Bioimpacts. 12(6):561-566.
doi: 10.34172/bi.2022.23825
Short Communication
Role of nanostructured lipid carriers in the expression alterations of ATP-binding cassette transporter genes in fluconazole-resistant Candida glabrata
Maryam Moazeni 1, 2, *  , Majid Saeedi 3
, Majid Saeedi 3  , Hamidreza Kelidari 3
, Hamidreza Kelidari 3  , Behrad Roohi 4, Mohammad T Hedayati 1, 2
, Behrad Roohi 4, Mohammad T Hedayati 1, 2  , Tahereh Shokohi 1, 2
, Tahereh Shokohi 1, 2  , Mojtaba Nabili 5
, Mojtaba Nabili 5  , Kofi Asare-Addo 6
, Kofi Asare-Addo 6  , Ali Nokhodchi 7, 8, *
, Ali Nokhodchi 7, 8, * 
Author information:
1Invasive Fungi Research Center, Communicable Diseases Institute, Mazandaran University of Medical Sciences, Sari, Iran
2Department of Medical Mycology, School of Medicine, Mazandaran University of Medical Sciences, Sari, Iran
3Department of Pharmaceutics, Faculty of Pharmacy, Mazandaran University of Medical Sciences, Sari, Iran
4Student Research Committee Center, Mazandaran University of Medical Sciences, Sari, Iran
5Faculty of Medicine, Sari Branch, Islamic Azad University, Sari, Iran
6Department of Pharmacy, University of Huddersfield, Queensgate, Huddersfield, HD1 3DH, UK
7Pharmaceutics Research Laboratory, School of Life Sciences, University of Sussex, Arundel Building, Brighton BN1 9QJ, UK
8Lupin Pharmaceutical Research Center, Coral Springs, Florida, USA
Abstract
Introduction:
This study was proposed to assess the potential role of efflux transporters in reversing fluconazole resistance in Candida glabrata isolates treated with fluconazole loaded nanostructured lipid carriers (FLZ-NLCs).
Methods:
The ultrasound technique was used to synthesize the FLZ-NLCs. Four fluconazole-resistant, as well as one susceptible standard C. glabrata isolates, were applied and exposed to FLZ/ FLZ-NLCs for 20 h at 37°C. Real-time PCRs were done to estimate the likely changes in ATP-binding cassette transporter genes.
Results:
Similar to the FLZ-exposed-susceptible standard strain which showed no alteration, the genes were not up-regulated significantly under the FLZ-NLCs treated condition. While they were over-expressed when the yeasts were treated with fluconazole.
Conclusion:
It is highly suggested that due to the nature of the NLCs which shields the whole conformation of the drug, FLZ is not recognized by the efflux transporter subunits and consequently the translocation would not happen.
Keywords: Fluconazole, Nanostructured lipid carriers, Candida glabrata, Resistance
Copyright and License Information
© 2022 The Author(s).
This work is published by BioImpacts as an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (
http://creativecommons.org/licenses/by-nc/4.0/). Non-commercial uses of the work are permitted, provided the original work is properly cited.
Introduction
Fluconazole (FLZ) is a safe, available, commonly used antifungal for both superficial and deep-seated candidiasis management.
1
This drug is a broad-spectrum antifungal which prevents cytochrome P450- dependent lanosterol 14-α-demethylase, a vital enzyme for ergosterol synthesis in fungal cells. The antifungal effect of FLZ acts through the accumulation of C14-methylated sterols which are thought to disrupt membrane structure and function of fungal cells.
2
Although it is strongly suggested for prophylaxis or treatment of candidiasis, some species exhibit lower or no sensitivity against FLZ. Among Candida species, Candida glabrata is often one of the most common causes of candidiasis
3,4
which shows intrinsic/acquired resistance to FLZ. The development of resistant C. glabrata strains is almost exclusively mediated by adenosine-5-triphosphate (ATP)-binding cassette (ABC) transporters as efflux pumps. Upregulation of these efflux pumps reduces the accumulation of effective concentrations of azole antifungal drugs in the cytoplasm. Three main transporter proteins encoded by C. glabrata CDR1 (CgCDR1), CgCDR2, and CgSNQ2 genes partake in the advancement of azole resistance.
5-7
The overexpression of CgCDR1, CgCDR2 and CgSNQ2 is related either with mutations in regulators such as transcription factors or mitochondrial deficiency. C. glabrata tends to intrinsically induce mitochondrial alteration which results in strong transcriptional modification comprising multidrug resistance genes.
