Nanostructures as Labels for Signal Amplification. J Electrochem Soc 2016; 163:B119-B124. doi: 10.1149/2.0241605jes [Crossref] [ Google Scholar]
- Ghorbani F, Abbaszadeh H, Dolatabadi JEN, Aghebati-Maleki L, Yousefi M. Application of various optical and electrochemical aptasensors for detection of human prostate specific antigen: A review. Biosens Bioelectron 2019; 142:111484. doi: 10.1016/j.bios.2019.111484 [Crossref] [ Google Scholar]
- Khoshroo A, Hosseinzadeh L, Adib K, Rahimi-Nasrabadi M, Ahmadi F. Earlier diagnoses of acute leukemia by a sandwich type of electrochemical aptasensor based on copper sulfide-graphene composite. Anal Chim Acta 2021; 1146:1-10. doi: 10.1016/j.aca.2020.12.007 [Crossref] [ Google Scholar]
- Kara P, Kerman K, Ozkan D, Meric B, Erdem A, Ozkan Z. Electrochemical genosensor for the detection of interaction between methylene blue and DNA. Electrochem Commun 2002; 4:705-709. doi: 10.1016/S1388-2481(02)00428-9 [Crossref] [ Google Scholar]
- Chen Y, Song J, Li D. Study on gene sensor based on primer extension. Sci China Ser C Life Sci 1997; 40:463-469. doi: 10.1007/BF03183583 [Crossref] [ Google Scholar]
- McConnell EM, Morrison D, Rincon MAR, Salena BJ, Li Y. Selection and applications of synthetic functional DNAs for bacterial detection. TrAC Trends Anal Chem 2020; 124:115785. doi: 10.1016/j.trac.2019.115785 [Crossref] [ Google Scholar]
- Mazloum-Ardakani M, Hosseinzadeh L, Heidari MM. Detection of the M268T Angiotensinogen A3B2 mutation gene based on screen-printed electrodes modified with a nanocomposite: application to human genomic samples. Microchim Acta 2016; 183:219-227. doi: 10.1007/s00604-015-1616-3 [Crossref] [ Google Scholar]
- Dehghani S, Nosrati R, Yousefi M, Nezami A, Soltani F, Taghdisi SM. Aptamer-based biosensors and nanosensors for the detection of vascular endothelial growth factor (VEGF): A review. Biosens Bioelectron 2018; 110:23-37. doi: 10.1016/j.bios.2018.03.037 [Crossref] [ Google Scholar]
- Appaturi JN, Pulingam T, Thong KL, Muniandy S, Ahmad N, Leo BF. Rapid and sensitive detection of Salmonella with reduced graphene oxide-carbon nanotube based electrochemical aptasensor. Anal Biochem 2020; 589:113489. doi: 10.1016/j.ab.2019.113489 [Crossref] [ Google Scholar]
- Hosseinzadeh L, Mazloum-Ardakani M. Chapter Six - Advances in aptasensor technology. Adv Clin Chem 2020; 99:237-279. doi: 10.1016/bs.acc.2020.02.010 [Crossref] [ Google Scholar]
- Sedighi-Khavidak S, Mazloum-Ardakani M, Rabbani Khorasgani M, Emtiazi G, Hosseinzadeh L. Detection of aflD gene in contaminated pistachio with Aspergillus flavus by DNA based electrochemical biosensor. Int J Food Prop 2017; 20:S119-S130. doi: 10.1080/10942912.2017.1291675 [Crossref] [ Google Scholar]
- Ranjbar S, Shahrokhian S. Design and fabrication of an electrochemical aptasensor using Au nanoparticles/carbon nanoparticles/cellulose nanofibers nanocomposite for rapid and sensitive detection of Staphylococcus aureus. Bioelectrochemistry 2018; 123:70-76. doi: 10.1016/j.bioelechem.2018.04.018 [Crossref] [ Google Scholar]
- Ranjbar S, Shahrokhian S, Nurmohammadi F. Nanoporous gold as a suitable substrate for preparation of a new sensitive electrochemical aptasensor for detection of Salmonella typhimurium. Sensors Actuators B Chem 2018; 255:1536-1544. doi: 10.1016/j.snb.2017.08.160 [Crossref] [ Google Scholar]
- Burrs SL, Bhargava M, Sidhu R, Kiernan-Lewis J, Gomes C, Claussen JC. A paper based graphene-nanocauliflower hybrid composite for point of care biosensing. Biosens Bioelectron 2016; 85:479-487. doi: 10.1016/j.bios.2016.05.037 [Crossref] [ Google Scholar]
- Abbaspour A, Norouz-Sarvestani F, Noori A, Soltani N. Aptamer-conjugated silver nanoparticles for electrochemical dual-aptamer-based sandwich detection of Staphylococcus aureus. Biosens Bioelectron 2015; 68:149-155. doi: 10.1016/j.bios.2014.12.040 [Crossref] [ Google Scholar]
- Jo N, Kim B, Lee SM, Oh J, Park IH, Jin Lim K, Shin JS. Aptamer-functionalized capacitance sensors for real-time monitoring of bacterial growth and antibiotic susceptibility. Biosens Bioelectron 2018; 102:164-170. doi: 10.1016/j.bios.2017.11.010 [Crossref] [ Google Scholar]
- Ozkan-Ariksoysal D, Kayran YU, Yilmaz FF, Ciucu AA, David IG, David V. DNA-wrapped multi-walled carbon nanotube modified electrochemical biosensor for the detection of Escherichia coli from real samples. Talanta 2017; 166:27-35. doi: 10.1016/j.talanta.2017.01.005 [Crossref] [ Google Scholar]
- Raveendran Raveendran, M M. Raveendran, M, Andrade AF and G-RSelective and sensitive electrochemical DNA biosensor for the detection of Bacillus anthracis. Int J Electrochem Sci 2016; 11:763-776. [ Google Scholar]
- Rahi A, Sattarahmady N, Heli H. Zepto-molar electrochemical detection of Brucella genome based on gold nanoribbons covered by gold nanoblooms. Sci Rep 2015; 5:18060. doi: 10.1038/srep18060 [Crossref] [ Google Scholar]
- Henihan G, Schulze H, Corrigan DK, Giraud G, Terry JG, Hardie A, Campbell CJ, Walton AJ, Crain J, Pethig R TK. Label-and amplification-free electrochemical detection of bacterial ribosomal RNA. Biosens Bioelectron 2016; 81:487-494. doi: 10.1016/j.bios.2016.03.037 [Crossref] [ Google Scholar]
- Hamidi-Asl E, Dardenne F, Pilehvar S, Blust R, De Wael K. Unique properties of core shell Ag@Au nanoparticles for the aptasensing of bacterial cells. Chemosensors 2016; 4:16. doi: 10.3390/chemosensors4030016 [Crossref] [ Google Scholar]
- Liébana S, Brandão D, Cortés P, Campoy S, Alegret S, Pividori MI. Electrochemical genosensing of Salmonella, Listeria and Escherichia coli on silica magnetic particles. Anal Chim Acta 2016; 904:1-9. doi: 10.1016/j.aca.2015.09.044 [Crossref] [ Google Scholar]
- Nguyet NT, Yen LTH, Doan VY, Hoang NL, Van Thu V, Lan H. A label-free and highly sensitive DNA biosensor based on the core-shell structured CeO2 -NR@Ppy nanocomposite for Salmonella detection. Mater Sci Eng C 2019; 96:790-797. doi: 10.1016/j.msec.2018.11.059 [Crossref] [ Google Scholar]
- Luo F, Li Z, Dai G, Lu Y, He P, Wang Q. Ultrasensitive biosensing pathogenic bacteria by combining aptamer-induced catalysed hairpin assembly circle amplification with microchip electrophoresis. Sensors Actuators B Chem 2020; 306:127577. [ Google Scholar]
- Janczuk M, Niedziółka-Jönsson J, Szot-Karpińska K. Bacteriophages in electrochemistry: A review. J Electroanal Chem 2016; 779:207-219. doi: 10.1016/j.jelechem.2016.05.019 [Crossref] [ Google Scholar]
- Shabani A, Marquette CA, Mandeville R, Lawrence MF. Magnetically-assisted impedimetric detection of bacteria using phage-modified carbon microarrays. Talanta 2013; 116:1047-1053. doi: 10.1016/j.talanta.2013.07.078 [Crossref] [ Google Scholar]