8
To overcome FLZ-resistant C. glabrata isolates, one of the promising approaches would be either a new antifungal agent with a new design or formulation or the utilization of innovative approaches. Nanostructured lipid transporters (NLCs) are a new type of colloidal approach made out of a blend of solid and fluid lipids and broadens the capability of the drug assembling capacity and delivery properties. These nano-transporters contain no harmful lipids in the submicron range (40–1000 nm).
9
The innovation has been broadly studied for the transmission of antifungals, particularly for FLZ.
10-13
Studies showed that NLCs can be used as a novel delivery system to improve the antifungal activity and reduce minimum inhibitory concentration (MIC) of FLZ in contrast to various Candida strains.
14
Although the efficacy of the carrier on Candida albicans and non-albicans Candida species was previously reported,
14
the mechanism responsible for the decrease in the MICs of FLZ when FLZ-NLCs is used has not yet been studied. This report, therefore, demonstrates that there are feasible alterations in the up-regulation of drug efflux transporter genes, which are vital elements in the enhancement of resistance to FLZ in C. glabrata isolates when treated with FLZ-NLCs.
Materials and Methods
Strains
In the current study, four FLZ-resistant C. glabrata isolates with MIC value of 64 µg/mL were used. These strains were isolated from diverse clinical samples collected from patients with different clinical types of candidiasis. Stock cultures were reserved in the reference culture assortment of the Invasive Fungi Research Center (IFRC, Sari, Iran). Also, one FLZ-susceptible strain (C. glabrata 90030, MIC: 16 µg/mL) was applied as the positive control in the following assays. Species identification of clinical isolates was previously performed to the species level by using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis on Internal Transcribed Spacer DNA (ITS rDNA) region using Msp1 restriction enzyme.
15
In addition, resistance to FLZ was proven for isolates with MICs ≥ 64 µg/mL against FLZ according to the broth microdilution reference method as released by the Clinical and Laboratory Standards Institute (CLSI) document M60.
16
Also, our previous in vitro study reported that the MIC for all selected isolates was 4 µg/mL when treated with FLZ-NLCs.
11
To get a large mass of fresh viable yeast cells, the isolates were grown on yeast extract peptone dextrose agar (YEPD) and incubated at 35°C for a period of 48 hours.
Fluconazole loaded nanostructured lipid carriers preparation
Fluconazole loaded nanostructured lipid carriers (FLZ-NLCs) were manufactured and optimized using the ultrasound technique as previously employed by Kelidari et al.
11
Briefly, 2.8 g of solid lipid (stearic acid, Merck, Darmstadt, Germany) in a mixture of 1.2 g of liquid lipid (oleic acid, Merck, Darmstadt, Germany), 2.5 g of lipophilic surfactant (Span 80, Merck, Darmstadt, Germany) and 1 g of FLZ (Arasto Pharmaceuticals Chemicals Inc., Tehran-Iran) were liquefied using a hot plate at 85 °C. The heated mixture of the lipid phase was diffused in 1/3 of the aqueous solution of hydrophilic surfactant (made by weighing out 0.84% w/w Tween 80) (Merck, Darmstadt, Germany) at 85°C using a probe sonicator (Bandelin Sonopuls, Berlin, Germany) for 5 minutes (Model HD 3200, Prob TT25, 50% power and 14.28 kJ continuous). As it is affirmed that the homogenization occurs at least 10°C above the melting point of the liquid lipid, the prime product was a nanoemulsion due to the liquid state of the lipid. After sonication, the mixture was diffused into the rest of the hydrophilic surfactant solution and preserved in an ice bath. Eventually, the whole solution was sonicated once more for 10 minutes (50% power and 43.21 kJ) whilst still submerged in the ice bath. The constituent and physicochemical characteristics of applied FLZ-loaded NLCs (% w/w) were selected from the best experimental formulation described previously to have a small particle size (126.4±15.2 nm), reasonably high zeta potential (35.1±3.0) with a good entrapment efficiency (%93.6±3.5).
15
NLC without FLZ was used as the placebo prepared by the same procedure as explained above with the excluding FLZ during the liquefication process.