- Tlili C, Sokullu E, Safavieh M, Tolba M, Ahmed MU, Zourob M. Bacteria screening, viability, and confirmation assays using bacteriophage-impedimetric/loop-mediated isothermal amplification dual-response biosensors. Anal Chem 2013; 85:4893-4901. [ Google Scholar]
- Piskin E, Zareie HM, Erdem A, Moghtader F, Congur G. Impedimetric detection of pathogenic bacteria with bacteriophages using gold nanorod deposited graphite electrodes. RSC Adv 2016; 6:97832-97839. doi: 10.1039/c6ra18884b [Crossref] [ Google Scholar]
- Wang X, Zhu P, Pi F, Jiang H, Shao J, Zhang Y. A Sensitive and simple macrophage-based electrochemical biosensor for evaluating lipopolysaccharide cytotoxicity of pathogenic bacteria. Biosens Bioelectron 2016; 81:349-357. doi: 10.1016/j.bios.2016.03.007 [Crossref] [ Google Scholar]
- Yan C, Zhang Y, Yang H, Yu J, Wei H. Combining phagomagnetic separation with immunoassay for specific, fast and sensitive detection of Staphylococcus aureus. Talanta 2017; 170:291-297. doi: 10.1016/j.talanta.2017.04.007 [Crossref] [ Google Scholar]
- Zhou Y, Marar A, Kner P, Ramasamy RP. Charge-Directed Immobilization of Bacteriophage on Nanostructured Electrode for Whole-Cell Electrochemical Biosensors. Anal Chem 2017; 89:5734-5741. doi: 10.1021/acs.analchem.6b03751 [Crossref] [ Google Scholar]
- Xu J, Zhao C, Chau Y, Lee Y-K. The synergy of chemical immobilization and electrical orientation of T4 bacteriophage on a micro electrochemical sensor for low-level viable bacteria detection via Differential Pulse Voltammetry. Biosens Bioelectron 2020; 151:111914. doi: 10.1016/j.bios.2019.111914 [Crossref] [ Google Scholar]
- Amani J, Khoshroo A, Rahimi-Nasrabadi M. Electrochemical immunosensor for the breast cancer marker CA 15–3 based on the catalytic activity of a CuS/reduced graphene oxide nanocomposite towards the electrooxidation of catechol. Microchim Acta 2017; 185:79. doi: 10.1007/s00604-017-2532-5 [Crossref] [ Google Scholar]
- Mazloum‐Ardakani M, Hosseinzadeh L, Khoshroo A. Ultrasensitive Electrochemical Immunosensor for Detection of Tumor Necrosis Factor‐α Based on Functionalized MWCNT‐Gold Nanoparticle/Ionic Liquid Nanocomposite. Electroanalysis 2015; 27:2518-2526. doi: 10.1002/elan.201500104 [Crossref] [ Google Scholar]
- Li Z, Chen G-Y. Current conjugation methods for immunosensors. Nanomaterials 2018; 8:278. doi: 10.3390/nano8050278 [Crossref] [ Google Scholar]
- Mazloum-Ardakani M, Hosseinzadeh L, Khoshroo A. Label-free electrochemical immunosensor for detection of tumor necrosis factor α based on fullerene-functionalized carbon nanotubes/ionic liquid. J Electroanal Chem 2015; 757:58-64. doi: 10.1016/j.jelechem.2015.09.006 [Crossref] [ Google Scholar]
- Felix FS, Angnes L. Electrochemical immunosensors – A powerful tool for analytical applications. Biosens Bioelectron 2018; 102:470-478. doi: 10.1016/j.bios.2017.11.029 [Crossref] [ Google Scholar]
- Viswanathan S, Rani C, Ho JAA. Electrochemical immunosensor for multiplexed detection of food-borne pathogens using nanocrystal bioconjugates and MWCNT screen-printed electrode. Talanta 2012; 94:315-319. doi: 10.1016/j.talanta.2012.03.049 [Crossref] [ Google Scholar]
- Mavaei M, Chahardoli A, Shokoohinia Y, Khoshroo A, Fattahi A. One-step Synthesized Silver nanoparticles Using isoimperatorin: evaluation of photocatalytic, and electrochemical Activities. Sci Rep 2020; 10:1-12. doi: 10.1038/s41598-020-58697-x [Crossref] [ Google Scholar]