FLZ-NLCs exposure and RNA extraction
Antifungal susceptibility testing assays (AFSTs) were performed in 24-well trays with the sub- MIC of 32 µg/mL and 8 µg/mL of FLZ for resistant and susceptible isolates, respectively. FLZ-NLCs with a concentration of 2 µg/mL was employed only for resistant isolates. Resistant C. glabrata isolates were exposed to both FLZ and FLZ-NLCs through separate assays and along with each assay. FLZ-sensitive C. glabrata ATCC 90030 isolate was evaluated as a positive control. The standard isolate was exposed only to FLZ. For the inoculum preparation, 24h-grown C. glabrata cultured on Sabouraud Dextrose Agar (SDA, Liofilchem, Italy) were used. The antifungal agent stock solutions and dilutions and the yeast inoculum were prepared as described by the M60 reference method.
16
Each plate was incubated for 20 hours at 37°C. Total RNA was extracted from mid-logarithmic phase cultures according to the RNX Plus Solution Kit instruction (Sina clone, Karaj, Iran). RNA properties such as concentration and purity were measured by a spectrophotometric technique (Biochrom WPA Biowave II, UK) where a dilution of 1:10 of RNA was evaluated. The RNA purity was assessed considering the optical density of extracted RNA at 260, 280 and 230 nm wavelength and also at the ratio of OD260/OD280. RNA concentration (µg/mL) was calculated using the following formula: 40 × OD260 × dilution factor. The extracted RNA was used for the synthesis of first-strand complementary DNAs (cDNAs) using the Prime Script RT reagent kit (Vivantis, Malaysia).
Quantitative real-time reverse transcription polymerase chain reaction
Real-time PCRs was performed to evaluate the alternations in the expression of ATP-binding cassette transporter genes. Primers were designed based on the published sequence of the related genes in C. glabrata (Table 1). Four serially diluted cDNAs were used for establishing standard curves for each gene. Real-time PCRs were done as recently explained utilizing the ABI step one real-time PCR framework (Applied Biosystem, USA).
7
The program for amplification was 95°C for 30 seconds as the underlying denaturation step followed by 40 cycles, every one of which comprises of two stages: 95 °C for 5 s and 60 °C for 30 s. The expression of all genes was standardized to the endogenous reference gene RDN5.8 and investigated by utilizing the REST® (2009) software. The program applies the comparative Ct method (ΔΔCt) to analyze data. To assess reproducibility, each test was repeated twice on two different days and performed in duplicate. Pairwise fixed reallocation randomization test which was applied by the REST® (2009) program itself was performed for statistical analysis. A P value < 0.05 was considered to be statistically significant.
Table 1.
Primer sequences for expression tests
|
Name
|
Reference gene accession no.
|
Primer’s sequence (5´→3´ )
|
PCR product length
|
CgCDR1-F
CgCDR1-R
|
AF109723.1 |
TACACGAACGTGGTGCTTTG
TTCTGCCACCTGGTTAAAGG
|
101 |
CgCDR2-F
CgCDR2-R
|
AF251023.1 |
GGTGGTAGCCCTCAAGTTGG
CCGGATGCACCCATTAAAGC
|
153 |
CgSNQ2-F
CgSNQ2-R
|
AM849042.1 |
CCTAGTGAAAATCCCGCTGA
CATACTTGGTTGGTGCATCG
|
196 |
RDN5.8-F
RDN5.8-R
|
AB032177.1 |
CTTGGTTCTCGCATCGATGA
GGCGCAATGTGCGTTCA
|
98 |
Results
In this study, the REST® (2009) was applied to denote the relative expression between treated and untreated (control) samples. REST® software is used to visualize gene expression data as an informative boxplot in which the smallest observation (sample minimum), lower quartile, median, upper quartile, and largest observation (sample maximum) are indicated. After normalizing the data to the selected reference gene (e.g., CgRDN ) underexpression and overexpression were implied by values between 0-1 and values higher than 1, respectively. Fig. 1 indicates the ABC transporter genes expression ratios when yeasts were exposed to the highest sub-inhibitory concentrations of FLZ as well as FLZ-NLCs compared to unexposed conditions.
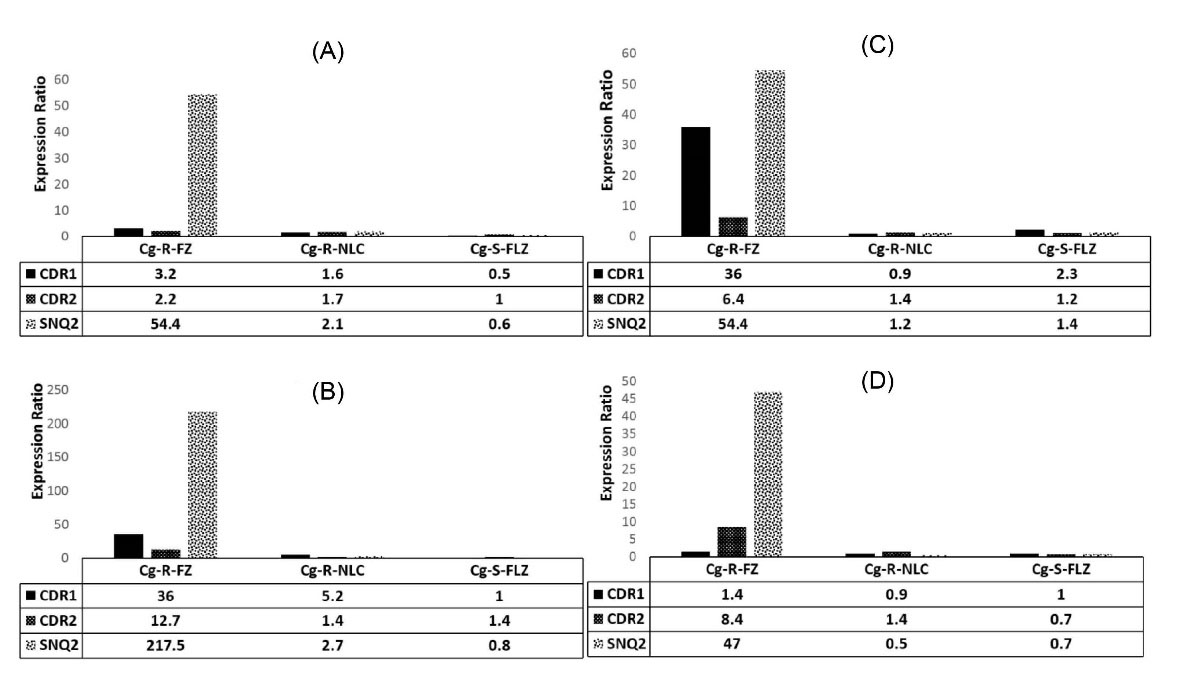
Fig. 1.
The CgCDR1, CgCDR2 and CgSNQ2 transporter genes expression ratios. Figure 1 indicates the ABC transporter genes expression ratios when yeasts were exposed to the sub-inhibitory concentration of fluconazole as well as fluconazole-NLCs compared to an unexposed condition. Along with a test performed for each resistant isolate, one FLZ-susceptible strain was applied in each run as positive control exposing to FLZ alone (FLZ-PC). Resistant isolates were as follow: isolate R1 (A), isolate R2 (B), isolate R3 (C) and isolate R4 (D).
.
The CgCDR1, CgCDR2 and CgSNQ2 transporter genes expression ratios. Figure 1 indicates the ABC transporter genes expression ratios when yeasts were exposed to the sub-inhibitory concentration of fluconazole as well as fluconazole-NLCs compared to an unexposed condition. Along with a test performed for each resistant isolate, one FLZ-susceptible strain was applied in each run as positive control exposing to FLZ alone (FLZ-PC). Resistant isolates were as follow: isolate R1 (A), isolate R2 (B), isolate R3 (C) and isolate R4 (D).
Based on the REST® output (Table 2), the expression of CgCDR1, CgCDR2, and CgSNQ2 were not constantly altered in all azole-resistant isolates exposed to applied antifungals. Under the highest sub-inhibitory concentrations of FLZ (32 µg/mL), meaningful and coordinated overexpression of all studiedgenes were observed only in isolate R3 (P < 0.05). The maximum expression variations was detected for CgSNQ2 (47- to 217-fold) when exposed to FLZ (overexpressed in all four isolates). CgCDR1 was over-expressed (3- to 36-fold) in three isolates, i.e. R1, R2, and R3. The expression of CgCDR2 was overexpressed significantly (8- to12-fold) only in isolates R3, and R4 (Fig. 1). CgCDR1 and CgSNQ2 were the genes that showed the least and the highest changes, respectively, when the strains were exposed to FLZ. Under the FLZ-NLCs treated conditions (2 µg/mL), although the studied genes were overexpressed in a non-coordinately manner, the observed overexpression was not significant. As it was expected, no significant overexpression in susceptible C. glabrata isolates was illustrated for the genes CgCDR1, CgCDR2, and CgSNQ2 when exposed to FLZ.
Table 2.