- Mazloum-Ardakani M, Brazesh B, Hosseinzadeh L, Khoshroo A. Graphene sheet for improving the electrocatalytic activity of a benzofuran derivative modified electrode for determination of epinephrine in the presence of serotonin. J Anal Chem 2017; 72:689-698. doi: 10.1134/S1061934817060119 [Crossref] [ Google Scholar]
- Mazloum-Ardakani M, Amin-sadrabadi E, Khoshroo A, Salavati-Niasari M. Electrocatalytic Properties of Vanadyl Complex in Graphite Nanocomposite and its Enhanced Electrochemical Catalysis Properties for Levodopa Oxidation. J Inorg Organomet Polym Mater 2015; 25:1576-1581. doi: 10.1007/s10904-015-0277-3 [Crossref] [ Google Scholar]
- Khoshroo A, Fattahi A. Electrochemical analysis of anionic analytes in weakly supported media using electron transfer promotion effect: a case study on nitrite. Sci Rep 2020; 10:1-9. doi: 10.1038/s41598-020-71365-4 [Crossref] [ Google Scholar]
- Xiang C, Li R, Adhikari B, She Z, Li Y, Kraatz HB. Sensitive electrochemical detection of Salmonella with chitosan-gold nanoparticles composite film. Talanta 2015; 140:122-127. doi: 10.1016/j.talanta.2015.03.033 [Crossref] [ Google Scholar]
- Mathelié-Guinlet M, Gammoudi I, Beven L, Moroté F, Delville MH, Grauby-Heywang C. Silica Nanoparticles Assisted Electrochemical Biosensor for the Detection and Degradation of Escherichia coli Bacteria. Procedia Eng 2016; 168:1048-1051. doi: 10.1016/j.proeng.2016.11.337 [Crossref] [ Google Scholar]
- Farka Z, Juřík T, Pastucha M, Kovář D, Lacina K, Skládal P. Rapid Immunosensing of Salmonella Typhimurium Using Electrochemical Impedance Spectroscopy: the Effect of Sample Treatment. Electroanalysis 2016; 28:1803-1809. doi: 10.1002/elan.201600093 [Crossref] [ Google Scholar]
- Altintas Z, Akgun M, Kokturk G, Uludag Y. A fully automated microfluidic-based electrochemical sensor for real-time bacteria detection. Biosens Bioelectron 2018; 100:541-548. doi: 10.1016/j.bios.2017.09.046 [Crossref] [ Google Scholar]
- Zhang X, Zhang F, Zhang H, Shen J, Han E, Dong X. Functionalized gold nanorod-based labels for amplified electrochemical immunoassay of E. coli as indicator bacteria relevant to the quality of dairy product. Talanta 2015; 132:600-605. doi: 10.1016/j.talanta.2014.10.013 [Crossref] [ Google Scholar]
- Zhong M, Yang L, Yang H, Cheng C, Deng W, Tan Y. An electrochemical immunobiosensor for ultrasensitive detection of Escherichia coli O157:H7 using CdS quantum dots-encapsulated metal-organic frameworks as signal-amplifying tags. Biosens Bioelectron 2019; 126:493-500. doi: 10.1016/j.bios.2018.11.001 [Crossref] [ Google Scholar]
- Krithiga N, Viswanath KB, Vasantha VS, Jayachitra A. Specific and selective electrochemical immunoassay for Pseudomonas aeruginosa based on pectin–gold nano composite. Biosens Bioelectron 2016; 79:121-129. doi: 10.1016/j.bios.2015.12.006 [Crossref] [ Google Scholar]
- Mao S, Yu K, Chang J, Steeber DA, Ocola LE, Chen J. Direct growth of vertically-oriented graphene for field-effect transistor biosensor. Sci Rep 2013; 3:33-36. doi: 10.1038/srep01696 [Crossref] [ Google Scholar]
-
Wu G, Meyyappan M, Lai KWC. Graphene field-effect transistors-based biosensors for Escherichia coli detection. 16th Int Conf Nanotechnol - IEEE NANO; 2016: 22-25. 10.1109/NANO.2016.7751414