Expression patterns of ABC transporter genes in four fluconazole-resistant Candida glabrata isolates
|
Isolate
|
Exposure condition
|
Gene
|
Type
|
Expression
|
Std. Error
|
95% CI
|
P
(H1)
a
|
Result
|
| FLZ-R* Cg1 |
FLZ |
CDR1 |
TRG |
3.182 |
2.550 - 4.169 |
2.098 - 4.893 |
0.000 |
UP |
| CDR2 |
TRG |
2.250 |
1.296 - 4.127 |
1.052 - 4.886 |
0.169 |
|
| SNQ2 |
TRG |
54.380 |
38.776 - 78.859 |
33.065 - 90.281 |
0.000 |
UP |
| RDN |
REF |
1.000 |
|
|
|
|
| FLZ-NLC |
CDR1 |
TRG |
1.597 |
1.051 - 2.456 |
0.957 - 2.673 |
0.341 |
|
| CDR2 |
TRG |
1.693 |
1.372 - 2.115 |
1.253 - 2.296 |
0.170 |
|
| SNQ2 |
TRG |
2.071 |
1.022 - 4.246 |
0.931 - 4.621 |
0.321 |
|
| RDN |
REF |
1.000 |
|
|
|
|
| FLZ-S Cg** |
FLZ |
CDR1 |
TRG |
0.518 |
0.393 - 0.684 |
0.372 - 0.720 |
0.000 |
DOWN |
| CDR2 |
TRG |
0.990 |
0.811 - 1.209 |
0.784 - 1.249 |
0.661 |
|
| SNQ2 |
TRG |
0.605 |
0.370 - 1.010 |
0.327 - 1.127 |
0.341 |
|
| RDN |
REF |
1.000 |
|
|
|
|
| FLZ-R Cg2 |
FLZ |
CDR1 |
TRG |
36.002 |
20.739 - 66.027 |
16.839 - 78.181 |
0.000 |
UP |
| CDR2 |
TRG |
6.364 |
2.592 - 16.50 |
2.105 - 19.545 |
0.169 |
|
| SNQ2 |
TRG |
54.380 |
38.776 - 78.859 |
33.065 - 90.281 |
0.000 |
UP |
| RDN |
REF |
1.000 |
|
|
|
|
| FLZ-NLC |
CDR1 |
TRG |
0.946 |
0.514 - 1.874 |
0.402 - 2.271 |
0.830 |
|
| CDR2 |
TRG |
1.419 |
0.986 - 2.160 |
0.798 - 2.564 |
0.336 |
|
| SNQ2 |
TRG |
1.227 |
0.738 - 2.065 |
0.672 - 2.247 |
0.661 |
|
| RDN |
REF |
1.000 |
|
|
|
|
| FLZ-S Cg |
FLZ |
CDR1 |
TRG |
2.321 |
1.563 - 3.705 |
1.227 - 4.482 |
0.339 |
|
| CDR2 |
TRG |
1.231 |
1.053 - 1.450 |
0.980 - 1.550 |
0.321 |
|
| SNQ2 |
TRG |
1.439 |
1.375 - 1.506 |
1.367 - 1.514 |
0.154 |
|
| RDN |
REF |
1.000 |
|
|
|
|
| FLZ-R Cg3 |
FLZ |
CDR1 |
TRG |
36.002 |
20.739 - 66.027 |
16.839 - 78.181 |
0.000 |
UP |
| CDR2 |
TRG |
12.729 |
10.201 - 16.675 |
8.393 - 19.572 |
0.000 |
UP |
| SNQ2 |
TRG |
217.519 |
155.105 - 315.437 |
132.262 - 361.122 |
0.000 |
UP |
| RDN |
REF |
1.000 |
|
|
|
|
| FLZ-NLC |
CDR1 |
TRG |
5.169 |
2.836 - 9.472 |
2.670 - 10.023 |
0.169 |
|
| CDR2 |
TRG |
1.419 |
0.986 - 2.160 |
0.798 - 2.564 |
0.336 |
|
| SNQ2 |
TRG |
2.704 |
1.490 - 8.463 |
0.689 - 12.748 |
0.341 |
|
| RDN |
REF |
1.000 |
|
|
|
|
| FLZ-S Cg |
FLZ |
CDR1 |
TRG |
0.973 |
0.648 - 1.527 |
0.538 - 1.780 |
0.830 |
|
| CDR2 |
TRG |
1.414 |
1.112 - 1.803 |
1.066 - 1.878 |
0.169 |
|
| SNQ2 |
TRG |
0.853 |
0.607 - 1.210 |
0.557 - 1.310 |
0.661 |
|
| RDN |
REF |
1.000 |
|
|
|
|
| FLZ-R Cg4 |
FLZ |
CDR1 |
TRG |
1.439 |
1.287 - 1.610 |
1.262 - 1.641 |
0.339 |
|
| CDR2 |
TRG |
8.427 |
6.887 - 10.318 |
6.752 - 10.519 |
0.000 |
UP |
| SNQ2 |
TRG |
47.013 |
28.304 - 79.309 |
25.463 - 87.179 |
0.000 |
UP |
| RDN |
REF |
1.000 |
|
|
|
|
| FLZ-NLC |
CDR1 |
TRG |
0.914 |
0.514 - 1.748 |
0.402 - 2.119 |
0.661 |
|
| CDR2 |
TRG |
1.371 |
0.735 - 2.585 |
0.672 - 2.807 |
0.830 |
|
| SNQ2 |
TRG |
0.478 |
0.223 - 1.066 |
0.187 - 1.234 |
0.491 |
|
| RDN |
REF |
1.000 |
|
|
|
|
| FLZ-S Cg |
FLZ |
CDR1 |
TRG |
1.003 |
0.588 - 1.720 |
0.557 - 1.811 |
1.000 |
|
| CDR2 |
TRG |
0.710 |
0.570 - 0.884 |
0.557 - 0.904 |
0.339 |
|
| SNQ2 |
TRG |
0.868 |
0.680 - 1.107 |
0.667 - 1.129 |
0.661 |
|
| RDN |
REF |
1.000 |
|
|
|
|
Note: FLZ-R Cg: fluconazole-resistant Candida glabrata; FLZ-R Cg: fluconazole-resistant Candida glabrata, TRG: target gene;REF: reference gene.
a
P(H1): the probability of the alternate hypothesis that variation between test and control groups is only accidental.
Discussion
Great attention has been drawn to C. glabrata due to the increasing trend in fungal infections caused by the species and also concerns regarding the development of FLZ-resistant strains. It has been shown that the acquired resistance to FLZ is the most common form of resistancein C. glabrata.
17
FLZ is still the most widely prescribed antifungal for prophylaxis for patients who are at great risk of acquiring a fungal infection and may be the reason for the emergence of FLZ-resistant C. glabrata strains.
18
It has been documented that ATP-binding cassette (ABC) transporters like efflux pumps (encoded by CgCDR1, CgCDR2, and CgSNQ2 genes) are responsible for FLZ-resistant development
7
. The result of a study conducted by Gohar et al indicated that translocation of drug-mediated by ATP-binding cassette transporters, particularly encoded by CgSNQ2 and CgCDR1 genes, is the major mechanism of FLZ resistance in C. glabrata.
7
Also, our previous study demonstrated that FLZ loaded on NLCs can be used as a novel approach to overcome FLZ-resistant C. glabrata isolates.
14
Similar to susceptible isolates, the obtained results revealed that the MIC values were lower than 32 µg/mL for resistant strains after being treated with FLZ-NLCs. One simple possible justification for obtaining this result may be the antifungal properties of the carrier components. However, this theory can be ruled out because the lack of antifungal properties of the carrier was proven when the yeasts were exposed to the placebo (NLCs without FLZ) in a pilot study (data not shown). An imaginable description for these antifungal susceptibility results is the up-regulation of plasma membrane transport proteins that translocate azoles across the cell membrane. The mechanism is recognized as the main route in charge of the drug resistance in important medical fungi especially C. glabrata.
14
Results of a study conducted by Bianchin et al showed that FLZ could reverse efflux pumps in FLZ-resistant Candida glabrata isolates.
19
However, the study was focused on the role of efflux transporters when the FLZ-resistant strain was exposed to FLZ alone using verapamil as transporter inhibitors. Hence, in the current study, the authors investigated the alterations in the expression of drug efflux transporter genes (CgCDR1, CgCDR2, and CgSNQ2) when FLZ-NLC was used. The results demonstrated that when the FLZ-NLCs were employed, significant overexpression of CgCDR1, CgCDR2, and CgSNQ2 genes was not observed. A probable explanation for this output is considering how the efflux transporters work. The clothes peg-like movement which results in an inward-open and outward-open conformation is the key point. This conformational alteration can provide an ability for the transporters to translocate the antifungal drug across the membrane through ATP binding and hydrolyzing.
20
Since the NLCs shield the whole conformation of the drug, the drug is not recognized by the efflux transporter subunits and consequently, the translocation does not occur.