- Cortina ME, Melli LJ, Roberti M, Mass M, Longinotti G, Tropea S. Electrochemical magnetic microbeads-based biosensor for point-of-care serodiagnosis of infectious diseases. Biosens Bioelectron 2016; 80:24-33. doi: 10.1016/j.bios.2016.01.021 [Crossref] [ Google Scholar]
- Wang D, Wang Z, Chen J, Kinchla AJ, Nugen SR. Rapid detection of Salmonella using a redox cycling-based electrochemical method. Food Control 2016; 62:81-88. doi: 10.1016/j.foodcont.2015.10.021 [Crossref] [ Google Scholar]
- Fei J, Dou W, Zhao G. Amperometric immunoassay for the detection of Salmonella pullorum using a screen - printed carbon electrode modified with gold nanoparticle-coated reduced graphene oxide and immunomagnetic beads. Microchim Acta 2016; 183:757-764. doi: 10.1007/s00604-015-1721-3 [Crossref] [ Google Scholar]
- Xu M, Wang R, Li Y. Xu M, Wang R, Li YAn electrochemical biosensor for rapid detection of: Ecoli O157:H7 with highly efficient bi-functional glucose oxidase-polydopamine nanocomposites and Prussian blue modified screen-printed interdigitated electrodes. Analyst 2016; 141:5441-5449. doi: 10.1039/c6an00873a [Crossref] [ Google Scholar]
- Guo Y, Wang Y, Liu S, Yu J, Wang H, Cui M. Electrochemical immunosensor assay (EIA) for sensitive detection of E. coli O157:H7 with signal amplification on a SG-PEDOT-AuNPs electrode interface. Analyst 2015; 140:551-559. doi: 10.1039/c4an01463d [Crossref] [ Google Scholar]
- Tufa LT, Oh S, Tran VT, Kim J, Jeong KJ, Park TJ. Electrochemical immunosensor using nanotriplex of graphene quantum dots, Fe3O4 and Ag nanoparticles for tuberculosis. Electrochim Acta 2018; 290:369-377. doi: 10.1016/j.electacta.2018.09.108 [Crossref] [ Google Scholar]
- Dastider SG, Mlaji Z, Shen Z, Almasri M, Liu J, El-Dweik M. An impedance biosensor for simultaneous detection of low concentration of Salmonella serogroups in poultry and fresh produce samples. Biosens Bioelectron 2018; 126:292-300. doi: 10.1016/j.bios.2018.10.065 [Crossref] [ Google Scholar]
- Cui F, Zhou Z, Zhou HS. Molecularly imprinted polymers and surface imprinted polymers based electrochemical biosensor for infectious diseases. Sensors 2020; 20:996. doi: 10.3390/s20040996 [Crossref] [ Google Scholar]
- Dickert FL, Lieberzeit P, Hayden O. Sensor strategies for microorganism detection—from physical principles to imprinting procedures. Anal Bioanal Chem 2003; 377:540-549. doi: 10.1007/s00216-003-2060-5 [Crossref] [ Google Scholar]
- Cohen T, Starosvetsky J, Cheruti U, Armon R. Whole cell imprinting in sol-gel thin films for bacterial recognition in liquids: Macromolecular fingerprinting. Int J Mol Sci 2010; 11:1236-1252. doi: 10.3390/ijms11041236 [Crossref] [ Google Scholar]
- Borovička J, Stoyanov SD, Paunov VN. Shape recognition of microbial cells by colloidal cell imprints. Nanoscale 2013; 5:8560-8568. doi: 10.1039/C3NR01893H [Crossref] [ Google Scholar]
- Harvey SD, Mong GM, Ozanich RM, Mclean JS, Goodwin SM, Valentine NB. Preparation and evaluation of spore-specific affinity-augmented bio-imprinted beads. Anal Bioanal Chem 2006; 386:211-219. doi: 10.1007/s00216-006-0622-z [Crossref] [ Google Scholar]
- Sharma PS, Pietrzyk-Le A, D’souza F, Kutner W. Electrochemically synthesized polymers in molecular imprinting for chemical sensing. Anal Bioanal Chem 2012; 402:3177-3204. doi: 10.1007/s00216-011-5696-6 [Crossref] [ Google Scholar]
- Jia M, Zhang Z, Li J, Ma X, Chen L, Yang X. Molecular imprinting technology for microorganism analysis. TrAC Trends Anal Chem 2018; 106:190-201. doi: 10.1016/j.trac.2018.07.011 [Crossref] [ Google Scholar]
- Hayden O, Dickert FL. Selective microorganism detection with cell surface imprinted polymers. Adv Mater 2001; 13:1480-1483. doi: 10.1002/1521-4095(200110)13:19 [Crossref] [ Google Scholar]
- Khan MAR, T T. Khan MAR, TCMoreira F, Riu J, FSales MGPlastic antibody for the electrochemical detection of bacterial surface proteins. Sens Actuators B Chem 2016; 233:697-704. doi: 10.1016/j.snb.2016.04.075 [Crossref] [ Google Scholar]
- Khan MAR, Cardoso ARA, Sales MGF, Merino S, Tomas JM, Rius FX. Artificial receptors for the electrochemical detection of bacterial flagellar filaments from Proteus mirabilis. Sens Actuators B Chem 2017; 244:732-741. doi: 10.1016/j.snb.2017.01.018 [Crossref] [ Google Scholar]
- Lahcen AA, Arduini F, Lista F, Amine A. Label-free electrochemical sensor based on spore-imprinted polymer for Bacillus cereus spore detection. Sens Actuators B Chem 2018; 276:114-120. doi: 10.1016/j.snb.2018.08.031 [Crossref] [ Google Scholar]
- Jiang H, Jiang D, Shao J, Sun X. Magnetic molecularly imprinted polymer nanoparticles based electrochemical sensor for the measurement of Gram-negative bacterial quorum signaling molecules (N-acyl-homoserine-lactones). Biosens Bioelectron 2016; 75:411-419. doi: 10.1016/j.bios.2015.07.045 [Crossref] [ Google Scholar]
- Golabi M, Kuralay F, Jager EWH, Beni V, Turner APF. Electrochemical bacterial detection using poly (3-aminophenylboronic acid)-based imprinted polymer. Biosens Bioelectron 2017; 93:87-93. doi: 10.1016/j.bios.2016.09.088 [Crossref] [ Google Scholar]
- Mugo SM, Lu W, Dhanjai D. A pathogen imprinted hybrid polymer capacitive sensor for selective Escherichia coli detection. Med Devices Sensors 2020; 3:e10071. doi: 10.1002/mds3.10071 [Crossref] [ Google Scholar]
- Idil N, Hedström M, Denizli A, Mattiasson B. Whole cell based microcontact imprinted capacitive biosensor for the detection of Escherichia coli. Biosens Bioelectron 2017; 87:807-815. doi: 10.1016/j.bios.2016.08.096 [Crossref] [ Google Scholar]
- Chen S, Chen X, Zhang L, Gao J, Ma Q. Electrochemiluminescence detection of Escherichia coli O157: H7 based on a novel polydopamine surface imprinted polymer biosensor. ACS Appl Mater Interfaces 2017; 9:5430-5436. doi: 10.1021/acsami.6b12455 [Crossref] [ Google Scholar]
- Wu J, Wang R, Lu Y, Jia M, Yan J, Bian X. Facile Preparation of a Bacteria Imprinted Artificial Receptor for Highly Selective Bacterial Recognition and Label-Free Impedimetric Detection. Anal Chem 2018; 91:1027-1033. doi: 10.1021/acs.analchem.8b04314 [Crossref] [ Google Scholar]
- Jafari H, Amiri M, Abdi E, Navid SL, Bouckaert J, Jijie R. Entrapment of uropathogenic E. coli cells into ultra-thin sol-gel matrices on gold thin films: A low cost alternative for impedimetric bacteria sensing. Biosens Bioelectron 2019; 124:161-166. doi: 10.1016/j.bios.2018.10.029 [Crossref] [ Google Scholar]
- Bhardwaj N, Bhardwaj SK, Mehta J, Mohanta GC, Deep A. Bacteriophage immobilized graphene electrodes for impedimetric sensing of bacteria (Staphylococcus arlettae). Anal Biochem 2016; 505:18-25. doi: 10.1016/j.ab.2016.04.008 [Crossref] [ Google Scholar]