21
More studies are needed to reveal the information on the interactions of the efflux transporter subunits with antifungals by modeling and molecular ducking.
Research Highlights
What is the current knowledge?
√ Upregulation of (ATP)-binding cassette ABC-transporters as efflux pumps is the main mechanism of FLZ resistance in Candida glabrata.
√ At least three transporters, encoded by Candida glabrata CDR1 (CgCDR1), CgCDR2, and CgSNQ2 genes participate in the development of FLZ resistance.
√ FLZ loaded NLCs can lower the MIC values against FLZ in resistant isolates.
What is new here?
√ For the first time, the possible mechanism of FLZ loaded NLC against FLZ-resistant strains was deciphered.
√ The CgSNQ2 transporter genes were over-expressed in all FLZ-resistant C. glabrata strains exposed to FLZ.
√ Upon exposure to FLZ loaded NLCs, none of ABC transporter genes was significantly over-expressed as occurs in susceptible strains exposed to FLZ.
√ The FLZ loaded NLCs may avoid FLZ to be recognized by transporters and consequently avoid drug translocation across the membrane.
Conclusion
The results of the current research concluded that using FLZ-NLCs instead of the conventional formulations of FLZ can potentially be a promising approach for the treatment of candidiasis caused by. C. glabrata and can play an effective role in decreasing the therapeutic dose and risk of adverse drug effects. It is highly suggested that due to the nature of the NLCs which shields the whole conformation of the drug, FLZ cannot be recognized by the efflux transporter subunits and consequently the translocation does not happen. This understanding will also contribute to the awareness of the proper shields to avoid drug-protein interactions.
Acknowledgment
We thank our funding corporation, Mazandaran University of Medical Sciences (MazUMS).
Funding sources
The current research fund was granted by the MazUMS grant (No. 6325) received by MM (the first author). MazUMS had neither role in study design, nor data collection and interpretation, or the decision to submit the work for publication.
Ethical statement
The study was approved by the Ethical Committee of Mazandaran University of Medical Sciences under ethic No. IR.MAZUMS.REC.1398.6325.
Competing interests
There is no conflict of interest to declare for this article.
Authors’ contribution
Conceptualization: MM; Methodology: MM, MS; Validation: MM, HK, AN; Formal Analysis: MM, HK; Investigation: MM, HK; Resources: MM, MTH, TSH; Data Curation: MM, MS; Writing—Original Draft Preparation: MM, MN; Writing—Review and Editing: BR, AN, KAA; Visualization: BR; Supervision: MM, AN; Project Administration: MM, MS; Funding Acquisition: MM.
References
- Debruyne D. Debruyne DClinical pharmacokinetics of fluconazole in superficial and systemic mycoses. Clin Pharmacokinet 1997; 33:52-77. doi: 10.2165/00003088-199733010-00005 [Crossref] [ Google Scholar]
- Cooper Jr CR, McGinnis MR. In vitro susceptibility of clinical yeast isolates to fluconazole and terconazole. Am J ObstetGynecol 1996; 175:1626-31. doi: 10.1016/S0002-9378(96)70116-3 [Crossref] [ Google Scholar]
- Deorukhkar S, Saini S. Virulence markers and antifungal susceptibility profile of Candida glabrata: an emerging pathogen. Microbiol Res J Int 2014; 4:39-49. doi: 10.9734/BMRJ/2014/5806 [Crossref] [ Google Scholar]
- Pfaller MA, Castanheira M, Lockhart SR, Jones RN. Candida glabrata: multidrug resistance and increased virulence in a major opportunistic fungal pathogen. Curr Fungal Infect Rep 2012; 6:154-64. doi: 10.1007/s12281-012-0091-0 [Crossref] [ Google Scholar]
- Ernst R, Kueppers P, Stindt J, Kuchler K, Schmitt L. Multidrug efflux pumps: Substrate selection in ATP‐binding cassette multidrug efflux pumps–first come, first served?. FEBS Lett 2010; 277:540-9. doi: 10.1111/j.1742-4658.2009.07485.x [Crossref] [ Google Scholar]
- Torelli R, Posteraro B, Ferrari S, La Sorda M, Fadda G, Sanglard D. The ATP‐binding cassette transporter–encoding gene CgSNQ2 is contributing to the CgPDR1‐dependent azole resistance of Candida glabrata. Mol Microbiol 2008; 68:186-201. doi: 10.1111/j.1365-2958.2008.06143.x [Crossref] [ Google Scholar]
- Gohar AA, Badali H, Shokohi T, Nabili M, Amirrajab N, Moazeni M. Expression patterns of ABC transporter genes in fluconazole-resistant Candida glabrata. Mycopathologia 2017; 182:273-84. doi: 10.1007/s11046-016-0074-8 [Crossref] [ Google Scholar]
- Sanglard D, Coste A, Ferrari S. Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res 2009; 9:1029-50. doi: 10.1111/j.1567-1364.2009.00578.x [Crossref] [ Google Scholar]
- Khosa A, Reddi S, Saha RN. Nanostructured lipid carriers for site-specific drug delivery. Biomed Pharmacother 2018; 103:598-613. doi: 10.1016/j.biopha.2018.04.055 [Crossref] [ Google Scholar]
- Moazeni M, Davari A, Shabanzadeh S, Akhtari J, Saeedi M, Mortyeza-Semnani K. In vitro antifungal activity of Thymus vulgaris essential oil nanoemulsion. J Herb Med 2021; 28:100452. doi: 10.1016/j.hermed.2021.100452 [Crossref] [ Google Scholar]
- Pardeike J, Weber S, Haber T, Wagner J, Zarfl H, Plank H. Development of an itraconazole-loaded nanostructured lipid carrier (NLC) formulation for pulmonary application. Int J Pharm 2011; 419:329-38. doi: 10.1016/j.ijpharm.2011.07.040 [Crossref] [ Google Scholar]
- Maheshwari RG, Tekade RK, Sharma PA, Darwhekar G, Tyagi A, Patel RP. Ethosomes and ultradeformable liposomes for transdermal delivery of clotrimazole: a comparative assessment. Saudi Pharm J 2012; 20:161-70. doi: 10.1016/j.jsps.2011.10.001 [Crossref] [ Google Scholar]
- Prajapati V, Jain A, Jain R, Sahu S, Kohli DV. Treatment of cutaneous candidiasis through fluconazole encapsulated cubosomes. Drug Deliv 2014; 4:400-8. doi: 10.1007/s13346-014-0202-2 [Crossref] [ Google Scholar]
- Kelidari HR, Moazeni M, Babaei R, Saeedi M, Akbari J, Parkoohi PI. Improved yeast delivery of fluconazole with a nanostructured lipid carrier system. Biomed Pharmacother 2017; 89:83-8. doi: 10.1016/j.biopha.2017.02.008 [Crossref] [ Google Scholar]
- Mirhendi H, Makimura K, Khoramizadeh M, Yamaguchi H. A one-enzyme PCR-RFLP assay for identification of six medically important Candida species. Nippon Ishinkin Gakkai Zasshi 2006; 47:225-9. doi: 10.3314/jjmm.47.225 [Crossref] [ Google Scholar]
- Clinical and Laboratory Standards Institue (CLSI). Performance Standards for Antifungal Susceptibility Testing of Yeasts; Approved Standard, CLSI Document M60. Wayne, PA: CLSI; 2017.
- Niemirowicz K, Durnaś B, Piktel E, Bucki R. Development of antifungal therapies using nanomaterials. Lond 2017; 12:1891-905. doi: 10.2217/nnm-2017-0052 [Crossref] [ Google Scholar]
- Ellis M, Clink H, Ernst P, Halim M, Padmos A, Spence D. Controlled study of fluconazole in the prevention of fungal infections in neutropenic patients with haematological malignancies and bone marrow transplant recipients. Eur J Clin Mivrobiol Infect Dis 1994; 13:3-11. doi: 10.1007/BF02026116 [Crossref] [ Google Scholar]
- Bianchin MD, Borowicz SM, Machado GdRM, Pippi B, Guterres SS, Pohlmann AR. Lipid core nanoparticles as a broad strategy to reverse fluconazole resistance in multiple Candida species. Colloids Surf B 2019; 175:523-9. doi: 10.1016/j.colsurfb.2018.12.011 [Crossref] [ Google Scholar]
- Du D, Wang-Kan X, Neuberger A, van Veen HW, Pos KM, Piddock LJ. Multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol 2018; 16:523-39. doi: 10.1038/s41579-018-0048-6 [Crossref] [ Google Scholar]
- Harris A, Wagner M, Du D, Raschka S, Nentwig L-M, Gohlke H. Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5. Nat Commun 2021; 12:1-14. doi: 10.1038/s41467-021-25574-8 [Crossref] [ Google Scholar]